Determination of Domain Structure of Proteins from X-Ray Solution Scattering
|
|
|
- Berniece Barker
- 8 years ago
- Views:
Transcription
1 2946 Biophysical Journal Volume 80 June Determination of Domain Structure of Proteins from X-Ray Solution Scattering Dmitri I. Svergun,* Maxim V. Petoukhov, and Michel H. J. Koch* *European Molecular Biology Laboratory, Hamburg Outstation, D Hamburg, Germany; Institute of Crystallography, Russian Academy of Sciences, Leninsky pr. 59, Moscow; and Physics Department, Moscow State University, Moscow , Russia ABSTRACT An ab initio method for building structural models of proteins from x-ray solution scattering data is presented. Simulated annealing is employed to find a chain-compatible spatial distribution of dummy residues which fits the experimental scattering pattern up to a resolution of 0.5 nm. The efficiency of the method is illustrated by the ab initio reconstruction of models of several proteins, with known and unknown crystal structure, from experimental scattering data. The new method substantially improves the resolution and reliability of models derived from scattering data and makes solution scattering a useful technique in large-scale structural characterization of proteins. INTRODUCTION Structural studies in molecular biology face the challenge of the post-genomic era, with vast numbers of genome sequences becoming available. There is presently an interest in large-scale expression and purification of proteins for subsequent structure determination using x-ray crystallography and nuclear magnetic resonance (NMR) (Edwards et al., 2000). Obviously, x-ray crystallography requires crystals of good diffraction quality, whereas the application of NMR to structure determination is limited to small proteins. It is therefore clear that a significant fraction of proteins could not be analyzed using the two methods. X-ray scattering from protein solutions is a structural method applicable to a broad range of conditions and sizes of macromolecules (Feigin and Svergun, 1987). In a scattering experiment, a dilute protein solution is exposed to x-rays, and the scattered intensity (I) is recorded as a function of the scattering angle. Because of the random positions and orientations of particles, the intensity is isotropic and proportional to the scattering from a single particle averaged over all orientations. The measured intensity after subtraction of solvent scattering is (Feigin and Svergun, 1987) I s A a s s A s s b A b s 2 (1) where A a (s), A s (s), and A b (s) are, respectively, the scattering amplitudes from the particle in vacuo, from the excluded volume, and from the hydration shell. The electron density of the bulk solvent, s, may differ from that of the hydration shell, b, yielding a non-zero contrast of the shell b ( b s ). The scattering vector is s (s, ), where s 4 sin / denotes the momentum transfer, 2 is the scattering Received for publication 17 January 2001 and in final form 21 March Address reprint requests to Dmitri Svergun, EMBL c/o DESY; Notkestrasse 85, D Hamburg, Germany. Tel: ; Fax: , svergun@embl-hamburg.de by the Biophysical Society /01/06/2946/08 $2.00 angle, the wavelength of the radiation, and stands for the average over the solid angle in reciprocal space. The main advantage of solution scattering is its ability to study the structure of native particles in nearly physiological conditions and to analyze structural changes in response to variations in external parameters. The price to pay is a dramatic loss of information caused by the spherical averaging in the scattering pattern. This has also led to the widespread opinion that solution scattering provides information only about overall size and anisometry of the solute particles. Traditionally, only limited portions of the scattering patterns were used to construct three-dimensional models. At low angles (resolution of 2 to 3 nm), x-rays are insensitive to the internal structure and the scattering is essentially determined by the particle shape. Low resolution shape models were thus constructed on a trial-and-error basis using a priori information from other methods. More recently, ab initio modeling approaches have been developed representing the shape either by an angular envelope function (Svergun and Stuhrmann, 1991; Svergun et al., 1996), or as an ensemble of densely packed beads (Chacon et al., 1998; Svergun, 1999; Walther et al., 1999), and employing nonlinear minimization to fit the scattering data. In spite of the limited resolution and the restricted range of application of these methods, a number of successful studies (Bada et al., 2000; Chacon et al., 2000; Svergun et al., 2000a, b) have shown that solution scattering curves provide sufficient information to restore particle shapes ab initio. In contrast, higher resolution x-ray scattering patterns from proteins are even rarely measured, partly for experimental reasons but mainly because of lack of adequate methods to interpret the data in terms of structural models. The level of structural detail provided by x-ray solution scattering can be qualitatively assessed by considering Fig. 1 presenting theoretical scattering curves from twenty-five proteins with different folds and molecular masses (MM) ranging from 10 to 300 kda. The high resolution structures of the proteins were taken from the Protein Data Bank (PDB) (Bernstein et al., 1977) and the curves were com-
2 Protein Structure from Solution Scattering 2947 spherically averaged scattering amplitudes from the amino acid residues were computed and weighted according to their abundance to yield the averaged residue form factor f(s) in Fig. 2. To represent the bound solvent, the protein was surrounded by a hydration layer of thickness r 0.3 nm as follows. A quasi-uniform grid of M N angular directions based on Fibonacci numbers was generated (Svergun, 1994). For each direction, the most distant residue was found and a dummy solvent atom was placed 0.5 nm outside the protein. The scattering intensity from the entire assembly of K N M centers with coordinates r i was calculated using the Debye formula (Debye, 1915) K I DR s i 1 K g i s g j s sin sr ij (2) j 1 sr ij FIGURE 1 Theoretical x-ray solution scattering curves computed from atomic models of 25 different proteins. The upper axis displays the spatial resolution 2 /s and the text labels indicate the levels of structure organization characteristic of this resolution. where g i (s) f(s) foradrandg i (s) (4 r i 2 /M) r b for a solvent atom, and r ij r i r j is the distance between the ith and jth point. At higher angles, the intensities (I DR (s)) were systematically lower than the theoretical ones (I(s)) because the internal structure of the residues is neglected in the DR model. The ratio c(s) I(s)/I DR (s) computed for the 25 proteins in Fig. 1 was used to evaluate the average correction factor c(s) (Fig. 2) such that the product c(s) I DR (s) yielded a good agreement with the theoretical scattering patterns up to s 15 nm 1 for numerous proteins tested. The use of the C positions allows to impose restrictions on the spatial arrangement of the DRs. In addition to the 0.38-nm separation along the chain, excluded volume effects and local interactions lead to a characteristic distribution of nearest neighbors. A histogram of the average number of C atoms in a 0.1-nm thick spherical shell surrounding a given C atom as a function of the shell radius N(R k ) for 0 R k 1 nm is presented in Fig. 3. It is clear that the histogram N DR (R k ) for a plausible chaincompatible DR model should be similar to N(R k ). puted using the program CRYSOL (Svergun et al., 1995) assuming the bound solvent to be 10% denser than the bulk ( b 30 e/nm 3 ; Svergun et al., 1995, 1998). The patterns differ significantly up to s 12 nm 1 but appear to converge at higher resolution. This suggests that solution scattering is not suitable for determination of secondary structure elements but also that the patterns contain information about the shape and fold of proteins up to a resolution of 0.5 nm. In the present paper, a method is proposed to build structural models of proteins accounting for the entire scattering curve and not just for its initial portion corresponding to the shape scattering. The advantages of the new technique are illustrated by its application to several proteins, with both known and unknown crystal structures. Minimization algorithm A DR model of the protein structure can be retrieved from the scattering data as follows. Given the number of residues N (usually known from the MATERIALS AND METHODS Chain-compatible dummy residues model Proteins typically consist of folded polypeptide chains composed of amino acid residues separated by 0.38 nm between adjacent C atoms in the primary sequence. At a resolution of 0.5 nm, a protein structure can be considered as an assembly of dummy residues (DR) centered at the C positions. A three-dimensional model of the protein may therefore be constructed from solution scattering data by finding a chain-compatible spatial arrangement of the DRs that fits the experimental scattering pattern. That such a model adequately describes scattering patterns of proteins was verified by simulations. A protein with a known atomic structure containing N residues was represented by its C coordinates. Solvent-corrected FIGURE 2 Averaged form factor of a residue (1) and the average correction factor (2). Dotted curves represent individual correction functions for the proteins in Fig. 1.
3 2948 Svergun et al. outside the search volume, repeat (2). (3) Positions of the solvent atoms are updated if necessary and a difference { E E(r ) E(r)} is computed. If E 0, the move is accepted; if E 0, it is accepted with a probability exp( E/T). (4) Steps (2 and 3) are repeated a sufficient number of times (N T ) to equilibrate the system, after which the temperature is lowered (T 0.9T). The system is cooled until no improvement in E(r) is observed. Computer program and testing FIGURE 3 Histogram of an average number of C atoms in 0.1 nm thick spherical shells around a given C atom. Smaller error bars: variation of the averaged values over all proteins; larger error bars: averaged variation within one protein. protein or translated DNA sequence), coordinates of DRs r i are found which minimize a goal function E(r) 2 P(r). Here, 2 is the discrepancy such that 2 1 n c s I DR s j ci exp s j n 1 s j j 1 2 (3) where I exp (s) is the experimental intensity specified at n points s j, j 1,... n, (s j ) is the correspondent standard deviation, and c is a scaling coefficient. The penalty P(r) has the form P r k W R k N DR R k N R k ) 2 G r max 0, r c r 0 2 (4) The first term in Eq. 4 imposes a protein-like nearest-neighbor distribution (the weights W(R k ) are inversely proportional to the variations of N(R k ) in Fig. 3). The second term G(r) ensures that the model is interconnected, i.e., each DR has at least one neighbor at a distance of 0.38 nm. All connected fragments (graphs) in the current DR model are found and G(r) is computed as ln(n G /N) 0, where N G is the length of the longest graph. The third term keeps the center of mass of the DR model r c close to the origin (r D max is the radius of a penalty-free zone and D max is the maximum size of the protein). The penalty weight 0 is selected such that P(r) yields a significant contribution ( 10 to 50%) to E(r) at the end of the minimization. The easiest way to construct a DR model would be to generate a random-walk C chain and let it fold to minimize E(r) (this option will be discussed later). However, a better convergence was obtained for a restrained condensation of a gas of DRs within a spherical search volume (which resembles to some extent an ab initio phasing approach (Subbiah, 1991)). The simulated annealing (Kirkpatrick et al., 1983) condensation protocol is simple: (1) N coordinates r i of the DRs are randomly generated within a sphere of radius R D max /2 r 0, and M dummy solvent atoms are added as described. A value of the goal function E(r) is computed and a high starting temperature (T 0 ) is selected. (2) A DR taken at random is relocated to an arbitrary point at a distance of 0.38 nm from another randomly selected DR (move from r to r ). If the second location falls The above algorithm was implemented in a computer program GASBOR and computations were performed on simulated examples to select values of the parameters (T ,N T 10 2 K, 10 2 ) ensuring its convergence. As usual with simulated annealing, millions of function evaluations are required and it takes a prohibitively long time to fully re-compute E(r) each time. Fortunately, both Debye s formula (Eq. 2) and the penalty (Eq. 4) are computed from the distances r ij. A table of off-diagonal distances {r ij,i j} is evaluated and saved at step (1) and later updated during steps (2 and 3). Moving one DR at a time greatly speeds up the computations (for example, on a 500 MHz Pentium III PC, the CPU time required to build the model of lysozyme below is reduced from 100hto 1 h). The program is also able to take into account particle symmetry by generating symmetry mates for the DRs in the asymmetric unit (point groups P2 to P6 and P222 to P62 are currently supported). In all tests on proteins presented below, the value of D max was determined directly from the experimental data using the orthogonal expansion program ORTOGNOM (Svergun, 1993), and the DR models were restored without any a priori information except for the number of residues. The results of 10 independent annealing runs were compared with each other and/or with the crystallographic model if the latter was available. Because the DR models had an arbitrary orientation and handedness, they were automatically aligned with the C coordinates in the crystal structures using the program SUPCOMB (Kozin and Svergun, 2001). This program minimizes a dissimilarity measure between two models as a normalized spatial discrepancy (NSD). For every point in the first model (DR or C ), the minimum value among the distances between the point and all points in the second model is found, and the same is done for the points in the second model. These distances are added and normalized against the average distances between the neighboring points for the two models. In the context of the work described here, the physical meaning of NSD for the best superposition is that, on average, for any C atom in the atomic structure there is a DR at a distance of about NSD 0.38 nm (and vice versa). Scattering experiments and data treatment The synchrotron radiation x-ray scattering data from hexokinase, yeast pyruvate decarboxylase and chitin-binding protein were collected following standard procedures using the 33 camera (Boulin et al., 1986, 1988) of the European Molecular Biology Laboratory at Deutsches Elektronen Synchrotron, (Hamburg) and multiwire proportional chambers with delay line readout (Gabriel and Dauvergne, 1982). The data were recorded at different protein concentrations (2 to 25 mg/ml) for two or three sampledetector distances (3.9, 2.5, and 1.4 m) and the scattering patterns were merged to yield the final composite curves. A shortened camera setup (sample-detector distance, 0.5 m) was designed for the additional wideangle measurements on lysozyme and bovine serum albumine. Details of the experimental procedures are given elsewhere (Koenig et al., 1993; Svergun et al., 2000a). The data processing (normalization, buffer subtraction, etc) involved statistical error propagation using the program SAPOKO (Svergun and Koch, unpublished). RESULTS After validation on simulated examples, the method was used to construct DR models of a number of proteins with
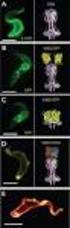
4 Protein Structure from Solution Scattering 2949 known and unknown crystal structure from experimental scattering data. For test purposes, wide-angle synchrotron x-ray scattering patterns were recorded from two readily available proteins, hen egg-white lysozyme (Merck Eurolab GmbH, Darmstadt, Germany) and bovine serum albumin (BSA; Sigma Chemical Corp., St. Louis, MO). These extensively characterized proteins are often used as model objects for structural and functional studies. The data were collected at protein concentrations ranging from 10 to 150 mg/ml and extrapolated to zero concentration of the solute protein for both small and wide angles to yield accurate scattering curves in the range up to s 13 nm 1 as illustrated in Fig. 4. The reconstructed models of lysozyme (MM 14 kda, 129 residues) superimposed with its atomic structure in the crystal (PDB entry 6lyz; Diamond, 1974) are presented in Fig. 5 A. The middle column displays the best (NSD 0.75), the right column the worst (NSD 0.85) solution out of 10 independent reconstructions. For comparison, the left column presents the low resolution shape of lysozyme restored ab initio using the program DAMMIN (Svergun, 1999). The latter model only fits the low angle portion of the scattering pattern (Fig. 4) whereas the new method neatly fits the entire curve and provides a significantly more detailed model of the protein structure. An interesting result was obtained for BSA (MW 67 kda, 583 residues). Initial attempts at modeling the structure failed to fit the experimental scattering pattern in Fig. 4. To further understand this discrepancy, theoretical scattering curves were computed from the coordinates of two FIGURE 5 Atomic models of lysozyme (A) and ligand-bound HSA (B) superimposed with DR models from solution scattering. The atomic models are displayed as C chains, DR models as semi-transparent spheres (yellow and cyan models yield best and worst agreement with the atomic model, respectively). For lysozyme, a low resolution shape obtained by DAMMIN (Svergun; 1999) is displayed in green (left column). The center and bottom rows are rotated counterclockwise by 90 0 around X and Y, respectively. All three-dimensional models were displayed using the program ASSA (Kozin et al., 1997). FIGURE 4 X-ray scattering from lysozyme (1) and BSA (2) (dots with error bars) and scattering from the DR models (full lines). For lysozyme, scattering from the low resolution shape model is also displayed (dashed line). available crystallographic models of the human homologue, human serum albumin (HSA), which shares 90% sequence homology with the BSA. One crystal structure is that of the unliganded form of HSA (PDB entry 1ao6; Sugio et al.,
5 2950 Svergun et al. 1999) while the other contains five fatty acids bound to the protein (entry 1bke; Curry et al., 1998) and reveals substantial conformational changes with respect to the former structure (r.m.s. displacement of C atoms, 0.46 nm; NSD 0.78). The scattering pattern computed using CRYSOL with coordinates from the ligand-bound form of HSA yielded a better fit to the experimental data compared with the ligandfree form. In both cases, however, a negative contrast of the hydration shell b 30 e/nm 3 must be assumed to fit the data. As a result, the density of the solvation layer surrounding the protein in solution was, somewhat unexpectedly, 10% below that of the bulk rather than 10% larger as is generally observed for globular proteins. Given that the fatty acids are 30% lighter than water, this result suggests that bound hydrophobic fatty acids may be exposed on the surface of this protein (which is a major fatty-acid transport protein in the circulatory system), leading to an apparently less dense hydration shell. In a dozen reconstructions with a negative contrast of the dummy solvent atoms b 30 e/nm, the entire BSA scattering pattern can be neatly fitted by that of a DR model (Fig. 4). The best and worst reconstructions (NSD 1.22 and 1.36, respectively) are superimposed in Fig. 5 B with the atomic model of the ligandbound HSA (Curry et al., 1998). Note that the DR models yielded somewhat worse agreement with the crystal structure of unliganded HSA (Sugio et al., 1999) (the NSD was, on average, 3 to 5% higher) indicating that the method is able to distinguish between the two conformations. In the following examples, DR models were constructed from scattering patterns recorded as part of ongoing research projects at the European Molecular Biology Laboratory, Hamburg, Germany. The data collection conditions were not optimized to yield high quality curves at wide angles, and the resolution was not better than 1 nm (Fig. 6). It was thus interesting to monitor the performance of the method against these data sets. The biologically active subunit of yeast hexokinase is a homodimer (a monomer with MW 53.8 kda has 485 residues). The scattering patterns from solutions of monomeric and dimeric hexokinase are presented in Fig. 6, curves 1 and 2, respectively. A crystal structure of the monomer is available (PDB entry 1hkg; Bennett and Steitz, 1980) but the quaternary structure of the dimeric enzyme in solution is uncertain. Theoretical curves computed from the crystallographic dimers, both symmetric and asymmetric (Bennett and Steitz, 1980; Steitz et al., 1976), failed to fit the experimental scattering of dimeric hexokinase. The scattering patterns in Fig. 6 were used to independently build DR models of the monomer and of the dimer (assuming P2 symmetry for the latter). Several reconstructions of the monomer yielded a good agreement with the atomic model in the crystal (NSD between 1.04 and 1.14). A typical superposition with NSD 1.08 is presented in Fig. 7 A, left panel. The symmetric model of the dimeric enzyme obtained without any assumption about the structure of the FIGURE 6 x-ray scattering patterns from monomeric (1) and dimeric (2) hexokinase, PDC (3) and CHB1 (4) (dots with error bars) and scattering from the reconstructed DR models (full lines). For chitin-binding protein, the scattering from a low resolution shape model is displayed (dashed line). The scattering patterns are displaced by one logarithmic unit for better visualization. monomer displays two distinct monomers and suggests a clear way of forming the dimer in solution (Fig. 7 A, right panel). Yeast pyruvate decarboxylase (PDC) provides another example of the use of symmetry restrictions. At low ph, PDC forms catalytically active tetramers with MM 236 kda and a total of 2148 residues (Koenig et al., 1993). The model of the tetrameric enzyme restored from the scattering pattern in Fig. 6 (curve 3) assuming P222 symmetry is presented in Fig. 7 B along with the crystal structure (PDB entry 1pvd; Arjunan et al., 1996). The comparison suggests that PDC in solution is more compact than in the crystal, partly as a result of an altered association between the dimers (corresponding to a relative tilt of about 10 with respect to the x axis in top orientation). This result is in an agreement with the model of tetrameric PDC in solution obtained earlier by rigid body refinement (Svergun et al., 2000c). The final example deals with a protein of unknown atomic structure, a chitin-binding protein CHB1 from Streptomyces (MM 18.7 kda, 201 residues). The low resolution shape of the protein recently obtained (Svergun et al., 2000a) from the scattering pattern in Fig. 6 (curve 4) is displayed in Fig. 7 C, left column. Ten independent DR models were generated, and the two solutions that differ
6 Protein Structure from Solution Scattering 2951 FIGURE 7 (A) Comparison of the atomic model of monomeric hexokinase and the DR models of the monomeric (left) and dimeric (right) hexokinase. (B) Atomic model of tetrameric PDC superimposed with the DR model. (C) Ab initio models of CHB1. Left column, low resolution envelope obtained by the program SASHA (Svergun et al., 1996); center and right columns, two most different DR models. The atomic models are displayed as C chains, DR models as spheres. The center and bottom rows are rotated as in Fig. 5. A space-filling representation of -chitin is presented in (C) (red) near the foot-like protuberance of CHB1. most from each other (NSD 0.95) are presented in Fig. 7 C, middle and right columns. Even these two most different models are fairly similar and, in particular, display a characteristic foot-like protuberance. Comparison with a spacefilling representation of the atomic structure of -chitin (taken from the Cambridge Crystallographic Database, suggests this protuberance as a plausible site for binding the chitin molecule. DISCUSSION It is clear that the method does not yield a unique solution (spatial distribution of DRs), but rather provides a manifold of configurations corresponding to virtually the same scattering pattern (e.g., yellow and cyan DR models in Fig. 5 A and B, and Fig. 7 C). Calculations on simulated and experimental scattering patterns indicate that the differences be-
7 2952 Svergun et al. tween the DR models are substantially smaller than those observed for the low resolution shape determination using densely packed beads (Svergun, 1999). This is not surprising given that the DR models use fewer independent parameters to fit much larger portions of the scattering pattern than do the bead modeling programs (Chacon et al., 1998; Svergun, 1999; Walther et al., 1999). The variations between the DR models preserve the domain structure of the protein, and, in analogy to NMR studies, an average (most probable) model can be generated by merging the results of independent simulated annealing runs. Could the method yield more than a detailed shape and domain structure of a protein? Although the DR models do not directly provide the tertiary structure, the map of approximate C positions can be incorporated as a constraint in protein folding prediction methods. We have also attempted to directly restore the protein fold from an x-ray scattering pattern. A C chain was folded accounting for the primary and secondary structure, backbone angles distribution (Kleywegt, 1997), hydrophobicity (Huang et al., 1995), and interaction potentials between residues (Miyazawa and Jernigan, 1999; Thomas and Dill, 1996). Following a random-walk simulated annealing protocol, various native-like folds could be generated fitting the data and satisfying all constraints (i.e., having a free energy much lower than that of the native protein). These results suggest, not unexpectedly, that there is insufficient information to reconstruct a protein fold described by C coordinates alone using a single x-ray pattern. Currently, the folding algorithm is extended to account for the centers of the side chains (Guo et al., 1995) and to incorporate additional information about the internal structure provided by contrast variation using isotopic H/D exchange in neutron scattering experiments. It should also be noted that the principle of DR modeling can be used to characterize the structure of unfolded or partially folded proteins and to construct low resolution ab initio models in protein crystallography. The DR modeling accounting for the complete scattering pattern already yields substantially more reliable and higher resolution models than previous methods, and the present approach has potential for future development. The new modeling technique makes x-ray scattering, which is free from major limitations of crystallography and NMR, a useful option for a large-scale structural characterization of proteins in solution.. The authors thank K. Brown for valuable comments on the manuscript, M. Malfois for assistance with measurements, V. Renkwitz for modifying the camera, H. Bartunik, S. Koenig and G. Grueber for providing the protein samples. The work was supported by International Association for the Promotion of Cooperation with Scientists from the Independent States of the Former Soviet Union, Grants and YSF The executable codes of the program GASBOR for IBM-PC and major UNIX platforms are available from ExternalInfo/Research/Sax. REFERENCES Arjunan, P., T. Umland, F. Dyda, S. Swaminathan, W. Furey, M. Sax, B. Farrenkopf, Y. Gao, D. Zhang, and F. Jordan Crystal structure of the thiamin diphosphate-dependent enzyme pyruvate decarboxylase from the yeast Saccharomyces cerevisiae at 2.3 A resolution. J. Mol. Biol. 256: Bada, M., D. Walther, B. Arcangioli, S. Doniach, and M. Delarue Solution structural studies and low-resolution model of the Schizosaccharomyces pombe sap1 protein. J. Mol. Biol. 300: Bennett, W. S., Jr., and T. A. Steitz Structure of a complex between yeast hexokinase A and glucose. II. Detailed comparisons of conformation and active site configuration with the native hexokinase B monomer and dimer. J. Mol. Biol. 140: Bernstein, F. C., T. F. Koetzle, G. J. B. Williams, E. F. Meyer, Jr., M. D. Brice, J. R. Rodgers, O. Kennard, T. Shimanouchi, and M. Tasumi The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 112: Boulin, C. J., R. Kempf, A. Gabriel, and M. H. J. Koch Data acquisition systems for linear and area X-ray detectors using delay line readout. Nucl. Instrum. Meth. A 269: Boulin, C., R. Kempf, M. H. J. Koch, and S. M. McLaughlin Data appraisal, evaluation and display for synchrotron radiation experiments: hardware and software. Nucl. Instrum. Meth. A 249: Chacon, P., J. F. Diaz, F. Moran, and J. M. Andreu Reconstruction of protein form with X-ray solution scattering and a genetic algorithm. J. Mol. Biol. 299: Chacon, P., F. Moran, J. F. Diaz, E. Pantos, and J. M. Andreu Low-resolution structures of proteins in solution retrieved from X-ray scattering with a genetic algorithm. Biophys. J. 74: Curry, S., H. Mandelkow, P. Brick, and N. Franks Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 5: Debye, P Zerstreuung von Roentgenstrahlen. Ann. Physik 46: Diamond, R Real-space refinement of the structure of hen egg-white lysozyme. J. Mol. Biol. 82: Edwards, A. M., C. H. Arrowsmith, D. Christendat, A. Dharamsi, J. D. Friesen, J. F. Greenblatt, and M. Vedadi Protein production: feeding the crystallographers and NMR spectroscopists. Nat. Struct. Biol. 7(Suppl): Feigin, L. A., and D. I. Svergun Structure Analysis by Small-Angle X-Ray and Neutron Scattering. New York: Plenum Press. pp 335 Gabriel, A., and F. Dauvergne The localization method used at EMBL. Nucl. Instrum. Meth. 201: Guo, D. Y., G. D. Smith, J. F. Griffin, and D. A. Langs Use of globic scattering factors for protein structures at low resolution. Acta Cryst. A51: Huang, E. S., S. Sibbiah, and M. Levitt Recognizing native folds by the arrangement of hydrophobic and polar residues. J. Mol. Biol. 252: Kirkpatrick, S., C. D. Gelatt, Jr., and M. P. Vecci Optimization by simulated annealing. Science 220: Kleywegt, G. J Validation of protein models from Calpha coordinates alone. J. Mol. Biol. 273: Koenig, S., D. Svergun, M. H. Koch, G. Hubner, and A. Schellenberger The influence of the effectors of yeast pyruvate decarboxylase (PDC) on the conformation of the dimers and tetramers and their ph-dependent equilibrium. Eur. Biophys. J. 22: Kozin, M. B., and D. I. Svergun Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34:33 41.
8 Protein Structure from Solution Scattering 2953 Kozin, M. B., V. V. Volkov, and D. I. Svergun ASSA: a program for three-dimensional rendering in solution scattering from biopolymers. J. Appl. Crystallogr. 30: Miyazawa, S., and R. L. Jernigan Self-consistent estimation of inter-residue protein contact energies based on an equilibrium mixture approximation of residues. Proteins. 34: Steitz, T. A., R. J. Fletterick, W. F. Anderson, and C. M. Anderson High resolution x-ray structure of yeast hexokinase, an allosteric protein exhibiting a non-symmetric arrangement of subunits. J. Mol. Biol. 104: Subbiah, S Low-resolution real-space envelopes: an approach to the ab initio macromolecular phase problem. Science. 252: Sugio, S., A. Kashima, S. Mochizuki, M. Noda, and K. Kobayashi Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 12: Svergun, D. I A direct indirect method of small-angle scattering data treatment. J. Appl. Crystallogr. 26: Svergun, D. I Solution scattering from biopolymers: advanced contrast variation data analysis. Acta Crystallogr. A50: Svergun, D. I Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76: Svergun, D. I., C. Barberato, and M. H. J. Koch CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28: Svergun, D. I., A. Becirevic, H. Schrempf, M. H. Koch, and G. Grueber. 2000a. Solution structure and conformational changes of the Streptomyces chitin-binding protein (CHB1). Biochemistry. 39: Svergun, D. I., M. Malfois, M. H. Koch, S. R. Wigneshweraraj, and M. Buck. 2000b. Low resolution structure of the sigma54 transcription factor revealed by X-ray solution scattering. J. Biol. Chem. 275: Svergun, D. I., M. V. Petoukhov, M. H. Koch, and S. Konig. 2000c. Crystal versus solution structures of thiamine diphosphate-dependent enzymes. J. Biol. Chem. 275: Svergun, D. I., S. Richard, M. H. Koch, Z. Sayers, S. Kuprin, and G. Zaccai Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. U. S. A. 95: Svergun, D. I., and H. B. Stuhrmann New developments in direct shape determination from small-angle scattering 1. Theory and model calculations. Acta Crystallogr. A47: Svergun, D. I., V. V. Volkov, M. B. Kozin, and H. B. Stuhrmann New developments in direct shape determination from small-angle scattering 2. Uniqueness. Acta Crystallogr. A52: Thomas, P. D., and K. A. Dill An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. U. S. A. 93: Walther, D., F. E. Cohen, and S. Doniach Reconstruction of lowresolution three-dimensional density maps from one-dimensional smallangle X-ray solution scattering data for biomolecules. J. Appl. Crystallogr. 33:
Phase determination methods in macromolecular X- ray Crystallography
 Phase determination methods in macromolecular X- ray Crystallography Importance of protein structure determination: Proteins are the life machinery and are very essential for the various functions in the
Phase determination methods in macromolecular X- ray Crystallography Importance of protein structure determination: Proteins are the life machinery and are very essential for the various functions in the
SASREF: Global Rigid Body Modelling
 SASREF: Global Rigid Body Modelling Starts from arbitrary initial positions and orientations of the subunits Employs simulated annealing to search interconnected arrangement of the subunits without clashes
SASREF: Global Rigid Body Modelling Starts from arbitrary initial positions and orientations of the subunits Employs simulated annealing to search interconnected arrangement of the subunits without clashes
Lecture 19: Proteins, Primary Struture
 CPS260/BGT204.1 Algorithms in Computational Biology November 04, 2003 Lecture 19: Proteins, Primary Struture Lecturer: Pankaj K. Agarwal Scribe: Qiuhua Liu 19.1 The Building Blocks of Protein [1] Proteins
CPS260/BGT204.1 Algorithms in Computational Biology November 04, 2003 Lecture 19: Proteins, Primary Struture Lecturer: Pankaj K. Agarwal Scribe: Qiuhua Liu 19.1 The Building Blocks of Protein [1] Proteins
Structure Determination
 5 Structure Determination Most of the protein structures described and discussed in this book have been determined either by X-ray crystallography or by nuclear magnetic resonance (NMR) spectroscopy. Although
5 Structure Determination Most of the protein structures described and discussed in this book have been determined either by X-ray crystallography or by nuclear magnetic resonance (NMR) spectroscopy. Although
ATSAS 2.1, a program package for small-angle scattering data analysis
 Journal of Applied Crystallography ISSN 0021-8898 Editor: Gernot Kostorz ATSAS 2.1, a program package for small-angle scattering data analysis Petr V. Konarev, Maxim V. Petoukhov, Vladimir V. Volkov and
Journal of Applied Crystallography ISSN 0021-8898 Editor: Gernot Kostorz ATSAS 2.1, a program package for small-angle scattering data analysis Petr V. Konarev, Maxim V. Petoukhov, Vladimir V. Volkov and
1 The water molecule and hydrogen bonds in water
 The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
CSC 2427: Algorithms for Molecular Biology Spring 2006. Lecture 16 March 10
 CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Structure Tools and Visualization
 Structure Tools and Visualization Gary Van Domselaar University of Alberta gary.vandomselaar@ualberta.ca Slides Adapted from Michel Dumontier, Blueprint Initiative 1 Visualization & Communication Visualization
Structure Tools and Visualization Gary Van Domselaar University of Alberta gary.vandomselaar@ualberta.ca Slides Adapted from Michel Dumontier, Blueprint Initiative 1 Visualization & Communication Visualization
(c) How would your answers to problem (a) change if the molecular weight of the protein was 100,000 Dalton?
 Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Structural Bioinformatics (C3210) Experimental Methods for Macromolecular Structure Determination
 Structural Bioinformatics (C3210) Experimental Methods for Macromolecular Structure Determination Introduction Knowing the exact 3D-structure of bio-molecules is essential for any attempt to understand
Structural Bioinformatics (C3210) Experimental Methods for Macromolecular Structure Determination Introduction Knowing the exact 3D-structure of bio-molecules is essential for any attempt to understand
FTIR Analysis of Protein Structure
 FTIR Analysis of Protein Structure Warren Gallagher A. Introduction to protein structure The first structures of proteins at an atomic resolution were determined in the late 1950 s. 1 From that time to
FTIR Analysis of Protein Structure Warren Gallagher A. Introduction to protein structure The first structures of proteins at an atomic resolution were determined in the late 1950 s. 1 From that time to
Structure Factors 59-553 78
 78 Structure Factors Until now, we have only typically considered reflections arising from planes in a hypothetical lattice containing one atom in the asymmetric unit. In practice we will generally deal
78 Structure Factors Until now, we have only typically considered reflections arising from planes in a hypothetical lattice containing one atom in the asymmetric unit. In practice we will generally deal
Myoglobin and Hemoglobin
 Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Functional Architecture of RNA Polymerase I
 Cell, Volume 131 Supplemental Data Functional Architecture of RNA Polymerase I Claus-D. Kuhn, Sebastian R. Geiger, Sonja Baumli, Marco Gartmann, Jochen Gerber, Stefan Jennebach, Thorsten Mielke, Herbert
Cell, Volume 131 Supplemental Data Functional Architecture of RNA Polymerase I Claus-D. Kuhn, Sebastian R. Geiger, Sonja Baumli, Marco Gartmann, Jochen Gerber, Stefan Jennebach, Thorsten Mielke, Herbert
PHYSIOLOGY AND MAINTENANCE Vol. II - On The Determination of Enzyme Structure, Function, and Mechanism - Glumoff T.
 ON THE DETERMINATION OF ENZYME STRUCTURE, FUNCTION, AND MECHANISM University of Oulu, Finland Keywords: enzymes, protein structure, X-ray crystallography, bioinformatics Contents 1. Introduction 2. Structure
ON THE DETERMINATION OF ENZYME STRUCTURE, FUNCTION, AND MECHANISM University of Oulu, Finland Keywords: enzymes, protein structure, X-ray crystallography, bioinformatics Contents 1. Introduction 2. Structure
Plate waves in phononic crystals slabs
 Acoustics 8 Paris Plate waves in phononic crystals slabs J.-J. Chen and B. Bonello CNRS and Paris VI University, INSP - 14 rue de Lourmel, 7515 Paris, France chen99nju@gmail.com 41 Acoustics 8 Paris We
Acoustics 8 Paris Plate waves in phononic crystals slabs J.-J. Chen and B. Bonello CNRS and Paris VI University, INSP - 14 rue de Lourmel, 7515 Paris, France chen99nju@gmail.com 41 Acoustics 8 Paris We
HYDRONMR and Fast-HYDRONMR
 File:hydronmr7c-pub.doc HYDRONMR and Fast-HYDRONMR Index 1. Introduction to HYDRONMR 2. Literature 3. Running HYDRONMR. Input data file 4. Output files 5. Notes and hints 7. Release notes Version 7c, September
File:hydronmr7c-pub.doc HYDRONMR and Fast-HYDRONMR Index 1. Introduction to HYDRONMR 2. Literature 3. Running HYDRONMR. Input data file 4. Output files 5. Notes and hints 7. Release notes Version 7c, September
Physics 9e/Cutnell. correlated to the. College Board AP Physics 1 Course Objectives
 Physics 9e/Cutnell correlated to the College Board AP Physics 1 Course Objectives Big Idea 1: Objects and systems have properties such as mass and charge. Systems may have internal structure. Enduring
Physics 9e/Cutnell correlated to the College Board AP Physics 1 Course Objectives Big Idea 1: Objects and systems have properties such as mass and charge. Systems may have internal structure. Enduring
X-ray Powder Diffraction Pattern Indexing for Pharmaceutical Applications
 The published version of this manuscript may be found at the following webpage: http://www.pharmtech.com/pharmtech/peer-reviewed+research/x-ray-powder-diffraction-pattern-indexing-for- Phar/ArticleStandard/Article/detail/800851
The published version of this manuscript may be found at the following webpage: http://www.pharmtech.com/pharmtech/peer-reviewed+research/x-ray-powder-diffraction-pattern-indexing-for- Phar/ArticleStandard/Article/detail/800851
Review of Chemical Equilibrium 7.51 September 1999. free [A] (µm)
![Review of Chemical Equilibrium 7.51 September 1999. free [A] (µm) Review of Chemical Equilibrium 7.51 September 1999. free [A] (µm)](/thumbs/40/21435474.jpg) Review of Chemical Equilibrium 7.51 September 1999 Equilibrium experiments study how the concentration of reaction products change as a function of reactant concentrations and/or reaction conditions. For
Review of Chemical Equilibrium 7.51 September 1999 Equilibrium experiments study how the concentration of reaction products change as a function of reactant concentrations and/or reaction conditions. For
Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert Karakaş, Phoebe L. Stewart, and Jens Meiler
 Structure 17 Supplemental Data EM-Fold: De Novo Folding of α-helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert
Structure 17 Supplemental Data EM-Fold: De Novo Folding of α-helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert
Nucleic Acid Purity Assessment using A 260 /A 280 Ratios
 Nucleic Acid Purity Assessment using A 260 /A 280 Ratios A common practice in molecular biology is to perform a quick assessment of the purity of nucleic acid samples by determining the ratio of spectrophotometric
Nucleic Acid Purity Assessment using A 260 /A 280 Ratios A common practice in molecular biology is to perform a quick assessment of the purity of nucleic acid samples by determining the ratio of spectrophotometric
Built from 20 kinds of amino acids
 Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
VTT TECHNICAL RESEARCH CENTRE OF FINLAND
 Figure from: http://www.embl.de/nmr/sattler/teaching Why NMR (instead of X ray crystallography) a great number of macromolecules won't crystallize) natural environmant (water) ligand binding and inter
Figure from: http://www.embl.de/nmr/sattler/teaching Why NMR (instead of X ray crystallography) a great number of macromolecules won't crystallize) natural environmant (water) ligand binding and inter
Consensus alignment server for reliable comparative modeling with distant templates
 W50 W54 Nucleic Acids Research, 2004, Vol. 32, Web Server issue DOI: 10.1093/nar/gkh456 Consensus alignment server for reliable comparative modeling with distant templates Jahnavi C. Prasad 1, Sandor Vajda
W50 W54 Nucleic Acids Research, 2004, Vol. 32, Web Server issue DOI: 10.1093/nar/gkh456 Consensus alignment server for reliable comparative modeling with distant templates Jahnavi C. Prasad 1, Sandor Vajda
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Nuclear Magnetic Resonance
 Nuclear Magnetic Resonance NMR is probably the most useful and powerful technique for identifying and characterizing organic compounds. Felix Bloch and Edward Mills Purcell were awarded the 1952 Nobel
Nuclear Magnetic Resonance NMR is probably the most useful and powerful technique for identifying and characterizing organic compounds. Felix Bloch and Edward Mills Purcell were awarded the 1952 Nobel
Chapter 3. Protein Structure and Function
 Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Peptide bonds: resonance structure. Properties of proteins: Peptide bonds and side chains. Dihedral angles. Peptide bond. Protein physics, Lecture 5
 Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Refinement of a pdb-structure and Convert
 Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Section I Using Jmol as a Computer Visualization Tool
 Section I Using Jmol as a Computer Visualization Tool Jmol is a free open source molecular visualization program used by students, teachers, professors, and scientists to explore protein structures. Section
Section I Using Jmol as a Computer Visualization Tool Jmol is a free open source molecular visualization program used by students, teachers, professors, and scientists to explore protein structures. Section
Indiana's Academic Standards 2010 ICP Indiana's Academic Standards 2016 ICP. map) that describe the relationship acceleration, velocity and distance.
 .1.1 Measure the motion of objects to understand.1.1 Develop graphical, the relationships among distance, velocity and mathematical, and pictorial acceleration. Develop deeper understanding through representations
.1.1 Measure the motion of objects to understand.1.1 Develop graphical, the relationships among distance, velocity and mathematical, and pictorial acceleration. Develop deeper understanding through representations
Quaternary structure
 Quaternary structure Assembly of multiple polypeptide chains in one integral structure The arrangement of the subunits gives rise to a stable structure Subunits may be identical or different A common shorthand
Quaternary structure Assembly of multiple polypeptide chains in one integral structure The arrangement of the subunits gives rise to a stable structure Subunits may be identical or different A common shorthand
Discrete representations of the protein C. chain Xavier F de la Cruz 1, Michael W Mahoney 2 and Byungkook Lee
 Research Paper 223 Discrete representations of the protein C chain Xavier F de la Cruz 1, Michael W Mahoney 2 and Byungkook Lee Background: When a large number of protein conformations are generated and
Research Paper 223 Discrete representations of the protein C chain Xavier F de la Cruz 1, Michael W Mahoney 2 and Byungkook Lee Background: When a large number of protein conformations are generated and
Lecture Overview. Hydrogen Bonds. Special Properties of Water Molecules. Universal Solvent. ph Scale Illustrated. special properties of water
 Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Automatic and Objective Measurement of Residual Stress and Cord in Glass
 Automatic and Objective Measurement of Residual Stress and Cord in Glass GlassTrend - ICG TC15/21 Seminar SENSORS AND PROCESS CONTROL 13-14 October 2015, Eindhoven Henning Katte, ilis gmbh copyright ilis
Automatic and Objective Measurement of Residual Stress and Cord in Glass GlassTrend - ICG TC15/21 Seminar SENSORS AND PROCESS CONTROL 13-14 October 2015, Eindhoven Henning Katte, ilis gmbh copyright ilis
Sizes, shapes and interactions of molecules in solution
 Sizes, shapes and interactions of molecules in solution Light Scattering Analytical Viscometry Ultracentrifugation Steve Harding, NCMH University of Nottingham 1. Molecular weight distribution analysis
Sizes, shapes and interactions of molecules in solution Light Scattering Analytical Viscometry Ultracentrifugation Steve Harding, NCMH University of Nottingham 1. Molecular weight distribution analysis
1 Peptide bond rotation
 1 Peptide bond rotation We now consider an application of data mining that has yielded a result that links the quantum scale with the continnum level electrostatic field. In other cases, we have considered
1 Peptide bond rotation We now consider an application of data mining that has yielded a result that links the quantum scale with the continnum level electrostatic field. In other cases, we have considered
Disaccharides consist of two monosaccharide monomers covalently linked by a glycosidic bond. They function in sugar transport.
 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
Peptide Bonds: Structure
 Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Chapter 8. Low energy ion scattering study of Fe 4 N on Cu(100)
 Low energy ion scattering study of 4 on Cu(1) Chapter 8. Low energy ion scattering study of 4 on Cu(1) 8.1. Introduction For a better understanding of the reconstructed 4 surfaces one would like to know
Low energy ion scattering study of 4 on Cu(1) Chapter 8. Low energy ion scattering study of 4 on Cu(1) 8.1. Introduction For a better understanding of the reconstructed 4 surfaces one would like to know
Carbohydrates, proteins and lipids
 Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Polymers: Introduction
 Chapter Outline: Polymer Structures Hydrocarbon and Polymer Molecules Chemistry of Polymer Molecules Molecular Weight and Shape Molecular Structure and Configurations Copolymers Polymer Crystals Optional
Chapter Outline: Polymer Structures Hydrocarbon and Polymer Molecules Chemistry of Polymer Molecules Molecular Weight and Shape Molecular Structure and Configurations Copolymers Polymer Crystals Optional
Subject Area(s) Biology. Associated Unit Engineering Nature: DNA Visualization and Manipulation. Associated Lesson Imaging the DNA Structure
 Subject Area(s) Biology Associated Unit Engineering Nature: DNA Visualization and Manipulation Associated Lesson Imaging the DNA Structure Activity Title Inside the DNA Header Image 1 ADA Description:
Subject Area(s) Biology Associated Unit Engineering Nature: DNA Visualization and Manipulation Associated Lesson Imaging the DNA Structure Activity Title Inside the DNA Header Image 1 ADA Description:
Helices From Readily in Biological Structures
 The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
Publication of small-unit-cell structures in Acta Crystallographica Michael Hoyland ECM28 University of Warwick, 2013
 Publication of small-unit-cell structures in Acta Crystallographica Michael Hoyland ECM28 University of Warwick, 2013 International Union of Crystallography 5 Abbey Square Chester CH1 2HU UK International
Publication of small-unit-cell structures in Acta Crystallographica Michael Hoyland ECM28 University of Warwick, 2013 International Union of Crystallography 5 Abbey Square Chester CH1 2HU UK International
4. Which carbohydrate would you find as part of a molecule of RNA? a. Galactose b. Deoxyribose c. Ribose d. Glucose
 1. How is a polymer formed from multiple monomers? a. From the growth of the chain of carbon atoms b. By the removal of an OH group and a hydrogen atom c. By the addition of an OH group and a hydrogen
1. How is a polymer formed from multiple monomers? a. From the growth of the chain of carbon atoms b. By the removal of an OH group and a hydrogen atom c. By the addition of an OH group and a hydrogen
DYNAMIC LIGHT SCATTERING COMMON TERMS DEFINED
 DYNAMIC LIGHT SCATTERING COMMON TERMS DEFINED Abstract: There are a number of sources of information that give a mathematical description of the terms used in light scattering. However, these will not
DYNAMIC LIGHT SCATTERING COMMON TERMS DEFINED Abstract: There are a number of sources of information that give a mathematical description of the terms used in light scattering. However, these will not
Disulfide Bonds at the Hair Salon
 Disulfide Bonds at the Hair Salon Three Alpha Helices Stabilized By Disulfide Bonds! In order for hair to grow 6 inches in one year, 9 1/2 turns of α helix must be produced every second!!! In some proteins,
Disulfide Bonds at the Hair Salon Three Alpha Helices Stabilized By Disulfide Bonds! In order for hair to grow 6 inches in one year, 9 1/2 turns of α helix must be produced every second!!! In some proteins,
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING
 CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
18.2 Protein Structure and Function: An Overview
 18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
Deposition and use of raw diffraction images
 Deposition and use of raw diffraction images By John.R. Helliwell Crystallographic Information and Data Management A Satellite Symposium to the 28th European Crystallographic Meeting University of Warwick,
Deposition and use of raw diffraction images By John.R. Helliwell Crystallographic Information and Data Management A Satellite Symposium to the 28th European Crystallographic Meeting University of Warwick,
7. DYNAMIC LIGHT SCATTERING 7.1 First order temporal autocorrelation function.
 7. DYNAMIC LIGHT SCATTERING 7. First order temporal autocorrelation function. Dynamic light scattering (DLS) studies the properties of inhomogeneous and dynamic media. A generic situation is illustrated
7. DYNAMIC LIGHT SCATTERING 7. First order temporal autocorrelation function. Dynamic light scattering (DLS) studies the properties of inhomogeneous and dynamic media. A generic situation is illustrated
Fundamentals of grain boundaries and grain boundary migration
 1. Fundamentals of grain boundaries and grain boundary migration 1.1. Introduction The properties of crystalline metallic materials are determined by their deviation from a perfect crystal lattice, which
1. Fundamentals of grain boundaries and grain boundary migration 1.1. Introduction The properties of crystalline metallic materials are determined by their deviation from a perfect crystal lattice, which
International Year of Light 2015 Tech-Talks BREGENZ: Mehmet Arik Well-Being in Office Applications Light Measurement & Quality Parameters
 www.led-professional.com ISSN 1993-890X Trends & Technologies for Future Lighting Solutions ReviewJan/Feb 2015 Issue LpR 47 International Year of Light 2015 Tech-Talks BREGENZ: Mehmet Arik Well-Being in
www.led-professional.com ISSN 1993-890X Trends & Technologies for Future Lighting Solutions ReviewJan/Feb 2015 Issue LpR 47 International Year of Light 2015 Tech-Talks BREGENZ: Mehmet Arik Well-Being in
LMB Crystallography Course, 2013. Crystals, Symmetry and Space Groups Andrew Leslie
 LMB Crystallography Course, 2013 Crystals, Symmetry and Space Groups Andrew Leslie Many of the slides were kindly provided by Erhard Hohenester (Imperial College), several other illustrations are from
LMB Crystallography Course, 2013 Crystals, Symmetry and Space Groups Andrew Leslie Many of the slides were kindly provided by Erhard Hohenester (Imperial College), several other illustrations are from
Assessing Checking the the reliability of protein-ligand structures
 Assessing Checking the the reliability of protein-ligand structures Aim A common task in structure-based drug design is the validation of protein-ligand structures. This process needs to be quick, visual
Assessing Checking the the reliability of protein-ligand structures Aim A common task in structure-based drug design is the validation of protein-ligand structures. This process needs to be quick, visual
A reinterpretation of the phase transitions in Na 2 CO 3
 Acta Crystallographica Section B Structural Science ISSN 0108-7681 Editor: Carolyn P. Brock A reinterpretation of the phase transitions in Na 2 CO 3 Alla Arakcheeva and Gervais Chapuis Copyright International
Acta Crystallographica Section B Structural Science ISSN 0108-7681 Editor: Carolyn P. Brock A reinterpretation of the phase transitions in Na 2 CO 3 Alla Arakcheeva and Gervais Chapuis Copyright International
http://faculty.sau.edu.sa/h.alshehri
 http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
DOING PHYSICS WITH MATLAB COMPUTATIONAL OPTICS RAYLEIGH-SOMMERFELD DIFFRACTION INTEGRAL OF THE FIRST KIND
 DOING PHYSICS WITH MATLAB COMPUTATIONAL OPTICS RAYLEIGH-SOMMERFELD DIFFRACTION INTEGRAL OF THE FIRST KIND THE THREE-DIMENSIONAL DISTRIBUTION OF THE RADIANT FLUX DENSITY AT THE FOCUS OF A CONVERGENCE BEAM
DOING PHYSICS WITH MATLAB COMPUTATIONAL OPTICS RAYLEIGH-SOMMERFELD DIFFRACTION INTEGRAL OF THE FIRST KIND THE THREE-DIMENSIONAL DISTRIBUTION OF THE RADIANT FLUX DENSITY AT THE FOCUS OF A CONVERGENCE BEAM
Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O.
 Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O. Melnyk b a National University of life and environmental sciences
Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O. Melnyk b a National University of life and environmental sciences
Lectures 2 & 3. If the base pair is imbedded in a helix, then there are several more angular attributes of the base pair that we must consider:
 Lectures 2 & 3 Patterns of base-base hydrogen bonds-characteristics of the base pairs How are double helices assembled?? Figure 13 Let us first examine the angular characteristics of base pairs. Figure
Lectures 2 & 3 Patterns of base-base hydrogen bonds-characteristics of the base pairs How are double helices assembled?? Figure 13 Let us first examine the angular characteristics of base pairs. Figure
JBS FUNDAMENT Thermofluor Screen
 Cat. No. CS-330 Amount 1 Kit For in vitro use only. Quality guaranteed for 3 months. Store at 4 C. Application Screen for thermal stability of proteins as a function of the FUNDAMENTAL variables ph and
Cat. No. CS-330 Amount 1 Kit For in vitro use only. Quality guaranteed for 3 months. Store at 4 C. Application Screen for thermal stability of proteins as a function of the FUNDAMENTAL variables ph and
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK
 Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
PyRy3D: a software tool for modeling of large macromolecular complexes MODELING OF STRUCTURES FOR LARGE MACROMOLECULAR COMPLEXES
 MODELING OF STRUCTURES FOR LARGE MACROMOLECULAR COMPLEXES PyRy3D is a method for building low-resolution models of large macromolecular complexes. The components (proteins, nucleic acids and any other
MODELING OF STRUCTURES FOR LARGE MACROMOLECULAR COMPLEXES PyRy3D is a method for building low-resolution models of large macromolecular complexes. The components (proteins, nucleic acids and any other
Part-Based Recognition
 Part-Based Recognition Benedict Brown CS597D, Fall 2003 Princeton University CS 597D, Part-Based Recognition p. 1/32 Introduction Many objects are made up of parts It s presumably easier to identify simple
Part-Based Recognition Benedict Brown CS597D, Fall 2003 Princeton University CS 597D, Part-Based Recognition p. 1/32 Introduction Many objects are made up of parts It s presumably easier to identify simple
Comparative crystal structure determination of griseofulvin: Powder X-ray diffraction versus single-crystal X-ray diffraction
 Article Analytical Chemistry October 2012 Vol.57 No.30: 3867 3871 doi: 10.1007/s11434-012-5245-5 SPECIAL TOPICS: Comparative crystal structure determination of griseofulvin: Powder X-ray diffraction versus
Article Analytical Chemistry October 2012 Vol.57 No.30: 3867 3871 doi: 10.1007/s11434-012-5245-5 SPECIAL TOPICS: Comparative crystal structure determination of griseofulvin: Powder X-ray diffraction versus
arxiv:cond-mat/9709083v1 [cond-mat.stat-mech] 6 Sep 1997
![arxiv:cond-mat/9709083v1 [cond-mat.stat-mech] 6 Sep 1997 arxiv:cond-mat/9709083v1 [cond-mat.stat-mech] 6 Sep 1997](/thumbs/40/21220062.jpg) Are Protein Folds Atypical? arxiv:cond-mat/97983v1 [cond-mat.stat-mech] 6 Sep 1997 Hao Li, Chao Tang, and Ned S. Wingreen NEC Research Institute, 4 Independence Way, Princeton, New Jersey 854 (August 29,
Are Protein Folds Atypical? arxiv:cond-mat/97983v1 [cond-mat.stat-mech] 6 Sep 1997 Hao Li, Chao Tang, and Ned S. Wingreen NEC Research Institute, 4 Independence Way, Princeton, New Jersey 854 (August 29,
AN EFFECT OF GRID QUALITY ON THE RESULTS OF NUMERICAL SIMULATIONS OF THE FLUID FLOW FIELD IN AN AGITATED VESSEL
 14 th European Conference on Mixing Warszawa, 10-13 September 2012 AN EFFECT OF GRID QUALITY ON THE RESULTS OF NUMERICAL SIMULATIONS OF THE FLUID FLOW FIELD IN AN AGITATED VESSEL Joanna Karcz, Lukasz Kacperski
14 th European Conference on Mixing Warszawa, 10-13 September 2012 AN EFFECT OF GRID QUALITY ON THE RESULTS OF NUMERICAL SIMULATIONS OF THE FLUID FLOW FIELD IN AN AGITATED VESSEL Joanna Karcz, Lukasz Kacperski
The process components and related data characteristics addressed in this document are:
 TM Tech Notes Certainty 3D November 1, 2012 To: General Release From: Ted Knaak Certainty 3D, LLC Re: Structural Wall Monitoring (#1017) rev: A Introduction TopoDOT offers several tools designed specifically
TM Tech Notes Certainty 3D November 1, 2012 To: General Release From: Ted Knaak Certainty 3D, LLC Re: Structural Wall Monitoring (#1017) rev: A Introduction TopoDOT offers several tools designed specifically
HiPer Ion Exchange Chromatography Teaching Kit
 HiPer Ion Exchange Chromatography Teaching Kit Product Code: HTC001 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 5-6 hours Storage Instructions: The kit is stable for
HiPer Ion Exchange Chromatography Teaching Kit Product Code: HTC001 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 5-6 hours Storage Instructions: The kit is stable for
Isomers Have same molecular formula, but different structures
 Isomers ave same molecular formula, but different structures Constitutional Isomers Differ in the order of attachment of atoms (different bond connectivity) Stereoisomers Atoms are connected in the same
Isomers ave same molecular formula, but different structures Constitutional Isomers Differ in the order of attachment of atoms (different bond connectivity) Stereoisomers Atoms are connected in the same
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
11.1. Objectives. Component Form of a Vector. Component Form of a Vector. Component Form of a Vector. Vectors and the Geometry of Space
 11 Vectors and the Geometry of Space 11.1 Vectors in the Plane Copyright Cengage Learning. All rights reserved. Copyright Cengage Learning. All rights reserved. 2 Objectives! Write the component form of
11 Vectors and the Geometry of Space 11.1 Vectors in the Plane Copyright Cengage Learning. All rights reserved. Copyright Cengage Learning. All rights reserved. 2 Objectives! Write the component form of
Atomic Force Microscope and Magnetic Force Microscope Background Information
 Atomic Force Microscope and Magnetic Force Microscope Background Information Lego Building Instructions There are several places to find the building instructions for building the Lego models of atomic
Atomic Force Microscope and Magnetic Force Microscope Background Information Lego Building Instructions There are several places to find the building instructions for building the Lego models of atomic
Incorporating Internal Gradient and Restricted Diffusion Effects in Nuclear Magnetic Resonance Log Interpretation
 The Open-Access Journal for the Basic Principles of Diffusion Theory, Experiment and Application Incorporating Internal Gradient and Restricted Diffusion Effects in Nuclear Magnetic Resonance Log Interpretation
The Open-Access Journal for the Basic Principles of Diffusion Theory, Experiment and Application Incorporating Internal Gradient and Restricted Diffusion Effects in Nuclear Magnetic Resonance Log Interpretation
State of Stress at Point
 State of Stress at Point Einstein Notation The basic idea of Einstein notation is that a covector and a vector can form a scalar: This is typically written as an explicit sum: According to this convention,
State of Stress at Point Einstein Notation The basic idea of Einstein notation is that a covector and a vector can form a scalar: This is typically written as an explicit sum: According to this convention,
Statistical Forecasting of High-Way Traffic Jam at a Bottleneck
 Metodološki zvezki, Vol. 9, No. 1, 2012, 81-93 Statistical Forecasting of High-Way Traffic Jam at a Bottleneck Igor Grabec and Franc Švegl 1 Abstract Maintenance works on high-ways usually require installation
Metodološki zvezki, Vol. 9, No. 1, 2012, 81-93 Statistical Forecasting of High-Way Traffic Jam at a Bottleneck Igor Grabec and Franc Švegl 1 Abstract Maintenance works on high-ways usually require installation
Lab 2 Biochemistry. Learning Objectives. Introduction. Lipid Structure and Role in Food. The lab has the following learning objectives.
 1 Lab 2 Biochemistry Learning Objectives The lab has the following learning objectives. Investigate the role of double bonding in fatty acids, through models. Developing a calibration curve for a Benedict
1 Lab 2 Biochemistry Learning Objectives The lab has the following learning objectives. Investigate the role of double bonding in fatty acids, through models. Developing a calibration curve for a Benedict
Organic Molecules of Life - Exercise 2
 Organic Molecules of Life - Exercise 2 Objectives -Know the difference between a reducing sugar and a non-reducing sugar. -Distinguish Monosaccharides from Disaccharides and Polysaccharides -Understand
Organic Molecules of Life - Exercise 2 Objectives -Know the difference between a reducing sugar and a non-reducing sugar. -Distinguish Monosaccharides from Disaccharides and Polysaccharides -Understand
Protein Dynamics Intro
 Protein Dynamics Intro From rigid structures to motions on energy landscapes Do you all remember Anfinsen? What concept now associated with his name made Anfinsen famous? Right, it is the concept that
Protein Dynamics Intro From rigid structures to motions on energy landscapes Do you all remember Anfinsen? What concept now associated with his name made Anfinsen famous? Right, it is the concept that
Chapter Outline. How do atoms arrange themselves to form solids?
 Chapter Outline How do atoms arrange themselves to form solids? Fundamental concepts and language Unit cells Crystal structures Simple cubic Face-centered cubic Body-centered cubic Hexagonal close-packed
Chapter Outline How do atoms arrange themselves to form solids? Fundamental concepts and language Unit cells Crystal structures Simple cubic Face-centered cubic Body-centered cubic Hexagonal close-packed
Molecular Spectroscopy
 Molecular Spectroscopy UV-Vis Spectroscopy Absorption Characteristics of Some Common Chromophores UV-Vis Spectroscopy Absorption Characteristics of Aromatic Compounds UV-Vis Spectroscopy Effect of extended
Molecular Spectroscopy UV-Vis Spectroscopy Absorption Characteristics of Some Common Chromophores UV-Vis Spectroscopy Absorption Characteristics of Aromatic Compounds UV-Vis Spectroscopy Effect of extended
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Measuring Protein Concentration through Absorption Spectrophotometry
 Measuring Protein Concentration through Absorption Spectrophotometry In this lab exercise you will learn how to homogenize a tissue to extract the protein, and then how to use a protein assay reagent to
Measuring Protein Concentration through Absorption Spectrophotometry In this lab exercise you will learn how to homogenize a tissue to extract the protein, and then how to use a protein assay reagent to
Phasing & Substructure Solution
 CCP4 Workshop Chicago 2011 Phasing & Substructure Solution Tim Grüne Dept. of Structural Chemistry, University of Göttingen June 2011 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de CCP4 Chicago 2011:
CCP4 Workshop Chicago 2011 Phasing & Substructure Solution Tim Grüne Dept. of Structural Chemistry, University of Göttingen June 2011 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de CCP4 Chicago 2011:
Topic 2: Energy in Biological Systems
 Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Structure Bioinformatics Course Basel 2004. Sergei V. Strelkov M.E. Mueller Institute for Structural Biology at Biozentrum Basel
 1 Structure Bioinformatics Course Basel 2004 Introduction to X-ray X crystallography Sergei V. Strelkov M.E. Mueller Institute for Structural Biology at Biozentrum Basel sergei-v.strelkov@unibas.ch 2 Intro
1 Structure Bioinformatics Course Basel 2004 Introduction to X-ray X crystallography Sergei V. Strelkov M.E. Mueller Institute for Structural Biology at Biozentrum Basel sergei-v.strelkov@unibas.ch 2 Intro
Steady Heat Conduction
 Steady Heat Conduction In thermodynamics, we considered the amount of heat transfer as a system undergoes a process from one equilibrium state to another. hermodynamics gives no indication of how long
Steady Heat Conduction In thermodynamics, we considered the amount of heat transfer as a system undergoes a process from one equilibrium state to another. hermodynamics gives no indication of how long
Nonlinear Iterative Partial Least Squares Method
 Numerical Methods for Determining Principal Component Analysis Abstract Factors Béchu, S., Richard-Plouet, M., Fernandez, V., Walton, J., and Fairley, N. (2016) Developments in numerical treatments for
Numerical Methods for Determining Principal Component Analysis Abstract Factors Béchu, S., Richard-Plouet, M., Fernandez, V., Walton, J., and Fairley, N. (2016) Developments in numerical treatments for
How To Check For Differences In The One Way Anova
 MINITAB ASSISTANT WHITE PAPER This paper explains the research conducted by Minitab statisticians to develop the methods and data checks used in the Assistant in Minitab 17 Statistical Software. One-Way
MINITAB ASSISTANT WHITE PAPER This paper explains the research conducted by Minitab statisticians to develop the methods and data checks used in the Assistant in Minitab 17 Statistical Software. One-Way
Organic Chemistry Calculations
 Organic Chemistry Calculations There are three basic units for measurement in the organic laboratory mass, volume, and number, measured in moles. Most of the other types of measurements are combinations
Organic Chemistry Calculations There are three basic units for measurement in the organic laboratory mass, volume, and number, measured in moles. Most of the other types of measurements are combinations
3D structure visualization and high quality imaging. Chimera
 3D structure visualization and high quality imaging. Chimera Vincent Zoete 2008 Contact : vincent.zoete@isb sib.ch 1/27 Table of Contents Presentation of Chimera...3 Exercise 1...4 Loading a structure
3D structure visualization and high quality imaging. Chimera Vincent Zoete 2008 Contact : vincent.zoete@isb sib.ch 1/27 Table of Contents Presentation of Chimera...3 Exercise 1...4 Loading a structure
Engineering Problem Solving and Excel. EGN 1006 Introduction to Engineering
 Engineering Problem Solving and Excel EGN 1006 Introduction to Engineering Mathematical Solution Procedures Commonly Used in Engineering Analysis Data Analysis Techniques (Statistics) Curve Fitting techniques
Engineering Problem Solving and Excel EGN 1006 Introduction to Engineering Mathematical Solution Procedures Commonly Used in Engineering Analysis Data Analysis Techniques (Statistics) Curve Fitting techniques
MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788
 MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788 ISSN 0340-6253 Three distances for rapid similarity analysis of DNA sequences Wei Chen,
MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788 ISSN 0340-6253 Three distances for rapid similarity analysis of DNA sequences Wei Chen,
AP1 Oscillations. 1. Which of the following statements about a spring-block oscillator in simple harmonic motion about its equilibrium point is false?
 1. Which of the following statements about a spring-block oscillator in simple harmonic motion about its equilibrium point is false? (A) The displacement is directly related to the acceleration. (B) The
1. Which of the following statements about a spring-block oscillator in simple harmonic motion about its equilibrium point is false? (A) The displacement is directly related to the acceleration. (B) The
Chem 106 Thursday Feb. 3, 2011
 Chem 106 Thursday Feb. 3, 2011 Chapter 13: -The Chemistry of Solids -Phase Diagrams - (no Born-Haber cycle) 2/3/2011 1 Approx surface area (Å 2 ) 253 258 Which C 5 H 12 alkane do you think has the highest
Chem 106 Thursday Feb. 3, 2011 Chapter 13: -The Chemistry of Solids -Phase Diagrams - (no Born-Haber cycle) 2/3/2011 1 Approx surface area (Å 2 ) 253 258 Which C 5 H 12 alkane do you think has the highest
Optical Design Tools for Backlight Displays
 Optical Design Tools for Backlight Displays Introduction Backlights are used for compact, portable, electronic devices with flat panel Liquid Crystal Displays (LCDs) that require illumination from behind.
Optical Design Tools for Backlight Displays Introduction Backlights are used for compact, portable, electronic devices with flat panel Liquid Crystal Displays (LCDs) that require illumination from behind.
