Residues Ser195. His57. structures of chymotrypsin (in blue, Asp102
|
|
|
- Janis Rich
- 7 years ago
- Views:
Transcription
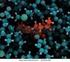
1 Enzyme Mechanisms Enzymes function by a wide variety of mechanisms. We will cover a few examples to illustrate the means that enzymes use to catalyze reactions. Serine proteases bond hydrolysis is a very common process. A wide variety of enzymes can perform proteolytic reactions. Members of one large family of protease are called serine proteases because of the important serine at the active site. All of the serine proteases contain three residues at their active site: a serine, a histidine, and an aspartate. These comprise the characteristic catalytic triad. In the numbering scheme for chymotrypsin (a numbering scheme which is typically used in studies of any of the mammalian serine proteases), the residues are Ser195, is57, and Asp102. ote that these residues are distributed throughout the 240 amino acid residues found in chymotrypsin, but are located in close proximity in the active three-dimensional conformation of the enzyme (see figure, below). The serine proteases are synthesized as larger, inactive, precursors. As an example: chymotrypsinogen is converted to chymotrypsin by two cleavage processes involving excision of residues and Interestingly the structures of chymotrypsinogen and chymotrypsin (including the catalytic residues) are almost completely superimposable; the conformational change involved in the conversion process appears to be fairly small. The implication is that relatively small structural changes can cause dramatic changes in activity. (ote: some changes in other aspects of the active site are observed. owever, because the catalytic triad residues are observed in their normal conformation, the zymogen forms of serine proteases usually exhibit low levels of catalytic activity.) Ser195 esidues This figure shows superimposed structures of chymotrypsin (in blue, from PDB ID 1A0) and chymotrypsinogen (in red, from PDB ID 2GA). ote the overall similarity in the structures, especially on the left side of the figure and in the position of the catalytic triad residues. The location of the excised residues (12-15 and ) removed in the conversion of chymotrypsinogen to chymotrypsin is shown in orange. is57 Asp102 esidues opyright Mark Brandt, Ph.D. 28
2 Some bacterial serine proteases (e.g., Streptomyces griseus Protease A) are structurally similar to the mammalian enzymes; the invention of the serine protease structure therefore occurred prior to the divergence of prokaryotes and eukaryotes. n the other hand, the Bacillus subtilus protease subtilisin, although it has the same catalytic residues, has a completely different structure, suggesting that serine protease mechanism has been invented more than once during evolution. (The figure below has a superimposition of bovine trypsin and subtilisin; note that the structures are totally different, but that Ser 195, is 57, and from trypsin have a very similar arrangement in three-dimensional space to Ser 221, is 64, and Asp 32 of subtilisin.) Bos taurus trypsin (pdb ID 2PT) Ser 195 is 57 Ser195 Ser221 is 64 Asp 32 Ser 221 Bacillus subtilis subtilisin (pdb ID 1GI) is 57 is 64 Asp 32 The serine proteases differ in their sequence and in their substrate specificity: the bacterial protease subtilisin (one of the major enzymes that resulted in the advertising slogan the cleaning power of enzymes! ) will cleave essentially any substrate, while one of the enzymes in the clotting cascade, Factor Xa, requires a four residue recognition sequence, Ile-Glu-Gly-Arg, in order to uniquely hydrolyze its polypeptide substrate after the arginine. Trypsin is specific for cleavage after lysine and arginine. Trypsin contains Asp 189 in the substrate side-chain binding pocket, and this residue forms an electrostatic interaction with basic residues. owever, the specificity is not completely dependent on a single residue; mutating Asp 189 to serine (the corresponding residue in chymotrypsin) converts trypsin into a non-specific protease, rather than into an enzyme with chymotrypsin-like specificity. (Even incorporating other mutations in nearby residues is only partially effective at producing a chymotrypsin-like enzyme; proteins are complex entities!) opyright Mark Brandt, Ph.D. 29
3 atalytic mechanism of serine proteases Serine proteases increase the rate of peptide bond hydrolysis by ~10 10 compared to the uncatalyzed reaction. A variety of structural features are responsible for the catalytic effectiveness of these enzymes. As mentioned above, the serine proteases all have three residues that are critical for catalysis: a serine, a histidine, and an aspartic acid. These are conserved in all of the serine proteases, and are superimposable in the crystal structures of these proteins. Steps in the catalytic process: 1) Substrate binding. ote that for the substrate peptide, the side-chain of the amino acid residue immediately before the scissile peptide bond can bind to the recognition site on the enzyme. Substrate side-chain recognition site Ser 195 is 57 2) ucleophilic attack. Serine 195 acts as a nucleophile, facilitated by istidine 57, which abstracts a proton from Ser 195. The result of the nucleophilic attack is a covalent bond between the Ser 195 side-chain oxygen and the substrate. The negative charge that develops on the peptide carbonyl oxygen is stabilized by hydrogen bonds formed from two protease backbone amide protons. This region of the protein is called the oxyanion hole, because it stabilizes the negative charge on the oxygen; the oxyanion hole is critical for catalysis. (ote: the oxyanion hole includes the backbone amide proton from Ser 195 ; the diagrams illustrating this discussion are thus somewhat distorted from the three-dimensional structure.) xyanion hole Ser 195 is 57 opyright Mark Brandt, Ph.D. 30
4 3) Protonation. is 57 donates a proton to the substrate amide nitrogen, allowing release of the -terminal part of the substrate as a free peptide. xyanion hole Ser 195 is 57 Ser 195 is 57 4) Ester hydrolysis. The final step is an attack by water on the ester bond between the peptide and the Ser 195 oxygen. This forms the second product peptide with a normal carboxyl group, and regenerates the serine hydroxyl. The second peptide then dissociates from the enzyme to allow another catalytic cycle to begin. 5) Ser 195 is 57 Ser 195 is 57 As is apparent in the series of drawings above, Ser 195 is the residue that performs the actual catalysis, while the other residues seem to be important for positioning of the serine and for stabilizing the intermediate states. Aspartate 102 is relatively far from the substrate; helps position is 57, and raises the pk a of is 57 to allow the histidine to act as a base. This is important to the catalytic process: mutation of to asparagine decreases the k cat by ~10,000-fold. The serine protease reaction mechanism involves a covalent intermediate between Ser 195 and the substrate. Although polypeptides do (very slowly) hydrolyze in aqueous solution, the hydrolysis reaction does not involve the formation of a covalent bond with a passing serine side-chain. Thus, the serine protease mechanism involves the use of an alternative pathway from that used in the uncatalyzed reaction. opyright Mark Brandt, Ph.D. 31
5 The serine protease mechanism does, however, involve stabilization of the transition state. This has been observed in mutant studies in which all of the catalytic triad residues were mutated. Although the reaction mechanism might be interpreted to predict that the loss of the active site serine would inactivate the enzyme, in fact, the triple mutant (where all three of the catalytic triad residues are changed to other residues) retains some ability to catalyze the reaction, and increases the rate of hydrolysis by ~5 x fold over the uncatalyzed reaction. This appears to be the result of the binding of the transition state to the enzyme, and in particular to the precise positioning of the oxyanion hole which allows the stabilization of the charged transition state. (Although all proteins contain peptide backbones, in general proteins lack the precise three-dimensional structure to catalyze reactions, which is why all proteins do not act as proteases.) Serine proteases are very useful as model enzymes for studying catalytic processes, because they use a variety of techniques for accelerating the reaction rate. They use acid-base catalysis (the histidine abstraction of the serine proton, and the histidine donation of the proton to allow release of the first peptide). Serine proteases also use charge stabilization (in particular, the oxyanion hole) and geometric factors (in particular the better fit of the tetrahedral alkoxide intermediate than the planar carbonyl structures) to lower the energy of the transition state, and use covalent catalysis to assist in the reaction. ow do we know all this? The serine proteases have been heavily studied. We will return to the serine protease mechanism later when we discuss inhibition, but one feature of research into the mechanism deserves some emphasis. The formation of the covalent enzymesubstrate intermediate was first proposed after a series of studies by B.S. artley and B.A. Kilby in They used a model substrate (p-nitrophenylacetate) to examine the kinetics of the reaction. hymotrypsin has a low k cat for p- nitrophenylacetate, which simplifies some of the analysis. The model substrate also has the advantage of forming a colored product p-nitrophenolate. They discovered that some p-nitrophenolate was released rapidly (burst phase kinetics) prior to the establishment of the steady state. [Product] ~1 mole product per mole enzyme Steady state Burst phase Time ormal enzyme kinetic measurements involve a brief period following addition of opyright Mark Brandt, Ph.D. 32
6 the enzyme, in which the enzyme-substrate complex forms. This pre-steady state condition is followed by the linear formation of product with time characteristic of steady state conditions. (The length of time required for steady state formation is somewhat exaggerated in the graph below). [Product] Steady state Time In the case of the serine proteases, a brief period of rapid product formation also occurs prior to establishment of steady state conditions. The amount of product formed in this burst phase corresponded to about 1 mole of product per mole of enzyme, suggesting that the enzyme rapidly formed a product, but then became trapped in a non-catalytic state that cycled back to the catalytic state more slowly. The mechanism that artley and Kilby proposed to explain the burst phase kinetics was that a covalent intermediate was formed between the enzyme and the acetate from the substrate, rapidly releasing the colored p-nitrophenolate. The acetylatedenzyme intermediate then slowly hydrolyzed to regenerate active enzyme. The initial burst of p-nitrophenolate production occurred at a high rate because it preceded the rate-limiting step for the reaction. nce the majority of the enzyme had become acetylated, the release of p-nitrophenolate became dependent on the rate-limiting step, which is the hydrolysis of the covalent bond formed to the enzyme. 3 hymotrypsin + p-nitrophenylacetate 2 2 p-nitrophenylate hydrolysis 3 hymotrypsin hymotrypsin + 3 When using normal polypeptide substrates, this burst phase kinetics can also occur. For most peptide substrates, however, the burst phase tends to be less obvious, because chymotrypsin cleaves polypeptides more effectively than it cleaves the model substrate p-nitrophenylacetate. These types of kinetic studies, when properly performed, allow insights into the enzyme mechanism, and have been the source of opyright Mark Brandt, Ph.D. 33
7 much of the available information regarding enzyme catalysis. The diagram below summarizes the reaction pathway for serine protease peptide bond cleavage. ote that the highest energy peak in the serine protease reaction pathway corresponds to the covalent intermediate hydrolysis process, which is the slowest step in the reaction. (X ) Uncatalyzed hydrolysis Energy E S ES E-S E-P P + 1 E-P 2 EP 2 Serine protease-catalyzed hydrolysis E + P Progress of eaction Aspartyl proteases Although serine proteases constitute a large family of enzymes with differing specificity, the serine-dependent catalytic triad is only one of many possible mechanisms for hydrolyzing peptide bonds. We will examine one other mechanism for proteolysis (a number of others exist). Aspartyl proteases comprise another family of proteolytic enzymes. This family includes the stomach digestive enzyme pepsin, the highly specific protease renin (the plasma enzyme that converts the circulating precursor angiotensinogen to the hormone angiotensin), and the IV protease (the enzyme that cleaves IV proteins during construction of the viral particle). Asp 25 IV Protease (from PDB ID 1V) Aspartyl proteases contain two aspartate residues at the active site; in some cases, as in the IV protease structure shown above, each monomer provides one of the opyright Mark Brandt, Ph.D. 34
8 two aspartate residues in the dimeric protein complex. While their catalytic mechanism differs considerably from that of the serine proteases, the end result, the hydrolysis of a peptide bond within the substrate to release two polypeptide products, is the same. atalytic mechanism The mechanism is different from that of the serine proteases. In the aspartyl proteases, the substrate never forms a covalent bond with the enzyme. Instead, the first step of the reaction (following substrate binding) is a nucleophilic attack by a water molecule on the substrate due to proton abstraction from the water by one of the aspartate from the enzyme. ne aspartate therefore acts as a base, while the water forms a covalent bond to the substrate. ote that while one aspartate removes a proton from the water molecule; the other aspartate acts as an acid, by releasing a proton to the carbonyl oxygen of the polypeptide substrate. In the next step, one aspartate residue acts as a base again, removing a proton from one of the hydroxyls that have been formed on the former carbonyl carbon. At the same time, the carbon-nitrogen bond breaks, with the nitrogen taking a proton from the other active site aspartate. This results in the formation of the carboxylic acid at one end of the new peptide, and the free amino group at the end of the other new peptide. The aspartyl protease mechanism requires that the environment of the protein opyright Mark Brandt, Ph.D. 35
9 result in the two aspartate residues being 50% protonated. Since the typical pk a of aspartate residues is about 4, this means that either the external p must be fairly low (as is the case for pepsin, a stomach enzyme that is most active at p of about 2), or the environment of the active site must alter the pk a of the aspartate residues enough to allow the partial protonation of the aspartate residues required for activity (as is the case for the renin and the IV protease, which are active at p of 7.4). Summary In order to recognize a substrate, the enzyme must have a structure that is capable of binding to the substrate. owever, many enzymes alter their structure upon substrate binding. Enzymes alter reaction rates by a variety of methods. These methods include increasing the local concentration of the substrate molecules by confining them to a small volume, positioning the substrates in appropriate orientations for the reaction to occur, inducing strain in the substrate, donating or abstracting protons, stabilizing charge development, and interacting covalently with the substrate. The variety of methods used result in enhanced reaction rates because of a decreased energy of the most energetic transition state in the pathway. This is possible because the enzyme binds to the transition state more tightly than to its substrate or products. Serine proteases, and to a lesser extent, aspartyl proteases, have been used as models for studying the effect of protein structure on specific catalytic processes. Serine proteases and aspartyl proteases catalyze peptide bond hydrolysis by different mechanisms; both mechanisms take advantage of acid-base catalysis, but the serine proteases form a covalent intermediate with their substrate, while the aspartyl proteases do not. opyright Mark Brandt, Ph.D. 36
ENZYMES. Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin. Principle of Enzyme Catalysis
 ENZYMES Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin Principle of Enzyme Catalysis Linus Pauling (1946) formulated the first basic principle of enzyme catalysis Enzyme increase the rate
ENZYMES Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin Principle of Enzyme Catalysis Linus Pauling (1946) formulated the first basic principle of enzyme catalysis Enzyme increase the rate
Lecture 15: Enzymes & Kinetics Mechanisms
 ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
Enzymes Enzyme Mechanism
 Mechanisms of Enzymes BCMB 3100 Chapters 6, 7, 8 Enzymes Enzyme Mechanism 1 Energy diagrams Binding modes of enzyme catalysis Chemical modes of enzyme catalysis Acid-Base catalysis Covalent catalysis Binding
Mechanisms of Enzymes BCMB 3100 Chapters 6, 7, 8 Enzymes Enzyme Mechanism 1 Energy diagrams Binding modes of enzyme catalysis Chemical modes of enzyme catalysis Acid-Base catalysis Covalent catalysis Binding
Biochemistry - I. Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture-11 Enzyme Mechanisms II
 Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture-11 Enzyme Mechanisms II In the last class we studied the enzyme mechanisms of ribonuclease A
Biochemistry - I Prof. S. Dasgupta Department of Chemistry Indian Institute of Technology, Kharagpur Lecture-11 Enzyme Mechanisms II In the last class we studied the enzyme mechanisms of ribonuclease A
Previous lecture: Today:
 Previous lecture: The energy requiring step from substrate to transition state is an energy barrier called the free energy of activation G Transition state is the unstable (10-13 seconds) highest energy
Previous lecture: The energy requiring step from substrate to transition state is an energy barrier called the free energy of activation G Transition state is the unstable (10-13 seconds) highest energy
How To Understand The Chemistry Of An Enzyme
 Chapt. 8 Enzymes as catalysts Ch. 8 Enzymes as catalysts Student Learning Outcomes: Explain general features of enzymes as catalysts: Substrate -> Product Describe nature of catalytic sites general mechanisms
Chapt. 8 Enzymes as catalysts Ch. 8 Enzymes as catalysts Student Learning Outcomes: Explain general features of enzymes as catalysts: Substrate -> Product Describe nature of catalytic sites general mechanisms
Enzymes reduce the activation energy
 Enzymes reduce the activation energy Transition state is an unstable transitory combination of reactant molecules which occurs at the potential energy maximum (free energy maximum). Note - the ΔG of the
Enzymes reduce the activation energy Transition state is an unstable transitory combination of reactant molecules which occurs at the potential energy maximum (free energy maximum). Note - the ΔG of the
Lecture 11 Enzymes: Kinetics
 Lecture 11 Enzymes: Kinetics Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 216-225 Key Concepts Kinetics is the study of reaction rates (velocities). Study of enzyme kinetics is useful for
Lecture 11 Enzymes: Kinetics Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 216-225 Key Concepts Kinetics is the study of reaction rates (velocities). Study of enzyme kinetics is useful for
A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys
 Questions- Proteins & Enzymes A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys Reaction of the intact peptide
Questions- Proteins & Enzymes A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys Reaction of the intact peptide
Enzymes: Introduction
 Enzymes: Introduction Firefly bioluminescence is produced by an oxidation reaction catalyzed by the enzyme firefly luciferase. The oxidized substrate (product of the reaction) is in an electronically excited
Enzymes: Introduction Firefly bioluminescence is produced by an oxidation reaction catalyzed by the enzyme firefly luciferase. The oxidized substrate (product of the reaction) is in an electronically excited
Chemistry 20 Chapters 15 Enzymes
 Chemistry 20 Chapters 15 Enzymes Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An
Chemistry 20 Chapters 15 Enzymes Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An
topics about enzyme function:
 6 Enzymes CHAPTER 6 Enzymes topics about enzyme function: Physiological significance of enzymes Origin of catalytic power of enzymes Chemical mechanisms of catalysis Mechanisms of chymotrypsin and lysozyme
6 Enzymes CHAPTER 6 Enzymes topics about enzyme function: Physiological significance of enzymes Origin of catalytic power of enzymes Chemical mechanisms of catalysis Mechanisms of chymotrypsin and lysozyme
CHM333 LECTURE 13 14: 2/13 15/13 SPRING 2013 Professor Christine Hrycyna
 INTRODUCTION TO ENZYMES Enzymes are usually proteins (some RNA) In general, names end with suffix ase Enzymes are catalysts increase the rate of a reaction not consumed by the reaction act repeatedly to
INTRODUCTION TO ENZYMES Enzymes are usually proteins (some RNA) In general, names end with suffix ase Enzymes are catalysts increase the rate of a reaction not consumed by the reaction act repeatedly to
Proteases: Hydrolysis of Peptide Bonds: Specificity and Mechanism
 Proteases: Hydrolysis of Peptide Bonds: Specificity and Mechanism C. Walsh Proteases (protein hydrolases) catalyze amide (peptide) bond hydrolysis in protein or peptide substrates: Cellular and physiologic
Proteases: Hydrolysis of Peptide Bonds: Specificity and Mechanism C. Walsh Proteases (protein hydrolases) catalyze amide (peptide) bond hydrolysis in protein or peptide substrates: Cellular and physiologic
Chapter 8: An Introduction to Metabolism
 Chapter 8: An Introduction to Metabolism Name Period Concept 8.1 An organism s metabolism transforms matter and energy, subject to the laws of thermodynamics 1. Define metabolism. The totality of an organism
Chapter 8: An Introduction to Metabolism Name Period Concept 8.1 An organism s metabolism transforms matter and energy, subject to the laws of thermodynamics 1. Define metabolism. The totality of an organism
Regulation of enzyme activity
 1 Regulation of enzyme activity Regulation of enzyme activity is important to coordinate the different metabolic processes. It is also important for homeostasis i.e. to maintain the internal environment
1 Regulation of enzyme activity Regulation of enzyme activity is important to coordinate the different metabolic processes. It is also important for homeostasis i.e. to maintain the internal environment
CHM333 LECTURE 13 14: 2/13 15/12 SPRING 2012 Professor Christine Hrycyna
 INTRODUCTION TO ENZYMES Enzymes are usually proteins (some RNA) In general, names end with suffix ase Enzymes are catalysts increase the rate of a reaction not consumed by the reaction act repeatedly to
INTRODUCTION TO ENZYMES Enzymes are usually proteins (some RNA) In general, names end with suffix ase Enzymes are catalysts increase the rate of a reaction not consumed by the reaction act repeatedly to
Lecture 10 Enzymes: Introduction
 Lecture 10 Enzymes: Introduction Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 205-217 (These pages in textbook are very important -- concepts of thermodynamics are fundamental to all of biochemistry.)
Lecture 10 Enzymes: Introduction Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 205-217 (These pages in textbook are very important -- concepts of thermodynamics are fundamental to all of biochemistry.)
ENZYMES FOR BIOCATALYSIS
 ENZYMES FOR BIOCATALYSIS for smarter chemical synthesis Graphical representation of alcalase molecule Rethink Tomorrow Proteases Biocatalysis Biocatalysis involves the implementation of natural catalysts,
ENZYMES FOR BIOCATALYSIS for smarter chemical synthesis Graphical representation of alcalase molecule Rethink Tomorrow Proteases Biocatalysis Biocatalysis involves the implementation of natural catalysts,
BNFO601 Introduction to Molecular Biology Protein
 BNFO601 Introduction to Molecular Biology Protein Outline: A. What can protein do? B. What are proteins? C. Structure and basis for catalysis D. Targeting protein E. Alteration of protein structure and
BNFO601 Introduction to Molecular Biology Protein Outline: A. What can protein do? B. What are proteins? C. Structure and basis for catalysis D. Targeting protein E. Alteration of protein structure and
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7. 4. Which of the following weak acids would make the best buffer at ph = 5.0?
 Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 4 ENZYMATIC CATALYSIS
 ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 4 ENZYMATIC CATALYSIS We will continue today our discussion on enzymatic catalysis
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 4 ENZYMATIC CATALYSIS We will continue today our discussion on enzymatic catalysis
The Citric Acid Cycle
 The itric Acid ycle February 14, 2003 Bryant Miles I. itrate Synthase + 3 SoA The first reaction of the citric acid cycle is the condensation of acetyloa and oxaloacetate to form citrate and oas. The enzyme
The itric Acid ycle February 14, 2003 Bryant Miles I. itrate Synthase + 3 SoA The first reaction of the citric acid cycle is the condensation of acetyloa and oxaloacetate to form citrate and oas. The enzyme
CHAPTER 6 AN INTRODUCTION TO METABOLISM. Section B: Enzymes
 CHAPTER 6 AN INTRODUCTION TO METABOLISM Section B: Enzymes 1. Enzymes speed up metabolic reactions by lowering energy barriers 2. Enzymes are substrate specific 3. The active site in an enzyme s catalytic
CHAPTER 6 AN INTRODUCTION TO METABOLISM Section B: Enzymes 1. Enzymes speed up metabolic reactions by lowering energy barriers 2. Enzymes are substrate specific 3. The active site in an enzyme s catalytic
Copyright 2000-2003 Mark Brandt, Ph.D. 54
 Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
CHAPTER 4: Enzyme Structure ENZYMES
 CHAPTER 4: ENZYMES Enzymes are biological catalysts. There are about 40,000 different enzymes in human cells, each controlling a different chemical reaction. They increase the rate of reactions by a factor
CHAPTER 4: ENZYMES Enzymes are biological catalysts. There are about 40,000 different enzymes in human cells, each controlling a different chemical reaction. They increase the rate of reactions by a factor
Carboxylic Acid Derivatives and Nitriles
 Carboxylic Acid Derivatives and itriles Carboxylic Acid Derivatives: There are really only four things to worry about under this heading; acid chlorides, anhydrides, esters and amides. We ll start with
Carboxylic Acid Derivatives and itriles Carboxylic Acid Derivatives: There are really only four things to worry about under this heading; acid chlorides, anhydrides, esters and amides. We ll start with
The Organic Chemistry of Amino Acids, Peptides, and Proteins
 Essential rganic Chemistry Chapter 16 The rganic Chemistry of Amino Acids, Peptides, and Proteins Amino Acids a-amino carboxylic acids. The building blocks from which proteins are made. H 2 N C 2 H Note:
Essential rganic Chemistry Chapter 16 The rganic Chemistry of Amino Acids, Peptides, and Proteins Amino Acids a-amino carboxylic acids. The building blocks from which proteins are made. H 2 N C 2 H Note:
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon. V. Polypeptides and Proteins
 IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
(c) How would your answers to problem (a) change if the molecular weight of the protein was 100,000 Dalton?
 Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
The peptide bond Peptides and proteins are linear polymers of amino acids. The amino acids are
 Introduction to Protein Structure Proteins are large heteropolymers usually comprised of 50 2500 monomer units, although larger proteins are observed 7. The monomer units of proteins are amino acids. The
Introduction to Protein Structure Proteins are large heteropolymers usually comprised of 50 2500 monomer units, although larger proteins are observed 7. The monomer units of proteins are amino acids. The
ENZYMES - EXTRA QUESTIONS
 ENZYMES - EXTRA QUESTIONS 1. A chemical reaction has a G o = -60 kj/mol. If this were an enzyme-catalyzed reaction what can you predict about the kinetics? A. It will exhibit very rapid kinetics. B. It
ENZYMES - EXTRA QUESTIONS 1. A chemical reaction has a G o = -60 kj/mol. If this were an enzyme-catalyzed reaction what can you predict about the kinetics? A. It will exhibit very rapid kinetics. B. It
ENZYMES (DR. NUGENT)
 ENZYMES (DR. NUGENT) I. INTRODUCTION TO ENZYMES/CATALYSTS a. Definition: enzyme = biological catalyst (catalyst = speeds rate of reaction) b. Features of enzymes: i. Enzymes speeds up the rate of a reaction
ENZYMES (DR. NUGENT) I. INTRODUCTION TO ENZYMES/CATALYSTS a. Definition: enzyme = biological catalyst (catalyst = speeds rate of reaction) b. Features of enzymes: i. Enzymes speeds up the rate of a reaction
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
CHAPTER 11 Mechanism of Enzyme Action
 CHAPTER 11 Mechanism of Enzyme Action 1. General properties of enzymes 2. Activation energy and the reaction coordinate 3. Catalytic mechanism 4. Lysozyme 5. Serine proteases Enzyme act with great speed
CHAPTER 11 Mechanism of Enzyme Action 1. General properties of enzymes 2. Activation energy and the reaction coordinate 3. Catalytic mechanism 4. Lysozyme 5. Serine proteases Enzyme act with great speed
Lecture 3: Enzyme kinetics
 Computational Systems Biology Lecture 3: Enzyme kinetics Fri 19 Jan 2009 1 Images from: D. L. Nelson, Lehninger Principles of Biochemistry, IV Edition, W. H. Freeman ed. A. Cornish-Bowden Fundamentals
Computational Systems Biology Lecture 3: Enzyme kinetics Fri 19 Jan 2009 1 Images from: D. L. Nelson, Lehninger Principles of Biochemistry, IV Edition, W. H. Freeman ed. A. Cornish-Bowden Fundamentals
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
General Protein Metabolism
 General Protein Metabolism Protein Digestion Dietary proteins are very large complex molecules that cannot be absorbed from the intestine. To be absorbed, dietary proteins must be digested to small simple
General Protein Metabolism Protein Digestion Dietary proteins are very large complex molecules that cannot be absorbed from the intestine. To be absorbed, dietary proteins must be digested to small simple
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Two Forms of Energy
 Module 2D - Energy and Metabolism Objective # 19 All living organisms require energy for survival. In this module we will examine some general principles about chemical reactions and energy usage within
Module 2D - Energy and Metabolism Objective # 19 All living organisms require energy for survival. In this module we will examine some general principles about chemical reactions and energy usage within
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK
 Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
green B 1 ) into a single unit to model the substrate in this reaction. enzyme
 Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
Enzymes. Enzyme Structure. Enzyme Classification. CHEM464/Medh, J.D. Reaction Rate and Enzyme Activity
 Enzymes Enzymes are biological catalysts They are not consumed or altered during the reaction They do not change the equilibrium, just reduce the time required to reach equilibrium. They increase the rate
Enzymes Enzymes are biological catalysts They are not consumed or altered during the reaction They do not change the equilibrium, just reduce the time required to reach equilibrium. They increase the rate
H H N - C - C 2 R. Three possible forms (not counting R group) depending on ph
 Amino acids - 0 common amino acids there are others found naturally but much less frequently - Common structure for amino acid - C, -N, and functional groups all attached to the alpha carbon N - C - C
Amino acids - 0 common amino acids there are others found naturally but much less frequently - Common structure for amino acid - C, -N, and functional groups all attached to the alpha carbon N - C - C
Energy & Enzymes. Life requires energy for maintenance of order, growth, and reproduction. The energy living things use is chemical energy.
 Energy & Enzymes Life requires energy for maintenance of order, growth, and reproduction. The energy living things use is chemical energy. 1 Energy exists in two forms - potential and kinetic. Potential
Energy & Enzymes Life requires energy for maintenance of order, growth, and reproduction. The energy living things use is chemical energy. 1 Energy exists in two forms - potential and kinetic. Potential
Part A: Amino Acids and Peptides (Is the peptide IAG the same as the peptide GAI?)
 ChemActivity 46 Amino Acids, Polypeptides and Proteins 1 ChemActivity 46 Part A: Amino Acids and Peptides (Is the peptide IAG the same as the peptide GAI?) Model 1: The 20 Amino Acids at Biological p See
ChemActivity 46 Amino Acids, Polypeptides and Proteins 1 ChemActivity 46 Part A: Amino Acids and Peptides (Is the peptide IAG the same as the peptide GAI?) Model 1: The 20 Amino Acids at Biological p See
Enzymes. Enzymes are characterized by: Specificity - highly specific for substrates
 Enzymes Enzymes are characterized by: Catalytic Power - rates are 10 6-10 12 greater than corresponding uncatalyzed reactions Specificity - highly specific for substrates Regulation - acheived in many
Enzymes Enzymes are characterized by: Catalytic Power - rates are 10 6-10 12 greater than corresponding uncatalyzed reactions Specificity - highly specific for substrates Regulation - acheived in many
Everything You Need to Know About Mechanisms. First rule: Arrows are used to indicate movement of electrons
 Everything You eed to Know About Mechanisms A) The orrect Use of Arrows to Indicate Electron Movement The ability to write an organic reaction mechanism properly is key to success in organic chemistry
Everything You eed to Know About Mechanisms A) The orrect Use of Arrows to Indicate Electron Movement The ability to write an organic reaction mechanism properly is key to success in organic chemistry
Carboxylic Acid Structure and Chemistry: Part 2
 Principles of Drug Action 1, pring 2005, Carboxylic Acids Part 2 Carboxylic Acid tructure and Chemistry: Part 2 Jack Deuiter IV. eactions of the Carboxylic Acid eactions Depending on their overall structure,
Principles of Drug Action 1, pring 2005, Carboxylic Acids Part 2 Carboxylic Acid tructure and Chemistry: Part 2 Jack Deuiter IV. eactions of the Carboxylic Acid eactions Depending on their overall structure,
BCH401G Lecture 39 Andres
 BCH401G Lecture 39 Andres Lecture Summary: Ribosome: Understand its role in translation and differences between translation in prokaryotes and eukaryotes. Translation: Understand the chemistry of this
BCH401G Lecture 39 Andres Lecture Summary: Ribosome: Understand its role in translation and differences between translation in prokaryotes and eukaryotes. Translation: Understand the chemistry of this
Enzymes. Enzyme Classification
 Enzyme Classification Simple Enzymes: composed of whole proteins Complex Enzymes: composed of protein plus a relatively small organic molecule holoenzyme = apoenzyme + prosthetic group / coenzyme A prosthetic
Enzyme Classification Simple Enzymes: composed of whole proteins Complex Enzymes: composed of protein plus a relatively small organic molecule holoenzyme = apoenzyme + prosthetic group / coenzyme A prosthetic
The Aerobic Fate of Pyruvate
 The Aerobic Fate of yruvate February 12, 2003 Bryant Miles I could tell that some of you were not impressed by the mere 2 ATs produced per glucose by glycolysis. The 2 AT s produced are only a small fraction
The Aerobic Fate of yruvate February 12, 2003 Bryant Miles I could tell that some of you were not impressed by the mere 2 ATs produced per glucose by glycolysis. The 2 AT s produced are only a small fraction
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Chapter 26 Biomolecules: Amino Acids, Peptides, and Proteins
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 26 Biomolecules: Amino Acids, Peptides, and Proteins Proteins Amides from Amino Acids Amino acids contain a basic amino group and an acidic carboxyl
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 26 Biomolecules: Amino Acids, Peptides, and Proteins Proteins Amides from Amino Acids Amino acids contain a basic amino group and an acidic carboxyl
Enzymes. A. a lipid B. a protein C. a carbohydrate D. a mineral
 Enzymes 1. All cells in multicellular organisms contain thousands of different kinds of enzymes that are specialized to catalyze different chemical reactions. Given this information, which of the following
Enzymes 1. All cells in multicellular organisms contain thousands of different kinds of enzymes that are specialized to catalyze different chemical reactions. Given this information, which of the following
Name Date Period. Keystone Review Enzymes
 Name Date Period Keystone Review Enzymes 1. In order for cells to function properly, the enzymes that they contain must also function properly. What can be inferred using the above information? A. Cells
Name Date Period Keystone Review Enzymes 1. In order for cells to function properly, the enzymes that they contain must also function properly. What can be inferred using the above information? A. Cells
Helices From Readily in Biological Structures
 The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
MCAT Organic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins
 MCAT rganic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins Question No. 1 of 10 Question 1. Which amino acid does not contain a chiral center? Question #01 (A) Serine (B) Proline (C)
MCAT rganic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins Question No. 1 of 10 Question 1. Which amino acid does not contain a chiral center? Question #01 (A) Serine (B) Proline (C)
PROTEINS THE PEPTIDE BOND. The peptide bond, shown above enclosed in the blue curves, generates the basic structural unit for proteins.
 Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
8/20/2012 H C OH H R. Proteins
 Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Chapter 2 Polar Covalent Bonds; Acids and Bases
 John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
Enzymes and Metabolic Pathways
 Enzymes and Metabolic Pathways Enzyme characteristics Made of protein Catalysts: reactions occur 1,000,000 times faster with enzymes Not part of reaction Not changed or affected by reaction Used over and
Enzymes and Metabolic Pathways Enzyme characteristics Made of protein Catalysts: reactions occur 1,000,000 times faster with enzymes Not part of reaction Not changed or affected by reaction Used over and
In addition to being shorter than a single bond, the double bonds in ethylene don t twist the way single bonds do. In other words, the other atoms
 In addition to being shorter than a single bond, the double bonds in ethylene don t twist the way single bonds do. In other words, the other atoms attached to the carbons (hydrogens in this case) can no
In addition to being shorter than a single bond, the double bonds in ethylene don t twist the way single bonds do. In other words, the other atoms attached to the carbons (hydrogens in this case) can no
Lecture 13-14 Conformation of proteins Conformation of a protein three-dimensional structure native state. native condition
 Lecture 13-14 Conformation of proteins Conformation of a protein refers to the three-dimensional structure in its native state. There are many different possible conformations for a molecule as large as
Lecture 13-14 Conformation of proteins Conformation of a protein refers to the three-dimensional structure in its native state. There are many different possible conformations for a molecule as large as
Enzymes and Metabolism
 Enzymes and Metabolism Enzymes and Metabolism Metabolism: Exergonic and Endergonic Reactions Chemical Reactions: Activation Every chemical reaction involves bond breaking and bond forming A chemical reaction
Enzymes and Metabolism Enzymes and Metabolism Metabolism: Exergonic and Endergonic Reactions Chemical Reactions: Activation Every chemical reaction involves bond breaking and bond forming A chemical reaction
Experiment 10 Enzymes
 Experiment 10 Enzymes Enzymes are proteins that act as catalysts for biological reactions. Enzymes, like all catalysts, speed up reactions without being used up themselves. They do this by lowering the
Experiment 10 Enzymes Enzymes are proteins that act as catalysts for biological reactions. Enzymes, like all catalysts, speed up reactions without being used up themselves. They do this by lowering the
1. A covalent bond between two atoms represents what kind of energy? a. Kinetic energy b. Potential energy c. Mechanical energy d.
 1. A covalent bond between two atoms represents what kind of energy? a. Kinetic energy b. Potential energy c. Mechanical energy d. Solar energy A. Answer a is incorrect. Kinetic energy is the energy of
1. A covalent bond between two atoms represents what kind of energy? a. Kinetic energy b. Potential energy c. Mechanical energy d. Solar energy A. Answer a is incorrect. Kinetic energy is the energy of
PHYSIOLOGY AND MAINTENANCE Vol. II Enzymes: The Biological Catalysts of Life - Pekka Mäntsälä and Jarmo Niemi
 ENZYMES: THE BIOLOGICAL CATALYSTS OF LIFE Pekka Mäntsälä and Jarmo Niemi University of Turku, Department of Biochemistry, Finland Keywords: enzymes, specificity, catalysis, cofactors, enzyme turnover,
ENZYMES: THE BIOLOGICAL CATALYSTS OF LIFE Pekka Mäntsälä and Jarmo Niemi University of Turku, Department of Biochemistry, Finland Keywords: enzymes, specificity, catalysis, cofactors, enzyme turnover,
Lecture 4: Peptides and Protein Primary Structure [PDF] Key Concepts. Objectives See also posted Peptide/pH/Ionization practice problems.
![Lecture 4: Peptides and Protein Primary Structure [PDF] Key Concepts. Objectives See also posted Peptide/pH/Ionization practice problems. Lecture 4: Peptides and Protein Primary Structure [PDF] Key Concepts. Objectives See also posted Peptide/pH/Ionization practice problems.](/thumbs/25/5655226.jpg) Lecture 4: Peptides and Protein Primary Structure [PDF] Reading: Berg, Tymoczko & Stryer, Chapter 2, pp. 34-37 Practice problems (peptide ionization) [PDF]; problems in textbook: chapter 2, pp. 63-64,
Lecture 4: Peptides and Protein Primary Structure [PDF] Reading: Berg, Tymoczko & Stryer, Chapter 2, pp. 34-37 Practice problems (peptide ionization) [PDF]; problems in textbook: chapter 2, pp. 63-64,
Role of Hydrogen Bonding on Protein Secondary Structure Introduction
 Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Chapter 3. Protein Structure and Function
 Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
What affects an enzyme s activity? General environmental factors, such as temperature and ph. Chemicals that specifically influence the enzyme.
 CH s 8-9 Respiration & Metabolism Metabolism A catalyst is a chemical agent that speeds up a reaction without being consumed by the reaction. An enzyme is a catalytic protein. Hydrolysis of sucrose by
CH s 8-9 Respiration & Metabolism Metabolism A catalyst is a chemical agent that speeds up a reaction without being consumed by the reaction. An enzyme is a catalytic protein. Hydrolysis of sucrose by
Computational Systems Biology. Lecture 2: Enzymes
 Computational Systems Biology Lecture 2: Enzymes 1 Images from: David L. Nelson, Lehninger Principles of Biochemistry, IV Edition, Freeman ed. or under creative commons license (search for images at http://search.creativecommons.org/)
Computational Systems Biology Lecture 2: Enzymes 1 Images from: David L. Nelson, Lehninger Principles of Biochemistry, IV Edition, Freeman ed. or under creative commons license (search for images at http://search.creativecommons.org/)
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
Chapter 8: Energy and Metabolism
 Chapter 8: Energy and Metabolism 1. Discuss energy conversions and the 1 st and 2 nd law of thermodynamics. Be sure to use the terms work, potential energy, kinetic energy, and entropy. 2. What are Joules
Chapter 8: Energy and Metabolism 1. Discuss energy conversions and the 1 st and 2 nd law of thermodynamics. Be sure to use the terms work, potential energy, kinetic energy, and entropy. 2. What are Joules
Chapter 16 Amino Acids, Proteins, and Enzymes
 Chapter 16 Amino Acids, Proteins, and Enzymes 1 Functions of Proteins Proteins in the body are polymers made from 20 different amino acids differ in characteristics and functions that depend on the order
Chapter 16 Amino Acids, Proteins, and Enzymes 1 Functions of Proteins Proteins in the body are polymers made from 20 different amino acids differ in characteristics and functions that depend on the order
Chapter 3: Biological Molecules. 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids
 Chapter 3: Biological Molecules 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids Elements in Biological Molecules Biological macromolecules are made almost entirely of just 6 elements: Carbon (C)
Chapter 3: Biological Molecules 1. Carbohydrates 2. Lipids 3. Proteins 4. Nucleic Acids Elements in Biological Molecules Biological macromolecules are made almost entirely of just 6 elements: Carbon (C)
How To Understand Enzyme Kinetics
 Chapter 12 - Reaction Kinetics In the last chapter we looked at enzyme mechanisms. In this chapter we ll see how enzyme kinetics, i.e., the study of enzyme reaction rates, can be useful in learning more
Chapter 12 - Reaction Kinetics In the last chapter we looked at enzyme mechanisms. In this chapter we ll see how enzyme kinetics, i.e., the study of enzyme reaction rates, can be useful in learning more
Regulation of the Citric Acid Cycle
 Regulation of the itric Acid ycle I. hanges in Free Energy February 17, 2003 Bryant Miles kj/mol 40 20 0 20 40 60 80 Reaction DGo' DG TA Free Energy hanges 1 2 3 4 5 6 7 8 9 1.) itrate Synthase 2.) Aconitase
Regulation of the itric Acid ycle I. hanges in Free Energy February 17, 2003 Bryant Miles kj/mol 40 20 0 20 40 60 80 Reaction DGo' DG TA Free Energy hanges 1 2 3 4 5 6 7 8 9 1.) itrate Synthase 2.) Aconitase
Preliminary MFM Quiz
 Preliminary MFM Quiz 1. The major carrier of chemical energy in all cells is: A) adenosine monophosphate B) adenosine diphosphate C) adenosine trisphosphate D) guanosine trisphosphate E) carbamoyl phosphate
Preliminary MFM Quiz 1. The major carrier of chemical energy in all cells is: A) adenosine monophosphate B) adenosine diphosphate C) adenosine trisphosphate D) guanosine trisphosphate E) carbamoyl phosphate
Copyright 2000-2003 Mark Brandt, Ph.D. 35
 Amino acid breakdown Amino acids comprise one of the three major energy sources for animals. They are an especially important energy source for carnivorous animals, and for all animals during early starvation
Amino acid breakdown Amino acids comprise one of the three major energy sources for animals. They are an especially important energy source for carnivorous animals, and for all animals during early starvation
Shu-Ping Lin, Ph.D. E-mail: splin@dragon.nchu.edu.tw
 Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute te of Biomedical Engineering ing E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ edu tw/pweb/users/splin/ Date: 10.13.2010
Amino Acids & Proteins Shu-Ping Lin, Ph.D. Institute te of Biomedical Engineering ing E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ edu tw/pweb/users/splin/ Date: 10.13.2010
4/18/2011. 9.8 Substituent Effects in Electrophilic Substitutions. Substituent Effects in Electrophilic Substitutions
 9.8 Substituent effects in the electrophilic substitution of an aromatic ring Substituents affect the reactivity of the aromatic ring Some substituents activate the ring, making it more reactive than benzene
9.8 Substituent effects in the electrophilic substitution of an aromatic ring Substituents affect the reactivity of the aromatic ring Some substituents activate the ring, making it more reactive than benzene
Patrick, An Introduction to Medicinal Chemistry 4e Chapter 13 Drug design: optimizing target interactions. Pyrrole ring N H
 Patrick, An Introduction to dicinal hemistry 4e hapter 13 Drug design: optimizing target interactions Answers to end-of-chapter questions 1) The pyrrole ring of DU 122290 serves to increase the rigidity
Patrick, An Introduction to dicinal hemistry 4e hapter 13 Drug design: optimizing target interactions Answers to end-of-chapter questions 1) The pyrrole ring of DU 122290 serves to increase the rigidity
LAB 3: DIGESTION OF ORGANIC MACROMOLECULES
 LAB 3: DIGESTION OF ORGANIC MACROMOLECULES INTRODUCTION Enzymes are a special class of proteins that lower the activation energy of biological reactions. These biological catalysts change the rate of chemical
LAB 3: DIGESTION OF ORGANIC MACROMOLECULES INTRODUCTION Enzymes are a special class of proteins that lower the activation energy of biological reactions. These biological catalysts change the rate of chemical
PRACTICAL 3: DIGESTIVE ENZYMES, SPECIFICITY AND ph
 PRACTICAL 3: DIGESTIVE ENZYMES, SPECIFICITY AND ph 3.1 Introduction The aims of this practical are: to illustrate the different ph dependence of gastric and pancreatic digestive proteases to illustrate
PRACTICAL 3: DIGESTIVE ENZYMES, SPECIFICITY AND ph 3.1 Introduction The aims of this practical are: to illustrate the different ph dependence of gastric and pancreatic digestive proteases to illustrate
Ionization of amino acids
 Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
1. Enzymes. Biochemical Reactions. Chapter 5: Microbial Metabolism. 1. Enzymes. 2. ATP Production. 3. Autotrophic Processes
 Chapter 5: Microbial Metabolism 1. Enzymes 2. ATP Production 3. Autotrophic Processes 1. Enzymes Biochemical Reactions All living cells depend on biochemical reactions to maintain homeostasis. All of the
Chapter 5: Microbial Metabolism 1. Enzymes 2. ATP Production 3. Autotrophic Processes 1. Enzymes Biochemical Reactions All living cells depend on biochemical reactions to maintain homeostasis. All of the
Amino Acids as Acids, Bases and Buffers:
 Amino Acids as Acids, Bases and Buffers: - Amino acids are weak acids - All have at least 2 titratable protons (shown below as fully protonated species) and therefore have 2 pka s o α-carboxyl (-COOH)
Amino Acids as Acids, Bases and Buffers: - Amino acids are weak acids - All have at least 2 titratable protons (shown below as fully protonated species) and therefore have 2 pka s o α-carboxyl (-COOH)
Recap. Lecture 2. Protein conformation. Proteins. 8 types of protein function 10/21/10. Proteins.. > 50% dry weight of a cell
 Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Keystone Review Practice Test Module A Cells and Cell Processes. 1. Which characteristic is shared by all prokaryotes and eukaryotes?
 Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Recognizing Organic Molecules: Carbohydrates, Lipids and Proteins
 Recognizing Organic Molecules: Carbohydrates, Lipids and Proteins Oct 15 8:05 PM What is an Organic Molecule? An Organic Molecule is a molecule that contains carbon and hydrogen and oxygen Carbon is found
Recognizing Organic Molecules: Carbohydrates, Lipids and Proteins Oct 15 8:05 PM What is an Organic Molecule? An Organic Molecule is a molecule that contains carbon and hydrogen and oxygen Carbon is found
Amino Acids and Proteins
 Amino Acids and Proteins Proteins are composed of amino acids. There are 20 amino acids commonly found in proteins. All have: N2 C α R COO Amino acids at neutral p are dipolar ions (zwitterions) because
Amino Acids and Proteins Proteins are composed of amino acids. There are 20 amino acids commonly found in proteins. All have: N2 C α R COO Amino acids at neutral p are dipolar ions (zwitterions) because
21.9 REDUCTION OF CARBOXYLIC ACID DERIVATIVES
 10 APTER 1 TE EMITRY F ARBXYLI AID DERIVATIVE TUDY GUIDE LIK 1.5 Esters and ucleophiles 1.17 Give the structure of the product in the reaction of each of the following esters with isotopically labeled
10 APTER 1 TE EMITRY F ARBXYLI AID DERIVATIVE TUDY GUIDE LIK 1.5 Esters and ucleophiles 1.17 Give the structure of the product in the reaction of each of the following esters with isotopically labeled
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Gene Regulation -- The Lac Operon
 Gene Regulation -- The Lac Operon Specific proteins are present in different tissues and some appear only at certain times during development. All cells of a higher organism have the full set of genes:
Gene Regulation -- The Lac Operon Specific proteins are present in different tissues and some appear only at certain times during development. All cells of a higher organism have the full set of genes:
Effect of temperature and ph on the enzymatic activity of salivary amylase
 Effect of temperature and ph on the enzymatic activity of salivary amylase Gae Khalil Rodillas, Nonia Carla Ysabel Samson, Raphael Jaime Santos* and Brylle Tabora Department of Biological Sciences, College
Effect of temperature and ph on the enzymatic activity of salivary amylase Gae Khalil Rodillas, Nonia Carla Ysabel Samson, Raphael Jaime Santos* and Brylle Tabora Department of Biological Sciences, College
Figure 5. Energy of activation with and without an enzyme.
 Biology 20 Laboratory ENZYMES & CELLULAR RESPIRATION OBJECTIVE To be able to list the general characteristics of enzymes. To study the effects of enzymes on the rate of chemical reactions. To demonstrate
Biology 20 Laboratory ENZYMES & CELLULAR RESPIRATION OBJECTIVE To be able to list the general characteristics of enzymes. To study the effects of enzymes on the rate of chemical reactions. To demonstrate
Lecture 4. Polypeptide Synthesis Overview
 Initiation of Protein Synthesis (4.1) Lecture 4 Polypeptide Synthesis Overview Polypeptide synthesis proceeds sequentially from N Terminus to C terminus. Amino acids are not pre-positioned on a template.
Initiation of Protein Synthesis (4.1) Lecture 4 Polypeptide Synthesis Overview Polypeptide synthesis proceeds sequentially from N Terminus to C terminus. Amino acids are not pre-positioned on a template.
