Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
|
|
|
- Emery Maxwell
- 8 years ago
- Views:
Transcription
1 Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
2 Introduction Molecular Mechanics or force-field methods use classical type models to predict the energy of a molecule as a function of its conformation. This allows predictions of Equilibrium geometries and transition states Relative energies between conformers or between different molecules Molecular mechanics can be used to supply the potential energy for molecular dynamics computations on large molecules. However, they are not appropriate (nor are most ab initio methods!) for bond-breaking reactions.
3 Stretching Interactions Express energy due to stretching bonds as a Taylor series about the equilibrium position R e : E(R) = k 2 (R R e ) 2 + k 3 (R R e ) 3 + (1) where R is a bond length. The first term should dominate. How do we get the parameters (k 2 and k 3, etc.)? Could get from experiment or by a full quantum mechanical calculation. The central idea of molecular mechanics is that these constants are transferrable to other molecules. Most C H bond lengths are 1.06 to 1.10 Å in just about any molecule, with stretching frequencies between 2900 and 3300 cm 1. This strategy is refined using different atom types.
4
5 Figure 1: Atom Types for MM2
6 The Force-Field Molecular mechanics expresses the total energy as a sum of Taylor series expansions for stretches for every pair of bonded atoms, and adds additional potential energy terms coming from bending, torsional energy, van der Waals energy, electrostatics, and cross terms: E = E str + E bend + E tors + E vdw + E el + E cross. (2) By separating out the van der Waals and electrostatic terms, molecular mechanics attempts to make the remaining constants more transferrable among molecules than they would be in a spectroscopic force field.
7 History D. H. Andrews (Phys. Rev., 1930) proposed extending spectroscopic force field ideas to doing molecular mechanics F. H. Westheimer (1940) performed the only molecular mechanics calculation done by hand to determine the transition state of a tetrasubstituted biphenyl J. B. Hendrickson (1961) performs conformational analysis of larger than 6 membered rings K. B. Wiberg (1965) publishes first general molecular mechanics type program with ability to find energy minimum N. L. Allinger [Adv. Phys. Org. Chem. 13, 1 (1976)]
8 publishes the first (MM1) in a series of highly popular force fields; the second, MM2, follows in 1977 Many other force field methods have been developed over the years
9 Stretch Energy The stretching potential for a bond between atoms A and B is given by the Taylor series E(R AB ) = k AB 2 (R AB R AB 0 ) 2 + k AB 3 (R AB R AB 0 ) 3 (3) + k AB 4 (R AB R AB 0 ) 4 + and different force field methods retain different numbers of terms in this expansion (frequently, only the first term is kept). Note that such expansions have incorrect limiting behavior at large distances.
10 Morse Potentials for Stretch Energy A simple function with correct limiting behavior is the Morse potential E str (R R 0 ) = D[1 e k/2d(r R 0 ) ] 2, (4) where D is the dissociation energy. However, this potential gives very small restoring forces for large R and therefore causes slow convergence in geometry optimization. For this reason, the truncated polynomial expansion is usually preferred.
11 Figure 2: Stretching Potential for CH 4 with exact, 2nd order, 4th order, and Morse potentials (Jensen, Introduction to Computational Chemistry)
12 Bend Energy Bending energy potentials are usually treated very similarly to stretching potentials; the energy is assumed to increase quadratically with displacement of the bond angle from equilibrium. E bend (θ ABC θ ABC 0 ) = k ABC (θ ABC θ ABC 0 ) 2 (5) An unusual thing happens for θ ABC = 180 o : the derivative of the potential needs to go to zero. This is sometimes enforced (Fig. 3).
13 Figure 3: Bending Potential for H 2 O (Jensen)
14 Out-of-Plane Bending The potential for moving an atom out of a plane is sometimes treated separately from bending (although it also involves bending). An out-of-plane coordinate (either χ or d) is displayed below. The potential is usually taken quadratic in this out-of-plane bend, E bend oop (χ B ) = k B (χ B ) 2. (6)
15 Torsional Energy The torsional energy term attempts to capture some of the steric and electrostatic nonbonded interactions between two atoms A and D connected through an intermediate bond B C. The torsional angle ω (also often denoted τ) is depicted below. It is the angle betwen the two planes defined by atoms A, B, and C and by B, C, and D.
16 Form of the Torsional Potential The torsional potential is not expanded as a Taylor series because the torsional angle can go far from equilibrium. Fourier series are used instead: E tors (ω ABCD ) = n=1 V ABCD n cos(nω ABCD ). (7) Often this is rewritten to make sure the energy is non-negative, and typically the number of terms is 3 (bad for octahedral!). E tors (ω ABCD ) = 1 2 V ABCD 1 [1 + cos(ω ABCD )] (8) V ABCD 2 [1 cos(2ω ABCD )] V ABCD 3 [1 + cos(3ω ABCD )].
17 For a molecule like ethylene, rotation about the C=C bond must be periodic by 180 o, so only even terms n = 2, 4,... can occur. For a molecule like ethane, only terms n = 3, 6, 9,... can occur.
18
19 Figure 4: Torsional Potentials for n = 3 and 2 (Jensen)
20 Figure 5: Example Torsional Potential for n = 1 (Jensen)
21 van der Waals Energy The van der Waals energy arises from the interactions between electron clouds around two nonbonded atoms. Short range: strongly repulsive Intermediate range: attractive Long range: goes to zero The attraction is due to electron correlation which results in dispersion or London forces (instantaneous multipole / induced multipole).
22 Form of the van der Waals Energy van der Waals energies are usually computed for atoms which are connected by no less than two atoms (e.g., 1-4 interactions between A and D in A-B-C-D and higher). Interactions betwen atoms closer than this are already accounted for by stretching and/or bending terms. At intermediate to long ranges, the attraction is proportional to 1/R 6. At short ranges, the repulsion is close to exponential. Hence, an appropriate model of the van der Waals interaction is E vdw (R AB ) = Ce DR E R6. (9)
23 Since the van der Walls interaction is long range, it becomes the dominant cost of a force-field computation. It can be speeded up substantially by a more economical expression, the Lennard-Jones potential E vdw (R AB ) = ǫ [ (R0 R ) 12 2 ( R0 R ) 6 ]. (10) The R 12 term is easier to compute than the exponential because no square roots need to be taken to get R. Figure 6 presents a comparison of various van der Waals potentials.
24 Figure 6: Example van der Waals Potentials (Jensen)
25 Electrostatic Energy Electrostatic terms describe the Coulomb interaction between atoms A and B with partial charges (e.g., in carbonyls), according to E el (R AB ) = QA Q B, (11) ǫrab where ǫ is an effective dialectric constant which is 1 in vacuum but higher when there are intermediate atoms or solvent. Usually ǫ is picked fairly arbitrarily; higher values or so-called distance-dependent dialectrics (ǫ = ǫ 0 R AB ) account for screening and kill off the electrostatic contributions faster, making them easier to compute.
26 Alternative Approaches to Electrostatics A slightly different approach to partial charges is to consider polarized bonds as dipoles, and compute the electrostatic interaction between these dipoles (e.g., MM2 and MM3): E el (R AB ) = µa µ B ǫ(r AB ) 3 (cosχ 3 cosα Acosα B ), (12) where χ is the angle between the dipoles and α A and α B are the angles each dipole makes with the line joining atoms A and B.
27 Fitting Atomic Charges Atoms don t have charges!! Quantum mechanics tells us electrons are smeared out, and their charge is shared among nearby atoms. No unique way to assign electrons to atoms. Nevertheless, atomic charges are a useful approximation. Most favored procedure is to perform a quantum computation and then choose charges that reproduce the quantum electrostatic potential. Restrained Electrostatic Potential (RESP) procedure used for AMBER force field. φ esp (r) = nuc A Z A Ψ R A r (r )Ψ(r ) dr (13) r r
28 Computational Cost of Electrostatic Terms Like van der Waals terms, electrostatic terms are typically computed for nonbonded atoms in a 1,4 relationship or further apart. They are also long range interactions and dominate the computation time. The number of nonbonded interactions grows quadratically with molecule size. The computation time can be reduced by cutting off the interactions after a certain distance. The van der Waals terms die off relatively quickly ( R 6 ) and can be cut off around 10 Å. The electrostatic terms die off slower ( R 1, although sometimes faster in practice), and are much harder to treat with cutoffs. This is a problem of interest to developers.
29 Improved Electrostatic Energies The point-charge model has serious deficiencies: (a) electrostatic potentials are not accurately reproduced; (b) simple models don t allow the charges to change as the molecular geometry changes, but they should; (c) only pairwise interactions are considered, but an electrostatic interaction can actually change by 10-20% in the presence of a third body (induction or polarization effects).
30 Improved Electrostatic Potentials Electrostatic potentials can be more accurately reproduced by: Allowing non-atom-centered charges Adding point dipoles, quadrupoles, etc. Both are employed in Anthony Stone s Distributed Multipole Analysis (DMA). However, this does not yet allow the electrostatic variables to change as a function of geometry or to respond to electrostatic potentials generated by nearby atoms. This requires polarizable force-fields such as the fluctuating charge model.
31 The Fluctuating Charge Model Consider the energy as a function of the number of electrons N: E = E 0 + E N N E + (14) 2 N 2( N)2 Electronetativity χ = E/ N. Chemical hardness η = 2 E/ N 2. Setting Q = N and expanding about the point with no net atomic charges, E = χq ηq2 + (15) Considering the electrostatic energy also depends on the external potential φ, and summing over all sites (usually
32 atomic centers), E el = A φ A Q A + A χ A Q A η ABQ A Q B. (16) In vector notation, and requiring that the energy be stationary with respect to the charges with and without an external potential, which yields E Q φ 0 = φ + χ + ηq = 0 (17) E Q φ=0 = χ + ηq 0 = 0 (18) Q = η 1 φ, (19) where φ depends on the charges at all sites (and thus the
33 equations must be solved iteratively), φ(r) = A Q A r R A. (20)
34 Other Polarizable Models The fluctuating charge model has some limitations. More general models incorporate explicit polarizability terms. A dipole moment can result from a polarization of the electronic charge as a result of an electric field (F = φ/ r): µ ind = αf (21) E pol el = 1 2 µ indf = 1 2 αf2 (22) It is common to break up the polarizability α into atomic polarizability contributions.
35 Cross Terms Cross terms are required to account for some interactions affecting others. For example, a strongly bent H 2 O molecule tends to stretch its O H bonds. This can be modeled by cross terms such as E str/bend = k ABC (θ ABC θ0 ABC ) [ (R AB R0 AB ) + (R BC R0 BC ) ] Other cross terms might include stretch-stretch, bend-bend, stretch-torsion, bend-torsion, etc. Force field models vary in what types of cross terms they use. (23)
36 Parameterizing the Force Fields Molecular mechanics requires many parameters, e.g., R AB 0, k AB, θ0 ABC, k ABC, Vn ABCD, etc. Assuming there are 30 atoms which form bonds with each other, there are 30 4 /2 torsional parameters for each Vn ABCD term, or parameters for V 1 through V 3! Only the 2466 most useful torsional parameters are present in MM2, meaning that certain torsions cannot be described. Lack of parameters is a serious drawback of all force field methods. Automatic guessing of unavailable parameters is sometimes done but is very dangerous.
37 Heats of Formation H f is the heat conent relative to the elements at standard state at 25 o C (g). This is a useful quantity for comparing the energies of two conformers of a molecule or two different molecules. Bond energy schemes estimate the overall H f by adding tabulated contributions from each type of bond. This works acceptably well for strainless systems. Molecular mechanics adds steric energy to the bond/structure increments to obtain better estimates of H f. The bare molecular mechanics energy is not a meaningful quantity for comparing different molecules.
38 Accuracy of Heats of Formation In principle, other corrections should be added (but usually are not): population increments (for low-lying conformers), torsional increments (for shallow wells), and corrections for low (< 7 kcal/mol) barriers other than methyl rotation (already included in group increment). The performance of the MM2 force field for typical molecules is given in Table 1. Overall, these results are rather good; however, unusual molecules can exhibit far larger errors.
39 Table 1: Average errors in heat of formations (kcal/mol) by MM2 a Compound type Avg error H f Hydrocarbons 0.42 Ethers and alcohols 0.50 Carbonyls 0.81 Aliphatic amines 0.46 Aromatic amines 2.90 Silanes 1.08 a Table 2.6 from Jensen s Introduction to Computational Chemistry.
40 Different Force Field Methods Class I Methods: Higher order terms and cross terms. Higher accuracy, used for small or medium sized molecules. Examples: Allinger s MM1-4, EFF, and CFF. Class II Methods: For very large molecules (e.g., proteins). Made cheaper by using only quadratic Taylor expansions and neglecting cross terms. Examples: AMBER, CHARMM, GROMOS, etc. Made even cheaper by using united atoms.
41 Hybrid Force Field/Electronic Structure Methods Treat uninteresting parts of the molecule by force field methods and interesting parts by high-accuracy electronic structure methods. This approach is useful for systems where part of the molecule is needed at high accuracy or for which no force field parameters exist (e.g., metal centers in metalloenzymes). Examples: Morokuma s ONIOM method; QM/MM methods.
42 QM/MM Approaches E total = E QM + E MM + E QM/MM (24) Mechanical embedding: Include bonded and steric interactions between QM and MM regions, and maybe assign partial charges to the QM atoms Electronic embedding: Also add partial charges on MM atoms to QM computation Polarizable embedding: Also allow QM atoms to polarize MM region (only applies to polarizable force fields or Gordon s effective fragment method) If the QM/MM boundary cuts covalent bonds, dangling bonds are terminated by so-called link atoms (typically hydrogen).
43 Morokuma s ONIOM Approach ONIOM: Our Own n-layered Integrated Molecular Orbital Molecular Mechanics Method E ONIOM (real system, high level) = E low level (real system) + E high level (model system) E low level (model system) Can also involve multiple layers. Originally used only mechanical embedding for the QM/MM interface, but more recently uses electronic embedding. Can also do as a QM/QM with high and low levels of quantum mechanics.
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Thomas W. Shattuck Department of Chemistry Colby College Waterville, Maine 04901
 Thomas W. Shattuck Department of Chemistry Colby College Waterville, Maine 04901 2 Colby College Molecular Mechanics Tutorial Introduction September 2008 Thomas W. Shattuck Department of Chemistry Colby
Thomas W. Shattuck Department of Chemistry Colby College Waterville, Maine 04901 2 Colby College Molecular Mechanics Tutorial Introduction September 2008 Thomas W. Shattuck Department of Chemistry Colby
The strength of the interaction
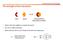 The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
INTERMOLECULAR FORCES
 INTERMOLECULAR FORCES Intermolecular forces- forces of attraction and repulsion between molecules that hold molecules, ions, and atoms together. Intramolecular - forces of chemical bonds within a molecule
INTERMOLECULAR FORCES Intermolecular forces- forces of attraction and repulsion between molecules that hold molecules, ions, and atoms together. Intramolecular - forces of chemical bonds within a molecule
H 2O gas: molecules are very far apart
 Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Chapter 2. Atomic Structure and Interatomic Bonding
 Chapter 2. Atomic Structure and Interatomic Bonding Interatomic Bonding Bonding forces and energies Primary interatomic bonds Secondary bonding Molecules Bonding Forces and Energies Considering the interaction
Chapter 2. Atomic Structure and Interatomic Bonding Interatomic Bonding Bonding forces and energies Primary interatomic bonds Secondary bonding Molecules Bonding Forces and Energies Considering the interaction
Use the Force! Noncovalent Molecular Forces
 Use the Force! Noncovalent Molecular Forces Not quite the type of Force we re talking about Before we talk about noncovalent molecular forces, let s talk very briefly about covalent bonds. The Illustrated
Use the Force! Noncovalent Molecular Forces Not quite the type of Force we re talking about Before we talk about noncovalent molecular forces, let s talk very briefly about covalent bonds. The Illustrated
Molecular Geometry and Chemical Bonding Theory
 Chapter 10 Molecular Geometry and Chemical Bonding Theory Concept Check 10.1 An atom in a molecule is surrounded by four pairs of electrons, one lone pair and three bonding pairs. Describe how the four
Chapter 10 Molecular Geometry and Chemical Bonding Theory Concept Check 10.1 An atom in a molecule is surrounded by four pairs of electrons, one lone pair and three bonding pairs. Describe how the four
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras
 Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
A pure covalent bond is an equal sharing of shared electron pair(s) in a bond. A polar covalent bond is an unequal sharing.
 CHAPTER EIGHT BNDING: GENERAL CNCEPT or Review 1. Electronegativity is the ability of an atom in a molecule to attract electrons to itself. Electronegativity is a bonding term. Electron affinity is the
CHAPTER EIGHT BNDING: GENERAL CNCEPT or Review 1. Electronegativity is the ability of an atom in a molecule to attract electrons to itself. Electronegativity is a bonding term. Electron affinity is the
An Introduction to Hartree-Fock Molecular Orbital Theory
 An Introduction to Hartree-Fock Molecular Orbital Theory C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology June 2000 1 Introduction Hartree-Fock theory is fundamental
An Introduction to Hartree-Fock Molecular Orbital Theory C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology June 2000 1 Introduction Hartree-Fock theory is fundamental
AP CHEMISTRY 2009 SCORING GUIDELINES
 AP CHEMISTRY 2009 SCORING GUIDELINES Question 6 (8 points) Answer the following questions related to sulfur and one of its compounds. (a) Consider the two chemical species S and S 2. (i) Write the electron
AP CHEMISTRY 2009 SCORING GUIDELINES Question 6 (8 points) Answer the following questions related to sulfur and one of its compounds. (a) Consider the two chemical species S and S 2. (i) Write the electron
Potential Energy Surfaces C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
 Potential Energy Surfaces C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Potential Energy Surfaces A potential energy surface is a mathematical function that gives
Potential Energy Surfaces C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Potential Energy Surfaces A potential energy surface is a mathematical function that gives
Basis Sets in Quantum Chemistry C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
 Basis Sets in Quantum Chemistry C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Basis Sets Generically, a basis set is a collection of vectors which spans (defines)
Basis Sets in Quantum Chemistry C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Basis Sets Generically, a basis set is a collection of vectors which spans (defines)
Determining the Structure of an Organic Compound
 Determining the Structure of an Organic Compound The analysis of the outcome of a reaction requires that we know the full structure of the products as well as the reactants In the 19 th and early 20 th
Determining the Structure of an Organic Compound The analysis of the outcome of a reaction requires that we know the full structure of the products as well as the reactants In the 19 th and early 20 th
Applications of Quantum Chemistry HΨ = EΨ
 Applications of Quantum Chemistry HΨ = EΨ Areas of Application Explaining observed phenomena (e.g., spectroscopy) Simulation and modeling: make predictions New techniques/devices use special quantum properties
Applications of Quantum Chemistry HΨ = EΨ Areas of Application Explaining observed phenomena (e.g., spectroscopy) Simulation and modeling: make predictions New techniques/devices use special quantum properties
Chapter 9. Chemical reactivity of molecules depends on the nature of the bonds between the atoms as well on its 3D structure
 Chapter 9 Molecular Geometry & Bonding Theories I) Molecular Geometry (Shapes) Chemical reactivity of molecules depends on the nature of the bonds between the atoms as well on its 3D structure Molecular
Chapter 9 Molecular Geometry & Bonding Theories I) Molecular Geometry (Shapes) Chemical reactivity of molecules depends on the nature of the bonds between the atoms as well on its 3D structure Molecular
KINETIC THEORY OF MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature
 1 KINETIC TERY F MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature TE STATES F MATTER 1. Gas a) ideal gas - molecules move freely - molecules have
1 KINETIC TERY F MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature TE STATES F MATTER 1. Gas a) ideal gas - molecules move freely - molecules have
Introduction, Noncovalent Bonds, and Properties of Water
 Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
Chapter 13 - LIQUIDS AND SOLIDS
 Chapter 13 - LIQUIDS AND SOLIDS Problems to try at end of chapter: Answers in Appendix I: 1,3,5,7b,9b,15,17,23,25,29,31,33,45,49,51,53,61 13.1 Properties of Liquids 1. Liquids take the shape of their container,
Chapter 13 - LIQUIDS AND SOLIDS Problems to try at end of chapter: Answers in Appendix I: 1,3,5,7b,9b,15,17,23,25,29,31,33,45,49,51,53,61 13.1 Properties of Liquids 1. Liquids take the shape of their container,
Chapter 2 Polar Covalent Bonds: Acids and Bases
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds: Acids and Bases Modified by Dr. Daniela R. Radu Why This Chapter? Description of basic ways chemists account for chemical
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds: Acids and Bases Modified by Dr. Daniela R. Radu Why This Chapter? Description of basic ways chemists account for chemical
Chapter 6 An Overview of Organic Reactions
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
2/10/2011. Stability of Cycloalkanes: Ring Strain. Stability of Cycloalkanes: Ring Strain. 4.3 Stability of Cycloalkanes: Ring Strain
 4.3 Stability of Cycloalkanes: Ring Strain Angle strain The strain induced in a molecule when bond angles are forced to deviate from the ideal 109º tetrahedral value (Adolf von Baeyer 1885) Stability of
4.3 Stability of Cycloalkanes: Ring Strain Angle strain The strain induced in a molecule when bond angles are forced to deviate from the ideal 109º tetrahedral value (Adolf von Baeyer 1885) Stability of
Chapter 2 Polar Covalent Bonds; Acids and Bases
 John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
Non-Covalent Bonds (Weak Bond)
 Non-Covalent Bonds (Weak Bond) Weak bonds are those forces of attraction that, in biological situations, do not take a large amount of energy to break. For example, hydrogen bonds are broken by energies
Non-Covalent Bonds (Weak Bond) Weak bonds are those forces of attraction that, in biological situations, do not take a large amount of energy to break. For example, hydrogen bonds are broken by energies
AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts
 AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts 8.1 Types of Chemical Bonds A. Ionic Bonding 1. Electrons are transferred 2. Metals react with nonmetals 3. Ions paired have lower energy
AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts 8.1 Types of Chemical Bonds A. Ionic Bonding 1. Electrons are transferred 2. Metals react with nonmetals 3. Ions paired have lower energy
Infrared Spectroscopy: Theory
 u Chapter 15 Infrared Spectroscopy: Theory An important tool of the organic chemist is Infrared Spectroscopy, or IR. IR spectra are acquired on a special instrument, called an IR spectrometer. IR is used
u Chapter 15 Infrared Spectroscopy: Theory An important tool of the organic chemist is Infrared Spectroscopy, or IR. IR spectra are acquired on a special instrument, called an IR spectrometer. IR is used
Mulliken suggested to split the shared density 50:50. Then the electrons associated with the atom k are given by:
 1 17. Population Analysis Population analysis is the study of charge distribution within molecules. The intention is to accurately model partial charge magnitude and location within a molecule. This can
1 17. Population Analysis Population analysis is the study of charge distribution within molecules. The intention is to accurately model partial charge magnitude and location within a molecule. This can
Symmetric Stretch: allows molecule to move through space
 BACKGROUND INFORMATION Infrared Spectroscopy Before introducing the subject of IR spectroscopy, we must first review some aspects of the electromagnetic spectrum. The electromagnetic spectrum is composed
BACKGROUND INFORMATION Infrared Spectroscopy Before introducing the subject of IR spectroscopy, we must first review some aspects of the electromagnetic spectrum. The electromagnetic spectrum is composed
Infrared Spectroscopy
 Infrared Spectroscopy 1 Chap 12 Reactions will often give a mixture of products: OH H 2 SO 4 + Major Minor How would the chemist determine which product was formed? Both are cyclopentenes; they are isomers.
Infrared Spectroscopy 1 Chap 12 Reactions will often give a mixture of products: OH H 2 SO 4 + Major Minor How would the chemist determine which product was formed? Both are cyclopentenes; they are isomers.
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.
 CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005
 Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Molecular Geometry & Polarity
 Name AP Chemistry Molecular Geometry & Polarity Molecular Geometry A key to understanding the wide range of physical and chemical properties of substances is recognizing that atoms combine with other atoms
Name AP Chemistry Molecular Geometry & Polarity Molecular Geometry A key to understanding the wide range of physical and chemical properties of substances is recognizing that atoms combine with other atoms
Why? Intermolecular Forces. Intermolecular Forces. Chapter 12 IM Forces and Liquids. Covalent Bonding Forces for Comparison of Magnitude
 1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
PCV Project: Excitons in Molecular Spectroscopy
 PCV Project: Excitons in Molecular Spectroscopy Introduction The concept of excitons was first introduced by Frenkel (1) in 1931 as a general excitation delocalization mechanism to account for the ability
PCV Project: Excitons in Molecular Spectroscopy Introduction The concept of excitons was first introduced by Frenkel (1) in 1931 as a general excitation delocalization mechanism to account for the ability
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Name Lab #3: Solubility of Organic Compounds Objectives: Introduction: soluble insoluble partially soluble miscible immiscible
 Lab #3: Solubility of rganic Compounds bjectives: - Understanding the relative solubility of organic compounds in various solvents. - Exploration of the effect of polar groups on a nonpolar hydrocarbon
Lab #3: Solubility of rganic Compounds bjectives: - Understanding the relative solubility of organic compounds in various solvents. - Exploration of the effect of polar groups on a nonpolar hydrocarbon
HOMEWORK PROBLEMS: IR SPECTROSCOPY AND 13C NMR. The peak at 1720 indicates a C=O bond (carbonyl). One possibility is acetone:
 HMEWRK PRBLEMS: IR SPECTRSCPY AND 13C NMR 1. You find a bottle on the shelf only labeled C 3 H 6. You take an IR spectrum of the compound and find major peaks at 2950, 1720, and 1400 cm -1. Draw a molecule
HMEWRK PRBLEMS: IR SPECTRSCPY AND 13C NMR 1. You find a bottle on the shelf only labeled C 3 H 6. You take an IR spectrum of the compound and find major peaks at 2950, 1720, and 1400 cm -1. Draw a molecule
Chapter 10 Molecular Geometry and Chemical Bonding Theory
 Chem 1: Chapter 10 Page 1 Chapter 10 Molecular Geometry and Chemical Bonding Theory I) VSEPR Model Valence-Shell Electron-Pair Repulsion Model A) Model predicts Predicts electron arrangement and molecular
Chem 1: Chapter 10 Page 1 Chapter 10 Molecular Geometry and Chemical Bonding Theory I) VSEPR Model Valence-Shell Electron-Pair Repulsion Model A) Model predicts Predicts electron arrangement and molecular
Application Note AN4
 TAKING INVENTIVE STEPS IN INFRARED. MINIATURE INFRARED GAS SENSORS GOLD SERIES UK Patent App. No. 2372099A USA Patent App. No. 09/783,711 World Patents Pending INFRARED SPECTROSCOPY Application Note AN4
TAKING INVENTIVE STEPS IN INFRARED. MINIATURE INFRARED GAS SENSORS GOLD SERIES UK Patent App. No. 2372099A USA Patent App. No. 09/783,711 World Patents Pending INFRARED SPECTROSCOPY Application Note AN4
Bonding & Molecular Shape Ron Robertson
 Bonding & Molecular Shape Ron Robertson r2 n:\files\courses\1110-20\2010 possible slides for web\00bondingtrans.doc The Nature of Bonding Types 1. Ionic 2. Covalent 3. Metallic 4. Coordinate covalent Driving
Bonding & Molecular Shape Ron Robertson r2 n:\files\courses\1110-20\2010 possible slides for web\00bondingtrans.doc The Nature of Bonding Types 1. Ionic 2. Covalent 3. Metallic 4. Coordinate covalent Driving
Chapter 7: Polarization
 Chapter 7: Polarization Joaquín Bernal Méndez Group 4 1 Index Introduction Polarization Vector The Electric Displacement Vector Constitutive Laws: Linear Dielectrics Energy in Dielectric Systems Forces
Chapter 7: Polarization Joaquín Bernal Méndez Group 4 1 Index Introduction Polarization Vector The Electric Displacement Vector Constitutive Laws: Linear Dielectrics Energy in Dielectric Systems Forces
Organic Chemistry Tenth Edition
 Organic Chemistry Tenth Edition T. W. Graham Solomons Craig B. Fryhle Welcome to CHM 22 Organic Chemisty II Chapters 2 (IR), 9, 3-20. Chapter 2 and Chapter 9 Spectroscopy (interaction of molecule with
Organic Chemistry Tenth Edition T. W. Graham Solomons Craig B. Fryhle Welcome to CHM 22 Organic Chemisty II Chapters 2 (IR), 9, 3-20. Chapter 2 and Chapter 9 Spectroscopy (interaction of molecule with
Vocabulary: VSEPR. 3 domains on central atom. 2 domains on central atom. 3 domains on central atom NOTE: Valence Shell Electron Pair Repulsion Theory
 Vocabulary: VSEPR Valence Shell Electron Pair Repulsion Theory domain = any electron pair, or any double or triple bond is considered one domain. lone pair = non-bonding pair = unshared pair = any electron
Vocabulary: VSEPR Valence Shell Electron Pair Repulsion Theory domain = any electron pair, or any double or triple bond is considered one domain. lone pair = non-bonding pair = unshared pair = any electron
Chemistry 1050 Chapter 13 LIQUIDS AND SOLIDS 1. Exercises: 25, 27, 33, 39, 41, 43, 51, 53, 57, 61, 63, 67, 69, 71(a), 73, 75, 79
 Chemistry 1050 Chapter 13 LIQUIDS AND SOLIDS 1 Text: Petrucci, Harwood, Herring 8 th Edition Suggest text problems Review questions: 1, 5!11, 13!17, 19!23 Exercises: 25, 27, 33, 39, 41, 43, 51, 53, 57,
Chemistry 1050 Chapter 13 LIQUIDS AND SOLIDS 1 Text: Petrucci, Harwood, Herring 8 th Edition Suggest text problems Review questions: 1, 5!11, 13!17, 19!23 Exercises: 25, 27, 33, 39, 41, 43, 51, 53, 57,
Polarity. Andy Schweitzer
 Polarity Andy Schweitzer What does it mean to be polar? A molecule is polar if it contains + and somewhere in the molecule. Remember: Protons can not move. So for a molecule to get a +/- it must somehow
Polarity Andy Schweitzer What does it mean to be polar? A molecule is polar if it contains + and somewhere in the molecule. Remember: Protons can not move. So for a molecule to get a +/- it must somehow
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Suggested solutions for Chapter 3
 s for Chapter PRBLEM Assuming that the molecular ion is the base peak (00% abundance) what peaks would appear in the mass spectrum of each of these molecules: (a) C5Br (b) C60 (c) C64Br In cases (a) and
s for Chapter PRBLEM Assuming that the molecular ion is the base peak (00% abundance) what peaks would appear in the mass spectrum of each of these molecules: (a) C5Br (b) C60 (c) C64Br In cases (a) and
CHAPTER 6 Chemical Bonding
 CHAPTER 6 Chemical Bonding SECTION 1 Introduction to Chemical Bonding OBJECTIVES 1. Define Chemical bond. 2. Explain why most atoms form chemical bonds. 3. Describe ionic and covalent bonding.. 4. Explain
CHAPTER 6 Chemical Bonding SECTION 1 Introduction to Chemical Bonding OBJECTIVES 1. Define Chemical bond. 2. Explain why most atoms form chemical bonds. 3. Describe ionic and covalent bonding.. 4. Explain
AP CHEMISTRY 2007 SCORING GUIDELINES. Question 6
 AP CHEMISTRY 2007 SCORING GUIDELINES Question 6 Answer the following questions, which pertain to binary compounds. (a) In the box provided below, draw a complete Lewis electron-dot diagram for the IF 3
AP CHEMISTRY 2007 SCORING GUIDELINES Question 6 Answer the following questions, which pertain to binary compounds. (a) In the box provided below, draw a complete Lewis electron-dot diagram for the IF 3
Molecular Geometry and VSEPR We gratefully acknowledge Portland Community College for the use of this experiment.
 Molecular and VSEPR We gratefully acknowledge Portland ommunity ollege for the use of this experiment. Objectives To construct molecular models for covalently bonded atoms in molecules and polyatomic ions
Molecular and VSEPR We gratefully acknowledge Portland ommunity ollege for the use of this experiment. Objectives To construct molecular models for covalently bonded atoms in molecules and polyatomic ions
EXPERIMENT 9 Dot Structures and Geometries of Molecules
 EXPERIMENT 9 Dot Structures and Geometries of Molecules INTRODUCTION Lewis dot structures are our first tier in drawing molecules and representing bonds between the atoms. The method was first published
EXPERIMENT 9 Dot Structures and Geometries of Molecules INTRODUCTION Lewis dot structures are our first tier in drawing molecules and representing bonds between the atoms. The method was first published
Chapter 8 Concepts of Chemical Bonding
 Chapter 8 Concepts of Chemical Bonding Chemical Bonds Three types: Ionic Electrostatic attraction between ions Covalent Sharing of electrons Metallic Metal atoms bonded to several other atoms Ionic Bonding
Chapter 8 Concepts of Chemical Bonding Chemical Bonds Three types: Ionic Electrostatic attraction between ions Covalent Sharing of electrons Metallic Metal atoms bonded to several other atoms Ionic Bonding
1 The water molecule and hydrogen bonds in water
 The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
GCE. Chemistry A. Mark Scheme for June 2012. Advanced Subsidiary GCE Unit F321: Atoms, Bonds and Groups. Oxford Cambridge and RSA Examinations
 GCE Chemistry A Advanced Subsidiary GCE Unit F321: Atoms, Bonds and Groups Mark Scheme for June 2012 Oxford Cambridge and RSA Examinations OCR (Oxford Cambridge and RSA) is a leading UK awarding body,
GCE Chemistry A Advanced Subsidiary GCE Unit F321: Atoms, Bonds and Groups Mark Scheme for June 2012 Oxford Cambridge and RSA Examinations OCR (Oxford Cambridge and RSA) is a leading UK awarding body,
Aspects of an introduction to photochemistry
 Aspects of an introduction to photochemistry Ground state reactants Excited state reactants Reaction Intermediates Ground state products Orbital occupancy Carbonyl photochemistry Vibrational structure
Aspects of an introduction to photochemistry Ground state reactants Excited state reactants Reaction Intermediates Ground state products Orbital occupancy Carbonyl photochemistry Vibrational structure
VAPORIZATION IN MORE DETAIL. Energy needed to escape into gas phase GAS LIQUID. Kinetic energy. Average kinetic energy
 30 VAPORIZATION IN MORE DETAIL GAS Energy needed to escape into gas phase LIQUID Kinetic energy Average kinetic energy - For a molecule to move from the liquid phase to the gas phase, it must acquire enough
30 VAPORIZATION IN MORE DETAIL GAS Energy needed to escape into gas phase LIQUID Kinetic energy Average kinetic energy - For a molecule to move from the liquid phase to the gas phase, it must acquire enough
1 Peptide bond rotation
 1 Peptide bond rotation We now consider an application of data mining that has yielded a result that links the quantum scale with the continnum level electrostatic field. In other cases, we have considered
1 Peptide bond rotation We now consider an application of data mining that has yielded a result that links the quantum scale with the continnum level electrostatic field. In other cases, we have considered
Section 3: Crystal Binding
 Physics 97 Interatomic forces Section 3: rystal Binding Solids are stable structures, and therefore there exist interactions holding atoms in a crystal together. For example a crystal of sodium chloride
Physics 97 Interatomic forces Section 3: rystal Binding Solids are stable structures, and therefore there exist interactions holding atoms in a crystal together. For example a crystal of sodium chloride
Chemistry Workbook 2: Problems For Exam 2
 Chem 1A Dr. White Updated /5/1 1 Chemistry Workbook 2: Problems For Exam 2 Section 2-1: Covalent Bonding 1. On a potential energy diagram, the most stable state has the highest/lowest potential energy.
Chem 1A Dr. White Updated /5/1 1 Chemistry Workbook 2: Problems For Exam 2 Section 2-1: Covalent Bonding 1. On a potential energy diagram, the most stable state has the highest/lowest potential energy.
Group Theory and Chemistry
 Group Theory and Chemistry Outline: Raman and infra-red spectroscopy Symmetry operations Point Groups and Schoenflies symbols Function space and matrix representation Reducible and irreducible representation
Group Theory and Chemistry Outline: Raman and infra-red spectroscopy Symmetry operations Point Groups and Schoenflies symbols Function space and matrix representation Reducible and irreducible representation
Orbits of the Lennard-Jones Potential
 Orbits of the Lennard-Jones Potential Prashanth S. Venkataram July 28, 2012 1 Introduction The Lennard-Jones potential describes weak interactions between neutral atoms and molecules. Unlike the potentials
Orbits of the Lennard-Jones Potential Prashanth S. Venkataram July 28, 2012 1 Introduction The Lennard-Jones potential describes weak interactions between neutral atoms and molecules. Unlike the potentials
Quantum Calculations on Hydrogen Bonds in Certain Water Clusters Show Cooperative Effects
 J. Chem. Theory Comput. 2007, 3, 103-114 103 Quantum Calculations on Hydrogen Bonds in Certain Water Clusters Show Cooperative Effects Vasiliy S. Znamenskiy and Michael E. Green* Department of Chemistry,
J. Chem. Theory Comput. 2007, 3, 103-114 103 Quantum Calculations on Hydrogen Bonds in Certain Water Clusters Show Cooperative Effects Vasiliy S. Znamenskiy and Michael E. Green* Department of Chemistry,
Exam 4 Practice Problems false false
 Exam 4 Practice Problems 1 1. Which of the following statements is false? a. Condensed states have much higher densities than gases. b. Molecules are very far apart in gases and closer together in liquids
Exam 4 Practice Problems 1 1. Which of the following statements is false? a. Condensed states have much higher densities than gases. b. Molecules are very far apart in gases and closer together in liquids
7.14 Linear triatomic: A-----B-----C. Bond angles = 180 degrees. Trigonal planar: Bond angles = 120 degrees. B < B A B = 120
 APTER SEVEN Molecular Geometry 7.13 Molecular geometry may be defined as the three-dimensional arrangement of atoms in a molecule. The study of molecular geometry is important in that a molecule s geometry
APTER SEVEN Molecular Geometry 7.13 Molecular geometry may be defined as the three-dimensional arrangement of atoms in a molecule. The study of molecular geometry is important in that a molecule s geometry
Section Activity #1: Fill out the following table for biology s most common elements assuming that each atom is neutrally charged.
 LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
Role of Hydrogen Bonding on Protein Secondary Structure Introduction
 Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
10.7 Kinetic Molecular Theory. 10.7 Kinetic Molecular Theory. Kinetic Molecular Theory. Kinetic Molecular Theory. Kinetic Molecular Theory
 The first scheduled quiz will be given next Tuesday during Lecture. It will last 5 minutes. Bring pencil, calculator, and your book. The coverage will be pp 364-44, i.e. Sections 0.0 through.4. 0.7 Theory
The first scheduled quiz will be given next Tuesday during Lecture. It will last 5 minutes. Bring pencil, calculator, and your book. The coverage will be pp 364-44, i.e. Sections 0.0 through.4. 0.7 Theory
Laboratory 11: Molecular Compounds and Lewis Structures
 Introduction Laboratory 11: Molecular Compounds and Lewis Structures Molecular compounds are formed by sharing electrons between non-metal atoms. A useful theory for understanding the formation of molecular
Introduction Laboratory 11: Molecular Compounds and Lewis Structures Molecular compounds are formed by sharing electrons between non-metal atoms. A useful theory for understanding the formation of molecular
VSEPR Model. The Valence-Shell Electron Pair Repulsion Model. Predicting Molecular Geometry
 VSEPR Model The structure around a given atom is determined principally by minimizing electron pair repulsions. The Valence-Shell Electron Pair Repulsion Model The valence-shell electron pair repulsion
VSEPR Model The structure around a given atom is determined principally by minimizing electron pair repulsions. The Valence-Shell Electron Pair Repulsion Model The valence-shell electron pair repulsion
Molecular Geometry and Hybrid Orbitals. Molecular Geometry
 Molecular Geometry and ybrid Orbitals + -- bond angle 90 o Molecular Geometry Why Should I are bout Molecular Geometry? Molecular geometry (shape) influences... 3 Physical properties: 3 3 3 3 3 Pentane
Molecular Geometry and ybrid Orbitals + -- bond angle 90 o Molecular Geometry Why Should I are bout Molecular Geometry? Molecular geometry (shape) influences... 3 Physical properties: 3 3 3 3 3 Pentane
Name: Class: Date: 3) The bond angles marked a, b, and c in the molecule below are about,, and, respectively.
 Name: Class: Date: Unit 9 Practice Multiple Choice Identify the choice that best completes the statement or answers the question. 1) The basis of the VSEPR model of molecular bonding is. A) regions of
Name: Class: Date: Unit 9 Practice Multiple Choice Identify the choice that best completes the statement or answers the question. 1) The basis of the VSEPR model of molecular bonding is. A) regions of
Chapter 4 Lecture Notes
 Chapter 4 Lecture Notes Chapter 4 Educational Goals 1. Given the formula of a molecule, the student will be able to draw the line-bond (Lewis) structure. 2. Understand and construct condensed structural
Chapter 4 Lecture Notes Chapter 4 Educational Goals 1. Given the formula of a molecule, the student will be able to draw the line-bond (Lewis) structure. 2. Understand and construct condensed structural
Benzene Benzene is best represented as a resonance hybrid:
 Electrophilic Aromatic Substitution (EAS) is a substitution reaction usually involving the benzene ring; more specifically it is a reaction in which the hydrogen atom of an aromatic ring is replaced as
Electrophilic Aromatic Substitution (EAS) is a substitution reaction usually involving the benzene ring; more specifically it is a reaction in which the hydrogen atom of an aromatic ring is replaced as
7. Gases, Liquids, and Solids 7.1 Kinetic Molecular Theory of Matter
 7. Gases, Liquids, and Solids 7.1 Kinetic Molecular Theory of Matter Kinetic Molecular Theory of Matter The Kinetic Molecular Theory of Matter is a concept that basically states that matter is composed
7. Gases, Liquids, and Solids 7.1 Kinetic Molecular Theory of Matter Kinetic Molecular Theory of Matter The Kinetic Molecular Theory of Matter is a concept that basically states that matter is composed
Order of Filling Subshells
 Bonding: General Concepts Ionic Bonds Sections 13.2-13.6 Covalent Bonds Section 13.7 Covalent Bond Energy & Chemical Reactions Section 13.8-13.9 Lewis Structures Sections 13.10-13.12 VSEPR Theory Section
Bonding: General Concepts Ionic Bonds Sections 13.2-13.6 Covalent Bonds Section 13.7 Covalent Bond Energy & Chemical Reactions Section 13.8-13.9 Lewis Structures Sections 13.10-13.12 VSEPR Theory Section
Ultraviolet Spectroscopy
 Ultraviolet Spectroscopy The wavelength of UV and visible light are substantially shorter than the wavelength of infrared radiation. The UV spectrum ranges from 100 to 400 nm. A UV-Vis spectrophotometer
Ultraviolet Spectroscopy The wavelength of UV and visible light are substantially shorter than the wavelength of infrared radiation. The UV spectrum ranges from 100 to 400 nm. A UV-Vis spectrophotometer
Chem 112 Intermolecular Forces Chang From the book (10, 12, 14, 16, 18, 20,84,92,94,102,104, 108, 112, 114, 118 and 134)
 Chem 112 Intermolecular Forces Chang From the book (10, 12, 14, 16, 18, 20,84,92,94,102,104, 108, 112, 114, 118 and 134) 1. Helium atoms do not combine to form He 2 molecules, What is the strongest attractive
Chem 112 Intermolecular Forces Chang From the book (10, 12, 14, 16, 18, 20,84,92,94,102,104, 108, 112, 114, 118 and 134) 1. Helium atoms do not combine to form He 2 molecules, What is the strongest attractive
A mutual electrical attraction between the nuclei and valence electrons of different atoms that binds the atoms together is called a(n)
 Chemistry I ATOMIC BONDING PRACTICE QUIZ Mr. Scott Select the best answer. 1) A mutual electrical attraction between the nuclei and valence electrons of different atoms that binds the atoms together is
Chemistry I ATOMIC BONDING PRACTICE QUIZ Mr. Scott Select the best answer. 1) A mutual electrical attraction between the nuclei and valence electrons of different atoms that binds the atoms together is
Physical Chemistry. Tutor: Dr. Jia Falong
 Physical Chemistry Professor Jeffrey R. Reimers FAA School of Chemistry, The University of Sydney NSW 2006 Australia Room 702 Chemistry School CCNU Tutor: Dr. Jia Falong Text: Atkins 9 th Edition assumed
Physical Chemistry Professor Jeffrey R. Reimers FAA School of Chemistry, The University of Sydney NSW 2006 Australia Room 702 Chemistry School CCNU Tutor: Dr. Jia Falong Text: Atkins 9 th Edition assumed
Molecular Structures. Chapter 9 Molecular Structures. Using Molecular Models. Using Molecular Models. C 2 H 6 O structural isomers: .. H C C O..
 John W. Moore onrad L. Stanitski Peter. Jurs http://academic.cengage.com/chemistry/moore hapter 9 Molecular Structures Stephen. oster Mississippi State University Molecular Structures 2 6 structural isomers:
John W. Moore onrad L. Stanitski Peter. Jurs http://academic.cengage.com/chemistry/moore hapter 9 Molecular Structures Stephen. oster Mississippi State University Molecular Structures 2 6 structural isomers:
INFRARED SPECTROSCOPY (IR)
 INFRARED SPECTROSCOPY (IR) Theory and Interpretation of IR spectra ASSIGNED READINGS Introduction to technique 25 (p. 833-834 in lab textbook) Uses of the Infrared Spectrum (p. 847-853) Look over pages
INFRARED SPECTROSCOPY (IR) Theory and Interpretation of IR spectra ASSIGNED READINGS Introduction to technique 25 (p. 833-834 in lab textbook) Uses of the Infrared Spectrum (p. 847-853) Look over pages
: : Solutions to Additional Bonding Problems
 Solutions to Additional Bonding Problems 1 1. For the following examples, the valence electron count is placed in parentheses after the empirical formula and only the resonance structures that satisfy
Solutions to Additional Bonding Problems 1 1. For the following examples, the valence electron count is placed in parentheses after the empirical formula and only the resonance structures that satisfy
Eðlisfræði 2, vor 2007
 [ Assignment View ] [ Pri Eðlisfræði 2, vor 2007 28. Sources of Magnetic Field Assignment is due at 2:00am on Wednesday, March 7, 2007 Credit for problems submitted late will decrease to 0% after the deadline
[ Assignment View ] [ Pri Eðlisfræði 2, vor 2007 28. Sources of Magnetic Field Assignment is due at 2:00am on Wednesday, March 7, 2007 Credit for problems submitted late will decrease to 0% after the deadline
for excitation to occur, there must be an exact match between the frequency of the applied radiation and the frequency of the vibration
 ! = 1 2"c k (m + M) m M wavenumbers! =!/c = 1/" wavelength frequency! units: cm 1 for excitation to occur, there must be an exact match between the frequency of the applied radiation and the frequency
! = 1 2"c k (m + M) m M wavenumbers! =!/c = 1/" wavelength frequency! units: cm 1 for excitation to occur, there must be an exact match between the frequency of the applied radiation and the frequency
Bonding Models. Bonding Models (Lewis) Bonding Models (Lewis) Resonance Structures. Section 2 (Chapter 3, M&T) Chemical Bonding
 Bonding Models Section (Chapter, M&T) Chemical Bonding We will look at three models of bonding: Lewis model Valence Bond model M theory Bonding Models (Lewis) Bonding Models (Lewis) Lewis model of bonding
Bonding Models Section (Chapter, M&T) Chemical Bonding We will look at three models of bonding: Lewis model Valence Bond model M theory Bonding Models (Lewis) Bonding Models (Lewis) Lewis model of bonding
Question 4.2: Write Lewis dot symbols for atoms of the following elements: Mg, Na, B, O, N, Br.
 Question 4.1: Explain the formation of a chemical bond. A chemical bond is defined as an attractive force that holds the constituents (atoms, ions etc.) together in a chemical species. Various theories
Question 4.1: Explain the formation of a chemical bond. A chemical bond is defined as an attractive force that holds the constituents (atoms, ions etc.) together in a chemical species. Various theories
Boyle s law - For calculating changes in pressure or volume: P 1 V 1 = P 2 V 2. Charles law - For calculating temperature or volume changes: V 1 T 1
 Common Equations Used in Chemistry Equation for density: d= m v Converting F to C: C = ( F - 32) x 5 9 Converting C to F: F = C x 9 5 + 32 Converting C to K: K = ( C + 273.15) n x molar mass of element
Common Equations Used in Chemistry Equation for density: d= m v Converting F to C: C = ( F - 32) x 5 9 Converting C to F: F = C x 9 5 + 32 Converting C to K: K = ( C + 273.15) n x molar mass of element
Peptide bonds: resonance structure. Properties of proteins: Peptide bonds and side chains. Dihedral angles. Peptide bond. Protein physics, Lecture 5
 Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Chapter 4. Electrostatic Fields in Matter
 Chapter 4. Electrostatic Fields in Matter 4.1. Polarization A neutral atom, placed in an external electric field, will experience no net force. However, even though the atom as a whole is neutral, the
Chapter 4. Electrostatic Fields in Matter 4.1. Polarization A neutral atom, placed in an external electric field, will experience no net force. However, even though the atom as a whole is neutral, the
A. Essentially the order of Pt II affinity for these ligands (their Lewis basicity or nucleophilicity)
 (c) Entering Ligand Effects 284 The second order rate constant k 2 in square planar complexes in the rate law below that we just discussed is strongly dependent on the nature of Y, the entering ligand
(c) Entering Ligand Effects 284 The second order rate constant k 2 in square planar complexes in the rate law below that we just discussed is strongly dependent on the nature of Y, the entering ligand
Lecture 24 - Surface tension, viscous flow, thermodynamics
 Lecture 24 - Surface tension, viscous flow, thermodynamics Surface tension, surface energy The atoms at the surface of a solid or liquid are not happy. Their bonding is less ideal than the bonding of atoms
Lecture 24 - Surface tension, viscous flow, thermodynamics Surface tension, surface energy The atoms at the surface of a solid or liquid are not happy. Their bonding is less ideal than the bonding of atoms
Raman Scattering Theory David W. Hahn Department of Mechanical and Aerospace Engineering University of Florida (dwhahn@ufl.edu)
 Introduction Raman Scattering Theory David W. Hahn Department of Mechanical and Aerospace Engineering University of Florida (dwhahn@ufl.edu) The scattering of light may be thought of as the redirection
Introduction Raman Scattering Theory David W. Hahn Department of Mechanical and Aerospace Engineering University of Florida (dwhahn@ufl.edu) The scattering of light may be thought of as the redirection
CHAPTER 10 THE SHAPES OF MOLECULES
 ATER 10 TE AE MLEULE 10.1 To be the central atom in a compound, the atom must be able to simultaneously bond to at least two other atoms. e,, and cannot serve as central atoms in a Lewis structure. elium
ATER 10 TE AE MLEULE 10.1 To be the central atom in a compound, the atom must be able to simultaneously bond to at least two other atoms. e,, and cannot serve as central atoms in a Lewis structure. elium
Kinetic Molecular Theory. Chapter 5. KE AVE and Average Velocity. Graham s Law of Effusion. Chapter 7. Real Gases
 hapter 5 1. Kinetic Molecular Theory. 2. Average kinetic energy and velocity. 3. Graham s Law of Effusion. 4. Real gases and the van der Waals equation. Kinetic Molecular Theory The curves below represent
hapter 5 1. Kinetic Molecular Theory. 2. Average kinetic energy and velocity. 3. Graham s Law of Effusion. 4. Real gases and the van der Waals equation. Kinetic Molecular Theory The curves below represent
EXPERIMENT 17 : Lewis Dot Structure / VSEPR Theory
 EXPERIMENT 17 : Lewis Dot Structure / VSEPR Theory Materials: Molecular Model Kit INTRODUCTION Although it has recently become possible to image molecules and even atoms using a high-resolution microscope,
EXPERIMENT 17 : Lewis Dot Structure / VSEPR Theory Materials: Molecular Model Kit INTRODUCTION Although it has recently become possible to image molecules and even atoms using a high-resolution microscope,
4.5 Physical Properties: Solubility
 4.5 Physical Properties: Solubility When a solid, liquid or gaseous solute is placed in a solvent and it seems to disappear, mix or become part of the solvent, we say that it dissolved. The solute is said
4.5 Physical Properties: Solubility When a solid, liquid or gaseous solute is placed in a solvent and it seems to disappear, mix or become part of the solvent, we say that it dissolved. The solute is said
Peptide Bonds: Structure
 Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
