Ubiquitin-conjugated degradation of Golden 2-Like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato
|
|
|
- Anabel Ramsey
- 8 years ago
- Views:
Transcription
1 New Phytologist Supporting Information Ubiquitin-conjugated degradation of Golden 2-Like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato Xiaofeng Tang, Min Miao, Xiangli Niu, Danfeng Zhang, Xulv Cao, Xichen Jin, Yunye Zhu, Youhong Fan, Hongtao Wang, Ying Liu, Yuan Sui, Wenjie Wang, Anquan Wang, Fangming Xiao, Jim Giovannoni and Yongsheng Liu Article acceptance date: 11 August 2015 The following Supporting Information is available for this article: Fig. S1 Prediction of ubiquitination sites in protein. Fig. S2 eal-time quantitative T-PC analysis of mna level in protoplasts prepared from tomato lines wild type (WT), high pigment-1 mutant (hp1), and high pigment-2 mutant (hp2). Fig. S3 eal-time quantitative T-PC analysis of virus-induced gene silencing (VIGS)- silenced N. benthamiana plants. Fig. S4 Subcellular localizations of SlDDB1 and proteins. Table S1 List of primers used in this study Methods S1 eal-time T-PC analysis.
2 Fig. S1 Prediction of ubiquitination sites in protein. Using UbPred ( two lysines K11 and K253 in protein were predicted as the possible ubiquitination sites with medium confidence.
3 Fig. S2 eal-time quantitative T-PC analysis of mna level in protoplasts prepared from tomato lines WT, high pigment-1 mutant (hp1), and high pigment-2 mutant (hp2). eal-time quantitative T-PC was analyzed for mna level in protoplasts derived from tomato lines including wild type (WT), high pigment-1 mutant (hp1), and high pigment-2 mutant (hp2), respectively. -Flag construct was transiently expressed in the prepared tomato protoplasts. MG132 was added at 1h after transformation. Total NAs were isolated at 12h after transformation and used to generate first-strand cdna. The Solanum lycopersicum UBI3 (GenBank accession No. X58253), served as an internal control for normalization. Values are means ± SE of three replicates.
4 Fig. S3 eal-time quantitative T-PC analysis of virus-induced gene silencing (VIGS)- silenced N. benthamiana plants. eal-time quantitative T-PC analysis of NbCUL4, NbDDB1 and NbDET1 mna level in NbCUL4-silenced, NbDDB1-silenced and NbDET1-silenced N. benthamiana plants, respectively. -Flag construct was transiently expressed in the VIGSsilenced plants. Total NAs were isolated after Agrobacterium infiltration and used to generate first-strand cdna. The N. benthamiana EF1α gene (accession No. AY ) served as an internal control for normalization. Values are means ± SE of three replicates.
5 Fig. S4 Subcellular localizations of SlDDB1 and proteins. (a) The PC-amplified GFP fragment, encoding green fluorescent protein, was cloned into Pst I and Sph I sites of ptex to generate ptex::gfp. The coding sequences of SlDDB1 and genes were PC amplified from tomato leaf cdna and cloned in the ptex::gfp vector to generate SlDDB1- GFP and -GFP constructs. Subcellular localization assay was carried out via transformation of tomato protoplasts. After DAPI staining the protoplasts was examined using confocal microscope to simultaneously capture DAPI and GFP signals. ptex::gfp constructs was included as controls. Left to right: green, GFP fluorescence; blue, nucleus stained with DAPI; red, chlorophyll autofluorescence; merged, combined fluorescence from GFP, chlorophyll
6 and DAPI. Bars, 5 μm. Primers used in generation of these constructs are listed in Table S1. (b) The YFP signal of root tips of DDB1-YFP transgenic tomato seedlings and (c) protoplasts prepared from transgenic DDB1-YFP line was determined by OLYMPUS FV1000-IX81 confocal microscopy. Bars: (b) 2 μm; (c) 5 μm.
7 Table S1 List of primers used in this study SacII-F ApaI- KpnI-F SalI- StuI- XbaI-F XhoI- K11-F K11- K253-F K253- SlCUL4 BamHI-F SlCUL4 SalI- SlCUL4 XbaI- SlCUL4 EcoI-F SlCUL4 XhoI- SlDDB1 KpnI-F SlDDB1 StuI- SlDDB1 SalI- SlDDB1 BamHI-F SlDDB1 SalI- Sequence (5-3 ) CCGCGGATGCTTGCTCTATCTT CATCATTGA GGGCCCTCAAGTTGGGGGTAT TTTGGTAA GGTACCATGCTTGCTCTATCTT CATCA GTCGACAGTTGGGGGTATTTT GGTAA AGGCCTAGTTGGGGGTATTTT GGTAA GCTCTAGAATGCTTGCTCTATC TTCATCA CCGCTCGAGAGTTGGGGGTAT TTTGGTAA TCTTCATCATTGAGCTACAGA AATGAAAGG TCTGTAGCTCAATGATGAAGA TAGAGCAAGC CAAGCCCTGCCATCATCAGAG TTGACACA TCTGATGATGGCAGGGCTTGG TGGGATGCC CGGGATCCATGAAGAAAGCTA AGTCACAAG CGCGTCGACAGCAAGGTAGTT GTATATTTGAG ATTTCTAGATTGATATAGATTT CCTCCCATT CAGAATTCATGAAGAAAGCTA AGTCAC ATTCTCGAGCTAAGCAAGGTA GTTGTATA GTCGGTACCATGAGTGTATGG AACTAC GAAAGGCCTATGCAACCTTGT CAACTC GAAGTCGACATGCAACCTTGT CAACTC GTCGGATCCATGAGTGTATGG AACTAC GATGTCGACATGCAACCTTGT CAACTC Usage Forward primer for peg202:: or pjg4-5:: everse primer for peg202:: or pjg4-5:: Forward primer for ptex::-flag, ptex::k11-flag, ptex::k253-flag, ptex::k11, 253-Flag or pbtex::-flag /HA constructs everse primer for ptex::-flag, ptex::k11-flag, ptex::k253-flag, ptex::k11, 253-Flag or pbtex::-flag constructs everse primer for pbtex::-ha construct Forward primer for puc-spyne:: construct everse primer for puc-spyne:: construct Forward primer for K11 mutant gene everse primer for K11 mutant gene Forward primer for K253 mutant gene everse primer for K253 mutant gene Forward primer for puc-spyce::slcul4 construct or ptex::slcul4- GFP,or pcb302:: CUL4-HA constructs everse primer for puc-spyce::slcul4 or ptex::slcul4-gfp constructs everse primer for generating pcb302:: SlCUL4-HA construct Forward primer for peg202::slcul4 or pjg4-5::slcul4 everse primer for peg202::slcul4 or pjg4-5::slcul4 Forward primer for generating pbtex::slddb1-ha /FLAG, pbtex::hp1-flag, pbtex::hp1 w -FLAG, ptex::slddb1-gfp constructs everse primer for generating pbtex::slddb1-ha construct everse primer for generating pbtex::slddb1-flag, pbtex::hp1- FLAG, pbtex::hp1 w -FLAG, ptex::slddb1-gfp constructs Forward primer for puc-spyce::slddb1 construct everse primer for puc-spyce::slddb1 construct
8 SlDDB1 SacII-F SlDDB1 ApaI- SlDET1 BamHI-F SlDET1 SalI- SlDET1 SacII-F SlDET1 ApaI- SlDET1(26-87) SacIIF SlDET1(26-87) ApaI SlDET1 KpnI-F SlDET1 StuI- SlDET1 SalI- NbDET1 XbaI-F NbDET1 BamHI- NbCUL4 EcoI-F NbCUL4 StuI- NbDDB1 EcoI-F NbDDB1 XbaI- NbCUL4 qt-f NbCUL4 qt- NbDDB1 qt-f NbDDB1 qt- NbDET1 qt-f NbDET1 qt- TCCCCGCGGATGAGTGTATGG AACTACGTGG TTCGGGCCCCTAATGCAACCT TGTCAACTC CGCGGATCCATGTTCAAAACT AACAATGTTACC ACGCGTCGACTCGACGAAAAT GGATATTTAC TCCCCGCGGATGTTCAAAACT AACAATGTTACC TTCGGGCCCTTATCGACGAAA ATGGATATT TCCCCGCGGCATCGTGCCAGA AGATTTTAT TTCGGGCCCATCTTCTTCTTTG CAGGAAAAT CGGGGTACCATGTTCAAAACT AACAATG CAAAGGCCTTCGACG AAAATGGATATT GACGTCGACTCGACG AAAATGGATATT TTCTCTAGAATGTTCAAAACT AACAATG GTCGGATCCAAACTTTCTAGC TTTCAAAGG GAAGAATTCCCATGAAGAAAG CTAAGTCAC GAAAGGCCTAGCAAGGTAGTT GTATATTTGAG CATGAATTCGATGTTGGGCAG ATACCC GATTCTAGAATGCAACCTTGT CAACTC CTTGATAAGGGTTTCACGCTG TT CAATGCTCTGACCAGTTCTTCG GCTTCACATCCGTTCAATACCC AAGCAGTCCCAACGCAATAAT A CAGTCTCAGAGTCCTTCGCCTT A ATCTTCTTAGTCCGCCCATCC Forward primer for peg202::slddb1 everse primer for peg202::slddb1 Forward primer for puc-spyce::sldet1 construct everse primer for puc-spyce::sldet1 construct Forward primer for peg202::sldet1 everse primer for peg202::sldet1 Forward primer for peg202::sldet aa everse primer for peg202::sldet aa Forward primer For pbtex::sldet1-flag/ha,pbtex::hp2-flag, pbtex::hp2 j -FLAG, pbtex::hp2 dg -FLAG, ptex::sldet1-gfp constructs everse primer for pbtex::sldet1-ha construct everse primer For pbtex::sldet1-flag, pbtex::hp2-flag, pbtex::hp2 j -FLAG, pbtex::hp2 dg -FLAG, ptex::sldet1-gfp constructs Forward primer for TV2::NbDET1 VIGS everse primer for TV2::NbDET1 VIGS Forward primer for TV2::NbCUL4 VIGS everse primer for TV2::NbCUL4 VIGS Forward primer for TV2::NbDDB1 VIGS everse primer for TV2::NbDDB1 VIGS Forward primer for realtime qt-pc everse primer for realtime qt-pc Forward primer for realtime qt-pc everse primer for realtime qt-pc Forward primer for realtime qt-pc everse primer for realtime qt-pc
9 NbEF1α qt-f NbEF1α qt- UBI3-F TGTGATTGATGCCCCTGGAC CCAAGGGTGAAAGCAAGCAAT AGGTTGATGACACTGGAAAGG TT Forward primer for realtime qt-pc everse primer for realtime qt-pc Forward primer for realtime qt-pc UBI3- AATCGCCTCCAGCCTTGTTGTA everse primer for realtime qt-pc hp1 w -F hp1 w - hp2 j -F hp2 j - hp2 dg -F hp2 dg - hp2-f hp2- GFP PstI-F GFP SphI- GATGCCAGAGGAAAATAAACC TACTAAG TATTTTCCTCTGGCATCACATA TGCA GTTCCTTCCTCTTCCACTCAAT ATTACC AGTGGAAGAGGAAGGAACAG ATCTTCTT CCAGAAGATTTTATGAGATCG TTGTACCAAG ATCTCATAAAATCTTCTGGCA CGATGGATG CTGAAATTCAAAATGAAGCCA GCTGGCAGCACAGATGGGCGA ACTAAATC TCGCCCATCTGTGCTGCCAGCT GGCTTCATTTTGAATTTCAGGA TATTGGG AAACTGCAGGGTGCCGGCGCA GGAGC ACATGCATGCTTACTTGTACA GCTCGTCCATGCCG Forward primer for hp1 w mutant gene everse primer for hp1 w mutant gene Forward primer for hp2 j mutant gene everse primer for hp2 j mutant gene Forward primer for hp2 dg mutant gene everse primer for hp2 dg mutant gene Forward primer for hp2 gene everse primer for hp2 gene Forward primer for ptex::gfp construct everse primer for ptex::gfp construct
10 Methods S1 eal-time T-PC analysis Total NAs were isolated with TIzol reagent (Invitrogen) and treated with DNase I. everse transcription was conducted using the SuperScriptII reverse transcriptase (Invitrogen, USA) and real-time PC analysis was performed on an ABI Prism 7700 sequence detection system using SYB Green reagents (Life Technologies, USA). The N. benthamiana EF1α gene (accession No. AY ) was used as an internal reference. elative expression ratios were determined using the EST software (Pfaffl et al., 2002). Primers used in the real-time T-PC analysis are listed in Supporting Information Table S1. eference Pfaffl MW, Horgan GW, Dempfle L elative expression software tool (EST) for group-wise comparison and statistical analysis of relative expression results in real-time PC. Nucleic Acids esearch 30: e36.
SUPPLEMENTARY DATA 1
 SUPPLEMENTARY DATA 1 Supplementary Figure S1. Overexpression of untagged Ago2 inhibits the nuclear transport of the dotted foci of GFP-signal of myc-gfp-tnrc6a-nes-mut. (A C) HeLa cells expressing myc-gfp-tnrc6a-nes-mut
SUPPLEMENTARY DATA 1 Supplementary Figure S1. Overexpression of untagged Ago2 inhibits the nuclear transport of the dotted foci of GFP-signal of myc-gfp-tnrc6a-nes-mut. (A C) HeLa cells expressing myc-gfp-tnrc6a-nes-mut
Mir-X mirna First-Strand Synthesis Kit User Manual
 User Manual Mir-X mirna First-Strand Synthesis Kit User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories, Inc.
User Manual Mir-X mirna First-Strand Synthesis Kit User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories, Inc.
Engineering of Yellow Mosaic Virus Resistance (YMVR) in Blackgram. Project ID: 1 April 2000 to 31 August 2004. Project Duration:
 Engineering of Yellow Mosaic Virus Resistance (YMVR) in Blackgram ID: Duration: Coordinator in Switzerland: PS1 1 April 2000 to 31 August 2004 Prof. Thomas Hohn University of Basel Botanisches Institut
Engineering of Yellow Mosaic Virus Resistance (YMVR) in Blackgram ID: Duration: Coordinator in Switzerland: PS1 1 April 2000 to 31 August 2004 Prof. Thomas Hohn University of Basel Botanisches Institut
Recombinant DNA & Genetic Engineering. Tools for Genetic Manipulation
 Recombinant DNA & Genetic Engineering g Genetic Manipulation: Tools Kathleen Hill Associate Professor Department of Biology The University of Western Ontario Tools for Genetic Manipulation DNA, RNA, cdna
Recombinant DNA & Genetic Engineering g Genetic Manipulation: Tools Kathleen Hill Associate Professor Department of Biology The University of Western Ontario Tools for Genetic Manipulation DNA, RNA, cdna
REAL TIME PCR USING SYBR GREEN
 REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
mircute mirna qpcr Detection Kit (SYBR Green)
 mircute mirna qpcr Detection Kit (SYBR Green) For detection of mirna using real-time RT-PCR (SYBR Green I) www.tiangen.com QP110302 mircute mirna qpcr Detection Kit (SYBR Green) Kit Contents Cat. no. FP401
mircute mirna qpcr Detection Kit (SYBR Green) For detection of mirna using real-time RT-PCR (SYBR Green I) www.tiangen.com QP110302 mircute mirna qpcr Detection Kit (SYBR Green) Kit Contents Cat. no. FP401
Recombinant DNA and Biotechnology
 Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
Supplemental Information. McBrayer et al. Supplemental Data
 1 Supplemental Information McBrayer et al. Supplemental Data 2 Figure S1. Glucose consumption rates of MM cell lines exceed that of normal PBMC. (A) Normal PBMC isolated from three healthy donors were
1 Supplemental Information McBrayer et al. Supplemental Data 2 Figure S1. Glucose consumption rates of MM cell lines exceed that of normal PBMC. (A) Normal PBMC isolated from three healthy donors were
ARABIDOPSIS. A Laboratory Manual DETLEF WEIGEL JANE GLAZEBROOK
 ARABIDOPSIS A Laboratory Manual DETLEF WEIGEL Salk Institute, Plant Biology Laboratory La Jolla, California Max Planck Institute for Developmental Biology Tubingen, Germany JANE GLAZEBROOK Torrey Mesa
ARABIDOPSIS A Laboratory Manual DETLEF WEIGEL Salk Institute, Plant Biology Laboratory La Jolla, California Max Planck Institute for Developmental Biology Tubingen, Germany JANE GLAZEBROOK Torrey Mesa
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
Real-time PCR: Understanding C t
 APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
Fig. S1. Identification of CRMs with activity in Ci-Tbx2/3 expression domains outside the notochord. B-F
 Fig. S1. Identification of CRMs with activity in Ci-Tbx2/3 expression domains outside the notochord. (A) Top: map of the Ci- Tbx2/3 locus. Teal rectangles and lines denote exons and introns, respectively.
Fig. S1. Identification of CRMs with activity in Ci-Tbx2/3 expression domains outside the notochord. (A) Top: map of the Ci- Tbx2/3 locus. Teal rectangles and lines denote exons and introns, respectively.
Recombinant DNA Unit Exam
 Recombinant DNA Unit Exam Question 1 Restriction enzymes are extensively used in molecular biology. Below are the recognition sites of two of these enzymes, BamHI and BclI. a) BamHI, cleaves after the
Recombinant DNA Unit Exam Question 1 Restriction enzymes are extensively used in molecular biology. Below are the recognition sites of two of these enzymes, BamHI and BclI. a) BamHI, cleaves after the
Notch 1 -dependent regulation of cell fate in colorectal cancer
 Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Transcriptional control of lifespan
 Transcriptional control of lifespan Enhanced plant productivity through control of lifespan Kiel, Germany 8-17 May Seite 1 von 7 Tuesday 8 May 2012 Bildungszentrum Tannenfelde 15:00 Diter von Wettstein
Transcriptional control of lifespan Enhanced plant productivity through control of lifespan Kiel, Germany 8-17 May Seite 1 von 7 Tuesday 8 May 2012 Bildungszentrum Tannenfelde 15:00 Diter von Wettstein
REAL TIME PCR SYBR GREEN
 REAL TIME PCR SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM QUANTITATION
REAL TIME PCR SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM QUANTITATION
First Strand cdna Synthesis
 380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
Relative Quantification of mirna Target mrnas by Real-Time qpcr. 1 Introduction. Gene Expression Application Note No. 4
 Gene Expression Application Note No. 4 Relative Quantification of mirna Target mrnas by Real-Time qpcr Ute Ernst, Jitao David Zhang, Anja Irsigler, Stefan Wiemann, Ulrich Tschulena Division: Molecular
Gene Expression Application Note No. 4 Relative Quantification of mirna Target mrnas by Real-Time qpcr Ute Ernst, Jitao David Zhang, Anja Irsigler, Stefan Wiemann, Ulrich Tschulena Division: Molecular
Biotechnology: DNA Technology & Genomics
 Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Investigating the role of a Cryptosporidium parum apyrase in infection
 Investigating the role of a Cryptosporidium parum apyrase in infection David Riccardi and Patricio Manque Abstract This project attempted to characterize the function of a Cryptosporidium parvum apyrase
Investigating the role of a Cryptosporidium parum apyrase in infection David Riccardi and Patricio Manque Abstract This project attempted to characterize the function of a Cryptosporidium parvum apyrase
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
RevertAid Premium First Strand cdna Synthesis Kit
 RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
SUGAR BEET (Beta vulgaris L.) PROMOTERS FOR DIRECTED TISSUE- SPECIFIC ROOT TRANSCRIPTION
 SUGAR BEET (Beta vulgaris L.) PROMOTERS FOR DIRECTED TISSUE- SPECIFIC ROOT TRANSCRIPTION Senthilkumar Padmanaban, Haiyan Li, David P. Puthoff and Ann C. Smigocki USDA-ARS Molecular Plant Pathology Laboratory,
SUGAR BEET (Beta vulgaris L.) PROMOTERS FOR DIRECTED TISSUE- SPECIFIC ROOT TRANSCRIPTION Senthilkumar Padmanaban, Haiyan Li, David P. Puthoff and Ann C. Smigocki USDA-ARS Molecular Plant Pathology Laboratory,
2.500 Threshold. 2.000 1000e - 001. Threshold. Exponential phase. Cycle Number
 application note Real-Time PCR: Understanding C T Real-Time PCR: Understanding C T 4.500 3.500 1000e + 001 4.000 3.000 1000e + 000 3.500 2.500 Threshold 3.000 2.000 1000e - 001 Rn 2500 Rn 1500 Rn 2000
application note Real-Time PCR: Understanding C T Real-Time PCR: Understanding C T 4.500 3.500 1000e + 001 4.000 3.000 1000e + 000 3.500 2.500 Threshold 3.000 2.000 1000e - 001 Rn 2500 Rn 1500 Rn 2000
DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
 Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2014 DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2014 DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna
 All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna All-in-One TM mirna First-Strand cdna Synthesis Kit AMRT-0020 (20 RT reactions), AMRT-0060 (60 RT reactions) Used in combination
All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna All-in-One TM mirna First-Strand cdna Synthesis Kit AMRT-0020 (20 RT reactions), AMRT-0060 (60 RT reactions) Used in combination
Mechanosensitive Neurons on the Internal Reproductive Tract Contribute to Egg-Laying- Induced Acetic Acid Attraction in Drosophila
 Cell Reports, Volume 9 Supplemental Information Mechanosensitive Neurons on the Internal Reproductive Tract Contribute to Egg-Laying- Induced Acetic Acid Attraction in Drosophila Bin Gou, Ying Liu, Ananya
Cell Reports, Volume 9 Supplemental Information Mechanosensitive Neurons on the Internal Reproductive Tract Contribute to Egg-Laying- Induced Acetic Acid Attraction in Drosophila Bin Gou, Ying Liu, Ananya
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual
 Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
ONLINE SUPPLEMENTAL MATERIAL. Allele-Specific Expression of Angiotensinogen in Human Subcutaneous Adipose Tissue
 ONLINE SUPPLEMENTAL MATERIAL Allele-Specific Expression of Angiotensinogen in Human Subcutaneous Adipose Tissue Sungmi Park 1, Ko-Ting Lu 1, Xuebo Liu 1, Tapan K. Chatterjee 2, Steven M. Rudich 3, Neal
ONLINE SUPPLEMENTAL MATERIAL Allele-Specific Expression of Angiotensinogen in Human Subcutaneous Adipose Tissue Sungmi Park 1, Ko-Ting Lu 1, Xuebo Liu 1, Tapan K. Chatterjee 2, Steven M. Rudich 3, Neal
Outline. 1. Experiment. 2. Sample analysis and storage. 3. Image analysis and presenting data. 4. Probemaker
 Tips and tricks Note: this is just an informative document with general recommendations. Please contact support@olink.com should you have any queries. Document last reviewed 2011-11-17 Outline 1. Experiment
Tips and tricks Note: this is just an informative document with general recommendations. Please contact support@olink.com should you have any queries. Document last reviewed 2011-11-17 Outline 1. Experiment
Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr
 User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
Real-time quantitative RT -PCR (Taqman)
 Real-time quantitative RT -PCR (Taqman) Author: SC, Patti Lab, 3/03 This is performed as a 2-step reaction: 1. cdna synthesis from DNase 1-treated total RNA 2. PCR 1. cdna synthesis (Advantage RT-for-PCR
Real-time quantitative RT -PCR (Taqman) Author: SC, Patti Lab, 3/03 This is performed as a 2-step reaction: 1. cdna synthesis from DNase 1-treated total RNA 2. PCR 1. cdna synthesis (Advantage RT-for-PCR
Supplementary Figure 1.
 Supplementary Figure 1. (A) MicroRNA 212 enhances IS from pancreatic β-cells. INS-1 832/3 β-cells were transfected with precursors for mirnas 212, 375, or negative control oligonucleotides. 48 hrs after
Supplementary Figure 1. (A) MicroRNA 212 enhances IS from pancreatic β-cells. INS-1 832/3 β-cells were transfected with precursors for mirnas 212, 375, or negative control oligonucleotides. 48 hrs after
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR
 Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
!!!!!!!!!!!!!!!!!!!!!!!!!!
 Figure S7 cdl-1 3'UTR gld-1 binding site (4..10) let-7 binding site (326..346) Figure Legends Figure S1. gld-1 genetically interacts with nhl-2 and vig-1 during germline development. (A) nhl-2(ok818) enhances
Figure S7 cdl-1 3'UTR gld-1 binding site (4..10) let-7 binding site (326..346) Figure Legends Figure S1. gld-1 genetically interacts with nhl-2 and vig-1 during germline development. (A) nhl-2(ok818) enhances
GENE CLONING AND RECOMBINANT DNA TECHNOLOGY
 GENE CLONING AND RECOMBINANT DNA TECHNOLOGY What is recombinant DNA? DNA from 2 different sources (often from 2 different species) are combined together in vitro. Recombinant DNA forms the basis of cloning.
GENE CLONING AND RECOMBINANT DNA TECHNOLOGY What is recombinant DNA? DNA from 2 different sources (often from 2 different species) are combined together in vitro. Recombinant DNA forms the basis of cloning.
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID papillary renal cell carcinoma (translocation-associated) PRCC Human This gene
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID papillary renal cell carcinoma (translocation-associated) PRCC Human This gene
Real-Time PCR Vs. Traditional PCR
 Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE Q5B
 INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
HCS604.03 Exercise 1 Dr. Jones Spring 2005. Recombinant DNA (Molecular Cloning) exercise:
 HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI,
 Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
Essentials of Real Time PCR. About Sequence Detection Chemistries
 Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS)
 Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS) A typical RNA Seq experiment Library construction Protocol variations Fragmentation methods RNA: nebulization,
Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS) A typical RNA Seq experiment Library construction Protocol variations Fragmentation methods RNA: nebulization,
GeneCopoeia Genome Editing Tools for Safe Harbor Integration in. Mice and Humans. Ed Davis, Liuqing Qian, Ruiqing li, Junsheng Zhou, and Jinkuo Zhang
 G e n e C o p o eia TM Expressway to Discovery APPLICATION NOTE Introduction GeneCopoeia Genome Editing Tools for Safe Harbor Integration in Mice and Humans Ed Davis, Liuqing Qian, Ruiqing li, Junsheng
G e n e C o p o eia TM Expressway to Discovery APPLICATION NOTE Introduction GeneCopoeia Genome Editing Tools for Safe Harbor Integration in Mice and Humans Ed Davis, Liuqing Qian, Ruiqing li, Junsheng
Chlamydomonas adapted Green Fluorescent Protein (CrGFP)
 Chlamydomonas adapted Green Fluorescent Protein (CrGFP) Plasmid pfcrgfp for fusion proteins Sequence of the CrGFP In the sequence below, all amino acids which have been altered from the wildtype GFP from
Chlamydomonas adapted Green Fluorescent Protein (CrGFP) Plasmid pfcrgfp for fusion proteins Sequence of the CrGFP In the sequence below, all amino acids which have been altered from the wildtype GFP from
Supplemental Fig. S1. The schematic diagrams of the expression constructs used in this study.
 1 Supplemental data Supplemental Fig. S1. The schematic diagrams of the expression constructs used in this study. Supplemental Fig. S2. Ingenuity Pathway Analysis (IPA) of the 56 putative caspase substrates
1 Supplemental data Supplemental Fig. S1. The schematic diagrams of the expression constructs used in this study. Supplemental Fig. S2. Ingenuity Pathway Analysis (IPA) of the 56 putative caspase substrates
by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains was previously described
 Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
Genetic Engineering and Biotechnology
 1 So, what is biotechnology?? The use of living organisms to carry out defined chemical processes for industrial or commercial application. The office of Technology Assessment of the U.S. Congress defines
1 So, what is biotechnology?? The use of living organisms to carry out defined chemical processes for industrial or commercial application. The office of Technology Assessment of the U.S. Congress defines
European Medicines Agency
 European Medicines Agency July 1996 CPMP/ICH/139/95 ICH Topic Q 5 B Quality of Biotechnological Products: Analysis of the Expression Construct in Cell Lines Used for Production of r-dna Derived Protein
European Medicines Agency July 1996 CPMP/ICH/139/95 ICH Topic Q 5 B Quality of Biotechnological Products: Analysis of the Expression Construct in Cell Lines Used for Production of r-dna Derived Protein
ab185916 Hi-Fi cdna Synthesis Kit
 ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
Data Analysis on the ABI PRISM 7700 Sequence Detection System: Setting Baselines and Thresholds. Overview. Data Analysis Tutorial
 Data Analysis on the ABI PRISM 7700 Sequence Detection System: Setting Baselines and Thresholds Overview In order for accuracy and precision to be optimal, the assay must be properly evaluated and a few
Data Analysis on the ABI PRISM 7700 Sequence Detection System: Setting Baselines and Thresholds Overview In order for accuracy and precision to be optimal, the assay must be properly evaluated and a few
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes.
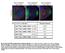 Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
MEF Starter Nucleofector Kit
 page 1 of 7 MEF Starter Nucleofector Kit for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the mice they are isolated
page 1 of 7 MEF Starter Nucleofector Kit for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the mice they are isolated
restriction enzymes 350 Home R. Ward: Spring 2001
 restriction enzymes 350 Home Restriction Enzymes (endonucleases): molecular scissors that cut DNA Properties of widely used Type II restriction enzymes: recognize a single sequence of bases in dsdna, usually
restriction enzymes 350 Home Restriction Enzymes (endonucleases): molecular scissors that cut DNA Properties of widely used Type II restriction enzymes: recognize a single sequence of bases in dsdna, usually
pcas-guide System Validation in Genome Editing
 pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
CompleteⅡ 1st strand cdna Synthesis Kit
 Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Nucleic Acid Techniques in Bacterial Systematics
 Nucleic Acid Techniques in Bacterial Systematics Edited by Erko Stackebrandt Department of Microbiology University of Queensland St Lucia, Australia and Michael Goodfellow Department of Microbiology University
Nucleic Acid Techniques in Bacterial Systematics Edited by Erko Stackebrandt Department of Microbiology University of Queensland St Lucia, Australia and Michael Goodfellow Department of Microbiology University
Global MicroRNA Amplification Kit
 Global MicroRNA Amplification Kit Store kit at -20 C on receipt (ver. 3-060901) A limited-use label license covers this product. By use of this product, you accept the terms and conditions outlined in
Global MicroRNA Amplification Kit Store kit at -20 C on receipt (ver. 3-060901) A limited-use label license covers this product. By use of this product, you accept the terms and conditions outlined in
Real-time monitoring of rolling circle amplification using aggregation-induced emission: applications for biological detection
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 215 Supplementary Information Real-time monitoring of rolling circle amplification using aggregation-induced
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 215 Supplementary Information Real-time monitoring of rolling circle amplification using aggregation-induced
Real-time qpcr Assay Design Software www.qpcrdesign.com
 Real-time qpcr Assay Design Software www.qpcrdesign.com Your Blueprint For Success Informational Guide 2199 South McDowell Blvd Petaluma, CA 94954-6904 USA 1.800.GENOME.1(436.6631) 1.415.883.8400 1.415.883.8488
Real-time qpcr Assay Design Software www.qpcrdesign.com Your Blueprint For Success Informational Guide 2199 South McDowell Blvd Petaluma, CA 94954-6904 USA 1.800.GENOME.1(436.6631) 1.415.883.8400 1.415.883.8488
Introduction to Bioinformatics 3. DNA editing and contig assembly
 Introduction to Bioinformatics 3. DNA editing and contig assembly Benjamin F. Matthews United States Department of Agriculture Soybean Genomics and Improvement Laboratory Beltsville, MD 20708 matthewb@ba.ars.usda.gov
Introduction to Bioinformatics 3. DNA editing and contig assembly Benjamin F. Matthews United States Department of Agriculture Soybean Genomics and Improvement Laboratory Beltsville, MD 20708 matthewb@ba.ars.usda.gov
Anti-ATF6 α antibody, mouse monoclonal (1-7)
 Anti-ATF6 α antibody, mouse monoclonal (1-7) 73-500 50 ug ATF6 (activating transcription factor 6) is an endoplasmic reticulum (ER) membrane-bound transcription factor activated in response to ER stress.
Anti-ATF6 α antibody, mouse monoclonal (1-7) 73-500 50 ug ATF6 (activating transcription factor 6) is an endoplasmic reticulum (ER) membrane-bound transcription factor activated in response to ER stress.
Optimized Protocol sirna Test Kit for Cell Lines and Adherent Primary Cells
 page 1 of 8 sirna Test Kit for Cell Lines and Adherent Primary Cells Kit principle Co-transfection of pmaxgfp, encoding the green fluorescent protein (GFP) from Pontellina p. with an sirna directed against
page 1 of 8 sirna Test Kit for Cell Lines and Adherent Primary Cells Kit principle Co-transfection of pmaxgfp, encoding the green fluorescent protein (GFP) from Pontellina p. with an sirna directed against
TransIT -2020 Transfection Reagent
 Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/5400 INTRODUCTION TransIT -2020 Transfection Reagent is a high-performance, animal-origin free, broad spectrum reagent
Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/5400 INTRODUCTION TransIT -2020 Transfection Reagent is a high-performance, animal-origin free, broad spectrum reagent
All-in-One mirna qrt-pcr Detection System Handbook
 All-in-One mirna qrt-pcr Detection System Handbook For quantitative detection of mature mirna All-in-One mirna First-Strand cdna Synthesis Kit Cat. No. AMRT-0020 (20 mirna reverse transcription reactions)
All-in-One mirna qrt-pcr Detection System Handbook For quantitative detection of mature mirna All-in-One mirna First-Strand cdna Synthesis Kit Cat. No. AMRT-0020 (20 mirna reverse transcription reactions)
QuantStudio 3D AnalysisSuite Software
 USER GUIDE QuantStudio 3D AnalysisSuite Software for use with QuantStudio 3D Digital PCR System Publication Number MAN0008161 Revision 1.0 For Research Use Only. Not for use in diagnostic procedures. For
USER GUIDE QuantStudio 3D AnalysisSuite Software for use with QuantStudio 3D Digital PCR System Publication Number MAN0008161 Revision 1.0 For Research Use Only. Not for use in diagnostic procedures. For
Beginner s Guide to Real-Time PCR
 Beginner s Guide to Real-Time PCR 02 Real-time PCR basic principles PCR or the Polymerase Chain Reaction has become the cornerstone of modern molecular biology the world over. Real-time PCR is an advanced
Beginner s Guide to Real-Time PCR 02 Real-time PCR basic principles PCR or the Polymerase Chain Reaction has become the cornerstone of modern molecular biology the world over. Real-time PCR is an advanced
J. Cell Sci. 128: doi:10.1242/jcs.164566: Supplementary Material
 Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Hepatitis B Virus. genesig Advanced Kit. DNA testing. Everything... Everyone... Everywhere... Core Protein Region. 150 tests.
 TM Primerdesign Ltd TM Primerdesign Ltd Hepatitis B Virus Core Protein Region genesig Advanced Kit 150 tests DNA testing Everything... Everyone... Everywhere... For general laboratory and research use
TM Primerdesign Ltd TM Primerdesign Ltd Hepatitis B Virus Core Protein Region genesig Advanced Kit 150 tests DNA testing Everything... Everyone... Everywhere... For general laboratory and research use
BaculoDirect Baculovirus Expression System Free your hands with the BaculoDirect Baculovirus Expression System
 BaculoDirect Baculovirus Expression System Free your hands with the BaculoDirect Baculovirus Expression System The BaculoDirect Baculovirus Expression System gives you: Unique speed and simplicity High-throughput
BaculoDirect Baculovirus Expression System Free your hands with the BaculoDirect Baculovirus Expression System The BaculoDirect Baculovirus Expression System gives you: Unique speed and simplicity High-throughput
All-in-One First-Strand cdna Synthesis Kit
 All-in-One First-Strand cdna Synthesis Kit For reliable first-strand cdna synthesis from all RNA sources Cat. No. AORT-0020 (20 synthesis reactions) Cat. No. AORT-0050 (50 synthesis reactions) User Manual
All-in-One First-Strand cdna Synthesis Kit For reliable first-strand cdna synthesis from all RNA sources Cat. No. AORT-0020 (20 synthesis reactions) Cat. No. AORT-0050 (50 synthesis reactions) User Manual
MystiCq microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C
 microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C Product Description The microrna cdna Synthesis Mix has been designed to easily convert micrornas into cdna templates for qpcr
microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C Product Description The microrna cdna Synthesis Mix has been designed to easily convert micrornas into cdna templates for qpcr
HENIPAVIRUS ANTIBODY ESCAPE SEQUENCING REPORT
 HENIPAVIRUS ANTIBODY ESCAPE SEQUENCING REPORT Kimberly Bishop Lilly 1,2, Truong Luu 1,2, Regina Cer 1,2, and LT Vishwesh Mokashi 1 1 Naval Medical Research Center, NMRC Frederick, 8400 Research Plaza,
HENIPAVIRUS ANTIBODY ESCAPE SEQUENCING REPORT Kimberly Bishop Lilly 1,2, Truong Luu 1,2, Regina Cer 1,2, and LT Vishwesh Mokashi 1 1 Naval Medical Research Center, NMRC Frederick, 8400 Research Plaza,
Understanding the immune response to bacterial infections
 Understanding the immune response to bacterial infections A Ph.D. (SCIENCE) DISSERTATION SUBMITTED TO JADAVPUR UNIVERSITY SUSHIL KUMAR PATHAK DEPARTMENT OF CHEMISTRY BOSE INSTITUTE 2008 CONTENTS Page SUMMARY
Understanding the immune response to bacterial infections A Ph.D. (SCIENCE) DISSERTATION SUBMITTED TO JADAVPUR UNIVERSITY SUSHIL KUMAR PATHAK DEPARTMENT OF CHEMISTRY BOSE INSTITUTE 2008 CONTENTS Page SUMMARY
Quantitative Real Time PCR Protocol. Stack Lab
 Quantitative Real Time PCR Protocol Stack Lab Overview Real-time quantitative polymerase chain reaction (qpcr) differs from regular PCR by including in the reaction fluorescent reporter molecules that
Quantitative Real Time PCR Protocol Stack Lab Overview Real-time quantitative polymerase chain reaction (qpcr) differs from regular PCR by including in the reaction fluorescent reporter molecules that
PrimePCR Assay Validation Report
 Gene Information Gene Name sorbin and SH3 domain containing 2 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SORBS2 Human Arg and c-abl represent the mammalian
Gene Information Gene Name sorbin and SH3 domain containing 2 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SORBS2 Human Arg and c-abl represent the mammalian
MEF Nucleofector Kit 1 and 2
 page 1 of 7 MEF Nucleofector Kit 1 and 2 for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the they are isolated
page 1 of 7 MEF Nucleofector Kit 1 and 2 for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the they are isolated
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
RT31-020 20 rxns. RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl
 Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
Mitotic Membrane Turnover Coordinates Differential Induction of the Heart Progenitor Lineage
 Developmental Cell Supplemental Information Mitotic Membrane Turnover Coordinates Differential Induction of the Heart Progenitor Lineage Christina D. Cota and Brad Davidson Supplemental Figures S1-S7 S1.
Developmental Cell Supplemental Information Mitotic Membrane Turnover Coordinates Differential Induction of the Heart Progenitor Lineage Christina D. Cota and Brad Davidson Supplemental Figures S1-S7 S1.
The heart specimens were fixed with formalin, embedded in paraffin, and sectioned at 6-
 Hsu et al. 2010/958033 R2 SUPPLEMENTAL MATERIALS Supplemental methods: Immunohistochemistry The heart specimens were fixed with formalin, embedded in paraffin, and sectioned at 6- μm thickness. Formalin-fixed
Hsu et al. 2010/958033 R2 SUPPLEMENTAL MATERIALS Supplemental methods: Immunohistochemistry The heart specimens were fixed with formalin, embedded in paraffin, and sectioned at 6- μm thickness. Formalin-fixed
Absolute Quantification Getting Started Guide
 5 cdna Reverse Primer Oligo d(t) or random hexamer Synthesis of 1st cdna strand 3 5 cdna Applied Biosystems 7300/7500/7500 Fast Real-Time PCR System Absolute Quantification Getting Started Guide Introduction
5 cdna Reverse Primer Oligo d(t) or random hexamer Synthesis of 1st cdna strand 3 5 cdna Applied Biosystems 7300/7500/7500 Fast Real-Time PCR System Absolute Quantification Getting Started Guide Introduction
protocol handbook 3D cell culture mimsys G hydrogel
 handbook 3D cell culture mimsys G hydrogel supporting real discovery handbook Index 01 Cell encapsulation in hydrogels 02 Cell viability by MTS assay 03 Live/Dead assay to assess cell viability 04 Fluorescent
handbook 3D cell culture mimsys G hydrogel supporting real discovery handbook Index 01 Cell encapsulation in hydrogels 02 Cell viability by MTS assay 03 Live/Dead assay to assess cell viability 04 Fluorescent
Transfection-Transfer of non-viral genetic material into eukaryotic cells. Infection/ Transduction- Transfer of viral genetic material into cells.
 Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
SureSilencing shrna Plasmids
 User Manual BIOMOL GmbH Waidmannstr. 35 22769 Hamburg info@biomol.de www.biomol.de Phone:+49-40-8532600 or 0800-2466651 (D) Fax: +49-40-85326022 or 0800-2466652 (D) SureSilencing shrna Plasmids Genome-Wide
User Manual BIOMOL GmbH Waidmannstr. 35 22769 Hamburg info@biomol.de www.biomol.de Phone:+49-40-8532600 or 0800-2466651 (D) Fax: +49-40-85326022 or 0800-2466652 (D) SureSilencing shrna Plasmids Genome-Wide
FastLine cell cdna Kit
 1. FastLine cell cdna Kit For high-speed preparation of first-strand cdna directly from cultured cells without RNA purification www.tiangen.com RT100701 FastLine cell cdna Kit Cat. no. KR105 Kit Contents
1. FastLine cell cdna Kit For high-speed preparation of first-strand cdna directly from cultured cells without RNA purification www.tiangen.com RT100701 FastLine cell cdna Kit Cat. no. KR105 Kit Contents
The cell lines used in this study were obtained from the American Type Culture
 Supplementary materials and methods Cell culture and drug treatments The cell lines used in this study were obtained from the American Type Culture Collection (ATCC) and grown as previously described.
Supplementary materials and methods Cell culture and drug treatments The cell lines used in this study were obtained from the American Type Culture Collection (ATCC) and grown as previously described.
Name Class Date. Figure 13 1. 2. Which nucleotide in Figure 13 1 indicates the nucleic acid above is RNA? a. uracil c. cytosine b. guanine d.
 13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
Hydroxynitrile lyase (Hnl)
 Hydroxynitrile lyase (Hnl) R 1 HCN R 1 OH R 2 C O R 2 C * CN S-selective: Hevea brasiliensis R-selective: Prunus spp. (S)-Hnl of Hevea brasiliensis and (R)-Hnl of Prunus amygdalus Hb_Hnl Type II Hnl intracellular
Hydroxynitrile lyase (Hnl) R 1 HCN R 1 OH R 2 C O R 2 C * CN S-selective: Hevea brasiliensis R-selective: Prunus spp. (S)-Hnl of Hevea brasiliensis and (R)-Hnl of Prunus amygdalus Hb_Hnl Type II Hnl intracellular
Path-ID Multiplex One-Step RT-PCR Kit
 USER GUIDE Path-ID Multiplex One-Step RT-PCR Kit TaqMan probe-based multiplex one-step real-time RT-PCR detection of RNA targets Catalog Numbers 4428206, 4428207, 4440022 Publication Part Number 4440907
USER GUIDE Path-ID Multiplex One-Step RT-PCR Kit TaqMan probe-based multiplex one-step real-time RT-PCR detection of RNA targets Catalog Numbers 4428206, 4428207, 4440022 Publication Part Number 4440907
RealStar HBV PCR Kit 1.0 11/2012
 RealStar HBV PCR Kit 1.0 11/2012 RealStar HBV PCR Kit 1.0 For research use only! (RUO) Product No.: 201003 96 rxns INS-201000-GB-02 Store at -25 C... -15 C November 2012 altona Diagnostics GmbH Mörkenstraße
RealStar HBV PCR Kit 1.0 11/2012 RealStar HBV PCR Kit 1.0 For research use only! (RUO) Product No.: 201003 96 rxns INS-201000-GB-02 Store at -25 C... -15 C November 2012 altona Diagnostics GmbH Mörkenstraße
Bacterial Transformation and Plasmid Purification. Chapter 5: Background
 Bacterial Transformation and Plasmid Purification Chapter 5: Background History of Transformation and Plasmids Bacterial methods of DNA transfer Transformation: when bacteria take up DNA from their environment
Bacterial Transformation and Plasmid Purification Chapter 5: Background History of Transformation and Plasmids Bacterial methods of DNA transfer Transformation: when bacteria take up DNA from their environment
SYBR Green Realtime PCR Master Mix -Plus-
 Instruction manual SYBR Green Realtime PCR Master Mix -Plus- 0810 F0925K SYBR Green Realtime PCR Master Mix -Plus- Contents QPK-212T 1mLx1 QPK-212 1mLx5 Store at -20 C, protected from light [1] Introduction
Instruction manual SYBR Green Realtime PCR Master Mix -Plus- 0810 F0925K SYBR Green Realtime PCR Master Mix -Plus- Contents QPK-212T 1mLx1 QPK-212 1mLx5 Store at -20 C, protected from light [1] Introduction
Integrated Protein Services
 Integrated Protein Services Custom protein expression & purification Version DC04-0012 Expression strategy The first step in the recombinant protein generation process is to design an appropriate expression
Integrated Protein Services Custom protein expression & purification Version DC04-0012 Expression strategy The first step in the recombinant protein generation process is to design an appropriate expression
TITRATION OF raav (VG) USING QUANTITATIVE REAL TIME PCR
 Page 1 of 5 Materials DNase digestion buffer [13 mm Tris-Cl, ph7,5 / 5 mm MgCl2 / 0,12 mm CaCl2] RSS plasmid ptr-uf11 SV40pA Forward primer (10µM) AGC AAT AGC ATC ACA AAT TTC ACA A SV40pA Reverse Primer
Page 1 of 5 Materials DNase digestion buffer [13 mm Tris-Cl, ph7,5 / 5 mm MgCl2 / 0,12 mm CaCl2] RSS plasmid ptr-uf11 SV40pA Forward primer (10µM) AGC AAT AGC ATC ACA AAT TTC ACA A SV40pA Reverse Primer
About Our Products. Blood Products. Purified Infectious/Inactivated Agents. Native & Recombinant Viral Proteins. DNA Controls and Primers for PCR
 About Our Products Purified Infectious/Inactivated Agents ABI produces a variety of specialized reagents, allowing researchers to choose the best preparations for their studies. Available reagents include
About Our Products Purified Infectious/Inactivated Agents ABI produces a variety of specialized reagents, allowing researchers to choose the best preparations for their studies. Available reagents include
RealLine HCV PCR Qualitative - Uni-Format
 Instructions for use PCR KIT FOR EXTRACTION OF RNA AND REAL TIME PCR DETECTION KIT FOR HEPATITIS C VIRUS RNA Research Use Only Qualitative Uni Format VBD0798 48 tests valid from: December 2013 Rev11122013
Instructions for use PCR KIT FOR EXTRACTION OF RNA AND REAL TIME PCR DETECTION KIT FOR HEPATITIS C VIRUS RNA Research Use Only Qualitative Uni Format VBD0798 48 tests valid from: December 2013 Rev11122013
DyNAmo cdna Synthesis Kit for qrt-pcr
 DyNAmo cdna Synthesis Kit for qrt-pcr Instruction manual F- 470S Sufficient for 20 cdna synthesis reactions (20 µl each) F- 470L Sufficient for 100 cdna synthesis reactions (20 µl each) Description...
DyNAmo cdna Synthesis Kit for qrt-pcr Instruction manual F- 470S Sufficient for 20 cdna synthesis reactions (20 µl each) F- 470L Sufficient for 100 cdna synthesis reactions (20 µl each) Description...
Production of antigens and antibodies in plants: alternative technology?
 Production of antigens and antibodies in plants: alternative technology? George Lomonossoff John Innes Centre Norwich, UK ECOPA, Alicante 29 th Sept. 2006 Why use Plants as Biofactories? Produce large
Production of antigens and antibodies in plants: alternative technology? George Lomonossoff John Innes Centre Norwich, UK ECOPA, Alicante 29 th Sept. 2006 Why use Plants as Biofactories? Produce large
RT-PCR: Two-Step Protocol
 RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
