Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome
|
|
|
- Jasmine Norris
- 7 years ago
- Views:
Transcription
1 Hum Genet (2003) 112 : DOI /s ORIGINAL INVESTIGATION Ivan Nasidze Tamara Sarkisian Azer Kerimov Mark Stoneking Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome Received: 9 August 2002 / Accepted: 23 October 2002 / Published online: 14 December 2002 Springer-Verlag 2002 Abstract A previous analysis of mtdna variation in the Caucasus found that Indo-European-speaking Armenians and Turkic-speaking Azerbaijanians were more closely related genetically to other Caucasus populations (who speak Caucasian languages) than to other Indo-European or Turkic groups, respectively. Armenian and Azerbaijanian therefore represent language replacements, possibly via elite dominance involving primarily male migrants, in which case genetic relationships of Armenians and Azerbaijanians based on the Y-chromosome should more closely reflect their linguistic relationships. We therefore analyzed 11 bi-allelic Y-chromosome markers in 389 males from eight populations, representing all major linguistic groups in the Caucasus. As with the mtdna study, based on the Y-chromosome Armenians and Azerbaijanians are more closely-related genetically to their geographic neighbors in the Caucasus than to their linguistic neighbors elsewhere. However, whereas the mtdna results show that Caucasian groups are more closely related genetically to European than to Near Eastern groups, by contrast the Y-chromosome shows a closer genetic relationship with the Near East than with Europe. Introduction I. Nasidze ( ) M. Stoneking Max Planck Institute for Evolutionary Anthropology, Inselstrasse 22, Leipzig, Germany Tel.: , Fax: , nasidze@eva.mpg.de T. Sarkisian Center of Medical Genetics, National Academy of Sciences of Republic of Armenia, 5/1 Zakyan Str., Yerevan, Armenia A. Kerimov Scientific-research Institute of Haematology and Transfusiology, Azerbaijan Republic Ministry of Health, Gashgay Street 87, Baku, Azerbaijan The Caucasus, the region between the Caspian and Black Seas, is characterized by extremely high linguistic diversity with four major families (South Caucasian, North Caucasian, Indo-European, and Altaic) spoken by more then 50 ethnic groups. In addition, a major geographic barrier, the Caucasus Mountain range, divides the region into the North and South Caucasus sub-regions. The Caucasus therefore offers the opportunity for studing the impact of linguistic diversity and geographic barriers on the genetic structure of human populations. In particular, the presence of populations who are linguistically-related to populations outside the Caucasus, such as the Armenians (who speak an Indo-European language) and the Azerbaijanians (who speak a Turkic language) raises the question as to whether language affiliation or geographic proximity best explains the genetic relationships of these Caucasian populations. We have previously addressed this question by studying sequence variation in the first hypervariable segment (HV1) of the mtdna control region and Alu insertion polymorphisms in Caucasian populations (Nasidze and Stoneking 2001; Nasidze et al. 2001). Both studies have shown that Armenians and Azerbaijanians are more closely related genetically to their geographic neighbors in the Caucasus and not to their linguistic neighbors elsewhere. In addition, both studies have demonstrated that Caucasian populations are genetically intermediate between European and Near Eastern populations, but that they are more closely related to European than to Near Eastern populations. The genetic results thus suggest that both the Armenian and Azerbaijanian languages represent language replacements in the Caucasus. The origins of the Armenian language are obscure, but the Azerbaijanian language was probably introduced in the 11 th century AD by central Asian nomads (Johanson 1998). A common mechanism of language replacement is elite dominance (Renfrew 1991), whereby the language of a small invading group is adopted by the larger resident population, either because it is imposed by force or because it is considered socially desirable to speak the language of the invaders. If the invading group is primarily male, then one might expect patterns of Y-chromosome variation to retain some trace of the invaders. To test this hypothesis of language re-
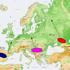
2 256 Fig. 1 Map of the Caucasus indicating the Y-SNP haplogroup frequencies in European, Near Eastern, Caucasian, and Central Asian populations (AB Abazinians, KA Kabardinians, IN Ingushians, CH Chechenians, DA Darginians, SV Svans, OS South Ossetians, K Georgians from Kazbegi, GE Georgians, AR Armenians, AZ Azerbaijanians, LE Lezginians). SV, K, and LE are from Wells et al. (2001) placement via elite dominance, we report here an analysis of Y-chromosomal bi-allelic markers in the same set of Caucasian populations previously analyzed for mtdna HV1 sequence variation (Nasidze and Stoneking 2001). Materials and methods Subjects A total of 389 samples (289 whole blood and 100 cheek cell swabs) from unrelated male individuals were collected in the following eight autochthonous populations (Fig. 1): Georgians (South Caucasian speakers), Armenians (Indo-European speakers), Azerbaijanians (Turkic speakers), and Abazinians, Kabardinians, Ingushians, Chechenians, and Darginians (North Caucasian speakers). DNA from the blood samples had been used previously for analyses of mtdna HV1 sequence variation and Alu insertion polymorphisms (Nasidze and Stoneking 2001; Nasidze et al. 2001); cheek cell swabs from additional males from Armenia and Azerbaijanwere collected in order to increase the sample size for Y-chromosome studies. Informed consent and information about the birthplace of the donor and the donor s parents and grandparents were obtained from all donors. Genomic DNA from blood samples was extracted by using an IsoQuick DNA extraction kit (Orca Research, Bothell, Wash., USA) or a conventional phenolchloroform method (Maniantis et al. 1982). DNA from cheek cell swabs was extracted by using a conventional salting-out procedure (Miller et al. 1988). Published Y-chromosome single nucleotide polymorphism (Y-SNP) data from the South Caucasus were used from 25 Svans (Georgia), 25 Georgians from Kazbegi (Georgia), 12 Lezgi (Azerbaijan), and 17 South Ossetians (Wells et al. 2001). Additional published data on 21 Azerbaijanians and 47 Armenians (Wells et al. 2001), and 63 Georgians (Semino et al. 2000; Wells et al. 2001), did not differ in any respect from our samples (data not shown) and were not included in the analysis to avoid weighting the results too heavily on these populations. Published Y-SNP data (Semino et al. 2000; Wells et al. 2001) for European, Near Eastern, and Central Asian populations were also included. Molecular analysis Ten SNP markers previously reported to be polymorphic in Europe and the Near East (Semino et al. 2000) were typed in all samples: RPS4Y (M130), M9, M89, M124, M45, M173, M17, M201, M170, and M172 (Underhill et al and references therein); the YAP Alu insertion polymorphism (Hammer and Horai 1995) was also typed. For all SNP markers except M130 (RPS4Y), Taqman (Applied Biosystems) assays were designed. Primers and dye-labeled probes were designed by using Primer Express (Version 1 for Macintosh Power PC; PE Biosystems). Primer and probe sequences are given in Table 1. Reaction mixes were prepared as described previously (Morin et al. 1999). M130 was typed by using the polymerase chain reaction/restriction fragment length polymorphism procedure described elsewhere (Kayser et al. 2000), whereas the YAP Alu insertion was typed as described previously (Hammer and Horai 1995). All samples were typed for all markers; no discrepancy was found between the hierarchical order of markers described by Underhill et al. (2000) and our results. The Y-SNP haplogroup nomenclature used here is according to the recent recommendations of the Y Chromosome Consortium (2002). The phylogenetic relationship of they-chromosome haplotypes, based on the 11 bi-allelic markers analyzed here, is shown in Fig. 2. Statistical analysis Haplogroup diversity and F st values were calculated with Arlequin (Schneider et al. 2000), which was also used to carry out Mantel tests for correlations between matrices. Multidimensional scaling (MDS) analysis (Kruskal 1964) of the F st values was carried out with STATISTICA (StatSoft). Programs in PHYLIP3.5c (Felsenstein 1993) were used to construct a neighbor-joining tree based on pairwise F st values.
3 257 Table 1 Primer and probe sequences for TaqMan assays with corresponding annealing temperatures (italic nucleotide in polymorphic site) SNP Forward primer Reverse primer Probes a Annealing marker temperature M9 CCTGAAATACAGAACTGCAAAGAA GACATTGAACGTTTGAACATGTCT CCTAAGATGGTTGAATCCTCTTTATTTTTCTTTAATTT 62 C CCTAAGATGGTTGAATGCTCTTTATTTTTCTTTAATTT M124 AATGCAAATTCCTGGGCAA TTCCTATAAAGCAAAGTTGAGGTTG AACAGGGAAGTCGGTTTAATAATACTGAGTTTGT 62 C AACAGGGAAGTTGGTTTAATAATACTGAGTTTGTG M45 AACAGTAACTCTAGGAGAGAGGATATCAA CTATCTCCTGGCCTGGACCT TGAAAAATTATAGATAGGCAAAAAGCTCCTTCTG 62 C CAGTGAAAAATTATAGATAAGCAAAAAGCTCCTTCT M173 TACTGTAACTTCCTAGAAAATTGGAAATAA CAGTTTTCCCAGATCCTGAAAA CAAGGGCATTTAGAACACTTTGTCATCTGTTA 62 C AGGGCATTTAGAACCCTTTGTCATCTGTT M17 AAATCAGATTCTGTCTACTCACCAGAG TCACAAAAATAGTTTGGCCACTT TTGCTGGTTGTTACGGGGTTTTTTTAA 60 C TTGCTGGTTGTTACGGG-TTTTTTTAAGTG M89 CCTGGATTCAGCTCTCTTCCT CCAGCAAAGGTAGCTGCAAC TTATGTACAAAAATCTCATCTCTCACTTTGCCTGA 58 C TTATGTACAAAAATCTTATCTCTCACTTTGCCTGA M201 GCAATAGTTACTACTTGAGTTACTATATTAGTGCAA CATCCTATCAGCTTCATCCAACA TCCAGTATCAACTGAGGGTTTTCGTAATAGGTACTTA 62 C ATCCAGTATCAACTGAGGTTTTTCGTAATAGGTACTTA M172 GGATTTTTCATTTTTATCCCCC CCAGGTACAGAGAAAGTTTGGACT AACCCATTTTGATGCTTTACTTAAAAGGTCTTC 60 C ACCCATTTTGATGCTTGACTTAAAAGGTC M170 TTTCATATTCTGTGCATTATACAAATTACTATT CCACACAAAAACAGGTCCTCA AAAAATCATTGTTCATTTTTTTCAGTGTGGGT 58 C AAAATCATTGTTCCTTTTTTTCAGTGTGGG a Ancestral probe is shown on the first line and the mutant probe on the second Results and discussion Y-SNP haplogroups in the Caucasus Eleven Y-SNP haplogroups (Fig. 1) were found in the Caucasus (Table 2, Fig. 1). The most frequent haplogroups were F*, G* and J2*; together the frequency of these three haplogroups was in all groups except for the Darginians. The Darginians had a high frequency of haplogroup I* (0.58), which elsewhere was found at a frequency of 0.1 or less. Svans were previously reported (Wells et al. 2001) to have a high frequency (0.92) of haplogroup F*, and Kazbegi to have a high frequency of haplogroup J2* (0.72). No other groups had any single haplogroup at a frequency greater than 0.5, with the exception of the small sample of 12 Lezgi, for which the frequency of haplogroup F* was The Darginians, Lezgi, Svans, and Kazbegi correspondingly had the lowest haplogroup diversities ( ), whereas for the other groups, the haplogroup diversity was Two of the three common Caucasus haplogroups (F* and J2*) are also common in Near Eastern populations Lebanese, Turks, Syrians (Semino et al. 2000), and Iranians (Wells et al. 2001), with average frequencies of and 0.28, respectively, but present in lower frequencies in Europe (average frequencies and 0.074, respectively). The third common Caucasus haplogroup, G*, is rare in Europe (frequency=0.061) and, in the Near East, has been reported only in the Turkish and Lebanese groups (Semino et al. 2000). Haplogroup R1*, which is common in Western and Central Europe, is observed in the South Caucasus at frequencies higher then 0.1, whereas in the North Caucasus, it is absent or nearly so. Haplogroup R1a1*, which is observed in high frequencies in the south-west Mediterranean region, Eastern Europe, and Central Asia, is present at a low frequency in the Caucasus, similar to the Near East. The other Caucasus Y-haplogroups occur at low frequency. The Darginian, Svan, and Kazbegi groups appear to be outliers compared with the other Caucasus groups. The F st value was highest between Svans and other Caucasus groups (average F st = 0.332), followed by the Kazbegi (average F st = 0.286) and Darginians (average F st = 0.25), whereas the average pairwise F st value among the remaining Caucasus groups was only These high F st values, coupled with the lower haplogroup diversity and reduced number of haplogroups (Table 2) in the Darginians, Svans, and Kazbegi, are most likely the result of genetic drift operating in small isolated populations. The correlation between the geographic and genetic (pairwise F st ) distances separating pairs of Caucasus populations was not statistically significant (Mantel test: Z = 0.113, P =0.671). Removal of the outliers (Svans, Kazbegi, and Darginians) resulted in a correlation that was higher but still non-significant (Z =0.301, P =0.134). To determine whether the Caucasus Mountains have an influence on the genetic structure of Caucasian populations, we examined the correlation between geographic and ge-
4 258 Fig. 2 Phylogenetic relationship of Y-chromosome haplogroups, based on 10 Y-SNPs and the YAP marker netic distance for the South and North Caucasus separately, but neither was statistically significant (North Caucasus: Z=0.331, P =0.133; South Caucasus: Z= 0.346, P = 0.65). Moreover, the average F st value between South and North Caucasus populations (excluding outliers) was 0.075, similar to that among North Caucasus populations (0.096) and that among South Caucasus populations (0.040). Therefore, the Caucasus Mountains appear not to have had a detectable influence on the genetic structure of Caucasus populations; instead, genetic drift operating in small isolated populations seems to have dominated the genetic structure of Caucasus populations. Comparison of Caucasian, European, and Near Eastern Y-haplogroups The haplogroup diversity in the Caucasus (average value: 0.797) is almost as high as that in Central Asia (average value: 0.824) and the Near East (average value: 0.769) and is significantly higher (t -test, P =0.024) than the haplogroup diversity in Europe (average value: 0.633). An MDS plot and neighbor-joining tree based on F st values (Fig. 3A, B) split European populations into Western and Eastern groups, as has been observed previously (Semino et al. 2000), with Central Asian populations falling in between the Western and Eastern European groups. The Caucasus populations are intermingled with Near Eastern populations. These patterns have further been confirmed by the pairwise F st comparisons; the mean pairwise F st value for the Caucasus vs Europe is 0.254, whereas the mean F st value for the Caucasus vs the Near East is 0.079, which is significantly lower (t -test based on average F st values jacknifed over populations, P <0.001). On average, South Caucasian populations are more similar to both Near Eastern and European populations (average F st = and 0.222, respectively) than are North Caucasian populations (average F st = and 0.303); however, both the South Caucasus and the North Caucasus are more similar to the Near East than to Europe with respect to Y-SNP haplogroups. The MDS and F st analyses included some groups from Wells et al. (2001) in which the M201 marker, which distinguishes haplogroup G* from haplogroup F* (Fig. 2), was not analyzed (Table 2). In the above analyses, these individuals were classified as haplogroup F*, although some unknown proportion could be haplogroup G*. To determine whether this inability to distinguish between haplogroups F* and G* for some groups influenced the results of the MDS and F st analyses, we classified all haplogroup G* individuals as haplogroup F* and repeated the analyses. The results (not shown) were essentially identical; thus, the inability to distinguish between haplogroups F* and G* in some groups does not influence our conclusions. In order to exclude any possible bias in our conclusions caused by the small size of the samples from some of the groups, we repeated all analyses after excluding all groups with sample size less then 25 (see Table 2). All conclusions remained the same (data not shown). Genetic relationship between Basques and Caucasians A common origin of Caucasian and Basque speakers has been proposed, based on (controversial) assertions of the similarity in their languages (Gamkrelidze and Ivanov 1995; Ruhlen 1991). A genetic relationship between Basques and Caucasians is not supported by the Y-SNP data. The
5 259 Table 2 Y chromosomal haplogroup frequencies and haplogroup diversities in the Caucasus. Haplogroup designations are according to the nomenclature proposed by the Y Chromosome consortium (2002) Population Number Haplogroups Haplo- Source group E* C* K* P1 P* R1* R1a1* F* G* J2* I* diversity Armenians Present study Azerbaijanians Present study Georgians Present study Kabardinians Present study Ingushians Present study Chechenians Present study Darginians Present study Abazinians Present study Georgians Semino et al Svans a Wells et al Kazbegi a Wells et al Ossetians a Wells et al Lezgi a Wells et al a Not typed for M201 (haplogroup G*) average pairwise F st value between Basques and Caucasian groups is much higher (F st = 0.563) than that between Basques and Indo-European groups (F st = 0.311). The Caucasus groups and Basques are not clustered together in either the neighbor-joining tree or the MDS plot (Fig. 3A, B). These results are in agreement with previous studies based on classical markers and mtdna HV1 sequence data (Bertorelle et al. 1995; Nasidze and Stoneking 2001). Language replacements and genetic relationships in the Caucasus The presence, in the Caucasus, of groups whose geographic neighbors are not their linguistic neighbors allows us to address the question as to which better explains the genetic relationships of these groups: geography or language? In particular, Azerbaijanians speak a Turkic language but are surrounded by non-turkic speakers, and Armenians speak an Indo-European language but are surrounded by non-indo-european speakers. Previous mtdna analyses have shown that both Azerbaijanians and Armenians are more closely related genetically to other Caucasus groups than to their respective linguistic neighbors (Nasidze and Stoneking 2001), which indicates that the Azerbaijanian and Armenian languages were introduced via language replacements. We have hypothesized that these language replacements have occurred via elite dominance (Renfrew 1991) in which a resident population adopts the language of a small group of migrants either by force or because speaking the language confers social prestige. If such were the case, and the migrants were primarily male, then Y-DNA analyses may more closely reflect the linguistic relationships of Azerbaijanians and Armenians than have mtdna analyses. Indeed, the Y-SNP haplogroups give similar results. The Turkic-speaking Azerbaijanians are genetically closer to their geographic neighbors in the Caucasus (average F st =0.047) than to other Turkic-speaking groups (average F st =0.105; t -test, P <0.001). Similarly, Indo-European-speaking Armenians are genetically closer to other Caucasus groups (average F st = 0.053) than to other Indo-European-speaking groups (average F st =0.146; t -test, P <0.001). This conclusion is also supported by the MDS plot and neighbor-joining tree (Fig. 3A, B), in which Armenians and Azerbaijanians both cluster with other Caucasian populations. Thus, in agreement with previous studies of mtdna diversity and Alu insertion polymorphisms (Nasidze and Stoneking 2001; Nasidze et al. 2001), Y-chromosome haplogroups indicate that Indo-European-speaking Armenians and Turkic-speaking Azerbaijanians are genetically more closely related to their geographic neighbors in the Caucasus than to their linguistic neighbors elsewhere. A similar observation has been reported recently in a study of Y-chromosomal markers in Armenian populations (Weale et al. 2001). Thus, all of the genetic evidence agrees that the Armenian and Azerbaijanian languages reflect language replacements, which occurred without any detectable genetic contribution of the original Indo-European and
6 260 Fig. 3A, B The phylogenetic relationship of Y-chromosome haplogroups, based on 10 Y-SNPs and the YAP marker. A MDS plot based on pairwise F st values, showing relationships among the Caucasus, European, Near Eastern, and Central Asian populations (open stars with boldface italic population names Caucasus groups, closed circles European populations, open squares Central Asian groups, closed diamonds Near Eastern populations). The stress value for the MDS plot is B Neighbor-joining tree based on pairwise F st values for the same populations (boldface Caucasus groups) Turkic groups, respectively. This may still reflect an elite dominance scenario, as presumably the original Indo-European/Turkic migrant groups were very small and/or did not mix extensively with the resident groups. In any event, the migrant groups had a negligible genetic impact on the resident groups. Currently, there are approximately 8million Azerbaijanians and 3.5 million Armenians, attesting to the remarkable rapidity of these language shifts; such large-scale shifts would have to be accomplished by cultural rather than merely biological means.
7 However, the Y-haplogroups contrast sharply with the previous mtdna and Alu insertion polymorphism studies (Nasidze and Stoneking 2001; Nasidze et al. 2001) in terms of the overall relationships of Caucasus populations. For the Y-chromosome, the Caucasus populations are more closely related to Near Eastern populations than to European populations. Evidence for this is provided by the higher diversity values and by the pairwise F st values and MDS plot. In particular, the Caucasus appears to be a break zone in the Y-chromosome genetic landscape of Eurasia; haplogroups such as R1a1* are at a high frequency in East Europe but at low frequency in the Caucasus and the Near East, whereas haplogroups such as F* and J2* are common in the Caucasus and the Near East, but rare in Europe (Table 2, Fig. 1). By contrast, both mtdna and Alu insertion polymorphisms group Caucasus populations with Europe rather than the Near East. How can these contrasting views on the relationship of Caucasus populations be reconciled? A possible explanation is that the Y-chromosome results reflect repeated invading migrations from the Near East, migrations that probably involved mostly males. Various Near Eastern groups invaded the Caucasus numerous times during the last two millennia including the occupation of Georgia by Arab caliphs after 654 AD (Muskhelishvili 1977), the Seljuc Turks invasion of the South Caucasus in the 11 th century (Muskhelishvili 1977), and repeated invasions by Turks and Persians (Muskhelishvili 1977 and references therein). Indeed, strong Y-chromosomal gene flow from the Near East to the Caucasus has been clearly demonstrated for the interpolated maps of admixture proportions in a recent study of Central Asian populations (Zerjal et al. 2002). Although these male-mediated invasions from the Near East might possibly explain why the Caucasian Y-haplogroups are of Near Eastern origin, they do not explain why mtdna and Alu insertion polymorphisms group the Caucasus with Europe. A possible explanation for this latter result would be a common ancestry of Caucasian and European populations. This hypothesized common ancestry could date back to pre-neolithic times, as suggested by Renfrew (1992) who considers Caucasian languages to reflect human dispersal before 15,000 years ago. On the other hand, it could reflect a route for Neolithic farmers from the Near East to Europe via the Caucasus; there are several securely dated Neolithic sites in the Caucasus that are 6,000 7,000 years old (Masson and Merpert 1982; Muskhelishvili 1977). Analyses of additional autosomal markers are needed to verify this hypothesized common ancestry of Caucasian and European populations. Acknowledgements We thank Manfred Kayser for useful discussion, Peter Underhill for control DNA samples, and Hiltrud Schaedlich for technical assistance. This research was supported by funding from the Max Planck Society. References Bertorelle G, Bertranpetit J, Calafell F, Nasidze I, Barbujani G (1995) Do Basque-speaking and Caucasian-speaking populations share non-indo-european ancestors. Eur J Hum Genet 3: Felsenstein J (1993) PHYLIP (phylogeny inference package) v. 3.5c. Department of Genetics, University of Washington, Seattle Gamkrelidze T, Ivanov V (1995) Indo-European and the Indo-Europeans. De Gruyter, Berlin Hammer M, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56: Johanson L (1998) The history of Turkic. In: Johanson L, Csato E (eds) The Turkic languages. Routledge, London, pp Kayser M, Brauer S, Weiss G, Underhill PA, Roewer L, Schiefenhover W, Stoneking M (2000) Melanesian origin of Polynesian Y chromosome. Curr Biol 10: Kruskal JB (1964) Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Pyschometrika 29:1 27 Maniatis T, Fritsh EF, Sambrook J (1982) Molecular clonning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY Masson VM, Merpert NJ (1982) Archaeology of USSR: Neolith of USSR. Nauka, Moscow, pp Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 Morin PA, Saiz R, Monjazeb A (1999) High-throughput single nucleotide polymorphism genotyping by fluorescent 5 exonuclease assay. BioTechniques 3: Muskhelishvili D (1977) The main problems of Georgian historicalgeography. Metsniereba, Tbilisi Nasidze I, Stoneking M (2001) Mitochondrial DNA variation and language replacements in the Caucasus. Proc R Soc Lond [Biol] 268: Nasidze I, Risch GM, Robichaux M, Sherry ST, Batzer MA, Stoneking M (2001) Alu insertion polymorphisms and the genetic structure of human populations from the Caucasus. Eur J Hum Genet 9: Renfrew C (1991) Before Babel: speculations on the origins of linguistic diversity. Cambridge Archaeol J 1:13 23 Renfrew C (1992) Archaeology, genetics, and linguistic diversity. Man 27: Ruhlen M (1991) A guide to the world s languages. Stanford University Press, Stanford Schneider S, Roessli D, Excoffier L (2000) Arlequin v. 2000: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, et al (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290: Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, et al (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26: Weale ME, Yepiskoposyan L, Jager RF, Hovhannisyan N, Khudoyan A, Burbage-Hall O, Bradman N, Thomas MG (2001) Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet 109: Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshimi S, Balakrishnan K, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98: Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2: Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C (2002) A genetic landscape reshaped by recent events: Y-chromosomal insights into Central Asia. Am J Hum Genet 71:
Report Concomitant Replacement of Language and mtdna in South Caspian Populations of Iran
 Current Biology 16, 668 673, April 4, 2006 ª2006 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2006.02.021 Report Concomitant Replacement of Language and mtdna in South Caspian Populations of Iran
Current Biology 16, 668 673, April 4, 2006 ª2006 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2006.02.021 Report Concomitant Replacement of Language and mtdna in South Caspian Populations of Iran
Genetic Variation and Human Evolution Lynn B. Jorde, Ph.D. Department of Human Genetics University of Utah School of Medicine.
 Genetic Variation and Human Evolution Lynn B. Jorde, Ph.D. Department of Human Genetics University of Utah School of Medicine. The past two decades have witnessed an explosion of human genetic data. Innumerable
Genetic Variation and Human Evolution Lynn B. Jorde, Ph.D. Department of Human Genetics University of Utah School of Medicine. The past two decades have witnessed an explosion of human genetic data. Innumerable
Y Chromosome Markers
 Y Chromosome Markers Lineage Markers Autosomal chromosomes recombine with each meiosis Y and Mitochondrial DNA does not This means that the Y and mtdna remains constant from generation to generation Except
Y Chromosome Markers Lineage Markers Autosomal chromosomes recombine with each meiosis Y and Mitochondrial DNA does not This means that the Y and mtdna remains constant from generation to generation Except
Gene Mapping Techniques
 Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Single Nucleotide Polymorphisms (SNPs)
 Single Nucleotide Polymorphisms (SNPs) Additional Markers 13 core STR loci Obtain further information from additional markers: Y STRs Separating male samples Mitochondrial DNA Working with extremely degraded
Single Nucleotide Polymorphisms (SNPs) Additional Markers 13 core STR loci Obtain further information from additional markers: Y STRs Separating male samples Mitochondrial DNA Working with extremely degraded
Matthew Kaplan and Taylor Edwards. University of Arizona Tucson, Arizona
 Matthew Kaplan and Taylor Edwards University of Arizona Tucson, Arizona Unresolved paternity Consent for testing Ownership of Samples Uncovering genetic disorders SRY Reversal / Klinefelter s Syndrome
Matthew Kaplan and Taylor Edwards University of Arizona Tucson, Arizona Unresolved paternity Consent for testing Ownership of Samples Uncovering genetic disorders SRY Reversal / Klinefelter s Syndrome
Elsevier Editorial System(tm) for Forensic Science International: Genetics Manuscript Draft
 Elsevier Editorial System(tm) for Forensic Science International: Genetics Manuscript Draft Manuscript Number: Title: A comment on the Paper: A comparison of Y-chromosomal lineage dating using either resequencing
Elsevier Editorial System(tm) for Forensic Science International: Genetics Manuscript Draft Manuscript Number: Title: A comment on the Paper: A comparison of Y-chromosomal lineage dating using either resequencing
Forensic DNA Testing Terminology
 Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
The sample is taken with a simple mouth swab no blood is involved. There will be instructions included on how to take the sample.
 DNA testing Thanks for your enquiry about DNA testing. I oversee the Scottish DNA Project on behalf of the University of Strathclyde, Glasgow and act as representative for Family Tree DNA who host our
DNA testing Thanks for your enquiry about DNA testing. I oversee the Scottish DNA Project on behalf of the University of Strathclyde, Glasgow and act as representative for Family Tree DNA who host our
Seeing Faces and History through Human Genome Sequences
 Seeing Faces and History through Human Genome Sequences CAS/MPG Partner Group on the Human Functional Genetic Variations Shanghai-Leipzig, 2011.2.1 2016.1.31 Prof. Dr. TANG Kun (middle) with his cooperator,
Seeing Faces and History through Human Genome Sequences CAS/MPG Partner Group on the Human Functional Genetic Variations Shanghai-Leipzig, 2011.2.1 2016.1.31 Prof. Dr. TANG Kun (middle) with his cooperator,
Development of two Novel DNA Analysis methods to Improve Workflow Efficiency for Challenging Forensic Samples
 Development of two Novel DNA Analysis methods to Improve Workflow Efficiency for Challenging Forensic Samples Sudhir K. Sinha, Ph.D.*, Anne H. Montgomery, M.S., Gina Pineda, M.S., and Hiromi Brown, Ph.D.
Development of two Novel DNA Analysis methods to Improve Workflow Efficiency for Challenging Forensic Samples Sudhir K. Sinha, Ph.D.*, Anne H. Montgomery, M.S., Gina Pineda, M.S., and Hiromi Brown, Ph.D.
Globally, about 9.7% of cancers in men are prostate cancers, and the risk of developing the
 Chapter 5 Analysis of Prostate Cancer Association Study Data 5.1 Risk factors for Prostate Cancer Globally, about 9.7% of cancers in men are prostate cancers, and the risk of developing the disease has
Chapter 5 Analysis of Prostate Cancer Association Study Data 5.1 Risk factors for Prostate Cancer Globally, about 9.7% of cancers in men are prostate cancers, and the risk of developing the disease has
Title:Global patterns of sex-biased migrations in humans
 Title:Global patterns of sex-biased migrations in humans Authors:Chuan-Chao Wang 1, Li Jin 1, 2, 3, Hui Li 1,* Affiliations: 1. State Key Laboratory of Genetic Engineering and MOE Key Laboratory of Contemporary
Title:Global patterns of sex-biased migrations in humans Authors:Chuan-Chao Wang 1, Li Jin 1, 2, 3, Hui Li 1,* Affiliations: 1. State Key Laboratory of Genetic Engineering and MOE Key Laboratory of Contemporary
D-Loop-BASE is online now Central European database of mitochondrial DNA
 International Congress Series 1239 (2003) 505 509 D-Loop-BASE is online now Central European database of mitochondrial DNA Holger Wittig a, *, Mike Koecke b, Kai-Uwe Sattler b, Dieter Krause a a Institute
International Congress Series 1239 (2003) 505 509 D-Loop-BASE is online now Central European database of mitochondrial DNA Holger Wittig a, *, Mike Koecke b, Kai-Uwe Sattler b, Dieter Krause a a Institute
Mitochondrial DNA Analysis
 Mitochondrial DNA Analysis Lineage Markers Lineage markers are passed down from generation to generation without changing Except for rare mutation events They can help determine the lineage (family tree)
Mitochondrial DNA Analysis Lineage Markers Lineage markers are passed down from generation to generation without changing Except for rare mutation events They can help determine the lineage (family tree)
From Africa to Aotearoa Part 1: Out of Africa
 From Africa to Aotearoa Part 1: Out of Africa The spread of modern humans out of Africa started around 65,000 years ago, and ended with the settlement of New Zealand 750 years ago. These PowerPoint presentations
From Africa to Aotearoa Part 1: Out of Africa The spread of modern humans out of Africa started around 65,000 years ago, and ended with the settlement of New Zealand 750 years ago. These PowerPoint presentations
Rethinking Polynesian Origins: Human Settlement of the Pacific
 LENScience Senior Biology Seminar Series Rethinking Polynesian Origins: Human Settlement of the Pacific Michal Denny, and Lisa Matisoo-Smith Our Polynesian ancestors are renowned as some of the world s
LENScience Senior Biology Seminar Series Rethinking Polynesian Origins: Human Settlement of the Pacific Michal Denny, and Lisa Matisoo-Smith Our Polynesian ancestors are renowned as some of the world s
Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group
 Hum Genet (2001) 109 :659 674 DOI 10.1007/s00439-001-0627-9 ORIGINAL INVESTIGATION Michael E. Weale Levon Yepiskoposyan Rolf F. Jager Nelli Hovhannisyan Armine Khudoyan Oliver Burbage-Hall Neil Bradman
Hum Genet (2001) 109 :659 674 DOI 10.1007/s00439-001-0627-9 ORIGINAL INVESTIGATION Michael E. Weale Levon Yepiskoposyan Rolf F. Jager Nelli Hovhannisyan Armine Khudoyan Oliver Burbage-Hall Neil Bradman
SECOND INTERNATIONAL CONFERENCE ON THE GREAT MIGRATIONS ASIA TO AMERICA
 The Permanent Delegation of Kazakhstan to the UNESCO The Embassy of the Republic of Kazakhstan to the United States of America The Harriman Institute, Columbia University East Central European Center,
The Permanent Delegation of Kazakhstan to the UNESCO The Embassy of the Republic of Kazakhstan to the United States of America The Harriman Institute, Columbia University East Central European Center,
HLA data analysis in anthropology: basic theory and practice
 HLA data analysis in anthropology: basic theory and practice Alicia Sanchez-Mazas and José Manuel Nunes Laboratory of Anthropology, Genetics and Peopling history (AGP), Department of Anthropology and Ecology,
HLA data analysis in anthropology: basic theory and practice Alicia Sanchez-Mazas and José Manuel Nunes Laboratory of Anthropology, Genetics and Peopling history (AGP), Department of Anthropology and Ecology,
Algorithms in Computational Biology (236522) spring 2007 Lecture #1
 Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Analysis of 16 Y STR loci in the Finnish population reveals a local reduction in the diversity of male lineages
 Forensic Science International 142 (2004) 37 43 Analysis of 16 Y STR loci in the Finnish population reveals a local reduction in the diversity of male lineages M. Hedman a, V. Pimenoff a, M. Lukka b, P.
Forensic Science International 142 (2004) 37 43 Analysis of 16 Y STR loci in the Finnish population reveals a local reduction in the diversity of male lineages M. Hedman a, V. Pimenoff a, M. Lukka b, P.
2. True or False? The sequence of nucleotides in the human genome is 90.9% identical from one person to the next. False (it s 99.
 1. True or False? A typical chromosome can contain several hundred to several thousand genes, arranged in linear order along the DNA molecule present in the chromosome. True 2. True or False? The sequence
1. True or False? A typical chromosome can contain several hundred to several thousand genes, arranged in linear order along the DNA molecule present in the chromosome. True 2. True or False? The sequence
DNA Genealogy, Mutation Rates, and Some Historical Evidences Written in Y-Chromosome. I. Basic Principles and the Method
 DNA Genealogy, Mutation Rates, and Some Historical Evidences Written in Y-Chromosome. I. Basic Principles and the Method Anatole A. Klyosov 1 Abstract Origin of peoples in a context of DNA genealogy is
DNA Genealogy, Mutation Rates, and Some Historical Evidences Written in Y-Chromosome. I. Basic Principles and the Method Anatole A. Klyosov 1 Abstract Origin of peoples in a context of DNA genealogy is
MCB41: Second Midterm Spring 2009
 MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
Molecular and Cell Biology Laboratory (BIOL-UA 223) Instructor: Ignatius Tan Phone: 212-998-8295 Office: 764 Brown Email: ignatius.tan@nyu.
 Molecular and Cell Biology Laboratory (BIOL-UA 223) Instructor: Ignatius Tan Phone: 212-998-8295 Office: 764 Brown Email: ignatius.tan@nyu.edu Course Hours: Section 1: Mon: 12:30-3:15 Section 2: Wed: 12:30-3:15
Molecular and Cell Biology Laboratory (BIOL-UA 223) Instructor: Ignatius Tan Phone: 212-998-8295 Office: 764 Brown Email: ignatius.tan@nyu.edu Course Hours: Section 1: Mon: 12:30-3:15 Section 2: Wed: 12:30-3:15
Table of Contents: Introduction. DNA Tribes Digest July 30, 2010
 DNA Tribes Digest July 30, 2010 Copyright 2010 DNA Tribes. All rights reserved. To request an email subscription to DNA Tribes Digest, email digest@dnatribes.com with the subject heading Subscribe. To
DNA Tribes Digest July 30, 2010 Copyright 2010 DNA Tribes. All rights reserved. To request an email subscription to DNA Tribes Digest, email digest@dnatribes.com with the subject heading Subscribe. To
Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis
 Hum Genet (2005) 117: 428 443 DOI 10.1007/s00439-005-1333-9 ORIGINAL INVESTIGATION Manfred Kayser Æ Oscar Lao Æ Katja Anslinger Christa Augustin Æ Grazyna Bargel Æ Jeanett Edelmann Sahar Elias Æ Marielle
Hum Genet (2005) 117: 428 443 DOI 10.1007/s00439-005-1333-9 ORIGINAL INVESTIGATION Manfred Kayser Æ Oscar Lao Æ Katja Anslinger Christa Augustin Æ Grazyna Bargel Æ Jeanett Edelmann Sahar Elias Æ Marielle
Online Y-chromosomal Short Tandem Repeat Haplotype Reference Database (YHRD) for U.S. Populations*
 Manfred Kayser, 1 Ph.D.; Silke Brauer; 1 Sascha Willuweit; 2 Hiltrud Schädlich, 1 Ph.D.; Mark A. Batzer, 3 Ph.D.; Jennifer Zawacki; 4 Mechthild Prinz, 4 M.D.; Lutz Roewer, 2 Ph.D.; and Mark Stoneking,
Manfred Kayser, 1 Ph.D.; Silke Brauer; 1 Sascha Willuweit; 2 Hiltrud Schädlich, 1 Ph.D.; Mark A. Batzer, 3 Ph.D.; Jennifer Zawacki; 4 Mechthild Prinz, 4 M.D.; Lutz Roewer, 2 Ph.D.; and Mark Stoneking,
DNA and Forensic Science
 DNA and Forensic Science Micah A. Luftig * Stephen Richey ** I. INTRODUCTION This paper represents a discussion of the fundamental principles of DNA technology as it applies to forensic testing. A brief
DNA and Forensic Science Micah A. Luftig * Stephen Richey ** I. INTRODUCTION This paper represents a discussion of the fundamental principles of DNA technology as it applies to forensic testing. A brief
DNA Tribes is on Facebook. Find us at http://facebook.com/dnatribes. DNA Tribes Digest February 1, 2013 Page 1 of 10
 Copyright 2013 DNA Tribes. All rights reserved. DNA Tribes Digest February 1, 2013 To request an email subscription to DNA Tribes Digest, email digest@dnatribes.com with the subject heading Subscribe.
Copyright 2013 DNA Tribes. All rights reserved. DNA Tribes Digest February 1, 2013 To request an email subscription to DNA Tribes Digest, email digest@dnatribes.com with the subject heading Subscribe.
Thesis Guidelines Handbook
 Thesis Guidelines Handbook For For MS Program COMSATS Institute of Information Technology Guidelines for MS Thesis These guidelines for MS thesis should be read in conjunction with the GHB. Please read
Thesis Guidelines Handbook For For MS Program COMSATS Institute of Information Technology Guidelines for MS Thesis These guidelines for MS thesis should be read in conjunction with the GHB. Please read
Some ancestors are difficult to trace. Adoptions illegitimacies, name changes and migrations can present brick walls in our research that seem
 Some ancestors are difficult to trace. Adoptions illegitimacies, name changes and migrations can present brick walls in our research that seem impregnable. In other cases, documentation that would shed
Some ancestors are difficult to trace. Adoptions illegitimacies, name changes and migrations can present brick walls in our research that seem impregnable. In other cases, documentation that would shed
Lab 2/Phylogenetics/September 16, 2002 1 PHYLOGENETICS
 Lab 2/Phylogenetics/September 16, 2002 1 Read: Tudge Chapter 2 PHYLOGENETICS Objective of the Lab: To understand how DNA and protein sequence information can be used to make comparisons and assess evolutionary
Lab 2/Phylogenetics/September 16, 2002 1 Read: Tudge Chapter 2 PHYLOGENETICS Objective of the Lab: To understand how DNA and protein sequence information can be used to make comparisons and assess evolutionary
RETRIEVING SEQUENCE INFORMATION. Nucleotide sequence databases. Database search. Sequence alignment and comparison
 RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
PREHISTORIC IBERIA. Genetics, Anthropology, and Linguistics
 PREHISTORIC IBERIA Genetics, Anthropology, and Linguistics PREHISTORIC IBERIA Genetics, Anthropology, and Linguistics Edited by Antonio Arnaiz-Villena Hospital "12 de Octubre" Universidad Complutense Madrid,
PREHISTORIC IBERIA Genetics, Anthropology, and Linguistics PREHISTORIC IBERIA Genetics, Anthropology, and Linguistics Edited by Antonio Arnaiz-Villena Hospital "12 de Octubre" Universidad Complutense Madrid,
The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations
 Ann. Hum. Genet. (2001), 65, 43 62 Printed in Great Britain 43 The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations P. A. UNDERHILL *, G. PASSARINO,, A.A.LIN,
Ann. Hum. Genet. (2001), 65, 43 62 Printed in Great Britain 43 The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations P. A. UNDERHILL *, G. PASSARINO,, A.A.LIN,
SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE
 AP Biology Date SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE LEARNING OBJECTIVES Students will gain an appreciation of the physical effects of sickle cell anemia, its prevalence in the population,
AP Biology Date SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE LEARNING OBJECTIVES Students will gain an appreciation of the physical effects of sickle cell anemia, its prevalence in the population,
Introduction to Bioinformatics 3. DNA editing and contig assembly
 Introduction to Bioinformatics 3. DNA editing and contig assembly Benjamin F. Matthews United States Department of Agriculture Soybean Genomics and Improvement Laboratory Beltsville, MD 20708 matthewb@ba.ars.usda.gov
Introduction to Bioinformatics 3. DNA editing and contig assembly Benjamin F. Matthews United States Department of Agriculture Soybean Genomics and Improvement Laboratory Beltsville, MD 20708 matthewb@ba.ars.usda.gov
Chapter 8: Recombinant DNA 2002 by W. H. Freeman and Company Chapter 8: Recombinant DNA 2002 by W. H. Freeman and Company
 Genetic engineering: humans Gene replacement therapy or gene therapy Many technical and ethical issues implications for gene pool for germ-line gene therapy what traits constitute disease rather than just
Genetic engineering: humans Gene replacement therapy or gene therapy Many technical and ethical issues implications for gene pool for germ-line gene therapy what traits constitute disease rather than just
( 1) Most human populations are a product of mixture of genetically distinct groups that intermixed within the last 4,000 years.
 Frequently asked questions about A Genetic Atlas of Human Admixture History G. Hellenthal, G.B.J. Busby, G. Band, J.F. Wilson, C. Capelli, D. Falush, S. Myers, Science (2014) SUMMARY What is your work
Frequently asked questions about A Genetic Atlas of Human Admixture History G. Hellenthal, G.B.J. Busby, G. Band, J.F. Wilson, C. Capelli, D. Falush, S. Myers, Science (2014) SUMMARY What is your work
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Lecture 13: DNA Technology. DNA Sequencing. DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology
 Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Annex to the Accreditation Certificate D-PL-13372-01-00 according to DIN EN ISO/IEC 17025:2005
 Deutsche Akkreditierungsstelle GmbH German Accreditation Body Annex to the Accreditation Certificate D-PL-13372-01-00 according to DIN EN ISO/IEC 17025:2005 Period of validity: 26.03.2012 to 25.03.2017
Deutsche Akkreditierungsstelle GmbH German Accreditation Body Annex to the Accreditation Certificate D-PL-13372-01-00 according to DIN EN ISO/IEC 17025:2005 Period of validity: 26.03.2012 to 25.03.2017
14.3 Studying the Human Genome
 14.3 Studying the Human Genome Lesson Objectives Summarize the methods of DNA analysis. State the goals of the Human Genome Project and explain what we have learned so far. Lesson Summary Manipulating
14.3 Studying the Human Genome Lesson Objectives Summarize the methods of DNA analysis. State the goals of the Human Genome Project and explain what we have learned so far. Lesson Summary Manipulating
CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA
 CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA Cytogenetics is the study of chromosomes and their structure, inheritance, and abnormalities. Chromosome abnormalities occur in approximately:
CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA Cytogenetics is the study of chromosomes and their structure, inheritance, and abnormalities. Chromosome abnormalities occur in approximately:
I Have the Results of My Genetic Genealogy Test, Now What?
 I Have the Results of My Genetic Genealogy Test, Now What? Version 2.1 1 I Have the Results of My Genetic Genealogy Test, Now What? Chapter 1: What Is (And Isn t) Genetic Genealogy? Chapter 2: How Do I
I Have the Results of My Genetic Genealogy Test, Now What? Version 2.1 1 I Have the Results of My Genetic Genealogy Test, Now What? Chapter 1: What Is (And Isn t) Genetic Genealogy? Chapter 2: How Do I
Data Analysis for Ion Torrent Sequencing
 IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
Essentials of Real Time PCR. About Sequence Detection Chemistries
 Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Population Genetics and Multifactorial Inheritance 2002
 Population Genetics and Multifactorial Inheritance 2002 Consanguinity Genetic drift Founder effect Selection Mutation rate Polymorphism Balanced polymorphism Hardy-Weinberg Equilibrium Hardy-Weinberg Equilibrium
Population Genetics and Multifactorial Inheritance 2002 Consanguinity Genetic drift Founder effect Selection Mutation rate Polymorphism Balanced polymorphism Hardy-Weinberg Equilibrium Hardy-Weinberg Equilibrium
Estimating Scandinavian and Gaelic Ancestry in the Male Settlers of Iceland
 Am. J. Hum. Genet. 67:697 717, 2000 Estimating Scandinavian and Gaelic Ancestry in the Male Settlers of Iceland Agnar Helgason, 1 Sigrún Sigurðardóttir, 3 Jayne Nicholson, 2 Bryan Sykes, 2 Emmeline W.
Am. J. Hum. Genet. 67:697 717, 2000 Estimating Scandinavian and Gaelic Ancestry in the Male Settlers of Iceland Agnar Helgason, 1 Sigrún Sigurðardóttir, 3 Jayne Nicholson, 2 Bryan Sykes, 2 Emmeline W.
Early modern human dispersal from Africa: genomic evidence for multiple waves of migration
 Tassi et al. Investigative Genetics (2015) 6:13 DOI 10.1186/s13323-015-0030-2 RESEARCH Early modern human dispersal from Africa: genomic evidence for multiple waves of migration Francesca Tassi 1, Silvia
Tassi et al. Investigative Genetics (2015) 6:13 DOI 10.1186/s13323-015-0030-2 RESEARCH Early modern human dispersal from Africa: genomic evidence for multiple waves of migration Francesca Tassi 1, Silvia
Worksheet - COMPARATIVE MAPPING 1
 Worksheet - COMPARATIVE MAPPING 1 The arrangement of genes and other DNA markers is compared between species in Comparative genome mapping. As early as 1915, the geneticist J.B.S Haldane reported that
Worksheet - COMPARATIVE MAPPING 1 The arrangement of genes and other DNA markers is compared between species in Comparative genome mapping. As early as 1915, the geneticist J.B.S Haldane reported that
Reduced-Median-Network Analysis of Complete Mitochondrial DNA Coding-Region Sequences for the Major African, Asian, and European Haplogroups
 Am. J. Hum. Genet. 70:1152 1171, 2002 Reduced-Median-Network Analysis of Complete Mitochondrial DNA Coding-Region Sequences for the Major African, Asian, and European Haplogroups Corinna Herrnstadt, 1
Am. J. Hum. Genet. 70:1152 1171, 2002 Reduced-Median-Network Analysis of Complete Mitochondrial DNA Coding-Region Sequences for the Major African, Asian, and European Haplogroups Corinna Herrnstadt, 1
Human Genome and Human Genome Project. Louxin Zhang
 Human Genome and Human Genome Project Louxin Zhang A Primer to Genomics Cells are the fundamental working units of every living systems. DNA is made of 4 nucleotide bases. The DNA sequence is the particular
Human Genome and Human Genome Project Louxin Zhang A Primer to Genomics Cells are the fundamental working units of every living systems. DNA is made of 4 nucleotide bases. The DNA sequence is the particular
Paternity Testing. Chapter 23
 Paternity Testing Chapter 23 Kinship and Paternity DNA analysis can also be used for: Kinship testing determining whether individuals are related Paternity testing determining the father of a child Missing
Paternity Testing Chapter 23 Kinship and Paternity DNA analysis can also be used for: Kinship testing determining whether individuals are related Paternity testing determining the father of a child Missing
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
STATISTICA. Clustering Techniques. Case Study: Defining Clusters of Shopping Center Patrons. and
 Clustering Techniques and STATISTICA Case Study: Defining Clusters of Shopping Center Patrons STATISTICA Solutions for Business Intelligence, Data Mining, Quality Control, and Web-based Analytics Table
Clustering Techniques and STATISTICA Case Study: Defining Clusters of Shopping Center Patrons STATISTICA Solutions for Business Intelligence, Data Mining, Quality Control, and Web-based Analytics Table
Summary. 16 1 Genes and Variation. 16 2 Evolution as Genetic Change. Name Class Date
 Chapter 16 Summary Evolution of Populations 16 1 Genes and Variation Darwin s original ideas can now be understood in genetic terms. Beginning with variation, we now know that traits are controlled by
Chapter 16 Summary Evolution of Populations 16 1 Genes and Variation Darwin s original ideas can now be understood in genetic terms. Beginning with variation, we now know that traits are controlled by
BASIC STATISTICAL METHODS FOR GENOMIC DATA ANALYSIS
 BASIC STATISTICAL METHODS FOR GENOMIC DATA ANALYSIS SEEMA JAGGI Indian Agricultural Statistics Research Institute Library Avenue, New Delhi-110 012 seema@iasri.res.in Genomics A genome is an organism s
BASIC STATISTICAL METHODS FOR GENOMIC DATA ANALYSIS SEEMA JAGGI Indian Agricultural Statistics Research Institute Library Avenue, New Delhi-110 012 seema@iasri.res.in Genomics A genome is an organism s
Bologna Seminar ALIGNING NATIONAL AGAINST EUROPEAN QUALIFICATION FRAMEWORKS: THE PRINCIPLES OF SELF CERTIFICATION
 Bologna Seminar ALIGNING NATIONAL AGAINST EUROPEAN QUALIFICATION FRAMEWORKS: THE PRINCIPLES OF SELF CERTIFICATION 27 28 November 2008, Tbilisi, Georgia Iv. Javakhishvili Tbilisi State University, Conference
Bologna Seminar ALIGNING NATIONAL AGAINST EUROPEAN QUALIFICATION FRAMEWORKS: THE PRINCIPLES OF SELF CERTIFICATION 27 28 November 2008, Tbilisi, Georgia Iv. Javakhishvili Tbilisi State University, Conference
A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups
 Resource A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups The Y Chromosome Consortium 1 The Y chromosome contains the largest nonrecombining block in the human genome. By virtue
Resource A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups The Y Chromosome Consortium 1 The Y chromosome contains the largest nonrecombining block in the human genome. By virtue
IV. DEMOGRAPHIC PROFILE OF THE OLDER POPULATION
 World Population Ageing 195-25 IV. DEMOGRAPHIC PROFILE OF THE OLDER POPULATION A. AGE COMPOSITION Older populations themselves are ageing A notable aspect of the global ageing process is the progressive
World Population Ageing 195-25 IV. DEMOGRAPHIC PROFILE OF THE OLDER POPULATION A. AGE COMPOSITION Older populations themselves are ageing A notable aspect of the global ageing process is the progressive
ABSTRACT. Promega Corporation, Updated September 2008. http://www.promega.com/pubhub. 1 Campbell-Staton, S.
 A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA... A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA from Shed Reptile Skin ABSTRACT
A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA... A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA from Shed Reptile Skin ABSTRACT
Journal of Statistical Software
 JSS Journal of Statistical Software October 2006, Volume 16, Code Snippet 3. http://www.jstatsoft.org/ LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria between Single Nucleotide
JSS Journal of Statistical Software October 2006, Volume 16, Code Snippet 3. http://www.jstatsoft.org/ LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria between Single Nucleotide
DNA as a Biometric. Biometric Consortium Conference 2011 Tampa, FL
 DNA as a Biometric Biometric Consortium Conference 2011 Tampa, FL September 27, 2011 Dr. Peter M. Vallone Biochemical Science Division National Institute of Standards and Technology Gaithersburg, MD 20899
DNA as a Biometric Biometric Consortium Conference 2011 Tampa, FL September 27, 2011 Dr. Peter M. Vallone Biochemical Science Division National Institute of Standards and Technology Gaithersburg, MD 20899
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs)
 Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
Y-STR haplotype diversity and population data for Central Brazil: implications for environmental forensics and paternity testing
 Short Communication Y-STR haplotype diversity and population data for Central Brazil: implications for environmental forensics and paternity testing T.C. Vieira 1,2,3,4, M.A.D. Gigonzac 2,3,4, D.M. Silva
Short Communication Y-STR haplotype diversity and population data for Central Brazil: implications for environmental forensics and paternity testing T.C. Vieira 1,2,3,4, M.A.D. Gigonzac 2,3,4, D.M. Silva
DnaSP, DNA polymorphism analyses by the coalescent and other methods.
 DnaSP, DNA polymorphism analyses by the coalescent and other methods. Author affiliation: Julio Rozas 1, *, Juan C. Sánchez-DelBarrio 2,3, Xavier Messeguer 2 and Ricardo Rozas 1 1 Departament de Genètica,
DnaSP, DNA polymorphism analyses by the coalescent and other methods. Author affiliation: Julio Rozas 1, *, Juan C. Sánchez-DelBarrio 2,3, Xavier Messeguer 2 and Ricardo Rozas 1 1 Departament de Genètica,
Technical Note. Roche Applied Science. No. LC 18/2004. Assay Formats for Use in Real-Time PCR
 Roche Applied Science Technical Note No. LC 18/2004 Purpose of this Note Assay Formats for Use in Real-Time PCR The LightCycler Instrument uses several detection channels to monitor the amplification of
Roche Applied Science Technical Note No. LC 18/2004 Purpose of this Note Assay Formats for Use in Real-Time PCR The LightCycler Instrument uses several detection channels to monitor the amplification of
Molecular typing of VTEC: from PFGE to NGS-based phylogeny
 Molecular typing of VTEC: from PFGE to NGS-based phylogeny Valeria Michelacci 10th Annual Workshop of the National Reference Laboratories for E. coli in the EU Rome, November 5 th 2015 Molecular typing
Molecular typing of VTEC: from PFGE to NGS-based phylogeny Valeria Michelacci 10th Annual Workshop of the National Reference Laboratories for E. coli in the EU Rome, November 5 th 2015 Molecular typing
GENETIC GENEALOGY AND DNA TESTING
 GENETIC GENEALOGY AND DNA TESTING by Ted Steele This publication may be ordered from: St. Louis Genealogical Society P. O. Box 432010 St. Louis, MO 63143 Copyright 2013 St. Louis Genealogical Society All
GENETIC GENEALOGY AND DNA TESTING by Ted Steele This publication may be ordered from: St. Louis Genealogical Society P. O. Box 432010 St. Louis, MO 63143 Copyright 2013 St. Louis Genealogical Society All
NORSE VIKING HERITAGE
 NORSE VIKING HERITAGE My mother's maiden name is WILLIAMSON. Her ancestors in the paternal line came from the Shetland Islands. The Shetland Islands were settled by Norse Vikings beginning before 800 AD.
NORSE VIKING HERITAGE My mother's maiden name is WILLIAMSON. Her ancestors in the paternal line came from the Shetland Islands. The Shetland Islands were settled by Norse Vikings beginning before 800 AD.
CCR Biology - Chapter 9 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
SNPbrowser Software v3.5
 Product Bulletin SNP Genotyping SNPbrowser Software v3.5 A Free Software Tool for the Knowledge-Driven Selection of SNP Genotyping Assays Easily visualize SNPs integrated with a physical map, linkage disequilibrium
Product Bulletin SNP Genotyping SNPbrowser Software v3.5 A Free Software Tool for the Knowledge-Driven Selection of SNP Genotyping Assays Easily visualize SNPs integrated with a physical map, linkage disequilibrium
Association between Dopamine Gene and Alcoholism in Pategar Community of Dharwad, Karnataka
 International Journal of Scientific and Research Publications, Volume 3, Issue 10, October 2013 1 Association between Dopamine Gene and Alcoholism in Pategar Community of Dharwad, Karnataka SOMASHEKHAR
International Journal of Scientific and Research Publications, Volume 3, Issue 10, October 2013 1 Association between Dopamine Gene and Alcoholism in Pategar Community of Dharwad, Karnataka SOMASHEKHAR
Real-Time PCR Vs. Traditional PCR
 Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Becker Muscular Dystrophy
 Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Quantifiler Human DNA Quantification Kit Quantifiler Y Human Male DNA Quantification Kit
 Product Bulletin Human Identification Quantifiler Human DNA Quantification Kit Quantifiler Y Human Male DNA Quantification Kit The Quantifiler kits produce reliable and reproducible results, helping to
Product Bulletin Human Identification Quantifiler Human DNA Quantification Kit Quantifiler Y Human Male DNA Quantification Kit The Quantifiler kits produce reliable and reproducible results, helping to
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR
 Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
RACE. By Michael J. Bamshad and Steve E. Olson
 DOES RACE EXIST By Michael J. Bamshad and Steve E. Olson IF RACES ARE DEFINED AS GENETICALLY DISCRETE GROUPS, NO. BUT RESEARCHERS CAN USE SOME GENETIC INFORMATION TO GROUP INDIVIDUALS INTO CLUSTERS WITH
DOES RACE EXIST By Michael J. Bamshad and Steve E. Olson IF RACES ARE DEFINED AS GENETICALLY DISCRETE GROUPS, NO. BUT RESEARCHERS CAN USE SOME GENETIC INFORMATION TO GROUP INDIVIDUALS INTO CLUSTERS WITH
DNA Fingerprinting. Unless they are identical twins, individuals have unique DNA
 DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
The Central Dogma of Molecular Biology
 Vierstraete Andy (version 1.01) 1/02/2000 -Page 1 - The Central Dogma of Molecular Biology Figure 1 : The Central Dogma of molecular biology. DNA contains the complete genetic information that defines
Vierstraete Andy (version 1.01) 1/02/2000 -Page 1 - The Central Dogma of Molecular Biology Figure 1 : The Central Dogma of molecular biology. DNA contains the complete genetic information that defines
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
DNA Sequence Analysis
 DNA Sequence Analysis Two general kinds of analysis Screen for one of a set of known sequences Determine the sequence even if it is novel Screening for a known sequence usually involves an oligonucleotide
DNA Sequence Analysis Two general kinds of analysis Screen for one of a set of known sequences Determine the sequence even if it is novel Screening for a known sequence usually involves an oligonucleotide
Original article: DXS10079, DXS10074 and DXS10075: new alleles and SNP occurrence
 EXCLI Journal 2007;6:177-182 ISSN 1611-2156 Received: December 14, 2006, accepted: June 21, 2007, published: June 26, 2007 Original article: DXS10079, DXS10074 and DXS10075: new alleles and SNP occurrence
EXCLI Journal 2007;6:177-182 ISSN 1611-2156 Received: December 14, 2006, accepted: June 21, 2007, published: June 26, 2007 Original article: DXS10079, DXS10074 and DXS10075: new alleles and SNP occurrence
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15
 Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
High-throughput sequencing and big data: implications for personalized medicine?
 High-throughput sequencing and big data: implications for personalized medicine? Dominick J. Lemas, PhD Postdoctoral Fellow in Pediatrics - Neonatology Mentor: Jacob E. (Jed) Friedman, PhD What is Big
High-throughput sequencing and big data: implications for personalized medicine? Dominick J. Lemas, PhD Postdoctoral Fellow in Pediatrics - Neonatology Mentor: Jacob E. (Jed) Friedman, PhD What is Big
New Technologies for Sensitive, Low-Input RNA-Seq. Clontech Laboratories, Inc.
 New Technologies for Sensitive, Low-Input RNA-Seq Clontech Laboratories, Inc. Outline Introduction Single-Cell-Capable mrna-seq Using SMART Technology SMARTer Ultra Low RNA Kit for the Fluidigm C 1 System
New Technologies for Sensitive, Low-Input RNA-Seq Clontech Laboratories, Inc. Outline Introduction Single-Cell-Capable mrna-seq Using SMART Technology SMARTer Ultra Low RNA Kit for the Fluidigm C 1 System
Comparison of Y-chromosomal lineage dating using either evolutionary
 Comparison of Y-chromosomal lineage dating using either evolutionary or genealogical Y-STR mutation rates Chuan-Chao Wang 1, Hui Li 1,* 1 State Key Laboratory of Genetic Engineering and MOE Key Laboratory
Comparison of Y-chromosomal lineage dating using either evolutionary or genealogical Y-STR mutation rates Chuan-Chao Wang 1, Hui Li 1,* 1 State Key Laboratory of Genetic Engineering and MOE Key Laboratory
Nucleic Acid Purity Assessment using A 260 /A 280 Ratios
 Nucleic Acid Purity Assessment using A 260 /A 280 Ratios A common practice in molecular biology is to perform a quick assessment of the purity of nucleic acid samples by determining the ratio of spectrophotometric
Nucleic Acid Purity Assessment using A 260 /A 280 Ratios A common practice in molecular biology is to perform a quick assessment of the purity of nucleic acid samples by determining the ratio of spectrophotometric
Prentice Hall World Geography: Building a Global Perspective 2003 Correlated to: Arkansas Social Studies Curriculum Frameworks (Grades 9-12)
 Arkansas Social Studies Curriculum Frameworks (Grades 9-12) Strand 1: Time, Continuity, and Change Content Standard 1: Students will demonstrate an understanding of the chronology and concepts of history
Arkansas Social Studies Curriculum Frameworks (Grades 9-12) Strand 1: Time, Continuity, and Change Content Standard 1: Students will demonstrate an understanding of the chronology and concepts of history
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
Real time and Quantitative (RTAQ) PCR. so I have an outlier and I want to see if it really is changed
 Real time and Quantitative (RTAQ) PCR or.. for this audience so I have an outlier and I want to see if it really is changed Nigel Walker, Ph.D. Laboratory of Computational Biology and Risk Analysis, Environmental
Real time and Quantitative (RTAQ) PCR or.. for this audience so I have an outlier and I want to see if it really is changed Nigel Walker, Ph.D. Laboratory of Computational Biology and Risk Analysis, Environmental
Milk protein genetic variation in Butana cattle
 Milk protein genetic variation in Butana cattle Ammar Said Ahmed Züchtungsbiologie und molekulare Genetik, Humboldt Universität zu Berlin, Invalidenstraβe 42, 10115 Berlin, Deutschland 1 Outline Background
Milk protein genetic variation in Butana cattle Ammar Said Ahmed Züchtungsbiologie und molekulare Genetik, Humboldt Universität zu Berlin, Invalidenstraβe 42, 10115 Berlin, Deutschland 1 Outline Background
Integrating DNA Motif Discovery and Genome-Wide Expression Analysis. Erin M. Conlon
 Integrating DNA Motif Discovery and Genome-Wide Expression Analysis Department of Mathematics and Statistics University of Massachusetts Amherst Statistics in Functional Genomics Workshop Ascona, Switzerland
Integrating DNA Motif Discovery and Genome-Wide Expression Analysis Department of Mathematics and Statistics University of Massachusetts Amherst Statistics in Functional Genomics Workshop Ascona, Switzerland
Protein Sequence Analysis - Overview -
 Protein Sequence Analysis - Overview - UDEL Workshop Raja Mazumder Research Associate Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center Topics Why do protein
Protein Sequence Analysis - Overview - UDEL Workshop Raja Mazumder Research Associate Professor, Department of Biochemistry and Molecular Biology Georgetown University Medical Center Topics Why do protein
Bob Jesberg. Boston, MA April 3, 2014
 DNA, Replication and Transcription Bob Jesberg NSTA Conference Boston, MA April 3, 2014 1 Workshop Agenda Looking at DNA and Forensics The DNA, Replication i and Transcription i Set DNA Ladder The Double
DNA, Replication and Transcription Bob Jesberg NSTA Conference Boston, MA April 3, 2014 1 Workshop Agenda Looking at DNA and Forensics The DNA, Replication i and Transcription i Set DNA Ladder The Double
