X-Chromosome Inactivation
|
|
|
- Melvyn Newton
- 10 years ago
- Views:
Transcription
1 Stanley M Gartler, University of Washington, Seattle, Washington, USA Michael A Goldman, San Francisco State University, San Francisco, California, USA In female mammals, one of the X-chromosomes is transcriptionally silenced by heterochromatin formation to bring about equal expression of X-linked genes in XX females and XY males. Introduction In mammals, the female has two large, gene-rich X chromosomes, whereas the male has a single X chromosome and a Y chromosome, which harbours few genes. If the X-linked genes were expressed equally in both sexes, females would produce approximately twice the level of X- linked gene products as would males. Since the great majority of genes on the X are not concerned with sex determination and reproduction, this difference would disturb metabolic balance in one sex or the other. In mammals and some other groups with XY sex determination, a system has evolved to bring about the equivalence in expression of X-linked genes in females and males. H. J. Muller, in 1931, dubbed this phenomenon dosage compensation (Muller, 1932). Different groups of organisms have evolved distinct strategies to achieve dosage compensation. In Drosophila, the transcription rate of the single X in males is higher than that of the two Xs in females. In Caenorhabditis elegans, the transcription rate is halved in the female s two Xs. In mammals, most of the genes on one of the Xs in the female are transcriptionally silenced in a process called X-chromosome inactivation (XCI), the focus of this entry. Muller described dosage compensation in Drosophila.It was not until 30 years after that the phenomenon was clearly recognized in mammals. Based on observations by several workers, Mary Lyon, in 1961, proposed that each cell of a mammal maintained one and only one active X chromosome, inactivating the other to produce a darklystaining mass called the sex chromatin or Barr body (Lyon, 1961). Morphological evidence for X-chromosome inactivation could be seen in the coat colours of various mammals, including the calico cat a female cat in which one X chromosome encodes one coat colour, the other a different one, and the cat s fur is a mosaic of patches having one coat colour or the other. This early model was, of course, incomplete, but the basic idea of a single active X chromosome in female cells has stood the test of time.. Introduction Secondary article Article Contents. X Inactivation in Somatic Cells. X Inactivation in the Germline. Random and Nonrandom X Inactivation. Stability of X Inactivation. Genes that Escape Inactivation. XIST and the X-Inactivation Centre. DNA Methylation. Replication Timing. Histone Acetylation. Inactive-X-Specific Proteins. A Model for X-Chromosome Inactivation. Perspective X Inactivation in Somatic Cells In the mouse, the two X chromosomes in the early female embryo are not differentiated, either cytologically or functionally; both X chromosomes are active. In the late morula or early blastocyst stages, the earliest evidence of XCI are seen: asynchrony of deoxyribonucleic acid (DNA) replication between the two Xs, differential gene expression and sex chromatin formation. These differences are first observed in the extraembryonic cell lineages that form the membranes surrounding the embryo. In these cells the paternal X is preferentially inactivated. At about the time of gastrulation, random X inactivation occurs in the epiblast, which contains the cells that will give rise to the embryo itself. When inactivation occurs in these cells, the paternal or maternal X is randomly chosen to be inactivated. However, once inactivation has occurred, all of the descendants of a cell will have the same X silent. This results in patches of cells having an inactive maternal X, and in patches having an inactive paternal X, explaining the mosaic pattern of allelic expression seen, for example, in the calico cat. X Inactivation in the Germline In contrast to somatic cells, in which X inactivation is extremely stable, inactivation in the germline is cyclic. In the female, one X is inactive during the oogonial or mitotic phase, while both X chromosomes are active in oogenesis. Having both Xs active may be necessary to permit pairing and recombination in meiosis, which may not be possible if one X is heterochromatic and highly condensed and the other is not. In the male, with a single X, inactivation does not occur in somatic cells; however, early in ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group / 1
2 spermatogenesis, the single X is condensed and inactivated, possibly to restrict pairing and recombination between the nonhomologous parts of the X and Y chromosomes. Random and Nonrandom X Inactivation X inactivation in somatic cells is generally random, with the maternal and paternal Xs having an equal probability of inactivation. Thus, in females heterozygous for X-linked traits, such as haemophilia, about half the cells of the liver should, on average, produce normal clotting factor. This is typically enough to ensure normal blood clotting. If the trait affects fur colour, as in the calico cat, about half the patches in the cat should exhibit one phenotype. Statistically, sometimes one X will be inactivated more frequently than the other, just as a series of coin tosses may sometimes yield something other than a 50:50 ratio of heads to tails. Thus, chance could lead to a female heterozygous for haemophilia in which little or no clotting factor is produced and the typical symptoms of this disease are observed. Such statistical extremes are uncommon. Aside from statistical fluctuation, preferential inactivation may occur, as in extraembryonic cells or in marsupials, where nonrandom inactivation of the paternal X is the rule (Graves, 1996). Genetic differences on the chromosome may render one X more likely to undergo inactivation. In addition, if one X has a mutant allele whose expression renders a cell inviable or inhibits its growth, cells inactivating the mutant allele will have a selective advantage over cells in which the normal allele is inactivated. Thus, although inactivation may occur randomly at first, cells that inactivate the wrong allele will be overgrown, and the outcome will be an organism in which all, or nearly all, cells have the same X inactivated. Stability of X Inactivation In placental mammals, the inactive state of the X is generally highly stable. As we discuss below, a number of factors collaborate to repress alleles on the inactive X. If all factors ensuring X inactivation operated independently and were functional throughout life, reactivation would probably not be possible. In the germline, for instance, the inactivation state is cyclic and DNA methylation, an important repressive factor in somatic cells (see below), is absent. DNA methylation also seems to be absent from the inactivation system in marsupials, where, even in somatic cells, the inactivation of the X is relatively unstable. Even in mouse somatic cells, some genes on the inactive X are reactivated at low, but significant levels as the animals age. We think that DNA methylation stabilizes the inactive state. Consistent with this notion, genes that exhibit agerelated reactivation typically do not utilize DNA methylation, while those genes that clearly utilize DNA methylation as a regulatory mechanism are not reactivated with age. In transformed cell lines, reactivation of some inactive X-linked genes can be induced with 5-azacytidine (5AC), a drug known to reverse DNA methylation. The same drug treatment does not reactivate X-linked genes in normal somatic cells. The reactivation of the X in transformed cells probably occurs because some of the redundant mechanisms maintaining the inactive state are not functioning in these cells, leaving methylation as one of the only repressive influences on X-linked gene expression (Hansen et al., 1998). Embryonal carcinoma (EC) cells are derived from gonadal tumours or embryonic tissues, and maintain their Xs in an active state as long as they remain undifferentiated. However, upon chemical-induced differentiation, the cells will go through X inactivation and one of the two X chromosomes will be silenced (Rastan and Robertson, 1985). When female mouse lymphocytes, having one active and one silent X, are fused with EC cells, the inactive X reactivates. This suggests that there are specific factors present in the EC cells that maintain or permit the active state of both X chromosomes. Genes that Escape Inactivation Once, every gene on the inactive X chromosome was believed to be transcriptionally silenced. We now know that a fair number of genes on human and mouse inactive Xs are not repressed (Disteche, 1995). One group of genes escaping X inactivation is found in the pseudoautosomal region (PAR) at the tip of the short arm of the human X, a region of homology between the X and Y chromosomes. Fertility in mammalian males requires pairing and recombination between the X and Y chromosomes within the PAR. Genes in the PAR are present on both the X and Y chromosomes, so no gene dosage difference between males and females exists; compensation is therefore not necessary and genes in the PAR escape X inactivation. In addition to the PAR at the tip of the X short arm, a second, smaller, PAR is present at the tip of the long arm in humans. Several genes have been detected there, although, surprisingly, one of these is inactivated. Several genes in humans and mice outside the PAR have functional Y- linked alleles and, as expected, their X-linked counterparts escape inactivation. In these latter cases the X- and Y- linked alleles may differ from one another, as no recombination between them occurs. Some X-linked genes that escape inactivation have nonfunctional Y-linked copies (pseudogenes), presumably due to accumulated inactivating mutations. Lastly, there are several reports of 2 ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group /
3 X-linked genes in humans that have no Y-chromosome counterparts, but still escape inactivation. Whether in the PAR or not, most genes that escape inactivation are organized in clusters or functional domains in which all of the genes escape inactivation. Such regulatory domains may contain sequence information specifying the spread or inhibition of an inactivation signal. Some that escape inactivation may have been repressed when X inactivation first occurs, but are reactivated later. Thus the entire X chromosome may be inactivated initially, but selected inactive domains may be inherently unstable and rapidly reversed. The mouse X appears to be more completely inactivated than is the human X, perhaps reflecting the occurrence of a longer evolutionary time for X inactivation in the mouse. Escape from inactivation of the SHOX gene in the human short arm PAR is apparently responsible for the short stature characteristic of Turner syndrome (XO) females. In mice, where XO is not associated with the phenotypic problems typical of Turner syndrome in humans, the SHOX homologue is not X linked. XIST and the X-Inactivation Centre A central feature of mammalian X inactivation is its chromosomal nature. With one exception (XIST, described below), all the inactivated genes are on one X, while the homologue contains the active alleles. This pattern is most easily explained by a single initiation site from which inactivation spreads along the rest of the chromosome. The evidence for the existence and location of an X-inactivation centre came from studies of X-autosomal translocations and deletions which interfered with the manifestation of X inactivation. Analysis of such translocations allows one to identify a short segment of the X, termed the X- inactivation centre or XIC, essential for X inactivation. Moreover, in mice, some Xs are more prone to inactivation than are others. The difference between them resides in a locus called Xce (or X-controlling element), which maps to about the same position as the XIC identified in translocation experiments. A single gene that maps to the XIC and that has properties suggesting that it might be the locus essential for the initiation of X inactivation has been designated X- inactive specific transcript or XIST. (Human genes are designated entirely in capital letters, such as XIST, whereas mouse genes are designated in capitals and lower-case letters, as in Xist. For convenience here we use a single acronym, XIST.) XIST is expressed only from the inactive X (Brown et al., 1991). The ribonucleic acid (RNA) transcript has no significant open reading frame and the product remains in the nucleus, coating the inactive X. This suggests that XIST is among those loci that produce a functional RNA molecule that is never translated into a protein. XIST expression is detected early in preimplantation development, often from both Xs, but well before any sign of X inactivation. However, this early expression is probably not functional, as the transcript is not stable, is unprocessed, and does not coat the X. In the mouse, the paternal X begins to accumulate transcripts in the early morula, apparently as a result of an imprint in the gametes that leads to the paternal nonrandom inactivation found in extraembryonic cells. Later, in the inner cell mass, unstable XIST is transcribed from both Xs, but the XIST transcript from one or the other X becomes stabilized and accumulates on that X. Random X inactivation follows and XIST transcription from the active X is silenced. Recent evidence suggests that alernative promoter utilization or an antisense transcript may be involved in XIST regulation. Although the properties and map location of XIST strongly suggest that it should be a key element in the X- inactivation process, further experimental evidence was required to show that this locus is necessary for inactivation of the X chromosome. This was shown through the use of various deletions of the promoter or exon region of the XIST gene that prevent production of full-sized XIST RNA. In these cases the chromosome bearing the deletion is not inactivated, showing that the XIST gene is required in cis for inactivation to take place. Further support for a critical role of XIST comes from experiments in which the XIST gene and varying amounts of surrounding sequence have been incorporated into an autosome in male embryonic stem cells. In some cases, where multiple copies of the XIST transgene have been tandemly incorporated, XIST RNA coats the autosome and represses transcription. Although other factors will probably be involved, these experiments indicate that XIST RNA appears to propagate inactivation by binding to the chromosome from which it is expressed. They also imply that specific X- linked sequences are not required for XIST RNA to coat a chromosome. DNA Methylation Methylation of the base cytosine occurs enzymatically after DNA synthesis and in mammals is restricted to the dinucleotide 5 -CpG-3 (CpG). About 7% of CpGs are present in clusters called CpG islands, which are usually located at the 5 ends of genes and have a relatively high density of CpGs. The remaining CpGs are dispersed throughout the genome, usually as singlets. Most CpG islands are unmethylated, but those near inactivated genes on the X chromosome, and those near some imprinted genes on autosomes, are methylated. Methylated CpG islands repress transcription, and the great majority of them in the genome are found on the inactive X in normal cells. They are important in maintaining the repressed state ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group / 3
4 of inactive X-linked genes and may also play a role in establishing X inactivation, as indicated by the fact that differential CpG island methylation occurs close to the time of inactivation of two X-linked genes that have been studied in detail. Interestingly, most of the dispersed CpGs throughout the genome tend to be methylated. There is limited evidence that the dispersed CpGs on the inactive X may be hypomethylated relative to the active X, suggesting a similarity in overall methylation levels on the active and inactive Xs. No evidence exists that the dispersed CpGs, methylated or unmethylated, play a role in gene silencing. A crucial role of methylation may be in the control of XIST expression, and therefore the initiation of X inactivation itself. Methylation analysis of a small cluster of CpGs in the XIST promoter shows that the silent allele on the active X becomes methylated while the expressed allele on the inactive X is hypomethylated. Developmental studies show that the sperm and oocyte are differentially methylated in part of the promoter region (sperm hypermethylated, oocyte hypomethylated) but these differences in methylation do not persist into preimplantation development (McDonald et al., 1998). Methylation may not be the factor responsible for turning XIST on or off in the first place, but it may function as an imprint and probably is crucial for keeping the silent copy of XIST stably repressed. Replication Timing The inactive X chromosome begins and ends DNA replication later than its active homologue. Replication timing studies of inactivated genes indicate that they replicate in the last half of S phase. The appearance of a late-replicating X is observed in early embryogenesis. In female embryonic stem cells, which can undergo X inactivation in culture, a late-replicating X is detectable 2 days after the onset of differentiation, well before the silencing of X-linked genes. Replication timing does not simply reflect whether a chromosome is active or inactive. For example, the blood clotting factor IX gene on the active X in nonexpressing cells replicates somewhat earlier than its allele on the inactive X and replicates even earlier in expressing cells. Thus, the time of replication appears to be related to the potential for transcription, whether determined by the chromosome activation state or the cell s differentiation phase. Studies with the demethylating agent 5AC suggest that methylation at controlling regions may play a role in altering timing of replication over domains of a megabase or more. Although it is possible that late replication may be merely an effect of the complicated inactivation process, its early appearance suggests a more significant role. There is evidence that late-replicating domains are compartmentalized and it is possible that late replication of the inactive X may play a role in the incorporation of the inactive X into a subnuclear compartment inhibiting transcription. Histone Acetylation Acetylation of histones occurs in all plant and animal forms. It occurs at specific lysine residues and is catalysed by histone acetyltransferases (HATs) and deacetylases (HDACs). An association between histone acetylation and gene expression has been considered for decades, and has attracted considerable attention recently with the discovery that HATs and HDACs are often identical to, or associated with, known regulators of transcription. In general, hyperacetylation is associated with gene expression, and hypoacetylation is associated with gene silencing. Using fluorescent antisera for individual acetylated lysine residues, cytological studies showed that the inactive X, with the exception of regions containing genes that escape inactivation, has a low level of the acetylated isoforms of histones H2A, H3 and H4 in humans, mice and marsupials (Keohane et al., 1998). That histone deacetylation is not an initiating event in X inactivation was shown by a study of the development of H4 deacetylation in female embryonic stem cells undergoing X inactivation. Increase in XIST expression, late replication onset and initial gene silencing occur some 2 days before H4 deacetylation of an X chromosome. Deacetylation of histones on the inactive X may be a consequence of other events associated with X inactivation. It seems possible that other histone modifications may also be associated with the activity state of the inactive X. For example, recently it has been shown that phosphorylation of histone H3 is associated with chromosome condensation in Tetrahymena. Inactive-X-Specific Proteins Although transcriptional proteins bind to the promoter regions of transcribed genes on the active X, all in vivo footprinting attempts to detect transcription inhibiting factors bound to promoter regions on the inactive X have failed. Recently, an in vitro binding assay identified a nuclear protein that recognizes the methylated region of the mouse XIST promoter on the active X. Although much work remains to be done, these results suggest that silencing proteins involved in repressing genes on the inactive X are more likely to function at a chromosomal level rather than at the level of individual genes. Probably XIST RNA, which coats the inactive X, does so by interacting with a DNA-binding protein, as suggested by a recent in vitro study showing significant interactions between heteronuclear proteins and XIST. 4 ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group /
5 The most exciting result in this area is the recent finding of a new family of histones, called macroh2a. One of these is macro histone 2A1(mH2A1) whose N-terminal third of the molecule is similar to H2A1, but the remainder of which is unrelated to any known histone (Costanzi and Pehrson, 1998). The unique part of mh2a1 was found to preferentially localize with the inactive X-chromosome during metaphase and interphase in a number of mammals, including humans. Surprisingly, other studies indicate that the nonhistone part of the protein is widely conserved, and that the gene may have originated before the appearance of eukaryotes. Although mh2a1 is enriched on the inactive X, it is found throughout the genome, in both males and females. It is conceivable that mh2a1 may interact with XIST RNA in the process of X inactivation. A Model for X-Chromosome Inactivation The study of X-chromosome inactivation is at a juncture in which models, grounded in recent data, can be used to direct future research. We can conveniently divide the X- inactivation process into three phases: initiation, spreading and maintenance. One of the key features of the initiation of XCI is the choice of which X is to remain active. In murine extraembryonic cells, where the paternal X is preferentially inactivated, choice may follow from the methylation imprint in the sperm XIST gene, the comparable oocyte region being unmethylated. In the epiblast where random X inactivation occurs, the earlier methylation imprint of the XIST gene has disappeared and both Xs are transcribing unstable XIST RNA. The popular idea has been that choice should involve selecting an X to remain active rather than the X or Xs to become inactive. This reasoning follows from the presumed difficulty of choice in cells with more than two Xs, where it should be simpler to choose one to be active rather than choosing two or more to become inactive. One proposal assumes a limiting amount of an autosomally produced blocking factor for which the XICs compete to remain active. A recent study has identified the 3 end of the XIST gene as a candidate region for the binding site of the blocking factor (Clerc and Avner, 1998). The requirement for stabilization of the XIST transcript has led to the proposal of XIST interacting factors affecting stability. A further complexity is the fact that mutations in different parts of the XIST gene can alter susceptibility to inactivation in different ways. It should not be surprising that there might be two or more mechanisms for ensuring that only one X remains active, as the presence of two active Xs is apparently lethal (Carrel and Willard, 1998; Panning and Jaenisch, 1998). XIST is certainly one of the keys to the initiation of X inactivation. How does it come to act only in cis, never affecting the other X-chromosome or the autosomes? One possibility is that the inactive X is compartmentalized within the nucleus. The inactive X appears isolated in a cell compartment in the form of sex chromatin, generally occupying a position near the periphery of the nucleus. A number of years ago it was proposed that compartmentalization might be an early event in X inactivation, helping target various repressive factors that modify the inactive X. This idea was based on the observation that the inactive X in somatic cell hybrids is relatively unstable and does not form a normal sex chromatin structure, suggesting that the inactive X does not occupy its traditional compartment within the nucleus. More recently it has been shown by high voltage electron microscopy and microscopic analysis of a protein (perichromin) normally covering chromosomes at mitosis, but present on the inactive X at interphase, that the sex chromatin has unique structural properties (Gartler et al., 1992). As mentioned earlier, late replication might signal this compartmentalization. How the inactivation signal spreads must also involve XIST. Since XIST RNA does not contact DNA, it seems likely that an RNA protein interaction might be involved in stabilization and propagation of the signal. It has been proposed that specific sequences along the X-chromosome could play an important role in signal spread, and, recently, long interspersed repetitive sequence elements (LINEs), which are present in an unusually high concentration on the X, have been proposed as a candidate (Lyon, 1998). LINEs occur throughout the genome, perhaps accounting for the fact that autosomal genes can be inactivated in X:autosome translocations. Moreover, LINEs vary subtly in sequence from one locus to another, possibly accounting for the fact that some loci on the X- chromosome escape X inactivation; such regions may simply lack the appropriate LINE sequence. However, the RNA protein interaction may be all that is necessary for the accumulation and spread of XIST RNA along the chromosome. How is the X-inactivation pattern maintained? Several observations on transformed cells indicate that if the XIST locus is deleted after X inactivation has occurred, the X- inactivation pattern is maintained. This is not surprising, as we know that a complex of repressive factors distinguish the inactive from the active X, and that promoter methylation, for one, can maintain the X-inactivation pattern in the absence of functioning XIST RNA. What we do not know is whether XIST is able to maintain inactivation in the absence of methylation or any combination of the other repressive factors. ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group / 5
6 Perspective During the first 20 years after the X-inactivation model was proposed, late replication and sex chromatin were the only physical features known to distinguish the two Xs. Methylation differences were detected in the 1980s and for a time it was believed to be the key process. XIST as a single explanation of X inactivation promised a solution in the 1990s, only to be modified by the more recent reports on histone acetylation differences between the two Xs. As this entry was being written, the first report of a protein (mh2a1) preferentially associated with the inactive X appeared. It is likely that that report is merely the first associating proteins with the inactivation process. References Brown CJ, Ballabio A, Rupert JL et al. (1991) A gene from the region of the human inactivation center is expressed exclusively from the inactive X chromosome. Nature 349: Carrel L and Willard HF (1998) Counting on Xist. Nature Genetics 19: Clerc P and Avner P (1998) Role of the region 3 to Xist exon 6 in the counting process of X-chromosome inactivation. Nature Genetics 19: Costanzi C and Pehrson JR (1998) Histone macroh2al is concentrated in the inactive X chromosome of female mammals. Nature 393: Disteche CM (1995) Escape from X inactivation in human and mouse. Trends in Genetics 11: Gartler SM, Dyer KA and Goldman MA (1992) Mammalian X chromosome inactivation. Molecular Genetic Medicine 2: Graves JAM (1996) Mammals that break the rules: genetics of marsupials and monotremes. Annual Review of Genetics. 30: Hansen RS, Canfield TK, Stanek AM, Keitges EA and Gartler SM (1998) Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proceedings of the National Academy of Sciences of the USA 95: Keohane AM, Lavender JS, O Neill LP and Turner BM (1998) Histone acetylation and X inactivation. Developmental Genetics 22: Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L). Nature 190: Lyon MF (1998) X-chromosome inactivation: a repeat hypothesis. Cytogenetics and Cell Genetics 80: McDonald LE, Paterson CA and Kay GF (1998) Bisulfite genomic sequencing-derived methylation profile of the Xist gene throughout early mouse development. Genomics 54: Muller HJ (1932) Further studies on the nature and causes of gene mutation. Proceedings of the 6th Congress of Genetics 1: Panning B and Jaenisch R (1998) RNA and the epigenetic regulation of X chromosome inactivation. Cell 93: Rastan S and Robertson EJ (1985) X-chromosome deletions in embryoderived (EK) cell lines associated with lack of X-chromosome inactivation. I. Embryology and Experimental Morphology 90: Further Reading Brockdorff N and Duthie SM (1998) X-chromosome inactivation and the Xist gene. Cellular and Molecular Life Sciences 54: Carrel L and Willard HF (1998) Counting on Xist. Nature Genetics 19: Goto T and Monk M (1998) Regulation of X-chromosome inactivation in development in mice and humans. Microbiology and Molecular Biology Reviews 62: Heard C, Clerc P and Avner P (1997) X-chromosome inactivation in mammals. Annual Review of Genetics 31: Panning B and Jaenisch R (1998) RNA and the epigenetic regulation of X chromosome inactivation. Cell 93: Spusta SC and Goldman MA (1999) XISTential wanderings: the role of XIST RNA in X-chromosome inactivation. Current Science 77: ENCYCLOPEDIA OF LIFE SCIENCES / & 2001 Nature Publishing Group /
Chromosomal Basis of Inheritance. Ch. 3
 Chromosomal Basis of Inheritance Ch. 3 THE CHROMOSOME THEORY OF INHERITANCE AND SEX CHROMOSOMES! The chromosome theory of inheritance describes how the transmission of chromosomes account for the Mendelian
Chromosomal Basis of Inheritance Ch. 3 THE CHROMOSOME THEORY OF INHERITANCE AND SEX CHROMOSOMES! The chromosome theory of inheritance describes how the transmission of chromosomes account for the Mendelian
17. A testcross A.is used to determine if an organism that is displaying a recessive trait is heterozygous or homozygous for that trait. B.
 ch04 Student: 1. Which of the following does not inactivate an X chromosome? A. Mammals B. Drosophila C. C. elegans D. Humans 2. Who originally identified a highly condensed structure in the interphase
ch04 Student: 1. Which of the following does not inactivate an X chromosome? A. Mammals B. Drosophila C. C. elegans D. Humans 2. Who originally identified a highly condensed structure in the interphase
CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE. Section B: Sex Chromosomes
 CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE Section B: Sex Chromosomes 1. The chromosomal basis of sex varies with the organism 2. Sex-linked genes have unique patterns of inheritance 1. The chromosomal
CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE Section B: Sex Chromosomes 1. The chromosomal basis of sex varies with the organism 2. Sex-linked genes have unique patterns of inheritance 1. The chromosomal
4 SEX CHROMOSOMES AND SEX DETERMINATION
 4 SEX CHROMOSOMES AND SEX DETERMINATION 4.1 Sex chromosomes and Sex Determination Sex- chromosomes. If present, sex chromosomes may not have the same size, shape, or genetic potential. In humans, females
4 SEX CHROMOSOMES AND SEX DETERMINATION 4.1 Sex chromosomes and Sex Determination Sex- chromosomes. If present, sex chromosomes may not have the same size, shape, or genetic potential. In humans, females
The correct answer is c A. Answer a is incorrect. The white-eye gene must be recessive since heterozygous females have red eyes.
 1. Why is the white-eye phenotype always observed in males carrying the white-eye allele? a. Because the trait is dominant b. Because the trait is recessive c. Because the allele is located on the X chromosome
1. Why is the white-eye phenotype always observed in males carrying the white-eye allele? a. Because the trait is dominant b. Because the trait is recessive c. Because the allele is located on the X chromosome
Fact Sheet 14 EPIGENETICS
 This fact sheet describes epigenetics which refers to factors that can influence the way our genes are expressed in the cells of our body. In summary Epigenetics is a phenomenon that affects the way cells
This fact sheet describes epigenetics which refers to factors that can influence the way our genes are expressed in the cells of our body. In summary Epigenetics is a phenomenon that affects the way cells
Influence of Sex on Genetics. Chapter Six
 Influence of Sex on Genetics Chapter Six Humans 23 Autosomes Chromosomal abnormalities very severe Often fatal All have at least one X Deletion of X chromosome is fatal Males = heterogametic sex XY Females
Influence of Sex on Genetics Chapter Six Humans 23 Autosomes Chromosomal abnormalities very severe Often fatal All have at least one X Deletion of X chromosome is fatal Males = heterogametic sex XY Females
Control of Gene Expression
 Home Gene Regulation Is Necessary? Control of Gene Expression By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Home Gene Regulation Is Necessary? Control of Gene Expression By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Chromosomes, Mapping, and the Meiosis Inheritance Connection
 Chromosomes, Mapping, and the Meiosis Inheritance Connection Carl Correns 1900 Chapter 13 First suggests central role for chromosomes Rediscovery of Mendel s work Walter Sutton 1902 Chromosomal theory
Chromosomes, Mapping, and the Meiosis Inheritance Connection Carl Correns 1900 Chapter 13 First suggests central role for chromosomes Rediscovery of Mendel s work Walter Sutton 1902 Chromosomal theory
Human Cloning The Science and Ethics of Nuclear Transplantation
 Human Cloning The Science and Ethics of Transplantation Rudolf Jaenisch, M.D. In addition to the moral argument against the use of somatic-cell nuclear for the creation of a child ( reproductive cloning
Human Cloning The Science and Ethics of Transplantation Rudolf Jaenisch, M.D. In addition to the moral argument against the use of somatic-cell nuclear for the creation of a child ( reproductive cloning
http://www.springer.com/3-540-23372-5
 http://www.springer.com/3-540-23372-5 Chromatin Remodeling Factors and DNA Replication Patrick Varga-Weisz Abstract Chromatin structures have to be precisely duplicated during DNA replication to maintain
http://www.springer.com/3-540-23372-5 Chromatin Remodeling Factors and DNA Replication Patrick Varga-Weisz Abstract Chromatin structures have to be precisely duplicated during DNA replication to maintain
Von Mäusen und Menschen E - 1
 Von Mäusen und Menschen E - 1 Mus musculus: Genetic Portrait of the House Mouse E - 3 Outline Mouse genome Mouse life cycle Transgenic protocols Addition of genes by nuclear injection Removal of genes
Von Mäusen und Menschen E - 1 Mus musculus: Genetic Portrait of the House Mouse E - 3 Outline Mouse genome Mouse life cycle Transgenic protocols Addition of genes by nuclear injection Removal of genes
Chapter 13: Meiosis and Sexual Life Cycles
 Name Period Chapter 13: Meiosis and Sexual Life Cycles Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know.
Name Period Chapter 13: Meiosis and Sexual Life Cycles Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know.
Complex multicellular organisms are produced by cells that switch genes on and off during development.
 Home Control of Gene Expression Gene Regulation Is Necessary? By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Home Control of Gene Expression Gene Regulation Is Necessary? By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Bio EOC Topics for Cell Reproduction: Bio EOC Questions for Cell Reproduction:
 Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
MCB41: Second Midterm Spring 2009
 MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
The world of non-coding RNA. Espen Enerly
 The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
Heredity - Patterns of Inheritance
 Heredity - Patterns of Inheritance Genes and Alleles A. Genes 1. A sequence of nucleotides that codes for a special functional product a. Transfer RNA b. Enzyme c. Structural protein d. Pigments 2. Genes
Heredity - Patterns of Inheritance Genes and Alleles A. Genes 1. A sequence of nucleotides that codes for a special functional product a. Transfer RNA b. Enzyme c. Structural protein d. Pigments 2. Genes
5. The cells of a multicellular organism, other than gametes and the germ cells from which it develops, are known as
 1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
www.njctl.org PSI Biology Mitosis & Meiosis
 Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
CHROMOSOME STRUCTURE CHROMOSOME NUMBERS
 CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells
 Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism )
 Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
GENE REGULATION. Teacher Packet
 AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes?
 Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
Histone modifications. and ChIP. G. Valle - Università di Padova
 Histone modifications and ChIP Histone acetylation It has been shown that the acetylation of lysines is highly dynamic and regulated by the opposing action of two families of enzymes, histone acetyltransferases
Histone modifications and ChIP Histone acetylation It has been shown that the acetylation of lysines is highly dynamic and regulated by the opposing action of two families of enzymes, histone acetyltransferases
Lecture 7 Mitosis & Meiosis
 Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
Biological Sciences Initiative. Human Genome
 Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
Cell Division CELL DIVISION. Mitosis. Designation of Number of Chromosomes. Homologous Chromosomes. Meiosis
 Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Genetics Module B, Anchor 3
 Genetics Module B, Anchor 3 Key Concepts: - An individual s characteristics are determines by factors that are passed from one parental generation to the next. - During gamete formation, the alleles for
Genetics Module B, Anchor 3 Key Concepts: - An individual s characteristics are determines by factors that are passed from one parental generation to the next. - During gamete formation, the alleles for
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Sex for the purposes of this class refers to 4 components
 Sex for the purposes of this class refers to 4 components Gonadal sex Gonads or where gametes are produced by meiosis Somatic sex Somatic cells are cells that undergo mitosis. They can be divided into
Sex for the purposes of this class refers to 4 components Gonadal sex Gonads or where gametes are produced by meiosis Somatic sex Somatic cells are cells that undergo mitosis. They can be divided into
The Human Genome Project
 The Human Genome Project Brief History of the Human Genome Project Physical Chromosome Maps Genetic (or Linkage) Maps DNA Markers Sequencing and Annotating Genomic DNA What Have We learned from the HGP?
The Human Genome Project Brief History of the Human Genome Project Physical Chromosome Maps Genetic (or Linkage) Maps DNA Markers Sequencing and Annotating Genomic DNA What Have We learned from the HGP?
The following chapter is called "Preimplantation Genetic Diagnosis (PGD)".
 Slide 1 Welcome to chapter 9. The following chapter is called "Preimplantation Genetic Diagnosis (PGD)". The author is Dr. Maria Lalioti. Slide 2 The learning objectives of this chapter are: To learn the
Slide 1 Welcome to chapter 9. The following chapter is called "Preimplantation Genetic Diagnosis (PGD)". The author is Dr. Maria Lalioti. Slide 2 The learning objectives of this chapter are: To learn the
Human Genome and Human Genome Project. Louxin Zhang
 Human Genome and Human Genome Project Louxin Zhang A Primer to Genomics Cells are the fundamental working units of every living systems. DNA is made of 4 nucleotide bases. The DNA sequence is the particular
Human Genome and Human Genome Project Louxin Zhang A Primer to Genomics Cells are the fundamental working units of every living systems. DNA is made of 4 nucleotide bases. The DNA sequence is the particular
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics 301 Sample Final Examination Spring 2003
 Genetics 301 Sample Final Examination Spring 2003 50 Multiple Choice Questions-(Choose the best answer) 1. A cross between two true breeding lines one with dark blue flowers and one with bright white flowers
Genetics 301 Sample Final Examination Spring 2003 50 Multiple Choice Questions-(Choose the best answer) 1. A cross between two true breeding lines one with dark blue flowers and one with bright white flowers
An Overview of Cells and Cell Research
 An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
BioBoot Camp Genetics
 BioBoot Camp Genetics BIO.B.1.2.1 Describe how the process of DNA replication results in the transmission and/or conservation of genetic information DNA Replication is the process of DNA being copied before
BioBoot Camp Genetics BIO.B.1.2.1 Describe how the process of DNA replication results in the transmission and/or conservation of genetic information DNA Replication is the process of DNA being copied before
Lecture 3: Mutations
 Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS
 LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS Los Angeles Mission College Biology 3 Name: Date: INTRODUCTION BINARY FISSION: Prokaryotic cells (bacteria) reproduce asexually by binary fission. Bacterial
LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS Los Angeles Mission College Biology 3 Name: Date: INTRODUCTION BINARY FISSION: Prokaryotic cells (bacteria) reproduce asexually by binary fission. Bacterial
Gene mutation and molecular medicine Chapter 15
 Gene mutation and molecular medicine Chapter 15 Lecture Objectives What Are Mutations? How Are DNA Molecules and Mutations Analyzed? How Do Defective Proteins Lead to Diseases? What DNA Changes Lead to
Gene mutation and molecular medicine Chapter 15 Lecture Objectives What Are Mutations? How Are DNA Molecules and Mutations Analyzed? How Do Defective Proteins Lead to Diseases? What DNA Changes Lead to
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
Chapter 5: Organization and Expression of Immunoglobulin Genes
 Chapter 5: Organization and Expression of Immunoglobulin Genes I. Genetic Model Compatible with Ig Structure A. Two models for Ab structure diversity 1. Germ-line theory: maintained that the genome contributed
Chapter 5: Organization and Expression of Immunoglobulin Genes I. Genetic Model Compatible with Ig Structure A. Two models for Ab structure diversity 1. Germ-line theory: maintained that the genome contributed
Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA
 Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Cell Division Mitosis and the Cell Cycle
 Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Name Date Period. 2. When a molecule of double-stranded DNA undergoes replication, it results in
 DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
Chapter 13: Meiosis and Sexual Life Cycles
 Name Period Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know. Define: gene locus gamete male gamete female
Name Period Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know. Define: gene locus gamete male gamete female
Forensic DNA Testing Terminology
 Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Biology 1406 Exam 4 Notes Cell Division and Genetics Ch. 8, 9
 Biology 1406 Exam 4 Notes Cell Division and Genetics Ch. 8, 9 Ch. 8 Cell Division Cells divide to produce new cells must pass genetic information to new cells - What process of DNA allows this? Two types
Biology 1406 Exam 4 Notes Cell Division and Genetics Ch. 8, 9 Ch. 8 Cell Division Cells divide to produce new cells must pass genetic information to new cells - What process of DNA allows this? Two types
Chapter 3. Chapter Outline. Chapter Outline 9/11/10. Heredity and Evolu4on
 Chapter 3 Heredity and Evolu4on Chapter Outline The Cell DNA Structure and Function Cell Division: Mitosis and Meiosis The Genetic Principles Discovered by Mendel Mendelian Inheritance in Humans Misconceptions
Chapter 3 Heredity and Evolu4on Chapter Outline The Cell DNA Structure and Function Cell Division: Mitosis and Meiosis The Genetic Principles Discovered by Mendel Mendelian Inheritance in Humans Misconceptions
Cell Growth and Reproduction Module B, Anchor 1
 Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Control of Gene Expression
 Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Germ cell formation / gametogenesis And Fertilisation
 Developmental Biology BY1101 P. Murphy Lecture 3 The first steps to forming a new organism Descriptive embryology I Germ cell formation / gametogenesis And Fertilisation Why bother with sex? In terms of
Developmental Biology BY1101 P. Murphy Lecture 3 The first steps to forming a new organism Descriptive embryology I Germ cell formation / gametogenesis And Fertilisation Why bother with sex? In terms of
Genetics Test Biology I
 Genetics Test Biology I Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Avery s experiments showed that bacteria are transformed by a. RNA. c. proteins.
Genetics Test Biology I Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Avery s experiments showed that bacteria are transformed by a. RNA. c. proteins.
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15
 Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
Chromosomes, Karyotyping, and Abnormalities (Learning Objectives) Learn the components and parts of a metaphase chromosome.
 Chromosomes, Karyotyping, and Abnormalities (Learning Objectives) Learn the components and parts of a metaphase chromosome. Define the terms karyotype, autosomal and sex chromosomes. Explain how many of
Chromosomes, Karyotyping, and Abnormalities (Learning Objectives) Learn the components and parts of a metaphase chromosome. Define the terms karyotype, autosomal and sex chromosomes. Explain how many of
CHAPTER 10 CELL CYCLE AND CELL DIVISION
 CHAPTER 10 CELL CYCLE AND CELL DIVISION Cell division is an inherent property of living organisms. It is a process in which cells reproduce their own kind. The growth, differentiation, reproduction and
CHAPTER 10 CELL CYCLE AND CELL DIVISION Cell division is an inherent property of living organisms. It is a process in which cells reproduce their own kind. The growth, differentiation, reproduction and
Trasposable elements: P elements
 Trasposable elements: P elements In 1938 Marcus Rhodes provided the first genetic description of an unstable mutation, an allele of a gene required for the production of pigment in maize. This instability
Trasposable elements: P elements In 1938 Marcus Rhodes provided the first genetic description of an unstable mutation, an allele of a gene required for the production of pigment in maize. This instability
Control of Gene Expression
 Control of Gene Expression (Learning Objectives) Explain the role of gene expression is differentiation of function of cells which leads to the emergence of different tissues, organs, and organ systems
Control of Gene Expression (Learning Objectives) Explain the role of gene expression is differentiation of function of cells which leads to the emergence of different tissues, organs, and organ systems
How To Understand How Gene Expression Is Regulated
 What makes cells different from each other? How do cells respond to information from environment? Regulation of: - Transcription - prokaryotes - eukaryotes - mrna splicing - mrna localisation and translation
What makes cells different from each other? How do cells respond to information from environment? Regulation of: - Transcription - prokaryotes - eukaryotes - mrna splicing - mrna localisation and translation
The Making of the Fittest: Evolving Switches, Evolving Bodies
 OVERVIEW MODELING THE REGULATORY SWITCHES OF THE PITX1 GENE IN STICKLEBACK FISH This hands-on activity supports the short film, The Making of the Fittest:, and aims to help students understand eukaryotic
OVERVIEW MODELING THE REGULATORY SWITCHES OF THE PITX1 GENE IN STICKLEBACK FISH This hands-on activity supports the short film, The Making of the Fittest:, and aims to help students understand eukaryotic
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
If and when cancer cells stop dividing, they do so at random points, not at the normal checkpoints in the cell cycle.
 Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Psychoonkology, Sept. 2010 lifestyle factors and epigenetics
 Psychoonkology, Sept. 2010 lifestyle factors and epigenetics Alexander G. Haslberger Dep. für Ernährungswissenschaften Univ. of Vienna Working group: Food, GI-Microbiology, Epigenetics Content Health:
Psychoonkology, Sept. 2010 lifestyle factors and epigenetics Alexander G. Haslberger Dep. für Ernährungswissenschaften Univ. of Vienna Working group: Food, GI-Microbiology, Epigenetics Content Health:
The cell cycle, mitosis and meiosis
 The cell cycle, mitosis and meiosis Learning objective This learning material is about the life cycle of a cell and the series of stages by which genetic materials are duplicated and partitioned to produce
The cell cycle, mitosis and meiosis Learning objective This learning material is about the life cycle of a cell and the series of stages by which genetic materials are duplicated and partitioned to produce
Chapter 9 Patterns of Inheritance
 Bio 100 Patterns of Inheritance 1 Chapter 9 Patterns of Inheritance Modern genetics began with Gregor Mendel s quantitative experiments with pea plants History of Heredity Blending theory of heredity -
Bio 100 Patterns of Inheritance 1 Chapter 9 Patterns of Inheritance Modern genetics began with Gregor Mendel s quantitative experiments with pea plants History of Heredity Blending theory of heredity -
LECTURE 6 Gene Mutation (Chapter 16.1-16.2)
 LECTURE 6 Gene Mutation (Chapter 16.1-16.2) 1 Mutation: A permanent change in the genetic material that can be passed from parent to offspring. Mutant (genotype): An organism whose DNA differs from the
LECTURE 6 Gene Mutation (Chapter 16.1-16.2) 1 Mutation: A permanent change in the genetic material that can be passed from parent to offspring. Mutant (genotype): An organism whose DNA differs from the
EPIGENETICS DNA and Histone Model
 EPIGENETICS ABSTRACT A 3-D cut-and-paste model depicting how histone, acetyl and methyl molecules control access to DNA and affect gene expression. LOGISTICS TIME REQUIRED LEARNING OBJECTIVES DNA is coiled
EPIGENETICS ABSTRACT A 3-D cut-and-paste model depicting how histone, acetyl and methyl molecules control access to DNA and affect gene expression. LOGISTICS TIME REQUIRED LEARNING OBJECTIVES DNA is coiled
DNA Fingerprinting. Unless they are identical twins, individuals have unique DNA
 DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
Cancer SBL101. James Gomes School of Biological Sciences Indian Institute of Technology Delhi
 Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs)
 Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
HUMAN CHROMOSOMES. Using this criterion, human chromosomes are divided in: metacentric, submetacentric, and acrocentric.
 HUMAN CHROMOSOMES Normal human somatic cells contain a diploid number of chromosomes (2n=46), so there are 23 pairs of chromosomes: - 22 pairs are identical in man and women and are called autosomes; -
HUMAN CHROMOSOMES Normal human somatic cells contain a diploid number of chromosomes (2n=46), so there are 23 pairs of chromosomes: - 22 pairs are identical in man and women and are called autosomes; -
Lecture 2: Mitosis and meiosis
 Lecture 2: Mitosis and meiosis 1. Chromosomes 2. Diploid life cycle 3. Cell cycle 4. Mitosis 5. Meiosis 6. Parallel behavior of genes and chromosomes Basic morphology of chromosomes telomere short arm
Lecture 2: Mitosis and meiosis 1. Chromosomes 2. Diploid life cycle 3. Cell cycle 4. Mitosis 5. Meiosis 6. Parallel behavior of genes and chromosomes Basic morphology of chromosomes telomere short arm
BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis
 BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis Introduction - Fields of Genetics To answer the following question, review the three traditional subdivisions of
BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis Introduction - Fields of Genetics To answer the following question, review the three traditional subdivisions of
How Cancer Begins???????? Chithra Manikandan Nov 2009
 Cancer Cancer is one of the most common diseases in the developed world: 1 in 4 deaths are due to cancer 1 in 17 deaths are due to lung cancer Lung cancer is the most common cancer in men Breast cancer
Cancer Cancer is one of the most common diseases in the developed world: 1 in 4 deaths are due to cancer 1 in 17 deaths are due to lung cancer Lung cancer is the most common cancer in men Breast cancer
LESSON 3.5 WORKBOOK. How do cancer cells evolve? Workbook Lesson 3.5
 LESSON 3.5 WORKBOOK How do cancer cells evolve? In this unit we have learned how normal cells can be transformed so that they stop behaving as part of a tissue community and become unresponsive to regulation.
LESSON 3.5 WORKBOOK How do cancer cells evolve? In this unit we have learned how normal cells can be transformed so that they stop behaving as part of a tissue community and become unresponsive to regulation.
Name: 4. A typical phenotypic ratio for a dihybrid cross is a) 9:1 b) 3:4 c) 9:3:3:1 d) 1:2:1:2:1 e) 6:3:3:6
 Name: Multiple-choice section Choose the answer which best completes each of the following statements or answers the following questions and so make your tutor happy! 1. Which of the following conclusions
Name: Multiple-choice section Choose the answer which best completes each of the following statements or answers the following questions and so make your tutor happy! 1. Which of the following conclusions
Genetic Mutations. Indicator 4.8: Compare the consequences of mutations in body cells with those in gametes.
 Genetic Mutations Indicator 4.8: Compare the consequences of mutations in body cells with those in gametes. Agenda Warm UP: What is a mutation? Body cell? Gamete? Notes on Mutations Karyotype Web Activity
Genetic Mutations Indicator 4.8: Compare the consequences of mutations in body cells with those in gametes. Agenda Warm UP: What is a mutation? Body cell? Gamete? Notes on Mutations Karyotype Web Activity
Chapter 8: Variation in Chromosome Structure and Number
 Chapter 8: Variation in Chromosome Structure and Number Student Learning Objectives Upon completion of this chapter you should be able to: 1. Know the principles and terminology associated with variations
Chapter 8: Variation in Chromosome Structure and Number Student Learning Objectives Upon completion of this chapter you should be able to: 1. Know the principles and terminology associated with variations
Appendix C DNA Replication & Mitosis
 K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257
 CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
REI Pearls: Pitfalls of Genetic Testing in Miscarriage
 The Skinny: Genetic testing of miscarriage tissue is controversial and some people question if testing is helpful or not. This summary will: 1) outline the arguments for and against genetic testing; 2)
The Skinny: Genetic testing of miscarriage tissue is controversial and some people question if testing is helpful or not. This summary will: 1) outline the arguments for and against genetic testing; 2)
Antibody Function & Structure
 Antibody Function & Structure Specifically bind to antigens in both the recognition phase (cellular receptors) and during the effector phase (synthesis and secretion) of humoral immunity Serology: the
Antibody Function & Structure Specifically bind to antigens in both the recognition phase (cellular receptors) and during the effector phase (synthesis and secretion) of humoral immunity Serology: the
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Structure and Function of DNA
 Structure and Function of DNA DNA and RNA Structure DNA and RNA are nucleic acids. They consist of chemical units called nucleotides. The nucleotides are joined by a sugar-phosphate backbone. The four
Structure and Function of DNA DNA and RNA Structure DNA and RNA are nucleic acids. They consist of chemical units called nucleotides. The nucleotides are joined by a sugar-phosphate backbone. The four
Name: Class: Date: ID: A
 Name: Class: _ Date: _ Meiosis Quiz 1. (1 point) A kidney cell is an example of which type of cell? a. sex cell b. germ cell c. somatic cell d. haploid cell 2. (1 point) How many chromosomes are in a human
Name: Class: _ Date: _ Meiosis Quiz 1. (1 point) A kidney cell is an example of which type of cell? a. sex cell b. germ cell c. somatic cell d. haploid cell 2. (1 point) How many chromosomes are in a human
CHROMOSOMES AND INHERITANCE
 SECTION 12-1 REVIEW CHROMOSOMES AND INHERITANCE VOCABULARY REVIEW Distinguish between the terms in each of the following pairs of terms. 1. sex chromosome, autosome 2. germ-cell mutation, somatic-cell
SECTION 12-1 REVIEW CHROMOSOMES AND INHERITANCE VOCABULARY REVIEW Distinguish between the terms in each of the following pairs of terms. 1. sex chromosome, autosome 2. germ-cell mutation, somatic-cell
MUTATION, DNA REPAIR AND CANCER
 MUTATION, DNA REPAIR AND CANCER 1 Mutation A heritable change in the genetic material Essential to the continuity of life Source of variation for natural selection New mutations are more likely to be harmful
MUTATION, DNA REPAIR AND CANCER 1 Mutation A heritable change in the genetic material Essential to the continuity of life Source of variation for natural selection New mutations are more likely to be harmful
The Somatic Cell Cycle
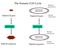 The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
Name Class Date. Figure 13 1. 2. Which nucleotide in Figure 13 1 indicates the nucleic acid above is RNA? a. uracil c. cytosine b. guanine d.
 13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
Lecture 11 The Cell Cycle and Mitosis
 Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
The Nucleus: DNA, Chromatin And Chromosomes
 The Nucleus: DNA, Chromatin And Chromosomes Professor Alfred Cuschieri Department of Anatomy, University of Malta. Objectives By the end of this unit the student should be able to: 1. List the major structural
The Nucleus: DNA, Chromatin And Chromosomes Professor Alfred Cuschieri Department of Anatomy, University of Malta. Objectives By the end of this unit the student should be able to: 1. List the major structural
Gene Therapy and Genetic Counseling. Chapter 20
 Gene Therapy and Genetic Counseling Chapter 20 What is Gene Therapy? Treating a disease by replacing, manipulating or supplementing a gene The act of changing an individual s DNA sequence to fix a non-functional
Gene Therapy and Genetic Counseling Chapter 20 What is Gene Therapy? Treating a disease by replacing, manipulating or supplementing a gene The act of changing an individual s DNA sequence to fix a non-functional
Transfection-Transfer of non-viral genetic material into eukaryotic cells. Infection/ Transduction- Transfer of viral genetic material into cells.
 Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
What is Cancer? Cancer is a genetic disease: Cancer typically involves a change in gene expression/function:
 Cancer is a genetic disease: Inherited cancer Sporadic cancer What is Cancer? Cancer typically involves a change in gene expression/function: Qualitative change Quantitative change Any cancer causing genetic
Cancer is a genetic disease: Inherited cancer Sporadic cancer What is Cancer? Cancer typically involves a change in gene expression/function: Qualitative change Quantitative change Any cancer causing genetic
