STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS
|
|
|
- Erica Riley
- 9 years ago
- Views:
Transcription
1 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS SECTION 1 The study of cell structure was revolutionised by the invention of the electron microscope. The best optical (light) microscopes can magnify specimens about 1,500 times under optimal conditions. With the electron microscope, magnifications of 100,000 times or more are possible, which enables detailed examination of sub-cellular components. This structural information, in conjunction with biochemical studies, has provided a wealth of information about how cells are constructed and how they function. 1.1 Features and ultrastructure of prokaryotic and eukaryotic cells The great variety that is found in different types of cell makes it difficult to present the average cell there is no such thing! However, there are things that most cells have in common the presence of genetic material, a cell membrane (plasma membrane), and some type of fluidbased matrix (the cytosol) that makes up much of the internal volume of the cell. We will consider bacterial, plant and animal cells, and try to illustrate the similarities and differences between them. Major aspects to consider are the arrangement of DNA in cells, whether or not subcellular organelles are present, the use of membranes within cells, and the organisation of the cytosol. The bacterial cell A generalised diagram of a bacterial cell is shown in Fig Bacteria are prokaryotic, and therefore lack a true membrane-bound nucleus. The DNA is present as a single circular molecule, usually termed the bacterial chromosome, although strictly speaking this term should be reserved for the more complex DNA:protein structures found in eukaryotic cells. The DNA is highly condensed or packaged by coiling and folding, and this produces a structure known as the nucleoid. Such packaging is needed because of the length of the DNA molecule a typical Escherichia coli cell is about 1 µm diameter 2 µm length, yet it contains about 1,400 µm of DNA. Fitting all this into the cell is only possible because DNA is a very long, thin molecule. Apart from the nucleoid, there is little internal structure evident in bacterial cell micrographs apart from a large number of ribosomes, BIOLOGY 1
2 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS essential for protein synthesis. The lack of internal structure means that the cytosol is effectively the site of all bacterial cell metabolism. This enables bacteria to adapt very quickly to changing nutritional conditions, but does mean that the regulation of genetic and metabolic activity has to be kept under tight control. Fig : Diagram of a typical bacterial cell. Not all of the structures shown may be present in all cells. Bacteria have cell walls that contain peptidoglycan, which is composed of linked disaccharide and peptide units. A major method of classifying bacteria into two groups is the Gram stain, which stains walls according to how thick the peptidoglycan layer is. Thus we have Gram-positive and Gram-negative bacteria, depending on whether or not the Gram stain is taken up by the cell wall. Lying outside the cell wall there may be a mucilaginous capsule, and there may also be projections from the cell surface known as pili or fimbriae. Longer flagella may also be present. Bacterial cells are often considered to be simple cells. Whilst this may be true in terms of the relatively few structural features that are present within the cell, it is certainly not true when you consider the complex chemical activity of the cell, all of which has to happen inside the bag of membrane-enclosed cytosol. Most bacterial cells exist as individual organisms, and need to be metabolically self-contained if they are to survive. Cells with a true nucleus and other membrane-bound organelles are known as eukaryotic cells, and are more complex than bacteria in terms of structure. Many eukaryotic cells are components of multicellular organisms, and therefore the requirements of the cell are somewhat different from those in bacteria. 2 BIOLOGY
3 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Plant cells A diagram of a typical plant cell is shown in Fig Plant cells show less diversity of form and function than animal cells, and have relatively rigid cell walls with cellulose as a major component. Thus connections are required if cell cell contact is to be achieved. These connecting structures are the plasmodesmata, which enable a continuous cytoplasmic link to be formed between cells. The region linking two adjacent cell walls is known as the middle lamella. Fig : Diagram of a cell from a higher plant. Cell sizes vary considerably in plants, but this would be around µm. Distinguishing features of such cells are the cellulose cell wall and the presence of a central vacuole and chloroplasts. (Copyright Philip Harris Education. Reproduced with permission.) Within most plant cells a central vacuole, which looks essentially empty, occupies a large proportion of the cell volume (up to 90% in some cases). The vacuole has a much more important role in the life of the plant cell than its apparent structural simplicity would suggest. The membrane surrounding the vacuole has an important function in controlling the movement of substances into and out of the vacuole, which can act as a storage reservoir for nutrients, waste products, enzymes and other metabolites. The vacuole is also important in maintaining cell water relations and thus cell turgor. BIOLOGY 3
4 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS The cytosol of plant cells contains numerous membrane-bound organelles and other structures that are not present in bacterial cells. The nucleus is of course the most notable, as this characterises the cell as being eukaryotic. The nuclear membrane (or nuclear envelope) is a double membrane structure, with nuclear pores and connections to a network of membrane-bound channels known as the endoplasmic reticulum (ER). This may be either Rough ER (ribosomes associated with the ER membranes) or Smooth ER, which has no ribosomes. The ribosomes function in the synthesis of proteins that are then transported via the ER channels to different parts of the cell. A derivative of the ER system is the Golgi apparatus (sometimes called the dictyosome in plants). This is named after Camillo Golgi, who first described it. The function of the Golgi apparatus is to modify and package materials such as proteins and polysaccharides. All cells carry out various functions that require energy. Energy conversion in eukaryotic cells involves two specialist organelles chloroplasts and mitochondria. Plants (both algae and higher plants) are the major primary producers in ecosystems, and have chloroplasts that localise the reactions of photosynthesis. The chloroplast has a complex arrangement of membranes arranged in stacks called grana. These increase the surface area inside the chloroplast and also provide the membranes on which electron transport proteins are localised. Chloroplast structure can be seen in Fig , which shows an electron micrograph of part of a plant cell. 4 BIOLOGY
5 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Fig : Electron micrograph of part of a plant cell, showing the chloroplast. Features labelled are as follows: A cell wall, B cell (plasma) membrane, C starch grain, D chloroplast stroma with ribosomes, E a granum (stack of thylakoid sacs), F DNA strands, G tonoplast or vacuolar membrane, H vacuole, I rough endoplasmic reticulum, J cytoplasmic ribosomes, K chloroplast envelope, L intercellular space. Photographed at 50,000 magnification. (Copyright A.W. Robards/ Philip Harris Education. Reproduced with permission.) The light reactions of photosynthesis provide energy by the photochemical splitting of water, which is used to drive the synthesis of ATP and NADPH 2. These compounds are then available for the dark (light-independent) reactions, in which carbon dioxide is reduced to carbohydrate. Energy capture in photosynthesis, and the consequent fixing of carbon dioxide into sugar, is the single most important set of reactions in biological systems. Without photosynthesis, there would be no carbohydrate available to be metabolised in respiration, and thus cells could not survive. In aerobic organisms glucose can be completely oxidised to carbon dioxide and water by the processes of glycolysis and the tricarboxylic acid (Krebs) cycle. The major energy yield in this process occurs during the re-oxidation of NADH 2 in mitochondria. These are in some ways similar to chloroplasts in that they have a folded membrane arrangement on which the electron transport proteins are arranged to enable ATP synthesis to be driven by proton pumping mechanisms. BIOLOGY 5
6 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Chloroplasts and mitochondria are essential parts of the biological energy chain, and both illustrate how membranes can be used within cell organelles to increase the area available for the localisation of biochemical reactions. A comparison of the structure of chloroplasts and mitochondria is shown in Fig Fig : Comparison of the internal structure of mitochondria and chloroplasts. The use of membranes to increase the surface area/volume ratio, and the presence of DNA, are common features of these two organelles. Animal cells We have already met most of the components of animal cells, as they are similar to those found in plant cells. There are two notable exceptions animal cells do not have chloroplasts or cell walls. In addition, any vacuoles present are usually very small (and transient) when compared to the plant cell vacuole. Animal cells show a much greater variation in structure and function than plant cells. This variation is however achieved by using the standard set of cell components in different ways, depending on the type of cell. A diagram of an animal cell is shown in Fig In addition to the main components such as the nucleus, Golgi apparatus, endoplasmic reticulum and mitochondria, animal cells may have microvilli on the cell surface. They also have centrioles, which assist in the organisation of spindle fibres during cell division. Some components of animal cells as revealed by electron microscopy are shown in Fig BIOLOGY
7 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Fig : Diagram of an animal cell. This cell would be approximately 20 µm in diameter. Note the lack of a cell wall, vacuole and chloroplasts. (Copyright Philip Harris Education. Reproduced with permission.) Fig : Electron micrograph of cells in rat liver. Features labelled are as follows: A rough endoplasmic reticulum, B mitochondria, C mitochondrial envelope (highlighted for clarity), D nuclear envelope, E nucleus (nucleoplasm), F part of the Golgi apparatus, G cell membrane separating two adjacent cells, H glycogen granules. Photographed at 25,000 magnification. (Copyright A.W. Robards/Philip Harris Education. Reproduced with permission.) BIOLOGY 7
8 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS In looking at plant and animal cells, we have seen that the use of membranes to provide internal structure is a central theme. The intracellular arrangement of membranes is often called the endomembrane system, and can be considered as a way of organising the cytosol so that the complex metabolism required for eukaryotic cell function can be controlled and regulated. In addition to the main subcellular structures already described for plant and animal cells, there are other organelles and structures found in eukaryotic cells. These include a variety of membrane-bound vesicles such as microbodies (also called peroxisomes) and lysosomes. The function of these vesicles is often to isolate specific reactions which might otherwise be harmful to the cell if they were not kept separate from the rest of the cytosol. Other structures of importance are the elements of the cytoskeleton, namely microtubules, intermediate filaments and microfilaments. We will consider the cytoskeleton in more detail elsewhere in this book. The organisation of DNA within the nucleus of a eukaryotic cell is more complex than the naked DNA found in prokaryotes. Eukaryotes have multiple chromosomes, and the packaging problem that is evident even in bacteria is more pronounced. In human cells, some 2m of DNA must be packed into a nucleus that is about 5µm in diameter. This is achieved by coiling the DNA around nucleosomes, which are made up of histone proteins. More extensive coiling of the nucleosome chain is required, particularly during cell division, and this produces the tightly packed structures that we recognise as eukaryotic chromosomes. 1.2 Cell growth and the cell cycle The cell theory states that cells can only arise by the division of existing cells. Cell growth and division is therefore a critical process in the life of cells and organisms. The reproduction of cells, from when the cell is produced by division of the mother cell until the new cell itself divides, is known as the cell cycle. The length of this cycle varies depending on the cell type. Bacterial cells can divide every 20 minutes when growth conditions are favourable, whereas human liver cells only divide about once a year. Mammalian cells in tissue culture have a cell cycle time of about 20 hours; frog embryo cells divide much more frequently with a cycle time of around 30 minutes. Despite the variation in the duration of the cell cycle, there are certain basic requirements for cell division. If two new cells are to be produced, 8 BIOLOGY
9 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS clearly the amount of cell material must double if the daughter cells are to remain the same size as the parent. This is particularly critical for the DNA of the cell, which must be copied exactly if the genetic integrity of the cell is to be maintained. This is achieved by the process of DNA replication, which is an important marker during the cell cycle. How has the cell cycle been studied? If you were to look at a mammalian cell under the microscope for a complete cell cycle, you would see very little evidence of cytological changes until the last hour of the process, when suddenly there is a flurry of activity as the chromosomes move apart and two new cells are formed. Thus early descriptions of the cycle separated the process into two parts, called interphase and division. The division process itself is more accurately called mitosis or the M phase of the cycle. Mitosis refers to the separation of the chromosomes, and this is followed by division of the cell during cytokinesis (CK). Interphase is sometimes called the resting phase, but in biochemical terms this is in fact a very active period of growth. A more accurate description of the cell cycle splits interphase into three parts. The period of DNA replication is known as the synthesis or S phase, and there are gaps known as G 1 and G 2. As our knowledge has increased, it has become clear that gap is perhaps not the best way to describe these two periods, as this suggests little activity, as does the term interphase. A diagram of the cell cycle is shown in Fig Fig : The cell cycle. Interphase is made up of phases G 1, S and G 2. During S phase the DNA is replicated, although no cytological changes are distinguished in the light microscope. The cell grows during interphase and enters the M phase (mitosis). The four sub-stages of M phase are prophase (P), metaphase (M), anaphase (A) and telophase (T). This is followed by cytokinesis (CK) which generates two new daughter cells, each of which enters G 1 to begin the cycle again. For animal cells in tissue culture, cycle duration is around 20 hours. BIOLOGY 9
10 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Mitosis The actual division of the cell requires a series of co-ordinated events that separate the replicated chromosomes and then split the cell into two, each half having received a complete set of chromosomes. After DNA replication the chromosomes are composed of two chromatids, held together at the centromere. Following separation during mitosis, the newly separated sister chromatids are known as daughter chromosomes. Although mitosis is a dynamic process, there are four stages that can be recognised easily in the light microscope. These are prophase, metaphase, anaphase and telophase. Movement of chromosomes is achieved by spindle fibres, which are microtubules. These are composed of alternating dimers of α and β tubulin (a protein). The spindle begins to form at prophase, and is organised by spindle poles or centrosomes at the two poles of the cell. During mitosis the spindle serves to guide the daughter chromosomes and pull them apart to opposite ends of the cell. Fig shows the events of mitosis in onion cells. Fig : Mitosis in onion cells (Allium sp.). Photographs represent cells seen under the light microscope at 100 magnification. The diagram and explanation describe the events in each stage. Due to the structural differences between plant and animal cells, cytokinesis is achieved by different mechanisms in these two cell types. 10 BIOLOGY
11 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS In animal cells, constriction of the cytoplasm by a contractile ring of actin and myosin produces a cleavage furrow, which essentially pinches the cell into two pieces. In plants the cell wall makes this impossible, and cytokinesis is achieved by the formation of a new cell wall to separate the daughter cells. Control of the cell cycle What controls cell division? This is a question that has occupied biologists for many years, and a wide variety of different organisms and cell types has been used to try to answer some of the main questions. Obviously it is important that DNA replication is complete before mitosis, and that the cell mass has increased sufficiently to enable two daughter cells to be formed. Also, the division processes of mitosis and cytokinesis have to be controlled in both temporal and spatial terms if success is to be achieved. In multicellular eukaryotes, additional constraints on cell division are required if cell numbers are to be controlled. To divide, these cells require specific growth factors, of which over 50 types have been isolated. In the absence of enough growth factor, cells stop at the G 1 checkpoint and enter a non-growing phase called G 0. Our current understanding of cell cycle control is that there is a central mechanism that is used to assess the status of the cell as it progresses through the cycle. This mechanism works through a series of three main checkpoints: G 1 checkpoint towards the end of G 1 phase, cell size is assessed. If sufficiently large to allow division, the cell enters S phase and DNA replication begins. G 2 checkpoint the success of DNA replication is monitored, and if all is well entry into mitosis is triggered. M checkpoint this occurs during metaphase and triggers the exit from mitosis and cytokinesis, and entry into the next G 1 phase in the daughter cells. The molecular mechanisms that control the cell cycle involve the interactions of many different genes and proteins to trigger the events of the cell cycle in their proper sequence. For example, a critical part of cell division is the entry into mitosis. This is triggered by a complex called mitosis promoting factor (MPF), which in turn is controlled by other intracellular cell cycle signals. BIOLOGY 11
12 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Abnormal cell division: cancer cells Cancer cells are those in which the normal control of cell proliferation has been lost, enabling these cells to form tumours. Although often not the only defective function, this loss of cell cycle control is a major feature of cancer cells. There are two main classes of genes that can generate abnormal cell division. Proliferation genes encode proteins that promote cell division, and any over-expression of these genes could result in excessive cell division. Genes in this category are called oncogenes when mutated (proto-oncogenes when normal). The second class of cancer-causing genes are known as antiproliferation genes, which normally help to restrict cell division by acting at cell cycle checkpoints. These genes are sometimes called tumour-suppressor genes. In diploid cells, only one copy of a proto-oncogene has to mutate into an oncogene to cause a problem, whereas both copies of a tumour-suppressor gene would have to become abnormal. Thus mutations in proto-oncogenes are dominant, those in tumoursuppressor genes are recessive. Our knowledge of how cancer cells arise has developed greatly over the past few years, and many types of protooncogene and tumour-suppressor gene have been identified. 1.3 Differentiation of cells into tissues and organs Although the cell is essentially a complete living system in its own right, in multicellular organisms cells are organised into tissues and systems for specific functions such as support, movement, nutrition, coordination and control. Much useful information about tissues and systems has been obtained by studying the final product liver, kidney, nerves, glands and so on but the key question is concerned with how these tissues and systems arise in the developing embryo. This is the area of developmental biology. When scientists began to isolate and study genes, a logical step was to try to understand how genes function in the control of developmental processes. Thus the field of developmental genetics has emerged as one of the central areas of modern biology. Despite great advances in our knowledge, development remains one of the most complex and astonishing branches of science. One way of looking at complex processes is to study simple organisms, to see if any functions correlate with those in higher organisms. Although there are not many developmental processes evident in bacteria, some aspects of how genes are expressed have become clearer by looking at adaptive responses such as expression of the lac operon. 12 BIOLOGY
13 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Multicellular plants provide relatively well-ordered systems, with much simpler developmental patterns than animals, yet development in plants is not as well understood as in animals. Development in animals begins with a single fertilised egg undergoing successive divisions. Cell proliferation is of course needed to make up the cell number required as the embryo develops an adult human has more than 1,000,000,000,000 cells! However, all these cells divide mitotically, and so have identical genomes. So how do different cells arise? And how do they end up in the correct place as the embryo grows? Differential gene expression in development To generate different cells from the same genetic information, there must be some sort of control over gene expression. This control can be both temporal (different genes expressed at certain specific times in development) and spatial (cells in different places in the embryo expressing different genes). Development is not a fixed series of genetic events, but rather a conversation between cells as the embryo develops. This conversation involves gene products, which influence cellular events, which in turn create patterns due to cell movement and differentiation. Unravelling all this complexity is an immense task, but it is becoming clear that similar processes operate in all animals. Studies on the fruit fly Drosophila melanogaster have enabled researchers to gain an almost complete understanding of how genes influence development in this organism. We will look at some examples to illustrate. Segmentation in Drosophila The body of a mature Drosophila has 17 segments. These are established early in development by the action of several genes in a hierarchical sequence where the action or effect of one set of genes depends on the effects of genes earlier in the developmental sequence. In Drosophila, the first set of genes produce gradients of concentration of gene product which determine the anterior/posterior and dorsal/ventral axes. The next set of genes respond to these gradients to divide the embryo into four main segments. The action of yet other genes further subdivides the embryo, and then sets up the final segmentation pattern. At this time regulatory (homeotic) genes control structural genes to determine the final fate of each of the segments by specifying the type of appendages and other structures that are specific for each segment. Gene action in Drosophila pattern formation is summarised in Fig BIOLOGY 13
14 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Fig : Differential gene expression during pattern formation in Drosophila. The developing embryo is divided into smaller and smaller segments by the sequential action of different sets of genes. Embryological development in vertebrates In addition to differential gene expression, other factors are important in development of the animal embryo. In vertebrates, the fertilised egg divides to produce a ball of cells called the blastula, which then folds in on itself during the process of gastrulation. This sets up the major body plan of the embryo, and by a conversation process similar to the Drosophila example, cells interact with each other, move and differentiate to give progressively higher degrees of order in the embryo. A schematic representation of this series of events in the mouse is shown in Fig BIOLOGY
15 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Fig : From egg to organism the sequence of events in the development of the mouse embryo. Vertebrate development involves a series of complex interactions involving differential gene expression, cell signalling, and cell movement. Developmental biology is a complex and exciting area of biology, that constantly provides new insights into how genes function to provide form and structure in the developing organism. It is clear that development involves molecular conversations between cells, and that it is therefore a dynamic and interactive process rather than a simple reading of the genetic instructions. 1.4 Cell and tissue culture The ability to grow cells in culture is essential for both pure and applied aspects of biology. The applications of cell culture are many and varied perhaps growing bacterial cells for a basic gene manipulation procedure, culturing mammalian cells for cancer studies, or producing new plants by using tissue culture techniques. Although there are marked differences in the techniques and growth media used with different cell BIOLOGY 15
16 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS types, there are certain things that are required for all cell culture procedures: a source of the cell material either freshly prepared, or from a stock of the cell line or bacterial culture. a suitable container for cell growth a simple flask may be sufficient, or a sophisticated fermenter with computer-controlled monitoring of culture conditions might be needed. a growth medium that provides all the required nutrients. opportunity for gas exchange (chiefly oxygen and carbon dioxide). control of factors such as temperature and ph. a method for measuring cell growth this might involve counting cell numbers in the culture, or measuring the optical density in a spectrophotometer. avoidance of contamination of the culture with unwanted microorganisms. Let s look at some aspects of cell and tissue culture using microorganisms and mammalian cell lines as examples. Micro-organisms Microbes inhabit many diverse ecological niches, and we might therefore expect that they should be adaptable and relatively easy to culture in the laboratory on a small scale (up to a few litres of culture). In addition, many types of microbe are used in the biotechnology industry in processes for the production of useful compounds. Industrial scale operations can involve culture volumes of up to tens or even hundreds of thousands of litres. The term fermentation is often used to describe any micro-organism growth procedure, although technically the term refers to anaerobic growth only. Unicellular algae have few requirements, and can be grown in simple mineral salts media. Bubbling with air (often enriched with carbon dioxide) increases the growth rate and yield of these photoautotrophs. Bacteria and yeasts need more complex media, as they are heterotrophic and therefore need an organic carbon source and other compounds such as amino acids. Micro-organisms can be grown in batch culture, where a culture is grown without dilution until maximum attainable density is reached. Batch cultures under appropriate conditions show a period of exponential growth, but this becomes limited by nutrient availability or cell density effects. Cultures then enter a stationary phase and eventually die if not sub-cultured. 16 BIOLOGY
17 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Exponential growth can be maintained in continuous culture if conditions are altered as growth continues. This can be achieved by continual addition of fresh medium and removal of an equal volume of the culture. Mammalian cell culture Animal cells are fragile and require more carefully controlled conditions than microbial cells if growth is to be maintained. Growth media are more complex, although the basic requirements are similar to those for bacterial cells. The minimum growth medium recipe consists of a balanced salt solution with amino acids, vitamins and glucose. Useful additions are a ph indicator and antibiotics to prevent bacterial growth, but the main additional requirement is for animal serum such as foetal bovine serum (FBS). This is difficult to define chemically, but many of the components appear to be essential for animal cell proliferation. Animal cell technologists have been trying to establish defined media for cell culture, but to date only a few cell types are supported by serumfree media. Growth media containing 5 10% FBS are often used for cell culture. Cells can be obtained by treating tissue samples with a proteolytic enzyme (such as trypsin) to separate cells from each other. This gives a primary cell culture, from which secondary cultures can be derived. However, one drawback is that normal cells only divide a finite number of times before they die, and thus long-term culturing of primary cell cultures is difficult. The most common cell lines used today have either been derived from tumours or have been transformed to produce immortalised cell lines. These cell lines are neoplastic, that is they produce cancers if transplanted into animals. Some common animal cell lines are shown in Table BIOLOGY 17
18 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Table 1.4.1: The origin and properties of some commonly used animal cell lines. Cells generally grow as monolayers in tissue culture flasks. Those that are able to grow in suspension culture conditions are indicated. Cell line Species of Tissue of Cell Growth in origin origin morphology suspension? 3T3 Mouse Connective Fibroblast No CHO Chinese hamster Ovary Epithelial Yes BHK21 Syrian hamster Kidney Fibroblast Yes HeLa Human Cervical Epithelial Yes carcinoma Cells in tissue culture generally grow as a monolayer adhering to the bottom of the plastic flask used as the culture vessel. Animal cells growing in culture are shown in Fig When the cells cover the available surface, they are said to be confluent, and proliferation stops until cells are sub-cultured into fresh medium. Fig : Animal cells growing in tissue culture. The cells form a flat sheet or monolayer on the bottom of the culture flask. One cell is outlined with a white border, showing the boundary of the cell (formed by the cell membrane) and the nucleus. (Courtesy of Dr Dajiang Li.) One of the main advantages of growing cells in culture is that they can be selected and cloned that is, a culture of identical cells can be derived from one isolated cell. This has been useful for the isolation of mutant cell lines, which can be used to understand normal cell growth 18 BIOLOGY
19 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS processes and may also be useful in biotechnological applications. In addition, cell lines can be fused to produce new hybrid cells which often have useful characteristics. 1.5 Plant tissue culture Plant cells can be grown in culture in a similar way to animal cells, although the growth requirements are simpler and defined media can be prepared more easily. One particularly useful trait of plant cells is their ability to regenerate complete plants under appropriate conditions. In theory, all somatic cells from multicellular organisms have this potential, as they have the same genome this is called nuclear totipotency. However, animals are much too complex to regenerate directly from somatic cells, and thus the applications of regeneration have been restricted to plants. Many commercially important ornamental plants are propagated in this way. Tissue culture is also important in the production of pathogen-free plants. Small pieces of plant tissue (explants) can be taken and grown on a medium containing plant growth regulators (plant hormones) such as auxin and cytokinin. Cell proliferation produces an undifferentiated mass of cells known as a callus. By sub-culturing and changing the balance of growth regulators, the callus tissue can be coaxed into differentiating into shoots and roots, and can be planted out to regenerate complete plants. Callus tissue with developing shoots can be seen in Fig Fig : Plant cell culture. This shows cells of potato growing on a petri dish in callus form. The cultures were derived from protoplasts of the potato cells. The formation of shoot tissue from some of the calli can be seen. (Courtesy of Dr Y Hamidoghli) BIOLOGY 19
20 STRUCTURE, FUNCTION AND GROWTH OF PROKARYOTIC AND EUKARYOTIC CELLS Hybrid plant cells can be generated using a technique known as protoplast fusion. A protoplast is a plant cell with the cell wall removed enzymatically. This enables cells to fuse together if conditions are favourable, and thus new combinations of cells can be produced. Protoplasts are also useful for gene manipulation procedures in plants, as the lack of a cell wall means that recombinant DNA can be readily taken up by the cell. Protoplast fusion is shown in Fig Fig : Fusion of protoplasts from two different species of potato. One unfused protoplast (U) is shown, lying on top of another. Compare this with the two heterokaryons shown (H). These are produced when two protoplasts join together during fusion. (Courtesy of Dr Y Hamidoghli) 20 BIOLOGY
21 STRUCTURE AND FUNCTION OF CELL COMPONENTS SECTION 2 Information about the ultrastructure of cell components, obtained by using the electron microscope, has provided much useful detail about how cells are organised. However, to get a full picture of how cells work, it is necessary to examine the biochemistry of the various molecules and structures found inside cells. The development of biochemical and molecular analysis has followed a similar path to that for microscopy, with more detailed information becoming available as more sophisticated techniques were developed. Much of the structural information about biological molecules has come from an understanding of their fundamental chemistry. This approach has been extended by studying how these molecules function in their biological roles, which has enabled the elucidation of the many metabolic pathways that are required for cells to function. Modern developments in molecular genetics can now provide information about how genes work at the molecular level, and thus in many cases the complete picture of how a particular molecular system functions at the cellular level is becoming clear. Many of the important discoveries of recent years in areas such as developmental biology and cancer have come from this approach, which is sometimes called molecular cell biology. In this section we consider the main groups of molecules that are found in cells carbohydrates, lipids, proteins and nucleic acids. We then consider two important cellular systems; cell membranes, and the cytoskeleton. Making and breaking the molecular architecture of cells One central theme runs throughout biochemistry the making and breaking of chemical bonds. Living systems are composed of a limited number of elements, with carbon, hydrogen, nitrogen, oxygen, phosphorus and sulphur (CHNOPS) making up around 99% of their mass. The carbon atom is of central importance, as it can form four covalent bonds with other atoms. This enables a variety of complex molecules to be constructed. In addition to the chemistry of carbon itself, there are many important functional groups associated with biological molecules. Some of these are shown in Fig BIOLOGY 21
22 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig. 2.1: Some functional chemical groups that are important in biological systems. Groups such as the carboxyl, amino and phosphate groups can be ionised, and thus carry a charge at neutral ph. The ionised forms of these groups are shown alongside the nonionised version. Many biologically important molecules are polymers, composed of smaller units called monomers linked together. Two monomers are joined by removing the elements of water. This reaction is a dehydration synthesis (a specific type of condensation reaction) and can be reversed by adding back the elements of water by hydrolysis. This feature of being able to construct and de-construct large molecules or macromolecules is one of the most important aspects of cell metabolism (see Fig. 2.2). Fig. 2.2: The monomer/polymer cycle. Monomeric units can be joined together by dehydration to give polymers. Hydrolysis reverses this and regenerates the monomers. Cyclical polymerisation/depolymerisation like this is important in many cellular processes. 22 BIOLOGY
23 STRUCTURE AND FUNCTION OF CELL COMPONENTS Making and breaking chemical bonds involves energy. A broad generalisation is that making more complex structures from simpler precursors requires energy (anabolic or biosynthetic reactions) whereas breaking bonds releases energy (catabolic reactions). Where there is little overall energy change, reactions are said to be reversible. The metabolism of a cell involves a complex mixture of these three types of reaction, all interacting with and responding to different concentrations of reactants and products, and the whole lot having to be very tightly controlled if energy chaos is to be avoided. The wonder of metabolism is that it works at all! In considering the structure of biological molecules, it is important to remember that these are three-dimensional arrangements of atoms. In fact, all the reactions in cells depend on the shapes of molecules, so this is a critical point. Whilst it is often difficult to represent 3-D structures in 2-D form (i.e. diagrams), there are a number of conventions that can be used. We will come across some of these as we look at the molecules and macromolecules of the cell. 2.1 Carbohydrates The carbohydrates are composed of carbon, hydrogen and oxygen in the ratio 1:2:1, giving a molecular formula of (CH 2 O) n for most simple carbohydrates. These are the monosaccharides or single sugars. There is considerable variation in monosaccharide structure, based on the number of carbon atoms and the arrangement of the hydrogen and oxygen atoms attached to them. We will examine the most common monosaccharide that is of central importance in biological systems glucose. Glucose (C 6 H 12 O 6 ) can be defined by its six carbons (making it a hexose sugar) and by the arrangement of the carbonyl (C=O) group at the terminus of the molecule. A different arrangement of the carbonyl group gives a different spatial arrangement of the atoms of a hexose sugar. All these variations of the same C 6 H 12 O 6 formula, known as isomers, make carbohydrate structure a complex topic. The simplest representation of glucose is the straight-chain form, shown in Fig By convention, if the OH group on carbon 5 (C5) projects to the right, the form is the D-form; if to the left, the L-form. Most sugars used in biological systems are the D-forms. Thus the representation in Fig is designated D-glucose. However, in solution glucose adopts a predominantly cyclical form, where C1 and C5 are linked through the BIOLOGY 23
24 STRUCTURE AND FUNCTION OF CELL COMPONENTS oxygen atom to give a ring structure. Depending on the arrangement of the hydroxyl group on C1, this generates two further variations known as the alpha and beta structures. In α-d-glucose the hydroxyl group attached to C1 is below the plane of the ring, in β-d-glucose the hydroxyl group is above the plane of the ring. In solution the equilibrium proportions of the three forms are approximately 38% α-d-glucose and 62% β-d-glucose, with at any given time only about 0.02% straight-chain form, with some other minor derivatives possible. Fig : The straight-chain form of glucose. The carbon atoms are numbered 1 6, with the carbonyl group at C1. As if trying to make sense of carbohydrate structure wasn t difficult enough, 3-dimensional representations give a better idea of what the molecule actually looks like. Fig (a) shows a full representation of the structure of α-d-glucose, with Fig (b) showing β-d-glucose in the standard shorthand version that is commonly used when drawing carbohydrate structures. 24 BIOLOGY
25 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : Haworth projections of the glucose molecule. (a) shows α-d-glucose with the carbons and hydrogens shown. The thicker lines show the 3-D effect. (b) shows β-d-glucose in the simplified form. Here, the carbons and hydrogens are not shown (unmarked corners represent carbon, unmarked line ends hydrogen). In addition, the 3-D line thickenings are not shown. It is assumed that the ring projects with the bottom edge towards the viewer. The OH groups that define the α and β forms are shaded. The glycosidic bond Two monosaccharides can be linked by a dehydration synthesis to give a disaccharide. These are defined by the component momomers and by the way in which the bond is arranged. If we consider two glucose monomers, the bond will be between the C1 of one molecule and the C4 of the other, giving either an α(1,4) linkage (maltose) or a β(1,4) linkage (cellobiose). These disaccharides are shown in Fig Other disaccharides include common table sugar sucrose (glucose and fructose), and the milk sugar lactose (glucose and galactose). BIOLOGY 25
26 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : Disaccharide structure. Two glucose monomers can be joined by an α(1,4) linkage to give maltose. If the linkage is a β(1,4) arrangement, this give cellobiose The H/OH configuration on the C1 on the right hand side of the diagrams is not specified, as this can exist in either the α or β arrangement. Polysaccharides Joining more monomeric units together produces larger polymers. If the repeating units are the same, we have a homopolymer, whereas different subunits give a heteropolymer. Long chains of simple sugars give the polysaccharides. There are many types of polysaccharide, including starch, glycogen and cellulose. These are all homopolymers made from glucose monomers linked together, but have markedly different properties and functions. This again illustrates how diversity of form and function can be generated by relatively simple variations in the chemistry of the molecules. The structure of these three examples is shown in Fig Fig : Polysaccharide structures found in starch (amylose, helical arrangement), glycogen (branched) and cellulose (parallel chain arrangement). Each of these is composed of a chain (or chains) of glucose monomers. 26 BIOLOGY
27 STRUCTURE AND FUNCTION OF CELL COMPONENTS Glycogen (animals) and starch (plants) are storage polysaccharides, readily hydrolysed to release the monomers for catabolic breakdown to provide energy. Cellulose is a much tougher arrangement of fibres that is ideally suited to its structural role in plants. It is the most abundant organic material on earth, yet most animals lack the enzyme cellulase that is needed to break it down into its component monomers. Other polysaccharides include chitin, a homopolysaccharide found in fungal cell walls and insect exoskeletons, and the glycosaminoglycans, which are heteropolysaccharides found in skin and connective tissues in vertebrates. 2.2 Lipids Lipids are important in cell membrane structure, and also as hormones and energy storage molecules. The common defining feature of lipids is that they are insoluble in water. Fats and oils are familiar lipids that we use every day, the distinction being a rather arbitrary one of the physical state of the molecule at room temperature. Although lipids are certainly smaller molecules than the large polysaccharides, proteins and nucleic acids, they are generally classed as one of the four groups of macromolecules. Three types of lipid are of particular importance in cells: triacylglycerols (or triglycerides), phospholipids and steroids. Triacylglycerols The constituents of triacylglycerols are a glycerol backbone and fatty acids. Glycerol is a 3-carbon alcohol; fatty acids are hydrocarbon chains ending with a carboxyl group. If all the available bonds are occupied by hydrogens, the fatty acid is said to be saturated. If there are carboncarbon double bonds in the molecule, this gives an unsaturated fatty acid. One structural consequence of this is that saturated fats pack closely together and tend to be solid, whereas in unsaturated fats kinks are introduced and the fatty acid chains do not fit together closely. This generally means that unsaturated fats are oils rather than hard fats. Most animal fats are saturated, those from plants tend to be unsaturated. Some common fatty acids are shown in Table BIOLOGY 27
28 STRUCTURE AND FUNCTION OF CELL COMPONENTS Table 2.2.1: Some common fatty acids found in lipids. Saturated fatty acids have no C=C double bonds. Oleic acid has one C=C double bond, linoleic acid has 3, and is therefore polyunsaturated. Fatty acid No. of Saturated/ Structure carbons unsaturated palmitic acid 16 saturated CH 3 (CH 2 ) 14 COOH stearic acid 18 saturated CH 3 (CH 2 ) 16 COOH oleic acid 18 unsaturated CH 3 (CH 2 ) 7 CH=CH(CH 2 ) 7 COOH linoleic acid 18 polyunsaturated CH 3 (CH 2 CH=CH) 3 (CH 2 ) 7 COOH Glycerol and fatty acids are joined together by dehydration (condensation) synthesis reactions between the hydroxyl and carboxyl groups, generating a triacylglycerol (triglyceride). This is shown in Fig Fig : Triglyceride structure. The glycerol molecule acts as the backbone to which three fatty acids are attached by ester linkages. This gives a triacylglycerol or triglyceride. The properties of triacylglycerols are determined by the properties of the fatty acids Phospholipids An important variant of the triacylglycerol structure is where one of the fatty acids is replaced by a phosphate group, which often has other groups attached. Usually one fatty acid is saturated, and one is unsaturated. The most abundant phospholipid in animal tisue is phosphatidylcholine (Fig ). 28 BIOLOGY
29 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : Phosphatidylcholine. This phospholipid has two fatty acid tails, one saturated (S) and one unsaturated (US). The polar head region is a choline/phosphate group linked to the glycerol backbone. As phospholipids have a non-polar fatty acid tail and a polar head, they are hydrophilic (water-loving) at the head and hydrophobic (water-hating) at the tail. This is a critical property in that it enables phospholipids to form bilayers, as shown in Fig As we shall see later, this is important in membrane structure. Fig : Phospholipids can form bilayers, with the polar heads on the outer (hydrophilic) surface, and the fatty acid tails forming a hydrophobic inner region. Steroids These lipids have a markedly different structure to that found in the glycerol-based triglycerides and phospholipids. Steroids are based on a four-ring structure, with associated side chain variations. The best known example of a steroid is cholesterol, which is found in cell membranes. Other steroids such as testosterone are hormones. The structure of cholesterol is shown in Fig BIOLOGY 29
30 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : The structure of cholesterol is based on a four-ring structure. Not all the atoms are labelled. Testosterone and other steroid hormones are similar, with different attached groups defining their chemical properties. 2.3 Proteins Protein molecules are heteropolymers made up of amino acids, of which there are 20 that are used in proteins. Variation in the length of the amino acid chain, and the order of the individual amino acids in it, theoretically enables an essentially unlimited variety of proteins to be constructed. This makes proteins the most diverse group of macromolecules in the cell, with many different roles to play in both structural and functional terms. Amino acid structure Amino acids are characterised by the amino (HN 2 ) group and the carboxylic acid (COOH) group. These are attached to the central or alpha carbon atom, which also carries a hydrogen atom and the variable part of the molecule (the R-group). Like carbohydrates, amino acids show isomerism, existing in both the D- and L-forms. The L-form is found exclusively in proteins. At neutral ph amino acids exist in the ionised form, although the charges on the amino and carboxylic acid groups disappear when the monomers are joined together. The simplest amino acid is glycine, which has a hydrogen atom as the R-group. A methyl (CH 3 ) group gives alanine, shown in Fig Fig : Amino acid structure. Amino acids have a hydrogen, and amino group and a carboxyl group attached to the central α-carbon atom. The fourth position is a variable side-chain or R-group. In this example the R-group is a methyl group, giving the amino acid alanine. 30 BIOLOGY
31 STRUCTURE AND FUNCTION OF CELL COMPONENTS As the variable part of amino acid structure, it is the R-group that gives each amino acid unique chemical properties. Interactions between R- groups also specify the particular shape that the protein has, and determine its overall properties. R-groups can be classed as acidic, basic, uncharged polar, and nonpolar. Some examples of the types of side chain found in these classes are shown in Table Table 2.3.2: Types of R-group found in amino acids. The number of amino acids (total 20) in each class is shown. The three-letter and single-letter abbreviations are shown as Asp / D etc. Class/No. amino acid abbreviation R-group amino acids Acidic aspartic acid Asp / D -CH 2 COOH (ionises to COO - ) (2) glutamic acid Glu / E -CH 2 CH 2 COOH (ionises to COO - ) Basic lysine Lys / K + -(CH 2 ) 4 NH 2 (ionises to NH 3 ) (3) Uncharged serine Ser / S -CH 2 OH polar (5) asparagine Asn / N -CH 2 C=O NH 2 (uncharged but polar) Nonpolar glycine Gly / G -H (10) alanine Ala / A -CH 3 cysteine Cys / C -CH 2 SH The peptide bond Proteins are made by joining amino acids together by an amide linkage called the peptide bond. A chain of amino acids is therefore known as a polypeptide. The peptide bond is formed by a dehydration synthesis reaction between the carboxylic acid group of one amino acid and the amino group of the next, as shown in Fig Amino acids joined together in this way are called residues. Although the peptide bond itself is a planar (flat) structure which does not allow rotation around BIOLOGY 31
32 STRUCTURE AND FUNCTION OF CELL COMPONENTS the C-N bond, the single bonds on either side of the peptide bond do allow rotation of the residues, which makes polypeptide chains very flexible structures. This is important in determining the way in which chains of amino acids can fold to generate the highly ordered structures found in complex proteins. Fig : The peptide bond links amino acids together by a dehydration synthesis between the carboxyl and amino groups of two different amino acid monomers. This gives the C-N linkage that is characteristic of the planar (flat) peptide bond. Levels of protein structure No group of biochemical molecules illustrates the concept of emergent properties better than the proteins. From the constituent atoms of the amino acids up to the final form of a large protein macromolecule, progressively more complex structural organisation enables specific form and function to be generated. To make sense of this complexity, protein chemists recognise four levels of protein structure, beginning with the sequence of amino acids in the polypeptide chain. Chemical bonding is obviously critical in determining protein shape, and different types of bonds are important at different levels. The covalent peptide bond that links amino acid residues together is a very strong bond. In higher-order protein structure weaker interactions are important. These involve non-covalent bonds such as hydrogen bonds and ionic bonds, van der Waals attractions, and hydrophobic interactions where the hydrophobic parts of R-groups tend to associate together. 32 BIOLOGY
33 STRUCTURE AND FUNCTION OF CELL COMPONENTS Primary (1 ) structure The primary structure of a protein is the sequence of amino acid residues in the polypeptide chain. The chain of amino acids has an amino terminus at one end and a carboxyl terminus at the other, reflecting the structure of the amino acid molecules. In writing out primary structures of proteins, the convention is to write from the amino terminus (N-terminus) on the left to the carboxyl terminus (Cterminus) on the right. In some cases the structure of the amino acid residues can be shown, although mostly the abbreviated names for the amino acids (either the three-letter abbreviations or the single-letter designations) are used. Representations of primary structure are shown in Fig Fig : Primary structure of a polypeptide. This could be written out in full, but is usually abbreviated to save space. In (a) a diagrammatic representation of the polypeptide is shown, with the N- and C-termini labelled. (b) shows the three-letter abbreviations for a short peptide sequence, and (c) shows the single-letter abbreviations for the same sequence. The N- and C-terminus labels may be omitted; if so the convention is that the N-terminus is on the left. Secondary (2 ) structure There are two main types of secondary structure. These are the α-helix and the β-sheet arrangements, which are generated from interactions between the atoms of the amino acid residues in the polypeptide chain. In the α-helix, as shown in Fig , the polypeptide chain is coiled into a right-handed helix by hydrogen bonds between the N-H group of a peptide bond and the C=O group of the peptide bond four residues away from it. In the β-sheet configuration, polypeptide chains are linked together in a side-by-side configuration, again by hydrogen bonding. Beta sheets can be either parallel or antiparallel, depending on the orientation of the constituent parts of the arrangement. The two types of β-sheet are shown in Fig BIOLOGY 33
34 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : The α-helix. This secondary structure is a right-handed helix with four residues per turn. Hydrogen bonds between oxygen and hydrogen atoms stabilise the helix. Not all H-bonds are shown. Fig : The β-sheet arrangement of secondary structure can be either parallel or antiparallel. In the parallel arrangement the polypeptide chains run in the same direction with respect to their N/C polarity. In the antiparallel arrangement, the chains run in opposing directions. Hydrogen bonds are shown by dotted lines. 34 BIOLOGY
35 STRUCTURE AND FUNCTION OF CELL COMPONENTS Tertiary (3 ) structure The α-helix and β-sheet arrangements are relatively simple ways to organise stretches of polypeptide chain. More complex structures are found when we look at the tertiary structure of proteins, which describes the way in which the polypeptide folds to give the final protein structure. Given that a polypeptide may be several hundred amino acid residues in length, and that both α-helix and β-sheet arrangements may be found in the same protein, it is easy to see that predicting and analysing protein structure is a difficult task. Modern computer graphics programmes enable scientists to look at protein structure in ways that were not possible a few years ago, and many new insights have come from computer modelling coupled with experimental techniques such as examining protein crystals by X-ray crystallography and nuclear magnetic resonance (NMR). The tertiary structure of a protein is determined largely by hydrophobic interactions, which place the non-polar R-groups in the centre of the molecule. In many proteins, an additional important type of covalent bond is the disulphide bond, which can form between the sulphydryl (SH) groups on cysteine residues in different parts of the polypeptide (or between cysteine residued in two different polypeptides). Within any tertiary structure, parts of the amino acid sequence may adopt an α- helix arrangement, others may be β-sheets or more complex arrangements of the β-sheet structure. A representation of the tertiary structure of myoglobin is shown in Fig BIOLOGY 35
36 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : Tertiary structure of myoglobin. This is a single polypeptide chain with several α-helices linked by non-helical regions. Folding of the protein produces a hydrophobic pocket for the haeme group, which contains the iron atom involved in binding oxygen. This representation is a ribbon plot generated by the widely used RasMol computer software package (copyright Roger Sayle, public domain). This programme enables the structure to be viewed in many different ways such as balland-stick or spacefilled, and also permits rotation of the structure to enable viewing from different angles. Software such as this is of great benefit to protein chemists. Folded proteins may have other non-protein groups associated with them. These are called prosthetic groups, and are often essential for the biological activity of the protein. Examples include the ironcontaining haeme group of myoglobin (see Fig ) and haemoglobin. Proteins may also have carbohydrate, lipid or nucleic acid groups associated with them; these conjugated proteins are known as glycoproteins, lipoproteins and nucleoproteins, respectively. The fact that proteins are relatively stable structures in the cellular environment is remarkable in that the weak forces that hold the structure together can be disrupted easily if the chemical environment changes, or if the sequence of the amino acids is altered. Proteins fold to give a structure with the lowest free energy, therefore each polypeptide chain will have its own preferred conformation. Thus, although in theory there are essentially unlimited possible protein structures, only a small number of these possibilities will fold to give a single stable conformation, which has enabled the evolution of protein structures that are stable and uniquely suited for a particular purpose. 36 BIOLOGY
37 STRUCTURE AND FUNCTION OF CELL COMPONENTS Quaternary (4 ) structure Many proteins are made up of a single polypeptide chain, which folds to a particular tertiary structure unique to that protein. Other proteins are composed of two or more polypeptide subunits, each of which has its own specific conformation. The organisation of the subunits in a multisubunit protein is known as the quaternary structure of the protein. A good example is the tetrameric protein haemoglobin, composed of two α- and two β-globin subunits, each with its own prosthetic haeme group. Protein motifs and domains As our knowledge of protein structure has improved, two additional elements have been recognised within the traditional 1 /2 /3 /4 classification. These are motifs and domains. Motifs are elements of secondary structure that form in particular ways. Examples include the β-α-β motif, which gives a fold or crease in the protein, and the β-barrel, which forms a tube-like arrangement within the protein. Domains are regions of the polypeptide chain that fold independently to give structurally distinct regions of tertiary structure, often with different roles to play in the complete protein. Function of proteins As might be expected from their structural complexity and diversity, proteins have an equally wide range of different roles in the cell. One common way of describing proteins is to group them as either fibrous (structural) proteins, or globular (functional) proteins. However, this is a rather simplistic classification, and it is better to classify proteins using a more specific system that takes into account the many different and highly specific roles that they carry out. Thus we have proteins that act as enzymes, those that are found as structural components of cells and tissues, receptor and signalling proteins, and many others. Some examples of the types of function that proteins carry out in the cell are shown in Table BIOLOGY 37
38 STRUCTURE AND FUNCTION OF CELL COMPONENTS Table 2.3.8: Some classes and functions of proteins in the cell. Class of Function Examples protein Enzyme Catalysis (breaking & Thousands of examples! Usually forming of covalent enzyme names end in -ase. Groups bonds). include proteases, lipases, polymerases, kinases, phosphatases, isomerases, dehydrogenases, etc. Structural Provide support to cells Collagen, elastin, tubulin, keratin, and tissues. actin. Receptor Detection and Insulin receptor, acetylcholine transmission of signals. receptor, rhodopsin. Signal Intercellular signalling. Insulin, other hormones & growth factors. Transport Carries small molecules In the bloodstream, haemoglobin or ions in bloodstream carries oxygen, serum albumin carries or in membranes. lipids, transferrin carries iron. Many transmembrane proteins act as pumps for transporting small molecules or ions (protons, calcium, glucose) across the membrane. Motor Generates movement. Myosin in muscle is the most obvious example, also dynein in cilia and flagella. Kinesin interacts with microtubules to move organelles. Storage Stores small molecules Ferretin stores iron in the liver. or ions. Ovalbumin (egg white) and casein (milk) are storage proteins. Regulatory Binds to DNA to regulate The lac repressor binding to the gene activity. operator of the lac operon to switch it off. Special Varied range of specialist Glue proteins to attach mussels to purpose functions. rocks, antifreeze proteins of Arctic and Antarctic fish, plus many other specialised examples. 38 BIOLOGY
39 STRUCTURE AND FUNCTION OF CELL COMPONENTS 2.4 Nucleic acids The information-carrying molecules of the cell are the nucleic acids DNA (deoxyribonucleic acid) and RNA (ribonucleic acid). When compared to the complexity of proteins, nucleic acids have a relatively simple chemical structure that is based on a sugar-phosphate backbone. The variable information-coding part of nucleic acids is made up of a set of four nitrogenous bases, which arrange themselves in pairs. One of the most fascinating aspects of molecular biology is how this very simple coding system enables the great diversity of protein molecules to be constructed. As we shall see, the answer is yet again another elegant example of one of our main themes that of emergent properties. Nucleotide structure The monomeric units of nucleic acids are the nucleotides. These are composed of a pentose sugar (ribose in RNA, deoxyribose in DNA) and a phosphate (PO 4 ) group that make up the constant structural part of the molecule, and a variable nitrogenous base. The bases are either purines (double-ring structures) or pyrimidines (single-ring structures). This difference has important consequences for the way in which the bases join together in the DNA molecule. The purine bases are adenine and guanine (A and G, found in DNA and RNA). The pyrimidines are cytosine (C, DNA and RNA), thymine (T, DNA only) and uracil (U, found in RNA instead of thymine). Nucleotide structure is shown in Fig Fig : Nucleotide structure. This is based around a 5-carbon sugar (shaded) with a phosphate and a nitrogenous base attached to the C5 and C1 positions respectively. In RNA the sugar is ribose, with an OH group at position X on C2. In DNA the sugar is deoxyribose, with X being a hydrogen. BIOLOGY 39
40 STRUCTURE AND FUNCTION OF CELL COMPONENTS The phosphodiester bond As with the other macromolecules, a dehydration synthesis reaction generates the bond that joins two nucleotides together. This phosphodiester bond is formed between the phosphate group of one nucleotide and a hydroxyl group on the sugar of the next nucleotide, as shown in Fig Fig : Nucleotides are joined together by a phosphodiester linkage between the C5 phosphate and the hydroxyl group on C3 (shaded area). This gives polynucleotides a defined polarity reflecting that of the component nucleotides. Polynucleotides and nucleic acid function Polynucleotide chains provide the structural and functional basis for the encoding and decoding of genetic information. The sugar/phosphate backbone of a polynucleotide carries the sequence of bases that makes up the genetic code as a series of triplet codons. The functional basis of the code is that of base-base recognition. Bases can fit together and join by hydrogen bonding, A with T (or U) and G with C, as shown in Fig Bases that fit together in this way are said to be complementary. 40 BIOLOGY
41 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : Base pairing between purines and pyrimidines in the DNA double helix. To fit the helix, one purine (A or G) must pair with one pyrimidine (T or C). This gives a stable arrangement of A:T and G:C base pairs, with two hydrogen bonds in an A:T bp and three in a G:C bp. Not all the atoms are shown or labelled in this representation. Whilst it may be rather too simplistic to say that one aspect of biology is more important than any other, without base pairing there could be no storage, replication, encoding or decoding of the information needed to make all the components and systems found in living organisms. Thus the A:T and G:C base pairs found in DNA have a significance far greater than that which their relatively simple chemistry would suggest. The DNA molecule is a double-stranded helix, the structure of which was proposed by James Watson and Francis Crick in This double helix (not to be confused with the α-helix of proteins!) is undoubtedly the biological structure that is most widely recognised by non-scientists. As with all the molecules and macromolecules that we have considered, it is an elegant example of the interdependence of structure and function. The double helix has two polynucleotide chains that run in different directions (known as an antiparallel arrangement). The bases fit across the centre of the right-handed helix, with one purine pairing with its complementary pyrimidine. The significance of the sizes of the purines and pyrimidines can now be appreciated, as the helix can accommodate only a purine:pyrimidine base pair if the hydrogen bonds are to be stable. The double helical arrangement of DNA is shown in Fig BIOLOGY 41
42 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : The double helix of DNA. Two polynucleotide chains are twisted into a right-handed helix. The chains are linked together by A:T and G:C base pairs. The chains are in an antiparallel arrangement (polarity indicated by arrows). The pitch of the helix is 3.4nm, diameter 2nm. The arrangement produces grooves on the surface of the helix. (Copyright D S T Nicholl/Cambridge University Press. Reproduced with permission.) The structure of RNA differs from that of DNA in a number of ways. The sugar in RNA is ribose, and the base uracil replaces thymine. Also, RNA molecules are generally single-stranded, as in messenger RNA (mrna), although they do fold to give the 3-D conformations seen in ribosomal RNA (rrna) and transfer RNA (trna). Complementary base pairing holds the key to the copying of genetic information in the processes of DNA replication and transcription, which are carried out by polymerase enzymes. The replication of DNA, where each DNA strand is used as a template for the synthesis of a complementary polynucleotide, is a critical part of the cell cycle. Replication enables a complete copy of the genome to be passed on to each daughter cell during mitosis. Transcription of genetic information (into RNA from a DNA template) provides the mechanism for the expression of genes. In addition to polymerases, many other types of enzyme are associated with nucleic acid biochemistry, both in the cell and as part of the genetic engineer s toolkit that makes it possible to manipulate DNA in the test tube. One example is DNA ligase, which forms phosphodiester bonds and can be used to join DNA molecules together to create recombinant DNA (rdna). 42 BIOLOGY
43 STRUCTURE AND FUNCTION OF CELL COMPONENTS 2.5 Cell membranes The cell membrane or plasma membrane is of fundamental importance, as it represents the barrier that separates the cell contents from the extracellular environment. Also, as we have already seen, eukaryotic cells use membranes to generate compartments within the cell, each with a specialised function. In discussing membranes we will concentrate on the plasma membrane (PM), although many of the features of the PM are shared by the other membranes in the cell. We can recognise several important functions of membranes, as follows: Providing selectively permeable barriers the PM prevents unrestricted movement of materials by acting as a barrier. However, it also enables substances to enter and leave the cell by being selectively permeable, a feature that is also critical for internal membrane function. Compartmentalisation membranes are used extensively in eukaryotic cells to form structures such as the nuclear envelope, endoplasmic reticulum, Golgi apparatus, mitochondria and chloroplasts. Localising reactions in the cell membranes provide the structural framework for organising many of the reactions in the cell as a consequence of compartmentalisation. Also, critical energytransducing mechanisms such as the light reactions of photosynthesis and the respiratory electron transport chain are closely associated with membranes. Transport of solutes in addition to their selective permeability, membranes have the machinery necessary for transporting solutes specifically across the membrane, often against a concentration gradient. Signal transduction receptors on the membrane surface recognise and respond to different stimulating molecules, enabling specific responses to be generated within the cell. Cell-cell recognition the external surface of the membrane is important in that it represents the cell s biochemical personality. In multicellular organisms this feature enables cells to recognise each other as similar or different, which is necessary for the correct association of cells during development. BIOLOGY 43
44 STRUCTURE AND FUNCTION OF CELL COMPONENTS Membrane structure The basic structural unit of membranes is the phospholipid bilayer, as outlined in Fig The proposal that this was the basis of membrane structure was put forward in 1925, following a simple calculation about the surface area (SA) covered by the lipids extracted from red blood cells. It was found that this was roughly twice that of the SA of the cell, and it therefore seemed likely that the lipids formed a double layer surrounding the cell. The currently accepted model for membrane structure is based on that of Singer and Nicolson, who proposed the fluid mosaic model in In this structure the phospholipid bilayer has proteins embedded within it (intrinsic or transmembrane proteins) or associated with its surface (extrinsic or peripheral proteins). Other components include cholesterol, glycoproteins and glycolipids. The membrane is not a static or rigid structure, but is a dynamic arrangement of lipids and proteins. Thus the components of membranes are able to drift laterally within the membrane, and therefore the term fluid mosaic is an accurate description of the nature of the membrane. The fluid mosaic model is shown in Fig Fig : The fluid mosaic model of membrane structure. The phospholipid bilayer contains partially- and fully-embedded proteins, cholesterol, glycolipids and glycoproteins. The structure is a dynamic or fluid structure, in which the proteins and other components are in a state of continuous lateral drift or motion. 44 BIOLOGY
45 STRUCTURE AND FUNCTION OF CELL COMPONENTS Function of membrane proteins Proteins make up around 50% of the mass of the PM, and can be classified into different groups according to either their arrangement in the membrane and/or their function. As shown in Fig , proteins may be embedded in the lipid bilayer or attached to the surface. The embedded (intrinsic) proteins may be transmembrane proteins or they may be linked to the lipids in one side of the bilayer only. The extrinsic or peripheral proteins are loosely attached to the membrane by noncovalent association with other proteins. When classifying membrane proteins according to function, several types can be recognised. Transport proteins, as the name suggests, are involved in transporting non-diffusible substances across the membrane. Transport proteins may be either channel proteins or carrier proteins. Channel proteins provide a pore across the membrane, through which substances (usually small charged molecules) can diffuse. Carrier proteins are more specific, with binding sites for one type of solute only. Both channel and carrier transport proteins can permit passive transport (with the concentration gradient, sometimes called facilitated diffusion). To transport molecules against the concentration gradient, special types of carrier proteins are needed that can harness energy to drive the transport process during active transport. In addition to transport functions, membrane proteins are important as enzymes and receptors. They have a role to play in cell adhesion and cell-cell recognition (glycoproteins; glycolipids are also important) and provide a structural support to the cell as part of the membrane-linked cytoskeleton. Membrane protein function is shown in Fig BIOLOGY 45
46 STRUCTURE AND FUNCTION OF CELL COMPONENTS Fig : Function of membrane proteins. (a) Channel protein. (b) Carrier protein, shown here as an active transport protein which requires ATP hydrolysis to transport molecules against the concentration gradient. (c) Membrane-bound enzyme, converting substrate X to product Y. (d) Receptor protein, which generates an intracellular response (R) to an extracellular signal (S). (e) Cell adhesion protein, linking two cells together. (f) Cell:cell recognition via a glycoprotein. (g) A membrane protein attached to the cytoskeleton. Sometimes a single protein may perform more than one of these functions. 2.6 The cytoskeleton The eukaryotic cell has an intricate network of thread-like filaments called the cytoskeleton, which supports the interior of the cell and the organelles within it. The cytoskeleton is made up of three components. In order of increasing diameter these are actin filaments (also known as microfilaments), intermediate filaments and microtubules. Microfilaments are composed of the protein actin, arranged as two strands of protein molecules twisted together to give a rope-like structure about 7nm in diameter. These are present throughout the cell, but are most highly concentrated just inside the plasma membrane. Microfilaments are important in cell movements. 46 BIOLOGY
47 STRUCTURE AND FUNCTION OF CELL COMPONENTS Intermediate filaments are about 10nm in diameter and are composed of tough fibrous protein strands twisted together. This means that they are very stable structures in the cell, and are particularly important for providing mechanical strength to animal cells, which lack the strong cell walls of plants. They may be anchored to the cell membrane to provide support. Microtubules are hollow tubes made up of the protein tubulin. Heterodimers (one α- and one β-tubulin subunit per dimer) are arranged as a set of 13 protofilaments to generate the final microtubule, which is a relatively rigid structure. Microtubules are important in cell division as part of the spindle fibre network, and are also involved in the movement of components within the cell. Microfilaments and microtubules can de-polymerise and re-polymerise very easily. Thus the cytoskeleton is not a static structure, but a dynamic one, which is continually changing to provide the basis of support and movement in the cell. The control of microtubule polymerisation and depolymerisation in animal cells is controlled by the centrosome or microtubule organiser. In interphase, the centrosome serves as the site for the production of cytoplasmic microtubules that make up part of the cytoskeleton. During mitosis, the microtubules are re-deployed as the spindle fibres that separate the chromatids to generate the daughter nuclei. BIOLOGY 47
48 48 BIOLOGY
49 SECTION 3 MOLECULAR INTERACTIONS IN CELL EVENTS In the previous sections we have looked at how cells are put together, concentrating mainly on the structure of components such as molecules and macromolecules, membranes and organelles. In this section we probe a little deeper into some examples of how the cell actually works as a molecular machine, by examining the interactions that make the link between structure and function. We will consider three examples: catalysis by enzymes, the sodium-potassium pump, and cell signalling. 3.1 Catalysis Enzymes are the biological catalysts of the cell. They increase the rate at which reactions happen, often by millions of times. Only by having this set of very specific catalysts can the cell carry out all the reactions that are necessary for metabolic processes to work effectively. Some of the types of enzymes were listed in Table 2.3.8, and it is now useful to describe a few of these in more detail. Enzymes that break things apart include: hydrolases these enzymes catalyse a hydrolytic cleavage reaction. phosphatases remove a phosphate from a molecule by a hydrolytic cleavage. proteases hydrolyse peptide bonds to break down proteins into amino acids. nucleases hydrolyse phosphodiester bonds to break down nucleic acids into nucleotides. ATPases hydrolyse ATP. Many proteins have an ATPase activity which is essential for their function. Enzymes that join things together include: kinases catalyse the transfer of a phosphate group onto a molecule such as a carbohydrate or a protein. synthases these enzymes join two molecules together by catalysing a dehydration synthesis reaction. BIOLOGY 49
50 MOLECULAR INTERACTIONS IN CELL EVENTS polymerases catalyse polymerisation reactions where molecules are added sequentially to a chain, as in DNA and RNA synthesis. Other types of enzyme are: oxidoreductases catalyse electron transfer oxidation/reduction reactions. Include the oxidases, reductases and dehydrogenases (found in, for example, respiration and photosynthesis). isomerases change the form of a molecule without altering its overall chemical composition. Most enzymes are now named according to their substrate and their function, although a few are named inconsistently due to historical reasons. Enzymes are also given a standard reference number (the EC number) to help characterise the 1,500 or so that are now known in detail. Examples of enzyme names are glucose oxidase, glyceraldehyde 3-phosphate dehydrogenase, citrate synthase, phosphofructokinase, and (the most important one on earth!) ribulose bis-phosphate carboxylase. Form and function Enzymes work by bringing the substrate(s) of a reaction close together in the active site region so that bond breakage or formation may occur at the atomic level. This is often facilitated by specific chemical effects such as the transfer of protons or the alteration of charge distribution around the target atoms. As might be expected, the substrate and enzyme must fit together very precisely. We can see this most easily by looking at an enzyme that has a large substrate (i.e. of a similar size to the enzyme itself). This is shown in Fig in this case, a restriction endonuclease and its substrate, which is a defined sequence in the DNA molecule. 50 BIOLOGY
51 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : The interaction of the enzyme BamHI (a restriction endonuclease) with its DNA substrate. The enzyme is shown as a RasMol ribbon plot with the DNA helix in black. In (a) the view is from the side, in (b) it is along the axis of the DNA helix. The way in which the enzyme wraps around the DNA is shown clearly. The catalytic cycle As enzymes are catalysts, they remain unchanged at the end of a reaction, and are available for the next interaction with substrate. This catalytic cycle can be represented as shown in Fig This shows the enzyme sucrase, which catalyses the hydrolysis of sucrose into its component monosaccharides, glucose and fructose. At the start of the cycle, enzyme (E) and substrate (S) are available. The molecular interaction of enzyme and substrate at the active site forms the enzyme:substrate (ES) complex. Catalysis occurs, forming the enzyme:product complex (not shown in the diagram) and the products are released, which frees the enzyme for the next substrate molecule. Fig : The catalytic cycle of sucrase. The enzyme hydrolyses the disaccharide sucrose, releasing glucose (G) and fructose (F). The enzyme remains unchanged at the end of the reaction cycle. BIOLOGY 51
52 MOLECULAR INTERACTIONS IN CELL EVENTS A common model for enzyme action is the lock and key model, which is often represented as shown in Fig However, this model is a little misleading in that it tends to give the impression that enzymes are rigid structures, whereas in fact they are quite flexible and can alter their conformation in response to the binding of other molecules. The currently accepted model for enzyme action is the induced fit model, in which conformational changes to the protein occur on binding of the substrate. We will look at the action of the enzyme hexokinase to illustrate the induced fit model. Hexokinase catalyses the transfer of a phosphate from ATP onto glucose. The structure of the enzyme is shown in Fig The active site and the two domains of the single polypeptide chain are clearly visible in this view of the backbone of the molecule. You might think of the protein about to close round the substrate in the active site in a similar way to your hand closing round a door handle. The effect of this is that glucose fits the active site more closely, and the binding of ATP is also enhanced. The catalytic cycle of hexokinase is shown in Fig It may help to think of the protein flexing during the binding of substrate. Fig : The structure of hexokinase. This is a RasMol plot of the α- carbon/peptide bond backbone (repeated C-C-N-C sequences) of the polypeptide. The protein is a single polypeptide chain, with two distinct domains that create the pocket where the active site is found. The domains are shown by the dotted line. 52 BIOLOGY
53 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : The catalytic cycle of hexokinase. (a) Enzyme and substrate are apart. (b) Substrate binds to active site region. (c) A conformational change in the protein closes the active site around the substrate (induced fit). Binding of ATP is enhanced, catalysis occurs and the phosphate is transferred to glucose. (d) Enzyme plus products. Control of enzyme activity The activity of enzymes must be regulated in some way if metabolic chaos is to be avoided. Regulation can be achieved by a number of different mechanisms. A major influence is the number of enzyme molecules in the cell, which is controlled at the level of gene expression. Compartmentalisation also enables the cell to keep sets of enzymes together and away from other enzymes, and temperature and ph also affect enzyme activity. Many enzymes also require cofactors to function. However, the most effective way of enabling a fine control of enzyme activity is to alter the shape of the enzyme itself, and thus cause a change in its catalytic efficiency. Examples of this type of metabolic control include inhibitors, allosteric effects, covalent modification, and end-product inhibition. Inhibitors can be either competitive or non-competitive. As the name suggests, competitive inhibitors compete for the active site, thus reducing the effectiveness of the enzyme. Competitive inhibitors are usually similar in structure to the substrate, and the enzyme is fooled into accepting the inhibitor, which blocks the active site. Noncompetitive inhibitors bind at a different location and change the BIOLOGY 53
54 MOLECULAR INTERACTIONS IN CELL EVENTS conformation of the protein, altering the active site and again reducing catalytic efficiency. Inhibition can be either reversible or nonreversible, depending on how the inhibitor binds to the enzyme. A diagram of enzyme inhibition is shown in Fig Fig : Enzyme inhibition. (a) shows competitive inhibition, with the inhibitor occupying the active site. In (b) non-competitive inhibition is shown, with the inhibitor binding to a site that is separate from the active site region. Allosteric enzymes are those that change form in response to binding of a regulating molecule (often called a modulator or effector). Allosteric modulators can be either positive or negative effectors of enzyme activity. They function by binding to allosteric sites that are distinct from the active site of the enzyme. As shown in Fig , noncompetitive inhibition is a form of allosteric regulation. In multi-subunit enzymes, the structure is more complex, and the enzyme often exists in two different conformational states (active and inactive). These can be stabilised by binding the modulator; positive modulators stabilise the active form of the enzyme, whilst negative modulators stabilise the inactive form. This is shown in Fig In addition to positive and negative modulators changing the activity of an allosteric enzyme, sometimes binding of the substrate itself to one active site enhances binding at the other active sites. This is known as cooperativity. 54 BIOLOGY
55 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : Allosteric effects shown for an enzyme with four subunits. There is an equilibrium between active and inactive forms of the enzyme (a), with the active form able to bind the substrate. In (b) the effect of binding a positive modulator is shown. This binds to an allosteric site and stabilises the active form of the enzyme. The effect of a negative modulator is shown in (c); in this case the inactive form is stabilised. Covalent modification of enzymes is another strategy that is used widely in metabolic regulation. One of the most common additions is a phosphate group, which can alter the shape of a protein by attracting positively charged R-groups (as phosphates carry two negative charges on the two single-bonded oxygen atoms). Protein kinases add phosphate groups, and phosphatases remove them, thus the effect can be reversed. Some proteins are activated by phosphorylation, others are inactivated (Fig ). Fig : General outline of protein phosphorylation effects. Kinase and phosphatase enzymes add or remove phosphates from enzymes. In (a) the effect of phosphorylation is to increase activity (shown by the + sign). In (b) the effect is to inactivate the enzyme ( sign). BIOLOGY 55
56 MOLECULAR INTERACTIONS IN CELL EVENTS An example of phosphorylation activating an enzyme is the skeletal muscle enzyme glycogen phosphorylase. This is used to generate glucose from glycogen when heavy demands are placed on muscle tissue. The enzyme is present as an inactive non-phosphorylated form, which is converted to the active form by the addition of a phosphate to a serine residue in the protein by phosphorylase kinase. When energy demand falls off, phosphorylase phosphatase removes the phosphate group and inactivates the enzyme. However, this is not the whole story glycogen phosphorylase is also regulated by an allosteric effect. Glucose and ATP act as negative modulators, and AMP acts as a positive modulator. This avoids having full activation by the phosphorylation alone, and provides a regulatory mechanism that is responsive to the ATP/AMP ratio in the cell. A further complication is that there is a hormonal control mechanism by epinephrine and glucagon. This illustrates very clearly the complexity of metabolic regulation, with often two or more distinct mechanisms used to control enzyme activity in a precise and responsive way. Yet another form of control by a covalent activating mechanism is proteolytic cleavage, as found in the enzyme trypsin (a protease, important in digestion). It is synthesised in the pancreas, but must not be made in its active form, or it would digest the pancreatic tissue itself! Thus it is synthesised as a slightly longer protein called trypsinogen), which is inactive (the general name zymogen is used to describe these inactive protease precursors). Activation occurs when trypsinogen is cleaved by a protease in the duodenum. Once active, trypsin can activate more trypsinogen molecules, resulting in an autocatalytic cascade that produces a large number of active trypsin molecules very rapidly. Trypsin also cleaves other zymogens such as chymotrypsinogen. Cascades like this are another important part of the overall metabolic regulatory strategy of the cell. So far we have built up a picture of the complex mechanisms used to control the activity of specific enzymes. However, metabolism is organised as a series of metabolic pathways, and control of these pathways is an important feature of cell biochemistry. One way in which control can be exercised is by end-product inhibition, as shown in Fig Regulating the output of a biochemical pathway according to the amount of its product is an efficient mechanism, as it avoids the wasteful conversion of the intermediates in the pathway. This is a form of negative feedback. 56 BIOLOGY
57 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : End-product inhibition in a metabolic pathway. A, B, C and D are the substrates/products of the enzymes (1, 2 and 3). When D increases in concentration, it can bind to the first enzyme in the pathway and reduce the efficiency of conversion of A to B. This in turn controls the whole pathway as the supply of intermediates is restricted. 3.2 The sodium-potassium pump The movement of solutes against a concentration gradient by active transport is an essential part of the cell s metabolism. One of the best examples of how this works is the sodium-potassium pump (Na + /K + pump) in animal cells. This transports sodium ions out of cells, and potassium ions in. The pump is driven by the hydrolysis of ATP. The overall result is that the intracellular concentration of Na + is kept low compared to the external concentration. For K + the opposite applies the intracellular concentration is high compared to the outside. Maintaining this imbalance of intracellular/extracellular Na + and K + concentrations is one of the cell s most critical functions, and it accounts for around 30% of the energy expenditure of a typical animal cell. A summary of the features of the Na + /K + pump is shown in Fig BIOLOGY 57
58 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : Summary of the sodium-potassium pump. The membranebound protein complex transports sodium ions to the extracellular environment, and potassium ions to the intracellular environment. The pump is driven by ATP-derived phosphorylation, which induces conformational changes in the protein. Three sodium and two potassium ions are exchanged for each cycle of operation. This results in the maintenance of Na + /K + concentration (electrochemical) gradients as shown. The pump is a very elegant example of our theme of structure and function being closely related. The protein complex is actually made up of four subunits; two large and two small. We will consider the protein complex as a single unit for simplicity. The key features of the pump are: it is a transmembrane carrier protein it has three binding sites for Na + ions it has two binding sites for K + ions there is a phosphorylation site to accept a phosphate from ATP two different conformations of the protein are possible (controlled by the phosphorylation state). As the protein works by phosphorylation, and ATP is the source of the phosphate group, the whole complex is often called the Na + /K + ATPase. The overall pattern is that the hydrolysis of one ATP molecule drives the export of 3 sodium ions and the import of 2 potassium ions. 58 BIOLOGY
59 MOLECULAR INTERACTIONS IN CELL EVENTS The way in which the pump works is shown in Fig Initially, the pump is open to the cytosol. Sodium ions can bind to the Na + binding sites, and the ATPase function phosphorylates the protein. This causes a conformational change, which makes the protein open to the extracellular environment. The Na + binding sites now have a lowered affinity for sodium, and the Na + ions are released. The K + binding sites (high affinity for K + ) can then be occupied; again, a conformational change occurs, with the release of the phosphate, which switches the protein back to the open to inside structure. The K + sites now have a lowered affinity for potassium, and the K + ions are released. This cycle is repeated, with as many as 300 sodium ions being transported per second. Fig : The mechanism of action of the sodium-potassium pump. The membrane is shaded, with the external environment at the top of each diagram. Sodium ions are black squares, potassium ions are black circles. (a) The protein is in the open to cytoplasm conformation, which allows sodium ions to bind. (b) Phosphorylation of the protein. (c) A conformational change releases sodium to the external environment. (d) Potassium binds to the binding sites now exposed to the external environment. (e) Dephosphorylation of the protein restores the open to inside conformation. (f) Potassium ions released; the pump is now ready for the next cycle of operation. BIOLOGY 59
60 MOLECULAR INTERACTIONS IN CELL EVENTS The Na + /K + pump is an excellent example of a molecular machine, in which two features work in concert. Firstly, the conformational change in protein structure caused by ion binding and phosphorylation/ dephosphorylation provides the mechanism of action of the pump. Secondly, the alteration of the affinity for Na + and K + ion binding enables the pump to capture and release the ions at the correct points in the pumping cycle. 3.3 Cell signalling Although cells are in many ways self-contained units, they do not exist in isolation. Even a unicellular organism must detect and respond to outside influences chemicals, light, nutrient availability, and the presence of other cells. In multicellular organisms, the organisation of cells into tissues and systems brings added complexity. It is therefore essential that cells are able to talk to each other by a set of processes that can be grouped under the general heading of cell-cell communication or cell signalling. In this section we will consider how this is achieved in animal cells. General principles Communication between cells is very much like any other form of communication it involves transmitting and receiving information. A signal is sent by a signalling cell and received by a target cell. Where a change in the form of the signal is required, this is signal transduction (faxing a letter involves signal transduction conversion of the printed form into the electronic form). Signal reception and transduction are the two areas of cell signalling that most is known about. Communication can be achieved by a number of different systems: endocrine secretion of a hormone into the bloodstream, which enables dispersal throughout the body. The signalling cell and the target cell can be far apart. paracrine works over a more restricted area than endocrine signalling. A local mediator is secreted, which can affect cells in the immediate area of the signalling cell. neuronal nerve cells or neurones elicit responses by the release of a neurotransmitter at synapses. Like hormones, the signal can cover long distances; in this case, through the network of nerve cells rather than in the bloodstream. 60 BIOLOGY
61 MOLECULAR INTERACTIONS IN CELL EVENTS contact-dependent signal molecules in the plasma membrane of the signal cell interact with membrane-bound receptors on the target cell. These signals are therefore restricted to cells that are in direct contact. Often similar types of molecules are used in these different forms of signalling. Examples include amino acid derivatives such as adrenaline (hormone), histamine (local mediator) and GABA (γ-aminobutyric acid, an inhibitory neurotransmitter). Many signal molecules are proteins, such as insulin (hormone) and EGF (epidermal growth factor, a local mediator). At the other end of the scale in terms of complexity is the local mediator nitric oxide (NO), which causes relaxation of smooth muscle cells in blood vessels, dilation and therefore increased blood flow. Another important group of signalling molecules are the steroid hormones, which are based on the cholesterol four-ring structure (as outlined in Fig ). In multicellular organisms, there may be hundreds of different signal molecules. Responses to these can be controlled by variations in the reception at the cell surface. In some cases, one signal/receptor is required, whilst in others multiple signals are involved. The signal can also be interpreted in different ways within the cell, often by a cascade system involving modulation of the signal, amplification, and the generation of different responses by different parts of the cell. Thus the signalling system can be controlled in a very precise way; this is obviously critical if the correct response is to be generated. Extracellular hydrophobic signalling molecules Some small hydrophobic molecules can cross the plasma membrane and enter the cell by diffusion. The best known classes are the steroid hormones (such as cortisol and testosterone) and the thyroid hormones (such as thyroxine; see Fig ). They work by activating gene regulatory proteins in the cell, which stimulate transcription of particular sets of genes in the nucleus. The hormones can diffuse across the plasma membrane and bind to receptor proteins that are located either in the cytosol or in the nucleus itself. The mode of action of cortisol is shown in Fig BIOLOGY 61
62 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : Structure of the thyroid hormone thyroxine. This is an amino acid derivative with two aromatic rings and four iodine atoms. Fig : Cortisol works by diffusing across the plasma membrane and binding to an intracellular receptor protein (RP). This enters the nucleus and binds to a regulatory site on the DNA, stimulating transcription. Extracellular hydrophilic signalling molecules In contrast to the hydrophobic signals outlined above, the majority of signalling molecules are either too large or too hydrophilic to cross the plasma membrane. The receptor proteins for these signals must therefore present a binding site to the extracellular environment, and elicit a response in the cytosol. There are three main classes of these cell surface transmembrane receptors: ion-channel-linked enzyme-linked G-protein-linked These types of receptor all bind extracellular signal molecules, but generate intracellular responses in different ways. A summary of their mode of action is shown in Fig BIOLOGY
63 MOLECULAR INTERACTIONS IN CELL EVENTS Fig : The three classes of receptor proteins. (a) Ion-channel-linked receptors. In this diagram the channel is closed and ions cannot pass through. Binding of the signal (S) causes a conformational change, opening the channel. (b) Enzyme-linked receptors, with the inactive enzyme converted to the active form on binding of the signal molecule. (c) G-protein-linked receptors (R) activate the G-protein (G) in response to signal binding. The G-protein acts on a target protein (T), which may be an enzyme or an ion-channel protein. This generates the intracellular response. Ion-channel-linked receptors (also known as chemically-gated ion channels) open pores through the protein in response to binding of the signal molecule. Ions flow through this gate, generating an electrical effect. This type of receptor is found in excitable cells such as nerve cells and muscle cells. Enzyme-linked and G-protein-linked receptors are found in all types of cells. The enzyme-linked receptors generate an enzyme activity (usually a kinase activity) on the cytoplasmic end of the protein. This kinase activity causes the phosphorylation of other intracellular proteins, thus activating them. G-protein-linked receptors activate a GTP-binding protein (the G-protein) that sets off a chain of events in the cell. This group of receptors is the largest known, and many different signals and responses can be associated with G- protein activity. Let s look at how G-protein-linked receptors work in a little more detail. Although many different variations exist, all G-protein-linked receptors have the same structural arrangement within the membrane. This is known as a seven-pass transmembrane protein, as shown in Fig Several hundred types of receptor are known, which bind signals as diverse as peptide hormones, amino acids, fatty acids, and neurotransmitters. On binding the signal, the G-protein is activated by binding of GTP. The activated protein diffuses away from the receptor protein site, and activates its target protein. This may be an ion-channel BIOLOGY 63
64 MOLECULAR INTERACTIONS IN CELL EVENTS protein, or an enzyme such as adenylate cyclase or phospholipase C. These enzymes catalyse the formation of small molecules known as second messengers, which trigger the intracellular response to the original signal transduction event at the cell surface. Adenylate cyclase activity generates cyclic AMP (camp), phospholipase C generates inositol triphosphate (IP 3 ). Second messengers are important parts of the signal transduction pathway, and can have many different effects. An outline of the camp pathway is shown in Fig Fig : Structure of the G-protein-linked receptor family. The sevenpass transmembrane protein crosses the membrane seven times, and has distinct binding sites for the extracellular signal molecule and the intracellular G-protein. Fig : The cyclic AMP (camp) signal transduction pathway. The G- protein-linked receptor binds the signal molecule and this causes activation of the G-protein (GDP is exchanged for GTP). The activated G-protein diffuses to its site of action in this case, adenylate cyclase (AC). This catalyses the formation of camp from ATP. The camp acts as a second messenger, triggering a variety of intracellular reactions that often form a cascade effect. 64 BIOLOGY
65 MOLECULAR INTERACTIONS IN CELL EVENTS Signal transduction is a complex topic. We have seen that signals can be of many different types, and can act either by diffusing across the plasma membrane (steroid hormones and nitric oxide) or by interacting with a receptor protein on the cell surface. The variety of signals, receptors and responses means that the system of signal reception and transduction can generate very specific effects in different types of cell. The response of a cell to a signal can involve ion flow, activation of specific proteins, or changes in gene expression. These effects can be short-lived, as in the case of the generation of an action potential, or they may be permanent alterations that control the developmental fate of the cell. It is therefore clear that the idea of a cell as a self-contained unit is in fact very far from the reality of the situation cells are constantly engaged in the exchange of information in the form of molecular signals, and it is this that enables cells in multicellular systems to function in an integrated way. BIOLOGY 65
66 66 BIOLOGY
67 SECTION 4 APPLICATIONS OF DNA TECHNOLOGY One of the defining features of modern biology is the extensive use of the technology of gene manipulation. The first recombinant DNA molecules were constructed at Stanford University in 1972, and developments since then have been truly staggering. It is now possible to isolate genes relatively easily, determine their DNA sequence, and assess their function. Genes can be modified in the test tube and replaced in the original organism or in a different host, and many types of transgenic plants and animals have been developed. In this final part of our look at cell and molecular biology, we will consider four of the areas in which gene manipulation technology is having (or will have) a major effect on our lives the human genome project, human therapeutics, forensic science, and agriculture. Although we will concentrate on these aspects, it is useful to remind ourselves that the great advances in recombinant DNA technology all depend on the simple rules of A:T and G:C base pairing. This is illustrated in Fig. 4.1, which shows the use of a radioactive probe in the identification of DNA fragments in two common applications. BIOLOGY 67
68 APPLICATIONS OF DNA TECHNOLOGY Fig 4.1: Detecting DNA sequences by hybridisation. In (a) a radioactive probe sequence (asterisks) is shown hybridising to its complementary sequence. This can be used to detect cloned sequences in bacterial colonies, as shown in (b). Clones with the desired sequence are identified by the positive response on X-ray film, caused by the probe binding to the complementary sequence on the filter replica of the bacterial colonies. In (c) a similar process is shown for DNA fragments in an electrophoresis gel. The fragments are transferred to a filter and the probe sequence binds to complementary DNA sequences. The pattern can be used to determine which fragments contain the desired sequence. 68 BIOLOGY
69 APPLICATIONS OF DNA TECHNOLOGY Recombinant DNA technology is now part of our culture, for both the scientist and for members of the public. Public concern about the use of gene technology is an area that poses perhaps the greatest challenge to the biological community over the next few years. If we are to gain from the benefits that gene technology offers, and avoid the pitfalls, we will all have to take an active part in the debates that lie ahead. 4.1 The human genome project The genome of an organism is its complete complement of genetic information. Most organisms have a DNA genome, although a few viruses have RNA as their genetic material. In animal cells, the nuclear genome is the major store of genetic information, with mitochondria having their own smaller genome (mitochondrial DNA or mtdna). In plants, the chloroplast also has its own genome (chloroplast DNA or cpdna). The big project in biology at the moment is the human genome project (HGP). The aim of this major international effort is to determine the complete DNA sequence of the human genome, with a projected completion date of In parallel with the human genome effort, many other organisms are being investigated, with some genome projects (such as E. coli and the yeast Saccharomyces cerevisiae) having already been completed. The obvious question in genome sequencing is why do it? The answer is that it will help us to understand the very nature of life itself, by studying the information that organisms require to function. This area is sometimes called bioinformatics, and there are many similarities between bioinformatics and the great technical advances that have been made in computing. Just as it is impossible to understand fully how a computer works without examining the software that drives it, genome projects aim to decipher the software that makes organisms what they are. As well as technical difficulties, this raises ethical problems. Many people are concerned that information about their individual genetic makeup could be used to discriminate against them in areas such as medical care and life assurance. In humans, the amount of information in the haploid nuclear genome is some base pairs (3,000,000,000 or 3 billion!). The structure of the genome is complex, and different classes of DNA can be recognised. About 40% of the total is either highly or moderately repetitive sequence DNA. Of the remaining 60%, which represents unique sequence and low copy number sequence elements, only around 3% is BIOLOGY 69
70 APPLICATIONS OF DNA TECHNOLOGY the actual coding sequence that makes up the estimated 60,000 80,000 genes that are needed to make a human being. The extra DNA is found between genes as spacer DNA, and also within gene sequences themselves. These intervening sequences or introns mean that eukaryotic genes are much larger than is necessary to code for the proteins that they specify. The RNA copy of the gene produced by transcription therefore has to be edited by a process known as RNA splicing or RNA processing. Whilst the exact role of introns is still not clear, it seems likely that they are important in regulating the expression of genes and in generating new protein molecules during the process of evolution. When we consider the 3 billion base pairs in the genome, it is difficult to know what this means in real terms. One example that is often used is to consider writing out the genome sequence. Fig shows a short stretch of DNA sequence. If we were to write out the human genome on a paper ribbon using this size of typeface, we would need around 5,000 kilometres to represent the information about the distance from Glasgow to New York! Imagine trying to make sense of this amount of information, with the only variation being the order of the four bases A, G, C and T, and you will begin to get some idea of the scale of the problem... Fig : The DNA sequence. If the human genome was written out like this, you would need about 5,000km of this ribbon to represent the entire sequence. Another analogy may help to set the HGP in context. If you wish to travel from Inverness to Plymouth, it is useful to follow a map. You would not usually buy street maps of all the towns and villages between Inverness and Plymouth, although in theory you could find your way by doing this and linking all the hundreds of maps together. A more sensible way would be to look at a road atlas of Britain, and see what major destinations you would aim for say Inverness, Glasgow, Carlisle, Manchester, Birmingham, Bristol, Plymouth. You would then fill in more detail as required Inverness, Aviemore, Perth, Stirling, Glasgow, and so 70 BIOLOGY
71 APPLICATIONS OF DNA TECHNOLOGY on. Your route would take you on particular roads the A9, M74, M6 etc., so you could keep track of the destinations and the routes between them. Sequencing the genome is a bit like constructing a road map. The first requirement is to pinpoint the major landmarks, keeping the information in order, and then completing the picture with the detailed sequence information. In the HGP this is achieved by three approaches: genetic mapping physical mapping DNA sequencing. One particular problem is the interface between the techniques particularly the link between genetic and physical maps, which are based on different criteria. Put simply, it is important to get the stretches of sequence information in the right order! Let s now consider how these techniques contribute to the HGP. Genetic linkage mapping Linkage mapping can be used to locate genes on particular chromosomes, and establish the order of these genes and the approximate distances between them by determining their recombination frequency. This approach relies on having genetic markers that are detectable sometimes these are genes that cause disease, traced in families by pedigree analysis. The marker alleles must be heterozygous so that meiotic recombination can be detected. If two genes are on different chromosomes, they are unlinked and will assort independently during meiosis. However, genes on the same chromosome are physically linked together, and a crossover between them during prophase I of meiosis can generate non-parental genotypes. The chance of this happening depends on how far apart they are if they are very close together, it is very unlikely that a crossover will occur between them. If far apart, they may behave as though they are essentially unlinked. By working out the recombination frequency it is therefore possible to produce a map of the relative locations of the marker genes. A summary of recombination possibilities is shown in Fig BIOLOGY 71
72 APPLICATIONS OF DNA TECHNOLOGY Fig : Genetic linkage. In (a) two maternal (M) and two paternal (P) chromosomes are shown (chromatids are not shown in this diagram). Gene A (alleles A & a) and B (alleles B & b) are on different chromosomes, and so will behave independently during meiosis. In (b) the two genes D and E are linked; the paternal chromosome has D and e alleles, the maternal has d and E. If a crossover (X) between non-sister chromatids occurs between the loci for D and E, recombination can generate non-parental genotypes in the gametes. The frequency of this event occurring between loci can be used as a measure of how far apart they are. The overall result of genetic mapping is to produce a picture of the locations of the marker loci on the chromosome a bit like establishing the order of the cities and large towns on the route from Inverness to Plymouth, but not yet knowing the precise distances between them, or the road numbers. Physical mapping is required to add some more of the detail. Physical mapping Until the early 1980s it was thought that a physical map of the human genome was unlikely to be achieved. As with genetic maps, construction of a physical map requires markers that can be mapped to a precise location on the DNA sequence. Physical maps of the genome can be constructed in a number of ways, all of which have the aim of generating a map in which the distances between markers are known with reasonable accuracy. The various methods include restriction mapping, in which fragments of DNA are generated by cutting with restriction enzymes (also known as restriction endonucleases). The recognition sequences for restriction enzymes are short (usually 4, 5 or 6 base pair) sequences that occur at defined positions in the DNA. By 72 BIOLOGY
73 APPLICATIONS OF DNA TECHNOLOGY using combinations of restriction enzymes and working out the sizes of the fragments, the puzzle can be pieced together to give the pattern of restriction enzyme recognition sites in the DNA. This is useful as defined fragments can then be identified, either by size or by using a specific DNA probe to bind to its complementary sequence. The technique of restriction mapping is shown in Table and Fig Table 4.1.3: Fragments produced from a 15 kbp DNA fragment digested with the restriction enzymes BamHI, EcoRI and PstI. Single, double and triple digests are shown. All fragments are in kbp. These data can be used to map the restriction sites as shown in Fig BamHI EcoRI PstI BamHI BamHI EcoRI BamHI EcoRI PstI PstI EcoRI PstI BIOLOGY 73
74 APPLICATIONS OF DNA TECHNOLOGY Fig : Determination of the restriction map for the fragments listed in Table In (a) the BamHI site cuts the 15 kbp fragment into 14 and 1 kbp fragments. In (b) the EcoRI fragments of 12 and 3 kbp can be orientated in two ways with respect to the BamHI site (i and ii). A double digest with BamHI and EcoRI generates fragments of 11, 3 and 1 kbp, so the correct orientation is (b)ii, giving the pattern shown in (c). Similar reasoning with the pattern of the PstI fragments shown in (d) enables the final map to be determined (e). (Copyright D S T Nicholl/ Cambridge University Press. Reproduced with permission.) Another method of identifying particular DNA sequences is to amplify the target sequence using the polymerase chain reaction (PCR). This technique has become very important in molecular biology, for both mapping and identification of genes, and in forensic analysis. The basis of the technique is shown in Fig In mapping, unique sequences in the genome can be amplified by PCR if suitable primers are available. This approach has become one of the most widely used for physical mapping of genome fragments. 74 BIOLOGY
75 APPLICATIONS OF DNA TECHNOLOGY Fig : The polymerase chain reaction (PCR). The DNA duplex is heat-denatured and two primers are annealed to the sequence. A DNA polymerase then copies the templates, doubling the number of copies. The denature-prime-copy cycle can be repeated automatically if a thermostable polymerase (such as Taq polymerase) is used. The PCR reaction generates a large number of copies of the target sequence in a short time. (Copyright D S T Nicholl/Cambridge University Press. Reproduced with permission.) The various methods used in genome mapping enabled the construction of a useful genetic and physical map of the human genome by The pieces of DNA that are actually used for sequencing are often cloned in large-capacity vectors that are essentially artificial chromosomes. These can then be restriction mapped and sequenced. The link between genetic, physical and restriction maps is shown in Fig BIOLOGY 75
76 APPLICATIONS OF DNA TECHNOLOGY Fig : Genome mapping. In (a) the genetic map is shown, with genetic markers assigned to positions on the chromosome. In (b) a section of the chromosome is shown, with the physical map of this region. Various methods may be used to assign physical markers to their chromosomal locations. In (c) the clone map of a section of the physical map is shown, with large overlapping DNA fragments. From this a more detailed restriction map and the DNA sequence itself can be determined. DNA sequencing The final stage of the genome project is to determine and assemble the actual DNA sequence itself. There are several critical requirements for this part the DNA fragments must be generated, the sequencing technology must be accurate and fast, and computer hardware and software must be available to analyse the sequence data. The technique used for sequencing is based on one of the original methods developed in the mid 1970s. This is called the dideoxy chaintermination method, and relies on making a copy of the DNA template to be sequenced. A DNA polymerase is used, along with a primer and the four dntps (datp, dgtp, dctp and dttp). With the correct biochemical conditions, including a radioactive label to enable the product to be detected, the polymerase can make a copy of the DNA by a process that is essentially the same as that used in DNA replication. The chain termination part is what makes the key difference. This involves setting up four separate reactions, each including one of the four dideoxy NTPs (ddatp, ddgtp, ddctp and ddttp). These modified nucleotides cannot form the next phosphodiester bond in the growing chain hence when a ddntp is incorporated into the copy, it terminates the process. The large number of fragments that are produced in the four reactions produce a set of sequences that differ in length by one 76 BIOLOGY
77 APPLICATIONS OF DNA TECHNOLOGY base, and end with a particular ddntp. These fragments can be separated by gel electrophoresis, and an image produced on X-ray film, which is darkened by the radioactive label in the copied DNA strand. The sequence is read from the film as shown in Fig Fig : Reading a sequencing gel. In (a), part of an X-ray film is shown. The film was produced from the electrophoresis gel that separated the DNA fragments in the direction of the arrow. In (b), a tracing of the film is shown. A separate lane is used for each dideoxy reaction, and the sequence can be read from the smallest fragment at the bottom. (Autoradiograph courtesy of Dr N Urwin. Copyright D S T Nicholl/Cambridge University Press. Reproduced with permission.) The standard chain-termination method of sequencing was adapted for genome sequencing by using fluorescent labelling instead of radioactive methods. In one method, the ddntps can be tagged with different labels, and one reaction carried out where all four ddntps are used together. The products are separated by gel electrophoresis, and the fluorescent labels detected as they come off the bottom of the gel. This gives a direct readout of the sequence. The process can be automated, and is much faster than conventional sequencing. It is summarised in Fig BIOLOGY 77
78 APPLICATIONS OF DNA TECHNOLOGY Fig : Automated DNA sequencing using fluorescent marker dyes. Each ddntp is tagged with a different dye as shown in (a). On separation in a single lane of a sequencing gel, the DNA fragments pass through a detector (b) and the fluorescent labels are monitored. A computer captures the data and displays the sequence as a series of peaks (c), from which the sequence is read as shown in (d). Comparative genome analysis As already mentioned, other genomes are being sequenced in addition to the human genome. Whilst only a few have been completely sequenced to date, the analysis of genomes has been carried out for a number of years. Genetic and physical mapping, hybridisation studies and sequencing of individual genes have provided a lot of useful information about genome size and organisation. In fact, the emphasis in molecular biology is changing as the amount of detailed information about genes increases we are now much more likely to consider the function of a gene as part of the genome, rather than in isolation. Some genome sizes are shown in Table Size and complexity generally increases with the increasing complexity of the organism, as might be expected. Gene structure in bacteria is simpler than that in eukaryotes. Comparing gene structure and organisation in E. coli, the yeast S. cerevisiae and humans puts the complexity in the expected order bacterium, yeast, human. However, there are some anomalies many plants have much larger genomes than humans, for example. Thus genome analysis is an area of research that is likely to give rise to many unexpected discoveries in the next few years. 78 BIOLOGY
79 APPLICATIONS OF DNA TECHNOLOGY Table 4.1.9: Genome size and estimated number of genes for some organisms. Genome sizes are given in Megabase pairs (1 Megabase = bases). Organism Genome size Number of (Mb) genes Escherichia coli (bacterium) 4.6 4,405 Saccharomyces cerevisiae (yeast) ,800 Drosophila melanogaster (fruit fly) ,000 Homo sapiens (man) 3,000 70,000 Nicotiana tabacum (tobacco) 4,500 43, Human therapeutics Congenital abnormalities are genetically-based diseases (often simply called genetic diseases) that are present at birth. Diseases caused by a single-gene defect are known as monogenic traits, and to date around 5,000 have been described. These are characterised as either autosomal dominant, autosomal recessive or X-linked. Most X-linked traits are recessive. Diseases that involve several genes are known as polygenic traits, and are usually more difficult to diagnose and treat than the single-gene defects. In this section we will consider how molecular genetics has improved the diagnosis and treatment of genetic disease, concentrating on two examples; cystic fibrosis and Duchenne muscular dystrophy. Detecting gene disorders The characterisation of a monogenic genetic disease usually begins with the presentation of the disease symptoms. Often a large amount of historical information is available about the disease and its effects before the definitive genetic cause is established. The first step is to trace the disease through family relationships by carrying out pedigree analysis to determine if the faulty gene is dominant, recessive or X-linked. This information can often be useful in advising prospective parents about the probability of their children inheriting the disease. This genetic BIOLOGY 79
80 APPLICATIONS OF DNA TECHNOLOGY counselling is an important part of preventive healthcare, and modern molecular techniques have greatly improved the diagnosis of defective alleles of the genes involved. Once the disease has been identified as a monogenic trait, the search for the gene defect itself can begin. This process can take many years to complete, and often involves collaboration between many groups of scientists specialising in the disease. The processes involved are similar to those employed for the human genome project. The defect is firstly mapped onto a chromosomal region by looking for genetic markers that are co-inherited with the disease. Meiotic recombination frequency gives an indication of how close the markers are to the target gene; the more often the gene and marker are co-inherited, the closer they are on the chromosome. When the chromosomal location of the gene is established, the more detailed analysis of the region can begin. By using a combination of genetic map information and physical mapping, the defective gene can be tracked down, characterised and sequenced, which can lead to the development of more accurate diagnostic procedures and potential new treatments. Cystic fibrosis (CF) is one disease that has been investigated in detail at the genetic and molecular levels. CF is an autosomal recessive monogenic trait that affects around 1 in 2,000 people. The CF gene codes for a membrane carrier protein of 1,480 amino acids, which has a complex structure with two transmembrane domains, two ATP-binding domains, and a regulatory region. Mutations in the CF gene result in a defective ion transport system which means that epithelial surfaces are not fully hydrated. This causes sticky mucus accumulation in the lungs. Major symptoms are inflammation of lung tissue and persistent bacterial infection. Other symptoms can include defects in pancreatic function. The disease is relatively easy to diagnose, as a symptom is the production of salty sweat. Affected individuals have a reduced quality of life and a life expectancy of about 30 years. The defective gene for CF has a carrier frequency of around 1 in 22. In cases where two carriers have children, there is a 1 in 4 chance of the child receiving both defective alleles (one from each parent) and thus being homozygous and suffering from the disease. The search for the CF gene was a major undertaking, involving several research groups at different stages. The gene was mapped onto chromosome 7, and linkage studies with the gene and two types of physical marker narrowed down the search. By an extensive programme of examining cloned DNA fragments, using the techniques of chromosome walking and chromosome jumping (see Fig ), the gene was eventually found in BIOLOGY
81 APPLICATIONS OF DNA TECHNOLOGY Fig 4.2.1: Chromosome walking and jumping. In (a) chromosome walking is shown. The end of each overlapping fragment is used in a hybridisation test to identify the next fragment. This is often used to walk from a marker gene towards the target gene. Chromosome jumping (b) is a similar technique but in this case a special cloning technique is used to isolate complementary fragments that are far apart. This enables a jump along the chromosome, which is useful if the marker gene is far from the target gene. Often a mixture of walks and jumps is needed to progress, as shown in (b). Once the gene had been identified, more and more details about the molecular biology of CF began to emerge. Some 550 mutations have been described, but the most common one by far is a deletion of three base pairs that removes one amino acid from the polypeptide. This is called the F508 mutation ( is deletion, F is phenylalanine and 508 is the position in the protein). The defective protein does not fold up properly, and does not reach its membrane location. A summary of the CF gene, protein and the F508 mutation is shown in Fig BIOLOGY 81
82 APPLICATIONS OF DNA TECHNOLOGY Fig 4.2.2: The cystic fibrosis F508 mutation. In (a) the gene is shown. Transcription produces the primary RNA transcript, that is converted into the functional mrna by removal of intervening sequences. On translation the CF transmembrane conductance regulator protein (CFTR) is produced. In (b) the normal and mutant proteins are shown. Normal CFTR has phenylalanine (F) at position 508. In the mutant form this is deleted, causing the protein to fold incorrectly. Duchenne muscular dystrophy (DMD) is an X-linked disease that affects around 1 in 3,300 boys. It causes progressive wasting of muscles, resulting in wheelchair confinement by the teenage years and a life expectancy similar to that for CF. There is a milder form of the disease called Becker muscular dystrophy (BMD). As with the CF gene, a search for the DMD gene was undertaken and it was found in The protein (called dystrophin) is 3,685 amino acids in length! Its normal function is to link the cytoskeleton with the sarcolemma (muscle cell membrane) in muscle cells. Molecular characterisation of genes enables accurate tests to be devised for diagnosis of the conditions. The F508 deletion in CF can be identified by amplifying a short (100 base pair) DNA fragment that spans the area of the deletion. The polymerase chain reaction is used for this, followed by separation of the DNA fragments on an electrophoresis gel. In homozygous normal cases the two DNA fragments produced (one from each allele) will be normal length and identical. The heterozygote will show one shorter band in addition to the normal one, and the homozygous recessive (two alleles with deletions) will show one band at the lower position on the gel (Fig ). Molecular tests can be devised 82 BIOLOGY
83 APPLICATIONS OF DNA TECHNOLOGY for most other gene mutations once the genes have been characterised, and this area of molecular genetics has had a great impact on the diagnosis of many types of inherited disease. Fig 4.2.3: A PCR-based test for the cystic fibrosis (CF) defective allele. A short sequence that spans the mutated region is amplified using the polymerase chain reaction. In lane 1 a normal homozygous pattern is shown (+/+). In lane 2 a carrier (heterozygous; +/ F508) shows two bands, one normal and one smaller by 3 nucleotides (one codon, representing the deletion of phenylalanine). In lane 3 a CF-affected individual (homozygous recessive; F508/ F508) shows one band at the lower position. In lanes 1 and 3 the DNA band contains sequences from both paternal and maternal chromosomes that run to the same position in the gel. It is only in the heterozygous case that the two bands are distinguished. Gene therapy When a gene defect has been identified and the gene cloned, there is the possibility of using the good copy of the gene to overcome the problem. This is known as gene therapy. The proposition is attractive as the cause of the disease is the target rather than just the symptoms. Gene therapy might also be used to kill abnormal cells, or to inhibit the spread of viruses by preventing DNA replication. Although still in the early stages, some success has been achieved. In 1990 the first gene therapy procedure was used with a 4-year-old girl patient who had the inherited disease adenosine deaminase (ADA) deficiency. The defect in this enzyme leads to the condition known as SCIDS (severe combined immunodeficiency syndrome), which means that the patient cannot fight infection. By treating cells from the immune system with a viral vector that carried the functional copy of the gene, and replacing the cells in the patient, the condition was improved. Cystic fibrosis and Duchenne MD are other obvious candidates for gene BIOLOGY 83
84 APPLICATIONS OF DNA TECHNOLOGY therapy. In both cases some success has been achieved using mice as model organisms, although an effective therapy for use in humans is still some way off due to the technical difficulties in getting the procedures to work reliably. When assessing if gene therapy is likely to be appropriate, several factors must be considered: the nature of the gene defect the gene must be available in cloned form, and the defective function characterised. the target cells in the patients obviously the therapy must act on the cells that are involved in the presentation of the disease. The cells can be treated outside the body (ex vivo) and replaced, or may have to be treated within the patient s body (in vivo). the method of delivery of the normal gene viral vectors can be used but are not ideal. Other methods with potential are liposomes that can fuse with cell membranes, or human artificial chromosomes. the expression and stability of the normal gene in the target cells the normal gene has to be expressed in the cells and the gene product has to function properly. One aspect of gene therapy that raises particular ethical questions concerns the type of cells that are the targets. Most scientists and observers accept that carrying out gene therapy on somatic cells (body cells) is not much different from taking an aspirin the chemical (DNA) is just a little more complex. However, it is also possible that germ cell gene therapy could be developed, in which the reproductive cells are the targets. This would effectively alter the gene pool of the species, as the alteration would be passed on to the next generation. At present, there are no plans to carry out genetic manipulation of germ-line cells, and most people agree that this should not be attempted. 4.3 Forensic uses In the legal profession, the use of DNA technology has become one of the most important tools for identifying individuals in both criminal cases and in disputes over whether people are related or not (paternity disputes and immigration cases are the most common). The original technique was called DNA fingerprinting. Improvements in the technology have increased the range of tests that can be carried out, and today the more general term DNA profiling is used to describe the methods available. The basis of all the techniques is that a sample of DNA 84 BIOLOGY
85 APPLICATIONS OF DNA TECHNOLOGY from a suspect (or person in a paternity or immigration dispute) can be matched with that of the reference sample (from the victim of a crime, or a relative in a civil case). In scene-of-crime investigations, the technique can be limited by the small amount of DNA available in forensic samples. Modern techniques use the polymerase chain reaction (see Fig ) to amplify and detect minute samples of DNA from bloodstains, skin fragments or hair roots. The original DNA fingerprinting technique is based on the fact that there are highly variable regions of the genome that are specific to each individual. These are minisatellite regions, which have a variable number of short repeated-sequence elements known as variable number tandem repeats (VNTRs). When digested with restriction enzymes, different sized fragments are produced (Fig ). The fragments can be detected using a probe that binds to the VNTR sequence. This generates a unique profile of the DNA from that person. Fig 4.3.1: Variation in restriction fragment lengths caused by different numbers of tandem repeat elements. A locus that is heterozygous for the tandem repeat length is shown. In the upper example, one allele with a 5-copy variable number tandem repeat (VNTR) is shown. In the lower example, the allele has a VNTR with 8 copies. If these DNA molecules are cut with a restriction enzyme that does not cut in the repeat sequence, but cuts frequently outside this, the VNTR sequences will show as different length fragments (labelled 1 and 2 in the diagram; restriction sites shown by arrows). A radiolabelled probe can then be used to identify the repeat sequences as part of a DNA fingerprinting (profiling) experiment. BIOLOGY 85
86 APPLICATIONS OF DNA TECHNOLOGY Detecting fragments and producing a DNA profile using the original techniques involves a number of stages: DNA isolation this is carried out on the sample (often a blood sample). If sufficient DNA is available it may be used directly in the profiling technique. restriction enzyme digestion this generates the DNA fragments that produce the profile, with fragments of different lengths produced from the variable regions of the DNA. gel electrophoresis this separates the DNA fragments according to length, and will produce the banding pattern that is used to compare the different samples. blotting the DNA onto a filter this enables the DNA fragment pattern to be transferred to a filter for the hybridisation stage which detects the target sequences. hybridisation with the probe this involves using a labelled nucleic acid probe (see Fig. 4.1c) which binds to a specific base sequence in the target fragments. The critical part is the selection of the appropriate probe sequence. There are two main classes of these multi-locus probes and singlelocus probes. Multi-locus probes bind to more than one site in the sample, and give complex profile patterns. This can make the results difficult to analyse, but it does decrease the possibility of a chance match. This is obviously important in cases where legal decisions are made on the strength of DNA fingerprint evidence. The odds against a chance match for varying numbers of bands in a DNA profile are shown in Table The use of multi-locus DNA profiling in a forensic case is shown in Fig In this example, blood from the victim is the reference sample. Samples from seven suspects were obtained and treated along with the sample from the victim. By matching the band patterns it is clear that suspect 5 is the guilty party. 86 BIOLOGY
87 APPLICATIONS OF DNA TECHNOLOGY Table 4.3.2: The odds against chance matches in a DNA fingerprint. The more bands present, the less likely it is that any match is due to chance. (Courtesy of Cellmark Diagnostics.) Number of bands in fingerprint Odds against a chance match : 1 6 4,000 : ,000 : million : million : million : ,300 million : ,000 million : million million : 1 BIOLOGY 87
88 APPLICATIONS OF DNA TECHNOLOGY Fig : A DNA profile prepared using a multi-locus probe. Samples of DNA from the victim (V; boxed) and seven suspects (1 7) were cut with a restriction enzyme and separated on an agarose gel. The fragments were blotted onto a filter and a radioactive probe added. The probe hybridises to the target sequences, producing a profile pattern on X-ray film. The band patterns from the victim and suspect 5 match. (Copyright Cellmark Diagnostics. Reproduced with permission.) Single-locus probes will bind to just one complementary sequence in the haploid genome. Thus, two bands will be visible in the resulting autoradiogram; one from the paternal chromosome and one from the maternal chromosome. This gives a simple profile that is often sufficient to demonstrate an unambiguous match between the suspect and the reference. The result of a paternity test using a single-locus probe is shown in Fig Sometimes two or more probes can be used to increase the number of bands in the profile. Single-locus probes are more sensitive multi-locus probes, and can detect much smaller amounts of DNA. Usually both single-locus and multi-locus probes are used in any given case, and the results combined. 88 BIOLOGY
89 APPLICATIONS OF DNA TECHNOLOGY Fig : A DNA profile prepared using a single-locus probe for paternity testing. Samples of DNA from the mother (M), 4 children (1 4) and the father (F) were prepared as in Fig A single-locus probe was used in this analysis. The band patterns show two maternal bands and two paternal bands. In the case of child 1, the paternal band is different from either of the two bands in lane F, indicating a different father (band labelled DF). This child was in fact born to the mother during a previous marriage. (Copyright Cellmark Diagnostics. Reproduced with permission.) In forensic analysis, the original DNA profiling technique shown in Fig has now been largely replaced by a PCR-based technique that amplifies parts of the DNA known as short tandem repeats (STRs, also known as microsatellites). The PCR reaction overcomes any problems associated with the tiny amounts of sample that are often found at the crime scene. By using fluorescent labels and automated DNA detection equipment (similar to the genome sequencing equipment shown in Fig ) a DNA profile can be generated quickly and accurately. To ensure that results from DNA profile analysis are admissible as evidence in legal cases, several important quality control steps must be in place. These include accurate recording of the samples as they arrive at the laboratory, and careful cross-checking of the procedures to make sure that the test is carried out properly and that the samples do not get mixed up. If PCR amplification is used as part of the procedure, great care must be taken to ensure that no trace of DNA contamination is present. A smear of the operator s sweat can often be enough to ruin a BIOLOGY 89
90 APPLICATIONS OF DNA TECHNOLOGY test, so strict operating procedures must be observed, and laboratories inspected and authorised to conduct the tests. This is essential if public confidence in the technique is to be maintained. 4.4 Agriculture Gene technology in agriculture has great potential, and can be applied to both plants and animals. The aim is usually to achieve genetic modification of the target organism to improve some aspect that would be of benefit to the producer or the consumer. Plants that are resistant to disease, drought, herbicides and pesticides increase yields and enable growth to be controlled more easily. Farm animals with increased yields of milk and meat would seem to be a positive development, as would increasing the shelf life of fruits and vegetables. However, not all developments are accepted by the consumer, which poses a problem for all concerned in the development, production and marketing of genetically modified plants and animals. Genetic modification usually involves inserting genes for the desired characteristic into the host s DNA to produce a transgenic organism, which carries the new genetic material in a stable form that is expressed and is also transmitted from generation to generation. An alternative use of DNA technology is to produce substances such as growth hormones by recombinant DNA methods, and use the product to affect the target organism. We will consider both of these approaches by looking firstly at transgenic plants, and then the production and use of bovine growth hormone. Transgenic plants There are several key stages in the production of a transgenic organism, but two requirements are of particular importance. These are (i) a suitable vector system that will enable the cloned DNA fragment to be inserted into the plant cell genome, and (ii) a mechanism for regenerating whole plants in which all the cells carry the transgene. If the latter is not achieved, a mosaic organism can arise, in which only some of the cells have been modified. In plants, the techniques of tissue culture can be used to regenerate whole plants from single cells (see Fig.1.5.1) and thus mosaics are not usually a problem. The most commonly used vector system for plant gene manipulation is the Ti plasmid. This is found in the soil bacterium Agrobacterium tumefaciens, and is responsible for causing crown gall disease in plants. 90 BIOLOGY
91 APPLICATIONS OF DNA TECHNOLOGY This disease produces tumours at the base of the plant stem. The Ti plasmid (Ti stands for Tumour inducing) is a large plasmid that carries a region of DNA called T-DNA that can integrate into the plant cell genome. This feature can be used to construct vectors that can be used to deliver the target gene into the plant cell. The Ti plasmid itself is too large to be used as a vector directly, as large plasmids are difficult to handle in vitro without fragmenting. Thus smaller vector plasmids with only the essential T-DNA region have been constructed. In many cases these plasmids lack the tumour-forming characteristics of the intact Ti plasmid, and thus do not cause tumours when regenerating the plant from the genetically engineered cells. This makes it easier to produce plants that are essentially normal apart from the inclusion of the transgene. A summary of the method for producing a transgenic plant is shown in Fig Fig : Generating a transgenic plant. In (a), the Ti-based vector is prepared from the host strain of Agrobacterium tumefaciens (although often E. coli is used as the host at this stage). The vector is cut with a restriction enzyme (RE). In (b), the gene of interest is isolated and inserted into the vector to generate a recombinant plasmid. This can be transferred into plant cells in culture, either by direct introduction or by using Agrobacterium infection to deliver the gene. Plants with the gene are regenerated from tissue culture, and can then be used in standard breeding procedures. An example of the use of transgenic plant technology is the construction of transgenic tomato plants that have been engineered to delay the ripening process. In the transgenic plant the production of ethylene (which causes ripening) is inhibited, and thus the fruit stays firmer for BIOLOGY 91
92 APPLICATIONS OF DNA TECHNOLOGY longer. This technology was used by the biotechnology company Calgene to produce the Flavr Savr (sic) tomato. Another example is the incorporation of insect resistance into plants by inserting a gene that produces an insecticidal protein. In one case a gene from the soil bacterium Bacillus thuringiensis was inserted into tomato and tobacco plants. When insect pests feed on the plants expressing the gene, the protein is converted into an insecticidal toxin by enzymes in the insect s gut. Other animals do not have this enzyme, and thus are not affected by the toxin. A major goal of plant genetic modification is the introduction of nitrogen fixation genes into non-leguminous crops, which lack the root nodules that contain the nitrogen-fixing bacteria Rhizobium spp. One of the problems is that the Ti-based vector system does not infect monocot plants such as cereals and grasses, many of which are the main target crops. However, other methods of introducing genes into these crops are being used with some success. One of the main controversies in plant genetic modification arose over the production of Roundup ready soybean plants by the biotechnology company Monsanto. These plants were modified to be resistant to the herbicide glyphosate, which is the active component of the commercial weedkiller Roundup. The benefits of this are not in question farmers can use this broad-spectrum herbicide to control weeds without affecting the modified crop plants. However, the main concern has centred on the containment of the modified plants. There are strict guidelines to regulate how such plants can be grown, and several cases have been reported where these guidelines were breached. Many people are concerned that herbicide resistance may spread to other species, with potentially serious consequences in the medium to long term. This is an area of active debate at present, and much more careful work is needed to reassure the public that genetically modified crops are safe for both consumption and in terms of their environmental impact. Bovine growth hormone Agricultural animals are also targets for genetic modification, and many types of transgenic animals have been constructed. As with plants, there are concerns about the use of this type of modification, both in terms of the technical aspects and also in the area of animal welfare, and this is again an area of heated debate. However, one application of gene technology in agriculture is already well established, and is different in that it involves a genetically engineered product that is administered to 92 BIOLOGY
93 APPLICATIONS OF DNA TECHNOLOGY farm animals. Examples include vaccines and antibodies, as well as growth hormones that can be used to increase growth and milk production in cattle. Tests have also been carried out with recombinant cellulase, which hydrolyses cellulose and therefore improves the digestibility of animal feed that includes plant material. The growth hormone bovine somatotrophin (BST) has been available for several years from Monsanto. The gene for BST has been cloned into a bacterial system where it is expressed during bacterial growth. The product is then purified and prepared for administration to cattle, either by injection or by including the protein in animal feed. The production of recombinant BST is outlined in Fig The effects of BST are to increase milk production by around 10%, which is obviously attractive to farmers. In strictly technical terms the result is not harmful to humans, as any ingested protein will be digested in the gut and will not have any effect on the consumer. However, as in the case of crop plants, there is widespread public concern over the use of this technology, with some countries refusing to import meat or milk from BST-treated cattle. Fig : The production of bovine somatotrophin (BST). In (a), the vector plasmid is prepared and cut with a restriction enzyme. In (b), the gene for BST is isolated from cattle and ligated into the vector to produce a recombinant. This is replaced into E. coli cells, where BST protein is synthesised. This can then be produced commercially using largescale fermentation processes as shown in (c). BIOLOGY 93
94 APPLICATIONS OF DNA TECHNOLOGY The future? Despite the many useful advances that have been made in applying gene manipulation techniques to agriculture, at the end of the 1990s a very strong feeling of unease developed in some parts of the scientific community and within the general public. This centred on the problems associated with transgenic crop plants for human consumption. Terms such as Frankenstein foods have been used, which tend to sensationalise the problems rather than address them objectively. Animal welfare issues associated with transgenic farm animals have also provoked heated debate, and many people have called for a halt to developments. Such caution is not new in DNA technology in the 1970s scientists were concerned about the spread of potentially harmful characteristics among bacterial populations. What is different is that now the public are more aware of the impact that gene manipulation is having on our lives and on the environment. Perhaps the greatest challenge for scientists in the new millennium is to use DNA technology in a way that is acceptable to everyone concerned. Towards the end of 1999 many supermarkets were claiming that they would not stock genetically modified foods, and this is a very real problem for the companies involved in the production and use of transgenic crop plants. In many ways the problems that lie ahead in genetic modification are the social and ethical aspects of the technology, rather than technical difficulties. What is certain is that the debates will continue, and that we need to increase public awareness of both the benefits and the possible problems associated with gene manipulation and its applications. 94 BIOLOGY
95 FURTHER READING Campbell, N A et al, Biology (5th edn), Menlo Park and Harlow: Addison-Wesley, 1999 Raven, P H and Johnson, G B, Biology (5th edn), Boston and London: WCB/McGraw-Hill, 1999 Two popular general biology texts that have proved their worth through numerous editions. Both provide excellent coverage of the topics in the Unit, and also cover many other aspects in the Advanced Higher Course. Excellent value for money. Additional educational aids are available. Alberts, B et al, Essential Cell Biology: An Introduction to the Molecular Biology of the Cell, New York and London: Garland Publishing, 1998 An excellent shorter version of the classic Molecular Biology of the Cell. Covers all elements of the Advanced Higher Biology Unit in Cell and Molecular Biology. Very clearly written and illustrated. Brown, T A, Genomes, Oxford: Bios Scientific Publishers, 1999 A new text which approaches the subject of molecular genetics from the perspective of the genome rather than a more traditional treatment. Detailed coverage of genome structure and function, including mapping and sequencing. Sudbery, P, Human Molecular Genetics, Harlow: Longman, 1998 A detailed text dealing with aspects of human genetics, including disease, mapping and genome sequencing. BIOLOGY 95
96 96 BIOLOGY
97 GLOSSARY α-helix A right-handed helical form of secondary structure in proteins. actin A protein found in muscle fibres and other contractile components such as microfilaments. active transport The movement of substances across a membrane against the concentration gradient. This process requires energy. ADA Adenosine deaminase, an enzyme involved in nucleotide metabolism. ADA deficiency causes SCIDS q.v. ADA deficiency was one of the first targets for gene therapy. adenine Nitrogenous base found in DNA and RNA. Forms A:T base pairs with thymine. adenylate cyclase An enzyme which catalyses the formation of cyclic adenosine monophosphate (camp) from ATP. Involved in signal transduction. Sometimes known as adenyl cyclase. adrenaline A hormone of the catecholamine group, which has a variety of effects such as speeding up heartbeat and stimulating the breakdown of glycogen to glucose these effects are sometimes called the fight or flight response. Adrenaline can also act as a neurotransmitter. Also known as epinephrine. Agrobacterium tumefaciens A bacterium that is used in gene manipulation of plants. It carries the plasmid responsible for crown gall disease. allosteric modulator (effector) An effector that binds to a site on an enzyme, usually distinct from the active site, to regulate its activity. amino acid The monomeric unit of proteins. Made up of an amino group, carboxyl group and a hydrogen atom attached to a carbon known as the α-carbon. The fourth group is an R-group, which gives the amino acid its characteristics. There are 20 amino acids found in proteins. amino terminus (N-terminus) The end of a polypeptide chain which has the free amino group. BIOLOGY 97
98 GLOSSARY anabolic reaction A building up or biosynthetic reaction. Opposite of catabolic. anticodon The triplet of bases on a trna molecule that is complementary to the codon in mrna. antiparallel Describes the orientation of the strands in a DNA duplex; these run in opposite directions with respect to their 5'-3' polarity. antiproliferation genes Also known as tumour-suppressor genes, these are involved in controlling (restricting) cell division activity. When antiproliferation genes are defective, tumours may form. ATP Adenosine triphosphate. Made up of the sugar ribose and the base adenine (the nucleoside adenosine) and three phosphate groups (the triphosphate). ATP is one of the most important molecules in cell metabolism, as it is used as an energy transfer compound. ATPase An enzyme that hydrolyses ATP to produce ADP and energy. autosome A chromosome that is not a sex-determining chromosome. We have 22 pairs of autosomes and one pair of sex chromosomes. β-sheet Type of secondary structure in proteins where the polypeptide chains are arranged as parallel sheets, unlike the α-helix. batch culture Method of growing cells in culture where the volume of the culture is fixed. bioinformatics Applies to the gathering, storage and analysis of biological information. Usually refers to DNA sequence information generated by genome sequencing. blastula The hollow ball of cells formed after several rounds of cell division following fertilisation. bovine somatotrophin (BST) Growth hormone from cattle. Produced by recombinant DNA technology and available for administration to cattle to improve yield. C-terminus See carboxyl terminus. callus An undifferentiated mass of cells growing in tissue culture. 98 BIOLOGY
99 GLOSSARY cancer cell A cell that has become transformed to grow in an unrestrained way, often due to defective cell cycle control processes. carbohydrates Important metabolic compounds made up of carbon, hydrogen and oxygen. Monomers are the monosaccharides q.v., from which large polysaccharides can be synthesised. carboxyl terminus (C-terminus) The end of a polypeptide chain which has the free carboxyl group. carrier protein A membrane-bound protein that can bind other ions or molecules and transport them across the membrane by either facilitated diffusion or active transport. catabolic reaction A breaking down reaction (opposite of anabolic). The breakdown of glucose in respiration is an example of an important catabolic pathway. catalytic cycle The bind-catalyse-release cycle of events that enable enzymes to perform their catalytic functions. cell cycle The period between the formation of a cell and its division to form two new cells. Has four stages: G1, S, G2 and M. cell signalling General term to cover the generation, reception, transduction and effect of various types of molecules that act as interor intracellular signals. cellobiose Disaccharide composed of two glucose molecules joined together by a β(1,4) linkage. cellulase An enzyme that hydrolyses cellulose. cellulose Polysaccharide made up of glucose monomers joined by β(1,4) linkages. Found in cell walls of plants, it is the most abundant organic compound in the biosphere. centriole Microtubule-based structure found mostly in animal cells as a pair of centrioles, important in organising spindle fibres during cell division. The centriole is also part of the structure of cilia and flagella. centromere The constricted region of a replicated chromosome where the two chromatids are held together. BIOLOGY 99
100 GLOSSARY centrosome Structure found in eukarytoic cells that is involved in the organisation of microtubules, particularly during cell division. Cf. microtubule organiser. channel protein Type of transport protein that forms a channel across a membrane, through which substances can pass. chitin Polysaccharide found in fungal cell walls and insect exoskeletons, composed of N-acetylglucosamine units. chloroplast Organelle found in plant and algal cells. Highly organised internal structure of membranes, on which the light reactions of photosynthesis are localised. The dark reactions occur in the nonmembrane region or stroma. cholesterol Steroid found in cell membranes. Also the basis of many other steroid hormones. chromatid Refers to the copy of a chromosome after DNA replication, prior to cell division. Each replicated chromosome is composed of two chromatids, joined at the centromere. chromosome A DNA molecule carrying genetic information. In prokaryotic cells there is a single circular chromosome. In eukaryotic cells there are multiple linear chromosomes (46 in man), associated with histone and non-histone proteins. codon Triplet of bases in DNA or mrna which specifies an amino acid or a stop signal. complementary Refers to pairs of strands in a double-stranded nucleic acid molecule that bind together by base pairing. condensation Refers to a reaction in which two molecules are joined together by the removal of specific groups. Dehydration is a form of condensation synthesis where water is removed. conjugated protein A protein which has a non-protein component associated with it. Examples include glycoproteins, lipoproteins and nucleoproteins. cortisol A steroid hormone that can diffuse across the plasma membrane and act on gene regulatory proteins to stimulate transcription. 100 BIOLOGY
101 GLOSSARY covalent modification A method of regulating enzyme activity by adding/removing groups such as a phosphate group. cyclic AMP (camp) Cyclical form of AMP, often formed from ATP by the action of adenylate cyclase. Acts as a signalling molecule or second messenger. cystic fibrosis (CF) Disease in which ion transport across membranes is affected in individuals who are homozygous for the defective allele. Some 550 mutations in the CF gene have been described. CF is the most common monogenic disorder in Western societies, with around 1 in 2,000 affected. Carrier frequency is approximately 1 in 22. cytokinesis The separation of the cytoplasm during cell division. cytoplasm The contents of a cell apart from the nucleus. Includes the fluid cytosol and the cell organelles. cytosine A nitrogenous base found in DNA and RNA. Forms G:C base pairs with guanine. cytoskeleton The network of structural fibres in the cell, often associated with the cell membrane. The cytoskeleton is important for the maintenance of cell shape. cytosol The fluid-based part of the cytoplasm, excluding the organelles. dehydration Removal of the elements of water from a reaction in which two molecules are joined together. A specific type of condensation reaction. dictyosome Name sometimes used to describe the Golgi apparatus q.v. in plant cells. disulphide bond Covalent bond between two sulphur atoms. In proteins, disulphide bonds are formed between the sulphydryl (-SH) groups of cysteine residues in the polypeptide chain. Important in stabilising the 3-D structure of proteins. DNA Deoxyribonucleic acid (infrequently deoxyribosenucleic acid is used). DNA is a polynucleotide consisting of two complementary strands, base paired A:T and G:C. The genetic material in most organisms. BIOLOGY 101
102 GLOSSARY DNA fingerprinting (profiling) Technique which enables identification of individuals based on variations in the pattern of restriction fragments or by polymerase chain reaction q.v. amplification of specific sequences. The original technique gives a bar code result that is unique to each individual. DNA ligase Enzyme which catalyses the formation of phosphodiester bonds between adjacent nucleotides in a polynucleotide. Used in DNA replication and repair processes in vivo, it is also an essential part of recombinant DNA technology, where it is used as molecular glue. DNA replication The copying of the genetic material prior to cell division. Each strand of the double helix is used as a template for the formation of a new strand; replication is therefore semi-conservative in that the products contain one original and one new strand of DNA. DNA replication occurs during the S (synthesis) phase of the cell cycle. DNA sequencing Determination of the order of bases in a DNA strand. Various methods are available, the most commonly used being the dideoxy chain termination method. Automated DNA sequencing has enabled great progress to be made in sequencing whole genomes. domain In proteins, a region of defined 3-D structure. A single polypeptide chain may contain one or more domains, each folding into a separate region of tertiary structure. double helix The term used for the double-stranded DNA molecule, usually to describe the right-handed B-form of the helix. Drosophila melanogaster The fruit fly, an organism that has been used extensively in genetic research for classical, molecular and developmental genetic studies. EGF Epidermal Growth Factor, a local mediator signalling molecule. Stimulates cell division in epidermal and other cell types. electron microscope Complex (and expensive!) microscope which uses electron beams as the illumination source. Due to the short wavelength, much higher resolution (and hence magnification) can be achieved compared with light (optical) microscopy. Electromagnetic lenses are used to focus the beam of electrons, and a vacuum is required in the microscope body. Specimens must be dead 102 BIOLOGY
103 GLOSSARY and cut very thinly for transmission electron microscopy. Scanning electron microscopes generate 3-D images of the specimen. endocrine system The system of glands (endocrine glands) which facilitates cell cell signalling by secretion of hormones into the bloodstream. Cf. paracrine system. endomembrane system The system of internal membranes in eukaryotic cells. Usually refers to the endoplasmic reticulum, Golgi apparatus and plasma membrane with associated membrane-bound vesicles such as lysosomes. Chloroplasts and mitochondria are not considered part of the endomembrane network in cells. endoplasmic reticulum (ER) Part of the endomembrane system in eukaryotic cells. Consists of flattened sacs of membrane, linked to give a complex network of channels. May be smooth (SER; lacks ribosomes) or rough (RER; has ribosomes associated with the outer surface of the membranes). end-product inhibition Inhibition of an enzyme early in a metabolic pathway q.v. by the final product of the pathway. A form of negative feedback q.v. enzyme A biological catalyst, specific for a particular reaction. Usually protein molecules, often with associated non-protein components such as metal ions. Some RNA molecules can act as enzymes, and are known as ribozymes. enzyme-linked receptor Type of receptor found in membranes, which has an enzyme activity (often a kinase activity) associated with the cytoplasmic part of the receptor. This generates a response in the cytosol when the signal molecule binds. epidermal growth factor (EGF) See EGF. Escherichia coli Bacterium that has been a major research organism in microbial genetics and biochemistry. Thousands of different strains of E. coli are available. eukaryote (eukaryotic) Applies to cells (or organisms) in which there is a membrane-bound nucleus. explant Piece of tissue (usually applies to plants) prepared for growth under tissue culture conditions. BIOLOGY 103
104 GLOSSARY extrinsic protein A protein associated with the outer surface of the plasma membrane. facilitated diffusion Transport of molecules or ions across a membrane by a carrier protein with the concentration gradient. Does not require energy. fatty acid A long-chain organic acid (functional group COOH) that is a component of triacylglycerols and phospholipids. The general formula is CH 3 (CH 2 ) n COOH for saturated fatty acids. fermentation Anaerobic carbohydrate catabolism. The term is often used to describe the production of carbon dioxide and ethanol by microorganisms such as yeast. fibrous protein A protein which forms fibres and has a structural rather than enzymatic function. Examples include keratin and collagen. Cf. globular protein. fimbriae Projections from the surface of a bacterial cell. Cf. pili. flagella Thread-like structures, longer than fimbriae, involved in motility. Found in bacteria and also unicellular eukaryotes. fluid mosaic model The accepted model for membrane structure, in which proteins are embedded or associated with a phospholipid bilayer. foetal bovine serum (FBS) Calf serum, used as a supplement in growth media for animal cells in tissue culture. functional group The defining group of a molecule, such as COOH, NH 2, C=O, etc. GABA γ-aminobutyric acid, an inhibitory neurotransmitter. gastrulation Stage in embryological development formed by invagination of the blastula. gene Unit of genetic information which generates an RNA molecule. This may be mrna (which is translated to give a protein) or may be trna or rrna. gene manipulation Term used to describe the methods of recombinant DNA technology. 104 BIOLOGY
105 GLOSSARY gene regulatory protein A protein which binds to DNA to regulate or control gene expression. gene therapy Use of a gene sequence as a therapeutic agent to attempt to overcome the effects of a defective gene, as in ADA deficiency and cystic fibrosis. genetic mapping Method of determining the relative positions of genes on a chromosome by analysing recombination frequencies. Cf. physical mapping. genetic modification Alternative term for gene manipulation, often used where the aim is to alter the genetic makeup of an organism rather than simply to clone a gene or other DNA sequence. genome The genetic material of a cell or organism. In eukaryotes, can be used to describe nuclear, mitochondrial and chloroplast DNA. germ cell A reproductive cell as opposed to a somatic or body cell. globular protein Protein which folds to give a complex 3-D tertiary structure. Usually part of an enzyme. Cf. fibrous protein. glucose Important hexose monosaccharide of the formula C 6 H 12 O 6. Glucose catabolism is a major source of energy generation in the cell. glycerol Sugar alcohol with 3 carbons, forming the backbone of triacylglycerols and phospholipids. glycogen A storage polysaccharide found in animal cells. Composed of branched chains of glucose molecules. glycolysis A major pathway of primary carbohydrate catabolism where glucose is broken down to pyruvate. The term literally means sugar splitting. glycoprotein A protein with sugars attached to specific amino acid residues. glycosaminoglycans Polysaccharides made up of repeating amino sugar disaccharides. glycosidic bond The covalent bond joining two sugars together. BIOLOGY 105
106 GLOSSARY Golgi apparatus or complex Structure in eukaryotic cells involved in modifying and packaging materials such as proteins. Consists of flattened sacs of membrane. G-protein-linked receptor Class of receptor based on the seven-pass transmembrane protein, which has a G-protein binding site on the intracellular side of the membrane. Gram stain Method of staining bacterial cells which distinguishes cells on the basis of their cell wall structure. granum (pl. grana) Structural component of chloroplasts, composed of a stack of flattened membrane sacs known as thylakoids. growth factor Applies to any substance, other than a carbon source, which is needed for growth. guanine A nitrogenous base found in DNA and RNA. Forms G:C base pairs with cytosine. haeme group Iron-containing prosthetic group found in proteins such as myoglobin and haemoglobin. Involved in the transport of oxygen. haemoglobin Blood protein involved in carrying oxygen. Composed of 4 subunits, 2 α and 2 β chains, each with its own haem prosthetic group. heteropolymer A polymer in which the monomeric units are different; the best examples are protein molecules, made up of 20 different amino acid monomers. heterotroph An organism that requires an organic carbon source. Cf. photoautotroph. histamine Amine derived from histidine. Acts as a local mediator and is involved in inflammation and allergy responses. homopolymer A polymer in which the monomeric units are all the same; e.g. polyphenylalanine. hormone Chemical signalling molecule that is made in one tissue or gland, and transported to its site of action (another tissue) in the bloodstream. 106 BIOLOGY
107 GLOSSARY human genome project The international project to determine the base sequence of the human genome. Scheduled to be almost completed by mid-2000, the entire sequence of the 3 billion base pairs should be known by hybrid Can be used to describe cells that are fused to generate a new cell line in tissue culture. hybridisation In molecular biology, refers to the annealing of complementary nucleic acid sequences. Used to identify genes and other DNA fragments. hydrolase An enzyme that catalyses a hydrolytic reaction. hydrolysis The splitting apart of two molecules by the addition of the elements of water; the reverse of dehydration synthesis. hydrophilic Literally water-loving ; refers to molecules or parts of molecules that are attracted to an aqueous environment, such as the phosphate/nitrogen group of phospholipids. hydrophobic Literally water-hating ; hydrophobic molecules, such as the fatty acid tails of phospholipids, avoid or exclude water. hydrophobic interactions Attractions between hydrophobic R-groups, important in determination of the tertiary structure of proteins. immortalised cell line A cell line that is able to continue cell division indefinitely when grown in tissue culture. induced fit Alteration of the shape of an enzyme on binding a substrate, which enhances the binding of the substrate and action of the enzyme. Cf. lock and key. inositol triphosphate Intracellular signalling molecule or second messenger, produced by the action of phospholipase C. insulin Hormone involved in the regulation of blood glucose levels. intermediate filaments Elements of the cyoskeleton (others are microfilaments and microtubules), composed of fibrous protein strands. intrinsic protein A protein that is fully embedded in a membrane; sometimes called a transmembrane protein. BIOLOGY 107
108 GLOSSARY intron (intervening sequence) Region of a gene that does not code for the protein. Found in eukaryotic genes, introns account for the very large size of some genes. The are removed by RNA processing after transcription of the gene region. ion-channel-linked receptor Class of receptor that forms a pore across a membrane. May be permanently open or gated, where opening the channel requires a specific stimulus. isomers Compounds with the same number and type of atoms but different structures. isomerase An enzyme that catalyses an isomerisation, causing a change to the structure of a molecule. kinase An enzyme that adds a phosphate group, derived from ATP, onto another molecule. Krebs cycle Biochemical pathway for the complete oxidation of pyruvate. Also known as the citric acid cycle or tricarboxylic acid cycle. lac operon The cluster of genes encoding the enzymes needed for lactose catabolism in bacterial cells. lactose A disaccharide composed of glucose and galactose. light microscope Microscope with glass lenses that uses visible light as the illumination source. Also known as an optical microscope. lipid Molecule, usually classed with the macromolecules, that is soluble in an organic solvent but insoluble in water. A diverse range of molecules including the triacylglycerols, phospholipids and steroids. lipoprotein A protein with a lipid molecule attached. lock and key Model of enzyme action in which the substrate fits exactly into a pocket in the enzyme. Although essentially correct, the induced fit model q.v. is a better representation of what actually happens. lysosome Sub-cellular membrane-bound organelle containing hydrolytic enzymes involved in intracellular digestion. 108 BIOLOGY
109 GLOSSARY macromolecule A molecule made up of monomeric units joined together by dehydration synthesis. Polysaccharides, proteins and nucleic acids are the three main types found in cells. Lipids, although smaller, are often classed as one of the four groups of intracellular macromolecules. maltose Disaccharide composed of two glucose molecules. Usually produced by the hydrolysis of starch, and further hydrolysed by maltase to give glucose. messenger RNA (mrna) The RNA molecule that carries information, in the form of codons, produced by transcription of DNA. Cf. ribosomal RNA, transfer RNA. metabolic pathway A sequence of biochemical reactions that produces a particular product or products. May be controlled by end-product inhibition q.v. microbodies Diverse class of small membrane-bound sub-cellular organelles, including peroxisomes and glyoxysomes. microfilaments One of the three types of filament found in the cytoskeleton. Composed of the protein actin. Cf. intermediate filaments, microtubules. microtubule organiser Structure in a eukaryotic cell that controls microtubule formation. Cf. centrosome. microtubules Filamentous hollow strands composed of the protein tubulin. With microfilaments and intermediate filaments they make up the cytoskeleton. Also involved in spindle formation for cell division. microvilli Projections from the surface of epithelial cells, particularly gut cells. middle lamella The layer between two adjacent plant cell walls. mitochondrion (pl. mitochondria) Sub-cellular membrane-bound organelle involved in respiration. The inner mitochondrial membrane is folded to increase the surface area available for the localisation of the reactions of the electron transport chain. mitosis Cell division process in which chromatids (components of a replicated chromosome) are separated prior to cytokinesis q.v. BIOLOGY 109
110 GLOSSARY mitosis promoting factor (MPF) Protein complex involved in controlling the entry of cells into mitosis during the cell cycle. Activation of MPF is triggered by a rise in the concentration of cyclin, another cell cycle protein. monogenic Applies to a genetic trait that is caused by a single gene, as opposed to a polygenic trait, in which there may be many genes involved. monolayer Refers to a layer of cells, one cell deep, growing in a flask or dish. monomer The building block of polymers; examples are amino acids (polymers are proteins) and nucleotides (polymers are nucleic acids). monosaccharide Literally single sugar. Monosaccharides have the formula (CH 2 O) n. The simplest group are the trioses, with three carbon atoms. mosaic An organism whose cells have two or more genotypes. In transgenic (q.v.) organisms, refers to the condition where not all the cells carry the transgene. motif A particular structural feature of proteins or nucleic acids. Important in regulation of gene or enzyme activity. mucilaginous capsule Outer layer of some bacterial cells, composed of polysaccharide-based sticky material. multi-locus probe In genetic fingerprinting, a probe that binds to more than one sequence in the sample. muscular dystrophy (MD) An X-linked disease affecting young boys (frequency around 1 in 3,300), causing muscle wasting. Two variants are Duchenne MD and Becker MD. myoglobin Oxygen-carrying haeme protein composed of a single polypeptide chain. N-terminus See amino terminus. NAD(H) Co-enzyme involved in oxidation/reduction. Important in energy transfer reactions in the cell, the reduced form can be exchanged for ATP (q.v.) in aerobic respiration via the electron transport chain. 110 BIOLOGY
111 GLOSSARY negative feedback General term for control of a process where detection of high levels of a product causes a reduction in its production. Occurs in homeostatic physiological mechanisms and also in biochemical pathways as end-product inhibition q.v. neoplastic Refers to cells or tissues that arise as a result of uncontrolled cell growth, often causing cancers in animals. neuronal Type of communication involving nerve cells and neurotransmitters. neurone A nerve cell. neurotransmitter A chemical substance that transmits an electrical signal across the synapse in nerve cells. Examples include acetylcholine and noradrenaline. nitrogenous base A nitrogen-containing base found in nucleic acids. Chemically can be either a purine (double-ring structure; e.g. adenine and guanine) or a pyrimidine (single-ring; e.g. cytosine, thymine and uracil). nuclear envelope The double membrane surrounding the eukaryotic nucleus. nuclear magnetic resonance (NMR) A technique for determining the 3-D structure of small proteins and other molecules. nuclear membrane See nuclear envelope. nuclease An enzyme that hydrolyses phosphodiester bonds in nucleic acids. May be an endonuclease (cuts within a nucleic acid strand) or an exonuclease (cuts from one end). Ribonucleases digest RNA, deoxyribonucleases digest DNA. The restriction endonucelases are one important group. nucleic acid General term used to describe DNA and RNA. nucleoid Region in a prokaryotic cell where the DNA is localised; not a true membrane-bound nucleus. nucleoprotein A complex of protein and nucleic acid. nucleoside A sugar joined to a nitrogenous base. BIOLOGY 111
112 GLOSSARY nucleosomes Structures composed of histone proteins that are involved in packaging DNA in chromosomes. Nucleosomes look like beads on a string when seen in the electron microscope. nucleotide A sugar, base and phosphate. nucleus Membrane-bound compartment in eukaryotic cells. The DNA is located in the nucleus, which is also the site of DNA replication and transcription. oncogene A gene involved in cancer. The normal version of an oncogene is called a proto-oncogene. optical microscope See light microscope. organelle Sub-cellular membrane-bound structure in eukaryotic cells. oxidoreductases A class of enzymes involved in oxidation/reduction reactions. paracrine system Intercellular signalling system involving short-range effects. Cf. endocrine system. passive transport Transport of substances across a membrane without the need for energy expenditure. pedigree analysis Method of retrospective genetic analysis. Involves tracing inherited traits through family trees. Can enable predictions to be made about the likelihood of parents passing on genetic defects to their children. peptide bond The C N bond linking two amino acids together. Formed by dehydration of the amino and carboxyl groups of adjacent amino acids. peptidoglycan Component of the bacterial cell wall, composed of polysaccharide chains linked by short peptides. peripheral protein A protein associated with the outer layer of the plasma membrane. Sometimes called an extrinsic protein q.v. peroxisomes Sub-cellular membrane-bound organelles which contain catalase and peroxidases. Sometimes called microbodies q.v. 112 BIOLOGY
113 GLOSSARY phosphatase Enzyme that removes phosphate groups from molecules. phosphatidylcholine A phospholipid which is the major component of cell membranes. phosphodiester bond The bond linking two nucleotides together via a 5'-3' linkage, formed between the 5' phosphate of one nucleotide and the 3' hydroxyl of the next. phospholipase C Enzyme that catalyses the formation of the second messengers (q.v.) inositol triphosphate (IP 3 ) and diacylglycerol (DAG) from membrane lipids. phospholipid Type of lipid found in cell membranes. Based on triacylglycerol structure, with one of the fatty acid chains being substituted by a phosphate. This gives the molecule a hydrophobic/ hydrophilic polarity. phospholipid bilayer Two layers of phospholipid arranged with the hydrophobic fatty acid tails together. This gives a layer with a hydrophobic interior and a hydrophilic surface, which is the basis of cell membrane structure. photoautotroph An organism that can synthesise its requirements from inorganic carbon (CO 2 ) and light. Cf. heterotroph. photosynthesis The use of light energy to fix and reduce carbon dioxide into sugars. Carried out in the chloroplasts of algae and plants, and also by certain bacteria. physical mapping Applies to a variety of techniques to determine the positions of genes on a chromosome by locating a physical marker. Cf. genetic mapping. pili Projections from the bacterial cell surface. Cf. fimbriae. plasma membrane The membrane surrounding the cell. Sometimes known as the cell membrane or plasmalemma. plasmodesmata Cytoplasmic connections across cell walls in plants, linking adjacent plant cells. pluripotent Having the potential to develop into a number of different cell types, but not all the types that are found in the adult organism. BIOLOGY 113
114 GLOSSARY polygenic Refers to a genetic trait that involves a number of genes. Cf. monogenic. polymer A large molecule (macromolecule) composed of monomeric units joined together, usually by dehydration synthesis. polymerase An enzyme that synthesises a polymer. Examples include DNA and RNA polymerases, which are template-dependent polymerases. polymerase chain reaction (PCR) Method of amplifying DNA sequences by repeatedly copying the region between two primers. polynucleotide Polymer composed of nucleotides linked by phosphodiester bonds. polypeptide Polymer composed of amino acids linked by peptide bonds. polysaccharide Polymer composed of monosaccharides linked by glycosidic bonds. primary cell culture Cell culture derived from cells taken directly from a tissue sample. primary structure In proteins, the sequence of amino acids in a polypeptide chain. prokaryote Cell without a membrane-bound nucleus. proliferation genes Genes that encode proteins which promote cell division. Cf. antiproliferation genes. prosthetic group Non-protein group, such as a metal atom or a haeme group, associated with a protein and essential for its biological function. proteases Enzymes that catalyse the hydrolysis of peptide bonds, thus digesting proteins. Also known as proteinases. protein Heteropolymer (q.v.) of amino acids linked by peptide bonds. May contain one or more polypeptide chains. protofilament Element of microtubule structure made up of tubulin dimer subunits. 114 BIOLOGY
115 GLOSSARY protoplast A plant cell with the cell wall removed, usually by treatment with enzymes. protoplast fusion The joining of two protoplasts to form a hybrid cell. purine Double-ring base found in nucleic acids, the most common being adenine and guanine. pyrimidine Single-ring bases such as cytosine, thymine and uracil. quaternary structure Level of protein structure in which two or more subunits, each with its own secondary and tertiary structure, are held together in a complex 3-D arrangement. R-group Used to describe the variable functional group in amino acids. Responsible for determining the properties of polypeptide chains. receptor Protein to which a signalling molecule binds to elicit an intracellular response. recombinant DNA (rdna) General term used to describe the techniques of gene manipulation which generate DNA molecules with sequences not found naturally. residue Applies to the element of a monomer that is incorporated into a polymer. Thus amino acids in proteins are called amino acid residues. restriction endonuclease A nuclease that recognises a specific base sequence in DNA (usually 4, 5 or 6 base pairs in length) and cuts within or close to that sequence. Rhizobium spp. Bacteria involved in nitrogen fixation, associated with root nodules in certain plants. ribosomal RNA (rrna) RNA molecule that is a component of ribosome structure. Occurs as different forms, characterised by their size. Cf. messenger RNA, transfer RNA. ribosome The jig that brings together mrna and charged trna molecules during protein synthesis. Composed of a complex arrangement of ribosomal RNA and ribosomal proteins RNA Ribonucleic acid (infrequently the term ribosenucleic acid is used). A polynucleotide, usually single-stranded, composed of BIOLOGY 115
116 GLOSSARY ribonucleotides with the bases adenine, guanine, cytosine and uracil. Occurs as different forms such as mrna, rrna and trna q.v. The genetic material in some viruses. sarcolemma The membranous sheath around a muscle fibre. SCIDS Severe combined immunodeficiency syndrome; a disease caused by a deficiency in the enzyme adenosine deaminase (ADA, q.v.). A target for gene therapy. second messenger A small molecule involved in signal transduction. Generates a specific intracellular response to a signal received by a transmembrane receptor. Examples of second messengers include cyclic AMP (camp), inositol triphosphate (IP 3 ) and diacylglycerol (DAG). secondary structure In proteins, refers to the α-helix and β-sheet arrangements of the polypeptide chain. May also be used for some nucleic acid structures. signal transduction The conversion of signals from one form into another, as in the conversion of an extracellular signal to an intracellular response, often via a receptor protein and a second messenger q.v. single-locus probe In DNA fingerprinting, refers to a probe that binds to a single sequence in the genome. Diploid organisms will usually show two bands in a profile, one of maternal and one of paternal origin. sodium-potassium pump Transmembrane protein complex that pumps sodium out of the cell and potassium into the cell in a 3:2 ratio, using ATP hydrolysis to drive the process. Also known as the Na + -K + ATPase. somatic cell Body cell, i.e. any cell in an organism apart from the reproductive cells. spindle fibres Microtubule-based fibres used to pull chromosomes or chromatids apart during cell division. spindle poles The ends of the spindle fibre structure not attached to the centromere of the chromosome. Cf. centrosome, microtubule organiser. 116 BIOLOGY
117 GLOSSARY starch Storage polysaccharide of plants, composed of glucose monomers joined by α(1,4) bonds. May be unbranched (amylose) or branched (amylopectin). steroids Large group of polycyclic lipids based on a 4-ring structure. Examples include cholesterol and testosterone. subunit A component of a multi-subunit complex such as a large protein or a ribosome. synthase (synthetase) An enzyme that catalyses the joining of two molecules. tertiary structure Higher-order structure in proteins, where the polypeptide is folded into a complex 3-D shape or conformation. testosterone Steroid hormone that is the main male sex hormone in mammals. thymine Nitrogenous base found in DNA but not in RNA, where it is replaced by uracil. Forms A:T base pairs with adenine. thyroxine Iodine-containing hormone produced by the thyroid gland. Derived from the amino acid tyrosine. Ti plasmid Plasmid of Agrobacterium used for gene manipulation in plants. totipotent Having the potential to develop into all the types of cell that are found in the adult organism. transcription The synthesis of an RNA copy of DNA by the enzyme RNA polymerase. transfer RNA (trna) Clover-leaf shaped RNA which binds a specific amino acid and has an anticodon which pairs with the codon in mrna. Cf. messenger RNA, ribosomal RNA. transgenic An organism that has been genetically modified to carry a foreign gene. translation The synthesis of proteins, involving ribosomes, trnas with their associated amino acids, and mrna. BIOLOGY 117
118 GLOSSARY transmembrane protein A protein that is embedded in a membrane so that one portion is on the outside and one on the inside. Often function as receptor proteins. triacylglycerol A lipid composed of glycerol plus three fatty acids. Often called triglycerides. tricarboxylic acid (TCA) cycle Biochemical pathway for the complete oxidation of pyruvate. Also known as the citric acid cycle or Krebs cycle. trypsin Protease, formed from its zymogen (q.v.) trypsinogen, involved in digestion. tubulin Protein component of microtubules q.v. Occurs as two forms, α and β. tumour-suppressor genes Also known as antiproliferation genes, these are involved in controlling (restricting) cell division activity. When defective, tumours may form. uracil Nitrogenous base found in RNA only, where it replaces thymine. Forms A:U base pairs with adenine. vacuole Fluid-filled membrane-bound organelle in plant cells which shows little structural detail. Often occupies a large proportion of the cell volume. X-ray crystallography Method of determining the structure of complex molecules by analysing the diffraction patterns of X-rays by crystals of the molecule. zymogen Inactive precursor of some enzymes. Converted to the active enzyme by cleavage of the polypeptide. 118 BIOLOGY
Disaccharides consist of two monosaccharide monomers covalently linked by a glycosidic bond. They function in sugar transport.
 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
Chapter 5. The Structure and Function of Macromolecule s
 Chapter 5 The Structure and Function of Macromolecule s Most Macromolecules are polymers: Polymer: (poly: many; mer: part) Large molecules consisting of many identical or similar subunits connected together.
Chapter 5 The Structure and Function of Macromolecule s Most Macromolecules are polymers: Polymer: (poly: many; mer: part) Large molecules consisting of many identical or similar subunits connected together.
Carbohydrates, proteins and lipids
 Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
A disaccharide is formed when a dehydration reaction joins two monosaccharides. This covalent bond is called a glycosidic linkage.
 CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
The Structure and Function of Macromolecules: Carbohydrates, Lipids & Phospholipids
 The Structure and Function of Macromolecules: Carbohydrates, Lipids & Phospholipids The FOUR Classes of Large Biomolecules All living things are made up of four classes of large biological molecules: Carbohydrates
The Structure and Function of Macromolecules: Carbohydrates, Lipids & Phospholipids The FOUR Classes of Large Biomolecules All living things are made up of four classes of large biological molecules: Carbohydrates
Chapter 3 Molecules of Cells
 Bio 100 Molecules of cells 1 Chapter 3 Molecules of Cells Compounds containing carbon are called organic compounds Molecules such as methane that are only composed of carbon and hydrogen are called hydrocarbons
Bio 100 Molecules of cells 1 Chapter 3 Molecules of Cells Compounds containing carbon are called organic compounds Molecules such as methane that are only composed of carbon and hydrogen are called hydrocarbons
Biochemistry of Cells
 Biochemistry of Cells 1 Carbon-based Molecules Although a cell is mostly water, the rest of the cell consists mostly of carbon-based molecules Organic chemistry is the study of carbon compounds Carbon
Biochemistry of Cells 1 Carbon-based Molecules Although a cell is mostly water, the rest of the cell consists mostly of carbon-based molecules Organic chemistry is the study of carbon compounds Carbon
Cell Structure & Function!
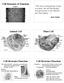 Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
How To Understand The Chemistry Of Organic Molecules
 CHAPTER 3 THE CHEMISTRY OF ORGANIC MOLECULES 3.1 Organic Molecules The chemistry of carbon accounts for the diversity of organic molecules found in living things. Carbon has six electrons, four of which
CHAPTER 3 THE CHEMISTRY OF ORGANIC MOLECULES 3.1 Organic Molecules The chemistry of carbon accounts for the diversity of organic molecules found in living things. Carbon has six electrons, four of which
THE HISTORY OF CELL BIOLOGY
 SECTION 4-1 REVIEW THE HISTORY OF CELL BIOLOGY Define the following terms. 1. cell 2. cell theory Write the correct letter in the blank. 1. One early piece of evidence supporting the cell theory was the
SECTION 4-1 REVIEW THE HISTORY OF CELL BIOLOGY Define the following terms. 1. cell 2. cell theory Write the correct letter in the blank. 1. One early piece of evidence supporting the cell theory was the
Biological molecules:
 Biological molecules: All are organic (based on carbon). Monomers vs. polymers: Monomers refer to the subunits that, when polymerized, make up a larger polymer. Monomers may function on their own in some
Biological molecules: All are organic (based on carbon). Monomers vs. polymers: Monomers refer to the subunits that, when polymerized, make up a larger polymer. Monomers may function on their own in some
The Molecules of Cells
 The Molecules of Cells I. Introduction A. Most of the world s population cannot digest milk-based foods. 1. These people are lactose intolerant because they lack the enzyme lactase. 2. This illustrates
The Molecules of Cells I. Introduction A. Most of the world s population cannot digest milk-based foods. 1. These people are lactose intolerant because they lack the enzyme lactase. 2. This illustrates
4. Which carbohydrate would you find as part of a molecule of RNA? a. Galactose b. Deoxyribose c. Ribose d. Glucose
 1. How is a polymer formed from multiple monomers? a. From the growth of the chain of carbon atoms b. By the removal of an OH group and a hydrogen atom c. By the addition of an OH group and a hydrogen
1. How is a polymer formed from multiple monomers? a. From the growth of the chain of carbon atoms b. By the removal of an OH group and a hydrogen atom c. By the addition of an OH group and a hydrogen
Review of the Cell and Its Organelles
 Biology Learning Centre Review of the Cell and Its Organelles Tips for most effective learning of this material: Memorize the names and structures over several days. This will help you retain what you
Biology Learning Centre Review of the Cell and Its Organelles Tips for most effective learning of this material: Memorize the names and structures over several days. This will help you retain what you
Cytology. Living organisms are made up of cells. Either PROKARYOTIC or EUKARYOTIC cells.
 CYTOLOGY Cytology Living organisms are made up of cells. Either PROKARYOTIC or EUKARYOTIC cells. A. two major cell types B. distinguished by structural organization See table on handout for differences.
CYTOLOGY Cytology Living organisms are made up of cells. Either PROKARYOTIC or EUKARYOTIC cells. A. two major cell types B. distinguished by structural organization See table on handout for differences.
Lecture Overview. Hydrogen Bonds. Special Properties of Water Molecules. Universal Solvent. ph Scale Illustrated. special properties of water
 Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
CELLS: PLANT CELLS 20 FEBRUARY 2013
 CELLS: PLANT CELLS 20 FEBRUARY 2013 Lesson Description In this lesson we will discuss the following: The Cell Theory Terminology Parts of Plant Cells: Organelles Difference between plant and animal cells
CELLS: PLANT CELLS 20 FEBRUARY 2013 Lesson Description In this lesson we will discuss the following: The Cell Theory Terminology Parts of Plant Cells: Organelles Difference between plant and animal cells
Anatomy and Physiology Placement Exam 2 Practice with Answers at End!
 Anatomy and Physiology Placement Exam 2 Practice with Answers at End! General Chemical Principles 1. bonds are characterized by the sharing of electrons between the participating atoms. a. hydrogen b.
Anatomy and Physiology Placement Exam 2 Practice with Answers at End! General Chemical Principles 1. bonds are characterized by the sharing of electrons between the participating atoms. a. hydrogen b.
Chapter 5: The Structure and Function of Large Biological Molecules
 Name Period Concept 5.1 Macromolecules are polymers, built from monomers 1. The large molecules of all living things fall into just four main classes. Name them. 2. Circle the three classes that are called
Name Period Concept 5.1 Macromolecules are polymers, built from monomers 1. The large molecules of all living things fall into just four main classes. Name them. 2. Circle the three classes that are called
Name: Hour: Elements & Macromolecules in Organisms
 Name: Hour: Elements & Macromolecules in Organisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight. All compounds
Name: Hour: Elements & Macromolecules in Organisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight. All compounds
Plasma Membrane hydrophilic polar heads
 The Parts of the Cell 3 main parts in ALL cells: plasma membrane, cytoplasm, genetic material this is about the parts of a generic eukaryotic cell Plasma Membrane -is a fluid mosaic model membrane is fluid
The Parts of the Cell 3 main parts in ALL cells: plasma membrane, cytoplasm, genetic material this is about the parts of a generic eukaryotic cell Plasma Membrane -is a fluid mosaic model membrane is fluid
1. The diagram below represents a biological process
 1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
Keystone Review Practice Test Module A Cells and Cell Processes. 1. Which characteristic is shared by all prokaryotes and eukaryotes?
 Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
I. Chapter 5 Summary. II. Nucleotides & Nucleic Acids. III. Lipids
 I. Chapter 5 Summary A. Simple Sugars (CH 2 O) n : 1. One C contains a carbonyl (C=O) rest contain - 2. Classification by functional group: aldoses & ketoses 3. Classification by number of C's: trioses,
I. Chapter 5 Summary A. Simple Sugars (CH 2 O) n : 1. One C contains a carbonyl (C=O) rest contain - 2. Classification by functional group: aldoses & ketoses 3. Classification by number of C's: trioses,
Carbon-organic Compounds
 Elements in Cells The living substance of cells is made up of cytoplasm and the structures within it. About 96% of cytoplasm and its included structures are composed of the elements carbon, hydrogen, oxygen,
Elements in Cells The living substance of cells is made up of cytoplasm and the structures within it. About 96% of cytoplasm and its included structures are composed of the elements carbon, hydrogen, oxygen,
Chapter 3: Biological Molecules. 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids
 Chapter 3: Biological Molecules 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids Elements in Biological Molecules Biological macromolecules are made almost entirely of just 6 elements: Carbon (C)
Chapter 3: Biological Molecules 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids Elements in Biological Molecules Biological macromolecules are made almost entirely of just 6 elements: Carbon (C)
Chapter 2: Cell Structure and Function pg. 70-107
 UNIT 1: Biochemistry Chapter 2: Cell Structure and Function pg. 70-107 Organelles are internal structures that carry out specialized functions, interacting and complementing each other. Animal and plant
UNIT 1: Biochemistry Chapter 2: Cell Structure and Function pg. 70-107 Organelles are internal structures that carry out specialized functions, interacting and complementing each other. Animal and plant
PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY
 Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Elements in Biological Molecules
 Chapter 3: Biological Molecules 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids Elements in Biological Molecules Biological macromolecules are made almost entirely of just 6 elements: Carbon (C)
Chapter 3: Biological Molecules 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids Elements in Biological Molecules Biological macromolecules are made almost entirely of just 6 elements: Carbon (C)
BIOLOGICAL MOLECULES OF LIFE
 BIOLOGICAL MOLECULES OF LIFE C A R B O H Y D R A T E S, L I P I D S, P R O T E I N S, A N D N U C L E I C A C I D S The Academic Support Center @ Daytona State College (Science 115, Page 1 of 29) Carbon
BIOLOGICAL MOLECULES OF LIFE C A R B O H Y D R A T E S, L I P I D S, P R O T E I N S, A N D N U C L E I C A C I D S The Academic Support Center @ Daytona State College (Science 115, Page 1 of 29) Carbon
Biology 101 Chapter 4 Cells as the Basic Unit of Life. The Cell Theory Major Contributors: Galileo = first observations made with a microscope
 Biology 101 Chapter 4 Cells as the Basic Unit of Life The Cell Theory Major Contributors: Galileo = first observations made with a microscope Robert Hooke = first to observe small compartments in dead
Biology 101 Chapter 4 Cells as the Basic Unit of Life The Cell Theory Major Contributors: Galileo = first observations made with a microscope Robert Hooke = first to observe small compartments in dead
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Cells & Cell Organelles
 Cells & Cell Organelles The Building Blocks of Life H Biology Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
Cells & Cell Organelles The Building Blocks of Life H Biology Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
Chapter 4: A Tour of the Cell. 1. Cell Basics. Limits to Cell Size. 1. Cell Basics. 2. Prokaryotic Cells. 3. Eukaryotic Cells
 Chapter 4: A Tour of the Cell 1. Cell Basics 2. Prokaryotic Cells 3. Eukaryotic Cells 1. Cell Basics Limits to Cell Size There are 2 main reasons why cells are so small: If cells get too large: 1) there
Chapter 4: A Tour of the Cell 1. Cell Basics 2. Prokaryotic Cells 3. Eukaryotic Cells 1. Cell Basics Limits to Cell Size There are 2 main reasons why cells are so small: If cells get too large: 1) there
3.1 AS Unit: Cells, Exchange and Transport
 3.1 AS Unit: Cells, Exchange and Transport Module 1: Cells 1.1.1 Cell Structure Candidates should be able to: (a) state the resolution and magnification that can be achieved by a light microscope, a transmission
3.1 AS Unit: Cells, Exchange and Transport Module 1: Cells 1.1.1 Cell Structure Candidates should be able to: (a) state the resolution and magnification that can be achieved by a light microscope, a transmission
Microscopes. Eukaryotes Eukaryotic cells are characterized by having: DNA in a nucleus that is bounded by a membranous nuclear envelope
 CH 6 The Cell Microscopy Scientists use microscopes to visualize cells too small to see with the naked eye. In a light microscope (LM), visible light is passed through a specimen and then through glass
CH 6 The Cell Microscopy Scientists use microscopes to visualize cells too small to see with the naked eye. In a light microscope (LM), visible light is passed through a specimen and then through glass
Given these characteristics of life, which of the following objects is considered a living organism? W. X. Y. Z.
 Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
If and when cancer cells stop dividing, they do so at random points, not at the normal checkpoints in the cell cycle.
 Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
The Cell: Organelle Diagrams
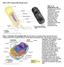 The Cell: Organelle Diagrams Fig 7-4. A prokaryotic cell. Lacking a true nucleus and the other membrane-enclosed organelles of the eukaryotic cell, the prokaryotic cell is much simpler in structure. Only
The Cell: Organelle Diagrams Fig 7-4. A prokaryotic cell. Lacking a true nucleus and the other membrane-enclosed organelles of the eukaryotic cell, the prokaryotic cell is much simpler in structure. Only
Cell and Membrane Practice. A. chromosome B. gene C. mitochondrion D. vacuole
 Name: ate: 1. Which structure is outside the nucleus of a cell and contains N?. chromosome. gene. mitochondrion. vacuole 2. potato core was placed in a beaker of water as shown in the figure below. Which
Name: ate: 1. Which structure is outside the nucleus of a cell and contains N?. chromosome. gene. mitochondrion. vacuole 2. potato core was placed in a beaker of water as shown in the figure below. Which
1. When applying the process of science, which of these is tested? a. an observation b. a result c. a hypothesis d. a question e.
 BCOR 11 Exam 1, 2004 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1. When applying the process of science, which of these is tested? a. an observation
BCOR 11 Exam 1, 2004 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1. When applying the process of science, which of these is tested? a. an observation
Plant and Animal Cells
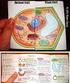 Plant and Animal Cells a. Explain that cells take in nutrients in order to grow, divide and to make needed materials. S7L2a b. Relate cell structures (cell membrane, nucleus, cytoplasm, chloroplasts, and
Plant and Animal Cells a. Explain that cells take in nutrients in order to grow, divide and to make needed materials. S7L2a b. Relate cell structures (cell membrane, nucleus, cytoplasm, chloroplasts, and
Lab 3 Organic Molecules of Biological Importance
 Name Biology 3 ID Number Lab 3 Organic Molecules of Biological Importance Section 1 - Organic Molecules Section 2 - Functional Groups Section 3 - From Building Blocks to Macromolecules Section 4 - Carbohydrates
Name Biology 3 ID Number Lab 3 Organic Molecules of Biological Importance Section 1 - Organic Molecules Section 2 - Functional Groups Section 3 - From Building Blocks to Macromolecules Section 4 - Carbohydrates
Organic Compounds. Essential Questions: What is Organic? What are the 4 major Organic Compounds? How are they made? What are they used for?
 Organic Compounds Essential Questions: What is Organic? What are the 4 major Organic Compounds? How are they made? What are they used for? Aristotle: Francesco Redi: What do we already know? Spontaneous
Organic Compounds Essential Questions: What is Organic? What are the 4 major Organic Compounds? How are they made? What are they used for? Aristotle: Francesco Redi: What do we already know? Spontaneous
Cell Structure and Function. Eukaryotic Cell: Neuron
 Cell Structure and Function Eukaryotic Cell: Neuron Cell Structure and Function Eukaryotic Cells: Blood Cells Cell Structure and Function Prokaryotic Cells: Bacteria Cell Structure and Function All living
Cell Structure and Function Eukaryotic Cell: Neuron Cell Structure and Function Eukaryotic Cells: Blood Cells Cell Structure and Function Prokaryotic Cells: Bacteria Cell Structure and Function All living
An Overview of Cells and Cell Research
 An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
Elements & Macromolecules in Organisms
 Name: Date: Per: Table # Elements & Macromolecules in rganisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight.
Name: Date: Per: Table # Elements & Macromolecules in rganisms Most common elements in living things are carbon, hydrogen, nitrogen, and oxygen. These four elements constitute about 95% of your body weight.
Introduction to the Cell: Plant and Animal Cells
 Introduction to the Cell: Plant and Animal Cells Tissues, Organs, and Systems of Living Things Cells, Cell Division, and Animal Systems and Plant Systems Cell Specialization Human Systems All organisms
Introduction to the Cell: Plant and Animal Cells Tissues, Organs, and Systems of Living Things Cells, Cell Division, and Animal Systems and Plant Systems Cell Specialization Human Systems All organisms
Comparing Plant And Animal Cells
 Comparing Plant And Animal Cells http://khanacademy.org/video?v=hmwvj9x4gny Plant Cells shape - most plant cells are squarish or rectangular in shape. amyloplast (starch storage organelle)- an organelle
Comparing Plant And Animal Cells http://khanacademy.org/video?v=hmwvj9x4gny Plant Cells shape - most plant cells are squarish or rectangular in shape. amyloplast (starch storage organelle)- an organelle
Carbon Hydrogen Oxygen Nitrogen
 Concept 1 - Thinking Practice 1. If the following molecules were to undergo a dehydration synthesis reaction, what molecules would result? Circle the parts of each amino acid that will interact and draw
Concept 1 - Thinking Practice 1. If the following molecules were to undergo a dehydration synthesis reaction, what molecules would result? Circle the parts of each amino acid that will interact and draw
cells - relatively simple cells - lack nuclear membrane and many organelles - bacteria and their relatives are all prokaryotic
 Cell Biology A cell is chemical system that is able to maintain its structure and reproduce. Cells are the fundamental unit of life. All living things are cells or composed of cells. 1 The interior contents
Cell Biology A cell is chemical system that is able to maintain its structure and reproduce. Cells are the fundamental unit of life. All living things are cells or composed of cells. 1 The interior contents
The Cell Teaching Notes and Answer Keys
 The Cell Teaching Notes and Answer Keys Subject area: Science / Biology Topic focus: The Cell: components, types of cells, organelles, levels of organization Learning Aims: describe similarities and differences
The Cell Teaching Notes and Answer Keys Subject area: Science / Biology Topic focus: The Cell: components, types of cells, organelles, levels of organization Learning Aims: describe similarities and differences
Preliminary MFM Quiz
 Preliminary MFM Quiz 1. The major carrier of chemical energy in all cells is: A) adenosine monophosphate B) adenosine diphosphate C) adenosine trisphosphate D) guanosine trisphosphate E) carbamoyl phosphate
Preliminary MFM Quiz 1. The major carrier of chemical energy in all cells is: A) adenosine monophosphate B) adenosine diphosphate C) adenosine trisphosphate D) guanosine trisphosphate E) carbamoyl phosphate
Compartmentalization of the Cell. Objectives. Recommended Reading. Professor Alfred Cuschieri. Department of Anatomy University of Malta
 Compartmentalization of the Cell Professor Alfred Cuschieri Department of Anatomy University of Malta Objectives By the end of this session the student should be able to: 1. Identify the different organelles
Compartmentalization of the Cell Professor Alfred Cuschieri Department of Anatomy University of Malta Objectives By the end of this session the student should be able to: 1. Identify the different organelles
Organelles and Their Functions
 Organelles and Their Functions The study of cell organelles and their functions is a fascinating part of biology. The current article provides a brief description of the structure of organelles and their
Organelles and Their Functions The study of cell organelles and their functions is a fascinating part of biology. The current article provides a brief description of the structure of organelles and their
Macromolecules 1 Carbohydrates, Lipids & Nucleic Acids
 VEA Bringing Learning to Life Program Support Notes Macromolecules 1 Carbohydrates, Lipids & Nucleic Acids Grades 10 - College 25mins Teacher Notes by Sue Wright, B. Sc., Dip. Ed. Produced by VEA Pty Ltd
VEA Bringing Learning to Life Program Support Notes Macromolecules 1 Carbohydrates, Lipids & Nucleic Acids Grades 10 - College 25mins Teacher Notes by Sue Wright, B. Sc., Dip. Ed. Produced by VEA Pty Ltd
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Cellular Structure and Function
 Chapter Test A CHAPTER 7 Cellular Structure and Function Part A: Multiple Choice In the space at the left, write the letter of the term or phrase that best answers each question. 1. Which defines a cell?
Chapter Test A CHAPTER 7 Cellular Structure and Function Part A: Multiple Choice In the space at the left, write the letter of the term or phrase that best answers each question. 1. Which defines a cell?
1.1.2. thebiotutor. AS Biology OCR. Unit F211: Cells, Exchange & Transport. Module 1.2 Cell Membranes. Notes & Questions.
 thebiotutor AS Biology OCR Unit F211: Cells, Exchange & Transport Module 1.2 Cell Membranes Notes & Questions Andy Todd 1 Outline the roles of membranes within cells and at the surface of cells. The main
thebiotutor AS Biology OCR Unit F211: Cells, Exchange & Transport Module 1.2 Cell Membranes Notes & Questions Andy Todd 1 Outline the roles of membranes within cells and at the surface of cells. The main
AP BIOLOGY 2006 SCORING GUIDELINES. Question 1
 AP BIOLOGY 2006 SCORING GUIDELINES Question 1 A major distinction between prokaryotes and eukaryotes is the presence of membrane-bound organelles in eukaryotes. (a) Describe the structure and function
AP BIOLOGY 2006 SCORING GUIDELINES Question 1 A major distinction between prokaryotes and eukaryotes is the presence of membrane-bound organelles in eukaryotes. (a) Describe the structure and function
7.2 Cells: A Look Inside
 CHAPTER 7 CELL STRUCTURE AND FUNCTION 7.2 Cells: A Look Inside Imagine a factory that makes thousands of cookies a day. Ingredients come into the factory, get mixed and baked, then the cookies are packaged.
CHAPTER 7 CELL STRUCTURE AND FUNCTION 7.2 Cells: A Look Inside Imagine a factory that makes thousands of cookies a day. Ingredients come into the factory, get mixed and baked, then the cookies are packaged.
Organic Molecules of Life - Exercise 2
 Organic Molecules of Life - Exercise 2 Objectives -Know the difference between a reducing sugar and a non-reducing sugar. -Distinguish Monosaccharides from Disaccharides and Polysaccharides -Understand
Organic Molecules of Life - Exercise 2 Objectives -Know the difference between a reducing sugar and a non-reducing sugar. -Distinguish Monosaccharides from Disaccharides and Polysaccharides -Understand
pathway that involves taking in heat from the environment at each step. C.
 Study Island Cell Energy Keystone Review 1. Cells obtain energy by either capturing light energy through photosynthesis or by breaking down carbohydrates through cellular respiration. In both photosynthesis
Study Island Cell Energy Keystone Review 1. Cells obtain energy by either capturing light energy through photosynthesis or by breaking down carbohydrates through cellular respiration. In both photosynthesis
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
Bacterial (Prokaryotic) Cell. Common features of all cells. Tour of the Cell. Eukaryotic Cell. Plasma Membrane defines inside from outside
 www.denniskunkel.com Tour of the Cell www.denniskunkel.com Today s Topics Properties of all cells Prokaryotes and Eukaryotes Functions of Major Cellular Organelles Information, Synthesis&Transport,, Vesicles
www.denniskunkel.com Tour of the Cell www.denniskunkel.com Today s Topics Properties of all cells Prokaryotes and Eukaryotes Functions of Major Cellular Organelles Information, Synthesis&Transport,, Vesicles
Lecture 4 Cell Membranes & Organelles
 Lecture 4 Cell Membranes & Organelles Structure of Animal Cells The Phospholipid Structure Phospholipid structure Encases all living cells Its basic structure is represented by the fluidmosaic model Phospholipid
Lecture 4 Cell Membranes & Organelles Structure of Animal Cells The Phospholipid Structure Phospholipid structure Encases all living cells Its basic structure is represented by the fluidmosaic model Phospholipid
Name Date Period. 2. When a molecule of double-stranded DNA undergoes replication, it results in
 DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
Electron Transport Generates a Proton Gradient Across the Membrane
 Electron Transport Generates a Proton Gradient Across the Membrane Each of respiratory enzyme complexes couples the energy released by electron transfer across it to an uptake of protons from water in
Electron Transport Generates a Proton Gradient Across the Membrane Each of respiratory enzyme complexes couples the energy released by electron transfer across it to an uptake of protons from water in
Cell Division Mitosis and the Cell Cycle
 Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cells. Structure, Function and Homeostasis
 Cells Structure, Function and Homeostasis Characteristics of Cells Basic unit of life anything alive is made of cells Plasma membrane (skin) that separates them from the environment. Skeletonsfor protection
Cells Structure, Function and Homeostasis Characteristics of Cells Basic unit of life anything alive is made of cells Plasma membrane (skin) that separates them from the environment. Skeletonsfor protection
Biology Chapter 7 Practice Test
 Biology Chapter 7 Practice Test Multiple Choice Write the letter that best answers the question or completes the statement on the line provided. 1. The work of Schleiden and Schwann can be summarized by
Biology Chapter 7 Practice Test Multiple Choice Write the letter that best answers the question or completes the statement on the line provided. 1. The work of Schleiden and Schwann can be summarized by
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257
 CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
7.2 Cell Structure. Lesson Objectives. Lesson Summary. Cell Organization Eukaryotic cells contain a nucleus and many specialized structures.
 7.2 Cell Structure Lesson Objectives Describe the structure and function of the cell nucleus. Describe the role of vacuoles, lysosomes, and the cytoskeleton. Identify the role of ribosomes, endoplasmic
7.2 Cell Structure Lesson Objectives Describe the structure and function of the cell nucleus. Describe the role of vacuoles, lysosomes, and the cytoskeleton. Identify the role of ribosomes, endoplasmic
Visualizing Cell Processes
 Visualizing Cell Processes A Series of Five Programs produced by BioMEDIA ASSOCIATES Content Guide for Program 3 Photosynthesis and Cellular Respiration Copyright 2001, BioMEDIA ASSOCIATES www.ebiomedia.com
Visualizing Cell Processes A Series of Five Programs produced by BioMEDIA ASSOCIATES Content Guide for Program 3 Photosynthesis and Cellular Respiration Copyright 2001, BioMEDIA ASSOCIATES www.ebiomedia.com
Cellular Reproduction
 9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
Examination One. Biology 101. Dr. Jaeson T. Fournier
 Examination One Biology 101 Dr. Jaeson T. Fournier Examination Instructions: Answers are to be indicated on a scantron. Keep your work protected! This helps prevent dishonesty. The instructor will not
Examination One Biology 101 Dr. Jaeson T. Fournier Examination Instructions: Answers are to be indicated on a scantron. Keep your work protected! This helps prevent dishonesty. The instructor will not
The Molecules of Life - Overview. The Molecules of Life. The Molecules of Life. The Molecules of Life
 The Molecules of Life - Overview The Molecules of Life The Importance of Carbon Organic Polymers / Monomers Functions of Organic Molecules Origin of Organic Molecules The Molecules of Life Water is the
The Molecules of Life - Overview The Molecules of Life The Importance of Carbon Organic Polymers / Monomers Functions of Organic Molecules Origin of Organic Molecules The Molecules of Life Water is the
MCAS Biology. Review Packet
 MCAS Biology Review Packet 1 Name Class Date 1. Define organic. THE CHEMISTRY OF LIFE 2. All living things are made up of 6 essential elements: SPONCH. Name the six elements of life. S N P C O H 3. Elements
MCAS Biology Review Packet 1 Name Class Date 1. Define organic. THE CHEMISTRY OF LIFE 2. All living things are made up of 6 essential elements: SPONCH. Name the six elements of life. S N P C O H 3. Elements
Worksheet 13.1. Chapter 13: Human biochemistry glossary
 Worksheet 13.1 Chapter 13: Human biochemistry glossary α-helix Refers to a secondary structure of a protein where the chain is twisted to form a regular helix, held by hydrogen bonds between peptide bonds
Worksheet 13.1 Chapter 13: Human biochemistry glossary α-helix Refers to a secondary structure of a protein where the chain is twisted to form a regular helix, held by hydrogen bonds between peptide bonds
Cell Division CELL DIVISION. Mitosis. Designation of Number of Chromosomes. Homologous Chromosomes. Meiosis
 Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
BSC 2010 - Exam I Lectures and Text Pages. The Plasma Membrane Structure and Function. Phospholipids. I. Intro to Biology (2-29) II.
 BSC 2010 - Exam I Lectures and Text Pages I. Intro to Biology (2-29) II. Chemistry of Life Chemistry review (30-46) Water (47-57) Carbon (58-67) Macromolecules (68-91) III. Cells and Membranes Cell structure
BSC 2010 - Exam I Lectures and Text Pages I. Intro to Biology (2-29) II. Chemistry of Life Chemistry review (30-46) Water (47-57) Carbon (58-67) Macromolecules (68-91) III. Cells and Membranes Cell structure
Biological cell membranes
 Unit 14: Cell biology. 14 2 Biological cell membranes The cell surface membrane surrounds the cell and acts as a barrier between the cell s contents and the environment. The cell membrane has multiple
Unit 14: Cell biology. 14 2 Biological cell membranes The cell surface membrane surrounds the cell and acts as a barrier between the cell s contents and the environment. The cell membrane has multiple
Unit I: Introduction To Scientific Processes
 Unit I: Introduction To Scientific Processes This unit is an introduction to the scientific process. This unit consists of a laboratory exercise where students go through the QPOE2 process step by step
Unit I: Introduction To Scientific Processes This unit is an introduction to the scientific process. This unit consists of a laboratory exercise where students go through the QPOE2 process step by step
WATER CHAPTER 3 - BIOCHEMISTRY "THE CHEMISTRY OF LIFE" POLARITY HYDROGEN BONDING
 CHAPTER 3 - BIOCHEMISTRY "THE CHEMISTRY OF LIFE" WATER Compare the body of the jellyfish with our own bodies. The jellyfish will die if it is removed from its water environment, yet we can live in the
CHAPTER 3 - BIOCHEMISTRY "THE CHEMISTRY OF LIFE" WATER Compare the body of the jellyfish with our own bodies. The jellyfish will die if it is removed from its water environment, yet we can live in the
10.1 The function of Digestion pg. 402
 10.1 The function of Digestion pg. 402 Macromolecules and Living Systems The body is made up of more than 60 % water. The water is found in the cells cytoplasm, the interstitial fluid and the blood (5
10.1 The function of Digestion pg. 402 Macromolecules and Living Systems The body is made up of more than 60 % water. The water is found in the cells cytoplasm, the interstitial fluid and the blood (5
CHROMOSOME STRUCTURE CHROMOSOME NUMBERS
 CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
3120-1 - Page 1. Name:
 Name: 1) Which series is arranged in correct order according to decreasing size of structures? A) DNA, nucleus, chromosome, nucleotide, nitrogenous base B) chromosome, nucleus, nitrogenous base, nucleotide,
Name: 1) Which series is arranged in correct order according to decreasing size of structures? A) DNA, nucleus, chromosome, nucleotide, nitrogenous base B) chromosome, nucleus, nitrogenous base, nucleotide,
Biology I. Chapter 7
 Biology I Chapter 7 Interest Grabber NOTEBOOK #1 Are All Cells Alike? All living things are made up of cells. Some organisms are composed of only one cell. Other organisms are made up of many cells. 1.
Biology I Chapter 7 Interest Grabber NOTEBOOK #1 Are All Cells Alike? All living things are made up of cells. Some organisms are composed of only one cell. Other organisms are made up of many cells. 1.
Investigating cells. Cells are the basic units of living things (this means that all living things are made up of one or more cells).
 SG Biology Summary notes Investigating cells Sub-topic a: Investigating living cells Cells are the basic units of living things (this means that all living things are made up of one or more cells). Cells
SG Biology Summary notes Investigating cells Sub-topic a: Investigating living cells Cells are the basic units of living things (this means that all living things are made up of one or more cells). Cells
Multiple Choice Questions
 Chapter 5 THE FUNDAMENTAL UNIT OF LIFE Multiple Choice Questions 1. Which of the following can be made into crystal? (a) A Bacterium (b) An Amoeba (c) A Virus (d) A Sperm 2. A cell will swell up if (a)
Chapter 5 THE FUNDAMENTAL UNIT OF LIFE Multiple Choice Questions 1. Which of the following can be made into crystal? (a) A Bacterium (b) An Amoeba (c) A Virus (d) A Sperm 2. A cell will swell up if (a)
Proteins and Nucleic Acids
 Proteins and Nucleic Acids Chapter 5 Macromolecules: Proteins Proteins Most structurally & functionally diverse group of biomolecules. : o Involved in almost everything o Enzymes o Structure (keratin,
Proteins and Nucleic Acids Chapter 5 Macromolecules: Proteins Proteins Most structurally & functionally diverse group of biomolecules. : o Involved in almost everything o Enzymes o Structure (keratin,
From DNA to Protein
 Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
Cell Unit Practice Test #1
 ell Unit Practice Test #1 Name: ate: 1. Which organelle is primarily concerned with the conversion of potential energy of organic compounds into suitable form for immediate use by the cell?. mitochondria.
ell Unit Practice Test #1 Name: ate: 1. Which organelle is primarily concerned with the conversion of potential energy of organic compounds into suitable form for immediate use by the cell?. mitochondria.
RAD 223. Radiography physiology. Lecture Notes. First lecture: Cell and Tissue
 RAD 223 Radiography physiology Lecture Notes First lecture: Cell and Tissue Physiology: the word physiology derived from a Greek word for study of nature. It is the study of how the body and its part work
RAD 223 Radiography physiology Lecture Notes First lecture: Cell and Tissue Physiology: the word physiology derived from a Greek word for study of nature. It is the study of how the body and its part work
The Cell Interior and Function
 The Cell Interior and Function 5 5.0 CHAPTER PREVIEW Investigate and understand the organization and function of the cell interior. Define the differences between eukaryotic and prokaryotic cell structure.
The Cell Interior and Function 5 5.0 CHAPTER PREVIEW Investigate and understand the organization and function of the cell interior. Define the differences between eukaryotic and prokaryotic cell structure.
Chapter 5 Organelles. Lesson Objectives List the organelles of the cell and their functions. Distinguish between plant and animal cells.
 Chapter 5 Organelles Lesson Objectives List the organelles of the cell and their functions. Distinguish between plant and animal cells. Check Your Understanding What is a cell? How do we visualize cells?
Chapter 5 Organelles Lesson Objectives List the organelles of the cell and their functions. Distinguish between plant and animal cells. Check Your Understanding What is a cell? How do we visualize cells?
AS Biology Unit 2 Key Terms and Definitions. Make sure you use these terms when answering exam questions!
 AS Biology Unit 2 Key Terms and Definitions Make sure you use these terms when answering exam questions! Chapter 7 Variation 7.1 Random Sampling Sampling a population to eliminate bias e.g. grid square
AS Biology Unit 2 Key Terms and Definitions Make sure you use these terms when answering exam questions! Chapter 7 Variation 7.1 Random Sampling Sampling a population to eliminate bias e.g. grid square
Chapter 3. Cellular Structure and Function Worksheets. 39 www.ck12.org
 Chapter 3 Cellular Structure and Function Worksheets (Opening image copyright by Sebastian Kaulitzki, 2010. Used under license from Shutterstock.com.) Lesson 3.1: Introduction to Cells Lesson 3.2: Cell
Chapter 3 Cellular Structure and Function Worksheets (Opening image copyright by Sebastian Kaulitzki, 2010. Used under license from Shutterstock.com.) Lesson 3.1: Introduction to Cells Lesson 3.2: Cell
CELL/ PHOTOSYNTHESIS/ CELLULAR RESPIRATION Test 2011 ANSWER 250 POINTS ANY WAY IN WHICH YOU WANT
 CELL/ PHOTOSYNTHESIS/ CELLULAR RESPIRATION Test 2011 ANSWER 250 POINTS ANY WAY IN WHICH YOU WANT Completion: complete each statement. (1 point each) 1. All cells arise from. 2. The basic unit of structure
CELL/ PHOTOSYNTHESIS/ CELLULAR RESPIRATION Test 2011 ANSWER 250 POINTS ANY WAY IN WHICH YOU WANT Completion: complete each statement. (1 point each) 1. All cells arise from. 2. The basic unit of structure
