for Detection of Multiple Pathogens and William C. Reeves 1
|
|
|
- Walter Alexander
- 7 years ago
- Views:
Transcription
1 Bioelectronic DNA Detection of Human Papillomaviruses Using esensor : A Model System for Detection of Multiple Pathogens Suzanne D. Vernon 1 (svernon@cdc.gov), Daniel H. Farkas 2* (dfarkas@bcm.tmc.edu), Elizabeth R. Unger 1 (eru0@cdc.gov), Vivian Chan 2* (vwyc@yahoo.com), Donna L. Miller 1 (dlm5@cdc.gov), Yin-Peng Chen 2 (yin.peng@motorola.com) Gary F. Blackburn 2 (gary.blackburn@motorola.com) and William C. Reeves 1 (wcr1@cdc.gov) 1 Division of Viral and Rickettsial Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia 30333, 2 Motorola Life Sciences, 757 South Raymond Avenue, Pasadena California *Current Affiliation of D.H.F. is Department of Pathology, Baylor College of Medicine, One Baylor Plaza, Houston, Texas 77030; of V.C. is Department of Pathology, Stanford University Medical Center, Stanford, California Abstract: 141 words Text: 2,016 words Running Head: Bioelectric Detection of Pathogens Address correspondence to: Suzanne D. Vernon, Ph.D. Centers for Disease Control and Prevention 1600 Clifton Road, Mail Stop A-15 Atlanta, GA Voice: FAX: svernon@cdc.gov 1
2 Abstract Background. We used human papillomaviruses (HPV) as a model system to evaluate the utility of a nucleic acid, hybridization-based bioelectronic DNA detection platform (esensor) in identifying multiple pathogens. Methods. Two chips were spotted with capture probes consisting of DNA oligonucleotide sequences specific for HPV types. Electrically conductive signal probes were synthesized to be complementary to a distinct region of the amplified HPV target DNA. A portion of the HPV L1 region that was amplified by using consensus primers served as target DNA. The amplified target was mixed with a cocktail of signal probes and added to a cartridge containing a DNA chip to allow for hybridization with complementary capture probes. Results. Two bioelectric chips were designed and successfully detected 86% of the HPV types contained in clinical samples. Conclusions. This model system demonstrates the potential of the esensor platform for rapid and integrated detection of multiple pathogens. 2
3 Background Global emergence of pathogenic infectious diseases by both natural and intentional means presents a formidable challenge to infectious disease surveillance and response, namely timely and efficient pathogen detection. Many laboratory methods exist for identifying pathogens, but most require exquisite care in sample handling and processing prior to characterization of a pathogen. In addition, costly and perishable reagents, equipment, and supplies are required for sensitive and specific detection. The ideal detection system would integrate sample processing and pathogen characterization into a single automated device that would eliminate laborious, and time consuming sample processing and costly detection. Bioelectronic detection of nucleic acids on a miniature solid support is one of the first steps toward development of such an integrated detection device. Bioelectronic DNA detection involves forming an electronic circuit mediated by nucleic acid hybridization and it serves as the basis for a DNA detection system called esensor [1-4]. This system uses low-density DNA chips containing electrodes coated with DNA capture probes. Target DNA present in the sample hybridizes specifically both to capture probes and ferrocene labeled signal probes in solution thereby generating an electric current. Current esensor DNA chips contain as many as 36 electrodes for simultaneous detection of multiple pathogens from a single sample. Many pathogens cause both acute and chronic disease at relatively low copy number and may be difficult or impossible to propagate in culture. Thus, most pathogen detection systems rely on nucleic acid amplification by using polymerase chain reaction (PCR). One highly effective amplification strategy targets conserved sequences among the family of organisms of interest. Such broad-range PCR strategies have been used to identify and characterize several known and previously uncharacterized bacteria [5, 6] and viruses [7, 8]. In order to maximize 3
4 the utility of these effective pathogen nucleic acid amplification systems, amplification needs to be coupled with rapid, sensitive, and specific detection. Bioelectronic DNA detection by use of the esensor chip might fulfill this need. Human papillomaviruses (HPV) serve as an ideal model system for determining the efficiency and feasibility of esensor DNA detection technology since there are at least 30 distinct genital HPV types that can be effectively amplified with broad-range consensus PCR primers [9]. We designed two esensor chips, each containing 14 probes specific for the conserved L1 region of the HPV genome. We evaluated clinical cervical cytology samples known to contain one or more HPV types. The esensor DNA detection platform successfully detected the correct HPV type in most of these clinical samples, demonstrating that the system provides a rapid, sensitive, specific, and economical approach for multiple-pathogen detection and identification from a single sample. 4
5 Methods esensor DNA Chip Design We used previously described printed circuit board technology to manufacture esensor chips with 16 gold electrodes, one reference electrode, and one auxiliary electrode [2-4]. Each electrode was wired to a connector at the edge of the chip. Capture probes were synthesized using an alkane chain of 16 residues terminated at the 3 -end with a disulfide which was spotted onto the gold electrode on the chips to form a self-assembled monolayer [10]. The chips were glued into a plastic housing with a port for sample introduction. Hybsimulator (RNAture, Inc., Irvine, CA) was used to design the HPV target mimics, and signal and capture probes. For these experiments, we designed capture (Table 1) and signal (Table 2) probes to hybridize specifically to the exon sequence of -globin and to HPV sequences within the 450-bp L1 region amplified by the PGMY primer set. Signal probes (Table 2) tagged with eight ferrocenes were synthesized with a modified adenine residue (N6) containing a ferrocene substitution on the ribose ring [11]. No signal probes were synthesized for HPV types 33, 39, 55, and 73. HPV target mimic oligonucleotides were synthesized for types 6, 11, 16, 18, 26, 31, 35, 40, 42, 45, 51, 52, 54, 56, 58, 59, 66, 68, 82, 83, and 84 (Table 3). Sample selection and DNA extraction Residual ThinPrep (Cytyc Corporation, Boxborough, MA) cervical samples previously tested for HPV using the Roche line blot assay [9] were used in this study. Twenty samples containing 24 of the 27 HPV types on the reverse line blot were selected for extraction and testing. (No samples were positive for HPV 53, 55, and 57.) Therefore a total of 21 HPV types could be evaluated (there were 22 HPV capture and signal probes minus no clinical sample containing HPV 53 leaves 21 possible HPV type answers). Samples were coded so that 5
6 investigators were blinded to the presence of multiple HPV types. DNA was extracted from 250 µl aliquots by using the QIAmp DNA Mini Kit (Qiagen Inc., Valencia, CA). The procedure provided with the kit was followed except that the proteinase K digestion was done overnight at 56 C. Following DNA elution from the column, DNA was concentrated in a Centricon Centrifugal Filter Device YM-100 (Millipore Corporation, Bedford MA) according to manufacturer s specifications. Each DNA sample was brought to 50 µl with the addition of ultra pure water. HPV PCR and Typing Five microliters of purified DNA from cytology samples was used in each PCR assay. HPV DNA was amplified with the PGMY09/PGMY11 primer system that uses a multiplex format to amplify both HPV and -globin in the same tube [9]. To allow for strand separation, only one PGMY and one -globin primer were biotinylated (Table 1). The use of one biotinylated primer to amplify the product was still sufficient to verify the amplified HPV type by reverse line blot. Amplification without DNA template was used to monitor contamination. Avidin-coated beads were used to harvest the biotin-free DNA strand from 50 µl of product, which was then used as the target in esensor chip assay. Briefly, the target mimic or target DNA was mixed with a signal probe cocktail (125 nm of each signal probe) in a hybridization buffer (proprietary). This mixture was injected into the esensor chip for hybridization at 40 o C for 2 to 8 hours. The chips were then scanned by an AC voltammetry technique built into the esensor detection device called the esensor 4800, from 50 to +550 millivolts, to detect and interpret the electrochemical signal generated during hybridization. Investigators recording esensor results remained blinded to prior or current HPV results. 6
7 Forty microliters of the amplified products was also independently tested for HPV using a reverse line probe assay (typing strip and reagents generously provided by Janet Kornegay, Roche Molecular Systems, Inc., Alameda, CA). In each case the HPV results on the strip were concordant with prior results. HPV detection and typing results from esensor and Roche were then directly compared. 7
8 Results The electrochemical signal is generated when the HPV amplicon hybridizes to the capture probe and ferrocene-labeled signal probe, thereby bringing the reporter molecule, ferrocene, into contact with the self-assembled monolayer on the gold electrode (Fig. 1). Many of the HPV target mimics tested resulted in a specific signal, with the exception of HPV types 16, 26, 35, 40, 42, 51, 52, and 82. This nonspecific hybridization signal generally resulted from a long stretch of complementary sequences (6-8 bases) among the HPV types due to their close genetic relatedness. Nonspecific hybridization was eliminated by introducing a mismatched base to the complementary region in the signal probes for HPV types 16, 26, 35, 42, and 82 and the capture probes for HPV types 40, 42, 51, 52, and 82. For instance, the HPV 16 signal probe hybridized with both the HPV 16 target mimic (Fig. 2, left panel) and the HPV 52 capture probe (Fig. 2, center panel). This cross-reactivity was eliminated by changing the second A base in the 5 -end of signal probe to a G base (Fig. 2, right panel). In addition, we demonstrated that one base modification of the signal probe (Fig. 2, left and right panels) generated minimal negative impact to the assay signal level. Applying this strategy, we were able to substantially reduce the cross-reactivity among the signal and capture probes for various HPV types without compromising their hybridization strengths. In most instances for the 20 samples tested, there was perfect concordance between the HPV type(s) detected by reverse line blot and the esensor system (Table 4). The esensor detected 18 (86%) of 21 HPV types. In samples with multiple HPV types amplified by the consensus primer system and typed by reverse line blot (but blinded to the individual conducting the esensor assay), the esensor successfully detected the multiple HPV targets in all six samples (Table 4). 8
9 Discussion We have presented a model application of a novel electronic detection platform [1-4]. We used HPV as a model infectious disease system because consensus primers are available that amplify at least 30 closely related genital HPV types. The resulting amplicons are extremely similar, and this relatedness challenges the ability of the esensor technology to distinguish accurately among these pathogens. The first optimization steps involved evaluation of electrochemical signal generation by using HPV target mimics. Cross-hybridization was observed between a subset of HPV target mimics and the capture and signal probes. This crosshybridization was easily overcome by the introduction of mismatched base pairs. The effectiveness of single-base mismatch discrimination of the esensor has been demonstrated by using wild type and mutant strains of human immunodeficiency virus [3]. We used the esensor DNA chip to determine the presence of single or multiple HPV types in cervical cytology samples. While the presence of the target HPV type was known, the presence of other HPV types in the same sample remained blinded (e.g., the HPV 42 in a clinical sample was identified but the presence of HPV 68 in the same sample was not known to those performing electrochemical detection). There was good concordance for the detection of the target HPV type by the esensor DNA Detection System. Eighteen (86%) of 21 target HPV types represented on the chip were readily detected by the esensor (Table 4); HPV 40, 45 and 54 were not detected. Some of the other discrepancies could be due to lack of rigorous optimization of sensitivity parameters for the esensor system, such as the weak HPV 54 result on reverse line blot and lack of detection on esensor. Low-level signals on the line probe assay have been validated by sequencing the amplified product [12]. For HPV 40 discrepancy, the capture probe design may not have been optimal to allow for hybridization with the HPV 40 target. These HPV types may be variant types that did not hybridize to the capture probe and subsequently could not be detected by the signal probe. Not only are there over 200 HPV types, but several 9
10 sequence variants of HPV types are known to exist [13]. Most hybridization reactions could be analyzed after 2 hours, although in several instances up to 8 hours of hybridization time was required. The intent of this study was to use HPV as a model system to assess the feasibility of electrochemical detection with the esensor chip of multiple pathogens, not to achieve perfect concordance of HPV detection with the reverse line blot. In this model assay, we have performed optimization of probe sequences and assay protocol by using synthesized target mimics. The resultant assay correctly identified nearly all HPV type in the samples. No attempt to further optimize the assay using PCR amplicons, rather than target mimics, was made and neither was the PCR process optimized for detection by using the esensor DNA detection system. This work demonstrates the ease with which a multi-analyte panel assay can be prototyped using the esensor DNA detection system. For clinical applications, the prototype assay can be readily fine-tuned by optimizing the conditions for PCR and hybridization to yield concordance levels approaching 100% (data not published). 10
11 Conclusions The bioelectronic detection platform described here includes disposable DNA chips and laboratory-based electronic readers and is a first-generation system that ultimately will facilitate the development of relatively low cost, arrayed nucleic acid-based tests. For widespread clinical laboratory adoption, these arrays must have clinically appropriate densities compatible with the needs of molecular diagnostics laboratories, (i.e., low cost and not labor-intensive). The system described could also be developed as a foundation for devices to be used for point-of-care testing for other infectious diseases. 11
12 Author s Contributions Author 1 (SDV) contributed to the conception and design of this study, the analysis of the results and drafted the manuscript. Author 2 (DHF) contributed to the design of the experimental approach using the bioelectric chip, implemented the experimental approach, and assisted in drafting the manuscript. Author 3 (ERU) contributed to the conception and design of this study, the analysis of the results and drafted the manuscript. Author 4 (VC) conducted the laboratory work at Clinical Microsensors. Author 5 (DLM) conducted all laboratory work involving sample preparation, amplification and typing of human papillomaviruses at CDC. Author 6 (YPC) conducted the laboratory work at Clinical Microsensors. Author 7 (GFB) supported the memorandum of understanding between Clinical Microsensors and CDC and edited the manuscript. Author 8 (WCR) contributed to the conception and design of this study, assisted in drafting the manuscript and supported the memorandum of understanding between Clinical Microsensors and CDC. All authors read and approved the final manuscript. 12
13 Acknowledgements Use of trade names is for identification only and does not imply endorsement by the Public Health Service or the U.S. Department of Health and Human Services. This research was supported in part by Memorandum of Understanding between the Viral Exanthems and Herpesvirus Branch in the Division of Viral and Rickettsial Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention and Clinical Microsensors, A Division of Motorola. 13
14 References 1. Farkas DH: Bioelectronic DNA chips for the clinical laboratory. Clin Chem 2001, 47: Umek RM, Lin SS, Chen Yp Y, Irvine B, Paulluconi G, Chan V, Chong Y, Cheung L, Vielmetter J, Farkas DH: Bioelectronic detection of point mutations using discrimination of the H63D polymorphism of the Hfe gene as a model. Mol Diagn 2000, 5: Umek RM, Lin SW, Vielmetter J, Terbrueggen RH, Irvine B, Yu CJ, Kayyem JF, Yowanto H, Blackburn GF, Farkas DH, Chen YP: Electronic detection of nucleic acids: versatile platform for molecular diagnostics. J Mol Diagn 2001, 3: Yu CJ, Wan Y, Yowanto H, Li J, Tao C, James MD, Tan CL, Blackburn GF, Meade TJ: Electronic detection of single-base mismatches in DNA with ferrocene-modified probes. J Am Chem Soc 2001, 123: Relman DA, Schmidt TM, MacDermott RP, Falkow S: Identification of the uncultured bacillus of Whipple's disease. N Engl J Med 1992, 327: Shukla SK, Vevea DN, Frank DN, Pace NR, Reed KD: Isolation and characterization of a black-pigmented Corynebacterium sp. from a woman with spontaneous abortion. J Clin Microbiol 2001, 39: Resnick RM, Cornelissen MT, Wright DK, Eichinger GH, Fox HS, ter Schegget J, Manos MM: Detection and typing of human papillomavirus in archival cervical cancer specimens by DNA amplification with consensus primers. J Natl Cancer Inst 1990, 82:
15 8. Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ: Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 1993, 262: Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ: Improved amplification of genital human papillomaviruses. J Clin Microbiol 2000, 38: Palegrosdemange C, Simon ES, Prime KL, Whitesides GM: Formation of selfassembled monolayers by chemisorption of derivatives of oligo(ethylene glycol) of structure HS(CH2)11(OCH2CH2)Meta-OH on gold. J Am Chem Soc 1991, 113: Yu CJ, Wan Y, Yowanto H, Li J, Tao C, James MD, Tan CL, Blackburn GF: Uridineconjugated ferrocene DNA oligonucleotides: unexpected cyclization reaction of the uridine base. J Am Chem Soc 2000,122: Vernon SD, Unger ER, Williams D: Comparison of human papillomavirus detection and typing by cycle sequencing, line blot and hybrid capture. J Clin Microbiol 2000, 38: Xi LF, Carter JJ, Galloway DA, Kuypers J, Hughes JP, Lee SK, Adam DE, Kiviat NB, Koutsky LA: Acquisition and natural history of human papillomavirus type 16 variant infection among a cohort of female university students. Cancer Epidemiol Biomarkers Prev 2002, 11:
16 Table 1. Human papillomavirus (HPV) capture probe and PCR primers sequences. 6 capture CAG AAT TGG TGT ATG TGG AAG A(N152) 11 capture TAA TCT GAA TTA GTG TAT GTA GCA GAT TTA GAC A(N152) 16 capture GTA GTT TCT GAA GTA GAT ATG G(N152) 18 capture TGG TAG CAT CAT ATT GCC CAG G(N152) 26 capture ATC AGA TGG TTT AAA TGG AGT GGA TGC(N152) 31 capture TAC TAC TTT TAA ATG TAG TAT CAC(N152) 35 capture ACT GTC ACT AGA AGA CAC AGC AGA ACA CA(N152) 40 capture GGG GGA CTG TGT GGC ACC A(N152) 42 capture AGC AGC TGT ATA TGT ATC ACC AGA TGT TGC AGT GGC TCA(N152) 45 capture CTT AGT AGG GTC ATA TGT ACT TGG C(N152) 51 capture TTG GGG AAA CCG CAG CAG TGG CAG GGC TA(N152) 52 capture TAT GTG CTT TCC TTT TTA ACC T(N152) 54 capture GTC AGA ATT ATT AAA GCT ATC CTG CG(N152) 56 capture TTT TCG TGC ATC ATA TTT ACT TA(N152) 58 capture GTA CCT TCC TTA GTT ACT TCA G(N152) 59 capture CTG GTA GGT GTG TAT ACA TTA G(N152) 66 capture CAC GGG CAT CAT ATT TAG TTA A(N152) 68 capture TTA AAT TTA TTA GGA TCA TAA ATA TTT GGT A(N152) 82 capture AAT GTT TGT GCA ACA GAT TGA GTA ACA GCT GTG(N152) 83 capture GTT AGA GGC TGT GTA TTC ATT AGC(N152) 84 capture GGT TTA TAT TCT GAT TCG GTG T(N152) globin capture AGC AAT AGA TGG CTC TGC CC(N152) globin primer GAA GAG CCA AGG ACA GGT AC globin rev primer BIOTIN CAA CTT CAT CCA CGT TCA CC HMB01 rev primer BIOTIN GCG ACC CAA TGC AAA TTG GT PGMY09F primer BIOTIN CGT CCC AAA GGA AAC TGA TC PGMY09G primer BIOTIN CGA CCT AAA GGA AAC TGA TC PGMY09H primer BIOTIN CGT CCA AAA GGA AAC TGA TC PGMY09I primer BIOTIN GCC AAG GGG AAA CTG ATC PGMY09J primer BIOTIN CGT CCC AAA GGA TAC TGA TC PGMY09K primer BIOTIN CGT CCA AGG GGA TAC TGA TC PGMY09L primer BIOTIN CGA CCT AAA GGG AAT TGA TC PGMY09M primer BIOTIN CGA CCT AGT GGA AAT TGA TC PGMY09N primer BIOTIN CGA CCA AGG GGA TAT TGA TC PGMY09P primer BIOTIN GCC CAA CGG AAA CTG ATC PGMY09Q primer BIOTIN CGA CCC AAG GGA AAC TGG TC PGMY09R primer BIOTIN CGT CCT AAA GGA AAC TGG TC PGMY11A primer GCA CAG GGA CAT AAC AAT GG PGMY11B primer GCG CAG GGC CAC AAT AAT GG PGMY11C primer GCA CAG GGA CAT AAT AAT GG PGMY11D primer GCC CAG GGC CAC AAC AAT GG PGMY11F primer GCT CAG GGT TTA AAC AAT GG 16
17 Table 2. HPV type and signal probe sequence. 6 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGT AGT TAC GGA TGC AC 11 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CAG ATG CAG ATA GTG TCA T 16 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CAG CGC ATA ATG ACA TAT TT 18 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TAC AGG AGA CTG TGT AG 26 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) AGA TGC TGT AGA TAA TGT AC 31 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGT TTG CAA TTG CAG CA 35 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CAG ACA TAT TTG TTC TAC GG 40 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TAA GGT TAA ATT AGT GCA ACG A 42 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CAA AGA CAT GTT AGT ACT AC 45 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) ACA GGA TTT TGT GTA GAG G 51 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) ATA GTT AAA TTT GTA CTT CTG G 52 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CAG CAC ATA AAG TCA TG 54 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGG ATG CTG TAG CAC AC 56 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) ACT GTT CTG TAG CAG TAC T 58 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGC ATA ATG TCA TAT TAG TG 59 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) GAA TAG AAG AAG TAG TAG AA 66 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGT GCT TTT AGC TGC AT 68 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CAG CTG ATT CAG TAG TAG TA 82 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CTA ATG GTT AAA TTG GTA GTT 83 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) CTG TGT AGC AGG AGC TGA AA 84 (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGG TAG CAG CAC TAA TA Globin (N6) C(N6) G(N6) C(N6) GCT TA(N6) C(N6) G(N6) C(N6) TGA CTT TTA TGC CCA GAC CTG G 17
18 Table 3. HPV type and target mimic oligonucleotide sequences 6 ACC ACA CGC AGT ACC AAC ATG ACA TTA TGT GCA TCC GTA ACT ACA TCT TCC ACA TAC ACC AAT TCT GAT TAT AAA 11 GTA CAA ATA TGA CAC TAT GTG CAT CTG TGT CTA AAT CTG CTA CAT ACA CTA ATT CAG ATT ATA AGG AAT ACA TGC G 16 ACT ACA CGC AGT ACA AAT ATG TCA TTA TGT GCT GCC ATA TCT ACT TCA GAA ACT ACA TAT AAA AAT ACT AAC TTT AA 18 ACC ACT CCC AGT ACC AAT TTA ACA ATA TGT GCT TCT ACA CAG TCT CCT GTA CCT GGG CAA TAT GAT GCT ACC AAA T 26 GTA CTA ACC TTA CCA TTA GTA CAT TAT CTG CAG CAT CTG CAT CCA CTC CAT TTA AAC CAT CTG ATT ATA AAC AAT T 31 ACC ACA CGT AGT ACC AAT ATG TCT GTT TGT GCT GCA ATT GCA AAC AGT GAT ACT ACA TTT AAA AGT AGT AAT TTT AA 35 TGT AGT TGA TAC AAC CCG TAG TAC AAA TAT GTC TGT GTG TTC TGC TGT GTC TTC TAG TGA CAG TAC ATA TAA AAA T 40 AGT TGT AGA CAC CAC TCG TAG CAC TAA TTT AAC CTT ATG TGC TGC CAC ACA GTC CCC CAC ACC AAC CCC ATA TAA T 42 ACT ACC CGT AGT ACT AAC ATG ACT TTG TGT GCC ACT GCA ACA TCT GGT GAT ACA TAT ACA GCT GCT AAT TTT AAG G 45 AAC ATT ATG TGC CTC TAC ACA AAA TCC TGT GCC AAG TAC ATA TGA CCC TAC TAA GTT TAA GCA GTA TAG TAG ACA T 51 GTT GAT ACT ACC AGA AGT ACA AAT TTA ACT ATT AGC ACT GCC ACT GCT GCG GTT TCC CCA ACA TTT ACT CCA AGT A 52 ACC ACT CGT AGC ACT AAC ATG ACT TTA TGT GCT GAG GTT AAA AAG GAA AGC ACA TAT AAA AAT GAA AAT TTT AAG 54 GTA CTA ACC TAA CAT TGT GTG CTA CAG CAT CCA CGC AGG ATA GCT TTA ATA ATT CTG ACT TTA GGG AGT ATA TTA G 56 GTA CTA ACA TGA CTA TTA GTA CTG CTA CAG AAC AGT TAA GTA AAT ATG ATG CAC GAA AAA TTA ATC AGT ACC TTA G 58 ACC ACT CGT AGC ACT AAT ATG ACA TTA TGC ACT GAA GTA ACT AAG GAA GGT ACA TAT AAA AAT GAT AAT TTT AAG 59 ACT ACT CGC AGC ACC AAT CTT TCT GTG TGT GCT TCT ACT ACT TCT TCT ATT CCT AAT GTA TAC ACA CCT ACC AGT 66 ACT ACC AGA AGC ACC AAC ATG ACT ATT AAT GCA GCT AAA AGC ACA TTA ACT AAA TAT GAT GCC CGT GAA ATC AAT 68 GTA CCA ATT TTA CTT TGT CTA CTA CTA CTG AAT CAG CTG TAC CAA ATA TTT ATG ATC CTA ATA AAT TTA AGG AAT A 82 ACT ACT AGA AGT ACC AAT TTA ACC ATT AGC ACT GCT GTT ACT CAA TCT GTT GCA CAA ACA TTT ACT CCA GCA AAC T 83 GTA CCA ATA TTA CTA TTT CAG CTG CTG CTA CAC AGG CTA ATG AAT ACA CAG CCT CTA ACT TTA AGG AAT ACC TCC G 84 ACC ACC CGC AGC ACC AAT TTT ACT ATT AGT GCT GCT ACC AAC ACC GAA TCA GAA TAT AAA CCT ACC AAT TTT AAG globin GGG AGG GCA GGA GCC AGG GCT GGG CAT AAA AGT CAG GGC AGA GCC ATC TAT TGC TTA CAT TTG CTT CTG ACA CAA C 18
19 Table 4. Comparison of HPV detection and typing results Target HPV Type(s) in clinical sample HPV Type(s) by esensor Comments on esensor , 35, 40 11, 35 HPV 40 ND , 26, 42 18, 26 HPV 42 ND , 84 weak 84 signal , 35, 40 11, 35 HPV 40 ND 42 42, 68 42, ND no globin signal weak 54 ND , 68 42, , 31, weak 51 82, 31 HPV 51 ND ND, not detected 19
20 Figure 1. A) Schematic represention of the esensor DNA Detection System. A self-assembled monolayer is generated on a gold electrode. For electrochemical detection to occur across the monolayer, two hybridization events must occur: the first between the capture probe (shown in black perpendicular to the electrode) and the target (for example, HPV DNA, shown in gray), and the second between an adjacent region of the target and ferrocene labeled signal probe (shown in black parallel to the monolayer). B) The esensor bioelectronic representative signal output. A fourth harmonic alternating current voltammogram is obtained from an HPV-positive sample (left panel), and one obtained from an HPV-negative sample (right panel). The faradic signal in the fourth harmonic closely fits the third derivative of a Gaussian curve. The peak current (100 na) in the fourth harmonic voltammogram is determined by averaging the two peaks that center around 130 mv. Fig. 2. HPV 16 Signaling Probe Optimization. The HPV esensor chip was decorated with 11 different HPV capture probes and an HIV capture probe as a negative control (shown in x-axis). An HPV 16 target mimic was detected by the esensor in the presence of HPV 16 specific signaling probe (left panel) in which the sample also reacted with HPV 52-specific capture probe. The signal was identified as being from the hybridization between HPV 16 signal probe and the HPV 52 capture probe as shown in the mid-panel in which HPV 16 signal probe was introduced into the chip without any target mimic. The HPV 16 signal probe was modified replacing the second A base in the 5 -end with a G base. The resulted new signal probe hybridized specifically with the HPV 16 target mimic and did not cross-react with the HPV 52 capture probe. 20
21 Figure 1 5 Fe Fe 5 Ferrocene Labels Fe 3 Gold electrode Self-Assembled Monolayer
22 Potential (mv) Potential (mv) Signal Output (na) Figure 2
23 HPV16.cp22 HPV18.cp22 HPV31.cp24 HPV52.cp22 HPV53.cp22 HPV58.cp22 HPV59.cp22 HPV66.cp22 HPV6b.cp22 HPVmm8.cp22 HPV31.cp22mm HIV.cp21.N152 HPV16.cp22 HPV18.cp22 HPV31.cp24 HPV52.cp22 HPV53.cp22 HPV58.cp22 HPV59.cp22 HPV66.cp22 HPV6b.cp22 HPVmm8.cp22 HPV31.cp22mm HIV.cp21.N152 HPV16.cp22 HPV18.cp22 HPV31.cp24 HPV52.cp22 HPV53.cp22 HPV58.cp22 HPV59.cp22 HPV66.cp22 HPV6b.cp22 HPVmm8.cp22 HPV31.cp22mm HIV.cp21.N HPV 16 target mimic with 1st generation signaling probe HPV 16 signaling probe only HPV target mimic with modified signaling probe Average Signal (nanoamps) Figure 3
GENEWIZ, Inc. DNA Sequencing Service Details for USC Norris Comprehensive Cancer Center DNA Core
 DNA Sequencing Services Pre-Mixed o Provide template and primer, mixed into the same tube* Pre-Defined o Provide template and primer in separate tubes* Custom o Full-service for samples with unknown concentration
DNA Sequencing Services Pre-Mixed o Provide template and primer, mixed into the same tube* Pre-Defined o Provide template and primer in separate tubes* Custom o Full-service for samples with unknown concentration
(http://genomes.urv.es/caical) TUTORIAL. (July 2006)
 (http://genomes.urv.es/caical) TUTORIAL (July 2006) CAIcal manual 2 Table of contents Introduction... 3 Required inputs... 5 SECTION A Calculation of parameters... 8 SECTION B CAI calculation for FASTA
(http://genomes.urv.es/caical) TUTORIAL (July 2006) CAIcal manual 2 Table of contents Introduction... 3 Required inputs... 5 SECTION A Calculation of parameters... 8 SECTION B CAI calculation for FASTA
DNA Sample preparation and Submission Guidelines
 DNA Sample preparation and Submission Guidelines Requirements: Please submit samples in 1.5ml microcentrifuge tubes. Fill all the required information in the Eurofins DNA sequencing order form and send
DNA Sample preparation and Submission Guidelines Requirements: Please submit samples in 1.5ml microcentrifuge tubes. Fill all the required information in the Eurofins DNA sequencing order form and send
(A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors.
 Legends of supplemental figures and tables Figure 1: Overview of study design and results. (A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors. After raw data gene expression
Legends of supplemental figures and tables Figure 1: Overview of study design and results. (A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors. After raw data gene expression
10 µg lyophilized plasmid DNA (store lyophilized plasmid at 20 C)
 TECHNICAL DATA SHEET BIOLUMINESCENCE RESONANCE ENERGY TRANSFER RENILLA LUCIFERASE FUSION PROTEIN EXPRESSION VECTOR Product: prluc-c Vectors Catalog number: Description: Amount: The prluc-c vectors contain
TECHNICAL DATA SHEET BIOLUMINESCENCE RESONANCE ENERGY TRANSFER RENILLA LUCIFERASE FUSION PROTEIN EXPRESSION VECTOR Product: prluc-c Vectors Catalog number: Description: Amount: The prluc-c vectors contain
Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation
 BDGP Resources Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation For recovery of sequences flanking PZ, PlacW and PEP elements E. Jay Rehm Berkeley Drosophila Genome Project I.
BDGP Resources Inverse PCR & Cycle Sequencing of P Element Insertions for STS Generation For recovery of sequences flanking PZ, PlacW and PEP elements E. Jay Rehm Berkeley Drosophila Genome Project I.
Table S1. Related to Figure 4
 Table S1. Related to Figure 4 Final Diagnosis Age PMD Control Control 61 15 Control 67 6 Control 68 10 Control 49 15 AR-PD PD 62 15 PD 65 4 PD 52 18 PD 68 10 AR-PD cingulate cortex used for immunoblot
Table S1. Related to Figure 4 Final Diagnosis Age PMD Control Control 61 15 Control 67 6 Control 68 10 Control 49 15 AR-PD PD 62 15 PD 65 4 PD 52 18 PD 68 10 AR-PD cingulate cortex used for immunoblot
SERVICES CATALOGUE WITH SUBMISSION GUIDELINES
 SERVICES CATALOGUE WITH SUBMISSION GUIDELINES 3921 Montgomery Road Cincinnati, Ohio 45212 513-841-2428 www.agctsequencing.com CONTENTS Welcome Dye Terminator Sequencing DNA Sequencing Services - Full Service
SERVICES CATALOGUE WITH SUBMISSION GUIDELINES 3921 Montgomery Road Cincinnati, Ohio 45212 513-841-2428 www.agctsequencing.com CONTENTS Welcome Dye Terminator Sequencing DNA Sequencing Services - Full Service
Molecular analyses of EGFR: mutation and amplification detection
 Molecular analyses of EGFR: mutation and amplification detection Petra Nederlof, Moleculaire Pathologie NKI Amsterdam Henrique Ruijter, Ivon Tielen, Lucie Boerrigter, Aafke Ariaens Outline presentation
Molecular analyses of EGFR: mutation and amplification detection Petra Nederlof, Moleculaire Pathologie NKI Amsterdam Henrique Ruijter, Ivon Tielen, Lucie Boerrigter, Aafke Ariaens Outline presentation
Supplementary Online Material for Morris et al. sirna-induced transcriptional gene
 Supplementary Online Material for Morris et al. sirna-induced transcriptional gene silencing in human cells. Materials and Methods Lentiviral vector and sirnas. FIV vector pve-gfpwp was prepared as described
Supplementary Online Material for Morris et al. sirna-induced transcriptional gene silencing in human cells. Materials and Methods Lentiviral vector and sirnas. FIV vector pve-gfpwp was prepared as described
pcas-guide System Validation in Genome Editing
 pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
Next Generation Sequencing
 Next Generation Sequencing 38. Informationsgespräch der Blutspendezentralefür Wien, Niederösterreich und Burgenland Österreichisches Rotes Kreuz 22. November 2014, Parkhotel Schönbrunn Die Zukunft hat
Next Generation Sequencing 38. Informationsgespräch der Blutspendezentralefür Wien, Niederösterreich und Burgenland Österreichisches Rotes Kreuz 22. November 2014, Parkhotel Schönbrunn Die Zukunft hat
Gene Synthesis 191. Mutagenesis 194. Gene Cloning 196. AccuGeneBlock Service 198. Gene Synthesis FAQs 201. User Protocol 204
 Gene Synthesis 191 Mutagenesis 194 Gene Cloning 196 AccuGeneBlock Service 198 Gene Synthesis FAQs 201 User Protocol 204 Gene Synthesis Overview Gene synthesis is the most cost-effective way to enhance
Gene Synthesis 191 Mutagenesis 194 Gene Cloning 196 AccuGeneBlock Service 198 Gene Synthesis FAQs 201 User Protocol 204 Gene Synthesis Overview Gene synthesis is the most cost-effective way to enhance
UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet
 1 UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam:.
1 UNIVERSITETET I OSLO Det matematisk-naturvitenskapelige fakultet Exam in: MBV4010 Arbeidsmetoder i molekylærbiologi og biokjemi I MBV4010 Methods in molecular biology and biochemistry I Day of exam:.
The p53 MUTATION HANDBOOK
 The p MUTATION HANDBOOK Version 1. /7 Thierry Soussi Christophe Béroud, Dalil Hamroun Jean Michel Rubio Nevado http://p/free.fr The p Mutation HandBook By T Soussi, J.M. Rubio-Nevado, D. Hamroun and C.
The p MUTATION HANDBOOK Version 1. /7 Thierry Soussi Christophe Béroud, Dalil Hamroun Jean Michel Rubio Nevado http://p/free.fr The p Mutation HandBook By T Soussi, J.M. Rubio-Nevado, D. Hamroun and C.
Mutations and Genetic Variability. 1. What is occurring in the diagram below?
 Mutations and Genetic Variability 1. What is occurring in the diagram below? A. Sister chromatids are separating. B. Alleles are independently assorting. C. Genes are replicating. D. Segments of DNA are
Mutations and Genetic Variability 1. What is occurring in the diagram below? A. Sister chromatids are separating. B. Alleles are independently assorting. C. Genes are replicating. D. Segments of DNA are
Introduction to Perl Programming Input/Output, Regular Expressions, String Manipulation. Beginning Perl, Chap 4 6. Example 1
 Introduction to Perl Programming Input/Output, Regular Expressions, String Manipulation Beginning Perl, Chap 4 6 Example 1 #!/usr/bin/perl -w use strict; # version 1: my @nt = ('A', 'C', 'G', 'T'); for
Introduction to Perl Programming Input/Output, Regular Expressions, String Manipulation Beginning Perl, Chap 4 6 Example 1 #!/usr/bin/perl -w use strict; # version 1: my @nt = ('A', 'C', 'G', 'T'); for
Hands on Simulation of Mutation
 Hands on Simulation of Mutation Charlotte K. Omoto P.O. Box 644236 Washington State University Pullman, WA 99164-4236 omoto@wsu.edu ABSTRACT This exercise is a hands-on simulation of mutations and their
Hands on Simulation of Mutation Charlotte K. Omoto P.O. Box 644236 Washington State University Pullman, WA 99164-4236 omoto@wsu.edu ABSTRACT This exercise is a hands-on simulation of mutations and their
Supplementary Information. Binding region and interaction properties of sulfoquinovosylacylglycerol (SQAG) with human
 Supplementary Information Binding region and interaction properties of sulfoquinovosylacylglycerol (SQAG) with human vascular endothelial growth factor 165 revealed by biosensor based assays Yoichi Takakusagi
Supplementary Information Binding region and interaction properties of sulfoquinovosylacylglycerol (SQAG) with human vascular endothelial growth factor 165 revealed by biosensor based assays Yoichi Takakusagi
NimbleGen SeqCap EZ Library SR User s Guide Version 3.0
 NimbleGen SeqCap EZ Library SR User s Guide Version 3.0 For life science research only. Not for use in diagnostic procedures. Copyright 2011 Roche NimbleGen, Inc. All Rights Reserved. Editions Version
NimbleGen SeqCap EZ Library SR User s Guide Version 3.0 For life science research only. Not for use in diagnostic procedures. Copyright 2011 Roche NimbleGen, Inc. All Rights Reserved. Editions Version
TITRATION OF raav (VG) USING QUANTITATIVE REAL TIME PCR
 Page 1 of 5 Materials DNase digestion buffer [13 mm Tris-Cl, ph7,5 / 5 mm MgCl2 / 0,12 mm CaCl2] RSS plasmid ptr-uf11 SV40pA Forward primer (10µM) AGC AAT AGC ATC ACA AAT TTC ACA A SV40pA Reverse Primer
Page 1 of 5 Materials DNase digestion buffer [13 mm Tris-Cl, ph7,5 / 5 mm MgCl2 / 0,12 mm CaCl2] RSS plasmid ptr-uf11 SV40pA Forward primer (10µM) AGC AAT AGC ATC ACA AAT TTC ACA A SV40pA Reverse Primer
Title : Parallel DNA Synthesis : Two PCR product from one DNA template
 Title : Parallel DNA Synthesis : Two PCR product from one DNA template Bhardwaj Vikash 1 and Sharma Kulbhushan 2 1 Email: vikashbhardwaj@ gmail.com 1 Current address: Government College Sector 14 Gurgaon,
Title : Parallel DNA Synthesis : Two PCR product from one DNA template Bhardwaj Vikash 1 and Sharma Kulbhushan 2 1 Email: vikashbhardwaj@ gmail.com 1 Current address: Government College Sector 14 Gurgaon,
Inverse PCR and Sequencing of P-element, piggybac and Minos Insertion Sites in the Drosophila Gene Disruption Project
 Inverse PCR and Sequencing of P-element, piggybac and Minos Insertion Sites in the Drosophila Gene Disruption Project Protocol for recovery of sequences flanking insertions in the Drosophila Gene Disruption
Inverse PCR and Sequencing of P-element, piggybac and Minos Insertion Sites in the Drosophila Gene Disruption Project Protocol for recovery of sequences flanking insertions in the Drosophila Gene Disruption
Event-specific Method for the Quantification of Maize MIR162 Using Real-time PCR. Protocol
 Event-specific Method for the Quantification of Maize MIR162 Using Real-time PCR Protocol 31 January 2011 Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics
Event-specific Method for the Quantification of Maize MIR162 Using Real-time PCR Protocol 31 January 2011 Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics
EU Reference Laboratory for E. coli Department of Veterinary Public Health and Food Safety Unit of Foodborne Zoonoses Istituto Superiore di Sanità
 Identification and characterization of Verocytotoxin-producing Escherichia coli (VTEC) by Real Time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated
Identification and characterization of Verocytotoxin-producing Escherichia coli (VTEC) by Real Time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated
Gene Finding CMSC 423
 Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would you decipher
Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would you decipher
pcmv6-neo Vector Application Guide Contents
 pcmv6-neo Vector Application Guide Contents Package Contents and Storage Conditions... 2 Product Description... 2 Introduction... 2 Production and Quality Assurance... 2 Methods... 3 Other required reagents...
pcmv6-neo Vector Application Guide Contents Package Contents and Storage Conditions... 2 Product Description... 2 Introduction... 2 Production and Quality Assurance... 2 Methods... 3 Other required reagents...
Taqman TCID50 for AAV Vector Infectious Titer Determination
 Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Coding sequence the sequence of nucleotide bases on the DNA that are transcribed into RNA which are in turn translated into protein
 Assignment 3 Michele Owens Vocabulary Gene: A sequence of DNA that instructs a cell to produce a particular protein Promoter a control sequence near the start of a gene Coding sequence the sequence of
Assignment 3 Michele Owens Vocabulary Gene: A sequence of DNA that instructs a cell to produce a particular protein Promoter a control sequence near the start of a gene Coding sequence the sequence of
Part ONE. a. Assuming each of the four bases occurs with equal probability, how many bits of information does a nucleotide contain?
 Networked Systems, COMPGZ01, 2012 Answer TWO questions from Part ONE on the answer booklet containing lined writing paper, and answer ALL questions in Part TWO on the multiple-choice question answer sheet.
Networked Systems, COMPGZ01, 2012 Answer TWO questions from Part ONE on the answer booklet containing lined writing paper, and answer ALL questions in Part TWO on the multiple-choice question answer sheet.
RT-PCR: Two-Step Protocol
 RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
ANALYSIS OF A CIRCULAR CODE MODEL
 ANALYSIS OF A CIRCULAR CODE MODEL Jérôme Lacan and Chrstan J. Mchel * Laboratore d Informatque de Franche-Comté UNIVERSITE DE FRANCHE-COMTE IUT de Belfort-Montbélard 4 Place Tharradn - BP 747 5 Montbélard
ANALYSIS OF A CIRCULAR CODE MODEL Jérôme Lacan and Chrstan J. Mchel * Laboratore d Informatque de Franche-Comté UNIVERSITE DE FRANCHE-COMTE IUT de Belfort-Montbélard 4 Place Tharradn - BP 747 5 Montbélard
Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians
 Vol. 44 No. 3 SCIENCE IN CHINA (Series C) June 2001 Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians KE Yuehai ( `º) 1, SU Bing (3 Á) 1 3, XIAO Junhua
Vol. 44 No. 3 SCIENCE IN CHINA (Series C) June 2001 Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians KE Yuehai ( `º) 1, SU Bing (3 Á) 1 3, XIAO Junhua
Module 6: Digital DNA
 Module 6: Digital DNA Representation and processing of digital information in the form of DNA is essential to life in all organisms, no matter how large or tiny. Computing tools and computational thinking
Module 6: Digital DNA Representation and processing of digital information in the form of DNA is essential to life in all organisms, no matter how large or tiny. Computing tools and computational thinking
Supplemental Data. Short Article. PPARγ Activation Primes Human Monocytes. into Alternative M2 Macrophages. with Anti-inflammatory Properties
 Cell Metabolism, Volume 6 Supplemental Data Short Article PPARγ Activation Primes Human Monocytes into Alternative M2 Macrophages with Anti-inflammatory Properties M. Amine Bouhlel, Bruno Derudas, Elena
Cell Metabolism, Volume 6 Supplemental Data Short Article PPARγ Activation Primes Human Monocytes into Alternative M2 Macrophages with Anti-inflammatory Properties M. Amine Bouhlel, Bruno Derudas, Elena
Chapter 9. Applications of probability. 9.1 The genetic code
 Chapter 9 Applications of probability In this chapter we use the tools of elementary probability to investigate problems of several kinds. First, we study the language of life by focusing on the universal
Chapter 9 Applications of probability In this chapter we use the tools of elementary probability to investigate problems of several kinds. First, we study the language of life by focusing on the universal
http://www.life.umd.edu/grad/mlfsc/ DNA Bracelets
 http://www.life.umd.edu/grad/mlfsc/ DNA Bracelets by Louise Brown Jasko John Anthony Campbell Jack Dennis Cassidy Michael Nickelsburg Stephen Prentis Rohm Objectives: 1) Using plastic beads, construct
http://www.life.umd.edu/grad/mlfsc/ DNA Bracelets by Louise Brown Jasko John Anthony Campbell Jack Dennis Cassidy Michael Nickelsburg Stephen Prentis Rohm Objectives: 1) Using plastic beads, construct
Marine Biology DEC 2004; 146(1) : 53-64 http://dx.doi.org/10.1007/s00227-004-1423-6 Copyright 2004 Springer
 Marine Biology DEC 2004; 146(1) : 53-64 http://dx.doi.org/10.1007/s00227-004-1423-6 Copyright 2004 Springer Archimer http://www.ifremer.fr/docelec/ Archive Institutionnelle de l Ifremer The original publication
Marine Biology DEC 2004; 146(1) : 53-64 http://dx.doi.org/10.1007/s00227-004-1423-6 Copyright 2004 Springer Archimer http://www.ifremer.fr/docelec/ Archive Institutionnelle de l Ifremer The original publication
ANALYSIS OF GROWTH HORMONE IN TENCH (TINCA TINCA) ANALÝZA RŮSTOVÉHO HORMONU LÍNA OBECNÉHO (TINCA TINCA)
 ANALYSIS OF GROWTH HORMONE IN TENCH (TINCA TINCA) ANALÝZA RŮSTOVÉHO HORMONU LÍNA OBECNÉHO (TINCA TINCA) Zrůstová J., Bílek K., Baránek V., Knoll A. Ústav morfologie, fyziologie a genetiky zvířat, Agronomická
ANALYSIS OF GROWTH HORMONE IN TENCH (TINCA TINCA) ANALÝZA RŮSTOVÉHO HORMONU LÍNA OBECNÉHO (TINCA TINCA) Zrůstová J., Bílek K., Baránek V., Knoll A. Ústav morfologie, fyziologie a genetiky zvířat, Agronomická
Cloning, sequencing, and expression of H.a. YNRI and H.a. YNII, encoding nitrate and nitrite reductases in the yeast Hansenula anomala
 Cloning, sequencing, and expression of H.a. YNRI and H.a. YNII, encoding nitrate and nitrite reductases in the yeast Hansenula anomala -'Pablo García-Lugo 1t, Celedonio González l, Germán Perdomo l, Nélida
Cloning, sequencing, and expression of H.a. YNRI and H.a. YNII, encoding nitrate and nitrite reductases in the yeast Hansenula anomala -'Pablo García-Lugo 1t, Celedonio González l, Germán Perdomo l, Nélida
2006 7.012 Problem Set 3 KEY
 2006 7.012 Problem Set 3 KEY Due before 5 PM on FRIDAY, October 13, 2006. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Which reaction is catalyzed by each
2006 7.012 Problem Set 3 KEY Due before 5 PM on FRIDAY, October 13, 2006. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Which reaction is catalyzed by each
APOT - Assay. Protocol for HPV16 and 18. Amplification of Papilloma Virus Oncogene Transcripts HPV. E6 E7 E1 Zelluläre DNA poly(a)
 E5 E2 E1 APOT - Assay Amplification of Papilloma Virus Oncogene Transcripts URR E6 E7 L1 HPV L2 E4 E6 E7 E1 Zelluläre DNA poly(a) Protocol for HPV16 and 18 Brief summary of the APOT assay Fig.1A shows
E5 E2 E1 APOT - Assay Amplification of Papilloma Virus Oncogene Transcripts URR E6 E7 L1 HPV L2 E4 E6 E7 E1 Zelluläre DNA poly(a) Protocol for HPV16 and 18 Brief summary of the APOT assay Fig.1A shows
Heraeus Sepatech, Kendro Laboratory Products GmbH, Berlin. Becton Dickinson,Heidelberg. Biozym, Hessisch Oldendorf. Eppendorf, Hamburg
 13 4. MATERIALS 4.1 Laboratory apparatus Biofuge A Centrifuge 5804R FACScan Gel electrophoresis chamber GPR Centrifuge Heraeus CO-AUTO-ZERO Light Cycler Microscope Motopipet Neubauer Cell Chamber PCR cycler
13 4. MATERIALS 4.1 Laboratory apparatus Biofuge A Centrifuge 5804R FACScan Gel electrophoresis chamber GPR Centrifuge Heraeus CO-AUTO-ZERO Light Cycler Microscope Motopipet Neubauer Cell Chamber PCR cycler
Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps and colorectal cancer risk
 DOI 10.1007/s10552-009-9438-4 ORIGINAL PAPER Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps and colorectal cancer risk Elisabeth Feik Æ Andreas Baierl Æ Barbara Hieger Æ Gerhard Führlinger
DOI 10.1007/s10552-009-9438-4 ORIGINAL PAPER Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps and colorectal cancer risk Elisabeth Feik Æ Andreas Baierl Æ Barbara Hieger Æ Gerhard Führlinger
The making of The Genoma Music
 242 Summary Key words Resumen Palabras clave The making of The Genoma Music Aurora Sánchez Sousa 1, Fernando Baquero 1 and Cesar Nombela 2 1 Department of Microbiology, Ramón y Cajal Hospital, and 2 Department
242 Summary Key words Resumen Palabras clave The making of The Genoma Music Aurora Sánchez Sousa 1, Fernando Baquero 1 and Cesar Nombela 2 1 Department of Microbiology, Ramón y Cajal Hospital, and 2 Department
On Covert Data Communication Channels Employing DNA Recombinant and Mutagenesis-based Steganographic Techniques
 On Covert Data Communication Channels Employing DNA Recombinant and Mutagenesis-based Steganographic Techniques MAGDY SAEB 1, EMAN EL-ABD 2, MOHAMED E. EL-ZANATY 1 1. School of Engineering, Computer Department,
On Covert Data Communication Channels Employing DNA Recombinant and Mutagenesis-based Steganographic Techniques MAGDY SAEB 1, EMAN EL-ABD 2, MOHAMED E. EL-ZANATY 1 1. School of Engineering, Computer Department,
Provincial Exam Questions. 9. Give one role of each of the following nucleic acids in the production of an enzyme.
 Provincial Exam Questions Unit: Cell Biology: Protein Synthesis (B7 & B8) 2010 Jan 3. Describe the process of translation. (4 marks) 2009 Sample 8. What is the role of ribosomes in protein synthesis? A.
Provincial Exam Questions Unit: Cell Biology: Protein Synthesis (B7 & B8) 2010 Jan 3. Describe the process of translation. (4 marks) 2009 Sample 8. What is the role of ribosomes in protein synthesis? A.
INTERPRETATION INFORMATION SHEET
 Creative Testing Solutions 2424 West Erie Dr. 2205 Highway 121 10100 Martin Luther King Jr. St. No. Tempe, AZ 85282 Bedford, TX 76021 St. Petersburg, FL 33716 INTERPRETATION INFORMATION SHEET Human Immunodeficiency
Creative Testing Solutions 2424 West Erie Dr. 2205 Highway 121 10100 Martin Luther King Jr. St. No. Tempe, AZ 85282 Bedford, TX 76021 St. Petersburg, FL 33716 INTERPRETATION INFORMATION SHEET Human Immunodeficiency
Molecular chaperones involved in preprotein. targeting to plant organelles
 Molecular chaperones involved in preprotein targeting to plant organelles Dissertation der Fakultät für Biologie der Ludwig-Maximilians-Universität München vorgelegt von Christine Fellerer München 29.
Molecular chaperones involved in preprotein targeting to plant organelles Dissertation der Fakultät für Biologie der Ludwig-Maximilians-Universität München vorgelegt von Christine Fellerer München 29.
DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108
 DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 THE INTERLEUKIN-10 FAMILY CYTOKINES GENE POLYMORPHISMS IN PLAQUE PSORIASIS KÜLLI KINGO TARTU
DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 DISSERTATIONES MEDICINAE UNIVERSITATIS TARTUENSIS 108 THE INTERLEUKIN-10 FAMILY CYTOKINES GENE POLYMORPHISMS IN PLAQUE PSORIASIS KÜLLI KINGO TARTU
Application Guide... 2
 Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
Transmembrane Signaling in Chimeras of the E. coli Chemotaxis Receptors and Bacterial Class III Adenylyl Cyclases
 Transmembrane Signaling in Chimeras of the E. coli Chemotaxis Receptors and Bacterial Class III Adenylyl Cyclases Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
Transmembrane Signaling in Chimeras of the E. coli Chemotaxis Receptors and Bacterial Class III Adenylyl Cyclases Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
Five-minute cloning of Taq polymerase-amplified PCR products
 TOPO TA Cloning Version R 8 April 2004 25-0184 TOPO TA Cloning Five-minute cloning of Taq polymerase-amplified PCR products Catalog nos. K4500-01, K4500-40, K4510-20, K4520-01, K4520-40, K4550-01, K4550-40,
TOPO TA Cloning Version R 8 April 2004 25-0184 TOPO TA Cloning Five-minute cloning of Taq polymerase-amplified PCR products Catalog nos. K4500-01, K4500-40, K4510-20, K4520-01, K4520-40, K4550-01, K4550-40,
2. The number of different kinds of nucleotides present in any DNA molecule is A) four B) six C) two D) three
 Chem 121 Chapter 22. Nucleic Acids 1. Any given nucleotide in a nucleic acid contains A) two bases and a sugar. B) one sugar, two bases and one phosphate. C) two sugars and one phosphate. D) one sugar,
Chem 121 Chapter 22. Nucleic Acids 1. Any given nucleotide in a nucleic acid contains A) two bases and a sugar. B) one sugar, two bases and one phosphate. C) two sugars and one phosphate. D) one sugar,
RealStar HBV PCR Kit 1.0 11/2012
 RealStar HBV PCR Kit 1.0 11/2012 RealStar HBV PCR Kit 1.0 For research use only! (RUO) Product No.: 201003 96 rxns INS-201000-GB-02 Store at -25 C... -15 C November 2012 altona Diagnostics GmbH Mörkenstraße
RealStar HBV PCR Kit 1.0 11/2012 RealStar HBV PCR Kit 1.0 For research use only! (RUO) Product No.: 201003 96 rxns INS-201000-GB-02 Store at -25 C... -15 C November 2012 altona Diagnostics GmbH Mörkenstraße
The DNA-"Wave Biocomputer"
 The DNA-"Wave Biocomputer" Peter P. Gariaev (Pjotr Garjajev)*, Boris I. Birshtein*, Alexander M. Iarochenko*, Peter J. Marcer**, George G. Tertishny*, Katherine A. Leonova*, Uwe Kaempf ***. * Institute
The DNA-"Wave Biocomputer" Peter P. Gariaev (Pjotr Garjajev)*, Boris I. Birshtein*, Alexander M. Iarochenko*, Peter J. Marcer**, George G. Tertishny*, Katherine A. Leonova*, Uwe Kaempf ***. * Institute
ncounter Gene Expression Assay Manual Total RNA and Cell Lysate Protocols
 ncounter Gene Expression Assay Manual Total RNA and Cell Lysate Protocols v.20090807 For research use only. Not for use in diagnostic procedures. Limited License Subject to the terms and conditions of
ncounter Gene Expression Assay Manual Total RNA and Cell Lysate Protocols v.20090807 For research use only. Not for use in diagnostic procedures. Limited License Subject to the terms and conditions of
Supporting Information
 Supporting Information Wiley-VCH 2007 69451 Weinheim, Germany Evolving a Thermostable DNA polymerase that Accurately Amplifies Highly-Damaged Templates Christian Gloeckner, Katharina B. M. Sauter, and
Supporting Information Wiley-VCH 2007 69451 Weinheim, Germany Evolving a Thermostable DNA polymerase that Accurately Amplifies Highly-Damaged Templates Christian Gloeckner, Katharina B. M. Sauter, and
ISTEP+: Biology I End-of-Course Assessment Released Items and Scoring Notes
 ISTEP+: Biology I End-of-Course Assessment Released Items and Scoring Notes Page 1 of 22 Introduction Indiana students enrolled in Biology I participated in the ISTEP+: Biology I Graduation Examination
ISTEP+: Biology I End-of-Course Assessment Released Items and Scoring Notes Page 1 of 22 Introduction Indiana students enrolled in Biology I participated in the ISTEP+: Biology I Graduation Examination
Mutation. Mutation provides raw material to evolution. Different kinds of mutations have different effects
 Mutation Mutation provides raw material to evolution Different kinds of mutations have different effects Mutational Processes Point mutation single nucleotide changes coding changes (missense mutations)
Mutation Mutation provides raw material to evolution Different kinds of mutations have different effects Mutational Processes Point mutation single nucleotide changes coding changes (missense mutations)
All commonly-used expression vectors used in the Jia Lab contain the following multiple cloning site: BamHI EcoRI SmaI SalI XhoI_ NotI
 2. Primer Design 2.1 Multiple Cloning Sites All commonly-used expression vectors used in the Jia Lab contain the following multiple cloning site: BamHI EcoRI SmaI SalI XhoI NotI XXX XXX GGA TCC CCG AAT
2. Primer Design 2.1 Multiple Cloning Sites All commonly-used expression vectors used in the Jia Lab contain the following multiple cloning site: BamHI EcoRI SmaI SalI XhoI NotI XXX XXX GGA TCC CCG AAT
Drosophila NK-homeobox genes
 Proc. Natl. Acad. Sci. USA Vol. 86, pp. 7716-7720, October 1989 Biochemistry Drosophila NK-homeobox genes (NK-1, NK-2,, and DNA clones/chromosome locations of genes) YONGSOK KIM AND MARSHALL NIRENBERG
Proc. Natl. Acad. Sci. USA Vol. 86, pp. 7716-7720, October 1989 Biochemistry Drosophila NK-homeobox genes (NK-1, NK-2,, and DNA clones/chromosome locations of genes) YONGSOK KIM AND MARSHALL NIRENBERG
were demonstrated to be, respectively, the catalytic and regulatory subunits of protein phosphatase 2A (PP2A) (29).
 JOURNAL OF VIROLOGY, Feb. 1992, p. 886-893 0022-538X/92/020886-08$02.00/0 Copyright C) 1992, American Society for Microbiology Vol. 66, No. 2 The Third Subunit of Protein Phosphatase 2A (PP2A), a 55- Kilodalton
JOURNAL OF VIROLOGY, Feb. 1992, p. 886-893 0022-538X/92/020886-08$02.00/0 Copyright C) 1992, American Society for Microbiology Vol. 66, No. 2 The Third Subunit of Protein Phosphatase 2A (PP2A), a 55- Kilodalton
Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541
 PRODUCT INFORMATION Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541 Lot _ Store at -20 C Expiry Date _ www.thermoscientific.com/onebio CERTIFICATE OF ANALYSIS The Phusion Site-Directed Mutagenesis
PRODUCT INFORMATION Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541 Lot _ Store at -20 C Expiry Date _ www.thermoscientific.com/onebio CERTIFICATE OF ANALYSIS The Phusion Site-Directed Mutagenesis
Data Analysis for Ion Torrent Sequencing
 IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
Essentials of Real Time PCR. About Sequence Detection Chemistries
 Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
BacReady TM Multiplex PCR System
 BacReady TM Multiplex PCR System Technical Manual No. 0191 Version 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI Detailed Experimental
BacReady TM Multiplex PCR System Technical Manual No. 0191 Version 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI Detailed Experimental
N-terminal Regulatory Domains of Phosphodiesterases 1, 4, 5 and 10 examined with an Adenylyl Cyclase as a Reporter
 N-terminal Regulatory Domains of Phosphodiesterases 1, 4, 5 and 10 examined with an Adenylyl Cyclase as a Reporter Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
N-terminal Regulatory Domains of Phosphodiesterases 1, 4, 5 and 10 examined with an Adenylyl Cyclase as a Reporter Dissertation der Mathematisch-Naturwissenschaftlichen Fakultät der Eberhard Karls Universität
PNA BRAF Mutation Detection Kit
 - PNA BRAF Mutation Detection Kit Catalog Number KA2102 50 tests/kit Version: 01 Intended for research use only www.abnova.com Introduction and Background Intended use The PNA BRAF Mutation Detection Kit
- PNA BRAF Mutation Detection Kit Catalog Number KA2102 50 tests/kit Version: 01 Intended for research use only www.abnova.com Introduction and Background Intended use The PNA BRAF Mutation Detection Kit
Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction
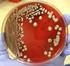 Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction Yogesh Chander 1 M. A. Ramakrishnan 1 Naresh Jindal 1 Kevan Hanson 2 Sagar M. Goyal 1 1 Department of Veterinary
Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction Yogesh Chander 1 M. A. Ramakrishnan 1 Naresh Jindal 1 Kevan Hanson 2 Sagar M. Goyal 1 1 Department of Veterinary
Synthetic Biology: DNA Digital Storage, Computation and the Organic Computer
 Synthetic Biology: DNA Digital Storage, Computation and the Organic Computer Alex Widdel University of Minnesota, Morris 1 / 27 Outline Overview of Synthetic Biology 1 Overview of Synthetic Biology 2 3
Synthetic Biology: DNA Digital Storage, Computation and the Organic Computer Alex Widdel University of Minnesota, Morris 1 / 27 Outline Overview of Synthetic Biology 1 Overview of Synthetic Biology 2 3
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
Molecular detection of Babesia rossi and Hepatozoon sp. in African wild dogs (Lycaon pictus) in South Africa
 Available online at www.sciencedirect.com Veterinary Parasitology 157 (2008) 123 127 Short communication Molecular detection of Babesia rossi and Hepatozoon sp. in African wild dogs (Lycaon pictus) in
Available online at www.sciencedirect.com Veterinary Parasitology 157 (2008) 123 127 Short communication Molecular detection of Babesia rossi and Hepatozoon sp. in African wild dogs (Lycaon pictus) in
Six Homeoproteins and a Iinc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
 Six Homeoproteins and a Iinc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype Iori Sakakibara 1,2,3, Marc Santolini 4, Arnaud Ferry 2,5, Vincent Hakim 4, Pascal Maire 1,2,3 * 1 INSERM U1016,
Six Homeoproteins and a Iinc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype Iori Sakakibara 1,2,3, Marc Santolini 4, Arnaud Ferry 2,5, Vincent Hakim 4, Pascal Maire 1,2,3 * 1 INSERM U1016,
Distribution of the DNA transposon family, Pokey in the Daphnia pulex species complex
 Eagle and Crease Mobile DNA (2016) 7:11 DOI 10.1186/s13100-016-0067-7 RESEARCH Open Access Distribution of the DNA transposon family, Pokey in the Daphnia pulex species complex Shannon H. C. Eagle and
Eagle and Crease Mobile DNA (2016) 7:11 DOI 10.1186/s13100-016-0067-7 RESEARCH Open Access Distribution of the DNA transposon family, Pokey in the Daphnia pulex species complex Shannon H. C. Eagle and
Accurate and sensitive mutation detection and quantitation using TaqMan Mutation Detection Assays for disease research
 PPLICTION NOTE Mutation Detection ssays ccurate and sensitive mutation detection and quantitation using Mutation Detection ssays for disease research In this research study, we addressed the feasibility
PPLICTION NOTE Mutation Detection ssays ccurate and sensitive mutation detection and quantitation using Mutation Detection ssays for disease research In this research study, we addressed the feasibility
Molecular Assessment of Dried Blood Spot Quality during Development of a Novel Automated. Screening
 Molecular Assessment of Dried Blood Spot Quality during Development of a Novel Automated in situ TREC qpcr Assay for SCID Screening J Bai, T Henry, J Benfer, S Berberich, T Kreman, and L DesJardin State
Molecular Assessment of Dried Blood Spot Quality during Development of a Novel Automated in situ TREC qpcr Assay for SCID Screening J Bai, T Henry, J Benfer, S Berberich, T Kreman, and L DesJardin State
Technical Note. Roche Applied Science. No. LC 18/2004. Assay Formats for Use in Real-Time PCR
 Roche Applied Science Technical Note No. LC 18/2004 Purpose of this Note Assay Formats for Use in Real-Time PCR The LightCycler Instrument uses several detection channels to monitor the amplification of
Roche Applied Science Technical Note No. LC 18/2004 Purpose of this Note Assay Formats for Use in Real-Time PCR The LightCycler Instrument uses several detection channels to monitor the amplification of
Introduction to Bioinformatics (Master ChemoInformatique)
 Introduction to Bioinformatics (Master ChemoInformatique) Roland Stote Institut de Génétique et de Biologie Moléculaire et Cellulaire Biocomputing Group 03.90.244.730 rstote@igbmc.fr Biological Function
Introduction to Bioinformatics (Master ChemoInformatique) Roland Stote Institut de Génétique et de Biologie Moléculaire et Cellulaire Biocomputing Group 03.90.244.730 rstote@igbmc.fr Biological Function
Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer s disease is this type 3 diabetes?
 Journal of Alzheimer s Disease 7 (2005) 63 80 63 IOS Press Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer s disease is this type 3 diabetes? Eric Steen,
Journal of Alzheimer s Disease 7 (2005) 63 80 63 IOS Press Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer s disease is this type 3 diabetes? Eric Steen,
Insulin Receptor Gene Mutations in Iranian Patients with Type II Diabetes Mellitus
 Iranian Biomedical Journal 13 (3): 161-168 (July 2009) Insulin Receptor Gene Mutations in Iranian Patients with Type II Diabetes Mellitus Bahram Kazemi 1*, Negar Seyed 1, Elham Moslemi 2, Mojgan Bandehpour
Iranian Biomedical Journal 13 (3): 161-168 (July 2009) Insulin Receptor Gene Mutations in Iranian Patients with Type II Diabetes Mellitus Bahram Kazemi 1*, Negar Seyed 1, Elham Moslemi 2, Mojgan Bandehpour
inhibition of mitosis
 The EMBO Journal vol.13 no.2 pp.425-434, 1994 cdt 1 is an essential target of the Cdc 1 O/Sct 1 transcription factor: requirement for DNA replication and inhibition of mitosis Johannes F.X.Hofmann and
The EMBO Journal vol.13 no.2 pp.425-434, 1994 cdt 1 is an essential target of the Cdc 1 O/Sct 1 transcription factor: requirement for DNA replication and inhibition of mitosis Johannes F.X.Hofmann and
Genetic Analysis. Phenotype analysis: biological-biochemical analysis. Genotype analysis: molecular and physical analysis
 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
Irina V Nesterova, Cecily A. Bennett, S. Sibel Erdem, Robert P. Hammer, Prescott L. Deininger, and Steven A. Soper
 ear-ir Single Fluorophore Quenching System Based on Phthalocyanine (Pc) Aggregation and its Application for Monitoring Inhibitor/Activator Action on a Therapeutic Target: L1-E Irina V esterova, Cecily
ear-ir Single Fluorophore Quenching System Based on Phthalocyanine (Pc) Aggregation and its Application for Monitoring Inhibitor/Activator Action on a Therapeutic Target: L1-E Irina V esterova, Cecily
a. Ribosomal RNA rrna a type ofrna that combines with proteins to form Ribosomes on which polypeptide chains of proteins are assembled
 Biology 101 Chapter 14 Name: Fill-in-the-Blanks Which base follows the next in a strand of DNA is referred to. as the base (1) Sequence. The region of DNA that calls for the assembly of specific amino
Biology 101 Chapter 14 Name: Fill-in-the-Blanks Which base follows the next in a strand of DNA is referred to. as the base (1) Sequence. The region of DNA that calls for the assembly of specific amino
The Arabinosyltransferase EmbC Is Inhibited by Ethambutol in Mycobacterium tuberculosis
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Oct. 2009, p. 4138 4146 Vol. 53, No. 10 0066-4804/09/$08.00 0 doi:10.1128/aac.00162-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. The
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, Oct. 2009, p. 4138 4146 Vol. 53, No. 10 0066-4804/09/$08.00 0 doi:10.1128/aac.00162-09 Copyright 2009, American Society for Microbiology. All Rights Reserved. The
360 Master Mix. , and a supplementary 360 GC Enhancer.
 Product Bulletin AmpliTaq Gold 360 Master Mix and 360 DNA Polymerase AmpliTaq Gold 360 Master Mix AmpliTaq Gold 360 DNA Polymerase 360 Coverage for a Full Range of Targets AmpliTaq Gold 360 Master Mix
Product Bulletin AmpliTaq Gold 360 Master Mix and 360 DNA Polymerase AmpliTaq Gold 360 Master Mix AmpliTaq Gold 360 DNA Polymerase 360 Coverage for a Full Range of Targets AmpliTaq Gold 360 Master Mix
Archimer http://archimer.ifremer.fr
 Please note that this is an author-produced PDF of an article accepted for publication following peer review. The definitive publisher-authenticated version is available on the publisher Web site Fish
Please note that this is an author-produced PDF of an article accepted for publication following peer review. The definitive publisher-authenticated version is available on the publisher Web site Fish
Interleukin-4 Receptor Signal Transduction: Involvement of P62
 Interleukin-4 Receptor Signal Transduction: Involvement of P62 Den Naturwissenschaftlichen Fakultäten der Friedrich Alexander Universität Erlangen Nürnberg zur Erlangung des Doktorgrades vorgelegt von
Interleukin-4 Receptor Signal Transduction: Involvement of P62 Den Naturwissenschaftlichen Fakultäten der Friedrich Alexander Universität Erlangen Nürnberg zur Erlangung des Doktorgrades vorgelegt von
TA Cloning Kit. Version V 7 April 2004 25-0024. Catalog nos. K2000-01, K2000-40, K2020-20, K2020-40, K2030-01 K2030-40, K2040-01, K2040-40
 TA Cloning Kit Version V 7 April 2004 25-0024 TA Cloning Kit Catalog nos. K2000-01, K2000-40, K2020-20, K2020-40, K2030-01 K2030-40, K2040-01, K2040-40 ii Table of Contents Table of Contents...iii Important
TA Cloning Kit Version V 7 April 2004 25-0024 TA Cloning Kit Catalog nos. K2000-01, K2000-40, K2020-20, K2020-40, K2030-01 K2030-40, K2040-01, K2040-40 ii Table of Contents Table of Contents...iii Important
Intended Use: The kit is designed to detect the 5 different mutations found in Asian population using seven different primers.
 Unzipping Genes MBPCR014 Beta-Thalassemia Detection Kit P r o d u c t I n f o r m a t i o n Description: Thalassemia is a group of genetic disorders characterized by quantitative defects in globin chain
Unzipping Genes MBPCR014 Beta-Thalassemia Detection Kit P r o d u c t I n f o r m a t i o n Description: Thalassemia is a group of genetic disorders characterized by quantitative defects in globin chain
Path-ID Multiplex One-Step RT-PCR Kit
 USER GUIDE Path-ID Multiplex One-Step RT-PCR Kit TaqMan probe-based multiplex one-step real-time RT-PCR detection of RNA targets Catalog Numbers 4428206, 4428207, 4440022 Publication Part Number 4440907
USER GUIDE Path-ID Multiplex One-Step RT-PCR Kit TaqMan probe-based multiplex one-step real-time RT-PCR detection of RNA targets Catalog Numbers 4428206, 4428207, 4440022 Publication Part Number 4440907
Directed-Mutagenesis and Deletion Generated through an Improved Overlapping-Extension PCR Based Procedure
 Research Article Directed-Mutagenesis and Deletion Generated through an Improved Overlapping-Extension PCR Based Procedure Wirojne Kanoksilapatham 1*, Juan M. González 2 and Frank T. Robb 3 1 Department
Research Article Directed-Mutagenesis and Deletion Generated through an Improved Overlapping-Extension PCR Based Procedure Wirojne Kanoksilapatham 1*, Juan M. González 2 and Frank T. Robb 3 1 Department
The Power of Next-Generation Sequencing in Your Hands On the Path towards Diagnostics
 The Power of Next-Generation Sequencing in Your Hands On the Path towards Diagnostics The GS Junior System The Power of Next-Generation Sequencing on Your Benchtop Proven technology: Uses the same long
The Power of Next-Generation Sequencing in Your Hands On the Path towards Diagnostics The GS Junior System The Power of Next-Generation Sequencing on Your Benchtop Proven technology: Uses the same long
Metabolic Engineering of Escherichia coli for Enhanced Production of Succinic Acid, Based on Genome Comparison and In Silico Gene Knockout Simulation
 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 2005, p. 7880 7887 Vol. 71, No. 12 0099-2240/05/$08.00 0 doi:10.1128/aem.71.12.7880 7887.2005 Copyright 2005, American Society for Microbiology. All Rights
APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Dec. 2005, p. 7880 7887 Vol. 71, No. 12 0099-2240/05/$08.00 0 doi:10.1128/aem.71.12.7880 7887.2005 Copyright 2005, American Society for Microbiology. All Rights
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR
 Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Characterization of cdna clones of the family of trypsin/a-amylase inhibitors (CM-proteins) in barley {Hordeum vulgare L.)
 Characterization of cdna clones of the family of trypsin/a-amylase inhibitors (CM-proteins) in barley {Hordeum vulgare L.) J. Paz-Ares, F. Ponz, P. Rodríguez-Palenzuela, A. Lázaro, C. Hernández-Lucas,
Characterization of cdna clones of the family of trypsin/a-amylase inhibitors (CM-proteins) in barley {Hordeum vulgare L.) J. Paz-Ares, F. Ponz, P. Rodríguez-Palenzuela, A. Lázaro, C. Hernández-Lucas,
Biopython Tutorial and Cookbook
 Biopython Tutorial and Cookbook Jeff Chang, Brad Chapman, Iddo Friedberg, Thomas Hamelryck, Michiel de Hoon, Peter Cock Last Update September 2008 Contents 1 Introduction 5 1.1 What is Biopython?.........................................
Biopython Tutorial and Cookbook Jeff Chang, Brad Chapman, Iddo Friedberg, Thomas Hamelryck, Michiel de Hoon, Peter Cock Last Update September 2008 Contents 1 Introduction 5 1.1 What is Biopython?.........................................
HBV Quantitative Real Time PCR Kit
 Revision No.: ZJ0002 Issue Date: Aug 7 th, 2008 HBV Quantitative Real Time PCR Kit Cat. No.: HD-0002-01 For Use with LightCycler 1.0/LightCycler2.0/LightCycler480 (Roche) Real Time PCR Systems (Pls ignore
Revision No.: ZJ0002 Issue Date: Aug 7 th, 2008 HBV Quantitative Real Time PCR Kit Cat. No.: HD-0002-01 For Use with LightCycler 1.0/LightCycler2.0/LightCycler480 (Roche) Real Time PCR Systems (Pls ignore
Chapter 5. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development
 Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development This chapter has been adapted from: W.K. Smits*, C.C. Eschevins*, K.A. Susanna, S. Bron, O.P.
Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development This chapter has been adapted from: W.K. Smits*, C.C. Eschevins*, K.A. Susanna, S. Bron, O.P.
