High-throughput RNAi screening by time-lapse imaging of live human cells
|
|
|
- Edith Mitchell
- 8 years ago
- Views:
Transcription
1 High-throughput RNAi screening by time-lapse imaging of live human cells 26 Nature Publishing Group h ttp:// e method s Beate Neumann 1,4, Michael Held 1,4, Urban Liebel 1,4, Holger Erfle 1,4, Phill Rogers 1, Rainer Pepperkok 2 & Jan Ellenberg 2,3 RNA interference (RNAi) is a powerful tool to study gene function in cultured cells. Transfected cell microarrays in principle allow high-throughput phenotypic analysis after gene knockdown by microscopy. But bottlenecks in imaging and data analysis have limited such high-content screens to endpoint assays in fixed cells and determination of global parameters such as viability. Here we have overcome these limitations and developed an automated platform for high-content RNAi screening by time-lapse fluorescence microscopy of live HeLa cells expressing histone-gfp to report on chromosome segregation and structure. We automated all steps, including printing transfection-ready small interfering RNA (sirna) microarrays, fluorescence imaging and computational phenotyping of digital images, in a high-throughput workflow. We validated this method in a pilot screen assaying cell division and delivered a sensitive, time-resolved phenoprint for each of the 49 endogenous genes we suppressed. This modular platform is scalable and makes the power of time-lapse microscopy available for genome-wide RNAi screens. After the completion of the human genome sequencing project 1, the task of functional genomics is to discover protein function genome-wide. Currently, RNAi is the method of choice to study loss-of-function phenotypes in human cells by specifically suppressing the expression of virtually any desired protein-coding gene. Indeed, several RNAi screens in human cells have already been reported 2. These time-consuming and expensive primary screens have typically been based on endpoint assays of cells transfected in microtiter plates 2. This allowed reasonable throughput but limited the information content of the phenotypic readout. This is problematic for two reasons. Endpoint assays capture only phenotypes for genes whose protein products are sufficiently reduced at the time the assay is performed, which depends on the stability of the targeted gene product. In a genome-wide screen this time varies between one and several days and endpoint assays are therefore typically scored late after RNAi transfection to detect as many phenotypes as possible. At late time points, however, most assays suffer from specificity problems, because for rapidly turned over proteins they can no longer differentiate between primary consequences of knockdown and secondary effects, and therefore lead to misinterpretation of phenotypes. In principle, these limitations of RNAi screening can be overcome using the power of time-resolved live cell imaging, a strategy that has been used very successfully but with an enormous labor effort in the model system Caenorhabditis elegans 6. Limitations in manual data acquisition of live movies as well as manual, and thus inherently biased and nonquantitative, data annotation have so far precluded the use of live-cell imaging for RNAi screening in vertebrate cell systems. Here we have overcome these bottlenecks by taking advantage of the miniaturized RNAi delivery offered by transfected cell microarrays, in which individually spotted sirna transfection mixes are directly taken up from the solid phase by cells seeded on top of the array 7,8. By spotting sirna microarrays directly in live-cell imaging chambers, we were able to perform time-lapse microscopy of HeLa cells on the arrays. By massively increasing the throughput of fluorescence imaging and developing computerized analysis of the phenotypes by digital image processing, we established a fully automated high-throughput and highcontent workflow of RNAi screening by time-lapse imaging (Fig. 1). We validated this method in a pilot experiment screening 49 endogenous human genes with a live-cell assay for chromosome segregation and structure in HeLa cells stably expressing the core histone 2B tagged with GFP. This platform is applicable to many assays that can be scored by in vivo (or fixed-cell) imaging, and is directly scalable to genome-wide RNAi screens. RESULTS sirna validation We chose to target 49 genes (Supplementary Table 1 online) known or predicted to have a function in chromosome segregation or nuclear structure by one chemically synthesized sirna each. For 43 of these sirnas, mrna knockdown efficiency could be measured in transfected HeLa cells by quantitative reverse-transcriptase PCR (qrt-pcr). We found that 74% (32) of the sirnas suppressed the mrna of the targeted gene by more than 7%, 88% (38) by at least 6% and all sirnas showed partial knockdown (Supplementary Fig. 1 online). In duplicate transfections of all 49 gene-specific sirnas and one scrambled sirna (randomized 1 MitoCheck Project Group, 2 Cell Biology/Biophysics and 3 Gene Expression Programmes, European Molecular Biology Laboratory, Meyerhofstrasse 1, D Heidelberg, Germany. 4 These authors contributed equally to this work. Correspondence should be addressed to J.E. (jan.ellenberg@embl.de). RECEIVED 6 JANUARY; ACCEPTED 22 MARCH; PUBLISHED ONLINE 2 APRIL 26; DOI:1.138/NMETH876 NATURE METHODS VOL.3 NO. MAY 26 38
2 26 Nature Publishing Group h ttp:// e method s Preparation of sirna transfection mix Spotting of sirna microarray Automated time-lapse image acquisition Automated image analysis 1 h 6 h 48 replicates t 44 h 9 µm 4 µm Cell seeding ~37, images ~1 Gb per microarray Figure 1 Workflow of high-throughput RNAi screening by time-lapse imaging. Flowchart of the steps, including time lines of each process based on one live-cell microarray containing 384 sirnas. RNA sequence not targeting any human gene), we scored phenotypes manually by fluorescence microscopy of HeLa cells stably expressing H2B-GFP (see below). For 8% (42) of the targeted genes, the sirna caused reproducible defects in nuclear structure or chromosome segregation, whereas cells transfected with the scrambled sirna had no detectable phenotype (data not shown). We thus estimated the overall hit rate of this sirna design as 74 8% and decided to use the validated sirnas (Supplementary Table 1) to develop the entire high-throughput workflow of live-cell RNAi screening by time-lapse imaging. sirna microarrays for live cell imaging Application of live-cell imaging to genome-wide RNAi screens requires increasing the throughput of fluorescence microscopy by miniaturizing and parallelizing the sirna transfection in a format compatible with imaging. We printed automatically prepared transfection cocktails for each of the sirnas directly in onewell live cell imaging chambers with coverslip bottoms 8,9 (Fig. 1). A 4-mm spot size and 9-mm inter-spot distance allowed us to observe B cells growing on each spot in a single field of view using a 1 microscope objective, and provided 384 spots per imaging chamber. The transfection efficiency of HeLa cells used in this study was 99% judged by uptake of fluorescently labeled scrambled sirna (Supplementary Protocol online). In addition, A49, RPE and U2OS cell lines as well as primary skin fibroblasts could be transfected with more than 9% transfection efficiency (Supplementary Protocol). sirna uptake in HeLa cells on microarrays led to depletion of the targeted gene product as judged by immunofluorescence for two proteins, TPX2 and COPB. In both cases, specific protein staining was abolished in all (TPX2) or the majority (for the much more stable protein COPB) of HeLa cells growing on the sirna spot (Supplementary Protocol). The sirna microarrays could be stored for more than 7 months without loss of transfection efficiency as judged by B7% of prometaphase-arrested HeLa cells after knockdown of PLK1 (Supplementary Protocol). This has the advantage that the automated production of microarrays (Supplementary Protocol) can be uncoupled from the phenotypic analysis of the cells by imaging, allowing the use of replicates of identical microarrays over long time intervals to control the reproducibility of cell-based assays or the multiplexing of different assays on the same set of targeted genes. Time-lapse imaging of chromosomes to assay cell division The biological problem we investigated in the pilot screen was chromosome segregation during cell division. Fluorescent labeling of chromosomes allows dynamic processes such as mitosis to be phenotyped in great detail by fluorescence microscopy 1. Mitosis is a rare event with typically less than % of a population of logarithmically growing HeLa cells undergoing division at one time. Furthermore, mitotic phenotypes such as chromosome segregation defects can lead to programmed cell death 11 and are often difficult to interpret in endpoint assays. We therefore decided to implement a time-lapse imaging assay for mitosis using a monoclonal HeLa cell line stably expressing the core histone 2B tagged with GFP 12, which highlights chromosomes and nuclei in all cell cycle stages. We imaged cells growing on scrambled sirna every 3 min for 44 h starting 2 h after seeding, long enough to observe at least one division for each of the B cells transfected on the spot. This provided the minimal temporal resolution to reliably score mitotic phases based on the changes in chromosome morphology (Fig. 2a). By starting the imaging 2 h after plating cells on sirna microarrays, the total observation period allowed the scoring of early as well as late phenotypes (Figs. 2b e and 3a; and below). High-throughput time-lapse microscopy of cell microarrays After establishing the assay conditions for a single spot, the next step was to image as many microarray spots as possible within a time lapse of 3 min for large-scale screening applications. By further developing previously established hardware and software methods 13, we implemented a fully automated and 37 1Cincubated wide-field epifluorescence screening microscope. After an initial autofocus of the entire microarray, we acquired single fluorescence images of each spot sequentially throughout the array with minimal illumination. Using real-time control of the fully motorized microscope hardware we could revisit up to 1,36 positions (four sirna microarrays) every 3 min. To our knowledge, this is the first time that such a throughput in live-cell imaging has been described and makes live-cell imaging feasible for genome-wide RNAi screening. We used this system to produce two-day time-lapse movies of HeLa H2B-GFP cells on triplicates of a custom microarray that contained one sirna spot for each of the 49 endogenous genes and three spots of a scrambled sirna as negative controls (data not shown). Notably, cells transfected by the scrambled sirna proliferated normally throughout the observation period (Supplementary Protocol), showing no toxicity by the repeated illumination and prolonged incubation on the screening microscope. Automatic and quantitative phenotype annotation After acquisition, the frames comprising each movie (89 per sirna) had to be scored for mitotic phenotypes. We developed an automatic method also for phenotyping for two reasons. First, 386 VOL.3 NO. MAY 26 NATURE METHODS
3 ARTICLES 26 Nature Publishing Group h ttp:// e method s a b c d e siincenp h Mitotic index siincenp Mitotic index sisyne2 Mitotic index 3. h 6. h siplk h. h. h 1. h 1. h 2. h 2. h sisyne2 Interphase Apoptosis 28. h Shape index Shape index Shape index Mitosis Shape 34. h 7. h only automatic annotation by image processing allows the quantitative, unbiased and reproducible analysis of phenotypes that can later be easily parameterized and statistically evaluated to compare phenotypic signatures of genes. Second, because genome-wide RNAi screens would produce tens of thousands of movies, it was clear that annotation would become the rate-limiting step in the workflow for large-scale screening experiments. The software we developed for automatic phenotyping proceeds in three major steps: (i) location of chromosome sets within single cells for each frame of the movie, (ii) classification of the chromosomes according to their morphology and (iii) detection of cell-cycle phenotypes based on the classification results for the entire movie. For the first step, we defined the boundary of fluorescent chromosomes by segmentation using optimized local adaptive thresholding 14, which identified both interphase nuclei and mitotic chromosome sets with more than 99% accuracy compared to manual image analysis (data not shown). In the second step we computed the texture and shape properties of each segmented set of chromosomes based on our past experience in identifying subcellular localization patterns 1 and used them to assign the cell to a biological class. The software distinguishes between interphase, mitosis, apoptosis and shape Max. penetrance Automatic nuclei classification siplk1 21. h 3. h 42. h Apoptotic index Apoptotic index Apoptotic index Figure 2 Imaging of cell microarrays and automatic phenotype analysis. (a) HeLa H2B-EGFP cells on a scrambled sirna spot (white circle represents spot diameter; left). Division of two cells (right, top row). Interphase and mitotic chromosomes recognized by the automatic classification are marked as indicated (right, bottom row; Supplementary Video 1). (b) Mitotic RNAi phenotypes followed by either binucleated arrested cells (INCENP sirna), or by apoptosis-like SYNE2 sirna and PLK1 sirna. Interphase, mitotic, apoptotic and binucleated ( shape ) cells are automatically recognized and labeled in green, yellow, red and blue, respectively (Supplementary Videos 2 4). (c e) Indices of mitosis, shape and apoptosis classes for three scrambled sirnas and one INCENP sirna movie are plotted over time. Data was smoothed and fitted with a thirdorder polygon by local regression (solid line). Divergence between the average scrambled sirna and the INCENP sirna was computed for all 89 time points. The earliest statistically significant divergence is marked as the first phenotype appearance (arrows) and the maximal divergence as the phenotype penetrance (double-headed arrows) for each index. (d,e) The same analysis is shown for exemplary SYNE2 (d) and PLK1 (e) sirna movies. cells by machine-learning using multiclass support vector machines (SVM; see Methods for details). The classification was 97% accurate compared with manual annotation. We carried out segmentation and classification analysis for two normal cell divisions on a scrambled sirna spot (Fig. 2a). The software correctly assigned interphase and mitotic chromosomes, the latter including all stages of M phase from late prophase to telophase. The correct identification of apoptotic cells was illustrated for the two distinct cell death morphologies resulting from suppression of the nuclear envelope proteins SYNE2 or PLK1 (Fig. 2b). By contrast, knockdown of the inner centromere protein INCENP resulted in bi- and multinucleated cells as a result of segregation defects that were correctly assigned to the shape class (Fig. 2b). Examples of the automatic classification for three additional gene knockdown experiments are shown (Supplementary Fig. 2 online). Finally we analyzed the classification results for an entire movie to decide whether the suppression of the targeted gene caused a chromosome segregation or nuclear structure phenotype. The software computes the number of cells in each biological class relative to the total number of cells within the spot area of a movie. We generated mitotic, shape and apoptotic index plots for the PLK1 knockdown experiment (Fig. 2e). We then compared the percentage of cells in a biological class to the three scrambled sirnas on the same cell microarray. If any of the three classes showed a statistically significant difference to the controls, the earliest time at which the difference became significant is defined as the phenotype appearance and the maximum difference as the phenotype penetrance. For example, suppression of the essential NATURE METHODS VOL.3 NO. MAY
4 26 Nature Publishing Group h ttp:// e method s a b c Figure 3 Clustering of genes by time-resolved phenoprints. (a) Heat map of Phenotype penetrance Mitosis None Weak Shape Apoptosis Penetrance: Strong First phenotype appearance Phenotype classes Mitosis Shape Apoptosis NUP17 NUMA1 NUP13 CBX3 SEH1L CCNB2 RANBP2 CBX1 CDC27 CDH1 CBX H2AFY SMC4L1 ANAPC11 ANAPC1 LMNA CENPH SUV39H2 EGFR POM121 FLT3 RAD21 CENPA PLK1 KIFF11 AURKB SYNE2 TPX2 CAPG CDC14A COPB TOP2A ANAPC2 MAD2L1 H2AFY2 CDC2C SEC13L1 NUP62 NUP37 ANAPC1 ANAPC CDK7 MAP2K4 KIF23 INCENP CDC16 LBR HIST1H1E H3F3A Scrambled Phenotype clustering mitotic kinase PLK1 led to prometaphase arrest 11 and consequently the mitotic index was elevated to 2% (compared to o% in the controls) at the beginning of the movie, and the first appearance of the mitosis phenotype was thus calculated as 2 h after seeding (Fig. 2e). The maximum mitotic index was reached at 27 h yielding a phenotype penetrance of 48% (Fig. 2e). The apoptotic index plot of the same movie reveals the long term consequence of the prometaphase arrest. Corresponding to the decrease in mitotic index after 27 h, the apoptotic index increased to a maximum penetrance of 44% at 4 h (Fig. 2e). As expected, the shape index indicative of binucleated cells was not affected, as cells lacking PLK1 do not initiate chromosome segregation but enter apoptosis directly from prometaphase 11 (Fig. 2e). We carried out an analogous analysis for INCENP and SYNE2 knockdown (Fig. 2c,d). In summary, time-resolved automatic phenotyping provides information on when the suppressed gene product becomes limiting and allowed us to distinguish between primary (for example, prometaphase arrest) and secondary (for example, apoptosis) phenotypes. Time-resolved phenoprints can cluster genes by phenotype We next used the quantitative image processing software to detect phenotypes in the entire pilot screen dataset. We benchmarked the automatic phenotyping against manual annotation of both mitotic as well as apoptotic phenotypes in the same dataset, which we the phenotype penetrances for the three phenotypic classes mitosis, shape and apoptosis automatically computed for all 49 targeted genes of the pilot screen and the scrambled sirna. Shown are mean penetrances of three movies for each sirna. The dynamic range of phenotype penetrance displayed in pseudocolor was scaled for every phenotypic class individually according to the maximal penetrance of each class, mitosis 3.3% (KIF11), shape 24.2% (INCENP) and apoptosis 37.% (COPB). (b) First appearance of a phenotype in each of the three classes mitosis (yellow), shape (blue) or apoptosis (red) plotted over time after cell seeding is shown for all 49 targeted genes and the scrambled sirna. (c) Genes were hierarchically clustered by penetrance and relative phenotype sequence shown in b. The dendrogram was reduced to four branches, yielding groups of genes related by their timeresolved phenotypic fingerprint. assumed to be 1% correct. For mitotic phenotypes, automatic and manual annotation agreed for 86% (42) of the targeted genes, with a false-positive rate of 14% (7) and no false negatives. Similarly for apoptosis, the automatic method had 88% (43) agreement with 1% () false positives and 2% (1) false negatives. Thus the automatic phenotyping was highly accurate and delivered unbiased and quantitative parameters about phenotype appearance and penetrance that allowed us to cluster this set of candidate genes according to phenotypic similarities. Genes were clustered by the kind of phenotype they showed (increased mitotic, shape or apoptotic index; Fig. 3a), by the mean maximal penetrance of each phenotype (ranging from 2 38%; Fig. 3a) and by the relative order in which multiple phenotypes, if present, appeared (Fig. 3b). This multiparameter clustering (Fig. 3c) revealed groups of genes related by their time-resolved phenotypic signature, or phenoprint. For example, a large group of genes that initially caused a mitotic phenotype followed later by apoptosis is represented by PLK1 (Fig. 2b,e) and CDC16 (Supplementary Fig. 2). Other groups of genes produced only apoptotic phenotypes (for example, NUMA1, Supplementary Fig. 2) or complex phenotypes in all classes, for example the gene SYNE2, encoding a nuclear envelope protein (Fig. 2d). It should be noted that the purpose of this study was to develop and apply a fully automated method for high-throughput RNAi screening by time-lapse imaging of live human cells and not to fully define the biological phenotype of each single gene in our pilot set. Because we only used one sirna per gene, we cannot exclude that some phenotypes are weak hypomorphs or may result at least in part from off-target effects. But the high specificity of our mitosis assay and the very good correspondence of our data with previously published phenotypes (see below) make it unlikely that off-target effects had a major contribution to the observed phenotypes. DISCUSSION RNAi screening by time-lapse imaging of living cells has key advantages over traditional endpoint assays. Most importantly, it allows one to discriminate the primary phenotype caused by gene suppression from secondary cellular responses. In addition it can detect early as well as late phenotypes resulting from different stabilities of the targeted gene product. Time-lapse imaging is also very sensitive and can detect hypomorphic phenotypes such as mitotic delays rather than arrest, which can occur with partial depletion by RNAi. But phenotypes caused by partial depletion can be very different from those caused by complete removal of a protein 16, and therefore a hit in a primary screening method as the 388 VOL.3 NO. MAY 26 NATURE METHODS
5 26 Nature Publishing Group h ttp:// e method s one presented here should be followed up by secondary experiments in which the depletion level of individual proteins can be quantitated. The three phenotypes shown (Fig. 2b) would all be misinterpreted or missed in a typical endpoint determination 48 h after sirna transfection. SYNE2 would already show more apoptotic than mitotic cells; PLK1 would be wrongly scored as an apoptotic phenotype, which is the secondary consequence of the earlier prometaphase arrest. It is worth noting that the HeLa cells we used in this study are known to have reduced p3 levels, which has recently been reported to enhance the apoptosis response caused by depletion of mitotic genes such as PLK1 (ref. 17). It is therefore likely that the high percentage of apoptotic phenotypes is at least in part due to this cell type. INCENP would be scored correctly as a binucleated cell phenotype, but the segregation defects causing this phenotype would not be detected because they already occur 2 h after seeding. The wide range of first phenotype appearances in our pilot screen 2 44 h after seeding (Fig. 3b) demonstrates the need to sample phenotypes at different times. In addition to classification and penetrance of the primary phenotype itself, the temporal sequence of phenotypes caused by suppression of a gene gives important clues about its function and can therefore be used to cluster genes into groups (Fig. 3b,c). We used only the relative timing of phenotypes for hierarchical clustering because the first appearance depends not only on stability of the targeted protein but also on the efficiency of the sirna knockdown. Nevertheless we expect that in genome-wide screens the first appearance will provide valuable, albeit indirect, clues about average protein stability, as we find that genes encoding shortlived cyclically regulated proteins, such as PLK1, AURKB and TPX2, cause phenotypes early, whereas those encoding stable proteins, such as CENPA and LMNA, cause late phenotypes (Fig. 3b). The automatic phenotyping software we developed matched manual annotation for 86 88% of the genes and correctly classified many phenotypes known from the literature (for example, mitotic phenotypes PLK1 (ref. 11), TPX2 (ref. 18), KIF11 (ref. ) and KIF23 (ref. 19)). Machine learning, however, also has limitations, because it will only correctly recognize phenotypes that were used to train the classification software. This explains the up to 14% falsepositive rate of the automatic annotation we observed in this pilot screen. An example of a false-positive mitotic recognition is the gene NUMA1 (Fig. 3a,b). In conclusion, we report for the first time an integrated highthroughput workflow that combines two powerful methods of functional genomics and cell biology: RNAi screening and live-cell time-lapse microscopy. This method delivers detailed time-resolved and quantitative phenoprints of human genes and is directly scalable to genome-wide screens. This method is not limited to HeLa cells, and can be applied to adherent cell lines derived from different tissues (Supplementary Protocol). It is worth noting, however, that a different cell line will require adaptation of the imaging conditions (slower cell cycle, for example, in primary skin fibroblasts, would require longer time-lapse sequences) and image processing (a different nuclear morphology, for example, in RPE cells, would have to be retrained for accurate classification). In general, this method can be adapted to any assay that can be detected by fluorescence microscopy and scored by computerized image processing, most prominently the host of GFP reporters available in the cell biology community for diverse cellular functions. METHODS High-throughput time-lapse imaging. We acquired images with an automated epifluorescence microscope (IX-81; Olympus Europe) as described 13 with the following new developments for time-lapse acquisition and increased throughput. We implemented stabilized light sources (MT2; Olympus Biosystems), firewire cameras (DB-H1, 1,3 1,24, Olympus Biosystems) and an in house modified version of the ScanR software (now commercially available with all new developments from Olympus Biosystems), EMBL environmental microscope incubator (European Molecular Biology Laboratory, GP 16) as well as a new objective (Plan, 1; numerical aperture (NA),.4; Olympus Europe) and filter sets for GFP (Chroma Inc.). We used an image-based autofocussing procedure 13 to focus on the maximum number of healthy cells (scoring size, intensity, contrast) in a field of view. The focus z coordinates of the transfected cell microarray were saved during the first round of imaging. The temperature stability of the incubated system allowed us to reuse the saved focus map for 44 h without further autofocussing between the 3-min imaging intervals. As cells needed on average B1 h to go through mitosis, we obtained B2 mitotic images per cell that enabled us to detect delays Z3 min, or a mitotic index change from % to 7.%. Higher time resolution would increase the sensitivity to subtle delays but reduce throughput and increase overall data volume. We reduced illumination of the specimen to the minimum necessary for sufficient signal-to-noise ratio for automated phenotyping (see below) with exposure times of 18 ms (for GFP). Stage movements thus limited throughput. Data was losslessly compressed on the fly and continuously saved on a Tb network-attached storage. We connected microscopes via a 2 Gbit/s network to the networkattached storage to sustain uninterrupted data flow. Automated phenotyping by image processing. We segmented chromosome sets by an optimized method of local adaptive thresholding, reducing computational costs by 9% compared to the original implementation 2. We extracted numerical features of shape and texture from each segmented set of chromosomes resulting in a vector of 214 features each. Automatic classification into biologically defined classes was by machine-learning using multiclass SVMs 14,1,21,22. Classes were interphase, mitotic, apoptotic (several cell death phenotypes with supercompacted or fragmented nuclei including, but not limited to, apoptosis), shape (nuclei of abnormal shape including, but not limited to, a high percentage of binucleated cells), and artifact (containing other objects). Feature selection was on B2 manually annotated examples per class. We trained the SVM classifier on the 4 bestranked features of all B1, manually labeled cells. The classifier achieved an overall accuracy of 97% on all samples measured by fivefold cross-validation. In each single frame of time-lapse movies for each spotting position, the classifier then automatically assigned cells to the classes (Fig. 2a) resulting in kinetic plots of the fraction of a given class relative to the total number of cells for the entire movie (class index; Fig. 2c e). Three classes mitosis, shape and apoptosis were mined individually for the presence of a cell-cycle phenotype by statistical analysis. To increase the robustness and to cope with occasional false classifications, the data of the knockdown experiment of interest was first smoothed by local regression, which provides a fitted value (Fig. 2e) and a confidence interval (data not shown) for the fit at each time point. We then compared NATURE METHODS VOL.3 NO. MAY
6 26 Nature Publishing Group h ttp:// e method s the fitted value to the mean value of the three regression fitted control experiments on the same microarray, taking into consideration the standard deviation (s.d.) of the controls. If the divergence between the class index of the RNAi experiment and the mean of the negative controls on the same cell array was above a certain level of significance ( mitosis 2 s.d., apoptosis 3 s.d., shape 2 s.d.), the earliest difference was calculated as the first appearance of the phenotype and the maximum difference as its maximal penetrance in the cell population (Fig. 2c e). Besides statistical significance, we also assessed biological significance by requiring each phenotypic class index to be above a certain absolute threshold ( mitosis and apoptosis at least 1%, shape at least 7%), which we determined empirically to remove false positives resulting from noise in the data (experimental and classification errors) that could not be removed by local regression smoothing. We excluded mitotic hits with an onset of more than 6 h after apoptosis based on apriori biological knowledge. We analyzed triplicates separately and averaged the results. We applied hierarchical clustering on relative phenotype sequence and maximum penetrance for these three phenotypic classes to the entire pilot screen dataset (Fig. 3c). We performed hierarchical cluster analysis on a dissimilarity structure, which was generated by the Euclidean distance between hextuples containing the maximum penetrance and the relative phenotype appearance for the three classes mitosis, shape and apoptosis. As there was no apriorireason to emphasize the categories penetrance and appearance or the classes mitosis, shape and apoptosis, all values of the hextuple were equally weighted for clustering. The automatic phenotyping software, which is coded in C++ and Python, is available upon request for academic researchers. Additional methods. See Supplementary Table 1 for sirna sequences and Supplementary Table 2 for reverse-transcriptase PCR primers used in this study; the Supplementary Protocol for description of the sirna transfection methodology and its performance in different cell lines; Supplementary Methods for details of qrt-pcr; and Supplementary Videos 1 7 for sample images of selected phenotypes. Note: Supplementary information is available on the Nature Methods website. ACKNOWLEDGMENTS We thank S. Narumiya (Kyoto University, Kyoto) and T. Hirota (Institute of Molecular Pathology; IMP; Vienna) for HeLa Kyoto cells; W. Huber (European Bioinformatics Institute; EBI; Hinxton) for advice on statistical analysis of kinetic data; O. Gruss (Zentrum für Molekulare Biologie Heidelberg; ZNBH; Heidelberg) for TPX2 antibody; J.-M. Peters (IMP; Vienna) for RPE cells; I. Hoffmann (Deutsches Krebsforschungszentrum; DKFZ; Heidelberg) for U2OS cells; H. Runz (Univ. Heidelberg) for primary human fibroblasts; Chroma Inc. for providing customized emission filter sets free of charge; EMBL s IT Services group (B. Kindler, M. Hemberger, R. Lück) for support; Olympus Biosystems, Hamilton and Bio-Rad for continuous support; Cenix BioScience GmbH for sirna design and for providing the A49 cells; and Ambion Europe, Ltd. for providing sirnas for validation. This project was funded by grants to J.E. within the MitoCheck consortium by the European Commission (FP6-3464) as well as in part by the Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (NGFN) (NGFN-2 SMP-RNAi, FKZ1GR43 to J.E. and NGFN-2 SMP-Cell FKZ1GR423, NGFN-1 FKZ1GR11, FKZ1KW13 to R.P.). COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests. Published online at Reprints and permissions information is available online at 1. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, (24). 2. Kittler, R. et al. An endoribonuclease-prepared sirna screen in human cells identifies genes essential for cell division. Nature 432, (24). 3. Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p3 pathway. Nature 428, (24). 4. Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, (2).. Zhu, C. et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell 16, (2). 6. Sonnichsen, B. et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, (2). 7. Ziauddin, J. & Sabatini, D.M. Microarrays of cells expressing defined cdnas. Nature 411, (21). 8. Erfle, H. et al. sirna cell arrays for high-content screening microscopy. Biotechniques 37, 44 48, 46, 462 (24). 9. Wheeler, D.B., Carpenter, A.E. & Sabatini, D.M. Cell microarrays and RNA interference chip away at gene function. Nat. Genet. 37 (Suppl.), S2 S3 (2). 1. Gerlich, D. & Ellenberg, J. 4D imaging to assay complex dynamics in live specimens. Nat. Cell Biol. 4 (Suppl.), S14 S19 (23). 11. Sumara, I. et al. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14, (24). 12. Kanda, T. & Wahl, G.M. The dynamics of acentric chromosomes in cancer cells revealed by GFP-based chromosome labeling strategies. J. Cell. Biochem. (Suppl.) 3, (2). 13. Liebel, U. et al. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 4, (23). 14. Huang, K. & Murphy, R.F. From quantitative microscopy to automated image understanding. J. Biomed. Opt. 9, (24). 1. Conrad, C. et al. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 14, (24). 16. Meraldi, P. & Sorger, P.K. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 24, (2). 17. Liu, X., Lei, M. & Erikson, L. Normal cells, but not cancer cells, survive severe plk1 depletion. Mol. Cell. Biol. 26, (26). 18. Gruss, O.J. et al. Chromosome-induced microtubule assembly mediated by Tpx2 is required for spindle formation in HeLA cells. Nat. Cell Biol. 4, (22). 19. Zhu, C., Bossy-Wetzel, E. & Jiang, W. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem. J. 389, (2). 2. Hirota, T. et al. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117, (24). 21. Seul, M., Lawrence, O. & Sammon, M. Practical Algorithms for Image Analysis (Cambridge Univ. Press, Cambridge, UK, 2). 22. Huang, K. & Murphy, R.F. Boosting accuracy of automated classification of fluorescence microscope images for location proteomics. BMC Bioinformatics, 78 (24). 39 VOL.3 NO. MAY 26 NATURE METHODS
by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains was previously described
 Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
J. Cell Sci. 128: doi:10.1242/jcs.164566: Supplementary Material
 Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Application Note No. 2 / July 2012. Quantitative Assessment of Cell Quality, Viability and Proliferation. System
 Application Note No. 2 / July 2012 Quantitative Assessment of Cell Quality, Viability and Proliferation System Quantitative Assessment of Cell Quality, Viability and Proliferation Introduction In vitro
Application Note No. 2 / July 2012 Quantitative Assessment of Cell Quality, Viability and Proliferation System Quantitative Assessment of Cell Quality, Viability and Proliferation Introduction In vitro
岑 祥 股 份 有 限 公 司 技 術 專 員 費 軫 尹 20100803
 技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
Cell Division Mitosis and the Cell Cycle
 Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Notch 1 -dependent regulation of cell fate in colorectal cancer
 Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Interphase (A) and mitotic (B) D16 cells were plated on concanavalin A and processed for
 E07-10-1069 Rogers SUPPLEMENTARY DATA Supplemental Figure 1. Interphase day-7 control and γ-tubulin23c RNAi-treated S2 cells stained for MTs (green), γ-tubulin (red), and DNA (blue). Scale, 5μm. Supplemental
E07-10-1069 Rogers SUPPLEMENTARY DATA Supplemental Figure 1. Interphase day-7 control and γ-tubulin23c RNAi-treated S2 cells stained for MTs (green), γ-tubulin (red), and DNA (blue). Scale, 5μm. Supplemental
EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES
 C GUGAAG EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES MicroRNA-adapted shrna (shrna mir ) for increased, specific and consistent knockdown. MicroRNA PROCESSING PATHWAY UTILIZED FOR shrna mir Developed
C GUGAAG EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES MicroRNA-adapted shrna (shrna mir ) for increased, specific and consistent knockdown. MicroRNA PROCESSING PATHWAY UTILIZED FOR shrna mir Developed
High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B. PEPperPRINT GmbH Heidelberg, 06/2014
 High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B PEPperPRINT GmbH Heidelberg, 06/2014 Introduction This report describes the epitope mapping of autoimmune sera
High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B PEPperPRINT GmbH Heidelberg, 06/2014 Introduction This report describes the epitope mapping of autoimmune sera
Supplemental Information. McBrayer et al. Supplemental Data
 1 Supplemental Information McBrayer et al. Supplemental Data 2 Figure S1. Glucose consumption rates of MM cell lines exceed that of normal PBMC. (A) Normal PBMC isolated from three healthy donors were
1 Supplemental Information McBrayer et al. Supplemental Data 2 Figure S1. Glucose consumption rates of MM cell lines exceed that of normal PBMC. (A) Normal PBMC isolated from three healthy donors were
Cell Discovery 360: Explore more possibilities.
 Cell Discovery 360: Explore more possibilities. Flexible solutions for the BioPharma laboratory Blood Banking Capillary Electrophoresis Centrifugation Flow Cytometry Genomics Lab Automation Lab Tools Particle
Cell Discovery 360: Explore more possibilities. Flexible solutions for the BioPharma laboratory Blood Banking Capillary Electrophoresis Centrifugation Flow Cytometry Genomics Lab Automation Lab Tools Particle
Introduction to Pattern Recognition
 Introduction to Pattern Recognition Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2009 CS 551, Spring 2009 c 2009, Selim Aksoy (Bilkent University)
Introduction to Pattern Recognition Selim Aksoy Department of Computer Engineering Bilkent University saksoy@cs.bilkent.edu.tr CS 551, Spring 2009 CS 551, Spring 2009 c 2009, Selim Aksoy (Bilkent University)
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
Cytell Cell Imaging System
 Cytell Cell Imaging System Simplify your cell analysis and start changing tomorrow today www.gelifesciences.com/cytell The Cytell Cell Imaging System is an intuitive cell analysis instrument that streamlines
Cytell Cell Imaging System Simplify your cell analysis and start changing tomorrow today www.gelifesciences.com/cytell The Cytell Cell Imaging System is an intuitive cell analysis instrument that streamlines
Biology 3A Laboratory MITOSIS Asexual Reproduction
 Biology 3A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Biology 3A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Cellular Reproduction
 9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
Molecular Genetics: Challenges for Statistical Practice. J.K. Lindsey
 Molecular Genetics: Challenges for Statistical Practice J.K. Lindsey 1. What is a Microarray? 2. Design Questions 3. Modelling Questions 4. Longitudinal Data 5. Conclusions 1. What is a microarray? A microarray
Molecular Genetics: Challenges for Statistical Practice J.K. Lindsey 1. What is a Microarray? 2. Design Questions 3. Modelling Questions 4. Longitudinal Data 5. Conclusions 1. What is a microarray? A microarray
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Example 2. What is meant by the cell cycle? Concept 12.1
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Example 2. What is meant by the cell cycle? Concept 12.1
PreciseTM Whitepaper
 Precise TM Whitepaper Introduction LIMITATIONS OF EXISTING RNA-SEQ METHODS Correctly designed gene expression studies require large numbers of samples, accurate results and low analysis costs. Analysis
Precise TM Whitepaper Introduction LIMITATIONS OF EXISTING RNA-SEQ METHODS Correctly designed gene expression studies require large numbers of samples, accurate results and low analysis costs. Analysis
SmartFlare RNA Detection Probes: Principles, protocols and troubleshooting
 Technical Guide SmartFlare RNA Detection Probes: Principles, protocols and troubleshooting Principles of SmartFlare technology RNA detection traditionally requires transfection, laborious sample prep,
Technical Guide SmartFlare RNA Detection Probes: Principles, protocols and troubleshooting Principles of SmartFlare technology RNA detection traditionally requires transfection, laborious sample prep,
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
Proposal Writing in a Nutshell
 Proposal Writing in a Nutshell Context Resources http://www.sci.utah.edu/~macleod/grants/ Find Your Purpose Who is the Audience? If you don t know, find out.?? Don t assume too much!! What are Review Criteria?
Proposal Writing in a Nutshell Context Resources http://www.sci.utah.edu/~macleod/grants/ Find Your Purpose Who is the Audience? If you don t know, find out.?? Don t assume too much!! What are Review Criteria?
Tutorial for proteome data analysis using the Perseus software platform
 Tutorial for proteome data analysis using the Perseus software platform Laboratory of Mass Spectrometry, LNBio, CNPEM Tutorial version 1.0, January 2014. Note: This tutorial was written based on the information
Tutorial for proteome data analysis using the Perseus software platform Laboratory of Mass Spectrometry, LNBio, CNPEM Tutorial version 1.0, January 2014. Note: This tutorial was written based on the information
Primetime for KNIME:
 Primetime for KNIME: Towards an Integrated Analysis and Visualization Environment for RNAi Screening Data F. Oliver Gathmann, Ph. D. Director IT, Cenix BioScience Presentation for: KNIME User Group Meeting
Primetime for KNIME: Towards an Integrated Analysis and Visualization Environment for RNAi Screening Data F. Oliver Gathmann, Ph. D. Director IT, Cenix BioScience Presentation for: KNIME User Group Meeting
A pretty picture, or a measurement? Retinal Imaging
 Big Data Challenges A pretty picture, or a measurement? Organelles Dynamics Cells Retinal Imaging Physiology Pathology Fundus Camera Optical coherence tomography Fluorescence Histology High Content Screening
Big Data Challenges A pretty picture, or a measurement? Organelles Dynamics Cells Retinal Imaging Physiology Pathology Fundus Camera Optical coherence tomography Fluorescence Histology High Content Screening
Quantitative proteomics background
 Proteomics data analysis seminar Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post transcriptional regulation of key cellular processes de Groot, M., Daran
Proteomics data analysis seminar Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post transcriptional regulation of key cellular processes de Groot, M., Daran
Analysis of gene expression data. Ulf Leser and Philippe Thomas
 Analysis of gene expression data Ulf Leser and Philippe Thomas This Lecture Protein synthesis Microarray Idea Technologies Applications Problems Quality control Normalization Analysis next week! Ulf Leser:
Analysis of gene expression data Ulf Leser and Philippe Thomas This Lecture Protein synthesis Microarray Idea Technologies Applications Problems Quality control Normalization Analysis next week! Ulf Leser:
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
Aiping Lu. Key Laboratory of System Biology Chinese Academic Society APLV@sibs.ac.cn
 Aiping Lu Key Laboratory of System Biology Chinese Academic Society APLV@sibs.ac.cn Proteome and Proteomics PROTEin complement expressed by genome Marc Wilkins Electrophoresis. 1995. 16(7):1090-4. proteomics
Aiping Lu Key Laboratory of System Biology Chinese Academic Society APLV@sibs.ac.cn Proteome and Proteomics PROTEin complement expressed by genome Marc Wilkins Electrophoresis. 1995. 16(7):1090-4. proteomics
Core Facility Genomics
 Core Facility Genomics versatile genome or transcriptome analyses based on quantifiable highthroughput data ascertainment 1 Topics Collaboration with Harald Binder and Clemens Kreutz Project: Microarray
Core Facility Genomics versatile genome or transcriptome analyses based on quantifiable highthroughput data ascertainment 1 Topics Collaboration with Harald Binder and Clemens Kreutz Project: Microarray
If and when cancer cells stop dividing, they do so at random points, not at the normal checkpoints in the cell cycle.
 Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Real-Time PCR Vs. Traditional PCR
 Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Call 2014: High throughput screening of therapeutic molecules and rare diseases
 Call 2014: High throughput screening of therapeutic molecules and rare diseases The second call High throughput screening of therapeutic molecules and rare diseases launched by the French Foundation for
Call 2014: High throughput screening of therapeutic molecules and rare diseases The second call High throughput screening of therapeutic molecules and rare diseases launched by the French Foundation for
Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA
 Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Thermo Scientific CellInsight CX5 HCS Platform. discover more uncover the possibilities
 Thermo Scientific CellInsight CX5 HCS Platform discover more uncover the possibilities high content screening Driving your Productivity with Innovated Technology What is High Content Screening (HCS)? High
Thermo Scientific CellInsight CX5 HCS Platform discover more uncover the possibilities high content screening Driving your Productivity with Innovated Technology What is High Content Screening (HCS)? High
Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS)
 Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS) A typical RNA Seq experiment Library construction Protocol variations Fragmentation methods RNA: nebulization,
Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS) A typical RNA Seq experiment Library construction Protocol variations Fragmentation methods RNA: nebulization,
Basic Analysis of Microarray Data
 Basic Analysis of Microarray Data A User Guide and Tutorial Scott A. Ness, Ph.D. Co-Director, Keck-UNM Genomics Resource and Dept. of Molecular Genetics and Microbiology University of New Mexico HSC Tel.
Basic Analysis of Microarray Data A User Guide and Tutorial Scott A. Ness, Ph.D. Co-Director, Keck-UNM Genomics Resource and Dept. of Molecular Genetics and Microbiology University of New Mexico HSC Tel.
Advances in scmos Camera Technology Benefit Bio Research
 Advances in scmos Camera Technology Benefit Bio Research scmos camera technology is gaining in popularity - Why? In recent years, cell biology has emphasized live cell dynamics, mechanisms and electrochemical
Advances in scmos Camera Technology Benefit Bio Research scmos camera technology is gaining in popularity - Why? In recent years, cell biology has emphasized live cell dynamics, mechanisms and electrochemical
Comparison of Non-linear Dimensionality Reduction Techniques for Classification with Gene Expression Microarray Data
 CMPE 59H Comparison of Non-linear Dimensionality Reduction Techniques for Classification with Gene Expression Microarray Data Term Project Report Fatma Güney, Kübra Kalkan 1/15/2013 Keywords: Non-linear
CMPE 59H Comparison of Non-linear Dimensionality Reduction Techniques for Classification with Gene Expression Microarray Data Term Project Report Fatma Güney, Kübra Kalkan 1/15/2013 Keywords: Non-linear
Cell Viability Assays: Microtitration (MTT) Viability Test Live/Dead Fluorescence Assay. Proliferation Assay: Anti-PCNA Staining
 Cell Viability Assays: Microtitration (MTT) Viability Test Live/Dead Fluorescence Assay Proliferation Assay: Anti-PCNA Staining Spring 2008 1 Objectives To determine the viability of cells under different
Cell Viability Assays: Microtitration (MTT) Viability Test Live/Dead Fluorescence Assay Proliferation Assay: Anti-PCNA Staining Spring 2008 1 Objectives To determine the viability of cells under different
RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial
 RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial Samuel J. Rulli, Jr., Ph.D. qpcr-applications Scientist Samuel.Rulli@QIAGEN.com Pathway Focused Research from Sample Prep to Data Analysis! -2-
RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial Samuel J. Rulli, Jr., Ph.D. qpcr-applications Scientist Samuel.Rulli@QIAGEN.com Pathway Focused Research from Sample Prep to Data Analysis! -2-
The Somatic Cell Cycle
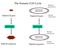 The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
Cell Growth and Reproduction Module B, Anchor 1
 Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
use. 3,5 Apoptosis Viability Titration Assays
 Introduction Monitoring cell viability can provide critical insights into the effectiveness of biological protocols and the stability of biological systems including: the efficacy of cell harvesting procedures,
Introduction Monitoring cell viability can provide critical insights into the effectiveness of biological protocols and the stability of biological systems including: the efficacy of cell harvesting procedures,
HiPerFect Transfection Reagent Handbook
 Fifth Edition October 2010 HiPerFect Transfection Reagent Handbook For transfection of eukaryotic cells with sirna and mirna Sample & Assay Technologies QIAGEN Sample and Assay Technologies QIAGEN is the
Fifth Edition October 2010 HiPerFect Transfection Reagent Handbook For transfection of eukaryotic cells with sirna and mirna Sample & Assay Technologies QIAGEN Sample and Assay Technologies QIAGEN is the
Mitosis in Onion Root Tip Cells
 Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
a measurable difference
 Thermo Scientific Cell Analysis Software and Informatics Products a measurable difference for everyone Thermo Scientific HCS Studio Cell Analysis Software Thermo Scientific Store Image and Database Management
Thermo Scientific Cell Analysis Software and Informatics Products a measurable difference for everyone Thermo Scientific HCS Studio Cell Analysis Software Thermo Scientific Store Image and Database Management
Standards, Guidelines and Best Practices for RNA-Seq V1.0 (June 2011) The ENCODE Consortium
 Standards, Guidelines and Best Practices for RNA-Seq V1.0 (June 2011) The ENCODE Consortium I. Introduction: Sequence based assays of transcriptomes (RNA-seq) are in wide use because of their favorable
Standards, Guidelines and Best Practices for RNA-Seq V1.0 (June 2011) The ENCODE Consortium I. Introduction: Sequence based assays of transcriptomes (RNA-seq) are in wide use because of their favorable
From DNA to Protein
 Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257
 CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
Cell Division CELL DIVISION. Mitosis. Designation of Number of Chromosomes. Homologous Chromosomes. Meiosis
 Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Smart Fluorescence and Dual Live Cell Imaging
 Smart Fluorescence and Dual Live Cell Imaging Smart Fluorescence and Dual Live Cell Imaging Compact Real-Time Cell Analyzer Specialized For Fluorescence-Expressing Cell Joo, Jae Yeon and An, Seong Soo
Smart Fluorescence and Dual Live Cell Imaging Smart Fluorescence and Dual Live Cell Imaging Compact Real-Time Cell Analyzer Specialized For Fluorescence-Expressing Cell Joo, Jae Yeon and An, Seong Soo
Z-Stacking and Z-Projection using a Scaffold-based 3D Cell Culture Model
 A p p l i c a t i o n N o t e Z-Stacking and Z-Projection using a Scaffold-based 3D Cell Culture Model Brad Larson and Peter Banks, BioTek Instruments, Inc., Winooski, VT Grant Cameron, TAP Biosystems
A p p l i c a t i o n N o t e Z-Stacking and Z-Projection using a Scaffold-based 3D Cell Culture Model Brad Larson and Peter Banks, BioTek Instruments, Inc., Winooski, VT Grant Cameron, TAP Biosystems
Cell Biology Questions and Learning Objectives
 Cell Biology Questions and Learning Objectives (with hypothetical learning materials that might populate the objective) The topics and central questions listed here are typical for an introductory undergraduate
Cell Biology Questions and Learning Objectives (with hypothetical learning materials that might populate the objective) The topics and central questions listed here are typical for an introductory undergraduate
Euro-BioImaging European Research Infrastructure for Imaging Technologies in Biological and Biomedical Sciences
 Euro-BioImaging European Research Infrastructure for Imaging Technologies in Biological and Biomedical Sciences WP11 Data Storage and Analysis Task 11.1 Coordination Deliverable 11.2 Community Needs of
Euro-BioImaging European Research Infrastructure for Imaging Technologies in Biological and Biomedical Sciences WP11 Data Storage and Analysis Task 11.1 Coordination Deliverable 11.2 Community Needs of
Inhibition of MicroRNA Sensitizes 3D Breast Cancer Microtissues to Radiation Therapy
 CASE STUDY High Content Image Analysis Inhibition of MicroRNA Sensitizes 3D Breast Cancer Microtissues to Radiation Therapy Dr. Nataša Anastasov leads the Personalized Radiation Therapy Group within the
CASE STUDY High Content Image Analysis Inhibition of MicroRNA Sensitizes 3D Breast Cancer Microtissues to Radiation Therapy Dr. Nataša Anastasov leads the Personalized Radiation Therapy Group within the
Thermo Scientific ArrayScan XTI High Content Analysis Reader. revolutionizing cell biology with the power of high content
 Thermo Scientific ArrayScan XTI High Content Analysis Reader revolutionizing cell biology with the power of high content learn more about your cells using high content technology Thermo Scientific High
Thermo Scientific ArrayScan XTI High Content Analysis Reader revolutionizing cell biology with the power of high content learn more about your cells using high content technology Thermo Scientific High
!!!!!!!!!!!!!!!!!!!!!!!!!!
 Figure S7 cdl-1 3'UTR gld-1 binding site (4..10) let-7 binding site (326..346) Figure Legends Figure S1. gld-1 genetically interacts with nhl-2 and vig-1 during germline development. (A) nhl-2(ok818) enhances
Figure S7 cdl-1 3'UTR gld-1 binding site (4..10) let-7 binding site (326..346) Figure Legends Figure S1. gld-1 genetically interacts with nhl-2 and vig-1 during germline development. (A) nhl-2(ok818) enhances
Intrusion Detection via Machine Learning for SCADA System Protection
 Intrusion Detection via Machine Learning for SCADA System Protection S.L.P. Yasakethu Department of Computing, University of Surrey, Guildford, GU2 7XH, UK. s.l.yasakethu@surrey.ac.uk J. Jiang Department
Intrusion Detection via Machine Learning for SCADA System Protection S.L.P. Yasakethu Department of Computing, University of Surrey, Guildford, GU2 7XH, UK. s.l.yasakethu@surrey.ac.uk J. Jiang Department
Appendix C DNA Replication & Mitosis
 K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
Frequently Asked Questions (FAQ)
 Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells
 Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
PREDA S4-classes. Francesco Ferrari October 13, 2015
 PREDA S4-classes Francesco Ferrari October 13, 2015 Abstract This document provides a description of custom S4 classes used to manage data structures for PREDA: an R package for Position RElated Data Analysis.
PREDA S4-classes Francesco Ferrari October 13, 2015 Abstract This document provides a description of custom S4 classes used to manage data structures for PREDA: an R package for Position RElated Data Analysis.
CCR Biology - Chapter 5 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 5 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. If a cell cannot move enough material
Name: Class: Date: CCR Biology - Chapter 5 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. If a cell cannot move enough material
Biomarker Discovery and Data Visualization Tool for Ovarian Cancer Screening
 , pp.169-178 http://dx.doi.org/10.14257/ijbsbt.2014.6.2.17 Biomarker Discovery and Data Visualization Tool for Ovarian Cancer Screening Ki-Seok Cheong 2,3, Hye-Jeong Song 1,3, Chan-Young Park 1,3, Jong-Dae
, pp.169-178 http://dx.doi.org/10.14257/ijbsbt.2014.6.2.17 Biomarker Discovery and Data Visualization Tool for Ovarian Cancer Screening Ki-Seok Cheong 2,3, Hye-Jeong Song 1,3, Chan-Young Park 1,3, Jong-Dae
Data Integration. Lectures 16 & 17. ECS289A, WQ03, Filkov
 Data Integration Lectures 16 & 17 Lectures Outline Goals for Data Integration Homogeneous data integration time series data (Filkov et al. 2002) Heterogeneous data integration microarray + sequence microarray
Data Integration Lectures 16 & 17 Lectures Outline Goals for Data Integration Homogeneous data integration time series data (Filkov et al. 2002) Heterogeneous data integration microarray + sequence microarray
School of Nursing. Presented by Yvette Conley, PhD
 Presented by Yvette Conley, PhD What we will cover during this webcast: Briefly discuss the approaches introduced in the paper: Genome Sequencing Genome Wide Association Studies Epigenomics Gene Expression
Presented by Yvette Conley, PhD What we will cover during this webcast: Briefly discuss the approaches introduced in the paper: Genome Sequencing Genome Wide Association Studies Epigenomics Gene Expression
Single-Cell Whole Genome Sequencing on the C1 System: a Performance Evaluation
 PN 100-9879 A1 TECHNICAL NOTE Single-Cell Whole Genome Sequencing on the C1 System: a Performance Evaluation Introduction Cancer is a dynamic evolutionary process of which intratumor genetic and phenotypic
PN 100-9879 A1 TECHNICAL NOTE Single-Cell Whole Genome Sequencing on the C1 System: a Performance Evaluation Introduction Cancer is a dynamic evolutionary process of which intratumor genetic and phenotypic
Cell Cycle in Onion Root Tip Cells (IB)
 Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
www.gbo.com/bioscience Tissue Culture 1 Cell/ Microplates 2 HTS- 3 Immunology/ HLA 4 Microbiology/ Bacteriology Purpose Beakers 5 Tubes/Multi-
 11 Cryo 5 Tubes/Multi 2 HTS 3 Immunology / Immunology Technical Information 3 I 2 96 Well ELISA 3 I 4 96 Well ELISA Strip Plates 3 I 6 8 Well Strip Plates 3 I 7 12 Well Strip Plates 3 I 8 16 Well Strip
11 Cryo 5 Tubes/Multi 2 HTS 3 Immunology / Immunology Technical Information 3 I 2 96 Well ELISA 3 I 4 96 Well ELISA Strip Plates 3 I 6 8 Well Strip Plates 3 I 7 12 Well Strip Plates 3 I 8 16 Well Strip
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY
 MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS
 BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS 1. The Technology Strategy sets out six areas where technological developments are required to push the frontiers of knowledge
BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS 1. The Technology Strategy sets out six areas where technological developments are required to push the frontiers of knowledge
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
The Huntington Library, Art Collections, and Botanical Gardens
 The Huntington Library, Art Collections, and Botanical Gardens Rooting for Mitosis Overview Students will fix, stain, and make slides of onion root tips. These slides will be examined for the presence
The Huntington Library, Art Collections, and Botanical Gardens Rooting for Mitosis Overview Students will fix, stain, and make slides of onion root tips. These slides will be examined for the presence
Application Note. Single Cell PCR Preparation
 Application Note Single Cell PCR Preparation From Automated Screening to the Molecular Analysis of Single Cells The AmpliGrid system is a highly sensitive tool for the analysis of single cells. In combination
Application Note Single Cell PCR Preparation From Automated Screening to the Molecular Analysis of Single Cells The AmpliGrid system is a highly sensitive tool for the analysis of single cells. In combination
Cancer SBL101. James Gomes School of Biological Sciences Indian Institute of Technology Delhi
 Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Lecture 7 Mitosis & Meiosis
 Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem
 FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem Elsa Bernard Laurent Jacob Julien Mairal Jean-Philippe Vert September 24, 2013 Abstract FlipFlop implements a fast method for de novo transcript
FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem Elsa Bernard Laurent Jacob Julien Mairal Jean-Philippe Vert September 24, 2013 Abstract FlipFlop implements a fast method for de novo transcript
MicroRNA Mike needs help to degrade all the mrna transcripts! Aaron Arvey ISMB 2010
 Target mrna abundance dilutes microrna and sirna activity MicroRNA Mike needs help to degrade all the mrna transcripts! Aaron Arvey ISMB 2010 Target mrna abundance dilutes microrna and sirna activity Erik
Target mrna abundance dilutes microrna and sirna activity MicroRNA Mike needs help to degrade all the mrna transcripts! Aaron Arvey ISMB 2010 Target mrna abundance dilutes microrna and sirna activity Erik
Instructions. Torpedo sirna. Material. Important Guidelines. Specifications. Quality Control
 is a is a state of the art transfection reagent, specifically designed for the transfer of sirna and mirna into a variety of eukaryotic cell types. is a state of the art transfection reagent, specifically
is a is a state of the art transfection reagent, specifically designed for the transfer of sirna and mirna into a variety of eukaryotic cell types. is a state of the art transfection reagent, specifically
Protease Peptide Microarrays Ready-to-use microarrays for protease profiling
 Protocol Protease Peptide Microarrays Ready-to-use microarrays for protease profiling Contact us: InfoLine: +49-30-97893-117 Order per fax: +49-30-97893-299 Or e-mail: peptide@jpt.com www: www.jpt.com
Protocol Protease Peptide Microarrays Ready-to-use microarrays for protease profiling Contact us: InfoLine: +49-30-97893-117 Order per fax: +49-30-97893-299 Or e-mail: peptide@jpt.com www: www.jpt.com
Automating Cell Biology Annual general meeting, September 7, 2015. Phase Holographic Imaging www.phiab.se
 Automating Cell Biology Annual general meeting, September 7, 2015 www.phiab.se 1 PHASE HOLOGRAPHIC IMAGING (PHI) Began as a research project at Lund University, Sweden, in 2000 Founded in 2004 Sales in
Automating Cell Biology Annual general meeting, September 7, 2015 www.phiab.se 1 PHASE HOLOGRAPHIC IMAGING (PHI) Began as a research project at Lund University, Sweden, in 2000 Founded in 2004 Sales in
Current Motif Discovery Tools and their Limitations
 Current Motif Discovery Tools and their Limitations Philipp Bucher SIB / CIG Workshop 3 October 2006 Trendy Concepts and Hypotheses Transcription regulatory elements act in a context-dependent manner.
Current Motif Discovery Tools and their Limitations Philipp Bucher SIB / CIG Workshop 3 October 2006 Trendy Concepts and Hypotheses Transcription regulatory elements act in a context-dependent manner.
Introduction to Proteomics
 Introduction to Proteomics Why Proteomics? Same Genome Different Proteome Black Swallowtail - larvae and butterfly Biological Complexity Yeast - a simple proteome 6,113 proteins = 344,855 tryptic peptides
Introduction to Proteomics Why Proteomics? Same Genome Different Proteome Black Swallowtail - larvae and butterfly Biological Complexity Yeast - a simple proteome 6,113 proteins = 344,855 tryptic peptides
Dr Alexander Henzing
 Horizon 2020 Health, Demographic Change & Wellbeing EU funding, research and collaboration opportunities for 2016/17 Innovate UK funding opportunities in omics, bridging health and life sciences Dr Alexander
Horizon 2020 Health, Demographic Change & Wellbeing EU funding, research and collaboration opportunities for 2016/17 Innovate UK funding opportunities in omics, bridging health and life sciences Dr Alexander
IncuCyte ZOOM Whole Well Imaging Overview
 IncuCyte ZOOM Whole Well Imaging Overview With the release of the 2013B software, IncuCyte ZOOM users will have the added ability to image the complete surface of select multi-well plates and 35 mm dishes
IncuCyte ZOOM Whole Well Imaging Overview With the release of the 2013B software, IncuCyte ZOOM users will have the added ability to image the complete surface of select multi-well plates and 35 mm dishes
Cellular Imaging Solutions Imaging with a vision
 Cellular Imaging Solutions Imaging with a vision www.moleculardevices.com High content screening (HCS) utilizes automated, high-resolution microscopy systems to assay and visualize phenotypic responses
Cellular Imaging Solutions Imaging with a vision www.moleculardevices.com High content screening (HCS) utilizes automated, high-resolution microscopy systems to assay and visualize phenotypic responses
How To Cluster
 Data Clustering Dec 2nd, 2013 Kyrylo Bessonov Talk outline Introduction to clustering Types of clustering Supervised Unsupervised Similarity measures Main clustering algorithms k-means Hierarchical Main
Data Clustering Dec 2nd, 2013 Kyrylo Bessonov Talk outline Introduction to clustering Types of clustering Supervised Unsupervised Similarity measures Main clustering algorithms k-means Hierarchical Main
LAB 09 Cell Division
 LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
EMBL Identity & Access Management
 EMBL Identity & Access Management Rupert Lück EMBL Heidelberg e IRG Workshop Zürich Apr 24th 2008 Outline EMBL Overview Identity & Access Management for EMBL IT Requirements & Strategy Project Goal and
EMBL Identity & Access Management Rupert Lück EMBL Heidelberg e IRG Workshop Zürich Apr 24th 2008 Outline EMBL Overview Identity & Access Management for EMBL IT Requirements & Strategy Project Goal and
Clonetics Conditionally Immortalized Human Cells. Relevant Cells for High Throughput Screening
 Clonetics Conditionally Immortalized Human Cells Relevant Cells for High Throughput Screening Clonetics Conditionally Immortalized Cell Strains Conditionally immortalized cell strains are primary cells
Clonetics Conditionally Immortalized Human Cells Relevant Cells for High Throughput Screening Clonetics Conditionally Immortalized Cell Strains Conditionally immortalized cell strains are primary cells
inform Advanced Image Analysis Software For Accurately Quantifying Biomarkers in Tissue Sections
 P R O D U C T N O T E inform Advanced Image Analysis Key Features Quantitative analysis of biomarker epression in tissue sections and TMAs Separation of weakly epressing and overlapping markers Cellular
P R O D U C T N O T E inform Advanced Image Analysis Key Features Quantitative analysis of biomarker epression in tissue sections and TMAs Separation of weakly epressing and overlapping markers Cellular
Cell Cycle Phase Determination Kit
 Cell Cycle Phase Determination Kit Item No. 10009349 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Safety
Cell Cycle Phase Determination Kit Item No. 10009349 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Safety
List, describe, diagram, and identify the stages of meiosis.
 Meiosis and Sexual Life Cycles In this topic we will examine a second type of cell division used by eukaryotic cells: meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis
Meiosis and Sexual Life Cycles In this topic we will examine a second type of cell division used by eukaryotic cells: meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis
5. The cells of a multicellular organism, other than gametes and the germ cells from which it develops, are known as
 1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
PRODUCT INFORMATION...
 VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
Thermo Scientific Dharmacon sirna Libraries
 Thermo Scientific Dharmacon sirna Libraries Scientific innovation in sirna potency, specificity and delivery Superior sirna in comprehensive collections for reliable screens Market leading products for
Thermo Scientific Dharmacon sirna Libraries Scientific innovation in sirna potency, specificity and delivery Superior sirna in comprehensive collections for reliable screens Market leading products for
Lecture 11 The Cell Cycle and Mitosis
 Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
