ND10 Protein PML Is Recruited to Herpes Simplex Virus Type 1 Prereplicative Sites and Replication Compartments in the Presence of Viral DNA Polymerase
|
|
|
- Ira Doyle
- 7 years ago
- Views:
Transcription
1 JOURNAL OF VIROLOGY, Dec. 1998, p Vol. 72, No X/98/$ Copyright 1998, American Society for Microbiology. All Rights Reserved. ND10 Protein PML Is Recruited to Herpes Simplex Virus Type 1 Prereplicative Sites and Replication Compartments in the Presence of Viral DNA Polymerase JENNIFER BURKHAM, 1 DONALD M. COEN, 2 AND SANDRA K. WELLER 1 * Department of Microbiology, University of Connecticut Health Center, Farmington, Connecticut 06030, 1 and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts Received 31 March 1998/Accepted 20 August 1998 Herpes simplex virus type 1 (HSV-1) infection results in the disruption of ND10 (also called nuclear bodies, PODs, or PML-associated bodies), which are nuclear matrix domains of unknown function present in mammalian cells. After ND10 disruption, viral transcription and DNA replication occur in globular nuclear domains called replication compartments. In this report we define four stages of infection by using antibodies to ICP8 (also called SSB and UL29) and the ND10 antigen PML. Immediately after infection, cells contain intact ND10 as detected by staining for PMLs (stage I); within 1 hour, however, ND10 are disrupted and cells begin to exhibit diffuse staining for the major viral DNA binding protein, ICP8 (stage II). After all ND10 have been disrupted, foci which resemble but are not equivalent to ND10 appear, containing both PML and ICP8 (stage III). Cells infected with mutants defective in the helicase-primase or origin binding protein are unable to form stage III foci. Cells infected with a mutant that is null for the polymerase catalytic subunit, however, form stage III-like ICP8 foci which do not contain PML. Thus, stage III foci recruit the cellular PML protein in the presence but not the absence of HSV polymerase. PML was recruited to stage III foci in some but not all cells infected with a mutant defective in the polymerase accessory protein, UL42. Thus, UL42 is not required for the recruitment of PML to viral foci. In wild-type infection, stage III cells are quickly replaced by cells containing replication compartments (stage IV). PML and ICP8 staining are both observed within replication compartments, indicating a potential role for PML in HSV-1 replication. Models for the role of ND10 proteins in the formation of replication compartments are discussed. * Corresponding author. Mailing address: Department of Microbiology, University of Connecticut Health Center, Farmington, CT Phone: (860) Fax: (860) Weller@nso2.uchc.edu. Several DNA viruses, including adenovirus, simian virus 40, and herpesviruses, have been shown to affect the partitioning of cellular antigens within ND10 (1, 4, 7, 19, 21, 32, 38, 44). These nuclear matrix-associated domains occur at an average of 10 per nucleus and are also known as PODs, Kr bodies, nuclear bodies, and PML-associated bodies (7, 15, 38). ND10 proteins have been associated with the control of cellular growth, transcription, apoptosis, and the life cycles of several viruses (5, 11, 14, 35). The most extensively studied ND10 protein, PML, is expressed as a fusion with the retinoic acid receptor alpha in individuals with acute promyelocytic leukemia (APL) (18). Expression of the PML-retinoic acid receptor alpha fusion protein in APL cells results in both the disruption of ND10 foci and the loss of growth control. Retinoic acid treatment of APL cell lines results in both the restoration of growth control and the reformation of ND10 (20). The observations that dispersal and reformation of ND10 occur during the cell cycle and in response to stress (34, 43) suggest that the partitioning of ND10 antigens into ND10 foci is linked to cellular growth control. PML has been shown to play a role in cellular growth control (46), and both PML and another ND10 protein, Sp100, have been implicated in transcriptional regulation (14, 49). Soon after infection with herpes simplex virus type 1 (HSV- 1), the viral transactivator ICP0 migrates to ND10 and is believed to initiate the dispersal of PML and other ND10 antigens (8, 10, 12, 31, 47). Everett et al. have recently reported that the disruption of ND10 correlates with the ICP0- and proteosome-dependent loss of several PML isoforms (9). DNA genomes have been observed at the periphery of ND10 and have been reported to preferentially initiate transcription at sites adjacent to ND10 (33). During infection, viral DNA replication proteins are found in large globular nuclear domains called replication compartments (40). Under some experimental conditions replication compartments have been shown to form at sites adjacent to ND10 (15, 26). Taken together, these results suggest that ND10 or sites in the cell close to ND10 play an important role in the establishment of a productive infection (32). One attractive model posits that ND10 antigens mark sites in the nucleus that are necessary for the establishment of a productive viral infection (30); whether these sites represent nuclear matrix attachment sites or some other relatively fixed domains within the nucleus remains to be determined. We and others have also observed that ND10 proteins are recruited to replication compartments in infected cells (24, 39), although the role of these proteins in viral replication is not clear. In this report, we explore the disruption of ND10 and the formation of replication compartments in cells infected with wild-type and mutant viruses. Based on staining patterns of the ND10 protein PML and the HSV-1 major DNA binding protein (SSB, ICP8, or UL29), we define four distinct stages during early infection leading to replication compartment formation. Stage I cells resemble uninfected cells in that they contain intact ND10 and show no ICP8 staining. Stage II cells show diffuse PML staining and no intact ND10. ICP8 staining is diffuse and nuclear during this stage. After the initial disrup
2 VOL. 72, 1998 PML IS RECRUITED TO HSV-1 LOCI tion of ND10, cells enter a stage (stage III) in which ICP8 and PML colocalize in punctate foci, which resemble one type of prereplicative site. Prereplicative sites in cells infected with wild-type virus in the presence of polymerase inhibitors and in cells infected with viruses lacking the viral DNA polymerase have previously been described (6, 40). It is now recognized that there are two types of prereplicative sites: (i) numerous, thought to represent foci of ICP8 and other viral proteins at cellular replication forks in S-phase cells (24), and (ii) ND10 associated, thought to represent actual intermediates in the formation of replication compartments (24, 45). The PMLcontaining ICP8 foci which appear in stage III cells are likely equivalent to the so-called ND10-associated prereplicative sites. In cells competent to carry out viral DNA synthesis, stage III cells are quickly replaced by cells containing replication compartments (stage IV). In this paper we report the requirements for formation of PML-containing prereplicative sites and replication compartments by studying mutants defective in various HSV replication proteins. The presence of PML in stage III prereplicative sites and in replication compartments implicates ND10 proteins in the establishment of viral infection. MATERIALS AND METHODS Cells and viruses. HEp-2 cells were obtained from the American Type Culture Collection (Manassas, Va.). They were maintained in Dulbecco s modified Eagle s medium (ICN Biomedicals Inc., Aurora, Ohio) supplemented with 10% fetal calf serum (Atlanta Biologicals) and 1% penicillin streptomycin solution (Sigma) at 37 C in a 7% CO 2 atmosphere. The KOS strain of HSV-1 was used as the wild-type strain. ICP6::lacZ insertion mutants in the helicase-primase genes, UL5 (hr99), UL8 (hr80), and UL52 (hr114) and the origin binding protein, UL9 (hr94) were previously described (3, 13, 28, 51). The polymerase null mutant HP66 and B10, in which the polymerase coding sequences were restored, were previously described (29, 50). The UL42 mutant Cgal 42 was generously provided by P. Johnson and D. Parris (17). Immunofluorescence and imaging. Cells were grown on glass coverslips and infected as described above. Cells on coverslips were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min, washed in PBS, and permeabilized in 1.0% Triton X-100 in PBS for 10 min. The coverslips were again washed in PBS and pretreated with 3% normal goat serum in PBS for several minutes. The polyclonal antibody used to detect the viral ICP8 was anti-icp8, a kind gift from Bill Ruyechan (State University of New York at Buffalo) (41). The monoclonal antibody used for detection of the ND10 protein, PML, was obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). All antibodies were used at a dilution of 1:200 in 3% normal goat serum. Cells were stained with the primary antibodies for 30 min. Coverslips were washed extensively with PBS between primary and secondary antibodies. The secondary antibodies, Texas red-conjugated goat anti-rabbit and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulins, were obtained from Cappel, Organon Teknika Corporation (Durham, N.C.) and were used at a dilution of 1:200 in PBS for the 30-min staining of the coverslips. Coverslips were then washed extensively in PBS and mounted in glycerol gelatin containing 2.5% diazibicyclooctane (DABCO) to retard bleaching. Imaging was performed on a Zeiss Axiovert 135 laser scanning microscope (confocal) equipped with an argon-krypton laser. Texas red was excited at 568 nm; fluorescein isothiocyanate was excited at 488 nm. Emissions were collected separately, and the channels were overlaid by computer for the dual images. Images were collected with a 63X Neofluar lens and arranged and labeled by using Adobe Photoshop 3.0. RESULTS Four stages of HSV infection. In this report we describe four stages of early HSV-1 infection based on the PML and ICP8 staining patterns observed during the first 6 h of infection. HEp2 cells were infected with KOS at a multiplicity of infection (MOI) of 2 PFU/cell and were harvested at 1-h intervals between 0 and 6 h postinfection. Cells were prepared for immunofluorescence microscopy and stained with a polyclonal antibody specific for ICP8 (41) as a marker for viral replication proteins and a monoclonal antibody specific for PML as a marker for ND10. As a control, cells were stained with one primary antibody and both secondary antibodies: in all cases only the expected staining was observed. This indicates that there was no cross-reactivity of antibodies and no leakage of light from either fluor into the inappropriate channel. A minimum of 100 cells on each coverslip were tallied according to their staining patterns, and the percentages of cells exhibiting each staining pattern are shown in the first column of Fig. 1. At time zero, almost all cells contained PML in intact ND10 and showed no ICP8 staining (Fig. 1, top row); this pattern was designated stage I. Within the first 2hofinfection, PML staining became diffuse and cells exhibited a gradient of ICP8 staining from no ICP8 to strong diffuse nuclear ICP8 staining; we defined this staining pattern stage II (Fig. 1, second row). After the disruption of ND10 and prior to the formation of replication compartments, we observed the unexpected reappearance of PML foci within a background of diffuse nuclear PML staining, peaking at about 3 h postinfection. These nuclear PML foci resembled intact ND10 of uninfected cells and colocalized with ICP8. Although Everett et al. (9) have reported the degradation of several isoforms of PML in cells infected with HSV, some isoforms remain. We suggest that it is these remaining isoforms of PML which appear to be recruited into PML-ICP8 foci. Cells exhibiting these foci were classified as stage III cells (Fig. 1, third row). Stage III appears to be transitory, never exceeding approximately 20% of the cells, and cells in stage III are quickly replaced by cells containing replication compartments (stage IV), which stained for both ICP8 and PML (Fig. 1, bottom row). By approximately 5 h postinfection, almost all cells exhibited replication compartments. In this experiment, some PML staining was seen in the cytoplasm of many cells, although some of this staining may have been perinuclear (Fig. 1). This observation is consistent with the previous report that PML has been shown to shuttle between the cytoplasm and nucleus; however, cells stained with secondary antibodies alone show faint cytoplasmic and/or perinuclear staining but no nuclear staining (data not shown). Therefore, at least some of the cytoplasmic staining is due to background staining with the secondary antibody. The PML staining observed in stage III foci and in the replication compartments in HEp2 cells is not due to cross-reactivity of the PML antibody with viral proteins, because no PML staining was observed in KOS-infected Vero cells (Fig. 2). Vero cells contain a version of PML that does not react with the PML antibody used. These results indicate that the nuclear PML staining observed in infected HEp2 cells is due to PML colocalization with viral replication compartments and not crossreactivity with viral antigens. In conclusion, we observe and define four different stages of viral infection with respect to PML and ICP8 antigens. Moreover, we confirm that ND10, as observed by PML staining, are disrupted early in viral infection and we suggest that at least some isoforms of the PML protein are present within HSV-1 replication compartments. The most surprising aspect of these experiments was the apparent reformation of PML foci (stage III) after the initial disruption of ND10 during stage II. Stage III foci may be equivalent to the previously described ND10-associated prereplicative sites. Stage III foci seen during wild-type infection resemble the previously described ND10-associated prereplicative sites observed in cells infected in the presence of polymerase inhibitors such as phosphonoacetic acid (PAA) (24, 45): the ICP8 and PML staining patterns are similar in both cases. The first row of Fig. 3 shows cells infected with wild-type virus. In the absence of PAA (Fig. 3A and B), replication compartments, which stain for both PML and ICP8, were observed. In the presence of PAA (Fig. 3C and D), both types of prereplicative sites were observed (23, 24). The upper cell (Fig. 3C and D) contains numerous prereplicative sites which are thought to represent ICP8 localization to cellular
3 FIG. 1. Early HSV-1 infection occurs in four distinct stages. HEp2 cells were infected with KOS and harvested for immunofluorescence at various time points between 0 and 6 h postinfection. Infected cells were stained with anti-pml monoclonal antibody and anti-icp8 polyclonal antibody. Over 100 cells for each time point were tallied according to the staining patterns of these two proteins, and four stages of infection were defined. The first column shows the percentage of infected cells in each stage at various time points. The photographs in the second, third, and fourth columns show representative fields of HEp2 cells infected with KOS. The column labeled Merge shows the merged image of staining patterns for PML (green) and ICP8 (red). The columns labeled PML and ICP8 show the single staining patterns of PML (green) and ICP8 (red), respectively. The different rows (graphs and three columns of photographs) represent cells in stages as follows: stage I, cells demonstrating intact ND10 by PML staining and no ICP8; stage II, cells exhibiting disrupted ND10 and diffuse ICP8; stage III, cells exhibiting PML-ICP8 foci against a diffuse nuclear background staining; stage IV, cells exhibiting PML and ICP8 staining in replication compartments. Marker bar 15 m
4 VOL. 72, 1998 PML IS RECRUITED TO HSV-1 LOCI FIG. 2. The anti-pml antibody does not cross-react with HSV-1 proteins. HEp2 (top row) and Vero (bottom row) cells were infected with KOS for 5 h, processed for immunofluorescence, and stained with anti-pml and anti-icp8 as indicated. Marker bar 15 m. replication forks (24); the majority of these numerous prereplicative sites do not colocalize with PML. The lower cell (Fig. 3C and D) contains ND10-associated prereplicative sites, which are thought to represent intermediates in the formation of replication compartments (23, 24). Note that the cells containing ND10-associated prereplicative sites resemble the stage III cells in Fig. 1, third row. The similarity between these staining patterns prompted us to undertake a more detailed analysis of the kinetics of formation of the ND10-associated prereplicative sites. In order to determine if stage III foci and ND10-associated prereplicative sites form with similar kinetics, we infected HEp2 cells with KOS in the absence and presence of PAA and harvested the infections at 1-h intervals to determine staining patterns of PML and ICP8. In the presence of PAA, cells progressed through stages I (intact ND10), II (disrupted ND10), and III (foci) with the same kinetics as were observed in the absence of PAA (data not shown). In the presence of PAA, however, cells did not progress to stage IV, consistent with the observation that PAA inhibits the formation of replication compartments (40). Since ND10-associated prereplicative sites resemble stage III foci in appearance and formation kinetics, we propose that ND10-associated prereplicative sites represent stage III foci which are not able progress to stage IV because of the presence of PAA. When ND10-associated prereplicative sites were first observed and named, it was assumed these foci appeared prior to the disruption of ND10. The results of the present study with cells infected for shorter periods indicate, however, that these foci formed after, not before, the disruption of ND10. Thus, the observation that ND10 are disrupted before the formation of ICP8-containing foci indicates that the term ND10-associated prereplicative sites may be inappropriate; hereafter in this report we refer to them as PML-containing prereplicative sites. Whether the PML-containing prereplicative sites form at the site of the original ND10 remains to be determined. HSV-1 mutants defective in DNA synthesis do not form PML-containing prereplicative sites. To better characterize stage III foci (PML-containing prereplicative sites) and understand the events leading to their formation, we assessed HSV-1 mutants defective in DNA synthesis for their ability to form the PML-containing prereplicative sites. Insertion mutants (hr99, hr80, and hr114) of the HSV-1 helicase-primase (UL5, UL8, and UL52, respectively) and an insertion mutant (hr94) of the origin binding protein (UL9) were previously described (3, 13, 28, 51). ICP8 staining patterns in cells infected with hr99, hr80, hr114, and hr94 were previously shown to be diffuse and nuclear in untreated cells and prereplicative sites in the presence of PAA (23, 25). Since the two types of prereplicative sites had not been distinguished at the time of those observations, we asked whether both types of prereplicative sites are present in HEp2 cells infected with these mutants in the presence and absence of PAA. At 5 h postinfection, cells were fixed for immunofluorescence and stained with antibodies to ICP8 and PML. Cells infected with mutants defective in helicase-primase or origin binding protein in the absence of PAA exhibited similar patterns of ICP8 staining: the untreated cells exhibited diffuse and somewhat granular ICP8 staining which does not colocalize with PML (Fig. 3E, F, I, J, M, N, Q, and R). Cells infected with these mutants in the presence of PAA were able to form the numerous prereplicative sites, but they did not form the PML-containing prereplicative sites, which are fewer in number and colocalize precisely with foci of PML (Fig. 3C, H, K, L, O, P, S, and T). Thus, in the presence of PAA, cells infected with helicase-primase or UL9 mutants exhibited either the diffuse granular pattern of ICP8 staining (as seen in the upper cell in Fig. 3G) or the numerous prereplicative site pattern of ICP8 staining (as seen in the lower cell in Fig. 3G). The fact that PML-containing prereplicative sites (stage III foci) did not form in cells infected with mutants defective in helicase-primase and UL9, proteins necessary for DNA replication, supports the proposal that this subset of prereplicative sites represents a functional intermediate in the formation of replication compartments (24, 45). Consistent with previous results (6, 40), cells infected with a mutant defective in the catalytic subunit of the HSV-1 polymerase (HP66) (29) were able to form both types of prereplicative sites in the presence and absence of PAA. Figure 4 shows not only the numerous prereplicative sites (Fig. 4B, bottom cell) but also the less numerous ICP8 foci, which resemble stage III foci (Fig. 4B, top cell; Fig. 4C, all three cells). To our surprise, however, the less numerous ICP8 foci did not stain with PML (Fig. 4A to D). This is in contrast to cells infected with wild-type virus, which exhibited colocalization of ICP8 and PML not only in the absence of PAA (Fig. 1, third row) but also in PML-containing prereplicative sites in the presence of PAA (Fig. 3C and D). The mutant staining pattern is due specifically to the mutation in the polymerase gene, as an HP66-derived virus, B10, in which the polymerase primary coding sequences were restored (29, 50), regains its ability to recruit PML and progress to replication compartment formation (Fig. 4E to H). The observation that PML is not present in the stage III-like foci in cells infected with the polymerase mutant suggests that PML is somehow recruited to these viral structures in the presence of polymerase. This observation raised a question about the apparent interaction of PML with viral foci: is the interaction between PML and (i) the polymerase, (ii) the polymerase accessory protein (UL42), or (iii) a viral complex that is dependent on the polymerase? Since polymerase is thought to recruit UL42 to viral replication foci (23), we hypothesized that the recruitment of PML to viral foci might be dependent on the presence of the polymerase accessory protein. We asked whether cells infected with a virus lacking the UL42 gene would show PML recruitment to viral foci. In HEp2 cells infected with the UL42 null
5 10104 BURKHAM ET AL. J. VIROL. FIG. 3. PML and ICP8 staining in cells infected with various HSV-1 mutants in the presence and absence of PAA. HEp2 cells were infected at an MOI of 10 PFU/cell with HSV-1 strain KOS or mutants lacking helicase-primase subunits or origin binding protein and were processed for immunofluorescence as described in the legend to Fig. 1 at 5 h postinfection. The first two columns represent cells infected with various viruses (as indicated) in the absence of polymerase inhibitors: the first column shows only PML staining, and the second column shows a merged image of the same cells exhibiting both PML and ICP8 staining. The third and fourth columns represent cells infected with various viruses in the presence of the polymerase inhibitor PAA (400 g/ml): the third column shows the merged image of PML and ICP8 staining, and the fourth column shows PML staining only of the same cells. (A to D) KOS (wild-type)-infected cells; (E to H) cells infected with hr99 lacking the helicase UL5; (I to L) cells infected with hr80 lacking the helicase-primase accessory protein UL8; (M to P) cells infected with hr114 lacking the primase UL52; (Q to T) cells infected with hr94 lacking the origin binding protein, UL9. Marker bar 15 m.
6 VOL. 72, 1998 PML IS RECRUITED TO HSV-1 LOCI FIG. 4. Viral foci in cells infected with polymerase and polymerase accessory mutant viruses. HEp2 cells were infected at an MOI of 10 PFU/cell with either HP66, a virus lacking the catalytic domain of the HSV-1 polymerase; B10, a virus derived from HP66 whose polymerase coding sequences have been restored by marker rescue; or Cgal 42, a virus lacking the polymerase accessory protein. The first two columns represent cells infected as indicated in the absence of polymerase inhibitors: the first column shows only PML staining, and the second column shows a merged image of the same cells exhibiting both PML and ICP8 staining. The third and fourth columns represent cells infected with various viruses in the presence of the polymerase inhibitor PAA (400 g/ml): the third column shows the merged image of PML and ICP8 staining, and the fourth column shows PML staining only of the same cells. (A to D) HP66-infected cells; (E to H) B10-infected cells showing a wild-type phenotype; (I to L) Cgal 42-infected cells. The strong cytoplasmic fluorescein staining seen in panels E to L is likely due to the secondary antibody used; control experiments show no secondary antibody staining of the nucleus. Marker bar 15 m. virus, Cgal 42 (17), PML was recruited to viral foci in some, but not all, of the cells showing viral foci (Fig. 4I to L). This intermediate phenotype suggests that the presence of UL42 is not essential for the recruitment of PML to stage III foci. In sum, viral mutants lacking the helicase-primase subunits or the origin binding protein cannot form stage III foci (PMLassociated prereplicative sites). The viral mutant lacking an active polymerase can form ICP8-containing foci resembling stage III foci; however, these foci do not associate with PML as they do during wild-type infection. DISCUSSION Several observations regarding the relationship between ND10 and viral infection are made in this report. (i) The early events of HSV-1 replication can be broken down into four stages as identified by confocal microscopy of ICP8 and PML staining patterns. PML appears to be recruited into stage III and stage IV foci after the initial disruption of ND10. (ii) Stage III foci do not form in cells infected with mutants lacking UL5, UL8, UL9, or UL52. (iii) The association of PML with the stage III-like ICP8 foci requires the HSV DNA polymerase catalytic subunit. (iv) UL42, the HSV-1 polymerase accessory protein, enhances but is not absolutely required for the recruitment of PML to viral foci. In this report we have confirmed that ND10 are disrupted almost immediately upon infection (stage II) (10, 31). An unexpected result, however, was the appearance of PML-containing foci after the initial disruption of ND10 (stage III). Stage III foci, which may represent the earliest replication compartments, resemble the cellular ND10 in their staining with PML antibodies, in their morphology, and in their approximate number per cell, but they form after most cells have undergone complete disruption of ND10 in stage II. The similarity between ND10 and stage III foci raises the question of whether stage III foci form at the sites originally occupied by ND10 or whether they form at unrelated sites in the nucleus. If stage III foci form at the same subnuclear sites which were previously occupied by ND10, it would indicate that ND10 mark sites within the nucleus which are important to the replication of the virus. The concept that ND10 marks important sites on the
7 10106 BURKHAM ET AL. J. VIROL. nuclear matrix was originally proposed by Maul (30). The observations that replication compartments form adjacent to ND10 in transfected cells (26) and in cells infected with an ICP0 mutant (16) support the notion that the PML-containing prereplicative sites form at or near the site previously occupied by ND10. We cannot rule out, however, that PML-containing stage III foci represent the nucleation or aggregation of PML protein at unrelated sites in the nucleus as it is recruited to viral foci after being released from cellular ND10. Müller et al. (36) and Sternsdorf et al. (42) recently showed that modification of the ND10 protein SUMO, also called PIC1 (2, 27, 37), affects its partitioning within the nucleus (36, 42). When the PML protein is modified by SUMO, it is observed in the nuclear matrix-insoluble cell fraction, or ND10 (36, 42). When, however, the protein is unmodified, the protein is found in the soluble nucleoplasm outside of ND10 (36, 42). The modification and movement appear to be reversible and may represent a nuclear response to influences such as heat shock, interferon treatment, and heavy metal treatment. More recently, Everett et al. have shown that HSV-1 infection has profound effects on the modification state of PML (9). Upon infection, SUMO-modified forms of PML appear to be degraded, leaving only the unmodified, soluble forms of PML in the nucleus (9). Further experiments will be required to determine the modification state of the PML which is recruited to HSV-1-infected cells during stages III and IV. It is possible that the PML observed in stage III and stage IV foci represents unmodified isoforms not targeted for degradation. In order to better understand the viral requirements for the formation of stage III foci and replication compartments, we studied the viral foci formed in cells infected with viruses defective in DNA replication genes. Since the previously identified ND10-associated prereplicative sites (now designated PML-containing prereplicative sites) resemble stage III foci both in their morphology and in the time of their appearance, we propose that PML-containing prereplicative sites are equivalent to these intermediates but are unable to progress to replication compartments because of polymerase inhibition. In previous studies Liptak et al. (23) and Lukonis and Weller (25) examined the requirements for the formation of prereplicative sites, using viral mutants lacking individual HSV replication proteins such as helicase-primase, origin binding protein, and viral polymerase. Both of these studies, however, were performed before the two types of prereplicative sites were identified. In this study we have examined the involvement of viral DNA replication genes in the formation of the PML-containing prereplicative sites. We show that stage III foci do not form in cells infected with mutants lacking the helicase-primase or the origin binding protein. Furthermore, in the presence of a polymerase inhibitor, the helicase-primase and origin binding protein mutant viruses form the numerous type of prereplicative sites, thought to represent localization of ICP8 to cellular replication forks of cells in S phase, but they do not form the PML-containing prereplicative sites which resemble stage III foci. This result indicates that the formation of PML-containing prereplicative sites (and stage III foci) requires the presence of each of the helicase-primase subunits and the origin binding protein. This supports the model proposed by Liptak et al. in which ICP8, UL5, UL8, UL52, and UL9 form a subassembly of viral replication proteins which later recruits the polymerase and polymerase accessory protein (23). In addition to containing each of the viral DNA replication proteins, replication compartments have been reported to contain both PML and Sp100 and another ND10 protein recognized by monoclonal antibody 138 (24, 26, 39). A surprising finding made in the present study is that PML is recruited to PML-containing prereplicative sites (stage III foci) and replication compartments in a manner which is dependent on the presence of the viral polymerase. ICP8 foci resembling stage III foci are observed in cells infected with a virus lacking the polymerase, but they do not contain PML. We considered several possible explanations for the inability of a polymerase null mutant to recruit PML into stage III foci. First, it was possible that UL42, the polymerase accessory factor, is responsible for the recruitment of PML to replication foci. However, in cells infected with a UL42 null virus, PML was detected in viral foci in some but not all of the cells. This result indicates that UL42 is not absolutely required for PML recruitment but may enhance PML recruitment in some way. Second, it was possible that the polymerase protein itself is required for PML recruitment. Experiments using small deletions and point mutations of the polymerase catalytic subunit will be required to answer this question. It is also possible that PML is recruited only to areas at which DNA synthetic machinery is intact, although it is interesting that the presence of a polymerase inhibitor, PAA, does not prevent PML recruitment to viral foci. Regardless of how PML is recruited to wild-type viral foci, the recruitment of PML to stage III foci and replication compartments raises questions about its role in these domains. Since PML has never been implicated directly in cellular DNA replication, it seems unlikely that it plays a direct role in DNA synthesis; however, it may play a role in the recruitment of other cellular proteins which are involved either directly or indirectly in DNA replication. The observation that several cellular proteins involved in DNA replication and repair are recruited to viral prereplicative sites and replication compartments (48) is consistent with this hypothesis. During HSV infection ICP0 is responsible for the selective degradation of some isoforms of PML as well as other cellular proteins including the large subunit of the DNA-dependent protein kinase (9, 22). The loss of the PML isoforms is correlated with the ND10 disruption (9). At least two models which are not mutually exclusive can be envisioned for how ND10 disruption may contribute to the establishment of a productive viral replication. (i) Everett et al. have proposed that the selective degradation of cellular proteins leads to a stimulation of viral gene expression (9). The positive effects on viral gene expression may increase the likelihood that viral infection will progress efficiently. (ii) Viral DNA replication may require association with nuclear sites that are marked by ND10 (30). The disruption of ND10 and the degradation of some isoforms of PML upon infection may expose underlying sites, allowing the viral genome or viral replication proteins to become associated. According to this model, the presence of intact ND10 may inhibit viral DNA replication by masking important nuclear attachment sites. ACKNOWLEDGMENTS We thank Bill Ruyechan for providing antisera and P. A. Johnson and D. S. Parris for providing the UL42 mutant virus, Cgal 42. We also thank the members of our laboratory and Mark Challberg for helpful comments. This investigation was supported by Public Health Service grants AI21747 to S.K.W. and AI19838 to D.M.C. REFERENCES 1. Ahn, J. H., and G. S. Hayward The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PMLassociated nuclear bodies at very early times in infected permissive cells. J. Virol. 71: Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:
8 VOL. 72, 1998 PML IS RECRUITED TO HSV-1 LOCI Carmichael, E. P., M. J. Kosovsky, and S. K. Weller Isolation and characterization of herpes simplex virus type 1 host range mutants defective in viral DNA synthesis. J. Virol. 62: Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131: Chen, G. Q., X. G. Shi, W. Tang, S. M. Xiong, J. Zhu, X. Cai, Z. G. Han, J. H. Ni, G. Y. Shi, P. M. Jia, M. M. Liu, K. L. He, C. Niu, J. Ma, P. Zhang, T. D. Zhang, P. Paul, T. Naoe, K. Kitamura, W. Miller, S. Waxman, Z. Y. Wang, H. de The, S. J. Chen, and Z. Chen Use of arsenic trioxide (As 2 O 3 )in the treatment of acute promyelocytic leukemia (APL): I. As 2 O 3 exerts dosedependent dual effects on APL cells. Blood 89: de Bruyn Kops, A., and D. M. Knipe Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55: Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10: Dyck, J. A., G. G. Maul, W. H. Miller, Jr., J. D. Chen, A. Kakizuka, and R. M. Evans A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76: Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72: Everett, R. D., and G. G. Maul HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13: Fagioli, M., F. Grignani, P. F. Ferrucci, M. Alcalay, A. Mencarelli, I. Nicoletti, F. Grignani, and P. G. Pelicci Effect of the acute promyelocytic leukemia PML/RAR alpha protein on differentiation and survival of myeloid precursors. Leukemia 8(Suppl.): Gelman, I. H., and S. Silverstein Co-ordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J. Mol. Biol. 191: Goldstein, D. J., and S. K. Weller An ICP6::lacZ insertional mutagen is used to demonstrate that the UL52 gene of herpes simplex virus type 1 is required for virus growth and DNA synthesis. J. Virol. 62: Guiochon-Mantel, A., J. F. Savouret, F. Quignon, K. Delabre, E. Milgrom, and H. de The Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol. Endocrinol. 9: Ishov, A. M., and G. G. Maul The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134: Ishov, A. M., R. M. Stenberg, and G. G. Maul Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138: Johnson, P. A., M. G. Best, T. Friedmann, and D. S. Parris Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J. Virol. 65: Kakizuka, A., W. H. Miller, Jr., K. Umesono, R. P. Warrell, Jr., S. R. Frankel, V. V. Murty, E. Dmitrovsky, and R. M. Evans Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66: Kelly, C., R. Van Driel, and G. W. Wilkinson Disruption of PMLassociated nuclear bodies during human cytomegalovirus infection. J. Gen. Virol. 76: Koken, M. H., F. Puvion-Dutilleul, M. C. Guillemin, A. Viron, G. Linares- Cruz, N. Stuurman, L. de Jong, C. Szostecki, F. Calvo, C. Chomienne, L. Degos, E. Puvion, and H. de The The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 13: Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229: Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70: Liptak, L. M., S. L. Uprichard, and D. M. Knipe Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70: Lukonis, C. J., J. Burkham, and S. K. Weller Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71: Lukonis, C. J., and S. K. Weller Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J. Virol. 70: Lukonis, C. J., and S. K. Weller Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71: Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88: Malik, A. K., R. Martinez, L. Muncy, E. P. Carmichael, and S. K. Weller Genetic analysis of the herpes simplex virus type 1 UL9 gene: isolation of a LacZ insertion mutant and expression in eukaryotic cells. Virology 190: Marcy, A. I., D. R. Yager, and D. M. Coen Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J. Virol. 64: Maul, G Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20: Maul, G. G., and R. D. Everett The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75: Maul, G. G., H. H. Guldner, and J. G. Spivack Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74: Maul, G. G., A. M. Ishov, and R. D. Everett Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217: Maul, G. G., E. Yu, A. M. Ishov, and A. L. Epstein Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell. Biochem. 59: Mu, Z. M., K. V. Chin, J. H. Liu, G. Lozano, and K. S. Chang PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell. Biol. 14: Müller, S., M. J. Matunis, and A. Dejean Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17: Okura, T., L. Gong, T. Kamitani, T. Wada, I. Okura, C. F. Wei, H. M. Chang, and E. T. Yeh Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. 157: Puvion-Dutilleul, F., M. K. Chelbi-Alix, M. Koken, F. Quignon, E. Puvion, and H. de The Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp. Cell Res. 218: Puvion-Dutilleul, F., L. Venturini, M. C. Guillemin, H. de The, and E. Puvion Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp. Cell Res. 221: Quinlan, M. P., L. B. Chen, and D. M. Knipe The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36: Shelton, L. S., A. G. Albright, W. T. Ruyechan, and F. J. Jenkins Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68: Sternsdorf, T., K. Jensen, and H. Will Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/ SUMO-1. J. Cell Biol. 139: Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel A monoclonal antibody recognizing nuclear matrixassociated nuclear bodies. J. Cell Sci. 101: Szekely, L., K. Pokrovskaja, W.-Q. Jiang, H. de The, N. Ringertz, and G. Klein The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J. Virol. 70: Uprichard, S. L., and D. M. Knipe Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229: Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi Role of PML in cell growth and the retinoic acid pathway. Science 279: Weis, K., S. Rambaud, C. Lavau, J. Jansen, T. Carvalho, M. Carmo-Fonseca, A. Lamond, and A. Dejean Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 76: Wilcock, D., and D. P. Lane Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349: Xie, K., E. J. Lambie, and M. Snyder Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol. Cell. Biol. 13: Yager, D. R., A. I. Marcy, and D. M. Coen Translational regulation of herpes simplex virus DNA polymerase. J. Virol. 64: Zhu, L., and S. K. Weller The UL5 gene of herpes simplex virus type 1: isolation of a lacz insertion mutant and association of the UL5 gene product with other members of the helicase-primase complex. J. Virol. 66:
Cross-reactivity of the anti-pml antibody PG-M3 with the herpes simplex virus type 1 immediate early protein ICP4
 Journal of General Virology (2000), 81, 1773 1777. Printed in Great Britain... SHORT COMMUNICATION Cross-reactivity of the anti-pml antibody PG-M3 with the herpes simplex virus type 1 immediate early protein
Journal of General Virology (2000), 81, 1773 1777. Printed in Great Britain... SHORT COMMUNICATION Cross-reactivity of the anti-pml antibody PG-M3 with the herpes simplex virus type 1 immediate early protein
Chapter 18: Applications of Immunology
 Chapter 18: Applications of Immunology 1. Vaccinations 2. Monoclonal vs Polyclonal Ab 3. Diagnostic Immunology 1. Vaccinations What is Vaccination? A method of inducing artificial immunity by exposing
Chapter 18: Applications of Immunology 1. Vaccinations 2. Monoclonal vs Polyclonal Ab 3. Diagnostic Immunology 1. Vaccinations What is Vaccination? A method of inducing artificial immunity by exposing
MUTATION, DNA REPAIR AND CANCER
 MUTATION, DNA REPAIR AND CANCER 1 Mutation A heritable change in the genetic material Essential to the continuity of life Source of variation for natural selection New mutations are more likely to be harmful
MUTATION, DNA REPAIR AND CANCER 1 Mutation A heritable change in the genetic material Essential to the continuity of life Source of variation for natural selection New mutations are more likely to be harmful
PROTOCOL. Immunocytochemistry (ICC) MATERIALS AND EQUIPMENT REQUIRED
 PROTOCOL Immunocytochemistry (ICC) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 11-07 MATERIALS AND EQUIPMENT REQUIRED Materials: MitoSciences primary monoclonal antibody/antibodies Fluorophore-conjugated
PROTOCOL Immunocytochemistry (ICC) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 11-07 MATERIALS AND EQUIPMENT REQUIRED Materials: MitoSciences primary monoclonal antibody/antibodies Fluorophore-conjugated
Cell Biology Questions and Learning Objectives
 Cell Biology Questions and Learning Objectives (with hypothetical learning materials that might populate the objective) The topics and central questions listed here are typical for an introductory undergraduate
Cell Biology Questions and Learning Objectives (with hypothetical learning materials that might populate the objective) The topics and central questions listed here are typical for an introductory undergraduate
4. DNA replication Pages: 979-984 Difficulty: 2 Ans: C Which one of the following statements about enzymes that interact with DNA is true?
 Chapter 25 DNA Metabolism Multiple Choice Questions 1. DNA replication Page: 977 Difficulty: 2 Ans: C The Meselson-Stahl experiment established that: A) DNA polymerase has a crucial role in DNA synthesis.
Chapter 25 DNA Metabolism Multiple Choice Questions 1. DNA replication Page: 977 Difficulty: 2 Ans: C The Meselson-Stahl experiment established that: A) DNA polymerase has a crucial role in DNA synthesis.
INTERPRETATION INFORMATION SHEET
 Creative Testing Solutions 2424 West Erie Dr. 2205 Highway 121 10100 Martin Luther King Jr. St. No. Tempe, AZ 85282 Bedford, TX 76021 St. Petersburg, FL 33716 INTERPRETATION INFORMATION SHEET Human Immunodeficiency
Creative Testing Solutions 2424 West Erie Dr. 2205 Highway 121 10100 Martin Luther King Jr. St. No. Tempe, AZ 85282 Bedford, TX 76021 St. Petersburg, FL 33716 INTERPRETATION INFORMATION SHEET Human Immunodeficiency
CHAPTER 6 ANTIBODY GENETICS: ISOTYPES, ALLOTYPES, IDIOTYPES
 CHAPTER 6 ANTIBODY GENETICS: ISOTYPES, ALLOTYPES, IDIOTYPES See APPENDIX: (3) OUCHTERLONY; (4) AFFINITY CHROMATOGRAPHY Human immunoglobulins are made up of LIGHT and HEAVY chains encoded by a total of
CHAPTER 6 ANTIBODY GENETICS: ISOTYPES, ALLOTYPES, IDIOTYPES See APPENDIX: (3) OUCHTERLONY; (4) AFFINITY CHROMATOGRAPHY Human immunoglobulins are made up of LIGHT and HEAVY chains encoded by a total of
Module 3 Questions. 7. Chemotaxis is an example of signal transduction. Explain, with the use of diagrams.
 Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
The Need for a PARP in vivo Pharmacodynamic Assay
 The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
2006 7.012 Problem Set 6 KEY
 2006 7.012 Problem Set 6 KEY ** Due before 5 PM on WEDNESDAY, November 22, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You create an artificial
2006 7.012 Problem Set 6 KEY ** Due before 5 PM on WEDNESDAY, November 22, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You create an artificial
PREPARED FOR: U.S. Army Medical Research and Materiel CommandFort Detrick, Maryland 21702 5012
 AD Award Number: W81XWH 07 1 0542 TITLE: Is Nuclear Structure Altered in Breast Cancer Cells? PRINCIPAL INVESTIGATOR: Han Htun, Ph.D. CONTRACTING ORGANIZATION: University of California Los Angeles, CA
AD Award Number: W81XWH 07 1 0542 TITLE: Is Nuclear Structure Altered in Breast Cancer Cells? PRINCIPAL INVESTIGATOR: Han Htun, Ph.D. CONTRACTING ORGANIZATION: University of California Los Angeles, CA
Transfection-Transfer of non-viral genetic material into eukaryotic cells. Infection/ Transduction- Transfer of viral genetic material into cells.
 Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
PART 3.3: MicroRNA and Cancer
 BIBM 2010 Tutorial: Epigenomics and Cancer PART 3.3: MicroRNA and Cancer Dec 18, 2010 Sun Kim at Indiana University Outline of Part 3.3 Background on microrna Role of microrna in cancer MicroRNA pathway
BIBM 2010 Tutorial: Epigenomics and Cancer PART 3.3: MicroRNA and Cancer Dec 18, 2010 Sun Kim at Indiana University Outline of Part 3.3 Background on microrna Role of microrna in cancer MicroRNA pathway
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Structure and Function of DNA
 Structure and Function of DNA DNA and RNA Structure DNA and RNA are nucleic acids. They consist of chemical units called nucleotides. The nucleotides are joined by a sugar-phosphate backbone. The four
Structure and Function of DNA DNA and RNA Structure DNA and RNA are nucleic acids. They consist of chemical units called nucleotides. The nucleotides are joined by a sugar-phosphate backbone. The four
Lecture Series 7. From DNA to Protein. Genotype to Phenotype. Reading Assignments. A. Genes and the Synthesis of Polypeptides
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Viral Infection: Receptors
 Viral Infection: Receptors Receptors: Identification of receptors has come from expressing the gene for the receptor in a cell to which a virus does not normally bind -OR- By blocking virus attachment
Viral Infection: Receptors Receptors: Identification of receptors has come from expressing the gene for the receptor in a cell to which a virus does not normally bind -OR- By blocking virus attachment
Viral Replication. Viral Replication: Basic Concepts
 Viral Replication Scott M. Hammer, M.D. Viral Replication: Basic Concepts Viruses are obligate intracellular parasites Viruses carry their genome (RNA or DNA) and sometimes functional proteins required
Viral Replication Scott M. Hammer, M.D. Viral Replication: Basic Concepts Viruses are obligate intracellular parasites Viruses carry their genome (RNA or DNA) and sometimes functional proteins required
Publikationsliste Claudia Götz
 Publikationsliste Claudia Götz 1. Reinhard,B., Götz, C., and Faillard, H.: Synthesis of N-Acetyl-9-Oacetylneuraminic acid α-p-aminophenylthioketoside and its application as ligand in the affinity chromatography
Publikationsliste Claudia Götz 1. Reinhard,B., Götz, C., and Faillard, H.: Synthesis of N-Acetyl-9-Oacetylneuraminic acid α-p-aminophenylthioketoside and its application as ligand in the affinity chromatography
Lecture 8. Protein Trafficking/Targeting. Protein targeting is necessary for proteins that are destined to work outside the cytoplasm.
 Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
The role of IBV proteins in protection: cellular immune responses. COST meeting WG2 + WG3 Budapest, Hungary, 2015
 The role of IBV proteins in protection: cellular immune responses COST meeting WG2 + WG3 Budapest, Hungary, 2015 1 Presentation include: Laboratory results Literature summary Role of T cells in response
The role of IBV proteins in protection: cellular immune responses COST meeting WG2 + WG3 Budapest, Hungary, 2015 1 Presentation include: Laboratory results Literature summary Role of T cells in response
岑 祥 股 份 有 限 公 司 技 術 專 員 費 軫 尹 20100803
 技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
Guidance for Industry
 Guidance for Industry Interpreting Sameness of Monoclonal Antibody Products Under the Orphan Drug Regulations U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation
Guidance for Industry Interpreting Sameness of Monoclonal Antibody Products Under the Orphan Drug Regulations U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation
Name Class Date. Figure 13 1. 2. Which nucleotide in Figure 13 1 indicates the nucleic acid above is RNA? a. uracil c. cytosine b. guanine d.
 13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE Q5B
 INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
Special report. Chronic Lymphocytic Leukemia (CLL) Genomic Biology 3020 April 20, 2006
 Special report Chronic Lymphocytic Leukemia (CLL) Genomic Biology 3020 April 20, 2006 Gene And Protein The gene that causes the mutation is CCND1 and the protein NP_444284 The mutation deals with the cell
Special report Chronic Lymphocytic Leukemia (CLL) Genomic Biology 3020 April 20, 2006 Gene And Protein The gene that causes the mutation is CCND1 and the protein NP_444284 The mutation deals with the cell
Exploiting science for engineering: BRCA2 targeted therapies
 20.109 MOD1 DNA ENGINEERING Fall 2010 Exploiting science for engineering: BRCA2 targeted therapies Orsi Kiraly Engelward lab Homologous recombination is important No HR chromosomal aberrations cell death
20.109 MOD1 DNA ENGINEERING Fall 2010 Exploiting science for engineering: BRCA2 targeted therapies Orsi Kiraly Engelward lab Homologous recombination is important No HR chromosomal aberrations cell death
Validation & Assay Performance Summary
 Validation & Assay Performance Summary GeneBLAzer RAR alpha DA Assay Kit GeneBLAzer RAR alpha-uas-bla GripTite Cells Cat. no. K1387, K1685 Target Description The retinoic acid receptor alpha (RAR alpha)
Validation & Assay Performance Summary GeneBLAzer RAR alpha DA Assay Kit GeneBLAzer RAR alpha-uas-bla GripTite Cells Cat. no. K1387, K1685 Target Description The retinoic acid receptor alpha (RAR alpha)
How To Understand How Gene Expression Is Regulated
 What makes cells different from each other? How do cells respond to information from environment? Regulation of: - Transcription - prokaryotes - eukaryotes - mrna splicing - mrna localisation and translation
What makes cells different from each other? How do cells respond to information from environment? Regulation of: - Transcription - prokaryotes - eukaryotes - mrna splicing - mrna localisation and translation
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
GENE REGULATION. Teacher Packet
 AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
Appendix C DNA Replication & Mitosis
 K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
Lesson 3 Reading Material: Oncogenes and Tumor Suppressor Genes
 Lesson 3 Reading Material: Oncogenes and Tumor Suppressor Genes Becoming a cancer cell isn t easy One of the fundamental molecular characteristics of cancer is that it does not develop all at once, but
Lesson 3 Reading Material: Oncogenes and Tumor Suppressor Genes Becoming a cancer cell isn t easy One of the fundamental molecular characteristics of cancer is that it does not develop all at once, but
Chapter 8. Summary and Perspectives
 Chapter 8 Summary and Perspectives 131 Chapter 8 Summary Overexpression of the multidrug resistance protein MRP1 confer multidrug resistance (MDR) to cancer cells. The contents of this thesis describe
Chapter 8 Summary and Perspectives 131 Chapter 8 Summary Overexpression of the multidrug resistance protein MRP1 confer multidrug resistance (MDR) to cancer cells. The contents of this thesis describe
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Biochemistry
 Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Biochemistry The Master Degree in Medical Laboratory Sciences /Clinical Biochemistry, is awarded by the Faculty of Graduate Studies
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Biochemistry The Master Degree in Medical Laboratory Sciences /Clinical Biochemistry, is awarded by the Faculty of Graduate Studies
Notch 1 -dependent regulation of cell fate in colorectal cancer
 Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
What is Cancer? Cancer is a genetic disease: Cancer typically involves a change in gene expression/function:
 Cancer is a genetic disease: Inherited cancer Sporadic cancer What is Cancer? Cancer typically involves a change in gene expression/function: Qualitative change Quantitative change Any cancer causing genetic
Cancer is a genetic disease: Inherited cancer Sporadic cancer What is Cancer? Cancer typically involves a change in gene expression/function: Qualitative change Quantitative change Any cancer causing genetic
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
RNA and Protein Synthesis
 Name lass Date RN and Protein Synthesis Information and Heredity Q: How does information fl ow from DN to RN to direct the synthesis of proteins? 13.1 What is RN? WHT I KNOW SMPLE NSWER: RN is a nucleic
Name lass Date RN and Protein Synthesis Information and Heredity Q: How does information fl ow from DN to RN to direct the synthesis of proteins? 13.1 What is RN? WHT I KNOW SMPLE NSWER: RN is a nucleic
From DNA to Protein. Proteins. Chapter 13. Prokaryotes and Eukaryotes. The Path From Genes to Proteins. All proteins consist of polypeptide chains
 Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Control of Gene Expression
 Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
The world of non-coding RNA. Espen Enerly
 The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
European Medicines Agency
 European Medicines Agency July 1996 CPMP/ICH/139/95 ICH Topic Q 5 B Quality of Biotechnological Products: Analysis of the Expression Construct in Cell Lines Used for Production of r-dna Derived Protein
European Medicines Agency July 1996 CPMP/ICH/139/95 ICH Topic Q 5 B Quality of Biotechnological Products: Analysis of the Expression Construct in Cell Lines Used for Production of r-dna Derived Protein
An Overview of Cells and Cell Research
 An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
4.1 3T12 and 312 are immortalized cell lines with transforming potential:
 DISCUSSION CHAPTER 4 4.1 3T12 and 312 are immortalized cell lines with transforming potential: Transformation of a normal cell with finite number of divisions into a tumorigenic cell of potentially infinite
DISCUSSION CHAPTER 4 4.1 3T12 and 312 are immortalized cell lines with transforming potential: Transformation of a normal cell with finite number of divisions into a tumorigenic cell of potentially infinite
DNA, RNA, Protein synthesis, and Mutations. Chapters 12-13.3
 DNA, RNA, Protein synthesis, and Mutations Chapters 12-13.3 1A)Identify the components of DNA and explain its role in heredity. DNA s Role in heredity: Contains the genetic information of a cell that can
DNA, RNA, Protein synthesis, and Mutations Chapters 12-13.3 1A)Identify the components of DNA and explain its role in heredity. DNA s Role in heredity: Contains the genetic information of a cell that can
Molecular Genetics. RNA, Transcription, & Protein Synthesis
 Molecular Genetics RNA, Transcription, & Protein Synthesis Section 1 RNA AND TRANSCRIPTION Objectives Describe the primary functions of RNA Identify how RNA differs from DNA Describe the structure and
Molecular Genetics RNA, Transcription, & Protein Synthesis Section 1 RNA AND TRANSCRIPTION Objectives Describe the primary functions of RNA Identify how RNA differs from DNA Describe the structure and
Blood-Based Cancer Diagnostics
 The Biotechnology Education Company Blood-Based Cancer Diagnostics EDVO-Kit 141 Store entire experiment at room temperature. EXPERIMENT OBJECTIVE: The objective of this experiment is to learn and understand
The Biotechnology Education Company Blood-Based Cancer Diagnostics EDVO-Kit 141 Store entire experiment at room temperature. EXPERIMENT OBJECTIVE: The objective of this experiment is to learn and understand
UNIVERSITY OF WROCŁAW
 BIO-IMAGing in research INnovation and Education UNIVERSITY OF WROCŁAW Faculty of Biotechnology Tamka 2, Przybyszewskiego 63/77, WROCŁAW, POLAND Legend I. Confocal system (LSM510 META - Zeiss) - Laboratory
BIO-IMAGing in research INnovation and Education UNIVERSITY OF WROCŁAW Faculty of Biotechnology Tamka 2, Przybyszewskiego 63/77, WROCŁAW, POLAND Legend I. Confocal system (LSM510 META - Zeiss) - Laboratory
Visualization of Myc/Max/Mad Family Dimers and the Competition for Dimerization in Living Cells
 MOLECULAR AND CELLULAR BIOLOGY, May 2004, p. 4294 4308 Vol. 24, No. 10 0270-7306/04/$08.00 0 DOI: 10.1128/MCB.24.10.4294 4308.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, May 2004, p. 4294 4308 Vol. 24, No. 10 0270-7306/04/$08.00 0 DOI: 10.1128/MCB.24.10.4294 4308.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved.
Viruses. Viral components: Capsid. Chapter 10: Viruses. Viral components: Nucleic Acid. Viral components: Envelope
 Viruses Chapter 10: Viruses Lecture Exam #3 Wednesday, November 22 nd (This lecture WILL be on Exam #3) Dr. Amy Rogers Office Hours: MW 9-10 AM Too small to see with a light microscope Visible with electron
Viruses Chapter 10: Viruses Lecture Exam #3 Wednesday, November 22 nd (This lecture WILL be on Exam #3) Dr. Amy Rogers Office Hours: MW 9-10 AM Too small to see with a light microscope Visible with electron
B Cell Generation, Activation & Differentiation. B cell maturation
 B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
Chapter 6 DNA Replication
 Chapter 6 DNA Replication Each strand of the DNA double helix contains a sequence of nucleotides that is exactly complementary to the nucleotide sequence of its partner strand. Each strand can therefore
Chapter 6 DNA Replication Each strand of the DNA double helix contains a sequence of nucleotides that is exactly complementary to the nucleotide sequence of its partner strand. Each strand can therefore
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology
 Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology The Master Degree in Medical Laboratory Sciences / Clinical Microbiology, Immunology or
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology The Master Degree in Medical Laboratory Sciences / Clinical Microbiology, Immunology or
Sickle cell anemia: Altered beta chain Single AA change (#6 Glu to Val) Consequence: Protein polymerizes Change in RBC shape ---> phenotypes
 Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Control of Gene Expression
 Control of Gene Expression (Learning Objectives) Explain the role of gene expression is differentiation of function of cells which leads to the emergence of different tissues, organs, and organ systems
Control of Gene Expression (Learning Objectives) Explain the role of gene expression is differentiation of function of cells which leads to the emergence of different tissues, organs, and organ systems
2.1.2 Characterization of antiviral effect of cytokine expression on HBV replication in transduced mouse hepatocytes line
 i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
The immune response Antibodies Antigens Epitopes (antigenic determinants) the part of a protein antigen recognized by an antibody Haptens small
 The immune response Antibodies Antigens Epitopes (antigenic determinants) the part of a protein antigen recognized by an antibody Haptens small molecules that can elicit an immune response when linked
The immune response Antibodies Antigens Epitopes (antigenic determinants) the part of a protein antigen recognized by an antibody Haptens small molecules that can elicit an immune response when linked
Recognition of T cell epitopes (Abbas Chapter 6)
 Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved
 microrna 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions of target mrnas Act as negative
microrna 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions of target mrnas Act as negative
Biotechnology and Recombinant DNA (Chapter 9) Lecture Materials for Amy Warenda Czura, Ph.D. Suffolk County Community College
 Biotechnology and Recombinant DNA (Chapter 9) Lecture Materials for Amy Warenda Czura, Ph.D. Suffolk County Community College Primary Source for figures and content: Eastern Campus Tortora, G.J. Microbiology
Biotechnology and Recombinant DNA (Chapter 9) Lecture Materials for Amy Warenda Czura, Ph.D. Suffolk County Community College Primary Source for figures and content: Eastern Campus Tortora, G.J. Microbiology
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA
 CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA INTRODUCTION DNA : DNA is deoxyribose nucleic acid. It is made up of a base consisting of sugar, phosphate and one nitrogen base.the
CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA INTRODUCTION DNA : DNA is deoxyribose nucleic acid. It is made up of a base consisting of sugar, phosphate and one nitrogen base.the
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
Activity 7.21 Transcription factors
 Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
CD3/TCR stimulation and surface detection Determination of specificity of intracellular detection of IL-7Rα by flow cytometry
 CD3/TCR stimulation and surface detection Stimulation of HPB-ALL cells with the anti-cd3 monoclonal antibody OKT3 was performed as described 3. In brief, antibody-coated plates were prepared by incubating
CD3/TCR stimulation and surface detection Stimulation of HPB-ALL cells with the anti-cd3 monoclonal antibody OKT3 was performed as described 3. In brief, antibody-coated plates were prepared by incubating
Guidance. 2. Definitions. 1. Introduction
 Work with naked DNA or RNA (including oligonucleotides, sirna, mirna, sequences that code for highly biologically active molecules and full length viral genomes) Guidance 1. Introduction The following
Work with naked DNA or RNA (including oligonucleotides, sirna, mirna, sequences that code for highly biologically active molecules and full length viral genomes) Guidance 1. Introduction The following
KMS-Specialist & Customized Biosimilar Service
 KMS-Specialist & Customized Biosimilar Service 1. Polyclonal Antibody Development Service KMS offering a variety of Polyclonal Antibody Services to fit your research and production needs. we develop polyclonal
KMS-Specialist & Customized Biosimilar Service 1. Polyclonal Antibody Development Service KMS offering a variety of Polyclonal Antibody Services to fit your research and production needs. we develop polyclonal
High Grade Gliomas: Update in Treatment and Care Ryan T. Merrell, M.D. Clinical Assistant Professor of Neurology NorthShore University HealthSystem
 High Grade Gliomas: Update in Treatment and Care Ryan T. Merrell, M.D. Clinical Assistant Professor of Neurology NorthShore University HealthSystem rmerrell@northshore.org Objectives Provide updates on
High Grade Gliomas: Update in Treatment and Care Ryan T. Merrell, M.D. Clinical Assistant Professor of Neurology NorthShore University HealthSystem rmerrell@northshore.org Objectives Provide updates on
Copyright 2000-2003 Mark Brandt, Ph.D. 54
 Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Liver Disease and Therapy of Hepatitis B Virus Infections
 Liver Disease and Therapy of Hepatitis B Virus Infections University of Adelaide Catherine Scougall Arend Grosse Huey-Chi Low Allison Jilbert Fox Chase Cancer Center Chunxiao Xu Carol Aldrich Sam Litwin
Liver Disease and Therapy of Hepatitis B Virus Infections University of Adelaide Catherine Scougall Arend Grosse Huey-Chi Low Allison Jilbert Fox Chase Cancer Center Chunxiao Xu Carol Aldrich Sam Litwin
FITC Annexin V/Dead Cell Apoptosis Kit with FITC annexin V and PI, for Flow Cytometry
 FITC Annexin V/Dead Cell Apoptosis Kit with FITC annexin V and PI, for Flow Cytometry Catalog no. V13242 Table 1. Contents and storage information. Material Amount Composition Storage* Stability FITC annexin
FITC Annexin V/Dead Cell Apoptosis Kit with FITC annexin V and PI, for Flow Cytometry Catalog no. V13242 Table 1. Contents and storage information. Material Amount Composition Storage* Stability FITC annexin
T Cell Maturation,Activation and Differentiation
 T Cell Maturation,Activation and Differentiation Positive Selection- In thymus, permits survival of only those T cells whose TCRs recognize self- MHC molecules (self-mhc restriction) Negative Selection-
T Cell Maturation,Activation and Differentiation Positive Selection- In thymus, permits survival of only those T cells whose TCRs recognize self- MHC molecules (self-mhc restriction) Negative Selection-
1) Aug 2005 In Vitro Chromosomal Aberration Study Cytotoxicity (File: TS39) Conclusion No cytotoxicity can be detected at concentrations up to 100%.
 Endotoxicity and Cytotoxicity Studies 1) Aug 2005 In Vitro Chromosomal Aberration Study Cytotoxicity (File: TS39) Chinese Hamster Ovary (CHO) cells were grown in monolayer with and without metabolic activation.
Endotoxicity and Cytotoxicity Studies 1) Aug 2005 In Vitro Chromosomal Aberration Study Cytotoxicity (File: TS39) Chinese Hamster Ovary (CHO) cells were grown in monolayer with and without metabolic activation.
LECTURE 6 Gene Mutation (Chapter 16.1-16.2)
 LECTURE 6 Gene Mutation (Chapter 16.1-16.2) 1 Mutation: A permanent change in the genetic material that can be passed from parent to offspring. Mutant (genotype): An organism whose DNA differs from the
LECTURE 6 Gene Mutation (Chapter 16.1-16.2) 1 Mutation: A permanent change in the genetic material that can be passed from parent to offspring. Mutant (genotype): An organism whose DNA differs from the
Complex multicellular organisms are produced by cells that switch genes on and off during development.
 Home Control of Gene Expression Gene Regulation Is Necessary? By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Home Control of Gene Expression Gene Regulation Is Necessary? By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Rubisco; easy Purification and Immunochemical Determination
 Rubisco; easy Purification and Immunochemical Determination Ulrich Groß Justus-Liebig-Universität Gießen, Institute of Plant Nutrition, Department of Tissue Culture, Südanlage 6, D-35390 Giessen e-mail:
Rubisco; easy Purification and Immunochemical Determination Ulrich Groß Justus-Liebig-Universität Gießen, Institute of Plant Nutrition, Department of Tissue Culture, Südanlage 6, D-35390 Giessen e-mail:
Custom Antibody Services
 prosci-inc.com Custom Antibody Services High Performance Antibodies and More Broad Antibody Catalog Extensive Antibody Services CUSTOM ANTIBODY SERVICES Established in 1998, ProSci Incorporated is a leading
prosci-inc.com Custom Antibody Services High Performance Antibodies and More Broad Antibody Catalog Extensive Antibody Services CUSTOM ANTIBODY SERVICES Established in 1998, ProSci Incorporated is a leading
Introduction to flow cytometry
 Introduction to flow cytometry Flow cytometry is a popular laser-based technology. Discover more with our introduction to flow cytometry. Flow cytometry is now a widely used method for analyzing the expression
Introduction to flow cytometry Flow cytometry is a popular laser-based technology. Discover more with our introduction to flow cytometry. Flow cytometry is now a widely used method for analyzing the expression
Activation and effector functions of HMI
 Activation and effector functions of HMI Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 August 2015 ว ตถ ประสงค หล งจากช วโมงบรรยายน แล วน กศ กษาสามารถ
Activation and effector functions of HMI Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 August 2015 ว ตถ ประสงค หล งจากช วโมงบรรยายน แล วน กศ กษาสามารถ
The Evaluation of Interferon-beta Levels in HPV-positive Cervical Cancer Cell Lines
 The Evaluation of Interferon-beta Levels in HPV-positive Cervical Cancer Cell Lines Olivia M. Ford and Jennifer T. Thomas, Ph.D. Human Papillomavirus (HPV) is the most common viral sexually transmitted
The Evaluation of Interferon-beta Levels in HPV-positive Cervical Cancer Cell Lines Olivia M. Ford and Jennifer T. Thomas, Ph.D. Human Papillomavirus (HPV) is the most common viral sexually transmitted
Trasposable elements: P elements
 Trasposable elements: P elements In 1938 Marcus Rhodes provided the first genetic description of an unstable mutation, an allele of a gene required for the production of pigment in maize. This instability
Trasposable elements: P elements In 1938 Marcus Rhodes provided the first genetic description of an unstable mutation, an allele of a gene required for the production of pigment in maize. This instability
Anti-ATF6 α antibody, mouse monoclonal (1-7)
 Anti-ATF6 α antibody, mouse monoclonal (1-7) 73-500 50 ug ATF6 (activating transcription factor 6) is an endoplasmic reticulum (ER) membrane-bound transcription factor activated in response to ER stress.
Anti-ATF6 α antibody, mouse monoclonal (1-7) 73-500 50 ug ATF6 (activating transcription factor 6) is an endoplasmic reticulum (ER) membrane-bound transcription factor activated in response to ER stress.
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes.
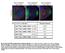 Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Recombinant DNA and Biotechnology
 Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
LEUKEMIA LYMPHOMA MYELOMA Advances in Clinical Trials
 LEUKEMIA LYMPHOMA MYELOMA Advances in Clinical Trials OUR FOCUS ABOUT emerging treatments Presentation for: Judith E. Karp, MD Advancements for Acute Myelogenous Leukemia Supported by an unrestricted educational
LEUKEMIA LYMPHOMA MYELOMA Advances in Clinical Trials OUR FOCUS ABOUT emerging treatments Presentation for: Judith E. Karp, MD Advancements for Acute Myelogenous Leukemia Supported by an unrestricted educational
a. Ribosomal RNA rrna a type ofrna that combines with proteins to form Ribosomes on which polypeptide chains of proteins are assembled
 Biology 101 Chapter 14 Name: Fill-in-the-Blanks Which base follows the next in a strand of DNA is referred to. as the base (1) Sequence. The region of DNA that calls for the assembly of specific amino
Biology 101 Chapter 14 Name: Fill-in-the-Blanks Which base follows the next in a strand of DNA is referred to. as the base (1) Sequence. The region of DNA that calls for the assembly of specific amino
Introduction to Flow Cytometry
 Introduction to Flow Cytometry presented by: Flow Cytometry y Core Facility Biomedical Instrumentation Center Uniformed Services University Topics Covered in this Lecture What is flow cytometry? Flow cytometer
Introduction to Flow Cytometry presented by: Flow Cytometry y Core Facility Biomedical Instrumentation Center Uniformed Services University Topics Covered in this Lecture What is flow cytometry? Flow cytometer
DNA Replication & Protein Synthesis. This isn t a baaaaaaaddd chapter!!!
 DNA Replication & Protein Synthesis This isn t a baaaaaaaddd chapter!!! The Discovery of DNA s Structure Watson and Crick s discovery of DNA s structure was based on almost fifty years of research by other
DNA Replication & Protein Synthesis This isn t a baaaaaaaddd chapter!!! The Discovery of DNA s Structure Watson and Crick s discovery of DNA s structure was based on almost fifty years of research by other
Ms. Campbell Protein Synthesis Practice Questions Regents L.E.
 Name Student # Ms. Campbell Protein Synthesis Practice Questions Regents L.E. 1. A sequence of three nitrogenous bases in a messenger-rna molecule is known as a 1) codon 2) gene 3) polypeptide 4) nucleotide
Name Student # Ms. Campbell Protein Synthesis Practice Questions Regents L.E. 1. A sequence of three nitrogenous bases in a messenger-rna molecule is known as a 1) codon 2) gene 3) polypeptide 4) nucleotide
Uses of Flow Cytometry
 Uses of Flow Cytometry 1. Multicolour analysis... 2 2. Cell Cycle and Proliferation... 3 a. Analysis of Cellular DNA Content... 4 b. Cell Proliferation Assays... 5 3. Immunology... 6 4. Apoptosis... 7
Uses of Flow Cytometry 1. Multicolour analysis... 2 2. Cell Cycle and Proliferation... 3 a. Analysis of Cellular DNA Content... 4 b. Cell Proliferation Assays... 5 3. Immunology... 6 4. Apoptosis... 7
Cancer SBL101. James Gomes School of Biological Sciences Indian Institute of Technology Delhi
 Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Comparative genomic hybridization Because arrays are more than just a tool for expression analysis
 Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
hydrocortisone (5 mg/ml), EGF (10 µg/ml) and Heparin (5000 U/ml). Antibodies against the N-terminal peptide of MEK1 (MEK1-N) and against Flotillin 1
 SUPPLEMENTARY METHODS Cells HUVEC (Human Umbilical Vein Endothelial Cells) were grown in complete Medium 199 (Gibco) supplemented with glutamax, 10% foetal calf serum, BBE (9 mg/ml), hydrocortisone (5
SUPPLEMENTARY METHODS Cells HUVEC (Human Umbilical Vein Endothelial Cells) were grown in complete Medium 199 (Gibco) supplemented with glutamax, 10% foetal calf serum, BBE (9 mg/ml), hydrocortisone (5
RETRIEVING SEQUENCE INFORMATION. Nucleotide sequence databases. Database search. Sequence alignment and comparison
 RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY
 MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
