How To Understand The Evolutionary Relationship Between Emperor Penguins And Other Species
|
|
|
- Kelly King
- 5 years ago
- Views:
Transcription
1 Biology 18 Spring 2008 Lab 2 - Illustrating Evolutionary Relationships Between Organisms: Emperor Penguins and Phylogenetic Trees Pre-Lab Reference Reading: Review pp and pp in Life by Sadava et al., 8 th edition, I. Introduction The ability to think broadly about the living world is rooted in an understanding of life's diversity. However, the large number of ways in which various organisms differ from one another can often mask their similarities. The diversity and unity of living organisms would seem to be contradictory concepts. Recall, though, that these were the major themes in Darwin's great work, in which he asked and answered the following question. Since life is so diverse, are there any unifying themes at all? By looking backwards in time, Darwin showed that the answer to this question is a resounding Yes! Darwin proposed both a pattern invoking common descent (as the basis for the unity of life) and a mechanism to explain how the relationships between species became fragmented over time (resulting in diversity). In addition, Darwin himself is responsible for the metaphor that is used today to illustrate this shared descent of living organisms: the Tree of Life (TOL). In fact, the diagram shown below was the only figure from his book On the Origin of Species by Natural Selection. Time...great tree of life...with its ever-branching and beautiful ramifications... (C. Darwin, 1859).
2 The ATOL (Assembling the Tree of Life) Project represents a collective effort of biologists working on different organisms to understand how the diversity of life fits together. Resolving evolutionary relationships that underlie the TOL is unquestionably one of the most important problems in biology today, for two major reasons. Because it is relevant to all organisms, with the tree potentially encompassing the 1.75 million living species characterized to date and an estimated tens of millions more to be discovered (not to mention the even larger number of species that existed and are now extinct). There are major applied consequences of understanding the evolutionary relationships between organisms with respect to human health and the environment. These include the origins and dynamics of disease-causing microbes, or the interactions between species that constitute a rich and self-sustaining ecosystem. Reconstructing an evolutionary tree (i.e., a phylogeny) is a powerful way to answer questions about the evolutionary relatedness of living organisms. In addition, modern tools of molecular biology are greatly advancing our analytical power, particularly in the use of DNA and protein sequence data to investigate the similarity/divergence between species. II. Overview of Activity Millions of people around the world have been captivated by the 2005 documentary movie The March of the Penguins. In this movie, viewers learn about the complex reproductive behaviors of Emperor penguins, which include a 70-mile march (by walking or belly gliding) to an inland breeding ground. (See Appendix for life cycle.) Strong selective pressures have likely been in force to establish reproductive behaviors that differ so greatly between Emperor penguins and other birds, thereby reinforcing the evolutionary independence and integrity of related species. A number of interesting biological questions come to mind while watching The March of the Penguins. For example, 1) What is the closest living relative of Emperor penguins? 2) Are all penguins living today the descendants of the same common ancestor? 3) If so, to which taxonomic group of birds are penguins most closely related? 4) What would the common ancestor of penguins and their closest bird relatives have looked like? 5) What is the evolutionary relationship between penguins (and birds in general) to other classes of vertebrates? To address these and other questions, students will use the National Center for Biotechnology Information (NCBI) Website to reconstruct a phylogeny, which will illustrate the evolutionary relationships between Emperor penguins and other living organisms. Students will use gene sequence databases to do this, and DNA sequence comparisons will be done behind the scenes using NCBI s BLAST search engine. BLAST stands for Basic Local Alignment Search Tool, and the program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches ( 2
3 A comparison of DNA or protein sequences from different species can reveal evolutionary relationships. This assumption is based on the idea that the nucleotide or amino acid sequence of a particular gene or the protein it encodes, respectively, mutates at a similar rate in different individuals. However, students should remember that the rate of mutation alone does not determine how fast a DNA sequence will change. The effects of mutation on survival and reproductive success (natural selection), as well as the generation time of the organism, also play important roles in the overall rate of genetic change in a population. To sum up, the overall goal of this lab is for students to examine the evidence held within genetic sequences to determine how living organisms are related to one another and to confirm that they share common ancestors. You will do so by using BLAST to measure the degree of gene sequence similarity between various types of organisms. Each pair of students should go through the exercises below with Emperor penguins, using the worksheet on pp to record your answers to questions (in bold) asked in the text. The completed worksheet is due at the beginning of lab next week. 3
4 III. Procedure 1. Go to the NCBI Home Page at This page contains links to dozens of different databases and other resources. Every page that is linked to the main NCBI page has a small icon somewhere in the upper-left corner of the screen. Clicking on this icon will immediately return the computer to the NCBI home page. 2. We will build a phylogenetic tree by using BLAST to compare the degree of sequence similarity in a gene between different individuals. Generally, the more divergent gene sequences are, the more time and evolutionary distance separate the two species. 3. To begin our analysis, we would ideally select a gene that is present within the genome of all the organisms that we wish to compare. Think about the shared characteristics of different groups of organisms and the gene products that may contribute to these characteristics. What types of genes would you expect to be present in all organisms? In all eukaryotes? In all plants (but not in animals or fungi)? In all animals (but not in plants or fungi)? In all vertebrates (but not in invertebrates)? 4
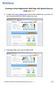
5 4. To identify possible gene candidates for the Emperor penguin analysis, type Emperor penguin in the for search box, and click on the Go button (as above). 5. A screen with all the NCBI-managed databases will appear next (partial screen shot below). Any database that contains Emperor penguin entries will have a number (representing the number of different entries in that database) in the small box to the left of the database name. 6. Click on the 10 in the box to the left of the CoreNucleotide database (highlighted in blue above). 7. The DNA sequence search results are listed on the next screen (partial screen shot on next page). Think about your answers to Question 3 above. Which of the ten gene entries, if selected, would enable you to build the most inclusive tree (i.e. containing the greatest number of organisms)? Why? Which entry would result in the least inclusive tree? Why? 5
6 8. We will build our first tree using a mitochondrial cytochrome gene. Scroll down to Entry 3 for the cytochrome b gene and click on the accession number DQ (highlighted in yellow above) to find out more about this gene. 9. The next screen (partial screenshot on the next page) contains a lot of useful information. First, note that the scientific name of Emperor penguins is Aptenodytes forsteri. Second, the classification hierarchy of Emperor penguins is listed below the ORGANISM name. Third, reference information for the paper in which this gene sequence was first published is next, followed by specific sequence information for the gene. Note that we will build our tree using a partial gene sequence of 1008 nucleotide base pairs (bp), which codes for a polypeptide chain of 336 amino acids. 6
7 10. If you click on the Aptenodytes forsteri link in the above screen, a new window opens that shows a more detailed taxonomic classification of that species. Also, a box will appear with the taxonomic rank (if applicable) of each entry in the lineage as one mouses over the entry. What is the class to which all birds belong? 11. Based on their morphological and behavioral characteristics, penguins have been grouped into a single family. What is the name of this family? Click on the family name, to see a taxonomic listing of the six genera of penguin species. Note: Print out this page or jot down the six genus names, as you will need this list later in the exercise. 12. Standard hierarchies such as this are based largely upon physical, easy-to-see characteristics. Phylogenies based on molecular sequence data often are consistent with the taxonomic classification system. However, as you will see shortly, there can be surprises with the molecular data. Thus, factors other than common descent must be responsible for why some organisms look very similar to one another. 7
8 13. To build our tree, we will now use BLAST to compare the cytochrome b gene from Emperor penguins to the same gene in a large number of other organisms. Click on the NCBI icon to go back to the NCBI homepage. 14. Type or paste the accession number for entry #2 (DQ137225) in the for search box and click on BLAST in the menu bar (purple highlight above). A screen that gives BLAST search options appears next, part of which is shown below. 15. Click on the nucleotide blast link following Choose a BLAST program to run in the Basic BLAST window (yellow highlight above). 8
9 16. In the BLASTN window that next appears (screenshot below), type the DQ accession number in the Enter Query Sequence box and select Others (nr etc.): next to Database in the Choose Search Set box. Note that Optimize for Highly similar sequences (megablast) is the default feature. Now, check the Show results in new window box, click on the BLAST button and prepare to wait! 17. The next window will tell you whether or not your request has been successfully submitted, and, if so, approximately how much time you will have to wait to get your results. The results will automatically appear in this same window once the BLAST search is completed. 9
10 18. The BLAST results (partial screenshot above) show the 100 best sequence matches (e.g., BLAST Hits) to the cytochrome b gene from the Emperor penguin. The results are in graphical form first, with the extent of sequence overlap shown as red bars. If you mouse over the first red bar, you will see that it is our query entry (e.g., the cytochrome b gene of Aptenodytes forsteri). 19. Now scroll down the screen to below the graphical output, to examine the table with the species names and BLAST scores of these 100 hits (see next partial screen shot). The species most closely-related to Emperor penguins are at the top of the table and have the highest scores. 10
11 20. Note that some species appear multiple times in the above list (due to multiple sequence database entries for the same gene and organism). Thus, the number of unique species in our data set is actually less than 100 species. 21. Now for the fun part! We will have BLAST reconstruct a phylogenetic tree to more easily see the evolutionary relationships between these 100 gene sequences (and the species of organisms from which the genes were sequenced!). To do so, simply click on the Distance tree of results link, which is between the graph and the table. 22. The top part of the resultant phylogeny is on the next page (you will have to use the arrow buttons to scroll down to see the entire tree). The entry for the gene from our query species (Aptenodytes forsteri) is highlighted in yellow near the bottom of the window. 23. In a molecular phylogeny, the closer two particular entries are among the branches of the tree, the more similar their DNA sequences are and, we assume, the shorter the evolutionary distance between the species from which the genes were obtained. Conversely, two species with a greater distance between their branches have less similar DNA sequences and are assumed to be less closely-related. 11
12 24. By default, the Sequence Title of each of the 100 hits is indicated on the above tree. This view is interesting, since it shows which gene sequences were used to build the tree, and you should spend a few minutes looking at the tree in this format. 25. For the rest of this exercise, you can simplify the tree to see just the species names by selecting Taxonomic Name (if available) in the drop-down menu under Sequence Label (highlighted in the screen shot above). 26. At this point, you are probably wondering what all the different animals are. By selecting BLAST Name (if available) in the same drop-down menu, you can find out what general class of organism each entry is. To which class of organisms do all the members of this phylogeny belong? (Note: one hint that they are all in the same class is that only birds appears in the BLAST names color map legend and each entry on the tree is preceded by the same blue circle.) 27. On the next page is a screen shot of the bottom half of the tree with Taxonomic Names selected. Using this tree you can now answer the first question on p. 2 of this exercise. What are the names (scientific and common) of the closest living relative of Emperor penguins? Note: make sure that you use the live Web site and not the printed screenshot to answer this and the following questions, as the tree may have changed since the lab manual was printed! 28. Does this tree support the standard NCBI taxonomy that you looked at earlier, which indicates that all penguins living today are in the same family (and thus share the same most-recent common ancestor)? Why or why not? 29. The standard NCBI taxonomy does not indicate the evolutionary relatedness amongst the different genera of penguins. What does your BLAST tree tell you about the evolutionary relationship of the genus Aptenodytes to the other penguin genera? Of the non-aptenodytes penguin genera to each other? 12
13 30. From the BLAST tree, what are the scientific and common names of the closest non-penguin relative(s) of the Emperor penguins? What do you think the common ancestor of Emperor penguins and the closest bird relative(s) would have looked like? (To obtain more information about the species and the families to which they belong, start by opening the Taxonomy Browser window of NCBI and typing the species name into the Search box. There are also many Web sites with detailed information, photographs, etc. of most living organisms, as well as reference bird atlases on the front table to flip through.) 31. Currently, there are conflicting interpretations of sequence data from the bird lineages in the scientific literature. Links to two such recently published papers are posted in the Laboratory Material area on Blackboard. See Figure 2 in the Van Tuinen et al., 2001 paper and Figs. 1 & 2 in the Watanabe et al., 2006 paper. What type of birds does each paper suggest are the closest living relatives to penguins? Does your BLAST tree agree with either of these trees? If not, which of the three trees do you think makes the most sense? Why? 32. To answer question 5 on page 2, you will need to build another tree that includes more distantly-related species. To do this, return to the BLAST Home page, open a new nucleotide BLAST window, type the DQ accession number (for the cytochrome b gene of A. forsteri) into the Enter Query Sequence box, and select 13
14 the button next to the Others (nr etc.): database. This time, however, select the Reference genomic sequences (refseq_genomic) database using the drop-down menu under the line of database selections. 33. Refseq refers to sequences that have been double checked and verified by a person at NCBI. Since there are fewer of these sequences in the database, this tree will include more distantly-related organisms (e.g., non-birds). Next, scroll down to the Program Selection box in the same window and next to Optimize for, select More dissimilar sequences (discontiguous megablast). Finally, click on Algorithm parameters, scroll down to the General Parameters box and select 250 next to Max target sequences. Now, BLAST away! 34. Repeat steps #17-26 above to analyze the tree created with this search. Once again, you will have to scroll down to find the query species A. forsteri. Examine the differences between this tree and the first phylogeny. What classes of organisms are represented in the second tree? Is there anything in common about all the organisms in the second phylogeny? Is there anything surprising about the taxonomic relationships in the second tree? 35. What do you think the common ancestor of all the animals in this tree looked like? Where do you think this common ancestor lived, how did it move, etc.? Be specific! 14
15 IV. Extension Exercises - The Circular Tree of Life 1. The Circular Tree of Life (Science 300: , 2003) is an alternative depiction of sequence relationships between extant species. This tree was created by David Hillis, Derrick Zwickl and Robin Gutell at the University of Texas, and their description follows. 2. About this Tree: This tree is from an analysis of small subunit rrna sequences sampled from about 3,000 species from throughout the Tree of Life. The species were chosen based on their availability, but we attempted to include most of the major groups, sampled very roughly in proportion to the number of known species in each group (although many groups remain over- or under-represented). The number of species represented is approximately the square-root of the number of species thought to exist on Earth (i.e., three thousand out of an estimated nine million species), or about 0.18% of the 1.7 million species that have been formally described and named. 3. The circular format of this phylogeny has some great advantages. First, one can easily visualize the relationships between thousands of species. Also, this format suppresses hierarchical thinking (such as the placement of humans at the top or far right of a tree), while at the same time emphasizing that all living species trace back to a common ancestor. 4. You can download the tree directly by going to the following link: 5. Using the Circular Tree of Life a. How is time represented in this tree? Where would extinct species lie on this tree? Where on the map is the last common ancestor of all living organisms? b. Using a meter stick and the wall poster of the Circular Tree, measure the distance to the center of the tree from the perimeter. What is this distance (in cm)? c. To see if a particular species is on the Circular Tree of Life, search (under the Edit tab in the menu bar) the pdf of this diagram, a link to which is posted on 15
16 Blackboard. Note: you will need to search for the scientific names of species of interest. You can instead randomly pick species from the perimeter of the tree. d. Once you have confirmed that a species is on the Circular Tree (and approximately which quadrant of the circle s circumference the species is in), examine the poster on display in lab to find the exact location of each species. e. Carefully mark each species with a sticky arrow on the wall poster. f. Which two species are you comparing? What is the distance (in cm) to the common ancestor of these two species from the perimeter? What percent is this distance relative to the distance to the center (which you measured for part b)? g. How many branch points are there from the common ancestor to each of the two species? If the number of branch points is different for the two, what is one possible explanation for this? h. Now, keep one of your two species the same but compare it to a third species. What is the distance in cm to the common ancestor of this new pair of species from the perimeter? What percent is this distance relative to the distance to the center (which you measured for part b)? Why is this distance different than the distance you measured for part (f)? i. Where on the map is the common ancestor of animals, plants and fungi? What might the common ancestor of these three groups of organisms have looked like? 16
17 Student Worksheet for Penguin Phylogeny Names Question III.3. What types of genes would you expect to be present in all organisms? In all eukaryotes? In all plants (but not in animals or fungi)? In all animals (but not in plants or fungi)? In all vertebrates (but not in invertebrates)? Question III.7 Which of the ten gene entries, if selected, would enable you to build the most inclusive tree (i.e. containing the greatest number of organisms)? Why? Which entry would result in the least inclusive tree? Why? Question III.10 What is the class to which all birds belong? Question III.11 What is the scientific name of the family to which all penguins belong? Question III.26 To which class of organisms do all the members of this phylogeny belong? 17
18 Question III.27 What are the names (scientific and common) of the closest living relative of Emperor penguins? Question III.28 Does this tree support the standard NCBI taxonomy that you looked at earlier, which indicates that all penguins living today are in the same family (and thus share the same, most-recent common ancestor)? Why or why not? Question III.29 What does your BLAST tree tell you about the evolutionary relationship of the genus Aptenodytes to the other penguin genera? Of the non-aptenodytes penguin genera to each other? Question III.30 From the BLAST tree, what are the scientific and common names of the closest non-penguin relative(s) of the Emperor penguins? What do you think the common ancestor of Emperor penguins and the closest bird relative(s) would have looked like? Question III.31 What type of birds does each paper suggest are the closest living relatives to penguins? 18
19 Does your BLAST tree agree with either of these trees? If not, which of the three trees do you think makes the most sense? Why? Question III.33 What classes of organisms are represented in the second tree? Is there anything in common about all the organisms in the second phylogeny? Is there anything surprising about the taxonomic relationships in the second tree? Question III.34 What do you think the common ancestor of all the animals in this tree looked like? Where do you think this common ancestor lived, how did it move, etc.? Be specific! 19
20 Question IV.5 a. How is time represented in this tree? Where would extinct species lie on this tree? Where on the map is the last common ancestor of all living organisms? b. What is this distance (in cm) to the center of the tree from the perimeter? f. Which two species are you comparing? What is the distance (in cm) to the common ancestor of these two species from the perimeter? What percent is this distance relative to the distance to the center (which you measured for part b)? g. How many branch points are there from the common ancestor to each of the two species? If the number of branch points is different for the two, what is one possible explanation for this? 20
21 h. Now, keep one of your two species the same but compare it to a third species. What is the distance in cm to the common ancestor of this new pair of species from the perimeter? What percent is this distance relative to the distance to the center (which you measured for part b)? Why is this distance different than the distance you measured for part (f)? i. Where on the map is the common ancestor of animals, plants and fungi? What might the common ancestor of these three groups of organisms have looked like? 21
22 Appendix: Emperor Penguin Life Cycle (as described in the 2005 documentary The March of the Penguins) 1. March (summer over): both sexes walk to inland breeding area (up to 70 miles away) 2. By May, courting and mating over. Days cold and dark - huddle together to stay warm 3. Eggs laid early June: transferred from Mom to Dad 4. Moms return to sea to feed (more ice/ longer journey), and Dads continue incubating eggs for > 2 months (go >125 days without food by end) 5. July: - Moms walk back to breeding ground - Chicks start hatching - Dads have a little food in throat pouch that can sustain chicks for 1-2 days 6. Mothers back: - Recognize mates by song - Chicks passed to Moms, regurgitate food for chick - Chick sings to Dad so he can recognize chick later - Dads walk back to sea - Chicks with Moms, chicks walking within a few days 7. Late August: - Moms leave chicks to feed in sea - Fathers return, recognize chicks by song, regurgitate food for chicks 8. Over next 2-3 months, parents take turns going to/from sea for food for themselves and chicks (shorter distances as ice melts & cracks w/ warmer summer temperatures); sometimes whole family together at breeding ground 9. November: Females and males have spent 9 mos. as a couple, now part ways & return to sea until following March (when will likely mate w/ someone else) 10. A few weeks later, chicks into the sea (which is very close by); live at sea for four years, then start at #1! 22
Analyzing A DNA Sequence Chromatogram
 LESSON 9 HANDOUT Analyzing A DNA Sequence Chromatogram Student Researcher Background: DNA Analysis and FinchTV DNA sequence data can be used to answer many types of questions. Because DNA sequences differ
LESSON 9 HANDOUT Analyzing A DNA Sequence Chromatogram Student Researcher Background: DNA Analysis and FinchTV DNA sequence data can be used to answer many types of questions. Because DNA sequences differ
Section 3 Comparative Genomics and Phylogenetics
 Section 3 Section 3 Comparative enomics and Phylogenetics At the end of this section you should be able to: Describe what is meant by DNA sequencing. Explain what is meant by Bioinformatics and Comparative
Section 3 Section 3 Comparative enomics and Phylogenetics At the end of this section you should be able to: Describe what is meant by DNA sequencing. Explain what is meant by Bioinformatics and Comparative
RETRIEVING SEQUENCE INFORMATION. Nucleotide sequence databases. Database search. Sequence alignment and comparison
 RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
Name Class Date. binomial nomenclature. MAIN IDEA: Linnaeus developed the scientific naming system still used today.
 Section 1: The Linnaean System of Classification 17.1 Reading Guide KEY CONCEPT Organisms can be classified based on physical similarities. VOCABULARY taxonomy taxon binomial nomenclature genus MAIN IDEA:
Section 1: The Linnaean System of Classification 17.1 Reading Guide KEY CONCEPT Organisms can be classified based on physical similarities. VOCABULARY taxonomy taxon binomial nomenclature genus MAIN IDEA:
COMPARING DNA SEQUENCES TO DETERMINE EVOLUTIONARY RELATIONSHIPS AMONG MOLLUSKS
 COMPARING DNA SEQUENCES TO DETERMINE EVOLUTIONARY RELATIONSHIPS AMONG MOLLUSKS OVERVIEW In the online activity Biodiversity and Evolutionary Trees: An Activity on Biological Classification, you generated
COMPARING DNA SEQUENCES TO DETERMINE EVOLUTIONARY RELATIONSHIPS AMONG MOLLUSKS OVERVIEW In the online activity Biodiversity and Evolutionary Trees: An Activity on Biological Classification, you generated
Algorithms in Computational Biology (236522) spring 2007 Lecture #1
 Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
MARCH OF THE PENGUINS
 MARCH OF THE PENGUINS S C I E N C E M O V I E N I G H T The Academic Support Center @ Daytona State College (Science 127 Page 1 of 33) The Academic Support Center @ Daytona State College (Science 127 Page
MARCH OF THE PENGUINS S C I E N C E M O V I E N I G H T The Academic Support Center @ Daytona State College (Science 127 Page 1 of 33) The Academic Support Center @ Daytona State College (Science 127 Page
SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE
 AP Biology Date SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE LEARNING OBJECTIVES Students will gain an appreciation of the physical effects of sickle cell anemia, its prevalence in the population,
AP Biology Date SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE LEARNING OBJECTIVES Students will gain an appreciation of the physical effects of sickle cell anemia, its prevalence in the population,
Bioinformatics Resources at a Glance
 Bioinformatics Resources at a Glance A Note about FASTA Format There are MANY free bioinformatics tools available online. Bioinformaticists have developed a standard format for nucleotide and protein sequences
Bioinformatics Resources at a Glance A Note about FASTA Format There are MANY free bioinformatics tools available online. Bioinformaticists have developed a standard format for nucleotide and protein sequences
Preparation. Educator s Section: pp. 1 3 Unit 1 instructions: pp. 4 5 Unit 2 instructions: pp. 6 7 Masters/worksheets: pp. 8-17
 ActionBioscience.org lesson To accompany the article by Lawrence M. Page, Ph.D.: "Planetary Biodiversity Inventories: A Response to the Taxonomic Crisis" (May 2006) http://www.actionbioscience.org/biodiversity/page.html
ActionBioscience.org lesson To accompany the article by Lawrence M. Page, Ph.D.: "Planetary Biodiversity Inventories: A Response to the Taxonomic Crisis" (May 2006) http://www.actionbioscience.org/biodiversity/page.html
Practice Questions 1: Evolution
 Practice Questions 1: Evolution 1. Which concept is best illustrated in the flowchart below? A. natural selection B. genetic manipulation C. dynamic equilibrium D. material cycles 2. The diagram below
Practice Questions 1: Evolution 1. Which concept is best illustrated in the flowchart below? A. natural selection B. genetic manipulation C. dynamic equilibrium D. material cycles 2. The diagram below
Lab 2/Phylogenetics/September 16, 2002 1 PHYLOGENETICS
 Lab 2/Phylogenetics/September 16, 2002 1 Read: Tudge Chapter 2 PHYLOGENETICS Objective of the Lab: To understand how DNA and protein sequence information can be used to make comparisons and assess evolutionary
Lab 2/Phylogenetics/September 16, 2002 1 Read: Tudge Chapter 2 PHYLOGENETICS Objective of the Lab: To understand how DNA and protein sequence information can be used to make comparisons and assess evolutionary
Name: Class: Date: Multiple Choice Identify the choice that best completes the statement or answers the question.
 Name: Class: Date: Chapter 17 Practice Multiple Choice Identify the choice that best completes the statement or answers the question. 1. The correct order for the levels of Linnaeus's classification system,
Name: Class: Date: Chapter 17 Practice Multiple Choice Identify the choice that best completes the statement or answers the question. 1. The correct order for the levels of Linnaeus's classification system,
Guide for Bioinformatics Project Module 3
 Structure- Based Evidence and Multiple Sequence Alignment In this module we will revisit some topics we started to look at while performing our BLAST search and looking at the CDD database in the first
Structure- Based Evidence and Multiple Sequence Alignment In this module we will revisit some topics we started to look at while performing our BLAST search and looking at the CDD database in the first
Introduction to Bioinformatics AS 250.265 Laboratory Assignment 6
 Introduction to Bioinformatics AS 250.265 Laboratory Assignment 6 In the last lab, you learned how to perform basic multiple sequence alignments. While useful in themselves for determining conserved residues
Introduction to Bioinformatics AS 250.265 Laboratory Assignment 6 In the last lab, you learned how to perform basic multiple sequence alignments. While useful in themselves for determining conserved residues
Biological Sciences Initiative. Human Genome
 Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
MASCOT Search Results Interpretation
 The Mascot protein identification program (Matrix Science, Ltd.) uses statistical methods to assess the validity of a match. MS/MS data is not ideal. That is, there are unassignable peaks (noise) and usually
The Mascot protein identification program (Matrix Science, Ltd.) uses statistical methods to assess the validity of a match. MS/MS data is not ideal. That is, there are unassignable peaks (noise) and usually
Evolution (18%) 11 Items Sample Test Prep Questions
 Evolution (18%) 11 Items Sample Test Prep Questions Grade 7 (Evolution) 3.a Students know both genetic variation and environmental factors are causes of evolution and diversity of organisms. (pg. 109 Science
Evolution (18%) 11 Items Sample Test Prep Questions Grade 7 (Evolution) 3.a Students know both genetic variation and environmental factors are causes of evolution and diversity of organisms. (pg. 109 Science
Mendelian and Non-Mendelian Heredity Grade Ten
 Ohio Standards Connection: Life Sciences Benchmark C Explain the genetic mechanisms and molecular basis of inheritance. Indicator 6 Explain that a unit of hereditary information is called a gene, and genes
Ohio Standards Connection: Life Sciences Benchmark C Explain the genetic mechanisms and molecular basis of inheritance. Indicator 6 Explain that a unit of hereditary information is called a gene, and genes
Talking About Penguins by Guy Belleranti
 Talking About Penguins Penguins are one of the world s most interesting birds. They waddle when they walk, and have flippers instead of wings. The bones in a penguin s flippers are heavier and more solid
Talking About Penguins Penguins are one of the world s most interesting birds. They waddle when they walk, and have flippers instead of wings. The bones in a penguin s flippers are heavier and more solid
Assign: Unit 1: Preparation Activity page 4-7. Chapter 1: Classifying Life s Diversity page 8
 Assign: Unit 1: Preparation Activity page 4-7 Chapter 1: Classifying Life s Diversity page 8 1.1: Identifying, Naming, and Classifying Species page 10 Key Terms: species, morphology, phylogeny, taxonomy,
Assign: Unit 1: Preparation Activity page 4-7 Chapter 1: Classifying Life s Diversity page 8 1.1: Identifying, Naming, and Classifying Species page 10 Key Terms: species, morphology, phylogeny, taxonomy,
Vector NTI Advance 11 Quick Start Guide
 Vector NTI Advance 11 Quick Start Guide Catalog no. 12605050, 12605099, 12605103 Version 11.0 December 15, 2008 12605022 Published by: Invitrogen Corporation 5791 Van Allen Way Carlsbad, CA 92008 U.S.A.
Vector NTI Advance 11 Quick Start Guide Catalog no. 12605050, 12605099, 12605103 Version 11.0 December 15, 2008 12605022 Published by: Invitrogen Corporation 5791 Van Allen Way Carlsbad, CA 92008 U.S.A.
Protein Sequence Analysis - Overview -
 Protein Sequence Analysis - Overview - UDEL Workshop Raja Mazumder Research Associate Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center Topics Why do protein
Protein Sequence Analysis - Overview - UDEL Workshop Raja Mazumder Research Associate Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center Topics Why do protein
Creating a Poster Presentation using PowerPoint
 Creating a Poster Presentation using PowerPoint Course Description: This course is designed to assist you in creating eye-catching effective posters for presentation of research findings at scientific
Creating a Poster Presentation using PowerPoint Course Description: This course is designed to assist you in creating eye-catching effective posters for presentation of research findings at scientific
Understanding by Design. Title: BIOLOGY/LAB. Established Goal(s) / Content Standard(s): Essential Question(s) Understanding(s):
 Understanding by Design Title: BIOLOGY/LAB Standard: EVOLUTION and BIODIVERSITY Grade(s):9/10/11/12 Established Goal(s) / Content Standard(s): 5. Evolution and Biodiversity Central Concepts: Evolution
Understanding by Design Title: BIOLOGY/LAB Standard: EVOLUTION and BIODIVERSITY Grade(s):9/10/11/12 Established Goal(s) / Content Standard(s): 5. Evolution and Biodiversity Central Concepts: Evolution
Building a phylogenetic tree
 bioscience explained 134567 Wojciech Grajkowski Szkoła Festiwalu Nauki, ul. Ks. Trojdena 4, 02-109 Warszawa Building a phylogenetic tree Aim This activity shows how phylogenetic trees are constructed using
bioscience explained 134567 Wojciech Grajkowski Szkoła Festiwalu Nauki, ul. Ks. Trojdena 4, 02-109 Warszawa Building a phylogenetic tree Aim This activity shows how phylogenetic trees are constructed using
Help Document for WWW.SAGIS.ORG. Step by step, how-to instructions for navigating and using the Savannah Area GIS viewer.
 Help Document for WWW.SAGIS.ORG Step by step, how-to instructions for navigating and using the Savannah Area GIS viewer. 1 SAGIS Savannah Area GIS is focused on providing access to Geospatial data in a
Help Document for WWW.SAGIS.ORG Step by step, how-to instructions for navigating and using the Savannah Area GIS viewer. 1 SAGIS Savannah Area GIS is focused on providing access to Geospatial data in a
Amazing DNA facts. Hands-on DNA: A Question of Taste Amazing facts and quiz questions
 Amazing DNA facts These facts can form the basis of a quiz (for example, how many base pairs are there in the human genome?). Students should be familiar with most of this material, so the quiz could be
Amazing DNA facts These facts can form the basis of a quiz (for example, how many base pairs are there in the human genome?). Students should be familiar with most of this material, so the quiz could be
Searching Nucleotide Databases
 Searching Nucleotide Databases 1 When we search a nucleic acid databases, Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from the forward strand and 3 reading frames
Searching Nucleotide Databases 1 When we search a nucleic acid databases, Mascot always performs a 6 frame translation on the fly. That is, 3 reading frames from the forward strand and 3 reading frames
Name: Date: Period: DNA Unit: DNA Webquest
 Name: Date: Period: DNA Unit: DNA Webquest Part 1 History, DNA Structure, DNA Replication DNA History http://www.dnaftb.org/dnaftb/1/concept/index.html Read the text and answer the following questions.
Name: Date: Period: DNA Unit: DNA Webquest Part 1 History, DNA Structure, DNA Replication DNA History http://www.dnaftb.org/dnaftb/1/concept/index.html Read the text and answer the following questions.
Visualization of Phylogenetic Trees and Metadata
 Visualization of Phylogenetic Trees and Metadata November 27, 2015 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com [email protected]
Visualization of Phylogenetic Trees and Metadata November 27, 2015 Sample to Insight CLC bio, a QIAGEN Company Silkeborgvej 2 Prismet 8000 Aarhus C Denmark Telephone: +45 70 22 32 44 www.clcbio.com [email protected]
Inside Blackboard Collaborate for Moderators
 Inside Blackboard Collaborate for Moderators Entering a Blackboard Collaborate Web Conference 1. The first time you click on the name of the web conference you wish to enter, you will need to download
Inside Blackboard Collaborate for Moderators Entering a Blackboard Collaborate Web Conference 1. The first time you click on the name of the web conference you wish to enter, you will need to download
PUSD High Frequency Word List
 PUSD High Frequency Word List For Reading and Spelling Grades K-5 High Frequency or instant words are important because: 1. You can t read a sentence or a paragraph without knowing at least the most common.
PUSD High Frequency Word List For Reading and Spelling Grades K-5 High Frequency or instant words are important because: 1. You can t read a sentence or a paragraph without knowing at least the most common.
Science 10-Biology Activity 14 Worksheet on Sexual Reproduction
 Science 10-Biology Activity 14 Worksheet on Sexual Reproduction 10 Name Due Date Show Me NOTE: This worksheet is based on material from pages 367-372 in Science Probe. 1. Sexual reproduction requires parents,
Science 10-Biology Activity 14 Worksheet on Sexual Reproduction 10 Name Due Date Show Me NOTE: This worksheet is based on material from pages 367-372 in Science Probe. 1. Sexual reproduction requires parents,
Excel -- Creating Charts
 Excel -- Creating Charts The saying goes, A picture is worth a thousand words, and so true. Professional looking charts give visual enhancement to your statistics, fiscal reports or presentation. Excel
Excel -- Creating Charts The saying goes, A picture is worth a thousand words, and so true. Professional looking charts give visual enhancement to your statistics, fiscal reports or presentation. Excel
Fry Phrases Set 1. TeacherHelpForParents.com help for all areas of your child s education
 Set 1 The people Write it down By the water Who will make it? You and I What will they do? He called me. We had their dog. What did they say? When would you go? No way A number of people One or two How
Set 1 The people Write it down By the water Who will make it? You and I What will they do? He called me. We had their dog. What did they say? When would you go? No way A number of people One or two How
PURPOSE OF GRAPHS YOU ARE ABOUT TO BUILD. To explore for a relationship between the categories of two discrete variables
 3 Stacked Bar Graph PURPOSE OF GRAPHS YOU ARE ABOUT TO BUILD To explore for a relationship between the categories of two discrete variables 3.1 Introduction to the Stacked Bar Graph «As with the simple
3 Stacked Bar Graph PURPOSE OF GRAPHS YOU ARE ABOUT TO BUILD To explore for a relationship between the categories of two discrete variables 3.1 Introduction to the Stacked Bar Graph «As with the simple
Auto Clicker Tutorial
 Auto Clicker Tutorial This Document Outlines Various Features of the Auto Clicker. The Screenshot of the Software is displayed as below and other Screenshots displayed in this Software Tutorial can help
Auto Clicker Tutorial This Document Outlines Various Features of the Auto Clicker. The Screenshot of the Software is displayed as below and other Screenshots displayed in this Software Tutorial can help
Phonics. High Frequency Words P.008. Objective The student will read high frequency words.
 P.008 Jumping Words Objective The student will read high frequency words. Materials High frequency words (P.HFW.005 - P.HFW.064) Choose target words. Checkerboard and checkers (Activity Master P.008.AM1a
P.008 Jumping Words Objective The student will read high frequency words. Materials High frequency words (P.HFW.005 - P.HFW.064) Choose target words. Checkerboard and checkers (Activity Master P.008.AM1a
When To Work Scheduling Program
 When To Work Scheduling Program You should receive your login information by the second week of May. Please login and follow the instructions included in this packet. You will be scheduled based upon the
When To Work Scheduling Program You should receive your login information by the second week of May. Please login and follow the instructions included in this packet. You will be scheduled based upon the
When you install Mascot, it includes a copy of the Swiss-Prot protein database. However, it is almost certain that you and your colleagues will want
 1 When you install Mascot, it includes a copy of the Swiss-Prot protein database. However, it is almost certain that you and your colleagues will want to search other databases as well. There are very
1 When you install Mascot, it includes a copy of the Swiss-Prot protein database. However, it is almost certain that you and your colleagues will want to search other databases as well. There are very
SolarEdge Monitoring Portal. User Guide 1.1. Table of Contents
 Table of Contents Table of Contents... 2 About This Guide... 3 Support and Contact Information... 4 Chapter 1 - Introducing the SolarEdge Monitoring Portal... 5 Chapter 2 - Using the SolarEdge Monitoring
Table of Contents Table of Contents... 2 About This Guide... 3 Support and Contact Information... 4 Chapter 1 - Introducing the SolarEdge Monitoring Portal... 5 Chapter 2 - Using the SolarEdge Monitoring
Table of Contents. Table of Contents
 Table of Contents Table of Contents Table of Contents... 2 About This Guide... 3 Support and Contact Information... 4 Chapter 1 - Introducing the SolarEdge Monitoring Portal... 5 Chapter 2 - Using the
Table of Contents Table of Contents Table of Contents... 2 About This Guide... 3 Support and Contact Information... 4 Chapter 1 - Introducing the SolarEdge Monitoring Portal... 5 Chapter 2 - Using the
WJEC AS Biology Biodiversity & Classification (2.1 All Organisms are related through their Evolutionary History)
 Name:.. Set:. Specification Points: WJEC AS Biology Biodiversity & Classification (2.1 All Organisms are related through their Evolutionary History) (a) Biodiversity is the number of different organisms
Name:.. Set:. Specification Points: WJEC AS Biology Biodiversity & Classification (2.1 All Organisms are related through their Evolutionary History) (a) Biodiversity is the number of different organisms
Geocortex HTML 5 Viewer Manual
 1 FAQ Nothing Happens When I Print? How Do I Search? How Do I Find Feature Information? How Do I Print? How can I Email A Map? How Do I See the Legend? How Do I Find the Coordinates of a Location? How
1 FAQ Nothing Happens When I Print? How Do I Search? How Do I Find Feature Information? How Do I Print? How can I Email A Map? How Do I See the Legend? How Do I Find the Coordinates of a Location? How
Exercises for the UCSC Genome Browser Introduction
 Exercises for the UCSC Genome Browser Introduction 1) Find out if the mouse Brca1 gene has non-synonymous SNPs, color them blue, and get external data about a codon-changing SNP. Skills: basic text search;
Exercises for the UCSC Genome Browser Introduction 1) Find out if the mouse Brca1 gene has non-synonymous SNPs, color them blue, and get external data about a codon-changing SNP. Skills: basic text search;
Guide To Creating Academic Posters Using Microsoft PowerPoint 2010
 Guide To Creating Academic Posters Using Microsoft PowerPoint 2010 INFORMATION SERVICES Version 3.0 July 2011 Table of Contents Section 1 - Introduction... 1 Section 2 - Initial Preparation... 2 2.1 Overall
Guide To Creating Academic Posters Using Microsoft PowerPoint 2010 INFORMATION SERVICES Version 3.0 July 2011 Table of Contents Section 1 - Introduction... 1 Section 2 - Initial Preparation... 2 2.1 Overall
Evolution by Natural Selection 1
 Evolution by Natural Selection 1 I. Mice Living in a Desert These drawings show how a population of mice on a beach changed over time. 1. Describe how the population of mice is different in figure 3 compared
Evolution by Natural Selection 1 I. Mice Living in a Desert These drawings show how a population of mice on a beach changed over time. 1. Describe how the population of mice is different in figure 3 compared
SNP Essentials The same SNP story
 HOW SNPS HELP RESEARCHERS FIND THE GENETIC CAUSES OF DISEASE SNP Essentials One of the findings of the Human Genome Project is that the DNA of any two people, all 3.1 billion molecules of it, is more than
HOW SNPS HELP RESEARCHERS FIND THE GENETIC CAUSES OF DISEASE SNP Essentials One of the findings of the Human Genome Project is that the DNA of any two people, all 3.1 billion molecules of it, is more than
Content Author's Reference and Cookbook
 Sitecore CMS 6.5 Content Author's Reference and Cookbook Rev. 110621 Sitecore CMS 6.5 Content Author's Reference and Cookbook A Conceptual Overview and Practical Guide to Using Sitecore Table of Contents
Sitecore CMS 6.5 Content Author's Reference and Cookbook Rev. 110621 Sitecore CMS 6.5 Content Author's Reference and Cookbook A Conceptual Overview and Practical Guide to Using Sitecore Table of Contents
Modifying Colors and Symbols in ArcMap
 Modifying Colors and Symbols in ArcMap Contents Introduction... 1 Displaying Categorical Data... 3 Creating New Categories... 5 Displaying Numeric Data... 6 Graduated Colors... 6 Graduated Symbols... 9
Modifying Colors and Symbols in ArcMap Contents Introduction... 1 Displaying Categorical Data... 3 Creating New Categories... 5 Displaying Numeric Data... 6 Graduated Colors... 6 Graduated Symbols... 9
Food Chains (and webs) Flow of energy through an ecosystem Grade 5 Austin Carter, Dale Rucker, Allison Hursey
 Food Chains (and webs) Flow of energy through an ecosystem Grade 5 Austin Carter, Dale Rucker, Allison Hursey References: Columbus Public Schools Curriculum Guide- Grade 5 GK-12 Biological Science Lesson
Food Chains (and webs) Flow of energy through an ecosystem Grade 5 Austin Carter, Dale Rucker, Allison Hursey References: Columbus Public Schools Curriculum Guide- Grade 5 GK-12 Biological Science Lesson
The Story of Human Evolution Part 1: From ape-like ancestors to modern humans
 The Story of Human Evolution Part 1: From ape-like ancestors to modern humans Slide 1 The Story of Human Evolution This powerpoint presentation tells the story of who we are and where we came from - how
The Story of Human Evolution Part 1: From ape-like ancestors to modern humans Slide 1 The Story of Human Evolution This powerpoint presentation tells the story of who we are and where we came from - how
The Art of the Tree of Life. Catherine Ibes & Priscilla Spears March 2012
 The Art of the Tree of Life Catherine Ibes & Priscilla Spears March 2012 from so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved. Charles Darwin, The
The Art of the Tree of Life Catherine Ibes & Priscilla Spears March 2012 from so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved. Charles Darwin, The
Creating Online Surveys with Qualtrics Survey Tool
 Creating Online Surveys with Qualtrics Survey Tool Copyright 2015, Faculty and Staff Training, West Chester University. A member of the Pennsylvania State System of Higher Education. No portion of this
Creating Online Surveys with Qualtrics Survey Tool Copyright 2015, Faculty and Staff Training, West Chester University. A member of the Pennsylvania State System of Higher Education. No portion of this
Differentiated Instruction Strategies
 Miss Taylor Brooke Stancil s Differentiated Instruction Strategies Choral Response: Choral response is a very simple technique in which the teacher asks questions to the class as a whole and the students
Miss Taylor Brooke Stancil s Differentiated Instruction Strategies Choral Response: Choral response is a very simple technique in which the teacher asks questions to the class as a whole and the students
Worksheet - COMPARATIVE MAPPING 1
 Worksheet - COMPARATIVE MAPPING 1 The arrangement of genes and other DNA markers is compared between species in Comparative genome mapping. As early as 1915, the geneticist J.B.S Haldane reported that
Worksheet - COMPARATIVE MAPPING 1 The arrangement of genes and other DNA markers is compared between species in Comparative genome mapping. As early as 1915, the geneticist J.B.S Haldane reported that
Helpful Links 8 Helpful Documents 8 Writing History 9 Pending Peer Reviews 9 Navigation Tabs 10 Changing Courses 10
 V7.30.15 2014 GETTING STARTED Table of Contents Welcome to WPP Online 3 WPP Online Welcome Page 3 Logging in to WPP Online 4 Changing your Password 5 Accessing your Courses 7 Selecting a Course 7 The Course
V7.30.15 2014 GETTING STARTED Table of Contents Welcome to WPP Online 3 WPP Online Welcome Page 3 Logging in to WPP Online 4 Changing your Password 5 Accessing your Courses 7 Selecting a Course 7 The Course
Creating and Using Links and Bookmarks in PDF Documents
 Creating and Using Links and Bookmarks in PDF Documents After making a document into a PDF, there may be times when you will need to make links or bookmarks within that PDF to aid navigation through the
Creating and Using Links and Bookmarks in PDF Documents After making a document into a PDF, there may be times when you will need to make links or bookmarks within that PDF to aid navigation through the
AP Biology 2015 Free-Response Questions
 AP Biology 2015 Free-Response Questions College Board, Advanced Placement Program, AP, AP Central, and the acorn logo are registered trademarks of the College Board. AP Central is the official online home
AP Biology 2015 Free-Response Questions College Board, Advanced Placement Program, AP, AP Central, and the acorn logo are registered trademarks of the College Board. AP Central is the official online home
MultiExperiment Viewer Quickstart Guide
 MultiExperiment Viewer Quickstart Guide Table of Contents: I. Preface - 2 II. Installing MeV - 2 III. Opening a Data Set - 2 IV. Filtering - 6 V. Clustering a. HCL - 8 b. K-means - 11 VI. Modules a. T-test
MultiExperiment Viewer Quickstart Guide Table of Contents: I. Preface - 2 II. Installing MeV - 2 III. Opening a Data Set - 2 IV. Filtering - 6 V. Clustering a. HCL - 8 b. K-means - 11 VI. Modules a. T-test
DCAD Website Instruction Manual
 DCAD Website Instruction Manual - 1-9/1/2010 INDEX PAGE Search Appraisal ---------------------------- 3-4 Owner Name ------------------------------ 5-6 Account Number ------------------------------ 7 Street
DCAD Website Instruction Manual - 1-9/1/2010 INDEX PAGE Search Appraisal ---------------------------- 3-4 Owner Name ------------------------------ 5-6 Account Number ------------------------------ 7 Street
Lecture 13: DNA Technology. DNA Sequencing. DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology
 Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Creating an Event Registration Web Page with Special Features using regonline Page 1
 Creating an Event Registration Web Page with Special Features using regonline 1. To begin, enter www.regonline.com in your browser s address bar. A red arrow on each screen shot shows you where to place
Creating an Event Registration Web Page with Special Features using regonline 1. To begin, enter www.regonline.com in your browser s address bar. A red arrow on each screen shot shows you where to place
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15
 Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
EDIT202 PowerPoint Lab Assignment Guidelines
 EDIT202 PowerPoint Lab Assignment Guidelines 1. Create a folder named LABSEC-CCID-PowerPoint. 2. Download the PowerPoint-Sample.avi video file from the course WebCT/Moodle site and save it into your newly
EDIT202 PowerPoint Lab Assignment Guidelines 1. Create a folder named LABSEC-CCID-PowerPoint. 2. Download the PowerPoint-Sample.avi video file from the course WebCT/Moodle site and save it into your newly
After you complete the survey, compare what you saw on the survey to the actual questions listed below:
 Creating a Basic Survey Using Qualtrics Clayton State University has purchased a campus license to Qualtrics. Both faculty and students can use Qualtrics to create surveys that contain many different types
Creating a Basic Survey Using Qualtrics Clayton State University has purchased a campus license to Qualtrics. Both faculty and students can use Qualtrics to create surveys that contain many different types
Phases of the Moon. Preliminaries:
 Phases of the Moon Sometimes when we look at the Moon in the sky we see a small crescent. At other times it appears as a full circle. Sometimes it appears in the daylight against a bright blue background.
Phases of the Moon Sometimes when we look at the Moon in the sky we see a small crescent. At other times it appears as a full circle. Sometimes it appears in the daylight against a bright blue background.
Lab 11: Budgeting with Excel
 Lab 11: Budgeting with Excel This lab exercise will have you track credit card bills over a period of three months. You will determine those months in which a budget was met for various categories. You
Lab 11: Budgeting with Excel This lab exercise will have you track credit card bills over a period of three months. You will determine those months in which a budget was met for various categories. You
Building Qualtrics Surveys for EFS & ALC Course Evaluations: Step by Step Instructions
 Building Qualtrics Surveys for EFS & ALC Course Evaluations: Step by Step Instructions Jennifer DeSantis August 28, 2013 A relatively quick guide with detailed explanations of each step. It s recommended
Building Qualtrics Surveys for EFS & ALC Course Evaluations: Step by Step Instructions Jennifer DeSantis August 28, 2013 A relatively quick guide with detailed explanations of each step. It s recommended
LESSON 9. Analyzing DNA Sequences and DNA Barcoding. Introduction. Learning Objectives
 9 Analyzing DNA Sequences and DNA Barcoding Introduction DNA sequencing is performed by scientists in many different fields of biology. Many bioinformatics programs are used during the process of analyzing
9 Analyzing DNA Sequences and DNA Barcoding Introduction DNA sequencing is performed by scientists in many different fields of biology. Many bioinformatics programs are used during the process of analyzing
Bitrix Site Manager 4.1. User Guide
 Bitrix Site Manager 4.1 User Guide 2 Contents REGISTRATION AND AUTHORISATION...3 SITE SECTIONS...5 Creating a section...6 Changing the section properties...8 SITE PAGES...9 Creating a page...10 Editing
Bitrix Site Manager 4.1 User Guide 2 Contents REGISTRATION AND AUTHORISATION...3 SITE SECTIONS...5 Creating a section...6 Changing the section properties...8 SITE PAGES...9 Creating a page...10 Editing
Online Tools Training Lesson Plan
 Online Tools Training Lesson Plan Reading grade 5 Assessment Development, OSPI, Washington State Table of Contents Purpose Statement... 2 Disclaimer... 2 Lesson Objectives... 2 Lesson Overview... 2 Lesson
Online Tools Training Lesson Plan Reading grade 5 Assessment Development, OSPI, Washington State Table of Contents Purpose Statement... 2 Disclaimer... 2 Lesson Objectives... 2 Lesson Overview... 2 Lesson
DNA Sequencing Overview
 DNA Sequencing Overview DNA sequencing involves the determination of the sequence of nucleotides in a sample of DNA. It is presently conducted using a modified PCR reaction where both normal and labeled
DNA Sequencing Overview DNA sequencing involves the determination of the sequence of nucleotides in a sample of DNA. It is presently conducted using a modified PCR reaction where both normal and labeled
Name: Date: Problem How do amino acid sequences provide evidence for evolution? Procedure Part A: Comparing Amino Acid Sequences
 Name: Date: Amino Acid Sequences and Evolutionary Relationships Introduction Homologous structures those structures thought to have a common origin but not necessarily a common function provide some of
Name: Date: Amino Acid Sequences and Evolutionary Relationships Introduction Homologous structures those structures thought to have a common origin but not necessarily a common function provide some of
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
Tutorial for Tracker and Supporting Software By David Chandler
 Tutorial for Tracker and Supporting Software By David Chandler I use a number of free, open source programs to do video analysis. 1. Avidemux, to exerpt the video clip, read the video properties, and save
Tutorial for Tracker and Supporting Software By David Chandler I use a number of free, open source programs to do video analysis. 1. Avidemux, to exerpt the video clip, read the video properties, and save
Tutorial Segmentation and Classification
 MARKETING ENGINEERING FOR EXCEL TUTORIAL VERSION 1.0.8 Tutorial Segmentation and Classification Marketing Engineering for Excel is a Microsoft Excel add-in. The software runs from within Microsoft Excel
MARKETING ENGINEERING FOR EXCEL TUTORIAL VERSION 1.0.8 Tutorial Segmentation and Classification Marketing Engineering for Excel is a Microsoft Excel add-in. The software runs from within Microsoft Excel
Lesson Overview. Biodiversity. Lesson Overview. 6.3 Biodiversity
 Lesson Overview 6.3 6.3 Objectives Define biodiversity and explain its value. Identify current threats to biodiversity. Describe how biodiversity can be preserved. THINK ABOUT IT From multicolored coral
Lesson Overview 6.3 6.3 Objectives Define biodiversity and explain its value. Identify current threats to biodiversity. Describe how biodiversity can be preserved. THINK ABOUT IT From multicolored coral
WINDOWS 7 EXPLORE INTERNET EXPLORER 8
 WINDOWS 7 EXPLORE INTERNET EXPLORER 8 Windows 7 Explore Internet Explorer 8 Last Edited: 2012-07-10 1 Explore changes in the UI... 3 Enhance with built-in Search engine... 3 Manage and Organize IE with
WINDOWS 7 EXPLORE INTERNET EXPLORER 8 Windows 7 Explore Internet Explorer 8 Last Edited: 2012-07-10 1 Explore changes in the UI... 3 Enhance with built-in Search engine... 3 Manage and Organize IE with
Microsoft PowerPoint 2010 Handout
 Microsoft PowerPoint 2010 Handout PowerPoint is a presentation software program that is part of the Microsoft Office package. This program helps you to enhance your oral presentation and keep the audience
Microsoft PowerPoint 2010 Handout PowerPoint is a presentation software program that is part of the Microsoft Office package. This program helps you to enhance your oral presentation and keep the audience
IDENTIFICATION OF ORGANISMS
 reflect Take a look at the pictures on the right. Think about what the two organisms have in common. They both need food and water to survive. They both grow and reproduce. They both have similar body
reflect Take a look at the pictures on the right. Think about what the two organisms have in common. They both need food and water to survive. They both grow and reproduce. They both have similar body
AP Biology 2013 Free-Response Questions
 AP Biology 2013 Free-Response Questions About the College Board The College Board is a mission-driven not-for-profit organization that connects students to college success and opportunity. Founded in 1900,
AP Biology 2013 Free-Response Questions About the College Board The College Board is a mission-driven not-for-profit organization that connects students to college success and opportunity. Founded in 1900,
Advanced Microsoft Excel 2010
 Advanced Microsoft Excel 2010 Table of Contents THE PASTE SPECIAL FUNCTION... 2 Paste Special Options... 2 Using the Paste Special Function... 3 ORGANIZING DATA... 4 Multiple-Level Sorting... 4 Subtotaling
Advanced Microsoft Excel 2010 Table of Contents THE PASTE SPECIAL FUNCTION... 2 Paste Special Options... 2 Using the Paste Special Function... 3 ORGANIZING DATA... 4 Multiple-Level Sorting... 4 Subtotaling
Learning Module 4 - Thermal Fluid Analysis Note: LM4 is still in progress. This version contains only 3 tutorials.
 Learning Module 4 - Thermal Fluid Analysis Note: LM4 is still in progress. This version contains only 3 tutorials. Attachment C1. SolidWorks-Specific FEM Tutorial 1... 2 Attachment C2. SolidWorks-Specific
Learning Module 4 - Thermal Fluid Analysis Note: LM4 is still in progress. This version contains only 3 tutorials. Attachment C1. SolidWorks-Specific FEM Tutorial 1... 2 Attachment C2. SolidWorks-Specific
Biological kinds and the causal theory of reference
 Biological kinds and the causal theory of reference Ingo Brigandt Department of History and Philosophy of Science 1017 Cathedral of Learning University of Pittsburgh Pittsburgh, PA 15260 E-mail: [email protected]
Biological kinds and the causal theory of reference Ingo Brigandt Department of History and Philosophy of Science 1017 Cathedral of Learning University of Pittsburgh Pittsburgh, PA 15260 E-mail: [email protected]
A Beginner s Guide to PowerPoint 2010
 A Beginner s Guide to PowerPoint 2010 I. The Opening Screen You will see the default opening screen is actually composed of three parts: 1. The Slides/Outline tabs on the left which displays thumbnails
A Beginner s Guide to PowerPoint 2010 I. The Opening Screen You will see the default opening screen is actually composed of three parts: 1. The Slides/Outline tabs on the left which displays thumbnails
Gexa Commercial Portal. User Guide. Version 1.8
 Gexa Commercial Portal User Guide Version 1.8 Table of Contents Accessing the Commercial Portal...2 Login Page...2 Account Registration...2 Verifying Your Account...3 Forgot Username and/or Password...4
Gexa Commercial Portal User Guide Version 1.8 Table of Contents Accessing the Commercial Portal...2 Login Page...2 Account Registration...2 Verifying Your Account...3 Forgot Username and/or Password...4
EHR: Scavenger Hunt I
 EHR: Scavenger Hunt I Metadata Author: Kay Folk Date Submitted: 03/21/07 Editor: Kathie Owens & Sandra Kersten Competency Domain(s) Health record data collection tools Competency Subdomain(s) IA. Health
EHR: Scavenger Hunt I Metadata Author: Kay Folk Date Submitted: 03/21/07 Editor: Kathie Owens & Sandra Kersten Competency Domain(s) Health record data collection tools Competency Subdomain(s) IA. Health
Intermediate PowerPoint
 Intermediate PowerPoint Charts and Templates By: Jim Waddell Last modified: January 2002 Topics to be covered: Creating Charts 2 Creating the chart. 2 Line Charts and Scatter Plots 4 Making a Line Chart.
Intermediate PowerPoint Charts and Templates By: Jim Waddell Last modified: January 2002 Topics to be covered: Creating Charts 2 Creating the chart. 2 Line Charts and Scatter Plots 4 Making a Line Chart.
Worksheet: The theory of natural selection
 Worksheet: The theory of natural selection Senior Phase Grade 7-9 Learning area: Natural Science Strand: Life and living Theme: Biodiversity, change and continuity Specific Aim 1: Acquiring knowledge of
Worksheet: The theory of natural selection Senior Phase Grade 7-9 Learning area: Natural Science Strand: Life and living Theme: Biodiversity, change and continuity Specific Aim 1: Acquiring knowledge of
FOR TEACHERS ONLY. The University of the State of New York REGENTS HIGH SCHOOL EXAMINATION LIVING ENVIRONMENT
 FOR TEACHERS ONLY LE The University of the State of New York REGENTS HIGH SCHOOL EXAMINATION LIVING ENVIRONMENT Tuesday, June 21, 2011 9:15 a.m. to 12:15 p.m., only SCORING KEY AND RATING GUIDE Directions
FOR TEACHERS ONLY LE The University of the State of New York REGENTS HIGH SCHOOL EXAMINATION LIVING ENVIRONMENT Tuesday, June 21, 2011 9:15 a.m. to 12:15 p.m., only SCORING KEY AND RATING GUIDE Directions
Training Manual. Version 6
 Training Manual TABLE OF CONTENTS A. E-MAIL... 4 A.1 INBOX... 8 A.1.1 Create New Message... 8 A.1.1.1 Add Attachments to an E-mail Message... 11 A.1.1.2 Insert Picture into an E-mail Message... 12 A.1.1.3
Training Manual TABLE OF CONTENTS A. E-MAIL... 4 A.1 INBOX... 8 A.1.1 Create New Message... 8 A.1.1.1 Add Attachments to an E-mail Message... 11 A.1.1.2 Insert Picture into an E-mail Message... 12 A.1.1.3
Quick Start to Family Tree
 Quick Start to Family Tree Family Tree is a new way to organize and record your genealogy online. It is free, is available to everyone, and provides an easy way to discover your place in history with free
Quick Start to Family Tree Family Tree is a new way to organize and record your genealogy online. It is free, is available to everyone, and provides an easy way to discover your place in history with free
Similarity Searches on Sequence Databases: BLAST, FASTA. Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Basel, October 2003
 Similarity Searches on Sequence Databases: BLAST, FASTA Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Basel, October 2003 Outline Importance of Similarity Heuristic Sequence Alignment:
Similarity Searches on Sequence Databases: BLAST, FASTA Lorenza Bordoli Swiss Institute of Bioinformatics EMBnet Course, Basel, October 2003 Outline Importance of Similarity Heuristic Sequence Alignment:
MICROSOFT POWERPOINT STEP BY STEP GUIDE
 IGCSE ICT SECTION 16 PRESENTATION AUTHORING MICROSOFT POWERPOINT STEP BY STEP GUIDE Mark Nicholls ICT Lounge Page 1 Contents Importing text to create slides Page 4 Manually creating slides.. Page 5 Removing
IGCSE ICT SECTION 16 PRESENTATION AUTHORING MICROSOFT POWERPOINT STEP BY STEP GUIDE Mark Nicholls ICT Lounge Page 1 Contents Importing text to create slides Page 4 Manually creating slides.. Page 5 Removing
Project Management within ManagePro
 Project Management within ManagePro This document describes how to do the following common project management functions with ManagePro: set-up projects, define scope/requirements, assign resources, estimate
Project Management within ManagePro This document describes how to do the following common project management functions with ManagePro: set-up projects, define scope/requirements, assign resources, estimate
Create a Poster Using Publisher
 Contents 1. Introduction 1. Starting Publisher 2. Create a Poster Template 5. Aligning your images and text 7. Apply a background 12. Add text to your poster 14. Add pictures to your poster 17. Add graphs
Contents 1. Introduction 1. Starting Publisher 2. Create a Poster Template 5. Aligning your images and text 7. Apply a background 12. Add text to your poster 14. Add pictures to your poster 17. Add graphs
