Supplementary Figure Legends
|
|
|
- Marjorie Cunningham
- 8 years ago
- Views:
Transcription
1 Supplementary Figure Legends All branches in the phylogenetic trees follow the same taxonomic colour- code: Metazoa- Black (e.g. Homo sapiens, Drosophila melanogaster, Daphnia pulex, Capitella teleta, Lottia gigantea, Nematostella vectensis, Amphimedon queenslandica), Choanoflagellata- Green (Monosiga brevicollis, Salpingoeca rosetta), Filasterea- Red (Capsaspora owczarzaki), Fungi- Orange (e.g. Laccaria bicolor, Saccharomyces cerevisiae, Allomyces macrogynus, Spizellomyces punctatus), Apusozoa- Yellow (Thecamonas trahens), Amoebozoa- Blue (Acanthamoeba castellanii, Dictyostelium discoideum), Bikonta- Grey (Arabidopsis thaliana, Ostreococcus taurii, Chlamydomonas reindhartii (Plantae), Trichomonas vaginalis, Naegleria gruberi (Excavata), Thalassiosira pseudonana, Tetrahymena thermophila, Toxoplasma gondii (Chromalveolata)). Fig. S1. Maximum likelihood phylogenetic tree of FERM- domain containing genes, including ERM, Merlin and FERM4A. Alignment is based on the FERM domain and contains 408 amino acid positions. The tree was inferred by RAxML using the WAG+Γ+I model of evolution. The tree is rooted using FERM4A as an outgroup. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and Bayesian Posterior Probabilities as inferred with MrBayes. PP. Both values are shown on key branches. Fig. S2. (A) Schematic alignment of ERM proteins. Pfam domains are highlighted. Key amino acids for binding to the membrane lipid phosphatidylinositol 4,5- bisphosphate (PI(4,5)P2), involved in regulation, are highlighted in blue. Key amino acids for actin- binding are highlighted in green. Key amino acids after Turunen et al and Niggli et al Taxa include Aq (Amphimedon queenslandica), Bf (Branchiostoma floridae), Co (Capsaspora owczarzaki), Dm (Drosophila melanogaster), Dp (Daphnia pulex), Hs (Homo sapiens), Mb (Monosiga brevicollis), Nv (Nematostella vectensis) and Sr (Salpingoeca rosetta). (B) Schematic alignment of fascin proteins, showing the four consecutive fascin protein domains. Key amino acids for protein folding are highlighted in blue. Key amino acids for actin- bundling activity are highlighted in green. Key amino acids after Sedeh et al. (2010) and Zanet et al. (2012).Taxa include Aq (Amphimedon queenslandica), Co (Capsaspora owczarzaki), Ct (Capitella teleta), Dm (Drosophila melanogaster), Dp (Daphnia pulex), Hs (Homo sapiens), Lg (Lottia gigantea), Mb (Monosiga brevicollis), Nv (Nematostella vectensis) and Sr (Salpingoeca rosetta). Fig. S3. Maximum likelihood phylogenetic tree of formin proteins. The alignment is based on the FH2 domain and comprises 308 amino acid positions. The tree was inferred by RAxML using the WAG+Γ+I model of evolution. The tree is rooted using the midpoint- root option. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and by BPP. Both values are shown on key branches. Domain architectures for each class are shown.taxa include Ac (Acanthamoeba castellanii), Am (Allomyces macrogynus), Aq (Amphimedon queenslandica), At (Arabidopsis thaliana), Co (Capsaspora owczarzaki), Cr (Chlamydomonas reindhartii), Ct (Capitella teleta), Dd (Dictyostelium discoideum), Dm (Drosophila melanogaster), Dp
2 (Daphnia pulex), Hs (Homo sapiens), Lb (Laccaria bicolor), Lg (Lottia gigantea), Mb (Monosiga brevicollis), Ng (Naegleria gruberi), Nv (Nematostella vectensis), Ot (Ostreococcus tauri), Sc (Saccharomyces cerevisiae), Sp (Spizellomyces punctatus), Sr (Salpingoeca rosetta), Tg (Toxoplasma gondii), Tp (Thalassiosira pseudonana), Tte (Tetrahymena thermophila),tt (Thecamonas trahens), Tv (Trichomonas vaginalis). Sequences used for this tree are reported in Table S1. Fig. S4. Schematic representation of the eukaryotic tree of life showing the distribution of the different formin classes. A black dot indicates the presence of clear homologs, whereas a dashed dot indicates the presence of putative or degenerate homologs. Absence of a dot indicates that an homolog is lacking in that taxon. *Choanoflagellate Diaphanous homologs lack a clear CRIB domain. Only two Naegleria gruberi proteins cluster with Diaphanous, but they don t have a canonical DRF structure (Drf_GBD- Drf_FH3- FH2- DAD). Among other eukaryotes, only the excavates Naegleria gruberi and Trichomonas vaginalis have bona fide DRF proteins. 4Defined as having the Drf_GBD- Drf_FH3- FH2- DAD domain architecture. Fig. S5. ML of gelsolin proteins. The alignment is based on the gelsolin domains and comprises 270 amino acid positions. The tree was inferred by RAxMlLusing the WAG+Γ+I model of evolution. The tree is rooted using the midpoint- root option. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and BPP. Both values are shown on key branches. Domain architectures for each class are shown.taxa include Ac (Acanthamoeba castellanii), Am (Allomyces macrogynus), Aq (Amphimedon queenslandica), At (Arabidopsis thaliana), Co (Capsaspora owczarzaki), Ct (Capitella teleta), Dd (Dictyostelium discoideum), Dm (Drosophila melanogaster), Dp (Daphnia pulex), Hs (Homo sapiens), Lg (Lottia gigantea), Mb (Monosiga brevicollis), Ng (Naegleria gruberi), Nv (Nematostella vectensis), Sp (Spizellomyces punctatus), Sr (Salpingoeca rosetta), Tp (Thalassiosira pseudonana), Tt (Thecamonas trahens). Sequences used are reported in Table S1. Fig. S6. Maximum likelihood phylogenetic tree of WASP proteins. The alignment comprises 139 amino acid positions. The tree was inferred by RAxML using the WAG+Γ+I model of evolution. The tree is rooted using the midpoint- root option. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and BPP. Both values are shown on key branches. Domain architectures for each class are shown. Sequences used are reported in Table S1. Fig. S7. Maximum likelihood phylogenetic tree of RhoGTPase proteins from unikont taxa. The alignment is based on the Ras domain and comprises 153 amino acid positions. The tree was inferred by RAxML using the WAG+Γ+I model of evolution. The tree is rooted using the MIRO GTPases as an outgroup. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and BPP. Both values are shown on key branches. Domain architectures for each class are shown. Taxa include Ac (Acanthamoeba castellanii), Am (Allomyces macrogynus), Aq (Amphimedon queenslandica), Co (Capsaspora owczarzaki), Ct (Capitella teleta), Dd (Dictyostelium discoideum), Dm (Drosophila melanogaster), Dp (Daphnia pulex), Hs (Homo sapiens), Lb (Laccaria bicolor), Lg (Lottia gigantea), Mb (Monosiga brevicollis), Nv (Nematostella
3 vectensis), Sc (Saccharomyces cerevisiae), Sp (Spizellomyces punctatus), Sr (Salpingoeca rosetta), Tt (Thecamonas trahens). Newly classified sequences are reported in Table S1. Fig. S8. Maximum likelihood phylogenetic tree of ablim and other LIM- domain containing proteins. The alignment comprises 239 amino acid positions. The tree was inferred by RAxML using the WAG+Γ+I model of evolution. The tree is rooted using the midpoint- root option. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and BPP. Both values are shown on key branches. Sequences used are reported in Table S1. Fig. S9. Maximum likelihood phylogenetic tree of IMD- domain containing proteins. The alignment is based on the IMD domain and comprises 180 amino acid positions. The tree was inferred by RAxML using the WAG+Γ+I model of evolution. The tree is rooted using the midpoint- root option. Statistical support was obtained by RAxML with 100 bootstrap replicates (BV) and BPP. Both values are shown on key branches. Domain architectures for each class are shown. Sequences used are reported in Table S1. Table S1. Sequences used in study.
4 Fig. S1 Merlin 76/ /1.00 Capitella teleta Lottia gigantea Capsaspora owczarzaki Radixin ERM Ezrin Moesin 40/0.83 Capitella teleta Lottia gigantea 50/0.92 Amphimedon queenslandica Monosiga brevicollis Salpingoeca rosetta FERM4A family Strongylocentrotus purpuratus Capitella teleta Amphimedon queenslandica Monosiga brevicollis Salpingoeca rosetta 0.4
5 A Hs_Ezrin MPKPINVRVTTMDAELEFAIQPNTTGKQLFDQVVKTIGLREVWYFGLHYVDNKGFPTWLKLDKKVSAQEVRKENPLQFKFRAKFYPEDVAEELIQDITQKLFFLQVKEGILSDEIYCPPETAVLLGSYAVQAKFGDYNKEVHKSGYLSSERLIPQRVMDQHKLTRDQWEDRIQVWHAEHRGMLKDNAMLEYLKIAQDLEMYGINYFEIKNKKGTDLWLGVDALGLNIYEKDDKLTPKIGF Hs_Moesin MPKTISVRVTTMDAELEFAIQPNTTGKQLFDQVVKTIGLREVWFFGLQYQDTKGFSTWLKLNKKVTAQDVRKESPLLFKFRAKFYPEDVSEELIQDITQRLFFLQVKEGILNDDIYCPPETAVLLASYAVQSKYGDFNKEVHKSGYLAGDKLLPQRVLEQHKLNKDQWEERIQVWHEEHRGMLREDAVLEYLKIAQDLEMYGVNYFSIKNKKGSELWLGVDALGLNIYEQNDRLTPKIGF Hs_Radixin MPKTISVRVTTMDAELEFAIQPNTTGKQLFDQVVKTIGLREVWFFGLQYQDTKGFSTWLKLNKKVTAQDVRKESPLLFKFRAKFYPEDVSEELIQDITQRLFFLQVKEGILNDDIYCPPETAVLLASYAVQSKYGDFNKEVHKSGYLAGDKLLPQRVLEQHKLNKDQWEERIQVWHEEHRGMLREDAVLEYLKIAQDLEMYGVNYFSIKNKKGSELWLGVDALGLNIYEQNDRLTPKIGF Bf_ERM ----VNVRVTTMDAELEFAIQSNTTGKQLFDQVVKTIGLREIWFFGLQYMDSKGYHTWLKLNKKVMSQDVRKENPLQFKFRAKFYPEDVSEELIQEITQRLFFLQVKDAILTDEVYCPPETSVLLASYAVQAKYGDHNKEVHKAGFLANDRLLPQRVIDQHKMTREQWEERITNWYAEHGGMLREDAMMEYLKIAQDLEMYGVNYFEIKNKKGTQLWLGVDALGLNIYEYDDKLTPKIGF Dm_ERM MPKALNVRVTTMDAELEFAIQSTTTGKQLFDQVVKTIGLREVWFFGLQYTDSKGDSTWIKLYKKVMNQDVKKENPLQFRFRAKFYPEDVAEELIQDITLRLFYLQVKNAILTDEIYCPPETSVLLASYAVQARHGDHNKTTHTAGFLANDRLLPQRVIDQHKMSKDEWEQSIMTWWQEHRSMLREDAMMEYLKIAQDLEMYGVNYFEIRNKKGTDLWLGVDALGLNIYEQDDRLTPKIGF Dp_ERM MDAELEFAIQPTTTGKQLFDQVVKTIGLREIWFFGLQYTDSKSYSTWLKLNKKVLNQDVKKESPLQFKFRAKFYPEDVAEELIQDITVRLFYLQVKNAILTDDIYCPPETSVLLASYAVQAKYGDHDKELHSSGYLANDRLLPQRVMDQHNLTREQWEERISTWHAEHGGMLREDAMMEYLKVAQDLEMYGVNYFEIKNKKGTQLWLGVDALGLNIYEKDDKLTPKIGF Nv_ERM MPKAINVRVTTMDAELEFAIQPSTTGKQLFDQVVKTIGLREIWFFGLQFTDNKGDASWLKLNKKVMSQDVKKETPLQFKFRVKFFPEDVAEELIQEVTQKMFFLQIKEAIFTSDVFCPPETTVLLASYMCQAKHGEYTDE--SAALIKDEKLLPDRVLDQHTLSKEQWDERIILWWKEHGRMAKEDAVIEYLKITQDLEMYGVNYFQIKNKKHTDLYLGVDALGLNIYDQNDKLTPKIGF Aq_ERM MPRPVTVRVTTMDAELEFAIQQSTTGKQLFDQVVKTIGLREVWWFGLQYEDDKGYITWLKLNKKVLAQDLPKETPLKFKFRVKFYPEDVQEELIQDVTQRLFYLQVKDSILTDEIYCPAETLVLLASYAVQAKYGDFNPDEHKPGFLTKDRLLPKRVYDTHKLTKEQWEERITTWHKEHKTMMREDAMLEYLKIAQDLEMYGVNYFEIKNKKGTDLYLGVDALGLNVYEKEDRLTPKIGF Mb_ERM MDAELEFAIQPATTGRQLFDQVVKTIGLREVWYFGLQFIDSKNLVSWLKMNKKVVSQDVRKEDPLQFKFRVKFYPEDVSEELVQDVTQRLFFLQVKEAVLNEDVYAPPETSVLLSSYAVQAKYGDYTPEIHRAGFLAHERLLPPRVLEQHRMTKSEWEGRITAWYSEHRGMLREDAMLEFLKVAQELEMYGVNYFAIKNRKGTDLWLGVDALGLNVYEKDDRLTPKVGF Sr_ERM MDCELEFAVLPATTGKQLFDQVVKTIGLREVWYFGLQYIDSKGLVTWLKMNKKVTAQDVRKETPLQFKFRAKFFPEDVSDELIQEITQRLFFLQVKEAILTEEYYCPPETSVLLASYAVQAKYGDYNPDVHKPGFLGHERLLPSRVLEQHRLSRAQWEERITNWYAEHRGMLREDAILEYLKIAQDLEMYGVNYFEIKNKKGTRLWLGVDALGLNVYEFEDRLTPKIGF FERM_N FERM_M Hs_Ezrin PWSEIRNISFNDKKFVIKPIDKKAPDFVFYAPRLRINKRILQLCMGNHELYMRRRKPDTIEVQQMKAQAREEKHQKQLERQQLETEKKRRETVEREKEQMMREKEELMLRLQDYEEKTKKAERELSEQIQRALQLEEERKRAQEEAERLEADRMAALRAKEELERQAVDQIKSQEQLAAELAEYTAKIALLEEARRRKEDEVEEWQHRAKEAQDDLVKTKEELHLVMTAPPPPPPPVYEP Hs_Moesin PWSEIRNISFNDKKFVIKPIDKKAPDFVFYAPRLRINKRILALCMGNHELYMRRRKPDTIEVQQMKAQAREEKHQKQMERAMLENEKKKREMAEKEKEKIEREKEELMERLKQIEEQTKKAQQELEEQTRRALELEQERKRAQSEAEKLAKERQEAEEAKEALLQASRDQKKTQEQLALEMAELTARISQLEMARQKKESEAVEWQQKAQMVQEDLEKTRAELKTAMSTP Hs_Radixin PWSEIRNISFNDKKFVIKPIDKKAPDFVFYAPRLRINKRILALCMGNHELYMRRRKPDTIEVQQMKAQAREEKHQKQMERAMLENEKKKREMAEKEKEKIEREKEELMERLKQIEEQTKKAQQELEEQTRRALELEQERKRAQSEAEKLAKERQEAEEAKEALLQASRDQKKTQEQLALEMAELTARISQLEMARQKKESEAVEWQQKAQMVQEDLEKTRAELKTAMSTP Bf_ERM PWSEIRNISFNDKKFVIKPIDKKAPDFQFFAPRLRINKRILALCMGNHELYMRRRKADTIEVQQMKAQAREEKHQKQMERAELARQKTLRERE ERQRLELEERLKRFEEEARQRQLELENMQTATHKMMEEKSRAEEEARVLERKRIEAEQERARLEELASREAAEKEEIARMQAALEEETRRLEQERLEKEEDSKKWQ--AERAIEEQQELEKKLVEATQTS Dm_ERM PWSEIRNISFSEKKFIIKPIDKKAPDFMFFAPRVRINKRILALCMGNHELYMRRRKPDTIDVQQMKAQAREEKNAKQQEREKLQLALAARERA EKKQQEYEDRLKQMQEDMERSQRDLLEAQDMIRRLEEQLKQLQAAKDELELRQKELQAMLQRLEEAKNMEAVEKLKLEEEIMAKQMEVQRIQDEVNAKDEETKRLQDEVEDARRKQAEAAAALLAASTTP Dp_ERM PWSEIRNISFNDRKFVIKPIDKKAPDFVFFAPRLRINKRILALCMGNHELYMRRRKPDTIEVQQMKAQAQEERRSKMKEREKLQREIQAREMA ERKQQEYADRLKSMQEEMERRQRELLEAQETIRRLEEQLRQLQKAKEELEVSQKELHNMMKRLEEAKEMEMAEKIKLEEEIRAKQVEVQRIAEEVQRKDDETRRLQEEVEDARRRQEEAAAALIAATTTP Nv_ERM PWSEIRNISFNDKKFCIKPIDKKAPDFIFFSPRLRSNKRILALCMGNHELYMRRRKPDTIEVQQMKAQAREEKVRREKERNALVKEAEARTKA ENDRKLLEEKLKQSEAEKEEMRAAQEEERRIKEELEKERKLIEQNRELLEKRVQEQEAETQRLQEERENAMHEKEELEEEKQAEEERRRRLEEEKIATQRELEELRLDQQRREEEFRMQEEMLVQSKMMP Aq_ERM PWSEIRNISFNDKKFVIKPIDKKAPDFVFNAPRLRINKRILALCMGNHELYLRRRKPDTIEVQQMKAQAREEKISKQSERVRFQKERQAREEA ERKTKDLQEKLQKVIEEEQRVRLEIEKKEQEQRDTAERLRREQAEKEELERKHEEA----RRAAEEKAQDAAEKKKLEEIAKEKEAAIAKANAEMEKMRKKMQELELE QSRLKKANETP Mb_ERM PWSEIRNISFSDKKFIIKPIDKKAPDFIFLATRLRINKRILALCMGNHELYMRRRRPDSIEVQQMKAQAREEKAIKHAERARLAREKTARVEA ERRQSEMAVRLKQYEDEFKQTMAALQESEDRSKKLAGKLEDMQTEAIRQQREHSQAMDSVRELQEQQSNNAEEREALERQRVEAESRAQRIRDEATQRDAEVQALKTQLVKAQEEQAANTRRLMAASAAP Sr_ERM PWSEIRNISFNDKKFIIKPNDKKAPDFVFYVSRLRINKRILALCMGNHELYLRRRRSDTIEVQQMKAQAREEKALRHAERAHLAREKQARMDA ERKRAELEKRVKEYEAEARRAMQALAQSEKTARDLEEKMKRVEAEAAERERLRLEAERLKRQAEESQTASEEEKRMILARTREAEEKAKQLEAEAAKRERDAEDLQAALMAAKKQQVEDAKALVNATSTD Hs_Ezrin VSYHVQES------LQDEGAEPTGYSAELSSEG-IRDDRNEEKRITEAEKNERVQRQLLTLSSELSQARDENKRTHNDIIHNENMRQGRDKYKTLRQIRQGNTKQRIDEFEAL Hs_Moesin ---HVAEP--AENEQDEQD-ENAEASADLRADA-MAKDRSEEERTTEAEKNERVQKHLKALTSELANARDESKKTANDMIHAENMRLGRDKYKTLRQIRQGNTKQRIDEFESM Hs_Radixin ---HVAEP--AENEQDEQD-ENAEASADLRADA-MAKDRSEEERTTEAEKNERVQKHLKALTSELANARDESKKTANDMIHAENMRLGRDKYKTLRQIRQGNTKQRIDEFESM Bf_ERM TALHVAEH--EEGEE NYTSLDLENNKDITNAGSEEDRVTFADKNKMMKDKLKELGKELEIAKDENKLTRNDQLNAENVKAGRDKYKTLKLIRQGNTKQRIDEFEAL Dm_ERM QHHHVAED---ENENEEELTNGGDVSRDLDTDEHIKDP--IEDRRTLAERNERLHDQLKALKQDLAQSRDETKETANDKIHRENVRQGRDKYKTLREIRKGNTKRRVDQFENM Dp_ERM QHQHVNEGDHDEGEDEDEDTPNNDLISESDINS-IRDP--VEDRLTLAEKNERLQNQLKMLKKDLAGTKDETKETAMDRLHKENVKQGRDKYKTLREIRKGNTKRRVDQFENM Nv_ERM AEHIPDK DEQ---GDRSSDLCHEG------IVGDRSEENRQPQNRSKALQDLSADLAAARDEKKQTHYDKLHDENARHGRDKYKTLKQIRQGNTKSRVDQFENL Aq_ERM AIALVTED--DHNEDDDKKTEG---TTDFSMEG-ASNVGSEMSRVHIAEKNKQMAQMLKTLTTELAESRDDTKATKLDQLHAENVKQGRDKYKTLRQIRQGNTKTRVEEFEHM Mb_ERM NLEASETDDVVETSQFSAELSH----ATLAAAG-RNWTSTAVDRDAVLQRSQRIRDQLAMLGRDLDAARDPTKVTSFDRLHSDNVVHGRDKFKTLQKIRSGNVRQRILEFEDM Sr_ERM KFHLMEANMNPDAMQHSQELGRTDLSAGDGKGG-PSWE--VGDRDAIAMKNDRIRQQLESLGAELDAAMDPEKVSAFDKLHRLNKSQGRNKFKTLQRIRAGNTRQRILDFESM Actin-binding region ERM FERM_C PI(4,5)P2 interaction Actin interaction B Hs MTANGTAEAVQ-----IQFGLIN-CGNKYLTAEAFGFKVNASASSLKKKQIWTLEQPPDEAGSAAVCLRSHLGRYLAADKDGNVTCEREVPGPDCR--FLIVAHDD--GRWSLQSEAHRRYFGGTEDRLSCF--AQT---VSPAEKWSVHIAMHPQVNIYSVTRKRYAHLSARPADEIAVDRDVPWGVDSLITLAF---Q-DQRYSVQTADHRFLRHDGRLVA-RPEPATGYTLEFRSGK---VAFRDCEGRYLAPSGPSGTLKAGKATKVGKDELFALE Dm --MNGQGCELGHSNGDIISQNQQKGWWTIGLIN-GQHKYMTAETFGFKLNANGASLKKKQLWTLE--PSNTGESIIYLRSHLNKYLSVDQFGNVLCESDERDAGSR--FQISISEDGSGRWALKNESRGYFLGGTPDKLVCT--AKT---PGASEFWTVHLAARPQVNLRSIGRKRFAHLSES-QDEIHVDANIPWGEDTLFTLEFRAEE-GGRYALHTCNNKYLNANGKLQV-VCNEDCLFSAEYHGGH---LALRDRQGQYLSPIGSKAVLK-SRSSSVTRDELFSLE Dp --MTGQN---GHSNGDVNPSDKPLTTWMVGLVN-SRLKYLTAETFGFKVNANGLLLKKKQLWTLE--AFGDSTDAVCLKSHLDKYLSVDQFGNVTCDAEEKDNGAR--FEIIVPESAAGKWAFRHPERGYFLGASADKLVCS--AKT---PTENELWFVHLAARPQVNLRSMGRRRFAHLSEQ-GDEIHVNANIPWGSTTLFTLEF-RDD-SRLYAIHTANNRYLSRDGRLLE-QCNPTCLFSLEYHAGA---LALRDTQGLYLAPLGSKAVLR-TRSTSVTKDELFTLE Lg --MNGVN---GHKNGGVNYHQ-----WKVGLLN-SSMKYLTAETFGFKINASGTALKKKQTWTLE---QDLTEEVVYIKSHLNRYMSADKYGNVTCEAEEKDEDPSQKFAIEYAKDGSGKWAIRNVMHGNYLSGDDDNVKCF--SKN---ISDKELWVGQLSIHPQVNLHNVNRKRYARLC---DDELQVTAVIPWGQESLIILHF---E-DGKYALKTCDNRFLHRDGHLVD-TLEDDAKFTLEIRSGANSGLAFRDSAGTYLTAVGATAVMK-GRNKSVGKDELFTLE Ct MSPNGVN---GQ--SGAETLQ-----WKVGLLN-NQGRYLTAEAFGFKINASGNVMRKKQVWVIE--HDTKEEDVVYIKSHLGRYLAGDKKGNVTCEHEERDPETK--FSIAYNPDGSGKWAIRNKVSTYYFGGSEDMLKVY--EKQ---PTASEWWTIRLGVHPQVNIRSIIRSRYARHHPE-NERIQFDELTPWGSDALITLEF---L-DGKYAVKASNNHYLKFDGTLVP-SPSQETLYTLEIRSGQYSGMALKDYRGKYLTTVGKDAVMQ-SRNNSFTKDELFKLE Nv MAEPSK-----WNIGLMNDNSSKYLTAETFGFKLNSDGVSLRKKQTWSLE----QVDGEAIYLKSHLGRYLTADPKGNLCCEAEERESNAK--FTVEIKD---GKWALKS-VHGAYFGANGDQLKCV--AK----IGKTELWTVHLAMHPQINLKSVLRKRYAHLDEE-DNDVKVSELIPWGADATITLKF---S-GGRYFLVTSNGKYVSASGDLQD-EPNDDCSFSVEFHQTS---IAFRDHKGKYLTAVG-TGKLQ-SKKETAGKDELFQLE Aq MAVASAKS-----WTVGLIN-SANKFLTAEKFQFKVNANGTSLRKKQYWILE----DAGSDMVAIKSSFGRYLGSDKDGKITADAEEVGDDNR--FILETRDD--GKVAIKSSKHGRYFGGTGDNLSGF--DKE---VSATSLWTIQLAIMPQLNLFNVNRKAYAHLDVT-NNEIRVNEVIPWGFDSTISIDF---H-EGKYSIRAANGSYLECSGALVE-KLNDNCLYTIVYRGTQ---VAFRDNAGKYLAGVGANAVLQ-SRKSSISKDELFTFE Sr MSSPNALE-----WTLGLKNRATGKYLTQEAFGNAVNVNGGSLRQKQTWTLI--SAEDGS--VHLRSYLGKYLYGDNDGNVKCDAESPSADTQ--WTIQPQPD--GTWALIS-AKNYYFHGTGTNISAFLEAKEGVAAPGDGLWVVHLAMHPQINLYNSMRKRYVHLS---GDELHCDEDIPWGDDALLNLIFFDDHPDGRYGIQSCNGKFLENSGRLVD-QPNANCQFLLGFHDNQ---VSFLDSDGKFLSCVGGSGVLK-TNKQKVTKDELFVIQ Sr_ MALPVHPGDSLP-----WALGFRD-VNGKYLTVENFGHRLNINGGSMKTKQIFQIE--QDEGAGSKVFIRTPHGRYLTAKPDGTWAADAETRGTNEE--FEIEVQPD--GRWALKS-AHGYYCGGAGEKIDAF--TKE---VAEDRLFIVNLAMHPQVCIWNVNRKTYLHLE---GEVINADEPIPWGADATITLTF-FDS-TGKYGLQACDGRFLKSDGTLTTVDTDPTIQYTLDMFGGQ---MAFRASNNMYLTCLGGKGTCRATKQGPPGKDELFVLE Mb MTMPTHPGQLLP-----WAFGLRN-HEGLYLTVEPFGNRINVSSKVMKNKQIFVFE--QPDGAGSTVALRTHQGRYLSATSDGRLESTAESIGENER--FELVPDDS--GRFALLS-SAGFYVGGHGEHVDAY--GRT---LAEDRYFHLNLALHPQMNIWNVNRKTFMHLNAD-GQRVTTDEVIPWGDDAVMSLQF-VEE-TNRYTIQAANGKFLDQSGSLVE-EAGPTAQYGLRLLGGQ---VAFQAANGRFLSSIGGDGVCKATKPGPPGKDELYVFE Mb_ MARNIS-----MTMGLRNASSGLYLTQETFGFAVNCNGKSLKKKQTLTLV--AQDGG---IHFKTHLNKFLYGDRDGNVKADADAPSAETQ--WTILPQAD--GTWALKS-AAGFFFHGTGDKLTAFVGGDD---VPADGRWVVHLAMHPQINLYNAMRKRYVHLST--DGELQCNEDVPWGEDALLNLIFFEEHPTGRYGLMACNGKYLENSGKLVD-APNGNCQFLLGFHDDH---ISLCDDEGRFLSCVGGKGTVR-VNKQKVTKDELFRIQ Co MSETLK-----WTLGLLN--NGKYLTQETFGNQMNISSPFLRKKQIFTLE---QNPGSSVVFFLTPNGKYLSAGPKGEFNGAAEEKGKNEE--WTVVPQAD--GRWALKS-THGYYFGGTEASLHAF--GRE---VGESEKWSIHLAMHPQVCIRNVNRKMYVHLA---GEELTADEAIPWGHDALVTFEF---K-GGKYALRASNNKFLDKSGKLVD-AVNDDSLYIIEFNDSQ---IALKANDGKYLQGYGPKGALQ-ARRTQVGKDELFELE Hs QSCAQ-VVLQAANERNVSTRQGMDLSANQD--EETDQ-ETFQLEIDRDTK-----KCAFRTHTGKYWTLTATGGVQSTASSKN-ASCYFDIEWR-DRRITLRASN-GKFVTSKKNGQLAASVETAGDSELFLMKLINRPIIVFRGEHGFIGCRKVTG-TLDANRSSYDVFQLEFND GAYNIKDSTGKYWTVGSD-SAVTSSGDTP-VDFFFEFCD-YNKVAIK----VG-GRYLKGDHAGVLKASAETVDPASLWEY Dm DSLPQASFIAGLNLRYVSVKQGVDVTANQD--EVGEN-ETFQLEYDWSAH-----RWALRTTQDRYWCLSAGGGIQATGNRRC-ADALFELIWHGDGSLSFRANN-GKFLATKRSGHLFATSESIEEIAKFYFYLINRPILVLKCEQGFVGYRTPGNLKLECNKATYETILVERAQ KGLVHLKAHSGKYWRIEGE-S-ISVDADAPSDGFFLELRE-PTRICIR----SQQGKYLGATKNGAFKLLDDGTDSATQWEF Dp DSLAQAVFVSMMNSRYVSVKQGVDVTANQD--EISDH-ETFQLEYDVSTG-----RWYIRTMGDRYWTLETAGGIQASSAKKS-SNALFQMEWQQDGSIGFRANN-GKFVGTKKSGHLFANTDSIDESSKFFFYLINRPMLVLKSEQGFVGYKSGSNCKLECNKANYEAIQVERGP LGLVYFKGQSGRYWTVSSD-G-ITADGDTP-EGFYLELRE-PSKLCIK----SGDGAYLMADKNGVFTVGSRDSELATRWEF Lg DTKPQ-VVIQAYSGKKVSIKQGQDVSANQE--EEEDETEIFQMEYDEHTE-----LWAFRTCSNKYWSLESSGGIMNVGKEIC-KNTLFNVEWQGDGTVTIKACN-NCYIFNKPTGCLFALSETATEKERFKIKMVNRPKIILRSDFGFVGIKSESKPEYICNKTGYENICLEPQE NGFYGFKGPNGKYWGLNAE-SFVMAEQEKP-VNFLLEFRK-QSQISIK----APNGKYLKGEQNGLFRARGDEIEAGSLWEY Ct DSHPQ-VFITAHNGKKVSIKQGIDLTANQD--ELSDR-ETFQCEFDKQSE-----KWRFRTTDNKYWSLESASGIQGVGNAQS-NTSLFGIEWHEDGHVNICGSN-GRYVTARMNGSLYAVSDAPTDKEKFYLTVINRPILVLKCDYGFIGFKLPNNPRIECNKASYDVITLEHNS GGSYFFKGQNGLYWALDDQ-QMINADSKDG-HPFIIELRG-LSRFAIK----APNGNYLKGEQNGIVSAKSSDLLRGTLWEF Nv DSHPQ-VSFIGHTNKRVSIKQHTDLRANQM--DMGDT-EIFQLEFSEDGK-----TVSLLGNNGMYWV--ANGPVSATEKTIT-PKAQFTMEYH-NNQVAFKAHT-GMYLTSNKQGQLLDGTPELEEAGKFSMEIVNRPLLILKGEFGFIGCKTSTN-RLECNRAAYDVFTLEVEEAESEEERGTWYKIGSEKGKYWRVEGD-GSISVSGEANSADLFEIFLERRNYLCIK----AKNGAYLRGQQNGVFNANGKSIAKDSLWEY Aq DTNPQ-ITLIAFNKKYVSQRQGVEVRANQT--DVTDD-ETFQIMAVDRSDLSGNVKWAVCNKKLKFWN--SESNLVAKSNDFSSPDCQFSIVWQ-GPMVLFQANN-GKYLKVTSNGQLSATGGVDDAECKFVLEIINHPILTLRSEFGFVGSKGASG-ILECNRSQYDVFRLKGNA GTYRFETSSGNQWKVEGD--NILANAPGG-SDFFIEFRA-HTRMCIV----APNGKCIQSAQNGDFKPVADDVTPATLWEF Sr DSQPQ-FIITDNRGKFVSVRNGFEVKADQR--EVTDF-ERFQMEITSEGN------VFLRSNKLKYWTVRPDGTIGAESDSPN-ADCTFTVDYSAGNRVRFIAQN-GNPVYVKPSGALMANAGGDPETTLFTLTIINRPTLLLRGQYGFVALKGASG-RVECNRATGNLFVLENKD GFYHLKTQDGKYWGVDVD-G-VTANSSAP-TDFVFEFVR-PTHALIKH---VESGKYLQGQQNGGFKAEGTSAGVNTLWEY Sr_2 DSHPQ-IKMTSWQGKKVSVRSSVEATANQT--DTTDA-ERFQIEIDDSGK------WNIRTNKNTFWYTSEDGTIMTDGASKNAANSKFTIEWL-NDSIALKASS-GKFVSVKKNGGLVAKDADIAAESTFVYELINRPLLVLRGQHGFIATTDKSK-QLMSTSAKAFTYNMHVKA GVCHISDPTKDFWNVASDASAVTVDGKQP-TKFFLEFVG-LSKIALKHMSEDGSSAYLRSHQNGSVTVDGDKVDASTTFEF Mb DSQPQ-IKMTAWFGKKVSVAVGTAILASQS--QTGDA-EQFQLEVGDDGR------WALRTHKNLFCYINGNGDLMGDSETKTQPEARFQLEFF-NEKLALRASN-GAYITAKKNGSMAANGHEPTEEALFVYEMTNRPRLVLRTKYGFLNMAGEETGNLMCNRAQPTVFHMHVKA GHCQIKGPNGRYFAPSDDYSKLAATSVEP-TSLFLEFVS-LSKFAIRVPGPAGETKYAKVHQNGAVSADATRIDESTLFEY Mb_2 DSEPQFIIRNIFKNLNVSARNGAEVKADQKLNDVQDT-ERFQFEV-ADGE------VHVMSDKLKYWAPRDDGSLAVETEKKS-ANTAFVVDYSAGNTVKFQHKASGKFLIAKPNGAMFATGDASDEAAAFELAIINRPTLILRGQYGFLGVKGASG-RVECNRSHPDVFNLVSKN GEYYLQ-KDGKYWTVDQD-G-VACVSSSP-VAFFFEFVQ-LSKALIKH---KESGKYIIGEQNGGFKANGDREETNTLWEF Co DSHPQ-FFLTGQNGKRVSIRQTEEIKAHQAIGDETDA-EIFQMELVGS KVAFRSNKGKYWT-SQDGAIKATSDKRG-DTELFDATWL-GHQVALKASN-GKFVAMKPNGSYAATGDEIDEIAKITLTLKNRPQLVLRCEYGFVATKAKEA--IVANGVEHEVLSLEANE GVYAFKSANGKYWKLEGD-GSFTATGAAP-EQFHFEFLL-HTRFAIKH---VASGKYIKGEHQGNFRATSTSVTKDELWEY Fascin Fascin Fascin Fascin Essential for protein folding Essential for bundling activity Fig. S2
6 Fig. S3 32/ / / / / / / / - 32/ / / / - 17/ / / / / / / / / / / / /1.00 Co 1 89/ /1.00 Ac 2 Ac 3 Ng 10 Dd 7 Nv 8 Am 2 Dd Ac 1 Ac 4 Tt 2 Dd 3 Hs 1 Hs 2 Ct 1 Nv 1 Aq 1 Dp 1 Ct 2 Hs 3 Aq 2 Nv 3 Hs 4 Hs 5 Ct 3 Dp 2 Dm 2 Nv 4 Co 2 Aq 3 Mb 2 Sr 2 Am 1 Sp 1 Dd 4 Dd 5 Dd 6 Dictyostelium Dia2 group CRIB motif Aq 4 Mb 3 Sr 3 Hs 6 Hs 7Hs 8 Dp 3 Dm 3 Ct 5 Aq 5 Mb 4 Sr 4 Lb Ng 6 Ng 7 Ng 4 Ng 8 Ng 9 Ng 5 Ng 11 Ng 3 Hs 9 Hs 10 Ct 6 Dp 4 Dm 4 Ct 7 Nv 6 Ct 8 Mb 6 Sr 6 Hs 11 Hs 12 Dm 5 Nv 7 Aq 7 Co 4 Co 5 Dd 8 Dd 9 Tt 3 Tt 4 Tg 1 At 1 At 2 At 3 At 4 At 5 At 6 At 7 At 8 Ot 1 Hs 13 Dp 5 Dm 6 Ct 9 Aq 9 Aq 10 Mb 7 Tv 1 Tv 2 Tv 4 Tv 3 Tt 5 Tt 8 Tt 9 Tt 10 Tp 1 Tp 2 Sr 1 Mb 5 Sr 5 Co 3 Nv 5 Sc 1 Sp 2 Tt 1 Ac 5 Ac 6 Dd 2 At 9 At 10 At 11 At 12 At 13 At 14 Tte 3 Tte 1 Tte 2 Hs 14 Tv 6 Tv Cr 5 Ac 1 7 Ot 2 An Mb 1 Tt 6 Tt 7 Dm 1 Am 3 Aq 6 Nv 2 Ct 4 Aq 8 Viridiplantae formins group 1 Tt 12 Tt 13 Tte 4 Cr 2 Sc 2 Ng 1 Ng 2 Tg 2 Diaphanous Daam Dictyostelium formins 1 Trichomonas formins Tt 11 Naegleria formins Delphilin FRL-like FRL PH domain formin Formin Fhos Viridiplantae formins group 2 INF Fungi DRFs related Thecamonas CAP_Gly domain formin DRFs Drf_GBD (Diaphanous GTPase binding domain) Drf_FH3 (Diaphanous FH3 domain) FH2 (Formin homology domain 2) DAD PDZ domain PH domain CAP_Gly domain PTEN C2 domain
7 Fig. S4 DRFs Diaphanous Daam FRL FRL-like Delphilin DRF-like 4 Formin Fhos INF PH domain formin Bilateria Metazoa Holozoa Opisthokonts Fungi Apusozoa Nematostella vectensis Amphimedon queenslandica Monosiga brevicollis Salpingoeca rosetta Capsaspora owczarzaki Other fungi Allomyces macrogynus Spizellomyces punctatus Thecamonas trahens Amoebozoa * * Other eukaryotes
8 47/ / / / / / /1.00 Co 1 Lg 3 Nv 1 Mb 1 Mb 2 Sr 1 Hs 1 Hs 2 Hs 3 Dp 1 Lg 1 Ct 1 Ac 1 Sp 1 Dp 2 Dp 3 Ct 2 Hs 4 Dm 1 51/0.97 Aq 3 Lg 5 Am Co 4 Nv 10 Aq 5 Ct 3 Lg 4 Aq 1 Aq 2 Nv 2 Nv 3 Nv 4 Nv 5 Nv 6 Co 2 Lb Dd 1 42/1.00 Ac 2 Tp Tt 1 Ng 1 Hs 5 Lg 6 Ct 4 Dp 4 Nv 7 Dm 3 98/1.00 Nv 8 53/0.93 Aq 4 Sp 2 34/0.93 Co 3 Dd 2 47/ /0.70 Ng 2 Sp 3 Dd 3 Ac 3 At 3 At 2 At 4 At 5 Ac 4 At 1 Ac 7 Dd 4 Dd 5 Ac 6 PBD Hs 6 Nv 9 Ct 5 Lg 7 Dm 4 Dp 5 Tt 2 PBD Ac 5 Dd 6 DUF1899 Dm 2 Lg 2 Plant Villin Sr 2 Ng 3 Severin Holozoan Villin Supervillin WD40 WD40 DUF1900 PH PH Mb 3 Villidin Amoebozoan GRP125 Amoebozoan Gelsolin Flightless PH PBD Gelsolin repeat Villin headpiece domain LRR domain (Leucine Rich Repeat) Pleckstrin homology domain p21-rho-binding domain Fig. S5
9 Fig. S Amphimedon queenslandica 1 Salpingoeca rosetta 1 Monosiga brevicollis 1 Amphimedon queenslandica 2 Capsaspora owczarzaki 1 Allomyces macrogynus 1 WAVE 45/0.86 Spizellomyces punctatus 1 Naegleria gruberi 1 Dictyostelium discoideum 1 Acanthamoeba castellanii 1 Thecamonas trahens Amphimedon queenslandica 3 Capsaspora owczarzaki 2 WH2 WASH 55/0.93 Salpingoeca rosetta 2 Monosiga brevicollis 2 Acanthamoeba castellanii 2 Dictyostelium discoideum 2 Naegleria gruberi 2 Thecamonas trahens 2 WASH_WHAD 3 3 WH2 WASP / Amphimedon queenslandica 4 Amphimedon queenslandica 6 Amphimedon queenslandica 5 Monosiga brevicollis 3 Salpingoeca rosetta 3 Capsaspora owczarzaki 3 Allomyces macrogynus 3 Allomyces macrogynus 2 Naegleria gruberi 3* Acanthamoeba castellanii 3 Acanthamoeba castellanii 4 Acanthamoeba castellanii 5 Tetrahymena thermophila* Trichomonas vaginalis* Saccharomyces cerevisiae Laccaria bicolor Spizellomyces punctatus 2 Dictyostelium discoideum 3 Thecamonas trahens 3 WH1 PBD WH2 (Except *)
10 Fig. S7 57/ / / / /0.82 Hs Cdc42 Aq 1 Nv 1 Co 1 Dp 1 Ct 1 Lg 1 Dm 1 Sr 1 70/0.99 Tt 1 Mb1 75/0.99 Spu 1 Sp Lb 1 Hs RhoQ Hs RhoJ 31/0.62 Hs RhoV Hs RhoU Lg 2 Dp 2 20/0.70 Ct 2 40/0.65 Ac 1 Co 2 Dd 1 Tt 2 Hs Rac1 Dp 3 Dm 3 53/1.00 Lg 3 Ct 3 Tt 3 Co Nv 3 Nv 4 Mb 2 Sr 2 Lb 2 Spu 2 Dd 3 Dd 2 Dd 4 Aq 4 Ac 2 Aq 5 Aq 6 Aq 7 Hs RhoB Aq 9 Dp 5 Dm 5 Hs RhoA Ct 5 Nv 6 Aq 8 Lg 5 Sr 3 Mb 3 Mb 4 Co 4 Lb 3 Spu 3 Spu 4 Sp 2 Sp 3 Dd 5 Lb 4 Lb 6 Lb 7 Lb 8 Lb 9 Lb 10 Lb 11 Lb 12 Lb 14 Sp 4 Lb 13 Sp 5 Hs RhoF/RIF Hs RhoD/RIF Hs Rnd2 Hs Rnd1 RhoQ/J Nv 7 Aq 3 Rac1 Cdc42 Nv 2 Hs RhoH Aq 2 Dm 4 Dp 4 Hs RhoBTB Lg 4 Ct 4 Nv 5 Lb 5 RhoU/V/H Dm 2 Rho BTB-like Rho A/B Hs Rnd3 Rho BTB Aq 10 Ras BTB Ras BTB RIF/Rnd Mitochondrial GTPase Ras EFhand MIRO 97/ / /
11 Fig. S8 97/1.00 ablim/unc-115 Caenorhabditis elegans Amphimedon queenslandica Capsaspora owczarzaki 3 Capsaspora owczarzaki 2 Capsaspora owczarzaki 1 Gallus gallus 2 2 Gallus gallus 1 Xenopus tropicalis 1 Xenopus tropicalis 2 1 PINCH 56/ / /0.91 Allomyces macrogynus Dictyostelium discoideum Dictyostelium purpureum Acanthamoeba castellanii 1 Acanthamoeba castellanii 2 Thecamonas trahens Xenopus tropicalis Capitella teleta Lottia gigantea Capsaspora owczarzaki Capitella teleta Lottia gigantea Amphimedon queenslandica Trichoplax adhaerens Capsaspora owczarzaki Batrachochytrium dendrobatidis Amphimedon queenslandica Trichoplax adhaerens Thecamonas trahens Allomyces macrogynus 1 Allomyces macrogynus 2 Monosiga brevicollis Spizellomyces punctatus Dictyostelium discoideum Acanthamoeba castellanii Paxillin FHL 0.4 Gallus gallus 2 1
12 Fig. S9 IMD SH3 Salpingoeca rosetta 2 IRSp53 Trichoplax adhaerens 82/0.89 Capsaspora owczarzaki 1 Thecamonas trahens Capitella teleta Lottia gigantea IMD Tuber melanosporum Pichia angusta Laccaria bicolor IMD Polysphondilium pallidum Dictyostelium discoideum Acanthamoeba castellanii 1 Acanthamoeba castellanii 2 83/1.00 Capsaspora owczarzaki 2 50/0.91 C apsaspora owczarzaki 4 78/0.98 Salpingoeca rosetta 1 Monosiga brevicollis 1 Capsaspora owczarzaki 3 MTSS Amphimedon queenslandica Salpingoeca rosetta 3 Monosiga brevicollis 1
Figure 1: Genome sizes of different organisms.
 How big are genomes? Genomes are now being sequenced at such a rapid rate that it is fair to say that it is becoming routine. As a result, there is a growing interest in trying to understand the meaning
How big are genomes? Genomes are now being sequenced at such a rapid rate that it is fair to say that it is becoming routine. As a result, there is a growing interest in trying to understand the meaning
Summary of Ph.D. thesis. Tamás Matusek. Supervisor: József Mihály Ph.D.
 Summary of Ph.D. thesis GENETIC, CELL BIOLOGICAL AND BIOCHEMICAL ANALYSIS OF THE TISSUE SPECIFIC FUNCTIONS OF A NEW DROSOPHILA FORMIN Tamás Matusek Supervisor: József Mihály Ph.D. Biological Research Center
Summary of Ph.D. thesis GENETIC, CELL BIOLOGICAL AND BIOCHEMICAL ANALYSIS OF THE TISSUE SPECIFIC FUNCTIONS OF A NEW DROSOPHILA FORMIN Tamás Matusek Supervisor: József Mihály Ph.D. Biological Research Center
Introduction to Genome Annotation
 Introduction to Genome Annotation AGCGTGGTAGCGCGAGTTTGCGAGCTAGCTAGGCTCCGGATGCGA CCAGCTTTGATAGATGAATATAGTGTGCGCGACTAGCTGTGTGTT GAATATATAGTGTGTCTCTCGATATGTAGTCTGGATCTAGTGTTG GTGTAGATGGAGATCGCGTAGCGTGGTAGCGCGAGTTTGCGAGCT
Introduction to Genome Annotation AGCGTGGTAGCGCGAGTTTGCGAGCTAGCTAGGCTCCGGATGCGA CCAGCTTTGATAGATGAATATAGTGTGCGCGACTAGCTGTGTGTT GAATATATAGTGTGTCTCTCGATATGTAGTCTGGATCTAGTGTTG GTGTAGATGGAGATCGCGTAGCGTGGTAGCGCGAGTTTGCGAGCT
Protein import complexes in the mitochondrial outer membrane of Amoebozoa representatives
 Buczek et al. BMC Genomics (2016) 17:99 DOI 10.1186/s12864-016-2402-2 RESEARCH ARTICLE Open Access Protein import complexes in the mitochondrial outer membrane of Amoebozoa representatives Dorota Buczek
Buczek et al. BMC Genomics (2016) 17:99 DOI 10.1186/s12864-016-2402-2 RESEARCH ARTICLE Open Access Protein import complexes in the mitochondrial outer membrane of Amoebozoa representatives Dorota Buczek
Phylogenetic Trees Made Easy
 Phylogenetic Trees Made Easy A How-To Manual Fourth Edition Barry G. Hall University of Rochester, Emeritus and Bellingham Research Institute Sinauer Associates, Inc. Publishers Sunderland, Massachusetts
Phylogenetic Trees Made Easy A How-To Manual Fourth Edition Barry G. Hall University of Rochester, Emeritus and Bellingham Research Institute Sinauer Associates, Inc. Publishers Sunderland, Massachusetts
Guide for Bioinformatics Project Module 3
 Structure- Based Evidence and Multiple Sequence Alignment In this module we will revisit some topics we started to look at while performing our BLAST search and looking at the CDD database in the first
Structure- Based Evidence and Multiple Sequence Alignment In this module we will revisit some topics we started to look at while performing our BLAST search and looking at the CDD database in the first
Protein Protein Interactions (PPI) APID (Agile Protein Interaction DataAnalyzer)
 APID (Agile Protein Interaction DataAnalyzer) 23 APID (Agile Protein Interaction DataAnalyzer) Integrates and unifies 7 DBs: BIND, DIP, HPRD, IntAct, MINT, BioGRID. Includes 51,873 proteins 241,204 interactions
APID (Agile Protein Interaction DataAnalyzer) 23 APID (Agile Protein Interaction DataAnalyzer) Integrates and unifies 7 DBs: BIND, DIP, HPRD, IntAct, MINT, BioGRID. Includes 51,873 proteins 241,204 interactions
Name: Class: Date: Multiple Choice Identify the choice that best completes the statement or answers the question.
 Name: Class: Date: Chapter 17 Practice Multiple Choice Identify the choice that best completes the statement or answers the question. 1. The correct order for the levels of Linnaeus's classification system,
Name: Class: Date: Chapter 17 Practice Multiple Choice Identify the choice that best completes the statement or answers the question. 1. The correct order for the levels of Linnaeus's classification system,
國 立 交 通 大 學. 同 源 蛋 白 質 - 蛋 白 質 交 互 作 用 之 研 究 A Study of Homologous Protein-protein Interactions 生 物 科 技 學 系 博 士 論 文
 國 立 交 通 大 學 生 物 科 技 學 系 博 士 論 文 同 源 蛋 白 質 - 蛋 白 質 交 互 作 用 之 研 究 A Study of Homologous Protein-protein Interactions 研 究 生 : 陳 俊 辰 Chen Chun-Chen 指 導 教 授 : 楊 進 木 博 士 Advisor: Yang Jinn-Moon, Ph. D. 中 華 民
國 立 交 通 大 學 生 物 科 技 學 系 博 士 論 文 同 源 蛋 白 質 - 蛋 白 質 交 互 作 用 之 研 究 A Study of Homologous Protein-protein Interactions 研 究 生 : 陳 俊 辰 Chen Chun-Chen 指 導 教 授 : 楊 進 木 博 士 Advisor: Yang Jinn-Moon, Ph. D. 中 華 民
Introduction to Bioinformatics AS 250.265 Laboratory Assignment 6
 Introduction to Bioinformatics AS 250.265 Laboratory Assignment 6 In the last lab, you learned how to perform basic multiple sequence alignments. While useful in themselves for determining conserved residues
Introduction to Bioinformatics AS 250.265 Laboratory Assignment 6 In the last lab, you learned how to perform basic multiple sequence alignments. While useful in themselves for determining conserved residues
ID of alternative translational initiation events. Description of gene function Reference of NCBI database access and relative literatures
 Data resource: In this database, 650 alternatively translated variants assigned to a total of 300 genes are contained. These database records of alternative translational initiation have been collected
Data resource: In this database, 650 alternatively translated variants assigned to a total of 300 genes are contained. These database records of alternative translational initiation have been collected
Online Resource 9. Figure S8: Amino acid alignments.
 Online Resource 9 This file contains amino acid alignments of the Long Oskar and Lasp-binding domains from 18 Drosophilids including the D. virilis allele from Webster et al. (1994) using the PRANK MSA,
Online Resource 9 This file contains amino acid alignments of the Long Oskar and Lasp-binding domains from 18 Drosophilids including the D. virilis allele from Webster et al. (1994) using the PRANK MSA,
*Corresponding author: Yoshitaka Nishiyama, E-mail: nishiyama@molbiol.saitama-u.ac.jp
 SUPPLEMENTAL DATA Elongation Factor G Is a Critical Target during Oxidative Damage to the Translation System of Escherichia coli Takanori Nagano 1, Kouji Kojima 1,7, Toru Hisabori 2, Hidenori Hayashi 3,
SUPPLEMENTAL DATA Elongation Factor G Is a Critical Target during Oxidative Damage to the Translation System of Escherichia coli Takanori Nagano 1, Kouji Kojima 1,7, Toru Hisabori 2, Hidenori Hayashi 3,
Bayesian Phylogeny and Measures of Branch Support
 Bayesian Phylogeny and Measures of Branch Support Bayesian Statistics Imagine we have a bag containing 100 dice of which we know that 90 are fair and 10 are biased. The
Bayesian Phylogeny and Measures of Branch Support Bayesian Statistics Imagine we have a bag containing 100 dice of which we know that 90 are fair and 10 are biased. The
An Overview of Cells and Cell Research
 An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
85748 Garching b. Munich, Germany 4 Scripps Institution of Oceanography, University of California San Diego, 9500 Gilman Drive,
 Assessing the root of bilaterian animals with scalable phylogenomic methods Andreas Hejnol 1, *, Matthias Obst 2, Alexandros Stamatakis 3, Michael Ott 3,GregW.Rouse 4, Gregory D. Edgecombe 5, Pedro Martinez
Assessing the root of bilaterian animals with scalable phylogenomic methods Andreas Hejnol 1, *, Matthias Obst 2, Alexandros Stamatakis 3, Michael Ott 3,GregW.Rouse 4, Gregory D. Edgecombe 5, Pedro Martinez
Title: Surveying Genome to Identify Origins of DNA Replication In Silico
 Title: Surveying Genome to Identify Origins of DNA Replication In Silico Abstract: DNA replication origins are the bases to realize the process of chromosome replication and analyze the progress of cell
Title: Surveying Genome to Identify Origins of DNA Replication In Silico Abstract: DNA replication origins are the bases to realize the process of chromosome replication and analyze the progress of cell
Functional Architecture of RNA Polymerase I
 Cell, Volume 131 Supplemental Data Functional Architecture of RNA Polymerase I Claus-D. Kuhn, Sebastian R. Geiger, Sonja Baumli, Marco Gartmann, Jochen Gerber, Stefan Jennebach, Thorsten Mielke, Herbert
Cell, Volume 131 Supplemental Data Functional Architecture of RNA Polymerase I Claus-D. Kuhn, Sebastian R. Geiger, Sonja Baumli, Marco Gartmann, Jochen Gerber, Stefan Jennebach, Thorsten Mielke, Herbert
Bioinformatics: Network Analysis
 Bioinformatics: Network Analysis Graph-theoretic Properties of Biological Networks COMP 572 (BIOS 572 / BIOE 564) - Fall 2013 Luay Nakhleh, Rice University 1 Outline Architectural features Motifs, modules,
Bioinformatics: Network Analysis Graph-theoretic Properties of Biological Networks COMP 572 (BIOS 572 / BIOE 564) - Fall 2013 Luay Nakhleh, Rice University 1 Outline Architectural features Motifs, modules,
Interaktionen von RNAs und Proteinen
 Sonja Prohaska Computational EvoDevo Universitaet Leipzig June 9, 2015 Studying RNA-protein interactions Given: target protein known to bind to RNA problem: find binding partners and binding sites experimental
Sonja Prohaska Computational EvoDevo Universitaet Leipzig June 9, 2015 Studying RNA-protein interactions Given: target protein known to bind to RNA problem: find binding partners and binding sites experimental
Protein Sequence Analysis - Overview -
 Protein Sequence Analysis - Overview - UDEL Workshop Raja Mazumder Research Associate Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center Topics Why do protein
Protein Sequence Analysis - Overview - UDEL Workshop Raja Mazumder Research Associate Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center Topics Why do protein
Bio-Informatics Lectures. A Short Introduction
 Bio-Informatics Lectures A Short Introduction The History of Bioinformatics Sanger Sequencing PCR in presence of fluorescent, chain-terminating dideoxynucleotides Massively Parallel Sequencing Massively
Bio-Informatics Lectures A Short Introduction The History of Bioinformatics Sanger Sequencing PCR in presence of fluorescent, chain-terminating dideoxynucleotides Massively Parallel Sequencing Massively
Multiple Losses of Flight and Recent Speciation in Steamer Ducks Tara L. Fulton, Brandon Letts, and Beth Shapiro
 Supplementary Material for: Multiple Losses of Flight and Recent Speciation in Steamer Ducks Tara L. Fulton, Brandon Letts, and Beth Shapiro 1. Supplementary Tables Supplementary Table S1. Sample information.
Supplementary Material for: Multiple Losses of Flight and Recent Speciation in Steamer Ducks Tara L. Fulton, Brandon Letts, and Beth Shapiro 1. Supplementary Tables Supplementary Table S1. Sample information.
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes.
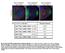 Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes
 Crisp et al. Genome Biology (2015) 16:50 DOI 10.1186/s13059-015-0607-3 RESEARCH Open Access Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes
Crisp et al. Genome Biology (2015) 16:50 DOI 10.1186/s13059-015-0607-3 RESEARCH Open Access Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes
Computational Systems Biology. Lecture 2: Enzymes
 Computational Systems Biology Lecture 2: Enzymes 1 Images from: David L. Nelson, Lehninger Principles of Biochemistry, IV Edition, Freeman ed. or under creative commons license (search for images at http://search.creativecommons.org/)
Computational Systems Biology Lecture 2: Enzymes 1 Images from: David L. Nelson, Lehninger Principles of Biochemistry, IV Edition, Freeman ed. or under creative commons license (search for images at http://search.creativecommons.org/)
4. Why are common names not good to use when classifying organisms? Give an example.
 1. Define taxonomy. Classification of organisms 2. Who was first to classify organisms? Aristotle 3. Explain Aristotle s taxonomy of organisms. Patterns of nature: looked like 4. Why are common names not
1. Define taxonomy. Classification of organisms 2. Who was first to classify organisms? Aristotle 3. Explain Aristotle s taxonomy of organisms. Patterns of nature: looked like 4. Why are common names not
Eukaryotes have organelles
 Energy-transducing Eukaryotes have organelles membrane systems An organelle is a discrete membrane bound cellular structure specialized functions. An organelle is to the cell what an organ is to the body
Energy-transducing Eukaryotes have organelles membrane systems An organelle is a discrete membrane bound cellular structure specialized functions. An organelle is to the cell what an organ is to the body
Supplementary Figures S1 - S11
 1 Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations Hang Yu 1,2, Klaus Schulten 1,2,3, 1 Beckman Institute, University of Illinois, Urbana, Illinois, USA 2 Center of Biophysics
1 Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations Hang Yu 1,2, Klaus Schulten 1,2,3, 1 Beckman Institute, University of Illinois, Urbana, Illinois, USA 2 Center of Biophysics
2011.008a-cB. Code assigned:
 This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the
The world of non-coding RNA. Espen Enerly
 The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
Table 1 Calcium regulation and signaling genes in Apicomplexans Function/structure
 Table Calcium regulation and signaling genes in Apicomplexans Function/structure P-type ATPase Toxoplasma gondii Most significant family/ a domain by InterProScan b (Accession No) TgA (AF,.m0000) IPR000
Table Calcium regulation and signaling genes in Apicomplexans Function/structure P-type ATPase Toxoplasma gondii Most significant family/ a domain by InterProScan b (Accession No) TgA (AF,.m0000) IPR000
Supporting Information. Fast and Efficient Fragment-Based Lead Generation. by Fully Automated Processing and Analysis of
 Supporting Information Fast and Efficient Fragment-Based Lead Generation by Fully Automated Processing and Analysis of Ligand-Observed NMR Binding Data Chen Peng, Alexandra Frommlet, Manuel Perez, Carlos
Supporting Information Fast and Efficient Fragment-Based Lead Generation by Fully Automated Processing and Analysis of Ligand-Observed NMR Binding Data Chen Peng, Alexandra Frommlet, Manuel Perez, Carlos
Evolution of microrna diversity and regulation in animals
 NON-CODING RNA Evolution of microrna diversity and regulation in animals Eugene Berezikov Abstract In the past decade, micrornas (mirnas) have been uncovered as key regulators of gene expression at the
NON-CODING RNA Evolution of microrna diversity and regulation in animals Eugene Berezikov Abstract In the past decade, micrornas (mirnas) have been uncovered as key regulators of gene expression at the
Building a phylogenetic tree
 bioscience explained 134567 Wojciech Grajkowski Szkoła Festiwalu Nauki, ul. Ks. Trojdena 4, 02-109 Warszawa Building a phylogenetic tree Aim This activity shows how phylogenetic trees are constructed using
bioscience explained 134567 Wojciech Grajkowski Szkoła Festiwalu Nauki, ul. Ks. Trojdena 4, 02-109 Warszawa Building a phylogenetic tree Aim This activity shows how phylogenetic trees are constructed using
Molecular Facts and Figures
 Nucleic Acids Molecular Facts and Figures DNA/RNA bases: DNA and RNA are composed of four bases each. In DNA the four are Adenine (A), Thymidine (T), Cytosine (C), and Guanine (G). In RNA the four are
Nucleic Acids Molecular Facts and Figures DNA/RNA bases: DNA and RNA are composed of four bases each. In DNA the four are Adenine (A), Thymidine (T), Cytosine (C), and Guanine (G). In RNA the four are
Introduction to Phylogenetic Analysis
 Subjects of this lecture Introduction to Phylogenetic nalysis Irit Orr 1 Introducing some of the terminology of phylogenetics. 2 Introducing some of the most commonly used methods for phylogenetic analysis.
Subjects of this lecture Introduction to Phylogenetic nalysis Irit Orr 1 Introducing some of the terminology of phylogenetics. 2 Introducing some of the most commonly used methods for phylogenetic analysis.
An ancestral MADS-box gene duplication occurred before the divergence of plants and animals
 An ancestral MADS-box gene duplication occurred before the divergence of plants and animals Elena R. Alvarez-Buylla*, Soraya Pelaz*, Sarah J. Liljegren*, Scott E. Gold*, Caroline Burgeff, Gary S. Ditta*,
An ancestral MADS-box gene duplication occurred before the divergence of plants and animals Elena R. Alvarez-Buylla*, Soraya Pelaz*, Sarah J. Liljegren*, Scott E. Gold*, Caroline Burgeff, Gary S. Ditta*,
Final Project Report
 CPSC545 by Introduction to Data Mining Prof. Martin Schultz & Prof. Mark Gerstein Student Name: Yu Kor Hugo Lam Student ID : 904907866 Due Date : May 7, 2007 Introduction Final Project Report Pseudogenes
CPSC545 by Introduction to Data Mining Prof. Martin Schultz & Prof. Mark Gerstein Student Name: Yu Kor Hugo Lam Student ID : 904907866 Due Date : May 7, 2007 Introduction Final Project Report Pseudogenes
Gene Models & Bed format: What they represent.
 GeneModels&Bedformat:Whattheyrepresent. Gene models are hypotheses about the structure of transcripts produced by a gene. Like all models, they may be correct, partly correct, or entirely wrong. Typically,
GeneModels&Bedformat:Whattheyrepresent. Gene models are hypotheses about the structure of transcripts produced by a gene. Like all models, they may be correct, partly correct, or entirely wrong. Typically,
Photosynthesis and Cellular Respiration. Stored Energy
 Photosynthesis and Cellular Respiration Stored Energy What is Photosynthesis? plants convert the energy of sunlight into the energy in the chemical bonds of carbohydrates sugars and starches. SUMMARY EQUATION:
Photosynthesis and Cellular Respiration Stored Energy What is Photosynthesis? plants convert the energy of sunlight into the energy in the chemical bonds of carbohydrates sugars and starches. SUMMARY EQUATION:
Genome Viewing. Module 2. Using Genome Browsers to View Annotation of the Human Genome
 Module 2 Genome Viewing Using Genome Browsers to View Annotation of the Human Genome Bert Overduin, Ph.D. PANDA Coordination & Outreach EMBL - European Bioinformatics Institute Wellcome Trust Genome Campus
Module 2 Genome Viewing Using Genome Browsers to View Annotation of the Human Genome Bert Overduin, Ph.D. PANDA Coordination & Outreach EMBL - European Bioinformatics Institute Wellcome Trust Genome Campus
IsoBase: a database of functionally related proteins across PPI networks
 D295 D300 doi:10.1093/nar/gkq1234 IsoBase: a database of functionally related proteins across PPI networks Daniel Park 1,2, Rohit Singh 1, Michael Baym 1,3,4, Chung-Shou Liao 5 and Bonnie Berger 1,4, *
D295 D300 doi:10.1093/nar/gkq1234 IsoBase: a database of functionally related proteins across PPI networks Daniel Park 1,2, Rohit Singh 1, Michael Baym 1,3,4, Chung-Shou Liao 5 and Bonnie Berger 1,4, *
PHYLOGENY OF THE TRAF/MATH DOMAIN
 PHYLOGENY OF THE TRAF/MATH DOMAIN Juan M. Zapata 1,2,3, Vanesa Martínez-García 1,2 and Sophie Lefebvre 1 1 Burnham Institute for Medical Research, 10901 N. Torrey Pines Road, La Jolla, CA 92037 and 2 Centro
PHYLOGENY OF THE TRAF/MATH DOMAIN Juan M. Zapata 1,2,3, Vanesa Martínez-García 1,2 and Sophie Lefebvre 1 1 Burnham Institute for Medical Research, 10901 N. Torrey Pines Road, La Jolla, CA 92037 and 2 Centro
Cellular Signalling 21 (2009) 1579 1585. Contents lists available at ScienceDirect. Cellular Signalling
 Cellular Signalling 21 (2009) 1579 1585 Contents lists available at ScienceDirect Cellular Signalling journal homepage: www.elsevier.com/locate/cellsig Phylogeny of the CDC25 homology domain reveals rapid
Cellular Signalling 21 (2009) 1579 1585 Contents lists available at ScienceDirect Cellular Signalling journal homepage: www.elsevier.com/locate/cellsig Phylogeny of the CDC25 homology domain reveals rapid
Hierarchical Bayesian Modeling of the HIV Response to Therapy
 Hierarchical Bayesian Modeling of the HIV Response to Therapy Shane T. Jensen Department of Statistics, The Wharton School, University of Pennsylvania March 23, 2010 Joint Work with Alex Braunstein and
Hierarchical Bayesian Modeling of the HIV Response to Therapy Shane T. Jensen Department of Statistics, The Wharton School, University of Pennsylvania March 23, 2010 Joint Work with Alex Braunstein and
MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788
 MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788 ISSN 0340-6253 Three distances for rapid similarity analysis of DNA sequences Wei Chen,
MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788 ISSN 0340-6253 Three distances for rapid similarity analysis of DNA sequences Wei Chen,
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
Victims Compensation Claim Status of All Pending Claims and Claims Decided Within the Last Three Years
 Claim#:021914-174 Initials: J.T. Last4SSN: 6996 DOB: 5/3/1970 Crime Date: 4/30/2013 Status: Claim is currently under review. Decision expected within 7 days Claim#:041715-334 Initials: M.S. Last4SSN: 2957
Claim#:021914-174 Initials: J.T. Last4SSN: 6996 DOB: 5/3/1970 Crime Date: 4/30/2013 Status: Claim is currently under review. Decision expected within 7 days Claim#:041715-334 Initials: M.S. Last4SSN: 2957
7 Answers to end-of-chapter questions
 7 Answers to end-of-chapter questions Multiple choice questions 1 B 2 B 3 A 4 B 5 A 6 D 7 C 8 C 9 B 10 B Structured questions 11 a i Maintenance of a constant internal environment within set limits i Concentration
7 Answers to end-of-chapter questions Multiple choice questions 1 B 2 B 3 A 4 B 5 A 6 D 7 C 8 C 9 B 10 B Structured questions 11 a i Maintenance of a constant internal environment within set limits i Concentration
The silicon cell. Ab initio prediction and folding impossible but for the smallest structures. Threading
 C E N T R E F O R I N T E G B I O I N F O R R M A A T T I I V C E S V U Homology searching (BLAST) Threading Sequence Structure Function Ab initio prediction and folding impossible but for the smallest
C E N T R E F O R I N T E G B I O I N F O R R M A A T T I I V C E S V U Homology searching (BLAST) Threading Sequence Structure Function Ab initio prediction and folding impossible but for the smallest
Dichotomic classes, correlations and entropy optimization in coding sequences
 Dichotomic classes, correlations and entropy optimization in coding sequences Simone Giannerini 1 1 Università di Bologna, Dipartimento di Scienze Statistiche Joint work with Diego Luis Gonzalez and Rodolfo
Dichotomic classes, correlations and entropy optimization in coding sequences Simone Giannerini 1 1 Università di Bologna, Dipartimento di Scienze Statistiche Joint work with Diego Luis Gonzalez and Rodolfo
A* with Partial Expansion for large branching factor problems
 A* with Partial Expansion for large branching factor problems Takayuki Yoshizumi, Teruhisa Miura and Toru Ishida Department of Social Informatics, Kyoto University, Kyoto 66-85, Japan fyosizumi, miura,
A* with Partial Expansion for large branching factor problems Takayuki Yoshizumi, Teruhisa Miura and Toru Ishida Department of Social Informatics, Kyoto University, Kyoto 66-85, Japan fyosizumi, miura,
Missing data and the accuracy of Bayesian phylogenetics
 Journal of Systematics and Evolution 46 (3): 307 314 (2008) (formerly Acta Phytotaxonomica Sinica) doi: 10.3724/SP.J.1002.2008.08040 http://www.plantsystematics.com Missing data and the accuracy of Bayesian
Journal of Systematics and Evolution 46 (3): 307 314 (2008) (formerly Acta Phytotaxonomica Sinica) doi: 10.3724/SP.J.1002.2008.08040 http://www.plantsystematics.com Missing data and the accuracy of Bayesian
High Throughput Network Analysis
 High Throughput Network Analysis Sumeet Agarwal 1,2, Gabriel Villar 1,2,3, and Nick S Jones 2,4,5 1 Systems Biology Doctoral Training Centre, University of Oxford, Oxford OX1 3QD, United Kingdom 2 Department
High Throughput Network Analysis Sumeet Agarwal 1,2, Gabriel Villar 1,2,3, and Nick S Jones 2,4,5 1 Systems Biology Doctoral Training Centre, University of Oxford, Oxford OX1 3QD, United Kingdom 2 Department
Photosynthesis (Life from Light)
 Photosynthesis Photosynthesis (Life from Light) Energy needs of life All life needs a constant input of energy o Heterotrophs (consumers) Animals, fungi, most bacteria Get their energy from other organisms
Photosynthesis Photosynthesis (Life from Light) Energy needs of life All life needs a constant input of energy o Heterotrophs (consumers) Animals, fungi, most bacteria Get their energy from other organisms
Sequence Analysis 15: lecture 5. Substitution matrices Multiple sequence alignment
 Sequence Analysis 15: lecture 5 Substitution matrices Multiple sequence alignment A teacher's dilemma To understand... Multiple sequence alignment Substitution matrices Phylogenetic trees You first need
Sequence Analysis 15: lecture 5 Substitution matrices Multiple sequence alignment A teacher's dilemma To understand... Multiple sequence alignment Substitution matrices Phylogenetic trees You first need
Copyright 1999 2003 by Mark Brandt, Ph.D.
 Central dogma of molecular biology The term central dogma of molecular biology is patterned after religious terminology. owever, it refers to a process that is subject to the changes in understanding that
Central dogma of molecular biology The term central dogma of molecular biology is patterned after religious terminology. owever, it refers to a process that is subject to the changes in understanding that
Genomes and SNPs in Malaria and Sickle Cell Anemia
 Genomes and SNPs in Malaria and Sickle Cell Anemia Introduction to Genome Browsing with Ensembl Ensembl The vast amount of information in biological databases today demands a way of organising and accessing
Genomes and SNPs in Malaria and Sickle Cell Anemia Introduction to Genome Browsing with Ensembl Ensembl The vast amount of information in biological databases today demands a way of organising and accessing
Bioinformatics Grid - Enabled Tools For Biologists.
 Bioinformatics Grid - Enabled Tools For Biologists. What is Grid-Enabled Tools (GET)? As number of data from the genomics and proteomics experiment increases. Problems arise for the current sequence analysis
Bioinformatics Grid - Enabled Tools For Biologists. What is Grid-Enabled Tools (GET)? As number of data from the genomics and proteomics experiment increases. Problems arise for the current sequence analysis
Chapter 3. Molecular cloning of tomato TCA cycle full-length. cdnas and characterisation α and β subunits of. succinyl CoA ligase
 Chapter 3 Molecular cloning of tomato TCA cycle full-length cdnas and characterisation α and β subunits of succinyl CoA ligase 3.1. Introduction The operation and the localization of the complete TCA cycle
Chapter 3 Molecular cloning of tomato TCA cycle full-length cdnas and characterisation α and β subunits of succinyl CoA ligase 3.1. Introduction The operation and the localization of the complete TCA cycle
Analysis of the Opsin Repertoire in the Tardigrade. Hypsibius dujardini Provides Insights into the Evolution of
 Genome Biology and Evolution Advance Access published September 4, 2014 doi:10.1093/gbe/evu193 Submission is intended as Research Article Analysis of the Opsin Repertoire in the Tardigrade Hypsibius dujardini
Genome Biology and Evolution Advance Access published September 4, 2014 doi:10.1093/gbe/evu193 Submission is intended as Research Article Analysis of the Opsin Repertoire in the Tardigrade Hypsibius dujardini
SEPTEMBER Unit 1 Page Learning Goals 1 Short a 2 b 3-5 blends 6-7 c as in cat 8-11 t 12-13 p
 The McRuffy Kindergarten Reading/Phonics year- long program is divided into 4 units and 180 pages of reading/phonics instruction. Pages and learning goals covered are noted below: SEPTEMBER Unit 1 1 Short
The McRuffy Kindergarten Reading/Phonics year- long program is divided into 4 units and 180 pages of reading/phonics instruction. Pages and learning goals covered are noted below: SEPTEMBER Unit 1 1 Short
The Central Dogma of Molecular Biology
 Vierstraete Andy (version 1.01) 1/02/2000 -Page 1 - The Central Dogma of Molecular Biology Figure 1 : The Central Dogma of molecular biology. DNA contains the complete genetic information that defines
Vierstraete Andy (version 1.01) 1/02/2000 -Page 1 - The Central Dogma of Molecular Biology Figure 1 : The Central Dogma of molecular biology. DNA contains the complete genetic information that defines
Unraveling protein networks with Power Graph Analysis
 Unraveling protein networks with Power Graph Analysis PLoS Computational Biology, 2008 Loic Royer Matthias Reimann Bill Andreopoulos Michael Schroeder Schroeder Group Bioinformatics 1 Complex Networks
Unraveling protein networks with Power Graph Analysis PLoS Computational Biology, 2008 Loic Royer Matthias Reimann Bill Andreopoulos Michael Schroeder Schroeder Group Bioinformatics 1 Complex Networks
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
PHYLOGENY AND COMPARATIVE METHODS SYMBIOMICS WORKSHOP
 PHYLOGENY AND COMPARATIVE METHODS SYMBIOMICS WORKSHOP March 4-7, 2013 Valencia, Spain Parc Cientific of the University of Valencia Goals The aim of this workshop is to provide the attendees with a broad
PHYLOGENY AND COMPARATIVE METHODS SYMBIOMICS WORKSHOP March 4-7, 2013 Valencia, Spain Parc Cientific of the University of Valencia Goals The aim of this workshop is to provide the attendees with a broad
Human Genome and Human Genome Project. Louxin Zhang
 Human Genome and Human Genome Project Louxin Zhang A Primer to Genomics Cells are the fundamental working units of every living systems. DNA is made of 4 nucleotide bases. The DNA sequence is the particular
Human Genome and Human Genome Project Louxin Zhang A Primer to Genomics Cells are the fundamental working units of every living systems. DNA is made of 4 nucleotide bases. The DNA sequence is the particular
Phylogeny of the Forgotten Cellular Slime Mold, Fonticula alba, Reveals a Key Evolutionary Branch within Opisthokonta
 Phylogeny of the Forgotten Cellular Slime Mold, Fonticula alba, Reveals a Key Evolutionary Branch within Opisthokonta Matthew W. Brown, Frederick W. Spiegel, and Jeffrey D. Silberman Department of Biological
Phylogeny of the Forgotten Cellular Slime Mold, Fonticula alba, Reveals a Key Evolutionary Branch within Opisthokonta Matthew W. Brown, Frederick W. Spiegel, and Jeffrey D. Silberman Department of Biological
Built from 20 kinds of amino acids
 Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Clustering Digital Data by Compression: Applications to Biology and Medical Images
 Clustering Digital Data by Compression: Applications to Biology and Medical Images BRUNO CARPENTIERI Dipartimento di Informatica Università di Salerno 84084 Fisciano (SA) ITALY bc@di.unisa.it Abstract:
Clustering Digital Data by Compression: Applications to Biology and Medical Images BRUNO CARPENTIERI Dipartimento di Informatica Università di Salerno 84084 Fisciano (SA) ITALY bc@di.unisa.it Abstract:
Formin-induced nucleation of actin filaments Sally H Zigmond
 Formin-induced nucleation of actin filaments Sally H Zigmond Formins are proteins best defined by the presence of the unique, highly conserved formin homology domain (). is necessary and sufficient to
Formin-induced nucleation of actin filaments Sally H Zigmond Formins are proteins best defined by the presence of the unique, highly conserved formin homology domain (). is necessary and sufficient to
Genomes with distinct function composition
 FEBS Letters 389 (1996) 96-101 FEBS 17138 Minireview Genomes with distinct function composition Javier Tamames a, Christos Ouzounis b, Chris Sander c,d, Alfonso Valencia a,* acentro Nacional de Biotecnologia
FEBS Letters 389 (1996) 96-101 FEBS 17138 Minireview Genomes with distinct function composition Javier Tamames a, Christos Ouzounis b, Chris Sander c,d, Alfonso Valencia a,* acentro Nacional de Biotecnologia
Efficient Parallel Execution of Sequence Similarity Analysis Via Dynamic Load Balancing
 Efficient Parallel Execution of Sequence Similarity Analysis Via Dynamic Load Balancing James D. Jackson Philip J. Hatcher Department of Computer Science Kingsbury Hall University of New Hampshire Durham,
Efficient Parallel Execution of Sequence Similarity Analysis Via Dynamic Load Balancing James D. Jackson Philip J. Hatcher Department of Computer Science Kingsbury Hall University of New Hampshire Durham,
Inferred thermophily of the last universal ancestor based on estimated
 1 Inferred thermophily of the last universal ancestor based on estimated amino acid composition Dawn J. Brooks and Eric A. Gaucher Address: Foundation for Applied Molecular Evolution, Gainesville, Florida
1 Inferred thermophily of the last universal ancestor based on estimated amino acid composition Dawn J. Brooks and Eric A. Gaucher Address: Foundation for Applied Molecular Evolution, Gainesville, Florida
PHYML Online: A Web Server for Fast Maximum Likelihood-Based Phylogenetic Inference
 PHYML Online: A Web Server for Fast Maximum Likelihood-Based Phylogenetic Inference Stephane Guindon, F. Le Thiec, Patrice Duroux, Olivier Gascuel To cite this version: Stephane Guindon, F. Le Thiec, Patrice
PHYML Online: A Web Server for Fast Maximum Likelihood-Based Phylogenetic Inference Stephane Guindon, F. Le Thiec, Patrice Duroux, Olivier Gascuel To cite this version: Stephane Guindon, F. Le Thiec, Patrice
PRODUCT INFORMATION...
 VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
T cell Epitope Prediction
 Institute for Immunology and Informatics T cell Epitope Prediction EpiMatrix Eric Gustafson January 6, 2011 Overview Gathering raw data Popular sources Data Management Conservation Analysis Multiple Alignments
Institute for Immunology and Informatics T cell Epitope Prediction EpiMatrix Eric Gustafson January 6, 2011 Overview Gathering raw data Popular sources Data Management Conservation Analysis Multiple Alignments
Name Class Date. binomial nomenclature. MAIN IDEA: Linnaeus developed the scientific naming system still used today.
 Section 1: The Linnaean System of Classification 17.1 Reading Guide KEY CONCEPT Organisms can be classified based on physical similarities. VOCABULARY taxonomy taxon binomial nomenclature genus MAIN IDEA:
Section 1: The Linnaean System of Classification 17.1 Reading Guide KEY CONCEPT Organisms can be classified based on physical similarities. VOCABULARY taxonomy taxon binomial nomenclature genus MAIN IDEA:
DNA Sequence formats
 DNA Sequence formats [Plain] [EMBL] [FASTA] [GCG] [GenBank] [IG] [IUPAC] [How Genomatix represents sequence annotation] Plain sequence format A sequence in plain format may contain only IUPAC characters
DNA Sequence formats [Plain] [EMBL] [FASTA] [GCG] [GenBank] [IG] [IUPAC] [How Genomatix represents sequence annotation] Plain sequence format A sequence in plain format may contain only IUPAC characters
Lecture Outline. Introduction to Databases. Introduction. Data Formats Sample databases How to text search databases. Shifra Ben-Dor Irit Orr
 Introduction to Databases Shifra Ben-Dor Irit Orr Lecture Outline Introduction Data and Database types Database components Data Formats Sample databases How to text search databases What units of information
Introduction to Databases Shifra Ben-Dor Irit Orr Lecture Outline Introduction Data and Database types Database components Data Formats Sample databases How to text search databases What units of information
A branch-and-bound algorithm for the inference of ancestral. amino-acid sequences when the replacement rate varies among
 A branch-and-bound algorithm for the inference of ancestral amino-acid sequences when the replacement rate varies among sites Tal Pupko 1,*, Itsik Pe er 2, Masami Hasegawa 1, Dan Graur 3, and Nir Friedman
A branch-and-bound algorithm for the inference of ancestral amino-acid sequences when the replacement rate varies among sites Tal Pupko 1,*, Itsik Pe er 2, Masami Hasegawa 1, Dan Graur 3, and Nir Friedman
From DNA to Protein. Proteins. Chapter 13. Prokaryotes and Eukaryotes. The Path From Genes to Proteins. All proteins consist of polypeptide chains
 Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
AP Bio Photosynthesis & Respiration
 AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
Probabilistic methods for post-genomic data integration
 Probabilistic methods for post-genomic data integration Dirk Husmeier Biomathematics & Statistics Scotland (BioSS) JMB, The King s Buildings, Edinburgh EH9 3JZ United Kingdom http://wwwbiossacuk/ dirk
Probabilistic methods for post-genomic data integration Dirk Husmeier Biomathematics & Statistics Scotland (BioSS) JMB, The King s Buildings, Edinburgh EH9 3JZ United Kingdom http://wwwbiossacuk/ dirk
A Step-by-Step Tutorial: Divergence Time Estimation with Approximate Likelihood Calculation Using MCMCTREE in PAML
 9 June 2011 A Step-by-Step Tutorial: Divergence Time Estimation with Approximate Likelihood Calculation Using MCMCTREE in PAML by Jun Inoue, Mario dos Reis, and Ziheng Yang In this tutorial we will analyze
9 June 2011 A Step-by-Step Tutorial: Divergence Time Estimation with Approximate Likelihood Calculation Using MCMCTREE in PAML by Jun Inoue, Mario dos Reis, and Ziheng Yang In this tutorial we will analyze
A first-use tutorial for the VIP Analysis software
 A first-use tutorial for the VIP Analysis software Luis C. Dias INESC Coimbra & School of Economics - University of Coimbra Av Dias da Silva, 165, 3004-512 Coimbra, Portugal LMCDias@fe.uc.pt The software
A first-use tutorial for the VIP Analysis software Luis C. Dias INESC Coimbra & School of Economics - University of Coimbra Av Dias da Silva, 165, 3004-512 Coimbra, Portugal LMCDias@fe.uc.pt The software
FINDING RELATION BETWEEN AGING AND
 FINDING RELATION BETWEEN AGING AND TELOMERE BY APRIORI AND DECISION TREE Jieun Sung 1, Youngshin Joo, and Taeseon Yoon 1 Department of National Science, Hankuk Academy of Foreign Studies, Yong-In, Republic
FINDING RELATION BETWEEN AGING AND TELOMERE BY APRIORI AND DECISION TREE Jieun Sung 1, Youngshin Joo, and Taeseon Yoon 1 Department of National Science, Hankuk Academy of Foreign Studies, Yong-In, Republic
PANTONE Solid to Process
 PANTONE Solid to Process PANTONE C:0 M:0 Y:100 K:0 Proc. Yellow PC PANTONE C:0 M:0 Y:51 K:0 100 PC PANTONE C:0 M:2 Y:69 K:0 106 PC PANTONE C:0 M:100 Y:0 K:0 Proc. Magen. PC PANTONE C:0 M:0 Y:79 K:0 101
PANTONE Solid to Process PANTONE C:0 M:0 Y:100 K:0 Proc. Yellow PC PANTONE C:0 M:0 Y:51 K:0 100 PC PANTONE C:0 M:2 Y:69 K:0 106 PC PANTONE C:0 M:100 Y:0 K:0 Proc. Magen. PC PANTONE C:0 M:0 Y:79 K:0 101
PROC. CAIRO INTERNATIONAL BIOMEDICAL ENGINEERING CONFERENCE 2006 1. E-mail: msm_eng@k-space.org
 BIOINFTool: Bioinformatics and sequence data analysis in molecular biology using Matlab Mai S. Mabrouk 1, Marwa Hamdy 2, Marwa Mamdouh 2, Marwa Aboelfotoh 2,Yasser M. Kadah 2 1 Biomedical Engineering Department,
BIOINFTool: Bioinformatics and sequence data analysis in molecular biology using Matlab Mai S. Mabrouk 1, Marwa Hamdy 2, Marwa Mamdouh 2, Marwa Aboelfotoh 2,Yasser M. Kadah 2 1 Biomedical Engineering Department,
CSC 2427: Algorithms for Molecular Biology Spring 2006. Lecture 16 March 10
 CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
Systematic assessment of cancer missense mutation clustering in protein structures
 Systematic assessment of cancer missense mutation clustering in protein structures Atanas Kamburov, Michael Lawrence, Paz Polak, Ignaty Leshchiner, Kasper Lage, Todd R. Golub, Eric S. Lander, Gad Getz
Systematic assessment of cancer missense mutation clustering in protein structures Atanas Kamburov, Michael Lawrence, Paz Polak, Ignaty Leshchiner, Kasper Lage, Todd R. Golub, Eric S. Lander, Gad Getz
Interaktionen von Nukleinsäuren und Proteinen
 Sonja Prohaska Computational EvoDevo Universitaet Leipzig June 9, 2015 DNA is never naked in a cell DNA is usually in association with proteins. In all domains of life there are small, basic chromosomal
Sonja Prohaska Computational EvoDevo Universitaet Leipzig June 9, 2015 DNA is never naked in a cell DNA is usually in association with proteins. In all domains of life there are small, basic chromosomal
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
A new type of Hidden Markov Models to predict complex domain architecture in protein sequences
 A new type of Hidden Markov Models to predict complex domain architecture in protein sequences Raluca Uricaru, Laurent Bréhélin and Eric Rivals LIRMM, CNRS Université de Montpellier 2 14 Juin 2007 Raluca
A new type of Hidden Markov Models to predict complex domain architecture in protein sequences Raluca Uricaru, Laurent Bréhélin and Eric Rivals LIRMM, CNRS Université de Montpellier 2 14 Juin 2007 Raluca
Pantone Matching System Color Chart PMS Colors Used For Printing
 Pantone Matching System Color Chart PMS Colors Used For Printing Use this guide to assist your color selection and specification process. This chart is a reference guide only. Pantone colors on computer
Pantone Matching System Color Chart PMS Colors Used For Printing Use this guide to assist your color selection and specification process. This chart is a reference guide only. Pantone colors on computer
Sequence Formats and Sequence Database Searches. Gloria Rendon SC11 Education June, 2011
 Sequence Formats and Sequence Database Searches Gloria Rendon SC11 Education June, 2011 Sequence A is the primary structure of a biological molecule. It is a chain of residues that form a precise linear
Sequence Formats and Sequence Database Searches Gloria Rendon SC11 Education June, 2011 Sequence A is the primary structure of a biological molecule. It is a chain of residues that form a precise linear
Integration of data management and analysis for genome research
 Integration of data management and analysis for genome research Volker Brendel Deparment of Zoology & Genetics and Department of Statistics Iowa State University 2112 Molecular Biology Building Ames, Iowa
Integration of data management and analysis for genome research Volker Brendel Deparment of Zoology & Genetics and Department of Statistics Iowa State University 2112 Molecular Biology Building Ames, Iowa
Crime Scene Investigation (Adopted from Forensics in the Classroom developed by Court TV)
 Crime Scene Investigation (Adopted from Forensics in the Classroom developed by Court TV) The case: Last night, the school cafeteria was vandalized; the kitchen area was broken into, several appliances
Crime Scene Investigation (Adopted from Forensics in the Classroom developed by Court TV) The case: Last night, the school cafeteria was vandalized; the kitchen area was broken into, several appliances
Hydroxynitrile lyase (Hnl)
 Hydroxynitrile lyase (Hnl) R 1 HCN R 1 OH R 2 C O R 2 C * CN S-selective: Hevea brasiliensis R-selective: Prunus spp. (S)-Hnl of Hevea brasiliensis and (R)-Hnl of Prunus amygdalus Hb_Hnl Type II Hnl intracellular
Hydroxynitrile lyase (Hnl) R 1 HCN R 1 OH R 2 C O R 2 C * CN S-selective: Hevea brasiliensis R-selective: Prunus spp. (S)-Hnl of Hevea brasiliensis and (R)-Hnl of Prunus amygdalus Hb_Hnl Type II Hnl intracellular
