Targeted Cancer Gene Analysis For Research Use Only - Not Intended for Clinical Use
|
|
|
- Rosalyn Murphy
- 10 years ago
- Views:
Transcription
1 Targeted Cancer Gene Analysis For Research Use Only - Not Intended for Clinical Use PGDX ID Sample ID Mutation Position Nucleotide (genomic) Amino Acid (protein) Mutation Type Consequence PGDX2163N_NotchCp SK100 chr9_ _c_t NA Substitution Splice site acceptor PGDX2115N_NotchCp SK51 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2115N_NotchCp SK51 chr9_ _c_t NA Substitution Splice site acceptor PGDX2115N_NotchCp SK51 chr9_ _c_t NA Substitution Splice site donor PGDX2116N_NotchCp SK52 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2116N_NotchCp SK52 chr9_ _c_t NA Substitution Splice site acceptor PGDX2117N_NotchCp SK53 chr9_ _a_c 819L>R Substitution Nonsynonymous coding PGDX2117N_NotchCp SK53 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2118N_NotchCp SK54 chr9_ _c_t NA Substitution Splice site acceptor PGDX2119N_NotchCp SK55 chr9_ _c_t NA Substitution Splice site acceptor PGDX2120N_NotchCp SK56 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2120N_NotchCp SK56 chr9_ _c_t NA Substitution Splice site acceptor PGDX2121N_NotchCp SK57 chr9_ _c_t NA Substitution Splice site acceptor PGDX2122N_NotchCp SK58 chr9_ _c_t NA Substitution Splice site acceptor PGDX2123N_NotchCp SK59 chr9_ _c_t NA Substitution Splice site acceptor PGDX2124N_NotchCp SK60 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2124N_NotchCp SK60 chr9_ _c_t NA Substitution Splice site acceptor PGDX2125N_NotchCp SK61 chr9_ _c_t NA Substitution Splice site acceptor PGDX2126N_NotchCp SK62 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2126N_NotchCp SK62 chr9_ _c_t NA Substitution Splice site acceptor PGDX2127N_NotchCp SK63 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2127N_NotchCp SK63 chr9_ _c_t NA Substitution Splice site acceptor PGDX2128N_NotchCp SK64 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2128N_NotchCp SK64 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2128N_NotchCp SK64 chr9_ _c_t NA Substitution Splice site acceptor PGDX2129N_NotchCp SK65 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2129N_NotchCp SK65 chr9_ _c_t NA Substitution Splice site acceptor PGDX2130N_NotchCp SK66 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2130N_NotchCp SK66 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2130N_NotchCp SK66 chr9_ _c_t NA Substitution Splice site acceptor PGDX2131N_NotchCp SK68 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding PGDX2131N_NotchCp SK68 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2131N_NotchCp SK68 chr9_ _c_t NA Substitution Splice site acceptor PGDX2132N_NotchCp SK69 chr9_ _c_t NA Substitution Splice site acceptor PGDX2133N_NotchCp SK70 chr9_ _t_g 65S>R Substitution Nonsynonymous coding PGDX2133N_NotchCp SK70 chr9_ _c_t NA Substitution Splice site acceptor PGDX2134N_NotchCp SK71 chr9_ _c_t NA Substitution Splice site acceptor PGDX2135N_NotchCp SK72 chr9_ _c_t NA Substitution Splice site acceptor PGDX2136N_NotchCp SK73 chr9_ _c_t NA Substitution Splice site acceptor PGDX2137N_NotchCp SK74 chr9_ _g_t 1531L>M Substitution Nonsynonymous coding PGDX2137N_NotchCp SK74 chr9_ _c_t NA Substitution Splice site acceptor PGDX2138N_NotchCp SK75 chr9_ _a_c 819L>R Substitution Nonsynonymous coding PGDX2138N_NotchCp SK75 chr9_ _c_t NA Substitution Splice site acceptor PGDX2139N_NotchCp SK76 chr9_ _c_t NA Substitution Splice site acceptor PGDX2140N_NotchCp SK77 chr9_ _c_t NA Substitution Splice site acceptor PGDX2141N_NotchCp SK78 chr9_ _c_t NA Substitution Splice site acceptor PGDX2142N_NotchCp SK79 chr9_ _c_t NA Substitution Splice site acceptor PGDX2143N_NotchCp SK80 chr9_ _c_t NA Substitution Splice site acceptor PGDX2144N_NotchCp SK81 chr9_ _c_t NA Substitution Splice site acceptor PGDX2145N_NotchCp SK82 chr9_ _g_a 1349A>V Substitution Nonsynonymous coding PGDX2145N_NotchCp SK82 chr9_ _c_t 245C>Y Substitution Nonsynonymous coding PGDX2145N_NotchCp SK82 chr9_ _c_t NA Substitution Splice site acceptor PGDX2146N_NotchCp SK83 chr9_ _t_c 1134Q>R Substitution Nonsynonymous coding PGDX2146N_NotchCp SK83 chr9_ _c_t NA Substitution Splice site acceptor PGDX2147N_NotchCp SK84 chr9_ _c_t NA Substitution Splice site acceptor PGDX2148N_NotchCp SK85 chr9_ _c_t NA Substitution Splice site acceptor PGDX2149N_NotchCp SK86 chr9_ _a_c 819L>R Substitution Nonsynonymous coding PGDX2149N_NotchCp SK86 chr9_ _c_t NA Substitution Splice site acceptor PGDX2150N_NotchCp SK87 chr9_ _c_t NA Substitution Splice site acceptor PGDX2151N_NotchCp SK88 chr9_ _t_c 1134Q>R Substitution Nonsynonymous coding PGDX2151N_NotchCp SK88 chr9_ _c_t NA Substitution Splice site acceptor PGDX2152N_NotchCp SK89 chr9_ _c_t NA Substitution Splice site acceptor PGDX2153N_NotchCp SK90 chr9_ _c_t NA Substitution Splice site acceptor PGDX2154N_NotchCp SK91 chr9_ _c_t NA Substitution Splice site acceptor PGDX2155N_NotchCp SK92 chr9_ _c_t NA Substitution Splice site acceptor PGDX2156N_NotchCp SK93 chr9_ _c_t NA Substitution Splice site acceptor PGDX2157N_NotchCp SK94 chr9_ _c_t NA Substitution Splice site acceptor PGDX2158N_NotchCp SK95 chr9_ _c_t NA Substitution Splice site acceptor PGDX2159N_NotchCp SK96 chr9_ _c_t NA Substitution Splice site acceptor PGDX2159N_NotchCp SK96 chr9_ _c_t NA Substitution Splice site donor PGDX2160N_NotchCp SK97 chr9_ _c_t NA Substitution Splice site acceptor PGDX2161N_NotchCp SK98 chr9_ _c_t NA Substitution Splice site acceptor PGDX2162N_NotchCp SK99 chr9_ _c_t NA Substitution Splice site acceptor Supplemental Table 6
2 Targeted Cancer Gene Analysis For Research Use Only - Not Intended for Clinical Use PGDX ID Sample ID Nucleotide (genomic) Mutation Position Amino Acid (protein) PGDX2040T_NotchCp SK1 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2040T_NotchCp SK1 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2040T_NotchCp SK1 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2040T_NotchCp SK1 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2048T_NotchCp SK10 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2087D_NotchCp SK101 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2087D_NotchCp SK101 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2088D_NotchCp SK102 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2088D_NotchCp SK102 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2089D_NotchCp SK103 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2089D_NotchCp SK103 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2090D_NotchCp SK104 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2090D_NotchCp SK104 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2091D_NotchCp SK105 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2091D_NotchCp SK105 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2092D_NotchCp SK106 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2092D_NotchCp SK106 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2093D_NotchCp SK107 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2094D_NotchCp SK108 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2094D_NotchCp SK108 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2095D_NotchCp SK109 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2095D_NotchCp SK109 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2049T_NotchCp SK11 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2049T_NotchCp SK11 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2049T_NotchCp SK11 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2049T_NotchCp SK11 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2096D_NotchCp SK110 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2096D_NotchCp SK110 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2097D_NotchCp SK111 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2097D_NotchCp SK111 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2098D_NotchCp SK112 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2098D_NotchCp SK112 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2069D_NotchCp SK117 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2069D_NotchCp SK117 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2070D_NotchCp SK118 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2070D_NotchCp SK118 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2071D_NotchCp SK119 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2071D_NotchCp SK119 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2072D_NotchCp SK120 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2072D_NotchCp SK120 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2073D_NotchCp SK121 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2073D_NotchCp SK121 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2074D_NotchCp SK122 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2075D_NotchCp SK123 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2075D_NotchCp SK123 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2076D_NotchCp SK124 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2077D_NotchCp SK125 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2078D_NotchCp SK126 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2079D_NotchCp SK127 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2080D_NotchCp SK128 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2081D_NotchCp SK129 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2050T_NotchCp SK13 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2050T_NotchCp SK13 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2050T_NotchCp SK13 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2082D_NotchCp SK130 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2083D_NotchCp SK131 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2084D_NotchCp SK132 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2085D_NotchCp SK133 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2086D_NotchCp SK134 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2099D_NotchCp SK135 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2099D_NotchCp SK135 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2099D_NotchCp SK135 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2100D_NotchCp SK136 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2101D_NotchCp SK137 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2102D_NotchCp SK138 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2103D_NotchCp SK139 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2104D_NotchCp SK140 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2105D_NotchCp SK141 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2106D_NotchCp SK142 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2107D_NotchCp SK143 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2108D_NotchCp SK144 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2108D_NotchCp SK144 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2108D_NotchCp SK144 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2109D_NotchCp SK145 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2110D_NotchCp SK146 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2111D_NotchCp SK147 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2112D_NotchCp SK148 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2113D_NotchCp SK149 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2114D_NotchCp SK150 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2051T_NotchCp SK19 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2051T_NotchCp SK19 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2051T_NotchCp SK19 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2051T_NotchCp SK19 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2041T_NotchCp SK2 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2041T_NotchCp SK2 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2041T_NotchCp SK2 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2041T_NotchCp SK2 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2052T_NotchCp SK22 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2052T_NotchCp SK22 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2052T_NotchCp SK22 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2052T_NotchCp SK22 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2053T_NotchCp SK23 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2053T_NotchCp SK23 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2053T_NotchCp SK23 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2053T_NotchCp SK23 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2054T_NotchCp SK24 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2054T_NotchCp SK24 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2054T_NotchCp SK24 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2054T_NotchCp SK24 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2055T_NotchCp SK25 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2055T_NotchCp SK25 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2055T_NotchCp SK25 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2055T_NotchCp SK25 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2056T_NotchCp SK27 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2056T_NotchCp SK27 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2056T_NotchCp SK27 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2057T_NotchCp SK28 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2057T_NotchCp SK28 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2057T_NotchCp SK28 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2057T_NotchCp SK28 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2058T_NotchCp SK29 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2058T_NotchCp SK29 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2058T_NotchCp SK29 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2058T_NotchCp SK29 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2042T_NotchCp SK3 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2042T_NotchCp SK3 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2042T_NotchCp SK3 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2042T_NotchCp SK3 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2059T_NotchCp SK30 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2059T_NotchCp SK30 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2059T_NotchCp SK30 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2059T_NotchCp SK30 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2060T_NotchCp SK32 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2060T_NotchCp SK32 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2060T_NotchCp SK32 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2060T_NotchCp SK32 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2061T_NotchCp SK34 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2061T_NotchCp SK34 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2061T_NotchCp SK34 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2061T_NotchCp SK34 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2062T_NotchCp SK38 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2062T_NotchCp SK38 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2062T_NotchCp SK38 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2062T_NotchCp SK38 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2043T_NotchCp SK4 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2043T_NotchCp SK4 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2043T_NotchCp SK4 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2043T_NotchCp SK4 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2063T_NotchCp SK40 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2063T_NotchCp SK40 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2063T_NotchCp SK40 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2063T_NotchCp SK40 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2064T_NotchCp SK41 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2064T_NotchCp SK41 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2064T_NotchCp SK41 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2064T_NotchCp SK41 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2065T_NotchCp SK42 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2065T_NotchCp SK42 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2065T_NotchCp SK42 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2068D_NotchCp SK46 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2068D_NotchCp SK46 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2068D_NotchCp SK46 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2068D_NotchCp SK46 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2044T_NotchCp SK5 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2044T_NotchCp SK5 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2044T_NotchCp SK5 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2044T_NotchCp SK5 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2066T_NotchCp SK50 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2066T_NotchCp SK50 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2066T_NotchCp SK50 chr9_ _c_t 245C>Y Substitution Nonsynonymous coding #N/A Yes PGDX2066T_NotchCp SK50 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2066T_NotchCp SK50 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2045T_NotchCp SK6 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2045T_NotchCp SK6 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2045T_NotchCp SK6 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2045T_NotchCp SK6 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2067T_NotchCp SK67 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2067T_NotchCp SK67 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2067T_NotchCp SK67 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2067T_NotchCp SK67 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2046T_NotchCp SK7 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2046T_NotchCp SK7 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2046T_NotchCp SK7 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2046T_NotchCp SK7 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes PGDX2047T_NotchCp SK8 chr9_ _t_g 2083H>P Substitution Nonsynonymous coding #N/A Yes PGDX2047T_NotchCp SK8 chr9_ _a_c 819L>R Substitution Nonsynonymous coding #N/A Yes PGDX2047T_NotchCp SK8 chr9_ _t_g 65S>R Substitution Nonsynonymous coding #N/A Yes PGDX2047T_NotchCp SK8 chr9_ _c_t NA Substitution Splice site acceptor Yes Yes Supplemental Table 7 Mutation Type Consequence In dbsnp In Normal Samples
3 Normals # Nonsynonymous Mutations in Normal Samples 73 # Unique Nonsyn Mutations in Normal Samples 49 Comment Unmatched Tumors # Nonsyn Mutations Also Present in Normal Samples 107 Includes 28 below as single mutation in dbsnp was also seen in normal samples # Nonsyn Mutations Also Present in dbsnp 28 # Nonsyn Mutations NOT Present in Normals/dbSNP 30 Previously reported Unmatched Dysplasia # Nonsyn Mutations Also Present in Normal Samples 71 Includes 47 below as single mutation in dbsnp was also seen in normal samples # Nonsyn Mutations Also Present in dbsnp 47 # Nonsyn Mutations NOT Present in Normals/dbSNP 50 Previously reported Supplemental Table 8
4 Sample ID Amino Acid (protein) Type Domain Exon # 1901E>K Missense Ankyrin repeats Ank_1 31 SK9 1662R>W Missense Heterodimerisation domain - N N>S Missense EGF-like domain (19) N>S Missense Ankyrin repeats Ank_1 31 SK12 952D>N Missense EGF-like domain (24) K>E Missense Next to Lin-12/Notch repeats (LNR)( ) 25 SK E>G Missense RBP-Jkappa-associated module (RAM) 28 SK D>G Missense RBP-Jkappa-associated module (RAM) P>S Missense Heterodimerisation domain - C 28 SK21 NA Indel SK G>S Missense RBP-Jkappa-associated module (RAM) C>Y Missense LNR D>N Missense Ankyrin repeats Ank_1 30/31 SK C>R Missense EGF-like domain (29) Q>R Missense PEST S>P Missense TAD S>N Missense RBP-Jkappa-associated module (RAM) 28 SK T>M Missense EGF-like domain (26) S>N Missense EGF-like domain (13) 9 NA Splice site acceptor 105A>V Missense EGF-like domain (3) 3 SK36 231G>D Missense EGF-like domain (6) 4 SK37 571D>A Missense EGF-like domain (15) 11 SK A>T Missense Ankyrin repeats Ank_ I>V Missense EGF-like domain (34) P>P Synonymous PEST 34 SK S>S Synonymous PEST L>L Synonymous RBP-Jkappa-associated module (RAM) 28 SK1 2429L>L Synonymous PEST P>P Synonymous EGF-like domain (23) 17 SK2 352D>Y Missense EGF-like domain (9) 6 SK5 1792V>A Missense RBP-Jkappa-associated module (RAM) H>H Synonymous EGF-like domain (30) 22 SK7 1627R>C Missense Heterodimerisation domain - N V>V Synonymous EGF-like domain (11) 7 SK8 1115L>P Missense EGF-like domain (29) 21 SK11 NA Indel SK13 _ISV-4> Splice site acceptor 1539H>P Missense LNR C>R Missense EGF-like domain (13) 9 SK R>Q Missense TAD P>P Synonymous EGF-like domain (20) 15 SK23 NA Splice site donor SK L>Q Missense LNR P>H Missense EGF-like domain (13) 9 SK N>D Missense EGF-like domain (32) G>G Synonymous EGF-like domain (25) 18 SK P>L Missense PEST K>R Missense EGF-like domain (21) 15 NA Splice site donor SK Y>X Nonsense TAD L>P Missense RBP-Jkappa-associated module (RAM) 29 SK P>P Synonymous TAD E>K Missense EGF-like domain (12) 8 SK L>P Missense LNR-2 26 SK S>P Missense Ankyrin repeats Ank_3 31 SK K>E Missense RBP-Jkappa-associated module (RAM) 29 SK G>G Synonymous Ankyrin repeats Ank_5 34 Song et al C1397F missense EGF-like domain (36) 25 SCC 1 P1641L missense Heterodimerisation domain - N 26 P1641S missense Heterodimerisation domain - N 26 SCC 4 C1218Y missense EGF-like domain (31) 23 SCC 5 C1133Y missense EGF-like domain (29) 21 Q1733 nonsense Heterodimerisation domain - C 28 R203C missense EGF-like domain (5) 4 R217W missense EGF-like domain (6) 4 R1762W missense RBP-Jkappa-associated module (RAM) 28 SCC 9 T232I missense EGF-like domain (6) 4 S1189F missense EGF-like domain (31) 22 N1347D missense EGF-like domain (34/35) 25 P1641L missense Heterodimerisation domain - N 26 S1389I missense EGF-like domain (36) 25 M1737I missense Trans-membrane region 28 SCC 10 P1641S missense Heterodimerisation domain - N 26 SCC 14 C1133Y missense EGF-like domain (29) 21 N1713S missense Heterodimerisation domain - C 27 SCC 15 C1133Y missense EGF-like domain (29) 21 SCC 19 E606 nonsense EGF-like domain (16) 11 SCC 21 N1489D missense LNR-3 25 SCC 22 Q1134R missense EGF-like domain (29) 21 SCC 23 Q1134R missense EGF-like domain (29) 21 SCC 26 E1084D missense EGF-like domain (28) 20 N1713S missense Heterodimerisation domain - C 27 SCC 28 Q1134R missense EGF-like domain (29) 21 SCC 31 N994K missense EGF-like domain (26) 19 P1641S missense Heterodimerisation domain - N 26 SCC 32 C1074Y missense EGF-like domain (28) 20 SCC 33 E424K missense EGF-like domain (11) 8 Q1080 nonsense EGF-like domain (28) 20 SCC 35 W1474 nonsense LNR-1 25 N1713S missense Heterodimerisation domain - C 27 SCC 42 R1107 nonsense EGF-like domain (28/29) 20 N548D missense EGF-like domain (14) 10 SCC 47 E606G missense EGF-like domain (16) 11 P1641L missense Heterodimerisation domain - N 26 P1641S missense Heterodimerisation domain - N 26 SCC 48 R1107 nonsense EGF-like domain (28/29) 20 W1474 nonsense LNR-1 25 SCC 50 E1084D missense EGF-like domain (28) 20 SCC 51 R1784Q missense RBP-Jkappa-associated module (RAM) 28 Supplemental Figure 9
5 Sample Amino acid (protein) Mutation type Domain Exon # HN14PT p.p391s Missense EGF-like domain (10) p.g484v Missense EGF-like domain (12) 9 HN 102 PT p.e450k Missense EGF-like domain (11) 8 HN 105 PT p.g812w Missense EGF-like domain (21) 15 HN 115 PT p.g1340a Missense EGF-like domain (34) 25 HN 117 PT p.g310r Missense EGF-like domain (8) 6 HN 117 PT p.c456r Missense EGF-like domain (12) 8 HN 142 PT p.r353h Missense EGF-like domain (9) 6 HN 142 PT p.m2011r Missense Ankyrin repeats Ank_4 32 HN 148 PT p.g1638v Missense Heterodimerisation domain - N 26 HN 148 PT p.p2272s Missense TAD 34 HN 208 PT p.r365c Missense EGF-like domain (9) 6 HN 208 PT p.r1280c Missense EGF-like domain (33) 23 HN 227 PT p.r365c Missense EGF-like domain (9) 6 HN 251 PT p.f1292l Missense EGF-like domain (33) 23 HN 255 PT p.v2039l Missense Ankyrin repeats Ank_5 33 HN 255 PT p.v2039e Missense Ankyrin repeats Ank_5 33 HN12PT p.w1843x Nonsense RBP-Jkappa-associated module (RAM) p.r1984x Nonsense Ankyrin repeats Ank_ Q290X Nonsense EGF-like domain (7) 6 HN 105 PT fs Indel HN 107 PT p.s402x Nonsense EGF-like domain (10) 7 HN 115 PT fs Indel HN 130 PT p.e949x Nonsense EGF-like domain (25) 18 HN 139 PT p.c554x Nonsense EGF-like domain (14) 10 HN 183 PT p.q1958x Nonsense Ankyrin repeats Ank_3 31 HN 194 PT fs Indel HN 245 PT fs Indel HN_63080-Tumor S953N Missense EGF-like domain (25) 18 HN_62318-Tumor D1517N Missense LRN repeat 2 25 HN_62755-Tumor Q1214* Nonsense EGF-like domain (31) 22 HN_63081-Tumor E719* Nonsense EGF-like domain (19) 13 HN_00076-Tumor Splice_Site HN_62863-Tumor G481S Missense EGF-like domain (12) 8 HN_62469-Tumor E424K Missense EGF-like domain (11) 8 HN_62995-Tumor P422S Missense EGF-like domain (11) 8 HN_62298-Tumor F357del Indel HN_62699-Tumor Splice_Site OSCJM-PT T G484V Missense EGF-like domain (12) 9 OSCJM-PT T S458L Missense EGF-like domain (12) 8 OSCJM-PT T Q290X Nonsense EGF-like domain (7) 6 OSCJM-PT T R1984X Nonsense Ankyrin repeats Ank_5 32 X39 R56* Nonsense EGF-like domain (1) 3 X17 Q154* Nonsense EGF-like domain (4) 4 X12 T1138I Missense EGF-like domain (29) 21 X48 E1294* Nonsense EGF-like domain (33) 23 X12 W1843* Nonsense RBP-Jkappa-associated module (RAM) 30 TCGA-BA-4076 G481C Missense EGF-like domain (12) 8 TCGA-BA-6868 C387Y Missense EGF-like domain (10) 7 TCGA-BB-4225 C461Y Missense EGF-like domain (12) 8 TCGA-CN-4723 D142H Missense EGF-like domain (4) 4 TCGA-CN-4729 C1085Y Missense EGF-like domain (28) 20 TCGA-CN-4736 C478S Missense EGF-like domain (12) 8 TCGA-CN-4742 P422S Missense EGF-like domain (11) 8 TCGA-CN-5369 Q1845* Nonsense RBP-Jkappa-associated module (RAM) 30 TCGA-CN-6011 E1305K Missense EGF-like domain (33) 24 TCGA-CN-6016 V1222M Missense EGF-like domain (32) 23 TCGA-CN-6989 C739* Nonsense EGF-like domain (19) 14 TCGA-CN-6994 P67T Missense EGF-like domain (2) 3 TCGA-CQ-5329 A2023T Missense Ankyrin repeats Ank_4 32 TCGA-CQ-5334 G1119* Nonsense EGF-like domain (29) 20/21 TCGA-CR-5243 C1085W Missense EGF-like domain (28) 20 TCGA-CR-7365 E1404K Missense EGF-like domain (36) 25 TCGA-CR-7376 G484V Missense EGF-like domain (12) 9 TCGA-CR-7377 E1450* Nonsense LRN repeat 1 25 TCGA-CR-7379 N1875K Missense Ankyrin repeats Ank_1 30 TCGA-CR-7380 C893S Missense EGF-like domain (23) 17 TCGA-CR-7390 Q1080* Nonsense EGF-like domain (28) 20 TCGA-CR-7394 Q1974* Nonsense Ankyrin repeats Ank_3 31 TCGA-CR-7395 P2064L Missense Ankyrin repeats Ank_6 34 TCGA-CV-5436 C478Y Missense EGF-like domain (12) 8 TCGA-CV-5971 R353C Missense EGF-like domain (9) 6 TCGA-CV-5973 R1984* Nonsense Ankyrin repeats Ank_5 32 TCGA-CV-6436 G310R Missense EGF-like domain (8) 6 TCGA-CV-6955 D1520H Missense LRN repeat 1 25 TCGA-CV-6955 N454T Missense EGF-like domain (12) 8 TCGA-CV-6959 R879W Missense EGF-like domain (23) 17 TCGA-CV-6962 C429Y Missense EGF-like domain (11) 8 TCGA-CV-7089 S385F Missense EGF-like domain (10) 7 TCGA-CV-7099 K1317N Missense EGF-like domain (34) 24 TCGA-CV-7101 T1996M Missense Ankyrin repeats Ank_4 32 TCGA-CV-7236 A465T Missense EGF-like domain (12) 8 TCGA-CV-7238 Q803* Nonsense EGF-like domain (21) 15 TCGA-CV-7410 E663K Missense EGF-like domain (17) 12 TCGA-CV-7410 E851Q Missense EGF-like domain (22) 16 TCGA-CV-7411 C1225* Nonsense EGF-like domain (32) 23 TCGA-CV-7427 W1089* Nonsense EGF-like domain (28) 20 TCGA-CV-7433 C440F Missense EGF-like domain (11) 8 TCGA-CX-7085 E455K Missense EGF-like domain (12) 8 TCGA-DQ-5625 D352V Missense EGF-like domain (9) 6 TCGA-DQ-5631 C440R Missense EGF-like domain (11) 8 TCGA-BA-4075 C449Y Missense EGF-like domain (11) 8 TCGA-CQ-6219 R353H Missense EGF-like domain (9) 6 TCGA-CV-5439 G95W Missense EGF-like domain (2) 3 TCGA-CN-5355 G1215_splice Splice_Site TCGA-CN-5363 F1977fs Indel TCGA-CN-6021 F357del IF-Indel TCGA-CQ-6220 S417fs Indel TCGA-BA-6873 P380fs Indel TCGA-CR-7365 K542fs Indel TCGA-CV-5441 A796fs Indel TCGA-CV-5966 T634fs Indel TCGA-CV-6938 E1148fs Indel TCGA-CV-6942 G684fs Indel TCGA-CV-6955 T123fs Indel TCGA-CV-7089 C1063fs Indel TCGA-BA-5149 Q513fs Indel TCGA-BB-7861 P2415del IF-Indel TCGA-BB-7863 G1380fs Indel TCGA-BB-7864 G115fs Indel TCGA-BB-7872 G736_splice Splice_Site IF - In Frame Supplemental Table 10
6 Sample ID Site HPV status HN14PT OC N 478 OC N HN 102 PT OC N HN 105 PT OC N HN 107 PT OC N HN 115 PT OC N HN 117 PT OC N HN 130 PT L N HN 139 PT OP N HN 142 PT OP P HN 148 PT OP N HN 183 PT OC N HN 194 PT OC N HN 208 PT OC N HN 227 PT HP N HN 245 PT OC N HN 251 PT OP P HN 255 PT OP N HN12PT OC N 385 OC N 600 OC N HN_63080-Tumor OC N HN_62318-Tumor OP P HN_62755-Tumor L N HN_63081-Tumor OP N HN_00076-Tumor OC N HN_62863-Tumor L N HN_62469-Tumor OC N HN_62995-Tumor OC N HN_62298-Tumor OC N HN_62699-Tumor HP N TCGA-BA-4076 L N TCGA-BA-5149 OC N TCGA-BA-6868 L N TCGA-BA-6873 OC P TCGA-BB-4225 OP P TCGA-CN-4723 L N TCGA-CN-4729 OC N TCGA-CN-4736 OC N TCGA-CN-4742 OC N TCGA-CN-5355 L N TCGA-CN-5363 L N TCGA-CN-5369 OC N TCGA-CN-6011 OC N TCGA-CN-6016 OC N TCGA-CN-6021 L N TCGA-CN-6989 L N TCGA-CN-6994 OC N TCGA-CQ-5329 OC N TCGA-CQ-5334 OC N TCGA-CQ-6220 OC N TCGA-CR-5243 OP P TCGA-CR-7365 OC N TCGA-CR-7376 OC N TCGA-CR-7377 OC N TCGA-CR-7379 OC N TCGA-CR-7380 OC N TCGA-CR-7390 OC N TCGA-CR-7394 OC N TCGA-CR-7395 OC N TCGA-CV-5436 OC N TCGA-CV-5441 L N TCGA-CV-5966 OC N TCGA-CV-5971 OC P TCGA-CV-5973 OC N TCGA-CV-6436 OC N TCGA-CV-6938 OC N TCGA-CV-6942 OC N TCGA-CV-6955 OC N TCGA-CV-6959 OC N TCGA-CV-6962 L N TCGA-CV-7089 L N TCGA-CV-7099 OC N TCGA-CV-7101 L N TCGA-CV-7236 OC N TCGA-CV-7238 OC N TCGA-CV-7410 L N TCGA-CV-7411 OC N TCGA-CV-7427 OC N TCGA-CV-7433 L N TCGA-CX-7085 OC N TCGA-DQ-5625 OC N TCGA-DQ-5631 OC N TCGA-BA-4075 OC ND TCGA-BB-7861 OP ND TCGA-BB-7863 OC ND TCGA-BB-7864 L ND TCGA-BB-7872 OC ND TCGA-CQ-6219 OC ND TCGA-CV-5439 OP N OSCJM-PT OC N OSCJM-PT OC N OSCJM-PT OC N OSCJM-PT OC ND X39 OC N X17 OC N X12 OC N X48 OC N OC - Oral Cavity OP - Oropharynx HP - Hypopharynx L - Larynx N - Negative P - Positive ND - Not defined Supplemental Table 11
plaque reduction assay, modified dye uptake assay including formazan test, dye uptake assay
 Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. 2015. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes
Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. 2015. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes
Domain Antivirals tested Test methods for phenotype References. Aciclovir (ACV), Foscarnet (FOS) ACV, FOS ACV,
 Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. 2015. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes
Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. 2015. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes
Victims Compensation Claim Status of All Pending Claims and Claims Decided Within the Last Three Years
 Claim#:021914-174 Initials: J.T. Last4SSN: 6996 DOB: 5/3/1970 Crime Date: 4/30/2013 Status: Claim is currently under review. Decision expected within 7 days Claim#:041715-334 Initials: M.S. Last4SSN: 2957
Claim#:021914-174 Initials: J.T. Last4SSN: 6996 DOB: 5/3/1970 Crime Date: 4/30/2013 Status: Claim is currently under review. Decision expected within 7 days Claim#:041715-334 Initials: M.S. Last4SSN: 2957
Long-QT syndrome (LQTS) is a cardiovascular disorder
 Spectrum of Mutations in Long-QT Syndrome Genes KVLQT1, HERG, SCN5A, KCNE1, and KCNE2 Igor Splawski, PhD; Jiaxiang Shen, MS; Katherine W. Timothy, BS; Michael H. Lehmann, MD; Silvia Priori, MD, PhD; Jennifer
Spectrum of Mutations in Long-QT Syndrome Genes KVLQT1, HERG, SCN5A, KCNE1, and KCNE2 Igor Splawski, PhD; Jiaxiang Shen, MS; Katherine W. Timothy, BS; Michael H. Lehmann, MD; Silvia Priori, MD, PhD; Jennifer
Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1
 www.sciencemag.org/cgi/content/full/science.1206923/dc1 Supporting Online Material for Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1 Nishant Agrawal,*
www.sciencemag.org/cgi/content/full/science.1206923/dc1 Supporting Online Material for Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1 Nishant Agrawal,*
Transient Voltage Suppressor SMBJ5.0 - SMBJ440CA
 Features: Glass passivated junction Low incremental surge resistance, excellent clamping capability 600W peak pulse power capability with a 10/1,000μs waveform, repetition rate (duty cycle): 0.01% Very
Features: Glass passivated junction Low incremental surge resistance, excellent clamping capability 600W peak pulse power capability with a 10/1,000μs waveform, repetition rate (duty cycle): 0.01% Very
The sequence of bases on the mrna is a code that determines the sequence of amino acids in the polypeptide being synthesized:
 Module 3F Protein Synthesis So far in this unit, we have examined: How genes are transmitted from one generation to the next Where genes are located What genes are made of How genes are replicated How
Module 3F Protein Synthesis So far in this unit, we have examined: How genes are transmitted from one generation to the next Where genes are located What genes are made of How genes are replicated How
CODES FOR PHARMACY ONLINE CLAIMS PROCESSING
 S FOR PHARMACY ONLINE CLAIMS PROCESSING The following is a list of error and warning codes that may appear when processing claims on the online system. The error codes are bolded. CODE AA AB AI AR CB CD
S FOR PHARMACY ONLINE CLAIMS PROCESSING The following is a list of error and warning codes that may appear when processing claims on the online system. The error codes are bolded. CODE AA AB AI AR CB CD
1. Oblast rozvoj spolků a SU UK 1.1. Zvyšování kvalifikace Školení Zapojení do projektů Poradenství 1.2. Financování 1.2.1.
 1. O b l a s t r o z v o j s p o l k a S U U K 1. 1. Z v y š o v á n í k v a l i f i k a c e Š k o l e n í o S t u d e n t s k á u n i e U n i v e r z i t y K a r l o v y ( d á l e j e n S U U K ) z í
1. O b l a s t r o z v o j s p o l k a S U U K 1. 1. Z v y š o v á n í k v a l i f i k a c e Š k o l e n í o S t u d e n t s k á u n i e U n i v e r z i t y K a r l o v y ( d á l e j e n S U U K ) z í
 C o a t i a n P u b l i c D e b tm a n a g e m e n t a n d C h a l l e n g e s o f M a k e t D e v e l o p m e n t Z a g e bo 8 t h A p i l 2 0 1 1 h t t pdd w w wp i j fp h D p u b l i c2 d e b td S t
C o a t i a n P u b l i c D e b tm a n a g e m e n t a n d C h a l l e n g e s o f M a k e t D e v e l o p m e n t Z a g e bo 8 t h A p i l 2 0 1 1 h t t pdd w w wp i j fp h D p u b l i c2 d e b td S t
SCO TT G LEA SO N D EM O Z G EB R E-
 SCO TT G LEA SO N D EM O Z G EB R E- EG Z IA B H ER e d it o r s N ) LICA TIO N S A N D M ETH O D S t DVD N CLUDED C o n t e n Ls Pr e fa c e x v G l o b a l N a v i g a t i o n Sa t e llit e S y s t e
SCO TT G LEA SO N D EM O Z G EB R E- EG Z IA B H ER e d it o r s N ) LICA TIO N S A N D M ETH O D S t DVD N CLUDED C o n t e n Ls Pr e fa c e x v G l o b a l N a v i g a t i o n Sa t e llit e S y s t e
Focusing on results not data comprehensive data analysis for targeted next generation sequencing
 Focusing on results not data comprehensive data analysis for targeted next generation sequencing Daniel Swan, Jolyon Holdstock, Angela Matchan, Richard Stark, John Shovelton, Duarte Mohla and Simon Hughes
Focusing on results not data comprehensive data analysis for targeted next generation sequencing Daniel Swan, Jolyon Holdstock, Angela Matchan, Richard Stark, John Shovelton, Duarte Mohla and Simon Hughes
1.- L a m e j o r o p c ió n e s c l o na r e l d i s co ( s e e x p li c a r á d es p u é s ).
 PROCEDIMIENTO DE RECUPERACION Y COPIAS DE SEGURIDAD DEL CORTAFUEGOS LINUX P ar a p od e r re c u p e ra r nu e s t r o c o rt a f u e go s an t e un d es a s t r e ( r ot u r a d e l di s c o o d e l a
PROCEDIMIENTO DE RECUPERACION Y COPIAS DE SEGURIDAD DEL CORTAFUEGOS LINUX P ar a p od e r re c u p e ra r nu e s t r o c o rt a f u e go s an t e un d es a s t r e ( r ot u r a d e l di s c o o d e l a
Accurate and sensitive mutation detection and quantitation using TaqMan Mutation Detection Assays for disease research
 PPLICTION NOTE Mutation Detection ssays ccurate and sensitive mutation detection and quantitation using Mutation Detection ssays for disease research In this research study, we addressed the feasibility
PPLICTION NOTE Mutation Detection ssays ccurate and sensitive mutation detection and quantitation using Mutation Detection ssays for disease research In this research study, we addressed the feasibility
Excel Invoice Format. SupplierWebsite - Excel Invoice Upload. Data Element Definition UCLA Supplier website (Rev. July 9, 2013)
 Excel Invoice Format Excel Column Name Cell Format Notes Campus* Supplier Number* Invoice Number* Order Number* Invoice Date* Total Invoice Amount* Total Sales Tax Amount* Discount Amount Discount Percent
Excel Invoice Format Excel Column Name Cell Format Notes Campus* Supplier Number* Invoice Number* Order Number* Invoice Date* Total Invoice Amount* Total Sales Tax Amount* Discount Amount Discount Percent
B I N G O B I N G O. Hf Cd Na Nb Lr. I Fl Fr Mo Si. Ho Bi Ce Eu Ac. Md Co P Pa Tc. Uut Rh K N. Sb At Md H. Bh Cm H Bi Es. Mo Uus Lu P F.
 Hf Cd Na Nb Lr Ho Bi Ce u Ac I Fl Fr Mo i Md Co P Pa Tc Uut Rh K N Dy Cl N Am b At Md H Y Bh Cm H Bi s Mo Uus Lu P F Cu Ar Ag Mg K Thomas Jefferson National Accelerator Facility - Office of cience ducation
Hf Cd Na Nb Lr Ho Bi Ce u Ac I Fl Fr Mo i Md Co P Pa Tc Uut Rh K N Dy Cl N Am b At Md H Y Bh Cm H Bi s Mo Uus Lu P F Cu Ar Ag Mg K Thomas Jefferson National Accelerator Facility - Office of cience ducation
ALLERGY/MOLD Gene & Variation rsid # Risk Allele Your Alleles and Results HLA rs7775228 C TT -/- HLA rs2155219 T GG -/-
 DETOX CYP1A1*2C A4889G rs1048943 C TT -/- CYP1A1*4 C2453A rs1799814 T GG -/- CYP1A2 C164A rs762551 C AA -/- CYP1B1 L432V rs1056836 C CC +/+ CYP1B1 N453S rs1800440 C TT -/- CYP1B1 R48G rs10012 C no call
DETOX CYP1A1*2C A4889G rs1048943 C TT -/- CYP1A1*4 C2453A rs1799814 T GG -/- CYP1A2 C164A rs762551 C AA -/- CYP1B1 L432V rs1056836 C CC +/+ CYP1B1 N453S rs1800440 C TT -/- CYP1B1 R48G rs10012 C no call
Next Generation Sequencing. mapping mutations in congenital heart disease
 Next Generation Sequencing mapping mutations in congenital heart disease AV Postma PhD Academic Medical Center Amsterdam, the Netherlands Overview talk Congenital heart disease and genetics Next generation
Next Generation Sequencing mapping mutations in congenital heart disease AV Postma PhD Academic Medical Center Amsterdam, the Netherlands Overview talk Congenital heart disease and genetics Next generation
Earthquake Hazard Zones: The relative risk of damage to Canadian buildings
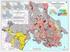 Earthquake Hazard Zones: The relative risk of damage to Canadian buildings by Paul Kovacs Executive Director, Institute for Catastrophic Loss Reduction Adjunct Research Professor, Economics, Univ. of Western
Earthquake Hazard Zones: The relative risk of damage to Canadian buildings by Paul Kovacs Executive Director, Institute for Catastrophic Loss Reduction Adjunct Research Professor, Economics, Univ. of Western
MUTATION, DNA REPAIR AND CANCER
 MUTATION, DNA REPAIR AND CANCER 1 Mutation A heritable change in the genetic material Essential to the continuity of life Source of variation for natural selection New mutations are more likely to be harmful
MUTATION, DNA REPAIR AND CANCER 1 Mutation A heritable change in the genetic material Essential to the continuity of life Source of variation for natural selection New mutations are more likely to be harmful
Concluding lesson. Student manual. What kind of protein are you? (Basic)
 Concluding lesson Student manual What kind of protein are you? (Basic) Part 1 The hereditary material of an organism is stored in a coded way on the DNA. This code consists of four different nucleotides:
Concluding lesson Student manual What kind of protein are you? (Basic) Part 1 The hereditary material of an organism is stored in a coded way on the DNA. This code consists of four different nucleotides:
 W Cisco Kompetanse eek end 2 0 0 8 SMB = Store Mu ll ii gg hh eter! Nina Gullerud ng ulleru@ c is c o. c o m 1 Vår E n t e r p r i s e e r f a r i n g... 2 S m å o g M e llo m s t o r e B e d r i f t e
W Cisco Kompetanse eek end 2 0 0 8 SMB = Store Mu ll ii gg hh eter! Nina Gullerud ng ulleru@ c is c o. c o m 1 Vår E n t e r p r i s e e r f a r i n g... 2 S m å o g M e llo m s t o r e B e d r i f t e
e4cash Configuration Reference PRELIMINARY
 e4cash Configuration Reference PRELIMINARY This document contains proprietary, privileged and preliminary information describing the e4cash multiple cash drawer network interface. This document and associated
e4cash Configuration Reference PRELIMINARY This document contains proprietary, privileged and preliminary information describing the e4cash multiple cash drawer network interface. This document and associated
A Quick Guide to Colleges. Offering Engineering Degrees
 Qk gs ffg gg Dgs Fby 2007 www.dgs.g Qk gs ffg gg Dgs Ts qk f g ps sy sg (by s) f U.S. gs ss ffg b s (4-y) gs gg 363 ss, spg 50 ss ps Ds f b R. Ts sy s s gg g pgs by f gg Tgy (T, gz f g sy pgs pp s, pg,
Qk gs ffg gg Dgs Fby 2007 www.dgs.g Qk gs ffg gg Dgs Ts qk f g ps sy sg (by s) f U.S. gs ss ffg b s (4-y) gs gg 363 ss, spg 50 ss ps Ds f b R. Ts sy s s gg g pgs by f gg Tgy (T, gz f g sy pgs pp s, pg,
CLASS TEST GRADE 11. PHYSICAL SCIENCES: CHEMISTRY Test 6: Chemical change
 CLASS TEST GRADE PHYSICAL SCIENCES: CHEMISTRY Test 6: Chemical change MARKS: 45 TIME: hour INSTRUCTIONS AND INFORMATION. Answer ALL the questions. 2. You may use non-programmable calculators. 3. You may
CLASS TEST GRADE PHYSICAL SCIENCES: CHEMISTRY Test 6: Chemical change MARKS: 45 TIME: hour INSTRUCTIONS AND INFORMATION. Answer ALL the questions. 2. You may use non-programmable calculators. 3. You may
BLADE 12th Generation. Rafał Olszewski. Łukasz Matras
 BLADE 12th Generation Rafał Olszewski Łukasz Matras Jugowice, 15-11-2012 Gl o b a l M a r k e t i n g Dell PowerEdge M-Series Blade Server Portfolio M-Series Blades couple powerful computing capabilities
BLADE 12th Generation Rafał Olszewski Łukasz Matras Jugowice, 15-11-2012 Gl o b a l M a r k e t i n g Dell PowerEdge M-Series Blade Server Portfolio M-Series Blades couple powerful computing capabilities
Advances in RainDance Sequence Enrichment Technology and Applications in Cancer Research. March 17, 2011 Rendez-Vous Séquençage
 Advances in RainDance Sequence Enrichment Technology and Applications in Cancer Research March 17, 2011 Rendez-Vous Séquençage Presentation Overview Core Technology Review Sequence Enrichment Application
Advances in RainDance Sequence Enrichment Technology and Applications in Cancer Research March 17, 2011 Rendez-Vous Séquençage Presentation Overview Core Technology Review Sequence Enrichment Application
Masters Mens Physique 45+
 C G x By, F, hysq, Bk Chpshps Ap, Cv Cy, Cf Mss Ms hysq + Fs Ls Css Ov Css s G MC Chpk M+ W M+/MC y Bs 9 8 9 9 8 B O'H 8 9 8 S Rs 8 8 9 h K 9 D Szwsk 8 9 8 9 9 G M+ h D Ly Iz M+ 8 M R : : C G x By, F,
C G x By, F, hysq, Bk Chpshps Ap, Cv Cy, Cf Mss Ms hysq + Fs Ls Css Ov Css s G MC Chpk M+ W M+/MC y Bs 9 8 9 9 8 B O'H 8 9 8 S Rs 8 8 9 h K 9 D Szwsk 8 9 8 9 9 G M+ h D Ly Iz M+ 8 M R : : C G x By, F,
 FORT WAYNE COMMUNITY SCHOOLS 12 00 SOUTH CLINTON STREET FORT WAYNE, IN 468 02 6:02 p.m. Ma r c h 2 3, 2 015 OFFICIAL P ROCEED ING S Ro l l Ca l l e a r d o f h o o l u e e o f t h e r t y m m u t y h o
FORT WAYNE COMMUNITY SCHOOLS 12 00 SOUTH CLINTON STREET FORT WAYNE, IN 468 02 6:02 p.m. Ma r c h 2 3, 2 015 OFFICIAL P ROCEED ING S Ro l l Ca l l e a r d o f h o o l u e e o f t h e r t y m m u t y h o
Payroll System Codes
 Page 1 of 6 Payroll System Codes Status Codes User Guide Section: 200-8 Screen: PF.PS.STAT.U A IA IC IN IP IW LP LR LS LW NA NE PA PG PP PR RP RW SC SP T TP Active Inactive for one tax quarter Inactive
Page 1 of 6 Payroll System Codes Status Codes User Guide Section: 200-8 Screen: PF.PS.STAT.U A IA IC IN IP IW LP LR LS LW NA NE PA PG PP PR RP RW SC SP T TP Active Inactive for one tax quarter Inactive
The Genetics of Hearing Loss. Heidi L. Rehm, Ph.D. Harvard Medical School Boston, MA
 The Genetics of Hearing Loss Heidi L. Rehm, Ph.D. Harvard Medical School Boston, MA Faculty Disclosure Information In the past 12 months, I have not had a significant financial interest or other relationship
The Genetics of Hearing Loss Heidi L. Rehm, Ph.D. Harvard Medical School Boston, MA Faculty Disclosure Information In the past 12 months, I have not had a significant financial interest or other relationship
Genomes and SNPs in Malaria and Sickle Cell Anemia
 Genomes and SNPs in Malaria and Sickle Cell Anemia Introduction to Genome Browsing with Ensembl Ensembl The vast amount of information in biological databases today demands a way of organising and accessing
Genomes and SNPs in Malaria and Sickle Cell Anemia Introduction to Genome Browsing with Ensembl Ensembl The vast amount of information in biological databases today demands a way of organising and accessing
FY 2013 Financial Status Report SOAR Operating Expenditures June 30, 2013. Table of Contents
 FY Financial Status Report SOAR Operating Expenditures 30, Table of Contents (A) Letter from the CFO... A 1 Key Increases (Decreases) in Local Funds Attachment A... A 3 (B) (C) District Summary by Percentage
FY Financial Status Report SOAR Operating Expenditures 30, Table of Contents (A) Letter from the CFO... A 1 Key Increases (Decreases) in Local Funds Attachment A... A 3 (B) (C) District Summary by Percentage
Recombinant DNA Technology
 Recombinant DNA Technology Stephen B. Gruber, MD, PhD Division of Molecular Medicine and Genetics November 4, 2002 Learning Objectives Know the basics of gene structure, function and regulation. Be familiar
Recombinant DNA Technology Stephen B. Gruber, MD, PhD Division of Molecular Medicine and Genetics November 4, 2002 Learning Objectives Know the basics of gene structure, function and regulation. Be familiar
Chem 115 POGIL Worksheet - Week 4 Moles & Stoichiometry Answers
 Key Questions & Exercises Chem 115 POGIL Worksheet - Week 4 Moles & Stoichiometry Answers 1. The atomic weight of carbon is 12.0107 u, so a mole of carbon has a mass of 12.0107 g. Why doesn t a mole of
Key Questions & Exercises Chem 115 POGIL Worksheet - Week 4 Moles & Stoichiometry Answers 1. The atomic weight of carbon is 12.0107 u, so a mole of carbon has a mass of 12.0107 g. Why doesn t a mole of
 How to Subnet a Network How to use this paper Absolute Beginner: Read all Sections 1-4 N eed a q uick rev iew : Read Sections 2-4 J ust need a little h elp : Read Section 4 P a r t I : F o r t h e I P
How to Subnet a Network How to use this paper Absolute Beginner: Read all Sections 1-4 N eed a q uick rev iew : Read Sections 2-4 J ust need a little h elp : Read Section 4 P a r t I : F o r t h e I P
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
Lessons Learned from Carrier Screening: Cystic Fibrosis The Parent s s Perspective
 Lessons Learned from Carrier Screening: Cystic Fibrosis The Parent s s Perspective Martin Kharrazi, Ph.D. Genetic Disease Screening Program California Department of Public Health Population-based Carrier
Lessons Learned from Carrier Screening: Cystic Fibrosis The Parent s s Perspective Martin Kharrazi, Ph.D. Genetic Disease Screening Program California Department of Public Health Population-based Carrier
Gene and Chromosome Mutation Worksheet (reference pgs. 239-240 in Modern Biology textbook)
 Name Date Per Look at the diagrams, then answer the questions. Gene Mutations affect a single gene by changing its base sequence, resulting in an incorrect, or nonfunctional, protein being made. (a) A
Name Date Per Look at the diagrams, then answer the questions. Gene Mutations affect a single gene by changing its base sequence, resulting in an incorrect, or nonfunctional, protein being made. (a) A
Role of Hydrogen Bonding on Protein Secondary Structure Introduction
 Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Exercises for the UCSC Genome Browser Introduction
 Exercises for the UCSC Genome Browser Introduction 1) Find out if the mouse Brca1 gene has non-synonymous SNPs, color them blue, and get external data about a codon-changing SNP. Skills: basic text search;
Exercises for the UCSC Genome Browser Introduction 1) Find out if the mouse Brca1 gene has non-synonymous SNPs, color them blue, and get external data about a codon-changing SNP. Skills: basic text search;
All answers must use the correct number of significant figures, and must show units!
 CHEM 10113, Quiz 2 September 7, 2011 Name (please print) All answers must use the correct number of significant figures, and must show units! IA Periodic Table of the Elements VIIIA (1) (18) 1 2 1 H IIA
CHEM 10113, Quiz 2 September 7, 2011 Name (please print) All answers must use the correct number of significant figures, and must show units! IA Periodic Table of the Elements VIIIA (1) (18) 1 2 1 H IIA
Para-Transit Pick Up Zones 2
 Para-Transit Pick Up Zones 2 Para-Transit Pick Up Locations Academic House (AH) Bowman-Oddy/Wolfe Hall (BO/WO) Carlson Library (CL) Carter Hall East/West (CE/CW) Center for Performing Arts (PA) Center
Para-Transit Pick Up Zones 2 Para-Transit Pick Up Locations Academic House (AH) Bowman-Oddy/Wolfe Hall (BO/WO) Carlson Library (CL) Carter Hall East/West (CE/CW) Center for Performing Arts (PA) Center
INTERACTIVE MAP EGYPT
 Multi-criteria Analysis for lanning Renewable Energy (MapRE) INTERACTIVE MA EGYT Of Southern and Eastern Africa Renewable Energy Zones (SEAREZs) This interactive DF map contains locations of high quality
Multi-criteria Analysis for lanning Renewable Energy (MapRE) INTERACTIVE MA EGYT Of Southern and Eastern Africa Renewable Energy Zones (SEAREZs) This interactive DF map contains locations of high quality
Vehicle Identification Numbering System 00.03
 Vehicle Identification Numbering System 00.03 IMPORTANT: See Subject 050 for the vehicle identification numbering system for vehicles built before May 1, 2000. Federal Motor Vehicle Safety Standard 115
Vehicle Identification Numbering System 00.03 IMPORTANT: See Subject 050 for the vehicle identification numbering system for vehicles built before May 1, 2000. Federal Motor Vehicle Safety Standard 115
Two River Views. A New Website at a New Home. Two River Community Bank Newsletter Fall 2015
 T Rv V T Rv mm Bk Nl Fll 2015 A N W N Hm. W l x mg l l. Dg, l lk m v vg, k ll v l ll kg. Pl, mz ml v, mll z, g mll l. Skg, l. W v l l -vl g l v, g kg m m --g. I, ll l mvg m TRv.Bk. Ol -v gz lgl.k, g l
T Rv V T Rv mm Bk Nl Fll 2015 A N W N Hm. W l x mg l l. Dg, l lk m v vg, k ll v l ll kg. Pl, mz ml v, mll z, g mll l. Skg, l. W v l l -vl g l v, g kg m m --g. I, ll l mvg m TRv.Bk. Ol -v gz lgl.k, g l
Rossmoor Website SEO Tracking Sheet 2012-2014 Updated: April 1, 2014
 As of 5/4/2012 As of 5/14/2012 active senior living no n/a active senior living no n/a adult golf community no n/a adult golf community no n/a 55+ community yes 8th 55+ community yes 8th retirement living
As of 5/4/2012 As of 5/14/2012 active senior living no n/a active senior living no n/a adult golf community no n/a adult golf community no n/a 55+ community yes 8th 55+ community yes 8th retirement living
Stephen du Toit Mathilda du Toit Gerhard Mels Yan Cheng. LISREL for Windows: SIMPLIS Syntax Files
 Stephen du Toit Mathilda du Toit Gerhard Mels Yan Cheng LISREL for Windows: SIMPLIS Files Table of contents SIMPLIS SYNTAX FILES... 1 The structure of the SIMPLIS syntax file... 1 $CLUSTER command... 4
Stephen du Toit Mathilda du Toit Gerhard Mels Yan Cheng LISREL for Windows: SIMPLIS Files Table of contents SIMPLIS SYNTAX FILES... 1 The structure of the SIMPLIS syntax file... 1 $CLUSTER command... 4
Disease gene identification with exome sequencing
 Disease gene identification with exome sequencing Christian Gilissen Dept. of Human Genetics Radboud University Nijmegen Medical Centre [email protected] Contents Infrastructure Exome sequencing
Disease gene identification with exome sequencing Christian Gilissen Dept. of Human Genetics Radboud University Nijmegen Medical Centre [email protected] Contents Infrastructure Exome sequencing
Vom Prototyp zum MVP. Peter Spisak Senior Lead Architect Online and VAS Development. Public Nicht vertraulich
 V Py z VP P Sk S Achc O VAS Dv Pbc Nch vch Py DSCOVER, DSRUPT, DEVER Pbc Nch vch x C DSCOVER, DSRUPT, DEVER 3 Pbc Nch vch DSCOVER, DSRUPT, DEVER 4 C - Dvb C C Pbc Nch vch DSCOVER, DSRUPT, DEVER 5 Dcb wh
V Py z VP P Sk S Achc O VAS Dv Pbc Nch vch Py DSCOVER, DSRUPT, DEVER Pbc Nch vch x C DSCOVER, DSRUPT, DEVER 3 Pbc Nch vch DSCOVER, DSRUPT, DEVER 4 C - Dvb C C Pbc Nch vch DSCOVER, DSRUPT, DEVER 5 Dcb wh
USI Master Policy Information
 Policy I.D. Gender A.B. (#327) M 86 United of Omaha $ 1,000,000.00 A.B. (#430) - (#436) M 86 Metlife $ 2,000,000.00 A.G. #1 (#371), (#610), (#624) M Conseco Life $ 3,125,000.00 10/Apr/10 A.G. #2 (#380),
Policy I.D. Gender A.B. (#327) M 86 United of Omaha $ 1,000,000.00 A.B. (#430) - (#436) M 86 Metlife $ 2,000,000.00 A.G. #1 (#371), (#610), (#624) M Conseco Life $ 3,125,000.00 10/Apr/10 A.G. #2 (#380),
Engineering. Installation Manager. Installations Manager (Gas Qualified) Whole House Assessors. Whole House Assessors (Gas Qualified)
 Apply via Engineering Role title BG12451 Installation Manager BG13779 Installations Manager (Gas Qualified) BG12450 Whole House Assessors BG13931 Whole House Assessors (Gas Qualified) BG12544 BMS Commissioning
Apply via Engineering Role title BG12451 Installation Manager BG13779 Installations Manager (Gas Qualified) BG12450 Whole House Assessors BG13931 Whole House Assessors (Gas Qualified) BG12544 BMS Commissioning
Chem 115 POGIL Worksheet - Week 4 Moles & Stoichiometry
 Chem 115 POGIL Worksheet - Week 4 Moles & Stoichiometry Why? Chemists are concerned with mass relationships in chemical reactions, usually run on a macroscopic scale (grams, kilograms, etc.). To deal with
Chem 115 POGIL Worksheet - Week 4 Moles & Stoichiometry Why? Chemists are concerned with mass relationships in chemical reactions, usually run on a macroscopic scale (grams, kilograms, etc.). To deal with
english parliament of finland
 gh f fd 213 P cvd f h f y f h g 4 Fby 213. E H (Sc Dcc Py) w -cd S, P Rv (N C Py) F Dy S d A Jh (Th F Py) Scd Dy S. Th g c c 5 Fby, wh Pd f h Rbc S Nö d P f h f fwg h c 212. P g dc c 12 Fby h b f P M c.
gh f fd 213 P cvd f h f y f h g 4 Fby 213. E H (Sc Dcc Py) w -cd S, P Rv (N C Py) F Dy S d A Jh (Th F Py) Scd Dy S. Th g c c 5 Fby, wh Pd f h Rbc S Nö d P f h f fwg h c 212. P g dc c 12 Fby h b f P M c.
Tools for human molecular diagnosis. Joris Vermeesch
 Tools for human molecular diagnosis Joris Vermeesch Chromosome > DNA Genetic Code Effect of point mutations/polymorphisms Effect of deletions/insertions Effect of splicing mutations IVS2-2A>G Normal splice
Tools for human molecular diagnosis Joris Vermeesch Chromosome > DNA Genetic Code Effect of point mutations/polymorphisms Effect of deletions/insertions Effect of splicing mutations IVS2-2A>G Normal splice
 G S e r v i c i o C i s c o S m a r t C a r e u ي a d e l L a b o r a t o r i o d e D e m o s t r a c i n R ل p i d a V e r s i n d e l S e r v i c i o C i s c o S m a r t C a r e : 1 4 ع l t i m a A c
G S e r v i c i o C i s c o S m a r t C a r e u ي a d e l L a b o r a t o r i o d e D e m o s t r a c i n R ل p i d a V e r s i n d e l S e r v i c i o C i s c o S m a r t C a r e : 1 4 ع l t i m a A c
Bioinformatics Resources at a Glance
 Bioinformatics Resources at a Glance A Note about FASTA Format There are MANY free bioinformatics tools available online. Bioinformaticists have developed a standard format for nucleotide and protein sequences
Bioinformatics Resources at a Glance A Note about FASTA Format There are MANY free bioinformatics tools available online. Bioinformaticists have developed a standard format for nucleotide and protein sequences
Lecture 3: Mutations
 Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
From Quantum to Matter 2006
 From Quantum to Matter 006 Why such a course? Ronald Griessen Vrije Universiteit, Amsterdam AMOLF, May 4, 004 vrije Universiteit amsterdam Why study quantum mechanics? From Quantum to Matter: The main
From Quantum to Matter 006 Why such a course? Ronald Griessen Vrije Universiteit, Amsterdam AMOLF, May 4, 004 vrije Universiteit amsterdam Why study quantum mechanics? From Quantum to Matter: The main
STAPLES: AFTERMARKET STAPLES CROSS-REFERENCE GUIDE BRAND OEM # MODELS / FINISHERS DESCRIPTION COPYLITE #
 C N O N S: FTERMRKET S CROSS-REFERENCE GUIDE CNON 0251001 CNON F23 5705 000 CNON E 1 CNON G 1 FINISHER D1, FINISHER D2, FINISHER F2 FINISHER G1, FINISHER H2, R E1 R E2, R E3, R F1 R H1, R H2, R J1 R K1,
C N O N S: FTERMRKET S CROSS-REFERENCE GUIDE CNON 0251001 CNON F23 5705 000 CNON E 1 CNON G 1 FINISHER D1, FINISHER D2, FINISHER F2 FINISHER G1, FINISHER H2, R E1 R E2, R E3, R F1 R H1, R H2, R J1 R K1,
A Parents Guide to Understanding. Reading
 A Parents Guide to Understanding Reading Dear Parents, After teaching first grade for many years, I was always faced with the same questions at the beginning of the year: What is Pathways to Reading? How
A Parents Guide to Understanding Reading Dear Parents, After teaching first grade for many years, I was always faced with the same questions at the beginning of the year: What is Pathways to Reading? How
tariff guide EFFECTIVE DATE 1 April 2013
 tariff guide EFFEIVE DATE 1 April 2013 Delivering peace of mind UCH Logistics is a dynamic, customer focused provider of specialist transport services to the airfreight industry. Having been established
tariff guide EFFEIVE DATE 1 April 2013 Delivering peace of mind UCH Logistics is a dynamic, customer focused provider of specialist transport services to the airfreight industry. Having been established
HR DEPARTMENTAL SUFFIX & ORGANIZATION CODES
 HR DEPARTMENTAL SUFFIX & ORGANIZATION CODES Department Suffix Organization Academic Affairs and Dean of Faculty, VP AA 1100 Admissions (Undergraduate) AD 1330 Advanced Ceramics, Colorado Center for--ccac
HR DEPARTMENTAL SUFFIX & ORGANIZATION CODES Department Suffix Organization Academic Affairs and Dean of Faculty, VP AA 1100 Admissions (Undergraduate) AD 1330 Advanced Ceramics, Colorado Center for--ccac
Gene Finding CMSC 423
 Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would you decipher
Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would you decipher
 Application Note: Cisco A S A - Ce r t if ica t e T o S S L V P N Con n e ct ion P r of il e Overview: T h i s a p p l i ca ti o n n o te e x p l a i n s h o w to co n f i g u r e th e A S A to a cco m
Application Note: Cisco A S A - Ce r t if ica t e T o S S L V P N Con n e ct ion P r of il e Overview: T h i s a p p l i ca ti o n n o te e x p l a i n s h o w to co n f i g u r e th e A S A to a cco m
Lecture Outline. Introduction to Databases. Introduction. Data Formats Sample databases How to text search databases. Shifra Ben-Dor Irit Orr
 Introduction to Databases Shifra Ben-Dor Irit Orr Lecture Outline Introduction Data and Database types Database components Data Formats Sample databases How to text search databases What units of information
Introduction to Databases Shifra Ben-Dor Irit Orr Lecture Outline Introduction Data and Database types Database components Data Formats Sample databases How to text search databases What units of information
QCC Human Resources Master Files Manual
 Quintessential School Systems QCC Human Resources Master Files Manual Quintessential School Systems (QSS), 2010 All Rights Reserved 867 American Street, 5Seconf Floor --- San Carlos, CA 94070 --- Voice
Quintessential School Systems QCC Human Resources Master Files Manual Quintessential School Systems (QSS), 2010 All Rights Reserved 867 American Street, 5Seconf Floor --- San Carlos, CA 94070 --- Voice
Government of the District of Columbia
 Paul Quer Deputy Mayor for Public Safety Justice Beatriz Otero Deputy Mayor for Health Human Vincent C. Gray Mayor Allen Y. Lew City Administrator Victor L. Hoskins Deputy Mayor for Planning Economic Development
Paul Quer Deputy Mayor for Public Safety Justice Beatriz Otero Deputy Mayor for Health Human Vincent C. Gray Mayor Allen Y. Lew City Administrator Victor L. Hoskins Deputy Mayor for Planning Economic Development
Montessori Academy of Owasso
 Montessori Academy of Owasso 5 & 6 Year-Old Curriculum Academic Area: Language Arts Category: Reading: Literature Subcategory: Key Ideas and Details Element 1:With prompting and support, ask and answer
Montessori Academy of Owasso 5 & 6 Year-Old Curriculum Academic Area: Language Arts Category: Reading: Literature Subcategory: Key Ideas and Details Element 1:With prompting and support, ask and answer
Form: Parental Consent for Blood Donation
 A R C Wt, C 20006 Ptl Ct f B i Ifi T f t y t ll f i y tl t q y t l A R C ly. Pl ll 1-800-RE-CROSS (1-800-733-2767) v. if y v q r t t i I iv t f yr,, t, y v t t: 1. Y y t t l i ly, 2. Y y t t t l i ( k
A R C Wt, C 20006 Ptl Ct f B i Ifi T f t y t ll f i y tl t q y t l A R C ly. Pl ll 1-800-RE-CROSS (1-800-733-2767) v. if y v q r t t i I iv t f yr,, t, y v t t: 1. Y y t t l i ly, 2. Y y t t t l i ( k
DNA Sequence Alignment Analysis
 Analysis of DNA sequence data p. 1 Analysis of DNA sequence data using MEGA and DNAsp. Analysis of two genes from the X and Y chromosomes of plant species from the genus Silene The first two computer classes
Analysis of DNA sequence data p. 1 Analysis of DNA sequence data using MEGA and DNAsp. Analysis of two genes from the X and Y chromosomes of plant species from the genus Silene The first two computer classes
Indiana Health Coverage Programs
 Indiana Health Coverage Programs Standard Companion Guide Transaction Information Instructions related to Transactions based on ASC X12 Implementation Guides, version 005010 Group Premium Payment for Insurance
Indiana Health Coverage Programs Standard Companion Guide Transaction Information Instructions related to Transactions based on ASC X12 Implementation Guides, version 005010 Group Premium Payment for Insurance
DATING YOUR GUILD 1952-1960
 DATING YOUR GUILD 1952-1960 YEAR APPROXIMATE LAST SERIAL NUMBER PRODUCED 1953 1000-1500 1954 1500-2200 1955 2200-3000 1956 3000-4000 1957 4000-5700 1958 5700-8300 1959 12035 1960-1969 This chart displays
DATING YOUR GUILD 1952-1960 YEAR APPROXIMATE LAST SERIAL NUMBER PRODUCED 1953 1000-1500 1954 1500-2200 1955 2200-3000 1956 3000-4000 1957 4000-5700 1958 5700-8300 1959 12035 1960-1969 This chart displays
Opis przedmiotu zamówienia - zakres czynności Usługi sprzątania obiektów Gdyńskiego Centrum Sportu
 O p i s p r z e d m i o t u z a m ó w i e n i a - z a k r e s c z y n n o c i f U s ł u i s p r z» t a n i a o b i e k t ó w G d y s k i e C eo n t r u m S p o r t us I S t a d i o n p i ł k a r s k i
O p i s p r z e d m i o t u z a m ó w i e n i a - z a k r e s c z y n n o c i f U s ł u i s p r z» t a n i a o b i e k t ó w G d y s k i e C eo n t r u m S p o r t us I S t a d i o n p i ł k a r s k i
Sickle cell anemia: Altered beta chain Single AA change (#6 Glu to Val) Consequence: Protein polymerizes Change in RBC shape ---> phenotypes
 Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
 2 1k 0 3k 2 0 1 4 S 5 7 P a s t w a c z ł o n k o w s k i e - Z a m ó w i e n i e p u b l i c z n e n a u s ł u g- i O g ł o s z e n i e o d o b r o w o l n e j p r z e j r z y s t o c i e x - a nnt e
2 1k 0 3k 2 0 1 4 S 5 7 P a s t w a c z ł o n k o w s k i e - Z a m ó w i e n i e p u b l i c z n e n a u s ł u g- i O g ł o s z e n i e o d o b r o w o l n e j p r z e j r z y s t o c i e x - a nnt e
H ig h L e v e l O v e r v iew. S te p h a n M a rt in. S e n io r S y s te m A rc h i te ct
 H ig h L e v e l O v e r v iew S te p h a n M a rt in S e n io r S y s te m A rc h i te ct OPEN XCHANGE Architecture Overview A ge nda D es ig n G o als A rc h i te ct u re O ve rv i ew S c a l a b ili
H ig h L e v e l O v e r v iew S te p h a n M a rt in S e n io r S y s te m A rc h i te ct OPEN XCHANGE Architecture Overview A ge nda D es ig n G o als A rc h i te ct u re O ve rv i ew S c a l a b ili
english parliament of finland
 gh f fd 2012 P cvd f h f y f h g Mdy, 6 Fby 2012. A h d MP, K T hd h ch h c f h S. E H (Sc Dcc Py) w -cd S, P Rv (N C Py) F Dy S d A Jh (F Py) Scd Dy S. Th g c c Tdy, 7 Fby, whch Pd f h Rbc Tj H d P f
gh f fd 2012 P cvd f h f y f h g Mdy, 6 Fby 2012. A h d MP, K T hd h ch h c f h S. E H (Sc Dcc Py) w -cd S, P Rv (N C Py) F Dy S d A Jh (F Py) Scd Dy S. Th g c c Tdy, 7 Fby, whch Pd f h Rbc Tj H d P f
d e f i n i c j i p o s t a w y, z w i z a n e j e s t t o m. i n. z t y m, i p o jі c i e t o
 P o s t a w y s p o і e c z e t s t w a w o b e c o s у b n i e p e і n o s p r a w n y c h z e s z c z e g у l n y m u w z g lb d n i e n i e m o s у b z z e s p o і e m D o w n a T h e a t t i t uodf
P o s t a w y s p o і e c z e t s t w a w o b e c o s у b n i e p e і n o s p r a w n y c h z e s z c z e g у l n y m u w z g lb d n i e n i e m o s у b z z e s p o і e m D o w n a T h e a t t i t uodf
Cikkszám Termék neve Fogyasztói bruttó árak
 Evobike 2014 Mounty termékcsalád Cikkszám Termék neve Fogyasztói bruttó árak 0713 MOUNTY LITE-PEDAL 10598 0713COMP MOUNTY COMP-PEDAL SCHWARZ/SILBER 16568 0713DH 0713DH nicht mehr lieferbar/11 13251 0713FREE
Evobike 2014 Mounty termékcsalád Cikkszám Termék neve Fogyasztói bruttó árak 0713 MOUNTY LITE-PEDAL 10598 0713COMP MOUNTY COMP-PEDAL SCHWARZ/SILBER 16568 0713DH 0713DH nicht mehr lieferbar/11 13251 0713FREE
Umm AL Qura University MUTATIONS. Dr Neda M Bogari
 Umm AL Qura University MUTATIONS Dr Neda M Bogari CONTACTS www.bogari.net http://web.me.com/bogari/bogari.net/ From DNA to Mutations MUTATION Definition: Permanent change in nucleotide sequence. It can
Umm AL Qura University MUTATIONS Dr Neda M Bogari CONTACTS www.bogari.net http://web.me.com/bogari/bogari.net/ From DNA to Mutations MUTATION Definition: Permanent change in nucleotide sequence. It can
DWC FORM-152 (Application for Attorney's Fees)
 DWC FORM-152 (Application for Attorney's Fees) The Division is authorized by the Texas Workers' Compensation Act, Texas Labor Code, Sections 408.221 and 408.222 to approve the fees paid to injured employees'
DWC FORM-152 (Application for Attorney's Fees) The Division is authorized by the Texas Workers' Compensation Act, Texas Labor Code, Sections 408.221 and 408.222 to approve the fees paid to injured employees'
Lung Cancer Research: From Prevention to Cure!
 Lung Cancer Research: From Prevention to Cure! Ravi Salgia, M.D, Ph.D Associate Professor of Medicine Director, Thoracic Oncology Research Program Department of Medicine Section of Hematology/Oncology
Lung Cancer Research: From Prevention to Cure! Ravi Salgia, M.D, Ph.D Associate Professor of Medicine Director, Thoracic Oncology Research Program Department of Medicine Section of Hematology/Oncology
BCD (ASCII) Arithmetic. Where and Why is BCD used? Packed BCD, ASCII, Unpacked BCD. BCD Adjustment Instructions AAA. Example
 BCD (ASCII) Arithmetic We will first look at unpacked BCD which means strings that look like '4567'. Bytes then look like 34h 35h 36h 37h OR: 04h 05h 06h 07h x86 processors also have instructions for packed
BCD (ASCII) Arithmetic We will first look at unpacked BCD which means strings that look like '4567'. Bytes then look like 34h 35h 36h 37h OR: 04h 05h 06h 07h x86 processors also have instructions for packed
Put the human back in Human Resources.
 Put the human back in Human Resources A Co m p l et e Hu m a n Ca p i t a l Ma n a g em en t So l u t i o n t h a t em p o w er s HR p r o f essi o n a l s t o m eet t h ei r co r p o r a t e o b j ect
Put the human back in Human Resources A Co m p l et e Hu m a n Ca p i t a l Ma n a g em en t So l u t i o n t h a t em p o w er s HR p r o f essi o n a l s t o m eet t h ei r co r p o r a t e o b j ect
RETRIEVING SEQUENCE INFORMATION. Nucleotide sequence databases. Database search. Sequence alignment and comparison
 RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
Payor Sheet for Medicare Part D/ PDP and MA-PD
 Payor Specification Sheet for MEDICARE PART D/PDP AND MA-PD PRIME THERAPEUTICS LLC CLIENTS JANUARY 1, 2006 (Page 1 of 8) BIN: PCN: See BINs on page 2 (in bold red type) See PCNs on page 2 (in bold red
Payor Specification Sheet for MEDICARE PART D/PDP AND MA-PD PRIME THERAPEUTICS LLC CLIENTS JANUARY 1, 2006 (Page 1 of 8) BIN: PCN: See BINs on page 2 (in bold red type) See PCNs on page 2 (in bold red
Future Trends in Airline Pricing, Yield. March 13, 2013
 Future Trends in Airline Pricing, Yield Management, &AncillaryFees March 13, 2013 THE OPPORTUNITY IS NOW FOR CORPORATE TRAVEL MANAGEMENT BUT FIRST: YOU HAVE TO KNOCK DOWN BARRIERS! but it won t hurt much!
Future Trends in Airline Pricing, Yield Management, &AncillaryFees March 13, 2013 THE OPPORTUNITY IS NOW FOR CORPORATE TRAVEL MANAGEMENT BUT FIRST: YOU HAVE TO KNOCK DOWN BARRIERS! but it won t hurt much!
Breast cancer and the role of low penetrance alleles: a focus on ATM gene
 Modena 18-19 novembre 2010 Breast cancer and the role of low penetrance alleles: a focus on ATM gene Dr. Laura La Paglia Breast Cancer genetic Other BC susceptibility genes TP53 PTEN STK11 CHEK2 BRCA1
Modena 18-19 novembre 2010 Breast cancer and the role of low penetrance alleles: a focus on ATM gene Dr. Laura La Paglia Breast Cancer genetic Other BC susceptibility genes TP53 PTEN STK11 CHEK2 BRCA1
How To Pay For An Ambulance Ride
 Chapter 9Ambulance 9 9.1 Enrollment........................................................ 9-2 9.2 Emergency Ground Ambulance Transportation.............................. 9-2 9.2.1 Benefits, Limitations,
Chapter 9Ambulance 9 9.1 Enrollment........................................................ 9-2 9.2 Emergency Ground Ambulance Transportation.............................. 9-2 9.2.1 Benefits, Limitations,
Genetic Testing in the Long QT Syndrome
 ORIGINAL CONTRIBUTION Genetic Testing in the Long QT Syndrome Development and Validation of an Efficient Approach to Genotyping in Clinical Practice Carlo Napolitano, MD, PhD Silvia G. Priori, MD, PhD
ORIGINAL CONTRIBUTION Genetic Testing in the Long QT Syndrome Development and Validation of an Efficient Approach to Genotyping in Clinical Practice Carlo Napolitano, MD, PhD Silvia G. Priori, MD, PhD
Overview of Eukaryotic Gene Prediction
 Overview of Eukaryotic Gene Prediction CBB 231 / COMPSCI 261 W.H. Majoros What is DNA? Nucleus Chromosome Telomere Centromere Cell Telomere base pairs histones DNA (double helix) DNA is a Double Helix
Overview of Eukaryotic Gene Prediction CBB 231 / COMPSCI 261 W.H. Majoros What is DNA? Nucleus Chromosome Telomere Centromere Cell Telomere base pairs histones DNA (double helix) DNA is a Double Helix
 Cisco Security Agent (CSA) CSA je v í c eúčelo v ý s o f t w a r o v ý ná s t r o j, k t er ý lze p o už í t k v ynuc ení r ů zný c h b ezp ečno s t ní c h p o li t i k. CSA a na lyzuje c h o v á ní a
Cisco Security Agent (CSA) CSA je v í c eúčelo v ý s o f t w a r o v ý ná s t r o j, k t er ý lze p o už í t k v ynuc ení r ů zný c h b ezp ečno s t ní c h p o li t i k. CSA a na lyzuje c h o v á ní a
Introduction to Genome Annotation
 Introduction to Genome Annotation AGCGTGGTAGCGCGAGTTTGCGAGCTAGCTAGGCTCCGGATGCGA CCAGCTTTGATAGATGAATATAGTGTGCGCGACTAGCTGTGTGTT GAATATATAGTGTGTCTCTCGATATGTAGTCTGGATCTAGTGTTG GTGTAGATGGAGATCGCGTAGCGTGGTAGCGCGAGTTTGCGAGCT
Introduction to Genome Annotation AGCGTGGTAGCGCGAGTTTGCGAGCTAGCTAGGCTCCGGATGCGA CCAGCTTTGATAGATGAATATAGTGTGCGCGACTAGCTGTGTGTT GAATATATAGTGTGTCTCTCGATATGTAGTCTGGATCTAGTGTTG GTGTAGATGGAGATCGCGTAGCGTGGTAGCGCGAGTTTGCGAGCT
