Recombinant Expressed Vector pet32a (+) S Constructed by Ligation Independent Cloning
|
|
|
- Stuart Chase
- 7 years ago
- Views:
Transcription
1 Molecules 2014, 19, ; doi: /molecules OPEN ACCESS molecules ISSN Article Recombinant Expressed Vector pet32a (+) S Constructed by Ligation Independent Cloning Yu Wang 1,, Guo-Hua Gong 1,, Cheng-Xi Wei 1, Long Liang 2, * and Bin Zhang 1,3, * 1 Medicinal Chemistry and Pharmacology Institute, Inner Mongolia University for the Nationalities, Tongliao , Inner Mongolia, China; s: shen348@126.com (Y.W.); gongguohua0211@163.com (G.-H.G.); weichengxi1224@163.com (C.-X.W.) 2 State Key Laboratory of Medical Microbiology and Biosafety, Academy of Military Medical Sciences, Biotechnology Institute of Beijing, Beijing , China 3 Institute of Mongolia and Western Medicinal Treatment, Affiliated Hospital of Inner Mongolia University for the Nationalities, Holin he Street 1742, Tongliao , Inner Mongolia, China These authors contributed equally to this work. * Authors to whom correspondence should be addressed; s: ll@bmi.ac.cn (L.L.); bzh9911@163.com (B.Z.); Tel./Fax: (B.Z.). External Editor: Derek J. McPhee Received: 5 August 2014; in revised form: 9 September 2014 / Accepted: 9 September 2014 / Published: 10 October 2014 Abstract: The aim of this work was to develop a new method for constructing vectors, named ligation-independent cloning (LIC) method. We constructed the S label expression vector and recombinant pet32a (+) S-phoN2 by LIC. The recombinant proteins were expressed in E. coli at a high level, and then the specificity of the recombinant proteins was identified by western blot. The target band was detected by S monoclonal antibody and Apyrase polyclonal antibodies but not Trx monoclonal antibody and HIS monoclonal antibody. Finally, we obtained protein Apyrase in E. coli (BL21), with a protein-only expression S tag. Collectively, our results demonstrated that LIC is effective for the construction of new vectors and recombinant plasmids. Free from the limitations of restriction enzyme sites and with a higher positive rate, LIC processes should find broad applications in molecular biology research.
2 Molecules 2014, Keywords: ligation-independent cloning; vector constructing; prokaryotic gene expression vector 1. Introduction Generally, the purpose of DNA molecular cloning is the insertion of a particular fragment into a vector to construct a recombinant plasmid. It is widely used in the medicine and biology field. The traditional cloning method involves acquisition of a particular fragment, digestion of a particular fragment and vector connection, transformation and selection. Traditional cloning methods are widely used in the construction of vectors and recombinant plasmids [1,2]. Some of them have even converted into kits, such as T-A Cloning [3] or Gateway [4,5]. These new technologies have greatly simplified the cloning process, and improved the efficiency of cloning, but some problems still exist. Traditional cloning techniques use restriction enzymes and ligation of DNA in vitro, which can be hampered by a lack of appropriate restriction-sites and inefficient enzymatic steps. For example, the restriction enzyme sites of particular fragments and vectors are considered, the particular fragment is treated by a restriction enzyme, and the ligation process needs DNA Ligase [6]. Using traditional cloning methods increases not only the cost of experiments and experiment duration, but also the risk of self-vector connection. In many cases, because of the properties of the original restriction sites of a particular fragment, the experiment has to use some rare restriction. This greatly increases the difficulty and cost of the experiment [7]. A way which is not subject to restriction sites could solve the problem above. Ligation independent cloning (LIC) has been developed as a new cloning approach which eliminates the use of restriction sites [8]. The principle of LIC is: first of all, the linearized vector and the insert fragment are amplified by PCR with primers containing homologous fragments; second, PCR products are treated with T4 DNA Polymerase (3'-5' exonuclease activity) to obtain the fragment containing 5'-cohesive ends; third, the intermediate is formed by denaturation-renaturation; finally, intermediate is transformed into Escherichia coli [9]. The LIC method has advantages over traditional cloning methods in vector and recombinant plasmid construction. LIC eliminates the use of restriction sites and DNA ligease [10]. Using the LIC method to construct vectors and recombinant plasmids not only shortens the experiments, but also reduces the cost of experiments. LIC uses constructed vectors [11], constructed recombinant proteins [12,13] and co-expression proteins [14,15]. LIC is particularly suitable for the construction of new vectors [16] and high molecular weight expression of recombinant proteins [17]. In this work, we report the development of an effective, inexpensive method named the LIC method to construct the expression vector pet32a (+) S and pet32a (+) S-phoN2, which can generate controllable overhangs.
3 Molecules 2014, Results and Discussion 2.1. Construction of a pet32a (+) S Expression Vector In order to construct the plasmid-only expressed S tag, we used the LIC method to construct pet32a (+) S. Cloning of expression vector pet32a (+) S via LIC was performed as described in Figure 1. After vector pet32a (+) was digested (Figure 1a), it was amplified with a pair of specific primers as described in the Experimental Section. The PCR product was characterized by electrophoresis through 1% agarose gels, and the 5900 bp band appeared as expected with the same size as pet32a (+) (Figure 1b). The transformants were acquired by use of E. coli DH5a (Figure 1c), the positive clones were identified by PCR (Figure 1d) and sequenced (Table 1). The result showed the constructed vector pet32a (+) S was not a mutant. Figure 1. Construction of expressed vector pet32a (+) S. (a) line A, MscI digested of vector pet32a (+), the 5900 bp bands were present. Line M, DL6000 Marker; (b) line B, PCR products of linear vector pet32a (+), the 5900 bp band was present. Line M, DL6000 Marker; (c) the result of colone; (d) PCR products of pet32a (+) S, the 604 bp band was present. Line M, DL6000 Marker.
4 Molecules 2014, Table 1. Sequence analysis of core of pet32a (+) S gene. CAA GAC CCG TTT AGA GGC CCC AAG GGG TTA TGC TAG TTA TTG CTC AGC GGT GGC AGC AGC CAA CTC AGC TTC CTT TCG GGC TTT GTT AGC AGC CGG ATC TCA GTG GTG GTG GTG GTG GTG CTC GAG TGC GGC CGC AAG CTT GAC GAC GGA GCT CGA ATT CGG ATC CGA TAT CAG CCA TGG CCT TGT CGT CGT CGT CGG TAC CCA GAT CTG GGC TGT CCA TGT GCT GGC GTT CGA ATT TAG CAG CAG CGG TTT CTT TAT GTA TAT CTC CTT CTT AAA GTT AAA CAA AAT TAT TTC TAG AGG GGA ATT GTT ATC CGC TCA CAA TTC CCC TAT AGT GAG TCG TAT TAA TTT CGC GGG ATC GAG ATC GAT CTC GAT CCT CTA CGC CGG ACG CAT CGT GGC CGG CAT CAC CGG CGC CAC AGG TGC GGT TGC TGG CGC CTAA TAT CGC CGA CAT CAC CGA TGG GGA AGA TCG GGC TCG CCA CTT CGG GCT CAT GAG CGC TTG TTT CGG CGT GGG TAT GGT GGC AGG CCC CGT GGC CGG GGG ACT GTT GGG CGC CAT CTC CTT GCA TGC ACC ATT CCT TGC GGC GGC GGT GCT CAA CGG CCT CAA CCT ACT ACT # Notes: # Length of the sequence was 604 bp; It contained bp of pet32a, where in addition to bp Construction of a pet32a (+) S-phoN2 Recombinant Plasmid The expression vector pet32a (+) S was amplified with a pair of specific primers as described in the Experimental Section 3.2. The PCR product was characterized by electrophoresis through 1% agarose gels, the 5502 bp band appeared as expected with the same size as pet32a (+) S (Figure 2a). The quality of the complete genome from Shigella flexneri M90T was characterized by electrophoresis through 1% agarose gels. On the basis of the sequence of phon2 in M90T, gene specific primers were designed. The PCR product was characterized by electrophoresis using 1% agarose gels, and the 741 bp band appeared as expected with the same size as phon2 (Figure 2b). The target fragment and linear vector were linked by T4 DNA Polymerase. The transformants were acquired by E. coli DH5a and identified by PCR (Figure 2c). The positive clones were sequenced. The result showed the constructed recombinant plasmid pet32a (+) S-phoN2 was not a mutant (Table 2). Figure 2. Construction of recombinant plasmid pet32a (+) S-phoN2. (a) Line D, the line of linear vector pet32a (+) S (5900 bp). Line M, DL6000 Marker; (b) Line E, the line of phon2 (741 bp). Line M, DL2000 Marker; (c) Line F, PCR products of recombinant plasmid pet32a (+) S-phoN2. Line M, DL2000 Marker.
5 Molecules 2014, Table 2. Sequence analysis of Apyrase gene and its encoding amino acids. ATG AAA ACC AAA AAC TTT CTT CTT TTT TGT ATT GCT ACA AAT ATG ATT TTT ATC CCC TCA M K T K N F L L F C I A T N M I F I P S GCA AAT GCT CTG AAG GCA GAA GGT TTT CTC ACT CAA CAA ACT TCA CCA GAC AGT TTG A N A L K A E G F L T Q Q T S P D S L TCA ATA CTT CCG CCG CCT CCG GCA GAG GAT TCA GTA GTA TTT CTG GCT GAC AAA GCT CAT S I L P P P P A E D S V V F L A D K A H TAT GAA TTC GGC CGC TCG CTC CGG GAT GCT AAT CGT GTA CGT CTC GCT AGC GAA GAT GCA Y E F G R S L R D A N R V R L A S E D A TAC TAC GAG AAT TTT GGT CTT GCA TTT TCA GAT GCT TAT GGC ATG GAT ATT TCA AGG GAA Y Y E N F G L A F S D A Y G M D I S R E AAT ACC CCA ATC TTA TAT CAG TTG TTA ACA CAA GTA CTA CAG GAT AGC CAT GAT TAC GCC N T P I L Y Q L L T Q V L Q D S H D Y A GTG CGT AAC GCC AAA GAA TAT TAT AAA AGA GTT CGT CCA TTC GTT ATT TAT AAA GAC GCA V R N A K E Y Y K R V R P F V I Y K D A ACC TGT ACA CCT GAT AAA GAT GAG AAA ATG GCT ATC ACT GGC TCT TAT CCC TCT GGT T C T P D K D E K M A I T G S Y P S G CAT GCA TCC TTT GGT TGG GCA GTA GCA CTG ATA CTT GCG GAG ATT AAT CCT CAA CGT AAA H A S F G W A V A L I L A E I N P Q R K GCG GAA ATA CTT CGA CGT GGA TAT GAG TTT GGA GAA AGT CGG GTC ATC TGC GGT GCG A E I L R R G Y E F G E S R V I C G A CAT TGG CAA AGC GAT GTA GAG GCT GGG CGT TTA ATG GGA GCA TCG GTT GTT GCA GTA H W Q S D V E A G R L M G A S V V A V CTT CAT AAT ACA CCT GAA TTT ACC AAA AGC CTT AGC GAA GCC AAA AAA GAG TTT GAA L H N T P E F T K S L S E A K K E F E GAA TTA AAT ACT CCT ACC AAT GAA CTG ACC CCA TAA E L N T P T N E L T P R Note: The full-length of the sequence was 741 bp and codes for 247 amino acid residues Recombinant Protein Expression The pet32a (+) S-Apyrase/BL21 was cultivated and OD600 was measured every hour. An S-type bacterial growth curve was observed, pet32a (+) S-Apyrase/BL21 was induced at 3 h when they were in the bacterial logarithmic growth phase (Figure 3). To determine the solubility of recombinant protein, pet32a (+) S-Apyrase/BL21 without induction, whole protein of pet32a (+) S-Apyrase/BL21 after induction, the supernatant of pet32a (+) S-Apyrase/BL21 and the pellet of pet32a (+) S-Apyrase/BL21 were analyzed by SDS-PAGE (Figure 4). The recombinant protein was identified in the induced bacteria but not the control. Comparing the content of ultrasound supernatant and ultrasound pellet, most of the fusion proteins were present in the supernatant, meaning that the fusion protein Apyrase-S was a soluble protein. Lastly, in order to check the activity of recombinant apyrase expression, Apyrase-S protein was purified (Figure 5).
6 Molecules 2014, Figure 3. Growth curve of pet32a (+) S-Apyrase/BL 21(DE3). OD600 reached 0.6 at 3 h. Figure 4. Identification and analysis of the fusion protein by SDS-PAGE. Line M: molecular weight marker: 175 kda, 80 kda, 58 kda, 46 kda, 30 kda and 23 kda; Line 1: whole bacteria of before induce; Line 2: whole bacteria of after induce; Line 3: supernatant of the ultrasound crushing; Line 4: precipitate of the ultrasound crushing. Figure 5. Purified protein Apyrase was identified by SDS-PAGE. Line M: molecular weight marker: 175 kda, 80 kda, 58 kda, 46 kda, 30 kda, 23 kda and 17 kda; Line A: the purified protein Aprase.
7 Molecules 2014, Detecting of the Specificity of Expressing Bacteria The induced bacteria were lysed, and the specificity of expressing bacteria was analyzed by western blot. The target band was detected by S monoclonal antibody and Apyrase polyclonal antibodies but not Trx monoclonal antibody and HIS monoclonal antibody (Figure 6). This means the expression of fusion protein pet32a (+) S-Apyrase/BL21 only contains S tag. The molecular weight of S tag is only 1.7 kda, which is too small to affect the structure of the target protein. Using the S tag, we can affinity purify the target protein and combine it with other tags for pull-down experiments. However, pet32a (+) contains two other tags in addition to the S tag. It was very inconvenient to construct a recombinant protein which only expressed S tag, so we constructed pet32a (+) S by LIC, which made the next experiment convenient. Figure 6. The specificity of expressing bacteria was detected by western blot. Line 1: whole bacteria of before induce; Line 2: whole bacteria of after induce; Line 3: supernatant of the ultrasound crushing; Line 4: precipitate of the ultrasound crushing. 3. Experimental Section 3.1. Materials E. coli DH5a and E. coli BL21(DE3) were purchased from the TIANGEN (Beijing, China); expression vector pet32a (+) was obtained from Novagen (Beijing, China); Antibodies for S, GAPDH, Trx and HIS were purchased from Sigma-Aldrich (Shanghai, China); PrimeSTAR HS DNA Polymerase, T4 DNA Polymerase and IPTG (isopropyl β-d-thiogalactoside) were purchased from the TaKaRa (Dalian, China) Vector Construction Construction of the expression vector pet32a (+) S is described in Figure 7. First of all, vector pet32a (+) was treated with MscI. LIC-specific primers were designed as shown below. The primers include homologous fragments. In the process of amplification, the bp (Trx tag site) of pet32a (+) was removed: SF: 5' GGTTTCTTTCATATGTATATCTCCTTCTTAAAGTTAAACA 3' SR: 5' ATGAAAGAAACCGCTGCTGCTAAATTCG 3'
8 Molecules 2014, Figure 7. Vector construct. The PCR was performed in 50 μl reaction mixes using the following cycles: initial denaturation at 98 C for 30 s; 25 cycles of denaturation at 98 C for 30 s and extension at 68 C for 6 min and 30 s; and a final extension at 68 C for 7 min. PCR products were analyzed by electrophoresis and purified with PCR cleanup kit (Beijing, China). The cloning reaction was performed by T4 DNA Ploymerase. There was 100 ng DNA model, 10 T4 DNA Polymerase buffers 1 μl, BSA 0.1 μl, T4 DNA Ploymerase 0.1 μl in the best 25 μl restriction system. The linear vector pet32a (+) was made using the following reaction: digestion at 37 C for 1 min; inactivation at 72 C for 20 min and annealing at 25 C. The annealing product was transformed into E. coli DH5α, and grown on LB agar plates (ampicillin) at 37 C for 16 h. LB medium was inoculated with bacterial colonies, the bacterial colonies were grown at 37 C, 200 rpm for h. Then, the bacteria was identified by primers: FSce: 5' CAAGACCCGTTTAGAGGC 3' (39 56 bp); Rsce: 5' AGTAGGTTGAGGCCGTTG 3' ( bp).
9 Molecules 2014, Construction of pet32a (+) S-Apyrase (Figure 8) Apyrase was amplified by PCR using AF (5'GCGGATCCATGAAAACCAAAAAC3') and AR (5'CTCGAGTTATGGGGTCAGTTCATTG3') primers. PCR products were analyzed by electrophoresis and purified with a PCR cleanup kit. The linear vector pet32a (+) S was amplified by primers SAF and SAR. The sequences are shown below; the upstream primers and downstream primers included homologous fragments of phon2: SAF: 5' GAAAGTTTTTGGTTTTCATGGATCCGATATCAGCCATGG 3' SAR: 5' CCAATGAACTGACCCCATAACTCGAGCACCACCACCACCA 3' The cloning reaction was performed by T4 DNA Polymerase. The annealing product was transformed into E. coli DH5α, and the bacteria were identified by primers AF and AR. Figure 8. Construction of pet32a (+) S-Apyrase Recombinant Protein Expression The recombinant pet32a (+) S-Ayrase was transformed into E. coli BL21 (DE3) and selected on LB agar (ampicillin) at 37 C overnight. The bacterial colonies were transferred into LB medium and
10 Molecules 2014, the bacteria were cultured at 37 C, 200 rpm overnight. In order to prepare the growth curve of the bacteria, the bacteria were sampled at 1, 2, 3, 4, 5, 6, 7 and 8 h. Then OD600 of each sample was measured. To determine the fusion of recombinant proteins, the bacteria were cultured at 37 C, 200 rpm for 4 h, until OD600 was 0.6. Then the bacteria were dissolved in PBS and lysed by sonication. The different samples were tested by SDS-PAGE Purification of Proteins The ps-apyrase protein was purified using S agarose and insoluble material was removed by centrifugation (12,000 r/min, 15 min, 4 C). Supernatants were filtered through 0.2 μm sterile Acrodisc syringe filters with membrane onto S agarose and rotated on a rocking platform at 4 C for 4 h. Beads were collected by centrifugation (3000 r/min, 5 min, 4 C) and washed three times in a wash buffer (20 mm Tris-HCl, 150 mm NaCl, 1%Triton (v/v) 100). The fusion proteins were stored in S agarose beads at 4 C. 4. Conclusions We have successfully constructed the expression vector pet32a (+) S and recombinant pet32a (+) S-Apyrase by LIC in just one week. Using LIC to construct vectors not only has a higher success rate, but also takes less time. The new method of construction of vectors and recombinant proteins provides an efficient and cost-effective parallel method and thus is applicable for the construction of different vectors, tags and proteins. Acknowledgments This study was supported by the National Natural Science Foundation of China (No ). Author Contributions Yu Wang did most of the experiments, Guo-Hua Gong corrected the manuscript, Cheng-Xi Wei provided help for Yu Wang and Guo-Hua Gong, and both Long Liang and Bin Zhang took up the idea and provided the important supervision of the research. Conflicts of Interest The authors declare no conflict of interest. References 1. Tenno, T.; Goda, N.; Tateishi, Y.; Tochio, H.; Mishima, M.; Hayashi, H.; Shirakawa, M.; Hiroaki, H. High-throughput construction method for expression vector of peptides for NMR study suited fo risotopic labeling. Protein Eng. Des. Sel. 2004, 17, Ohsugi, T.; Kumasaka, T.; Urano, T. Construction of a full-length human T cell leukemia virus type I genome from MT-2 cells containing multiple defective proviruses using overlapping polymerase chain reaction. Anal. Biochem. 2004, 329,
11 Molecules 2014, Holton, T.A.; Graham, M.W. A simple and efficient method for direct cloning of PCR products using ddt-tailed vectors. Nucleic Acids Res. 1991, 19, Katzen, F. Gateway recombinational cloning: A biological operating system. Expert Opin. Drug Discov. 2007, 2, Hartley, J.L.; Temple, G.F.; Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000, 10, Cohen, S.N.; Chang, A.C.; Boyer, H.W.; Helling, R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 1973, 70, Haun, R.S.; Serventi, I.M.; Moss, J. Rapid, reliable ligation-independent cloning of PCR products using modified plasmid vectors. Biotechniques 1992, 13, Aslanidis, C.; de Jong, P.J. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990, 18, Barry, E.M.; Pasetti, M.F.; Sztein, M.B.; Fasano, A.; Karen, L.; Levine, K.M.M. Progress and pitfalls in Shigella vaccine research. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, Haun, R.S.; Moss, J. Ligation-independent cloning of glutathione Stransferase fusion genes for expression in Escherichia coli. Gene 1992, 112, Stevenson, J.; Krycer, J.R.; Phan, L.; Brown, A.J. A practical comparison of ligation-independent cloning techniques. PLoS One 2013, 8, e Weeks, S.D.; Drinker, M.; Loll, P.J. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr. Purif. 2007, 53, Lee, J.; Kim, S.H. High-throughput T7 LIC vector for introducing Cterminal poly-histidine tags with variable lengths without extra sequences. Protein Expr. Purif. 2009, 63, Chanda, P.K.; Edris, W.A.; Kennedy, J.D. A set of ligation-independent expression vectors for co-expression of proteins in Escherichia coli. Protein Expr. Purif. 2006, 47, Qin, H.; Hu, J.; Hua, Y.Z.; Challa, S.V.; Cross, T.A.; Gao, F.P. Construction of a series of vectors for high throughput cloning and expression screening of membrane proteins from Mycobacterium tuberculosis. BMC Biotechnol. 2008, 8, Kato, T.; Thompson, J.R.; Park, E.Y. Construction of new ligation-independent cloning vectors for the expression and furifcation of recombinant proteins in silkworms using BmNPV bacmid system. PLoS One 2013, 8, e Wang, R.Y.; Shi, Z.-Y.; Guo, Y.-Y.; Chen, J.-C.; Chen, G.-Q. DNA fragments assembly based on nicking enzyme system. PLoS One 2013, 8, e Sample Availability: Samples of the compounds are available from the authors by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (
GENEWIZ, Inc. DNA Sequencing Service Details for USC Norris Comprehensive Cancer Center DNA Core
 DNA Sequencing Services Pre-Mixed o Provide template and primer, mixed into the same tube* Pre-Defined o Provide template and primer in separate tubes* Custom o Full-service for samples with unknown concentration
DNA Sequencing Services Pre-Mixed o Provide template and primer, mixed into the same tube* Pre-Defined o Provide template and primer in separate tubes* Custom o Full-service for samples with unknown concentration
DNA Sample preparation and Submission Guidelines
 DNA Sample preparation and Submission Guidelines Requirements: Please submit samples in 1.5ml microcentrifuge tubes. Fill all the required information in the Eurofins DNA sequencing order form and send
DNA Sample preparation and Submission Guidelines Requirements: Please submit samples in 1.5ml microcentrifuge tubes. Fill all the required information in the Eurofins DNA sequencing order form and send
(http://genomes.urv.es/caical) TUTORIAL. (July 2006)
 (http://genomes.urv.es/caical) TUTORIAL (July 2006) CAIcal manual 2 Table of contents Introduction... 3 Required inputs... 5 SECTION A Calculation of parameters... 8 SECTION B CAI calculation for FASTA
(http://genomes.urv.es/caical) TUTORIAL (July 2006) CAIcal manual 2 Table of contents Introduction... 3 Required inputs... 5 SECTION A Calculation of parameters... 8 SECTION B CAI calculation for FASTA
10 µg lyophilized plasmid DNA (store lyophilized plasmid at 20 C)
 TECHNICAL DATA SHEET BIOLUMINESCENCE RESONANCE ENERGY TRANSFER RENILLA LUCIFERASE FUSION PROTEIN EXPRESSION VECTOR Product: prluc-c Vectors Catalog number: Description: Amount: The prluc-c vectors contain
TECHNICAL DATA SHEET BIOLUMINESCENCE RESONANCE ENERGY TRANSFER RENILLA LUCIFERASE FUSION PROTEIN EXPRESSION VECTOR Product: prluc-c Vectors Catalog number: Description: Amount: The prluc-c vectors contain
pcas-guide System Validation in Genome Editing
 pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation
 BDGP Resources Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation For recovery of sequences flanking PZ, PlacW and PEP elements E. Jay Rehm Berkeley Drosophila Genome Project I.
BDGP Resources Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation For recovery of sequences flanking PZ, PlacW and PEP elements E. Jay Rehm Berkeley Drosophila Genome Project I.
UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet
 1 UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam:.
1 UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam:.
(A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors.
 Legends of supplemental figures and tables Figure 1: Overview of study design and results. (A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors. After raw data gene expression
Legends of supplemental figures and tables Figure 1: Overview of study design and results. (A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors. After raw data gene expression
pcmv6-neo Vector Application Guide Contents
 pcmv6-neo Vector Application Guide Contents Package Contents and Storage Conditions... 2 Product Description... 2 Introduction... 2 Production and Quality Assurance... 2 Methods... 3 Other required reagents...
pcmv6-neo Vector Application Guide Contents Package Contents and Storage Conditions... 2 Product Description... 2 Introduction... 2 Production and Quality Assurance... 2 Methods... 3 Other required reagents...
SERVICES CATALOGUE WITH SUBMISSION GUIDELINES
 SERVICES CATALOGUE WITH SUBMISSION GUIDELINES 3921 Montgomery Road Cincinnati, Ohio 45212 513-841-2428 www.agctsequencing.com CONTENTS Welcome Dye Terminator Sequencing DNA Sequencing Services - Full Service
SERVICES CATALOGUE WITH SUBMISSION GUIDELINES 3921 Montgomery Road Cincinnati, Ohio 45212 513-841-2428 www.agctsequencing.com CONTENTS Welcome Dye Terminator Sequencing DNA Sequencing Services - Full Service
Gene Synthesis 191. Mutagenesis 194. Gene Cloning 196. AccuGeneBlock Service 198. Gene Synthesis FAQs 201. User Protocol 204
 Gene Synthesis 191 Mutagenesis 194 Gene Cloning 196 AccuGeneBlock Service 198 Gene Synthesis FAQs 201 User Protocol 204 Gene Synthesis Overview Gene synthesis is the most cost-effective way to enhance
Gene Synthesis 191 Mutagenesis 194 Gene Cloning 196 AccuGeneBlock Service 198 Gene Synthesis FAQs 201 User Protocol 204 Gene Synthesis Overview Gene synthesis is the most cost-effective way to enhance
Table S1. Related to Figure 4
 Table S1. Related to Figure 4 Final Diagnosis Age PMD Control Control 61 15 Control 67 6 Control 68 10 Control 49 15 AR-PD PD 62 15 PD 65 4 PD 52 18 PD 68 10 AR-PD cingulate cortex used for immunoblot
Table S1. Related to Figure 4 Final Diagnosis Age PMD Control Control 61 15 Control 67 6 Control 68 10 Control 49 15 AR-PD PD 62 15 PD 65 4 PD 52 18 PD 68 10 AR-PD cingulate cortex used for immunoblot
Supplementary Online Material for Morris et al. sirna-induced transcriptional gene
 Supplementary Online Material for Morris et al. sirna-induced transcriptional gene silencing in human cells. Materials and Methods Lentiviral vector and sirnas. FIV vector pve-gfpwp was prepared as described
Supplementary Online Material for Morris et al. sirna-induced transcriptional gene silencing in human cells. Materials and Methods Lentiviral vector and sirnas. FIV vector pve-gfpwp was prepared as described
Mutations and Genetic Variability. 1. What is occurring in the diagram below?
 Mutations and Genetic Variability 1. What is occurring in the diagram below? A. Sister chromatids are separating. B. Alleles are independently assorting. C. Genes are replicating. D. Segments of DNA are
Mutations and Genetic Variability 1. What is occurring in the diagram below? A. Sister chromatids are separating. B. Alleles are independently assorting. C. Genes are replicating. D. Segments of DNA are
Supplementary Information. Binding region and interaction properties of sulfoquinovosylacylglycerol (SQAG) with human
 Supplementary Information Binding region and interaction properties of sulfoquinovosylacylglycerol (SQAG) with human vascular endothelial growth factor 165 revealed by biosensor based assays Yoichi Takakusagi
Supplementary Information Binding region and interaction properties of sulfoquinovosylacylglycerol (SQAG) with human vascular endothelial growth factor 165 revealed by biosensor based assays Yoichi Takakusagi
Introduction to Perl Programming Input/Output, Regular Expressions, String Manipulation. Beginning Perl, Chap 4 6. Example 1
 Introduction to Perl Programming Input/Output, Regular Expressions, String Manipulation Beginning Perl, Chap 4 6 Example 1 #!/usr/bin/perl -w use strict; # version 1: my @nt = ('A', 'C', 'G', 'T'); for
Introduction to Perl Programming Input/Output, Regular Expressions, String Manipulation Beginning Perl, Chap 4 6 Example 1 #!/usr/bin/perl -w use strict; # version 1: my @nt = ('A', 'C', 'G', 'T'); for
Molecular analyses of EGFR: mutation and amplification detection
 Molecular analyses of EGFR: mutation and amplification detection Petra Nederlof, Moleculaire Pathologie NKI Amsterdam Henrique Ruijter, Ivon Tielen, Lucie Boerrigter, Aafke Ariaens Outline presentation
Molecular analyses of EGFR: mutation and amplification detection Petra Nederlof, Moleculaire Pathologie NKI Amsterdam Henrique Ruijter, Ivon Tielen, Lucie Boerrigter, Aafke Ariaens Outline presentation
Next Generation Sequencing
 Next Generation Sequencing 38. Informationsgespräch der Blutspendezentralefür Wien, Niederösterreich und Burgenland Österreichisches Rotes Kreuz 22. November 2014, Parkhotel Schönbrunn Die Zukunft hat
Next Generation Sequencing 38. Informationsgespräch der Blutspendezentralefür Wien, Niederösterreich und Burgenland Österreichisches Rotes Kreuz 22. November 2014, Parkhotel Schönbrunn Die Zukunft hat
Inverse PCR and Sequencing of P-element, piggybac and Minos Insertion Sites in the Drosophila Gene Disruption Project
 Inverse PCR and Sequencing of P-element, piggybac and Minos Insertion Sites in the Drosophila Gene Disruption Project Protocol for recovery of sequences flanking insertions in the Drosophila Gene Disruption
Inverse PCR and Sequencing of P-element, piggybac and Minos Insertion Sites in the Drosophila Gene Disruption Project Protocol for recovery of sequences flanking insertions in the Drosophila Gene Disruption
Hands on Simulation of Mutation
 Hands on Simulation of Mutation Charlotte K. Omoto P.O. Box 644236 Washington State University Pullman, WA 99164-4236 omoto@wsu.edu ABSTRACT This exercise is a hands-on simulation of mutations and their
Hands on Simulation of Mutation Charlotte K. Omoto P.O. Box 644236 Washington State University Pullman, WA 99164-4236 omoto@wsu.edu ABSTRACT This exercise is a hands-on simulation of mutations and their
Title : Parallel DNA Synthesis : Two PCR product from one DNA template
 Title : Parallel DNA Synthesis : Two PCR product from one DNA template Bhardwaj Vikash 1 and Sharma Kulbhushan 2 1 Email: vikashbhardwaj@ gmail.com 1 Current address: Government College Sector 14 Gurgaon,
Title : Parallel DNA Synthesis : Two PCR product from one DNA template Bhardwaj Vikash 1 and Sharma Kulbhushan 2 1 Email: vikashbhardwaj@ gmail.com 1 Current address: Government College Sector 14 Gurgaon,
The p53 MUTATION HANDBOOK
 The p MUTATION HANDBOOK Version 1. /7 Thierry Soussi Christophe Béroud, Dalil Hamroun Jean Michel Rubio Nevado http://p/free.fr The p Mutation HandBook By T Soussi, J.M. Rubio-Nevado, D. Hamroun and C.
The p MUTATION HANDBOOK Version 1. /7 Thierry Soussi Christophe Béroud, Dalil Hamroun Jean Michel Rubio Nevado http://p/free.fr The p Mutation HandBook By T Soussi, J.M. Rubio-Nevado, D. Hamroun and C.
Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION
 Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION Susan Carson Heather B. Miller D.Scott Witherow ELSEVIER AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN
Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION Susan Carson Heather B. Miller D.Scott Witherow ELSEVIER AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN
Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541
 PRODUCT INFORMATION Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541 Lot _ Store at -20 C Expiry Date _ www.thermoscientific.com/onebio CERTIFICATE OF ANALYSIS The Phusion Site-Directed Mutagenesis
PRODUCT INFORMATION Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541 Lot _ Store at -20 C Expiry Date _ www.thermoscientific.com/onebio CERTIFICATE OF ANALYSIS The Phusion Site-Directed Mutagenesis
Gene Finding CMSC 423
 Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would you decipher
Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would you decipher
NimbleGen SeqCap EZ Library SR User s Guide Version 3.0
 NimbleGen SeqCap EZ Library SR User s Guide Version 3.0 For life science research only. Not for use in diagnostic procedures. Copyright 2011 Roche NimbleGen, Inc. All Rights Reserved. Editions Version
NimbleGen SeqCap EZ Library SR User s Guide Version 3.0 For life science research only. Not for use in diagnostic procedures. Copyright 2011 Roche NimbleGen, Inc. All Rights Reserved. Editions Version
EU Reference Laboratory for E. coli Department of Veterinary Public Health and Food Safety Unit of Foodborne Zoonoses Istituto Superiore di Sanità
 Identification and characterization of Verocytotoxin-producing Escherichia coli (VTEC) by Real Time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated
Identification and characterization of Verocytotoxin-producing Escherichia coli (VTEC) by Real Time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated
Cloning Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems
 Promega Notes Number 71, 1999, p. 10 Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems By Kimberly Knoche, Ph.D., and Dan Kephart, Ph.D. Promega Corporation Corresponding
Promega Notes Number 71, 1999, p. 10 Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems By Kimberly Knoche, Ph.D., and Dan Kephart, Ph.D. Promega Corporation Corresponding
Expression and Purification of Recombinant Protein in bacteria and Yeast. Presented By: Puspa pandey, Mohit sachdeva & Ming yu
 Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Event-specific Method for the Quantification of Maize MIR162 Using Real-time PCR. Protocol
 Event-specific Method for the Quantification of Maize MIR162 Using Real-time PCR Protocol 31 January 2011 Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics
Event-specific Method for the Quantification of Maize MIR162 Using Real-time PCR Protocol 31 January 2011 Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics
Five-minute cloning of Taq polymerase-amplified PCR products
 TOPO TA Cloning Version R 8 April 2004 25-0184 TOPO TA Cloning Five-minute cloning of Taq polymerase-amplified PCR products Catalog nos. K4500-01, K4500-40, K4510-20, K4520-01, K4520-40, K4550-01, K4550-40,
TOPO TA Cloning Version R 8 April 2004 25-0184 TOPO TA Cloning Five-minute cloning of Taq polymerase-amplified PCR products Catalog nos. K4500-01, K4500-40, K4510-20, K4520-01, K4520-40, K4550-01, K4550-40,
Module 6: Digital DNA
 Module 6: Digital DNA Representation and processing of digital information in the form of DNA is essential to life in all organisms, no matter how large or tiny. Computing tools and computational thinking
Module 6: Digital DNA Representation and processing of digital information in the form of DNA is essential to life in all organisms, no matter how large or tiny. Computing tools and computational thinking
Marine Biology DEC 2004; 146(1) : 53-64 http://dx.doi.org/10.1007/s00227-004-1423-6 Copyright 2004 Springer
 Marine Biology DEC 2004; 146(1) : 53-64 http://dx.doi.org/10.1007/s00227-004-1423-6 Copyright 2004 Springer Archimer http://www.ifremer.fr/docelec/ Archive Institutionnelle de l Ifremer The original publication
Marine Biology DEC 2004; 146(1) : 53-64 http://dx.doi.org/10.1007/s00227-004-1423-6 Copyright 2004 Springer Archimer http://www.ifremer.fr/docelec/ Archive Institutionnelle de l Ifremer The original publication
TITRATION OF raav (VG) USING QUANTITATIVE REAL TIME PCR
 Page 1 of 5 Materials DNase digestion buffer [13 mm Tris-Cl, ph7,5 / 5 mm MgCl2 / 0,12 mm CaCl2] RSS plasmid ptr-uf11 SV40pA Forward primer (10µM) AGC AAT AGC ATC ACA AAT TTC ACA A SV40pA Reverse Primer
Page 1 of 5 Materials DNase digestion buffer [13 mm Tris-Cl, ph7,5 / 5 mm MgCl2 / 0,12 mm CaCl2] RSS plasmid ptr-uf11 SV40pA Forward primer (10µM) AGC AAT AGC ATC ACA AAT TTC ACA A SV40pA Reverse Primer
TIANquick Mini Purification Kit
 TIANquick Mini Purification Kit For purification of PCR products, 100 bp to 20 kb www.tiangen.com TIANquick Mini Purification Kit (Spin column) Cat no. DP203 Kit Contents Contents Buffer BL Buffer PB Buffer
TIANquick Mini Purification Kit For purification of PCR products, 100 bp to 20 kb www.tiangen.com TIANquick Mini Purification Kit (Spin column) Cat no. DP203 Kit Contents Contents Buffer BL Buffer PB Buffer
TA Cloning Kit. Version V 7 April 2004 25-0024. Catalog nos. K2000-01, K2000-40, K2020-20, K2020-40, K2030-01 K2030-40, K2040-01, K2040-40
 TA Cloning Kit Version V 7 April 2004 25-0024 TA Cloning Kit Catalog nos. K2000-01, K2000-40, K2020-20, K2020-40, K2030-01 K2030-40, K2040-01, K2040-40 ii Table of Contents Table of Contents...iii Important
TA Cloning Kit Version V 7 April 2004 25-0024 TA Cloning Kit Catalog nos. K2000-01, K2000-40, K2020-20, K2020-40, K2030-01 K2030-40, K2040-01, K2040-40 ii Table of Contents Table of Contents...iii Important
ANALYSIS OF A CIRCULAR CODE MODEL
 ANALYSIS OF A CIRCULAR CODE MODEL Jérôme Lacan and Chrstan J. Mchel * Laboratore d Informatque de Franche-Comté UNIVERSITE DE FRANCHE-COMTE IUT de Belfort-Montbélard 4 Place Tharradn - BP 747 5 Montbélard
ANALYSIS OF A CIRCULAR CODE MODEL Jérôme Lacan and Chrstan J. Mchel * Laboratore d Informatque de Franche-Comté UNIVERSITE DE FRANCHE-COMTE IUT de Belfort-Montbélard 4 Place Tharradn - BP 747 5 Montbélard
Transmembrane Signaling in Chimeras of the E. coli Chemotaxis Receptors and Bacterial Class III Adenylyl Cyclases
 Transmembrane Signaling in Chimeras of the E. coli Chemotaxis Receptors and Bacterial Class III Adenylyl Cyclases Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
Transmembrane Signaling in Chimeras of the E. coli Chemotaxis Receptors and Bacterial Class III Adenylyl Cyclases Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
Directed-Mutagenesis and Deletion Generated through an Improved Overlapping-Extension PCR Based Procedure
 Research Article Directed-Mutagenesis and Deletion Generated through an Improved Overlapping-Extension PCR Based Procedure Wirojne Kanoksilapatham 1*, Juan M. González 2 and Frank T. Robb 3 1 Department
Research Article Directed-Mutagenesis and Deletion Generated through an Improved Overlapping-Extension PCR Based Procedure Wirojne Kanoksilapatham 1*, Juan M. González 2 and Frank T. Robb 3 1 Department
Coding sequence the sequence of nucleotide bases on the DNA that are transcribed into RNA which are in turn translated into protein
 Assignment 3 Michele Owens Vocabulary Gene: A sequence of DNA that instructs a cell to produce a particular protein Promoter a control sequence near the start of a gene Coding sequence the sequence of
Assignment 3 Michele Owens Vocabulary Gene: A sequence of DNA that instructs a cell to produce a particular protein Promoter a control sequence near the start of a gene Coding sequence the sequence of
Recombinant DNA Unit Exam
 Recombinant DNA Unit Exam Question 1 Restriction enzymes are extensively used in molecular biology. Below are the recognition sites of two of these enzymes, BamHI and BclI. a) BamHI, cleaves after the
Recombinant DNA Unit Exam Question 1 Restriction enzymes are extensively used in molecular biology. Below are the recognition sites of two of these enzymes, BamHI and BclI. a) BamHI, cleaves after the
Heraeus Sepatech, Kendro Laboratory Products GmbH, Berlin. Becton Dickinson,Heidelberg. Biozym, Hessisch Oldendorf. Eppendorf, Hamburg
 13 4. MATERIALS 4.1 Laboratory apparatus Biofuge A Centrifuge 5804R FACScan Gel electrophoresis chamber GPR Centrifuge Heraeus CO-AUTO-ZERO Light Cycler Microscope Motopipet Neubauer Cell Chamber PCR cycler
13 4. MATERIALS 4.1 Laboratory apparatus Biofuge A Centrifuge 5804R FACScan Gel electrophoresis chamber GPR Centrifuge Heraeus CO-AUTO-ZERO Light Cycler Microscope Motopipet Neubauer Cell Chamber PCR cycler
2006 7.012 Problem Set 3 KEY
 2006 7.012 Problem Set 3 KEY Due before 5 PM on FRIDAY, October 13, 2006. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Which reaction is catalyzed by each
2006 7.012 Problem Set 3 KEY Due before 5 PM on FRIDAY, October 13, 2006. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Which reaction is catalyzed by each
Rapid and Simple Amplification of Genomic DNA Sequences Flanking Transposon
 MICROBIOLOGY INDONESIA, April 2007, p 1-4 Volume. 1, Number 1 ISSN 1978-3477 Rapid and Simple Amplification of Genomic DNA Sequences Flanking Transposon ARIS TRI WAHYUDI Department of Biology, Institut
MICROBIOLOGY INDONESIA, April 2007, p 1-4 Volume. 1, Number 1 ISSN 1978-3477 Rapid and Simple Amplification of Genomic DNA Sequences Flanking Transposon ARIS TRI WAHYUDI Department of Biology, Institut
RT-PCR: Two-Step Protocol
 RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
Supplemental Data. Short Article. PPARγ Activation Primes Human Monocytes. into Alternative M2 Macrophages. with Anti-inflammatory Properties
 Cell Metabolism, Volume 6 Supplemental Data Short Article PPARγ Activation Primes Human Monocytes into Alternative M2 Macrophages with Anti-inflammatory Properties M. Amine Bouhlel, Bruno Derudas, Elena
Cell Metabolism, Volume 6 Supplemental Data Short Article PPARγ Activation Primes Human Monocytes into Alternative M2 Macrophages with Anti-inflammatory Properties M. Amine Bouhlel, Bruno Derudas, Elena
ANALYSIS OF GROWTH HORMONE IN TENCH (TINCA TINCA) ANALÝZA RŮSTOVÉHO HORMONU LÍNA OBECNÉHO (TINCA TINCA)
 ANALYSIS OF GROWTH HORMONE IN TENCH (TINCA TINCA) ANALÝZA RŮSTOVÉHO HORMONU LÍNA OBECNÉHO (TINCA TINCA) Zrůstová J., Bílek K., Baránek V., Knoll A. Ústav morfologie, fyziologie a genetiky zvířat, Agronomická
ANALYSIS OF GROWTH HORMONE IN TENCH (TINCA TINCA) ANALÝZA RŮSTOVÉHO HORMONU LÍNA OBECNÉHO (TINCA TINCA) Zrůstová J., Bílek K., Baránek V., Knoll A. Ústav morfologie, fyziologie a genetiky zvířat, Agronomická
On Covert Data Communication Channels Employing DNA Recombinant and Mutagenesis-based Steganographic Techniques
 On Covert Data Communication Channels Employing DNA Recombinant and Mutagenesis-based Steganographic Techniques MAGDY SAEB 1, EMAN EL-ABD 2, MOHAMED E. EL-ZANATY 1 1. School of Engineering, Computer Department,
On Covert Data Communication Channels Employing DNA Recombinant and Mutagenesis-based Steganographic Techniques MAGDY SAEB 1, EMAN EL-ABD 2, MOHAMED E. EL-ZANATY 1 1. School of Engineering, Computer Department,
Recombinant DNA & Genetic Engineering. Tools for Genetic Manipulation
 Recombinant DNA & Genetic Engineering g Genetic Manipulation: Tools Kathleen Hill Associate Professor Department of Biology The University of Western Ontario Tools for Genetic Manipulation DNA, RNA, cdna
Recombinant DNA & Genetic Engineering g Genetic Manipulation: Tools Kathleen Hill Associate Professor Department of Biology The University of Western Ontario Tools for Genetic Manipulation DNA, RNA, cdna
NimbleGen DNA Methylation Microarrays and Services
 NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
Chapter 9. Applications of probability. 9.1 The genetic code
 Chapter 9 Applications of probability In this chapter we use the tools of elementary probability to investigate problems of several kinds. First, we study the language of life by focusing on the universal
Chapter 9 Applications of probability In this chapter we use the tools of elementary probability to investigate problems of several kinds. First, we study the language of life by focusing on the universal
Part ONE. a. Assuming each of the four bases occurs with equal probability, how many bits of information does a nucleotide contain?
 Networked Systems, COMPGZ01, 2012 Answer TWO questions from Part ONE on the answer booklet containing lined writing paper, and answer ALL questions in Part TWO on the multiple-choice question answer sheet.
Networked Systems, COMPGZ01, 2012 Answer TWO questions from Part ONE on the answer booklet containing lined writing paper, and answer ALL questions in Part TWO on the multiple-choice question answer sheet.
Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians
 Vol. 44 No. 3 SCIENCE IN CHINA (Series C) June 2001 Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians KE Yuehai ( `º) 1, SU Bing (3 Á) 1 3, XIAO Junhua
Vol. 44 No. 3 SCIENCE IN CHINA (Series C) June 2001 Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians KE Yuehai ( `º) 1, SU Bing (3 Á) 1 3, XIAO Junhua
Cloning, sequencing, and expression of H.a. YNRI and H.a. YNII, encoding nitrate and nitrite reductases in the yeast Hansenula anomala
 Cloning, sequencing, and expression of H.a. YNRI and H.a. YNII, encoding nitrate and nitrite reductases in the yeast Hansenula anomala -'Pablo García-Lugo 1t, Celedonio González l, Germán Perdomo l, Nélida
Cloning, sequencing, and expression of H.a. YNRI and H.a. YNII, encoding nitrate and nitrite reductases in the yeast Hansenula anomala -'Pablo García-Lugo 1t, Celedonio González l, Germán Perdomo l, Nélida
were demonstrated to be, respectively, the catalytic and regulatory subunits of protein phosphatase 2A (PP2A) (29).
 JOURNAL OF VIROLOGY, Feb. 1992, p. 886-893 0022-538X/92/020886-08$02.00/0 Copyright C) 1992, American Society for Microbiology Vol. 66, No. 2 The Third Subunit of Protein Phosphatase 2A (PP2A), a 55- Kilodalton
JOURNAL OF VIROLOGY, Feb. 1992, p. 886-893 0022-538X/92/020886-08$02.00/0 Copyright C) 1992, American Society for Microbiology Vol. 66, No. 2 The Third Subunit of Protein Phosphatase 2A (PP2A), a 55- Kilodalton
HCS604.03 Exercise 1 Dr. Jones Spring 2005. Recombinant DNA (Molecular Cloning) exercise:
 HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
Provincial Exam Questions. 9. Give one role of each of the following nucleic acids in the production of an enzyme.
 Provincial Exam Questions Unit: Cell Biology: Protein Synthesis (B7 & B8) 2010 Jan 3. Describe the process of translation. (4 marks) 2009 Sample 8. What is the role of ribosomes in protein synthesis? A.
Provincial Exam Questions Unit: Cell Biology: Protein Synthesis (B7 & B8) 2010 Jan 3. Describe the process of translation. (4 marks) 2009 Sample 8. What is the role of ribosomes in protein synthesis? A.
PrimeSTAR HS DNA Polymerase
 Cat. # R010A For Research Use PrimeSTAR HS DNA Polymerase Product Manual Table of Contents I. Description...3 II. III. IV. Components...3 Storage...3 Features...3 V. General Composition of PCR Reaction
Cat. # R010A For Research Use PrimeSTAR HS DNA Polymerase Product Manual Table of Contents I. Description...3 II. III. IV. Components...3 Storage...3 Features...3 V. General Composition of PCR Reaction
Cloning GFP into Mammalian cells
 Protocol for Cloning GFP into Mammalian cells Studiepraktik 2013 Molecular Biology and Molecular Medicine Aarhus University Produced by the instructors: Tobias Holm Bønnelykke, Rikke Mouridsen, Steffan
Protocol for Cloning GFP into Mammalian cells Studiepraktik 2013 Molecular Biology and Molecular Medicine Aarhus University Produced by the instructors: Tobias Holm Bønnelykke, Rikke Mouridsen, Steffan
Improved methods for site-directed mutagenesis using Gibson Assembly TM Master Mix
 CLONING & MAPPING DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Improved methods for site-directed mutagenesis using Gibson Assembly TM Master Mix LIBRARY PREP FOR NET GEN SEQUENCING PROTEIN
CLONING & MAPPING DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Improved methods for site-directed mutagenesis using Gibson Assembly TM Master Mix LIBRARY PREP FOR NET GEN SEQUENCING PROTEIN
APOT - Assay. Protocol for HPV16 and 18. Amplification of Papilloma Virus Oncogene Transcripts HPV. E6 E7 E1 Zelluläre DNA poly(a)
 E5 E2 E1 APOT - Assay Amplification of Papilloma Virus Oncogene Transcripts URR E6 E7 L1 HPV L2 E4 E6 E7 E1 Zelluläre DNA poly(a) Protocol for HPV16 and 18 Brief summary of the APOT assay Fig.1A shows
E5 E2 E1 APOT - Assay Amplification of Papilloma Virus Oncogene Transcripts URR E6 E7 L1 HPV L2 E4 E6 E7 E1 Zelluläre DNA poly(a) Protocol for HPV16 and 18 Brief summary of the APOT assay Fig.1A shows
Bacterial Transformation and Plasmid Purification. Chapter 5: Background
 Bacterial Transformation and Plasmid Purification Chapter 5: Background History of Transformation and Plasmids Bacterial methods of DNA transfer Transformation: when bacteria take up DNA from their environment
Bacterial Transformation and Plasmid Purification Chapter 5: Background History of Transformation and Plasmids Bacterial methods of DNA transfer Transformation: when bacteria take up DNA from their environment
CLONING IN ESCHERICHIA COLI
 CLONING IN ESCHERICHIA COLI Introduction: In this laboratory, you will carry out a simple cloning experiment in E. coli. Specifically, you will first create a recombinant DNA molecule by carrying out a
CLONING IN ESCHERICHIA COLI Introduction: In this laboratory, you will carry out a simple cloning experiment in E. coli. Specifically, you will first create a recombinant DNA molecule by carrying out a
Molecular Cloning, Product Brochure
 , Product Brochure Interest in any of the products, request or order them at Bio-Connect. Bio-Connect B.V. T NL +31 (0)26 326 44 50 T BE +32 (0)2 503 03 48 Begonialaan 3a F NL +31 (0)26 326 44 51 F BE
, Product Brochure Interest in any of the products, request or order them at Bio-Connect. Bio-Connect B.V. T NL +31 (0)26 326 44 50 T BE +32 (0)2 503 03 48 Begonialaan 3a F NL +31 (0)26 326 44 51 F BE
N-terminal Regulatory Domains of Phosphodiesterases 1, 4, 5 and 10 examined with an Adenylyl Cyclase as a Reporter
 N-terminal Regulatory Domains of Phosphodiesterases 1, 4, 5 and 10 examined with an Adenylyl Cyclase as a Reporter Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
N-terminal Regulatory Domains of Phosphodiesterases 1, 4, 5 and 10 examined with an Adenylyl Cyclase as a Reporter Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS
 STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS THESIS SUBMITTED FOR THE DEGREB OF DOCTOR OF PHILOSOPHY (SCIENCE) OF THE UNIVERSITY OF CALCUTTA 1996 NRISINHA DE, M.Sc DEPARTMENT OF BIOCHEMISTRY
STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS THESIS SUBMITTED FOR THE DEGREB OF DOCTOR OF PHILOSOPHY (SCIENCE) OF THE UNIVERSITY OF CALCUTTA 1996 NRISINHA DE, M.Sc DEPARTMENT OF BIOCHEMISTRY
Bio 102 Practice Problems Recombinant DNA and Biotechnology
 Bio 102 Practice Problems Recombinant DNA and Biotechnology Multiple choice: Unless otherwise directed, circle the one best answer: 1. Which of the following DNA sequences could be the recognition site
Bio 102 Practice Problems Recombinant DNA and Biotechnology Multiple choice: Unless otherwise directed, circle the one best answer: 1. Which of the following DNA sequences could be the recognition site
http://www.life.umd.edu/grad/mlfsc/ DNA Bracelets
 http://www.life.umd.edu/grad/mlfsc/ DNA Bracelets by Louise Brown Jasko John Anthony Campbell Jack Dennis Cassidy Michael Nickelsburg Stephen Prentis Rohm Objectives: 1) Using plastic beads, construct
http://www.life.umd.edu/grad/mlfsc/ DNA Bracelets by Louise Brown Jasko John Anthony Campbell Jack Dennis Cassidy Michael Nickelsburg Stephen Prentis Rohm Objectives: 1) Using plastic beads, construct
Mutation. Mutation provides raw material to evolution. Different kinds of mutations have different effects
 Mutation Mutation provides raw material to evolution Different kinds of mutations have different effects Mutational Processes Point mutation single nucleotide changes coding changes (missense mutations)
Mutation Mutation provides raw material to evolution Different kinds of mutations have different effects Mutational Processes Point mutation single nucleotide changes coding changes (missense mutations)
All commonly-used expression vectors used in the Jia Lab contain the following multiple cloning site: BamHI EcoRI SmaI SalI XhoI_ NotI
 2. Primer Design 2.1 Multiple Cloning Sites All commonly-used expression vectors used in the Jia Lab contain the following multiple cloning site: BamHI EcoRI SmaI SalI XhoI NotI XXX XXX GGA TCC CCG AAT
2. Primer Design 2.1 Multiple Cloning Sites All commonly-used expression vectors used in the Jia Lab contain the following multiple cloning site: BamHI EcoRI SmaI SalI XhoI NotI XXX XXX GGA TCC CCG AAT
Taqman TCID50 for AAV Vector Infectious Titer Determination
 Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Characterization of cdna clones of the family of trypsin/a-amylase inhibitors (CM-proteins) in barley {Hordeum vulgare L.)
 Characterization of cdna clones of the family of trypsin/a-amylase inhibitors (CM-proteins) in barley {Hordeum vulgare L.) J. Paz-Ares, F. Ponz, P. Rodríguez-Palenzuela, A. Lázaro, C. Hernández-Lucas,
Characterization of cdna clones of the family of trypsin/a-amylase inhibitors (CM-proteins) in barley {Hordeum vulgare L.) J. Paz-Ares, F. Ponz, P. Rodríguez-Palenzuela, A. Lázaro, C. Hernández-Lucas,
Biotechnology: DNA Technology & Genomics
 Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
The making of The Genoma Music
 242 Summary Key words Resumen Palabras clave The making of The Genoma Music Aurora Sánchez Sousa 1, Fernando Baquero 1 and Cesar Nombela 2 1 Department of Microbiology, Ramón y Cajal Hospital, and 2 Department
242 Summary Key words Resumen Palabras clave The making of The Genoma Music Aurora Sánchez Sousa 1, Fernando Baquero 1 and Cesar Nombela 2 1 Department of Microbiology, Ramón y Cajal Hospital, and 2 Department
Protein Synthesis Simulation
 Protein Synthesis Simulation Name(s) Date Period Benchmark: SC.912.L.16.5 as AA: Explain the basic processes of transcription and translation, and how they result in the expression of genes. (Assessed
Protein Synthesis Simulation Name(s) Date Period Benchmark: SC.912.L.16.5 as AA: Explain the basic processes of transcription and translation, and how they result in the expression of genes. (Assessed
TransformAid Bacterial Transformation Kit
 Home Contacts Order Catalog Support Search Alphabetical Index Numerical Index Restriction Endonucleases Modifying Enzymes PCR Kits Markers Nucleic Acids Nucleotides & Oligonucleotides Media Transfection
Home Contacts Order Catalog Support Search Alphabetical Index Numerical Index Restriction Endonucleases Modifying Enzymes PCR Kits Markers Nucleic Acids Nucleotides & Oligonucleotides Media Transfection
How To Clone Into Pcdna 3.1/V5-His
 pcdna 3.1/V5-His A, B, and C Catalog no. V810-20 Rev. date: 09 November 2010 Manual part no. 28-0141 MAN0000645 User Manual ii Contents Contents and Storage... iv Methods... 1 Cloning into pcdna 3.1/V5-His
pcdna 3.1/V5-His A, B, and C Catalog no. V810-20 Rev. date: 09 November 2010 Manual part no. 28-0141 MAN0000645 User Manual ii Contents Contents and Storage... iv Methods... 1 Cloning into pcdna 3.1/V5-His
First Strand cdna Synthesis
 380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
Supporting Information
 Supporting Information Wiley-VCH 2007 69451 Weinheim, Germany Evolving a Thermostable DNA polymerase that Accurately Amplifies Highly-Damaged Templates Christian Gloeckner, Katharina B. M. Sauter, and
Supporting Information Wiley-VCH 2007 69451 Weinheim, Germany Evolving a Thermostable DNA polymerase that Accurately Amplifies Highly-Damaged Templates Christian Gloeckner, Katharina B. M. Sauter, and
Integrated Protein Services
 Integrated Protein Services Custom protein expression & purification Version DC04-0012 Expression strategy The first step in the recombinant protein generation process is to design an appropriate expression
Integrated Protein Services Custom protein expression & purification Version DC04-0012 Expression strategy The first step in the recombinant protein generation process is to design an appropriate expression
Chromatin Immunoprecipitation
 Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Integrated Protein Services
 Integrated Protein Services Custom protein expression & purification Last date of revision June 2015 Version DC04-0013 www.iba-lifesciences.com Expression strategy The first step in the recombinant protein
Integrated Protein Services Custom protein expression & purification Last date of revision June 2015 Version DC04-0013 www.iba-lifesciences.com Expression strategy The first step in the recombinant protein
The Arabinosyltransferase EmbC Is Inhibited by Ethambutol in Mycobacterium tuberculosis
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Oct. 2009, p. 4138 4146 Vol. 53, No. 10 0066-4804/09/$08.00 0 doi:10.1128/aac.00162-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. The
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Oct. 2009, p. 4138 4146 Vol. 53, No. 10 0066-4804/09/$08.00 0 doi:10.1128/aac.00162-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. The
Drosophila NK-homeobox genes
 Proc. Natl. Acad. Sci. USA Vol. 86, pp. 7716-7720, October 1989 Biochemistry Drosophila NK-homeobox genes (NK-1, NK-2,, and DNA clones/chromosome locations of genes) YONGSOK KIM AND MARSHALL NIRENBERG
Proc. Natl. Acad. Sci. USA Vol. 86, pp. 7716-7720, October 1989 Biochemistry Drosophila NK-homeobox genes (NK-1, NK-2,, and DNA clones/chromosome locations of genes) YONGSOK KIM AND MARSHALL NIRENBERG
Molecular chaperones involved in preprotein. targeting to plant organelles
 Molecular chaperones involved in preprotein targeting to plant organelles Dissertation der Fakultät für Biologie der Ludwig-Maximilians-Universität München vorgelegt von Christine Fellerer München 29.
Molecular chaperones involved in preprotein targeting to plant organelles Dissertation der Fakultät für Biologie der Ludwig-Maximilians-Universität München vorgelegt von Christine Fellerer München 29.
HiPer RT-PCR Teaching Kit
 HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
Rapid GST Inclusion Body Solubilization and Renaturation Kit
 Product Manual Rapid GST Inclusion Body Solubilization and Renaturation Kit Catalog Number AKR-110 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bacteria are widely used for His
Product Manual Rapid GST Inclusion Body Solubilization and Renaturation Kit Catalog Number AKR-110 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Bacteria are widely used for His
Recombinant DNA Technology
 Recombinant DNA Technology Dates in the Development of Gene Cloning: 1965 - plasmids 1967 - ligase 1970 - restriction endonucleases 1972 - first experiments in gene splicing 1974 - worldwide moratorium
Recombinant DNA Technology Dates in the Development of Gene Cloning: 1965 - plasmids 1967 - ligase 1970 - restriction endonucleases 1972 - first experiments in gene splicing 1974 - worldwide moratorium
Interleukin-4 Receptor Signal Transduction: Involvement of P62
 Interleukin-4 Receptor Signal Transduction: Involvement of P62 Den Naturwissenschaftlichen Fakultäten der Friedrich Alexander Universität Erlangen Nürnberg zur Erlangung des Doktorgrades vorgelegt von
Interleukin-4 Receptor Signal Transduction: Involvement of P62 Den Naturwissenschaftlichen Fakultäten der Friedrich Alexander Universität Erlangen Nürnberg zur Erlangung des Doktorgrades vorgelegt von
Identification and characterisation of Verocytotoxinproducing Escherichia coli (VTEC) by PCR amplification of virulence genes
 CRL_Method 01 28_04_2008 Pag 1 of 10 Identification and characterisation of Verocytotoxinproducing Escherichia coli (VTEC) by PCR amplification of virulence genes CRL_Method 01 28_04_2008 Pag 2 of 10 INDEX
CRL_Method 01 28_04_2008 Pag 1 of 10 Identification and characterisation of Verocytotoxinproducing Escherichia coli (VTEC) by PCR amplification of virulence genes CRL_Method 01 28_04_2008 Pag 2 of 10 INDEX
MultiSite Gateway Pro
 MultiSite Gateway Pro Using Gateway Technology to simultaneously clone multiple DNA fragments Catalog nos. 12537-102, 12537-103, 12537-104, 12537-100 Version B 3 October 2006 25-0942 Design your MultiSite
MultiSite Gateway Pro Using Gateway Technology to simultaneously clone multiple DNA fragments Catalog nos. 12537-102, 12537-103, 12537-104, 12537-100 Version B 3 October 2006 25-0942 Design your MultiSite
Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction
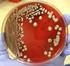 Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction Yogesh Chander 1 M. A. Ramakrishnan 1 Naresh Jindal 1 Kevan Hanson 2 Sagar M. Goyal 1 1 Department of Veterinary
Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction Yogesh Chander 1 M. A. Ramakrishnan 1 Naresh Jindal 1 Kevan Hanson 2 Sagar M. Goyal 1 1 Department of Veterinary
2. The number of different kinds of nucleotides present in any DNA molecule is A) four B) six C) two D) three
 Chem 121 Chapter 22. Nucleic Acids 1. Any given nucleotide in a nucleic acid contains A) two bases and a sugar. B) one sugar, two bases and one phosphate. C) two sugars and one phosphate. D) one sugar,
Chem 121 Chapter 22. Nucleic Acids 1. Any given nucleotide in a nucleic acid contains A) two bases and a sugar. B) one sugar, two bases and one phosphate. C) two sugars and one phosphate. D) one sugar,
DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108
 DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 THE INTERLEUKIN-10 FAMILY CYTOKINES GENE POLYMORPHISMS IN PLAQUE PSORIASIS KÜLLI KINGO TARTU
DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 THE INTERLEUKIN-10 FAMILY CYTOKINES GENE POLYMORPHISMS IN PLAQUE PSORIASIS KÜLLI KINGO TARTU
pbad/his A, B, and C pbad/myc-his A, B, and C
 pbad/his A, B, and C pbad/myc-his A, B, and C Vectors for Dose-Dependent Expression of Recombinant Proteins Containing N- or C-Terminal 6 His Tags in E. coli Catalog nos. V430-01, V440-01 Version J 29
pbad/his A, B, and C pbad/myc-his A, B, and C Vectors for Dose-Dependent Expression of Recombinant Proteins Containing N- or C-Terminal 6 His Tags in E. coli Catalog nos. V430-01, V440-01 Version J 29
DNA Sequencing of the eta Gene Coding for Staphylococcal Exfoliative Toxin Serotype A
 Journal of General Microbiology (1988), 134, 71 1-71 7. Printed in Great Britain 71 1 DNA Sequencing of the eta Gene Coding for Staphylococcal Exfoliative Toxin Serotype A By SUSUMU SAKURA, HTOSH SUZUK
Journal of General Microbiology (1988), 134, 71 1-71 7. Printed in Great Britain 71 1 DNA Sequencing of the eta Gene Coding for Staphylococcal Exfoliative Toxin Serotype A By SUSUMU SAKURA, HTOSH SUZUK
2.1.2 Characterization of antiviral effect of cytokine expression on HBV replication in transduced mouse hepatocytes line
 i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
 Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2014 DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2014 DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
HighPure Maxi Plasmid Kit
 HighPure Maxi Plasmid Kit For purification of high pure plasmid DNA with high yields www.tiangen.com PP120109 HighPure Maxi Plasmid Kit Kit Contents Storage Cat.no. DP116 Contents RNaseA (100 mg/ml) Buffer
HighPure Maxi Plasmid Kit For purification of high pure plasmid DNA with high yields www.tiangen.com PP120109 HighPure Maxi Plasmid Kit Kit Contents Storage Cat.no. DP116 Contents RNaseA (100 mg/ml) Buffer
CCR Biology - Chapter 9 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
