X-tra! X-tra! News from the Mouse X Chromosome
|
|
|
- Alberta Ford
- 8 years ago
- Views:
Transcription
1 Developmental Biology 298 (2006) Review X-tra! X-tra! News from the Mouse X Chromosome Joanne L. Thorvaldsen, Raluca I. Verona, Marisa S. Bartolomei Department of Cell and Developmental Biology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, USA Received for publication 10 April 2006; revised 5 July 2006; accepted 11 July 2006 Available online 15 July 2006 Abstract X chromosome inactivation (XCI) is the phenomenon through which one of the two X chromosomes in female mammals is silenced to achieve dosage compensation with males. XCI is a highly complex, tightly controlled and developmentally regulated process. The mouse undergoes two forms of XCI: imprinted, which occurs in all cells of the preimplantation embryo and in the extraembryonic lineage, and random, which occurs in somatic cells after implantation. This review presents results and hypotheses that have recently been proposed concerning important aspects of both imprinted and random XCI in mice. We focus on how imprinted XCI occurs during preimplantation development, including a brief discussion of the debate as to when silencing initiates. We also discuss regulation of random XCI, focusing on the requirement for Tsix antisense transcription through the Xist locus, on the regulation of Xist chromatin structure by Tsix and on the effect of Tsix regulatory elements on choice and counting. Finally, we review exciting new data revealing that X chromosomes co-localize during random XCI. To conclude, we highlight other aspects of X-linked gene regulation that make it a suitable model for epigenetics at work Elsevier Inc. All rights reserved. Keywords: X inactivation; Xist; Tsix; Imprinted; Random; Choice Introduction X chromosome inactivation (XCI) is the silencing mechanism used by eutherian mammals to equalize the expression of X- linked genes between males and females (Lyon, 1961). The process results in the silencing of a majority of genes on one of the two X chromosomes in females. To achieve transcriptional silencing of genes on one X chromosome, each cell of a female mammal must undergo a highly orchestrated set of events consisting of four stages: counting, choice, initiation and maintenance. First a cell must count the number of X chromosomes it contains. If more than one X is counted, the cell must then choose to inactive a specific X to ensure that only one remains active. The cell then initiates and propagates chromosome-wide silencing of the chosen X, and finally maintains this inactive state throughout subsequent cell divisions (Avner and Heard, 2001; Boumil and Lee, 2001). The study of XCI in mice has been critical for understanding how this process unravels during the development of the organism and has revealed important details about its mechanism. Corresponding author. Fax: address: bartolom@mail.med.upenn.edu (M.S. Bartolomei). The mouse undergoes two forms of XCI; random, which occurs in somatic cells, and imprinted, which occurs in the extraembryonic lineage (trophectoderm and primitive endoderm). In addition, recent work has shown that all cells of the preimplantation embryo exhibit the imprinted form of XCI, with the inner cell mass (ICM) of the late blastocyst reactivating the inactive X (Chaumeil et al., 2004; Huynh and Lee, 2003; Mak et al., 2004). The random form of X inactivation subsequently initiates in the epiblast and is thought to be complete by dpc (Gardner and Lyon, 1971; Hoppe and Whitten, 1972; Nesbitt, 1971; Rastan, 1982; Rastan et al., 1980; Takagi, 1974; Takagi et al., 1982; Tam et al., 1994; Tan et al., 1993). Both imprinted and random XCI require a specific region of the X chromosome, designated the X inactivation center (Xic). The Xic harbors distinct genetic elements that function in the silencing pathway, including the X inactive-specific transcript (Xist) and Tsix genes and the genetically defined X-controlling element (Xce). In particular, the Xist gene plays a vital role in inactivation (Borsani et al., 1991; Brown et al., 1991). Xist is expressed from the inactive X (Xi) and the RNA gradually spreads to coat the entire chromosome. Subsequent to this step, the Xi acquires distinctive chromatin features that are believed to be important for transcriptional silencing /$ - see front matter 2006 Elsevier Inc. All rights reserved. doi: /j.ydbio
2 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) Here we review the mechanism of XCI in the mouse by examining how imprinted XCI occurs during preimplantation development and how random XCI is initiated following implantation (Fig. 1). We also briefly recap other local and chromosome-wide aspects of X-linked gene regulation. Additional reviews are recommended for a discussion of X chromosome regulation in other mammals (Chow et al., 2005; Grutzner and Graves, 2004). X inactivation during preimplantation development During preimplantation development, cells of female embryos undergo imprinted XCI, in which the paternal X (Xp) chromosome is always inactivated. This silencing is dependent upon the Xist non-coding RNA and the subsequent recruitment of an ordered set of chromatin modifications. When does imprinted XCI begin? Until recently, the prevailing view was that XCI initiates upon embryo implantation. Yet, recent studies demonstrated that imprinted XCI occurs much earlier in the preimplantation embryo. The question of when the Xp is silenced during preimplantation has become a controversial issue, with two competing models attempting to resolve the debate (Fig. 1). Although these hypotheses are based largely on similar experimental approaches, their conclusions are conflicting. The Pre-inactivation hypothesis proposes that the Xp is inherited from the father in an already silent state, whereas the de novo model advocates that silencing is not initiated until after fertilization. According to the Pre-inactivation hypothesis, the predisposition of the Xp to be inactivated during preimplantation originates from passage of this chromosome through the male germline (Huynh and Lee, 2001). In meiotic spermatocytes, the sex chromosomes form the XY body and are silenced through a process known as meiotic sex chromosome inactivation (MSCI) (Handel, 2004). This mechanism of silencing is not fully understood, but differs from XCI in that it is Xist-independent and is characterized by several distinct chromatin modifications including the histone variant H2AX (Fernandez-Capetillo et al., 2003; Turner et al., 2002). The Xp has been proposed to persist in a silent state through the end of spermatogenesis and enter the egg in a pre-inactivated state (Huynh and Lee, 2001, 2005). Huynh and Lee have employed an RNA fluorescence in situ hybridization (FISH) approach using a Cot-1 probe that detects nascent RNA, thus marking regions of active transcription. In support of the Pre-inactivation model, they reported that at the 2-cell stage one X chromosome does not stain with a FISH Cot-1 probe, suggesting that it lacks active transcription (Huynh and Lee, 2003). As MSCI and XCI differ in many respects, the pre- Fig. 1. Life cycle of X chromosome inactivation and reactivation in mice. The state of an X chromosome (i.e. whether X-linked genes are actively transcribed) is depicted throughout development. The asterisk in the 1 cell embryo shows that the inactivation is reflective of the silent state of the entire genome and is not X chromosome specific. In the 2-cell embryo the conflict in the literature as to whether or not the paternal X remains inactive or is reactivated is illustrated. As indicated by the curved arrows, escape from XCI could occur before random XCI (and possibly imprinted XCI) is established or be a consequence of the failure to maintain XCI post-implantation.
3 346 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) inactivated Xp would have to convert to Xist-dependent silencing and acquire the correct chromatin modifications following fertilization. The Pre-inactivation model is attractive in that it eliminates the need for multiple rounds of inactivation and reactivation in the germline and zygote (Huynh and Lee, 2001, 2005). Two recent studies have shown that some transcriptional repression of the X chromosome persists after completion of meiosis in the male germline (Namekawa et al., 2006; Turner et al., 2006). However, other data support the idea that silencing does not persist after meiosis. First, some germ cell specific genes are reactivated in postmeiotic spermatocytes (Wang et al., 2005). Second, the presence of specific euchromatic histone modifications as well as the association of phosphorylated RNA Polymerase II with the X chromosome during these stages suggest the X may have reverted to an active state (Khalil et al., 2004). An alternative, de novo model proposes that the Xp is active at fertilization and is silenced during subsequent stages of development. In support of this hypothesis, Okamoto et al. report that the Xp is active in 2-cell embryos, using RNA Polymerase II (Pol II) and Cot-1 staining as markers of transcriptional activity. In addition, assays using transcript-specific RNA FISH showed signals from both chromosomes, in accordance with the hypothesis that the paternal X is active at this stage (Okamoto et al., 2004). Furthermore, inactivation of autosomal Xist transgenes transmitted through the male can occur independently of MSCI, suggesting that silencing during male meiosis is not a prerequisite for XCI (Okamoto et al., 2005). However, as this latter study is based on transgenes, it remains to be determined whether this holds true for genes on the X chromosome. It is largely unclear if experimental conditions (such embryo fixation) or other factors account for the conflicting results on the transcriptional status of the Xp in the zygote. Perhaps the key to reconciling these two models lies in a more detailed examination of the transcriptional activity of genes along the X chromosome early after fertilization using transcript-specific FISH rather than the more global Cot-1 and Pol II probes. The paternal X becomes inactivated Xist expression initiates at the 2-cell stage, when the zygotic genome is activated. Xist is imprinted in the early embryo, with exclusive expression from the Xp. It remains unclear how Xist is initially silenced on the maternal X (Xm) chromosome and how its imprinting is controlled. However, at later stages of development, maternal Xist expression is negatively regulated by the overlapping gene, Tsix. Expression of the Tsix non-coding RNA is detected after the 8-cell stage and is oppositely imprinted to Xist, with exclusive expression from the maternal allele (Sado et al., 2001). Using FISH analysis, Xist RNA can be detected in 2-cell embryos as a small punctate signal. Starting at the 4-cell stage, Xist transcripts begin to accumulate on the Xp and appear as a larger RNA domain. The accumulation of Xist RNA is the initiating event in the silencing process. Subsequently, the Xist RNA domain spreads in cis, eventually coating the X chromosome. An ordered set of chromatin modifications follows Xist accumulation during pre-implantation, reflecting the progressive silencing of the X chromosome. The initial chromatin changes, which occur between the 8- and 32-cell stages, are defined by the loss of euchromatin marks: the X exhibits hypoacetylation of histone H3-Lysine 9 (H3K9) and hypomethylation of histone H3-Lysine 4 (Mak et al., 2004; Okamoto et al., 2004). Subsequently, enrichment of the EED/EZH2 Polycomb Group complex has been shown to mediate the early epigenetic mark of histone H3-Lysine 27 (H3K27) methylation on the Xi chromosome from the 16-cell stage to the early blastocyst. Finally, H3K9 methylation follows on the Xi (Heard, 2004; Mak et al., 2004; Okamoto et al., 2004). These epigenetic modifications are thought to act cooperatively on the Xi to mediate transcriptional silencing. However, although the kinetics of these chromatin changes are becoming well characterized, it remains to be determined which alterations function in establishing the inactivated state and which play a role in its maintenance. For example, chromatin changes that appear towards the end of preimplantation (after transcriptional silencing has occurred) are unlikely to function in establishment of silencing. Conversely, the H3K27 methylation epigenetic mark mediated by the Polycomb Group proteins is evident at an earlier stage, but it is still unclear whether it is required for the establishment of XCI. X-linked gene expression Analysis of the transcriptional status of X-linked genes in 8 16-cell embryos revealed a gradient of silencing along the chromosome, with an inverse correlation between the degree of silencing and distance from the Xic (Huynh and Lee, 2003). Thus, most genes within 10 cm of the Xic were predominantly silent on the Xp, whereas those further away were more likely to be expressed. These data suggest that silencing is progressive and reflects the degree of spreading of Xist RNA in these embryos. These results support a model in which silencing is first established at the Xic and then proceeds to spread bidirectionally toward the ends of the chromosomes. Reactivation of the X chromosome in the ICM In early blastocysts, all cells exhibit the hallmark signs of XCI on the Xp: a large domain of Xist RNA, indicating its coating of the chromosome, association of the Polycomb Group proteins, H3K27 methylation and transcriptional silencing (Mak et al., 2004; Okamoto et al., 2004; Plath et al., 2003). However, at this point, cells of the ICM undergo a reversal of XCI. This reversal is characterized by loss of the factors that associated with the X chromosome earlier during development. Thus, in the ICM of expanding or hatching blastocysts, Xist RNA is dispersed or absent, no EED/EZH2 domain is present, and some cells also lack H3K27 methylation, presumably as a result of dissociation of the EED/EZH2 complex from the chromosome. These results highlight once again that during preimplantation, X-inactivation is strictly Xist-dependent, such that when Xist is downregulated, the silencing modifications are also lost.
4 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) In contrast to the ICM, cells of the trophectoderm maintain their imprinted XCI and the Xp remains silent following implantation. Recently, transient association of EED with the X chromosome in the trophectoderm progenitor cells was proposed to play a role in the maintenance of transcriptional silencing during subsequent differentiation (Kalantry et al., 2006). Thus, differentiated eed / cells exhibit reactivation of the X chromosome (Wang et al., 2001). However, in undifferentiated eed / mutant trophectoderm stem (TS) cells, the Xp is not reactivated despite the absence of Xist RNA coating, H3K27 methylation, macroh2a and H4-1mK20 (Kalantry et al., 2006). These apparently conflicting results suggest that our knowledge of how X chromosome silencing occurs in the trophectoderm lineage is incomplete. Distinct sequence requirements for imprinted versus random XCI? Transgenic approaches have been used to study which DNA sequences are important for XCI. Xist/Tsix spanning transgenes present at more than one copy at autosomal loci can induce Xistdependent silencing in cis at ectopic loci when the cell undergoes random XCI; these transgenes range in size from 80 kb in line πjl1.4.1 (Lee et al., 1999b) to 460 kb in line L412 (Heard et al., 1999) (Fig. 2B). Recently, transgenic lines containing a single copy insertion of X sequences onto autosomes were examined for their ability to undergo Xist-induced silencing in cis during preimplantation, i.e. imprinted XCI (Okamoto et al., 2005). Specifically, in transgenic line 53BL (Fig. 2B), a 460 kb transgenic fragment containing Xist and Tsix conferred silencing in cis upon paternal transmission and exhibited the characteristic chromatin modifications normally seen on the Xi during this time. Thus, this transgene contains sufficient X chromosome sequence for Xist-induced silencing in cis and therefore provides a useful system to identify minimal sequences required for imprinted XCI. Conversely, transgenic line L412, which has 2 copies of the same transgene and was earlier shown to exhibit random XCI, did not confer silencing during preimplantation. While transgenic studies initially suggested that distinct elements may be required for imprinted versus random XCI, the answer may be slightly more complex since these studies may be confounded by position effects, transgene copy number, introduction of foreign sequence and integrity of transgenic DNA. Random XCI As previously introduced, random XCI is a multi-step process that initiates in the embryo-proper upon implantation and is considered to be complete in the 6.5 dpc postimplantation embryo (Rastan, 1982; Takagi et al., 1982). First, X chromosome counting and choice ensure that only one X remains active. In males, a defect in counting would result in the inactivation of the single X. In females a defect in choice is evident by loss of random XCI, while a defect in counting might result in the inactivation of both or none of the X chromosomes. Once choice is made, initiation is then evident by the downregulation of Tsix and upregulation of the Xist transcription on the future Xi (Lee et al., 1999a). In contrast to earlier reports, it is unlikely that this latter step involves the stabilization of Xist RNA (Panning and Jaenisch, 1998; Sheardown et al., 1997; Sun et al., 2006). The Xist RNA coats the Xi in cis followed by a similar sequence of chromatin modifications observed during preimplantation. In addition, DNA hypermethylation is a late step in this process. These epigenetic modifications eventually serve to maintain Xi repression in a Xist-independent manner, although Xist continues to be highly expressed and coat the Xi even after XCI is fully established. Since random XCI occurs upon differentiation of female embryonic stem (ES) cells, which are derived from the blastocyst ICM, ES cells have served as a system to elucidate molecular mechanisms of random XCI that are difficult to access in vivo. Other reviews nicely present the specific chromatin modifications leading to the establishment of random XCI in both ES cells and embryos (Heard, 2005; Plath et al., 2002). Here we focus on recent developments in deducing how Tsix regulates Xist during early events of random XCI, and then we discuss some exciting news on nuclear organization of the X chromosomes during this process, as well as the identification of elements essential for XCI. The complex dependence of Xist on Tsix Within the Xic chromosomal region, Xist and Tsix are essential to mediate the early steps in XCI. Xist deletions confirm that Xist is necessary for silencing of the X chromosome in cis (Penny et al., 1996; Marahrens et al., 1997). The choice of which X to inactivate, however, appears to involve a complex interplay between Xist and Tsix. While the analyses of Tsix (major promoter) deletion alleles Tsix ΔCpG (Fig. 2B) (Lee and Lu, 1999) and Tsix AA2Δ1.7 (Sado et al., 2001) show that Tsix is critical for repressing Xist on the active X (Xa), it is not known how Tsix exerts its effects. Here we highlight three developments in our understanding of how Tsix mediates the control of XCI: the requirement for Tsix antisense transcription through Xist, the regulation of Xist chromatin structure by Tsix and the effect of Tsix regulatory elements on choice and counting. To discriminate between a role for the Tsix RNA itself or antisense transcription through Xist, two groups independently generated Tsix mutant alleles that express a truncated Tsix RNA that terminates before it overlaps with Xist-coding sequences (Luikenhuis et al., 2001; Shibata and Lee, 2004). The resulting mutant alleles are always chosen as the Xi, implying a role for antisense transcription through Xist. Furthermore, knocking in the Tsix cdna in cis cannot complement this phenotype (Shibata and Lee, 2004), emphasizing that transcription through the overlapping region is required for the ability of Tsix to repress Xist (Shibata and Lee, 2003). While analyses of Tsix mutant alleles have demonstrated that Tsix is essential for regulating choice of XCI in females, a Tsix-independent mechanism appears to regulate counting of random XCI in males, since viable males can inherit a Tsix deletion on their single X chromosome (Lee and Lu, 1999; Sado et al., 2001; Ohhata et al., 2006). While the interplay between Tsix and Xist remains incompletely understood, recent studies suggest that Tsix regulates
5 348 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) Fig. 2. Initiation of random XCI. (A) Co-localization of X chromosomes occurs early in differentiating ES cells. The X chromosomes are shown in the nucleus of differentiating ES cells and terminally differentiated MEFs. The Xics are depicted as rectangles. Red and green rectangles indicate Xist and Tsix expression, respectively, and the white box represents the absence of Tsix expression from the established Xa in MEFs. The blue triangle denotes a hypothetical anchor point for the co-localization of the X chromosomes. (B) Deletion and transgenic ES cell lines suggest sequence requirements for Xic Xic pairing. Schematic of the Xist/Tsix locus is shown at the top, with arrows denoting the start and direction of transcription. Transcripts that initiate from the Xite region also coincide with minor Tsix transcripts (Ogawa and Lee, 2003; Sado et al., 2001). For summary of transcripts at this locus see Rougeulle and Avner (2004). Tsx, the closest protein-coding gene, is indicated by a red box. ES cell lines analyzed by Bacher et al. (2006) are in bold; the other lines were assayed by Xu et al. (2006). The asterisk designates lines from which mice have been generated. For deletion lines, cross-hatched boxes indicated deleted sequence with size of deletions denoted in parentheses next to the line name. For line c.16.1 the gray box indicates the 16 kb of reinserted sequence. For autosomal transgenic lines (names in parentheses) yellow boxes designate the region of Xist/Tsix sequence that was used to generate the transgene. Estimated transgene copy numbers are designated. Pairing between two Xs (in female lines) or an X and A (in transgenic lines) is indicated to the right. Delayed shows that pairing occurred later than normal during differentiation of ES cells while reduced indicates pairing occurred less often than expected. Whether or not random XCI occurred is noted for deletion lines (random XCI). Partial corresponds to skewing of XCI for the deletion allele. For male transgenic lines (XY+TgA) the ability of the transgene to induce Xist-dependent silencing was noted (cis-silencing). For female transgenic lines the ability of cell to undergo XCI was noted. Xist transcription, at least in part, through chromatin remodeling at the Xist locus. Sado et al. modified the Xist locus in the presence or absence of a Tsix deletion in cis such that a truncated nonfunctional Xist was under the influence of the endogenous Xist promoter and determined the consequence in E13.5 tissues and mouse embryonic fibroblasts (MEFs) (Sado et al., 2005).
6 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) Here, the nonfunctional Xist RNA was expressed in the absence of Tsix transcription in cis. In addition, the active chromatin modification H3-2meK4, was observed on the Xist promoter of the nonfunctional Xist allele in the absence but not in the presence of Tsix expression in cis, suggesting that Tsix functions to prevent the recruitment of activating marks. In another study, Navarro et al. profiled the chromatin at the Xist and Tsix loci in TS cells that undergo imprinted XCI, in MEFs where random XCI is well established and in undifferentiated ES cells, where random XCI has not yet occurred (Navarro et al., 2005). Active Xist loci were enriched for H3-2meK4. Comparison of wild-type and Tsix mutant ES cells indicated that recruitment of H3-2meK4 throughout the Xist transcription unit (but not the promoter) is dependent upon Tsix expression. Sun et al. have performed a similar analysis in undifferentiated and differentiated ES cells, supporting the idea that Tsix-dependent chromatin modifications are important for controlling random XCI (Sun et al., 2006). These studies suggest that Tsix is affecting Xist local chromatin structure during random and imprinted XCI. While Sado et al propose from their analyses of cells in which random XCI is complete that Tsix represses Xist transcription by recruiting repressive chromatin modifications at the Xist promoter on the Xa, other groups suggest that Tsix causes the Xist gene body to become euchromatic in ES cells, which have not undergone XCI (Navarro et al., 2005; Sun et al., 2006). This latter activity may reflect an additional role for Tsix in priming the Xist locus for activation or inactivation (Navarro et al., 2005). Deletion and transgenic analyses of Tsix regulatory elements indicate that they are key to determining both the future Xa and Xi and may control choice and counting during random XCI (Fig. 2B). Initially, a 65-kb deletion of sequence distal to Xist (X Δ65 kb, Fig. 2B), which removed all identified antisense transcripts to Xist as well as other genes, perturbed choice in XX female cells and counting in XY and XO cells (Clerc and Avner, 1998; Rougeulle and Avner, 2004). In contrast, genetic manipulations that perturb Tsix expression from its major promoter lead to nonrandom XCI but counting in XY cells appeared unaffected (Lee, 2000; Lee and Lu, 1999; Luikenhuis et al., 2001; Morey et al., 2001; Sado et al., 2001). Subsequently, the X-intergenic transcription element, Xite, was characterized upstream of the major Tsix promoter and was shown to be the region from which multiple transcripts were initiating (Ogawa and Lee, 2003) [partly coincident with minor Tsix promoters (Sado et al., 2001)]. Deletion of the Xite region in ES cells (Xite ΔL, Fig. 2B) skewed random XCI, leading to the idea that Xite is affecting choice and may be the elusive Xce. While XX cells heterozygous for deletions of sequence 3 of Xist have identified sequences that regulate XCI choice, homozygous deletion of the Tsix major promoter in ES cells and mice (Tsix ΔCpG, Fig. 2B) led to failure of random XCI, indicating that the deleted sequence is also required for counting in females. The observed failure in XCI was termed chaotic choice since three possible outcomes were observed: two Xa's, two Xi's or, the most rare event, one Xi and one Xa. This indicates that the Tsix deleted sequence is also important for counting in female ES cells as well as choosing both the future Xi and Xa (Lee, 2002, 2005). In addition, when XX ES cells with high copy number transgenes of the Tsix major promoter (p3.7) or the Xite region (pxite), were differentiated, the cells exhibited a defect in growth and there was no evidence of XCI, suggesting that an excess of these elements was inhibiting counting and choice (Fig. 2B) (Lee, 2005). While this outcome could be due to the transgenic sequences sequestering transacting factors essential for Tsix expression away from the endogenous X chromosome, leading to its inactivation, Lee proposes that random XCI requires two factors, a blocking factor for the future Xa and a competence factor to enable inactivation of the future Xi (Lee and Lu, 1999). This model contrasts with an earlier proposal that the Xa requires a blocking factor to protect it from being inactivated and the Xi arises by default (Rastan, 1983). Before departing from a discussion of Tsix mediated control of XCI, it should be noted that while many of the experiments cited here rely on the assumption that the ES cell system recapitulates random X inactivation in the mouse embryo, there may be some potential problems with this assumption. For example, a recent study reported that XX female ES cells were globally hypomethylated relative to XY and XO ES cells (Zvetkova et al., 2005). The authors propose that the hypomethylated state is a disadvantage to XX ES cells and therefore the reason why these cells tend to be unstable. Consequently, it is critical to determine whether the ICM of female blastocysts is also hypomethylated. Although female ES cells represent a powerful tool to study random XCI, these results are a reminder that, when possible, findings should be verified in the embryo. Initiating random XCI Whereas sequences conferring choice and initiation of XCI are being delimited, less is known about the mechanism through which these elements may function. Recent evidence suggests that these sequences may be important to bring the X chromosomes together. The Xics of two X chromosomes were shown to transiently co-localize within interphase nuclei during the early stages of ES cell differentiation (Fig. 2) and this transient pairing appears to precede the upregulation of Xist expression from one of the X chromosomes and the subsequent accumulation of characteristic repressive chromatin modifications (Bacher et al., 2006; Xu et al., 2006). [For reviews see (Carrel, 2006; Morey and Bickmore, 2006).] Thus, X chromosome pairing appears to be important for choice and initiation of XCI. Interestingly, Bacher et al. also demonstrate that the Xs colocalize at the nuclear periphery, suggesting that pairing may also be due to localization within a common nuclear subcompartment. Deletion of 65 kb sequence 3 to Xist that is required for counting abolished X chromosome pairing (X Δ65 kb, Fig. 2B). However, reintroducing sequence 16 kb of spanning the Tsix regulatory elements into the 65 kb deletion restores colocalization but does not restore random XCI (X Δ65 kb + 16 kb, Fig. 2B), indicating that X chromosome pairing is necessary but not sufficient for choice/counting (Clerc and Avner, 1998; Morey et al., 2001). The deletion and transgenic analyses in
7 350 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) both studies, as summarized in Fig. 2B, suggest that elements regulating choice are required for X X pairing, and, when present at autosomal loci, can interfere with this process (Bacher et al., 2006; Xu et al., 2006). Additional proof that this process has a role in counting will come from X colocalization tests in cells with more than two X chromosomes or with increased autosomal ploidy (e.g. >2N) (Morey and Bickmore, 2006). Finally, the co-localization of Xs reported in these studies is consistent with the proposal that choice/ counting/initiation involves transvection of X chromosomes (Marahrens, 1999). At this time, transvection (defined as the pairing of homologous chromosomes) has been only shown to regulate transcription in Drosophila (Duncan, 2002). In addition, observations of Bacher et al. may also be consistent with an early hypothesis that XCI involves co-localization of Xs to the nuclear periphery (Comings, 1968). When is choice of the future Xa and Xi determined? Contrary to the established view, Williams and Wu have hypothesized that XCI does not even involve choice between two X chromosomes (Williams and Wu, 2004) and suggest that the future Xa and Xi are determined prior to developmental events that trigger the initiation of XCI (such as differentiation of ES cells). In general, they propose that only one of the two X chromosomes is susceptible to epigenetic modification designating the future Xa or Xi, while the other X is destined for the opposite outcome, apparently by default. While allele-specific chromatin and DNA modification are apparent after XCI initiates, they have not been reported in undifferentiated ES cells. However, using a DNA FISH procedure that preserves nuclear architecture in ES cells Panning and colleagues observe differences in the two X chromosomes prior to XCI (Mlynarczyk-Evans et al., 2006), thus supporting the hypothesis by Williams and Wu. These data suggest that the X chromosomes in ES cells have two distinct states, one more accessible to the X chromosome DNA probes than the other, perhaps correlating with yet-to-be defined epigenetic differences between the future Xa or Xi. Defining elements required for random XCI It is a challenge to define the minimal cis sequences and trans elements required for the initial XCI events of choice and counting. With regards to choice, Cattanach and colleagues characterized a putative X-linked element in the mouse, designated the Xce. Different Xce alleles skew X chromosome inactivation, resulting in a nonrandom X-linked phenotype (Cattanach and Isaacson, 1965, 1967). Though Xce has been mapped to the distal region of the mouse Xic (Chadwick et al., 2006; Simmler et al., 1993), it has eluded molecular identification and its mode of action remains only phenotypically defined. It is not known how the distinction of Xce alleles occurs in an Xce heterozygous animal, although characteristic epigenetic modifications and chromatin structure may play a role (Avner et al., 1998; Chao et al., 2002; Percec and Bartolomei, 2002; Prissette et al., 2001; Simmler et al., 1993). Notably, the Xce effect appears to be limited to tissues of the embryo proper, as the paternally imprinted inactivation characteristic of mouse extraembryonic tissues is not perturbed in Xce heterozygous females (Rastan, 1983). Lee and colleagues proposed that the regulatory element defined as Xite functions as the Xce (Ogawa and Lee, 2003). Ultimately, genetically engineered swapping of candidate Xce elements will define the Xce. Two approaches have been used to identify trans factors involved in random XCI. First, an ENU mutagenesis screen has identified multiple autosomal mutant loci that perturbed the Xce effect (Percec et al., 2002, 2003) and may harbor candidates for the blocking factor, a hypothetical complex proposed to function on the Xa (Lyon, 1971; Rastan, 1983). Fine mapping of the autosomal loci are required to elucidate whether or not these regions harbor elements that affect Xce, although preliminary data have indicated that the effect is more complex and not defined by a single locus [personal observation, (Chadwick and Willard, 2005)]. Second, a recent SAGE screen identified multiple genes that are differentially expressed in males and female 6.5 dpc embryos and, consequently, may include candidate genes for regulating XCI (Bourdet et al., 2006). For now, the minimal cis and trans elements required for XCI remain elusive, especially since no single copy Xist/Tsix containing transgene can recapitulate random XCI. Other processes of X-linked gene regulation X chromosome reactivation in PGCs and cloned embryos Once random XCI is established in somatic cells, it is maintained in subsequent cell divisions. At a specific time in developing primordial germ cells (PGCs) both X chromosomes are active once again. This could occur either if PGCs develop from a population of cells in which X reactivation occurs, or if they arise by expansion of a stem cell population in which XCI has never occurred. Studies favor the former hypothesis and indicate that the reactivation of the Xi occurs through the erasure of the marks that distinguish the Xi and Xa (McLaren, 2003). A similar process occurs at autosomal imprinting loci at this time, where the somatic imprinted marks are also being erased (Hajkova et al., 2002). Not much is known about the mechanism of X chromosome reactivation in PGCs. For example, is this process dependent upon X-linked genes? Is it independent of the Xic? To some extent, X chromosome reactivation in the PGCs is mimicked when somatic nuclei are transferred into oocytes during mouse cloning procedures (Bao et al., 2005; Eggan et al., 2000; Nolen et al., 2005). Marks associated with the Xi are erased, although incompletely, and XCI becomes randomized again in cloned embryos. Undoubtedly, this will provide a useful system to begin understanding the mechanism through which the X chromosome is reactivated. Exceptions in X-linked gene regulation We end this review with three additional anomalies of X- linked gene regulation for which the mechanisms remain to be resolved. First, a few genes escape XCI in mice, as opposed to
8 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) Table 1 Questions addressing inactivation and reactivation of the X chromosome Question 1 When is imprinted XCI established? 2 What are the cis elements required for imprinted XCI? 3 When is Tsix first required for imprinted XCI? 4 How does Tsix affect Xist chromatin structure during imprinted and random XCI? 5 Are Tsix and Xite regulatory sequences exclusively responsible for choice (and counting) in random XCI? 6 How is the Xce affecting choice in skewed/nonrandom XCI? 7 What are the trans factors involved in choice during random XCI? 8 What are the cis elements required for X chromosome pairing during early steps of random X inactivation? 9 Is X chromosome pairing during early steps of random XCI occurring at a fixed location in the nuclear periphery? 10 Is the hypomethylated state reported in female ES cells also present in the ICM of female blastocysts? 11 Are the steps of random XCI observed in differentiating ES cells recapitulating the steps of random XCI that occur in the embryo? 12 How do X chromosomal imprinted genes escape random XCI? (Is escape a consequence of the inability to maintain random XCI?) 13 Do genes that escape random XCI also escape imprinted XCI? 14 How are X-linked imprinted genes regulated in the midst of imprinted and random XCI? the 15 20% that escape XCI in humans (Carrel and Willard, 2005; Disteche et al., 2002). While escape has been proposed to be dependent upon the vertebrate insulator factor CTCF controlling boundary elements present between escapees and non-escapees (Filippova et al., 2005), many questions remain. For example, it is still unclear whether the genes that escape XCI do so from the very beginning of the X inactivation process or if they are initially inactivated and then become reactivated (Fig. 1). The latter mechanism was reported for mouse escapee Smcx (Lingenfelter et al., 1998). Furthermore, it also remains to be determined if genes can escape imprinted XCI. Secondly, in addition to the genes that escape XCI, there are new reports of imprinted X-linked genes, adding even greater complexity to X- linked gene expression (Davies et al., 2005; Kobayashi et al., 2006; Raefski and O'Neill, 2005). The neuronal specific expression of imprinted genes from the Xm may serve as a mouse model for uncovering complexities of Turner Syndrome. The Rhox5/Pem gene was recently reported to be imprinted, with expression from the Xp during implantation and from the Xm during postimplantation. As hinted by Eakin and Hadjantonakis, paternal-specific expression detected in preimplantation embryo may be a consequence of failure to completely inactivate the distally located X-linked Rhox5 that is expressed in sperm (Eakin and Hadjantonakis, 2006). Finally, X-linked gene expression levels on Xa are reported to be elevated relative to autosomal gene expression levels, such that expression levels from a single X corresponds to expression levels from two autosomes. Thus, not only is the Xi unique in its silencing, but the Xa is also unique in its elevated gene expression levels (Nguyen and Disteche, 2006). Elucidating the mechanisms behind these X-linked anomalies will further our understanding of the requirements for establishing and maintaining random XCI. Conclusion We have presented a brief overview of the complex stages of X chromosome inactivation and reactivation, focusing on recent exciting studies that will encourage investigators to reevaluate and broaden approaches to study these dynamic processes. We conclude in Table 1 with questions that undoubtedly will be tackled with the many pioneering and creative approaches that have and will continue to unravel the complexities of X chromosome regulation throughout development. Acknowledgments We apologize to those whose work we could not mention due to space constraints. This work was supported by the National Institutes of Health (GM74768). We thank Drs. Barbara Panning and Edith Heard for sharing results prior to publication. We thank Drs. Nora Engel and Jesse Mager for helpful comments. References Avner, P., Heard, E., X-chromosome inactivation: counting, choice and initiation. Nat. Rev. 2, Avner, P., Prissette, M., Arnaud, D., Courtier, B., Cecchi, C., Heard, E., Molecular correlates of the murine Xce locus. Genet. Res. 72, Bacher, C.P., Guggiari, M., Brors, B., Augui, S., Clerc, P., Avner, P., Eils, R., Heard, E., Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8, Bao, S., Miyoshi, N., Okamoto, I., Jenuwein, T., Heard, E., Azim Surani, M., Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep. 6, Borsani, G., Tonlorenzi, R., Simmler, M.C., Dandalo, L., Arnaud, D., Capra, V., Grompe, M., Pizzati, A., Muzay, D., Lawrence, C., Willard, H.F., Avner, P., Ballabio, A., Characterisation of a murine gene expressed from the inactive X chromosome. Nature 351, Boumil, R.M., Lee, J.T., Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 10, Bourdet, A., Ciaudo, C., Zakin, L., Elalouf, J.M., Rusniol, C., Weissenbach, J., Avner, P., A SAGE approach to identifying novel trans-acting factors involved in the X inactivation process. Cytogenet, Genome Res. 113, Brown, C.J., Ballabio, A., Rupert, J.L., Lafreniere, R.G., Grompe, M., Tonlorenzi, R., Willard, H.F., A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, Carrel, L., Molecular biology. X -rated chromosomal rendezvous. Science 311, Carrel, L., Willard, H.F., X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, Cattanach, B.M., Isaacson, J.H., Genetic control over the inactivation of autosomal genes attached to the X-chromosome. Z. Vererbungsl. 96, Cattanach, B.M., Isaacson, J.H., Controlling elements in the mouse X chromosome. Genetics 57, Chadwick, L.H., Willard, H.F., Genetic and parent-of-origin influences on X chromosome choice in Xce heterozygous mice. Mamm. Genome 16, Chadwick, L.H., Pertz, L., Broman, K.W., Bartolomei, M.S., Willard, H.F., Genetic control of X chromosome inactivation in mice: definition of the Xce candidate interval. Genetics (Electronic publication ahead of print). Chao, W., Huynh, K.D., Spencer, R.J., Davidow, L.S., Lee, J.T., CTCF, a
9 352 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) candidate trans-acting factor for X-inactivation choice. Science 295, Chaumeil, J., Okamoto, I., Heard, E., X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 376, Chow, J.C., Yen, Z., Ziesche, S.M., Brown, C.J., Silencing of the mammalian X chromosome. Annu. Rev. Genomics Hum. Genet. 6, Clerc, P., Avner, P., Role of the region 3 to Xist exon 6 in the counting process of X-chromosome inactivationtsix. Nat. Genet. 19, Comings, D.E., The rationale for an ordered arrangement of chromatin in the interphase nucleus. Am. J. Hum. Genet. 20, Davies, W., Isles, A., Smith, R., Karunadasa, D., Burrmann, D., Humby, T., Ojarikre, O., Biggin, C., Skuse, D., Burgoyne, P., Wilkinson, L., Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat. Genet. 37, Disteche, C.M., Filippova, G.N., Tsuchiya, K.D., Escape from X inactivation. Cytogenet. Genome Res. 99, Duncan, I.W., Transvection effects in Drosophila. Annu. Rev. Genet. 36, Eakin, G.S., Hadjantonakis, A.K., Sex-specific gene expression in preimplantation mouse embryos. Genome Biol. 7, Eggan, K., Akutsu, H., Hochedlinger, K., Rideout 3rd, W., Yanagimachi, R., Jaenisch, R., X-Chromosome inactivation in cloned mouse embryos. Science 290, Fernandez-Capetillo, O., Mahadevaiah, S.K., Celeste, A., Romanienko, P.J., Camerini-Otero, R.D., Bonner, W.M., Manova, K., Burgoyne, P., Nussenzweig, A., H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4, Filippova, G.N., Cheng, M.K., Moore, J.M., Truong, J.P., Hu, Y.J., Nguyen, D.K., Tsuchiya, K.D., Disteche, C.M., Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell 8, Gardner, R.L., Lyon, M.F., X chromosome inactivation studied by injection of a single cell into the mouse blastocyst. Nature 231, Grutzner, F., Graves, J.A., A platypus' eye view of the mammalian genome. Curr. Opin. Genet. Dev. 14, Hajkova, P., Erhardt, S., Lane, N., Haff, T., El-Maarri, O., Reik, W., Walter, J., Surani, A.H., Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, Handel, M.A., The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 296, Heard, E., Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16, Heard, E., Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 15, Heard, E., Mongelard, F., Arnaud, D., Avner, P., Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol. Cell. Biol. 19, Hoppe, P.C., Whitten, W.K., Does X chromosome inactivation occur during mitosis of first cleavage? Nature 239, 520. Huynh, K.D., Lee, J.T., Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr. Opin. Cell Biol. 13, Huynh, K.D., Lee, J.T., Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426, Huynh, K.D., Lee, J.T., X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat. Rev., Genet. 6, Kalantry, S., Mills, K.C., Yee, D., Otte, A.P., Panning, B., Magnuson, T., The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat. Cell Biol. 8, Khalil, A.M., Boyar, F.Z., Driscoll, D.J., Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc. Natl. Acad. Sci. 101, Kobayashi, S., Isotani, A., Mise, N., Yamamoto, M., Fujihara, Y., Kaseda, K., Nakanishi, T., Ikawa, M., Hamada, H., Abe, K., Okabe, M., Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr. Biol. 16, Lee, J.T., Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103, Lee, J.T., Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat. Genet. 32, Lee, J.T., Regulation of X-chromosome counting by Tsix and Xite sequences. Science 309, Lee, J.T., Lu, N., Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99, Lee, J.T., Davidow, L.S., Warshawsky, D., 1999a. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21, Lee, J.T., Lu, N., Han, Y., 1999b. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc. Natl. Acad. Sci. 96, Lingenfelter, P.A., Adler, D.A., Posllinski, D., Thomas, S., Elliott, R.W., Chapman, V.M., Disteche, C.M., Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 18, Luikenhuis, S., Wutz, A., Jaenisch, R., Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 21, Lyon, M.F., Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, Lyon, M.F., Possible mechanisms of X chromosome inactivation. Nat., New Biol. 232, Mak, W., Nesterova, T.B., de Napoles, M., Appanah, R., Yamanaka, S., Otte, A.P., Brockdorff, N., Reactivation of the paternal X chromosome in early mouse embryos. Science 303, Marahrens, Y., X-inactivation by chromosomal pairing events. Genes Dev. 13, Marahrens, Y., Panning, B., Dausman, J., Strauss, W., Jaenisch, R., Xistdeficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, McLaren, A., Primordial germ cells in the mouse. Dev. Biol. 262, Mlynarczyk-Evans, S., Royce-Tolland, M.E., Alexander, M.K., Andersen, A.A., Kalantry, S., Gribnau, J., Panning, B., X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 4, e159. Morey, C., Bickmore, W., Sealed with a X. Nat. Cell Biol. 8, Morey, C., Arnaud, D., Avner, P., Clerc, P., Tsix-mediated repression of Xist accumulation is not sufficient for normal random X inactivation. Hum. Mol. Genet. 10, Namekawa, S.H., Park, P.J., Zhang, L., Shima, E.J., McCarrey, J.R., Griswold, M.D., Lee, J.T., Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16, Navarro, P., Pichard, S., Ciaudo, C., Avner, P., Rougeulle, C., Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 19, Nesbitt, M.N., X chromosome inactivation mosaicism in the mouse. Dev. Biol. 26, Nguyen, D.K., Disteche, C.M., Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38, Nolen, L.D., Gao, S., Han, Z., Mann, M.R., Gie Chung, Y., Otte, A.P., Bartolomei, M.S., Latham, K.E., X chromosome reactivation and regulation in cloned embryos. Dev. Biol. 279, Ogawa, Y., Lee, J.T., Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell 11, Ohhata, T., Hoki, Y., Sasaki, H., Sado, T., Tsix-deficient X chromosome does not undergo inactivation in the embryonic lineage in males: implications for Tsix-independent silencing of Xist. Cytogenet. Genome Res. 113, Okamoto, I., Otte, A.P., Allis, C.D., Reinberg, D., Heard, E., Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, Okamoto, I., Arnaud, D., Le Baccon, P., Otte, A.P., Disteche, C.M., Avner, P., Heard, E., Evidence for de novo imprinted X-chromosome
10 J.L. Thorvaldsen et al. / Developmental Biology 298 (2006) inactivation independent of meiotic inactivation in mice. Nature 438, Panning, B., Jaenisch, R., RNA and the epigenetic regulation of X chromosome inactivation. Cell 93, Penny, G.D., Kay, G.F., Sheardown, S.A., Rastan, S., Brockdorff, N., Requirement for Xist in X chromosome inactivation. Nature 379, Percec, I., Bartolomei, M.S., Do X chromosomes set boundaries? Science 295, Percec, I., Plenge, R.M., Nadeau, J.H., Bartolomei, M.S., Willard, H.F., Autosomal dominant mutations affecting X inactivation choice in the mouse. Science 296, Percec, I., Thorvaldsen, J.L., Plenge, R.M., Krapp, C.J., Nadeau, J.H., Willard, H.F., Bartolomei, M.S., An N-ethyl-N-nitrosourea mutagenesis screen for epigenetic mutations in the mouse. Genetics 164, Plath, K., Mlynarczyk-Evans, S., Nusinow, D.A., Panning, B., Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 36, Plath, K., Fang, J., Mlynarczyk-Evans, S.K., Cao, R., Worringer, K.A., Wang, H., de la Cruz, C.C., Otte, A.P., Panning, B., Zhang, Y., Role of histone H3 lysine 27 methylation in X inactivation. Science 300, Prissette, M., El-Maarri, O., Arnaud, D., Walter, J., Avner, P., Methylation profiles of DXPas34 during the onset of X-inactivation. Hum. Mol. Genet. 10, Raefski, A.S., O'Neill, M.J., Identification of a cluster of X-linked imprinted genes in mice. Nat. Genet. 37, Rastan, S., Timing of X-chromosome inactivation in postimplantation mouse embryos. J. Embryol. Exp. Morphol. 71, Rastan, S., Non-random X-chromosome inactivation in mouse X-autosome translocation embryos-location of the inactivation centre. J. Embryol. Exp. Morphol. 78, Rastan, S., Kaufman, M.H., Handyside, A.H., Lyon, M.F., X-chromosome inactivation in extra-embryonic membranes of diploid parthenogenetic mouse embryos demonstrated by differential staining. Nature 228, Rougeulle, C., Avner, P., The role of antisense transcription in the regulation of X-inactivation. Curr. Top. Dev. Biol. 63, Sado, T., Wang, Z., Sasaki, H., Li, E., Regulation of imprinted X- chromosome inactivation in mice by Tsix. Development 128, Sado, T., Hoki, Y., Sasaki, H., Tsix silences Xist through modification of chromatin structure. Dev. Cell 9, Sheardown, S.A., Duthie, S.M., Johnston, C.M., Newall, A.E.T., Formstone, E.J., Arkell, R.M., Nesterova, T.B., Alghisi, G.-C., Rastan, S., Brockdorff, N., Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91, Shibata, S., Lee, J.T., Characterization and quantitation of differential Tsix transcripts: implications for Tsix function. Hum. Mol. Genet. 12, Shibata, S., Lee, J.T., Tsix transcription-versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr. Biol. 14, Simmler, M.C., Cattanach, B.M., Rasberry, C., Rougeulle, C., Avner, P., Mapping the murine Xce locus with (CA)n repeats. Mamm. Genome 4, Sun, B.K., Deaton, A.M., Lee, J.T., A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21, Takagi, N., Differentiation of X chromosomes in early female mouse embryos. Exp. Cell Res. 86, Takagi, N., Sugawara, O., Sasaki, M., Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma 85, Tam, P.P.L., Williams, E.A., Tan, S.-S., Expression of an X-linked HMG-lacZ transgene in mouse embryos: implication of chromosomal imprinting and lineage-specific X-chromosome activity. Dev. Genet. 15, Tan, S.-S., Williams, E.A., Tam, P.P.L., X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat. Genet. 3, Turner, J.M., Mahadevaiah, S.K., Elliott, D.J., Garchon, H.J., Pehrson, J.R., Jaenisch, R., Burgoyne, P.S., Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J. Cell Sci. 115, Turner, J.M., Mahadevaiah, S.K., Ellis, P.J., Mitchell, M.J., Burgoyne, P.S., Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev. Cell 10, Wang, J., Mager, J., Chen, Y., Schneider, E., Cross, J.C., Nagy, A., Magnuson, T., Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28, Wang, P.J., Page, D.C., McCarrey, J.R., Differential expression of sexlinked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 14, Williams, B.R., Wu, C.T., Does random X-inactivation in mammals reflect a random choice between two X chromosomes? Genetics 167, Xu, N., Tsai, C.L., Lee, J.T., Transient homologous chromosome pairing marks the onset of X inactivation. Science 311, Zvetkova, I., Apedaile, A., Ramsahoye, B., Mermoud, J.E., Crompton, L.A., John, R., Feil, R., Brockdorff, N., Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 37,
X-Chromosome Inactivation
 Stanley M Gartler, University of Washington, Seattle, Washington, USA Michael A Goldman, San Francisco State University, San Francisco, California, USA In female mammals, one of the X-chromosomes is transcriptionally
Stanley M Gartler, University of Washington, Seattle, Washington, USA Michael A Goldman, San Francisco State University, San Francisco, California, USA In female mammals, one of the X-chromosomes is transcriptionally
Von Mäusen und Menschen E - 1
 Von Mäusen und Menschen E - 1 Mus musculus: Genetic Portrait of the House Mouse E - 3 Outline Mouse genome Mouse life cycle Transgenic protocols Addition of genes by nuclear injection Removal of genes
Von Mäusen und Menschen E - 1 Mus musculus: Genetic Portrait of the House Mouse E - 3 Outline Mouse genome Mouse life cycle Transgenic protocols Addition of genes by nuclear injection Removal of genes
Human Cloning The Science and Ethics of Nuclear Transplantation
 Human Cloning The Science and Ethics of Transplantation Rudolf Jaenisch, M.D. In addition to the moral argument against the use of somatic-cell nuclear for the creation of a child ( reproductive cloning
Human Cloning The Science and Ethics of Transplantation Rudolf Jaenisch, M.D. In addition to the moral argument against the use of somatic-cell nuclear for the creation of a child ( reproductive cloning
17. A testcross A.is used to determine if an organism that is displaying a recessive trait is heterozygous or homozygous for that trait. B.
 ch04 Student: 1. Which of the following does not inactivate an X chromosome? A. Mammals B. Drosophila C. C. elegans D. Humans 2. Who originally identified a highly condensed structure in the interphase
ch04 Student: 1. Which of the following does not inactivate an X chromosome? A. Mammals B. Drosophila C. C. elegans D. Humans 2. Who originally identified a highly condensed structure in the interphase
The correct answer is c A. Answer a is incorrect. The white-eye gene must be recessive since heterozygous females have red eyes.
 1. Why is the white-eye phenotype always observed in males carrying the white-eye allele? a. Because the trait is dominant b. Because the trait is recessive c. Because the allele is located on the X chromosome
1. Why is the white-eye phenotype always observed in males carrying the white-eye allele? a. Because the trait is dominant b. Because the trait is recessive c. Because the allele is located on the X chromosome
4 SEX CHROMOSOMES AND SEX DETERMINATION
 4 SEX CHROMOSOMES AND SEX DETERMINATION 4.1 Sex chromosomes and Sex Determination Sex- chromosomes. If present, sex chromosomes may not have the same size, shape, or genetic potential. In humans, females
4 SEX CHROMOSOMES AND SEX DETERMINATION 4.1 Sex chromosomes and Sex Determination Sex- chromosomes. If present, sex chromosomes may not have the same size, shape, or genetic potential. In humans, females
CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE. Section B: Sex Chromosomes
 CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE Section B: Sex Chromosomes 1. The chromosomal basis of sex varies with the organism 2. Sex-linked genes have unique patterns of inheritance 1. The chromosomal
CHAPTER 15 THE CHROMOSOMAL BASIS OF INHERITANCE Section B: Sex Chromosomes 1. The chromosomal basis of sex varies with the organism 2. Sex-linked genes have unique patterns of inheritance 1. The chromosomal
Trasposable elements: P elements
 Trasposable elements: P elements In 1938 Marcus Rhodes provided the first genetic description of an unstable mutation, an allele of a gene required for the production of pigment in maize. This instability
Trasposable elements: P elements In 1938 Marcus Rhodes provided the first genetic description of an unstable mutation, an allele of a gene required for the production of pigment in maize. This instability
Stability and flexibility of epigenetic gene regulation in mammalian development Wolf Reik 1
 NATURE Vol 447 24 May 2007 doi:10.1038/nature05918 INSIGHT REVIEW Stability and flexibility of epigenetic gene regulation in mammalian development Wolf Reik 1 During development, s start in a pluripotent
NATURE Vol 447 24 May 2007 doi:10.1038/nature05918 INSIGHT REVIEW Stability and flexibility of epigenetic gene regulation in mammalian development Wolf Reik 1 During development, s start in a pluripotent
Chromosomal Basis of Inheritance. Ch. 3
 Chromosomal Basis of Inheritance Ch. 3 THE CHROMOSOME THEORY OF INHERITANCE AND SEX CHROMOSOMES! The chromosome theory of inheritance describes how the transmission of chromosomes account for the Mendelian
Chromosomal Basis of Inheritance Ch. 3 THE CHROMOSOME THEORY OF INHERITANCE AND SEX CHROMOSOMES! The chromosome theory of inheritance describes how the transmission of chromosomes account for the Mendelian
Influence of Sex on Genetics. Chapter Six
 Influence of Sex on Genetics Chapter Six Humans 23 Autosomes Chromosomal abnormalities very severe Often fatal All have at least one X Deletion of X chromosome is fatal Males = heterogametic sex XY Females
Influence of Sex on Genetics Chapter Six Humans 23 Autosomes Chromosomal abnormalities very severe Often fatal All have at least one X Deletion of X chromosome is fatal Males = heterogametic sex XY Females
Fact Sheet 14 EPIGENETICS
 This fact sheet describes epigenetics which refers to factors that can influence the way our genes are expressed in the cells of our body. In summary Epigenetics is a phenomenon that affects the way cells
This fact sheet describes epigenetics which refers to factors that can influence the way our genes are expressed in the cells of our body. In summary Epigenetics is a phenomenon that affects the way cells
The world of non-coding RNA. Espen Enerly
 The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
Chromosomes, Mapping, and the Meiosis Inheritance Connection
 Chromosomes, Mapping, and the Meiosis Inheritance Connection Carl Correns 1900 Chapter 13 First suggests central role for chromosomes Rediscovery of Mendel s work Walter Sutton 1902 Chromosomal theory
Chromosomes, Mapping, and the Meiosis Inheritance Connection Carl Correns 1900 Chapter 13 First suggests central role for chromosomes Rediscovery of Mendel s work Walter Sutton 1902 Chromosomal theory
MCB41: Second Midterm Spring 2009
 MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
The following chapter is called "Preimplantation Genetic Diagnosis (PGD)".
 Slide 1 Welcome to chapter 9. The following chapter is called "Preimplantation Genetic Diagnosis (PGD)". The author is Dr. Maria Lalioti. Slide 2 The learning objectives of this chapter are: To learn the
Slide 1 Welcome to chapter 9. The following chapter is called "Preimplantation Genetic Diagnosis (PGD)". The author is Dr. Maria Lalioti. Slide 2 The learning objectives of this chapter are: To learn the
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
The Making of the Fittest: Evolving Switches, Evolving Bodies
 OVERVIEW MODELING THE REGULATORY SWITCHES OF THE PITX1 GENE IN STICKLEBACK FISH This hands-on activity supports the short film, The Making of the Fittest:, and aims to help students understand eukaryotic
OVERVIEW MODELING THE REGULATORY SWITCHES OF THE PITX1 GENE IN STICKLEBACK FISH This hands-on activity supports the short film, The Making of the Fittest:, and aims to help students understand eukaryotic
Bio EOC Topics for Cell Reproduction: Bio EOC Questions for Cell Reproduction:
 Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
How to construct transgenic mice
 How to construct transgenic mice Sandra Beer-Hammer Autumn School 2012 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
How to construct transgenic mice Sandra Beer-Hammer Autumn School 2012 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
Control of Gene Expression
 Home Gene Regulation Is Necessary? Control of Gene Expression By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Home Gene Regulation Is Necessary? Control of Gene Expression By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
www.njctl.org PSI Biology Mitosis & Meiosis
 Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Heredity - Patterns of Inheritance
 Heredity - Patterns of Inheritance Genes and Alleles A. Genes 1. A sequence of nucleotides that codes for a special functional product a. Transfer RNA b. Enzyme c. Structural protein d. Pigments 2. Genes
Heredity - Patterns of Inheritance Genes and Alleles A. Genes 1. A sequence of nucleotides that codes for a special functional product a. Transfer RNA b. Enzyme c. Structural protein d. Pigments 2. Genes
The Plant Cell, March 2016, www.plantcell.org 2016 American Society of Plant Biologists. All rights reserved.
 Epigenetics (TTPB4) Outline and Study Guide Overview Epigenetic modifications of DNA and chromatin affect the activity of genes and transposons. Epigenetic controls affect processes as diverse as time-of-flowering,
Epigenetics (TTPB4) Outline and Study Guide Overview Epigenetic modifications of DNA and chromatin affect the activity of genes and transposons. Epigenetic controls affect processes as diverse as time-of-flowering,
GeneCopoeia Genome Editing Tools for Safe Harbor Integration in. Mice and Humans. Ed Davis, Liuqing Qian, Ruiqing li, Junsheng Zhou, and Jinkuo Zhang
 G e n e C o p o eia TM Expressway to Discovery APPLICATION NOTE Introduction GeneCopoeia Genome Editing Tools for Safe Harbor Integration in Mice and Humans Ed Davis, Liuqing Qian, Ruiqing li, Junsheng
G e n e C o p o eia TM Expressway to Discovery APPLICATION NOTE Introduction GeneCopoeia Genome Editing Tools for Safe Harbor Integration in Mice and Humans Ed Davis, Liuqing Qian, Ruiqing li, Junsheng
Nuclear transplantation, embryonic stem cells and the potential for cell therapy
 (2004) 5, S114 S117 & 2004 The European Hematology Association All rights reserved 1466-4680/04 $30.00 www.nature.com/thj STEM CELLS Nuclear transplantation, embryonic stem cells and the potential for
(2004) 5, S114 S117 & 2004 The European Hematology Association All rights reserved 1466-4680/04 $30.00 www.nature.com/thj STEM CELLS Nuclear transplantation, embryonic stem cells and the potential for
Chapter 13: Meiosis and Sexual Life Cycles
 Name Period Chapter 13: Meiosis and Sexual Life Cycles Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know.
Name Period Chapter 13: Meiosis and Sexual Life Cycles Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know.
Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice
 Manuscript EMBO-2012-82888 Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice Saya Kagiwada, Kazuki Kurimoto, Takayuki Hirota, Masashi Yamaji, and Mitinori Saitou
Manuscript EMBO-2012-82888 Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice Saya Kagiwada, Kazuki Kurimoto, Takayuki Hirota, Masashi Yamaji, and Mitinori Saitou
5. The cells of a multicellular organism, other than gametes and the germ cells from which it develops, are known as
 1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
Answer Key Problem Set 5
 7.03 Fall 2003 1 of 6 1. a) Genetic properties of gln2- and gln 3-: Answer Key Problem Set 5 Both are uninducible, as they give decreased glutamine synthetase (GS) activity. Both are recessive, as mating
7.03 Fall 2003 1 of 6 1. a) Genetic properties of gln2- and gln 3-: Answer Key Problem Set 5 Both are uninducible, as they give decreased glutamine synthetase (GS) activity. Both are recessive, as mating
Imprinted X chromosome inactivation: evolution of mechanisms in distantly related mammals
 Manuscript submitted to: Volume 2, Issue 2, 110-126. AIMS Genetics DOI: 10.3934/genet.2015.2.110 Received date 3 November 2014, Accepted date 1 March 2015, Published date 4 March 2015 Review Imprinted
Manuscript submitted to: Volume 2, Issue 2, 110-126. AIMS Genetics DOI: 10.3934/genet.2015.2.110 Received date 3 November 2014, Accepted date 1 March 2015, Published date 4 March 2015 Review Imprinted
Complex multicellular organisms are produced by cells that switch genes on and off during development.
 Home Control of Gene Expression Gene Regulation Is Necessary? By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Home Control of Gene Expression Gene Regulation Is Necessary? By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA
 Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
How to construct transgenic mice
 How to construct transgenic mice Sandra Beer-Hammer Autumn School 2011 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
How to construct transgenic mice Sandra Beer-Hammer Autumn School 2011 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains was previously described
 Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
CONTRACTING ORGANIZATION: University of Alabama at Birmingham Birmingham, AL 35294
 AD Award Number: W81XWH-08-1-0030 TITLE: Regulation of Prostate Cancer Bone Metastasis by DKK1 PRINCIPAL INVESTIGATOR: Gregory A. Clines, M.D., Ph.D. CONTRACTING ORGANIZATION: University of Alabama at
AD Award Number: W81XWH-08-1-0030 TITLE: Regulation of Prostate Cancer Bone Metastasis by DKK1 PRINCIPAL INVESTIGATOR: Gregory A. Clines, M.D., Ph.D. CONTRACTING ORGANIZATION: University of Alabama at
(1-p) 2. p(1-p) From the table, frequency of DpyUnc = ¼ (p^2) = #DpyUnc = p^2 = 0.0004 ¼(1-p)^2 + ½(1-p)p + ¼(p^2) #Dpy + #DpyUnc
 Advanced genetics Kornfeld problem set_key 1A (5 points) Brenner employed 2-factor and 3-factor crosses with the mutants isolated from his screen, and visually assayed for recombination events between
Advanced genetics Kornfeld problem set_key 1A (5 points) Brenner employed 2-factor and 3-factor crosses with the mutants isolated from his screen, and visually assayed for recombination events between
1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes?
 Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
In developmental genomic regulatory interactions among genes, encoding transcription factors
 JOURNAL OF COMPUTATIONAL BIOLOGY Volume 20, Number 6, 2013 # Mary Ann Liebert, Inc. Pp. 419 423 DOI: 10.1089/cmb.2012.0297 Research Articles A New Software Package for Predictive Gene Regulatory Network
JOURNAL OF COMPUTATIONAL BIOLOGY Volume 20, Number 6, 2013 # Mary Ann Liebert, Inc. Pp. 419 423 DOI: 10.1089/cmb.2012.0297 Research Articles A New Software Package for Predictive Gene Regulatory Network
Chapter 5: Organization and Expression of Immunoglobulin Genes
 Chapter 5: Organization and Expression of Immunoglobulin Genes I. Genetic Model Compatible with Ig Structure A. Two models for Ab structure diversity 1. Germ-line theory: maintained that the genome contributed
Chapter 5: Organization and Expression of Immunoglobulin Genes I. Genetic Model Compatible with Ig Structure A. Two models for Ab structure diversity 1. Germ-line theory: maintained that the genome contributed
Practice Problems 4. (a) 19. (b) 36. (c) 17
 Chapter 10 Practice Problems Practice Problems 4 1. The diploid chromosome number in a variety of chrysanthemum is 18. What would you call varieties with the following chromosome numbers? (a) 19 (b) 36
Chapter 10 Practice Problems Practice Problems 4 1. The diploid chromosome number in a variety of chrysanthemum is 18. What would you call varieties with the following chromosome numbers? (a) 19 (b) 36
Psychoonkology, Sept. 2010 lifestyle factors and epigenetics
 Psychoonkology, Sept. 2010 lifestyle factors and epigenetics Alexander G. Haslberger Dep. für Ernährungswissenschaften Univ. of Vienna Working group: Food, GI-Microbiology, Epigenetics Content Health:
Psychoonkology, Sept. 2010 lifestyle factors and epigenetics Alexander G. Haslberger Dep. für Ernährungswissenschaften Univ. of Vienna Working group: Food, GI-Microbiology, Epigenetics Content Health:
Chapter 7. Physical interaction between Jamb and Jamc is necessary for myoblast fusion. Summary
 Chapter 7 Physical interaction between Jamb and Jamc is necessary for myoblast fusion Summary In this chapter I describe a series of transplant experiments designed to further elucidate the function and
Chapter 7 Physical interaction between Jamb and Jamc is necessary for myoblast fusion Summary In this chapter I describe a series of transplant experiments designed to further elucidate the function and
Lecture 3: Mutations
 Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
http://www.springer.com/3-540-23372-5
 http://www.springer.com/3-540-23372-5 Chromatin Remodeling Factors and DNA Replication Patrick Varga-Weisz Abstract Chromatin structures have to be precisely duplicated during DNA replication to maintain
http://www.springer.com/3-540-23372-5 Chromatin Remodeling Factors and DNA Replication Patrick Varga-Weisz Abstract Chromatin structures have to be precisely duplicated during DNA replication to maintain
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
4.1 3T12 and 312 are immortalized cell lines with transforming potential:
 DISCUSSION CHAPTER 4 4.1 3T12 and 312 are immortalized cell lines with transforming potential: Transformation of a normal cell with finite number of divisions into a tumorigenic cell of potentially infinite
DISCUSSION CHAPTER 4 4.1 3T12 and 312 are immortalized cell lines with transforming potential: Transformation of a normal cell with finite number of divisions into a tumorigenic cell of potentially infinite
1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells
 Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Prof Brian McStay Wellcome Trust Senior Investigator Award April 2015- March 2020
 Prof Brian McStay Wellcome Trust Senior Investigator Award April 2015- March 2020 Career History BA (Genetics) Trinity College Dublin PhD University of Edinburgh (with Adrian Bird) Post-Doc Fred Hutchinson
Prof Brian McStay Wellcome Trust Senior Investigator Award April 2015- March 2020 Career History BA (Genetics) Trinity College Dublin PhD University of Edinburgh (with Adrian Bird) Post-Doc Fred Hutchinson
CHROMOSOME STRUCTURE CHROMOSOME NUMBERS
 CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
GENE REGULATION. Teacher Packet
 AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
Name: Class: Date: ID: A
 Name: Class: _ Date: _ Meiosis Quiz 1. (1 point) A kidney cell is an example of which type of cell? a. sex cell b. germ cell c. somatic cell d. haploid cell 2. (1 point) How many chromosomes are in a human
Name: Class: _ Date: _ Meiosis Quiz 1. (1 point) A kidney cell is an example of which type of cell? a. sex cell b. germ cell c. somatic cell d. haploid cell 2. (1 point) How many chromosomes are in a human
Cell Division CELL DIVISION. Mitosis. Designation of Number of Chromosomes. Homologous Chromosomes. Meiosis
 Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
AP BIOLOGY 2012 SCORING GUIDELINES
 AP BIOLOGY 2012 SCORING GUIDELINES Question 1 Note: At least 1 point must be earned from each of parts (a), (b), (c), and (d) in order to earn a maximum score of 10. The ability to reproduce is a characteristic
AP BIOLOGY 2012 SCORING GUIDELINES Question 1 Note: At least 1 point must be earned from each of parts (a), (b), (c), and (d) in order to earn a maximum score of 10. The ability to reproduce is a characteristic
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
Cancer SBL101. James Gomes School of Biological Sciences Indian Institute of Technology Delhi
 Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Cancer SBL101 James Gomes School of Biological Sciences Indian Institute of Technology Delhi All Figures in this Lecture are taken from 1. Molecular biology of the cell / Bruce Alberts et al., 5th ed.
Chromosomes, Karyotyping, and Abnormalities (Learning Objectives) Learn the components and parts of a metaphase chromosome.
 Chromosomes, Karyotyping, and Abnormalities (Learning Objectives) Learn the components and parts of a metaphase chromosome. Define the terms karyotype, autosomal and sex chromosomes. Explain how many of
Chromosomes, Karyotyping, and Abnormalities (Learning Objectives) Learn the components and parts of a metaphase chromosome. Define the terms karyotype, autosomal and sex chromosomes. Explain how many of
guides BIOLOGY OF AGING STEM CELLS An introduction to aging science brought to you by the American Federation for Aging Research
 infoaging guides BIOLOGY OF AGING STEM CELLS An introduction to aging science brought to you by the American Federation for Aging Research WHAT ARE STEM CELLS? Stem cells are cells that, in cell cultures
infoaging guides BIOLOGY OF AGING STEM CELLS An introduction to aging science brought to you by the American Federation for Aging Research WHAT ARE STEM CELLS? Stem cells are cells that, in cell cultures
Julie Chaumeil, Sandrine Augui, Jennifer C. Chow, and Edith Heard
 Chapter 18 Combined Immunofluorescence, RNA Fluorescent In Situ Hybridization, and DNA Fluorescent In Situ Hybridization to Study Chromatin Changes, Transcriptional Activity, Nuclear Organization, and
Chapter 18 Combined Immunofluorescence, RNA Fluorescent In Situ Hybridization, and DNA Fluorescent In Situ Hybridization to Study Chromatin Changes, Transcriptional Activity, Nuclear Organization, and
Embryonic & and induced pluripotent Stem Cells. May 2010 Dipl. Biol. Dr. Kurt Pfannkuche
 Embryonic & and induced pluripotent Stem Cells May 2010 Dipl. Biol. Dr. Kurt Pfannkuche Seite 2 What makes a stem cell? open chromatin structure plasticity! Seite 3 self-renewal Plasticity Seite 4 Culture
Embryonic & and induced pluripotent Stem Cells May 2010 Dipl. Biol. Dr. Kurt Pfannkuche Seite 2 What makes a stem cell? open chromatin structure plasticity! Seite 3 self-renewal Plasticity Seite 4 Culture
Support structure for genetic material
 Support structure for genetic material 1 Making proteins in the RER Making copies of humans 2 Making copies of cells Making copies of genetic material 3 Making copies of genetic material Making copies
Support structure for genetic material 1 Making proteins in the RER Making copies of humans 2 Making copies of cells Making copies of genetic material 3 Making copies of genetic material Making copies
What is Cancer? Cancer is a genetic disease: Cancer typically involves a change in gene expression/function:
 Cancer is a genetic disease: Inherited cancer Sporadic cancer What is Cancer? Cancer typically involves a change in gene expression/function: Qualitative change Quantitative change Any cancer causing genetic
Cancer is a genetic disease: Inherited cancer Sporadic cancer What is Cancer? Cancer typically involves a change in gene expression/function: Qualitative change Quantitative change Any cancer causing genetic
Should Stem Cells Be Used To Treat Human Diseases?
 SAMPLE ESSAY C Should Stem Cells Be Used To Treat Human Diseases? Stem cells can be defined as undifferentiated cells that are generated during the development of the embryo. There are two functions ascribed
SAMPLE ESSAY C Should Stem Cells Be Used To Treat Human Diseases? Stem cells can be defined as undifferentiated cells that are generated during the development of the embryo. There are two functions ascribed
Epigenetic reprogramming in mammals
 Human Molecular Genetics, 2005, Vol. 14, Review Issue 1 doi:10.1093/hmg/ddi114 R47 R58 Epigenetic reprogramming in mammals Hugh D. Morgan, Fátima Santos, Kelly Green, Wendy Dean and Wolf Reik* Laboratory
Human Molecular Genetics, 2005, Vol. 14, Review Issue 1 doi:10.1093/hmg/ddi114 R47 R58 Epigenetic reprogramming in mammals Hugh D. Morgan, Fátima Santos, Kelly Green, Wendy Dean and Wolf Reik* Laboratory
Biological Sciences Initiative. Human Genome
 Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
CHARACTERISTIC FEATURES OF STEM CELLS. CLONING TECHNOLOGIES
 CHARACTERISTIC FEATURES OF STEM CELLS. CLONING TECHNOLOGIES 1 Science is discovering the unknown Stem cell field is still in its infancy Human embryonic stem cell research is a decade old, adult stem cell
CHARACTERISTIC FEATURES OF STEM CELLS. CLONING TECHNOLOGIES 1 Science is discovering the unknown Stem cell field is still in its infancy Human embryonic stem cell research is a decade old, adult stem cell
Head of College Scholars List Scheme. Summer Studentship. Report Form
 Head of College Scholars List Scheme Summer Studentship Report Form This report should be completed by the student with his/her project supervisor. It should summarise the work undertaken during the project
Head of College Scholars List Scheme Summer Studentship Report Form This report should be completed by the student with his/her project supervisor. It should summarise the work undertaken during the project
CCR Biology - Chapter 7 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 7 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. A person who has a disorder caused
Name: Class: Date: CCR Biology - Chapter 7 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. A person who has a disorder caused
!!!!!!!!!!!!!!!!!!!!!!!!!!
 Figure S7 cdl-1 3'UTR gld-1 binding site (4..10) let-7 binding site (326..346) Figure Legends Figure S1. gld-1 genetically interacts with nhl-2 and vig-1 during germline development. (A) nhl-2(ok818) enhances
Figure S7 cdl-1 3'UTR gld-1 binding site (4..10) let-7 binding site (326..346) Figure Legends Figure S1. gld-1 genetically interacts with nhl-2 and vig-1 during germline development. (A) nhl-2(ok818) enhances
Genetics 301 Sample Final Examination Spring 2003
 Genetics 301 Sample Final Examination Spring 2003 50 Multiple Choice Questions-(Choose the best answer) 1. A cross between two true breeding lines one with dark blue flowers and one with bright white flowers
Genetics 301 Sample Final Examination Spring 2003 50 Multiple Choice Questions-(Choose the best answer) 1. A cross between two true breeding lines one with dark blue flowers and one with bright white flowers
The Somatic Cell Cycle
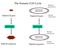 The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
Becker Muscular Dystrophy
 Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Control of Gene Expression
 Control of Gene Expression (Learning Objectives) Explain the role of gene expression is differentiation of function of cells which leads to the emergence of different tissues, organs, and organ systems
Control of Gene Expression (Learning Objectives) Explain the role of gene expression is differentiation of function of cells which leads to the emergence of different tissues, organs, and organ systems
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15
 Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
Biology 1406 - Notes for exam 5 - Population genetics Ch 13, 14, 15 Species - group of individuals that are capable of interbreeding and producing fertile offspring; genetically similar 13.7, 14.2 Population
Germ cell formation / gametogenesis And Fertilisation
 Developmental Biology BY1101 P. Murphy Lecture 3 The first steps to forming a new organism Descriptive embryology I Germ cell formation / gametogenesis And Fertilisation Why bother with sex? In terms of
Developmental Biology BY1101 P. Murphy Lecture 3 The first steps to forming a new organism Descriptive embryology I Germ cell formation / gametogenesis And Fertilisation Why bother with sex? In terms of
Chapter 13: Meiosis and Sexual Life Cycles
 Name Period Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know. Define: gene locus gamete male gamete female
Name Period Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know. Define: gene locus gamete male gamete female
STEM CELL FACTS. The ISSCR is an independent, nonproft organization providing a global forum for stem cell research and regenerative medicine.
 STEM CELL FACTS The ISSCR is an independent, nonproft organization providing a global forum for stem cell research and regenerative medicine. WHAT ARE STEM CELLS? Stem cells are the foundation cells for
STEM CELL FACTS The ISSCR is an independent, nonproft organization providing a global forum for stem cell research and regenerative medicine. WHAT ARE STEM CELLS? Stem cells are the foundation cells for
How To Understand How Gene Expression Is Regulated
 What makes cells different from each other? How do cells respond to information from environment? Regulation of: - Transcription - prokaryotes - eukaryotes - mrna splicing - mrna localisation and translation
What makes cells different from each other? How do cells respond to information from environment? Regulation of: - Transcription - prokaryotes - eukaryotes - mrna splicing - mrna localisation and translation
Genetics Module B, Anchor 3
 Genetics Module B, Anchor 3 Key Concepts: - An individual s characteristics are determines by factors that are passed from one parental generation to the next. - During gamete formation, the alleles for
Genetics Module B, Anchor 3 Key Concepts: - An individual s characteristics are determines by factors that are passed from one parental generation to the next. - During gamete formation, the alleles for
Yasunobu Mano 1,2, Tetsuya J. Kobayashi 3,4, Jun-ichi Nakayama 5,6, Hiroyuki Uchida 1, Masaya Oki 1,4,7 * Abstract. Introduction
 Single Cell Visualization of Yeast Gene Expression Shows Correlation of Epigenetic Switching between Multiple Heterochromatic Regions through Multiple Generations Yasunobu Mano 1,2, Tetsuya J. Kobayashi
Single Cell Visualization of Yeast Gene Expression Shows Correlation of Epigenetic Switching between Multiple Heterochromatic Regions through Multiple Generations Yasunobu Mano 1,2, Tetsuya J. Kobayashi
Gene Mapping Techniques
 Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
An Introduction to Stem Cell Biology. Michael L. Shelanski, MD,PhD Professor of Pathology and Cell Biology Columbia University
 An Introduction to Stem Cell Biology Michael L. Shelanski, MD,PhD Professor of Pathology and Cell Biology Columbia University Figures adapted from ISSCR. Presentations of Drs. Martin Pera (Monash University),
An Introduction to Stem Cell Biology Michael L. Shelanski, MD,PhD Professor of Pathology and Cell Biology Columbia University Figures adapted from ISSCR. Presentations of Drs. Martin Pera (Monash University),
PSYC G4498 -- Behavioral Epigenetics Dr. Marija Kundakovic Fall 2012
 PSYC G4498 -- Behavioral Epigenetics Dr. Marija Kundakovic Fall 2012 I. Bulletin description II. Full course description III. Rationale for giving the course IV. Weekly outline of topics and readings V.
PSYC G4498 -- Behavioral Epigenetics Dr. Marija Kundakovic Fall 2012 I. Bulletin description II. Full course description III. Rationale for giving the course IV. Weekly outline of topics and readings V.
Gene mutation and molecular medicine Chapter 15
 Gene mutation and molecular medicine Chapter 15 Lecture Objectives What Are Mutations? How Are DNA Molecules and Mutations Analyzed? How Do Defective Proteins Lead to Diseases? What DNA Changes Lead to
Gene mutation and molecular medicine Chapter 15 Lecture Objectives What Are Mutations? How Are DNA Molecules and Mutations Analyzed? How Do Defective Proteins Lead to Diseases? What DNA Changes Lead to
Mechanism of Mouse Germ Cell Specification: A Genetic Program Regulating Epigenetic Reprogramming
 Mechanism of Mouse Germ Cell Specification: A Genetic Program Regulating Epigenetic Reprogramming M.A. SURANI, K. ANCELIN, P. HAJKOVA, U.C. LANGE, B. PAYER, P. WESTERN, * AND M. SAITOU Wellcome Trust Cancer
Mechanism of Mouse Germ Cell Specification: A Genetic Program Regulating Epigenetic Reprogramming M.A. SURANI, K. ANCELIN, P. HAJKOVA, U.C. LANGE, B. PAYER, P. WESTERN, * AND M. SAITOU Wellcome Trust Cancer
Title: Genetics and Hearing Loss: Clinical and Molecular Characteristics
 Session # : 46 Day/Time: Friday, May 1, 2015, 1:00 4:00 pm Title: Genetics and Hearing Loss: Clinical and Molecular Characteristics Presenter: Kathleen S. Arnos, PhD, Gallaudet University This presentation
Session # : 46 Day/Time: Friday, May 1, 2015, 1:00 4:00 pm Title: Genetics and Hearing Loss: Clinical and Molecular Characteristics Presenter: Kathleen S. Arnos, PhD, Gallaudet University This presentation
LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS
 LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS Los Angeles Mission College Biology 3 Name: Date: INTRODUCTION BINARY FISSION: Prokaryotic cells (bacteria) reproduce asexually by binary fission. Bacterial
LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS Los Angeles Mission College Biology 3 Name: Date: INTRODUCTION BINARY FISSION: Prokaryotic cells (bacteria) reproduce asexually by binary fission. Bacterial
DNA Fingerprinting. Unless they are identical twins, individuals have unique DNA
 DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
Chapter 9 Patterns of Inheritance
 Bio 100 Patterns of Inheritance 1 Chapter 9 Patterns of Inheritance Modern genetics began with Gregor Mendel s quantitative experiments with pea plants History of Heredity Blending theory of heredity -
Bio 100 Patterns of Inheritance 1 Chapter 9 Patterns of Inheritance Modern genetics began with Gregor Mendel s quantitative experiments with pea plants History of Heredity Blending theory of heredity -
Site-Directed Nucleases and Cisgenesis Maria Fedorova, Ph.D.
 Site-Directed Nucleases and Cisgenesis Maria Fedorova, Ph.D. Regulatory Strategy Lead Enabling Technologies DuPont-Pioneer, USA 1 New Plant Breeding Techniques 2007 New Techniques Working Group established
Site-Directed Nucleases and Cisgenesis Maria Fedorova, Ph.D. Regulatory Strategy Lead Enabling Technologies DuPont-Pioneer, USA 1 New Plant Breeding Techniques 2007 New Techniques Working Group established
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs)
 Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Control of Gene Expression
 Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
Cat caryotype (38 chromosomes)
 CAT GENETICS Cat caryotype (38 chromosomes) D Dense pigment d dilute pigment L short hair dominant l long hair monohybrid dihybrid Cat Genetics and Mosaicism The Calico phenotype reflects transcriptional
CAT GENETICS Cat caryotype (38 chromosomes) D Dense pigment d dilute pigment L short hair dominant l long hair monohybrid dihybrid Cat Genetics and Mosaicism The Calico phenotype reflects transcriptional
Reprogramming hierarchy in mouse liver cell subpopulations depends on cell intrinsic factors including BAF complex members.
 Reprogramming hierarchy in mouse liver cell subpopulations depends on cell intrinsic factors including BAF complex members. A dissertation submitted to the faculty of natural sciences for the requirement
Reprogramming hierarchy in mouse liver cell subpopulations depends on cell intrinsic factors including BAF complex members. A dissertation submitted to the faculty of natural sciences for the requirement
Analysis and Integration of Big Data from Next-Generation Genomics, Epigenomics, and Transcriptomics
 Analysis and Integration of Big Data from Next-Generation Genomics, Epigenomics, and Transcriptomics Christopher Benner, PhD Director, Integrative Genomics and Bioinformatics Core (IGC) idash Webinar,
Analysis and Integration of Big Data from Next-Generation Genomics, Epigenomics, and Transcriptomics Christopher Benner, PhD Director, Integrative Genomics and Bioinformatics Core (IGC) idash Webinar,
Genetic Mutations. Indicator 4.8: Compare the consequences of mutations in body cells with those in gametes.
 Genetic Mutations Indicator 4.8: Compare the consequences of mutations in body cells with those in gametes. Agenda Warm UP: What is a mutation? Body cell? Gamete? Notes on Mutations Karyotype Web Activity
Genetic Mutations Indicator 4.8: Compare the consequences of mutations in body cells with those in gametes. Agenda Warm UP: What is a mutation? Body cell? Gamete? Notes on Mutations Karyotype Web Activity
NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS
 NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS Orly Alter (a) *, Gene H. Golub (b), Patrick O. Brown (c)
NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS Orly Alter (a) *, Gene H. Golub (b), Patrick O. Brown (c)
Plant Growth & Development. Growth Stages. Differences in the Developmental Mechanisms of Plants and Animals. Development
 Plant Growth & Development Plant body is unable to move. To survive and grow, plants must be able to alter its growth, development and physiology. Plants are able to produce complex, yet variable forms
Plant Growth & Development Plant body is unable to move. To survive and grow, plants must be able to alter its growth, development and physiology. Plants are able to produce complex, yet variable forms
Lecture 2: Mitosis and meiosis
 Lecture 2: Mitosis and meiosis 1. Chromosomes 2. Diploid life cycle 3. Cell cycle 4. Mitosis 5. Meiosis 6. Parallel behavior of genes and chromosomes Basic morphology of chromosomes telomere short arm
Lecture 2: Mitosis and meiosis 1. Chromosomes 2. Diploid life cycle 3. Cell cycle 4. Mitosis 5. Meiosis 6. Parallel behavior of genes and chromosomes Basic morphology of chromosomes telomere short arm
