Accepted 19 July, 2012
|
|
|
- Magnus Carter
- 9 years ago
- Views:
Transcription
1 African Journal of Microbiology Research Vol. 6(30), pp , 9 August, 2012 Available online at DOI: /AJMR ISSN Academic Journals Full Length Research Paper Comparison of bacterial diversity in large intestine of Xiangxi yellow cattle (Bos taurus) associated with different diet: Fresh Miscanthus sinensis and mixed forage Yadan Li 1,2, Lijuan Hu 1,2, Gaoshang Xue 1,2, Huhu Liu 1,2, Hui Yang 1,2, Yunhua Zhu 1,2,3, Yun Tian 1,2 * and Xiangyang Lu 1,2 * 1 College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha , China. 2 Hunan Agricultural Bioengineering Research Institute, Changsha , China. 3 College of Pharmacy and Life Science, Nanhua University, Hengyang , China. Accepted 19 July, 2012 Ruminants harbour a complicate ecosystem consisting mostly of microorganisms which have the ability to digest the fibrous cell walls of plant materials from host's food. To discover the novel enzyme from ruminant's digestive tract for exploration of the new biofuel plant-miscanthus sinensis, we comparatively analyzed the distribution of microbiome in the large intestine of two groups of local cattle fed with either M. sinensis [M. sinensis (+) group] or mixed forage [M. sinensis (-) group]. Two libraries of 16S rrna gene from intestinal microbiome of two groups of local cattle were constructed respectively, and subjected to restriction fragment length polymorphism (RFLP) and sequence analysis. After analyzing the sequences of 16S rrna genes, our results indicated that the intestinal bacteria of M. sinensis (+) group were composed of Firmicutes (48.89%), Bacteroidetes (6.67%), rumen bacteria (10%) and uncultured bacteria (34.44%), and the intestinal bacteria of M. sinensis (-) group were composed of Firmicutes (52.87%), Bacteroidetes (1.27%), rumen bacteria (9.55%), uncultured rumen bacteria (4.46%) and uncultured bacteria (31.21%). As expected, we found that five species of potential cellulolytic becteria can only be detected in the library of M. sinensis (+). Besides, through phylogenetic tree analysis, we found that the ratio of uncultured genus which were lack of recognized sequences in M. sinensis (+) library and M. sinensis (-) library were and 51.56%, respectively, indicating that the cattle fed on M. sinensis will produce more novel uncultured genus which probably have specific metabolic effect on decomposition of M. sinensis. Key words: Cattle, bacterial diversity, large intestine, Miscanthus sinensis, 16S rrna gene library, phylogenetic analysis. INTRODUCTION Cattle is a functionally important ruminant animal that was able to digest plant materials with high fiber concentration, as in its gastrointestinal tract there was a complex community of fibrolytic aerophilic and anaerobic microbes (Pandya et al., 2010). Most of the previous *Corresponding author. [email protected]., [email protected]. studies for investigating the microbial community in digestive system of ruminants usually focused on rumen, the largest and most important stomach compartment (Tajima et al., 1999, 2000; Pitta et al., 2010). However, the large intestine, especially the caecum and colon, has also received a lot of attention since it is also an important organ for nutrition supply and absorption of ruminant. Some studies suggested that the large intestine provide an active fermentation condition similar to the reticulo-rumen (Schwarz, 2001). Previous studies indicated
2 5966 Afr. J. Microbiol. Res. that the large intestine of cattle is an important organ for digesting glucose and soluble α-glucoside (Huntington et al., 2006; Noziere et al., 2005; Warner et al., 1972). In addition, cellulose and hemicellulose have also been reported to be largely digested in the large intestine of cattle, as the digestion of cellulose accounted for 18% to 27% of total digested cellulose, and the hemicellulose digested in the large intestine accounted for 30 to 40% of the total hemicellulose digestion (Hoover, 1978; Warner et al., 1972). It is reasonable that the large intestine has a strong digestive ability for cellulose and hemicellulose, as cecal bacteria in large intestine produce some special cellulolytic enzymes that could hydrolyze pentosans and hemicellulose (Bailey and MacRea, 1970). Thus, to identify novel fibrolytic enzymes with strong digestive ability for cellulose from the large intesteine s microorganism of cattle would be feasible and significant. Miscanthus sinensis, as an abundant source of biomass for biopower or biofules (Ge et al., 2011; Paul et al., 2011), has been of great interest to a lot of European countries. Retroactively, in 1992, European Miscanthus Network has been established (Vega et al., 1997). A kind of Chinese M. sinensis, called Chinese silvergrass, has been found to be extensively distributed in China. With high cellulose content and high yield, Chinese M. sinensis would be a gold plant for providing energy by biotransformation or chemical conversion or combination of both approaches (Shumny et al., 2011). Due to the complexity of lignocellulose s frame, this plant is very difficult to be degraded (Vega et al., 1997). However, the ruminant animal such as cattle would be able to digest M. sinensis as the result of numerous cellulolytic microorganisms habiting in its digestive tract (Maas and Glass, 1991; Ogura, 2011), which could be a potential source for identifying bio-enzyme. In consideration of this, we proposed that M. sinensis-fed cattle would have an increased population of cellulolytic microorganisms in rumen and large intesteine which provide much higher possibility for us to identify novel fibrolytic enzymes. Our final goal is to identify novel high efficient fibrolytic enzyme from cellulolytic microorganisms for exploitation of the new biofuel plant-m. sinensis. To start this project, it is of significance for us to first understand the complexity and diversity of microorganisms in the digestive system of cattle which were fed on M. sinensis, which would not only increase our chances to identify novel fibrolytic enzyme, but also provide more knowledge for understanding M. sinensis-fed cattle s physiology and nutrition. Although many investigations have been carried out to study microbial diversity in rumen of cattle (Bekele et al., 2010, 2011; Chen et al., 2011), and our investigation on microbial diversity in rumen of M. sinensis-fed cattle are also ongoing, the phylogenetic diversity in the large intestine of cattle, especially for M. sinensis-fed cattle, has rarely been studied. In the present study, we extracted the total microorganism s DNA from large intestines of M. sinensis-fed Xiangxi Yellow Cattle (Bos taurs) as well as mixed forage-fed cattle respectively, and then followed by polymerase chain reaction (PCR), cloning, and sequencing of 16S rrna genes. We found that the distribution of large intestinal microbiome varied with the diets. Five species of potential cellulolytic becteria can only be detected in the library M. sinensis (+) group. Moreover, we also found that a more significant part of unidentified sequences showed in phylogenetic tree analysis of M. sinensis (+) library, indicating that the cattle fed on M. sinensis can produce more novel uncultured genus which probably have specific metabolic effect on decomposition of M. sinensis, and these sequences data were previously unidentified. MATERIALS AND METHODS Animal diet, rumen content collection Six male cattle sampled were all from the same colony maintained in Liu Yang (Hunan provice, eastern region of China), belonging to Xiangxi Yellow Cattle (Bos taurs), the most widely distributed cattle in Hunan Province. They were separated into two groups. One group including three cattle were always fed on common feedstuff, including vinasse, bean dregs, dry straw (3:3:1), while the other group including three cattle were only given fresh M. sinensis diet from 2.5 years old for 18 months. All of the cattle in this experiment were the same age and their weight were 100±3 kg. Each diet was fed for 18 months and large intestinal samples were derived from each group of animals. Briefly, the whole contents in large guts of each group of cattle were extracted by hand with sterile gloves and were thoroughly mixed, then these pooled samples were separated in small aliquots (50±1 g) and were frozen by liquid nitrogen. Samples were stored at -80 C until DNA extraction. Isolation and purification of genomic DNA The genomic DNA was extracted from the large gut content of both treatments using the modified method as below. Briefly, 10 g sample was suspended completely in 30 ml PBS (ph 7.4) by beadbeating for min, then centrifuged at 600 g for 10 min at 4 C. The supernatant was collected and centrifuged again at 8000 g for 10 min to obtain the cell pellet. 6.3 ml DNA extraction buffer [1500 mm NaCl, 100 mm Tris-HCl (ph 8.0), 100 mm PBS, 100 mm EDTA, 2% CTAB], 2% (w/v) PVPP, 50 μl lysozyme (100 μg ml -1 ) were added to the pellet, then incubated at 37 C for 30 min. Furthermore, the mixture was added with proteinase K (50 μg ml -1 ) and incubated at 37 C for 30 min followed by added with 0.7 ml 20% SDS and incubated at 65 C for another 90 min. After that, the whole mixture was centrifuged at 8000 g for 15 min and the supernatant was transferred to a clean tube. The liquid was then purified twice with chloroform: isoamyl alcohol (24:1). Two volumes of ethanol were used to precipitate DNA. Primers and PCR conditions The PCR reaction was conducted using the 27F forward primer (5 - AGAGTTTGATCCTGGCTCAG-3 ) and 1492R reverse primer (5 - GGTTACCTTGTTACGACTT-3 ) (Pandya et al., 2010). Ex Taq polymerase (TaKaRa, Dalian, China) was used for PCR thermocycling. Each 50 μl PCR reaction contained: 30 ng of purified bacterial genomic DNA, 200 μmol of dntp mixture, 5 μl of
3 Li et al PCR Buffer, 10 pmol of each primer. PCR amplification was performed using the following program: denaturing at 95 C for 5 min, followed by 30 cycles of 30 s of denaturing at 95 C, 30 s of annealing at 58 C and 2 min of elongation at 72 C, with a final extension at 72 C for 10 min. 16S rrna gene library construction The purified PCR products were cloned into pmd-18 T vecter (TaKaRa, Dalian, China), and recombinant plasmids were transformed into competent Escherichia coli DH5α cells. Positive clones grew after incubation overnight at 37 C. The 16S rrna gene library from Chinese silvergrass group was named as M. sinensis (+) library (S) and another 16S rrna gene library from mixed forage group was M. sinensis (-) library (D). Restriction fragment length polymorphism (RFLP) screening of 16S rrna gene clones Inserted rrna genes from recombinant clones were reamplified by PCR (common primers-m13). Aliquots of crude reamplified rrna gene PCR products were digested twice with 0.5U of Mspl and Afal (TaKaRa, Dalian, China) in a final volume of 15 μl, for 3 h at 37 C. The operational taxonmic units (OTUs) was initially classified based on the RFLP patterns. Sequencing and phylogenetic analysis Positive clones were sequenced by company (BGI, Beijing Genomics Institute, China). The sequences were automatically aligned using CLUSTAL X, and phylogenetic trees were constructed by neighbor-joining distance matrix methods with the programs in the software package Mega, version 4.0. Bootstrap resampling analysis for 1000 replicates was performed to estimate the confidence of tree topologies. Good's coverage was calculated as [1-(n/N)] 100%, where n is the number of single-clone OTUs and N is the library size, that is, the total number of clones for the analyzed sample (Schloss and Handelsman, 2005). uncultured bacteria (34.44%) (Table 3), including a diverse array of bacterial species: Clostridium, Acetivibrio, Butyrivibrio, Eubacterium, Sporobacter, Oscillospira, Lachnospira, Ruminococcus, Anaerostipes, Marinilabilia. Moreover, eight OTUs (S-2, S-5, S-18, S- 20; S-33; S-12, S-29; S-14) could only be grouped at the family level (Lachnospiraceae, Ruminococcaceae, Rikenellaceae or Clostridiaceae); two OTUs (S-16, S-22) could only be identified at the order level (Clostridiales); and an unique OUT (S-9) could only be labeled at phylum level (Bacteroidetes); others could only be classified to bacteria. Among all of the analyzed sequences, the most abundant OTU was S-37, which comprised 9.17% of the analyzed clones, belonging to uncultured bacterium (Table 1). However, in M. sinensis (-) library, about 48.44% clones (62 clones) had <97% similarity and 51.56% clones (66 clones) had 97% similarity to the known 16S rrna gene sequences of NCBI database ( These corresponded to Firmicutes (52.87%), Bacteroidetes (1.27%), rumen bacteria (9.55%), uncultured rumen bacteria (4.46%) and uncultured bacteria (31.21%) (Table 3), including Christensenella, Eubacterium, Acetomicrobium, Clostridium, Butyricicoccus, Oscillospira, Acetivibrio, Sporobacter, Oscillibacter, Lachnospira, Ruminococcus, Hydrogenoanaero bacterium. And seven OTUs (D-2, D- 22; D-18; D-10, D-13, D-14, D-16) could only be ascribed at the family level (Lachnospiraceae, Clostridiaceae or Ruminococcaceae); five OTUs (D-5) could only be identified at the order level (Clostridiales). Besides, D-22 was showed as the most abundant OTU, including 9 clones (7.03%) of all, which was considered as Lachnospiraceae bacterium (Table 2). RESULTS Bacterial community structure in the large intestine of M. sinensis (+) group and M. sinensis (-) group To analyze the differences of bacterial community structure in the large intestine between M. sinensis (+) group (M. sinensis-fed cattle) and M. sinensis (-) group (mixed forage-fed cattle), two 16S rrna gene libraries were constructed. A total of S rdna clones, grouped into 41 OTUs, from M. sinensis (+) library and S rdna clones, typed into 43 OTUs, from M. sinensis (-) library were analyzed. The OTUs based on RFLP and the accession numbers of the closest relative sequences were categorized in Tables 1 and 2, respectively. As a result, in M. sinensis (+) library, about 58.72% clones (64 clones) had <97% similarity and 31.28% clones (45 clones) had 97% similarity to 16S rrna gene data sequences from GeneBank by BLAST. All the sequences consisted of Firmicutes (48.89%), Bacteroidetes (6.67%), rumen bacteria (10%) and Phylogenetic analysis of M. sinensis (+) group and M. sinensis (-) group The phylogenetic tree was constructed to further investigate bacteria s taxonomic affiliation. BLAST searching tool had been used to find the neighbors of these sequences, and 2-3 the nearest relative sequences were chosen to construct the phylogenetic tree together with the representative sequenced clones of each OTUs. In the library of the M. sinensis (+), all 109 sequences underwent in a phylogenetic analysis and could be classified into 41 OTUs. As shown in Table 1, almost 1/3 of the representive clones (13 OTUs) were close to uncultured bacteria. Furthermore, within the phylum Firmicutes, Lachnospiraceae bacterium (S-2, S-5, S-18, S-20) was the prevalent bacterial genera, whose proportion was 14.45% in the whole library, followed by uncultured Ruminococcaceae (5.56%), while the abundance of Bacteroidetese was 6.67%. The total library of the M. sinensis (+) contained 9 single-clone OTUs, and the Good's coverage of this library was
4 5968 Afr. J. Microbiol. Res. Table 1. Similarity values of operational taxonomic units (OTUs) based on RFLP of 16S rrna gene sequences derived from M. sinensis (+) library. OTU Length (bp) No. of clones Sequence identity (%) Nearest valid relative (GeneBank accession no.) S UB (EU466462) S Lachnospiraceae bacterium (AF550610) S Acetivibrio sp. (AB596889) S T.aceticus (Z49863) S Lachnospiraceae bacterium (HM099641) S UB (FJ848445) S UB (FJ825506) S Eubacterium siraeum (EU266550) S Bacteroidetes bacterium (GU409274) S UB (EU469073) S UB (EU774692) S Rikenellaceae bacterium (EU774692) S UB (DQ799972) S Clostridiaceae bacterium (AB298755) S Rumen bacterium (AB239481) S Clostridiales bacterium (AB477432) S Clostridium sp. (AB596885) S Lachnospiraceae bacterium (DQ789118) S Oscillospira guilliermondii (AB040497) S Lactobacillales bacterium (AY581272) S Rumen bacterium (GU324401) S Clostridiales bacterium (HQ ) S Paenibacillus campinasensis (EU ) S Rumen bacterium (GU324364) S UB (FJ951884) S Lachnospira pectinoschiza (AY169414) S Butyrate-producing bacterium (AY305315) S UB(FJ ) S Rikenellaceae bacterium (AB298736) S B.crossotus (X89981) S Ruminococcus flavefaciens (AF104841) S UB (EU466127) S Uncultured Ruminococcaceae (EU466127) S UB(FJ681013) S Uncultured Ruminococcaceae (EU794113) S UB (GQ451212) S UB (FJ825523) S Marinilabilia salmonicolor (AB ) S Anaerostipes caccae (AB243986) S Lactobacillales bacterium (EU728724) S UB (EU475425) UB=uncultured bacteria, URB=uncultured rumen bacteria, S: Miscanthus sinensis (+) %. That indicated the sequences identified in M. sinensis (+) library represent the majority of bacterial diversity present in the large gut. Through phylogenetic analysis of these OTUs, we found there was a relative big part of clade having no recognized sequences. It was named clade II, including 24 OTUs, 61 clones (Figure 1). In M. sinensis (-) library, as the data summarized in Table 2, 11 OTUs, nearly 25.6% clones were grouped to uncultured bacteria. Moreover, among the phylum Firmicutes, uncultured Ruminococcaceae (D-10, D-13, D-
5 Li et al Table 2. Similarity values of operational taxonomic units (OTUs) based on RFLP of 16S rrna gene sequences derived from M. sinensis (-) library. OTU Length (bp) No. of clones Sequence identity (%) Nearest valid relative (GeneBank accession no.) D UB (EU464240) D Lachnospiraceae bacterium (AF550610) D UB (FJ848449) D URB (GU304334) D Clostridiales bacterium (DQ168656) D UB (EU774974) D UB (GQ448460) D Rumen bacterium (AB239481) D UB (EU776277) D Uncultured Ruminococcaceae bacterium (HQ132395) D UB (EU466343) D Acetivibrio sp. (AB596889) D Uncultured Ruminococcaceae bacterium (EU794228) D Uncultured Ruminococcaceae bacterium (EU794227) D UB (EU465566) D Uncultured Ruminococcaceae bacterium (EU794158) D Hydrogenoanaero bacterium (EU170433) D Clostridiaceae bacterium (AB298755) D T.aceticus (Z49863) D UB (EU464238) D UB (FJ680663) D Lachnospiraceae bacterium (DQ789118) D Bacterium (DQ286651) D Lactobacillales bacterium (AY581272) D Eubacterium oxidoreducens (FR733672) D Ruminococcus sp. (EU815223) D Clostridium sp. (AB093546) D Rumen bacterium (GU324408) D Christensenella minuta (AB490809) D Lachnospira pectinoschiza (AY699282) D Acetomicrobium faecale (FR749983) D UB (FJ848445) D Oscillibacter sp. (HM626173) D Oscillospira guilliermondii (AB040498) D Eubacterium siraeum (L34625) D UB (EU773761) D Rumen bacterium (GU324404) D Butyricicoccus pullicaecorum (EU410376) D Uncultured Ruminococcaceae bacterium (EU794237) D URB (GU304068) D Lactococcus lactis subsp. (EU337112) D Blautia schinkii (X94964) D UB (FJ848445) UB=uncultured bacteria, URB=uncultured rumen bacteria, D: Miscanthus sinensis (-). 14, D-16) accounted for 15% of the total sequences, Lachnospiraceae bacterium (D-2, D-22) accounted for 10.19%, only one OTU D-222 affiliated to the CFB phylum, which had relatively remote similarity with Acetomicrobium faecale. Besides, the Good's coverage of this library was 95.32% owing to 6 single-clone OTUs
6 5970 Afr. J. Microbiol. Res. Table 3. Estimated composition of species of bacteria identified from the two libraries. Proportion (%) S/No Taxon M. sinensis (+)S M. sinensis (-) D firmicutes 48.89(21OTUs) 52.87(24OTUs) 1 Lachnospiraceae bacterium 14.45(4OTUs) (2OTUs) 2 Uncultured Ruminococcaceae bacterium 5.56(1OTU) 15(4OTUs) 3 Ruminococcus flavefaciens 2.22(1OTU) 0 4 Ruminococcus sp (1OTU) 5 Clostridiaceae bacterium 2.22(1OTU) 5.1(1OTU) 6 Clostridium sp. 3.33(1OTU) 5.1(1OTU) 7 Clostridiales bacterium 3.33(2OTUs) 2.55(2OTUs) 8 Lactobacillales bacterium 2.22(1OTU) 1.27(1OTU) 9 Lachnospira pectinoschiza 1.11(1OTU) 0.64(1OTU) 10 Hydrogenoanaero bacterium (1OTU) 11 Eubacterium oxidoreducens (1OTU) 12 Eubacterium siraeum 3.33(1OTU) 3.82(1OTU) 13 Oscillospira guilliermondii 1.11(1OTU) 1.27(1OTU) 14 Oscillibacter sp (1OTU) 15 Butyrate-producing bacterium 1.11(1OTU) 0 16 B.crossotus 1.11(1OTU) 0 17 Blautia schinkii (1OTU) 18 Butyricicoccus pullicaecorum (1OTU) 19 Paenibacillus campinasensis 1.11(1OTU) 0 20 Anaerostipes caccae 1.11(1OTU) 0 21 Acetivibrio sp. 3.33(2OTUs) 3.18(1OTU) 22 T. aceticus 1.11(1OTU) 2.55(1OTU) 23 Lactococcus lactis subsp (1OTU) 24 Christensenella minuta (1OTU) CFB phylum (Bacteroidetes) 6.67(5OTUs) 1.27(1OTU) 1 Uncultured Bacteroidales 2.22(1OTU) 0 2 Unclassified Bacteroidetes bacterium 1.11(1OTU) 0 3 Rikenellaceae bacterium 2.22(2OTUs) 0 4 Marinilabilia salmonicolor 1.11(1OTU) 0 5 Acetomicrobium faecale (1OTU) URB (2OTUs) rumen bacteria 10(2OTUs) 9.55(3OTUs) unidentified bacteria (1OTU) UB 34.44(13OTUs) 31.21(11OTUs) UB=uncultured bacteria, URB=uncultured rumen bacteria, S: Miscanthus sinensis (+), D: Miscanthus sinensis (-). in this library. This value also means the chosen clones was representative. Through the phylogenetic analysis, we also could find clade II, including 25 OTUs, 66 clones (Figure 2). DISCUSSION In the present study, we for the first time comparatively analyzed the bacterial diversity in large intestine of two
7 Li et al Figure 1. Phylogenetic tree derived from 16S rrna sequence data recovered from Bos taurus (M. sinensis (+)) large intestine. The tree was constructed depending on neighbour-joining analysis of a distance matrix obtained from a multiple sequence alignment. Bootstrap value was expressed as a percentage of 1000 trees. groups of cattle raised with/without M. sinensis. Important experimental findings from the present work were summarized below. Sequence analysis of 16S rrna gene clone libraries constructed using specific PCR primers have been frequently used to analyze diversity of microorganism community (Tajima et al., 1999, 2000, 2007). Based on this method, in this study we found that there are changed diversity profiles and previously rarely detected OTUs in the bacterial community of M. sinensis (+) group compared with that of M. sinensis (-) group. It was worthy of noting that compared with species in M. sinensis (-) library, a few species such as Ruminococcus flavefaciens, Butyrate-producing bacterium, Paenibacillus campinasensis, Anaerostipes caccae could only been identified in M. sinensis (+) library. R. flavefaciens was a cellulolytic ruminal bacterium that could utilize cellobiose but not glucose as a substrate for growth (Helaszek and White, 1991), and played an important role in the digestion of hemicellulose and cellulose in plant cell walls. It has been reported that they inherented in the pony and donkey cecum (Julliand et al., 1999). Butyrateproducing bacterium was essential for digestion of plant polysaccharides (Paggi and Fay, 2004), and was found in digestive tract of human (Barcenilla et al., 2000).
8 5972 Afr. J. Microbiol. Res. Figure 2. Phylogenetic tree derived from 16S rrna sequence data recoverd from Bos taurus (M. sinensis (-)) large intesine. The tree was constructed depending on neighbour-joining analysis of a distance matrix obtained from a multiple sequence alignment. Bootstrap value was expressed as a percentage of 1000 trees.
9 Li et al Moreover, previous study indicated high-fiber intake increased the amounts of Butyrate-producing bacterium (Mrazek et al., 2006). Additionally, Paenibacillus campinasensis has drawn much attention recently. P. campinasensis BL11, which produces multiple extracellular polysaccharide-degrading enzymes, including one xylanase, three cellulases, one pectinase and one cyclodextrin glucanotransferase, has a strong potential for plant material degradation and bioresource utilization (Ko et al., 2007). Anaerostipes caccae has been identified as a new saccharolytic, acetate-utilising, butyrate-producing bacterium (Schwiertz et al., 2002). As these species of Firmicutes were crucial for the host in decomposition of plant-derived material, our result thus implied that the intake of M. sinensis was possibly responsible for increasing fibrolytic or cellulolytic bacteria. Another interesting finding is that the ratio of Lachnospiraceae was significantly increased (4.26%) in the large intestine of M. sinensis-fed cattle compared with mixed forage-fed cattle (Table 1). Lachnospiraceae bacterium has been reported as a ubiquitous and important bacteria which could be often detected in the digestive tract of cattle (Dowd et al., 2008). Previous study implied that Lachnospiraceae bacterium could potentially involve in the degradation of cellulose, as Pope et al. (2010) have identified some genes that are homologous to genes present in the genomes of Lachnospiraceae and could express products with carboxymethylcellulase or xylanase activity. Our result also implied that the changes caused by M. sinensis might be due in part to variations in Lachnospiraceae. Further research is necessary to evaluate the potential role of Lachnospiraceae bacterium in the degradation of M. sinensis. It is noteworthy that total CFB clones number was much higher in the large intestine of M. sinensis fed cattle compared to mixed diet-fed cattle (Table 3). Bacteroidetes has been demonstrated to be associated with hydrolysis of polysaccharides in intestine (Mariat et al., 2009; Martens et al., 2009), and high concentration of fiber diet might trigger the increasing trend of Bacteroides in the digestive tract of ruminant. For example, Yuhei et al. (2005) reported that only 14.4% clones belonged to the Bacteroides fragilis group in the fecal sample when cattle were fed with forage containing 41% corns, 23% wheat bran, 12% corn byproducts, 8% rapeseed meal, 7% soybean meal. While, Weidong et al. (2007) indicated that 42.2% clones could be assigned to the phylum CFB in gayals (B. frontails) when cattle were fed with a diet composed of fresh bamboo leaves and twigs. Pitta et al. (2010) also revealed that bermudagrass with a high fiber content could significantly increase the population of Bacteroides in the digestive tract of cattle. Consistent with previous findings, M. sinensis with high concentration of cellulose in our study could result in an increase of Bacteroides. The exact function of Bacteroides in the degradation of M. sinensis needs further investigation. Moreover, from the taxonomic information generated by phylogenetic tree analysis, we found that the ratio of uncultured genus having no recognized relatives (clade II) in M. sinensis (+) library and M. sinensis (-) library were and 51.56%, respectively, indicating that the cattle fed on M. sinensis will produce more novel uncultured genus which probably have specific metabolic effect on decomposition of M. sinensis. However, these bacteria needed to be further identified and their roles also needed to be deeply explored. The diversity of micropopulation existing in digestive tract of ruminants has already been ascribed to breed, age, dietary composition, nutrient density and environmental conditions (Leng et al., 2012). Base on the finding of Dowd et al. (2008) they used 16S rdna bacterial tag-encoded FLX amplicon pyrosequencing (btefap) to study the diversity of cattle faeces, and identified Clostridium, Bacteroides, Porpyhyromonas, Ruminococcus, Alistipes, Lachnospiraceae, Prevotella, Lachnospira, Enterococcus, Oscillospira, Cytophaga, Anaerotruncus, and Acidaminococcus spp as the ubiquitous bacteria. However, in our study, only 6 out of 13 species mentioned above were detected: Clostridium, Bacteroides, Ruminococcus, Lachnospiraceae, Lachnospira, Oscillospira (Table 3). The reason for failing to detect other classification might be: Firstly, the size of our library was relatively small [M. sinensis (+) library and M. sinensis (-) library had 109 and 128 clones, respectively]. Secondly, experimental PCR biases might be unavoidable, like the specificity of the primers. It is been reported that DNA extracted from Fibrobacter succinogenes was amplified less efficiently by PCR in comparison to other gut bacteria (Reysenbach et al., 1992; Suzuki and Giovannoni, 1996). Thirdly, Xiangxi Yellow Cattle might form unique feature of their symbiotic microbe during the long-term evolution for better adaption to local environment. In conclusion, our present study for the first time comparatively analyze the bacterial diversity in the large intestine of Xiangxi Yellow Cattle which were fed on M. sinensis or mixed forage. Our results indicated that the dietary transition from mixed forage to M. sinensis significantly altered the diversity of bacteria including R. flavefaciens, Butyrate-producing bacterium, B. crossotus, P. campinasensis, A. caccae in the phylum Firmicute and Rikenellaceae bacterium, Marinilabilia salmonicolor in the phylum Bacteroidete. In the large intestine of cattle, these species may play roles in the process of degradation of M. sinensis. Therefore, M. sinensis appeared to successfully increase the population of fibrolytic bacteria in the intestinal system of cattle. In this case, we would have a novel way to investigate the degradation of M. sinensis for replaceable energy. Undoubtedly, the information of symbiotic microbiota s diversity of cattle's large intestine adapting to M. sinensis diet will help us better understand the distribution of complex community
10 5974 Afr. J. Microbiol. Res. in cattle s digestion system, will benefit our discovery of bio-enzyme based on this system, and will provide the theoretical support for future work. ACKNOWLEDGEMENTS This research was supported by the National Key Project of Scientific and Technical Supporting Programs Funded by Ministry of Science and Technology of China (NO. 2008BADC4B08), the International Scientific and Technological Cooperation Project (2010DFA62510) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT0963). REFERENCES Bailey RW, MacRea MJ (1970). The hydrolysis by rumen and caecal microbiol enzyme of hemicellulose in plant and degesta particles. J. Agric. Sci. 75:321. Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ (2000). Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66: Bekele AZ, Koike S, Kobayashi Y (2010). Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rrna gene-based analysis. FEMS Microbiol. Lett. 305: Bekele AZ, Koike S, Kobayashi Y (2011). Phylogenetic diversity and dietary association of rumen Treponema revealed using groupspecific 16S rrna gene-based analysis. FEMS Microbiol. Lett. 316: Chen Y, Penner GB, Li M, Oba M, Guan LL (2011). Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 77: Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rdna bacterial tag-encoded FLX amplicon pyrosequencing (btefap). BMC Microbiol. 8:125. Ge X, Burner DM, Xu J, Phillips GC, Sivakumar G (2011). Bioethanol production from dedicated energy crops and residues in Arkansas, USA. Biotechnol. J. 6: Helaszek CT, White BA (1991). Cellobiose uptake and metabolism by Ruminococcus flavefaciens. Appl. Environ. Microbiol. 57: Hoover WH (1978). Digestion and absorption in the hindgut of ruminants. J. Anim. Sci. 46: Huntington GB, Harmon DL, Richards CJ (2006). Sites, rates, and limits of starch digestion and glucose metabolism in growing cattle. J. Anim. Sci. 84:E Julliand V, Vaux A, Millet L, Fonty G (1999). Identification of Ruminococcus flavefaciens as the predominant cellulolytic bacterial species of the equine cecum. Appl. Environ. Microbiol. 65: Ko CH, Chen WL, Tsai CH, Jane WN, Liu CC, Tu J (2007). Paenibacillus campinasensis BL11: a wood material-utilizing bacterial strain isolated from black liquor. Bioresour. Technol. 98: Leng J, Cheng YM, Zhang CY, Zhu RJ, Yang SL, Gou X, Deng WD, Mao HM (2012). Molecular diversity of bacteria in Yunnan yellow cattle (Bos taurs) from Nujiang region, China. Mol. Biol. Rep. 39: Maas LK, Glass TL (1991). Cellobiose uptake by the cellulolytic ruminal anaerobe Fibrobacter (Bacteroides) succinogenes. Can. J. Microbiol. 37: Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, Corthier G, Furet JP (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9:123. Martens EC, Koropatkin NM, Smith TJ, Gordon JI (2009). Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284: Mrazek J, Tepsic K, Avgustin G, Kopecny J (2006). Diet-dependent shifts in ruminal butyrate-producing bacteria. Folia Microbiol. 51: Noziere P, Remond D, Lemosquet S, Chauveau B, Durand D, Poncet C (2005). Effect of site of starch digestion on portal nutrient net fluxes in steers. Br. J. Nutr. 94: Ogura S (2011). Diet selection and foraging behavior of cattle on species-rich, Japanese native grasslands. JIFS 8: Paggi RA, Fay JP (2004). Effect of short-chain acids on the carboxymethylcellulase activity of the ruminal bacterium Ruminococcus albus. Folia Microbiol. 49: Pandya PR, Singh KM, Parnerkar S, Tripathi AK, Mehta HH, Rank DN, Kothari RK, Joshi CG (2010). Bacterial diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rdna analysis. J. Appl. Genet. 51: Paul RM, John CB, Iain D (2011). Phenotypic variation in senescence in Miscanthus: towards optimising biomass quality and quantity. Bioenerg. Res. 5: Pitta DW, Pinchak E, Dowd SE, Osterstock J, Gontcharova V, Youn E, Dorton K, Yoon I, Min BR, Fulford JD, Wickersham TA, Malinowski DP (2010). Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb. Ecol. 59: Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, McHardy AC, Cheng JF, Hugenholtz P, McSweeney CS, Morrison M (2010). Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc. Natl. Acad. Sci. USA. 107: Reysenbach AL, Giver LJ, Wickham GS, Pace NR (1992). Differential amplification of rrna genes by polymerase chain reaction. Appl. Environ. Microbiol. 58: Schloss PD, Handelsman J (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71: Schwarz WH (2001). The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56: Schwiertz A, Hold GL, Duncan SH, Gruhl B, Collins MD, Lawson PA, Flint HJ, Blaut M (2002). Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 25: Shumny VK, Veprev SG, Nechiporenko NN, Goryachkovskaya TN, Slynko NM, Kolchanov NA, Peltek SE (2011). A New Variety of Chinese Silver Grass (Miscanthus sinensis Anderss.): A promising source of cellulose containing raw material. Russian J. Genet. Appl. Res. 1: Suzuki MT, Giovannoni SJ (1996). Bias caused by template annealing in the amplification of mixtures of 16S rrna genes by PCR. Appl. Environ. Microbiol. 62: Tajima K, Aminov RI, Nagamine T, Ogata K, Nakamura M, Matsui H, Benno Y (1999). Rumen bacterial diversity as determined by sequence analysis of 16S rdna libraries. FEMS Microbiol. Ecol. 29: Tajima K, Nonaka I, Higuchi K, Takusari N, Kurihara M, Takenaka A, Mitsumori M, Kajikawa H, Aminov RI (2007). Influence of high temperature and humidity on rumen bacterial diversity in Holstein heifers. Anaerobe 13: Tajima K, ShozoArai, Ogata K, Nagamine T, Matsui H, Nakamura M, Aminov RI, Benno Y (2000). Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe 6: Vega A, Bao M, Rodríguez JL (1997). Fractionating lignocellulose of Miscanthus sinensis with aqueous phenol in acidic medium. Eur. J. Wood Wood Prod. 55: Warner RL, Mitchell GE, Little CO (1972). Post-ruminal digestion of cellulose in wehters and steers. J. Anim. Sci. 34: Weidong D, Matha W, Songcheng M, Jing C, Dongmei X, Tianbao H, Zhifang Y, Huaming M (2007). Phylogenetic analysis of 16S rdna sequences mainfest rumen batcerial diversity in Gayals (Bos frontallis) fed fresh bamboo leaves and twigs (Sinarumdinaria). Asian- Aust. J. Anim. Sci. 20: Yuhei OHH, Mitsuo S, Hisao I, Yoshimi B (2005). Culture-independent analysis of fecal microbiota in cattle. Biosci. Biotechnol. Biochem. 69:
MagExtractor -Genome-
 Instruction manual MagExtractor-Genome-0810 F0981K MagExtractor -Genome- NPK-101 100 preparations Store at 4 C Contents [1] Introduction [2] Components [3] Materials required [4] Protocol 1. Purification
Instruction manual MagExtractor-Genome-0810 F0981K MagExtractor -Genome- NPK-101 100 preparations Store at 4 C Contents [1] Introduction [2] Components [3] Materials required [4] Protocol 1. Purification
DIGESTION is the physical and
 Digestion DIGESTION is the physical and chemical breakdown of feeds as they pass through the gastrointestinal tract. The structures of the gastrointestinal tract include the mouth, the esophagus, the stomach,
Digestion DIGESTION is the physical and chemical breakdown of feeds as they pass through the gastrointestinal tract. The structures of the gastrointestinal tract include the mouth, the esophagus, the stomach,
AmphoraNet: Taxonomic Composition Analysis of Metagenomic Shotgun Sequencing Data
 Csaba Kerepesi, Dániel Bánky, Vince Grolmusz: AmphoraNet: Taxonomic Composition Analysis of Metagenomic Shotgun Sequencing Data http://pitgroup.org/amphoranet/ PIT Bioinformatics Group, Department of Computer
Csaba Kerepesi, Dániel Bánky, Vince Grolmusz: AmphoraNet: Taxonomic Composition Analysis of Metagenomic Shotgun Sequencing Data http://pitgroup.org/amphoranet/ PIT Bioinformatics Group, Department of Computer
Understanding Feed Analysis Terminology
 Understanding Feed Analysis Terminology One of the most important steps in developing a ration suitable for dairy animals is feed testing. It is essential to have a starting point in order to formulate
Understanding Feed Analysis Terminology One of the most important steps in developing a ration suitable for dairy animals is feed testing. It is essential to have a starting point in order to formulate
Application Guide... 2
 Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
TIANquick Mini Purification Kit
 TIANquick Mini Purification Kit For purification of PCR products, 100 bp to 20 kb www.tiangen.com TIANquick Mini Purification Kit (Spin column) Cat no. DP203 Kit Contents Contents Buffer BL Buffer PB Buffer
TIANquick Mini Purification Kit For purification of PCR products, 100 bp to 20 kb www.tiangen.com TIANquick Mini Purification Kit (Spin column) Cat no. DP203 Kit Contents Contents Buffer BL Buffer PB Buffer
First Strand cdna Synthesis
 380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 [email protected] A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 [email protected] A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
Genomic DNA Clean & Concentrator Catalog Nos. D4010 & D4011
 Page 0 INSTRUCTION MANUAL Catalog Nos. D4010 & D4011 Highlights Quick (5 minute) spin column recovery of large-sized DNA (e.g., genomic, mitochondrial, plasmid (BAC/PAC), viral, phage, (wga)dna, etc.)
Page 0 INSTRUCTION MANUAL Catalog Nos. D4010 & D4011 Highlights Quick (5 minute) spin column recovery of large-sized DNA (e.g., genomic, mitochondrial, plasmid (BAC/PAC), viral, phage, (wga)dna, etc.)
HighPure Maxi Plasmid Kit
 HighPure Maxi Plasmid Kit For purification of high pure plasmid DNA with high yields www.tiangen.com PP120109 HighPure Maxi Plasmid Kit Kit Contents Storage Cat.no. DP116 Contents RNaseA (100 mg/ml) Buffer
HighPure Maxi Plasmid Kit For purification of high pure plasmid DNA with high yields www.tiangen.com PP120109 HighPure Maxi Plasmid Kit Kit Contents Storage Cat.no. DP116 Contents RNaseA (100 mg/ml) Buffer
NimbleGen DNA Methylation Microarrays and Services
 NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
Genomic DNA Extraction Kit INSTRUCTION MANUAL
 Genomic DNA Extraction Kit INSTRUCTION MANUAL Table of Contents Introduction 3 Kit Components 3 Storage Conditions 4 Recommended Equipment and Reagents 4 Introduction to the Protocol 4 General Overview
Genomic DNA Extraction Kit INSTRUCTION MANUAL Table of Contents Introduction 3 Kit Components 3 Storage Conditions 4 Recommended Equipment and Reagents 4 Introduction to the Protocol 4 General Overview
PrimeSTAR HS DNA Polymerase
 Cat. # R010A For Research Use PrimeSTAR HS DNA Polymerase Product Manual Table of Contents I. Description...3 II. III. IV. Components...3 Storage...3 Features...3 V. General Composition of PCR Reaction
Cat. # R010A For Research Use PrimeSTAR HS DNA Polymerase Product Manual Table of Contents I. Description...3 II. III. IV. Components...3 Storage...3 Features...3 V. General Composition of PCR Reaction
TOOLS FOR T-RFLP DATA ANALYSIS USING EXCEL
 TOOLS FOR T-RFLP DATA ANALYSIS USING EXCEL A collection of Visual Basic macros for the analysis of terminal restriction fragment length polymorphism data Nils Johan Fredriksson TOOLS FOR T-RFLP DATA ANALYSIS
TOOLS FOR T-RFLP DATA ANALYSIS USING EXCEL A collection of Visual Basic macros for the analysis of terminal restriction fragment length polymorphism data Nils Johan Fredriksson TOOLS FOR T-RFLP DATA ANALYSIS
Chromatin Immunoprecipitation (ChIP)
 Chromatin Immunoprecipitation (ChIP) Day 1 A) DNA shearing 1. Samples Dissect tissue (One Mouse OBs) of interest and transfer to an eppendorf containing 0.5 ml of dissecting media (on ice) or PBS but without
Chromatin Immunoprecipitation (ChIP) Day 1 A) DNA shearing 1. Samples Dissect tissue (One Mouse OBs) of interest and transfer to an eppendorf containing 0.5 ml of dissecting media (on ice) or PBS but without
Mammalian digestive tracts
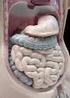 Mammalian digestive tracts Mouth: mastication, some digestive enzymes Esophagus: simple transport tube Stomach: most initial digestion, some physical processing Small intestine: digestion continues, some
Mammalian digestive tracts Mouth: mastication, some digestive enzymes Esophagus: simple transport tube Stomach: most initial digestion, some physical processing Small intestine: digestion continues, some
Biotechnology and Recombinant DNA (Chapter 9) Lecture Materials for Amy Warenda Czura, Ph.D. Suffolk County Community College
 Biotechnology and Recombinant DNA (Chapter 9) Lecture Materials for Amy Warenda Czura, Ph.D. Suffolk County Community College Primary Source for figures and content: Eastern Campus Tortora, G.J. Microbiology
Biotechnology and Recombinant DNA (Chapter 9) Lecture Materials for Amy Warenda Czura, Ph.D. Suffolk County Community College Primary Source for figures and content: Eastern Campus Tortora, G.J. Microbiology
Cloning Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems
 Promega Notes Number 71, 1999, p. 10 Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems By Kimberly Knoche, Ph.D., and Dan Kephart, Ph.D. Promega Corporation Corresponding
Promega Notes Number 71, 1999, p. 10 Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems By Kimberly Knoche, Ph.D., and Dan Kephart, Ph.D. Promega Corporation Corresponding
RevertAid Premium First Strand cdna Synthesis Kit
 RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
FAO Symposium on. The role of agricultural biotechnologies for production of bio-energy in developing countries"
 FAO Symposium on The role of agricultural biotechnologies for production of bio-energy in developing countries" ETHANOL PRODUCTION VIA ENZYMATIC HYDROLYSIS OF SUGAR-CANE BAGASSE AND STRAW Elba P. S. Bon
FAO Symposium on The role of agricultural biotechnologies for production of bio-energy in developing countries" ETHANOL PRODUCTION VIA ENZYMATIC HYDROLYSIS OF SUGAR-CANE BAGASSE AND STRAW Elba P. S. Bon
Summary. Keywords: methanol, glycerin, intake, beef cattle. Introduction
 Effect of Methanol Infusion on Intake and Digestion of a Grain-based Diet by Beef Cattle K.N. Winsco, N.M. Kenney, R.O. Dittmar, III, J.A. Coverdale, J.E. Sawyer, and T.A. Wickersham Texas A & M University,
Effect of Methanol Infusion on Intake and Digestion of a Grain-based Diet by Beef Cattle K.N. Winsco, N.M. Kenney, R.O. Dittmar, III, J.A. Coverdale, J.E. Sawyer, and T.A. Wickersham Texas A & M University,
Ruminant Digestive System
 Ruminant Digestive System Complex structure with four compartments Source: Animal Feeding and Nutrition (Jurgens) Ruminant Characteristics Primarily herbivores Cattle, sheep, goats, deer, elk Camelids
Ruminant Digestive System Complex structure with four compartments Source: Animal Feeding and Nutrition (Jurgens) Ruminant Characteristics Primarily herbivores Cattle, sheep, goats, deer, elk Camelids
ab185916 Hi-Fi cdna Synthesis Kit
 ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
BacReady TM Multiplex PCR System
 BacReady TM Multiplex PCR System Technical Manual No. 0191 Version 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI Detailed Experimental
BacReady TM Multiplex PCR System Technical Manual No. 0191 Version 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI Detailed Experimental
What a re r Lipids? What a re r Fatty y Ac A ids?
 2010 - Beef Cattle In-Service Training Inclusion of Lipids into Beef Cattle Diets Reinaldo F. Cooke, Ph. D. Oregon State University EOARC, Burns What are Lipids? Organic compounds Plant and animal compounds
2010 - Beef Cattle In-Service Training Inclusion of Lipids into Beef Cattle Diets Reinaldo F. Cooke, Ph. D. Oregon State University EOARC, Burns What are Lipids? Organic compounds Plant and animal compounds
DNA ligase. ATP (or NAD+)
 DNA Ligase enzyme catalysing formation of phosphodiesteric bound between group 3 -OH of one end of DNA molecule and group 5 -phosphate of the second end of DNA DNA ligase ATP (or NAD+) Ligase cofactors
DNA Ligase enzyme catalysing formation of phosphodiesteric bound between group 3 -OH of one end of DNA molecule and group 5 -phosphate of the second end of DNA DNA ligase ATP (or NAD+) Ligase cofactors
Online Supporting Material
 Supplemental Table 1 Bacterial pure cultures used in this study for optimization of the Lactobacillus group qpcr Species Strain Clostridial Phylogenetic affiliation according to cluster 1 NCBI 2 taxonomy
Supplemental Table 1 Bacterial pure cultures used in this study for optimization of the Lactobacillus group qpcr Species Strain Clostridial Phylogenetic affiliation according to cluster 1 NCBI 2 taxonomy
How To Make A Tri Reagent
 TRI Reagent For processing tissues, cells cultured in monolayer or cell pellets Catalog Number T9424 Store at room temperature. TECHNICAL BULLETIN Product Description TRI Reagent is a quick and convenient
TRI Reagent For processing tissues, cells cultured in monolayer or cell pellets Catalog Number T9424 Store at room temperature. TECHNICAL BULLETIN Product Description TRI Reagent is a quick and convenient
Protocols. Internal transcribed spacer region (ITS) region. Niklaus J. Grünwald, Frank N. Martin, and Meg M. Larsen (2013)
 Protocols Internal transcribed spacer region (ITS) region Niklaus J. Grünwald, Frank N. Martin, and Meg M. Larsen (2013) The nuclear ribosomal RNA (rrna) genes (small subunit, large subunit and 5.8S) are
Protocols Internal transcribed spacer region (ITS) region Niklaus J. Grünwald, Frank N. Martin, and Meg M. Larsen (2013) The nuclear ribosomal RNA (rrna) genes (small subunit, large subunit and 5.8S) are
Analysis of the DNA Methylation Patterns at the BRCA1 CpG Island
 Analysis of the DNA Methylation Patterns at the BRCA1 CpG Island Frédérique Magdinier 1 and Robert Dante 2 1 Laboratory of Molecular Biology of the Cell, Ecole Normale Superieure, Lyon, France 2 Laboratory
Analysis of the DNA Methylation Patterns at the BRCA1 CpG Island Frédérique Magdinier 1 and Robert Dante 2 1 Laboratory of Molecular Biology of the Cell, Ecole Normale Superieure, Lyon, France 2 Laboratory
Production of prebiotics from PATRICK ADLERCREUTZ, DIV. OF BIOTECHNOLOGY
 Production of prebiotics from hemicellulose PATRICK ADLERCREUTZ, DIV. OF BIOTECHNOLOGY Gut microbiota Gut microbiota microorganisms in our gastrointestinal tract The gut microbiota has a large influence
Production of prebiotics from hemicellulose PATRICK ADLERCREUTZ, DIV. OF BIOTECHNOLOGY Gut microbiota Gut microbiota microorganisms in our gastrointestinal tract The gut microbiota has a large influence
TransformAid Bacterial Transformation Kit
 Home Contacts Order Catalog Support Search Alphabetical Index Numerical Index Restriction Endonucleases Modifying Enzymes PCR Kits Markers Nucleic Acids Nucleotides & Oligonucleotides Media Transfection
Home Contacts Order Catalog Support Search Alphabetical Index Numerical Index Restriction Endonucleases Modifying Enzymes PCR Kits Markers Nucleic Acids Nucleotides & Oligonucleotides Media Transfection
Enzymes: Amylase Activity in Starch-degrading Soil Isolates
 Enzymes: Amylase Activity in Starch-degrading Soil Isolates Introduction This week you will continue our theme of industrial microbiologist by characterizing the enzyme activity we selected for (starch
Enzymes: Amylase Activity in Starch-degrading Soil Isolates Introduction This week you will continue our theme of industrial microbiologist by characterizing the enzyme activity we selected for (starch
IMBB 2013. Genomic DNA purifica8on
 IMBB 2013 Genomic DNA purifica8on Why purify DNA? The purpose of DNA purifica8on from the cell/8ssue is to ensure it performs well in subsequent downstream applica8ons, e.g. Polymerase Chain Reac8on (PCR),
IMBB 2013 Genomic DNA purifica8on Why purify DNA? The purpose of DNA purifica8on from the cell/8ssue is to ensure it performs well in subsequent downstream applica8ons, e.g. Polymerase Chain Reac8on (PCR),
DP419 RNAsimple Total RNA Kit. RNAprep pure Series. DP501 mircute mirna Isolation Kit. DP438 MagGene Viral DNA / RNA Kit. DP405 TRNzol Reagent
 Overview of TIANGEN Products DP419 RNAsimple Total RNA Kit DP430 RNAprep pure Kit(For Cell/Bacteria) DP315/DP315-R TIANamp Virus DNA/RNA Kit DP431 RNAprep pure Kit (For Tissue) Silica-membrane Technology
Overview of TIANGEN Products DP419 RNAsimple Total RNA Kit DP430 RNAprep pure Kit(For Cell/Bacteria) DP315/DP315-R TIANamp Virus DNA/RNA Kit DP431 RNAprep pure Kit (For Tissue) Silica-membrane Technology
DOE Office of Biological & Environmental Research: Biofuels Strategic Plan
 DOE Office of Biological & Environmental Research: Biofuels Strategic Plan I. Current Situation The vast majority of liquid transportation fuel used in the United States is derived from fossil fuels. In
DOE Office of Biological & Environmental Research: Biofuels Strategic Plan I. Current Situation The vast majority of liquid transportation fuel used in the United States is derived from fossil fuels. In
Molecular and Cell Biology Laboratory (BIOL-UA 223) Instructor: Ignatius Tan Phone: 212-998-8295 Office: 764 Brown Email: ignatius.tan@nyu.
 Molecular and Cell Biology Laboratory (BIOL-UA 223) Instructor: Ignatius Tan Phone: 212-998-8295 Office: 764 Brown Email: [email protected] Course Hours: Section 1: Mon: 12:30-3:15 Section 2: Wed: 12:30-3:15
Molecular and Cell Biology Laboratory (BIOL-UA 223) Instructor: Ignatius Tan Phone: 212-998-8295 Office: 764 Brown Email: [email protected] Course Hours: Section 1: Mon: 12:30-3:15 Section 2: Wed: 12:30-3:15
DNA Isolation Kit for Cells and Tissues
 DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
IMCAS-BRC: toward better management and more efficient exploitation of microbial resources
 IMCAS-BRC: toward better management and more efficient exploitation of microbial resources Xiuzhu Dong Biological Resources Center Institute of Microbiology, Chinese Academy of Sciences Challenges Global
IMCAS-BRC: toward better management and more efficient exploitation of microbial resources Xiuzhu Dong Biological Resources Center Institute of Microbiology, Chinese Academy of Sciences Challenges Global
The impact of antibiotics on the gut microbiota
 The impact of antibiotics on the gut microbiota Dr Paul Cotter [email protected] Teagasc Food Research Centre, Moorepark & Alimentary Pharmabiotic Centre (APC), University College Cork Cork, Ireland
The impact of antibiotics on the gut microbiota Dr Paul Cotter [email protected] Teagasc Food Research Centre, Moorepark & Alimentary Pharmabiotic Centre (APC), University College Cork Cork, Ireland
PyroPhage 3173 DNA Polymerase, Exonuclease Minus (Exo-)
 PyroPhage 3173 DNA Polymerase, Exonuclease Minus (Exo-) FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608)
PyroPhage 3173 DNA Polymerase, Exonuclease Minus (Exo-) FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608)
FEEDING THE DAIRY COW DURING LACTATION
 Department of Animal Science FEEDING THE DAIRY COW DURING LACTATION Dairy Cattle Production 342-450A Page 1 of 8 Feeding the Dairy Cow during Lactation There are main stages in the lactation cycle of the
Department of Animal Science FEEDING THE DAIRY COW DURING LACTATION Dairy Cattle Production 342-450A Page 1 of 8 Feeding the Dairy Cow during Lactation There are main stages in the lactation cycle of the
Biorefinery concepts in the paper industry
 Biorefinery concepts in the paper industry Graziano Elegir, Tullia Maifreni, Daniele Bussini Innovhub, Paper Division Azienda Speciale della Camera di Commercio di Milano OUTLINE General aspects on the
Biorefinery concepts in the paper industry Graziano Elegir, Tullia Maifreni, Daniele Bussini Innovhub, Paper Division Azienda Speciale della Camera di Commercio di Milano OUTLINE General aspects on the
Molecular Analysis of Phytase Gene Cloned. from Bacillus subtilis
 Advanced Studies in Biology, Vol. 3, 2011, no. 3, 103 110 Molecular Analysis of Phytase Gene Cloned from Bacillus subtilis R. Bawane 1, K. Tantwai 1*, L.P.S. Rajput 1, M. Kadam-Bedekar 2 S. Kumar 1, I.
Advanced Studies in Biology, Vol. 3, 2011, no. 3, 103 110 Molecular Analysis of Phytase Gene Cloned from Bacillus subtilis R. Bawane 1, K. Tantwai 1*, L.P.S. Rajput 1, M. Kadam-Bedekar 2 S. Kumar 1, I.
HiPer RT-PCR Teaching Kit
 HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
RT-PCR: Two-Step Protocol
 RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
Caecotrophy in Rabbits
 Caecotrophy In Rabbits Amy E. Halls, M.Sc. Monogastric Nutritionist Shur Gain, Nutreco Canada Inc. January 2008 Caecotrophy in Rabbits Amy E. Halls, M.Sc. Monogastric Nutritionist Shur-Gain, Nutreco Canada
Caecotrophy In Rabbits Amy E. Halls, M.Sc. Monogastric Nutritionist Shur Gain, Nutreco Canada Inc. January 2008 Caecotrophy in Rabbits Amy E. Halls, M.Sc. Monogastric Nutritionist Shur-Gain, Nutreco Canada
Chair of Chemistry of Biogenic Resources TU München - R&D activities -
 Chair of Chemistry of Biogenic Resources TU München - R&D activities - Chair of Chemistry of Biogenic Resources Schulgasse 18, 94315 Straubing, Germany [email protected] www.rohstoffwandel.de Locations Freising
Chair of Chemistry of Biogenic Resources TU München - R&D activities - Chair of Chemistry of Biogenic Resources Schulgasse 18, 94315 Straubing, Germany [email protected] www.rohstoffwandel.de Locations Freising
Hepatitis B Virus Genemer Mix
 Product Manual Hepatitis B Virus Genemer Mix Primer Pair for amplification of HBV Specific DNA Fragment Includes Internal Negative Control Primers and Template Catalog No.: 60-2007-12 Store at 20 o C For
Product Manual Hepatitis B Virus Genemer Mix Primer Pair for amplification of HBV Specific DNA Fragment Includes Internal Negative Control Primers and Template Catalog No.: 60-2007-12 Store at 20 o C For
The Techniques of Molecular Biology: Forensic DNA Fingerprinting
 Revised Fall 2011 The Techniques of Molecular Biology: Forensic DNA Fingerprinting The techniques of molecular biology are used to manipulate the structure and function of molecules such as DNA and proteins
Revised Fall 2011 The Techniques of Molecular Biology: Forensic DNA Fingerprinting The techniques of molecular biology are used to manipulate the structure and function of molecules such as DNA and proteins
PCR and Sequencing Reaction Clean-Up Kit (Magnetic Bead System) 50 preps Product #60200
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] PCR and Sequencing Reaction Clean-Up Kit (Magnetic Bead System)
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] PCR and Sequencing Reaction Clean-Up Kit (Magnetic Bead System)
Protein and Energy Supplementation to Beef Cows Grazing New Mexico Rangelands
 Protein and Energy Supplementation to Beef Cows Grazing New Mexico Rangelands Cooperative Extension Service Circular 564 College of Agriculture and Home Economics CONTENTS General ruminant nutrition...
Protein and Energy Supplementation to Beef Cows Grazing New Mexico Rangelands Cooperative Extension Service Circular 564 College of Agriculture and Home Economics CONTENTS General ruminant nutrition...
360 Master Mix. , and a supplementary 360 GC Enhancer.
 Product Bulletin AmpliTaq Gold 360 Master Mix and 360 DNA Polymerase AmpliTaq Gold 360 Master Mix AmpliTaq Gold 360 DNA Polymerase 360 Coverage for a Full Range of Targets AmpliTaq Gold 360 Master Mix
Product Bulletin AmpliTaq Gold 360 Master Mix and 360 DNA Polymerase AmpliTaq Gold 360 Master Mix AmpliTaq Gold 360 DNA Polymerase 360 Coverage for a Full Range of Targets AmpliTaq Gold 360 Master Mix
Forage Crises? Extending Forages and Use of Non-forage Fiber Sources. Introduction
 Forage Crises? Extending Forages and Use of Non-forage Fiber Sources Mike Allen and Jennifer Voelker Michigan State University Dept. of Animal Science Introduction Forage availability is sometimes limited
Forage Crises? Extending Forages and Use of Non-forage Fiber Sources Mike Allen and Jennifer Voelker Michigan State University Dept. of Animal Science Introduction Forage availability is sometimes limited
MEASURING PHYSICALLY EFFECTIVE FIBER ON-FARM TO PREDICT COW RESPONSE
 MEASURING PHYSICALLY EFFECTIVE FIBER ON-FARM TO PREDICT COW RESPONSE K. W. Cotanch and R. J. Grant W. H. Miner Agricultural Research Institute Chazy, NY INTRODUCTION This paper is a follow-up to our 2005
MEASURING PHYSICALLY EFFECTIVE FIBER ON-FARM TO PREDICT COW RESPONSE K. W. Cotanch and R. J. Grant W. H. Miner Agricultural Research Institute Chazy, NY INTRODUCTION This paper is a follow-up to our 2005
Data Analysis for Ion Torrent Sequencing
 IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
PicoMaxx High Fidelity PCR System
 PicoMaxx High Fidelity PCR System Instruction Manual Catalog #600420 (100 U), #600422 (500 U), and #600424 (1000 U) Revision C Research Use Only. Not for Use in Diagnostic Procedures. 600420-12 LIMITED
PicoMaxx High Fidelity PCR System Instruction Manual Catalog #600420 (100 U), #600422 (500 U), and #600424 (1000 U) Revision C Research Use Only. Not for Use in Diagnostic Procedures. 600420-12 LIMITED
Aurora Forensic Sample Clean-up Protocol
 Aurora Forensic Sample Clean-up Protocol 106-0008-BA-D 2015 Boreal Genomics, Inc. All rights reserved. All trademarks are property of their owners. http://www.borealgenomics.com [email protected]
Aurora Forensic Sample Clean-up Protocol 106-0008-BA-D 2015 Boreal Genomics, Inc. All rights reserved. All trademarks are property of their owners. http://www.borealgenomics.com [email protected]
QUANTITATIVE RT-PCR. A = B (1+e) n. A=amplified products, B=input templates, n=cycle number, and e=amplification efficiency.
 QUANTITATIVE RT-PCR Application: Quantitative RT-PCR is used to quantify mrna in both relative and absolute terms. It can be applied for the quantification of mrna expressed from endogenous genes, and
QUANTITATIVE RT-PCR Application: Quantitative RT-PCR is used to quantify mrna in both relative and absolute terms. It can be applied for the quantification of mrna expressed from endogenous genes, and
ZR DNA Sequencing Clean-up Kit
 INSTRUCTION MANUAL ZR DNA Sequencing Clean-up Kit Catalog Nos. D40 & D4051 Highlights Simple 2 Minute Bind, Wash, Elute Procedure Flexible 6-20 µl Elution Volumes Allow for Direct Loading of Samples with
INSTRUCTION MANUAL ZR DNA Sequencing Clean-up Kit Catalog Nos. D40 & D4051 Highlights Simple 2 Minute Bind, Wash, Elute Procedure Flexible 6-20 µl Elution Volumes Allow for Direct Loading of Samples with
GRS Plasmid Purification Kit Transfection Grade GK73.0002 (2 MaxiPreps)
 1 GRS Plasmid Purification Kit Transfection Grade GK73.0002 (2 MaxiPreps) (FOR RESEARCH ONLY) Sample : Expected Yield : Endotoxin: Format : Operation Time : Elution Volume : 50-400 ml of cultured bacterial
1 GRS Plasmid Purification Kit Transfection Grade GK73.0002 (2 MaxiPreps) (FOR RESEARCH ONLY) Sample : Expected Yield : Endotoxin: Format : Operation Time : Elution Volume : 50-400 ml of cultured bacterial
SOLIDscript Solid Phase cdna Synthesis Kit Instruction Manual
 Toll Free: 866-252-7771 752A Lincoln Blvd. Phone: 732-469-7771 Fax: 732-469-7782 Middlesex, NJ 08846 Web: www.purebiotechllc.com SOLIDscript Solid Phase cdna Synthesis Kit Instruction Manual Product: SOLIDscript
Toll Free: 866-252-7771 752A Lincoln Blvd. Phone: 732-469-7771 Fax: 732-469-7782 Middlesex, NJ 08846 Web: www.purebiotechllc.com SOLIDscript Solid Phase cdna Synthesis Kit Instruction Manual Product: SOLIDscript
EU Reference Laboratory for E. coli Department of Veterinary Public Health and Food Safety Unit of Foodborne Zoonoses Istituto Superiore di Sanità
 Identification and characterization of Verocytotoxin-producing Escherichia coli (VTEC) by Real Time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated
Identification and characterization of Verocytotoxin-producing Escherichia coli (VTEC) by Real Time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated
Energy in the New Dairy NRC. Maurice L. Eastridge 1 Department of Animal Sciences The Ohio State University
 Energy in the New Dairy NRC Maurice L. Eastridge 1 Department of Animal Sciences The Ohio State University Introduction Energy is vital to the function of all cells, and thus physiologically, it is vital
Energy in the New Dairy NRC Maurice L. Eastridge 1 Department of Animal Sciences The Ohio State University Introduction Energy is vital to the function of all cells, and thus physiologically, it is vital
Induction of Enzyme Activity in Bacteria:The Lac Operon. Preparation for Laboratory: Web Tutorial - Lac Operon - submit questions
 Induction of Enzyme Activity in Bacteria:The Lac Operon Preparation for Laboratory: Web Tutorial - Lac Operon - submit questions I. Background: For the last week you explored the functioning of the enzyme
Induction of Enzyme Activity in Bacteria:The Lac Operon Preparation for Laboratory: Web Tutorial - Lac Operon - submit questions I. Background: For the last week you explored the functioning of the enzyme
Intended Use: The kit is designed to detect the 5 different mutations found in Asian population using seven different primers.
 Unzipping Genes MBPCR014 Beta-Thalassemia Detection Kit P r o d u c t I n f o r m a t i o n Description: Thalassemia is a group of genetic disorders characterized by quantitative defects in globin chain
Unzipping Genes MBPCR014 Beta-Thalassemia Detection Kit P r o d u c t I n f o r m a t i o n Description: Thalassemia is a group of genetic disorders characterized by quantitative defects in globin chain
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual
 Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
IIID 14. Biotechnology in Fish Disease Diagnostics: Application of the Polymerase Chain Reaction (PCR)
 IIID 14. Biotechnology in Fish Disease Diagnostics: Application of the Polymerase Chain Reaction (PCR) Background Infectious diseases caused by pathogenic organisms such as bacteria, viruses, protozoa,
IIID 14. Biotechnology in Fish Disease Diagnostics: Application of the Polymerase Chain Reaction (PCR) Background Infectious diseases caused by pathogenic organisms such as bacteria, viruses, protozoa,
A simplified universal genomic DNA extraction protocol suitable for PCR
 A simplified universal genomic DNA extraction protocol suitable for PCR T.Y. Wang, L. Wang, J.H. Zhang and W.H. Dong Department of Biochemistry and Molecular Biology, Xinxiang Medical University, Henan,
A simplified universal genomic DNA extraction protocol suitable for PCR T.Y. Wang, L. Wang, J.H. Zhang and W.H. Dong Department of Biochemistry and Molecular Biology, Xinxiang Medical University, Henan,
CORN BY-PRODUCTS IN DAIRY COW RATIONS
 CORN BY-PRODUCTS IN DAIRY COW RATIONS Dennis Lunn, Ruminant Nutritionist Shur-Gain, Nutreco Canada Inc. CORN BY-PRODUCTS IN DAIRY COW RATIONS Dennis Lunn, Ruminant Nutritionist Shur-Gain, Nutreco Canada
CORN BY-PRODUCTS IN DAIRY COW RATIONS Dennis Lunn, Ruminant Nutritionist Shur-Gain, Nutreco Canada Inc. CORN BY-PRODUCTS IN DAIRY COW RATIONS Dennis Lunn, Ruminant Nutritionist Shur-Gain, Nutreco Canada
How To Understand The Human Body
 Introduction to Biology and Chemistry Outline I. Introduction to biology A. Definition of biology - Biology is the study of life. B. Characteristics of Life 1. Form and size are characteristic. e.g. A
Introduction to Biology and Chemistry Outline I. Introduction to biology A. Definition of biology - Biology is the study of life. B. Characteristics of Life 1. Form and size are characteristic. e.g. A
Identification of the VTEC serogroups mainly associated with human infections by conventional PCR amplification of O-associated genes
 Identification of the VTEC serogroups mainly associated with human infections by conventional PCR amplification of O-associated genes 1. Aim and field of application The present method concerns the identification
Identification of the VTEC serogroups mainly associated with human infections by conventional PCR amplification of O-associated genes 1. Aim and field of application The present method concerns the identification
Quantitative Real Time PCR Protocol. Stack Lab
 Quantitative Real Time PCR Protocol Stack Lab Overview Real-time quantitative polymerase chain reaction (qpcr) differs from regular PCR by including in the reaction fluorescent reporter molecules that
Quantitative Real Time PCR Protocol Stack Lab Overview Real-time quantitative polymerase chain reaction (qpcr) differs from regular PCR by including in the reaction fluorescent reporter molecules that
Plant Genomic DNA Extraction using CTAB
 Plant Genomic DNA Extraction using CTAB Introduction The search for a more efficient means of extracting DNA of both higher quality and yield has lead to the development of a variety of protocols, however
Plant Genomic DNA Extraction using CTAB Introduction The search for a more efficient means of extracting DNA of both higher quality and yield has lead to the development of a variety of protocols, however
Table of Contents. I. Description... 2. II. Kit Components... 2. III. Storage... 2. IV. 1st Strand cdna Synthesis Reaction... 3
 Table of Contents I. Description... 2 II. Kit Components... 2 III. Storage... 2 IV. 1st Strand cdna Synthesis Reaction... 3 V. RT-PCR, Real-time RT-PCR... 4 VI. Application... 5 VII. Preparation of RNA
Table of Contents I. Description... 2 II. Kit Components... 2 III. Storage... 2 IV. 1st Strand cdna Synthesis Reaction... 3 V. RT-PCR, Real-time RT-PCR... 4 VI. Application... 5 VII. Preparation of RNA
HiPer Ion Exchange Chromatography Teaching Kit
 HiPer Ion Exchange Chromatography Teaching Kit Product Code: HTC001 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 5-6 hours Storage Instructions: The kit is stable for
HiPer Ion Exchange Chromatography Teaching Kit Product Code: HTC001 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 5-6 hours Storage Instructions: The kit is stable for
Biotechnology: DNA Technology & Genomics
 Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Introduction. Preparation of Template DNA
 Procedures and Recommendations for DNA Sequencing at the Plant-Microbe Genomics Facility Ohio State University Biological Sciences Building Room 420, 484 W. 12th Ave., Columbus OH 43210 Telephone: 614/247-6204;
Procedures and Recommendations for DNA Sequencing at the Plant-Microbe Genomics Facility Ohio State University Biological Sciences Building Room 420, 484 W. 12th Ave., Columbus OH 43210 Telephone: 614/247-6204;
Nucleic Acid Techniques in Bacterial Systematics
 Nucleic Acid Techniques in Bacterial Systematics Edited by Erko Stackebrandt Department of Microbiology University of Queensland St Lucia, Australia and Michael Goodfellow Department of Microbiology University
Nucleic Acid Techniques in Bacterial Systematics Edited by Erko Stackebrandt Department of Microbiology University of Queensland St Lucia, Australia and Michael Goodfellow Department of Microbiology University
PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI,
 Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
1. The diagram below represents a biological process
 1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
Lecture 13: DNA Technology. DNA Sequencing. DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology
 Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Recombinant DNA and Biotechnology
 Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
1/12 Dideoxy DNA Sequencing
 1/12 Dideoxy DNA Sequencing Dideoxy DNA sequencing utilizes two steps: PCR (polymerase chain reaction) amplification of DNA using dideoxy nucleoside triphosphates (Figures 1 and 2)and denaturing polyacrylamide
1/12 Dideoxy DNA Sequencing Dideoxy DNA sequencing utilizes two steps: PCR (polymerase chain reaction) amplification of DNA using dideoxy nucleoside triphosphates (Figures 1 and 2)and denaturing polyacrylamide
TRI Reagent Solution. A. Product Description. RNA / DNA / Protein Isolation Reagent Part Number AM9738 100 ml
 TRI Reagent Solution RNA / DNA / Protein Isolation Reagent Part Number AM9738 100 ml A. Product Description TRI Reagent * solution is a complete and ready-to-use reagent for the isolation of total RNA
TRI Reagent Solution RNA / DNA / Protein Isolation Reagent Part Number AM9738 100 ml A. Product Description TRI Reagent * solution is a complete and ready-to-use reagent for the isolation of total RNA
50 g 650 L. *Average yields will vary depending upon a number of factors including type of phage, growth conditions used and developmental stage.
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
restriction enzymes 350 Home R. Ward: Spring 2001
 restriction enzymes 350 Home Restriction Enzymes (endonucleases): molecular scissors that cut DNA Properties of widely used Type II restriction enzymes: recognize a single sequence of bases in dsdna, usually
restriction enzymes 350 Home Restriction Enzymes (endonucleases): molecular scissors that cut DNA Properties of widely used Type II restriction enzymes: recognize a single sequence of bases in dsdna, usually
Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity
 Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity Today you analyze the results of your bacterial transformation from last week and determine the efficiency
Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity Today you analyze the results of your bacterial transformation from last week and determine the efficiency
BEC Feed Solutions. Steve Blake BEC Feed Solutions
 BEC Feed Solutions Presenter: Steve Blake BEC Feed Solutions Nutritional Role of Phosphorus Phosphorus (P) is present in all cells in the body Essential for many digestive and metabolic processes, including
BEC Feed Solutions Presenter: Steve Blake BEC Feed Solutions Nutritional Role of Phosphorus Phosphorus (P) is present in all cells in the body Essential for many digestive and metabolic processes, including
Genolution Pharmaceuticals, Inc. Life Science and Molecular Diagnostic Products
 Genolution Pharmaceuticals, Inc. Revolution through genes, And Solution through genes. Life Science and Molecular Diagnostic Products www.genolution1.com TEL; 02-3010-8670, 8672 Geno-Serum Hepatitis B
Genolution Pharmaceuticals, Inc. Revolution through genes, And Solution through genes. Life Science and Molecular Diagnostic Products www.genolution1.com TEL; 02-3010-8670, 8672 Geno-Serum Hepatitis B
How To Get Rid Of Small Dna Fragments
 AxyPrep TM Mag FragmentSelect-I Protocol (Fragment Size Selection for Illumina Genome Analyzer and Life Technologies SoLiD) Introduction The AxyPrep Mag FragmentSelect-I purification kit utilizes a unique
AxyPrep TM Mag FragmentSelect-I Protocol (Fragment Size Selection for Illumina Genome Analyzer and Life Technologies SoLiD) Introduction The AxyPrep Mag FragmentSelect-I purification kit utilizes a unique
ZR-96 DNA Sequencing Clean-up Kit Catalog Nos. D4052 & D4053
 INSTRUCTION MANUAL ZR-96 DNA Sequencing Clean-up Kit Catalog Nos. D4052 & D4053 Highlights Simple 10 Minute Bind, Wash, Elute Procedure Flexible 15-20 µl Elution Volumes Allow for Direct Loading of Samples
INSTRUCTION MANUAL ZR-96 DNA Sequencing Clean-up Kit Catalog Nos. D4052 & D4053 Highlights Simple 10 Minute Bind, Wash, Elute Procedure Flexible 15-20 µl Elution Volumes Allow for Direct Loading of Samples
AxyPrep TM Mag PCR Clean-up Protocol
 AxyPrep TM Mag PCR Clean-up Protocol Intro The AxyPrep Mag PCR Clean-up kit utilizes a unique paramagnetic bead technology for rapid, high-throughput purification of PCR amplicons. Using this kit, PCR
AxyPrep TM Mag PCR Clean-up Protocol Intro The AxyPrep Mag PCR Clean-up kit utilizes a unique paramagnetic bead technology for rapid, high-throughput purification of PCR amplicons. Using this kit, PCR
RT31-020 20 rxns. RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl
 Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
Effect of Flaxseed Inclusion on Ruminal Fermentation, Digestion and Microbial Protein Synthesis in Growing and Finishing Diets for Beef Cattle
 Effect of Flaxseed Inclusion on Ruminal Fermentation, Digestion and Microbial Protein Synthesis in Growing and Finishing Diets for Beef Cattle T.C. Gilbery, G.P. Lardy, D.S. Hagberg and M.L. Bauer NDSU
Effect of Flaxseed Inclusion on Ruminal Fermentation, Digestion and Microbial Protein Synthesis in Growing and Finishing Diets for Beef Cattle T.C. Gilbery, G.P. Lardy, D.S. Hagberg and M.L. Bauer NDSU
DNA Core Facility: DNA Sequencing Guide
 DNA Core Facility: DNA Sequencing Guide University of Missouri-Columbia 216 Life Sciences Center Columbia, MO 65211 http://biotech.missouri.edu/dnacore/ Table of Contents 1. Evaluating Sequencing Data..
DNA Core Facility: DNA Sequencing Guide University of Missouri-Columbia 216 Life Sciences Center Columbia, MO 65211 http://biotech.missouri.edu/dnacore/ Table of Contents 1. Evaluating Sequencing Data..
Chromatin Immunoprecipitation
 Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Tribuna Académica. Overview of Metagenomics for Marine Biodiversity Research 1. Barton E. Slatko* Metagenomics defined
 Tribuna Académica 117 Overview of Metagenomics for Marine Biodiversity Research 1 Barton E. Slatko* We are in the midst of the fastest growing revolution in molecular biology, perhaps in all of life science,
Tribuna Académica 117 Overview of Metagenomics for Marine Biodiversity Research 1 Barton E. Slatko* We are in the midst of the fastest growing revolution in molecular biology, perhaps in all of life science,
Terra PCR Direct Polymerase Mix User Manual
 Clontech Laboratories, Inc. Terra PCR Direct Polymerase Mix User Manual Cat. Nos. 639269, 639270, 639271 PT5126-1 (031416) Clontech Laboratories, Inc. A Takara Bio Company 1290 Terra Bella Avenue, Mountain
Clontech Laboratories, Inc. Terra PCR Direct Polymerase Mix User Manual Cat. Nos. 639269, 639270, 639271 PT5126-1 (031416) Clontech Laboratories, Inc. A Takara Bio Company 1290 Terra Bella Avenue, Mountain
