SYNOPSIS APPENDIX A.
|
|
|
- Egbert Burns
- 3 years ago
- Views:
Transcription
1 SYNOPSIS Even after illuminating most of the deep mysteries of brain, there is still a lot to know about this miraculous organ. The brain is a complex of pathways performing several different functions at a time. The molecular mechanism of these pathways needs insight to understand the complexity of the organ. The Olfactory Transduction is one such pathway which neuroscientists have wondered for its specificity to organisms. The pre-eminent objective of this project was to understand the molecular mechanism of this pathway by analysing different molecules involved in the pathway, their dominant functions and to study their evolutionary relationship among different vertebrates. The central idea involved in this comparative analysis was to correlate the molecular evolution with phylogenetic evolution. For the comparative analysis study, a comparison of 15 molecules involved in OT pathway namely; CALM (calmodulin), PKA(protein kinase A), CAMK2 (Calcium/calmodulindependent protein kinase), GNAL (guanine nucleotide binding proein), CNG (Cyclic nucleotide gated channel) A3, A4, B1, CLCA(calcium activated chloride channel) 1,2,3,4, PRKG(Protein kinase cgmp-dependent), ADCY3 (Adenylate cyclase 3), PDC( Phosducin), GUCA1(Guanylate cyclate activator 1) was done among eight different organisms namely; Human, Chimpanzee, Cow, Dog, Mouse, Chicken, Xenopus laevis and Zebrafish. Comparitive analysis showed that there is no significant relationship between molecular evolution and phylogenetic evolution. The OT pathway shows molecular evolution where number of components within the pathway varies among organisms which is specific to their ability to smell. Apart from analysing OT pathway, another objective was to collect the brain molecules involved in OT pathway for different organisms and to update the available database, i.e. Molecular Database of Brain (MDB) freely available at with these molecules. 390 molecules were collected for OT pathway. Currently, this database has 1251 brain molecules related to LTP, PT and OT pathway having the details of their gene, proteins and ligands. In this short period of time, an additional objective was to study chimeric molecule of Outer membrane protein F (OmpF) and endolysin gene as a way for enzyme presentation by a molecule fixed to membrane. This idea would further help in bioengineering of molecular presentation systems. This is included in APPENDIX A. 1
2 In APPENDIX B, is also reported the crystallography of lysozyme crystallization which was done as a part of learning process as it is a best model to study crystallization and is also comfortable to work with. Different concentrations of lysozyme were tried to study its effect on morphology of crystals. Besides, this trehalose, a cryoprotectant was also used to study its interaction with lysozyme to understand the whole mechanism behind this property of trehalose. The inherent property of trehalose acting as a heat-stress compound was exploited to collect data to make the process more cost effective. Co-crystals of lysozyme with trehalose were obtained at varying concentrations of trehalose from 5%, 10%, 15%, 20%, 25%, 30% and 35%. Though after data processing the ligand, trehalose was not found to be bound with lysozyme protein. 2
3 1. INTRODUCTION 3
4 1.1 Olfaction Sense of smell Olfaction or Olfactory perception is one of the most exquisite senses to communicate with the environment. It is the phenomenon of sensing smell, allowing detection of food, predators, mates, provides sensual pleasure of odour of flowers and perfumes and also gives warning alarms like that of chemical dangers and spoiled food. The sensory system used for olfaction is Olfactory System, which is one of the most pre-eminent senses, having remarkable capacity of detecting odorant molecules. These odorant molecules are volatile chemical molecules which may be air-borne molecules, detected by vertebrates, including humans, or water-soluble molecules detected by aquatic animals. Olfaction has also been reported in invertebrates like various insects including Drosophilla melanogaster [1], moths and honeybees and also in worms like C. elegans [2]. Reports are also there for olfaction in plants as their tendrils are sensitive to air-borne volatile compounds [3, 4]. In comparison to dogs and rhodents, though humans have poor olfactory ability, still they can specifically detect a wide range of odorant molecules. Olfaction is a type of chemosensory process in which volatile odorant molecule binds to specific receptors present on olfactory epithelium in nose leading to a series of chemical reactions and finally transducing the signal to the brain. The brain not only analyses the signal and categorizes the odorant molecule but also memorizes the variety of odours each time it comes across. In 1991, Richard Axel and Linda Buck were honoured by the Nobel Prize in the field of Physiology and Medicine, for their aesthetic research on the interpretation of odorant patterns by brain where the discovered that brain can detect and memorize different odorant molecules because of the special receptors, called olfactory receptors present in the nasal epithelium [5]. Olfaction is also associated with some disorders like Anosmia- inability to smell, Hyperosmia- having acute sense of smell, Hyposmia-decreased sense of smell and Dysosmiasmelling things differently Olfactory Receptors The ability of sensing millions of odorant molecules present in a different way is due to the presence of olfactory receptors. Olfactory receptors are largest multigene family in vertebrates and are chiefly located in the form of clusters in the posterior part of nasal cavity forming an olfactory epithelium, which is the primary sensing structure [6]. 4
5 In insects, it is located on their third segment of antennae and other chemosensory organs [7]. There has also been a report suggesting that olfactory receptors are present on sperms, which detects a chemical called Bourgeonal, released by egg cell by chemotactic movement [8, 9, 10]. Basically, these olfactory receptors (ORs) belong to heptahelical G-protein coupled receptors (GPCRs) superfamily that is invariably seven transmembrane domain (7TM) proteins. Figure 1.1 Structure of Olfactory Receptors showing seven transmembrane domain proteins Types of Olfactory Receptors ORs can be classified into two types of classes based on their amino acid sequences and phylogenetic analysis: Class I ORs referred as fish-like ORs specialized in recognizing water-soluble odorants, and, Class II ORs referred as mammalian-like ORs recognizing volatile odorants. The olfactory repertoire genes has been estimated to be 650 to 900 in humans [11, 12], 1200 to 1500 in mice [13, 14, 15], 1700 to 2000 in rats [16] and 1300 in dogs [17, 18,]. 5
6 This may attribute to the reason why dogs and mice have extraordinary smelling power which humans don t have [19] Nomenclature of Olfactory Receptors To avoid confusion in naming of these large numbers of olfactory genes, a nomenclature system has also been constructed for olfactory gene receptor family [20]. The name of the individual family are given in the format ORnXmY, where, OR is the root name indicating Olfactory Receptor superfamily n is an integer representing a family whose members have greater than 40% sequence identity X is a single letter denoting a subfamily, m is an integer representing an individual family member, and, Y is an integer denoting that it is a pseudogene Olfactory Transduction (OT) Although every day we come across different types of odour which may be familiar or unfamiliar to us, but still we are able to detect it. So, the question arises that how the brain can identify and categorize every odorant molecule. The answer to this question lies in the cellular composition of the olfactory epithelium and specificity of the olfactory receptors to a particular class of odorant molecule. Odorants are small volatile organic molecules of molecular weight less than 400 Da varying in size, shape, functional groups and charge [21]. These can be alcohols, aldehydes, ketones, esters or amines or can be aromatic, alicyclic, polycyclic or heterocyclic ring structures or can be substituted compounds of these types. Differences in the structure of an odorant can lead to modification in its quality [22]. 6
7 1.1.4 (a) Anatomical Organization of Olfactory Epithelium The olfactory system is well adapted structurally to perform its specific function. The olfactory epithelium present in the nasal cavity which is the primary sensory structure is basically made up of three types of cells: Olfactory receptor neurons (ORNs) Supporting sustentacular cells Basal cells. ORN is a bipolar neuron whose axons extends to the olfactory bulb present in the forebrain while its apical dendrites extend to the surface of neural epithelia forming cilia and detects odour [23, 24, 25, 26] Figure 1.2 Structure of Olfactory epithelium showing olfactory receptor neurons, supporting cell and basal cells (b) Mechanism Of Olfactory Transduction Though the mechanism is well known in vertebrates, it is still not well understood in invertebrates. The main mechanism behind olfaction involves cascade of events of signal transduction called Olfactory Transduction, where the chemical signal is transformed into electrical signals and sent to the brain which perceives it as smells [24, 25]. The whole event of signal 7
8 transduction involves the binding of odorant molecule to specific ORs leading to the activation of G-protein signal transduction [27]. Odorous ligands bind to specific olfactory receptors expressed on ORNs which subsequently gets coupled to an olfactory specific G- protein (G olf ) and activates adenylyl cyclase type III. This reaction transduces chemical information into electrical involving amplification of signal. Activation of adenylyl cyclase increases intracellular camp (cyclic AMP) levels leading to the opening of cyclic nucleotide gated ion channel (CNG). This allows cations, mainly, calcium (Ca 2+ ) and sodium (Na + ) ion to flow down their electrochemical gradients into the cell leading to the depolarization of the cell membrane ORNs. The Ca 2+ entering through CNG channel are also able to activated calcium activated chloride ion channel (CLCA) which is permeable to negatively charged chloride ion (Cl - ), thus allowing it to flow out of the cell, leading to increase in depolarization. Furthermore, increase in Ca 2+ concentration also adapts a negative feedback loop pathway which leads to two important steps: Inhibition of adenylyl cyclase activation by calmodulin-dependent protein kinase (CAMK) Downregulation of the affinity of CNG channel to camp. Figure1.3 Flowchart showing the mechanism of Olfactory Transduction. 8
9 Figure 1.4 Diagrammatic representation of the mechanism of olfactory transduction pathway showing molecules involved in the pathway. Biochemical studies have also shown that long stimulation to odorant molecules can lead to the elevation of cgmp levels [28, 29, 30]. Several Ca 2+ ion independent mechanism have been proposed to mediate odour adaptation, including odour dependent phosphorylation of protein kinase A (PKA) and G-protein couples receptor kinase 3 (GSK3) [31, 32] Molecules involved in OT Pathway Broadly, there are 16 molecules present in olfactory transduction pathway. 1. Calmodulin (CALM) CALcium MODULated protein is abbreviated as calmodulin which is abundantly expressed in cytoplasm of all eukaryotic cells and functions as a calcium-binding messenger protein. It mediates the control of a large number of enzymes, ion channels and other proteins by Ca 2+ and can stimulate number of protein kinase enzymes and phosphatases. Calmodulin also helps in the activation of adenylate cyclase in a calcium dependent manner and independent of direct stimulation of odorant molecules [33, 34]. 9
10 2. Olfactory receptors (ORs) These are G-protein coupled receptors (GPCRs) having seven transmembrane domains and act as primary sensory structure to detect odorant molecule. Binding of an odorant molecule then mediates olfactory transduction [35]. 3. Protein kinase A (PKA) Also known as camp-dependent protein kinase performs several functions in phosphorylating large number of substrates in the cytoplasm and nucleus [36]. It is a family consisting of four components, namely, PRKACA, PRKACB having catalytic alpha, beta subunit respectively and X-linked PRKX. 4. Calcium/calmodulin-dependent protein kinase (CAMK2) Consists of four different subunits, CAMK2A, CAMK2B, CAMK2D, CAMK2G having catalytic subunits alpha, beta, delta and gamma. These molecules perform several functions like extracellular signal-regulated kinase and regulation of Ca 2+ homeostasis [37]. 5. Guanine nucleotide binding protein alpha subunit (GNAL) These are basically adenylate cyclase stimulating G-alpha protein which are involved in modulating transmembrane signalling systems and mediates signal transduction within olfactory epithelium and basal ganglia [38]. 6. Cyclic nucleotide gated channel (CNG) It also consists of three subunits, namely, CNGA3, CNGA4 and CNGB1 having alpha3, alpha4 and beta1 catalytic subunits. It has ligand-gated ion channel activity and plays a central role in the transduction of odorant molecules [39, 40]. 7. Calcium activated chloride channel (CLCA) Its family consists of four different molecules: CLCA1, CLCA2, CLCA3, and CLCA4. It mainly has chloride channel activity and hydrogen ion transporting ATPase activity [41]. 10
11 8. Protein Kinase cgmp dependent (PRKG) It consists of two isoforms: PRKG1 and PRKG2 and is strongly expressed in smooth muscles, platelets and hippocampal cells. It mainly phosphorylates many cellular proteins and has ATP binding, cgmp binding activity. 9. Adenylate cyclase 3 (ADCY3) It catalyzes the formation of the secondary messenger, camp. 10. Phosducin (PDC) It is a phosphoprotein which mainly participates in regulation of visual phototransduction. It also shows phospholipase inhibitory activity. An odorant-induced rise of camp levels would activate protein kinase A leading to phosphorylation of phosducin which triggers the membrane targeting of GRK3 and phosphorylation of the receptor protein [42]. 11. Guanylate cyclase activator 1 (GUCA) It has two isoforms GUCA1A and GUCA1B and plays an important role in regulation of guanylyl cyclase by stimulating guanylyl cyclase1 when Ca 2+ concentration is low and inhibits when free Ca 2+ concentration is elevated [43]. 11
12 Figure 1.5 Diagrammatic representation of all the molecules involved in OT pathway. 1.2 MOLECULAR DATABASE OF BRAIN Of all the objects in the universe, human brain is the most complex. Despite of the recent advances in the science of the brain and mind, the mystery of the brain is still unrevealed. For better understanding of the brain, many databases have been developed for brain which has made the research easier. Nowadays, several databases of brain are available like Brain web: simulated Brain Database The BRAINnet database Brain Resource Database PMOD Normal Brain Database Sense lab which are freely accessible for the researches and help them in understanding of various brain disorders and genetics and functional analysis of brain. None of these databases provides molecular information of the brain. The Molecular Database of Brain (MDB) is one such novel database which deals with 12
13 the molecular aspects of brain. MDB is a unique database that has molecules related to different pathways of brain having details of all the genes, proteins and ligand involved in that pathway in a range of number of organisms like Humans, Chimpanzee, Cow, Dog, Mouse, Chick etc. MDB already has 317 molecules involved in Long Term Potentiation Pathway (LTP) [44] and Phototransduction Pathway (PT) [45]. The attempt was to update MDB with molecules related to Olfactory Transduction Pathway (OT). At present there are few databases for Olfactory Receptors (OR) namely, ORDBhttp://senselab.med.yale.edu/OrDB/,OdorDBhttp://senselab.med.yale.edu/OdorDB/, OdorMapDBhttp://senselab.med.yale.edu/OdorMapDB/,HORDEhttp://genome.weizmann.ac.il/hor de/, but all of them has information related to only ORs. MDB consists of all the molecules that are involved in the pathway. Kyoto Encyclopedia of Genes and Genomes (KEGG) database ( was used to retrieve all the information related to Olfactory Transduction Pathway. About 390 molecules related to olfactory transduction pathway were collected. An attempt was made to understand olfactory perception process in detail by phylogenetic analysis of molecules involved in the pathway among eight organisms. Presently, MDB has 1251 brain molecules related to different pathways. MDB is also being updated with the structural information of brain molecules which contains three categories: structure, model and Structural Genomics Initiative (SGI). This will again proof to be a great help for the neuroscientists for the functional analysis of brain molecules [46]. 13
14 2. MATERIALS AND METHODS 14
15 2.1 Analysis of Olfactory Transduction (OT) Pathway Retrieval of sequences of molecules of OT from different organisms For the analysis of OT pathway, 15 molecules were chosen- 1. CALM (Calmodulin) 2. PKA (Protein Kinase A) 3. CAMK2 (Calcium/calmodulin-dependent protein kinase) 4. GNAL (Guanine nucleotide binding protein alpha type) 5. CNGA3 (Cyclic nucleotide gated channel alpha 3) 6. CNGA4 (Cyclic nucleotide gated channel alpha 4) 7. CNGB1 (Cyclic nucleotide gated channel beta 1) 8. CLCA1 (Calcium-activated chloride channel alpha 1) 9. CLCA2 (Calcium-activated chloride channel alpha 2) 10. CLCA3 (Calcium-activated chloride channel alpha 3) 11. CLCA4 (Calcium-activated chloride channel alpha 4) 12. PRKG (Protein kinase cgmp dependent) 13. ADCY3 (Adenylase cyclase 3) 14. PDC (Phosducin) 15. GUCA1 (Guanylate cyclase activator 1) These molecules were phlylogenetically compared among eight different classes of vertebrates. I. Class: Mammalia Order: Primates Humans (Homo sapiens) II. Class: Mammalia Order: Primates Chimpanzee (Pan troglodytes) III. Class: Mammalia Order: Artiodacyla Cow (Bos taurus) IV. Class: Mammalia Order: Carnivora Dog (Canis familiaris) V. Class: Mammalia Order: Rhodentia Mouse (Mus musculus) VI. Class: Aves Order: Galliformes Chicken (Gallus gallus) VII. Class: Amphibia Order: Anura African clawed frog (Xenopus laevis) VIII. Class: Actinopterygii Order: Cypriniformes Zebra fish (Danio rerio) 15
16 2.1.2 Phylogenetic Analysis of Pathway Multiple sequence alignment (MSA) of eight organisms was done using ClustalW ( Phylogenetic trees were constructed using neighbour joining method as implemented in MEGA ( for 15 molecules in the pathway. The Neighbour joining method gave tree topologies with minimum branch length. It resulted in producing un-rooted, bifurcating trees [31]. The clustering in the constructed tree were then validated using the Bootstrap (a statistical method which provides the repeatability of clustering observed in randomized trials), where the number of trials was set to 100. Nodes with bootstrap value greater than 50% were used for analysis. 2.2 Molecular Database of Brain (MDB) Data collection and Updating the Database Different databases were accessed like KEGG Pathway Database ( NCBI-PubMed ( Uniprot/Swissprot (web.expasy.org/docs/swiss-prot_guideline.html), Protein Data Bank (PDB- ) for collection of molecules and various details like gene name, gene symbol, chromosome location, functions etc. The relational database management system MySQL is used for updating the database. HyperText Markup Language (HTML) and Cascading Styling Sheets (CSS) were used for styling and designing. PHP:Hypertext Preprocessor was used as the interface. This helped in providing a user friendly resource to access information on brain related molecules. 16
17 2.2.2 Sequence Alignment Studies Jalview ( was used for alignment studies. The alignment score using nucleotide sequences was calculated according to the following formula: Alignment Score = Percentage Identity/ Alignment length. Dividing the percentage identity by alignment length ensures that, small alignments with high identity do not bias the result. Percentage identity was also calculated using jalview which was used for making scatter plot of average % identity of molecules involved in the pathway among human, chimpanzee, dog, mouse and fish. Standard deviation (SD) was calculated for average percentage identity of the molecules involved in the pathway among these organisms. Then the range of standard deviation was calculated by adding and subtracting the standard deviation value to obtain significant range of variation among the molecules. Maximum Deviation = Mean value + SD Minimum Deviation = Mean value SD Scatter plot was made to plot the graph between molecules involved in the pathway against Average % identity. The plot signifies the conservation or variation of a molecule among different organisms. 17
18 3. RESULTS AND DISCUSSION 18
19 3.1Analysis of the olfactory transduction (OT) pathway The OT pathway was understood based on: Molecular analysis Phylogenetic analysis Molecular Analysis of OT Pathway The OT pathway is comprised of different molecules which work as a cascade to help in olfactory perception. These molecules consist of different components which vary in different organisms of different families or within the same family. The OT pathway can actually be referred as DUAL SECONDARY MESSENGER PATHWAY, as it involves two different secondary messengers, i.e. camp and cgmp for signal transduction, which are triggered by different concentration of odorant molecules. The first secondary messenger pathway involves camp while the second secondary messenger pathway involves cgmp which is triggered at high concentration of odorant molecules after its long exposure. The overall OT pathway is shown as in flowchart diagram. Figure3.1. The molecules and their different components involved in OT pathway were analysed among eight organisms, namely, Human, Chimpanzee, Cow, Dog, Mouse, Chicken, Frog and Fish. The molecules were analysed along the pathway for both First secondary-messenger pathway and Second secondary-messenger pathway. After the analysis of OT pathway in above selected organisms, it was found that no orthologs are present in chimpanzee for the molecule CNGA4 and CAMK2B. No ortholog of CAMK2B was found in dogs also. CLCA2 and CLCA4 are not present in frog and fishes. Most of the molecules involved in OT pathway in other organisms are as per the KEGG Database, are not present for frogs. 19
20 FLOWCHART DIAGRAM 20
21 Molecular analysis of the components involved in the pathway is shown as in Table 3.1. MOLECULES OLFACTORY RECEPTOR ORGANISMS HUMANS CHIMPANZEE COW DOG MOUSE CHICKEN FROG FISH FIRST SECONDARY- MESSENGER (camp) PATHWAY GNAL ADCY CNGA CNGA CNGB CALM CALM CALM CAMK2A CAMK2B CAMK2D CAMK2G CLCA CLCA CLCA SECOND SECONDARY-MESSENGER (cgmp) PATHWAY GUCA1A GUCA1B GUCA1C PRKG PRKG PRKX PRKACA PRKACB PDC Table 3.1 Dual secondary messenger pathway analysis of the molecules involved along the pathway. As per the pathway databases, the molecules involved in the pathway are denoted as + while the molecules absent are denoted as - sign. The molecules highlighted in red and pink are discussed further. 21
22 3.1.2 Sequence Alignment studies The alignment scores were calculated for looking the conservation or variation of a particular molecule involved along the pathway which is shown here in the Table 3.2, 3.3 and 3.4. Only those molecules were considered for calculating the alignment scores for which sequences are present. Similarly only those organisms were considered for sequence alignment studies, which has sequences of all the molecules considered above. Then the average scores were taken for these molecules involved in the pathway, which were then denoted graphically as a plot of average alignment score for each molecule along the pathway which is shown in Figure 3.2 and 3.3. ALIGNMENT SCORE ORGANISMS GNAL ADCY3 CNGA3 CALM1 CALM2 CALM3 Human & Chimpanzee Human & Dog Human & Mouse Human & Fish Chimpanzee & Dog Chimpanzee & Mouse Chimpanzee & Fish Dog & Mouse Dog & Fish Mouse & Fish AVERAGE Table 3.2 Alignment scores of molecules involved in First secondary-messenger pathway of Olfactory Transduction 22
23 Alignment score ALIGNMENT SCORE ORGANISMS CAMK2A CAMK2D CAMK2G CLCA1 Human & Chimpanzee Human & Dog Human & Mouse Human & Fish Chimpanzee & Dog Chimpanzee & Mouse Chimpanzee & Fish Dog & Mouse Dog & Fish Mouse & Fish AVERAGE Table 3.3 Alignment scores of molecules involved in First secondary-messenger pathway of Olfactory Transduction GNAL ADCY3 CNGA3 CALM1 CALM2 CALM3 CAMK2A CAMK2D CAMK2G CLCA1 Molecules involved along the pathway Figure 3.2 Sequence conservation and variation in molecules along the First secondarymessenger pathway of olfactory transduction. 23
24 Alignment Score ALIGNMENT SCORE ORGANISMS GUCA1A GUCA1B PRKG1 PRKX PRKACA PRKACB PDC Human & Chimpanzee Human & Dog Human & Mouse Human & Fish Chimpanzee & Dog Chimpanzee & Mouse Chimpanzee & Fish Dog & Mouse Dog & Fish Mouse & Fish AVERAGE Table 3.4 Alignment scores of molecules involved in Second secondary-messenger pathway of Olfactory Transduction GUCA1A GUCA1B PRKG1 PRKX PRKACA PRKACB PDC Molecules involved in the pathway Figure 3.3 Sequence showing conservation and variation in molecules along the Second secondary-messenger pathway of olfactory transduction. 24
25 The alignment score and the graphical plot denoted that the molecules having higher alignment score are more conserved and molecules having less alignment score shows variation. In the first secondary-messenger pathway, sequences of molecules like CALM 1, 2, 3 were found to be conserved while others shows some variation. Similarly, in second secondarymessenger pathway molecules like GUCA1A, GUCA1B were showing conservation while others were showing some variation. The significance of the variation is measured in terms of standard deviation and was depicted in the scatter plots. Two scatter plots were made for both secondary messenger pathways. It shows the molecules that are conserved or varying along the pathway among different organisms. Scatter plots are shown in Figure 3.4 and 3.5. It was found that in first secondary messenger pathway, sequences of CALM1, 2, 3 and CAMK2A, 2D, 2G were conserved while GNAL, ADCY3, CNGA3 and CLCA1 were varying among organisms. Similarly, in second secondary messenger pathway, it was observed that the molecules like GUCA1A, GUCA1B, PRKACA, PRKACB and PRKG1 were found to be conserved while PRKX and PDC were showing significant variation among organisms. 25
26 Figure 3.4 Plot showing the conservation and variation of molecules along the First Secondary messenger (camp) pathway. The numbering on X-axis indicates the molecules involved along the pathway (1-GNAL, 2-ADCY3, 3-CNGA3, 4-CALM1, 5-CALM2, 6- CALM3, 7-CAMK2A, 8-CAMK2D, 9-CAMK2G, and 10-CLCA1) and Y-axis denotes the average percentage identity. The pink line indicates the Mean value and the blue line indicate the range of standard deviation (SD), i.e. Maximum deviation (Mean + SD) and Minimum deviation (Mean SD). The blue circle indicates the molecules that are conserved along the pathway among organisms and red boxes indicate molecules that show variation among the organisms. 26
27 Figure3.5 Plot showing the conservation and variation of molecules along the Second Secondary messenger (cgmp) pathway. The numbering on X-axis indicates the molecules involved along the pathway (1-GUCA1A, 2-GUCA1B, 3-PRKG1, 4-PRKX, 5-PRKACA, 6- PRKACB and 7-PDC) and Y-axis denotes the average percentage identity. The pink line indicates the Mean value and the blue line indicate the range of standard deviation (SD), i.e. Maximum deviation (Mean + SD) and Minimum deviation (Mean SD). The blue circle indicates the molecules that are conserved along the pathway among organisms and red boxes indicate molecules that show variation among the organisms. 27
28 3.1.3 Phylogenetic Analysis of the Olfactory Transduction Pathway The relationship of different molecules involved in the pathway among different class of organism was studied by constructing a Phylogenetic tree. From the phylogenetic tree, those branching pattern was considered significant which had the bootstrap value greater than 50%. Phylogenetic trees of molecules involved along the pathway are shown in Figure 3.7. Also clustering of molecules was done based on the phylogenetic tree which is shown in Figure 3.8. Mouse Dog Figure 3.6 Standard taxonomic Tree 1. GNAL GNAL 2. ADCY3 ADCY3 28
29 3. CNGA3 CNGA3 4. CNGA4 CNGA4 5. CNBG1 CNBG1 29
30 6. CALM1 CALM1 7. CALM2 CALM2 8. CALM3 CALM3 30
31 9. CAMK2A CAMK2A 10. CAMK2B CAMK2B 11. CAMK2D CAMK2D 12. CAMK2G CAMK2G 31
32 13. CLCA1 CLCA1 14. CLCA3 CLCA3 15. CLCA4 CLCA4 16. PDC PDC 32
33 17. GUCA1A GUCA1A 18. GUCA1B GUCA1B 19. PRKG1 PRKG1 20. PRKG2 PRKG2 33
34 21. PRKX PRKX 22. PRKACA PRKACA 23. PRKACB PRKACB Figure 3.7 Evolutionary Trees generated using NJ method. Bootstrap values greater than 50% only given. (Mammals: Human, Chimpanzee, Dog, Cow, Mouse, Aves: Chicken, Amphibia: Xenopus laevis, Fishes: Zebrafish). Components of same molecule are shown in same colour. 34
35 Molecules GNAL Organisms Human Chimp Mouse Dog Cow Chick Frog Fish ADCY3 CALM1 CALM2 CALM3 CAMK2A CAMK2B CAMK2D CAMK2G 35
36 Molecules Organisms Human Chimp Mouse Dog Cow Chick Frog Fish CNGA3 CNGA4 CNBG1 CLCA1 CLCA2 CLCA4 GUCA1A GUCA1B PRKG1 36
37 Molecules Organisms Human Chimp Mouse Dog Cow Chick Frog Fish PRKG2 PRKX PRKACA PRKACB PDC Figure3.8 Based on the phylogenetic trees, clustering of the organisms has been made for those having bootstrap value greater than 50%. The figure compares the clustered obtained with the standard taxonomic relationship. Thus from the Figure 3.8 it is shown that there is no reasonable comparison between the clustering of molecules based on their sequences and the standard taxonomic clustering Molecular Database of Brain (MDB) Molecular database of brain gives information on both functional and structural part of the selected molecules. Gene, protein and ligand details of a particular pathway of brain can be easily accessed by simply giving the molecule name in the search bar. Figures 3.9, 3.10, 3.11 and 3.12 present the home page, view mode and search mode screenshots of MDB. 37
38 Figure 3.9 Home page of MDB Figure 3.10 View mode of MDB 38
39 Figure 3.11 Search mode of MDB Figure 3.12 Search mode (results) of MDB. 39
40 3.3 Analysis of the Olfactory Transduction pathway Molecular analysis of the OT pathway shows that though same molecules are involved in the pathway among different vertebrates, the number of components of these molecules vary from organism to organism which may contribute towards their specific olfaction power. The absence of many molecules in amphibians suggests that the function of these molecules in olfactory transduction might have been lost though these molecules may be present in frogs performing different functions. Molecular analysis of the components also shows that amphibians only have second secondary messenger pathway and the first secondary messenger pathway is not developed in them. The components of the molecules like CLCA2 and CLCA4 which are only present in terrestrial vertebrates signifies that these are terrestrial modifications and are not necessary for olfaction in aquatic life. Sequence alignment studies shows that the initial molecules and final molecules in first secondary- messenger pathway shows variation among organisms while the molecules in the middle of the pathway are conserved. Though this pattern is not observed in second secondary-messenger pathway. Only molecules like PRKX and PDC showed significant variation among organisms. The molecules showing variation can be attributed as specific molecules evolved for olfactory transduction. Comparison of phylogenetic tree of different molecules with standard taxonomic tree by clustering them suggested that evolution of the components of the olfactory transduction pathway does not follow the whole organism phylogenetic relationship. It is as if the molecules have evolved independently as per the need of time and function. The pathway has not evolved as one unit. The modification in the molecular components of the OT pathway in vertebrates has no significant match with their taxonomic relationship. 40
41 4. CONCLUSIONS 41
42 The olfactory transduction pathway was analysed on both molecular basis and phylogenetically among different classes of vertebrates and it was observed that there is no significant relationship between molecular evolution and phylogenetic evolution. The OT pathway showed molecular evolution where number of components within the pathway varied among organisms probably conferring specificity to their ability to smell. Molecular Database of Brain (MDB) available at the URL has been updated with 390 molecules involved in olfactory transduction pathway. Presently, this database has 1251 brain molecules related to three pathways namely, Long Term Potentiation (LTP), Phototransduction (PT) and Olfactory Transduction (OT). 42
43 REFERENCES 1. Benjamin Kaupp U. (2010). Olfactory signalling in vertebrates and insects: differences and commonalities. Nature Reviews Neuroscience 11: Cornelia I. Bargmann, Hartweig E., Hortvit H. R. (1993). Odorant-selective genes and neurons mediate olfaction in C.elegans. Cell 74: Wayne A., Fromm H. (2001). Calmodulin as a versatile calcium signal tranducer in plants. New Phytologist 151: Heil M., Karban R. (2010). Explaining evolution of plant communication by airborne signals. Trends in ecology and evolution 25: Buck L, Axel R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65 (1): Rinaldi A. (2007). The scent of life. The exquisite complexity of the sense of smell in animals and humans. EMBO Rep. 8 (7): Hallem EA, Dahanukar A, Carlson JR. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51: Spehr M., Schwane K., Riffell JA., Zimmer RK, Hatt H. (2006). Odorant receptors and olfactory-like signalling mechanisms in mammalian sperms. Mol. Cell Endocrinol. 250 (1-2): Olsson P., Laska M. (2010). Human male superiority in Olfactory Sensitvity to the sperm attractant odorant, Bougeonal. Chemical senses 10: MarcSpehr., Katlen S., Jeffrey A. Riffell, Jon Barbour, RichardK.Zimmer, Eva M. Neuhaus., Hanns H. (2004). Particulate Adenylate Cyclase Plays a Key Role in Human Sperm Olfactory Receptor-mediated Chemotaxis. Journal of Biochemistry 279:
44 11. Malnic B., Godfrey PA., Buck LB. (2004). The human olfactory receptor gene family. Proc Natl Acad Sci USA 101: Glusman G., Yanai I., Rubin I., Lancet D. (2001). The complete human olfactory subgenome. Genome Res 11: Zhang X., Firestein S. (2002). The olfactory receptor gene superfamily of the mouse. Nat. Neuroscience 5: Godfrey PA., Malnic B., Buck LB. (2004). The mouse olfactory receptor gene family. Proc Natl Acad Sci USA 101: Young JM., Friedmam C., Williams EM., Ross JA., Tonnes-Priddy L., Trask BJ. (2002). Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Human Mol. Genet. 11: Richard A. Gibbs, George. MW. (2004). Rat Genome Sequencing Project Consortium: Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: Quignon P., Kirkness E., Cadieu E., Touleimat N., Guyon R., Renier C., Hitte C., Andre C., Fraser C., Galibert F. (2003). Comparison of the canine and human olfactory receptor gene repertoires. Genome Biol 4:R Olender T, Fuchs T, Linhart C, Shamir R, Adams M, Kalush F, Khen M, Lancet D (2004). The canine olfactory subgenome. Genomics 83: Pascale Q., Mathieu G., Maud R., Patricia L., Sandrine Tache E M. (2005). The dog and rat olfactory receptor repertoires, Genome Biology 2 6:R Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D (2000). The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm. Genome 11 (11):
45 21. Amoore J. E., Hautala E. (1983). Odor as an aid to chemical safety: odor thresholds compared with threshold limit values and volatilities for 214 industrial chemicals in air and water dilution. J. Appl. Toxicol. 3: Gaillard I., Rouquier S., Giorgi D. (2003). Olfactory Receptors. Cellular and Molecular Life Sciences 61: Stuart F. (2001). How the olfactory system makes sense of scents. Nature 413: Frank Zufall, Trese Lienders Zufall (2000). The Cellular and Molecular basis of Odor Adaptation. Chemical Senses 25: David.T.Moran, Carter Rowley J, Bruce W. Jafek, Mark A.Lowell (1982). The fine structure of olfactory mucosa in man. Journal of neurocytology 11: Gabriele V. Ronnett1 and Cheil Moon (2002). G-proteins and olfactory signal transduction. Annual Review of Physiology 64: Breer H, Shepherd G.M (1993). Implications of the NO/cGMP system for olfaction, Trends in Neurosciences 16: Kroner C., Boekoeff.I, Lohmann S.M, Breer H (1996). Regulation of olfactory signalling via cgmp dependent protein kinase. European Journal of Biochemistry 236: Moon C., Jaberi. P,Otto Bruc A., Baehr W., Ronett. G.V. (1998). Calcium sensitive particulate guanyl cyclase as modulator camp in olfactory receptor neurons. Journal of Neurosciences 18: Dawson T.M., Arriza J.L., Jaworsky D.E., Borisy F.F., Ronett G.V. (1993). β- adrenargic receptor kinase 2 and β-arrestin 2 as mediators of odorant induced desensitization. Science 259:
46 31. Peppel K., Boekhoff I., Mc Donald P., Breer H., Caron M.G. (1997). G-protein coupled receptor kinase 3 (GSK3) gene disruption leads to loss of odorant receptor desensitization. The Journal of Biological Chemistry 272: Robert R. H. Anholt, Ann M. Rivers (1990). Olfactory transduction: cross-talk between second-messenger systems. 29 (17): Jonathan B., Wolfgang B., King-Wai Y., Stephan F. (2004). Calmodulin permanently associates with rat olfactory CNG channels under native conditions. Nature Neruoscience 7: Jones DT., Reed RR (1989). Golf: an olfactory neuron specific-g protein involved in odorant signal transduction. Science 244: Robert RH., Susanne M. Mumby, Doris A. Stoffers, Peggy R. Girard, JF. Kuo, Solomon H. Snyder (1987). Transduction proteins of olfactory receptor cells: identification of guanine nucleotide binding proteins and protein kinase C. Biochemistry 26 (3): Jia Wei, Allan Z. Zhao, Guy CK. Chan, Lauren P. Baker, Soren Impey, Joseph A. Beavo, Daniel R. Storm (1998). Phosphorylation and Inhibition of Olfactory Adenylyl Cyclase by CaM Kinase II in Neurons: a Mechanism for Attenuation of Olfactory Signals. Neuron 29: Allen MS.(1987). Signal transduction by guanine nucleotide binding proteins. Molecular and cellular endocrinology 49: Zagotta W.N, Siegelbaum SA. (1996). Structure and Function of Cyclic Nucleotide- Gated Channels. Annual Review of Neuroscience 19: Lisa J Brunet, Geoffrey G., John N. (1996). General Anosmia Caused by a Targeted Disruption of the Mouse Olfactory Cyclic Nucleotide Gated Cation Channel. Neuron 17:
47 40. Aaron B. Stephan, Eleen Y. Shum, Sarah Hirsh, Katherine D. Cygnar, Johannes Reisert, and Haiqing Zhao (2009). ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. PNAS 106 : Boekhoff I, Touhara K, Danner S, Inglese J, Lohse MJ, Breer H, Lefkowitz RJ. (1997). Phosducin, potential role in modulation of olfactory signalling. The Journal of Biological Chemistry 272(7): Trese Leinders-Zufall, Renee E. Cockerham, Stylianos Michalakis, Martin Biel, David L. Garbers, Randall R. Reed, Frank Zufall, and Steven D. Munger (2007). Contribution of the receptor guanylyl cyclase GC-D to chemosensory function in the olfactory epithelium. PNAS 104 (36): Pathania M. (2012). Molecular Understanding of Brain : Analysis of Long TermPotentiation(LTP ). M.Sc dissertation submitted to Madurai Kamaraj University Ramamoorthy (2012). Molecular Understanding of the Brain: Analysis of Phototransduction pathway. M.Sc dissertation submitted to Madurai Kamaraj University Mulakrishanan A. (2013). Structural genomics initiative for brain molecules and enhancing molecular database of brain. M.Sc dissertation submitted to Madurai Kamaraj University
48 SUPPLEMENTARY TABLE ORGANISMS MOLECULES KEGG Accession No. GNAL 2774 ADCY3 109 CNGA CNGA CNGB CALM1 801 CALM2 805 CALM3 808 CAMK2A 815 CAMK2B 816 CAMK2D 817 CAMK2G 818 CLCA Homo sapiens CLCA CLCA GUCA1A 2978 GUCA1B 2979 GUCA1C 9626 PRKG PRKG PRKX 5613 PRKACA 5566 PRKACB 5567 PDC 5132 GNAL ADCY CNGA CNGA CNGB Pan troglodytes CALM CALM CALM CAMK2A CAMK2D CAMK2G CLCA CLCA CLCA GUCA1A GUCA1B GUCA1C PRKG
49 PRKG PRKX PRKACA PRKACB PDC Mus musculus Canis familiaris GNAL ADCY CNGA CNGA CNGB CALM CALM CALM CAMK2A CAMK2B CAMK2D CAMK2G CLCA CLCA CLCA GUCA1A GUCA1B PRKG PRKG PRKX PRKACA PRKACB PDC GNAL ADCY CNGA CNGA CALM CALM CALM CAMK2A CAMK2D CAMK2G CLCA CLCA CLCA GUCA1A GUCA1B GUCA1C PRKG PRKG
50 PRKX PRKACA PRKACB PDC Bos taurus Gallus gallus GNAL ADCY CNGA CNGA CNGB CALM CALM CALM CAMK2A CAMK2B CAMK2D CAMK2G CLCA CLCA CLCA GUCA1A GUCA1B PRKG PRKG PRKX PRKACA PRKACB PDC GNAL ADCY CNGA CNGA CNGB CALM CALM CAMK2A CAMK2B CAMK2D CAMK2G CLCA CLCA GUCA1A GUCA1B GUCA1C PRKG PRKG PRKX
51 PRKACB PDC Xenopus laevis Danio rerio GNAL CALM CALM CAMK2A CAMK2B CAMK2D CAMK2G GUCA1A PRKG PRKACA PRKACB GNAL ADCY CNGA CNGA CNGB CALM CALM CALM CAMK2A CAMK2B CAMK2D CAMK2G CLCA GUCA1A GUCA1B PRKG PRKG PRKACA PRKACB PDC Supplementary Table shows KEGG accession number for nucleotide sequences of the molecules considered for the pathway analysis for different organisms. 51
52 APPENDIX A Cloning of endolysin (ybcs) gene in Loop 5 of Outer Membrane Protein F (OmpF) 52
53 A1 INTRODUCTION A1.1 Membrane Proteins Biological membranes exist as a dynamic structure allowing the cells to communicate with its environments. It has two essential components, i.e. lipids and proteins. Membrane proteins are interesting scientifically because of their key roles in various biological processes of life. They control cell adhesion to form tissues, help in development of plants and animals, control important metabolic processes including salt balance, energy production and transmission, and photosynthesis. Nowadays, they are also targets of most of the pharmaceuticals and becoming important across medicine and agriculture. An insight into the protein database indicates that approximately 20% of all the genes in most of the genomes encodes for membrane proteins [1]. Broadly, membrane proteins are classified as: Intergral membrane proteins Peripheral membrane proteins. Peripheral membrane proteins are attached to the lipid bilayer either by electrostatic or Van der Waals interactions or by covalent interactions [2]. Integral membrane proteins are permanently attached to the membrane and span the membrane [3]. Basically, integral membrane proteins are also of two types- α-helical β-barrels. α-helical membrane proteins are most abundantly found in all types of biological membranes and contain one or several α-helices. The helices are hydrophobic in nature. β-barrel proteins are mainly found in outer membranes of Gram-negative bacteria, lipid rich cell walls of some Gram-positive bacteria and in outer membranes of mitochondria and chloroplasts. They consist of β-sheets and form a β-barrel like structure. The residues alternatively face outwards towards the lipids or inwards towards the proteins such that they form a pattern having alternatively hydrophobic and polar. Thus a porous channel is formed through which water-soluble molecules can cross. It has been estimated that about 2-3% of all the genes in Gram-negative bacteria encodes for β-barrel membrane proteins [4]. 53
54 A1.2 Porins Porins belong to the β-barrel class of membrane proteins which were discovered in 1976 by T. Nakae in Salmonella as protein complexes that forms trans-membrane channels through which molecules can diffuse. These are large channels that are specific to different types of molecules. These are present in Gram-negative bacteria, Gram-positive bacteria of the group Mycolata and in mitochondria and chloroplasts of eukaryotes [5]. Generally, porins are present as oligomers mostly trimers [6]. OmpF. OmpC and PhoE (phosphorin) are generally referred as Classical porins. They show a general preference for charge and size of solute, OmpC and OmpF preferring cations over anions and PhoE preferring anions [7]. X-ray crystallization studies have shown that porins consists of antiparallel β- strands, the number of which differs in different porins. A1.3 OmpF The outer membrane protein F (OmpF) is one of the three major porins found in Gramnegative bacteria. It is a non specific transport channel that allows passive diffusion of small, polar molecules like water, ions, glucose and waste products, of Da in size through the cell s outer membrane [8, 9]. OmpF porins functions to regulate osmotic pressure between cell and its surroundings. The OmpF porin gene contains 1086 nucleotides which correspond to 362 amino acids. Out of these 362 amino acids, 22 amino acids make up the signal peptide ad form a precursor sequence to OmpF porins which are eventually cleaved off [10]. OmpF of Salmonella typhii has 37KDa molecular weight and it is made up of 371 amino acids. It shows 57.4% identity with E.coli OmpF. The structure of S.typhi OmpF was solved and submitted to PDB and the PDB ID is 3NSG [11]. OmpF has 16 antiparallel β-strands and 8 loops that connect β-helices to each other. Contact among the monomers is stabilized by hydrophobic and polar interaction, where Loop 2 (known as the latching loop ) tends to bend over the wall of the barrel of the neighbouring 54
55 subunit, playing a significant role in stabilization. The pore size is however reduced by the insertion of Loop 3 into the barrel, forming the eyelet [12]. Figure A1 Structure of OmpF monomer. porin 55
56 Figure A2 Structure of OmpF porin of Salmonella typhii as a trimer where red regions shows loop 5 of OmpF. A1.4 Clinical and Industrial manifestations of Porins The exposed external loops of porins are known to interact with antibodies, phages and colicins. OmpF, along with LPS, is the receptor for phage K20 [13, 14]. Porins also acts as potential target as an antigen that can further used for vaccination. Porins predominantly induce the production of IgG and IgM antibodies. It has been reported that OmpC is one of the major antibody inducer and confers a stronger long lasting bactericidal activity than OmpF [15]. It has also been showed that E.coli porins can be used as bioadsorbents as Yeast metallothioneins can be effectively expressed in E.coli as attachments to LamB protein which enhances E.coli natural capability to cadmium ions to about fold [16]. Bacterial outer membrane porins can also be used as efficient drug delivery system. OmpF can help in diffusion of a prodrug into vesicles acting as functional nanoreactor [17]. 56
57 A1.5 Endolysin Endolysin, generally termed as lysins are double stranded DNA bacteriophage encoding enzymes produced during the late phase of gene expression in the lytic cycle. It degrades the peptidoglycan layer which is the main component of bacterial cell wall, enabling the liberation of progeny virus [18]. Endolysins lack secretory signals, thus their access to peptidoglycans from inside the cells is dependent on small hydrophobic proteins called, Holins, which enable endolysin molecule to cross the inner membrane. Holins themselves do not have any enzymatic activity and are localized to the cytoplasmic membrane. The endolysin gene used for the study is ybcs which is found in defective prophage, DLP12 of E.coli K-12 as a part of two component lysis cassette along with essd, a putative holin. Endolysin structure of Bacillus anthracis is known and it contains an N-terminal amidase and a C-terminal membrane binding site. Some contain an N-terminal SAR (Signal Arrest Release) domain, that aids in holin independent release of the protein into the periplasmic space [18]. 57
58 A2. Materials and Methods A2.1 Plasmid isolation by Alkaline Lysis Method Requirements- Solution I 50 mm glucose 25 mm tris chloride (ph 8.0) 10 mm EDTA (ph 8.0) This solution was filter sterilized and stored at 4 C. Solution II 0.2N NaOH (freshly diluted from 10N stock) 1% (w/v) SDS This solution was freshly prepared and used at room temperature. Solution III 3M sodium acetate (ph 5.2, adjusted using glacial acetic acid) This solution was autoclaved and stored at 4 C. Procedure Inoculation of bacterial culture was done in LB broth (10ml) with their respective antibiotics i.e. Kanamycin (50µg/ml) for pokompfl5h+e and ampicillin (100µg/ml). Overnight culture was then pelleted out at rpm for 5 minutes. The pellet was then suspended in 200µl of ice-cold Solution I by vigorous vortexing. Then 400µl of freshly prepared Solution II was added and the contents were mixed gently by inversion, and incubated on ice for 5 minutes. 58
59 Then 300µl of ice-cold Solution III was added, mixed by inversion and stored on ice for 5 minutes. Then after centrifugation at rpm for 10 minutes, supernatant was collected in a fresh eppendorf. DNAse free RNAse was then added at a concentration of 2µg/ml and incubated at 37 0 C for 45 minutes. Then equal volume of chloroform was added and mixed by inversion. It was then spun at rpm for 5 minutes and the upper aqueous layer was collected in a fresh tube. After this, equal volume of isopropanol was added and the tubes were incubated for 10 minutes at room temperature. It was again centrifuged at rpm for 15 minutes. The pellet was then washed with 70% ethanol and the mixture was again centrifuged for 5 minutes at rpm. After then, the pellet was kept for air drying. Finally, the pellet was resuspended in 20µl of MilliQ and then stored at C. A2.2 Agarose Gel Electrophoresis The isolated plasmid was then checked by using Agarose Gel Electrophoresis. Running buffer (1X TAE) Tris : M EDTA : 1 mm The ph was adjusted to 8.0 using acetic acid. Sample buffer (10X) Glycerol : 50% Bromophenol blue : 0.0% EDTA : 0.01 M 59
60 1 % Agarose gel was casted with the addition of ethidium bromide (1μl/mL) in 1 X TAE buffer. Samples were loaded with the loading dye. Electrophoresis was carried out at 100V till the dye front reaches the end of gel. After electrophoresis the gel is visualized under UV transilluminator. A2.3 Restriction Digestion Double digestion and sequential digestion were performed and the digested products were analyzed by agarose gel electrophoresis. Sequential Digestion Digestion mixture was prepared by adding DNA, enzyme I, buffer and MilliQ and the mixture was then spun at rpm for 1 minutes for completely mixing of the contents. The digestion mix was kept in water bath for 3 hours at 37 0 C. After then, one-tenth volume of sodium acetate (ph 5.2) and twice the volume of isopropanol were added. It was then incubated at C for one hour. Then it was centrifuged at rpm for 10 minutes. The supernatant was discarded and the mixture was washed with 80% ethanol. It was again spun for rpm for 10 minutes. The supernatant was removed and the pellet was air dried completely. The pellet was again resuspended in Enzyme II, buffer and MilliQ according to the reaction volume and then the contents were again mixed by spinning at rpm for 1 minute. The mixture was again incubated in water bath at 37 0 C for 3 hours. The digestion mixture was finally run in agarose gel to analyze the digested products. A2.4 Gel Elution The agarose block containing the desired DNA fragment was taken and the DNA was eluted by using SIGMA GelElute TM Gel Extraction Kit as per the manufacturer s instructions. 60
61 A2.5 Ligation Reactions were set up with 5 Weiss Unit of T4 DNA Ligase (Fermentas). Ligation was allowed to take place at 4 0 C over. A2.6 Competent Cell Preparation by Calcium Chloride Method A single colony of E. coli DH5α, pet20b+ or BL21DE3 was inoculated into 2 ml LB broth and kept in a shaker at 180 rpm at 37 0 C overnight. Starter culture (1%) was then subcultured into 50ml of LB medium and was grown again at 37 0 C for two hours. When O.D. reaches to 0.6. the culture is taken out and kept in ice for 10 minutes. Then the culture is spun at 5000rpm for5 minutes at 4 0 C. The supernatant was then discarded and the pellet was suspended into ice-cold 1ml 100mM calcium chloride followed by gentle shaking. Then the volume was made upto 7ml by adding more calcium chloride. Then it was again centrifuged at 4000 rpm for 5 minutes at 4 0 C. After discarding the supernatant again 1ml of calcium chloride was added to the pellet and was mixed by gentle shaking. The volume was made upto 7ml by adding more calcium chloride, It was then incubated for half an hour on ice and then centrifuged at 3000 rpm for 5 minutes at 4 0 C. Finally, the pellet was resuspended into 500µl of calcium chloride. A2.7 Transformation 100μl of competent cells were taken in a 1.5 ml eppendorf. To this 100ng of plasmid DNA to be transformed was added or the ligation mixture can be added. The mixture was then incubated in ice for 30. The heat shock was then given for 90 seconds at 42 C in a water bath and immediately the tubes are transferred in ice. 900µl of LB media was taken and transformation mix was added to it. 61
62 It was then left in the shaker for expression for 90 minutes at 37 0 C at 180 rpm. The cells were then plated on LB-agar plates with suitable selection conditions and incubated at 37 0 C in an incubator overnight. A2.8 Quick screening An overnight culture was taken in 1.5 ml eppendorf and pelleted at 12000rpm for 5 minutes. The pellet was then suspended into 20µl of MilliQ by vortexing. Then 50µl each of DNA loading dye and phenol was added to the contents. The mixture was again vortexed strongly. It was again centrifuged at rpm for 10 minutes. After this three layers will be formed. The uppermost layer contains plasmid DNA was taken and check by Agarose Gel Electrophoresis. A2.9 Protein expression SDS-PAGE Separating gel (10 ml) - 12% 30% acrylamide bisacrylamide : 4 ml Tris HCl (ph 8.8 and 1.5M) SDS 10% APS 10% : 2.5 ml : 0.1 ml : 0.1 ml TEMED : 4 µl Distilled water : 3.4 ml 62
63 Stacking Gel (10 ml) - 5% 30% acrylamide bisacrylamide : 1.7 ml Tris HCl (ph 6.8 and 1M) SDS 10% APS 10% : 2.5 ml : 0.1 ml : 0.1 mg TEMED : 8 µl Distilled water : 5.7 ml Sample loading Buffer (2X) Tris-HCl (ph 6.25) : Bromophenol blue : 0.05% SDS : 4% Glycerol : 20% β-mercaptoethanol : 10% Running Buffer (ph 8.3) Glycine : M Tris-HCl : SDS : 0.1% Staining Solution (0.1%) Coomassive Brilliant Blue (G 250) : 0.25% Methanol : 50% Acetic acid : 10% Destaining Solution Acetic acid 10% 63
64 The SDS PAGE was performed with 12% separating gel and 5% stacking gel. A separating gel was cast in the gel assembly system and a small column of stacking gel was poured over it and an appropriate comb was placed and kept for polymerization. Samples were prepared by boiling with an equal volume of the sample buffer for 5 minutes. Electrophoresis was done at 100V, till the dye front reached bottom of the gel. After electrophoresis, the gel was kept in staining solution (Coomassie staining) for 1 hr and the excess stain was removed by keeping in the destaining solution till the bands were clear. 64
65 A3. RESULTS AND DISCUSSION A3.1 Introduction of endolysin (ybcs) into loop 5 of OmpF In this study, engineering of ybcs (endolysin) gene into loop 5 of OmpF was done. Loop 5 is one of the loops of OmpF that is away from the trimer interface. So, it is taken for engineering studies as it will not affect in trimer formation. Further, endolysin is an unstable enzyme, so its engineering into loop 5 will provide stability to its structure which can be further be utilized for various applications including endolysins as an antimicrobial agent. ybcs was obtained as plasmid from Genobase-ASKA library. It has an internal EcoRV site in its reading frame which was removed by inverse PCR mutating the restriction site and keeping the codon intact [19, 20]. It was cloned into TA cloning vector with HindIII at 5 end and EcoRV site at 3 end. After sequencing TA clone showed deletion mutation of two CC nucleotides within the endolysin which was again corrected by inverse PCR. Now the TAendolysin CC corrected has 549 bp endolysin as insert which is released after HindIII and EcoRV digestion. pokompf-yfp in loop F construct was used as backbone. So, pta-endolysincc corrected and pokompfl5-yfp was used for this part of study. Figure A3 Cloning Strategy of endolysn gene in OmpFL5. 65
66 A3.1.2 Isolation of plasmid DNA To isolate endolysin from TA vector, plasmid DNA of ptaendolysincc corrected was isolated. For the engineering of endolysin into OmpF, plasmid DNA of pokompfl5-yfp was also isolated. The isolated plasmids are shown in Figure A Lane 1: ptaendolysincc corrected Lane 2: pokompfl5-yfp Figure A4 Plasmid isolation. A3.1.3 Restriction Digestion pta-endolysincccorrected has HindIII and EcoRV restriction sites engineered at its 5 and 3 end of endolysin gene. TA vector also has two HindIII sites. So, sequential digestion was performed for pta-endolysincc corrected. The expected bands were 2.7 kb for TA backbone and 549 bp of endolysin. pokompfl5-yfp also has HindIII and EcoRV sites engineered in its loop 5. So, to obtain backbone for the insertion of endolysin vector, double digestion was performed for pokompfl5-yfp. After digestion the backbone of pokompfl5 was expected to give band of 3.2 bp length. 66
67 Both the restriction digestion results gave the expected band patterns which are shown in Figure A Lane 1: Undigested pta-endolysincc corrected Lane2: pta-endolysincc corrected digested with HindIII & EcoRV Lane 3: 1 Kb ladder Lane 4: pokompfl5-yfp digested with HindIII and EcoRV Lane 4: Undigested pokompfl5-yfp Figure A5 Restriction digestion products. 67
68 A3.1.4 Elution of pokompfl5 and ptaendolysincc corrected To isolate both backbone of pokompfl5 and vector i.e. endolysin, bands of required length were cut and eluted from the gel. The eluted DNA was again check by Agarose Gel Electrophoresis. Both endolysin and pokompfl5 were showing expected band length which is shown here in the Figure A (3.2 kb) (549 bp) Figure A6 Eluted products of vector and insert. A3.1.5 Ligation and Trasformation After elution, both vector and backbone were ligated using T4 DNA ligase (Fermentas) and then the ligation mix was transformed into DH5α. The mixture was then plated on LB media containing suitable antibiotics, i.e. Kanamycin (50µg/ml). 68
69 A3.1.6 Quick Screening All the observed colonies after transformation were screened by following quick screening protocol. Samples of the plasmid DNA of the colonies along with pokompf alone as control were loaded on gel. The transformants containing insert were expected to show band shift. As expected, band shift was observed in one of the colony as shown in Figure A Figure A7 Quick screening of the transformed colonies showing band shift in Lane 9 which was expected to be the correct clone. A3.1.7 Confirmation of the Clone The clone was then used for further confirmation and analysis. The clone was confirmed after plasmid isolation and restriction digestion by HindIII and EcoRV which gave the expected band patterns of 3.2 kb and 549 bp. The clone was also confirmed by restriction digestion by XbaI and EcoRI enzymes which was expected to give band patterns of 1.6 kb of OmpFendolysin CC corrected insert and 2.1 kb of pok backbone. 69
70 The clone was confirmed as it was showing band pattern which is shown in the Figure A kb pokompfl5 backbone 2.1 kb pok backbone 1.6 kb OmpF-endolysin insert 549 bp endolysin insert Lane 1: Undigested pokompfl5-endolysin. Lane 2: pokompfl5-endolysin digested with HindIII and EcoRV. Lane 3: pokompfl5-endolysin digested with XbaI and EcoRI. Lane 4: 1Kb ladder Figure A8 Confirmation of clone pokompfl5-endolysin. Then the pokompfl5-endolysin construct was sent for sequencing while the work was carried on further. 70
71 A3.1.8 Subcloning of OmpFL5-endolysin in pet20b(+) vector Double digestion with XbaI and EcoRI were performed with the clone for the isolation of OmpFL5-endolysin construct which was then eluted from the gel. Then subcloning was done into pet20b(+) vector which already has XbaI and EcoRI restriction sites engineered into it. Ligation with T4 DNA Ligase enzyme was done and then the ligation mix was transformed into DH5α. A3.1.9 Clone confirmation The plasmid DNA was further isolated for the observed colonies and the clones were confirmed by performing restriction digestion with XbaI and EcoRI. The clones were expected to give 3.7 kb length for pet20b+ backbone and 1.6 kb for OmpFL5-endolysin insert. The clone confirmation is shown as in Figure A Figure A9 Clone confirmation of pet-ompfl5-endolysin construct. 71
72 A Expression of OmpFL5-endolysin construct After confirmation of clones, one of the clones was taken for further experiment. Plasmid DNA was isolated for the clone petfendo which was then transformed into BL21DE3 for checking expression of the protein and the colonies were plated on LB medium having Ampicillin antibiotic (100µg/ml). The transformed colonies were induced by different concentration of IPTG, i.e. 0.1mM, 0.2mM, 0.3mM, 0.4mM and 0.5mM. The samples were loaded on SDS-PAGE to check the expression of the protein. As the expected size of the protein was around 60 KDa, so we used BSA of molecular weight 66.5 KDa as control Lane 1: BSA (66.5 KDa) Lane 2-6: Induced petfendo construct (0.1, 0.2, 0.3, 0.4, 0.5mM) Lane 7: Uninduced petfendo construct Lane 8: OmpFL7Δ6, 8-YFP (68 KDa) Figure A10 12% SDS-PAGE stained with coomassie stain showing petfendo expression. The protein got expressed at all concentrations of IPTG, though it came as a truncated protein. 72
73 Since the uninduced sample was also showing some leaky expression, so to check whether this expression was coming from BL21DE3, another SDS-PAGE was run with both induced (0.5 mm of IPTG) and uninduced BL21DE3 devoid of clone plasmid KDa 66 KDa 43 KDa 29 KDa 20 KDa Truncated protein 14.3 KDa Lane 1: Uninduced BL21DE3 Lane 2 : Induced BL21DE3 (0.5mM IPTG) Lane 3 : Protein Marker Lane 4 : Endolysin protein (20 KDa) Lane 5: Induced petfendo construct (0.5 mm IPTG) Lane 6: Uninduced petfendo construct. Figure A11. 12% SDS-PAGE confirming that the truncated protein construct petfendo is not endolysin. 73
74 The result confirmed that there is no leaky expression due to BL21DE3 and only OmpFL5- endolysin protein was showing leaky expression. Also the protein observed was truncated. The plasmid DNA was sent for sequencing. The reason for truncation can be further analysed only after sequencing results are obtained. A3.2 Cloning of endolysin in Loop5 of OmpF Endolysin (ybcs) gene of 549 bp was isolated from TA vector and successfully cloned into pokompfl5. Sub-cloning of pokompfl5-endo was also done in pet20b+ expression vector which also gave expected band pattern after restriction digestion. But the expression of petfendo construct came as a truncated protein. It was also confirmed that the truncated protein is neither endolysin nor OmpF alone. The reason for truncation is still to be validated which can be done only after the sequencing results are obtained. 4. CONCLUSIONS Endolysin gene was engineered in loop 5 of OmpF and was successfully cloned and subcloned in pet20b+ vector. The petfendo construct showed truncation after expression. The truncation can be corrected only after the sequencing results are obtained. 74
75 APPENDIX B LYSOZYME CRYSTALLIZATION AND ITS CO-CRYSTALLIZATION WITH TREHALOSE 75
76 B1. INTRODUCTION B1.1 CRYSTALLIZATION Crystallization refers to a natural or artificial process of formation of crystals from homogeneous solutions. It is a solid-liquid separation technique, in which mass transfer of a solute from the liquid solution to pure solid crystalline phase occurs. Crystallization is therefore an aspect of precipitation, obtained through a variation of the solubility conditions of the solute in the solvent, as compared to precipitation due to chemical reaction. To determine protein structure accurately and to analyse their interactions, X-ray crystallography is regarded as the best method. It is not only used for understanding the molecular structure of a substance but also for determining the drugs and their potential targets. It is more suitable method for solving the structure of larger proteins. The process of crystallization can be differentiated into two steps: 1. The nucleation process 2. Crystal growth. In order to produce crystals, all the systems need to reach nucleation zone after which they enter into metastable zone and where it grows and then finally arrive at solubility curve. Only at the nucleation stage atoms arrange in a definite and periodic manner forming crystal structure. The phase separation stage results in the nucleation of crystals. Figure B1. represents the process of crystallization by a phase diagram which has four distinct zones, namely, Unsaturated or solubility zone Metastable zone Labile zone or nucleation zone Precipitation zone. Low protein and /or precipitant concentration will result in undersaturation and no protein crystals will be formed at this zone. The labile zone (or nucleation zone) is where crystal nucleate and growth starts. As the crystals are formed the protein concentration decreases 76
77 resulting in the movement from labile zone to metastable zone. Precipitation will occur when the protein comes out of solution as an aggregate. Figure B1 Phase Diagram showing hoe protein crystals are formed. Basically, for the study of interactions of protein two types of studies can be done Co-crystallization. Soaking. B1.2 Co-crystallization It is regarded as a process of developing crystals made up of two or more components like protein ligand complexes in a definite stoichiometric ratio, where each component is defined as an atom, ion or molecule. Co-crystals exhibit long-range order and the components interact via non-covalent interactions such as hydrogen bonding, ionic interactions and van der Waals interactions. The intermolecular interactions and resulting crystal structures can generate physical and chemical properties that differ from the properties of the individual components [1]. It is widely used for structure-aided drug design [2]. 77
78 B1.3 Soaking In soaking approach, preformed crystals of the target protein are incubated in the compound. The compounds in which crystals were soaked may bind at some functional binding site of the protein, such as enzyme binding site or functional regulatory site [1]. B1.4 Lysozyme In our study, lysozyme was chosen as a model to study as it is easy to crystallize and to work with. It has also been reported that lyzozyme aggregates are found in the serum of the Alzheimer s patients [3]. Lysozyme protein was found to be accumulated in neurons which are associated with Age-related dementias [4]. B1.5 Lysozyme with Trehalose Trehalose, is a disaccharide and non-reducing sugar formed from two glucose unit joined by 1-1 α-bond forming α-d-glucopyronosyl-(1,1)-α-d-glucopyronoside. It is ubiquitous in nature and is found in animals, plants and microorganisms. Figure B2 Chemical Structure of trehalose. Recently, trehalose has been implicated as a stress-responsive factor synthesizing molecule. It has been found that when the organisms especially inicellular organisms are subjected to various stresses like heat, cold, oxidation or dessication, they adapt the changing environment by synthesizing huge amounts of trehalose to maintain their cellular integrity [5]. Sugars are often used to enhance the stabilty of a protein [6, 7, 8]. Recently, trehalose has came up as a molecule acting as a BIOPROTECTANT. Trehalose is one of the osmolytes that work as a stabilizer for biomolecules. It has many inherent properties like prevention of starch retrogradation and stabilization of lipids and proteins has made it useful cryoproetctant in many industries [9]. It is generally believed that the stabilization of the protein 78
79 confirmation is achieved by their preferential exclusion from the protein surface and the consequent hydration of the protein [10]. Generally, sugars like glucose, sucrose, lactose, raffinose, trehalose etc. has been reported to exert cryoprotectant properties [11] among which, trehalose has the most superior effect of cryoprotectant [11]. Although, the exact mechanism of how trehalose protects biomolecules is not yet understood, but still there are some theories that shed light on its property. When trehalose is added to water, it affects both the structure and dyanamics of water molecules relative to each other. Trehalose takes apat the tetrahedral hydrogen bond network of water, preventing the water from crystallizing into ice. It entraps the proteins in a protective cage which slows down the water molecules directly adjacent to the protein, preventing ice crystallization in its immediate vicinity [12]. Recently, it has been also been reported that immiscible oils can be used in the combination of lower concentration of cryoprotectant to prevent destructive ice formation during flash cooling of macromolecular crystals for cryocrytallography [13]. Oils like paraffin oils has been widely used to prevent ice formation which does not require the growth or soaking of crystals in solvents [14]. B1.6 Methods of Crystallization Broadly, there are four types of methods of crystallization, namely Microbatch method Vapour Diffusion method. Free-interface diffusion Dialysis. Vapour diffusion method is widely used method of crystallization. There are many other methods like of vapour diffusion: Hanging Drop, Sitting Drop, and Sandwich Drop. The vapour diffusion method of crystal formation is based on the principle of evaporation due to difference in concentration of protein and precipitant in the drop and in reservoir solution in the well. Water vapour diffuses into the reservoir solution in the well, resulting in gradual increase in the concentration of both protein and precipitant. This is the phase separation stage which results in the nucleation of crystals. The crystal will grow as a result the concentration of protein in the solution depletes. Once equilibrium is established between the crystals and the protein solution, the growth of the crystal stops 79
80 B2. Materials and methods B2.1 Lysozyme Crystallization B2.1.1 By Hanging Drop method Chicken egg-white lysozyme (Sigma) protein was used for crystallization studies using hanging drop method. Hanging Drop technique It is one of the commonly used techniques to grow crystals. In this method, protein solution is mixed with some concentration of reservoir solution containing the precipitants. A drop of mixture is then put on the siliconized coverslip kept upside down covering the reservoir solution for growth of the crystals. As the protein/precipitant mixture in the drop is less concentrated than the reservoir solution, water evaporates from the drop into the reservoir. As a result the concentration of both protein and precipitant in the drop slowly increases and crystals are formed. Figure B3 Hanging Drop technique of crystallization. (Adapted from Bernhard Rupp.BIOMOLECULAR CRYYSTALLOGRAPHY.Garland Science 2010) 80
81 B2.1.2 Preparation of solutions Reservoir solution 8% sodium chloride (NaCl) as precipitant 0.1M sodium acetate (CH 3 COONa) as buffer having ph 4.6. Different concentrations of trehalose i.e. 5%, 10%, 15%, 20%, 25%, 30% and 35% solution were made with reservoir solution. Protein solution 50 mg/ml lysozyme solution was made in 0.1M sodium acetate (ph 4.6). The contents were centrifuged at 14000rpm for 10 minutes at 25 0 C. After then the supernatant was collected in fresh eppendorf for further use. The crystallization of lysozyme was carried out in various ratios of lysozyme and reservoir solutions which shown below in Figure B4 and in Table B1. 5µl P + 5µl RS 4µl P + 2µl RS 2µl P + 4µl RS 3µl P + 3µl RS 4µl P + 2µl RS 2µl P + 4µl RS Figure B4 Various ratios of lysozyme protein (P) and reservoir solution (RS) were carried out at different drop size to obtain lysozyme crystals. 81
82 1 (Protein + Reservoir solution) 2 (Protein + Reservoir solution) 3 (Protein + Reservoir solution) 4 (Protein + Reservoir solution) 5 (Protein + Reservoir solution) 6 (Protein + Reservoir solution) A 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl B 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl C 4μl + 2μl 2μl + 4μl 4μl + 2μl 2μl + 4μl 4μl + 2μl 2μl + 4μl 4μl + 2μl 2μl + 4μl 4μl + 2μl 2μl + 4μl 4μl + 2μl 2μl + 4μl D 3μl + 3μl 4μl + 2μl 2μl + 4μl 3μl + 3μl 4μl + 2μl 2μl + 4μl 3μl + 3μl 4μl + 2μl 2μl + 4μl 3μl + 3μl 4μl + 2μl 2μl + 4μl 3μl + 3μl 4μl + 2μl 2μl + 4μl 3μl + 3μl 4μl + 2μl 2μl + 4μl Table B1 Various conditions of lysozyme crystallization. B2.2 Co-crystallization of Lysozyme with Trehalose Co-crystallization of lysozyme protein was tried at various concentration of trehalose in the reservoir solution. The different ratios and drop size tried to obtain co-crystals which are shown here in Table B2. 82
83 A 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl 5μl + 5μl (Protein + Reservoir solution + 5% Trehalose) (Protein + Reservoir solution + 10% Trehalose) (Protein + Reservoir solution + 15% Trehalose) (Protein + Reservoir solution + 20% Trehalose) (Protein + Reservoir solution + 25% Trehalose) (Protein + Reservoir solution + 30% Trehalose) B 5μl + 5μl 5μl + 5μl (Protein + Reservoir Solution + 40% Trehalose) (Protein + Reservoir solution + 40% Trehalose) Table B2 Different condition of co-crystallization of Lysozyme with Trehalose. Preparation of Cryo Free Solution (CFS) A CFS is a solution that is used for collecting data for crystallization without any use of liquid nitrogen. Liquid nitrogen is mainly used for collecting data during X-ray crystallography as it does not allows the crystals to freeze or degenerate during the collection. Looking into the high cost of liquid nitrogen, CFS was prepared by using common vegetable oil and petroleum jelly. Oils and jelly will not allow the crystals to degrade during the data collection. CFS was made by trying mixing various concentrations of almond oil and petroleum jelly. The CFS solution used for the present study was made by mixing 80% almond oil and 20% petroleum jelly. Along with these co-crystals of the lysozyme protein was obtained with trehalose. Trehalose acting as a heat-stress will further help in maintaining the integrity of the crystals. Thus it will the overall process cost effective. 83
84 B2.3. Data Collection and Processing The co-crystals obtained were then mounted onto the mounting capillary along with Cryo Free Solution (CFS). It was then put on the MARdb for data collection. Then different frames of the crystals were collected to get the diffraction pattern. The cocrystals were then used for diffraction studies using rotating anode X-ray source.. X-ray beam falls on the crystal and causes diffraction, which can be used for building the electron density map. Data processing was done using Collaborative Computational Project No. 4 interface (CCP4ihttp:// ) which is software for macromolecular X-ray Crystallography and Coot 7 which is a visualization tool. The crystal was exposed to X-ray for 30 seconds. Diffraction pattern for every 1 o was collected. 180 frames of crystals were collected. 84
85 B3 Results and Discussion B3.1.1 Lysozyme Crystallization Crystals were obtained in all the conditions after 24 hours, though the size of the crystals varied from microcrystals to macrocrystals. Only 5:5 ratio of the protein: precipitant gave crystals of good size whereas other ratios gave crystals of varying size. Good, big crystals of optimum size, sharp edges and smooth faces formed in condition with equal volume (5μl+5μl condition) and bigger drop size (10μl). But small crystals formed in smaller drop size (6 μl), though the protein and precipitant are in equal volume (3μl+3μl). The crystals obtained at different conditions are shown here in Figure B5. A B C D Figure B5 Crystals of lysozyme: A) 5μl of reservoir solution and 5μl of lysozyme. B) 3μl of reservoir solution and 3μl of lysozyme C) 2μl of reservoir solution and 4μl of lysozyme D) 4μl of Reservoir solution and 2μl of lysozyme. 85
86 B3.1.2 Co-crystallization of Lysozyme with Trehalose Co-crystals were obtained after 2 days for most of the conditions though they varied in their size. It was found that on increasing the concentration of trehalose from 5% to 35%, the size of the crystals decreases. For co-crystals at 40% trehalose, nucleation started after 20 days. Crystals with good size were obtained at 10% Trehalose which were further used for data collection and processing to check whether lysozyme is bound to trehalose. The co crystals are shown in the Figure B6 (i) and B6 (ii). A B C D FFigure B6 (i) Co-crystallization of lysozyme with different concentrations of trehalose, A] 5% trehalose B] 10% trehalose C] 15% trehalose D] 20% trehalose. 86
87 E F G H FFigure B6 (ii) Co-crystallization of lysozyme with different concentrations of trehalose E] 25% trehalose F] 30% trehalose G] 35% trehalose H] 40% trehalose. B3.1.2 [A] Data Collection Crystals of Lysozyme grown with 10% Trehalose was then soaked in CFS solution and transferred to capillary and mounted for data collection. The crystals diffracted at high resolution of angstrom showing more number of spots. Data was collected for which w as then used for processing. One such diffraction pattern obtained is shown as in Figure B7. 87
88 Figure B7 Diffraction pattern of lysozyme crystals, co-crystallized with 10% trehalose. B3.1.2 [B] Data Processing Processing of 180 frames of the diffraction pattern was done by using CCP4i software. Tetragonal form of lysozyme was obtained having P space group. But no significant electron density was found to fit trehalose. Following parameters were observed after processing as shown in Table B3. Parameters Lysozyme with 10% Trehalose Space Group P Parameters a (Å) b (Å) c (Å) α ( ο ) 90 β ( ο ) 90 γ ( ο ) 90 Resolution (Å) Completeness (%) 98.9 R- merge Table B3 Parameters of data processing. 88
89 Data processing of lysozyme crystal structure is done. The geometry of the crystal is tetragonal, whose space group was P Using Molecular Replacement method, the structure of lysozyme is elucidated.1h87 is the PDB_ID of the template used for Molecular Replacement. The elucidated structure of lysozyme has a resolution of Å and its R-free, R-factor values are and respectively. Unfortunately no trehalose is found in the co-crystallized lysozyme (Figure B8). Figure B8 Structure of Lysozyme (1.734 Å resolution) elucidated using X-ray crystallography method. B3.2. In lysozyme crystallization, on either increasing or decreasing the concentration of protein or precipitant, the microcrystals were obtained indicating that good size crystals are obtained only at optimum concentration of protein and precipitant and drop size. Equal drop ratio, drop size is also crucial to obtain good, big crystals of optimum size, sharp edges and smooth faces. Similar, results were obtained in co-crystallization of lysozyme with trehalose where on increasing the concentration of trehalose the size of the crystals was decreasing. Trehalose though acts as heat-stress compound for lysozyme, but significantly does not binds with it at 10% concentration. 89
AP Biology 2015 Free-Response Questions
 AP Biology 2015 Free-Response Questions College Board, Advanced Placement Program, AP, AP Central, and the acorn logo are registered trademarks of the College Board. AP Central is the official online home
AP Biology 2015 Free-Response Questions College Board, Advanced Placement Program, AP, AP Central, and the acorn logo are registered trademarks of the College Board. AP Central is the official online home
Keystone Review Practice Test Module A Cells and Cell Processes. 1. Which characteristic is shared by all prokaryotes and eukaryotes?
 Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
The Lipid Bilayer Is a Two-Dimensional Fluid
 The Lipid Bilayer Is a Two-Dimensional Fluid The aqueous environment inside and outside a cell prevents membrane lipids from escaping from bilayer, but nothing stops these molecules from moving about and
The Lipid Bilayer Is a Two-Dimensional Fluid The aqueous environment inside and outside a cell prevents membrane lipids from escaping from bilayer, but nothing stops these molecules from moving about and
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT
 BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
CELL MEMBRANES, TRANSPORT, and COMMUNICATION. Teacher Packet
 AP * BIOLOGY CELL MEMBRANES, TRANSPORT, and COMMUNICATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production
AP * BIOLOGY CELL MEMBRANES, TRANSPORT, and COMMUNICATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production
Transmembrane proteins span the bilayer. α-helix transmembrane domain. Multiple transmembrane helices in one polypeptide
 Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
PART I: Neurons and the Nerve Impulse
 PART I: Neurons and the Nerve Impulse Identify each of the labeled structures of the neuron below. A. B. C. D. E. F. G. Identify each of the labeled structures of the neuron below. A. dendrites B. nucleus
PART I: Neurons and the Nerve Impulse Identify each of the labeled structures of the neuron below. A. B. C. D. E. F. G. Identify each of the labeled structures of the neuron below. A. dendrites B. nucleus
Actions of Hormones on Target Cells Page 1. Actions of Hormones on Target Cells Page 2. Goals/ What You Need to Know Goals What You Need to Know
 Actions of Hormones on Target Cells Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com) Page 1. Actions of Hormones on Target Cells Hormones
Actions of Hormones on Target Cells Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com) Page 1. Actions of Hormones on Target Cells Hormones
Mechanisms of Hormonal Action Bryant Miles
 Mechanisms of ormonal Action Bryant Miles Multicellular organisms need to coordinate metabolic activities. Complex signaling systems have evolved using chemicals called hormones to regulate cellular activities.
Mechanisms of ormonal Action Bryant Miles Multicellular organisms need to coordinate metabolic activities. Complex signaling systems have evolved using chemicals called hormones to regulate cellular activities.
1.1.2. thebiotutor. AS Biology OCR. Unit F211: Cells, Exchange & Transport. Module 1.2 Cell Membranes. Notes & Questions.
 thebiotutor AS Biology OCR Unit F211: Cells, Exchange & Transport Module 1.2 Cell Membranes Notes & Questions Andy Todd 1 Outline the roles of membranes within cells and at the surface of cells. The main
thebiotutor AS Biology OCR Unit F211: Cells, Exchange & Transport Module 1.2 Cell Membranes Notes & Questions Andy Todd 1 Outline the roles of membranes within cells and at the surface of cells. The main
An Overview of Cells and Cell Research
 An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
BSC 2010 - Exam I Lectures and Text Pages. The Plasma Membrane Structure and Function. Phospholipids. I. Intro to Biology (2-29) II.
 BSC 2010 - Exam I Lectures and Text Pages I. Intro to Biology (2-29) II. Chemistry of Life Chemistry review (30-46) Water (47-57) Carbon (58-67) Macromolecules (68-91) III. Cells and Membranes Cell structure
BSC 2010 - Exam I Lectures and Text Pages I. Intro to Biology (2-29) II. Chemistry of Life Chemistry review (30-46) Water (47-57) Carbon (58-67) Macromolecules (68-91) III. Cells and Membranes Cell structure
CHAPTER 5.1 5.2: Plasma Membrane Structure
 CHAPTER 5.1 5.2: Plasma Membrane Structure 1. Describe the structure of a phospholipid molecule. Be sure to describe their behavior in relationship to water. 2. What happens when a collection of phospholipids
CHAPTER 5.1 5.2: Plasma Membrane Structure 1. Describe the structure of a phospholipid molecule. Be sure to describe their behavior in relationship to water. 2. What happens when a collection of phospholipids
3) There are different types of extracellular signaling molecules. 4) most signaling molecules are secreted by exocytosis
 XIV) Signaling. A) The need for Signaling in multicellular organisms B) yeast need to signal to respond to various factors C) Extracellular signaling molecules bind to receptors 1) most bind to receptors
XIV) Signaling. A) The need for Signaling in multicellular organisms B) yeast need to signal to respond to various factors C) Extracellular signaling molecules bind to receptors 1) most bind to receptors
Carbohydrates, proteins and lipids
 Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Todays Outline. Metabolism. Why do cells need energy? How do cells acquire energy? Metabolism. Concepts & Processes. The cells capacity to:
 and Work Metabolic Pathways Enzymes Features Factors Affecting Enzyme Activity Membrane Transport Diffusion Osmosis Passive Transport Active Transport Bulk Transport Todays Outline -Releasing Pathways
and Work Metabolic Pathways Enzymes Features Factors Affecting Enzyme Activity Membrane Transport Diffusion Osmosis Passive Transport Active Transport Bulk Transport Todays Outline -Releasing Pathways
Biological Membranes. Impermeable lipid bilayer membrane. Protein Channels and Pores
 Biological Membranes Impermeable lipid bilayer membrane Protein Channels and Pores 1 Biological Membranes Are Barriers for Ions and Large Polar Molecules The Cell. A Molecular Approach. G.M. Cooper, R.E.
Biological Membranes Impermeable lipid bilayer membrane Protein Channels and Pores 1 Biological Membranes Are Barriers for Ions and Large Polar Molecules The Cell. A Molecular Approach. G.M. Cooper, R.E.
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
Ions cannot cross membranes. Ions move through pores
 Ions cannot cross membranes Membranes are lipid bilayers Nonpolar tails Polar head Fig 3-1 Because of the charged nature of ions, they cannot cross a lipid bilayer. The ion and its cloud of polarized water
Ions cannot cross membranes Membranes are lipid bilayers Nonpolar tails Polar head Fig 3-1 Because of the charged nature of ions, they cannot cross a lipid bilayer. The ion and its cloud of polarized water
Six major functions of membrane proteins: Transport Enzymatic activity
 CH 7 Membranes Cellular Membranes Phospholipids are the most abundant lipid in the plasma membrane. Phospholipids are amphipathic molecules, containing hydrophobic and hydrophilic regions. The fluid mosaic
CH 7 Membranes Cellular Membranes Phospholipids are the most abundant lipid in the plasma membrane. Phospholipids are amphipathic molecules, containing hydrophobic and hydrophilic regions. The fluid mosaic
Copyright 2000-2003 Mark Brandt, Ph.D. 54
 Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Parts of the Nerve Cell and Their Functions
 Parts of the Nerve Cell and Their Functions Silvia Helena Cardoso, PhD [ 1. Cell body] [2. Neuronal membrane] [3. Dendrites] [4. Axon] [5. Nerve ending] 1. Cell body The cell body (soma) is the factory
Parts of the Nerve Cell and Their Functions Silvia Helena Cardoso, PhD [ 1. Cell body] [2. Neuronal membrane] [3. Dendrites] [4. Axon] [5. Nerve ending] 1. Cell body The cell body (soma) is the factory
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Cell Biology - Part 2 Membranes
 Cell Biology - Part 2 Membranes The organization of cells is made possible by membranes. Membranes isolate, partition, and compartmentalize cells. 1 Membranes isolate the inside of the cell from the outside
Cell Biology - Part 2 Membranes The organization of cells is made possible by membranes. Membranes isolate, partition, and compartmentalize cells. 1 Membranes isolate the inside of the cell from the outside
UNRAVELING THE SENSE OF SMELL
 UNRAVELING THE SENSE OF SMELL Nobel Lecture, December 8, 2004 by Linda B. Buck Howard Hughes Medical Institute, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109 1024,
UNRAVELING THE SENSE OF SMELL Nobel Lecture, December 8, 2004 by Linda B. Buck Howard Hughes Medical Institute, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109 1024,
Helices From Readily in Biological Structures
 The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
4. Biology of the Cell
 4. Biology of the Cell Our primary focus in this chapter will be the plasma membrane and movement of materials across the plasma membrane. You should already be familiar with the basic structures and roles
4. Biology of the Cell Our primary focus in this chapter will be the plasma membrane and movement of materials across the plasma membrane. You should already be familiar with the basic structures and roles
Total body water ~(60% of body mass): Intracellular fluid ~2/3 or ~65% Extracellular fluid ~1/3 or ~35% fluid. Interstitial.
 http://www.bristol.ac.uk/phys-pharm/teaching/staffteaching/sergeykasparov.htmlpharm/teaching/staffteaching/sergeykasparov.html Physiology of the Cell Membrane Membrane proteins and their roles (channels,
http://www.bristol.ac.uk/phys-pharm/teaching/staffteaching/sergeykasparov.htmlpharm/teaching/staffteaching/sergeykasparov.html Physiology of the Cell Membrane Membrane proteins and their roles (channels,
Preliminary MFM Quiz
 Preliminary MFM Quiz 1. The major carrier of chemical energy in all cells is: A) adenosine monophosphate B) adenosine diphosphate C) adenosine trisphosphate D) guanosine trisphosphate E) carbamoyl phosphate
Preliminary MFM Quiz 1. The major carrier of chemical energy in all cells is: A) adenosine monophosphate B) adenosine diphosphate C) adenosine trisphosphate D) guanosine trisphosphate E) carbamoyl phosphate
Chapter-21b: Hormones and Receptors
 1 hapter-21b: Hormones and Receptors Hormone classes Hormones are classified according to the distance over which they act. 1. Autocrine hormones --- act on the same cell that released them. Interleukin-2
1 hapter-21b: Hormones and Receptors Hormone classes Hormones are classified according to the distance over which they act. 1. Autocrine hormones --- act on the same cell that released them. Interleukin-2
PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY
 Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Cells & Cell Organelles
 Cells & Cell Organelles The Building Blocks of Life H Biology Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
Cells & Cell Organelles The Building Blocks of Life H Biology Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
Hormones & Chemical Signaling
 Hormones & Chemical Signaling Part 2 modulation of signal pathways and hormone classification & function How are these pathways controlled? Receptors are proteins! Subject to Specificity of binding Competition
Hormones & Chemical Signaling Part 2 modulation of signal pathways and hormone classification & function How are these pathways controlled? Receptors are proteins! Subject to Specificity of binding Competition
Nerve Cell Communication
 Nerve Cell Communication Core Concept: Nerve cells communicate using electrical and chemical signals. Class time required: Approximately 2 forty minute class periods Teacher Provides: For each student
Nerve Cell Communication Core Concept: Nerve cells communicate using electrical and chemical signals. Class time required: Approximately 2 forty minute class periods Teacher Provides: For each student
Chapter 3. Cellular Structure and Function Worksheets. 39 www.ck12.org
 Chapter 3 Cellular Structure and Function Worksheets (Opening image copyright by Sebastian Kaulitzki, 2010. Used under license from Shutterstock.com.) Lesson 3.1: Introduction to Cells Lesson 3.2: Cell
Chapter 3 Cellular Structure and Function Worksheets (Opening image copyright by Sebastian Kaulitzki, 2010. Used under license from Shutterstock.com.) Lesson 3.1: Introduction to Cells Lesson 3.2: Cell
Biological cell membranes
 Unit 14: Cell biology. 14 2 Biological cell membranes The cell surface membrane surrounds the cell and acts as a barrier between the cell s contents and the environment. The cell membrane has multiple
Unit 14: Cell biology. 14 2 Biological cell membranes The cell surface membrane surrounds the cell and acts as a barrier between the cell s contents and the environment. The cell membrane has multiple
A disaccharide is formed when a dehydration reaction joins two monosaccharides. This covalent bond is called a glycosidic linkage.
 CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
2006 7.012 Problem Set 6 KEY
 2006 7.012 Problem Set 6 KEY ** Due before 5 PM on WEDNESDAY, November 22, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You create an artificial
2006 7.012 Problem Set 6 KEY ** Due before 5 PM on WEDNESDAY, November 22, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You create an artificial
Quick Hit Activity Using UIL Science Contests For Formative and Summative Assessments of Pre-AP and AP Biology Students
 Quick Hit Activity Using UIL Science Contests For Formative and Summative Assessments of Pre-AP and AP Biology Students Activity Title: Quick Hit Goal of Activity: To perform formative and summative assessments
Quick Hit Activity Using UIL Science Contests For Formative and Summative Assessments of Pre-AP and AP Biology Students Activity Title: Quick Hit Goal of Activity: To perform formative and summative assessments
7 Answers to end-of-chapter questions
 7 Answers to end-of-chapter questions Multiple choice questions 1 B 2 B 3 A 4 B 5 A 6 D 7 C 8 C 9 B 10 B Structured questions 11 a i Maintenance of a constant internal environment within set limits i Concentration
7 Answers to end-of-chapter questions Multiple choice questions 1 B 2 B 3 A 4 B 5 A 6 D 7 C 8 C 9 B 10 B Structured questions 11 a i Maintenance of a constant internal environment within set limits i Concentration
CSC 2427: Algorithms for Molecular Biology Spring 2006. Lecture 16 March 10
 CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
The Molecules of Cells
 The Molecules of Cells I. Introduction A. Most of the world s population cannot digest milk-based foods. 1. These people are lactose intolerant because they lack the enzyme lactase. 2. This illustrates
The Molecules of Cells I. Introduction A. Most of the world s population cannot digest milk-based foods. 1. These people are lactose intolerant because they lack the enzyme lactase. 2. This illustrates
Fight or Flight Response: Play-by-Play
 One of the most remarkable examples of cell communication is the fight or flight response. When a threat occurs, cells communicate rapidly to elicit physiological responses that help the body handle extraordinary
One of the most remarkable examples of cell communication is the fight or flight response. When a threat occurs, cells communicate rapidly to elicit physiological responses that help the body handle extraordinary
Anatomy and Physiology Placement Exam 2 Practice with Answers at End!
 Anatomy and Physiology Placement Exam 2 Practice with Answers at End! General Chemical Principles 1. bonds are characterized by the sharing of electrons between the participating atoms. a. hydrogen b.
Anatomy and Physiology Placement Exam 2 Practice with Answers at End! General Chemical Principles 1. bonds are characterized by the sharing of electrons between the participating atoms. a. hydrogen b.
Nerves and Nerve Impulse
 Nerves and Nerve Impulse Terms Absolute refractory period: Period following stimulation during which no additional action potential can be evoked. Acetylcholine: Chemical transmitter substance released
Nerves and Nerve Impulse Terms Absolute refractory period: Period following stimulation during which no additional action potential can be evoked. Acetylcholine: Chemical transmitter substance released
MULTIPLE CHOICE QUESTIONS
 MULTIPLE CHOICE QUESTIONS 1. Most components of energy conversion systems evolved very early; thus, the most fundamental aspects of energy metabolism tend to be: A. quite different among a diverse group
MULTIPLE CHOICE QUESTIONS 1. Most components of energy conversion systems evolved very early; thus, the most fundamental aspects of energy metabolism tend to be: A. quite different among a diverse group
Given these characteristics of life, which of the following objects is considered a living organism? W. X. Y. Z.
 Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
AP BIOLOGY 2008 SCORING GUIDELINES
 AP BIOLOGY 2008 SCORING GUIDELINES Question 1 1. The physical structure of a protein often reflects and affects its function. (a) Describe THREE types of chemical bonds/interactions found in proteins.
AP BIOLOGY 2008 SCORING GUIDELINES Question 1 1. The physical structure of a protein often reflects and affects its function. (a) Describe THREE types of chemical bonds/interactions found in proteins.
Cell Membrane Structure (and How to Get Through One)
 Cell Membrane Structure (and How to Get Through One) A cell s membrane is a wall of sorts that defines the boundaries of a cell. The membrane provides protection and structure for the cell and acts as
Cell Membrane Structure (and How to Get Through One) A cell s membrane is a wall of sorts that defines the boundaries of a cell. The membrane provides protection and structure for the cell and acts as
Molecular Cell Biology. Prof. D. Karunagaran. Department of Biotechnology. Indian Institute of Technology Madras. Module 7 Cell Signaling Mechanisms
 Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 7 Cell Signaling Mechanisms Lecture 2 GPCR Signaling Receptors - G protein coupled receptors
Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 7 Cell Signaling Mechanisms Lecture 2 GPCR Signaling Receptors - G protein coupled receptors
MCAS Biology. Review Packet
 MCAS Biology Review Packet 1 Name Class Date 1. Define organic. THE CHEMISTRY OF LIFE 2. All living things are made up of 6 essential elements: SPONCH. Name the six elements of life. S N P C O H 3. Elements
MCAS Biology Review Packet 1 Name Class Date 1. Define organic. THE CHEMISTRY OF LIFE 2. All living things are made up of 6 essential elements: SPONCH. Name the six elements of life. S N P C O H 3. Elements
Chapter 5: The Structure and Function of Large Biological Molecules
 Name Period Concept 5.1 Macromolecules are polymers, built from monomers 1. The large molecules of all living things fall into just four main classes. Name them. 2. Circle the three classes that are called
Name Period Concept 5.1 Macromolecules are polymers, built from monomers 1. The large molecules of all living things fall into just four main classes. Name them. 2. Circle the three classes that are called
7. A selectively permeable membrane only allows certain molecules to pass through.
 CHAPTER 2 GETTING IN & OUT OF CELLS PASSIVE TRANSPORT Cell membranes help organisms maintain homeostasis by controlling what substances may enter or leave cells. Some substances can cross the cell membrane
CHAPTER 2 GETTING IN & OUT OF CELLS PASSIVE TRANSPORT Cell membranes help organisms maintain homeostasis by controlling what substances may enter or leave cells. Some substances can cross the cell membrane
REVIEW SHEET EXERCISE 3 Neurophysiology of Nerve Impulses Name Lab Time/Date. The Resting Membrane Potential
 REVIEW SHEET EXERCISE 3 Neurophysiology of Nerve Impulses Name Lab Time/Date ACTIVITY 1 The Resting Membrane Potential 1. Explain why increasing extracellular K + reduces the net diffusion of K + out of
REVIEW SHEET EXERCISE 3 Neurophysiology of Nerve Impulses Name Lab Time/Date ACTIVITY 1 The Resting Membrane Potential 1. Explain why increasing extracellular K + reduces the net diffusion of K + out of
Basic Scientific Principles that All Students Should Know Upon Entering Medical and Dental School at McGill
 Fundamentals of Medicine and Dentistry Basic Scientific Principles that All Students Should Know Upon Entering Medical and Dental School at McGill Students entering medical and dental training come from
Fundamentals of Medicine and Dentistry Basic Scientific Principles that All Students Should Know Upon Entering Medical and Dental School at McGill Students entering medical and dental training come from
Date: Student Name: Teacher Name: Jared George. Score: 1) A cell with 1% solute concentration is placed in a beaker with a 5% solute concentration.
 Biology Keystone (PA Core) Quiz Homeostasis and Transport - (BIO.A.4.1.1 ) Plasma Membrane, (BIO.A.4.1.2 ) Transport Mechanisms, (BIO.A.4.1.3 ) Transport Facilitation Student Name: Teacher Name: Jared
Biology Keystone (PA Core) Quiz Homeostasis and Transport - (BIO.A.4.1.1 ) Plasma Membrane, (BIO.A.4.1.2 ) Transport Mechanisms, (BIO.A.4.1.3 ) Transport Facilitation Student Name: Teacher Name: Jared
Modes of Membrane Transport
 Modes of Membrane Transport Transmembrane Transport movement of small substances through a cellular membrane (plasma, ER, mitochondrial..) ions, fatty acids, H 2 O, monosaccharides, steroids, amino acids
Modes of Membrane Transport Transmembrane Transport movement of small substances through a cellular membrane (plasma, ER, mitochondrial..) ions, fatty acids, H 2 O, monosaccharides, steroids, amino acids
Chapter 3 Molecules of Cells
 Bio 100 Molecules of cells 1 Chapter 3 Molecules of Cells Compounds containing carbon are called organic compounds Molecules such as methane that are only composed of carbon and hydrogen are called hydrocarbons
Bio 100 Molecules of cells 1 Chapter 3 Molecules of Cells Compounds containing carbon are called organic compounds Molecules such as methane that are only composed of carbon and hydrogen are called hydrocarbons
Cell and Membrane Practice. A. chromosome B. gene C. mitochondrion D. vacuole
 Name: ate: 1. Which structure is outside the nucleus of a cell and contains N?. chromosome. gene. mitochondrion. vacuole 2. potato core was placed in a beaker of water as shown in the figure below. Which
Name: ate: 1. Which structure is outside the nucleus of a cell and contains N?. chromosome. gene. mitochondrion. vacuole 2. potato core was placed in a beaker of water as shown in the figure below. Which
Biological molecules:
 Biological molecules: All are organic (based on carbon). Monomers vs. polymers: Monomers refer to the subunits that, when polymerized, make up a larger polymer. Monomers may function on their own in some
Biological molecules: All are organic (based on carbon). Monomers vs. polymers: Monomers refer to the subunits that, when polymerized, make up a larger polymer. Monomers may function on their own in some
Lecture 8. Protein Trafficking/Targeting. Protein targeting is necessary for proteins that are destined to work outside the cytoplasm.
 Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Chapter 8. Movement across the Cell Membrane. AP Biology
 Chapter 8. Movement across the Cell Membrane More than just a barrier Expanding our view of cell membrane beyond just a phospholipid bilayer barrier phospholipids plus Fluid Mosaic Model In 1972, S.J.
Chapter 8. Movement across the Cell Membrane More than just a barrier Expanding our view of cell membrane beyond just a phospholipid bilayer barrier phospholipids plus Fluid Mosaic Model In 1972, S.J.
ANIMATED NEUROSCIENCE
 ANIMATED NEUROSCIENCE and the Action of Nicotine, Cocaine, and Marijuana in the Brain Te a c h e r s G u i d e Films for the Humanities & Sciences Background Information This program, made entirely of
ANIMATED NEUROSCIENCE and the Action of Nicotine, Cocaine, and Marijuana in the Brain Te a c h e r s G u i d e Films for the Humanities & Sciences Background Information This program, made entirely of
Mechanism of hormone action
 Mechanism of hormone action ผศ.ดร.พญ.ส ว ฒณ ค ปต ว ฒ ภาคว ชาสร รว ทยา คณะแพทยศาสตร ศ ร ราชพยาบาล Aims What is hormone receptor Type of hormone receptors - cell surface receptor - intracellular receptor
Mechanism of hormone action ผศ.ดร.พญ.ส ว ฒณ ค ปต ว ฒ ภาคว ชาสร รว ทยา คณะแพทยศาสตร ศ ร ราชพยาบาล Aims What is hormone receptor Type of hormone receptors - cell surface receptor - intracellular receptor
http://faculty.sau.edu.sa/h.alshehri
 http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
Chapter 2 Phosphorus in the Organic Life: Cells, Tissues, Organisms
 Chapter 2 Phosphorus in the Organic Life: Cells, Tissues, Organisms As already mentioned (see Chap. 1 ), in the living cell phosphorus plays a decisive role in three different essential structures: In
Chapter 2 Phosphorus in the Organic Life: Cells, Tissues, Organisms As already mentioned (see Chap. 1 ), in the living cell phosphorus plays a decisive role in three different essential structures: In
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Regulation of enzyme activity
 1 Regulation of enzyme activity Regulation of enzyme activity is important to coordinate the different metabolic processes. It is also important for homeostasis i.e. to maintain the internal environment
1 Regulation of enzyme activity Regulation of enzyme activity is important to coordinate the different metabolic processes. It is also important for homeostasis i.e. to maintain the internal environment
Chapter 8: An Introduction to Metabolism
 Chapter 8: An Introduction to Metabolism Name Period Concept 8.1 An organism s metabolism transforms matter and energy, subject to the laws of thermodynamics 1. Define metabolism. The totality of an organism
Chapter 8: An Introduction to Metabolism Name Period Concept 8.1 An organism s metabolism transforms matter and energy, subject to the laws of thermodynamics 1. Define metabolism. The totality of an organism
Disaccharides consist of two monosaccharide monomers covalently linked by a glycosidic bond. They function in sugar transport.
 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
Expression and Purification of Recombinant Protein in bacteria and Yeast. Presented By: Puspa pandey, Mohit sachdeva & Ming yu
 Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Chemical Basis of Life Module A Anchor 2
 Chemical Basis of Life Module A Anchor 2 Key Concepts: - Water is a polar molecule. Therefore, it is able to form multiple hydrogen bonds, which account for many of its special properties. - Water s polarity
Chemical Basis of Life Module A Anchor 2 Key Concepts: - Water is a polar molecule. Therefore, it is able to form multiple hydrogen bonds, which account for many of its special properties. - Water s polarity
B Cell Generation, Activation & Differentiation. B cell maturation
 B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
green B 1 ) into a single unit to model the substrate in this reaction. enzyme
 Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
What affects an enzyme s activity? General environmental factors, such as temperature and ph. Chemicals that specifically influence the enzyme.
 CH s 8-9 Respiration & Metabolism Metabolism A catalyst is a chemical agent that speeds up a reaction without being consumed by the reaction. An enzyme is a catalytic protein. Hydrolysis of sucrose by
CH s 8-9 Respiration & Metabolism Metabolism A catalyst is a chemical agent that speeds up a reaction without being consumed by the reaction. An enzyme is a catalytic protein. Hydrolysis of sucrose by
008 Chapter 8. Student:
 008 Chapter 8 Student: 1. Some bacteria are strict aerobes and others are strict anaerobes. Some bacteria, however, are facultative anaerobes and can live with or without oxygen. If given the choice of
008 Chapter 8 Student: 1. Some bacteria are strict aerobes and others are strict anaerobes. Some bacteria, however, are facultative anaerobes and can live with or without oxygen. If given the choice of
4. Which carbohydrate would you find as part of a molecule of RNA? a. Galactose b. Deoxyribose c. Ribose d. Glucose
 1. How is a polymer formed from multiple monomers? a. From the growth of the chain of carbon atoms b. By the removal of an OH group and a hydrogen atom c. By the addition of an OH group and a hydrogen
1. How is a polymer formed from multiple monomers? a. From the growth of the chain of carbon atoms b. By the removal of an OH group and a hydrogen atom c. By the addition of an OH group and a hydrogen
Introduction to Proteins and Enzymes
 Introduction to Proteins and Enzymes Basics of protein structure and composition The life of a protein Enzymes Theory of enzyme function Not all enzymes are proteins / not all proteins are enzymes Enzyme
Introduction to Proteins and Enzymes Basics of protein structure and composition The life of a protein Enzymes Theory of enzyme function Not all enzymes are proteins / not all proteins are enzymes Enzyme
CELLS IN THE NERVOUS SYSTEM
 NEURONS AND GLIA CELLS IN THE NERVOUS SYSTEM Glia Insulates, supports, and nourishes neurons Neurons Process information Sense environmental changes Communicate changes to other neurons Command body response
NEURONS AND GLIA CELLS IN THE NERVOUS SYSTEM Glia Insulates, supports, and nourishes neurons Neurons Process information Sense environmental changes Communicate changes to other neurons Command body response
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
Introduction to Animals
 Introduction to Animals Unity and Diversity of Life Q: What characteristics and traits define animals? 25.1 What is an animal? WHAT I KNOW SAMPLE ANSWER: Animals are different from other living things
Introduction to Animals Unity and Diversity of Life Q: What characteristics and traits define animals? 25.1 What is an animal? WHAT I KNOW SAMPLE ANSWER: Animals are different from other living things
Cells and Their Housekeeping Functions Cell Membrane & Membrane Potential
 Cells and Their Housekeeping Functions Cell Membrane & Membrane Potential Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/
Cells and Their Housekeeping Functions Cell Membrane & Membrane Potential Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/
Cell Structure & Function!
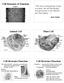 Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
ID of alternative translational initiation events. Description of gene function Reference of NCBI database access and relative literatures
 Data resource: In this database, 650 alternatively translated variants assigned to a total of 300 genes are contained. These database records of alternative translational initiation have been collected
Data resource: In this database, 650 alternatively translated variants assigned to a total of 300 genes are contained. These database records of alternative translational initiation have been collected
Chemistry 20 Chapters 15 Enzymes
 Chemistry 20 Chapters 15 Enzymes Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An
Chemistry 20 Chapters 15 Enzymes Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An
Disulfide Bonds at the Hair Salon
 Disulfide Bonds at the Hair Salon Three Alpha Helices Stabilized By Disulfide Bonds! In order for hair to grow 6 inches in one year, 9 1/2 turns of α helix must be produced every second!!! In some proteins,
Disulfide Bonds at the Hair Salon Three Alpha Helices Stabilized By Disulfide Bonds! In order for hair to grow 6 inches in one year, 9 1/2 turns of α helix must be produced every second!!! In some proteins,
Unit 2: Cells, Membranes and Signaling CELL MEMBRANE. Chapter 5 Hillis Textbook
 Unit 2: Cells, Membranes and Signaling CELL MEMBRANE Chapter 5 Hillis Textbook HOW DOES THE LAB RELATE TO THE NEXT CHAPTER? SURFACE AREA: the entire outer covering of a cell that enables materials pass.
Unit 2: Cells, Membranes and Signaling CELL MEMBRANE Chapter 5 Hillis Textbook HOW DOES THE LAB RELATE TO THE NEXT CHAPTER? SURFACE AREA: the entire outer covering of a cell that enables materials pass.
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Cell Membrane & Tonicity Worksheet
 NAME ANSWER KEY DATE PERIOD Cell Membrane & Tonicity Worksheet Composition of the Cell Membrane & Functions The cell membrane is also called the PLASMA membrane and is made of a phospholipid BI-LAYER.
NAME ANSWER KEY DATE PERIOD Cell Membrane & Tonicity Worksheet Composition of the Cell Membrane & Functions The cell membrane is also called the PLASMA membrane and is made of a phospholipid BI-LAYER.
Review of the Cell and Its Organelles
 Biology Learning Centre Review of the Cell and Its Organelles Tips for most effective learning of this material: Memorize the names and structures over several days. This will help you retain what you
Biology Learning Centre Review of the Cell and Its Organelles Tips for most effective learning of this material: Memorize the names and structures over several days. This will help you retain what you
Chapter 5. The Structure and Function of Macromolecule s
 Chapter 5 The Structure and Function of Macromolecule s Most Macromolecules are polymers: Polymer: (poly: many; mer: part) Large molecules consisting of many identical or similar subunits connected together.
Chapter 5 The Structure and Function of Macromolecule s Most Macromolecules are polymers: Polymer: (poly: many; mer: part) Large molecules consisting of many identical or similar subunits connected together.
Module 3 Questions. 7. Chemotaxis is an example of signal transduction. Explain, with the use of diagrams.
 Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
Ion Channels. Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com)
 Ion Channels Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com) ** There are a number of ion channels introducted in this topic which you
Ion Channels Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com) ** There are a number of ion channels introducted in this topic which you
Biology Slide 1 of 38
 Biology 1 of 38 2 of 38 35-2 The Nervous System What are the functions of the nervous system? 3 of 38 35-2 The Nervous System 1. Nervous system: a. controls and coordinates functions throughout the body
Biology 1 of 38 2 of 38 35-2 The Nervous System What are the functions of the nervous system? 3 of 38 35-2 The Nervous System 1. Nervous system: a. controls and coordinates functions throughout the body
AP Bio Photosynthesis & Respiration
 AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
How To Understand The Chemistry Of Organic Molecules
 CHAPTER 3 THE CHEMISTRY OF ORGANIC MOLECULES 3.1 Organic Molecules The chemistry of carbon accounts for the diversity of organic molecules found in living things. Carbon has six electrons, four of which
CHAPTER 3 THE CHEMISTRY OF ORGANIC MOLECULES 3.1 Organic Molecules The chemistry of carbon accounts for the diversity of organic molecules found in living things. Carbon has six electrons, four of which
Membrane Structure and Function
 Membrane Structure and Function -plasma membrane acts as a barrier between cells and the surrounding. -plasma membrane is selective permeable -consist of lipids, proteins and carbohydrates -major lipids
Membrane Structure and Function -plasma membrane acts as a barrier between cells and the surrounding. -plasma membrane is selective permeable -consist of lipids, proteins and carbohydrates -major lipids
Cell Transport and Plasma Membrane Structure
 Cell Transport and Plasma Membrane Structure POGIL Guided Inquiry Learning Targets Explain the importance of the plasma membrane. Compare and contrast different types of passive transport. Explain how
Cell Transport and Plasma Membrane Structure POGIL Guided Inquiry Learning Targets Explain the importance of the plasma membrane. Compare and contrast different types of passive transport. Explain how
