Supporting Information
|
|
|
- Regina Ball
- 7 years ago
- Views:
Transcription
1 Supporting Information Nature of Molecular Interactions of Peptides with Gold, Palladium, and Pd-Au Bimetal Surfaces in Aqueous Solution Hendrik Heinz, 1 * Barry L. Farmer, 2 Ras B. Pandey, 3 Joseph M. Slocik, 2 Soumya S. Patnaik, 2 Ruth Pachter, 2 and Rajesh R. Naik 2 1 Department of Polymer Engineering, University of Akron, Akron, Ohio Air Force Research Laboratory, Materials and Manufacturing Directorate, AFRL/RX, Wright- Patterson AFB, Ohio Department of Physics and Astronomy, University of Southern Mississippi, Hattiesburg, MS * Corresponding author: hendrik.heinz@uakron.edu 1 of 16
2 Report Documentation Page Form Approved OMB No Public reporting burden for the collection of information is estimated to average 1 hour per response, including the time for reviewing instructions, searching existing data sources, gathering and maintaining the data needed, and completing and reviewing the collection of information. Send comments regarding this burden estimate or any other aspect of this collection of information, including suggestions for reducing this burden, to Washington Headquarters Services, Directorate for Information Operations and Reports, 1215 Jefferson Davis Highway, Suite 1204, Arlington VA Respondents should be aware that notwithstanding any other provision of law, no person shall be subject to a penalty for failing to comply with a collection of information if it does not display a currently valid OMB control number. 1. REPORT DATE REPORT TYPE 3. DATES COVERED to TITLE AND SUBTITLE Nature of Molecular Interactions of Peptides with Gold, Palladium, and Pd-Au Bimetal Surfaces in Aqueous Solution 5a. CONTRACT NUMBER 5b. GRANT NUMBER 5c. PROGRAM ELEMENT NUMBER 6. AUTHOR(S) 5d. PROJECT NUMBER 5e. TASK NUMBER 5f. WORK UNIT NUMBER 7. PERFORMING ORGANIZATION NAME(S) AND ADDRESS(ES) Department of Polymer Engineering,University of Akron,Akron,OH, PERFORMING ORGANIZATION REPORT NUMBER 9. SPONSORING/MONITORING AGENCY NAME(S) AND ADDRESS(ES) 10. SPONSOR/MONITOR S ACRONYM(S) 12. DISTRIBUTION/AVAILABILITY STATEMENT Approved for public release; distribution unlimited 13. SUPPLEMENTARY NOTES 14. ABSTRACT 11. SPONSOR/MONITOR S REPORT NUMBER(S) 15. SUBJECT TERMS 16. SECURITY CLASSIFICATION OF: 17. LIMITATION OF ABSTRACT a. REPORT unclassified b. ABSTRACT unclassified c. THIS PAGE unclassified Same as Report (SAR) 18. NUMBER OF PAGES 16 19a. NAME OF RESPONSIBLE PERSON Standard Form 298 (Rev. 8-98) Prescribed by ANSI Std Z39-18
3 S1. Further Background on Simulation Methods In summary, we employ a classical atomistic molecular dynamics approach to analyze specific peptide binding to metal surfaces in comparison with thermochemical, IR, NMR, and TEM measurements (Figure 1). The availability of suitable parameters for biomolecules and recent improvements in the accuracy of parameters for inorganic components by more than one order of magnitude 29,37 41 are thereby very helpful Moreover, modeling at all levels, molecular, coarse-grain, and bioinformatics, is ultimately important in identifying the most suitable peptide sequences for controlled binding and detachment (Figure 1) as the huge number of possible sequences exceeds the number of available sequences in commercial phage libraries by more than a million. Quantum-mechanical approaches indicate approximate trends of the interaction of peptide fragments and a few solvent molecules with parts of a surface, and have shown that covalent interactions with metal surfaces are modest to small. 21,22,24,26,28,31 However, such approaches cannot fully explain the mechanisms of binding due to limitations to static calculations including a few hundred atoms, and neglect of thousands of needed solvent molecules, ions, and dynamics at significant time scales. Some ab-initio results also disagree with each other due to known difficulties with the treatment of van-der-waals interactions (only in wave-function based techniques) and accurate electrostatics. 29,38,40 A limitation of ab-initio and classical atomistic methods is also the difficulty to include polarization effects on metal surfaces; though estimates suggest modest contributions on even surfaces. Coarse-grain approaches can be computationally ~10 3 times more efficient and include certain specific peptide-surface interactions. 30 A critical role can also be attributed to bioinformatics approaches such as simple numerical screening functions on the basis of molecular-level insight to help eliminate sequences of undesirable binding strength. The success of such higher-level 2 of 16
4 approaches, however, yet depends on understanding the nature of the molecular interactions (Figure 1). S2. Efficient Computation of Adsorption Energies The quantitative computation of adsorption energies requires the inclusion of explicit water molecules and ionic solutes due to the formation of spatially oriented hydrogen bonds which change their orientation when peptides (or other adsorbates) move from the solution to the surface. The calculation of adsorption energies also requires an accurate summation of Coulomb interactions on the order of 0.1 kcal/mol uncertainty for the entire system (see section S3.3). Under these conditions, we have developed and tested two methods to compute the changes in energy, entropy, and free energy upon adsorption (Figure S1). S2.1. Method 1 Simple Approach. A conceptually simple approach involves the simulation of the adsorbate on the surface (simulation 1) and in solution (simulation 2) using the same box size and box content (Figure S1a). The box needs to be large enough to avoid contact of the adsorbate with the surface during the solution run. However, large boxes result in high values and large fluctuations of the total energy which require extra simulation time to compute the relatively small energy difference E = E 1 E2 between simulation 1 and simulation 2 with statistically acceptable errors (Figure S1a). The time dependence on the number of particles N is typically on the order N 2 for summations of Coulomb interactions using the Ewald method and for summations of van-der-waals interactions using spherical cutoffs, and on the order N lnn for summation of Coulomb interactions using the PPPM method. Method 1 is less efficient due to the inclusion of many water molecules in the surface run, and due to the inclusion of the surface atoms and surrounding water molecules in the 3 of 16
5 solution run (Figure S1b). This introduces particular computational overhead when the adsorption of a series of molecules on the same surface is of interest. In the presence of usual periodic boundary conditions, these issues of method 1 can be circumvented by taking the additivity of intermolecular interactions into account. As shown earlier, 41 the interaction of polar but overall charge neutral molecules or crystals becomes almost negligible at distances greater than 3 nm so that larger simulation boxes in Figure S1a can be split into multiple boxes of ~5 nm height as shown in Figure S1b. This partition of the computation boxes leads to method 2. S2.2. Method 2 Efficient Approach. In the second scheme (Figure S1c), a total of four calculations with smaller boxes are required which is approximately twice as fast as two calculations with larger boxes using Ewald procedures or spherical cutoffs for Lennard-Jones interactions ( t ( N) ~ N 2 ). For computational screening of additional adsorbates on the same surface, only two additional calculations per adsorbate are required to obtain E Surf+A+H2O and E A+H2O while the values for E H2O and E A+H2O remain the same. Assuming a dependence of simulation time t(n) on the number of atoms N, a box size of 2N in Fig. S1a, and about half the box size N in Fig. S1c, the computation of adsorption energies for k peptides on the same surface takes the relative time 2k+ 2 t( N). For example, method 2 will screen 20 peptides in 26% of 2k t(2n) the time compared to method 1 in a simulation with 2 t ( N) ~ N (Ewald or spherical cutoffs), and in less than 50% of the time compared to method 1 in a simulation with t( N) ~ N ln N (PPPM summations). A further advantage of method 2 over method 1 is the reduction of the total energy and its standard deviation, which lowers the standard deviation of computed adsorption energies on an absolute scale. Possible interferences from the surface in the solution run in method 1 are also 4 of 16
6 eliminated, and method 2 equally provides access to thermodynamic quantities including energies, entropies, and free energies of adsorption. The computation of entropy S (using A= U TS or other method) and free energy A is more time-consuming than that of the energies, however, because very well resolved histograms of the energy distribution are needed or multiple simulations for each system to facilitate thermodynamic integration, respectively. The application of either method, provided the box size is large enough to eliminate finite size effects and simulations are allowed to reach equilibrium, leads to identical results. Simulations are best performed using the NVT ensemble and additive molecular volumina, i.e., predefined densities for each constituent such as metal atoms, water molecules, and peptides. Then, for example, using the same cross section (A = xy) of the box, the box height (z) is incrementally additive for every component added. To compute differences in thermodynamic quantities, the simulation temperature must be constant or corrected to the same value using the heat capacity of each box (as derived in the simulation). 5 of 16
7 (a) Method 1 (b) Decomposition as H 2 O Peptide (adsorbate) E 1 E 2 Surface EH 2 O ESurf + A+ H 2O EA+ H 2O ESurf + H 2O Surface Solution E = E 1 E 2 (c) Method 2 E Surf+A+H2O E A+H2O E H2O E Surf+H2O + Constant term = E SL using E SL = E = E Surf+A+H2O E A+H2O + E H2O E Surf+H2O S, G analogous 6 of 16
8 Fig. S1. Computation of adsorption energies under periodic boundary conditions (illustrations in 2D for simplicity). (a) Method 1 involves the calculation of average energies in two simulation boxes with the adsorbate on the surface and in solution. (b) Decomposition into smaller boxes for reduced computation time and lower fluctuation in computed energy differences. (c) Method 2 involves four smaller boxes and is more efficient than method 1, particularly for screening many molecules on the same surface. 7 of 16
9 S3. Computational Details S3.1. Models of the Peptides, Water, and the Metallic Substrates. Atomistic Models of the peptides were build from scratch using the amino acid sequences and protonation state at ph = 7 outlined in Table 1. In addition to the possible presence of counterions (Flg-Na 3, Flgd- Na 2, Pd2-Cl, Pd4-Cl), all peptides are used in their zwitterionic form. Models of the peptides in different backbone conformations (α-helix, extended, random coil) were prepared with the help of the Hyperchem visualizer, 47 as well as by manual drawings (random coil) using the Materials Studio builder 48 and simple energy minimization. For water, we employ the standard SPC model implemented in the consistent valence force field (CVFF). 49 Models of the fcc metals were constructed from the unit cell parameter a 0 and the atom positions (0,0,0), (1/2,1/2,0), (1/2, 0, 1/2), (0, 1/2, 1/2). 35 The fcc unit cell and related supercells yield {100} surfaces along the Cartesian coordinate axes. To construct {111} surfaces, an equivalent larger cell was build as a 0 a0 3a0 with the atom positions (0, 0, 0), 2 2 (1/2, 1/2, 0), (0, 1/3, 1/3), (1/2, 5/6, 1/3), (1/2, 1/6, 2/3), (0, 2/3, 2/3). {111} surfaces are then obtained perpendicular to the z axis. 29 The Pd-Au {111} bimetal interface was constructed starting with the Au supercell and replacement of half the atoms by Pd. The lattice constants are very similar ( Å vs Å) so that the scale of the simulation box (3 nm) does not lead to dislocations; a fractional offset of the Pd lattice positions from the nominal Au lattice positions was observed in the course of the simulation. Complete start structures consist of the inorganic substrate, the peptide, and water, or subsets thereof with three-dimensional periodic boundary conditions (see Figure S1c). The structures for each set of simulations were assembled from slabs of the metal substrate, preequilibrated assemblies of 1000 water molecules (2000 water molecules using method 1 in 8 of 16
10 Figure S1a), and one peptide chain using the Materials Studio graphical interface. 48 The initial conformations of the peptide chains were obtained from the build of various conformations, the result of the solution run, as well as from pre-equilibrated peptide conformation on the metal surface in vacuum (without solvent). The setup of the hybrid structures assumes a density of 1000 kg/m 3 for all liquid phases such as water and peptide solutions, and the exact density of the metal consistent with unit cell parameters. The box dimensions are typically 3 nm 3 nm z whereby the box height z amounts to 1.2 nm for metal substrates, 3.5 nm for water, and 3.5 nm for the aqueous solutions of the peptides. The size was chosen such that 25% to 40% of the surface area is covered by the peptide, close to experimental conditions. This comparatively low surface coverage for a single peptide chain in the simulation box also reduces finite size effects to a tolerable level. The peptides can be initially superimposed onto the water slab; subsequent energy minimization removes close contacts with only modest distortions of the initial secondary structure. The concept of additive molecular volumes and the choice of a benchmark density for every component is required for NVT simulations according to Figure S1; computed adsorption properties are not affected if benchmark densities are changed consistently in all simulations in a range of ±1%. S3.2. Force Field Parameters for Peptides and Metals. At the classical atomistic level, several force fields with good parameters for peptides and water are available, e.g., AMBER, CHARMM, CVFF, OPLS-AA, PCFF, and compatible parameters for inorganic structures have been recently developed. 29,37 41 The improvement of Lennard-Jones parameters for fcc metals leads to better than 10% agreement of surface and interface properties with experiment for fcc metals 29,32 36 which is essential to understand sensitive interfacial adsorption processes. In contrast, earlier metal parameters in force fields were associated with deviations in excess of 9 of 16
11 100% and result in wide gaps between computed adsorption properties and actual interfacial interactions in experiment. 3,23 27 In this paper, we employ the consistent valence force field (CVFF) 48,49 extended for accurate Lennard-Jones parameters for fcc metals (Ag, Al, Au, Cu, Ni, Pb, Pd, Pt). 29 This extended version of CVFF, therefore, contains the existing parameters for the peptides and water 48,49 as well as new, accurate parameters for fcc metals. 29 The potential energy expression with no cross-terms and no morse potential is employed: E + pot = Ebond + Eangle + Etorsion + Eout of plane + ECoulomb EvdW (S1) = ij bonded + E K oop r, ij ( r ij r 1 + 4πε ε 0 0, ij ) 2 + θ, ijk ijk bonded r ij nonbonded (1,2 and1,3excl) K q q i r ij j ( θ + ijk θ 0, ijk ) ij nonbonded (1,2 and 1, 3excl) 2 + φ, ijkl ijkl bonded r0, ε ij rij ij V 12 [1+ cos( nφ r0, 2 rij ij 6 ijkl φ 0, ijkl )] Equation (S1) shows the additive energy terms for quadratic bond stretching E bond, quadratic angle bending E angle, a one-term torsion potential torsion E, quadratic out-of-plane deformation Eout of plane, electrostatic energy Coulomb E, and a 12-6 Lennard-Jones potential E vdw for van-der- Waals interactions. Nonbond interactions between 1,4 covalently bonded atoms are included to 100% and geometric combination rules apply for Lennard-Jones parameters between different atom types. A critical advantage over earlier models is the accurate 12 6 Lennard-Jones potential for fcc metals. Computed cell parameters for the metals approach experimental values within 0.2% deviation, computed surface tensions for the {111} surfaces of the metal fall within 1-5% of the experimental values at 298 K (equal to the uncertainty in experiment), and the computed surface energy anisotropy between the {111} and {100} metal surfaces compares favorably with experimental data. 29,33,34 Computed metal-water interfacial tensions are on average 10% to 15% 10 of 16
12 lower than expected from experimental data according to the Young equation. 29 A lower interface tension might have some justification, however, due to attractive polarization effects as the Young equation only provides an upper bound. In detail, polarization will be discussed in a separate contribution. In summary, computed surface energies using metal parameters in earlier semiempirical force fields were associated with errors on the order of 100% so that the present semi-quantitative treatment represents an improvement of between one and two orders of magnitude in accuracy. Remaining sources of uncertainties are (1) polarization effects, (2) the approximate nature of the energy model, specifically limitations of the SPC water model and of the peptide parameters, (3) geometric combination rules of the 12-6 Lennard-Jones parameters, (4) and the inability of the force field to consider covalent contributions, such as major shifts in electron density, to adsorption. S3.3. Simulation Protocol. The programs Discover 48 and LAMMPS 50 were used to carry out molecular dynamics (MD) simulations. Typically, we started with a short energy minimization for 200 steps using the conjugate gradient method, and then carried out NVT molecular dynamics. A time step of 1 fs, the Verlet integrator, the Anderson or the Nose thermostat at K, a van-der-waals cutoff at 1.2 nm, Ewald summation of Coulomb interactions with high accuracy (10 6 kcal/mol), or PPPM summation of Coulomb interactions with high accuracy (10 6 kcal/mol) were employed. The analysis of adsorption energies and chain conformations followed as a post-processing operation after typical simulation times of 5 ns to 10 ns, corresponding to 5 to 10 million time steps. For the analysis, the first 1 ns or more of the trajectory was discarded to take into account only the parts of the trajectory with steady energies and thermodynamically significant conformations. We note thermodynamic averages, 11 of 16
13 as well as the visual and statistical inspection of complete trajectories are required to understand conformational changes so that representative snapshots are intended for guidance only. A bottleneck is the generation of a Boltzmann average of equilibrium conformations of the peptides on the surface. While the modest length of the peptides up to 12 amino acids does not excessively complicate this goal, several start conformations that are distinct from each other, such as helix, outstretched, random coil, and optimized structures in vacuum were required. Solution structures converge to consistent results, however, on the metal surfaces with particularly strong adhesion, we cannot be certain that the global minimum energy has been reached in every case as relaxation times exceed 10 ns. However, the deviation in adsorption energies is mostly in a range of ±5 kcal/mol for different start structures in independent simulations which is less than 10% of the strongest adsorption energies seen. Besides, reference to coarse-grain models and Monte Carlo algorithms to pre-equilibrate the structures in a sequential, reversible two-scale approach may not result in major improvements in sampling as the competition of water and peptide on the surface and specific polar chemistries of side groups are difficult to capture in simplified models. An important aspect in the computation of adsorption energies was the adjustment of the average total energy for each simulation to the same reference temperature ( K) using the heat capacity of the simulation box. Even small temperature differences (<0.1 K) between individual calculations (Figure S1) cause noticeable energy differences (<2 kcal/mol) and could introduce errors in computed adsorption energies. 12 of 16
14 S4. Additional Evidence on the Binding Mechanism from Previous Simulations Additional evidence for the proposed binding mechanism by earlier computational results at various length scales is described in the following. S4.1. Ab-initio methods. Ab-initio methods have been employed to study peptide fragments such as amino acids or water in contact with fcc metals at the smallest level without significant dynamics or solvation. 21,22,24,26,28 In a benchmark study, Haftel et al. 21 compared the reliability of tight-binding (TB), electron density functional methods (DFT), embedded atom models (EAM), and modified embedded atom models (MEAM) to compute surface tensions of several fcc metals (Rh, Ir, Ni, Pd, Pt, Au). The methods exhibit scatter by a factor of two among each other, and typical deviations from experiment are ~20% in each method with individual deviations up to 50% both above and below experimental values. The reliability is therefore a problem and must be taken into account in the discussion of DFT results, as well as difficulties to combine DFT methods with classical force fields to simulate the dynamics of peptides with explicit solvent molecules in solution. Water adsorption and layering on transition metal surfaces was studied by Vassilev et al. 22 using Born-Oppenheimer DFT. On Rh {111} and Pt {111} surfaces, a planar orientation of water molecules is found, and the adsorption energy of single water molecules in vacuum at low surface coverage was computed as 8.6 and 6.9 kcal/mol, at a higher coverage with a water bilayer, 13.1 kcal/mol and 13.1 kcal/mol for Rh and Pt surfaces. Schravendijk et al. 24 also found a planar coordination of water molecules on the metal surfaces and computed slightly higher adsorption energies of single water molecules of 9.7 and 8.1 kcal/mol on Rh {111} and Pt {111} surfaces, as well as 2.3 and 7.6 kcal/mol on Au {111} and Pd {111} surfaces. Classical MD simulation (section S3) agrees with the preference for a planar orientation and adsorption 13 of 16
15 energies of 4.4 and 5.4 kcal/mol are computed on the Au {111} and on the Pd {111} surface at <5% surface coverage (calculation as in Figure S1a with one water molecule as adsorbate in vacuum). 29 These values agree with lower surface tensions of Au and Pd versus Rh and Pt; 32 the gap in DFT calculations between the Au {111} and Pd {111} surface yet remains to be explained on physical grounds. 24 Experimental measurements of contact angles indicate 0º and, therefore, hydrophilic properties of both clean Au and Pd surfaces, 36 in agreement with attraction of water to the metal surfaces in the computation. Further, dual-scale simulations of benzene adsorption on Ni {111} and Au {111} surfaces including a few explicit water molecules were carried out by DFT and Car-Parrinello ab-initio MD molecular dynamics as well as specialized Lennard-Jones and Morse potentials for a classical description of the interaction with the metal. Taking into account competition between a few water molecules and the benzene solute, the adsorption energy of benzene in solution on the Ni {111} surface was computed as 9.6 kcal/mol, slightly stronger adsorption was found on the Pd {111} surface, and a preference for desorption was predicted on the Au {111} surface. However, the surface tension of the metals was not validated in these models and the few included water molecules are a crude approximation of a solvation shell. Nevertheless, the binding energy agrees with the order of magnitude for aromatic amino acids (Table 2 and Table 4), and a weaker attraction of benzene (in Phe) to the Au {111} surface is also observed in our study. Our results indicate still adsorption in agreement with experimental data (section 5.1). 4,7,8 It may be alternatively possible that phenyl rings are positioned flat on the surface only in order to provide sufficient contact area for neighboring parts of the molecule which are factually involved in an adsorption process. Schravendijk s method 24 was further extended into a highly specialized force field description for the interaction of selected hydrated amino acids with a Ni {111} surface. 26 A new extensive set of parameters would be required for 14 of 16
16 every additional amino acid and every additional metal. Meanwhile, the validation of the metal surface energy and the physical justification of the numerous parameters remain unclear, and incompatibility with existing biomolecular force renders the approach less practical. Therefore, atomistic simulation with extended force field engines as presented in this paper yields results of the same, if not better, quality, and is simpler, faster, and more widely applicable. Ghiringhelli et al. 28 studied Phe adsorption in vacuum on {111} surfaces of fcc metals using DFT. Strong adsorption of 30 to 25 kcal/mol (Pt, Ni, Pd) and weaker adsorption of 12 to 7 kcal/mol (Cu, Ag, Au) was found. For Cu, Ag, Au, the phenyl ring is not in close contact with the surface. Although water was absent in this study, the range of values is similar to those inferred from the force-field based simulation (Tables 2 and Table 4); 30 a lesser attraction of the phenyl ring to the Au {111} surface compared to the Pd {111} surface is observed in our method as well. Hong et al. 31 have shown by further DFT calculations in vacuum that charge transfer is possible on Au and Pd surfaces. Up to 0.65e could be transferred per negatively charged residue such as CO 2 of Asp over numerous electron accepting superficial Au atoms in vacuum (Table 3). In aqueous solution, however, the presence of water molecules as a solvent of similar polarity diminishes the amount of charge transfer from a single CO 2 group which would be more challenging to analyze using this method. Among Asp, Lys, Arg, Ser, Pro, and Val, the strongest binding residue in vacuum has been Asp by a considerable margin ( 57 kcal/mol), followed by Lys ( 27 kcal/mol) and Arg ( 19 kcal/mol), and almost no binding was found for Pro and Ser. While the values will be substantially smaller in aqueous solution, this ranking coincides with the force-field based simulation (Tables 2 and 4). S4.2. Force-field based simulations. Sarikaya et al. 3,S1 carried out classical molecular dynamics simulations of large proteins (>40 amino acids) on Au {111} and {112} surfaces with 15 of 16
17 a number of heavy approximations which weigh on the reliability of the results: (1) The proteins were placed in a dielectric continuum so that explicit water molecules and hydrogen bonds are absent. (2) The surface properties of Au according to the chosen Lennard-Jones parameters were not validated which adds uncertainty in excess of 100% to adsorption energies and affects the molecular mechanism of adsorption of the peptide. 29 (3) The thickness of the surfaces was restricted to only 3 atomic layers (6 Å) which is shorter than the range of interactions between the metal atoms (at least Å). Comparisons relative to experiment are rather difficult under these conditions, and further simulations of peptides on metal surfaces using this approach lead to a number of questionable specific results. 23 An interesting outcome, however, is the response of peptides to gross surface features such as ridges in {110} surfaces. A subsequent study by Kantarci et al. 25 extended the approach using an all atom representation of peptides and water on Pt {111} surfaces. This represents a more realistic model, however, the Lennard-Jones parameters for the metal according to the unmodified CVFF overestimate the Pt surface tension by 101% relative to experiment and modify the interfacial structures and adsorption processes; 29 the thin metal layer also introduced residual interactions between water molecules above and below the metal slab. Under these severe approximations, Ser, Thr, Arg, and Pro were found in proximity to the metal surface (Pt). Overall, earlier all atom models may not have been reliable enough for quantitative or firm qualitative conclusions. References S1 Braun, R.; Sarikaya, M.; Schulten, K. S. J. Biomater. Sci. 2002, 13, of 16
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
The strength of the interaction
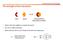 The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
H 2O gas: molecules are very far apart
 Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Peptide bonds: resonance structure. Properties of proteins: Peptide bonds and side chains. Dihedral angles. Peptide bond. Protein physics, Lecture 5
 Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Section 3: Crystal Binding
 Physics 97 Interatomic forces Section 3: rystal Binding Solids are stable structures, and therefore there exist interactions holding atoms in a crystal together. For example a crystal of sodium chloride
Physics 97 Interatomic forces Section 3: rystal Binding Solids are stable structures, and therefore there exist interactions holding atoms in a crystal together. For example a crystal of sodium chloride
Chapter 2. Atomic Structure and Interatomic Bonding
 Chapter 2. Atomic Structure and Interatomic Bonding Interatomic Bonding Bonding forces and energies Primary interatomic bonds Secondary bonding Molecules Bonding Forces and Energies Considering the interaction
Chapter 2. Atomic Structure and Interatomic Bonding Interatomic Bonding Bonding forces and energies Primary interatomic bonds Secondary bonding Molecules Bonding Forces and Energies Considering the interaction
Explain the ionic bonds, covalent bonds and metallic bonds and give one example for each type of bonds.
 Problem 1 Explain the ionic bonds, covalent bonds and metallic bonds and give one example for each type of bonds. Ionic Bonds Two neutral atoms close to each can undergo an ionization process in order
Problem 1 Explain the ionic bonds, covalent bonds and metallic bonds and give one example for each type of bonds. Ionic Bonds Two neutral atoms close to each can undergo an ionization process in order
Advanced Micro Ring Resonator Filter Technology
 Advanced Micro Ring Resonator Filter Technology G. Lenz and C. K. Madsen Lucent Technologies, Bell Labs Report Documentation Page Form Approved OMB No. 0704-0188 Public reporting burden for the collection
Advanced Micro Ring Resonator Filter Technology G. Lenz and C. K. Madsen Lucent Technologies, Bell Labs Report Documentation Page Form Approved OMB No. 0704-0188 Public reporting burden for the collection
OPTICAL IMAGES DUE TO LENSES AND MIRRORS *
 1 OPTICAL IMAGES DUE TO LENSES AND MIRRORS * Carl E. Mungan U.S. Naval Academy, Annapolis, MD ABSTRACT The properties of real and virtual images formed by lenses and mirrors are reviewed. Key ideas are
1 OPTICAL IMAGES DUE TO LENSES AND MIRRORS * Carl E. Mungan U.S. Naval Academy, Annapolis, MD ABSTRACT The properties of real and virtual images formed by lenses and mirrors are reviewed. Key ideas are
Chapter 2 Polar Covalent Bonds; Acids and Bases
 John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK
 Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Data Visualization for Atomistic/Molecular Simulations. Douglas E. Spearot University of Arkansas
 Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
CHAPTER 26 ELECTROSTATIC ENERGY AND CAPACITORS
 CHAPTER 6 ELECTROSTATIC ENERGY AND CAPACITORS. Three point charges, each of +q, are moved from infinity to the vertices of an equilateral triangle of side l. How much work is required? The sentence preceding
CHAPTER 6 ELECTROSTATIC ENERGY AND CAPACITORS. Three point charges, each of +q, are moved from infinity to the vertices of an equilateral triangle of side l. How much work is required? The sentence preceding
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras
 Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Role of Hydrogen Bonding on Protein Secondary Structure Introduction
 Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Using the Advancement Degree of Difficulty (AD 2 ) as an input to Risk Management
 Using the Advancement Degree of Difficulty (AD 2 ) as an input to Risk Management James W. Bilbro JB Consulting International Huntsville, AL Multi-dimensional Assessment of Technology Maturity Technology
Using the Advancement Degree of Difficulty (AD 2 ) as an input to Risk Management James W. Bilbro JB Consulting International Huntsville, AL Multi-dimensional Assessment of Technology Maturity Technology
Lecture 15: Enzymes & Kinetics Mechanisms
 ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005
 Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING
 CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
18.2 Protein Structure and Function: An Overview
 18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
Name Class Date. In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question.
 Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Chapter 13 - LIQUIDS AND SOLIDS
 Chapter 13 - LIQUIDS AND SOLIDS Problems to try at end of chapter: Answers in Appendix I: 1,3,5,7b,9b,15,17,23,25,29,31,33,45,49,51,53,61 13.1 Properties of Liquids 1. Liquids take the shape of their container,
Chapter 13 - LIQUIDS AND SOLIDS Problems to try at end of chapter: Answers in Appendix I: 1,3,5,7b,9b,15,17,23,25,29,31,33,45,49,51,53,61 13.1 Properties of Liquids 1. Liquids take the shape of their container,
What is molecular dynamics (MD) simulation and how does it work?
 What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
Why? Intermolecular Forces. Intermolecular Forces. Chapter 12 IM Forces and Liquids. Covalent Bonding Forces for Comparison of Magnitude
 1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
KINETIC MOLECULAR THEORY OF MATTER
 KINETIC MOLECULAR THEORY OF MATTER The kinetic-molecular theory is based on the idea that particles of matter are always in motion. The theory can be used to explain the properties of solids, liquids,
KINETIC MOLECULAR THEORY OF MATTER The kinetic-molecular theory is based on the idea that particles of matter are always in motion. The theory can be used to explain the properties of solids, liquids,
Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
 Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Introduction Molecular Mechanics or force-field methods use classical type models
Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Introduction Molecular Mechanics or force-field methods use classical type models
Asset Management- Acquisitions
 Indefinite Delivery, Indefinite Quantity (IDIQ) contracts used for these services: al, Activity & Master Plans; Land Use Analysis; Anti- Terrorism, Circulation & Space Management Studies; Encroachment
Indefinite Delivery, Indefinite Quantity (IDIQ) contracts used for these services: al, Activity & Master Plans; Land Use Analysis; Anti- Terrorism, Circulation & Space Management Studies; Encroachment
Simulation of Air Flow Through a Test Chamber
 Simulation of Air Flow Through a Test Chamber by Gregory K. Ovrebo ARL-MR- 0680 December 2007 Approved for public release; distribution unlimited. NOTICES Disclaimers The findings in this report are not
Simulation of Air Flow Through a Test Chamber by Gregory K. Ovrebo ARL-MR- 0680 December 2007 Approved for public release; distribution unlimited. NOTICES Disclaimers The findings in this report are not
Monitoring of Arctic Conditions from a Virtual Constellation of Synthetic Aperture Radar Satellites
 DISTRIBUTION STATEMENT A. Approved for public release; distribution is unlimited. Monitoring of Arctic Conditions from a Virtual Constellation of Synthetic Aperture Radar Satellites Hans C. Graber RSMAS
DISTRIBUTION STATEMENT A. Approved for public release; distribution is unlimited. Monitoring of Arctic Conditions from a Virtual Constellation of Synthetic Aperture Radar Satellites Hans C. Graber RSMAS
Use the Force! Noncovalent Molecular Forces
 Use the Force! Noncovalent Molecular Forces Not quite the type of Force we re talking about Before we talk about noncovalent molecular forces, let s talk very briefly about covalent bonds. The Illustrated
Use the Force! Noncovalent Molecular Forces Not quite the type of Force we re talking about Before we talk about noncovalent molecular forces, let s talk very briefly about covalent bonds. The Illustrated
Topic 2: Energy in Biological Systems
 Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Chemical Bonds and Groups - Part 1
 hemical Bonds and Groups - Part 1 ARB SKELETS arbon has a unique role in the cell because of its ability to form strong covalent bonds with other carbon atoms. Thus carbon atoms can join to form chains.
hemical Bonds and Groups - Part 1 ARB SKELETS arbon has a unique role in the cell because of its ability to form strong covalent bonds with other carbon atoms. Thus carbon atoms can join to form chains.
Introduction to Chemistry. Course Description
 CHM 1025 & CHM 1025L Introduction to Chemistry Course Description CHM 1025 Introduction to Chemistry (3) P CHM 1025L Introduction to Chemistry Laboratory (1) P This introductory course is intended to introduce
CHM 1025 & CHM 1025L Introduction to Chemistry Course Description CHM 1025 Introduction to Chemistry (3) P CHM 1025L Introduction to Chemistry Laboratory (1) P This introductory course is intended to introduce
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon. V. Polypeptides and Proteins
 IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
Language: English Lecturer: Gianni de Fabritiis. Teaching staff: Language: English Lecturer: Jordi Villà i Freixa
 MSI: Molecular Simulations Descriptive details concerning the subject: Name of the subject: Molecular Simulations Code : MSI Type of subject: Optional ECTS: 5 Total hours: 125.0 Scheduling: 11:00-13:00
MSI: Molecular Simulations Descriptive details concerning the subject: Name of the subject: Molecular Simulations Code : MSI Type of subject: Optional ECTS: 5 Total hours: 125.0 Scheduling: 11:00-13:00
(c) How would your answers to problem (a) change if the molecular weight of the protein was 100,000 Dalton?
 Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Introduction, Noncovalent Bonds, and Properties of Water
 Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
Chapter 6 An Overview of Organic Reactions
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
Chapter 5: Diffusion. 5.1 Steady-State Diffusion
 : Diffusion Diffusion: the movement of particles in a solid from an area of high concentration to an area of low concentration, resulting in the uniform distribution of the substance Diffusion is process
: Diffusion Diffusion: the movement of particles in a solid from an area of high concentration to an area of low concentration, resulting in the uniform distribution of the substance Diffusion is process
Overview Presented by: Boyd L. Summers
 Overview Presented by: Boyd L. Summers Systems & Software Technology Conference SSTC May 19 th, 2011 1 Report Documentation Page Form Approved OMB No. 0704-0188 Public reporting burden for the collection
Overview Presented by: Boyd L. Summers Systems & Software Technology Conference SSTC May 19 th, 2011 1 Report Documentation Page Form Approved OMB No. 0704-0188 Public reporting burden for the collection
Molecular Modelling of DNA. Charlie Laughton University of Nottingham
 Molecular Modelling of DNA Charlie Laughton University of Nottingham Why model DNA? Sequences/structures you can t crystallise or get by NMR Drug design Understanding protein-dna recognition Quantitating
Molecular Modelling of DNA Charlie Laughton University of Nottingham Why model DNA? Sequences/structures you can t crystallise or get by NMR Drug design Understanding protein-dna recognition Quantitating
Metamaterials and Transformation Optics
 AFRL-AFOSR-UK-TR-2011-0021 Metamaterials and Transformation Optics John Pendry Imperial College London Physics Department South Kensington Campus Exhibition Road London, United Kingdom SW7 2AZ EOARD GRANT
AFRL-AFOSR-UK-TR-2011-0021 Metamaterials and Transformation Optics John Pendry Imperial College London Physics Department South Kensington Campus Exhibition Road London, United Kingdom SW7 2AZ EOARD GRANT
Elements in the periodic table are indicated by SYMBOLS. To the left of the symbol we find the atomic mass (A) at the upper corner, and the atomic num
 . ATOMIC STRUCTURE FUNDAMENTALS LEARNING OBJECTIVES To review the basics concepts of atomic structure that have direct relevance to the fundamental concepts of organic chemistry. This material is essential
. ATOMIC STRUCTURE FUNDAMENTALS LEARNING OBJECTIVES To review the basics concepts of atomic structure that have direct relevance to the fundamental concepts of organic chemistry. This material is essential
Recap. Lecture 2. Protein conformation. Proteins. 8 types of protein function 10/21/10. Proteins.. > 50% dry weight of a cell
 Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Ultraviolet Spectroscopy
 Ultraviolet Spectroscopy The wavelength of UV and visible light are substantially shorter than the wavelength of infrared radiation. The UV spectrum ranges from 100 to 400 nm. A UV-Vis spectrophotometer
Ultraviolet Spectroscopy The wavelength of UV and visible light are substantially shorter than the wavelength of infrared radiation. The UV spectrum ranges from 100 to 400 nm. A UV-Vis spectrophotometer
CHAPTER 7 DISLOCATIONS AND STRENGTHENING MECHANISMS PROBLEM SOLUTIONS
 7-1 CHAPTER 7 DISLOCATIONS AND STRENGTHENING MECHANISMS PROBLEM SOLUTIONS Basic Concepts of Dislocations Characteristics of Dislocations 7.1 The dislocation density is just the total dislocation length
7-1 CHAPTER 7 DISLOCATIONS AND STRENGTHENING MECHANISMS PROBLEM SOLUTIONS Basic Concepts of Dislocations Characteristics of Dislocations 7.1 The dislocation density is just the total dislocation length
2054-2. Structure and Dynamics of Hydrogen-Bonded Systems. 26-27 October 2009. Hydrogen Bonds and Liquid Water
 2054-2 Structure and Dynamics of Hydrogen-Bonded Systems 26-27 October 2009 Hydrogen Bonds and Liquid Water Ruth LYNDEN-BELL University of Cambridge, Department of Chemistry Lensfield Road Cambridge CB2
2054-2 Structure and Dynamics of Hydrogen-Bonded Systems 26-27 October 2009 Hydrogen Bonds and Liquid Water Ruth LYNDEN-BELL University of Cambridge, Department of Chemistry Lensfield Road Cambridge CB2
Structure Check. Authors: Eduard Schreiner Leonardo G. Trabuco. February 7, 2012
 University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
: : Solutions to Additional Bonding Problems
 Solutions to Additional Bonding Problems 1 1. For the following examples, the valence electron count is placed in parentheses after the empirical formula and only the resonance structures that satisfy
Solutions to Additional Bonding Problems 1 1. For the following examples, the valence electron count is placed in parentheses after the empirical formula and only the resonance structures that satisfy
Bonding & Molecular Shape Ron Robertson
 Bonding & Molecular Shape Ron Robertson r2 n:\files\courses\1110-20\2010 possible slides for web\00bondingtrans.doc The Nature of Bonding Types 1. Ionic 2. Covalent 3. Metallic 4. Coordinate covalent Driving
Bonding & Molecular Shape Ron Robertson r2 n:\files\courses\1110-20\2010 possible slides for web\00bondingtrans.doc The Nature of Bonding Types 1. Ionic 2. Covalent 3. Metallic 4. Coordinate covalent Driving
Chapter Outline. Diffusion - how do atoms move through solids?
 Chapter Outline iffusion - how do atoms move through solids? iffusion mechanisms Vacancy diffusion Interstitial diffusion Impurities The mathematics of diffusion Steady-state diffusion (Fick s first law)
Chapter Outline iffusion - how do atoms move through solids? iffusion mechanisms Vacancy diffusion Interstitial diffusion Impurities The mathematics of diffusion Steady-state diffusion (Fick s first law)
2. Spin Chemistry and the Vector Model
 2. Spin Chemistry and the Vector Model The story of magnetic resonance spectroscopy and intersystem crossing is essentially a choreography of the twisting motion which causes reorientation or rephasing
2. Spin Chemistry and the Vector Model The story of magnetic resonance spectroscopy and intersystem crossing is essentially a choreography of the twisting motion which causes reorientation or rephasing
PROTEINS THE PEPTIDE BOND. The peptide bond, shown above enclosed in the blue curves, generates the basic structural unit for proteins.
 Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
Report Documentation Page
 (c)2002 American Institute Report Documentation Page Form Approved OMB No. 0704-0188 Public reporting burden for the collection of information is estimated to average 1 hour per response, including the
(c)2002 American Institute Report Documentation Page Form Approved OMB No. 0704-0188 Public reporting burden for the collection of information is estimated to average 1 hour per response, including the
Non-Autoclave (Prepreg) Manufacturing Technology
 Non-Autoclave (Prepreg) Manufacturing Technology Gary G. Bond, John M. Griffith, Gail L. Hahn The Boeing Company Chris Bongiovanni, Jack Boyd Cytec Engineered Materials 9 September 2008 Report Documentation
Non-Autoclave (Prepreg) Manufacturing Technology Gary G. Bond, John M. Griffith, Gail L. Hahn The Boeing Company Chris Bongiovanni, Jack Boyd Cytec Engineered Materials 9 September 2008 Report Documentation
100% ionic compounds do not exist but predominantly ionic compounds are formed when metals combine with non-metals.
 2.21 Ionic Bonding 100% ionic compounds do not exist but predominantly ionic compounds are formed when metals combine with non-metals. Forming ions Metal atoms lose electrons to form +ve ions. Non-metal
2.21 Ionic Bonding 100% ionic compounds do not exist but predominantly ionic compounds are formed when metals combine with non-metals. Forming ions Metal atoms lose electrons to form +ve ions. Non-metal
FIRST IMPRESSION EXPERIMENT REPORT (FIER)
 THE MNE7 OBJECTIVE 3.4 CYBER SITUATIONAL AWARENESS LOE FIRST IMPRESSION EXPERIMENT REPORT (FIER) 1. Introduction The Finnish Defence Forces Concept Development & Experimentation Centre (FDF CD&E Centre)
THE MNE7 OBJECTIVE 3.4 CYBER SITUATIONAL AWARENESS LOE FIRST IMPRESSION EXPERIMENT REPORT (FIER) 1. Introduction The Finnish Defence Forces Concept Development & Experimentation Centre (FDF CD&E Centre)
Vacuum Evaporation Recap
 Sputtering Vacuum Evaporation Recap Use high temperatures at high vacuum to evaporate (eject) atoms or molecules off a material surface. Use ballistic flow to transport them to a substrate and deposit.
Sputtering Vacuum Evaporation Recap Use high temperatures at high vacuum to evaporate (eject) atoms or molecules off a material surface. Use ballistic flow to transport them to a substrate and deposit.
Express Introductory Training in ANSYS Fluent Lecture 1 Introduction to the CFD Methodology
 Express Introductory Training in ANSYS Fluent Lecture 1 Introduction to the CFD Methodology Dimitrios Sofialidis Technical Manager, SimTec Ltd. Mechanical Engineer, PhD PRACE Autumn School 2013 - Industry
Express Introductory Training in ANSYS Fluent Lecture 1 Introduction to the CFD Methodology Dimitrios Sofialidis Technical Manager, SimTec Ltd. Mechanical Engineer, PhD PRACE Autumn School 2013 - Industry
KINETIC THEORY OF MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature
 1 KINETIC TERY F MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature TE STATES F MATTER 1. Gas a) ideal gas - molecules move freely - molecules have
1 KINETIC TERY F MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature TE STATES F MATTER 1. Gas a) ideal gas - molecules move freely - molecules have
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
UMass at TREC 2008 Blog Distillation Task
 UMass at TREC 2008 Blog Distillation Task Jangwon Seo and W. Bruce Croft Center for Intelligent Information Retrieval University of Massachusetts, Amherst Abstract This paper presents the work done for
UMass at TREC 2008 Blog Distillation Task Jangwon Seo and W. Bruce Croft Center for Intelligent Information Retrieval University of Massachusetts, Amherst Abstract This paper presents the work done for
MOLECULAR DYNAMICS INVESTIGATION OF DEFORMATION RESPONSE OF THIN-FILM METALLIC NANOSTRUCTURES UNDER HEATING
 NANOSYSTEMS: PHYSICS, CHEMISTRY, MATHEMATICS, 2011, 2 (2), P. 76 83 UDC 538.97 MOLECULAR DYNAMICS INVESTIGATION OF DEFORMATION RESPONSE OF THIN-FILM METALLIC NANOSTRUCTURES UNDER HEATING I. S. Konovalenko
NANOSYSTEMS: PHYSICS, CHEMISTRY, MATHEMATICS, 2011, 2 (2), P. 76 83 UDC 538.97 MOLECULAR DYNAMICS INVESTIGATION OF DEFORMATION RESPONSE OF THIN-FILM METALLIC NANOSTRUCTURES UNDER HEATING I. S. Konovalenko
Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert Karakaş, Phoebe L. Stewart, and Jens Meiler
 Structure 17 Supplemental Data EM-Fold: De Novo Folding of α-helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert
Structure 17 Supplemental Data EM-Fold: De Novo Folding of α-helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert
Matter, Materials, Crystal Structure and Bonding. Chris J. Pickard
 Matter, Materials, Crystal Structure and Bonding Chris J. Pickard Why should a theorist care? Where the atoms are determines what they do Where the atoms can be determines what we can do Overview of Structure
Matter, Materials, Crystal Structure and Bonding Chris J. Pickard Why should a theorist care? Where the atoms are determines what they do Where the atoms can be determines what we can do Overview of Structure
1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA
 1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA Tomáš Kubař Institute of Organic Chemistry and Biochemistry Praha, Czech Republic Thermodynamic Integration Alchemical change
1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA Tomáš Kubař Institute of Organic Chemistry and Biochemistry Praha, Czech Republic Thermodynamic Integration Alchemical change
Surface Area and Porosity
 Surface Area and Porosity 1 Background Techniques Surface area Outline Total - physical adsorption External Porosity meso micro 2 Length 1 Å 1 nm 1 µm 1 1 1 1 1 mm macro meso micro metal crystallite 1-1
Surface Area and Porosity 1 Background Techniques Surface area Outline Total - physical adsorption External Porosity meso micro 2 Length 1 Å 1 nm 1 µm 1 1 1 1 1 mm macro meso micro metal crystallite 1-1
Fundamentals of grain boundaries and grain boundary migration
 1. Fundamentals of grain boundaries and grain boundary migration 1.1. Introduction The properties of crystalline metallic materials are determined by their deviation from a perfect crystal lattice, which
1. Fundamentals of grain boundaries and grain boundary migration 1.1. Introduction The properties of crystalline metallic materials are determined by their deviation from a perfect crystal lattice, which
Refinement of a pdb-structure and Convert
 Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Section Activity #1: Fill out the following table for biology s most common elements assuming that each atom is neutrally charged.
 LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
Adsorption at Surfaces
 Adsorption at Surfaces Adsorption is the accumulation of particles (adsorbate) at a surface (adsorbent or substrate). The reverse process is called desorption. fractional surface coverage: θ = Number of
Adsorption at Surfaces Adsorption is the accumulation of particles (adsorbate) at a surface (adsorbent or substrate). The reverse process is called desorption. fractional surface coverage: θ = Number of
CHAPTER 6 AN INTRODUCTION TO METABOLISM. Section B: Enzymes
 CHAPTER 6 AN INTRODUCTION TO METABOLISM Section B: Enzymes 1. Enzymes speed up metabolic reactions by lowering energy barriers 2. Enzymes are substrate specific 3. The active site in an enzyme s catalytic
CHAPTER 6 AN INTRODUCTION TO METABOLISM Section B: Enzymes 1. Enzymes speed up metabolic reactions by lowering energy barriers 2. Enzymes are substrate specific 3. The active site in an enzyme s catalytic
Lecture Overview. Hydrogen Bonds. Special Properties of Water Molecules. Universal Solvent. ph Scale Illustrated. special properties of water
 Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Copyright 2011 Casa Software Ltd. www.casaxps.com. Centre of Mass
 Centre of Mass A central theme in mathematical modelling is that of reducing complex problems to simpler, and hopefully, equivalent problems for which mathematical analysis is possible. The concept of
Centre of Mass A central theme in mathematical modelling is that of reducing complex problems to simpler, and hopefully, equivalent problems for which mathematical analysis is possible. The concept of
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.
 CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
Chapter 2 Polar Covalent Bonds: Acids and Bases
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds: Acids and Bases Modified by Dr. Daniela R. Radu Why This Chapter? Description of basic ways chemists account for chemical
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds: Acids and Bases Modified by Dr. Daniela R. Radu Why This Chapter? Description of basic ways chemists account for chemical
Multiple Network Marketing coordination Model
 REPORT DOCUMENTATION PAGE Form Approved OMB No. 0704-0188 The public reporting burden for this collection of information is estimated to average 1 hour per response, including the time for reviewing instructions,
REPORT DOCUMENTATION PAGE Form Approved OMB No. 0704-0188 The public reporting burden for this collection of information is estimated to average 1 hour per response, including the time for reviewing instructions,
Material Deformations. Academic Resource Center
 Material Deformations Academic Resource Center Agenda Origin of deformations Deformations & dislocations Dislocation motion Slip systems Stresses involved with deformation Deformation by twinning Origin
Material Deformations Academic Resource Center Agenda Origin of deformations Deformations & dislocations Dislocation motion Slip systems Stresses involved with deformation Deformation by twinning Origin
RT 24 - Architecture, Modeling & Simulation, and Software Design
 RT 24 - Architecture, Modeling & Simulation, and Software Design Dennis Barnabe, Department of Defense Michael zur Muehlen & Anne Carrigy, Stevens Institute of Technology Drew Hamilton, Auburn University
RT 24 - Architecture, Modeling & Simulation, and Software Design Dennis Barnabe, Department of Defense Michael zur Muehlen & Anne Carrigy, Stevens Institute of Technology Drew Hamilton, Auburn University
Transmembrane proteins span the bilayer. α-helix transmembrane domain. Multiple transmembrane helices in one polypeptide
 Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Ionization of amino acids
 Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Chapter 8. Low energy ion scattering study of Fe 4 N on Cu(100)
 Low energy ion scattering study of 4 on Cu(1) Chapter 8. Low energy ion scattering study of 4 on Cu(1) 8.1. Introduction For a better understanding of the reconstructed 4 surfaces one would like to know
Low energy ion scattering study of 4 on Cu(1) Chapter 8. Low energy ion scattering study of 4 on Cu(1) 8.1. Introduction For a better understanding of the reconstructed 4 surfaces one would like to know
Studying an Organic Reaction. How do we know if a reaction can occur? And if a reaction can occur what do we know about the reaction?
 Studying an Organic Reaction How do we know if a reaction can occur? And if a reaction can occur what do we know about the reaction? Information we want to know: How much heat is generated? How fast is
Studying an Organic Reaction How do we know if a reaction can occur? And if a reaction can occur what do we know about the reaction? Information we want to know: How much heat is generated? How fast is
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT
 BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
Covalent Bonding & Molecular Orbital Theory
 Covalent Bonding & Molecular Orbital Theory Chemistry 754 Solid State Chemistry Dr. Patrick Woodward Lecture #16 References - MO Theory Molecular orbital theory is covered in many places including most
Covalent Bonding & Molecular Orbital Theory Chemistry 754 Solid State Chemistry Dr. Patrick Woodward Lecture #16 References - MO Theory Molecular orbital theory is covered in many places including most
PCV Project: Excitons in Molecular Spectroscopy
 PCV Project: Excitons in Molecular Spectroscopy Introduction The concept of excitons was first introduced by Frenkel (1) in 1931 as a general excitation delocalization mechanism to account for the ability
PCV Project: Excitons in Molecular Spectroscopy Introduction The concept of excitons was first introduced by Frenkel (1) in 1931 as a general excitation delocalization mechanism to account for the ability
INTERMOLECULAR FORCES
 INTERMOLECULAR FORCES Intermolecular forces- forces of attraction and repulsion between molecules that hold molecules, ions, and atoms together. Intramolecular - forces of chemical bonds within a molecule
INTERMOLECULAR FORCES Intermolecular forces- forces of attraction and repulsion between molecules that hold molecules, ions, and atoms together. Intramolecular - forces of chemical bonds within a molecule
AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts
 AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts 8.1 Types of Chemical Bonds A. Ionic Bonding 1. Electrons are transferred 2. Metals react with nonmetals 3. Ions paired have lower energy
AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts 8.1 Types of Chemical Bonds A. Ionic Bonding 1. Electrons are transferred 2. Metals react with nonmetals 3. Ions paired have lower energy
The Properties of Water
 1 Matter & Energy: Properties of Water, ph, Chemical Reactions EVPP 110 Lecture GMU Dr. Largen Fall 2003 2 The Properties of Water 3 Water - Its Properties and Its Role in the Fitness of Environment importance
1 Matter & Energy: Properties of Water, ph, Chemical Reactions EVPP 110 Lecture GMU Dr. Largen Fall 2003 2 The Properties of Water 3 Water - Its Properties and Its Role in the Fitness of Environment importance
Molecular Models in Biology
 Molecular Models in Biology Objectives: After this lab a student will be able to: 1) Understand the properties of atoms that give rise to bonds. 2) Understand how and why atoms form ions. 3) Model covalent,
Molecular Models in Biology Objectives: After this lab a student will be able to: 1) Understand the properties of atoms that give rise to bonds. 2) Understand how and why atoms form ions. 3) Model covalent,
Introduction to Principal Components and FactorAnalysis
 Introduction to Principal Components and FactorAnalysis Multivariate Analysis often starts out with data involving a substantial number of correlated variables. Principal Component Analysis (PCA) is a
Introduction to Principal Components and FactorAnalysis Multivariate Analysis often starts out with data involving a substantial number of correlated variables. Principal Component Analysis (PCA) is a
A GPS Digital Phased Array Antenna and Receiver
 A GPS Digital Phased Array Antenna and Receiver Dr. Alison Brown, Randy Silva; NAVSYS Corporation ABSTRACT NAVSYS High Gain Advanced GPS Receiver (HAGR) uses a digital beam-steering antenna array to enable
A GPS Digital Phased Array Antenna and Receiver Dr. Alison Brown, Randy Silva; NAVSYS Corporation ABSTRACT NAVSYS High Gain Advanced GPS Receiver (HAGR) uses a digital beam-steering antenna array to enable
DEFENSE CONTRACT AUDIT AGENCY
 DEFENSE CONTRACT AUDIT AGENCY Fundamental Building Blocks for an Acceptable Accounting System Presented by Sue Reynaga DCAA Branch Manager San Diego Branch Office August 24, 2011 Report Documentation Page
DEFENSE CONTRACT AUDIT AGENCY Fundamental Building Blocks for an Acceptable Accounting System Presented by Sue Reynaga DCAA Branch Manager San Diego Branch Office August 24, 2011 Report Documentation Page
Integrated Force Method Solution to Indeterminate Structural Mechanics Problems
 NASA/TP 2004-207430 Integrated Force Method Solution to Indeterminate Structural Mechanics Problems Surya N. Patnaik Ohio Aerospace Institute, Brook Park, Ohio Dale A. Hopkins and Gary R. Halford Glenn
NASA/TP 2004-207430 Integrated Force Method Solution to Indeterminate Structural Mechanics Problems Surya N. Patnaik Ohio Aerospace Institute, Brook Park, Ohio Dale A. Hopkins and Gary R. Halford Glenn
Headquarters U.S. Air Force
 Headquarters U.S. Air Force I n t e g r i t y - S e r v i c e - E x c e l l e n c e Air Force Technology Readiness Assessment (TRA) Process for Major Defense Acquisition Programs LtCol Ed Masterson Mr
Headquarters U.S. Air Force I n t e g r i t y - S e r v i c e - E x c e l l e n c e Air Force Technology Readiness Assessment (TRA) Process for Major Defense Acquisition Programs LtCol Ed Masterson Mr
Molecular Geometry and VSEPR We gratefully acknowledge Portland Community College for the use of this experiment.
 Molecular and VSEPR We gratefully acknowledge Portland ommunity ollege for the use of this experiment. Objectives To construct molecular models for covalently bonded atoms in molecules and polyatomic ions
Molecular and VSEPR We gratefully acknowledge Portland ommunity ollege for the use of this experiment. Objectives To construct molecular models for covalently bonded atoms in molecules and polyatomic ions
Universitätsstrasse 1, D-40225 Düsseldorf, Germany 3 Current address: Institut für Festkörperforschung,
 Lane formation in oppositely charged colloidal mixtures - supplementary information Teun Vissers 1, Adam Wysocki 2,3, Martin Rex 2, Hartmut Löwen 2, C. Patrick Royall 1,4, Arnout Imhof 1, and Alfons van
Lane formation in oppositely charged colloidal mixtures - supplementary information Teun Vissers 1, Adam Wysocki 2,3, Martin Rex 2, Hartmut Löwen 2, C. Patrick Royall 1,4, Arnout Imhof 1, and Alfons van
Finite Element Formulation for Plates - Handout 3 -
 Finite Element Formulation for Plates - Handout 3 - Dr Fehmi Cirak (fc286@) Completed Version Definitions A plate is a three dimensional solid body with one of the plate dimensions much smaller than the
Finite Element Formulation for Plates - Handout 3 - Dr Fehmi Cirak (fc286@) Completed Version Definitions A plate is a three dimensional solid body with one of the plate dimensions much smaller than the
