Horseradish peroxidase: a modern view of a classic enzyme
|
|
|
- Barbara Elliott
- 8 years ago
- Views:
Transcription
1 Phytochemistry 65 (2004) Molecules of interest Horseradish peroxidase: a modern view of a classic enzyme Nigel C. Veitch* Jodrell Laboratory, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3DS, UK Abstract Horseradish peroxidase is an important heme-containing enzyme that has been studied for more than a century. In recent years new information has become available on the three-dimensional structure of the enzyme and its catalytic intermediates, mechanisms of catalysis and the function of specific amino acid residues. Site-directed mutagenesis and directed evolution techniques are now used routinely to investigate the structure and function of horseradish peroxidase and offer the opportunity to develop engineered enzymes for practical applications in natural product and fine chemicals synthesis, medical diagnostics and bioremediation. A combination of horseradish peroxidase and indole-3-acetic acid or its derivatives is currently being evaluated as an agent for use in targeted cancer therapies. Physiological roles traditionally associated with the enzyme that include indole-3-acetic acid metabolism, cross-linking of biological polymers and lignification are becoming better understood at the molecular level, but the involvement of specific horseradish peroxidase isoenzymes in these processes is not yet clearly defined. Progress in this area should result from the identification of the entire peroxidase gene family of Arabidopsis thaliana, which has now been completed. # 2003 Elsevier Ltd. All rights reserved. Keywords: Horseradish peroxidase; Armoracia rusticana; Arabidopsis thaliana; Cruciferae; Heme; Hydrogen peroxide; Indole-3-acetic acid; Protein engineering 1. Introduction The horseradish (Armoracia rusticana P.Gaertn., B.Mey. & Scherb.; Cruciferae) is a hardy perennial herb cultivated in temperate regions of the world mainly for the culinary value of its roots (Fig. 1). These are also a rich source of peroxidase, a heme-containing enzyme that utilises hydrogen peroxide to oxidise a wide variety of organic and inorganic compounds. Production of peroxidase from horseradish roots occurs on a relatively large scale because of the commercial uses of the enzyme, for example as a component of clinical diagnostic kits and for immunoassays. Although the term horseradish peroxidase is used somewhat generically, the root of the plant contains a number of distinctive peroxidase isoenzymes of which the C isoenzyme (HRP C) is the most abundant. This has been the subject of much of the published work on horseradish peroxidase, which comprises many thousands of papers in the scientific literature. Some major advances in our understanding of the structure and function of HRP C have * Corresponding author. Tel.: ; fax: address: n.veitch@rbgkew.org.uk (N.C. Veitch). been achieved relatively recently, and were initiated by the successful production of recombinant enzyme (Smith et al., 1990). From this work came the longawaited solution of the three-dimensional structure of HRP C by X-ray crystallography and more recently, a high resolution description of the intermediates in the catalytic cycle of the enzyme (Gajhede et al., 1997; Berglund et al., 2002). Many physiological roles have been assigned to horseradish peroxidase isoenzymes including indole-3-acetic acid metabolism, lignification, crosslinking of cell wall polymers, suberin formation and resistance to infection. It is surprising therefore that so little is known about their specific functions in the plant. New insights into this problem may result from comparative studies of the peroxidases of the model plant, Arabidopsis thaliana (L.) Heynh., which is also a member of the Cruciferae. Two groups have now described the entire peroxidase gene family of A. thaliana, which comprises no less than 73 full-length genes, the majority of which are predicted to code for stable enzymes (Tognolli et al., 2002; Welinder et al., 2002). It seems therefore, that each plant species contains a suite of peroxidase isoenzymes with the potential to carry out a range of different functions. This is one of the issues that will be highlighted in the present paper, which aims to provide an overview of recent advances in our /$ - see front matter # 2003 Elsevier Ltd. All rights reserved. doi: /j.phytochem
2 250 N.C. Veitch / Phytochemistry 65 (2004) Fig. 1. An early illustration of the horseradish plant, from the 1633 edition of Gerard s celebrated Herball, or generall historie of plantes. According to Gerard the root is long and thick, white of colour, in taste sharpe, and very much biting the tongue like mustard. These properties are due to volatile isothiocyanates released by the hydrolysis of constituent glucosinolates when the roots are crushed, a reaction that is catalysed by myrosinase. The same roots are also one of the most important sources of the heme-containing enzyme, peroxidase. Reproduced with permission. Copyright 2003, Royal Botanic Gardens, Kew. understanding of horseradish peroxidase and a summary of its most important characteristics and uses. 2. Historical perspectives The first recorded observation of a reaction catalysed by horseradish peroxidase appears in a note published by Louis Antoine Planche ( ) describing the analysis of jalap resin imported to France for medicinal use, probably from the Carribean (Planche, 1810). A series of tests was carried out to determine whether a particular consignment was adulterated with gaiac resin (also known as guaiacum, the powdered heartwood of the Carribean trees, Guaiacum officinale and G. sanctum). During these investigations Planche noted that a tincture of gaiac resin became a beautiful blue colour when a piece of fresh horseradish root was placed in it. The reaction observed was probably the peroxidase-catalysed oxidation of 2,5-di-(4-hydroxy-3- methoxyphenyl)-3,4-dimethylfuran (a minor constituent of the resin and formerly referred to as a-guaiaconic acid) to give the corresponding bis-methylenequinone, a blue product also known as guaiacum blue (Kratochvil et al., 1971). Although the term peroxidase came into use towards the end of the nineteenth century it was the work of Robert Chodat ( ) and Alexei Nikolaevich Bach ( ) at the University of Geneva during the early years of the twentieth century that initiated further research on peroxidase from the horseradish root and other plant sources (Bach and Chodat, 1903). Some of their observations stimulated a vigorous debate on the nature of oxidases and peroxidases which was later recounted in a book by David Keilin ( ), another distinguished contributor to the peroxidase field (Keilin, 1966). Although Bach and Chodat obtained a crude preparation of horseradish peroxidase it was through the subsequent studies of Richard Willsta tter ( ) and Hugo Theorell ( ) that pure enzyme was finally isolated. Other advances in the period from approximately 1920 to 1945 included the demonstration of heme and carbohydrate as components of horseradish peroxidase, the first observation of the catalytic intermediates known as compounds I and II and the first kinetic analysis of the reaction with hydrogen peroxide. Detailed commentaries on the history and development of peroxidase are available in the literature (Saunders et al., 1964; Paul, 1986). These emphasise the important contribution that research on horseradish peroxidase has made to our understanding of peroxidase enzymes in general. 3. Description of the enzyme 3.1. General features Horseradish peroxidase isoenzyme C comprises a single polypeptide of 308 amino acid residues, the sequence of which was determined by Welinder (1976). The N- terminal residue is blocked by pyroglutamate and the C- terminus is heterogenous, with some molecules lacking the terminal residue, Ser308. There are 4 disulphide bridges between cysteine residues 11 91, 44 49, and , and a buried salt bridge between Asp99 and Arg123. Nine potential N-glycosylation sites can be recognised in the primary sequence from the motif Asn- X-Ser/Thr (where X represents an amino acid residue) and of these, eight are occupied. A branched heptasaccharide accounts for 75 to 80% of the glycans (Table 1) but the carbohydrate profile of HRP C is heterogeneous, and many minor glycans have also been characterised (Yang et al., 1996). These invariably contain two terminal GlcNAc and several mannose residues. A further complication is the variation in the type of glycan present at any of the glycosylation sites. The total carbohydrate content of HRP C is somewhat dependent on the source of the enzyme and values of between 18 and 22% are typical.
3 N.C. Veitch / Phytochemistry 65 (2004) Table 1 Anatomy of a peroxidase. Some essential structural features of horseradish peroxidase isoenzyme C (HRP C) Heme (i) (ii) (iii) (iv) His170 forms coordinate bond to heme iron atom. Asp247 carboxylate side-chain helps to control imidazolate character of His170 ring. His170Ala mutant undergoes heme degradation when H 2 O 2 added and compounds I and II are not detected; k 1 = M 1 s 1. Imidazoles can bind to heme iron in the artificially created cavity but full catalytic activity is not restored because the His170Ala-imidazole complex does not maintain a five-coordinate state (His42 also binds to Fe). Aromatic substrates are oxidized at the exposed heme edge but do not bind to heme iron. Calcium Distal O-donors Proximal O-donors (i) Distal and proximal Ca 2+ ions are both seven-coordinate. Asp43, Asp50, Ser52 Thr171, Asp222, (ii) On calcium loss enzyme activity decreases by 40%. (side-chain) Asp43, Val46, Gly48 (carbonyl) 1 structural water Thr225, Asp230 (side-chain) Thr171, Thr225, Ile228 (carbonyl) (iii) Structural water of distal Ca site hydrogen bonded to Glu64 which is itself hydrogen bonded to Asn70 and thus connects to the distal heme pocket. Carbohydrate (i) (ii) Sites of glycosylation are in loop regions of the structure, at Asn13, Asn57, Asn158, Asn186, Asn198, Asn214, Asn255 and Asn268. The major glycan is shown here; there are also minor glycans of the form Man m GlcNAc 2 (m = 4 to 7) and (Xyl) x Man m (Fuc) f GlcNAc 2 (m =2,4,5,6;f = 0 or 1; x = 0 or 1). Amino acid residues Arg38 Essential roles in (i), the formation and stabilization of compound I, (ii) binding and stabilization of ligands and aromatic substrates (e.g. benzhydroxamic acid, ferulate etc.). Arg38Leu mutant; k 1 = M 1 s 1 ; K d (BHA) mm. Phe41 Prevents substrate access to the ferryl oxygen of compound I. Phe41Ala/Leu/Thr mutants show improved rates of thioanisole oxidation (oxygen-transfer chemistry). His42 Asn70 Pro139 Essential roles in (i), compound I formation (accepts proton from H 2 O 2 ), (ii) binding and stabilization of ligands and aromatic substrates. Maintains basicity of His42 side-chain through Asn70-His42 couple (hydrogen bond from Asn70 amide oxygen to His42 imidazole NH). Part of a structural motif, -Pro-X-Pro- (Pro139-Ala140-Pro141 in HRP C), which is conserved in plant peroxidases. His42Leu mutant; k 1 = M 1 s 1 ; K d (BHA) mm. Asn70Val mutant; k 1 = M 1 s 1. Crystallographic evidence indicates role for Pro139 in substrate oxidation and binding. Note that residues Ala140, Phe142 and Phe143 are also part of the ferulate binding site shown in Fig. 5. Original references for the results described in this table can be found in Veitch and Smith (2001). The rate constant for compound I formation (k 1 ) for wild-type enzyme is M 1 s 1. The apparent dissociation constant (K d ) for complex formation between resting state HRP C and benzhydroxamic acid is mm (ph 7.0, 25 C).
4 252 N.C. Veitch / Phytochemistry 65 (2004) HRP C contains two different types of metal centre, iron(iii) protoporphyrin IX (usually referred to as the heme group ) and two calcium atoms (Fig. 2 and Table 1). Both are essential for the structural and functional integrity of the enzyme. The heme group is attached to the enzyme at His170 (the proximal histidine residue) by a coordinate bond between the histidine side-chain NE2 atom and the heme iron atom. The second axial coordination site (on the so-called distal side of the heme plane) is unoccupied in the resting state of the enzyme but available to hydrogen peroxide during enzyme turnover (Fig. 3). Small molecules such as carbon monoxide, cyanide, fluoride and azide bind to the heme iron atom at this distal site giving six-coordinate peroxidase complexes. Some bind only in their protonated forms, which are stabilized through hydrogen bonded interactions with the distal heme pocket amino acid side-chains of Arg38 (the distal arginine) and His42 (the distal histidine) (Fig. 3). The two calcium binding sites are located at positions distal and proximal to the heme plane and are linked to the heme-binding region by a network of hydrogen bonds. Each calcium site is seven-coordinate with oxygen-donor ligands provided by a combination of amino acid side-chain carboxylates (Asp), hydroxyl groups (Ser, Thr), backbone carbonyls and a structural water molecule (distal site only) (Table 1). Loss of calcium results in decreases to both enzyme activity and thermal stability (Haschke and Friedhoff, 1978) and to subtle changes in the heme environment that can be detected spectroscopically (Howes et al., 2001) Three-dimensional structure The first solution of the three-dimensional structure of HRP C using X-ray crystallography appeared in the literature relatively recently (Gajhede et al., 1997). The recombinant enzyme used as the source of crystals and heavy atom derivatives was produced by expression in Escherichia coli in non-glycosylated form (Smith et al., 1990). Previous attempts to obtain suitable crystals for diffraction were frustrated partly by the heterogeneity of plant HRP C preparations comprising multiple glycoforms. The structure of the enzyme is largely a-helical although there is also a small region of b-sheet (Fig. 2). There are two domains, the distal and proximal, between which the heme group is located. These domains probably originated as a result of gene duplication, a proposal supported by their common calcium binding sites and other structural elements (Welinder and Gajhede, 1993). Fig. 2. Three-dimensional representation of the X-ray crystal structure of horseradish peroxidase isoenzyme C (Brookhaven accession code 1H5A). The heme group (coloured in red) is located between the distal and proximal domains which each contain one calcium atom (shown as blue spheres). a-helical and b-sheet regions of the enzyme are shown in purple and yellow, respectively. The F 0 and F 00 a-helices appear in the bottom right-hand quadrant of the molecule. Fig. 3. Key amino acid residues in the heme-binding region of HRP C. The heme group and heme iron atom are shown in red, the remaining residues in atom colours. His170, the proximal histidine residue, is coordinated to the heme iron atom whereas the corresponding distal coordination site above the plane of the heme is vacant. Specific functions for the amino acid residues shown here are given in Table 1.
5 N.C. Veitch / Phytochemistry 65 (2004) HRP and the plant peroxidase superfamily Horseradish peroxidase isoenzymes belong to class III ( classical secretory plant peroxidases) of the plant peroxidase superfamily, which includes peroxidases of bacterial, fungal and plant origin (Welinder, 1992a). The remaining two classes comprise yeast cytochrome c peroxidase, gene-duplicated bacterial peroxidases and ascorbate peroxidases (class I), and fungal peroxidases (class II). This classification was based originally on comparisons of amino acid sequence but is well-supported by more recent three-dimensional structural data obtained for enzymes from each of the classes. Many structural elements are conserved among peroxidases of the three classes leading to the definition of a core peroxidase fold. The structures of HRP C and of other class III plant peroxidases contain three a-helices that are additional to this core peroxidase fold (Gajhede et al., 1997). Two of these (helices F 0 and F 00 ) are located in a long insertion that shows great variation in both sequence and number of residues (Fig. 2). The overall integrity of the structure in this region is maintained by the disulphide linkage from Cys177 to Cys209. What is particularly interesting in the case of HRP C is the location of residues in helix F 0 that are involved in substrate access and binding. Some authors have speculated that this structural region is important for stabilization or retention of radical species produced in reactions catalysed by plant peroxidases (Gajhede et al., 1997). A comparison of structural and functional characteristics of peroxidases from the three classes of the plant peroxidase superfamily has been published (Smith and Veitch, 1998). 4. Mechanism of action ascorbate peroxidase (Apx1) was demonstrated (Pnueli et al., 2003). An interesting observation in the knockout-apx1 plants was the induction of transcripts encoding for two class III plant peroxidases, presumably to compensate for the reduction in peroxidescavenging ability. The formation of radical products in HRP-catalysed reactions gives an indication of some possible in vivo functions in the plant. These may involve cross-linking reactions such as the formation of diferulate linkages from polymer-attached ferulate groups of polysaccharides or pectins, the formation of dityrosine linkages, the cross-linking of phenolic monomers in the formation of suberin and the oxidative coupling of phenolic compounds as part of the biosynthesis of lignin. Peroxidases that catalyse cross-linking reactions may be expressed in response to external factors such as the wounding of plant tissue. Water loss and invasion by pathogens can thus be limited by the formation of a protective polymeric barrier such as suberin. Although in vivo roles for HRP isoenzymes have not yet been determined, it is known that peroxidase activity is induced when horseradish leaves are wounded (Kawaoka et al., 1994) Catalytic mechanism The mechanism of catalysis of horseradish peroxidase and in particular, the C isoenzyme, has been investigated extensively (reviewed in Dunford, 1991, 1999; Veitch and Smith, 2001). Some important features of the catalytic cycle are illustrated in Fig. 4 with ferulic acid as reducing substrate. The generation of radical species in the two one-electron reduction steps can result in a complex profile of reaction products, including dimers, 4.1. Functional roles Most reactions catalysed by HRP C and other horseradish peroxidase isoenzymes can be expressed by the following equation, in which AH 2 and AH represent a reducing substrate and its radical product, respectively. Typical reducing substrates include aromatic phenols, phenolic acids, indoles, amines and sulfonates. HRP C H 2 O 2 þ 2AH 2! 2H2 O þ 2AH ð1þ The conversion of hydrogen peroxide to water is not the primary function of class III plant peroxidases such as HRP C. Other enzymes, including ascorbate peroxidase (class I), are used by plants to regulate levels of intracellular hydrogen peroxide (Mittler, 2002; Shigeoka et al., 2002). Little is known about the ascorbate peroxidases of horseradish, but in a recent study the accumulation of H 2 O 2 in Arabidopsis thaliana plants deficient in cytosolic Fig. 4. The catalytic cycle of horseradish peroxidase (HRP C) with ferulate as reducing substrate. The rate constants k 1, k 2 and k 3 represent the rate of compound I formation, rate of compound I reduction and rate of compound II reduction, respectively.
6 254 N.C. Veitch / Phytochemistry 65 (2004) trimers and higher oligomers that may themselves act as reducing substrates in subsequent turnovers. The first step in the catalytic cycle is the reaction between H 2 O 2 and the Fe(III) resting state of the enzyme to generate compound I, a high oxidation state intermediate comprising an Fe(IV) oxoferryl centre and a porphyrin-based cation radical. A transient intermediate (compound 0) formed prior to compound I has been detected in reactions between HRP C and H 2 O 2 at low temperatures and described as an Fe(III)-hydroperoxy complex. Molecular dynamics simulations of these peroxide-bound complexes have been carried out (Filizola and Loew, 2000). In formal terms, compound I is two oxidising equivalents above the resting state. The first one-electron reduction step requires the participation of a reducing substrate and leads to the generation of compound II, an Fe(IV) oxoferryl species that is one oxidising equivalent above the resting state. Both compound I and compound II are powerful oxidants, with redox potentials estimated to be close to +1 V. The second one-electron reduction step returns compound II to the resting state of the enzyme. Reaction of excess hydrogen peroxide with resting state enzyme gives compound III, which can also be prepared by several other routes (Dunford, 1999). This intermediate is best described as a resonance hybrid of iron(iii)-superoxide and iron(ii)-dioxygen complexes. A high-resolution crystal structure of 95% pure compound III published recently shows dioxygen bound to heme iron in a bent conformation (Berglund et al., 2002). Models for the irreversible inactivation of HRP C by peroxides have been developed in studies with m-chloroperoxybenzoic acid (Rodriguez-Lopez et al., 1997). Detailed mechanisms showing the role of the distal heme pocket residues Arg38 and His42 (conserved in all members of the plant peroxidase superfamily) in the formation of compound I have been described in the literature but are beyond the scope of this review. High resolution crystal structures of the oxidised intermediates of HRP C confirm the importance of Arg38 and His42 for peroxide catalysis (Berglund et al., 2002). For example, the ferryl oxygen of compound I is hydrogen bonded to Arg38 NEH and a water molecule that is also hydrogen bonded to both Arg38 and His42. A mechanism for the reduction of compounds I and II by ferulate has been proposed based on new information from the crystal structure of a ternary complex formed between ferulic acid and cyanide-ligated HRP C (Henriksen et al., 1999) Aromatic substrate binding sites Substrate oxidation by HRP C occurs at the exposed heme edge, a region comprising the heme methyl C18 and heme meso C20 protons (Table 1). This interaction site was identified in enzyme inactivation experiments with alkyl- and phenylhydrazines, sodium azide and other reactive agents (Ortiz de Montellano, 1992). The free radical species generated when these are incubated with HRP C and H 2 O 2 give adducts such as C18- hydroxymethyl and C20-meso-phenyl heme derivatives. What is particularly interesting is the contrast between the behaviour of HRP C and other heme proteins such as the globins and cytochromes P450, where modification to the heme iron atom and pyrrole nitrogens occurs. Substrate access to the oxoferryl centre of HRP C appears to be hindered by the local protein environment, an example of closed heme architecture. This is one reason why peroxidases are much less effective as catalysts of oxygen-transfer reactions than cytochromes P450. Attempts to improve this aspect of the chemistry of HRP C have been made by site-directed mutagenesis. In these experiments, substrate access to the oxoferryl centre of compound I was increased by substituting the distal residues Phe41 and His42 with smaller amino acid residues (Newmyer and Ortiz de Montellano, 1995; Ozaki and Ortiz de Montellano, 1995). Stable, reversible 1:1 complexes with a variety of aromatic molecules are formed both by resting state HRP C and some of its ligand-bound derivatives (e.g. cyanide-ligated HRP C) in the absence of H 2 O 2. Spectroscopic and crystallographic studies have revealed a detailed picture of the site where these aromatic substrates are located (Veitch and Smith, 2001). For example, when ferulic acid binds to cyanide-ligated HRP C its aromatic ring is located in a distal site close to the exposed heme edge and its phenolic acid side-chain is oriented towards the entrance of the binding site (Henriksen et al., 1999). There are hydrogen-bonded interactions between Arg38 NZH 2 and the phenolic and methoxyl oxygen atoms of ferulic acid and between the phenolic oxygen and an active site water molecule. The latter is also hydrogen bonded to the backbone carbonyl of Pro139 (an invariant residue in the plant peroxidase superfamily) and the nitrogen atom of the cyanide ligand. There are hydrophobic interactions with amino acid side-chains (Phe68, Gly69, Pro139, Ala140, Phe142 and Phe179) as well as heme methyl C18H 3 and the heme meso proton C20H (Fig. 5). The structure and dynamics of benzhydroxamic acid complexes of HRP C have also been investigated in detail with site-directed mutants used to explore the contribution of binding site residues to substrate affinity (Veitch and Smith, 2001). 5. The isoenzyme question; horseradish and Arabidopsis peroxidases 5.1. The horseradish peroxidase isoenzyme family Fifteen peroxidase isoenzymes have been isolated from horseradish root using classical protein purification
7 N.C. Veitch / Phytochemistry 65 (2004) neutral isoenzyme (HRP N) that has not yet been isolated from the plant (Bartonek-Roxa and Holm, 1999) Arabidopsis thaliana peroxidases Fig. 5. The ferulic acid binding site of horseradish peroxidase (HRP C). This representation is taken from the X-ray crystal structure of the ternary complex of ferulic acid (FA) and cyanide-ligated HRP C (Brookhaven accession code 7ATJ). The heme group is shown in red and ferulic acid, the cyanide ligand (bound to heme iron at the vacant distal coordination site) and the catalytic residues Arg38, His42 and His170 are in atom colours. Several phenylalanine side-chains that contribute to the binding site are shown in blue. Note that the ferulic acid side-chain is located towards the entrance of the binding site. methods and are generally referred to by codes based on their isoelectric point value as HRP A1 A3 ( acidic ), B1 B3 and C1 C2 ( neutral or neutral-basic ) and D and E1 E6 ( basic ). An even greater number of peroxidase isoforms can be detected by isoelectric focusing but their identity has proved controversial as some may be artifacts produced by deamidation, cross-linking or other processes. The fact that amino acid sequences determined for HRP A2 and E5 show only 54 and 70% identity, respectively, to HRP C, is one piece of evidence for the existence of distinct peroxidase isoenzymes in horseradish root (Welinder, 1992b). Four genomic DNA clones encoding neutral and neutral-basic HRP isoenzymes have been isolated and characterised from cultured cells of Armoracia rusticana (Fujiyama et al., 1988, 1990). Two of these peroxidase genes (prxc1a and prxc1b) are expressed both in stems and roots and two mainly in roots (prxc2 and prxc3). All consist of four exons and three introns and have identical splice site positions, a feature common to other plant peroxidase genes. Three highly homologous cdnas (prxc1a, prxc1b and prxc1c) have also been isolated using an oligonucleotide probe corresponding to the primary sequence region from His40 to Ala51 of HRP C (Fujiyama et al., 1988). The amino acid sequence deduced from prxc1a is identical to that of HRP C but also contains a hydrophobic leader sequence as well as a C-terminal extension which may be responsible for vacuolar targeting. Expression of the prxc2 gene is induced by wounding and its transcriptional regulation has been investigated in detail (Kaothien et al., 2000). One further cdna has been cloned and sequenced from horseradish corresponding to a The sequencing of the genome of the model plant, Arabidopsis thaliana, and the availability of expressed sequence tag (EST) databases of cdna clones has provided the opportunity to investigate an entire family of class III peroxidases from one plant for the first time, with striking results. According to recent studies there are 73 full-length genes for class III plant peroxidases in the Arabidopsis genome (Tognolli et al., 2002; Welinder et al., 2002). Of these, 71 are predicted to encode for stable enzymes folded similarly to HRP C (Welinder et al., 2002). Amino acid sequence identities among the predicted peroxidases range from 28 to 94%. Alignment of these sequences has been used as a basis for comparisons with other plant peroxidases and the identification of potentially similar enzymes. For example, expression of an acidic peroxidase isoenzyme from A. thaliana (AtPA2) with 95% amino acid sequence identity to HRP A2 (a horseradish peroxidase isoenzyme of unknown function) was found to coincide with lignification (Østergaard et al., 2000). The three-dimensional structure solved for recombinant AtPA2 by X- ray crystallography reveals a substrate binding site that can accommodate monolignols such as p-coumaryl and coniferyl alcohols (Nielsen et al., 2001). This structure is an excellent model for HRP A2, which has not yielded crystals suitable for crystallographic analysis. Both enzymes will catalyse the oxidation of monolignols and phenolic acids although small differences in their reactivity can be detected. These may have their origin in the levels of glycosylation of the two enzymes; recombinant AtPA2 (as expressed in E. coli) is non-glycosylated, whereas plant HRP A2 is highly glycosylated with one bulky glycan attached close to residues at the entrance of the substrate binding site. The presence of similar peroxidase isoenzymes in taxa from two genera of the same plant family may not seem surprising, but analysis of Arabidopsis peroxidases indicates that enzymes from species in quite unrelated plant families may also share high amino acid sequence identity, for example, Arabidopsis peroxidase AtP1 (Cruciferae) and a peroxidase from Gossypium hirsutum (Malvaceae). Duroux and Welinder (2003) have used the peroxidase gene family of A. thaliana as a basis for a comprehensive survey of evolutionary relationships among class III plant peroxidases from angiosperms, gymnosperms, ferns, mosses and liverworts. They suggest that the class III plant peroxidase gene family appeared when plants colonised the land and that adaptive advantages were conferred through peroxidase functionality (e.g. in cell-wall metabolism and defence). Phylogenetic analysis confirms that the class III peroxidases of A.
8 256 N.C. Veitch / Phytochemistry 65 (2004) thaliana represent a highly diverse gene family. Peroxidases from eudicots cluster with the different groups present in Arabidopsis, indicating that peroxidase diversity was extant before the radiation of eudicots. In contrast, peroxidases from monocots cluster in specific groups that have not been found in eudicots. It appears therefore that some groups of peroxidases evolved independently in monocots and eudicots or that some were lost. What is remarkable is the inference that each plant species contains a relatively large number of distinctive peroxidase enzymes. The challenge for the future lies in the determination of their specific functions. 6. Protein engineering and directed evolution of HRP C Several expression systems for the production of recombinant HRP C have been developed since the early 1990s and are compared in a recent review (Veitch and Smith, 2001). Active recombinant HRP C in yields of 5 10 mg/l can be produced in a baculovirus system although the enzyme is highly glycosylated. Yields are lower (typically 2 4 mg/l) when Escherichia coli is used as the expression system because the enzyme must be recovered from inclusion bodies. Essential steps in this process are the solubilization of the inclusion bodies, controlled reoxidation of the reduced, denatured enzyme and in vitro refolding in the presence of heme and calcium (Smith et al., 1990). One advantage of E. coli expression is that active recombinant enzyme is obtained in non-glycosylated form, a factor crucial to the success of subsequent crystallisation studies and the solution of the three-dimensional structure of the enzyme. Site-directed mutants of the enzyme have been generated in both baculovirus and E. coli expression systems. Areas of interest addressed in these studies include the role of distal heme pocket residues in peroxide binding and catalysis, the identity of structural determinants of substrate oxidation, characterisation of small molecule and substrate binding sites, the significance of calcium binding sites and the rational design of artificial binding sites. A comprehensive survey of work published on site-directed mutants of HRP C up to the end of 1999 is available (Veitch and Smith, 2001) and some results are summarised in Table 1. Directed evolution techniques have been applied to HRP C more recently in an attempt to improve qualities of the enzyme that are important for biotechnological and diagnostic applications, such as thermal stability and resistance to inactivation by peroxides. In these studies the host chosen for expression of HRP C was Saccharomyces cerevisiae rather than E. coli, as active enzyme can be produced without the need for in vitro refolding. However, expression levels in S. cerevisiae are low (approximately 50 mg/l) and the secreted enzyme is hyperglycosylated. A 40-fold increase in the HRP C activity of S. cerevisiae culture supernatant was achieved in three rounds of directed evolution using random point mutagenesis and screening (Morawski et al., 2000). The majority of the mutations were to amino acid residues in either loop or surface regions of the enzyme. Although activity was improved, thermal stability showed a small decrease with respect to wildtype enzyme. However, the combination of an HRP C mutant evolved for higher activity with a single site mutation at Asn175 (N175S) gave significant improvements in thermal stability (Morawski et al., 2001). Expression of this mutant in Pichia pastoris (giving sufficient amounts of the enzyme for purification and characterisation) yielded recombinant enzyme showing a 3-fold improvement in thermal stability at 60 C and ph 7.0 and increased resistance to inactivation by hydrogen peroxide and other additives. This recombinant enzyme ( HRP13A7-N175S ) comprised one synonymous mutation (A85) and three amino acid substitutions (N175S, N212D and Q223L). In contrast, directed evolution of a fungal peroxidase from Coprinus cinereus yielded site-directed mutants with up to 190- fold increases in thermal stability and 100 times greater oxidative stability than the wild-type enzyme (Cherry et al., 1999). These dramatic improvements must be considered in the light of the greater thermal and oxidative stability already possessed by wild-type HRP C as compared to Coprinus peroxidase and the much larger number of mutants screened for the latter. 7. Horseradish peroxidase and indole-3-acetic acid 7.1. Mechanisms of oxidation One of the most interesting reactions of HRP C occurs with the plant hormone, indole-3-acetic acid (IAA). In contrast to most peroxidase catalysed reactions, this takes place without added hydrogen peroxide, hence the use of the term indole acetic acid oxidase to describe this activity of HRP C in the older literature. More recent studies of the reaction at neutral ph indicate that it is not an oxidase mechanism that operates, but rather a peroxidase mechanism coupled to a very efficient branched-chain process in which organic peroxide is formed (Dunford, 1999). The reaction is initiated when a trace of the indole-3-acetate cation radical is produced. Major products of indole-3-acetic acid oxidation include indole-3-methanol, indole-3- aldehyde and 3-methylene-2-oxindole, the latter most probably as a result of the non-enzymatic conversion of indole-3-methylhydroperoxide. Conflicting theories have been proposed to explain the mechanism of reaction at lower ph (reviewed by Dunford, 1999), in which
9 N.C. Veitch / Phytochemistry 65 (2004) the formation of the ferrous enzyme, compound III and hydroperoxyl radicals must also be accounted for. The physiological significance of IAA metabolism by HRP C and other plant peroxidases is still an area of active debate. For example, studies of the expression of an anionic peroxidase in transgenic tobacco plants indicate that while overproduction of the enzyme favours defensive strategies (such as resistance to disease, physical damage and insect attack) it has a negative impact on growth because of increased IAA degradation activity (Lagrimini, 1996). Thus peroxidase expression in plant tissues at different stages of development must reflect a balance between the priorities of defence and growth Targeted cancer therapy Use of indole-3-acetic acid and HRP C in combination may offer new potential for targeted cancer therapy, according to recent work by Wardman and colleagues (Folkes and Wardman, 2001; Greco et al., 2001; Wardman, 2002). These authors observed that IAA is cytotoxic towards mammalian cells, including human tumour cells, in the presence of HRP C. The primary mechanism of toxicity is thought to involve 3- methylene-2-oxindole, a known product of the reaction between HRP C and IAA that shows high reactivity towards cellular nucleophiles such as glutathione and the thiol groups of proteins or histone (Fig. 6). However, the fact that 2-methylindole-3-acetic acid shows greater cytotoxicity in combination with HRP C than IAA suggests that there may be other mechanisms of toxicity which do not involve oxindole intermediates (Wardman, 2002). Many other substituted indole-3- acetic acid derivatives have been tested for cytotoxicity in combination with HRP C in an attempt to place relationships between structure and activity on a predictive level. No simple correlation was found between levels of cytotoxicity of indole derivatives and their reactivity towards compound I; for example 5-fluoroindole-3-acetic acid is more cytotoxic towards tumour cells than IAA but less effective as a reductant of compound I (Folkes et al., 2002). Other factors such as the pk a of the indolyl radical cation and rates of decarboxylation and radical fragmentation may also be significant. One of the most cytotoxic indoles identified from in vitro screens is 6-chloroindole-3-acetic acid, a derivative with potential as a prodrug for targeted cancer therapies mediated by HRP C (Rossiter et al., 2002). The challenge now is to develop strategies to evaluate and implement this promising system in vivo. Indeed the combination of HRP C and indole-3-acetic acid or its derivatives offers several advantages for future antibody-, gene- or polymer-directed enzyme:prodrug therapies (Folkes and Wardman, 2001; Wardman, 2002). Among the favourable properties of HRP C are its good stability at 37 C, high activity at neutral ph, lack of toxicity and the ease with which it can be conjugated to antibodies and polymers. Furthermore, evidence available at present suggests that IAA does not show any adverse side-effects in humans. The fact that peroxide is not required as a cosubstrate for the reaction with HRP C is also a significant advantage. 8. Applications overview Horseradish peroxidase (predominantly HRP C) is used as a reagent for organic synthesis and biotransformation as well as in coupled enzyme assays, chemiluminescent assays, immunoassays and the treatment of waste waters (for reviews see Ryan et al., 1994; Veitch and Smith, 2001; Krieg and Halbhuber, 2003). Improvements to desirable qualities of the enzyme such as its relatively good stability in aqueous and nonaqueous solvent systems are actively sought as an outcome of chemical modification, site-directed mutagenesis and directed evolution studies (O Fa ga in, 2003). Some applications of HRP C in small-scale organic synthesis include N- and O-dealkylation, oxidative coupling, selective hydroxylation and oxygen-transfer reactions. Site-directed mutagenesis at Phe41 and His42 of HRP C has been used to improve the enantioselectivity of arylmethylsulfide oxidations (Newmyer and Ortiz de Montellano, 1995; Ozaki and Ortiz de Montellano, 1995). However, Van de Velde et al. (2001) make the general point that scale-up of peroxidase- Fig. 6. A mechanism proposed for the formation of 3-methylene-2-oxindole from horseradish peroxidase (HRP C) and indole-3-acetic acid (after Folkes et al., 2002). R represents a cellular nucleophile (e.g. sulphydryl groups of enzymes or histone).
10 258 N.C. Veitch / Phytochemistry 65 (2004) catalysed enantioselective oxidations to industrial level will require a substantial reduction in the price of enzyme per unit product. Solutions to this problem may include better process management of hydrogen peroxide to avoid enzyme inactivation and use of engineered enzymes with improved stability and catalytic efficiency. Several examples of the use of HRP C in natural product synthesis or biotransformation have appeared in the literature. For example, peroxidase-catalysed oxidative coupling of methyl-(e)-sinapate with the syringyl lignin-model compound, 1-(4-hydroxy-3,5- dimethoxyphenyl)ethanol yielded a novel spirocyclohexadienone together with a dimerization side-product (Seta la et al., 1999). Coupling of catharanthine and vindoline to yield a-3 0,4 0 -anhydrovinblastine is a reaction catalysed by HRP C of potential interest as a semisynthetic step in the production of the anti-cancer drugs vinblastine and vincristine from Catharanthus roseus (Sottomayor et al., 1997). An enzyme with a- 3 0,4 0 -anhydrovinblastine synthase activity that shows many features characteristic of a plant peroxidase has now been purified from C. roseus leaves (Sottomayor et al., 1998). 9. Bibliography of horseradish peroxidase Few plant enzymes are represented so widely in the scientific and patent literature as horseradish peroxidase. This has limited the number of primary sources that can be cited in the present report and a bias towards more recent papers has been adopted. Review articles with more extensive lists of references are quoted where appropriate. Two important monographs on peroxidase enzymes have been published by Dunford (1999) and Saunders, Holmes-Siedle and Stark (1964). The latter contains approximately 1500 references covering the period from 1810 to Two volumes of reviews and literature compilations on peroxidase covering the decades and were published by the University of Geneva and contain more than 5000 references (Gaspar et al., 1982, 1992). Other general reviews on peroxidase are listed in Veitch and Smith (2001). Two review articles that deal specifically with horseradish peroxidase are available in the literature (Dunford, 1991; Veitch and Smith, 2001). Acknowledgements The author thanks Andrew Smith, Karen Gjesing Welinder and Claude Penel for their generous assistance during the preparation of this review. References Bach, A., Chodat, R., Untersuchungen u ber die Rolle der Peroxyde in der Chemie der lebenden Zelle. IV. Ueber Peroxydase. Ber. Deutsch. Chem. Gesell. 36, Bartonek-Roxå, E., Holm, C., Production of catalytically active horseradish peroxidase-n in the insect cell-baculovirus expression system. Biotech. Techniques 13, Berglund, G.I., Carlsson, G.H., Smith, A.T., Szo ke, H., Henriksen, A., Hajdu, J., The catalytic pathway of horseradish peroxidase at high resolution. Nature 417, Cherry, J.R., Lamsa, M.H., Schneider, P., Vind, J., Svendsen, A., Jones, A., Pedersen, A.H., Directed evolution of a fungal peroxidase. Nature Biotechnol. 17, Dunford, H.B., Horseradish peroxidase: Structure and kinetic properties. In: Everse, J., Everse, K.E., Grisham, M.B. (Eds.), Peroxidases in chemistry and biology, Vol. 2. CRC Press, Boca Raton, Florida, pp Dunford, H.B., Heme peroxidases. Wiley VCH, New York. Duroux, L., Welinder, K.G., The peroxidase gene family in plants: a phylogenetic overview. J. Mol. Evol. 57, Filizola, M., Loew, G.H., Role of protein environment in horseradish peroxidase compound I formation: molecular dynamics simulations of horseradish peroxidase HOOH complex. J. Am. Chem. Soc. 122, Folkes, L.K., Wardman, P., Oxidative activation of indole-3- acetic acids to cytotoxic species-a potential new role for plant auxins in cancer therapy. Biochem. Pharmacol. 61, Folkes, L.K., Greco, O., Dachs, G.U., Stratford, M.R.L., Wardman, P., Fluoroindole-3-acetic acid: a prodrug activated by a peroxidase with potential for use in targeted cancer therapy. Biochem. Pharmacol. 63, Fujiyama, K., Takemura, H., Shibayama, S., Kobayashi, K., Choi, J.-K., Shinmyo, A., Takano, M., Yamada, Y., Okada, H., Structure of the horseradish peroxidase isozyme C genes. Eur. J. Biochem. 173, Fujiyama, K., Takemura, H., Shinmyo, A., Okada, H., Takano, M., Genomic DNA structure of two new horseradish-peroxidaseencoding genes. Gene 89, Gajhede, M., Schuller, D.J., Henriksen, A., Smith, A.T., Poulos, T.L., Crystal structure of horseradish peroxidase C at 2.15 angstrom resolution. Nature Struct. Biol. 4, Gaspar, T., Penel, C., Thorpe, T., Greppin, H., Peroxidases A Survey of their Biochemical and Physiological Roles in Higher Plants. University of Geneva, Geneva. Gaspar, T., Penel, C., Greppin, H., Plant Peroxidases Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. University of Geneva, Geneva. Greco, O., Rossiter, S., Kanthou, C., Folkes, L.K., Wardman, P., Tozer, G.M., Dachs, G.U., Horseradish peroxidase-mediated gene therapy: choice of prodrugs in oxic and anoxic tumor conditions. Mol. Cancer Therapeutics 1, Haschke, R.H., Friedhoff, J.M., Calcium-related properties of horseradish peroxidase. Biochem. Biophys. Res. Commun. 80, Henriksen, A., Smith, A.T., Gajhede, M., The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates. J. Biol. Chem. 274, Howes, B.D., Feis, A., Raimondi, L., Indiani, C., Smulevich, G., The critical role of the proximal calcium ion in the structural properties of horseradish peroxidase. J. Biol. Chem. 276, Kaothien, P., Shimokawatoko, Y., Kawaoka, A., Yoshida, K., Shinmyo, A., A cis-element containing PAL-box functions in the expression of the wound-inducible peroxidase gene of horseradish. Plant Cell Rep. 19,
11 N.C. Veitch / Phytochemistry 65 (2004) Kawaoka, A., Kawamoto, T., Ohta, H., Sekine, M., Takano, M., Shinmyo, A., Wound-induced expression of horseradish peroxidase. Plant Cell Rep. 13, Keilin, D., The History of Cell Respiration and Cytochrome. Cambridge University Press, Cambridge. Kratochvil, J.F., Burris, R.H., Seikel, M.K., Harkin, J.M., Isolation and characterization of a-guaiaconic acid and the nature of guaiacum blue. Phytochemistry 10, Krieg, R., Halbhuber, K.-J., Recent advances in catalytic peroxidase histochemistry. Cellular Mol. Biol. 49, Lagrimini, L.M., The role of the tobacco anionic peroxidase in growth and development. In: Obinger, C., Burner, U., Ebermann, R., Penel, C., Greppin, H. (Eds.), Plant Peroxidases, Biochemistry and Physiology. University of Geneva, Geneva, pp Mittler, R., Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, Morawski, B., Lin, Z., Cirino, P., Joo, H., Bandara, G., Arnold, F.H., Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Protein Eng. 13, Morawski, B., Quan, S., Arnold, F.H., Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae. Biotechnol. Bioeng. 76, Newmyer, S.L., Ortiz de Montellano, P.R., Horseradish peroxidase His-42!Ala, His-42!Val, and Phe-41!Ala mutants. Histidine catalysis and control of substrate access to the heme iron. J. Biol. Chem. 270, Nielsen, K.L., Indiani, C., Henriksen, A., Feis, A., Becucci, M., Gajhede, M., Smulevich, G., Welinder, K.G., Differential activity and structure of highly similar peroxidases. Spectroscopic, crystallographic, and enzymatic analyses of lignifying Arabidopsis thaliana peroxidase A2 and horseradish peroxidase A2. Biochemistry 40, Ó Fágáin, C., Enzyme stabilization recent experimental progress. Enzyme Microb. Technol. 33, Ortiz de Montellano, P.R., Catalytic sites of hemoprotein peroxidases. Annu. Rev. Pharmacol. Toxicol. 32, Østergaard, L., Teilum, K., Mirza, O., Mattson, O., Petersen, M., Welinder, K.G., Mundy, J., Gajhede, M., Henriksen, A., Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 44, Ozaki, S., Ortiz de Montellano, P.R., Molecular engineering of horseradish peroxidase; thioether sulfoxidation and styrene epoxidation by Phe-41 leucine and threonine mutants. J. Am. Chem. Soc. 117, Paul, K.G., Peroxidases: historical background. In: Greppin, H., Penel, C., Gaspar, T. (Eds.), Molecular and Physiological Aspects of Plant Peroxidases. University of Geneva, Geneva, pp Planche, L.A., Note sur la sophistication de la résine de jalap et sur les moyens de la reconnaître, etc. Bull. Pharmacie 2, Pnueli, L., Liang, H., Rozenberg, M., Mittler, R., Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)- deficient Arabidopsis plants. Plant J. 34, Rodriguez-Lopez, J.N., Herna ndez-ruiz, J., Garcia-Cánovas, F., Thorneley, R.N.F., Acosta, M., Arnao, M.B., The inactivation and catalytic pathways of horseradish peroxidase with m- chloroperoxybenzoic acid. J. Biol. Chem. 272, Rossiter, S., Folkes, L.K., Wardman, P., Halogenated indole-3- acetic acids as oxidatively activated prodrugs with potential for targeted cancer therapy. Bioorg. Med. Chem. Lett. 12, Ryan, O., Smyth, M.R., Ó Fa gáin, C., Horseradish peroxidase: the analyst s friend. In: Ballou, D.K. (Ed.), Essays in Biochemistry, Vol. 28. Portland Press, London, pp Seta lä, H., Pajunen, A., Rummakko, P., Sipilä, J., Brunow, G., A novel type of spiro compound formed by oxidative cross coupling of methyl sinapate with a syringyl lignin model compound. A model system for the b-1 pathway in lignin biosynthesis. J. Chem. Soc. Perkin Trans. 1, Saunders, B.C., Holmes-Siedle, A.G., Stark, B.P., Peroxidase: The Properties and Uses of a Versatile Enzyme and Some Related Catalysts. Butterworths, London. Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., Yoshimura, K., Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Biol. 53, Smith, A.T., Veitch, N.C., Substrate binding and catalysis in heme peroxidases. Curr. Opin. Chem. Biol. 2, Smith, A.T., Santama, N., Dacey, S., Edwards, M., Bray, R.C., Thorneley, R.N.F., Burke, J.F., Expression of a synthetic gene for horseradish peroxidase C in Escherichia coli and folding and activation of the recombinant enzyme with Ca 2+ and heme. J. Biol. Chem. 265, Sottomayor, M., DiCosmo, F., Ros Barcelo, A.R., On the fate of catharanthine and vindoline during the peroxidase-mediated enzymatic synthesis of a-3 0,4 0 -anhydrovinblastine. Enzyme Microb. Technol. 21, Sottomayor, M., Lo pez-serrano, M., DiCosmo, F., Ros Barcelo, A.R., Purification and characterization of a-3 0,4 0 -anhydrovinblastine synthase (peroxidase-like) from Catharanthus roseus (L.). G. Don. FEBS Lett. 428, Tognolli, M., Penel, C., Greppin, H., Simon, P., Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288, Van de Velde, F., Van Rantwijk, F., Sheldon, R.A., Improving the catalytic performance of peroxidases in organic synthesis. Trends Biotechnol. 19, Veitch, N.C., Smith, A.T., Horseradish peroxidase. Adv. Inorg. Chem. 51, Wardman, P., Indole-3-acetic acids and horseradish peroxidase: A new prodrug/enzyme combination for targeted cancer therapy. Current Pharmaceutical Design 8, Welinder, K.G., Covalent structure of the glycoprotein horseradish peroxidase. FEBS Lett. 72, Welinder, K.G., 1992a. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 2, Welinder, K.G., 1992b. Plant peroxidases: structure-function relationships. In: Penel, C., Gaspar, T., Greppin, H. (Eds.), Plant Peroxidases Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. University of Geneva, Geneva, pp Welinder, K.G., Gajhede, M., Structure and evolution of peroxidases. In: Welinder, K.G., Rasmussen, S.K., Penel, C., Greppin, H. (Eds.), Plant Peroxidases: Biochemistry and Physiology. University of Geneva, Geneva, pp Welinder, K.G., Justesen, A.F., Kjærsgård, I.V.H., Jensen, R.B., Rasmussen, S.K., Jespersen, H.M., Duroux, L., Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 269, Yang, B.Y., Gray, J.S.S., Montgomery, R., The glycans of horseradish peroxidase. Carbohydrate Res. 287,
Myoglobin and Hemoglobin
 Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon. V. Polypeptides and Proteins
 IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK
 Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Peptide bonds: resonance structure. Properties of proteins: Peptide bonds and side chains. Dihedral angles. Peptide bond. Protein physics, Lecture 5
 Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Recap. Lecture 2. Protein conformation. Proteins. 8 types of protein function 10/21/10. Proteins.. > 50% dry weight of a cell
 Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
18.2 Protein Structure and Function: An Overview
 18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
Oxygen-Binding Proteins
 Oxygen-Binding Proteins Myoglobin, Hemoglobin, Cytochromes bind O 2. Oxygen is transported from lungs to various tissues via blood in association with hemoglobin In muscle, hemoglobin gives up O 2 to myoglobin
Oxygen-Binding Proteins Myoglobin, Hemoglobin, Cytochromes bind O 2. Oxygen is transported from lungs to various tissues via blood in association with hemoglobin In muscle, hemoglobin gives up O 2 to myoglobin
http://faculty.sau.edu.sa/h.alshehri
 http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys
 Questions- Proteins & Enzymes A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys Reaction of the intact peptide
Questions- Proteins & Enzymes A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys Reaction of the intact peptide
Biochemistry 462a Hemoglobin Structure and Function Reading - Chapter 7 Practice problems - Chapter 7: 1-6; Proteins extra problems
 Biochemistry 462a Hemoglobin Structure and Function Reading - Chapter 7 Practice problems - Chapter 7: 1-6; Proteins extra problems Myoglobin and Hemoglobin Oxygen is required for oxidative metabolism
Biochemistry 462a Hemoglobin Structure and Function Reading - Chapter 7 Practice problems - Chapter 7: 1-6; Proteins extra problems Myoglobin and Hemoglobin Oxygen is required for oxidative metabolism
Lecture 15: Enzymes & Kinetics Mechanisms
 ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
Shu-Ping Lin, Ph.D. E-mail: splin@dragon.nchu.edu.tw
 Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute te of Biomedical Engineering ing E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ edu tw/pweb/users/splin/ Date: 10.13.2010
Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute te of Biomedical Engineering ing E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ edu tw/pweb/users/splin/ Date: 10.13.2010
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Previous lecture: Today:
 Previous lecture: The energy requiring step from substrate to transition state is an energy barrier called the free energy of activation G Transition state is the unstable (10-13 seconds) highest energy
Previous lecture: The energy requiring step from substrate to transition state is an energy barrier called the free energy of activation G Transition state is the unstable (10-13 seconds) highest energy
PHYSIOLOGY AND MAINTENANCE Vol. II - On The Determination of Enzyme Structure, Function, and Mechanism - Glumoff T.
 ON THE DETERMINATION OF ENZYME STRUCTURE, FUNCTION, AND MECHANISM University of Oulu, Finland Keywords: enzymes, protein structure, X-ray crystallography, bioinformatics Contents 1. Introduction 2. Structure
ON THE DETERMINATION OF ENZYME STRUCTURE, FUNCTION, AND MECHANISM University of Oulu, Finland Keywords: enzymes, protein structure, X-ray crystallography, bioinformatics Contents 1. Introduction 2. Structure
Part A: Amino Acids and Peptides (Is the peptide IAG the same as the peptide GAI?)
 ChemActivity 46 Amino Acids, Polypeptides and Proteins 1 ChemActivity 46 Part A: Amino Acids and Peptides (Is the peptide IAG the same as the peptide GAI?) Model 1: The 20 Amino Acids at Biological p See
ChemActivity 46 Amino Acids, Polypeptides and Proteins 1 ChemActivity 46 Part A: Amino Acids and Peptides (Is the peptide IAG the same as the peptide GAI?) Model 1: The 20 Amino Acids at Biological p See
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Protein Physics. A. V. Finkelstein & O. B. Ptitsyn LECTURE 1
 Protein Physics A. V. Finkelstein & O. B. Ptitsyn LECTURE 1 PROTEINS Functions in a Cell MOLECULAR MACHINES BUILDING BLOCKS of a CELL ARMS of a CELL ENZYMES - enzymatic catalysis of biochemical reactions
Protein Physics A. V. Finkelstein & O. B. Ptitsyn LECTURE 1 PROTEINS Functions in a Cell MOLECULAR MACHINES BUILDING BLOCKS of a CELL ARMS of a CELL ENZYMES - enzymatic catalysis of biochemical reactions
Chapter 26 Biomolecules: Amino Acids, Peptides, and Proteins
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 26 Biomolecules: Amino Acids, Peptides, and Proteins Proteins Amides from Amino Acids Amino acids contain a basic amino group and an acidic carboxyl
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 26 Biomolecules: Amino Acids, Peptides, and Proteins Proteins Amides from Amino Acids Amino acids contain a basic amino group and an acidic carboxyl
Amino Acids, Peptides, Proteins
 Amino Acids, Peptides, Proteins Functions of proteins: Enzymes Transport and Storage Motion, muscle contraction Hormones Mechanical support Immune protection (Antibodies) Generate and transmit nerve impulses
Amino Acids, Peptides, Proteins Functions of proteins: Enzymes Transport and Storage Motion, muscle contraction Hormones Mechanical support Immune protection (Antibodies) Generate and transmit nerve impulses
Pipe Cleaner Proteins. Essential question: How does the structure of proteins relate to their function in the cell?
 Pipe Cleaner Proteins GPS: SB1 Students will analyze the nature of the relationships between structures and functions in living cells. Essential question: How does the structure of proteins relate to their
Pipe Cleaner Proteins GPS: SB1 Students will analyze the nature of the relationships between structures and functions in living cells. Essential question: How does the structure of proteins relate to their
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Biochemistry - I. Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture-11 Enzyme Mechanisms II
 Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture-11 Enzyme Mechanisms II In the last class we studied the enzyme mechanisms of ribonuclease A
Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture-11 Enzyme Mechanisms II In the last class we studied the enzyme mechanisms of ribonuclease A
The Organic Chemistry of Amino Acids, Peptides, and Proteins
 Essential rganic Chemistry Chapter 16 The rganic Chemistry of Amino Acids, Peptides, and Proteins Amino Acids a-amino carboxylic acids. The building blocks from which proteins are made. H 2 N C 2 H Note:
Essential rganic Chemistry Chapter 16 The rganic Chemistry of Amino Acids, Peptides, and Proteins Amino Acids a-amino carboxylic acids. The building blocks from which proteins are made. H 2 N C 2 H Note:
The peptide bond is rigid and planar
 Level Description Bonds Primary Sequence of amino acids in proteins Covalent (peptide bonds) Secondary Structural motifs in proteins: α- helix and β-sheet Hydrogen bonds (between NH and CO groups in backbone)
Level Description Bonds Primary Sequence of amino acids in proteins Covalent (peptide bonds) Secondary Structural motifs in proteins: α- helix and β-sheet Hydrogen bonds (between NH and CO groups in backbone)
H H N - C - C 2 R. Three possible forms (not counting R group) depending on ph
 Amino acids - 0 common amino acids there are others found naturally but much less frequently - Common structure for amino acid - C, -N, and functional groups all attached to the alpha carbon N - C - C
Amino acids - 0 common amino acids there are others found naturally but much less frequently - Common structure for amino acid - C, -N, and functional groups all attached to the alpha carbon N - C - C
Amino Acids as Acids, Bases and Buffers:
 Amino Acids as Acids, Bases and Buffers: - Amino acids are weak acids - All have at least 2 titratable protons (shown below as fully protonated species) and therefore have 2 pka s o α-carboxyl (-COOH)
Amino Acids as Acids, Bases and Buffers: - Amino acids are weak acids - All have at least 2 titratable protons (shown below as fully protonated species) and therefore have 2 pka s o α-carboxyl (-COOH)
Ionization of amino acids
 Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
CNAS ASSESSMENT COMMITTEE CHEMISTRY (CH) DEGREE PROGRAM CURRICULAR MAPPINGS AND COURSE EXPECTED STUDENT LEARNING OUTCOMES (SLOs)
 CNAS ASSESSMENT COMMITTEE CHEMISTRY (CH) DEGREE PROGRAM CURRICULAR MAPPINGS AND COURSE EXPECTED STUDENT LEARNING OUTCOMES (SLOs) DEGREE PROGRAM CURRICULAR MAPPING DEFINED PROGRAM SLOs Course No. 11 12
CNAS ASSESSMENT COMMITTEE CHEMISTRY (CH) DEGREE PROGRAM CURRICULAR MAPPINGS AND COURSE EXPECTED STUDENT LEARNING OUTCOMES (SLOs) DEGREE PROGRAM CURRICULAR MAPPING DEFINED PROGRAM SLOs Course No. 11 12
(c) How would your answers to problem (a) change if the molecular weight of the protein was 100,000 Dalton?
 Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Lecture Overview. Hydrogen Bonds. Special Properties of Water Molecules. Universal Solvent. ph Scale Illustrated. special properties of water
 Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Chapter 12 - Proteins
 Roles of Biomolecules Carbohydrates Lipids Proteins 1) Catalytic 2) Transport 3) Regulatory 4) Structural 5) Contractile 6) Protective 7) Storage Nucleic Acids 12.1 -Amino Acids Chapter 12 - Proteins Amino
Roles of Biomolecules Carbohydrates Lipids Proteins 1) Catalytic 2) Transport 3) Regulatory 4) Structural 5) Contractile 6) Protective 7) Storage Nucleic Acids 12.1 -Amino Acids Chapter 12 - Proteins Amino
1. The diagram below represents a biological process
 1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
8/20/2012 H C OH H R. Proteins
 Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Invariant residue-a residue that is always conserved. It is assumed that these residues are essential to the structure or function of the protein.
 Chapter 6 The amino acid side chains have polar and nonpolar properties, and the relative hydrophobicity of the amino acid side chains is critical for the folding and stability of a protein. The more hydrophobic
Chapter 6 The amino acid side chains have polar and nonpolar properties, and the relative hydrophobicity of the amino acid side chains is critical for the folding and stability of a protein. The more hydrophobic
Expression and Purification of Recombinant Protein in bacteria and Yeast. Presented By: Puspa pandey, Mohit sachdeva & Ming yu
 Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Chapter 3. Protein Structure and Function
 Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Copyright 2000-2003 Mark Brandt, Ph.D. 54
 Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Helices From Readily in Biological Structures
 The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
ENZYMES. Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin. Principle of Enzyme Catalysis
 ENZYMES Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin Principle of Enzyme Catalysis Linus Pauling (1946) formulated the first basic principle of enzyme catalysis Enzyme increase the rate
ENZYMES Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin Principle of Enzyme Catalysis Linus Pauling (1946) formulated the first basic principle of enzyme catalysis Enzyme increase the rate
Disaccharides consist of two monosaccharide monomers covalently linked by a glycosidic bond. They function in sugar transport.
 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
Chapter 3 Molecules of Cells
 Bio 100 Molecules of cells 1 Chapter 3 Molecules of Cells Compounds containing carbon are called organic compounds Molecules such as methane that are only composed of carbon and hydrogen are called hydrocarbons
Bio 100 Molecules of cells 1 Chapter 3 Molecules of Cells Compounds containing carbon are called organic compounds Molecules such as methane that are only composed of carbon and hydrogen are called hydrocarbons
Chapter 9 Mitochondrial Structure and Function
 Chapter 9 Mitochondrial Structure and Function 1 2 3 Structure and function Oxidative phosphorylation and ATP Synthesis Peroxisome Overview 2 Mitochondria have characteristic morphologies despite variable
Chapter 9 Mitochondrial Structure and Function 1 2 3 Structure and function Oxidative phosphorylation and ATP Synthesis Peroxisome Overview 2 Mitochondria have characteristic morphologies despite variable
Methods of Grading S/N Style of grading Percentage Score 1 Attendance, class work and assignment 10 2 Test 20 3 Examination 70 Total 100
 COURSE: MIB 303 Microbial Physiology and Metabolism (3 Units- Compulsory) Course Duration: Three hours per week for 15 weeks (45 hours). Lecturer: Jimoh, S.O. B.Sc., M.Sc, Ph.D Microbiology (ABU, Zaria)
COURSE: MIB 303 Microbial Physiology and Metabolism (3 Units- Compulsory) Course Duration: Three hours per week for 15 weeks (45 hours). Lecturer: Jimoh, S.O. B.Sc., M.Sc, Ph.D Microbiology (ABU, Zaria)
ATOMS AND BONDS. Bonds
 ATOMS AND BONDS Atoms of elements are the simplest units of organization in the natural world. Atoms consist of protons (positive charge), neutrons (neutral charge) and electrons (negative charge). The
ATOMS AND BONDS Atoms of elements are the simplest units of organization in the natural world. Atoms consist of protons (positive charge), neutrons (neutral charge) and electrons (negative charge). The
BOC334 (Proteomics) Practical 1. Calculating the charge of proteins
 BC334 (Proteomics) Practical 1 Calculating the charge of proteins Aliphatic amino acids (VAGLIP) N H 2 H Glycine, Gly, G no charge Hydrophobicity = 0.67 MW 57Da pk a CH = 2.35 pk a NH 2 = 9.6 pi=5.97 CH
BC334 (Proteomics) Practical 1 Calculating the charge of proteins Aliphatic amino acids (VAGLIP) N H 2 H Glycine, Gly, G no charge Hydrophobicity = 0.67 MW 57Da pk a CH = 2.35 pk a NH 2 = 9.6 pi=5.97 CH
博 士 論 文 ( 要 約 ) A study on enzymatic synthesis of. stable cyclized peptides which. inhibit protein-protein interactions
 博 士 論 文 ( 要 約 ) 論 文 題 目 A study on enzymatic synthesis of stable cyclized peptides which inhibit protein-protein interactions ( 蛋 白 質 間 相 互 作 用 を 阻 害 する 安 定 な 環 状 化 ペプチドの 酵 素 合 成 に 関 する 研 究 ) 氏 名 張 静 1
博 士 論 文 ( 要 約 ) 論 文 題 目 A study on enzymatic synthesis of stable cyclized peptides which inhibit protein-protein interactions ( 蛋 白 質 間 相 互 作 用 を 阻 害 する 安 定 な 環 状 化 ペプチドの 酵 素 合 成 に 関 する 研 究 ) 氏 名 張 静 1
Carbohydrates, proteins and lipids
 Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Structure of proteins
 Structure of proteins Primary structure: is amino acids sequence or the covalent structure (50-2500) amino acids M.Wt. of amino acid=110 Dalton (56 110=5610 Dalton). Single chain or more than one polypeptide
Structure of proteins Primary structure: is amino acids sequence or the covalent structure (50-2500) amino acids M.Wt. of amino acid=110 Dalton (56 110=5610 Dalton). Single chain or more than one polypeptide
CSC 2427: Algorithms for Molecular Biology Spring 2006. Lecture 16 March 10
 CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CHAPTER 6 AN INTRODUCTION TO METABOLISM. Section B: Enzymes
 CHAPTER 6 AN INTRODUCTION TO METABOLISM Section B: Enzymes 1. Enzymes speed up metabolic reactions by lowering energy barriers 2. Enzymes are substrate specific 3. The active site in an enzyme s catalytic
CHAPTER 6 AN INTRODUCTION TO METABOLISM Section B: Enzymes 1. Enzymes speed up metabolic reactions by lowering energy barriers 2. Enzymes are substrate specific 3. The active site in an enzyme s catalytic
Reactive oxygen species in leaves and how to catch these
 Reactive oxygen species in leaves and how to catch these Éva Hideg Molecular Stress- & Photobiology Group, BRC Szeged EPPN Summer School 2013 Outline How to prove that ROS are involved? The central role
Reactive oxygen species in leaves and how to catch these Éva Hideg Molecular Stress- & Photobiology Group, BRC Szeged EPPN Summer School 2013 Outline How to prove that ROS are involved? The central role
Disulfide Bonds at the Hair Salon
 Disulfide Bonds at the Hair Salon Three Alpha Helices Stabilized By Disulfide Bonds! In order for hair to grow 6 inches in one year, 9 1/2 turns of α helix must be produced every second!!! In some proteins,
Disulfide Bonds at the Hair Salon Three Alpha Helices Stabilized By Disulfide Bonds! In order for hair to grow 6 inches in one year, 9 1/2 turns of α helix must be produced every second!!! In some proteins,
Your partner in immunology
 Your partner in immunology Expertise Expertise Reactivity Reactivity Quality Quality Advice Advice Who are we? Specialist of antibody engineering Covalab is a French biotechnology company, specialised
Your partner in immunology Expertise Expertise Reactivity Reactivity Quality Quality Advice Advice Who are we? Specialist of antibody engineering Covalab is a French biotechnology company, specialised
A disaccharide is formed when a dehydration reaction joins two monosaccharides. This covalent bond is called a glycosidic linkage.
 CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
Figure 5. Energy of activation with and without an enzyme.
 Biology 20 Laboratory ENZYMES & CELLULAR RESPIRATION OBJECTIVE To be able to list the general characteristics of enzymes. To study the effects of enzymes on the rate of chemical reactions. To demonstrate
Biology 20 Laboratory ENZYMES & CELLULAR RESPIRATION OBJECTIVE To be able to list the general characteristics of enzymes. To study the effects of enzymes on the rate of chemical reactions. To demonstrate
Peptide Bonds: Structure
 Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Table of contents. Bibliografische Informationen http://d-nb.info/1006571213. digitalisiert durch
 1. Research scope: The role of structure rigid'tf'ication in nature and chemistry 1 2. Establishing a Dha=Tap backbone scan in order to elucidate structural properties of the N-terminusofNPY 5 2.1 Introduction.
1. Research scope: The role of structure rigid'tf'ication in nature and chemistry 1 2. Establishing a Dha=Tap backbone scan in order to elucidate structural properties of the N-terminusofNPY 5 2.1 Introduction.
5.111 Principles of Chemical Science
 MIT OpenCourseWare http://ocw.mit.edu 5.111 Principles of Chemical Science Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. 5.111 Principles
MIT OpenCourseWare http://ocw.mit.edu 5.111 Principles of Chemical Science Fall 2008 For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms. 5.111 Principles
KMS-Specialist & Customized Biosimilar Service
 KMS-Specialist & Customized Biosimilar Service 1. Polyclonal Antibody Development Service KMS offering a variety of Polyclonal Antibody Services to fit your research and production needs. we develop polyclonal
KMS-Specialist & Customized Biosimilar Service 1. Polyclonal Antibody Development Service KMS offering a variety of Polyclonal Antibody Services to fit your research and production needs. we develop polyclonal
Chapter 8. Summary and Perspectives
 Chapter 8 Summary and Perspectives 131 Chapter 8 Summary Overexpression of the multidrug resistance protein MRP1 confer multidrug resistance (MDR) to cancer cells. The contents of this thesis describe
Chapter 8 Summary and Perspectives 131 Chapter 8 Summary Overexpression of the multidrug resistance protein MRP1 confer multidrug resistance (MDR) to cancer cells. The contents of this thesis describe
Lecture 13-14 Conformation of proteins Conformation of a protein three-dimensional structure native state. native condition
 Lecture 13-14 Conformation of proteins Conformation of a protein refers to the three-dimensional structure in its native state. There are many different possible conformations for a molecule as large as
Lecture 13-14 Conformation of proteins Conformation of a protein refers to the three-dimensional structure in its native state. There are many different possible conformations for a molecule as large as
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
Control of Gene Expression
 Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
--not necessarily a protein! (all proteins are polypeptides, but the converse is not true)
 00Note Set 5b 1 PEPTIDE BONDS AND POLYPEPTIDES OLIGOPEPTIDE: --chain containing only a few amino acids (see tetrapaptide, Fig 5.9) POLYPEPTIDE CHAINS: --many amino acids joined together --not necessarily
00Note Set 5b 1 PEPTIDE BONDS AND POLYPEPTIDES OLIGOPEPTIDE: --chain containing only a few amino acids (see tetrapaptide, Fig 5.9) POLYPEPTIDE CHAINS: --many amino acids joined together --not necessarily
Enzymes reduce the activation energy
 Enzymes reduce the activation energy Transition state is an unstable transitory combination of reactant molecules which occurs at the potential energy maximum (free energy maximum). Note - the ΔG of the
Enzymes reduce the activation energy Transition state is an unstable transitory combination of reactant molecules which occurs at the potential energy maximum (free energy maximum). Note - the ΔG of the
The Need for a PARP in vivo Pharmacodynamic Assay
 The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
Papers listed: Cell2. This weeks papers. Chapt 4. Protein structure and function
 Papers listed: Cell2 During the semester I will speak of information from several papers. For many of them you will not be required to read these papers, however, you can do so for the fun of it (and it
Papers listed: Cell2 During the semester I will speak of information from several papers. For many of them you will not be required to read these papers, however, you can do so for the fun of it (and it
Recognizing Organic Molecules: Carbohydrates, Lipids and Proteins
 Recognizing Organic Molecules: Carbohydrates, Lipids and Proteins Oct 15 8:05 PM What is an Organic Molecule? An Organic Molecule is a molecule that contains carbon and hydrogen and oxygen Carbon is found
Recognizing Organic Molecules: Carbohydrates, Lipids and Proteins Oct 15 8:05 PM What is an Organic Molecule? An Organic Molecule is a molecule that contains carbon and hydrogen and oxygen Carbon is found
Structure and properties of proteins. Vladimíra Kvasnicová
 Structure and properties of proteins Vladimíra Kvasnicová Chemical nature of proteins biopolymers of amino acids macromolecules (M r > 10 000) Classification of proteins 1) by localization in an organism
Structure and properties of proteins Vladimíra Kvasnicová Chemical nature of proteins biopolymers of amino acids macromolecules (M r > 10 000) Classification of proteins 1) by localization in an organism
AMINO ACIDS & PEPTIDE BONDS STRUCTURE, CLASSIFICATION & METABOLISM
 AMINO ACIDS & PEPTIDE BONDS STRUCTURE, CLASSIFICATION & METABOLISM OBJECTIVES At the end of this session the student should be able to, recognize the structures of the protein amino acid and state their
AMINO ACIDS & PEPTIDE BONDS STRUCTURE, CLASSIFICATION & METABOLISM OBJECTIVES At the end of this session the student should be able to, recognize the structures of the protein amino acid and state their
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7. 4. Which of the following weak acids would make the best buffer at ph = 5.0?
 Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Ann.wellhouse@TouchStoneScience.net 1. Enzyme Function
 Ann.wellhouse@TouchStoneScience.net 1 Enzyme Function National Science Standards Science as Inquiry: Content Standard A: As a result of activities in grades 9-12, all students should develop: Abilities
Ann.wellhouse@TouchStoneScience.net 1 Enzyme Function National Science Standards Science as Inquiry: Content Standard A: As a result of activities in grades 9-12, all students should develop: Abilities
Interaktionen von RNAs und Proteinen
 Sonja Prohaska Computational EvoDevo Universitaet Leipzig June 9, 2015 Studying RNA-protein interactions Given: target protein known to bind to RNA problem: find binding partners and binding sites experimental
Sonja Prohaska Computational EvoDevo Universitaet Leipzig June 9, 2015 Studying RNA-protein interactions Given: target protein known to bind to RNA problem: find binding partners and binding sites experimental
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
Refinement of a pdb-structure and Convert
 Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Biochemistry of Cells
 Biochemistry of Cells 1 Carbon-based Molecules Although a cell is mostly water, the rest of the cell consists mostly of carbon-based molecules Organic chemistry is the study of carbon compounds Carbon
Biochemistry of Cells 1 Carbon-based Molecules Although a cell is mostly water, the rest of the cell consists mostly of carbon-based molecules Organic chemistry is the study of carbon compounds Carbon
Structures of Proteins. Primary structure - amino acid sequence
 Structures of Proteins Primary structure - amino acid sequence Secondary structure chain of covalently linked amino acids folds into regularly repeating structures. Secondary structure is the result of
Structures of Proteins Primary structure - amino acid sequence Secondary structure chain of covalently linked amino acids folds into regularly repeating structures. Secondary structure is the result of
CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA
 CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA INTRODUCTION DNA : DNA is deoxyribose nucleic acid. It is made up of a base consisting of sugar, phosphate and one nitrogen base.the
CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA INTRODUCTION DNA : DNA is deoxyribose nucleic acid. It is made up of a base consisting of sugar, phosphate and one nitrogen base.the
Chapter 16 Amino Acids, Proteins, and Enzymes
 Chapter 16 Amino Acids, Proteins, and Enzymes 1 Functions of Proteins Proteins in the body are polymers made from 20 different amino acids differ in characteristics and functions that depend on the order
Chapter 16 Amino Acids, Proteins, and Enzymes 1 Functions of Proteins Proteins in the body are polymers made from 20 different amino acids differ in characteristics and functions that depend on the order
Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism )
 Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
Metabolism Dr.kareema Amine Al-Khafaji Assistant professor in microbiology, and dermatologist Babylon University, College of Medicine, Department of
 Metabolism Dr.kareema Amine Al-Khafaji Assistant professor in microbiology, and dermatologist Babylon University, College of Medicine, Department of Microbiology. Metabolism sum of all chemical processes
Metabolism Dr.kareema Amine Al-Khafaji Assistant professor in microbiology, and dermatologist Babylon University, College of Medicine, Department of Microbiology. Metabolism sum of all chemical processes
Built from 20 kinds of amino acids
 Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Chemistry 20 Chapters 15 Enzymes
 Chemistry 20 Chapters 15 Enzymes Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An
Chemistry 20 Chapters 15 Enzymes Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An
Proteins the primary biological macromolecules of living organisms
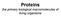 Proteins the primary biological macromolecules of living organisms Protein structure and folding Primary Secondary Tertiary Quaternary structure of proteins Structure of Proteins Protein molecules adopt
Proteins the primary biological macromolecules of living organisms Protein structure and folding Primary Secondary Tertiary Quaternary structure of proteins Structure of Proteins Protein molecules adopt
Peptides: Synthesis and Biological Interest
 Peptides: Synthesis and Biological Interest Therapeutic Agents Therapeutic peptides approved by the FDA (2009-2011) 3 Proteins Biopolymers of α-amino acids. Amino acids are joined by peptide bond. They
Peptides: Synthesis and Biological Interest Therapeutic Agents Therapeutic peptides approved by the FDA (2009-2011) 3 Proteins Biopolymers of α-amino acids. Amino acids are joined by peptide bond. They
Chemical Bonds and Groups - Part 1
 hemical Bonds and Groups - Part 1 ARB SKELETS arbon has a unique role in the cell because of its ability to form strong covalent bonds with other carbon atoms. Thus carbon atoms can join to form chains.
hemical Bonds and Groups - Part 1 ARB SKELETS arbon has a unique role in the cell because of its ability to form strong covalent bonds with other carbon atoms. Thus carbon atoms can join to form chains.
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
Ch18_PT MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Ch18_PT MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) All of the following can be classified as biomolecules except A) lipids. B) proteins. C)
Ch18_PT MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) All of the following can be classified as biomolecules except A) lipids. B) proteins. C)
How To Understand The Chemistry Of Organic Molecules
 CHAPTER 3 THE CHEMISTRY OF ORGANIC MOLECULES 3.1 Organic Molecules The chemistry of carbon accounts for the diversity of organic molecules found in living things. Carbon has six electrons, four of which
CHAPTER 3 THE CHEMISTRY OF ORGANIC MOLECULES 3.1 Organic Molecules The chemistry of carbon accounts for the diversity of organic molecules found in living things. Carbon has six electrons, four of which
The Molecules of Cells
 The Molecules of Cells I. Introduction A. Most of the world s population cannot digest milk-based foods. 1. These people are lactose intolerant because they lack the enzyme lactase. 2. This illustrates
The Molecules of Cells I. Introduction A. Most of the world s population cannot digest milk-based foods. 1. These people are lactose intolerant because they lack the enzyme lactase. 2. This illustrates
Biochemistry. Entrance Requirements. Requirements for Honours Programs. 148 Bishop s University 2015/2016
 148 Bishop s University 2015/2016 Biochemistry The Biochemistry program at Bishop s is coordinated through an interdisciplinary committee of chemists, biochemists and biologists, providing students with
148 Bishop s University 2015/2016 Biochemistry The Biochemistry program at Bishop s is coordinated through an interdisciplinary committee of chemists, biochemists and biologists, providing students with
BIOLOGICAL MOLECULES OF LIFE
 BIOLOGICAL MOLECULES OF LIFE C A R B O H Y D R A T E S, L I P I D S, P R O T E I N S, A N D N U C L E I C A C I D S The Academic Support Center @ Daytona State College (Science 115, Page 1 of 29) Carbon
BIOLOGICAL MOLECULES OF LIFE C A R B O H Y D R A T E S, L I P I D S, P R O T E I N S, A N D N U C L E I C A C I D S The Academic Support Center @ Daytona State College (Science 115, Page 1 of 29) Carbon
How To Make A Drug From A Peptide
 MODERN PERSPECTIVES ON PEPTIDE SYNTHESIS INTRODUCTION WHITEPAPER www.almacgroup.com The complexity of synthetic peptide products, whether as reagents used in research or as therapeutic APIs, is increasing.
MODERN PERSPECTIVES ON PEPTIDE SYNTHESIS INTRODUCTION WHITEPAPER www.almacgroup.com The complexity of synthetic peptide products, whether as reagents used in research or as therapeutic APIs, is increasing.
Patrick, An Introduction to Medicinal Chemistry 4e Chapter 13 Drug design: optimizing target interactions. Pyrrole ring N H
 Patrick, An Introduction to dicinal hemistry 4e hapter 13 Drug design: optimizing target interactions Answers to end-of-chapter questions 1) The pyrrole ring of DU 122290 serves to increase the rigidity
Patrick, An Introduction to dicinal hemistry 4e hapter 13 Drug design: optimizing target interactions Answers to end-of-chapter questions 1) The pyrrole ring of DU 122290 serves to increase the rigidity
MCAT Organic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins
 MCAT rganic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins Question No. 1 of 10 Question 1. Which amino acid does not contain a chiral center? Question #01 (A) Serine (B) Proline (C)
MCAT rganic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins Question No. 1 of 10 Question 1. Which amino acid does not contain a chiral center? Question #01 (A) Serine (B) Proline (C)
green B 1 ) into a single unit to model the substrate in this reaction. enzyme
 Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
