Rethinking synchronization of mammalian cells for cell cycle analysis
|
|
|
- Jonas Kelley
- 7 years ago
- Views:
Transcription
1 CMLS, Cell. Mol. Life Sci. 60 (2003) X/03/ Birkhäuser Verlag, Basel, 2003 CMLS Cellular and Molecular Life Sciences Review Rethinking synchronization of mammalian cells for cell cycle analysis S. Cooper Department of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, Michigan (USA), Fax , cooper@umich.edu Received 16 September 2002; received after revision 13 November 2002; accepted 22 November 2002 Abstract. An analysis of different classes of forced or batch synchronization methods reveals why these methods, in theory, do not produce synchronized cultures. Cells may be aligned for a particular property after specific treatments, but these aligned cells do not correspond to any particular cell age during the normal cell cycle. The experimental methods analyzed are those that arrest cells with a G1 phase amount of DNA, those that inhibit DNA synthesis, and those that arrest cells at mitosis. Release of arrested cells from inhibition does not produce cells reflecting cells during the normal division cycle. Thus, cells produced by batch or forcing methods are not experimental models for analysis of the normal cell cycle. Key words. Cell cycle; synchronization; double-thymidine block, G1 phase arrest; nocodazole; mitotic arrest. Introduction Genome-wide analysis of gene expression during the eukaryotic division cycle has been examined by a number of groups in a number of organisms using microarray technology. This type of analysis has been carried out in yeast [1, 2], primary human fibroblasts [3, 4], mouse fibroblasts [5], and human HeLa cells [6, 7]. From such studies, the overall and general conclusion is that there are a large number of genes expressed at particular times during the division cycle. For example, in the budding yeast, Saccharomyces cerevisiae, 800 genes were found to be periodically expressed [1]; in mouse fibroblasts, 578 such genes were identified [5]; in primary human fibroblasts, 700 periodically expressed genes were proposed to exist [4], and, most recently, more than 850 periodically expressed genes were identified in HeLa cells [7]. Problems with the large-scale gene analysis with microarrays to identify cell-cycle-specific expression patterns have been described [8]. More significant, detailed statistical analyses of the raw microarray data on gene expression during the division cycle of primary human fibroblasts [9] and yeast cells [10] have revealed problems with the proposal that there exist a large number of cell cycle specifically regulated genes. These reanalyses were stimulated by a proposal that the cell cycle is regulated by the continuous accumulation of material and not by sequential activation of specific genes [11 14]. The published analysis of the microarray data on fibroblasts serves as an instructive example [9]. A statistical reanalysis of the original data produced three principal findings. (i) Randomized data exhibit periodic patterns of similar or greater strength than the experimental data. This suggested that all apparent cyclicities in the expression measurements may arise from chance fluctuations. (ii) The presence of cyclicity and the timing of peak cyclicity in a given gene are not reproduced in two replicate experiments. This suggests that there is an uncontrolled source of experimental variation that is stronger than the innate variation of gene expression in cells over time. (iii) The amplitude of peak expression in the second cycle is not consistently smaller than the corresponding amplitude in the first cycle. This finding places doubt on CMLS 2253/M
2 2 S. Cooper Synchronization of mammalian cells the assumption that the cells are actually synchronized. The conclusion from this analysis was that the microarray results do not support the proposal that there are numerous cell cycle specifically expressed genes in human cells. Many batch treatments (i.e., treatments that affect all cells equally) are widely believed to produce synchronized cultures of eukaryotic cells. For example, serum starvation has been widely used as a synchronization method. Serum starvation or low serum concentration is believed to arrest cells at a particular point in the cell cycle. The arrested cells with a G1 phase amount of DNA are proposed to be arrested at this point in the G1 phase or in what is generally called the G0 phase. Upon restoration of serum, these arrested cells are assumed to pass synchronously through the cell cycle. Many methods related to serum starvation may be grouped as methods that arrest cells at a specific point in the G1 phase, sometimes referred to as the restriction point. Other synchronization methods such as the double-thymidine block method or hydroxyurea inhibition affect DNA synthesis and are proposed to arrest cells in S phase. A third class of batch synchronization methods arrests cells at mitosis. A reappraisal of these different synchronization methods is presented here. For the batch synchronization methods, theoretical problems argue against their use for understanding the normal cell cycle. Cells treated to starvation or inhibition methods are neither synchronized nor representative of cells at any particular stage of the cell cycle. Cell growth during the division cycle A synchronized culture is one where cells pass through the division cycle as a relatively uniform cohort and represent, at different time points, cells of different cell cycle ages. In passing through the cell cycle a newborn eukaryotic cell first passes through the G1 phase (absence of DNA synthesis), then S phase (period of DNA synthesis), then G2 phase (absence of DNA synthesis), with division occurring at M phase (mitosis). If many cells in a culture approximate this pattern as a group, these cells would be called a synchronized culture. A culture does not have to have perfect synchrony to be called synchronized. There is stochastic variation in interdivision times. If a cell in exponential growth had a measured doubling time of 20 h (i.e., an average interdivision time of 20 h), it might be expected that in a wellsynchronized culture started with a pure collection of newborn cells, a large majority of cells would divide between 18 and 22 h. Even a lower-resolution synchrony, with most of the cells dividing between 17 and 23 h might be acceptable for some purposes. So the discussion here is not related to whether the synchrony is good, or good enough, but rather to whether the methods, in theory, can produce a synchronized culture. A single cell passing through the cell cycle has two attributes. At each time point during the division cycle a cell has a particular DNA content and a particular cell mass. During steady-state exponential growth, the cell mass must increase during the division cycle, as a mother cell must have, on average, twice as much mass as a newborn cell. As cytoplasm is made in all phases of the cell cycle, it follows that the cell mass varies throughout the cell cycle. At each time point during synchronized growth, a synchronized culture should (on average) reflect in individual cells the cell mass and the DNA content of unperturbed cells. Beside the average DNA content and mass, there should be a relatively narrow spread in these cell characteristics so that cells in the synchronized culture can be considered equivalent to some particular cell at some particular cell cycle age in a normal culture. The concept of continuous change during the cell cycle is not restricted to cell mass. One can substitute a particular trigger molecule, or any molecule, or group of molecules for the generalized term mass. The proposal here is not that mass per se is the initiating or controlling factor of the cell cycle. It may be. But if a synchronized culture does not reflect the normal, unperturbed DNA-massmolecule synthetic pattern during the cell cycle, the burden of proof that such a culture is normal rests on those studying that synchronized culture. As will be shown below, merely aligning cells with some particular property (e.g., all cells with a G1 phase DNA content, or all cells arrested in mitosis) is not equivalent to producing a synchronized culture where the cells reflect the normal physiology of a growing cell. Underlying the analysis presented here is the assumption that the purpose of synchronization is not merely to obtain a set of starting cells that have some cell characteristic uniformly present in all cells (e.g., all cells with a G1 phase DNA content). Rather, the purpose of synchronization is to obtain a culture of cells mimicking the normal, unperturbed cell cycle. Expanding on this idea, the analysis below proposes that batch procedures where all cells in an exponentially growing culture are treated identically cannot, in theory, synchronize cells. Put another way, even cells that have one characteristic in common are not synchronized if this common feature was produced by a batch treatment (i.e., all cells treated identically). One of the most common observations regarding batch methods of synchronization is that the synchrony rapidly decays, so that a second cycle is rarely studied or observed. The general explanation for this lack of a second cycle is that interdivision time variability leads to a rapid decay in synchrony. The development of the membraneelution synchronization method, and the results from that method, presented in the following section, suggest that a
3 CMLS, Cell. Mol. Life Sci. Vol. 60, 2003 Review Article 3 more appropriate conclusion from the lack of a second cycle of synchrony in batch-synchronized cells is that the cells were not really synchronized. Membrane-elution: a new standard for synchronization The recent development of a new method for cell synchronization emphasizes the problems inherent in the batch synchronization methods. This method was developed by Charles Helmstetter after many years of effort [15]. The history of this development has been described [16]. In membrane-elution, exponentially growing cells are bound to a membrane. Cells grow normally on the membrane. At division, one of the daughter cells remains attached to the membrane while the other daughter cell is released into the eluate. Temperature and medium are maintained constant throughout the procedure. In a typical membrane-elution synchronized culture, one can produce three clear synchronized divisions with clear plateaus between the doublings in cell number. DNA content distributions as well as size distributions indicate that the cells eluted from the membrane-elution apparatus reflect the normal pattern of growth during the division cycle. The newborn cells are appropriately small (equivalent in size to the cells at the low-size end of the size distribution of the exponential culture) and these newborn cells have a G1 phase DNA content. The membrane-elution method therefore shows that without any starvation, one can have a narrowing in the size distribution and a narrow DNA content that reflects the normal cell cycle. The phrase narrow size distribution, must be clarified. If a normal culture has cell sizes ranging from size 1 to 2, it is not a narrowing of size distribution to produce a culture that has a size distribution from 0.75 to 1.5. Although in the normal culture, the absolute difference is 1.0 between the largest and smallest and only 0.75 for the synchronized culture, what really counts is the ratio of the largest to the smallest cells in a culture. A narrowing would occur only if there was a significant decrease in the ratio of largest to smallest cell sizes from 2.0 to something like 1.1 or 1.2. In what follows, the theoretical expectations of three different batch methods are compared for DNA contents and expected size distributions. The results obtained with other methods will be compared to those cells produced by the membrane-elution method. Analysis of batch/forcing synchronization The analysis of synchronization methods is presented in a series of four tables. Table 1 describes the DNA and cell mass configurations of cells during steady-state growth without any synchronization or inhibition. One purpose of this first table is to explain how to read the events as the initial cells move through time that is, down each column. The subsequent three tables then go through the different classes of synchronization illustrating why these methods do not work. Analysis of the methods is presented primarily in the table legends, with the text merely putting the analysis in perspective. Following the analysis of the tables, a diagrammatic summary of tables 1 4 is presented in figure 1. Steady-state growth The pattern of mass increase, DNA replication, and cell division for an exponentially growing culture is presented in table 1. Each of the initial cells (top row is time zero) should be followed down each column to follow the history of each initial cell. (The attribution of size 0.07 to the youngest cell in the first line is due to the use of the last size in the age bin as the size for the bin. Thus, for 14 cells in the first line, the sizes of the youngest cells go from 0.0 to It should not be considered that the first line are cells at some later time, because there is an instant at which the experimental starvation starts.) The top line in table 1 represents the initial cells at various stages during the division cycle. By following each cell down each column, one can see what happens to each individual cell after time zero. When a cell divides, the mother cell mass is apportioned equally to the two daughter cells. Cells divide at mass 2.00 but the daughter cells are not illustrated immediately after this division but 0.07 (one-fourteenth) of a cell cycle later. At this time, during steady-state growth, the daughter cells are of size During steady-state growth (table 1), across any row, the DNA pattern as well as the cell mass pattern are invariant. There are always five cells with a G1 phase amount of DNA, four in S phase, four in G2 phase, and one in M phase. Mass increases within each row from 1.05 to 2.00, although the columnar location of the largest and smallest cells varies during the period of steady-state growth. G1 phase arrest The consequences of inhibition of mass increase is considered in table 2. Although the analysis presented in table 2 is phrased in terms of mass increase, it can apply to any method that is proposed to arrest cells with a G1 phase amount of DNA or, as usually phrased, in G1 phase. Thus, alpha factor arrest in yeast, arrest at the restriction point, and even quiescence or G(0) arrest are considered in this analysis. The detailed analysis is presented in the accompanying legend.
4 4 S. Cooper Synchronization of mammalian cells Table 1. Steady-state growth. G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 G G G G G S 1.41 S 1.49 S 1.56 G G G G M 2.00 G G G G G S 1.35 S 1.49 S 1.56 G G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.56 G G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.49 G G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G M 2.00 G G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G L H Each vertical column illustrates the stage of the cell cycle (i.e., G1, S, G2, or M) and the relative cell mass (the number) for cells growing exponentially without inhibition. The cell at the upper left just started G1 phase a short time earlier. It moves through G1, S, and G2 phases (moving down the column) to divide at mitosis (M phase) to produce a cell with a G1 phase amount of DNA (lower left). The first row across the top of the table shows the stage of the cell cycle (as in the vertical column, with G1, S, G2, and M indicated) and the relative cell masses for cells throughout the cell cycle at the start of the analysis. These cells have the same DNA contents and cell masses presented horizontally as in the first column presented vertically. The cells across the first row each represent a cell during one-fourteenth of the cell cycle. At the start of the analysis (first row across top of table), the first five cells are in G1 phase, the next four cells are in S phase, four cells follow in G2 phase, and one cell represents the final cell at division, in M phase. The numerical values accompanying the DNA descriptions indicate the cytoplasmic mass of cells with the mass increasing exponentially during the cell cycle. A newborn cell has a mass of 1.0, a dividing cell has a mass of 2.00, and a cell of age 0.07 (one-fourteenth of the cell cycle time) has a mass of Each of these initial cells (top row) can now be followed down each column during a period of time equivalent to one interdivision time. Each of the cells increases in mass, the cells pass through the DNA phases of the cell cycle, and at M phase, the cells divide. The first cell (upper left) goes through G1 phase, enters S phase at size 1.35, enters G2 phase at size 1.64, and divides at size 2.00 to produce a cell with a G1 phase amount of DNA and size 1.05 after passing through a short period (0.07) of the next cell cycle. The bold, underlined values indicate that a newborn cell has been produced. The mass in these cells is halved as two daughter cells are produced at mitosis. The cell masses are given on a per cell basis. Thus, in the first column, the cell in M phase divides at size 2.00 producing two cells of size 1.00; after 0.07 of a cell division cycle, the two cells each have a size of Below the first column is an L and below the last column is an H. These letters indicate in this and subsequent tables, the highest and lowest masses present in the cells after a period equivalent to one full interdivision time. At the end of one interdivision time, and during the entire period of growth, the cell masses spread over a factor of 2.00, just as in the original culture. The smallest cell is always 1.05 (note, this cell is age 0.07) and the largest (age 1.00) is mass The assumption on which the analysis in table 2 is based is that initiation of S phase requires the accumulation of some cellular element or elements to a certain level. In the steady-state condition (table 1), this value is If a cell does not reach this value by continued mass increase, S phase initiation is prevented. Again, mass per se is not proposed to initiate S phase. It may be the accumulation of some specific molecule that reflects cell mass or is a constant proportion of cell mass. When mass increase ceases, the cells in S, G2, and M phases continue through these phases to divide and produce two daughter cells. As mass did not increase, we get some cells that are relatively small compared to the initial culture (lowest row, table 2). In this idealized example, all cells in S, G2, and M phases are assumed to proceed to division. The definition of a culture arrested with a G1 phase amount of DNA (or as usually phrased, arrested in G1 phase ), means that all cells in S, G2, and M phases must proceed to division. It has not been proposed that cells with greater than a G1 phase amount of DNA regress backwards through the cell cycle, reducing their DNA content to arrive at the G1 phase amount of DNA. One can substitute for mass any process that accumulates continuously through the cell cycle. The numbers in the column may refer to the relative amount of some trigger molecule. One can inhibit a process that is independent of the cell cycle (e.g., mass increase) and get cells all with a G1 phase amount of DNA. Are these cells in G1 phase? The answer given here is no. The cells all have different cell masses, and the spread in cell mass ranges over a factor of 2.0. For a culture to be synchronized, the cell masses should have a narrower range than the cell mass range in the original culture. Inhibition of DNA synthesis Table 3 illustrates inhibition of DNA replication as a synchronizing method. During inhibition of DNA synthesis, mass increases allowing cells to pass through the G1, G2, and M phases of the cell cycle to produce cells that are ar-
5 CMLS, Cell. Mol. Life Sci. Vol. 60, 2003 Review Article 5 Table 2. Inhibition of mass synthesis, or arrest with a G1 phase amount of DNA. G G G G G S 1.35 S 1.41 S 1.49 G G G G M 1.90 G G G G G G S 1.35 S 1.41 G G G G M 1.81 G G G G G G G S 1.35 G G G G M 1.72 G G G G G G G G G G G G M 1.64 G G G G G G G G G G G G M 1.56 G G G G G G G G G G G G M 1.49 G G G G G G G G G G G G M 1.41 G G G G G G G G G G G G M 1.35 G G G G G G G G H L The top row shows (as in table 1) cells in all phases of the division cycle at the start of the experiment. When mass synthesis is inhibited. This means that the cell masses in the first five columns do not change throughout the period of analysis. Furthermore, these cells do not start S phase as they never reach size 1.35, the size associated with the initiation of S phase or DNA synthesis. The cells are thus fixed in size and DNA content for the period of analysis. In each column, we observe what is happening to the cells in the initial culture. All cells do not react identically to the mass inhibition. Cells in columns 6 14 can eventually divide. In these columns, the total mass, as in columns 1 5, does not increase. However, cells can divide after passing through an S, G2, and M phase. At division there is a halving of the available mass (newborn cells again being indicated by bold and underlining). At the end of the experiment, all cells have a G1 phase amount of DNA, but the sizes of the cells are not equivalent. As in table 1, an H and an L indicate the highest and lowest cell masses after starvation for one full interdivision time. The masses in the bottom row range from 0.67 to Two points should be noted. The spread in cell masses in the bottom row is twofold, equivalent to the spread in cell masses in the original cells (sizes 1.0 to 2.0). Second, there are cells that are smaller than any of the G1 phase cells in the original culture (sizes 0.67 to 0.95 in the bottom row). Upon resumption of mass synthesis, the cell in column 5 (mass 1.28) would be expected to achieve mass 1.35 (the start of S phase mass) before other cells. With time, cells with lower masses (e.g., 1.22, 1.16, 1.10, 1.05, 1.00, 0.95, 0.91, 0.86, , 0.74, 0.71, and finally 0.67) would achieve a mass equivalent to that required for initiation of S phase (mass 1.35). Thus, the initiation of S phase would not be synchronous but would be spread over a time equivalent to that of one full doubling time. This is because one full doubling time would be needed for the cell of mass 0.67 to reach a mass of 1.35, which is associated with initiation of S phase. Table 3. Inhibition of DNA synthesis. G G G G S 1.35 S 1.41 S 1.49 S 1.56 S 1.64 G G G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 S 1.64 S 1.72 G G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 S 1.64 S 1.72 S 1.81 G M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 S 1.64 S 1.72 S 1.81 S 1.90 M 2.00 G G G G S 1.35 S 1.41 S 1.49 S 1.56 S 1.64 S 1.72 S 1.81 S 1.90 S 2.00 G G G G G S 1.41 S 1.49 S 1.56 S 1.64 S 1.72 S 1.81 S 1.90 S 2.00 S 2.10 G G G G S 1.35 S 1.49 S 1.56 S 1.64 S 1.72 S 1.81 S 1.90 S 2.00 S 2.10 S 2.21 G G G S 1.35 S 1.41 S 1.56 S 1.64 S 1.72 S 1.81 S 1.90 S 2.00 S 2.10 S 2.21 S 2.32 G G S 1.35 S 1.41 S 1.49 S 1.64 S 1.72 S 1.81 S 1.90 S 2.00 S 2.10 S 2.21 S 2.32 S 2.44 G S 1.35 S 1.41 S 1.49 S 1.56 S 1.72 S 1.81 S 1.90 S 2.00 S 2.10 S 2.21 S 2.32 S 2.44 S 2.56 S 1.35 S 1.41 S 1.49 S 1.56 S 1.64 S 1.81 S 1.90 S 2.00 S 2.10 S 2.21 S 2.32 S 2.44 S 2.56 S 2.69 S 1.41 S 1.49 S 1.56 S 1.64 S 1.72 S 1.90 S 2.00 S 2.10 S 2.21 S 2.32 S 2.44 S 2.56 S 2.69 S 2.83 S 1.49 S 1.56 S 1.64 S 1.72 S 1.81 S 2.00 S 2.10 S 2.21 S 2.32 S 2.44 S 2.56 S 2.69 S 2.83 S 2.97 S 1.56 S 1.64 S 1.72 S 1.81 S 1.90 S 2.10 S 2.21 S 2.32 S 2.44 S 2.56 S 2.69 S 2.83 S 2.97 S 3.12 S 1.64 S 1.72 S 1.81 S 1.90 S 2.00 H L The cells in a growing culture (top row) are subjected to inhibition of DNA replication for a period equivalent to one interdivision time. In this case, we assume some condition is imposed that specifically inhibits DNA synthesis from occurring. As in tables 1 and 2, the top row shows the starting cells with a given pattern of DNA contents and cell mass increasing exponentially during the division cycle. Cells in S phase remain in S phase although mass increases. These cells do not divide, as they never complete S phase. Cells in G1 phase increase mass and then presumably initiate S phase although no DNA replication occurs because DNA synthesis, by definition of the experiment, is inhibited. In this case, the initiation of S phase without subsequent DNA synthesis means that the cells have achieved the ability to initiate DNA replication. Cells at the start of S phase (in this nascent synthetic state) are indicated by italics. Cells in the G2 phase complete the G2 phase, perform mitosis, divide, pass through G1 phase, and then arrive at the start of S phase (again indicated by italics). At the end of the period of DNA inhibition, all cells are shown in S phase or at the start of S phase, but these cells all have different cell masses, ranging from 1.64 to (The H and L indicate the smallest and largest cells in the culture.) Upon release from inhibition of DNA replication, there may be a rapid synthesis of DNA, but subsequent initiations of DNA replication would be spread out in time (asynchronous) because of the variation in cell masses at the time that DNA replication was permitted to occur.
6 6 S. Cooper Synchronization of mammalian cells rested either at the start of S phase (italicized S in table 3), or in mid S phase for those cells caught in S phase at the moment of inhibition of DNA replication. By following each of the initial cells (top row) down each column, one can see that after a period of inhibition, a collection of cells is produced with varied sizes. The spread in cell sizes is no narrower than that in the original culture (from 1.64 to 3.12, compared to 1.05 to 2.00). Thus, as with the analysis in table 2, no synchronization has taken place. Mitotic arrest Table 4 shows the results obtained after an inhibition of mitosis. The classical inhibitor for such a process is nocodazole. As cells are arrested in M phase, a collection of cells is obtained with varying sizes spread over a factor of 2.00 (from 2.10 to 4.00). None of these cells after a period of inhibition reflects any of the normal cells in a culture. Upon arrest of cells with condensed chromosomes in mitosis, mass synthesis may cease. In this eventuality, cells would be arrested with a common DNA configuration (condensed chromosomes) and a common mass (size 2.0 as indicated in table 4 for cells in mitosis). In this case, if one starved cells for one cell interdivision time, the first cell arriving at mitosis would be inhibited for one cell interdivision time, and the last cell arriving at mitosis would be inhibited for essentially no time. Thus the history of the cells with apparent physical similarities is quite different, with cells having major differences in residence in arrest conditions. The burden of proof rests on those that use mitotic inhibition to demonstrate that cells inhibited in mitosis are not affected by this variable history and that they are actually synchronized and exhibit synchronized divisions. The results in tables 1 4 are illustrated in figure 1. According to the analyses in tables 2 4, the synchronization methods analyzed do not produce cells that have a narrow size distribution and a narrow DNA content distribution. One can contrast this result with the theoretical results from a selective method. Uses of cells arrested with a particular cell property There may be reasons to use some inhibition method to produce a particular set of cells. For example, one may wish to get a group of cells with chromosomes condensed so that there can be isolated. There is nothing wrong with this. Some cell fusion/cloning experiments ( Dolly -like cloning) may work best with cells with a particular DNA content. What is argued here is that the methods used to produce a culture of cells that have a particular property in common should be restricted to the study of that property. These cells should not be called synchronized. Similarly, cells arrested by a particular batch procedure may produce periodic expression patterns following re- Table 4. Inhibition of mitosis or entry into mitosis. G G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 M 2.10 G G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 M 2.10 M 2.21 G G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 M 2.10 M 2.21 M 2.32 G S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 S 1.35 S 1.41 S 1.49 S 1.56 G G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 S 1.41 S 1.49 S 1.56 G G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 S 1.49 S 1.56 G G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 S 1.56 G G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 G G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 M 3.12 G G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 M 3.12 M 3.28 G G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 M 3.12 M 3.28 M 3.45 G M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 M 3.12 M 3.28 M 3.45 M 3.62 M 2.00 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 M 3.12 M 3.28 M 3.45 M 3.62 M 3.81 M 2.10 M 2.21 M 2.32 M 2.44 M 2.56 M 2.69 M 2.83 M 2.97 M 3.12 M 3.28 M 3.45 M 3.62 M 3.81 M 4.00 L H Assume that some compound inhibits completion of mitosis or entry into mitosis. This is added to the initial cells (first row). Because cell division is inhibited, and in the case illustrated here we assume that mass synthesis increases unabated, the cells pass through the phases of the cell cycle (G1 to S to G2 to M), and arrest with an M phase DNA configuration. Mass increases continuously and exponentially and as there is no cell division, there is no point at which mass is halved between two daughter cells. At the end of the incubation period, the cells all have an M phase DNA configuration and the cell masses range from 2.10 to 4.0. Again, the spread in cell masses ranges over a factor of 2.0, similar to the range in mass sizes of the initial culture (1.05 to 2.0). Upon release from inhibition, one may envision a synchronous cell division if all cells complete M phase at the same time, but the next initiation would vary over time as the cell masses after the cell divisions would vary from 1.05 to 2.0. Some cells would have a mass greater than that required for initiation of S phase (1.35), while others would have to increase mass to reach Thus there would not be a synchronous initiation of S phase after the mitotic period. Also observe that the cells in M phase have masses between 2.10 and 4.00 which is not like the masses of 2.0 of the original cells (upper right corner).
7 CMLS, Cell. Mol. Life Sci. Vol. 60, 2003 Review Article 7 Figure 1. Diagrammatic representation of the results derived in tables 1 4. (A) The final result from table 1, steady-state growth. (B) The final result from table 2, inhibition of mass increase leading to cells arrested with a G1 phase amount of DNA. (C) The final result in table 3, arrest of cells by inhibition of DNA replication. (D) The final result from table 4, arrest of cells at mitosis. lease from inhibition. But these patterns of gene expression should be stated as being related to this or that treatment. Whether or not the periodicity reflects the normal pattern of gene expression is a separate question. Supporting experimental results Experimental support of the proposal made here is primarily a pattern of omission of successful synchronization results. For all of the hundreds or thousands of experiments where cells are synchronized using batch or forcing methods, a cell-synchrony curve is rarely pictured. Some of these synchrony experiments present FACS analyses of cellular DNA contents following release from growth arrest. Yet even these are generally not supportive of synchronization. However, there are occasional papers that present experimental data supporting the proposal made here, that batch synchronization methods do not actually synchronize cells. An experiment with cells arrested with a G1 phase amount of DNA (described as synchronization by the authors [17]) did not show the expected DNA patterns after release from arrest [17, 18]. An analysis of an experiment using a double-thymidine block (DNA inhibition) [4] also did not support the proposal of synchronization [9]. The use of lovastatin to synchronize cells [19] and to arrest cells in G1 phase [20, 21] was shown to have an alternative explanation, with the cells neither synchronized nor arrested in G1 phase [22]. An analysis of the classical restriction point proposal also revealed problems with the proposal that cells are arrested at a particular point in the G1 phase, generally called the restriction point [23]. Thus there is support for the proposals made here. Because negative results where cells after treatment are not observed to be synchronized are usually not published, there is a bias against finding non-synchronization results in the literature. That there are many such reports suggests that the inability to synchronize cells may be more common than the literature would suggest. The recent study of gene expression during the division cycle of HeLa cells [7] contains what may arguably be the best evidence in support of the proposals made here. Two of the methods used, a double-thymidine block and a thymidine-nocodazole block were analyzed by FACS determination of DNA contents of cells after release from the arrest procedures. Although the text of the paper proposes that the cells were synchronized, the results presented in their figure 1 suggest otherwise. The doublethymidine block has the initial cells with what appears to be an excess of DNA (8n compared to 4n normally found at later times), and the DNA patterns, while suggestive, are not clearly indicative of unperturbed synchronization. Similarly for the thymidine-nocodazole blocked cells where at 12 h there were cells from all phases of the cell cycle present, rather than cells from only one or at most two phases of the cell cycle as expected. A comparison of the HeLa results to synchronization using the membrane-
8 8 S. Cooper Synchronization of mammalian cells elution method [15] clearly indicates that the HeLa cells were not synchronized. Criteria for successful synchronization The proposal here is that there is no arrest or batch synchronization procedure that can bring all aspects of the cell into a common state. If one process of a cell is arrested, other processes continue. This conclusion is most evident in the literature on arrest with a G1 phase amount of DNA. Starvation of the cell for a required nutrient that presumably arrests mass synthesis (e.g., starvation for an amino acid stops protein synthesis) can lead to cells with a G1 phase amount of DNA. This requires that cells in S phase or G2/M phase at the time of arrest complete these processes and divide to produce cells with a G1 phase amount of DNA. Similarly for inhibition of DNA synthesis, where inhibition of DNA replication need not affect protein or mass synthesis. This produces a population of cells that may have one property in common, but all other aspects of the cell are as spread out as in the original culture. The proposal made here is best summarized in three criteria for a successful synchronization. One criterion is that the DNA pattern or content in the initial cell population should be uniform in the initial cells. The second criterion is that the size distribution of the initial cells should be narrower than the size distribution of the initial cells. And finally, and most important, the third criterion is that the synchronized culture should exhibit synchronized cell divisions. Unless one can theoretically describe a system to produce cells that have a narrow size distribution and a narrow DNA distribution and synchronized divisions, it is not clear that one can synchronize cells by non-selective (i.e., batch) methods. I do understand that many researchers believe that cells arrested with a common cell property (i.e., all cells with a G1 phase amount of DNA) are the same. But this is merely a belief. Until the methods are shown to work to produce a synchronously dividing culture, one cannot say that any batch synchronization method actually synchronizes cells. 1 Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B. et al. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 9: Cho R. J., Campbell M. J., Winzeler E. A., Steinmetz L., Conway A., Wodicka L. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 2: Iyer V. R., Eisen M. B., Ross D. T., Schuler G., Moore T., Lee J. C. et al. (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283: Cho R. J., Huang M., Campbell M. J., Dong H., Steinmetz L., Sapinoso L. et al. (2001) Transcriptional regulation and function during the human cell cycle. Nat. Genet. 27: Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M. et al. (2001) Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21: Crawford D. F. and Piwnica-Worms H. (2001) The G(2) DNA damage checkpoint delays expression of genes encoding mitotic regulators. J. Biol. Chem. 276: Whitfield M., Sherlock G., Saldanha A., Murray J. I., Ball C. A., Alexnder K. E. et al. (2002) Identification of Genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13: Cooper S. (2002) Cell cycle analysis and microarrays. Trends Genet. 18: Shedden K. and Cooper S. (2002) Analysis of cell-cycle-specific gene expression in human cells as determined by microarrays and double-thymidine block synchronization. Proc. Natl. Acad. Sci. USA 99: Shedden K. and Cooper S. (2002) Analysis of cell-cycle-specific gene exresssion in Saccharomyces cerevisiae as determined by microarrays and multiple synchronization methods. Nucleic. Acids. Res. 30: Cooper S. (2000) The continuum model and G1-control of the mammalian cell cycle. Prog. Cell Cycle Res. 4: Cooper S. and Shayman J. A. (2001) Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell. Mol. Life Sci. 58: Cooper S. (1979) A unifying model for the G1 period in prokaryotes and eukaryotes. Nature 280: Cooper S. (1981) The continuum model: application to G1-arrest and G(O). In: Cell Growth, pp , Nicolini C. (ed.), Plenum, New York 15 Thornton M., Eward K. L. and Helmstetter C. E. (2002) Production of minimally disturbed synchronous cultures of hematopoietic cells. Biotechniques 32: Cooper S. (2002) Minimally disturbed, multi-cycle, and reproducible synchrony using a eukaryotic baby machine. Bioessays 24: Di Matteo G., Fuschi P., Zerfass K., Moretti S., Ricordy R., Cenciarelli C. et al. (1995) Transcriptional control of the Htf9- A/RanBP-1 gene during the cell cycle. Cell Growth Differ. 6: Cooper S. (1998) Mammalian cells are not synchronized in G1- phase by starvation or inhibition: considerations of the fundamental concept of G1-phase synchronization. Cell Prolif. 31: Keyomarsi K., Sandoval L., Band V. and Pardee A. B. (1991) Synchronization of tumor and normal cells from G1 to multiple cell cycles by lovastatin. Cancer Res. 51: Rao S., Lowe M., Herliczek T. W. and Keyomarsi K. (1998) Lovastatin mediated G1 arrest in normal and tumor breast cells is through inhibition of CDK2 activity and redistribution of p21 and p27, independent of p53. Oncogene 17: Rao S., Porter D. C., Chen X., Herliczek T., Lowe M. and Keyomarsi K. (1999) Lovastatin-mediated G1 arrest is through inhibition of the proteasome, independent of hydroxymethyl glutaryl-coa reductase. Proc. Natl. Acad. Sci. USA 96: Cooper S. (2002) Reappraisal of G1-phase arrest and synchronization by lovastatin. Cell Biol. Int. 26: Coper S. (2003) Reappraisal of serum starvation, the restriction point, GO, and G1-phase arrest points. FASEB J. 17:
Checkpoints and Restriction Points in Bacteria and Eukaryotic Cells
 Checkpoints and Restriction Points in Bacteria and Eukaryotic Cells Stephen Cooper Department of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, Michigan 481090620; e-mail:
Checkpoints and Restriction Points in Bacteria and Eukaryotic Cells Stephen Cooper Department of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, Michigan 481090620; e-mail:
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Example 2. What is meant by the cell cycle? Concept 12.1
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Example 2. What is meant by the cell cycle? Concept 12.1
CCR Biology - Chapter 5 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 5 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. If a cell cannot move enough material
Name: Class: Date: CCR Biology - Chapter 5 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. If a cell cannot move enough material
The chip mold was constructed by patterning photoresist (SU8, Clariant Corp.) on silicon wafer in
 SUPPLEMENTARY INFORMATION Cell Cycle Synchronization by Nutrient Modulation Yuan Tian 1, 2, Chunxiong Luo 1, 2, Yuheng Lu 3, Chao Tang 2, 4 1, 2, Qi Ouyang Microfluidic Chip Fabrication The chip mold was
SUPPLEMENTARY INFORMATION Cell Cycle Synchronization by Nutrient Modulation Yuan Tian 1, 2, Chunxiong Luo 1, 2, Yuheng Lu 3, Chao Tang 2, 4 1, 2, Qi Ouyang Microfluidic Chip Fabrication The chip mold was
1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells
 Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Division Mitosis and the Cell Cycle
 Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
www.njctl.org PSI Biology Mitosis & Meiosis
 Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Appendix C DNA Replication & Mitosis
 K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
If and when cancer cells stop dividing, they do so at random points, not at the normal checkpoints in the cell cycle.
 Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes?
 Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
Cell Growth and Reproduction Module B, Anchor 1
 Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Lecture 11 The Cell Cycle and Mitosis
 Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
Given these characteristics of life, which of the following objects is considered a living organism? W. X. Y. Z.
 Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
Mitosis in Onion Root Tip Cells
 Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Chapter 10. Key Ideas Correlation, Correlation Coefficient (r),
 Chapter 0 Key Ideas Correlation, Correlation Coefficient (r), Section 0-: Overview We have already explored the basics of describing single variable data sets. However, when two quantitative variables
Chapter 0 Key Ideas Correlation, Correlation Coefficient (r), Section 0-: Overview We have already explored the basics of describing single variable data sets. However, when two quantitative variables
Title: Surveying Genome to Identify Origins of DNA Replication In Silico
 Title: Surveying Genome to Identify Origins of DNA Replication In Silico Abstract: DNA replication origins are the bases to realize the process of chromosome replication and analyze the progress of cell
Title: Surveying Genome to Identify Origins of DNA Replication In Silico Abstract: DNA replication origins are the bases to realize the process of chromosome replication and analyze the progress of cell
Every time a cell divides the genome must be duplicated and passed on to the offspring. That is:
 DNA Every time a cell divides the genome must be duplicated and passed on to the offspring. That is: Original molecule yields 2 molecules following DNA replication. Our topic in this section is how is
DNA Every time a cell divides the genome must be duplicated and passed on to the offspring. That is: Original molecule yields 2 molecules following DNA replication. Our topic in this section is how is
GENE REGULATION. Teacher Packet
 AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
Lecture 7 Mitosis & Meiosis
 Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
From DNA to Protein
 Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
BME 42-620 Engineering Molecular Cell Biology. Lecture 02: Structural and Functional Organization of
 BME 42-620 Engineering Molecular Cell Biology Lecture 02: Structural and Functional Organization of Eukaryotic Cells BME42-620 Lecture 02, September 01, 2011 1 Outline A brief review of the previous lecture
BME 42-620 Engineering Molecular Cell Biology Lecture 02: Structural and Functional Organization of Eukaryotic Cells BME42-620 Lecture 02, September 01, 2011 1 Outline A brief review of the previous lecture
Module 3 Questions. 7. Chemotaxis is an example of signal transduction. Explain, with the use of diagrams.
 Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
12.1 The Role of DNA in Heredity
 12.1 The Role of DNA in Heredity Only in the last 50 years have scientists understood the role of DNA in heredity. That understanding began with the discovery of DNA s structure. In 1952, Rosalind Franklin
12.1 The Role of DNA in Heredity Only in the last 50 years have scientists understood the role of DNA in heredity. That understanding began with the discovery of DNA s structure. In 1952, Rosalind Franklin
Keystone Review Practice Test Module A Cells and Cell Processes. 1. Which characteristic is shared by all prokaryotes and eukaryotes?
 Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
LAB 09 Cell Division
 LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
Drawing a histogram using Excel
 Drawing a histogram using Excel STEP 1: Examine the data to decide how many class intervals you need and what the class boundaries should be. (In an assignment you may be told what class boundaries to
Drawing a histogram using Excel STEP 1: Examine the data to decide how many class intervals you need and what the class boundaries should be. (In an assignment you may be told what class boundaries to
Modélisation. lisation,, simulation et analyse qualitatives de réseaux d'interactions, application au cycle cellulaire mammifère
 Modélisation lisation,, simulation et analyse qualitatives de réseaux d'interactions, application au cycle cellulaire mammifère Claudine Chaouiya chaouiya@tagc.univ-mrs.fr Technologies Avancées pour le
Modélisation lisation,, simulation et analyse qualitatives de réseaux d'interactions, application au cycle cellulaire mammifère Claudine Chaouiya chaouiya@tagc.univ-mrs.fr Technologies Avancées pour le
MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788
 MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788 ISSN 0340-6253 Three distances for rapid similarity analysis of DNA sequences Wei Chen,
MATCH Communications in Mathematical and in Computer Chemistry MATCH Commun. Math. Comput. Chem. 61 (2009) 781-788 ISSN 0340-6253 Three distances for rapid similarity analysis of DNA sequences Wei Chen,
NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS
 NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS Orly Alter (a) *, Gene H. Golub (b), Patrick O. Brown (c)
NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS Orly Alter (a) *, Gene H. Golub (b), Patrick O. Brown (c)
In this chapter, you will learn improvement curve concepts and their application to cost and price analysis.
 7.0 - Chapter Introduction In this chapter, you will learn improvement curve concepts and their application to cost and price analysis. Basic Improvement Curve Concept. You may have learned about improvement
7.0 - Chapter Introduction In this chapter, you will learn improvement curve concepts and their application to cost and price analysis. Basic Improvement Curve Concept. You may have learned about improvement
Essentials of Human Anatomy & Physiology 11 th Edition, 2015 Marieb
 A Correlation of Essentials of Human Anatomy Marieb To the Next Generation Science Standards Life A Correlation of, HS-LS1 From Molecules to Organisms: Structures and Processes HS-LS1-1. Construct an explanation
A Correlation of Essentials of Human Anatomy Marieb To the Next Generation Science Standards Life A Correlation of, HS-LS1 From Molecules to Organisms: Structures and Processes HS-LS1-1. Construct an explanation
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION. Professor Bharat Patel Office: Science 2, 2.36 Email: b.patel@griffith.edu.
 Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: b.patel@griffith.edu.au What is Gene Expression & Gene Regulation? 1. Gene Expression
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: b.patel@griffith.edu.au What is Gene Expression & Gene Regulation? 1. Gene Expression
CHROMOSOME STRUCTURE CHROMOSOME NUMBERS
 CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
Bob Jesberg. Boston, MA April 3, 2014
 DNA, Replication and Transcription Bob Jesberg NSTA Conference Boston, MA April 3, 2014 1 Workshop Agenda Looking at DNA and Forensics The DNA, Replication i and Transcription i Set DNA Ladder The Double
DNA, Replication and Transcription Bob Jesberg NSTA Conference Boston, MA April 3, 2014 1 Workshop Agenda Looking at DNA and Forensics The DNA, Replication i and Transcription i Set DNA Ladder The Double
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs)
 Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Name Class Date. Figure 13 1. 2. Which nucleotide in Figure 13 1 indicates the nucleic acid above is RNA? a. uracil c. cytosine b. guanine d.
 13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
13 Multiple Choice RNA and Protein Synthesis Chapter Test A Write the letter that best answers the question or completes the statement on the line provided. 1. Which of the following are found in both
Experimental Analysis
 Experimental Analysis Instructors: If your institution does not have the Fish Farm computer simulation, contact the project directors for information on obtaining it free of charge. The ESA21 project team
Experimental Analysis Instructors: If your institution does not have the Fish Farm computer simulation, contact the project directors for information on obtaining it free of charge. The ESA21 project team
2006 7.012 Problem Set 6 KEY
 2006 7.012 Problem Set 6 KEY ** Due before 5 PM on WEDNESDAY, November 22, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You create an artificial
2006 7.012 Problem Set 6 KEY ** Due before 5 PM on WEDNESDAY, November 22, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You create an artificial
AP: LAB 8: THE CHI-SQUARE TEST. Probability, Random Chance, and Genetics
 Ms. Foglia Date AP: LAB 8: THE CHI-SQUARE TEST Probability, Random Chance, and Genetics Why do we study random chance and probability at the beginning of a unit on genetics? Genetics is the study of inheritance,
Ms. Foglia Date AP: LAB 8: THE CHI-SQUARE TEST Probability, Random Chance, and Genetics Why do we study random chance and probability at the beginning of a unit on genetics? Genetics is the study of inheritance,
Simple Regression Theory II 2010 Samuel L. Baker
 SIMPLE REGRESSION THEORY II 1 Simple Regression Theory II 2010 Samuel L. Baker Assessing how good the regression equation is likely to be Assignment 1A gets into drawing inferences about how close the
SIMPLE REGRESSION THEORY II 1 Simple Regression Theory II 2010 Samuel L. Baker Assessing how good the regression equation is likely to be Assignment 1A gets into drawing inferences about how close the
Replication Study Guide
 Replication Study Guide This study guide is a written version of the material you have seen presented in the replication unit. Self-reproduction is a function of life that human-engineered systems have
Replication Study Guide This study guide is a written version of the material you have seen presented in the replication unit. Self-reproduction is a function of life that human-engineered systems have
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
The cell cycle, mitosis and meiosis
 The cell cycle, mitosis and meiosis Learning objective This learning material is about the life cycle of a cell and the series of stages by which genetic materials are duplicated and partitioned to produce
The cell cycle, mitosis and meiosis Learning objective This learning material is about the life cycle of a cell and the series of stages by which genetic materials are duplicated and partitioned to produce
Name Date Period. 2. When a molecule of double-stranded DNA undergoes replication, it results in
 DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
Cellular Reproduction
 9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
CHAPTER 5 SIGNALLING IN NEURONS
 5.1. SYNAPTIC TRANSMISSION CHAPTER 5 SIGNALLING IN NEURONS One of the main functions of neurons is to communicate with other neurons. An individual neuron may receive information from many different sources.
5.1. SYNAPTIC TRANSMISSION CHAPTER 5 SIGNALLING IN NEURONS One of the main functions of neurons is to communicate with other neurons. An individual neuron may receive information from many different sources.
2. The number of different kinds of nucleotides present in any DNA molecule is A) four B) six C) two D) three
 Chem 121 Chapter 22. Nucleic Acids 1. Any given nucleotide in a nucleic acid contains A) two bases and a sugar. B) one sugar, two bases and one phosphate. C) two sugars and one phosphate. D) one sugar,
Chem 121 Chapter 22. Nucleic Acids 1. Any given nucleotide in a nucleic acid contains A) two bases and a sugar. B) one sugar, two bases and one phosphate. C) two sugars and one phosphate. D) one sugar,
Algorithms in Computational Biology (236522) spring 2007 Lecture #1
 Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Forensic DNA Testing Terminology
 Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Workshop: Cellular Reproduction via Mitosis & Meiosis
 Workshop: Cellular Reproduction via Mitosis & Meiosis Introduction In this workshop you will examine how cells divide, including how they partition their genetic material (DNA) between the two resulting
Workshop: Cellular Reproduction via Mitosis & Meiosis Introduction In this workshop you will examine how cells divide, including how they partition their genetic material (DNA) between the two resulting
to 370C in a matter of seconds. Materials and Methods.-Yeast spheroplasts were prepared, resuspended to a concentration
 A MUTANT OF YEAST APPARENTLY DEFECTIVE IN THE INITIATION OF PROTEIN SYNTHESIS* BT LELAND H. HARTWELLt AND CALVIN S. MCLAUGHLIN DEPARTMENT OF MOLECULAR AND CELL BIOLOGY, UNIVERSITY OF CALIFORNIA (IRVINE)
A MUTANT OF YEAST APPARENTLY DEFECTIVE IN THE INITIATION OF PROTEIN SYNTHESIS* BT LELAND H. HARTWELLt AND CALVIN S. MCLAUGHLIN DEPARTMENT OF MOLECULAR AND CELL BIOLOGY, UNIVERSITY OF CALIFORNIA (IRVINE)
LAB : THE CHI-SQUARE TEST. Probability, Random Chance, and Genetics
 Period Date LAB : THE CHI-SQUARE TEST Probability, Random Chance, and Genetics Why do we study random chance and probability at the beginning of a unit on genetics? Genetics is the study of inheritance,
Period Date LAB : THE CHI-SQUARE TEST Probability, Random Chance, and Genetics Why do we study random chance and probability at the beginning of a unit on genetics? Genetics is the study of inheritance,
Cell Cycle in Onion Root Tip Cells (IB)
 Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Session 7 Bivariate Data and Analysis
 Session 7 Bivariate Data and Analysis Key Terms for This Session Previously Introduced mean standard deviation New in This Session association bivariate analysis contingency table co-variation least squares
Session 7 Bivariate Data and Analysis Key Terms for This Session Previously Introduced mean standard deviation New in This Session association bivariate analysis contingency table co-variation least squares
Transmission Line and Back Loaded Horn Physics
 Introduction By Martin J. King, 3/29/3 Copyright 23 by Martin J. King. All Rights Reserved. In order to differentiate between a transmission line and a back loaded horn, it is really important to understand
Introduction By Martin J. King, 3/29/3 Copyright 23 by Martin J. King. All Rights Reserved. In order to differentiate between a transmission line and a back loaded horn, it is really important to understand
CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA
 CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA INTRODUCTION DNA : DNA is deoxyribose nucleic acid. It is made up of a base consisting of sugar, phosphate and one nitrogen base.the
CHAPTER 6: RECOMBINANT DNA TECHNOLOGY YEAR III PHARM.D DR. V. CHITRA INTRODUCTION DNA : DNA is deoxyribose nucleic acid. It is made up of a base consisting of sugar, phosphate and one nitrogen base.the
Comparative genomic hybridization Because arrays are more than just a tool for expression analysis
 Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
A and B are not absolutely linked. They could be far enough apart on the chromosome that they assort independently.
 Name Section 7.014 Problem Set 5 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68-120 by 5:00pm on Friday
Name Section 7.014 Problem Set 5 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68-120 by 5:00pm on Friday
Translation Study Guide
 Translation Study Guide This study guide is a written version of the material you have seen presented in the replication unit. In translation, the cell uses the genetic information contained in mrna to
Translation Study Guide This study guide is a written version of the material you have seen presented in the replication unit. In translation, the cell uses the genetic information contained in mrna to
Chapter 19a Oxidative Phosphorylation and Photophosphorylation. Multiple Choice Questions
 Chapter 19a Oxidative Phosphorylation and Photophosphorylation Multiple Choice Questions 1. Electron-transfer reactions in mitochondria Page: 707 Difficulty: 1 Ans: E Almost all of the oxygen (O 2 ) one
Chapter 19a Oxidative Phosphorylation and Photophosphorylation Multiple Choice Questions 1. Electron-transfer reactions in mitochondria Page: 707 Difficulty: 1 Ans: E Almost all of the oxygen (O 2 ) one
Guided Notes: Chapter 9 Cellular Reproduction
 Guided Notes: Cellular Reproduction When do cells divide? Cells grow and function normally until they become too. Cell size is because increases faster than This means that there is not enough area on
Guided Notes: Cellular Reproduction When do cells divide? Cells grow and function normally until they become too. Cell size is because increases faster than This means that there is not enough area on
Bio EOC Topics for Cell Reproduction: Bio EOC Questions for Cell Reproduction:
 Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
Cell Cycle Vignettes Irreversible Transitions in the Cell Cycle
 Cell Cycle Vignettes Irreversible Transitions in the Cell Cycle John J. Tyson & Bela Novak Department of Biological Sciences Virginia Polytechnic Institute & State University Blacksburg, Virginia USA &
Cell Cycle Vignettes Irreversible Transitions in the Cell Cycle John J. Tyson & Bela Novak Department of Biological Sciences Virginia Polytechnic Institute & State University Blacksburg, Virginia USA &
Transfection-Transfer of non-viral genetic material into eukaryotic cells. Infection/ Transduction- Transfer of viral genetic material into cells.
 Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
II. DISTRIBUTIONS distribution normal distribution. standard scores
 Appendix D Basic Measurement And Statistics The following information was developed by Steven Rothke, PhD, Department of Psychology, Rehabilitation Institute of Chicago (RIC) and expanded by Mary F. Schmidt,
Appendix D Basic Measurement And Statistics The following information was developed by Steven Rothke, PhD, Department of Psychology, Rehabilitation Institute of Chicago (RIC) and expanded by Mary F. Schmidt,
First-year Statistics for Psychology Students Through Worked Examples
 First-year Statistics for Psychology Students Through Worked Examples 1. THE CHI-SQUARE TEST A test of association between categorical variables by Charles McCreery, D.Phil Formerly Lecturer in Experimental
First-year Statistics for Psychology Students Through Worked Examples 1. THE CHI-SQUARE TEST A test of association between categorical variables by Charles McCreery, D.Phil Formerly Lecturer in Experimental
Exercise with Gene Ontology - Cytoscape - BiNGO
 Exercise with Gene Ontology - Cytoscape - BiNGO This practical has material extracted from http://www.cbs.dtu.dk/chipcourse/exercises/ex_go/goexercise11.php In this exercise we will analyze microarray
Exercise with Gene Ontology - Cytoscape - BiNGO This practical has material extracted from http://www.cbs.dtu.dk/chipcourse/exercises/ex_go/goexercise11.php In this exercise we will analyze microarray
Lab 3: Testing Hypotheses about Mitosis
 Lab 3: Testing Hypotheses about Mitosis Why do cells divide? Lab today focuses on cellular division, also known as cellular reproduction. To become more familiar with why cells divide, the types of cell
Lab 3: Testing Hypotheses about Mitosis Why do cells divide? Lab today focuses on cellular division, also known as cellular reproduction. To become more familiar with why cells divide, the types of cell
Stochastic Gene Expression in Prokaryotes: A Point Process Approach
 Stochastic Gene Expression in Prokaryotes: A Point Process Approach Emanuele LEONCINI INRIA Rocquencourt - INRA Jouy-en-Josas Mathematical Modeling in Cell Biology March 27 th 2013 Emanuele LEONCINI (INRIA)
Stochastic Gene Expression in Prokaryotes: A Point Process Approach Emanuele LEONCINI INRIA Rocquencourt - INRA Jouy-en-Josas Mathematical Modeling in Cell Biology March 27 th 2013 Emanuele LEONCINI (INRIA)
Years after 2000. US Student to Teacher Ratio 0 16.048 1 15.893 2 15.900 3 15.900 4 15.800 5 15.657 6 15.540
 To complete this technology assignment, you should already have created a scatter plot for your data on your calculator and/or in Excel. You could do this with any two columns of data, but for demonstration
To complete this technology assignment, you should already have created a scatter plot for your data on your calculator and/or in Excel. You could do this with any two columns of data, but for demonstration
Biology 3A Laboratory MITOSIS Asexual Reproduction
 Biology 3A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Biology 3A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
UNDERSTANDING THE TWO-WAY ANOVA
 UNDERSTANDING THE e have seen how the one-way ANOVA can be used to compare two or more sample means in studies involving a single independent variable. This can be extended to two independent variables
UNDERSTANDING THE e have seen how the one-way ANOVA can be used to compare two or more sample means in studies involving a single independent variable. This can be extended to two independent variables
CYCLIN DEPENDENT KINASES AND CELL CYCLE CONTROL
 CYCLIN DEPENDENT KINASES AND CELL CYCLE CONTROL Nobel Lecture, December 9, 2001 by PAUL M. NURSE Cancer Research UK, PO Box 123, 61 Lincoln s Inn Fields, London WC2A 3PK, United Kingdom. The cell is the
CYCLIN DEPENDENT KINASES AND CELL CYCLE CONTROL Nobel Lecture, December 9, 2001 by PAUL M. NURSE Cancer Research UK, PO Box 123, 61 Lincoln s Inn Fields, London WC2A 3PK, United Kingdom. The cell is the
8.7 Exponential Growth and Decay
 Section 8.7 Exponential Growth and Decay 847 8.7 Exponential Growth and Decay Exponential Growth Models Recalling the investigations in Section 8.3, we started by developing a formula for discrete compound
Section 8.7 Exponential Growth and Decay 847 8.7 Exponential Growth and Decay Exponential Growth Models Recalling the investigations in Section 8.3, we started by developing a formula for discrete compound
1.5 page 3 DNA Replication S. Preston 1
 AS Unit 1: Basic Biochemistry and Cell Organisation Name: Date: Topic 1.5 Nucleic Acids and their functions Page 3 l. DNA Replication 1. Go through PowerPoint 2. Read notes p2 and then watch the animation
AS Unit 1: Basic Biochemistry and Cell Organisation Name: Date: Topic 1.5 Nucleic Acids and their functions Page 3 l. DNA Replication 1. Go through PowerPoint 2. Read notes p2 and then watch the animation
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE Q5B
 INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
Copyright 2000-2003 Mark Brandt, Ph.D. 54
 Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology
 Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology The Master Degree in Medical Laboratory Sciences / Clinical Microbiology, Immunology or
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology The Master Degree in Medical Laboratory Sciences / Clinical Microbiology, Immunology or
MCAS Biology. Review Packet
 MCAS Biology Review Packet 1 Name Class Date 1. Define organic. THE CHEMISTRY OF LIFE 2. All living things are made up of 6 essential elements: SPONCH. Name the six elements of life. S N P C O H 3. Elements
MCAS Biology Review Packet 1 Name Class Date 1. Define organic. THE CHEMISTRY OF LIFE 2. All living things are made up of 6 essential elements: SPONCH. Name the six elements of life. S N P C O H 3. Elements
Lecture 11 Enzymes: Kinetics
 Lecture 11 Enzymes: Kinetics Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 216-225 Key Concepts Kinetics is the study of reaction rates (velocities). Study of enzyme kinetics is useful for
Lecture 11 Enzymes: Kinetics Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 216-225 Key Concepts Kinetics is the study of reaction rates (velocities). Study of enzyme kinetics is useful for
CALCULATIONS & STATISTICS
 CALCULATIONS & STATISTICS CALCULATION OF SCORES Conversion of 1-5 scale to 0-100 scores When you look at your report, you will notice that the scores are reported on a 0-100 scale, even though respondents
CALCULATIONS & STATISTICS CALCULATION OF SCORES Conversion of 1-5 scale to 0-100 scores When you look at your report, you will notice that the scores are reported on a 0-100 scale, even though respondents
Cell Cycle Phase Determination Kit
 Cell Cycle Phase Determination Kit Item No. 10009349 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Safety
Cell Cycle Phase Determination Kit Item No. 10009349 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Safety
Exploring the new world of the genome. with DNA microarrays
 Exploring the new world of the genome with DNA microarrays Patrick O. Brown 1,3 & David Botstein 2 Departments of 1 Biochemistry and 2 Genetics, and the 3 Howard Hughes Medical Institute, Stanford University
Exploring the new world of the genome with DNA microarrays Patrick O. Brown 1,3 & David Botstein 2 Departments of 1 Biochemistry and 2 Genetics, and the 3 Howard Hughes Medical Institute, Stanford University
CCR Biology - Chapter 9 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Integrating DNA Motif Discovery and Genome-Wide Expression Analysis. Erin M. Conlon
 Integrating DNA Motif Discovery and Genome-Wide Expression Analysis Department of Mathematics and Statistics University of Massachusetts Amherst Statistics in Functional Genomics Workshop Ascona, Switzerland
Integrating DNA Motif Discovery and Genome-Wide Expression Analysis Department of Mathematics and Statistics University of Massachusetts Amherst Statistics in Functional Genomics Workshop Ascona, Switzerland
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
List, describe, diagram, and identify the stages of meiosis.
 Meiosis and Sexual Life Cycles In this topic we will examine a second type of cell division used by eukaryotic cells: meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis
Meiosis and Sexual Life Cycles In this topic we will examine a second type of cell division used by eukaryotic cells: meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis
AS Biology Unit 2 Key Terms and Definitions. Make sure you use these terms when answering exam questions!
 AS Biology Unit 2 Key Terms and Definitions Make sure you use these terms when answering exam questions! Chapter 7 Variation 7.1 Random Sampling Sampling a population to eliminate bias e.g. grid square
AS Biology Unit 2 Key Terms and Definitions Make sure you use these terms when answering exam questions! Chapter 7 Variation 7.1 Random Sampling Sampling a population to eliminate bias e.g. grid square
BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis
 BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis Introduction - Fields of Genetics To answer the following question, review the three traditional subdivisions of
BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis Introduction - Fields of Genetics To answer the following question, review the three traditional subdivisions of
Association Between Variables
 Contents 11 Association Between Variables 767 11.1 Introduction............................ 767 11.1.1 Measure of Association................. 768 11.1.2 Chapter Summary.................... 769 11.2 Chi
Contents 11 Association Between Variables 767 11.1 Introduction............................ 767 11.1.1 Measure of Association................. 768 11.1.2 Chapter Summary.................... 769 11.2 Chi
CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA
 CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA Cytogenetics is the study of chromosomes and their structure, inheritance, and abnormalities. Chromosome abnormalities occur in approximately:
CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA Cytogenetics is the study of chromosomes and their structure, inheritance, and abnormalities. Chromosome abnormalities occur in approximately:
Appendix A: Science Practices for AP Physics 1 and 2
 Appendix A: Science Practices for AP Physics 1 and 2 Science Practice 1: The student can use representations and models to communicate scientific phenomena and solve scientific problems. The real world
Appendix A: Science Practices for AP Physics 1 and 2 Science Practice 1: The student can use representations and models to communicate scientific phenomena and solve scientific problems. The real world
Summary. 16 1 Genes and Variation. 16 2 Evolution as Genetic Change. Name Class Date
 Chapter 16 Summary Evolution of Populations 16 1 Genes and Variation Darwin s original ideas can now be understood in genetic terms. Beginning with variation, we now know that traits are controlled by
Chapter 16 Summary Evolution of Populations 16 1 Genes and Variation Darwin s original ideas can now be understood in genetic terms. Beginning with variation, we now know that traits are controlled by
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY
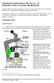 MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
Lecture 3: Mutations
 Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
Overview of Violations of the Basic Assumptions in the Classical Normal Linear Regression Model
 Overview of Violations of the Basic Assumptions in the Classical Normal Linear Regression Model 1 September 004 A. Introduction and assumptions The classical normal linear regression model can be written
Overview of Violations of the Basic Assumptions in the Classical Normal Linear Regression Model 1 September 004 A. Introduction and assumptions The classical normal linear regression model can be written
BCOR101 Midterm II Wednesday, October 26, 2005
 BCOR101 Midterm II Wednesday, October 26, 2005 Name Key Please show all of your work. 1. A donor strain is trp+, pro+, met+ and a recipient strain is trp-, pro-, met-. The donor strain is infected with
BCOR101 Midterm II Wednesday, October 26, 2005 Name Key Please show all of your work. 1. A donor strain is trp+, pro+, met+ and a recipient strain is trp-, pro-, met-. The donor strain is infected with
