Cost-based Optimization of Graph Queries in Relational Database Management Systems
|
|
|
- Chester Jenkins
- 10 years ago
- Views:
Transcription
1 Cost-based Optimization of Graph Queries in Relational Database Management Systems D I S S E R T A T I O N zur Erlangung des akademischen Grades Dr. rer nat. im Fach Informatik eingereicht an der Mathematisch-Naturwissenschaftlichen Fakultät II Humboldt-Universität zu Berlin von Dipl.-Ing. (FH) Silke Trißl M.Sc. Präsident der Humboldt-Universität zu Berlin: Prof. Dr. Jan-Hendrik Olbertz Dekan der Mathematisch-Naturwissenschaftlichen Fakultät II: Prof. Dr. Elmar Kulke Gutachter: 1. Prof. Dr. Ulf Leser 2. Prof. Johann-Christoph Freytag, Ph.D. 3. Prof. Dr. Thorsten Grust eingereicht am: Tag der mündlichen Prüfung:
2
3 Alles hat ein Ende nur die Wurst hat zwei. Stephan Remmler Acknowledgement This thesis would not have been possible without the help, support, and encouragement of many people. First of all, I would like to thank my supervisor Prof. Ulf Leser. He gave me the opportunity to start my PhD and provided a welcoming and pleasant working environment at Humboldt-Universität zu Berlin. I am greatly indebted to him for his patience, encouragement, and guidance during all these years with ups and downs. I could not have imagined a more motivated or dedicated advisor for my PhD study. I am grateful to all who gave me the opportunity to partly finance my PhD by teaching. I met committed and inquiring students in the courses and exercises I taught for Prof. Ulf Leser at HU Berlin and Prof. Felix Naumann at HPI Potsdam. Dr. Márta Gutsche and the Frauenförderung at HU Berlin gave me the opportunity to spark interest in girls to study computer science. I thank Prof. Louiqa Raschid at University of Maryland who invited me for a research exchange to the US. I am also grateful to the BMBF who supported my research. I would not have finished this PhD thesis without the help and support of many colleagues and friends. Thanks to Jörg, Timo, and Philippe who shared an office with me. Thanks to Jens, Melanie, Jana, Long, Roger, and Samira who also accompanied me for a long time during my thesis. I want to acknowledge all researchers and students from the groups WBI and DBIS at HU, Informationssysteme at HPI, and Genetik und Biometrie at FBN. Many thanks for constructive criticism and helpful suggestions. I am greatly indebted to all colleagues who tried to cheer me up during common lunch and coffee breaks. I acknowledge some students, who I met during my time in Berlin. Raphael and Philipp did a lot of programming in my first project Columba. Johannes, Christoph, Florian, and André used some ideas of GRIPP in their Studien- or Diplomarbeiten and gave feedback on the algorithm. Last but not least, würde ich mich gerne bei meiner Familie bedanken, die während der gesamten Zeit Freud und Leid mit mir geteilt hat. Meine Eltern hatten und haben immer ein offenes Ohr für meine Sorgen und Nöte von ganzem Herzen vielen Dank dafür. Also, many thanks to my sister. Whenever I needed to discuss a problem, she listened patiently and gave me good advice.
4
5 Abstract Graphs occur in many areas of life. We are interested in graphs in biology, where nodes are chemical compounds, enzymes, reactions, or interactions, which are connected by either directed or undirected edges. Efficiently querying these graphs is a challenging task. In this thesis we present GRIcano, a system that efficiently executes graph queries. For GRIcano we assume that graphs are stored and queried using relational database management systems (RDBMS). We use an extended version of the Pathway Query Language PQL to express graph queries, for which we describe the syntax and semantics in this work. We employ ideas from RDBMS to improve the performance of query execution. Thus, the core of GRIcano is a cost-based query optimizer, which is created using the Volcano optimizer generator. This thesis makes contributions to all three required components of the optimizer, the relational algebra, implementations, and cost model. Relational algebra operators alone are not sufficient to express graph queries. Thus, we first present new operators to rewrite PQL queries to algebra expressions. We propose the reachability φ, distance Φ, path length ψ, and path operator Ψ. In addition, we provide rewrite rules for the newly proposed operators in combination with standard relational algebra operators. Secondly, we present implementations for each proposed operator. The main contribution is GRIPP, an index structure that allows us to execute reachability queries on very large graphs containing directed edges. GRIPP has advantages over other existing index structures, which we review in this work. In addition, we show how to employ GRIPP and the recursive query strategy as implementation for all four proposed operators. The third component of GRIcano is the cost model, which requires cardinality estimates for the proposed operators and cost functions for the implementations. Based on extensive experimental evaluation of the proposed implementations we present functions to estimate the cardinality of the φ, Φ, ψ, and Ψ operator and the cost of executing a query. The novelty of our approach is that these functions only use key figures of the graph. We finally present the effectiveness of GRIcano using exemplary graph queries on real biological networks. v
6
7 Zusammenfassung Graphen sind in vielen Bereichen des Lebens zu finden, wobei wir speziell an Graphen aus der Biologie interessiert sind. Knoten in solchen Graphen sind chemische Komponenten, Enzyme, Reaktionen oder Interaktionen, die durch gerichtete oder ungerichtete Kanten miteinander verbunden sind. Eine effiziente Ausführung von Graphanfragen ist eine Herausforderung. In dieser Arbeit präsentieren wir GRIcano, ein System, das das effiziente Ausführen von Graphanfragen erlaubt. Wir nehmen an, dass die Graphen in relationalen Datenbankmanagementsystemen (RDBMS) gespeichert sind und darin auch angefragt werden. Als Graphanfragesprache schlagen wir eine erweiterte Version der Pathway Query Language (PQL) vor. Der Hauptbestandteil von GRIcano ist ein kostenbasierter Anfrageoptimierer, der mit Hilfe des Optimierergenerators Volcano erzeugt wird. Diese Arbeit enthält Beiträge zu allen drei benötigten Komponenten des Optimierers, der relationalen Algebra, Implementierungen und Kostenmodellen. Die Operatoren der relationalen Algebra alleine sind nicht ausreichend, um PQL Anfragen auszudrücken. Daher stellen wir zuerst die neuen Operatoren Erreichbarkeits- φ, Distanz- Φ, Pfadlängen- ψ und Pfadoperator Ψ vor. Zusätzlich geben wir Regeln für die Umformung von Ausdrücken an, die die neuen Operatoren zusammen mit den Standardoperatoren der relationalen Algebra enthalten. Des Weiteren präsentieren wir Implementierungen für jeden vorgeschlagenen Operatoren. Der Hauptbeitrag dabei ist GRIPP, eine Indexstruktur, die die effiziente Ausführung von Erreichbarkeitsanfragen auf sehr großen Graphen mit gerichteten Kanten erlaubt. Wir zeigen, wie GRIPP und die rekursive Anfragestrategie genutzt werden können, um Implementierungen für alle vorgeschlagenen Operatoren bereitzustellen. Die dritte Komponente von GRIcano ist das Kostenmodell, das Kardinalitätsabschätzungen für die vorgeschlagenen Operatoren und Kostenmodelle für die Implementierungen benötigt. Basierend auf umfangreichen Experimenten schlagen wir Funktionen für die Abschätzung der Kardinalitäten der Operatoren φ, Φ, ψ und Ψ vor. Zusätzlich leiten wir Funktionen für die Abschätzung der Kosten für die Ausführung von Graphanfragen ab. Der neue Ansatz der Kostenmodelle ist, dass die Funktionen nur Kennzahlen der Graphen verwenden. Abschließend zeigen wir die Wirkungsweise von GRIcano mit Beispielanfragen auf echten biologischen Netzwerken. vii
8
9 Contents 1. Introduction Queries on Graphs Motivation Contribution Structure of this Work Definitions and Terminology Graphs Definitions Storage and Traversal Relational Algebra Algebra and Relations Operators Equivalence Rules Cost-Based Query Optimization Query Processing Implementation of Operators Cost Function and Query Optimization Volcano Graph Queries Data Model Graph Queries Query Graph Evaluation of Graph Queries Pathway Query Language Graphs in PQL Syntax PQL and Non-graph Relations PQL Semantics Semantics of Node Conditions Semantics of Path Conditions Semantics of HAVING Conditions Semantic of the Subgraph Specification Conversion to Relational Algebra Related Work ix
10 Contents 4. Operators for Graph Queries Operators for Nodes Operators for Paths Path Operator, Ψ Reachability operator, φ Path Length Operator, ψ Distance Operator, Φ Summary Related Work Implementations for Operators GRIPP Index Structure Reachability Queries Distance Queries Path Length and Path Queries Other Index Structures Transitive Closure Dual Labeling Label + SSPI RDBMS Capabilities Recursive Strategies Summary Related Work Performance of GRIPP Experimental Setup Generated Graphs Real-world Graphs Implementation Details Index Creation Query Performance Reachability Queries Distance Queries Path Length Queries Path Queries Comparison of Query Types Summary GRIcano Cardinality Estimates Reachability Operator Distance Operator Path Length Operator x
11 Contents Path Operator Validation on Real World Graphs Cost Functions Reachability Queries Distance Queries Path Length Queries Path Queries Validation on Real World Graphs GRIcano Experimental Evaluation Related Work Cardinality and Cost Estimates Rule-based Query Optimization Cost-based Query Optimization Conclusion and Outlook Summary Future Work A. Strongly Connected Component 151 A.1. Kosaraju s Algorithm B. Rewrite Rules for Operators 153 B.1. Path Operator B.1.1. Restriction on Start and End Node B.1.2. Path Operator and Other Operators B.2. Path Length Operator B.2.1. Restriction on Start and End Node B.2.2. From Path Operator Ψ to Path Length Operator ψ B.2.3. Path Length Operator and Other Operators B.3. Distance Operator B.3.1. Restriction on Start and End Node B.3.2. From Path Operator Ψ to Distance Operator Φ B.3.3. Distance Operator and Other Operators B.4. Reachability Operator B.4.1. Restriction on Start and End Node B.4.2. From Path Operator Ψ to Reachability Operator φ B.4.3. Reachability Operator and Other Operators C. Additional Algorithms for GRIPP 161 C.1. Relational Schema for Storing GRIPP C.2. Stop Node List for GRIPP C.3. Reachability for Sets of Nodes xi
12 Contents D. Graph Properties 165 E. Model Specification for Volcano 167 F. Cost and Cardinality Functions for Volcano 173 G. Exemplary Queries for GRIcano 179 xii
13 1. Introduction The topic of this work is cost-based optimization of graph queries in relational database management systems. In Section 1.1 we first introduce the kind of graphs that led us to this topic, before we proceed in Section 1.2 with the motivation for our approach. In Section 1.3 we summarize our contribution in the area of cost-based optimization of graph queries. Finally, in Section 1.4 we give an overview of this work Queries on Graphs Graphs occur in many areas of life. Examples are public transport plans, road maps, the World Wide Web (WWW), or social networks. Common to all these graphs is that they consist of nodes and edges. Nodes are stations, junctions, web pages, or people. Edges in such networks are tracks, roads, links, or personal relationships. All these graphs have interesting features but we are interested in graphs in biology. To understand the content of these graphs we first make a short digression to cell biology. For a more comprehensive introduction we refer the reader to Alberts et al. [AJW + 08]. All biological cells are built in similar fashion, though there exist differences in the structure of cells between the three major groups, prokaryotes, eukaryotes, and archaea. All have in common that they contain a cell membrane as boundary to the outside and a genome, which holds information for building and maintaining the cell. In eukaryotes the genome is contained inside the nucleus, while in prokaryotes and archaea the genome is free in the cytoplasm. The genome is comprised of long stretches of DNA, the chromosomes. Genes are short regions of the genome that code for a functional product in the cell. During the transcription process genes are read and transcribed into RNA. Either the RNA itself is the functional product or the RNA, possibly with some modifications, is translated to proteins. Proteins in a cell are the workhorses as they catalyze reactions, process signals, or transport molecules. One class of proteins, the enzymes, catalyze chemical reactions, such as the degradation of sugar or the production of essential amino acids. Another class, the membrane proteins, reside inside the cell membrane and react to outer stimuli or facilitate the transport of substances in and out of a cell. When an outer stimuli occurs membrane proteins may activate or inactivate proteins inside the cell to enhance or suppress reactions. There exist other protein groups such as histones, which are concerned with packing the DNA in the nucleus of eukaryotes, collagens, which occur mainly in muscle cells, or antibodies, which are required in higher organisms for the immune response. 1
14 1. Introduction To give an impression of the complexity of the problem, every human has about 250,000 different proteins in his or her body, according to current estimates. Each protein may interact with numerous other proteins or some of the hundreds of thousands organic and inorganic substances. Biologists have studied these complex interactions involving proteins and other substances. Their knowledge is stored as graphs in publicly available data sources. Biological graphs may roughly be divided into three categories, metabolic networks, signaling pathways, and protein-protein interaction networks 1. For a review on different biological networks see [BN05]. Metabolic networks are graphs, which represent the conversion of substances in a cell. Nodes in these networks are proteins, other molecules such as sugars or fatty acids, or reactions. Edges in such graphs are usually directed and indicate that a molecule participates in a reaction. The most familiar conversion is the glycolysis. In the glycolysis glucose is converted to pyruvate, which produces energy during the conversion. Proteins and reactions participating in this conversion are said to be in the glycolysis pathway. In general, pathways in metabolic networks are subgraphs that stand for specific conversions defined by researchers. The pathways may overlap, i.e., they may share proteins or reactions. Data sources for metabolic networks are KEGG [KGK + 04], BioCyc [KOMK + 05], and Reactome [JTGV + 05] for instance. Figure 1.1 shows the glycolysis given by KEGG. Circles are molecules that are converted, rectangular boxes on edges stand for reactions catalyzed by enzymes that are identified by their EC number, and the boxes with rounded corners represent other pathways. Signaling pathways are graphs that capture the information flow in a cell. Nodes in these graphs are usually proteins or reactions, while edges represent the flow of information. For example, Figure 1.2 shows the activation of protein kinase A (PKA) by an outer stimuli as given by BioCarta [htt11b]. The activated form of PKA regulates several reactions, including one reaction of the glycolysis presented in Figure 1.1. Depending on the outer stimuli glucose PKA phosphorylates or dephosphorylates the complex of the two enzymes phosphofructokinase 2 and fructose-2,6-bisphosphatase. The phosphorylation status influences the reaction rate of the glycolysis. The third group of biological graphs are protein-protein interaction networks. In these graphs nodes are proteins, while edges represent interactions between proteins and they are usually undirected. Figure 1.3 shows known interactions for the protein complex phosphofructokinase 2 and fructose-2,6-bisphosphatase (PFKFB1) as given by String [vmjs + 05], a data source for protein-protein interactions. The red node in the center is PFKFB1. It interacts with protein kinase A (PKACA) and several other proteins. The different colors of the edges code for different evidences, e.g., interactions found in other data sources are represented by blue edges, while interactions derived using text mining methods are shown by light green edges. Other data sources that contain data about protein-protein interactions are for in- 1 See Pathguide: the pathway resource list for a list on data sources 2
15 1.1. Queries on Graphs Figure 1.1.: The glycolysis as given by KEGG. The circles are molecules that are converted, rectangular boxes on edges stand for reactions catalyzed by enzymes that are identified by their EC number, and the boxes with rounded corners stand for other pathways. 3
16 1. Introduction Figure 1.2.: The activation of PKA through an outer stimuli from BioCarta. stance DIP [XSD + 02], BIND [BBH03], Intact [XSD + 02], and PubGene [JLKH01] Motivation The examples in the last section show only small parts of different biological graphs. Table 1.1 shows the number of nodes and edges of selected data sources. For example, KEGG contains 42,002 nodes and 51,450 edges in its reference pathway as of March The reference pathway is a summarization of the pathways of all species. In contrast, BioCyc stores an individual metabolic network for each of the roughly 400 species. In addition, in contrast to KEGG BioCyc also represents relationships between genes and proteins. Biologist use specialized graph viewing tools to display those graphs. For a review on the tools see Suderman & Hallett [SH07]. The tools usually display parts of the entire graph, e.g., a single pathway of a metabolic network, possibly with links to other pathways as shown in Figure 1.1. With such tools a biologist is only able to navigate through graphs. Consider the question How many steps does a cell require to produce the amino acid lysine given the substrate glucose. A biologist may use the metabolic network of KEGG, 4
17 1.2. Motivation Figure 1.3.: Known protein-protein interactions for the protein complex PFKFB1 in humans. The different colors of edges stand for different evidences, e.g., interactions found in other data sources are represented by blue edges, while interactions derived using text mining methods are shown by light green edges. where she has to start at glucose in the glycolysis pathway, follow the link to the pathway of the citrate cycle, and then follow the link to the pathway of the lysine biosynthesis. This way, she will count that there are 25 steps required to produce lysine from the substrate glucose. Clearly, when manually navigating through the images of pathways a biologist might not find the shortest path or occasionally even no path at all although there exists one. Thus, tools are required that allow users to pose queries such as the one presented above and return an answer to the user. In [HNM + 00] van Helden and colleagues identified several other questions that are interesting for biologists: Get all reactions catalyzed by a given gene product. Find all metabolic pathways that convert compound A into compound B in less than X steps. Retrieve all genes whose expression is directly or indirectly affected by a given compound. Find all compounds that can be synthesized from a given precursor in less than X steps. Currently, researchers have to write specialized programs to traverse the graphs to 5
18 1. Introduction Biological graph Number of nodes Number of edges Metabolic networks KEGG [KGK + 04] 42,002 51,450 BioCyc A. thaliana [KOMK + 05] 10,951 23,649 Reactome [JTGV + 05] 11,795 23,649 Signaling pathways BioCarta [htt11b] only images NetPath TGF-β [KMR + 10] TransPath [KPV + 06] > 100,000 >240,000 Protein-protein interaction networks String [vmjs + 05] > 2,500,000 > 50,000,000 DIP [XSD + 02] 23,201 71,276 Intact 50, ,044 Table 1.1.: Sizes of biological graphs (in March 2011). answer such queries. Whenever they want to pose a new query these programs need to be adjusted. In this work we present GRIcano to overcome this problem Contribution In this work we present GRIcano, a novel tool that efficiently retrieves answers to graph queries. In GRIcano we employ ideas from query optimization in relational database management systems (RDBMS) and carry these ideas over to graph query optimization. In the following chapters we target several aspects of graph queries. We specifically make the following contributions: Extend the existing query language PQL. We present and extend the Pathway Query Language (PQL) [Les05a], which was developed to express graph queries. Using PQL a user may express conditions of a graph query as predicates. In Chapter 3 we describe the syntax as well as the semantics of PQL. Define relational operators to express PQL queries. In order to optimize a graph query we want to be able to alter the order in which predicates of the query are evaluated. We may achieve this by rewriting the PQL query to an algebraic expression and apply rewrite rules for transformation. As standard operators from relational algebra are not sufficient for expressing PQL queries, which we discuss in Chapter 4, we develop new and novel operators in this thesis. We define the path Ψ, path length ψ, distance Φ, and reachability operator φ to express predicates of graphs queries and provide rewrite rules for the exchange of operators. Propose and experimentally evaluate implementations for operators. For each proposed operator we have to provide implementations to compute the result. Thus, in Chapter 5 we discuss implementations to answer reachability, 6
19 1.4. Structure of this Work distance, path length, and path queries. We may use GRIPP, our newly developed index structure, for answering all four types of graph queries. Chapter 6 shows that we are able to compute the GRIPP index even for very large graphs, for which the transitive closure cannot be created. In addition, we are able to answer reachability queries on average in almost constant time regardless the size and shape of the graph using GRIPP. Develop functions to estimate cardinality of operators and cost of implementations. For cost-based query optimization we require cardinality estimates for the different operators and cost functions for each implementation. In Chapter 7 we develop equations that are based on key figures of the graph, which is to our knowledge a novel approach. Using our cost functions we correctly predict on generated as well as on real-world graphs the result sizes and fastest implementations. Present and evaluate a prototypical implementation of GRIcano. In Chapter 7 we present GRIcano, the first system that performs cost-based query optimization for graph queries. The underlying cost-based query optimizer is generated using the Volcano framework [GM93]. Volcano requires as input the available operators and rewrite rules of the algebra, the available implementations for the different operators, and the equations for the cardinality and cost estimates. We show the effect of GRIcano using exemplary queries Structure of this Work In Chapter 2 we introduce basic notation on graphs, relational algebra, and cost-based query optimization. Chapter 3 is devoted to a data model for storing graphs, graph queries, and PQL, a language to express graph queries. In Chapter 4 we first argue that PQL queries should be executed like standard SQL queries, i.e., first transforming them to an algebraic expression. We induce the necessity of new operators for the algebra and introduce the path operator, Ψ, path length operator, ψ, distance operator Φ, and reachability operator φ. We also provide rewrite rules for exchanging operators. In Chapter 5 we provide implementations for the operators proposed in Chapter 4. We present GRIPP, an index structure to efficiently answer reachability queries even on large graphs. In Chapter 6 we experimentally evaluate the presented implementations. In Chapter 7 we devise functions to estimate cardinality for the four newly defined operators and cost functions for the different implementations. In that chapter we also introduce GRIcano, our graph query optimizer. We show the capabilities of GRIcano using selected queries. Chapter 8 concludes the work. 7
20
21 2. Definitions and Terminology This chapter introduces basic notation on graphs, relational algebra, and query optimization. In Section 2.1 we formally define graphs and properties of graphs. Section 2.2 introduces fundamental concepts behind relational algebra. In Section 2.3 we present an introduction to cost-based query optimization in relational database management systems Graphs This work mostly deals with graph structured data. We therefore formally introduce graphs. For this purpose we adopt notation from Cormen et al. [CLR01] Definitions Definition 2.1 (Graph) A graph G = (V (G), E(G)) is a tuple consisting of a set of nodes V (G) and a set of edges E(G), with E(G) V (G) V (G). Whenever the context of the graph is clear we may write G = (V, E). There exist two types of graphs, directed and undirected graphs. Directed graphs have ordered pairs of nodes in E. In contrast, in undirected graphs the set E contains unordered pairs of nodes. Consider (u, v) E with u, v V and u v. In a directed graph only v is adjacent to u, while in an undirected graph the relation is symmetric, i.e., (u, v) is the same as (v, u). If (u, v) E in a directed graph we say node u has the outgoing edge (u, v) and therefore u is start node of (u, v). In analogy (u, v) is an incoming edge of node v and therefore v is target node of (u, v). We call u parent of v and v child of u. Definition 2.2 (Size of a graph) Let G = (V, E). The size of G is the number of nodes V plus the number of edges E in G, i.e., G = V + E. Based on the ratio between edges and nodes, which is called the density of a graph, we are able to divide graphs into two groups sparse and dense graphs. The literature does not provide a clear distinction between the two types. As rule of thumb, if the number of edges E is close to V 2 the graphs are called dense, otherwise if E V 2 they are sparse. 9
22 2. Definitions and Terminology e f a d b c Figure 2.1.: A directed graph. Circles represent nodes; arrows between nodes represent edges. Nodes in this example are uniquely labeled. The size of the graph is 14 (6 nodes plus 8 edges). For example, the degree of node b is deg(b) = 3. To describe the shape of a graph we look at the distribution of node degrees. To do so, we first define the degree of a node. Definition 2.3 (Degree of a node) Given a graph G = (V, E). The degree of node v V deg(v) is the number of edges in which v participates. If G is directed we may distinguish between an indegree deg in (v) and an outdegree deg out (v) of a node v. The indegree is the number of edges with v as target node and, in analogy, the outdegree is the number of edges with v as start node. Based on the distribution of the node degree we distinguish between different graph topologies. The distribution of the node degrees of random graphs follows a binomial distribution. Graphs where the distribution of the node degrees follows a power-law are called scale-free. Barabási and Oltvai describe in [BO04] these topologies. Nodes and edges are often labeled. Therefore we define a label function for nodes and edges of a graph. Definition 2.4 (Label function, φ) Let L be a set of labels. A label function φ assigns labels to nodes and edges, φ(v, L) : V L and φ(e, L) : E L. In this work we assume each label l L consists of a type and a value. Graphs also contain paths. Definition 2.5 (Path and path length) Let G = (V, E). A path p is a sequence of nodes v 0, v 1, v 2,..., v k, v i V such that (v i 1, v i ) E for i = 1, 2,..., k. The length of the path is the number of edges in the path. If there exists a path p from u to w we say w is reachable from u, written as u w. 10
23 2.1. Graphs A path is simple if all nodes in p are distinct, otherwise p is said to contain a cycle. Formally, a cycle is a path in a directed graph with nodes v 0, v 1, v 2,..., v k, where v 0 = v k and k 2. In this work we only consider simple paths. A directed graph G without cycles is called a directed acyclic graph (DAG). In a DAG a node may have many parent nodes. If a graph G contains no cycles and every node v V has at most one parent it is a tree. Definition 2.6 (Distance) Given a graph G = (V, E). The distance between two nodes u, w V is the length of the shortest path p between nodes u and w. If no path exists, the distance is. For a pair of nodes u, w V there may exist several paths p i with the same path length. Example 2.1 (Paths in graphs). Consider nodes a and e in Figure 2.1. Node e is reachable from node a as there exist several paths from a to e. One path is p 1 = a, b, c, f, e with length 4, another p 2 = a, b, c, e with length 3. As there exists no other simple path between both nodes we know the distance between the two nodes is 3. Path b, c, e, b forms a cycle. After introducing paths we now define ancestors and descendants of a node. Definition 2.7 (Ancestors and descendants of a node) Let G = (V, E). A node u V is ancestor of v V if a path u v in G exists. In analogy all nodes w V are descendants of node v V for which v w in G exists. We are able to represent all ancestor-descendant relationships in the transitive closure. Definition 2.8 (Transitive closure) Let G = (V, E) with v, w V. The transitive closure is the graph G = (V, E ) with E = {(v, w) v w in G}. When answering graph queries we are interested in certain nodes and edges of a given graph. We thus formally introduce the notation of a subgraph in the following. Definition 2.9 (Subgraph and induced subgraph) Let G = (V, E). A graph G = (V, E ) is a subgraph of G, written G G, iff V V and E E. The graph G = (V, E ) is an induced subgraph of G = (V, E) iff G is a subgraph of G and E = {(u, v) (u, v) E, u, v V }. Graphs and subgraphs may have special properties, for example they may be connected. We distinguish between undirected and directed graphs for the definition of connected and strongly connected graphs. 11
24 2. Definitions and Terminology Definition 2.10 (Connected and strongly connected graph) Given an undirected graph G = (V, E). G is connected iff for every pair of nodes u, v V a path u v exists. Given a directed graph G. G is strongly connected iff every two nodes u, v V are reachable from each other, i.e., u v and v u. Given an undirected graph G. A connected component G of G is a subgraph of G that is connected and can not be extended. In analogy for a directed graph a strongly connected component is a subgraph of G that is strongly connected and can not be extended. Example 2.2 (Strongly connected components). Consider the graph in Figure 2.1. The graph has one strongly connected component of size > 1, which contains nodes b, c, e, and f. Nodes a and d do not belong to this strongly connected component. Erdös and Rényi investigated random graphs and discovered that random directed graphs with certain properties contain one giant strongly connected component [ER60]. They showed that undirected graphs with n nodes and m = c n/2 edges contain for c < 1 many components, which are no larger than O(log n) nodes. In contrast, graphs with c > 1 already have with high probability one giant component of the size Θ(n) nodes. For graphs with directed edges the situation is more complicated [CF04]. Let d = i,j ij l i,j c n be the average directed degree, with l i,j being the number of vertices with indegree i and outdegree j and c > 0. Note, the shape of the graph (random or scale-free) and c influence i and j and thus l i,j. For d < 1 the graph contains strongly connected components of the size O(log n) nodes. Otherwise, if d > 1 the graph contains with high probability one giant strongly connected component. For a more detailed analysis of random graphs and other graph models see Bornholdt & Schuster [BS03] Storage and Traversal So far, we only considered properties of graphs. We now discuss how to store and traverse graphs. Representation of Graphs To represent graphs we may use two different formats, an adjacency matrix or an adjacency list. The adjacency matrix M is of size V V. The entry on position M(i, j) is 1 if an edge between nodes v i, v j with 1 i, j V exists, otherwise 0. The matrix representation of a graph is useful for dense graphs. The adjacency list contains one entry for every edge in the graph. Therefore this list is of size E. That representation is good for sparse graphs. As we mostly deal with sparse graphs, we use adjacency lists. 12
25 2.1. Graphs Graph Traversal To determine relationships between nodes in a graph we may use graph traversal. Common algorithms use breadth-first or depth-first search. Algorithm 2.1 shows the function breadth-first to traverse a graph G in breadth-first order starting from node u. The algorithm requires two lists, queue, which contains all nodes that still have to be considered, and traversed, which stores all descendant nodes of u. Initially, queue only contains u. In the while loop the first node of queue is removed and its child nodes are considered. If the child node has not been encountered before (element of traversed) it is added to both lists. The while loop is executed as long as queue contains nodes. The runtime of algorithms that perform a breadth-first search to find all descendants of u V is O( V + E ). Algorithm 2.1: Algorithm to perform a breadth-first search on G starting at u to find all nodes reachable from u. Data: graph G, node u 1 traversed 2 queue 3 FUNCTION breadth-first(u) 4 push(queue, u) 5 traversed traversed u 6 while queue do 7 u pop(queue) 8 foreach v children(u) do 9 if v / traversed then 10 push(queue, v) 11 traversed traversed v 12 end 13 end 14 end 15 end We may modify the breadth-first search algorithm to return the distance between u and all descendants of u. Thus, the breadth-first search algorithm may be used to target the single-source shortest-path problem [CLR01]. Another algorithm to traverse a graph is depth-first search. Algorithm 2.2 shows the function depth-first to perform a depth-first search in G starting from u. The characteristics of a depth-first search is that the algorithm searches deeper in the graph whenever possible [CLR01]. The algorithm starts at node u and considers all child nodes v of u. If v is not in list traversed, which stores all descendants of u and u itself, the algorithm calls the function depth-first again with v as new start node. As soon as a node u has no further untraversed child nodes the algorithm backtracks. As for the breadth-first search, the depth-first search reaches all nodes reachable from u in G. The runtime of algorithms that perform depth-first search to find all descendants of u V is O( V + E ). 13
26 2. Definitions and Terminology Algorithm 2.2: Algorithm to perform a depth-first search on G starting at u to find all nodes reachable from u. Data: graph G, node u 1 traversed 2 FUNCTION depth-first(u) 3 traversed traversed u 4 foreach v children(u) do 5 if v / traversed then 6 depth-first(v) 7 end 8 end 9 end Several algorithms use depth-first search as basis. For example, it is possible to assign every node in a graph a pre- and postorder value. This labeling scheme has been proposed by Dietz and Sleator [DS87] as indexing method. This scheme was used to label nodes in trees [GvKT04] and DAGs [ABJ89, TL05]. In this thesis we apply it to arbitrary graphs containing cycles in Chapter 5. Definition 2.11 (Pre- and postorder value of a node v) Let G = (V, E). Assume we start a depth-first search at u and reach v V. The preorder value, v pre is assigned as soon as node v is traversed during a depth-first search of G. The postorder value, v post is assigned after all descendant nodes of v have been traversed. After each assignment the counter for the pre- and postorder value is increased. Algorithm 2.3 shows the functions to assign every node in G a pre- and postorder value. Function pre-post ensures that every node in G will receive a pre- and postorder value. The function depth-first given in Algorithm 2.2 is extended to also assign these values. The order in which nodes of G are traversed may be determined in function pre-post. Note, the algorithm does not properly treat cycles. We apply Algorithm 2.3 to a tree T. Starting at its root node each node receives exactly one pre- and postorder value. A table of all nodes with their assigned pre- and postorder values forms an index with which reachability queries may be answered with a single query. If v is reachable from u, v must have a higher preorder and lower postorder value than w, i.e., v pre > u pre v post < u post [DS87]. This condition only holds for trees. We developed an algorithm in [TL07], called GRIPP, that uses a modified form of the pre- and postorder labeling and a specific query strategy to also apply pre- and postorder labeling on general graphs. We introduce GRIPP in Section 5.1. A further application of depth-first search is to determine which nodes of a graph are contained in a strongly connected component. There exist several algorithms to solve the problem, e.g., Kosaraju s algorithm, Tarjan s algorithm, Gabow s algorithm, or Karp s algorithm [Sed04]. We provide Kosaraju s algorithm in Appendix A. 14
27 2.2. Relational Algebra Algorithm 2.3: Algorithm to assign every node in G a pre- and postorder value. Data: graph G 1 traversed 2 pp_count = 0 3 FUNCTION pre-post 4 foreach u V do 5 if u / traversed then 6 depth-first(u) 7 end 8 end 9 end 10 FUNCTION depth-first(u) 11 traversed traversed u 12 u pre pp_count foreach v children(u) do 14 if v / traversed then 15 depth-first(v) 16 end 17 end 18 u post pp_count end 2.2. Relational Algebra This work presents a tool for the optimization of graph queries in relational database management systems (RDBMS). RDBMS use relational algebra during the evaluation of queries. In this section we briefly introduce and define terms and notation of the relational algebra as presented by Date in [Dat03] Algebra and Relations To understand relational algebra we first define algebra following Hamilton [Ham82]. Definition 2.12 (Algebra) An algebra consists of a set A together with a set of operators applied on A. An algebra allows users to build expressions by applying operators to A and combine those. Parentheses are used to group expressions, sets, and operators. In addition an algebra may define rules for its operators. Any rule may be applied to an expression, as long as the result of the expression is not changed. Therefore, not every rule is applicable to every operator. Such rules allow the exchange of operations performed in an expression. The relational algebra is one example of an algebra. The set A of this algebra consists of relations defined according to Maier [Mai83]. The notation for relational algebra used in this work is taken from Garcia-Molina et al. [GMUW02]. 15
28 2. Definitions and Terminology Definition 2.13 (Relation) Let a relation scheme S be a finite set of attributes A = {a 1, a 2,..., a n }. Corresponding to each attribute a i is a set D i, 1 i n, called the domain of values of a i. Let D = D 1 D 2 D n. A relation R on relation scheme S is a finite set of mappings {t 1, t 2,..., t j } from S to D with the restriction that for each mapping t R, t(a i ) must be in D i. The mappings are called tuples. The value of attribute a i A in tuple t R is t(a i ). A tuple t R is represented as t = {(a 1, t(a 1 )),..., (a n, t(a n ))} Operators Operators of relational algebra apply to relations or expressions. Expressions of the relational algebra construct new relations using operators. We distinguish between operators for single and for multiple relations. There exist different standard operators applied to a single relation: Selection (σ) eliminates tuples of a relation. Projection (π) eliminates attributes of a relation. Renaming (ρ) does not change the data of a relation, but changes the name of the relation or the name of attributes in the relation. Operators for multiple relations are: Set operators (,, \) the known set operators union, intersection, and difference applied to two relations. Cartesian Product ( ) produces all possible combinations of tuples from two relations. Join operators ( ) selectively join tuples from the two input relations based on a condition. In the following we formally introduce some of the operators. Selection The selection operator is applied to a single relation R. It produces a new relation R, such that R contains all tuples of R that fulfill a given conditional expression. Definition 2.14 (Conditional expression, C) A conditional expression C consists of conditions c 1,..., c i, i 1 combined by Boolean operators AND, OR, and NOT. 16
29 2.2. Relational Algebra Every condition c i of the conditional expression must evaluate to true or false. Combining these single conditions using Boolean operators returns true or false for the entire conditional expression. We now define the selection operator for a single condition c. Definition 2.15 (Selection operator, σ) Given a relation R with attributes A, and given a condition c of the form c = a θ b or c = a θ k, with a, b A, k = constant, and θ {<,, =,,, >}. The selection operator σ applied to R with condition c produces a relation as follows σ a θ b (R) = {t t R, t(a) θ t(b)} σ a θ k (R) = {t t R, t(a) θ k} When applying the selection operator σ to the relation R using a conditional expression C the resulting relation will contain all tuples t R for which the conditional expression evaluates to true. Projection The projection operator is used to produce a new relation R from a given relation R, such that R only has a subset of attributes of R. Definition 2.16 (Projection operator, π) Given a relation R with attributes A = {a 1,..., a n } and a set of attributes A with A A. The projection operator π applied to relation R produces a relation as follows π A (R) = {t[a ] t R}, with t[a ] = {(a i, t(a i )) (a i, t(a i )) t, a i A }. Join operators In a cartesian product all possible combinations of tuples from both relations occur. More often we want to join only those tuples that match in some way. The natural join operator and the theta join provide this behavior. The natural join combines tuples of two relations only if the values in common attributes are equal. The theta join relaxes this requirement by adding a conditional expression C to the join. We now define the theta join for a single condition c. Definition 2.17 (Theta join, R c S) Given two relations, R with attributes A and S with attributes B, and a condition c in the form c = aθb, with a A, b B, and θ {<,, =,,, >}. Applying the theta join operator to the two relations results in R c S = {r s r R, s S, r(a) θ s(b)}. 17
30 2. Definitions and Terminology The theta join only joins tuples for which condition c is true. If the theta join contains a conditional expression, only tuples are joined for which the conditional expression evaluates to true. In R c S we may write R.a and S.a for an attribute a contained in both relations R and S to distinguish the different origins of the attribute. This is different to the natural join, where common attributes occur only once. Additional operators There exist additional operators, which we just mention, that are required for working with relations. In commercial database systems relations are not sets of entities, but may be bags of entities, i.e., a tuple might occur more than once in a relation [GMUW02]. Duplicate-elimination operator (δ) turns a bag into a set. Grouping operator (γ) groups tuples in a relation according to their values in an attribute or a set of attributes Equivalence Rules In relational algebra we may form expressions that contain multiple relations and operators. We are able to change the order of operators in an expression applying rules. We only introduce some relevant equivalence rules. For a more comprehensive overview see [GMUW02]. Selection and Join Operator We consider the combination of join operators with selection operators. Note, we only show this rewrite rule for the natural join, for the theta join the same applies. The expression σ c (R S), with c = a θ k and k = const may be rewritten as Equations 2.1 and 2.2 show. σ c (R S) = σ c (R) S iff a A (2.1) σ c (R S) = R σ c (S) iff a B (2.2) When the selection operator contains a conditional expression we have to split the conditional expression and apply the rules described above for every condition. Example 2.3 (Reformulating a query). Given two relations Protein and Type and the query Return the names of proteins that bind DNA (stated in Type) and whose sequence starts with M. The relational algebra expression for this query is π name (σ sequence= M.* (Protein) σ description= Bind DNA (Type)). We are able to represent every expression from relational algebra as expression tree. Figure 2.2 shows two possible expression trees for the combination of a selection and projection operator on a single relation. The root of the expression tree is the result of 18
31 2.3. Cost-Based Query Optimization the expression, the leafs are the original relations, and inner nodes are intermediate relations resulting from applying operators. Using this representation we say the selection operators are pushed down in Figure 2.2(a). An alternative is to first join both relations, then select the desired tuples, and then project the required attribute. Figure 2.2(b) shows this possibility. Both expressions are equivalent, i.e., they will return the same result. When giving both expressions to a relational database system returning the answer might take longer in the second case as intermediate relations may be larger. In the next section we introduce cost-based query optimization. The knowledge of the possibilities to rewrite relational algebra expressions is important for cost-based query optimization. πname πname σ sequence= M.* AND description= Bind DNA. σ sequence= M.* σ description= Bind DNA. Protein Type (a) First select entries and then join these. Protein Type (b) First join the two relations and then select. Figure 2.2.: Two possible expression trees to answer the query Give me the names of proteins that bind DNA and whose sequence starts with M Cost-Based Query Optimization Many relational database management systems (RDBMS) use cost-based query optimization [GMUW02]. One of the aims of cost-based query optimization is to minimize the time for the execution of a given query. As we propose to use these ideas for optimizing graph queries in [Tri07] we first describe query processing and query optimization in an RDBMS in general Query Processing We now provide a short introduction to cost-based query optimization in RDBMS. For more elaborate reviews on this topic see [Ioa96, Cha98]. Figure 2.3 shows the typical workflow of query processing in an RDBMS. A user poses a query to the database system. The user usually does not enter a relational algebra expression but a query written in a query language. The most common query language for relational database systems is the structured query language (SQL) [DD97]. We assume familiarity of the reader with SQL. 19
32 2. Definitions and Terminology Query Optimization Query Relational Algebra Implementation Cost Model Result Query Parser Query Rewriter Query Planner Code Generator Query Processor Figure 2.3.: Query processing in RDBMS, including cost-based query optimization. The first step in query processing is to parse the query and rewrite it to a relational algebra expression. For query optimization the query planner uses three different sources of information. First, rules for rewriting a relational algebra expression as given in Section 2.2. Second, the RDBMS considers available implementations of operators and produces possible execution plans. We discuss implementations of operators in Section Third, the execution of a plan produces costs. These costs are accumulated over every step of the plan. Thus, every implementation requires a cost model to produce the cost of execution for this step. We discuss cost models in Section Finally, the query optimizer chooses the execution plan with lowest estimated cost. For this plan the query processor generates the code and executes the query. The result is then returned to the user Implementation of Operators The operators presented in Section 2.2 are only abstract representations of an actual process to handle data in an RDBMS. Consider the selection operator applied on a relation. To evaluate the expression the database system first has to access the data and then find tuples that fulfill the conditional expression given in the selection operator. For each step the database system requires implementations. We briefly discuss implementations for selected operators in the following. For all implementations we assume we only need to read the data of relations from disk, while the result of an operation fits into main-memory or is immediately returned to the user. Access to Relations An RDBMS stores a relation in blocks on secondary storage devices (disks). Thus, for the evaluation of an expression we have to access these blocks from disk and bring them to main-memory. RDBMS provide two possibilities to access relations, i.e., read the entire relation from disk or use indexes of a relation first. To read the relation from disk and bring it to main-memory the RDBMS has to locate the first block of the relation on disk. Every block contains a pointer to its next block. The RDBMS has to follow these pointers to retrieve the remaining blocks of the relation until all block are read. If the main-memory is not able to accommodate all blocks 20
33 2.3. Cost-Based Query Optimization of the relation some blocks have to be removed from main-memory before the entire relation is read. This will affect the choice of implementations for operators as discussed in following sections. Alternatively the RDBMS may access indexes of the relation first. Initially, an index is created for a single attribute or a set of attributes from one relation. As the index usually does not contain all attributes its space consumption is smaller than that of the relation itself. Thus, it may fit entirely into main-memory and can possibly even be kept there. The index stores a set of pointers for each unique value of the relation pointing to the location where this tuple is stored on disk. This way an index helps to efficiently find the location of tuples of interest on disk. For one-dimensional data the most common indexes are B-tree indexes [Bay72] or hash tables. For multi-dimensional data the most common indexes are grid files or R-tree indexes [Gut84]. For a description on the index structures mentioned as well as for other index structures see Database Systems: The Complete Book by Garcia-Mollina et al. [GMUW02]. Selection Operator An implementation of the selection operator must return tuples that fulfill the conditional expression given by the operator. To compute the result the RDBMS either accesses the relation from disk or employs a suitable index of the relation. In the first case the implementation sequentially reads tuples from disk and keeps or outputs those for which the conditional expression evaluates to true. The second case only applies when there exists an index on attributes given in the conditional expression. In this case the implementation may first use the index to find the tuples that fulfill the conditional expression and then specifically access these tuples on disk. Even if an index exists the RDBMS has to estimate if it is cheaper to read the entire relation sequentially from disk or to access the index and then perform a random read of the required tuples. Cartesian Product and Join Operators The cartesian product operator as well as join operators work with two relations whose tuples are combined in some way. For now, we assume at least one relation fits into main memory. If this is the case the implementations first read one relation into main-memory and then sequentially read the tuples from the second relation. Implementation of the cartesian product operator, natural join operator, and theta join operator are similar, except for the tuples that are joined. Implementations are nestedloop join, which uses two loops for the two relations, sort-merge join, where the relations are sorted on the attributes involved in the join, and hash join, where tuples are sorted into buckets and then joined. In case none of the relations fits into main-memory there exist variations of the algorithms. For a thorough description of the algorithms we refer the reader to [GMUW02]. Clearly, different implementations of operators require different amount of time to execute. To decide which implementation is best for the given relational operator we have to assign a cost function for each implementation. We describe the cost function 21
34 2. Definitions and Terminology more closely in the next section Cost Function and Query Optimization A cost function tries to correlate the cost of executing an implementation of a relational algebra operator with the time required. For the cost function several aspects are considered, which we mention in the following. First, the location of the data, i.e., is it necessary to read the input distributed in blocks from disk or is it already present in main-memory. Second, it takes the size of the relations that are input to operators and the sizes of intermediate results into account. Third, the complexity of the algorithm, e.g., the choice of the sorting or join algorithm. There exist many more factors, such as buffer size, disk seek time, layout of data on disk, available main-memory, or statistics such as the approximate number and frequency of values for an attribute that may be considered for a cost function. Access to Data The cost of accessing data depends on the location of the data. Accessing data in secondary storage devices is about 10 5 times slower than accessing data residing in main-memory [GMUW02]. Consider relation R stored in B(R) blocks on disk. To read these blocks the system initially has to locate the first block on disk, where the time depends on the disk seek time. If the blocks are continuous on disk, the system requires time proportional to the number of blocks of the relation on disk. Otherwise, more seek steps need to be performed. Depending on the number of blocks and the available main-memory (for this operation) a relation may or may not fit into main-memory. Indexes of a relation also require space, but usually not as much as the entire relation. Although it would be preferable to keep indexes permanently in main-memory it is not always possible. Thus the system must load indexes from disk to main-memory if necessary. If an index is actually used is down to the estimated costs. Size and Shape of Relations For the size of relations we have to differentiate between the size of the relation on disk, i.e., the number of blocks it occupies on disk, which is B(R) and the number of tuples in the relation T (R), also called cardinality. The number of tuples that fit into one block depends on the number and type of attributes. To estimate the cost of an implementation we require B(R) as well as T (R). Additionally, the RDBMS must know or must be able to estimate the cost for retrieving a block from disk (IO-cost) or handle a tuple in main-memory (CUP-cost). For relations stored on disk both parameters may be gathered by the RDBMS in advance. The RDBMS has also to estimate the size of the output of an operator. For some operators the size of the output is clear, e.g., for the projection operator, cartesian product, or sorting operator. For others such as the selection or join operator the RDBMS is only able to estimate the size of the result, e.g., by gathering statistics of the relation. These statistics may include the number of different values for an attribute but 22
35 2.3. Cost-Based Query Optimization also the distribution of values for which histograms are employed. For a review on this topic see Chaudhuri [Cha98]. Computational Complexity Consider the sorting operator. We assume we want to sort relation R, which fits into main-memory. For reading we have costs of B(R) IO-cost. As algorithm for sorting we use bucket sort, which has the computational complexity of O(n log(n)) [CLR01]. In the cost function we take this complexity into account by adding T (R) log(t (R)) CPUcost. Thus in total the cost for sorting R is B(R) IO-cost +T (R) log(t (R)) CPU-cost. The proposed equation to estimate the cost of sorting using bucket sort is a simplified assumption, but may be sufficient for cost-based query optimization. For cost functions of other operators we refer the reader to [GMUW02]. Query Optimizer The query optimizer receives a relational algebra expression and produces execution plans. The chosen plan is the plan with lowest estimated cost of execution. This cost is the sum of the costs for executing every single step in the execution plan (when no parallelization is applied). To find the plan with lowest cost the query optimizer considers three different sources of information as shown in Figure 2.3 on page 20. First, it uses relational algebra and the given rewrite rules to reformulate expressions. Second, there may exist multiple implementations for each operator. And third, for each implementation a cost function is provided. To find the best execution plan the query optimizer could compute for a given SQL query equivalent relational algebra expressions. It then could try all possible implementations for every operator and accumulate the cost of executing the plans. This strategy is called exhaustive search strategy. It results in the best plan given the costs, but generating all possible plans might be very time consuming. Thus, the query optimizer uses strategies to reduce the time spent on hopefully finding the best plan. Strategies a RDBMS may use are heuristic selection, branch-and-bound plan enumeration, hill climbing, or dynamic programming [GMUW02]. These strategies take less time than the exhaustive search strategy with the trade-off that they might not find the plan with lowest cost. Such strategies were first proposed by Selinger and colleagues in [SAC + 79] who developed System R, one of the first cost-based query optimizers. It was later improved by Haas et al. in [HFLP89]. Graefe and McKenna developed the Volcano framework [GM93], a query optimizer generator, which we use in this work. The optimizer generator requires the definition of rules for relational algebra expressions, knowledge about implementations for operators and their cost functions as well as their requirements. The Volcano optimizer uses dynamic programming to find the best execution plan for the query. In the following section we describe Volcano in more detail. 23
36 2. Definitions and Terminology 2.4. Volcano In this work we use Volcano, the optimizer generator [GM93], as basis for GRIcano. Figure 2.4 gives a schematic overview of Volcano. In general, to create the optimizer a database implementor has to provide the model specification and compile the optimizer. A database user may present a query to the optimizer, which returns an optimized query plan together with the appropriate algorithms and property information. In the following we describe the system in more detail. Operator Algorithm Enforcer Implementation Rewrite System property Logical property Physical property Cost function Applicability Model specification Original query SELECT cond Optimizer generator Optimizer Source Code Optimized query prop hash JOIN join_cond JOIN join_cond Compiler Optimizer prop filter SELECT cond R2 attr R1 attr R2 attr R1 attr Figure 2.4.: Schematic overview of Volcano. The database implementor specifies the capabilities of the database in advance and generates the optimizer. Given a query the optimizer returns the optimized query plan together with the appropriate algorithms and property information. Initially, a database implementor has provide the model specification, i.e., which operators does the database support, which algorithms are implemented, and which enforcers are required. Additionally, he has to specify which algorithms are implementations for which operators and define rewrite rules for operators. Example 2.4 (Operators and algorithms in Volcano). We assume the database system is able to perform join and selection. We specify operators JOIN and SELECT and as algorithms hash, merge, and filter. As merge join expects its input sorted on the join attributes we also have to define an enforcer SORT. We specify that algorithms hash and merge are implementations for JOIN and filter for SELECT. In addition we specify rewrite rules. For example, it is not relevant in which order the two input relations are used for the join operator as shown in Equation
37 2.4. Volcano Only the join condition needs to be adapted. JOIN join_cond (R1 R2) = JOIN join_cond (R2 R1) (2.3) Rewrite rules may also contain conditions as when to apply this rule. For each operator system properties, which are cardinality and record width, and logical properties, which are the schema of the resulting relation and selectivity estimates for attributes, need to be defined. These properties are independent of the algorithm applied. Algorithm dependent properties are physical properties, such as the sorting of a relation or the distribution of data. In addition, a cost function must be specified that estimates the cost for applying an algorithm. An algorithm may not always be applicable, which must also be specified. Having specified all necessary components, the optimizer generator creates optimizer source code, which is then compiled to produce the optimizer. Example 2.5 (Properties for operators and algorithms). To illustrate the different properties we use JOIN and its two implementations. Cardinality and record width are system properties. The record width of the resulting relation is the sum of the record widths of the two input relations. For the cardinality of the resulting relation the join condition and the selectivity of the join attributes needs to be considered and the size estimated. The schema of the resulting relation is the logical property. For JOIN it contains all attributes from the first relation together with the attributes from the second relation. hash and merge are two implementations for JOIN. As physical property hash produces an unsorted, while merge a sorted relation. In addition, merge may only be applied if both input relations are sorted on the join attribute, requiring a sorting step first, which causes additional cost. The two algorithms also have different cost functions. The cost function for hash is 3 (B(R1 ) + B(R2 )). In contrast, merge requires (B(R1 ) + B(R2 )), if the input relations are already sorted on the join attributes. The generated optimizer is then used to optimize a given user query. The original query only contains operators required together with their conditions. In addition, knowledge about the input relations is also required, such as the attributes, width, and cardinality of the relation. The Volcano optimizer uses, what the authors call, a directed dynamic programming approach to optimize the query. The optimizer does not create all possible equivalent expressions and plans, but only those that actually participate in sub-queries of the query. For a sub-query Volcano stores the best plan for each set of properties (sort order, distribution of data). Using this knowledge sub-plans with identical properties, but worse cost are immediately discarded. According to the authors this pruning strategy efficiently reduces the number of different plans that need to be considered. The optimizer produces as output the optimized plan, which contains not only the order and type of operators that must be applied, but also the appropriate algorithm and additional properties, such as estimated cardinality and cost, for each step. 25
38
39 3. Graph Queries This chapter describes a model for representing graphs, graph queries, languages to express graph queries, and the evaluation of graph queries. Consider the query Find all paths leading from a node with label name= Arginine to a node with label name= D-Proline whose path length is shorter than 6 in KEGG. To answer this query we first have to define the model to represent biological graphs before we proceed to answer the query. In Section 3.1 we describe a model for representing graphs. In Section 3.2 we define queries on graphs and the result of a graph query. Section 3.3 introduces PQL, a language to express graph queries. After introducing the query language we present the semantics of a graph query in Section 3.4. This chapter concludes with an overview of related work on graph models and graph query languages Data Model In this work we consider data that are structured as graphs, e.g., biological networks, social networks, or web graphs. Nodes for example are proteins, persons, or web pages. Every node may have additional information attached, such as the sequence of a protein, the name of a person, or the content of a web page. Nodes are connected by binary edges. Edges represent, for example, the participation of a protein in a reaction, two persons knowing each other, or a link on a web page pointing to another page. An edge might also have information attached. We describe our model to store such graphs in the following. Figure 3.1 depicts the schema for representing graphs. For every graph we may include additional information such as the name of the graph, its origin, or creation date. Node +node_id:integer +external_id:string Edge +edge_id:integer +from_id:node +to_id:node 0..n 0..n has has 0..n 0..n Label +label_id:integer +label_type:labelset +value:string Figure 3.1.: UML schema to represent a graph. 27
40 3. Graph Queries Nodes A node of the graph, identified by its external_id, is represented as one object in class Node. In our representation, each node receives a unique node-id. The labels of nodes are objects in class Label, which is discussed below. Each node may have many labels and each label may be used for many nodes or edges. In the implementation this many-to-many relationship between the two classes is resolved. Edges Each edge in the graph is represented as object in class Edge. This class resembles the adjacency list of a graph. The class Edge contains three attributes, edge_id, from_id, and to_id. The attribute edge_id allows us to uniquely identify an edge, while the other two attributes point to an object in class Node. We store edge labels in object Label. Each edge may have many labels and each label may be used for many nodes or edges. Labels Labels of nodes and edges are stored in object Label. This object contains attributes label_id, label_type, and value. Labels are stored according to Definition 2.4 on page 10, where each label contains a type and a value. Thus, in relation Label the type of the label is not a primitive data type, but a fixed collection of label types, such as name or sequence. Nodes and Edges in Biological Graphs In biological graphs it is not clear per se, which objects are nodes and edges. There is a common agreement that molecules, which are consumed or produced by a reaction, are nodes in a graph. The situation for proteins and reactions is more complicated. Consider KEGG [KGK + 04] for example. Only molecules consumed or produced by a reaction are nodes. Reactions themselves are represented as hyperedges, i.e., edges with many start and end nodes. These edges are labeled with names of enzymes that are responsible for the reaction. In contrast, in Reactome [JTGV + 05] molecules, enzymes, as well as reactions are modeled as nodes. Edges represent the fact that a molecule participates in a reaction. Both representations may be transformed into each other by either adding nodes for reactions and enzymes to KEGG and creating new edges or by representing reactions in Reactome as labeled hyperedges. Our model complies to the second representation. In a review by Schaefer [Sch04] a more detailed discussion of different graph types may be found Graph Queries After describing the data model that we use to represent graphs we proceed to describing queries on graphs and their evaluation. The goal of querying graphs is to retrieve certain 28
41 3.2. Graph Queries nodes and edges of the graph. To achieve this we want to pose a graph query. In the query we must be able to specify the nodes in which we are interested in. In addition, we also must be able to specify relationships between these nodes. As the query consists of nodes and relationships between nodes we consider the query as graph itself. Thus, in the following we first introduce query graphs intuitively and informally, before we proceed to the evaluation of graph queries. In Section 3.3 we introduce a query language that allows users to express graph queries. In this section we also formally specify the semantics of a graph query Query Graph We start with an example to illustrate query graphs, before we introduce in Definition 3.1 the term query graph more formally. Example 3.1 (First graph query). Assume a biologist wants to know if an organism produces D-Proline given the amino-acid Arginine in less than 6 steps. Figure 3.2 shows the query as graph. Nodes in the query graph are placeholders for nodes in a given graph. Our example contains two nodes, whose possible instantiations are restricted by the conditions name= Arginine and name= D-Proline. Edges in a query graph are placeholders for paths in the given graph. In Figure 3.2 the edge contains the restriction that only paths may be mapped that start at a node with label name= Arginine, end at a node with label name= D-Proline, and whose path length is shorter than 6. A path.length<6 B name= Arginine name= D-Proline Figure 3.2.: A query graph to find a path between Arginine and D-Proline. Definition 3.1 (Query graph) A query graph Q = (V, E, C, φ C ) is a quad-tuple consisting of a set of nodes V, a set of edges E, with E V V, a set of conditions C, and a condition function φ C that assigns conditions to nodes and edges in Q. According to Definition 3.1 a graph query also forms a graph. To each node and edge in Q conditions may be assigned. In Section 3.3 we present these conditions Evaluation of Graph Queries We now consider the evaluation of a query graph given a data graph G D. The data graph is the collection of graphs that we use for the evaluation of a query. To illustrate the evaluation of a graph query we first consider the result of the query given above on the data graph KEGG. 29
42 3. Graph Queries Example 3.2 (Evaluation of the first graph query). Figure 3.3 shows the Arginine and Proline metabolism as given by KEGG. We find one node with a label of type name whose value is Arginine and one node with the label of type name whose value is D- Proline. Figure 3.4 shows two different paths, which exist in the data graph between the two nodes that are shorter than 6. One path leads from Ornithine over L-Glutamate 5- Semialdehyde and L-1-Pyrroline 5-Carboxylate to L-Proline, while another goes directly from Ornithine to L-Proline. Apart from the two paths presented there exist other paths in the data graph, e.g., one leading over Citrulline. Figure 3.3.: The Arginine and Proline metabolism as given by KEGG. When querying a graph we search for a subgraph of G D that fulfills certain criteria. In Section 3.3 we specify possible criteria, while in Section 3.4 we give the semantics of these criteria. In general, given a data graph G D and a query graph Q the subgraph search problem is to find all subgraphs of G D for which nodes, edges, and paths of the subgraph of G D fulfill the conditions given by the query graph Q. Up to now, we have not defined what we mean by fulfill the conditions of Q. For now, let us assume every edge in Q stands for one edge in G D. Solving the subgraph 30
43 3.2. Graph Queries (a) Graph 1. (b) Graph 2. Figure 3.4.: Two result graphs for the graph query evaluated on KEGG. search problem would then essentially be equivalent to solving the subgraph isomorphism problem [CLR01]. Definition 3.2 (Graph isomorphism) Given graphs G 1 = (V, E) and G 2 = (V, E ). An isomorphism of G 1 and G 2 is a bijection f : V V such that for every (u, v) E there exists (f(u), f(v)) E. Finding subgraphs of G D that are isomorphic to Q is NP-complete [CLR01]. In the case when nodes or edges in G D have labels and Q contains conditions on those labels the problem may be solved more easily as shown by Koyutürk and colleagues in [KGS04]. The reason is that for a given condition on a node we may find only a few nodes in G D, depending on the selectivity of the condition. Thus, not all possible subgraphs of G D that are isomorphic to Q are of interest, but only those that contain specific nodes or edges. Using this knowledge we may start the search at this node or edge. Solving the graph isomorphism problem would be sufficient if we only allow edges in Q to stand for edges in G D. In this work we assume edges in Q may also stand for paths in G D. Thus finding an isomorphic subgraph to the query graph is not sufficient, but we want to find homeomorphic subgraphs of Q in G D [LR78]. Definition 3.3 (Graph homeomorphism) Let G 1 = (V, E) and G 2 = (V, E ). An homeomorphism of G 1 and G 2 is a bijection f : V V such that for every (u, v) E there exists a path f(u),..., f(v) in G 2. Definition 3.3 defines graph homeomorphism in the general case. This definition basically means an edge in G 1 may stand for an edge or a path in G 2. Thus, in our case, given a query graph Q we try to find all homeomorphic subgraphs in the data graph 31
44 3. Graph Queries G D. In its general case, i.e., if no bindings of nodes in Q to nodes in G D is known, the subgraph homeomorphism problem is also NP-complete [LR78]. Like Koyutürk and colleagues in [KGS04] we assume nodes and paths in the data graphs have labels and the query graph contains conditions on those labels. The conditions and their semantics are introduced in the following section when we introduce the query language for graph queries. For one query graph Q multiple homeomorphic subgraphs whose labels or properties comply to the conditions of Q in G D may exist. We therefore introduce the match graph of Q in G D. Definition 3.4 (Match graph G M (Q)) Given a query graph Q and a data graph G D. Let G = {G 1,..., G n } be the set of all homeomorphic subgraphs of Q in G D whose labels or properties comply to the conditions of Q. The match graph for Q in G D, written G M (Q) is defined as G M (Q) = G i G i G We give an example for a match graph in the following section, where we introduce the pathway query language, PQL, as syntax to express query graphs Pathway Query Language The syntax and semantics of the Pathway Query Language (PQL) was first described 2005 by Leser [Les05a]. A more detailed description may be found in [Les05b]. Two diploma theses suggested extensions of the language [Ahl06, Lis07]. In this work we enhance PQL in the following points: Ability to query multiple graphs simultaneously using the FROM clause Introducing the LET clause to declare variables for nodes and paths Possibility to use ordinary relations in the FROM clause Graphs in PQL In the previous section we introduced the data graph G D, the query graph Q, and the match graph G M. In this work we search for homeomorphic graphs of Q in G D, which form G M. A user may only be interested in parts of G M, such as nodes. Thus, we allow users to specify a select graph G S, which is a subgraph of Q, that specifies the parts of G M, which should be returned. When we restrict G M based on the specifications of G S we get the result graph G R. PQL allows users to specify Q and G S as we show in this section. In addition, we formally specify which nodes and edges are contained in G M and G S based on Q and G S. 32
45 3.3. Pathway Query Language To summarize, evaluating a PQL query conceptually requires five different graphs, namely data graph(s) G D query graph Q match graph G M, with G M G D select graph G S, with G S Q result graph G R, with G R G M Example 3.3 (Graphs for the query from Example 3.1). To illustrate the different types of graphs we use the same query as in the last section, where we wanted to find all paths in KEGG between a node with label name= Arginine and a node with label name= D-Proline that are shorter than 6. Figure 3.5(c) shows the query graph Q for this query. It contains nodes A and B, with conditions name= Arginine and name= D-Proline, respectively. In addition, it contains an edge with the condition that a directed, simple path between nodes A and B whose path length is shorter than 6 exists. The data graph G D is a small part of the metabolic network of KEGG shown in Figure 3.5(a). Figure 3.5(b) shows the match graph for Q in G M. We bind the node with label Arginine in G D to node A of Q and the node with label D-Proline to node B. For the path we find four bindings in G D. There exist other paths between these two nodes, but these are longer than 6. In a PQL query we may also specify the subgraph that is returned to the user. This subgraph is the select graph G S, which is a subgraph of Q. Figure 3.5(d) shows the G S. In this case we are only interested in nodes of G D bound to A and B of Q. Figure 3.5(e) shows the result graph G R of the query. In this case we get only one binding for each node variable as the conditions on the node variables are very restrictive. Clearly, for other queries we might find more bindings as following examples show Syntax In this section we specify the syntax of PQL. We start with the general syntax of a PQL query, before we specify the different parts separately. General Syntax and Examples The syntax of PQL is closely related to the syntax of SQL. A PQL query consists of five parts a SELECT, FROM, LET, WHERE, and HAVING clause. Figure 3.6 shows the syntax of a PQL query. To give a first impression of the query language we give two examples. 33
46 3. Graph Queries (a) Data graph G D: metabolic network of KEGG. (b) Match graph G M (Q). A path.length < 6 B name= Arginine name= L-Proline (c) Query graph Q: node A, node B, and path P. A B (d) Subgraph specification G S: node A, node B. (e) Result graph G R. Figure 3.5.: The five different graphs of a PQL query. Example 3.4 (PQL statement for graph query from Example 3.1). In the following we provide the PQL statement for the query graph from Example 3.1. SELECT A, B FROM Kegg k LET node A IN k, node B IN k, path P IN k WHERE A.name = Arginine AND B.name = D-Proline AND P.path = A[->]B AND P.length < 6; 34
47 3.3. Pathway Query Language SELECT, subgraph-specification,, FROM graph, relation, LET variable-type variable-name IN graph WHERE where-condition HAVING having-condition Figure 3.6.: PQL query syntax. Example 3.4 shows that the data graph Kegg is specified in the FROM clause. In the LET clause we may state node and path variables. In the WHERE and HAVING clause a user may specify conditions on node and path variables. Finally, the SELECT clause specifies the select graph. In the following we formally specify the syntax of the different parts of a PQL query. We first specify the FROM and LET clause, before we specify conditions on nodes and paths in the WHERE and HAVING clause. We conclude the language specification with the SELECT clause. To illustrate the clauses we introduce a second example for which the query is built during the introduction of the syntax of PQL. Example 3.5 (Running example). Consider Figure 1.1 on page 3. The conversion of α-d-glucose to pyruvate involves 22 enzymes. Assume a biologist is interested in all proteins that interact with one of those enzymes present in mammals and the direct interaction partners of those enzymes. No single data source for biological graphs supplies all required data. Thus, we use two different sources, namely KEGG for the conversion of substances and Intact for protein-protein interactions. FROM clause Let G D be the set of graphs consisting of G 1,..., G n, n 1, which are input to the graph query. We may specify this set of graphs in the FROM clause as Figure 3.6 shows. FROM clause := FROM graph, (graph)* graph := graph_name (alias)? alias := ( A... Z a... z )+ Every data graph in the database may be referred in the FROM clause by its name. Multiple graphs are separated by colons. Every graph in the FROM clause may optionally 35
48 3. Graph Queries be associated to an alias for this graph. The alias of the graph must be unique in the query. LET clause The LET clause specifies the node and path variables of the query graph Q. In addition, it allows a user to state of which graph nodes and paths must be bound to node and path variables in the query. Every node and path variable must be assigned to exactly one data graph. LET clause variable variable_type variable_name graph := LET variable (, variable)* := variable_type variable_name (IN graph)? := node path := ( A... Z a... z )+ := graph or graph alias specified in FROM clause A variable in the LET clause stands either for a node or an edge in the query graph Q. Thus, a variable in the LET clause may be of two different types, node and path. The variable_name is the name of the variable, by which it is referred to in other clauses. The variable_name must be unique within the query. The last part of the variable declaration consists of the specification from which graph nodes or paths must be bound to this variable. We may only use graphs or graph aliases specified in the FROM clause. If the FROM clause contains only one graph this last part may be omitted. WHERE clause The WHERE clause specifies conditions on variables in Q. All conditions must be connected by AND or OR 1. WHERE clause condition connector := WHERE condition (connector condition)* := node_condition path_condition := AND OR Node conditions We first describe conditions on node variables of the query graph Q. The node variables of Q are given in the LET clause. In the WHERE clause we may pose conditions on nodes in Q, e.g., the node has a specific label. Given two variables A and B that are specified as node variables in the LET clause, i.e.,... LET node A, node B.... We specify the following conditions on nodes. 1 Note, PQL currently does not support negation or parenthesis. 36
49 3.3. Pathway Query Language A = B A!= B A.type op n A.type text_op string A.type op B.type A HAS (ontology hierarchy, node-concept) type := any label type of G D (in out)degree op := =!= < > <= >= text_op := op LIKE n := numeric value In the following we give an example for conditions on node variables in the WHERE clause. Example 3.6 (WHERE clause with node conditions). In our running example we use six node variables in the LET clause. The following PQL statement shows the conditions for those node variables.... FROM Kegg k, Intact i LET node A IN k, node B IN k, node C IN k, node D IN k, path O IN k, path P IN k, path Q IN k, node X IN i, node Y IN i, path R IN i WHERE A.name = α-d glucose AND B.name = Pyruvate AND C HAS (Type, Reaction ) AND D HAS (Type, Enzyme ) AND D HAS (NCBI Taxonomy, Mammal ) AND X.name = D.name AND Y HAS (Type, Protein )... In Section 3.4 we formally define the semantics of the conditions. For now we describe the result intuitively. On node variable A we bind nodes from KEGG that have a label with type name whose value equals α-d Glucose. In similar fashion, on node variable B we bind nodes from KEGG that have a label with type name whose value equals Pyruvate. For node variable C we consider a Type hierarchy that contains concepts for node types, which biological graphs use, such as reaction, protein, enzyme, or catalytic enzyme, and their relationships. We may bind all nodes of KEGG that have a label of type Type whose value is either Reaction or the name of a successor concept of Reaction in the 37
50 3. Graph Queries Type hierarchy, e.g., catalyzed and self-catalyzed. Similarly on node variable D we may bind all nodes of KEGG that have a label of type Type whose value is Enzyme or a successor concept of Enzyme in the Type hierarchy. In addition these nodes must have a label of type NCBI Taxonomy whose value is Mammal or a successor concept of Mammal, such as Human, Mouse, or Chimp, in the NCBI Taxonomy. To node variables X and Y we may only bind nodes of the data graph Intact. For node variable Y we may bind nodes that have a label of type Type with the value Protein or a successor concept of Protein in the Type hierarchy. For node variable X the case is more complicated. We may bind to X all nodes that have a label of type name whose value is equal to the value of a label of type name from a node bound to node variable D. Note, these are not the same nodes as they originate from different graphs. Those two nodes must share the same label. Path conditions Until now, PQL queries are not much more powerful than searching keywords in a database of node labels. Thus, we now introduce path conditions. Given variables A and B specified as nodes and P and R specified as paths in the LET clause, i.e.,... LET node A, node B, path P, path R.... We specify the following conditions for paths: P.path = A[- -> <->]B P.start = A P.end = B P.edge = [- -> <->] P.length num_op n num_op := =!= < > <= >= n := positive integer For every path variable in the LET clause we only bind directed paths to this variable, unless otherwise specified in the WHERE clause using P.path or P.edge. The direction of edges in a path is specified by providing one of the options from [- -> <-> <-]. For - we consider all edges, regardless their direction. If -> is given the path may only contain directed edges, while for <-> each edge in the path must be bi-directional. Example 3.7 (WHERE clause with path conditions). In our running example we have path variables O, P, Q, and R. On O, P, and Q we may only bind paths from KEGG, while on R only paths from Intact may be bound. On O we bind paths that start at a node bound to A and end at a node bound to C, while for P the paths must start at a node bound to C and end at a node B. On Q we bind paths of KEGG that start at a node bound to C leading to a node bound to D, and whose length equals one. In addition all edges on this path must be bi-directional, i.e., for an adjacent node pair u, v there must exist both directed edges (u, v) and (v, u). 38
51 3.3. Pathway Query Language To find all interacting proteins for the enzymes in Intact we require the path variable R. We bind all paths from Intact regardless of the direction of the edges to paths that start at a node bound to X and end at a node bound to Y, whose length is shorter or equal to two.... FROM Kegg k, Intact i LET node A IN k, node B IN k, node C IN k, node D IN k, path O IN k, path P IN k, path Q IN k, node X IN i, node Y IN i, path R IN i WHERE... AND O.path = A[->]C AND P.path = C[->]B AND Q.path = C[<->]D AND Q.length = 1 AND R.path = X[-]Y AND R.length <= 2... HAVING clause Assume in our running example a user is interested in the result only if there exists exactly one binding for node variable A. This means, there must exist only one node with label (name, α-d glucose ) in KEGG. This example shows we want to pose a condition on the number of bindings for a node or path variable. To specify this we introduce the HAVING clause for PQL. HAVING clause := HAVING condition (connector condition)* connector := AND The conditions on nodes or paths filter the result according to the number of existing bindings. Given two variables A and P that are specified as node and path in the LET clause, i.e.,... LET node A, path P.... We specify the following conditions: A.count P.count num_op n num_op n num_op := =!= < > <= >= n := positive integer 39
52 3. Graph Queries Example 3.8 (HAVING clause). Consider our running example again. A user might only be interested in an answer to the query if only one node with label ( name, α- D glucose ) and one with label ( name, pyruvate ) exists. In addition the number of different paths between both nodes should not exceed five. The following HAVING clause expresses this.... FROM Kegg k, Intact i LET node A IN k, node B IN k, node C IN k, node D IN k, path O IN k, path P IN k, path Q IN k, node X IN i, node Y IN i, path R IN i WHERE... HAVING A.count = 1 AND B.count = 1 AND P.count <= 5... SELECT clause Until now we only specified the query graph Q and its bindings to nodes and paths in the data graph G D. The query graph may contain node and path variables in which a user is not interested in. Thus, PQL offers a possibility to state which node and path variables should occur in the result using the SELECT clause. SELECT clause := SELECT subgraph-specification (, subgraph-specification)* Given two variables A and P that are specified as node and path variable in the LET clause, i.e.,... LET node A, path P.... We specify the following subgraph-specifications in the SELECT clause: * A P P.start P.end n := positive integer Example 3.9 (Complete PQL statement for the running example). We complete the PQL statement of our running example by adding the SELECT clause. For Example 3.5 we are only interested in the proteins that interact with the enzymes and the shortest paths between them. Thus, we give the complete PQL statement in the following. 40
53 3.3. Pathway Query Language SELECT X, Y, R FROM Kegg k, Intact i LET node A IN k, node B IN k, node C IN k, node D IN k, path O IN k, path P IN k, path Q IN k, node X IN i, node Y IN i, path R IN i WHERE A.name = α-d glucose AND B.name = Pyruvate AND C HAS (Type, Reaction ) AND D HAS (Type, Enzyme ) AND D HAS (NCBI Taxonomy, Mammal ) AND X.name = D.name AND Y HAS (Type, Protein ) AND O.path = A[->]C AND P.path = C[->]B AND Q.path = C[<->]D AND Q.length = 1 AND R.path = X[-]Y AND R.length <= 2 HAVING A.count = 1 AND B.count = 1 AND P.count <= 5 To illustrate this PQL statement Figure 3.7(a) shows the query graph. Nodes and edges contain conditions given in the WHERE clause of the PQL query. Figure 3.7(b) shows the select graph G S. From the entire result we are only interested in nodes bound to node variables X and Y and paths bound to path variable R PQL and Non-graph Relations So far, we only allowed graphs to be input to a PQL query. This might not always be sufficient as the following example shows. Example 3.10 (Introducing non-graph relations to PQL). In Example 3.9 we provide the complete PQL statement to identify all proteins that interact directly or via another component with an enzyme that participates in the conversion of α-d glucose to pyruvate. Assume we also have a relation called Microarray_experiment that stores information about microarray experiments [THC + 99], i.e., gene name, fold change for this gene, and experiment number. We are now only interested in proteins that directly or indirectly interact with enzymes if both, the protein as well as the enzyme with which it interacts with, is up-regulated in the same microarray experiment. Although, we might have stored all relevant data in the same database, i.e., graphs and other relations, we have no opportunity to express a query such as the one of 41
54 3. Graph Queries name=d.name R.length <= 2 Intact X Y HAS (Type, Protein ) KEGG Q.length = 1 D HAS (Type, Enzyme ) HAS (NCBI Taxonomy, Mammal ) P.count <= 5 A C B name= α-d glucose count = 1 HAS (Type, Reaction ) name= Pyruvate (a) Query graph Q. R Intact X Y (b) Select graph G S. Figure 3.7.: The query and select graph specified in the PQL query for Example 3.5. Example 3.10 in a PQL statement. Thus, we extend PQL to also use relations that are not part of the graph model (see Section 3.1) as input to a PQL query. Extension to the FROM clause The FROM clause of PQL contains the graphs that are input to the graph query. It is a natural extension to also state all non-graph relations in the FROM clause of a PQL query as well. Thus, the FROM clause of a PQL query is as follows. FROM clause := FROM graph, (graph relation)* graph relation := graph_name (alias)? := relation_name (alias)? alias := ( A... Z a... z )+ As for the alias for the graph the alias or tuple variable of the relations must be unique within the query. 42
55 3.4. PQL Semantics Extension to the WHERE clause To use non-graph relations we also have to extend the syntax of the WHERE clause. This extension is straight forward. If an expression only affects attributes of a relation the same rules apply as in an SQL statement. There is only a difference when we want to pose a condition on node variables involving a relation. In this case the definition of the semantics of node conditions must be extended. Given a node variable A specified in the LET clause and relation R with attribute attr specified in the FROM clause. We define the following condition for nodes. A.type op R.attr op := =!= < > <= >= Example 3.11 (PQL query with non-graph relations). To illustrate the usage of relations in a PQL query we extend the PQL statement from Example 3.9 to also account for the expression level of different enzymes and proteins as given in Example SELECT X, Y, R.shortest FROM Kegg k, Intact i, Microarray_experiment ma, Microarray_experiment mb LET node A IN k, node B IN k, node C IN k, node D IN k, path O IN k, path P IN k, path Q IN k, node X IN i, node Y IN i, path R IN i WHERE... AND D.name = ma.name AND ma.fold_change > 2 AND mb.fold_change > 2 AND mb.name = Y.name AND ma.exp_no = mb.exp_no HAVING PQL Semantics There may be many interpretations on the meaning of a PQL query with multiple graphs, relations, node and path variables. We first start by giving the semantics of node conditions, path conditions, HAVING conditions, and the subgraph specification Semantics of Node Conditions Let A and B be two node variables specified in the LET clause. A node condition in the WHERE clause is interpreted as Boolean function. Given a mapping of node variables A and B to nodes in G D a condition evaluates to TRUE or FALSE. A = B returns TRUE if A and B are mapped to the same node in G D, and otherwise FALSE. 43
56 3. Graph Queries A!= B returns TRUE if A and B are not mapped to the same node in G D, and otherwise FALSE. A.type op n returns TRUE if the node of G D mapped to node variable A in Q contains a label of type type, whose value is equal to, not equal to, smaller than, or larger than (depending on op) the given numeric value, and otherwise FALSE. A.type text_op string returns TRUE if the node of G D mapped to node variable A in Q contains a label of type type, whose value is equal to, not equal to, smaller than, larger than, or LIKE (depending on text_op) the given string, and otherwise FALSE. A.type op B.type returns TRUE if the node of G D mapped to node variable A contains a label of type type whose value is equal to, not equal to, smaller than, or larger than (depending on op) the value of the label of the node mapped to B whose type equals type, and otherwise FALSE. A HAS (ontology hierarchy, node-concept) returns TRUE if the node of G D mapped to A has a label of type ontology hierarchy with the value node-concept or any successor of node-concept in the ontology or hierarchy specified by ontology hierarchy, and otherwise FALSE. A.type op R.attr returns TRUE if the value of label with type type of the node mapped to A is equal to, not equal to, smaller than, or larger than (depending on op) the value of tuple t for attribute attr, t(attr) and otherwise FALSE Semantics of Path Conditions Let A and B be node variables and P be a path variable specified in the LET clause. A path condition is interpreted as a Boolean function. Given an assignment of path variables P to paths in G D a condition evaluates to TRUE or FALSE. P.path = A[- -> <->]B returns TRUE if the simple path assigned to P = v 0, v 1,..., v k with i = 1, 2,..., k has v 0 set of nodes bound to A, v k set of nodes bound to B, and (v i 1, v i ) E(G D ) (v i, v i 1 ) E(G D ) for A[-]B (v i 1, v i ) E(G D ) for A[->]B (v i 1, v i ) E(G D ) (v i, v i 1 ) E(G D ) for A[<->]B otherwise FALSE. P.start = A returns TRUE if the simple path assigned to P begins at a node bound to A, and otherwise FALSE. P.end = B returns TRUE if the simple path assigned to P ends at a node bound to B, and otherwise FALSE. P.edge = [- -> <->] returns TRUE if the simple path assigned to P = v 0, v 1,..., v k with i = 1, 2,..., k only contains edges such that (v i 1, v i ) E(G D ) (v i, v i 1 ) E(G D ) for [-] 44
57 3.4. PQL Semantics (v i 1, v i ) E(G D ) for [->] (v i 1, v i ) E(G D ) (v i, v i 1 ) E(G D ) for [<->] for all i = 1, 2,..., k, and otherwise FALSE. P.length num_op n returns TRUE if the simple path assigned to P has a length equal to, not equal to, shorter than, or longer than (depending on num_op) n, otherwise FALSE Semantics of HAVING Conditions Let A be a node variable and P be a path variable specified in the LET clause. A node condition in the HAVING clause is interpreted as Boolean function. Given an assignment of node variable A to nodes in G D or a path variable P to paths in G D a condition evaluates to TRUE or FALSE. A.count num_op n returns TRUE if the number of different nodes from G D assigned to node A equals, not equals, is smaller, or is larger than (depending on num_op) the numeric value n, and otherwise FALSE. P.count num_op n returns TRUE if the number of different paths from G D assigned to path P equals, not equals, is smaller, or is larger than (depending on num_op) the numeric value n, and otherwise FALSE Semantic of the Subgraph Specification Let A be one of the node variables and P be one of the path variables specified in the LET clause. To construct the result graph G R all subgraph-specification elements in the SELECT clause are evaluated successively. If the element is * all unique nodes and paths bound to any node or path variable of the LET clause are added to G R. If the element is A then all unique nodes bound to A are added to G R. If the element is P then all unique paths (nodes and edges) bound to P are added to G R. If the element is P.start then all unique start nodes of paths bound to P are added to G R. If the element is P.end then all unique end nodes of paths bound to P are added to G R Conversion to Relational Algebra The syntax of PQL is closely related to the syntax of SQL. Thus, we also convert PQL statements to relational algebra in the same way as SQL statements are converted. To convert an SQL statement into a relational algebra expression we require the following steps [GMUW02]: 45
58 3. Graph Queries Start with the tuple variables given in the FROM clause and create the cartesian product of their tuples. If the WHERE clause contains conditions, apply the selection operator σ on this product with the conditions of the WHERE clause. Apply the projection operator π with the attributes in the SELECT clause. The expression created this way may then be transformed using the equivalence rules for relational algebra operators given in Section 2.2. In the same spirit we want to convert a PQL statement to a relational algebra expression. The problem is, a PQL query may contain multiple graphs, node and path variables, and relations with their tuple variables. We have to consider the node and path variables as well as the tuple variables for the conversion of a PQL query. The first step of the conversion is to bind all tuples of a relation to its tuple variable, bind all nodes and paths of the graph to node and path variables given in the LET clause. Thus, the evaluation of a PQL query is as follows: Start with the node and path variables given in the LET clause and the tuple variables given in the FROM clause and create the cartesian product of all nodes bound to node variables all paths bound to path variables all tuples bound to tuple variables If the WHERE clause contains conditions apply the selection operator σ on this product with the conditions of the WHERE clause. Apply the projection operator π on the result with the node and path variables stated in the SELECT clause. If the PQL statement contains an aggregate function we also have to use an aggregation operator. Thus, converting a PQL query to an algebra expression conceptually follows the same principle as in SQL. The only difference is that PQL also uses node and path variables. So far, we have not defined what node and path variables are. We will do this in the following. Binding Nodes and Paths to Variables To store graphs we map our graph data model to the relational data model. In addition, we also translate PQL statements into an algebra expression. Thus, node and path variables must also comply to the relational data model. While tuple variables in a PQL statement are treated in the same was as in an SQL statement, we have to specify the data model for node and path variables. We define a virtual relation for each of the node and path variables given in a PQL statement. Figure 3.8 shows the two virtual relations that represent nodes and paths of a graph. We discuss in Chapter 4 how and when these virtual relations are filled. Depending on the PQL statement and the condition on nodes and paths these virtual relations may contain only few nodes or paths of the graph. 46
59 3.4. PQL Semantics Nodes node_id external_id label_id label_type value (a) Node view Paths path_id start end length node_id position (b) Path view Figure 3.8.: Virtual relations Nodes and Paths. The view Nodes holds all nodes together with their annotation, while the view Paths may hold information for every cycle-free path of the graph. Example 3.12 (Expression tree for PQL statement). To show the translation of a PQL statement to a relational algebra expression we consider the PQL statement from Example 3.4. SELECT A, B FROM Kegg k LET node A IN k, node B IN k, path P IN k WHERE A.name = Arginine AND B.name = D-Proline AND P.path = A[->]B AND P.length < 6; When converting this PQL statement into an algebra expression we require twice the virtual relation Nodes and once Paths. Figure 3.9 shows two possible expression trees for the given PQL statement. The tree in Figure 3.9(a) shows the tree that is initially produced, while Figure 3.9(b) shows a tree, which is transformed according to relational algebra rules. Note, the views that stand for node and path variables are not resolved. How to resolve these views and rules for rewriting are described in Chapter 4. Evaluation of the HAS function The HAS function is a special construct in the WHERE clause of a PQL query. It takes as argument the name and one concept of a hierarchy or an ontology. We consider a hierarchy or an ontology as graph as well. Thus, to retrieve all successor nodes of a given concept we pose a PQL statement. Example 3.13 (PQL query to retrieve successor concepts of a given concept). Assume we want to retrieve all successor concepts of macromolecule from the Type hierarchy. If we consider the Type hierarchy as graph we use the following PQL statement to get an answer to this question. 47
60 3. Graph Queries π A, B π A, B σ P.path = A[->]B σ A.name= Arginine AND B.name= D-Proline AND P.path = A[->]B AND P.length < 6 Paths P σ A.name= Arginine σ P.length < 6 σ B.name= D-Proline Nodes B Nodes A Nodes B (a) Expression tree 1. Nodes A Paths P (b) Expression tree 2. Figure 3.9.: Possible expression trees for the PQL statement. SELECT B FROM Type t LET node A IN t, node B IN t, path P IN t WHERE A.concept = macromolecule AND P.path = A[->]B This basically means the construct HAS is an abbreviation for the type of query given in Example More specifically, the following holds: HAS (hierarchy, value ) = SELECT B FROM hierarchy LET node A, node B, path P WHERE A.concept_name = value AND P.path = A[->]B When converting a PQL statement to a relational algebra expression we thus expand the HAS condition. To find all successor concepts of an hierarchy or ontology the same algorithms and optimizations are applicable as for optimizing graph queries Related Work In this section we discuss related work on graph queries. Angles and Gutierrez present a survey on models to represent graphs and graph query languages in [AG05]. In the following we also discuss the works presented there, with main focus on the following points. First, the data model and its ability to store graphs as well as non-graph data. Second, we discuss the capabilities, usage, and optimization possibilities of existing languages. Third, we discuss if the data models together with their respective query languages are implemented in existing systems and if these systems are already used for storing biological graphs. 48
61 3.5. Related Work Deductive Databases Several works discuss the possibility to represent graph structured data using first order logic. Bancilhon and Ramakrishnan introduce in [BR86] Datalog as possibility to represent and query graphs. Datalog is a logical query language [GMUW02], which is inspired by Prolog. Nodes and edges in Datalog are represented by predicates. To access data in other data sources these data also must be modeled as predicates. In Datalog a graph query is expressed using rules on predicates. Using these predicates it is possible to form complex queries that may contain cycles or regular expressions over paths, i.e., the path should not contain a node with a specific label. This is certainly an advantage over PQL, but on the other hand, the formulation of graph queries using rules is quite complex. Rules may be combined to form programs as presented for G-Log by Jaredaens and colleagues in [PPT95]. Writing rule-based programs might be difficult for scientists used to programming languages and SQL, thus Consens and Mendelzon presented in [CM90] GraphLog, a graphical query language to express graph queries. This query language is an extension to G and G+ presented by Mendelzon and Wood in [MW89] and may be led back to Datalog. As the complexity of evaluating arbitrary combination of rules is high, GraphLog does not allow queries that contain cycles for performance reasons. Object-oriented Graph Representation In the object-oriented model nodes, edges, and paths are represented as objects of specific classes. Every class may contain further attributes, e. g., the class for nodes may contain the attribute name. Güting presents in [Güt94] GraphDB, a database that uses such a data model. This data model allows the storage of any type of graph. One drawback is, for any new attribute the classes need to be rewritten. Accessing data from other data sources is possible if a class for these data exists and the relationship between nodes or edges and these data is defined. To query GraphDB a scientist may use the proposed query language. The query language allows the specification of the subgraph of interest, which may also contain paths or regular expressions over path labels. This clearly is an advantage over PQL, but GraphDB never left the stadium of a proposal for a data model and query language. In the same style Amann and Scholl propose in [AS92] a graph data model and the query language GRAM. The model assumes that there exist classes for nodes and edges. The query language is based on regular expressions over these classes to find paths in a graph. Sheng and colleagues present in [SÖÖ99] an object-oriented graph model and the query language GOQL. To query graphs they introduce new capabilities to OQL, the standard query language for object-oriented databases. This extension is capable to express graph queries, including regular expressions over path labels. Object-oriented database system and the capabilities of the proposed query languages make these systems interesting for storing and querying graphs, but implementations of the query languages never left the status of prototypes. For example Sheng and colleagues [SÖÖ99] implemented GOQL to VStore, an object-oriented database system. 49
62 3. Graph Queries VStore is based in the o-algebra, an algebra similar to relational algebra, but adapted for object oriented database systems. In their work the authors introduce new operators for the o-algebra to capture the capabilities of GOQL, but they do not give rewrite rules or special implementation for path expressions as we introduce for PQL in Section 4. In addition, research on object-oriented databases is currently much less popular than for relational database systems. Thus, in our work we prefer to store and query graphs in an RDBMS and not in an object-oriented environment. Semi-structured Data and XML Abiteboul describes in [Abi97] the main aspects of semi-structured data and argues for the need of query languages for this type of data. The structure of these data is irregular and only implicitly given by the data. Intuitively, semi-structured data may be considered as graphs (in most cases they are trees or DAGs). A special case of semi-structured data are data that comply to the Extensible Markup Language (XML) [BPSM + 06]. XML data usually form a tree, where nodes are elements of the XML document and edges are given by the nested structure of the document. In addition, there exists an order between sibling nodes. The tree structure is broken (then forming DAGs or graphs) when XPointer [DJG + 02] or XLink [DMO01] elements are used in the document. The World Wide Web Consortium (W3C) recommends XML Query (XQuery) [DFF + 07] as query language. This language is based on FLWOR expressions (FOR, LET, WHERE, ORDER BY, and RETURN), which is similar to SQL and PQL. A subpart of XQuery is XPath, which allows the specification of path structured query graphs to retrieve parts of an XML document. The main drawback of XQuery compared to PQL is that it is only capable to query tree structured XML documents, thus making it inapplicable for storing and querying biological networks. Prior to XQuery other query languages have been proposed to query semi-structured data that form general graphs. In [AQM + 97] the group around Abiteboul proposes the query language Lorel. This language is specifically designed to cope with schema heterogeneity and additionally allows regular path expressions. The authors briefly introduce the implementation in the Lore system, which also performs query optimization. In [BDHS96] Buneman and colleagues present a query language for unstructured and semi-structured data called UnQL. They also introduce new operators to rewrite the queries to a calculus. The group around Shasha present in [GS02] GraphGrep, a method for querying graphs. They introduce the query language Glide, which combines features from XPath and Smart, a language to query chemical molecules. To evaluate graph queries efficiently they index the data, which we discuss more closely in Section 5.6. Most systems proposed above never left the status of prototypical implementations. Current systems to store XML data are native XML databases such as exist [htt08] or XML extensions to RDBMS. These systems provide XQuery as query language. As much research is put into optimization of queries on XML documents as well as on index structures for XML documents we discuss some works in Section 4.3 and in Section 5.6, respectively. Several groups use XML to store biological data. The most prominent exchange for- 50
63 3.5. Related Work mats for biological graphs are the Systems Biology Markup Language (SBML) and BioPAX. Both exchange formats are based on XML with XLink elements. For an overview over the predominant formats SBML, PSI, MI, and BioPAX we refer the reader to Strömbäck and Lambrix [SL05]. Although XML allows the storage of biological graphs, querying the data using XQuery is not possible as especially XLink elements cannot be resolved. Thus, making the result of an XQuery possibly incomplete. Sohler and colleagues present in [SHZ04] ToPNet, a tool for storing and querying biological graphs. They query ToPNet using XML-based templates. In [SZ05] they give algorithms to find matching subgraphs to such an XML template. They only allow users to query for subgraphs that are isomorphic to the XML template. In PQL we also allow the search for homeomorphic subgraphs. Semantic Web and RDF The semantic web aims to provide semantic information about resources on the web. A W3C recommendation for the representation of information is the Resource Description Framework (RDF) [Bec04]. RDF allows the representation of semantic information as graphs. As for semi-structured data several query languages have been proposed. Karvounarakis et al. present in [KAC + 02] the query language RQL, which is inspired by OQL. For an RQL query graph the matching subgraphs may be found by subgraph isomorphism, but not by subgraph homeomorphism. The W3C also provides a recommendation for querying RDF graphs, called SPARQL [PS08]. As for RQL this language only allows the specification of query graphs in which the edges are mapped to edges in the data graph, but not to paths in the data graph. Several groups presented extensions to the query language SPARQL that allow regular path expressions. Kochut & Janik introduce in [KJ07] SPARQLeR, Alkhateeb and colleagues present in [ABE09] PSPARQL, and Detwiler et al. describe in [DSB08] GLEEN. For the efficient execution of SPARQL queries indexing techniques for RDF graphs have been proposed. Heese et al. [HLQR07] suggested to index the RDF graph for graph patterns in analogy to index frequently queried attributes in RDBMS. In addition, Hartig and Heese presented in [HH07] operators to logically represent a SPARQL query. They also present transformation rules to optimize such a query. We discuss both in Section 4.3 more closely. We may not be able to transfer knowledge gained from these works to the evaluation of PQL queries. The reason is we intend to find homeomorphic subgraphs, i.e., where an edge in the query may stand for a path in the data graph, which is more complex than to find isomorphic subgraphs, the intention of SPARQL. In contrast, the authors of PSPARQL present algorithms that find homeomorphic subgraphs. We discuss these algorithms in Section 5.6. Relational Database Management Systems In RDBMS graphs may be stored in relations. Due to the high flexibility in schema design RDBMS are well suited to store graphs. In this work we propose in Figure 3.1 (on page 27) a model to store any type of graph. Dar and Agrawal use a similar model 51
64 3. Graph Queries to store graphs in an RDBMS and extend in [DA93] SQL to query graphs. They propose a syntax where the closure of the graph is stated in the FROM clause. In the WHERE clause it is possible to restrict paths by their start or end node, while a subquery on the paths is necessary to restrict paths by their properties, such as path length. This proposed extension to SQL allows users to pose all types of graph queries, but as conditions may be stated in two different location its syntax is difficult to understand. In [DNR09] Dries and colleagues describe a graph model for relational database systems and a query language. The graph model consists of objects and relations between objects. Thus, in their model nodes and edges of a graph are objects and the connection between an edge and a node is given by the relationship between the objects. Their proposed query language is a mixture of SQL and query languages developed for deductive databases, i.e., it contains a SELECT, FROM, and WHERE clause as in SQL and allows the specification of regular path expressions as in query language for deductive databases. In contrast to PQL only one data graph at a time may be used and the query language is not able to query normal relations in parallel. Moreover, the authors only propose the query language, but state that the development of an efficient and scalable implementation remains an important issue. He and Singh present in [HS08] GraphQL, a graph query language. As this language is similar to Datalog, the specification of the query graph is not as straightforward as in PQL. In addition to the query language the authors also present a graph algebra to rewrite a graph query into a graph algebra. Moreover they present algorithms to answer graph queries and optimization methods. We discuss both in Section 7.4. For biological graphs different database schemas have been developed, an overview may be found in Schaefer [Sch04]. For the metabolic network amaze the database schema may be found in Lemer et al. [LAC + 04] and for Reactome in Vastrik et al. [VDS + 07]. In addition RDBMS also allow users to access further data stored as relations. This is a great advantage over other systems, as many data are available for RDBMS. Some RDBMS also store and query graphs differently. In [EB06] Eckman and Brown propose to store graphs as first-class SQL data type within the database. This extension is called the Systems Biology Graph Extender (SBGE), which performs various functions on the data type graph, such as graph set operations, shortest path computation, and subgraph isomorphism tests. One drawback of their approach is that the graph must be loaded entirely in main-memory for efficient query execution. The predominant query language in RDBMS is the Structured Query Language (SQL). Since the ANSI SQL-99 standard it is possible to express recursive queries using the syntax WITH RECURSIVE [GP99]. Database systems have adopted this standard by now [PBBS10]. Some database system use their own syntax, e.g., Oracle10g uses CONNECT BY PRIOR. The current specification only targets edge disjunct paths using the additional condition NO CYCLES, but we are interested in node disjunct paths, which is not possible using the given syntax. Many researchers work on cost-based query optimization in RDBMS [Cha98]. The insights of this research have gone into commercial as well as open-source RDBMS. To our knowledge current RDBMS do not optimize the recursive part of an SQL query. Therefore, the evaluation of an SQL statement that contains a recursive part may be 52
65 3.5. Related Work prohibitively slow as we show in Section 6. One reason for missing optimization is that known operators from relational algebra are not sufficient to express recursion [Agr88]. We target this problem in the next chapter. 53
66
67 4. Operators for Graph Queries This chapter discusses the possibility to express graph queries using algebra expressions. In Section 4.1 we show that existing relational algebra operators are sufficient for binding nodes to node variables. In Section 4.2 we then motivate the need for new operators to express PQL statements. We introduce four new operators, the path operator Φ, path length operator φ, distance operator Ψ, and reachability operator ψ. Finally in Section 4.3 we present related work in the area of algebraic operators for graph query execution Operators for Nodes In Section 3.4 we introduced view Nodes. This view is basically composed of information that is already present in the model for storing graphs, which is given in Figure 3.1 (on page 27). Assume, we create a relation for each class present in the model. We first show how view Nodes is defined and then explain how to evaluate node conditions on this view. Figure 4.1 gives the SQL statement to declare view Nodes. Equation 4.1 shows the relational algebra expression to resolve it. We see, relational algebra operators presented in Section 2.2 are sufficient to resolve this view. CREATE VIEW Nodes AS SELECT Node.node_id, Node.external_id, Label.label_id, Label.label_type, Label.value FROM Node, Node_label, Label WHERE Node.node_id = Node_label.node_id AND Node_label.label_id = Label.label_id; Figure 4.1.: View definition for view Nodes. Nodes = (Node Node_label Label) (4.1) Example 4.1 (Sample graph). To illustrate views Nodes and Paths we provide a small exemplary graph. Figure 4.2 shows a simplified subgraph of the Arginine and Proline metabolism shown in Figure 3.3 (on page 30). We store the data of the graph according to the model presented in Figure 3.1 (on page 27). Figure 4.3 shows the relations for the exemplary graph. 55
68 4. Operators for Graph Queries Figure 4.2.: Exemplary subgraph of the Arginine and Proline metabolism as given by KEGG. Reactions together with their catalyzing enzymes are omitted. node_id external_id 1 Arginine 2 Ornithine 3 Urea 4 Citrulline 5 L-Argininosuccinate 6 Aspartate (a) NodeKegg node_id label_id (d) NodeKegg_label edge_id from_id to_id (b) EdgeKegg label_id label_type value edge_id name Arginine 2 name Ornithine 3 name Urea 4 name Citrulline 5 name L-Argininosuccinate 6 name Aspartate 20 Type compound 99 null null (e) LabelKegg label_id (c) EdgeKegg_label Figure 4.3.: Relations to represent the exemplary graph from Figure 4.2. Selection on Nodes In PQL it is possible to select specific nodes from the graph using type op n or type text_op s in the WHERE clause. Equation 4.2 defines the rewrite rule for the selection condition type op n. The equation also shows the selection condition on the resolved view. The rewrite rule for selection condition type text_op s is easy to deduce from 56
69 4.2. Operators for Paths Equation 4.2 and thus omitted. σ type op n Nodes = σ label_type=type AND value op n Nodes = σ label_type=type AND value op n (Node Node_label Label) (4.2) Equality Condition on Nodes The WHERE clause may also contain equality conditions for nodes, i.e., A = B. Equation 4.3 shows how to rewrite the expression to be applicable on view Nodes. We are able to transform the original expression, which contains a cartesian product, to an expression with a theta join by applying relational algebra rules. σ A=B (Nodes A Nodes B) = σ A.node_id=B.node_id (Nodes A Nodes B) = Nodes A A.node_id=B.node_id Nodes B (4.3) Label Comparison for Nodes The WHERE clause may also contain the condition A.type op B.type. Equation 4.4 shows how to rewrite the expression to be applicable on two views Nodes. σ A.type op B.type (Nodes A Nodes B) = σ A.label_type=type AND B.label_type=type AND A.value op B.value(Nodes A Nodes B) (4.4) 4.2. Operators for Paths If we resolve the Paths view using an SQL statement that joins relation Edge n times we face two problems. First, we run into cycles, i.e., nodes occur more than once in a path. Second, even if we join Edge of cycle-free graphs we do not know how long the longest path in the graph is, and thus how many joins are required. In relational algebra we are only able to express that we want to join Edge exactly n times. This means, classical operators of relational algebra are not sufficient to express path queries. Therefore, we have to introduce new operators Path Operator, Ψ To resolve the Paths view we introduce the path operator Ψ. This operator uses relation Edge as input and takes the orientation of edges in the path and possibly conditions on the length of the path as parameters. Definition 4.1 (Path operator, Ψ) Given relation Edge, which stores all edges E of a graph G as {(edge_id, e), (from_id, v m ), (to_id, v n )}, with v n, v m being node_ids of nodes in G. Given condi- 57
70 4. Operators for Graph Queries tions on paths, cond := ([->] [-] [<->]), (length_cond)? and length_cond := length op n (AND length op n)* 1. The path operator Ψ cond applied to Edge produces relation Paths such that Ψ cond (Edge) = {(path_id, j), (start, u), (end, w), (length, length(p j )), (node_id, v i ), (position, i)} v i p j = v 0,..., v i,..., v n, 0 h, i < n, u = v 0, w = v n, v i v h for i h and (e k, v i, v i+1 ) Edge if cond = [->] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge, k l if cond = [-] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge, k l if cond = [<->] and length(p j ) fulfills length_cond If no length condition is given, all paths regardless their length are of interest. Example 4.2 (The Paths relation). To illustrate the idea of the path operator Ψ and the content of view Paths we use nodes and edges from Example 4.1 to produce Ψ [ >] (EdgeKegg). Figure 4.4 shows all directed paths for the graph from Figure 4.2. path_id start end length node_id position (a) Paths path_id start end length node_id position (b) Paths (cont.) Figure 4.4.: Ψ [ >] (EdgeKegg) for the graph in Figure 4.2. Tuples displayed in bold are the result of the query given in Example 4.3. Path Operator with Restriction on Start and End Nodes In a PQL query we may state that paths bound to a path variable start or end at a node bound to a node variable. Consider the following example. Example 4.3. Assume we want to find all paths in KEGG that start at a node that has as label (name, Arginine ). We express the query using the following PQL statement. 1 We do not support disjunction inside the operator. This needs to be resolved differently, like by enhancing the PQL query capabilities. 58
71 4.2. Operators for Paths SELECT * FROM Kegg LET node A, path P WHERE A.name = Arginine AND P.start = A; Converting the given PQL statement to an algebra expression results in σ P.start=A.node_id AND A.name= Arginine (NodesKegg A Ψ [ >](EdgeKegg) P). A naive strategy to evaluate this statement would be to first resolve the Paths view using Ψ, create the cartesian product with the nodes view, before restricting the tuples to those whose start nodes are nodes bound to node variable A and whose label has the value (name, Arginine ). Actually computing the result would require to first compute all paths of a graph, before possibly discarding many unnecessary tuples. A more reasonable step would be to only compute paths that start at a node bound to A. Using Ψ we are not able not express this fact. The example above motivates the introduction of three more operators, called Ψ start, Ψ end, and Ψ both, which are derived from Ψ. Equations show the definition of these operators (cond stands for [->], (length_cond)?, [-], (length_cond)?, or [<->], (length_cond)? ). Ψ start cond, start=a.node_id(nodes A, Edge) := σ start=a.node_id (Nodes A Ψ cond (Edge)) = Nodes A A.node_id=start Ψ cond (Edge) (4.5) Ψ end cond, end=b.node_id(edge, Nodes B) := σ end=b.node_id (Ψ cond (Edge) Nodes B) = Ψ cond (Edge) end=b.node_id Nodes B (4.6) Ψ both cond, start=a.node_id AND end=b.node_id(nodes A, Edge, Nodes B) := σ start=a.node_id AND end=b.node_id (Nodes A Ψ cond (Edge) Nodes B) = Nodes A A.node_id=start Ψ cond (Edge) end=b.node_id Nodes B (4.7) Rewrite Rules for Path Operators We also define rewrite rules for path operators in combination with other operators from relational algebra as rewriting expressions is an essential step in query optimization. No rewrite rules for operator Ψ cond (Edge) are necessary. Equation 4.8 shows the rewrite rule for operator Ψ start cond(nodes, Edge) together with a selection operator. This rewrite rule may only be applied if the selection operator contains only attributes from relation 59
72 4. Operators for Graph Queries Nodes. σ sel_cond (Ψ start cond(nodes, Edge)) = Ψ start cond(σ sel_cond (Nodes), Edge) if sel_cond contains only conditions on attributes of Nodes (4.8) Clearly, we may also push the selection operator to relation Nodes for operators Ψ end and Ψ both. The according rewrite rules may be found in Appendix B. We also give rewrite rules for path operators and the join operator. Equation 4.9 shows the equation for Ψ start. This equation shows that we may push a join down to the Nodes view, if the join condition only involves an attribute from the Nodes view and one from the second relation. For Ψ end and Ψ both the equations are also given in Appendix B. R R.attr op A.attr Ψ start cond(nodes A, Edge)) = (Ψ start cond((r R.attr op A.attr Nodes A), Edge) (4.9) Example 4.4. The PQL statement from Example 4.3 may be converted to a relational algebra expression. We picture this statement as expression tree in Figure 4.5(a). We are able to transform this tree into the expression tree given in Figure 4.5(b). Clearly, we may first split the selection operator and then push the operator with the condition A.name= Arginine down to the Nodes view. σ A.name= Arginine AND start = A.node_id Nodes A Ψ [->] Ψ start [->], start = A.node_id σ A.name= Arginine EdgeKegg EdgeKegg (a) Original expression tree. Nodes A (b) Transformed tree. expression Figure 4.5.: Possible expression trees for the PQL statement from Example Reachability operator, φ In the examples so far we required the entire information contained in view Paths, such as intermediate nodes or path lengths, for the result. Sometimes we also want to answer simpler queries, such as the one given in Example 4.5. Example 4.5 (Reachability query). Assume we want are interested in all nodes of type compound that are reachable from a node with label (name, Arginine ), i.e., for 60
73 4.2. Operators for Paths which a path between the two nodes exists. The following PQL statement expresses this query. SELECT B FROM Kegg k LET node A, node B, path P WHERE A.name = Arginine AND B HAS (type, compound ) AND P.path = A[->]B This query retrieves only nodes bound to node variable B, not actual paths bound to path variable P. More generally, assume, we have a PQL statement that contains a path variable P, which is not stated in the SELECT clause and only contains conditions on its start and/or end node in the WHERE clause. Such a query only retrieves distinct node pairs between which paths exist. Converting this expression to our algebra using only path, selection, and duplicate-elimination operators results in δ(π start,end (Ψ cond (Edge))). This statement requires to first resolve the Paths view before projecting attributes start and end and finally de-duplicating the node pairs. A more efficient way is to only compute reachability information, which is faster than computing paths (see Chapter 6). Therefore, we introduce the reachability operator φ to be able to use dedicated algorithms for answering reachability queries. Definition 4.2 (Reachability operator, φ) Given relation Edge, which stores all edges E of a graph G as {(edge_id, e), (from_id, v m ), (to_id, v n )}, with v n, v m being node_ids of nodes in G. And given conditions on paths, cond := ([->] [-] [<->]). The reachability operator φ cond applied to relation Edge produces φ cond (Edge) = {(start, u), (end, w) p j = v 0,..., v i,..., v n, 0 h, i < n, u = v 0, w = v n, v i v h for i h and (e k, v i, v i+1 ) Edge if cond = [->] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge if cond = [-] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge if cond = [<->] } Example 4.6 (Result for Example 4.5). Figure 4.6 shows φ [ >] (EdgeKegg) from Example 4.1. Consider again Example 4.5. To answer the query we have to know the node with label (name, Arginine ) has node_id 1 and nodes with node_id 2, 4, and 5 are compounds. Thus, the answer to the query are node_ids 2, 4, and 5. 61
74 4. Operators for Graph Queries start end Figure 4.6.: The reachability operator φ applied on relation EdgeKegg. The tuples in bold are required for the PQL statement in Example 4.5. Reachability Operator with Start and End Nodes Consider Example 4.5, where we have a path variable P on which only paths may be bound that start at a node bound to A and end at a node bound to B. For the path operator, Ψ we defined Equations , which are also valid in modified form for φ. For example, Equation 4.10 is the modified form of Equation 4.7. The equations for the remaining operators φ start and φ end are easy to deduce from Equations 4.5 and 4.6 and are thus omitted. σ start=a.node_id AND end=b.node_id (Nodes A φ cond (Edge) Nodes B) = Nodes A A.node_id=start φ cond (Edge) end=b.node_id Nodes B = φ both cond, start=a.node_id AND end=b.node_id(nodes A, Edge, Nodes B) (4.10) From Path Operator Ψ to Reachability Operator φ So far, we only defined the reachability operator. We gave no rules how to rewrite an expression containing a path operator to an expression containing a reachability operator. Equation 4.11 shows this rewrite rule for Ψ to φ. δ(π A (Ψ cond (Edge))) = π A (φ cond (Edge)) if A = {start, end} and cond = [->] [-] [<->] (4.11) Using Equation 4.11 and the rewrite rules from φ to φ start, φ end, or φ both, shown for φ both in Equation 4.10, we are able to deduce the rewrite rules for Ψ start to φ start, Ψ end to φ end, and Ψ both to φ both. Rewrite Rules for Reachability Operators The reachability operator is defined in the same style as the path operator. Thus, the rewrite rules defined for Ψ in Equations may be used. As rewriting the equations for the reachability operator, φ is trivial, the equations are omitted. Basically, 62
75 4.2. Operators for Paths we push down a selection operator to one of the participating relations of φ start, φ end, and φ both. We also push down a join operator together with its join partner to the respective relation Path Length Operator, ψ In a PQL statement we may also pose length conditions on paths, as Example 4.7 shows. Example 4.7 (Path length query). Assume, we are still interested in all nodes of type compound that are reachable from a node with label (name, Arginine ), as stated in Example 4.5. Now we also require the path length to be equal to 2. The following PQL statement expresses this query. SELECT B FROM Kegg k LET node A, node B, path P WHERE A.name = Arginine AND B HAS (type, compound ) AND P.path = A[->]B AND P.length = 2 We are only interested in nodes bound to node variable B. Thus, we only require information about start and end node of a path and path lengths for paths between the two nodes. Assume, the WHERE clause contains a length restriction or the HAVING clause a restriction on the number of paths bound to P. There may also conditions on the start and end node, but P is not stated in the SELECT clause. Thus, we are only interested in attributes path_id, start node, end node, and length. A possible expression using our algebra results in δ(π path_id,start,end,length (Ψ cond (Edge))). Figure 4.7 shows δ(π path_id,start,end,length (Ψ [ >] (EdgeKegg)) for the graph from Figure 4.2. path_id start end length Figure 4.7.: δ(π path_id,start,end,length (Ψ [ >] (EdgeKegg)) for the graph in Figure 4.2. Tuples displayed in bold are required for the result of the query given in Example 4.7. Compare the tables in Figure 4.4 and 4.7. As one may see, the latter contains fewer tuples and attributes. In Chapter 6 we show, immediately computing the result of 63
76 4. Operators for Graph Queries δ(π path_id,start,end,length (Ψ cond (Edge))) is more efficient than to first compute all paths of a graph, before discarding some information. To give this opportunity to the query optimizer we need a third operator, the path length operator ψ. Definition 4.3 (Path length operator, ψ) Given relation Edge, which stores all edges E of a graph G as {(edge_id, e), (from_id, v m ), (to_id, v n )}, with v n, v m being node_ids of nodes in G. Given conditions on paths, cond := ([->] [-] [<->]), (length_cond)? and length_cond := length op n (AND length op n)* 2. The path length operator ψ cond applied to Edge produces ψ cond (Edge) = {(path_id, j), (start, u), (end, w), (length, length(p j ))} p j = v 0,..., v i,..., v n, 0 h, i < n, u = v 0, w = v n, v i v h for i h and (e k, v i, v i+1 ) Edge if cond = [->] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge, k l if cond = [-] (e k, v i, v i+1 )Edge (e l, v i+1, v i ) Edge, k l if cond = [<->] and length(p j ) fulfills length_cond Please note, in the result set of the path length operator we might find a pair of nodes with the same path length multiple times. The reason is that multiple paths of the same length between these two nodes may exist. The tuples are only distinguished by their path_id. Path Length Operator with Start and End Node For the path operator Ψ we defined Ψ start, Ψ end, and Ψ both in Equations These equations are also valid in modified form for ψ. The equations are given in Appendix B. From Path Operator Ψ to Path Length Operator ψ We also have to define rules to rewrite an expression containing a path operator Ψ to an expression containing a path length operator ψ. Equations 4.12 and 4.13 state the rewrite rules. δ(π A (Ψ cond (Edge))) = π A (ψ cond (Edge)) if A = {path_id, start, end, length} (4.12) 2 We also do not support disjunction inside this operator. 64
77 4.2. Operators for Paths δ(γ A,count(path_id) (Ψ cond (Edge))) = γ A,count(path_id) (ψ cond (Edge)) if A = {path_id, start, end, length} (4.13) In the same style we define rewrite rules for ψ start, ψ end, and ψ both, which are given in Appendix B. Rewrite Rules for Path Length Operators The path length operator ψ is almost identical to the path operator Ψ. Thus, Equations 4.8 and 4.9 may be modified for ψ (equations given in Appendix B) Distance Operator, Φ The path length operator may always be applied when a length restriction on a path is given and when the actual path is not required in the SELECT clause. In some cases, not all lengths of paths are required. Consider Example 4.8. Example 4.8 (Distance query). Remember, in the last example (Example 4.7) we required that the length of the path between nodes with label (name, Arginine ) and nodes of type compound must be greater or equal to 2. We now require that the length of the path is less than or equal to 2. To answer this query it is sufficient to know for each node pair that there exists at least one path that is shorter than 2, i.e., the distance (the length of the shortest path) between these nodes must be lower or equal to 2. Definition 4.4 (Distance operator, Φ) Given relation Edge, which stores all edges E of a graph G as {(edge_id, e), (from_id, v m ), (to_id, v n )}, with v n, v m being node_ids of nodes in G. And given conditions on paths, cond := ([->] [-] [<->]), (length < n)?. The distance operator Φ cond applied to relation Edge produces Φ cond (Edge) = {(start, u), (end, w), (length, length(p j )) p j = v 0,..., v i,..., v n and p l = v 0,..., v i,..., v m with length(p l ) < length(p j ), 0 h, i < m < n, u = v 0, w = v n = v m, v i v h for i h, and (e k, v i, v i+1 ) Edge if cond = [->] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge if cond = [-] (e k, v i, v i+1 ) Edge (e l, v i+1, v i ) Edge if cond = [<->] } Consider the graph given in Figure 4.2 again. Φ [ >] (EdgeKegg) produces the relation given in Figure 4.8. As one may see, this relation contains fewer attributes and, in this 65
78 4. Operators for Graph Queries example also fewer tuples, than ψ [ >] (EdgeKegg) given in Figure 4.7. Please note, while the path length operator may produce a node pair with the same length multiple times, only differentiated by the path_id, the distance operator produces each node pair only once with its distance. start end length Figure 4.8.: Φ [ >] (EdgeKegg) for the graph in Figure 4.2. Tuples displayed in bold are required for the result of the query given in Example 4.8. Distance Operator with Start and End Node As for the path operator Ψ, the path length operator ψ, and the reachability operator φ we define Φ start, Φ end, and Φ both according to Equations (given in Appendix B). From Path Operator Ψ to Distance Operator Φ In Equation 4.14 we also provide rewrite rules to rewrite an algebra expression containing a path operator to one containing a distance operator. π A (Ψ cond (Edge)) = π A (φ cond (Edge)) if A = {start, end} and cond = [->] [-] [<->], (length_cond)? with length_cond := length op n (AND length op n)* and op := < (4.14) In the same style we define the rewrite rules for Φ start, Φ end, and Φ both (see Appendix B). Rewrite Rules for Distance Operators The rewrite rules for the distance operator Φ are similar to those for the remaining three operators (see Appendix B) Summary The path operator Ψ would be sufficient to convert all possible PQL statement to relational algebra expressions. For query optimization we introduced three more operators, 66
79 4.3. Related Work the reachability operator φ, the path length operator ψ, and the distance operator Φ. For each of these operators we gave rewrite rules as when to transform an expression containing a path operator to an expression containing one of the other operators. In this section we summarize which operators are applicable in which situation. For example, the PQL statement in Example 4.5 expresses a reachability query for which we are interested in the end nodes of paths. Thus, in our extended algebra we may apply the path operator Ψ, but we may also transform the expression to contain either the path length operator ψ, the distance operator Φ, or the reachability operator φ. In Table 4.1 the applicability of operators for different combinations of projection attributes as well as select conditions is displayed. The PQL statements always have the form of SELECT...FROM...LET node A, node B, path P (WHERE)?...(HAVING)?..., i.e., with or without a WHERE and HAVING clause. Take for example the third column and the third row of Table 4.1. This combination represents the following PQL query without HAVING clause. SELECT A, B FROM... LET node A, node B, path P WHERE P.path = A[->]B AND P.length < n For this query we may use the path operator Ψ, path length operator ψ, or the distance operator Φ to express the query in relational algebra as Table 4.1 shows. The reachability operator φ is not applicable as a length restriction is applied. Table 4.1 shows some of the conditions possible in the WHERE clause of a PQL statement. Further conditions are A.name= x, B HAS (Type, y), or P.start=A. Any select condition on nodes may be directly applied to the Nodes view and does not influence the choice of operator to assess paths. The remaining conditions P.start = A and P.end = B are semantically the same as P.path = A[->]B and the same operators may be applied Related Work In this section we discuss related work on operators for extending the relational algebra to handle path queries. Our focus lies on three main points. First, the capability of the introduced operators to answer graph queries. We discuss, which type of queries are supported, such as reachability, distance, path length, or path queries. Second, we discuss rewrite rules for the operators and their applicability. Finally, as outlook to Chapters 5, 6 and 7 we consider if there exist implementations for the operators including cost functions and cardinality estimates, as these are important for efficient query execution. 67
80 4. Operators for Graph Queries SELECT A B A, B P A, P B, P * WHERE HAVING φ, Φ, ψ, Ψ φ, Φ, ψ, Ψ φ, Φ, ψ, Ψ Ψ Ψ Ψ Ψ WHERE P.path = A[->]B HAVING φ, Φ, ψ, Ψ φ, Φ, ψ, Ψ φ, Φ, ψ, Ψ Ψ Ψ Ψ Ψ WHERE P.path = A[->]B AND P.length < n HAVING Φ, ψ, Ψ Φ, ψ, Ψ Φ, ψ, Ψ Ψ Ψ Ψ Ψ WHERE P.path = A[->]B AND P.length = n HAVING WHERE P.path = A[->]B HAVING A.count op m WHERE P.path = A[->]B HAVING P.count op m ψ, Ψ ψ, Ψ ψ, Ψ Ψ Ψ Ψ Ψ φ, Φ, ψ, Ψ φ, Φ, ψ, Ψ φ, Φ, ψ, Ψ Ψ Ψ Ψ Ψ ψ, Ψ ψ, Ψ ψ, Ψ Ψ Ψ Ψ Ψ Table 4.1.: Applicability of operators path (Φ), path length (φ), distance (Ψ), and reachability (ψ) for different PQL statements. The PQL statements always have the form of SELECT... FROM... LET node A, node B, path P (WHERE)?... (HAVING)?... A is meant as start node, B as end node. Path Operators for RDBMS Rosenthal et al. propose in [RHDM86] new operators for relational algebra to answer graph queries. They state a path enumeration operator is needed, but do neither define its input nor its output. They also state that in the case of distance or reachability queries a grouping and projection is necessary, but they do not explicitly introduce new operators to resolve these combinations. They present some rewrite rules for the new operator by example, but no formal introduction is given. In [Agr88] Agrawal proposes the α-operator as additional operator for relational algebra to handle recursive queries. Applying the α-operator to a relation of edges results in a new relation that stores node pairs and as additional attribute all cycle-free paths between the pairs as an enumeration. As the attribute for the path is not in first normal form, it may not be directly accessed using relational algebra operators. Thus, the author proposes special predicates to work with paths stored in this attribute. The α- operator is similar to our proposed path operator Ψ, yet no rewrite rules, especially for the situation with a restriction on the start or end node, are given. In [DAJ91] Dar and colleagues extend the idea given in [Agr88] to overcome this shortage. They state that in case a restriction on the start node or on nodes or edges in the path is given, this selection should be pushed into the path computation. In addition they state in some cases nodes and edges of a path are not required and thus do not 68
81 4.3. Related Work have to be computed. Our proposed operators and rewrite rules follow the same idea. In [DA93] Dar and Agrawal refine their previous approach and propose an extension to SQL to provide a syntax to query graphs as discussed in Section 3.5. Ordonez presents in [Ord05] an extension of the database system Teradata to express recursive views. Using these views it is possible to express recursive queries. The author also presents query optimization strategies for evaluating the recursive views when conditions are applied. One problem of this approach is that recursive views are not able to handle cycles, i.e., an execution will not terminate unless a termination condition is explicitly given in the view definition. Eckman & Brown follow in [EB06] a different approach. They store biological graphs as data object in an RDBMS. They provide several methods that may be applied to the data object, which perform various analyses. A disadvantage is that the graph is stored as single object and thus, for efficient execution of a query should fit together with is result into main-memory. Operators for Graph Queries in Other Areas Query optimization is also important in other areas than RDBMS, for example for semistructured data or data complying to the RDF data model. For semi-structured data Buneman and colleagues present in [BDHS96] a query language called UnQL. They also introduce new operators to rewrite the queries to lambda calculus. Although the operators are not expressed the same way, the capability and optimization strategies are similar to those proposed for our path operator Ψ, but defined only for DAGs and trees. In [BFS00] the authors refine their proposed language and operators, but they do not consider cases where only path length or reachability is required. They also state in the paper that future work is needed to combine these (calculus and rewriting rules) with a cost model and an optimization algorithm. Beeri and Tzaban present in [BT99] SAL, an algebra for semi-structured data. They propose several operators to convert queries on XML documents to an algebraic expression, but this algebra lacks an equivalent to our proposed path operator. Although selection of nodes and edges, projection of subtrees, and join of trees are possible, the algebra is not capable to express paths of arbitrary length, which we find necessary. In [BMR99] Beech and colleagues overcome this problem. Their follow operator takes a collection of nodes as input and in its basic form selects edges of a specific type starting at nodes in the input, or, if a Kleene star is added, allows arbitrary paths. Thus, their follow operator is similar to our proposed path operator Ψ. In the paper the authors do not give any rewrite rules for optimization nor introduce algorithms to efficiently execute the queries. In [FHP02] Frasincar and colleagues take up the operators proposed by Beech et al. and give rewrite rules involving these operators. Neither reachability queries nor path length queries can be expressed using their proposed algebra. In addition, they do not give algorithms for evaluating their version of the path operator. Fernandez and colleagues present in [FSW00] an algebra for XML Query. Apart from selection, projection, and join they also introduce a structural recursion operator, which basically resembles our path operator. They also give equivalence and rewrite rules for 69
82 4. Operators for Graph Queries query optimization, but their proposed operators are only applicable for tree structured XML documents. In addition reachability or path length are not supported as these are not necessary for querying XML documents. In [CCS00] the authors follow a different approach. They propose the bind operator to convert queries on XML document to relational algebra expressions. This operator takes as input a collection of trees, contains as argument a tree pattern, and returns variable bindings between the pattern and the input trees. To rewrite the expression they split a bind operator, i.e., split the pattern of the argument, to produce an expression containing two bind operators. In addition, they give rewrite rules to transform expressions containing the bind operator together with selection, projection, join, and map. Following the same concept, Jagadish and colleagues propose TAX [JLST01], an algebra for XML. They propose four operators (selection, projection, product, and grouping) that take as input a collection of trees. The selection operator, similar to the bind operator proposed in [CCS00], returns those trees that satisfy the selection condition, given as pattern tree. In the same line He and Singh [HS08] define their selection operator. Their proposed operator may also be used for general graphs as input and returns isomorphic subgraphs to the input pattern. Thus, it is not comparable to our path operator as we return homeomorphic subgraphs. In addition, all three operators are not capable to express reachability or path length queries. Also, for the first two the authors do not give any algorithms for the proposed operators, no cardinality estimates, or cost functions. Only He and Singh provide in [HS08] algorithms and cost functions, which we discuss in Section 7.4. Wu and colleagues present in [WPJ03a] an optimization strategy for queries on XML documents. They introduce the structural join to determine ancestor-descendant relationships of XML elements in a document. In this work the authors only introduce a strategy that is similar to our proposed reachability operator φ, while path lengths or paths are not considered, as they are not part of the XQuery language. Apart from the introduction of the structural join, they also investigate the best order for a query that contains multiple structural joins. In the area of the semantic web Hartig and Heese presented in [HH07] the SPARQL query graph model for the execution of SPARQL queries. This model is based on relational algebra and contains operators and transformation rules to rewrite a SPARQL query. Although RDF data are graph structured, the original query language SPARQL does not allow for queries that contain paths of arbitrary length. Thus, the operators and rewrite rules proposed by the authors may not be applicable in our setting. In the works on extending SPARQL for regular path expressions the authors only introduce the language and some algorithms for execution, but no explicit operator or algebra [ABE09]. Concluding, optimization of graph queries in a RDBMS still requires attention, as current proposals either lack the ability to handle cycles [Ord05], are not capable to express path queries, or lack an efficient implementation for the proposed operators [DA93]. 70
83 5. Implementations for Operators In this chapter we present implementations for the reachability, distance, path length, and path operator presented in the previous chapter. We assume that the data are stored in an RDBMS. Our goal is that the entire path computation occurs inside this system. In an RDBMS we find tuples either by scanning the entire data at query time or by using a precomputed index, which according to [GMUW02] is a data structure that makes it more efficient to find... tuples. Conveying this fact to graph queries means, we may either scan relation Edge multiple times at query time or precompute and store index structures. There exist several index structures, which may be used for answering different kinds of queries. In this chapter we present four index structures more closely, namely our newly developed index structure, called GRIPP, the transitive closure, DualLabeling [WHY + 06], and Label+SSPI [CGK05]. In Section 5.1 we present GRIPP. We describe how to create the index structure itself before we present different query strategies. We provide implementations for all four types of operators that use the GRIPP index. In Section 5.2 we present the remaining three index structures. In contrast to GRIPP, these index structures may only be used for answering reachability and distance queries. We also presents two methods that traverse the graph at query time. For these methods no precomputed index is necessary. In Section 5.3 we describe the built-in function of an RDBMS to answer graph queries, while in Section 5.4 we show algorithms that recursively traverse a graph. In Section 5.5 we summarize which algorithms are applicable for which operators. All algorithms are experimentally evaluated in Chapter 6. This chapter concludes in Section 5.6 with a reflection on related work in the area of index structures and algorithms to answer graph queries GRIPP In this section we present the theoretic foundations of GRIPP. We first describe the GRIPP index structure itself before we present the query strategies to answer reachability, distance, path length, and path queries Index Structure The GRIPP index structure, which we propose in [TL07], is based on pre- and postorder labeling of nodes in a graph as explained in Section 2.1 on page 14. In GRIPP every node in the graph receives at least one pair of pre- and postorder values. As nodes may have multiple parents one pair is not sufficient to encode the entire graph structure. Therefore, some nodes are assigned more than one value pair. 71
84 5. Implementations for Operators For now, we assume the graph has exactly one root node, i.e., one node without incoming edges. We also assume an arbitrary, yet fixed order among child nodes, e.g., given by the ID of the node. Later in this section we explain how to deal with graphs with multiple or no root nodes. For the creation of the GRIPP index we start at the root node of G. During a depth-first traversal of G we assign pre- and postorder values. We traverse child nodes according to their order. A node v with n > 1 incoming edges is reached n times on edges e i, with 1 i n. Edge e i on which we reach v for the first time is called a tree edge. We assign a preorder value to v and proceed the depth-first traversal. After all child nodes have a value pair, v receives its postorder value. We reach v n 1 times again. Assume we reach v over edge e j, with e j e i. We call e j a non-tree edge and assign a pre- and postorder value to v, but do not traverse child nodes of v. We store the pre- and postorder values of tree- and non-tree instances together with the node identifier, depth information, and edge_id as node instances in an index table, IND(G). The depth of an instance is its distance from the root instance. Every node has as many instances in IND(G) as it has incoming edges in G. Analogously to the distinction of tree and non-tree edges we distinguish between tree and non-tree instances in IND(G). Definition 5.1 (Tree and non-tree instances) Let IND(G) be the index table of graph G. Let v V be a node of G and v be an instance of v in IND(G). v is a tree instance of v, iff it was the first instance created for v in IND(G). Otherwise v is a non-tree instance of v. The GRIPP index structure resembles a rooted tree, which we call the order tree, O(G). Definition 5.2 (Order tree) Let G = (V, E) and let IND(G) be its index table. The order tree, O(G), is a tree that contains all instances of IND(G) as nodes and all edges of G as edges between the nodes in O(G). Every non-tree instance in O(G) is a leaf node, while tree instances may be inner or leaf nodes. Note, the shape of O(G) depends on the order with which G is traversed. In Section we shall explain how we determine an order that is well suited for our purpose. Example 5.1 (IND(G) and O(G)). Figure 5.1 shows a graph and Figure 5.2(a) shows its index table resulting from a traversal in lexicographical order of node labels. Nodes A and B have two instances in IND(G) because they have two incoming edges in G. In Figure 5.2(b) the instances of IND(G) shown in Figure 5.2(a) are plotted using pre- and postorder values as coordinates. Nodes A and B occur twice in O(G) as they have two instances in IND(G). The space requirement to store the GRIPP index table is O(n + m), i.e., linear in the size of the graph. More precisely IND(G) has as many entries as G has edges plus one 72
85 5.1. GRIPP r A B C D E F G H Figure 5.1.: A graph G node pre post depth type r tree A tree B tree E tree F tree C tree D tree G tree B non-tree H tree A non-tree (a) Index table, IND(G). post 20 r A B F E C D G B H A pre (b) Order tree O(G). Figure 5.2.: GRIPP index table IND(G) and its order tree O(G). Non-tree instances in O(G) are displayed in gray, tree instances in black. entry for every root node. To create the GRIPP index structure we perform a depth-first traversal, requiring O(n + m) time. Algorithm for Index Creation Traversal Order We choose a traversal order of nodes to optimize reachability queries. Based on considerations presented in Section we found that the degree of nodes is important. We thus traverse nodes in descending order of their degrees. Graphs with Multiple or no Root Nodes So far, we only explained the creation of the GRIPP index structure for graphs with a single root node. Other kinds of graphs may be treated as follows. We first add a virtual root node r to the graph. We add an edge between r and the node with the highest degree among all nodes. We then traverse and label nodes starting from r and using child nodes in the order of their degree. In general, some nodes will not be reached during this traversal, i.e., nodes without incoming edges 73
86 5. Implementations for Operators or nodes in not connected subgraphs. We identify these nodes and add another edge from r to the node with highest degree. This is repeated until all nodes have at least one instance in the index table. This way, we uniformly handle graphs with none, one, or multiple root nodes. Algorithm 5.1 shows the algorithm to compute the GRIPP index table IND(G). To answer distance, path length, and path queries we also compute and store the depth of an instance, i.e., the distance of the instance to the root instance in O(G). To store traversed nodes we use the global variable seen. Algorithm 5.1: The GRIPP algorithm to compute IND(G) 1 pre_post 0; seen 2 PROCEDURE compute_gripp() 3 while empty(node \ seen) do 4 pre_node pre_post 5 pre_post pre_post next_node next(node \ seen) // order by degree 7 8 PPtraversal(next_node, 0 ) 9 GRIPP GRIPP (next_node, pre_node, pre_post, 0, T) 10 pre_post pre_post end 12 end 13 PROCEDURE PPtraversal(next_node, cur_dist) 14 seen seen next_node 15 while child next(children(next_node)) // order by degree 16 do 17 pre_node pre_post 18 pre_post pre_post if child / seen then 20 node_inst T 21 PPtraversal(child, cur_dist +1) 22 else 23 node_inst N 24 end 25 GRIPP GRIPP (child, pre_node, pre_post, cur_dist +1, node_inst) 26 pre_post pre_post end 28 end Reachability Queries In the following section we show how to use the GRIPP index to efficiently answer reachability queries for a fixed pair of nodes. Recall from Section 2.1 reachability queries in pre- and postorder labeled trees are answered with a single lookup as all reachable nodes of u have a preorder value that is contained within the borders given by u pre and u post. When we try to query the GRIPP index structure in this way, we face two problems. First, u may have multiple instances in IND(G), each with its individual 74
87 5.1. GRIPP pre- and postorder value. Second, in the preorder range of an instance u we only find instances of nodes that are reachable from u in O(G). Nodes reachable from u in G but not from u in O(G) are missed. Thus, to find all reachable nodes in G, we have to extend the search, using the hop technique. To evaluate if v is reachable from u we use the index table IND(G). Observe, u may have many instances in IND(G), but only one of them is a tree instance. Every non-tree instance of u in IND(G) is a leaf node in O(G) and therefore has no successors in O(G). Let u be the tree instance of u. If u is an inner node in O(G) then it has reachable instances v in O(G) such that u pre < v pre < u post. Those are retrieved with a single query. We call this set of instances reachable instance set of u. Definition 5.3 (Reachable instance set) Let u V be a node of graph G and u IND(G) its tree instance. The reachable instance set of u, written RIS(u), is the set of all nodes that have instances reachable from u in O(G), i.e., which have a preorder value in [u pre, u post]. To determine if v is reachable from u we proceed as follows. We first search for the tree instance u of u and retrieve its reachable instance set. If v RIS(u), we finish and return true, otherwise we have to extend the search. If RIS(u) contains non-tree instances of nodes, their child nodes might not have an instance in RIS(u), i.e., these nodes are reachable from u in G, but not from u in O(G). To account for this, we have to examine all non-tree instances of nodes in RIS(u). We call those nodes hop nodes. Definition 5.4 (Hop node) Let u, h V and h be a non-tree instance of h. If h RIS(u) then h is called a hop node for u. Example 5.2 (RIS(u) and non-tree instances). In Figure 5.3(a) the reachable instance set of node D is shown. It contains instances of nodes G, B, H, and A. Two instances in RIS(D) are non-tree instance, namely B and A, i.e., both are hop nodes for D. Every hop node in RIS(u) has a reachable instance set in O(G). The nodes in this set are reachable from u in G, but not from u in O(G). We need to check if v is in one of those. Therefore, we identify all hop nodes and recursively check their reachable instance sets by performing a depth-first search over O(G) using hop nodes in ascending order of their preorder values. We stop traversing O(G) if we find node v in some reachable instance set or if there exists no further non-traversed hop node. In IND(G) exist (m n) non-tree instances, each of which may be a hop node. Thus, querying GRIPP to answer if v is reachable from u requires in worst case (m n) queries. However, in the following we show pruning strategies that allow us to query graphs on average in almost constant time regardless the size and shape of the graph, as shown in Section 6. 75
88 5. Implementations for Operators post r 20 A B F E C D G B H A pre (a) RIS(D) post r 20 A C B F E D G B H A pre (b) RIS(D) and RIS(B) in dark gray; RIS(A) in light gray. Figure 5.3.: The example shows the evaluation if r is reachable from D on the GRIPP index structure from Figure 5.2(a). Nodes A and B are hop nodes for D. Pruning Strategies Example 5.3 (Query evaluation using GRIPP). Consider Figure 5.3(b) and the question if r is reachable from D. We find non-tree instances of nodes B and A in RIS(D). If we first use node A as hop node, we find non-tree instances of A and B in RIS(A). Clearly, we do not need to use A as hop node again. Therefore, we next use B as hop node. The tree instance of B is successor of the tree instance of A in O(G). This implies RIS(B) is contained in RIS(A), i.e., we do not find new instances in RIS(B) that are not already contained in RIS(A). Therefore, using B to retrieve RIS(B) is not necessary; B is pruned from the list of hop nodes. In general we want to avoid posing queries for preorder ranges which we have already checked. During our search we keep a list U of all nodes that have been used to retrieve a reachable instance set. Now assume we have found a new hop node h. The decision whether we need to consider the reachable instance set of h entirely, partly, or not at all depends on the location of the tree instance h of h relative to the tree instances of nodes in U. There are four possible locations of h in relation to the tree instance u of a node u U in O(G). These are shown in Figure 5.4. h either is (a) equal to, (b) a successor of, (c) an ancestor of, or (d) a sibling to u. Given that fact, we may consider all nodes in U for pruning, it results in four possible cases: (a) h is equal to the tree instance of some node in U; (b) h is successor of the tree instance of some node in U; (c) h is ancestor to all tree instances of nodes in U; (d) h is sibling to all tree instances of nodes in U. Note, by construction, the pre- and postorder ranges of two instances never overlap. They are either disjoint or one is entirely contained in the other. In the first case (Figure 5.4(a)), we skip h entirely because a non-tree instance of h has already been used as hop node and thus RIS(h) has already been checked. 76
89 h = u (a) h equals u u h (b) h successor of u 5.1. GRIPP h u h u (c) h ancestor of u (d) h sibling to u Figure 5.4.: Possible locations of h of hop node h relative to u, u U. In the second case, we also skip h. In this case (see Figure 5.4(b)) there exists u U such that h is successor of u, i.e., h RIS(u) in O(G). Thus, RIS(h) RIS(u). In the third case (Figure 5.4(c)) we have to be more careful. Consider Figure 5.3(b) and the query reach(d, r). Assume, we have retrieved RIS(D) and RIS(B) and expand the search using A as hop node. RIS(A) contains the tree instance of B and D and therefore also contains RIS(B) and RIS(D). Thus, when we consider RIS(A) we skip the pre- and postorder range of RIS(B) and RIS(D). In the last case (Figure 5.4(d)), no pruning is possible and we have to consider the entire reachable instance set of h, as there exists no previous reachable instance set that covers instances in RIS(h). Skip Strategy We first assume only one u exists that is successor of h. Thus, RIS(u) RIS(h). This situation is displayed in Figure 5.4(c). Considering the entire reachable instance set of h leads to duplication of work. To avoid this work we use the skip strategy working as follows. For every node u U we stored the pre- and postorder value, i.e., the borders of RIS(u). In this range all instances are covered by RIS(u) and we are able to skip the preorder range without missing instances. We only have to consider instances from RIS(h) whose preorder values lie outside the pre- and postorder range of u. If there is more than one successor node of h in U, the situation is slightly more complicated. Essentially, we skip all their ranges when searching RIS(h). We could optimize the search merging ranges iteratively, thus reducing the number of necessary interval operations. However, as we search U only a few times during a reachability query (see also Section 6) we believe the cost to merge ranges does not account for the gain of merging. Therefore, if multiple u exist in RIS(h) each of their ranges is considered separately for skipping. Stop Strategy When querying graphs for reachability between nodes u and v we stop extending the search as soon as we have found an instance of v in the reachable instance set of the current hop node h. If v / RIS(h) we must check every hop node in RIS(h) and start a recursive search. It would be advantageous if we knew in advance that in RIS(h) no hop node exists that extends the search. In this case we do not have to query for the tree instances of hop nodes. We now show cases where this property may be precomputed. 77
90 5. Implementations for Operators Recall, a hop node for node s is a node h that has a non-tree instance in RIS(s). h is not used as hop node if the tree instance of h is in RIS(s) (Figures 5.4(a) and 5.4(b)). We are able to precompute a list of nodes S for which all hop nodes have this property. We call these nodes stop nodes as their reachable instance sets do not extend the search. Definition 5.5 (Stop node) Let s V be a node of graph G and let RIS(s) be its reachable instance set in O(G). s is called a stop node iff all non-tree instances in RIS(s) also have their corresponding tree instances in RIS(s). Intuitively, a stop node s is a node in G for which a corresponding tree instance for every non-tree instance in RIS(s) exists in the same set. Thus, all nodes reachable from s in G are reachable from s in O(G), i.e., have an instance in RIS(s). Clearly, nodes reachable from s in G may also have non-tree instances in other reachable instance sets than in RIS(s). Algorithm C.1 in Appendix C.2 shows the function to compute stop nodes. When we reach the tree instance of a stop node s during the search we immediately know we do not need to extend the search using hop nodes of RIS(s). Example 5.4 (Stop nodes). The GRIPP index structure in Figure 5.2 contains several stop nodes, namely nodes r, A, B, E, F, and C. As heuristic, during the search we prefer stop nodes as hop nodes over non-stop nodes. Algorithms In this section we present the algorithm to answer reachability queries using the GRIPP index. We show strategies to answer reachability queries for a single pair of nodes. In addition, we briefly describe how to find all reachable nodes for a given start node. Pair of Nodes To answer if v is reachable from u Algorithm 5.2 starts by testing v RIS(u) with a query over the index. If v RIS(u) the algorithm stops. Otherwise it adds u to the list U of used nodes. If u is a stop node, the algorithm returns. Next, we perform a depth-first search considering non-tree instances in RIS(u) in ascending order of their preorder rank as hop nodes, unless RIS(u) contains a non-tree instance of a stop node, which is preferentially used. In the next step we select all hop nodes from RIS(u) which are not already covered by another reachable instance set. For every hop node h we determine the location of its tree instance h and test if RIS(h) is completely or partly covered by nodes in U. If not, we proceed, using h as next hop node. We stop once we find an instance of v or if there are no more non-traversed hop nodes. Set of Reachable Nodes In some cases we want to find all nodes that are reachable from a given node u. In this case, we could use Algorithm 5.2 and test for all nodes in G if they are reachable from u. A more efficient way is to use a different function, called reachability_set(u) that receives as input u and returns all nodes that are reachable from u in G. This function is a slight variation of Algorithm 5.2 and is given in Appendix C.3. 78
91 5.1. GRIPP Algorithm 5.2: Function to answer if v is reachable from u using GRIPP. 1 used_hops ; used_stops 2 FUNCTION reachability(u, v) RETURNS boolean 3 if v RIS(u) then 4 return true 5 else 6 used_hops used_hops (u) 7 if u STOP_NODES then 8 used_stops used_stops (u) 9 return false 10 else 11 while non_tree_inst nextstop(ris(u)) do 12 tree_inst gettreeinst(non_tree_inst) 13 if reachability(tree_inst, v) then return true 14 end 15 if isinris(u, used_stops) then return false 16 H 1... H n getusedhopsinris(u) // skip ranges 17 non_tree_instances getnontreeinst(ris(u) \ RIS(H 1) \... \ RIS(H n)) 18 foreach non_tree_inst non_tree_instances do 19 tree_inst gettreeinst(non_tree_inst) 20 if!haschildren(tree_inst) then continue // if new hop has been used as hop 21 if tree_inst used_hops then continue // if new hop is in a RIS of a used hop 22 if isinris(tree_inst, used_hops) then continue // otherwise call recursively 23 if reachability(tree_inst, v) then return true 24 if isinris(u, used_stops) then return false 25 end 26 return false 27 end 28 end 29 end Theoretic considerations We would like to estimate how many recursive calls are required to answer u w. We know, in worst case we require (m n) calls, as (m n) non-tree instances are in IND(G). But on average we need less calls as the following considerations show. In every graph one may identify strongly connected components C 1... C k in linear time using Algorithm A.1 on page 152. Each component is collapsed into a representative node (see Figure 5.5(a)). The reachability information for nodes within one component are identical (this obvious optimization is used by many graph indexing strategies, such as [ABJ89] or [WHY + 06]). Therefore, we divide the problem in two separate parts. First, estimate the number of calls if u and w are within one strongly connected component, and second, if they belong to different components of the component graph. To estimate the number of recursive calls if u and w are in the same component we have to consider the traversal order for nodes within one strongly connected component 79
92 5. Implementations for Operators r r a 1 a n a 1 a n c C 1 c s n C 2 s 1 s n (a) Structure of a graph. Solid lines indicate edges, dotted lines paths. The gray area mark strongly connected components of the graph. s 1 (b) Optimal GRIPP index structure. Circles indicate tree instances, squares non-tree instances. The double circled node is the stop node, the double squared nodes are its non-tree instances. In gray is the area of instances of the giant strong component. Figure 5.5.: General idea of an optimal index structure for GRIPP. C. Assume, during index creation we reach node c C. We add the tree instance of c to IND(G). If no other node of C has been traversed before, we traverse all remaining nodes of C all are reachable from c since C is a strongly connected component. Thus, every node in C will have a tree instance in RIS(c) and we are able to answer if w is reachable from u for u = c and w C with a single lookup. If u c, but u C the situation is different. Suppose RIS(u) contains a non-tree instance of c and suppose we use c as first hop node. Then we are able to answer if w is reachable from u (with w C) with two recursive calls, i.e., one to retrieve RIS(u) and one for RIS(c). To achieve this for every u C, we have to find a traversal order such that for every node u C, RIS(u) contains a non-tree instance of c. We therefore must solve the following problem: Find a node c C such that we may divide C in partitions P 1,..., P n with n deg in (c). For every P i, 1 i n compute a Hamilton path starting at node u and ending at node c, with u = c or u child node of a node in P j, j i. If we create GRIPP along those Hamilton paths we ensure for every node u C that RIS(u) contains at least one non-tree instance of c. Now suppose, we have not traversed any successor node of c in G when we traverse c, i.e., we have not traversed any nodes of C or any nodes in successor components of C. We traverse nodes in C along Hamilton paths [BS03] and also traverse all nodes in successor components of C. This means all reachable nodes of c in G have a tree instance in RIS(c). In addition, every non-tree instance in RIS(c) must also have its corresponding tree instance in RIS(c), i.e., c is a stop node. In Figure 5.5(b) the tree instance of c, c is shown as double circled node in the gray area. Given u C we may answer if w is reachable from u for any node w G with at most two calls, one initial call to test RIS(u), finding a non-tree instance of c (or possibly already an instance of 80
93 5.1. GRIPP w), and a second call using c as hop node to test RIS(c). As c is a stop node we do not have to use any further hop node, regardless if RIS(c) contains an instance of w or not. Next, we have to consider the case when both nodes are in different components of the component graph. In case the start node is in C and the end node in a successor component of C we have to ensure component C is traversed before any of its successor components. Clearly, this is not possible for any C, but the problem is alleviated (for some graphs) by the following observation. Erdös and Rényi [ER60] proved that directed random graphs with more edges than nodes contain one giant strongly connected component C. The size of C depends on the graph density. The experimental results given in [Wag06] show it is also true for scale-free graphs (but we are not aware of a formal proof). Therefore, graphs of a certain density usually appear as shown in Figure 5.5(a), with one component being very large (giant) and all other components being small. In this setting, it is only important to traverse the giant component before any of its successor components. Recall, for nodes in a component C that has been traversed before any of its successor components we may answer if w is reachable from u, with u C and w G, with two recursive calls. We may also estimate the number of recursive calls if both nodes are not in C. If RIS(u) contains no non-tree instance we immediately return false using one call. Otherwise, we have to query GRIPP recursively, but we use at most m n recursive calls with m number of edges and n number of nodes in the component graph. In some cases this number may even be reduced. Consider the case where u is sibling to nodes in C and RIS(u) only contains non-tree instances of nodes in C and possibly of nodes in successor components of C. Suppose we first use a node from C as hop node. We then need at most three recursive calls to answer if w is reachable from u. One call to retrieve RIS(u), finding the non-tree instance h of a node h C and using h as hop node, one call to retrieve RIS(h), which contains a non-tree instance of c, and one call to test RIS(c). If we ensure this order of hop nodes we may also answer reachability queries for such cases with a constant number of calls. Clearly, these considerations also have implications for a good traversal order of the graph. An algorithm to determine a good traversal order would proceed as follows. First identify all strongly connected components and build the component graph. Using Kosaraju s algorithm (Appendix A) this takes O(n + m) time. Second, determine the traversal order of components in the component graph by computing the size of the successor sets of all k components, which basically means to compute the transitive closure over the components and thus requires O(k 3 ) time. Third, compute a good order for nodes within every component C by first identifying a node c and then computing Hamilton paths as described above. As finding Hamilton paths in graphs is NP-complete [CLR01], especially this step is not feasible for practical application. Thus we use a heuristic when creating the GRIPP index, which we present in the following. Heuristic for Traversal Order During index creation, we want to ensure to traverse nodes from the giant strongly connected component before any other nodes. We assume nodes with a high degree have many successor nodes and may be reached by many nodes, 81
94 5. Implementations for Operators thus they are very likely member of the giant strongly connected component. Choosing the node with the highest degree as first node to traverse during index creation has the additional advantage that it also has many incoming edges and therefore will get many non-tree instances in IND(G). Thus, it is likely to find a non-tree instance of this node in the reachable instance set of other nodes, and recall, this node is a stop node. During graph traversal we try to traverse the child node with the largest reachable instance set first as this node covers a large part of the remaining graph. We use the heuristic that a node with a high degree is likely to have a larger reachable instance set than a node with a lower degree. Therefore, we prefer child nodes with a high degree, i.e., we traverse child nodes according to their degree. In Chapter 6 we show this using these heuristics we reach an almost constant query time over different sizes and shapes of graphs Distance Queries In this section we show how to answer distance queries using GRIPP. Please remember, for reachability queries it was sufficient to find one path between a given pair of nodes. For distance queries it is not the case as we want to find a specific path. We first consider a strategy for computing the distance between a given pair of nodes. We use a bidirectional breadth-first traversal for which the basic idea is shown in Figure 5.6. In a bidirectional search we start the search from the start node in forward direction and from the end node in reverse direction. In each step we expand both sets of reachable nodes. The search terminates when we find a node that is common in both sets or no set may be expanded further. In case we find a common node in both sets, the sum of the distances between the start and common node and common and end node is the distance between the two nodes. start end Figure 5.6.: Bidirectional search in a directed graph. The search proceeds in forward direction from the start node and in backward direction from the end node. The different colors represent the steps 1 3. The bidirectional search to answer distance queries for a given pair of nodes u, w using GRIPP works as follows. We first retrieve the tree instance u of u from IND(G). Next, we retrieve RIS(u). For each reachable instance v RIS(u) we may compute v depth u depth. Note, the difference in depth values does not reflect the distance between the two nodes u and v. It is possible that we reach v starting from u on a shorter way by using hop nodes. This has two implications for the algorithm. First, we know 82
95 5.1. GRIPP the distance between u and v only after all hop nodes with lower depth difference have been traversed. Second, we have to use more hop nodes than for answering reachability queries. After retrieving RIS(u) we want to retrieve RIS(w), which are the ancestors of w. We could retrieve some ancestors from IND(G), i.e., those that are on a path from the virtual root of O(G) to instances of w. This is neither complete nor efficient. Instead, we compute a second index called GRIPPreverse. This index is built in the same fashion as the original GRIPP index, which we now call GRIPPforward, but on the reverse edges of the graph. Thus, using GRIPPreverse we retrieve all ancestors for w the same way as we retrieve successors from GRIPPforward for u. The next step in the bidirectional search is to compare the instances in RIS(u) and RIS(w). If we do not find instances of the same node in both sets we have to proceed the search using the hop node with lowest difference of depth values. This instance may either be in RIS(u) or RIS(w). Otherwise, if we find instances of v in both sets, i.e., v RIS(u) and v RIS(w), we have to check if there exists an instance x RIS(u) such that x depth u depth < v depth u depth. If this is the case we have to proceed using hop nodes as we have not yet determined the distance between u and v. In analogy we perform this step for v. We have to continue until no further hop nodes are available or we have found a node that has instances in both sets and no further hop nodes with shorter depth difference can be used. Example 5.5 (Distance query evaluation). Consider again Figure 5.3 on page 76 and the GRIPP index structure in Figure 5.2(a) on page 73, which now becomes GRIPPforward. We are interested in the distance between nodes D and E. For simplicity we only describe a breadth-first search in forward direction. In RIS(D) we find no instance of E, thus we have to use a hop node. We may use A or B as hop nodes as both hop nodes have the path length of 4 2 = 2. We select A as next hop node. In RIS(A) we find an instance of E with path length = 4. In case of a reachability query we could stop here, but for the distance query we need to explore all hop nodes with shorter path length than the currently found shortest path length. Thus, we also have to use B as hop node. In RIS(B) we also find an instance of E with path length of = 3. As there are no further unexplored hop nodes we know the distance between D and E is 3. Pruning Strategies For answering distance queries we may only prune in two cases. Consider again Figure 5.4 (page 77). In the first case (Figure 5.4(a)), we skip h entirely because a non-tree instance of h has already been used as hop node and therefore the reachable instance set of the tree instance of h has already been checked, no shorter path may be found. In the second case (see Figure 5.4(b)), we cannot skip h when answering distance queries. There exists u U such that h is successor of u, i.e., h RIS(u) in O(G). Although, we have seen the entire reachable instance set of hop node h, we 83
96 5. Implementations for Operators may find shorter path lengths to instances in RIS(h). Thus, we have to use h as hop node. In the third case (Figure 5.4(c)) we again have be careful. We skip the pre- and postorder range of RIS(h) as we have already found h and thus all instances in RIS(h) with a shorter path length during the breadth-first search. We only have to consider the remaining instances of RIS(u). In the last case (Figure 5.4(d)), again, no pruning is possible. Algorithms We compute the distance between nodes using the GRIPP index. As for reachability we differentiate between computing the distance for a pair of nodes and the distance for all reachable nodes given a start node. In the following we describe the more complicated algorithm for determining the distance for a pair of nodes. We briefly describe the set-based algorithm later in this section. Pair of Nodes Algorithm 5.3 shows the function to compute the distance for a given pair of nodes. For the start and end node we have two sets each, one set for possible hop nodes (poss_hop_start and poss_hop_end) and one set for reached nodes (found_start and found_end). Initially, poss_hop_start and found_start contain only the start node. For each node in both sets we also store the length of the shortest path between the start node and this node found so far. For the start node to itself this length is 0. We do the same for the end node. In contrast to a bidirectional breadth-first search, where each step increases the distance by exactly one in each direction, in GRIPP we may increase the distance by more than one. Thus, we first check, in which direction we should expand the search. If the distance from the start node is smaller or equal to the distance from the end node, we expand the search in forward direction (line 7) by using function search. In this function we first consider all possible hop nodes from poss_hop_start to find the next hop node (lines 23 32). We basically use the hop node, whose path length to the start node is lowest (called next). In addition, in this step we delete all possible hop nodes for which we have found a tree instance with shorter path length to the start node (line 25). If we are not able to identify a next hop node we return (line 44). Otherwise, we use next to expand the search (lines 35 42). We add the node of next to found and delete it from poss_hop. In the next step we retrieve all reachable instances of next. For each instance we check if we have already seen an instance of this node. If we have not seen this node or seen this node with a longer path length to the start, we adjust found for this node. In addition, if it is a non-tree instance, we have to add it to the set of possible hop nodes. Otherwise, we delete it from this set if it exists. Before we return from search we adjust the distance for which we have already checked all hop nodes (line 45). After expanding either set we have to check if found_start and found_end contain instances of the same nodes (lines 11 17). Note, even if both sets have nodes in common, we may not have found the distance between the nodes. In found_start we only 84
97 5.1. GRIPP know the distance from the start node for those nodes, whose path length is shorter than or equal to the minimum path length in poss_hop_start. We return the distance if the path length for both nodes is shorter than distance_start and distance_end, respectively, and the sum of both shorter than the distance found so far. Distance for Sets For a given start node we may compute the distance to all reachable nodes. We use a variation of Algorithm 5.3. In this algorithm, we only require the search in forward direction starting from the start node. We terminate the search only if no further possible hop nodes are available. The set found_start contains all reachable nodes together with their distances to the start node. Distance with Length Restrictions In case we have a restriction on the maximal length of paths we have to add a condition. Whenever we compute distance_start or distance_end we check if they exceed the given path length. If this is the case, we return Path Length and Path Queries We may use GRIPP to answer path length and path queries. The strategy for GRIPP is similar to the approach for the recursive query strategy, but we may utilize the knowledge from the order tree O(G). Example 5.6 (Path query evaluation). Consider the GRIPP order tree, O(G) in Figure 5.2(b). We are interested in all paths between D and E. In RIS(D) exists a path from D over H to A. We may use A as hop node and find a path from A over B to E. Thus, the entire path is p 1 = D, H, A, B, E with length 4. We also find a second path p 2 = D, G, B, E with length 3 when we use B as hop node. Example 5.6 shows that we have to consider the GRIPP index structure only three times to find all paths between the two nodes. In comparison, for the recursive query strategy we would have to consider relation Edge six times. Pruning Strategies For path length and path queries we may use the same pruning strategies as for distance queries. Algorithms In this subsection we describe the function to compute actual paths using GRIPP. The computation of path lengths is a slight variation and thus described later in this subsection. 85
98 5. Implementations for Operators Algorithm 5.3: Function to find the distance between two given nodes u and v using the GRIPP index. 1 distance = ; dist_start = 0; dist_end = 0; continue = true 2 poss_hop_start [u] = dist_start; poss_hop_end [v] = dist_end 3 found_start [u] = dist_start; found_end [v] = dist_end 4 FUNCTION distance(u, v) RETURNS integer 5 repeat 6 if dist_start dist_end then 7 continue = search(poss_hop_start, found_start, dist_start, RIS_fw) 8 else 9 continue = search(poss_hop_end, found_end, dist_end, RIS_rv) 10 end 11 foreach n ( found_start found_end) do 12 if found_start [n] dist_start AND found_end [n] dist_end 13 AND found_start [n]+ found_end [n] distance then 14 distance = found_start [n]+ found_end [n] 15 continue = false 16 end 17 end 18 until continue = false 19 return distance 20 end 21 FUNCTION search(poss_hop, found, dist, RIS_dir) RETURNS boolean 22 min_distance = ; next = null 23 foreach hop poss_hop do 24 if poss_hop [hop] > found [hop] then 25 poss_hop [hop ].delete 26 else 27 if min_distance poss_hop [hop] then 28 dist = min_distance = poss_hop [hop] 29 next = hop 30 end 31 end 32 end 33 if next null then 34 found [next] = dist; poss_hop [next].delete 35 foreach i RIS_dir(next) do 36 d = dist + depth(i) - depth(next) 37 if (! found [i].exists) OR ( found [i].exists AND found [i] > d) then 38 found [i] = d 39 if inst(i) = non_tree_inst then poss_hop [i] = d 40 else if poss_hop [i].exists then poss_hop [i].delete 41 end 42 end 43 else return false 44 dist = getminval(poss_hop); return true 45 end Paths for Sets Algorithm 5.4 shows the function to compute paths given a single start node. We decided not to use the bidirectional search because we are only interested 86
99 5.1. GRIPP in cycle-free paths. Computing paths in forward and reverse direction would require to check each forward path if it contains nodes from the reverse path. As this is time consuming we decided to compute all paths in forward direction and then restrict them to those containing an end node. In Algorithm 5.4 we basically perform a depth-first traversal of the GRIPP index, during which we store nodes of the path that we are currently exploring in list path_nodes. The algorithm starts by retrieving RIS(u) ordered by their preorder values. For each instance inst we first compute the length to the start node, before we check if we have already seen the node of inst in the path we are currently expanding. If this is not the case, we add the node of inst to path_nodes at position len, which is the length to the start node. As we are interested in all paths, including their intermediate nodes, we loop through the nodes of the current path (lines 13 15) and add all nodes together with the remaining information as new tuples to relation Paths, which is returned in the end (line 22). If inst is a non-tree instance, we use this node as hop node by calling function paths. We continue using hop nodes until no further hop node may be used. If an end node or a set of end nodes is given, we restrict the returned paths to those containing as end node one of the nodes in the given set. Algorithm 5.4: Function to find all paths that start at a given node u using the GRIPP index. 1 path_nodes (0) = u 2 start = u 3 FUNCTION paths(u, length) RETURNS Paths 4 skip_to = 0 // RIS ordered by preorder value 5 foreach inst RIS(u) do 6 len = length- u depth + inst depth 7 if inst pre skip_to then 8 if inst node path_nodes then 9 skip_to = inst post 10 continue 11 end 12 path_nodes (len) = inst node 13 for n = 0; n < path_nodes.size; n ++ do 14 Paths Paths (start, inst node, len, path_nodes (n), inst edge_id, n) 15 end 16 if inst inst = nontree then 17 Paths Paths paths(inst node, len) 18 end 19 path_nodes (len) = null 20 end 21 end 22 return Paths 23 end The algorithm for computing all path lengths is a slight variation of Algorithm 5.4. Instead of returning paths, we are only interested in node pairs with their path length. This means, instead of looping through all nodes of the current path in lines of 87
100 5. Implementations for Operators Algorithm 5.4 we only return the start node, the current instance and the length of the path. The rest of the algorithm is the same. Paths and Path Lengths Queries with Length Restrictions If we have restrictions on the length of paths we add in line 16 of Algorithm 5.4 the condition that we only call the function paths(u, length) again if the path length does not exceed the given maximal path length Other Index Structures There exist several other index structures to support reachability and distance queries. In the following we present three of them, namely the transitive closure, Dual Labeling [WHY + 06], and Label+SSPI [CGK05]. The latter two may only be used to answer reachability queries. The transitive closure may be modified to also store distance values for each node pair Transitive Closure The transitive closure of a graph was formally defined in Definition 2.8 (page 11). The transitive closure for a directed graph contains one entry for each node pair (u, v) for which a directed path from u to v exists. The computation of the transitive closure may easily be extended to also contain the distance between the two nodes. Theorem 5.1. Computing the transitive closure using the Floyd-Warshall algorithm requires O(n 3 ) time and O(n 2 ) space [CLR01]. For a proof of this theorem see Cormen et al. [CLR01]. Index Creation Several algorithms [AJ87, Ioa86, War75, War62] were developed to compute the transitive closure inside an RDBMS. We use the semi-naive algorithm presented by Lu in [Lu87], as it is well suited for the implementation in an RDBMS. For this algorithm we give experimental results in Section 6.2. Reachability and Distance Queries We use the transitive closure to answer reachability and distance queries. For path length and path queries the transitive closure cannot be applied as this information is not present in the index structure. To answer reachability or distance queries a single lookup of relation TC is sufficient. 88
101 5.2. Other Index Structures Dual Labeling In [WHY + 06] Wang et al. emphasize that the size of the transitive closure grows quadratic in the number of nodes in the graph. Their conclusion is that the transitive closure is not applicable for large graphs. In their work they propose a different approach, called Dual Labeling, which we briefly present here. Index Creation The algorithm to create the Dual Labeling index consists of three steps. Given a graph G, the first step is to compute all strongly connected components of G. After identifying the components they construct the component graph G C, which forms a DAG and is usually smaller than G. The second step is to label a spanning tree of G C using pre- and postorder values as given in Algorithm 2.3 on page 15. To also encode the reachability information induced by non-tree edges of G C they create a transitive link table, which basically represents the transitive closure over non-tree edges of G C. The resulting index structure has the size O(n+t 2 ), where n is the number of nodes in G and t is the number of nodes in G C. As G C is usually smaller than G, the index created by Dual Labeling also generally is smaller than the transitive closure. An advantage is it may be queried for reachability in constant time as we explain in the following. Reachability Queries The algorithm for querying the index structure generated by Dual Labeling proceeds in three steps. Given a pair of nodes u, w and the question whether w is reachable from u. The algorithm first consider if both nodes are in the same connected component. If this is the case it returns true, otherwise it considers the pre- and postorder encoded spanning tree. If u is in a component that has a lower preorder and higher postorder number than the component in which w is, the algorithm returns true, otherwise it proceeds to query the transitive link table. Using this table and the pre- and postorder values of the components of u and w it is possible to determine whether w is reachable from u over non-tree edges. For further details on the index structure and query method we refer the reader to the original publication [WHY + 06]. Concluding, querying the index of Dual Labeling requires at most three queries and thus the query time is constant Label + SSPI For Dual Labeling it is still necessary to compute a transitive closure. In [CGK05] Chen et al. present an index structure to answer reachability queries where no such computation is necessary. They call their index Label + SSPI. Index Creation To create Label + SSPI the first two steps are identical to the first steps of Dual Labeling. They also start by identifying connected components, compute the component 89
102 5. Implementations for Operators graph G C, and then label a spanning tree using pre- and postorder values. In the third step they only store the non-tree edges of G C plus adjacent edges in the Surrogate & Surplus Predecessor Index (SSPI). For more details please refer to the original publication [CGK05]. Reachability Queries The query processing for Label + SSPI is similar to the reachability query processing of Dual Labeling. To answer the query whether w is reachable from u Label + SSPI also first considers if the two nodes are contained in the same component. If not, the reachability information encoded in the pre- and postorder labels is considered. If the two components are not reachable via a tree edge, they might be reachable via a combination of tree and non-tree edges. Thus, the algorithm has to consider the SSPI index. As this index only stores the non-tree edges plus some additional information, but no transitive relationships, it has to recursively traverse the SSPI index to answer the query. Thus, this method does not have a constant query time, but it depends linearly on the number of nodes and edges in the component graph RDBMS Capabilities The SQL 99 standard [GW02] defines WITH as syntax to answer recursive queries using SQL. Several database systems have implemented this standard so far [PBBS10]. The advantage of using recursive SQL is that no specialized, precomputed index is required to answer recursive queries inside an RDBMS. Oracle s implementation 1 does not comply to the SQL 99 standard as they use the syntax CONNECT BY PRIOR [Pri04]. In Section 6 we use Oracle 10g to answer reachability and distance queries. We cannot use recursive SQL for path and path length queries as it does not produce node disjunct paths. Instead it produces edge disjunct paths, which means, a node may occur multiple times in a path. This does not comply to the requirement given in Section 3.3 that paths returned from a PQL query must be simple, i.e., no node may occur multiple times in a path. Answering reachability and distance queries using Oracle s syntax requires as input relation Edge. This relation is then joined multiple times to itself using the condition given by CONNECT BY PRIOR... To ensure that the statement terminates on a graph containing cycles the additional keyword NOCYCLE has to be included. It is also possible to add a start node using START WITH. We may state the end node inside the WHERE clause. To also answer distance queries Oracle provides the attribute level, which is the number of edges traversed starting from the start node. Example 5.7 (Distance query using RDBMS capabilities). Using the following SQL statement in Oracle we find all nodes that are reachable from node D, including the distance from node D. The edges of the graph are stored in relation Edge
103 5.4. Recursive Strategies SELECT D AS start_node, to_id AS end_node, min(level) AS distance FROM Edge WHERE to_id <> D START WITH from_id = D CONNECT BY NOCYCLE PRIOR to_id = from_id GROUP BY D, to_id 5.4. Recursive Strategies We may also answer graph queries using user defined functions that implement the algorithms presented in Section 2.1. We either use breadth-first traversal strategies as presented by Algorithm 2.1 on page 13 or depth-first traversal strategies as presented by Algorithm 2.2 on page 14. The advantage of recursive query strategies is they require no precomputed index structure. In the following we omit any algorithms as they are only slight variations of the breadth-first and depth-first traversal given in Section 2.1. For each operator we only state the type of search and the alterations one has to make in comparison to the original algorithms. Reachability Queries For reachability queries we use depth-first traversal. For a given pair of nodes we start at the start node and stop as soon as we have found the target node. If only a start node is given, we continue the traversal until we have no more untraversed nodes available and return all traversed nodes. Distance Queries For distance queries we apply a breadth-first traversal. In case we are looking for distances for a pair of nodes we apply the bi-directional breadth-first traversal. We describe the principle in Section If just a start node is given, we use a simple breadth-first search starting from the start node. Path Length and Path Queries To answer path length queries we perform a depth-first traversal, during which we store all nodes of the current path. The traversal stops as soon as we have traversed all paths. This may require to traverse nodes multiple times when these nodes lie on different paths. For path length queries we only return the node pair together with their path lengths, while for path queries we also return the entire path for the each pair of nodes. 91
104 5. Implementations for Operators 5.5. Summary Table 5.1 summarizes the available implementations for the different operators for directed paths. For paths where the direction of the edges is irrelevant and for cases where we require the edges to be bidirectional we may use a specialized form of the recursive query strategies, which is not discussed in this work. GRIPP recursive TC SQL Dual Label Label +SSPI Reachability Distance no no Path length no no no no Path no no no no Table 5.1.: Summary of the available implementations for the different operators ( means the implementation is available) Related Work The aim of this thesis is to query large biological graphs fast and efficiently. Whenever possible we want to employ index structures for graphs. Many groups investigated index structures for different types of graphs to answer a variety of graph queries. We first consider strategies to index graphs for answering reachability and distance queries. We survey methods to compute the transitive closure and then proceed to various node labeling techniques, before we present methods that combine both. In [YC10] Yu and Cheng also review several of the methods presented in the following. In the area of path queries we present index structures that are based on common substructures, before we describe query strategies that do not rely on precomputed index structures, but traverse the graph at query-time. For all methods presented we discuss their applicability on typical biological graphs. As presented in Section 1.2 biological graphs have sizes between a few thousand nodes and edges and millions of nodes and edges. Table 5.2 summarizes the indexing times, index sizes, and query times for different index structures. In addition, in that table we present the maximum number of nodes and edges for which the index structure was created. Transitive Closure Based Methods Using the transitive closure (TC) we may answer reachability and distance queries. Many researchers have developed and improved algorithms to compute the transitive closure. The earliest ideas were to compute TC using an adjacency matrix. Warshal presented in [War62] this idea first. In [War75] Warren improved the algorithm given by Warshal. Both algorithms are based on three nested loops over all nodes in the graph. Agrawal and 92
105 5.6. Related Work Jagadish revisited the ideas and presented in [AJ87] algorithms to reduce the amount of main-memory consumption by successively looping over specific blocks of the adjacency matrix. In an RDBMS it is advantageous to use an adjacency list instead of an adjacency matrix as input. In [Lu87] Lu thus presents an algorithm that is based on the idea from Warren, which works on adjacency lists. Lu et al. show this algorithm only is advantageous if the intermediate results fit into main-memory [LMR87]. The naive, semi-naive, and logarithmic algorithm presented by Ioannidis in [Ioa86] are better suited to compute TC if this is not the case. In this work we use the semi-naive algorithm, which takes the tuples in TC with the highest distance at each step and produces new node pairs by joining those with the edge table. In contrast, the naive algorithm joins all tuples in TC with the edge table, thus producing more new tuples, but also more irrelevant tuples. The logarithmic algorithm takes all tuples with highest distance in TC and joins these with relation TC producing in step n node pairs for paths with length up to 2 n 1. Thus, this algorithm requires only log n steps to produce TC. The transitive closure has a great disadvantage for large graphs. It requires O(n 3 ) time to compute and O(n 2 ) space to store. Therefore, other index structures have been developed. Cohen et al. present the 2-Hop-Cover [CHKZ03], an index structure, which may be much smaller than the transitive closure. The idea of 2-Hop-Cover is the following. Determine a node h over which many paths lead. Use this node and find all nodes that reach h (stored in L in (h)) and which h reaches (stored in L out (h)). Repeat finding new hop nodes h until all node pairs of TC are covered. To answer a reachability query one has to join the sets L in (h) and L out (h) on the same h. To compute the 2-Hop-Cover we still require the transitive closure first. In [STW04] Schenkel et al. proposed HOPI, a method that is based on graph partitioning. They first compute 2-Hop-Covers for partitions of the graph, which is advantageous, as the 2-Hop-Cover for a partition is smaller than for the entire graph. To also cover cross-partition edges they have to do a postprocessing step. In [CYL + 08] Cheng et al. also propose to partition the graph for constructing the 2-Hop-Cover. In contrast to Schenkels approach they partition the graph hierarchically, i.e., they keep splitting the graph into two subgraphs. In their algorithm called MaxCardinality-I they try to create splits that are optimal for the 2- Hop-Cover. In [CY09] they use this strategy to compute a distance-aware 2-Hop-Cover, i.e., an index structure with which distance queries may be answered. On the downside, the size of this distance-aware 2-Hop-Cover is much larger than that of the reachability only encoding. The disadvantage of 2-Hop-Cover is that the transitive closure either for the entire graph or for partitions has to be computed, which may be impossible for typical biological graphs. In [JXRF09] Jin et al. extend the 2-Hop-Cover to a 3-Hop-Cover for answering reachability queries. They follow the idea presented by Bast et al. [BFM06] to create the 3-Hop-Cover. Bast et al. produce an index structure for road networks, which stores for each node a set of nearby hop nodes. The idea is as follows. For a sufficiently long distance regardless where you start you have to pass one close-by hop node to reach your destination, most likely on a highway. Likewise, before you reach your destina- 93
106 5. Implementations for Operators tion you have to pass through a hop node close to the destination. The challenge is to identify these hop nodes for each node. Bast et al. limit the complexity by applying a grid to the road network and searching only within a cell for those hop nodes. For the 3-Hop-Cover this method is not applicable as the graphs do not have 2-dimensional coordinates. In [JXRF09] the authors use a chain decomposition of a DAG. The chains may be viewed as highways, on which to identify hop nodes. For each reachability query they have to hop from a start node to a chain and find a hop from this chain to the end node, thus 3-Hop-Cover. Although the 3-Hop-Cover reduces the size of the 2-Hop-Cover as their experiments show, this index structure is still bigger than GRIPP. Node Labeling Based Methods The transitive closure or the 2-Hop-Label have the disadvantage that both have to store many node pairs. Index schemes that are based on node labeling only provide labels for each node, but may also be used to answer reachability queries. In [DS87] Dietz and Sleator present the pre- and postorder labeling. This method is heavily used in the area of XML documents to label elements of a document [Gru02, GvKT03, GvKT04]. Grust et al. present the Staircase join in [GvKT03]. They use preand postorder values to retrieve ancestor, successor, and sibling concepts efficiently. In [BKS02] Bruno et al. use this scheme to efficiently answer path and twig queries on XML documents. The drawback of pre- and postorder labeling is, it is only applicable to trees. In a DAG some nodes have to get more than one pre- and postorder label to cover the entire reachability information. In [TL05] we solve the problem by traversing and labeling each node as often as this node has a path to the root node. This leads to an exponential growth for the number of pre- and postorder labels in the number of additional edges. In [ABJ89] Agrawal et al. present a method to label a DAG using preand postorder labels, but they propagate intervals of a child node with multiple parent nodes to all their ancestor nodes. When appropriate, they merge intervals of ancestor nodes. Using this technique they avoid an exponential growth, but the construction requires O(n 2 ) time. In [CGK05] Chen et al. propose to index a DAG-structured XML document by first indexing a spanning tree and encode the remaining non-tree edges using a special index structure (SSPI, see 5.2.3). Yildirim et al. present in [YCZ10] GRAIL an indexing method for DAGs to answer reachability queries. In GRAIL they label each node with d postorder values, generated by d depth-first traversals using a random order of child nodes in each of the traversals. This way they quickly answer that w is not reachable from u by comparing the postorder values, but to verify that w is reachable from u they need to do a depth-first traversal. Thus, this method is only well suited for very sparse graphs, where few node pairs are reachable from each other, while for denser graphs other indexing methods should be employed. Jin et al. present in [JXRW08] a different method to label nodes of a DAG. They determine a traversal order based on a path decomposition of the DAG, where nodes on the same path are traversed before nodes of other paths. Each node receives a postorder value and a path identifier. The edges that are not covered by this index structure need to be traversed 94
107 5.6. Related Work recursively utilizing the information given by the postorder value and the path identifier, just as for GRIPP. Another node labeling scheme is the Dewey encoding. In [TVB + 02] Tatarinov et al. and in [GV07] Georgiadis & Vassalos applied this scheme to XML documents to answer XPath and XQuery queries. As with the pre- and postorder labeling the Dewey encoding is only applicable for tree structured graphs. Hybrid Methods Both indexing schemes, the transitive closure as well as node label based methods, provide advantages. Thus, several groups have developed indexes that combine these two schemes. In [HWYY05] He et al. present HLSS. The algorithm first computes strongly connected components and merges all nodes of a component to get the component graph. Next, they index a spanning tree using pre- and postorder labeling, and encode the non-tree edges using 2-hop-labeling. This achieves a constant query time. In [CYL + 06] Cheng et al. use the same idea as He et al., but in their algorithm called MaxCardinality- H they compute the 2-Hop-Cover differently. Wang et al. present in [WHY + 06] Dual Labeling, where they compute the transitive closure for non-tree edges as described in Section All hybrid methods reduce the number of edges, which need to be indexed, by first identifying strongly connected components and then indexing a spanning tree separately. Over the remaining non-tree edges the transitive closure or the 2-Hop-Cover needs to be computed and their complexities remain. Thus, for huge graphs those methods might not be applicable. In addition, all methods that reduce the graph by identifying the strongly connected components are not suitable to also index the distance between nodes. Subgraph Based Methods Queries on semi-structured or XML documents may also be considered as graph queries. For this type of queries several index structures have been developed, which we briefly discuss here for the sake of completeness. Most of these index structures rely on graphs that contain only a few distinct labels, which is not the case for typical biological networks. In [GW97] Goldman and Widom present DataGuides, an index structure that summarizes the structure of a semi-structured document, such as an XML document. The actual data values of leaf nodes are omitted. This index structure assumes that there exist few different labels on inner nodes, which result in only few different paths and thus in a small DataGuide compared to the original document. Milo & Suciu present in [MS99] a similar structure, which they call 1-index. They also present an index called T-index that is based on indexing paths of semi-structured data. The index of Giugno & Shasha, called GraphGrep [GS02], indexes all paths of a semi-structured document. As this index might grow quite considerably, this group presents in [SWG02] an index that holds all paths up to a specific length. The authors of [CMS02] also want to reduce the number of indexed paths. They analyze the workload and index only those paths 95
108 5. Implementations for Operators that frequently occur in results to queries. Kaushik et al. follow in [KSBG02] a different idea. They merge nodes in a DataGuide to produce the A(k) index, in which k stands for the length of the paths that must be preserved. This way, the DataGuide may be condensed. Yang and Sze describe in [YS07] a method to extract meaningful pathways from biological networks. Given a graph query q. Their aim is to find similar subgraphs in the data graph G. They present an algorithm that either maps an edge in q to an edge in G or that allows for up to n inserted, deleted, or mismatched edges between any two nodes in q. To achieve this, the authors compute the distance between selected node pairs and build and index of these at query time. In [ZL07] the authors argue this procedure is time consuming, especially for greater n and thus present an index structure, which enhances the performance of subgraph matching. Their index only allows the search for isomorphic subgraphs, i.e., an edge in q is mapped to an edge in G. Although interesting, both methods suffer from the fact that a user has to specify a concrete subgraph, of which isomorphic or very similar subgraphs in a data graph are retrieved. A researcher may find such similar subgraphs also by browsing the data graph. Indexing subgraphs is not just of interest for semi-structured data, but also in the following scenario. Given a database D that contains k graphs. Given a query Q, find the graphs of D for which the query graph is a subgraph. One option is to index all paths up to a specified length as proposed in [SWG02]. Several groups [KK04, YYH04, YYH05, HS06, CYY + 07, CKNL07, ZHY07] show it is better for this scenario to not just index paths, but build an index on frequent subgraphs. All these methods differ in their choice of frequent subgraphs and how to use the knowledge gained from these subgraphs for a query. These methods are interesting, but not applicable for PQL, as they only try to find graphs that are isomorphic to the query graph, which is not sufficient in our scenario. In addition, all methods that summarize the structure of the data or that are based on frequent subgraphs assume that many nodes in the graph share the same label. In biological graphs every node may have a different set of labels and each node must be considered independently. Thus, biological graphs provide no simple possibility to summarize the structure or to find frequent subgraphs. Querying Without Indexing Rosenthal et al. describe query strategies to answer reachability, distance, and path queries [RHDM86]. Their methods are solely based on graph traversal, no precomputed index is used. Clearly, this method has some advantages (no computation and storage of an index, no update problems), but on the downside, recursively traversing the graph at query-time requires lots of time as we show in Section 6. In [MW89] Mendelzon & Wood assume that they have a graph that has labeled nodes and edges. They also assume the query is a regular expression over node and edge labels, which may also include Kleene stars. They suggest to solve the query by using a finite state automaton, for which they present an algorithm that is based on graph traversal at query time. Kim et al. propose in [KYHJ02] a strategy to find the distance between a node pair in scale-free graphs with directed, unlabeled edges. Given a start node they traverse the 96
109 5.6. Related Work child nodes in order of their degree. The assumption is that the shortest path for many node pairs leads over the node with highest degree called hub in a scale-free graph. The experiments show the returned distance for a node pair is only about 50 % worse than distance of the exact algorithm. Zwick proposed in [Zwi98] also an exact and an almost exact algorithm to determine the distance for a pair of nodes. Their proposed algorithms only work on graphs with weighted ( 1), directed edges. In [ABE09] Alkhateeb et al. want find matching homeomorphic subgraphs in an RDF dataset given a PSPARQL query containing a regular expression. They use a nondeterministic finite automaton to represent the given regular expression pattern and then try to find instantiations of it in the RDF data set using a variation of the recursive search. Koschmieder [Kos09] et al. also answer graph queries containing regular path expressions. Both methods differ only in the usage of the automaton. As PQL does not allow for regular path expressions, it is not necessary to build an automaton for its execution. 97
110 5. Implementations for Operators Algorithm Index Time Index Size Query time Technique Largest Graphs GRIPP [TL07] O(n + m) O(n + m) O(m n) (worst), O(1) (average) Transitive Closure [Lu87] O(n 3 ) O(n 2 ) O(1) TC Dual Labeling [WHY + 06] O(n + m + n 3 ) O(n + m + n 2 ) O(1) PP labeling Component Graph + PP labeling +TC over m non-tree edges Label+SSPI [CGK05] O(n + m) O(n + m) O(m n) PP labeling (graph) 2-Hop-Cover [CHKZ03] O(n 3 ) O(nm 1/2 ) O(1) 2-Hop-Cover HOPI [STW04] O(n 3 ) O(nm 1/2 ) O(1) MaxCardinality- I [CYL + 08] O(n 3 ) O(nm 1/2 ) O(1) 3-Hop-Cover [JXRF09] O(kn 2 ) not given O(log n + k) PP labeling in XML [GvKT04] PP labeling in DAGs [ABJ89] GRAIL [YCZ10] O(d(n + m)) O(d(n)) Graph partitioning + 2-Hop-Cover Graph partitioning + 2-Hop-Cover Path decomposition + Partial TC O(n + m) O(n) (tree) O(1) (tree) PP labeling (tree) O(n 2 ) O(n 2 ) (DAG) O(1) (DAG) PP labeling (DAG) O(n + m) (worst), O(d) (best) Path tree [JXRW08] O(m + n log n) not given O(1) (DAG) HLSS [HWYY05] O(n + m + m 3 ) O(n + m + n m 1/2 ) O(1) MaxCardinality- G [CYL + 06] O(n + m + m 3 ) O(n + m + n m 1/2 ) O(1) multiple (d) PP labelings Path decomposition + PP labeling Component Graph + PP labeling + 2-Hop-Cover over m non-tree edges Component Graph + PP labeling + 2-Hop-Cover over m non-tree edges 5,000,000 nodes; 10,000,000 edges 10,000 nodes; 20,000 edges 13,969 nodes; 17,694 edges 400,000 nodes; 720,000 edges 900 nodes; 1,740 edges 5,244,872 nodes; 5,308,087 edges 500,000 nodes; 700,000 edges 10,000 nodes; 20,000 edges 5,077,531 nodes; 5,077,530 edges 1,000 nodes; 10,000 edges 100,000,000 nodes; 500,000,000 edges 100,000 nodes; 200,000 edges 1,000 nodes; 3,000 edges 336,244 nodes; 397,713 edges Table 5.2.: Overview of methods described in literature to answer reachability queries. 98
111 6. Performance of GRIPP In this chapter we evaluate the performance of the algorithms proposed in Chapter 5. In Section 6.1 we first describe the graphs used for our experiments, before we present the actual experimental setup. In Section 6.2 we show index times and index sizes for GRIPP in comparison to other methods, such as the transitive closure, Dual Labeling, and Label + SSPI. In Section 6.3 we show the query performance of GRIPP and compare it to other methods. For reachability and distance queries we compare GRIPP to the recursive query strategy, the transitive closure, and built-in recursive query capabilities of the RDBMS. For path length and path queries we compare GRIPP only to the recursive query strategy. From these experiments we shall deduce cost functions and cardinality estimates in Chapter Experimental Setup Generated Graphs To systematically evaluate our approach we use synthetic data. We created random and scale-free graphs in the size of 25 to 2,500,000 nodes and average outdegree between 1 and 10 using the methods described in [BA99]. The degree distribution in scale-free graphs follows a power law with an exponent γ = 2.7. More information about the graphs may be found in Appendix D Real-world Graphs We also use real-world data for evaluation. We use data of signal transduction pathways from NetPath [KMR + 10] and metabolic networks provided by KEGG [KGK + 04], Reactome [JTGV + 05], and BioCyc [KOMK + 05]. Nodes represent enzymes, chemical compounds, or reactions, while edges represent the participation of an enzyme or compound in a reaction. The degree distribution in these networks follows a power law with exponent γ = 3.0, i.e., they are also scale-free. Properties of these data set are provided in Table Implementation Details GRIPP as well as all competitive methods (based on the original code kindly provided by their authors and reimplemented for RDBMS) are implemented as stored procedures in Oracle 10g. We performed the tests on a DELL dual Xeon machine with 4 GB RAM. Queries were run without rebooting the database or emptying the cache. 99
112 6. Performance of GRIPP Database No. nodes No. edges Max. degree No. degree zero Signal transduction pathways NetPath KitReceptor NetPath Notch NetPath TGF-β NetPath TNF-α 809 1, Metabolic pathways KEGG 42,002 51,470 1,745 25,028 Reactome 11,795 19, BioCyc A. thaliana 10,951 23, BioCyc E. coli 3,844 8, BioCyc H. sapiens 8,816 18, Table 6.1.: Number of nodes and edges in biological graphs. Indexing is performed by stored procedures entirely inside the database system. The resulting index structure is stored in relations. The indexing times are averaged over five different graphs for every graph type (random and scale-free), number of nodes, and number of edges. All query functions are implemented as stored procedures as well. Therefore, we are able to compute the result entirely inside the RDBMS. Depending on the type of query (path, path length, distance, or reachability query) the procedure returns relation Paths given in Figure 3.8(b) on page 47 or a relation containing a subset of the attributes of Paths. For path length queries these are path_id, start, end, and path_length, for distance queries attribute path_id is omitted from this set and for reachability queries only attributes start and end are returned. For each query type, i.e., path, path length, distance, and reachability, different methods are available to query the graph. These methods are pair, seta, setb, setab, and all. For example, for reachability queries the method pair expects as input a pair of nodes and uses the algorithms presented in Chapter 6 to return the node pair if they are reachable from each other. Methods seta and setb expect as input a set of start nodes (A) or a set of end nodes (B) and use for each node in the set the algorithm for a start node to find all reachable nodes either on forward edges (seta) or on reverse edges (setb). The method setab expects as input a set of start and end nodes and uses for each pair of input nodes the algorithm for pairs of nodes. Method all uses the algorithm for a start node to find all end nodes to process each node of the graph. These five methods are implemented for each operator and each query type. Method pair is repeated 1,000 times on each graph, i.e., giving an average over 5,000 queries on 5 graphs. The methods setab, seta, setb, and all are repeated 2 times on each graph, i.e., giving an average over 10 queries on 5 different graphs. 100
113 6.2. Index Creation 6.2. Index Creation For indexing we compare GRIPP to the Transitive Closure (TC), the Dual Labeling approach by Wang et al. [WHY + 06], and the Labeling+SSPI approach by Chen et al. [CGK05]. The latter two algorithms are only able to index directed, acyclic graphs (DAG). Therefore we first identify strongly connected components (SCC) of G and collapse each component into one node. This step requires O(n + m) using Kosaraju s algorithm [CLR01] presented in Appendix A.1. Table 6.2 (page 114) shows the average time required to create the different index structures for scale-free graphs with 100 to 2.5 million nodes and twice the number of edges. The data show that GRIPP scales roughly linear with the number of nodes for a fixed density. For example, we computed the GRIPP index table for a scale-free graph with 2,500,000 nodes and 5,000,000 edges in less than three hours. Thus, we may compute the GRIPP index even for very large graphs. In contrast, the time to compute the TC grows roughly quadratic in the number of nodes. Therefore, we did not compute the transitive closure for graphs with more than 10,000 nodes. For example, computing the transitive closure for graphs with 10,000 nodes and 20,000 edges requires about 30,000 seconds or 8.5 hours. In comparison, computing GRIPP for the same graphs takes less than 35 seconds. The time to create the list of stop nodes for GRIPP always takes less than five seconds. We computed the SCC and thus Dual Labeling and Label+SSPI only for graphs with 25,000 nodes or less. The reason is that Dual Labeling does not scale well for index creation and Label+SSPI for query times. While the indexing times for Label + SSPI grow linear similar to GRIPP, the indexing time for Dual Labeling grows quadratic. Indexing times for random graphs are similar and thus omitted. Table 6.3 shows the average number of tuples for the different index structures for scale-free graphs with 100 % more edges than nodes. The figures show that the size of the GRIPP index table (figures are only shown for forward, figures for reverse are similar) grows linear with the size of the graph. The GRIPP index table of a scalefree graph with 10,000 nodes and 20,000 edges contains about 22,000 instances (20,000 instances for the edges, 2,000 instances for nodes without incoming edges, root nodes of subgraphs). In contrast, the transitive closure for the same graph contains more than 60 million node pairs. For graphs with twice the number of edges than nodes we always identify exactly one stop node. The figures for random graphs behave similar and are therefore omitted. Identifying strongly connected components and building the component graph reduces the number of nodes and edges. The component graph for graphs with average outdegree 2 contains about 45 % of the initial number of nodes and about 30 % of the initial number of edges. Dual Labeling as well as Label+SSPI use this component graph for indexing. Both index structures require three index tables, one to map nodes in the original graph to nodes in the component graph (contains as many tuples as there are nodes in the original graph), one for the pre- and postorder labeling of the spanning tree (contains as many tuples as there are nodes in the component graph) and either the transitive links for Dual Labeling or the Surrogate and Surplus predecessor index (SSPI) for Label+SSPI. 101
114 6. Performance of GRIPP The figures show that the transitive link table also grows quadratic in the number of the nodes in the graph, while the SSPI index grows linear with the number of edges in the component graph. Table 6.4 shows the time required to index graphs with 5,000 nodes and an increasing number of edges. The indexing times for GRIPP (times only shown for GRIPP forward, times for GRIPP reverse are similar) and the computation of the strongly connected components grow linear with increasing number of edges in the graph. Note, the indexing time for GRIPP is faster than the time for computing the strongly connected components. This behavior has two reasons, first in GRIPP we require only one pass of the graph, while for SCC we require two, and second constructing the component graph also takes time. The indexing times for the transitive closure also grow with increasing number of edges. In comparison, for Dual Labeling and Label+SSPI the indexing times drop. This behavior is due to size of the component graph. Table 6.5 shows that the component graph shrinks with increasing number of edges in the graph. While the component graph of a graph with 5,000 nodes and 5,000 edges contains almost the same number of nodes and edges, a graph with 5,000 nodes and 20,000 edges contains only about 200 nodes and 220 edges. Clearly, the time required to index a small component graph is much less compared to index a large one. Table 6.5 also shows that the transitive closure for graphs with 5,000 edges contains only few pairs of nodes. In contrast, in graphs with 5,000 nodes and 15,000 edges almost all nodes are reachable from each other, as the transitive closure contains almost the maximal number of (5, 000) 2 entries. For GRIPP, the number of instances is dependent on the number of nodes and edges in the graph. The table also shows that only graphs with very low density have more than one stop node for GRIPP Query Performance We now show a systematic evaluation of the performance of the algorithms presented in Section 5 using synthetic graphs. We provide average result sizes and average query times to answer different types of queries, namely reachability, distance, path length, and path queries. In addition, we show for each algorithm the performance of the different query methods, pair, all, seta, setb, and setab Reachability Queries We compare querying GRIPP to answer reachability queries with recursive query strategies using depth-first search, querying the transitive closure, Dual Labeling, and Label+SSPI. We also tested Oracle s 10g implementation of recursive SQL. We first show the performance for answering reachability queries for a given pair of nodes. Given an end node we stop the search in all algorithms as soon as we have found this end node. Figures 6.1(a) and 6.1(b) show the average number of tuples returned for answering reachability queries on different graphs. The average number of tuples for different number of nodes with fixed density of 2 remains almost constant at 0.6, i.e., about
115 6.3. Query Performance Average number of tuples for method pair on different graphs Average number of tuples for method pair on different graphs 1.4 random scale-free 1.4 random scale-free Avg.number of tuples Avg.number of tuples Number of nodes Number of edges (a) Increasing number of nodes. (b) Increasing number of edges. Figure 6.1.: Average number of tuples returned for answering reachability queries for pairs of nodes on different graphs. % of the randomly selected node pairs are reachable from each other. There are only minor differences between random and scale-free graphs. For graphs with 5,000 nodes and 5,000 edges Figure 6.1(b) shows that for a given node pair it is very unlikely (0.4 %) that the end node is reachable from the start node. With increasing graph density this probability increases. For graphs with 5,000 nodes and 30,000 edges almost all node pairs are reachable from each other. Avg. number of recursive calls Average number of recursive calls for method pair on scale-free graphs 1000 recursive GRIPP TC Dual SSPI Avg. query time (sec) Query times for method pair on scale-free graphs recursive GRIPP TC Dual SSPI Number of nodes Number of nodes (a) Number of calls. (b) Query times. Figure 6.2.: Average number of calls and average query time for answering reachability queries for pairs of nodes on different graph sizes (25 2,500,000 nodes) of scale-free graphs with average outdegree of 2. Figure 6.2(a) shows the average number of recursive calls for the different query strategies on scale-free graphs with 25 to 2,500,000 nodes and 100 % more edges than nodes. The figures for random graphs are similar and thus omitted. Clearly, for TC and Dual Labeling we need only one lookup to answer reachability queries. The number of recursive calls for the recursive query strategy and for Label+SSPI depends on the size of the 103
116 6. Performance of GRIPP graph. For graphs with 1,000 nodes and 2,000 edges we require on average 27 calls for the recursive query strategy, ranging from 1 call for a node without child nodes to 704 calls in worst case. When querying graphs using GRIPP the number of recursive calls remains almost constant at 2.2 over different sizes of graphs, with the maximum number of calls ranges from 4 to 8. This fact indicates that using our heuristics to create the GRIPP index produces an index structure that is close to the optimal index structure described in Section Thus, answering reachability queries requires almost constant time. The query times shown in Figure 6.2(b) for GRIPP, the recursive query strategy, and Label+SSPI correspond well with the number of recursive calls. The increase in query time for GRIPP on graphs with 2.5 million nodes and 5.0 million edges could not be explained. For random graphs this increase could even be seen for smaller graphs (500,000 nodes). A reason could not be detected and requires further investigation. Please note, querying GRIPP is only about three times slower than querying TC, while the recursive query strategy is up to over an order of magnitude slower than using GRIPP, depending on the graph size. The figures for Oracle s implementation of recursive SQL are not shown, as they are very slow even for small graphs. A single query on a graph with 100 nodes and 200 edges took more than 10 seconds, on a graph with 1,000 nodes and 2,000 edges it did not complete within 5 hours. The reason seems to be that Oracle enumerates all paths in the graph beginning from the start node. For tree structured data, as found for example in parts hierarchies, this poses no problem as only few paths will be found, whereas for general graphs the number of paths grows exponentially with an increasing number of nodes and edges. Avg. number of recursive calls Average number of recursive calls for method pair on scale-free graphs 100 recursive GRIPP TC Dual SSPI 10 Avg. query time (sec) Query times for method pair on scale-free graphs recursive GRIPP TC Dual SSPI Number of edges (a) Number of calls Number of edges (b) Query times. Figure 6.3.: Average number of recursive calls and average query time for answering reachability queries for pairs of nodes on different graph densities for graphs with 5,000 nodes and 5,000 50,000 edges. Figure 6.3 shows the average number of calls and average query time on scale-free graphs with 5,000 nodes and 5,000 to 50,000 edges. The figures for random graphs are similar and therefore not given. For GRIPP, the recursive query strategy, and La- 104
117 6.3. Query Performance bel+sspi the number of recursive calls peaks at 7,500 10,000 edges and then drops. This behavior is expected as for graphs with 5,000 edges the number of nodes without outgoing edges is high. Thus, we may start at such a node or hit such a node quite early during the search and return. For graphs with 7,500 or 10,000 edges this number is already much lower and thus, we have to explore more nodes on the way. In denser graphs the recursive query strategy more likely finds the target node during the search, whereas for GRIPP it is more likely to find a non-tree instance of a stop node in a RIS. For Label+SSPI the reason is different, as with increasing number of edges the size of the component graph decreases and thus the size of the giant strongly component increases. If a node pair is contained in the strongly connected component no recursive calls on SSPI are necessary Query times for method set A on graphs with 1000 nodes recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) Query times for method set AB on scale-free graphs with 1,000 nodes recursive (scale-free) GRIPP (scale-free) TC (scale-free) Avg. query time (sec) Size Set A Size Set B Size set A (a) Query times. (b) Query times. Figure 6.4.: Average number of tuples and average query time for answering reachability queries for a given set of start nodes on graphs with 1,000 nodes and 2,000 edges. Figure 6.4(a) shows average query times for the three different methods to answer reachability queries on graphs with 1,000 nodes and 2,000 edges for an increasing number of start nodes to return all reachable end nodes. In this case, querying the transitive closure is even slower than querying GRIPP, which is surprising. An explanation may be found in the fact that we implemented all query strategies as stored procedures. They take the set of start nodes as input. In case of the transitive closure, this set cannot directly be joined with TC, but for each node in the input set TC must be queried and the result appended to the output, just as for GRIPP. Thus, if the the size of the input set is 100 we have to perform 100 queries on TC, which has over 600,000 tuples, compared to about 260 queries on the GRIPP index, which has some 2,200 tuples. It should be faster if transitive closure is directly joined with the input set. Figure 6.4(b) shows average query times for varying sizes of start and end nodes on scale-free graphs with 1,000 nodes and 2,000 edges. The figures show that the time to answer reachability queries using recursive query strategies is much slower than using either TC or GRIPP. In comparison, querying TC is about ten times faster for 100 start and end nodes than GRIPP, which in turn is also about ten times faster than using the 105
118 6. Performance of GRIPP recursive query strategy. Concluding, the time to answer reachability queries using GRIPP is almost constant over different number of nodes and different densities of graphs for a given pair of nodes. Using TC is fastest to answer reachability queries, but we are not able to compute it for graphs with more than 10,000 nodes in reasonable time. The time required to answer reachability queries using the recursive query strategy grows steeply with increasing number of nodes. The query time of Dual Labeling is also constant, but its index structure cannot be computed in reasonable time for graphs with more than 25,000 nodes. The disadvantage of Label+SSPI is that its query time grows with increasing number of nodes in the graph. Thus, out of the five presented methods the only practical method to answer reachability queries in large graphs is GRIPP Distance Queries To answer distance queries we may use GRIPP, the transitive closure, if it also contains distance information, recursive query strategies, and DB-built-in recursive query functions. Average number of recursive calls for method pair on graphs Query times for method pair on graphs 100 recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) 1 recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) Avg. number of recursive calls 10 Avg. query time (sec) Number nodes (a) Number of calls Number nodes (b) Query times. Figure 6.5.: Average number of recursive calls and average query time for answering distance queries for pairs of nodes on different graph sizes (25 5,000 nodes) and average outdegree of 2. Distance queries return the same number of tuples as reachability queries, thus these figures are omitted. Figure 6.5 shows the average number of recursive calls and average query time to answer distance queries for a given pair of nodes on graphs with increasing number of nodes. As expected, the query time for the transitive closure is almost constant for different sizes of graphs as we require only one lookup in TC. In contrast, the number of recursive calls for the recursive query strategy and for GRIPP increases with increasing number of nodes in the graph. Although the recursive query strategy requires more recursive calls than querying GRIPP, the recursive query strategy is faster. The reason is that GRIPP performs many additional steps in each call to find the next hop node and to update distance values of nodes found, which is not necessary in the re- 106
119 6.3. Query Performance cursive query strategy. For graphs with increasing average degree GRIPP also performs worse than the recursive query strategy and querying TC. The difference between the query times for random and scale-free graphs is due to the difference in the average distance between two nodes (data not shown). The average distance for scale-free graphs is smaller than for random graphs. Thus, less calls and also less time is required to answer distance queries for a given pair of nodes on scale-free graphs. Query times for method pair on graphs with 2500 nodes and 5000 edges Query times for method pair and pair low on scale-free graphs Avg. query time (sec) recursive (random) GRIPP (random) TC (random) Oracle (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) Oracle (scale-free) Avg. query time (sec) recursive GRIPP TC recursive (low 10) GRIPP (low 10) TC (low 10) Maximum path length Number nodes (a) Length restriction. (b) With and without length restriction. Figure 6.6.: Average query time for distance queries with a given pair of nodes and length restriction on the path length on graphs with 2,500 nodes and 5,000 edges. If a length restriction in the query is given we may use this knowledge while computing the distance between a pair of nodes. Figure 6.6(a) shows the average query time for the four different methods on scale-free and random graphs with 2,500 nodes and 5,000 edges and increasing maximal path length. While the query time for TC remains almost constant for different path lengths, GRIPP, the recursive query strategy, and the DBbuilt-in recursive query function increases with increasing path length. The DB-built-in recursive query function shows the steepest increase. It is similar to the query times for path length and path queries given in the next section, which leads us to the assumption that the implementation enumerates all paths in the graph. Figure 6.6(b) compares the query times with and without length restriction. The figure shows that only GRIPP benefits from the length restriction, as it has to update the distance for fewer nodes Path Length Queries We may answer path length queries using either GRIPP or the recursive query strategy. For the evaluation we always restrict the maximal length of the paths. This has two reasons. First, in a large graph the length of the longest path between two nodes may be as long as the number of nodes in the graph. This path is most likely not of interest for a biologist, but shorter paths are. Second, with increasing path length the size of the intermediate and final results grows exponentially. Thus, the system quickly runs out of memory for larger maximal path length. Clearly, this is a limitation of the algorithms 107
120 6. Performance of GRIPP and the system presented and may arouse the discussion if the setting inside a classical RDBMS is up to date. Number of tuples for method pair on random graphs Number of tuples for method pair on scale-free graphs size 25/50 size 50/100 size 100/200 size 250/500 size 500/1000 size 1000/ size 25/50 size 50/100 size 100/200 size 250/500 size 500/1000 size 1000/2000 Avg. number of tuples Avg. number of tuples Maximum path length Maximum path length (a) Random graphs. (b) Scale-free graphs. Figure 6.7.: Average number of tuples as answer to path length queries for pairs of nodes on different graph sizes ( nodes) with average outdegree of 2 and increasing maximal path length. Query times for method pair, GRIPP on random graphs Query times for method pair, GRIPP on scale-free graphs 1 time 25/50 time 50/100 time 100/200 time 250/500 time 500/1000 time 1000/ time 25/50 time 50/100 time 100/200 time 250/500 time 500/1000 time 1000/2000 Avg. query time (sec) Avg. query time (sec) Maximum path length Maximum path length (a) Random graphs. (b) Scale-free graphs. Figure 6.8.: Average query time for path length queries for pairs of nodes on different graph sizes ( nodes) with average outdegree of 2 and increasing maximal path length. Figure 6.7 shows the average number of tuples for different graph sizes for the method pair. The figures show that with increasing maximal path length the average number of tuples returned increases. Remember, reachability queries return on average about 0.6 tuples for a given pair of nodes, regardless the graph type. In contrast, path length queries with the length restriction 10 return on average 24 tuples per pair on scale-free graphs with 500 nodes. Recall, for every unique path between start and end node the path length is returned. Thus, we may return the node pair with length l multiple times. In both graph types the average number of tuples is dependent on the graph size. For random graphs the curves remain almost parallel for different graph sizes over length, 108
121 6.3. Query Performance with larger numbers for smaller graphs. The reason is that in smaller graphs it is more likely to find the end node in a path of length l in comparison to larger graphs. In scalefree graphs these curves cross at a maximal path length of about 7. The reason is that the degree distribution, which is binomial in random graphs and follows a power law in scale-free graphs. While in random graphs the probability to find the target node in a path remains almost constant over the path length, in scale-free graphs this probability increases with increasing length as we might find a hub node, i.e., a node with a high outdegree. This node is connected to many other nodes, thus, it opens many paths, possibly some to the target node. In Chapter 7 we show that with increasing graph size the maximum outdegree of this hub grows. As this outdegree is similar in both graph types for small graphs (25 and 50 nodes) their behavior is similar. Only for larger graphs the maximum outdegree differs more considerably as Table 7.1 in Chapter 7 (page 118) shows. Figure 6.8 shows the average query time for randomly selected node pairs on different graph sizes using GRIPP. The query times for the recursive query strategy are slightly slower than for GRIPP (data not shown). In both cases, the query times are approximately proportional to the number of recursive calls for each method (data also not shown) Query times for method all on graphs with maximum path length less than 8 recursive random GRIPP random recursive scale-free GRIPP scale-free Query times for method all on graphs with 100 nodes and maximum path length less than 8 recursive (random) GRIPP (random) recursive (scale-free) GRIPP (scale-free) Avg. query time (sec) 10 Avg. query time (sec) Number nodes Number edges (a) Increasing number of nodes. (b) Increasing average outdegree. Figure 6.9.: Average query time to answer path length queries with method all on different graphs. Figures 6.9(a) and 6.9(b) show the time required to get the results for path length queries for all possible node pairs of a graph. The query times are proportional to the number of tuples returned (data not shown). The figures also show that GRIPP is slightly faster than the recursive query strategy. Using knowledge from the order tree O(G) does not boost the performance significantly. In addition, this speed increase gets lost for denser graphs, as Figure 6.9(b) shows. The reason is that the more additional edges are in the graph, the more often GRIPP needs to recursively query the index structure. Figure 6.10(a) shows the average number of tuples returned for varying set sizes of start and end nodes for random graphs. As expected the larger both sets are, the more 109
122 6. Performance of GRIPP Number of tuples for method set AB on random graphs with 100 nodes and maximum path length less than 8 Comparison of forward and reverse methods for GRIPP with increasing set sizes Avg. number of tuples Size Set A random Size Set B Avg. query time Size Set A GRIPP fw (random) GRIPP rv (random) Size Set B (a) Number of Tuples. (b) Query time. Figure 6.10.: Average number of tuples and average query time to answer path length queries for a varying number of start and end nodes on random graphs with 100 nodes, an average outdegree of 2, and given maximum path length of less than 8. tuples are returned, resulting in a maximum when both sets contain all nodes. Figure 6.10(b) shows the query times to answer path length queries for increasing sizes of the sets of start and end nodes (setab). We may utilize two different strategies. First, starting at the nodes in seta and search in forward direction (orange). Second, starting at the end nodes of setb and search in backward direction (green). The colored bottom indicates which method is advantageous for which number of nodes. In the orange area the reverse search strategy is advantageous, in the white area both strategies are almost equal, while in the green area the forward strategy is better. This indicates which method to use for answering path length queries when a set of start and end nodes is given Path Queries To answer path queries we may use GRIPP or the recursive query strategy. We again restrict the maximal length of paths. The figures are very similar to the figures for path queries, only the absolute number of tuples returned is higher for path than for path length queries. The query times are just slightly higher (figures not shown) as all nodes of the path must be returned. Concluding, for path length and path queries it is only feasible to return the result sets for a maximum path length as otherwise the number of tuples grows exponentially. When executing path and path length queries GRIPP has only marginal advantages over recursive query strategies Comparison of Query Types Figure 6.11 shows the average number of tuples returned to answer different types of graph queries on random and scale-free graphs for a given pair of nodes. While in 110
123 6.3. Query Performance both graphs the number of tuples returned for reachability and distance queries remains almost constant at 0.6, the numbers for path length and path queries vary. In random graphs the number decreases, as with increasing size of the graph it is less likely to find an end node on a path of length less than 10. In contrast, this number is hardly influenced in scale-free graphs, as we may find a hub node on this path, which opens many new paths. Number of tuples for method pair on random graphs Number of tuples for method pair on scale-free graphs 100 Path less 8 Path Length less 8 Distance Reachability 100 Path less 8 Path Length less 8 Distance Reachability Avg. Number of Tuples 10 Avg. Number of Tuples e e+06 Number of Nodes Number of Nodes (a) Random graphs. (b) Scale-free graphs. Figure 6.11.: Comparison of result set sizes to answer different types of queries given a pair of nodes (graphs with average outdegree of 2). For path and path length queries the maximum path length is restricted to less than 8. Figure 6.12 compares the average query times for method pair to answer queries for the four different query types, namely reachability, distance, path length, and path. Figures 6.12(a) and 6.12(b) show the comparison for random graphs, while Figures 6.12(c) and 6.12(d) do this for scale-free graphs. We only show the figures for GRIPP and the recursive query strategy as only those are able to process all four types of queries. Figures 6.12(a) and 6.12(c) show almost no difference between the depth-first search used to answer reachability queries and the bi-directional breadth-first search used for distance queries in the graphs under consideration. In contrast, for GRIPP a difference between both query types (reachability and distance) is clearly visible. The figures also show that on scale-free graphs the recursive query strategy and GRIPP are orders of magnitude faster for answering reachability and distance queries compared to answering path or path length queries. Note, there is an increase in query time to answer reachability queries for GRIPP with increasing graph size. Although the number of recursive calls remains almost constant over the different sizes of graphs the query time increases. We are not able to explain this behavior, but believe this is due to buffer size. More investigation is needed to look into this behavior. Figure 6.13 compares the query times to answer reachability, distance, and path length queries for a given pair of nodes on real-world graphs. Figure 6.13(b) shows that the query time to answer reachability queries using GRIPP is almost constant over the different graph types, which verifies our findings on synthetic graphs. In comparison, 111
124 6. Performance of GRIPP Query times for method pair, recursive on random graphs Query times for method pair, GRIPP on random graphs 1 Path less 8 Path Length less 8 Distance Reachability 1 Path less 8 Path Length less 8 Distance Reachability Avg. query time [sec] Avg. query time [sec] e e+06 Number of Nodes Number of Nodes (a) Recursive on random graphs. (b) GRIPP on random graphs. Query times for method pair, recursive on scale-free graphs Query times for method pair, GRIPP on scale-free graphs 1 Path less 8 Path Length less 8 Distance Reachability 1 Path less 8 Path Length less 8 Distance Reachability Avg. query time [sec] Avg. query time [sec] e e+06 Number of Nodes Number of Nodes (c) Recursive on scale-free graphs. (d) GRIPP on scale-free graphs. Figure 6.12.: Comparison of query times to answer different types of queries given a pair of nodes on graphs with average outdegree of 2. answering reachability queries using the recursive query strategy is dependent on the size of the graph as Figure 6.13(a) shows. For the small graphs from NetPath we may answer reachability queries very fast, while we require more time for the larger graphs KEGG and Reactome. The figure also shows that the query times for answering reachability and distance queries using the recursive query strategy are similar. In comparison, for GRIPP these times differ greatly, as we already showed for synthetic graphs. The query times to answer path length queries differ greatly for the different graphs. Both algorithms are very fast for the small and very sparse graphs from NetPath. For the metabolic networks of KEGG, Reactome, and BioCyc they require orders of magnitude more time than for answering reachability or distance queries Summary Reachability and distance queries return fewer tuples than path length queries, which in turn return fewer tuples than path queries. Only for path length and path queries a difference between random and scale-free graphs is observable. 112
125 6.3. Query Performance Avg. query time (sec) Query times for method pair, recursive on real world graphs Reachability Distance Path Length less 6 Avg. query time (sec) Query times for method pair, GRIPP on real world graphs Reachability Distance Path Length less NetPath TGF-beta NetPath Notch NetPath KitReceptor Reactome KEGG NetPath TNF-alpha BioCyc Hsapiens BioCyc Ecoli BioCyc Athaliana NetPath TGF-beta NetPath Notch NetPath KitReceptor Reactome KEGG NetPath TNF-alpha BioCyc Hsapiens BioCyc Ecoli BioCyc Athaliana Datasources (a) Recursive on real-world graphs. Datasources (b) GRIPP on real-world graphs. Figure 6.13.: Comparison of query times to answer different types of queries given a pair of nodes on real-world graphs. In terms of query times, for reachability and distance queries the TC would be the first choice, if it is possible to compute the TC. Otherwise, for reachability queries GRIPP is best, regardless the size or shape of the graph, while for distance queries a bi-directional breadth-first search is best. Answering path length and path queries is orders of magnitude slower than answering distance or reachability queries. For both query types GRIPP has minor advantages over the recursive query strategy. In the next chapter we use the knowledge gained in this chapter to develop cardinality estimates for the different types of queries and cost functions for the different algorithms. 113
126 6. Performance of GRIPP Table 6.2.: Average time in seconds and standard deviation for different indexing methods on synthetic scale-free graphs with 100 % more edges than nodes. The indexing times for GRIPP reverse are almost identical to GRIPP forward. (TC = transitive closure, SSC = strongly connected components) GRIPP forward TC SCC Dual Labeling Label + SSPI No. Nodes Index Stop nodes ± ± ± ± ± ± ± ± ± ± ± ± , ± ± ± ± ± ± , ± ± ,001.1 ± ± ± ± , ± ± ± ,771.9 ± ± , ± ± ± ,284.2 ± 1, ± , ± ± , ± ± ,000 1,140.7 ± ± ,000,000 2,811.9 ± ± ,500,000 9,061.7 ± ± Table 6.3.: Average number of tuples and standard deviation for different indexing methods on synthetic scale-free graphs with 100 % more edges than nodes. For these graphs we always identify just one stop node. The index sizes for GRIPP reverse are similar to the ones for GRIPP forward and thus omitted. (TC = transitive closure, SSC = strongly connected components) GRIPP forward TC SCC Dual Labeling Label + SSPI No. Nodes Index Nodes Edges transitive links SSPI ± 4.6 6,361 ± ± ± ± ± ,091 ± ,421 ± 6, ± ± ,461 ± ± ,000 2,183 ± ,728 ± 18, ± ± ,647 ± 2, ± ,000 10,941 ± ,080,968 ± 289, ± ± ,354 ± 24, ,834 ± ,000 21,810 ± ,310 ± ,143 ± ,534 ± 18, ,575 ± ,000 54,572 ± ,945 ± ,621 ± ,835,822 ± 142, ,193 ± , ,178 ± , ,433 ± ,000 1,091,998 ± ,000,000 2,184,202 ± ,500,000 5,465,794 ± 2,610,
127 6.3. Query Performance Table 6.4.: Average time in seconds and standard deviation for different indexing methods on synthetic scale-free graphs with 5,000 nodes and increasing number of edges. The indexing times for GRIPP reverse are similar to the times for GRIPP forward. (TC = transitive closure, SSC = strongly connected components) No. GRIPP forward TC SCC Dual Labeling Label + SSPI Edges Index Stop nodes 5, ± ± ± ± ± ± , ± ± ,365.5 ± ± ± ± , ± ± ,001.1 ± ± ± ± , ± ± ,002.9 ± 3, ± ± ± , ± ± ,001.3 ± ± ± ± , ± ± ± ± ± , ± ± ± ± ± , ± ± ± ± ± , ± ± ± ± ± , ± ± ± ± ± 0.01 Table 6.5.: Average number of tuples and standard deviation for different indexing methods on synthetic scale-free graphs with 5,000 nodes and increasing number of edges. The number of tuples for GRIPP reverse are similar to the number for GRIPP forward. (TC = transitive closure, SSC = strongly connected components) No. GRIPP forward TC SCC Dual Labeling Label + SSPI Edges Index Stop nodes Nodes Edges transitive links SSPI 5,000 7,481 ± ± ,383 ± 11, ,999 ± 2.2 4,998 ± 2.7 1,413,956 ± 43, ,798 ± ,500 9,109 ± ± 0.0 8,957,335 ± 313, ,586 ± ,513 ± ,916 ± 46, ,977 ± ,000 10,941 ± ± ,080,968 ± 289, ,202 ± ,128 ± ,354 ± 24, ,834 ± ,500 13,119 ± ± ,823,289 ± 8,280, ,109 ± ,446 ± ,548 ± 4, ,349 ± ,000 15,363 ± ± ,202,338 ± 212, ± ± ± 2, ± ,000 20,141 ± ± ± ± ± ± ,000 25,069 ± ± ± ± ± ± ,000 30,027 ± ± ± ± ± ± ,000 40,006 ± ± ± ± ± ± ,000 50,003 ± ± ± ± ± ±
128
129 7. GRIcano In this chapter we describe GRIcano, a query optimizer for PQL queries. The optimizer supports the operators and rewrite rules given in Chapter 4 and uses knowledge about implementations presented in Chapter 5. Our aim is to perform a cost-based query optimization. Therefore, cardinality estimates for approximating the size of intermediate result sets and cost functions for estimating the cost of each step are required. In Section 7.1 we develop cardinality estimates for the operators presented in Chapter 4. In Section 7.2 we provide cost functions for the different implementations of reachability, distance, path length, and path queries given in Chapter 5. In Section 7.3 we show how we use the cardinality estimates and cost functions to create GRIcano, a query optimizer for PQL queries, which is based on Volcano, the query optimizer generator presented in Section 2.4. We show the effect of graph query optimization using exemplary queries. In Section 7.4 we discuss related work on costbased graph query optimization Cardinality Estimates In this section we develop functions to estimate the cardinality, i.e., the size of the result set for the four operators presented in Section 4. The cardinality for the different operators is defined as follows. Definition 7.1 (Cardinality of operators) Let G = (V, E) be a graph. Let S V be the set of start nodes and T V be the set of target nodes, with u S, w T, s = S, and t = T. The cardinality of the reachability operator, φ(s, t, G), is the number of distinct node pairs u, w for which u w holds in G. The cardinality of the distance operator, Φ(s, t, G), is equal to φ(s, t, G). The cardinality of the path length operator, ψ(s, t, G) is the number of unique simple paths between any pair u, w in G. The cardinality of the path operator, Ψ(s, t, G) is the number of nodes in all unique simple paths between any u and w in G. Cost-based query optimization in RDBMS uses statistical data of relations to estimate cardinalities of predicates and sub-queries [Cha98]. These statistical data include the size of a relation and the distribution of values of attributes, which are stored in histograms [Ioa03]. We may consider graphs as input relations. Thus, we use key figures of 117
130 7. GRIcano the graph to estimate cardinality for graph queries. These properties should be easy to determine and significant. To our knowledge no cardinality estimates for graph queries have been described that are based on key figures of the graph. We found the number of nodes and edges, the outdegree of the node with highest outdegree, and the number of nodes without outgoing edges are properties that differentiate well between different graph sizes and shapes [TL10]. This knowledge may easily be gained while loading the graph. No. nodes No. edges Random graphs Scale-free graphs Max. degree Zero degree Max. degree Zero degree , ,000 2, ,000 10, ,000 20, , , , , , , , , , , ,000 1,000, , , ,000,000 2,000, , , ,000 5, , , ,000 10, ,000 15, ,000 20, ,000 25, ,000 30, ,000 40, ,000 50, Table 7.1.: Key figures for different random and scale-free graphs (Max. degree = outdegree of the node with highest outdegree; Zero degree = number of nodes with outdegree 0). The figures are averaged over five generated graphs for each graph type. Table 7.1 shows values of these key figures for different graphs. In Appendix D a more complete list, including the standard deviation, is given. The outdegree of the node with maximum outdegree for random graphs increases only slightly with growing number of nodes, whereas in scale-free graphs a much steeper increase is visible. In contrast, the increase in the number of nodes without outgoing edges increases in both graph types almost identically. For a constant number of nodes and increasing number of edges a clear difference between random and scale-free graphs in terms of the highest outdegree is also observable Reachability Operator Assume we want to estimate the cardinality of the reachability operator φ for a given set of start and end nodes. Clearly, the larger both sets are, the more tuples are returned on average. The size of the result set is also influenced by the density of the graph. With increasing average node degree we may expect an increase in the probability of 118
131 7.1. Cardinality Estimates u w until a saturation occurs. We use the saturation function 1 e ( x ) as it( reflects ) our experimental data better than other saturation functions, such as a b x or a x b+x (data not shown). Variable x is a placeholder for a function that contains the key figures of the graph. Our experimental evaluation in Chapter 6 shows that only the average degree influences the result size. Our experiments show that the fraction between additional edges (m n) and number of nodes n provides a good approximation. Equation 7.1 shows φ(s, t, G) to estimate the cardinality of reachability queries. φ(s, t, G) = s t (1 e m n n ) (7.1) Experimental Validation We validate Equation 7.1 using the data from Section 6. Figure 7.1 shows that the estimated cardinalities corresponds well with the experimentally determined result sizes. Please note, the shape of the graph, i.e., if it is random or scale-free, has no influence on the cardinality of the result set as we already discussed in Section Average number of tuples for method pair on different graphs Average number of tuples for method pair on different graphs random scale-free card_reach(s, t, G) random scale-free card_reach(s, t, G) Avg. number of tuples Avg. number of tuples Number of nodes Number of edges (a) Pairs of nodes, increasing number of nodes, avg. outdegree = 2. (b) Pairs of nodes, increasing avg. outdegree, n = 5,000. Figure 7.1.: Average number of tuples and estimated cardinality of reachability queries Distance Operator The cardinality of the distance operator is equal to the cardinality of the reachability operator as for each node pair only one distance value is returned. Thus, Equation 7.2 holds. Φ(s, t, G) = φ(s, t, G) (7.2) 119
132 7. GRIcano Distance with Length Restriction Using the distance operator we may restrict the result to those node pairs, whose distance is shorter than l. The upper bound of the cardinality of the distance operator with length restriction, Φ(low)(s, t, G, l) is Φ(s, t, G) for infinite l. Given a finite value for l we have to estimate the number of unique nodes that may be reached within path length l from a given start node. Assume we start at a single start node on a graph with average outdegree of m n = 2. At path length l = 1 we find on average 1 2 = 2 nodes, at length l = 2 we find 2 2 = 4 nodes, and at length l = 3 we find 4 2 = 8 new nodes, which results in = 15 nodes seen at length l. Equation 7.3 provides the function to calculate the number of nodes seen at length l, which is the number of nodes seen in the last step plus the number of newly found nodes. We may estimate the number of newly found unique nodes by multiplying the number of nodes found at length l 1 with the average outdegree. This equation would only be true on infinite graphs with a constant outdegree. For the graphs under consideration we have to regard several side effects, such as the degree of nodes or duplicate nodes in sets. In Equation 7.4 we account for these side effects by applying a correction factor corr_factor_dist(g, l) when computing the number of newly found unique nodes. We develop the correction factor in the following. For now assume d x (G) = m n. We provide an extended function for d x (G) in Equation 7.10 later in this section. { 1 l = 0 seen_nodes(g, l) = (7.3) seen_nodes(g, l 1) + found_nodes(g, l) l > 0 { 1 l = 0 found_nodes(g, l) = found_nodes(g, l 1) d x (G) corr_factor_dist(g, l) l > 0 (7.4) Assume at length l 1 we find k new nodes. Computing the number of nodes for l would result in k d x (G) new nodes, ignoring the correction factor. This figure is only valid, if all k nodes have outgoing edges. Table 7.1 shows this may not be true for the graphs under consideration, as about 15 % of nodes in scale-free graphs have an outdegree of 0. Thus, we have to consider the probability of a node having outdegree 0 to compute the number of nodes found at step l. If we assume equal distribution of selected nodes in the set of k nodes we may use Equation 7.5 to estimate this probability. pzero(g) = z n (7.5) Equation 7.5 estimates the probability for a node to have outdegree of 0 by dividing the number of nodes with outdegree 0 (z) with the number of nodes n. We use (1 pzero(g)), which is the probability of a node to have outgoing edges, to estimate the number of nodes found at length l. Applying the correction factor provided in Equation 7.5 results at length l in a set of k d x (G) (1 pzero(g)) new nodes, as only the fraction of 120
133 7.1. Cardinality Estimates (1 pzero(g)) nodes have outgoing edges. Equation 7.6 provides the general equation for estimating the number of newly found nodes. new_nodes(g, l) = found_nodes(g, l 1) d x (G) (1 pzero(g)) (7.6) Within this set we may find the same node multiple times over different edges. For distance queries we are only interested in finding each node once. To calculate the number of newly found unique nodes we have to estimate the probability of finding a node more than once. We may use the urn model known from statistics to develop an equation. Consider an urn with 1 red ball and n 1 black balls. We draw b times and (( ) always put back the ball. The probability to draw the red ball exactly c times is c 1 n (1 1 n )b c (b c) ) [KRBV06]. For our problem we may consider nodes of G as balls and new_nodes(g, l) as number of draws. ( ( ) 1 0 ( pintra(g, l) = ( ) new_nodes(g,l) 0 new_nodes(g, l) n n) 0 ( ) 1 1 ( ( new_nodes(g,l) 1 new_nodes(g, l) n n) 1 ( ( = ) new_nodes(g,l) n ( ( n) n) 1 new_nodes(g,l) 1 ) new_nodes(g, l) ) ) (7.7) In Equation 7.7 we estimate the probability of finding duplicate nodes within the set of newly found nodes, pintra(g, l) by estimating the probabilities of finding a node exactly 0 or 1 time. The sum of both probabilities is subtracted from 1 to estimate the probability of finding a node more than once. We may not only find duplicate nodes within the set of newly found nodes. At length l we may find nodes in the new set that have already been found in a previous step. We account for these duplicates by estimating the probability for a node to be part of the set of all nodes seen up to length l 1. Equation 7.8 shows the equation to compute the probability that a node has been found before. pinter(g, l) = seen_nodes(g, l) n (7.8) We use the three presented probabilities pzero(g), pintra(g, l), and pinter(g, l) for corr_factor_dist(g, l) in Equation 7.9. (1 pzero(g)) is the fraction of nodes with outgoing edges, which may expand the search. Using (1 pintra(g, l)) allows us to estimate the fraction of non-duplicate nodes in the set of newly found nodes. Finally, (1 pinter(g, l)) is the fraction of nodes that may not have been encountered before. Applying the correction factor in Equation 7.4 we may estimate the number of newly found unique nodes. 121
134 7. GRIcano corr_factor_dist(g, l) = (1 pzero(g, l)) (1 pintra(g, l)) (1 pinter(g, l)) (7.9) So far, the presented equations do not account for different graph types, i.e., if they are random or scale-free. The main difference between random and scale-free graphs is the outdegree of the node with highest degree x as Table 7.1 shows. Assume, we hit a node with high outdegree during the search. This node leads to many newly found nodes. Thus, we assume the higher the highest outdegree, the more nodes may be found in each step. So far we assume d x (G) = m n. We found the average outdegree being too low to account for the figures, as nodes with outdegree 0 are already handled by pzero(g, l). On the other hand, using d x (G) = x would be too high. We found that 1/2 (m/n + x) reflects our data best as Equation 7.10 shows. d x (G) = 1 ( m 2 n x ) (7.10) In Equation 7.11 we use Equations 7.2 and 7.3 to estimate the cardinality of distance queries with length restrictions given a set of start and end nodes. Φ(low)(s, t, G, l) = Φ(s, t, G) seen_nodes(g, l) n (7.11) Experimental Validation We validate Equation 7.11 using the experimentally determined result sizes from Section 6. Apart from the experimental values and the cardinality estimates based on Equation 7.11 we also show in Figure 7.2 the influence of the different terms. Using d x (G) = m n and corr_factor_dist = 1 (labeled found * d) we see an exponential growth of the cardinality, and an underestimation of the cardinality in scale-free graphs for small l. Taking the number of nodes with outdegree 0 into account by applying (1 pzero(g)) already decreases the exponential growth (found * d * (1-pzero)). We already observe a curve similar to the experimentally determined values when adding the correction factors for duplicate nodes within as well as between sets. Finally, using Equation 7.10 for d x (G) results in Φ(low)(s, t, G, l). The figures show we are able to predict the experimentally determined cardinality well. Figure 7.3 verifies this finding for constant l and average degree but increasing number of edges. The proposed function in Equation 7.11 captures the general trend for all graph sizes and types well, which is important. It is necessary to know if the size of the result set for s start and t target nodes is more likely close to (s t) or to
135 7.1. Cardinality Estimates Average number of tuples for method pair low on random graphs with 1,000 nodes Average number of tuples for method pair low on scale-free graphs with 1,000 nodes Avg.number of tuples size 1000/2000 card_reach(s, t, G) found*d found*d*(1-pzero) found*d*(1-pzero)*(1-pintra)*(1-pinter) card_dist_low(s,t,g,l) Avg.number of tuples size 1000/2000 card_reach(s, t, G) found*d found*d*(1-pzero) found*d*(1-pzero)*(1-pinter)*(1-pintra) card_dist_low(s,t,g,l) Maximum path length (a) Random graph Maximum path length (b) Scale-free graph. Figure 7.2.: Average number of tuples and estimated cardinality to answer distance queries with length restriction for pairs of nodes on graphs with 1,000 nodes and 2,000 edges and increasing path length Path Length Operator Next, we estimate the cardinality of the result when applying ψ. This operator returns the path lengths for all paths between a given pair of nodes. If a length restriction is given, the path lengths of all paths shorter than the given length restriction are returned. We have to estimate the number of paths between a given start and end node to estimate the cardinality of the result of the path length operator. Estimating the number of paths of length up to l is similar to estimating the numbers of nodes seen for distance queries with length restriction given in Equation 7.3. Thus, we may use similar considerations to estimate seen_paths(g, l). The number of paths seen at length l is the number of paths seen at length l 1 plus the newly found paths as Equation 7.12 shows. To estimate the number of newly found paths we may again assume to start the search at a single start node on a graph with average outdegree of 2. At length l = 1 we find on average 1 2 = 2 new paths and at l = = 4 new paths. Equation 7.13 shows the equation to estimate the number of new paths found at length l. As for estimating the number of nodes found in distance queries we have to consider correction factors to estimate the number of paths found at length l, which we discuss in the following. { 0 l = 0 seen_paths(g, l) = (7.12) seen_paths(g, l 1) + found_paths(g, l) l > 0 { 1 l = 0 found_paths(g, l) = found_paths(g, l 1) d x (G) corr_factor_path(g, l) l > 0 (7.13) Assume, at length l 1 we found k paths. We now have to estimate how many of 123
136 7. GRIcano Average number of tuples for method pair low with l<6 Average number of tuples for method pair low with l< random (exp) scale-free(exp) card_dist_low, random card_dist_low, scale-free random (exp) scale-free(exp) card_dist_low, random card_dist_low, scale-free Avg.number of tuples Avg.number of tuples Number of nodes Number of nodes (a) Path length < 6. (b) Path length < 8. Figure 7.3.: Average number of tuples and estimated cardinality to answer distance queries with length restriction for pairs of nodes on graphs with varying graph sizes and constant average outdegree of 2. these paths may be expanded to length l. There exist two conditions that prohibit the extension of a path. First, we clearly cannot extend the path if the end node of the path has no outgoing edges. Second, we stop extending the path if we find a node more than once in a path. We may account for both conditions using the correction factor for paths. We already provided Equation 7.5 that allows us to estimate the fraction of paths that contain end nodes without outgoing. For estimating the fraction of paths in which we may find a node multiple times we use the urn model presented in the last section again. The nodes of the graph again are the balls. As we want to estimate the probability of a node to occur more than once in a path the number of draws is l. Thus, we may compute the probability of not finding a node exactly 0 or 1 time in a path as Equation 7.14 shows. ( ( 1 ppathdupl(g, l) = 1 n = 1 ( ( 1 1 n ) 0 ( 1 1 n) l 0 ) l + 1 n ( 1 1 n) l 1 l ( ) ( ) l 1 1 ( l 1 0 n n) ) ( )) l 1 (7.14) In Equation 7.15 we take the probability of not finding a node with outdegree 0, pzero(g) and the probability of not finding a node more than once, ppathdupl(g, l) into account for corr_factor_path(g, l) to estimate the number of newly found paths at length l. corr_factor_path(g, l) = (1 pzero(g)) (1 ppathdupl(g, l)) (7.15) Using seen_paths(g, l) given in Equation 7.12 we may estimate the number of paths 124
137 7.1. Cardinality Estimates found for a single start node of length up to l, regardless their end nodes. To estimate the cardinality for a given pair of start and end nodes we have to divide seen_paths(g, l) by the number of nodes of the graph as Equation 7.16 shows. ψ(low)(s, t, G, l) = s t seen_paths(g, l) n (7.16) Experimental Validation To validate Equation 7.16 we use the experimentally determined values from Chapter 6. Figure 7.4(a) shows the results for varying number of nodes, constant average outdegree of 2, and maximum path length of less than 8. As the figures show, the cardinality estimates work well. Figure 7.4(b) shows the numbers for an increasing average outdegree but constant number of nodes. Although the estimates are not perfect, they provide a good approximation for the size of the result set. Figures 7.4(c) and 7.4(d) show the influence of the length of the path and the sizes of the sets of start and end nodes. As one may see, in both cases the estimated cardinalities correspond well with the observed values Path Operator Finally, we deduce the cardinality estimates when applying a path operator Ψ. In contrast to the path length operator, which only returns one tuple for each path, the path operator returns l + 1 tuples for each path. As the basic principle of the path operator and the path length operator is the same, we use the same considerations. Equation 7.17 shows the equation to estimate the number of tuples returned for a single start node. Remember, in Equation 7.12 we computed seen_paths(g, l). Equation 7.17 uses the same functions, but multiplies the number of newly found paths with their length to return the number of tuples. path_tuples(g, l) = { 0 l = 0 path_tuples(g, l 1) + (l + 1) found_paths(g, l) l > 0 (7.17) Using Equation 7.17 we estimate the cardinality of the path operator in Equation As for the path length operator, we assume we find an end node with equal probability and thus divide the number of tuples to represent all paths starting at a start node by the number of nodes in the graph. Ψ(low)(s, t, G, l) = s t path_tuples(g, l) n (7.18) Experimental Validation We validate Equation 7.18 using again the experimentally determined result sizes of the generated graphs from Chapter 6. The figures (data not shown here) show a similar 125
138 7. GRIcano Avg. number tuples Number of tuples for method all on graphs with maxium path length less than 8 1e+07 random scale-free card_path_len(s, t, G, l) random card_path_len(s, t, G, l) scale-free 1e Number of tuples for method all on graphs with 100 nodes and maxium path length less than 8 1e+08 random scale-free 1e+07 card_path_len(s, t, G, l) random card_path_len(s, t, G, l) scale-free Avg. number tuples 1e Number nodes Number edges (a) Increasing number of nodes, avg. outdegree = 2. (b) Increasing avg. outdegree, n = 100. Number of tuples for method all on graphs with 250 nodes and 500 edges 1e+07 random scale-free card_path_len(s, t, G, l) random 1e+06 card_path_len(s, t, G, l) scale-free Number of nodes for method set AB on scale-free graphs with 100 nodes and max. path length less than 8 scale-free card_path_len(s, t, G, l) Avg. number tuples Maximum path length Size Set A Size Set B (c) Increasing path length, n = 250, m = 500. (d) Varying sizes of sets, n = 100, m = 200. Figure 7.4.: Average number of tuples and estimated cardinality of path length queries. Figures 7.4(a) and 7.4(b) show the figures for all possible node pairs of different graphs using a given maximal path length of 8. Figure 7.4(c) shows the numbers for varying path lengths on graphs with 250 nodes and 500 edges, averaged over all node pairs of a graph. Figure 7.4(d) shows the number of tuples for varying set sizes. behavior as for path length queries. They confirm the cardinality estimates work well Validation on Real World Graphs We validate our cardinality estimates postulated for synthetic graphs using the biological graphs given in Table 6.1 (page 100). Figure 7.5 shows the experimentally determined result sizes and estimated cardinalities on different real-world graphs to answer queries for a given pair of nodes. Figure 7.5(a) shows that our estimates work well for reachability queries on metabolic networks (KEGG, Reactome, BioCyc), while for the smaller graphs from NetPath we severely overestimate the size of the result set. This behavior may be caused by the average degree, which is close to 1 for graphs from NetPath. Figure 7.5(b) shows the cardinality estimates for path length queries also work well for metabolic 126
139 7.2. Cost Functions networks, while we again overestimate the number of paths for graphs from NetPath. Determined and estimated cardinality for pairs of nodes on real world graphs 1 Exp. determined Estimated 0.8 Determined and estimated cardinality for pairs of nodes (l<6) on real world graphs 100 Exp. determined Estimated 10 Cardinality Cardinality NetPath KitReceptor NetPath Notch NetPath TNF-alpha NetPath TGF-beta KEGG Reactome BioCyc Athaliana BioCyc Ecoli BioCyc Hsapiens NetPath KitReceptor NetPath Notch NetPath TNF-alpha NetPath TGF-beta KEGG Reactome BioCyc Athalia BioCyc Ecoli BioCyc Hsapiens Datasources Datasources (a) Reachability queries. (b) Path length queries (length < 6). Figure 7.5.: Average number of tuples and estimated cardinality to answer queries for pairs of nodes on real-world graphs Cost Functions The goal of cost-based query optimization in RDBMS is to execute queries as efficiently as possible [Cha98]. For query optimization possible plans receive a cost value, which is essentially the sum of the predicted cost values of all individual steps. The plan with lowest predicted cost is used for execution. In this work we consider the time required to answer a query as sole efficiency criteria. Thus, our cost functions should roughly emulate the time required to compute the result. The cost functions depend on the algorithm used to compute the result. For the reachability and distance operator we provide cost functions for the recursive query strategy, GRIPP, and the transitive closure in Section and For the path length and path operator only the first two possibilities may be used, for which we provide cost functions in Section and For the two index-based methods, GRIPP and TC, the query optimizer requires apart from the cost function itself also the information if the indexes have been created to judge if they may be used. Cost functions generally require formulas to estimate CPU and IO costs [Cha98]. In this work we consider these two factors in the following equations, but we also use some simplifications. We assume uniform access cost for relations, i.e., accessing a small relation is as expensive as accessing a large one. In addition we do not take caching or sequential IO into account Reachability Queries To answer reachability queries we provide cost functions for three different algorithms, namely for TC, GRIPP, and recursive query strategy. Equations show the cost functions for a given set of start and end nodes. 127
140 7. GRIcano Cost φ(tc) (s, t, G) = s t IO_cost (7.19) Cost φ(gripp) (s, t, G) = s t 2.2 IO_cost + s t 2.2 CPU _cost (7.20) Cost φ(recursive) (s, t, G) = s t ( m n n + 1) IO_cost m (7.21) The terms IO_cost and CPU_cost depend on the system configuration and may be determined experimentally. For all equations we do not model the size of the input relation or access mode, as we assume uniform access cost. Thus, our cost model is merely based on the complexity of the algorithms. In Equation 7.19 the cost for querying TC is given. For each pair of nodes we have to access the transitive closure once and perform a lookup if the node pair is present in TC. No further processing is required. Answering reachability queries using GRIPP requires on average 2.2 recursive calls, regardless the size or shape of the graph (see Figure 6.2(a)). During each of these calls we have to consider the reachable instance set and determine the next hop node to use. As we assume this computation to occur in main memory we capture the cost of this additional computation by the second summand in Equation For the recursive query strategy the situation is more complicated. We assume the query time and thus the cost is proportional to the number of recursive calls. This number may range from one call for a node without outgoing edges to (n 1) calls on a graph with n nodes. We could argue on average we require (n/2) recursive calls to answer u w for any node pair u, w. Based on the experimentally determined values presented in Figure 6.2(a) on page 103 we found the number of recursive calls to be proportional to m n for graphs with constant outdegree. Figure 6.3(a) on page 104 shows that the degree is also dependent on the average outdegree. Thus, we add the second factor (n/m) in Equation 7.21, which reflects the fact that the higher the degree of a graph is, the less calls are required. Set of Start Nodes To evaluate reachability queries we also have implementations that return all reachable nodes given a set of start nodes. As these implementations are slightly different from the ones described by the cost functions above we have to define three more cost functions given in Equations 7.22, 7.23, and Cost φ(tcsingle) (s, G) = s IO_cost (7.22) Cost φ(grippsingle) (s, G) = s 3 IO_cost + s 3 CPU _cost (7.23) Cost φ(recursivesingle) (s, G) = s ( m n + 1) IO_cost m (7.24) For the transitive closure we have to query TC once for each node in the set of start nodes. Thus, Equation 7.22 holds. For GRIPP we have to perform an exhaustive search for which no longer 2.2 calls on average are sufficient. We require on average 3.0 recursive 128
141 7.2. Cost Functions calls as experimental data from Section 6.3 show. For the recursive query strategy we also require more calls for the exhaustive search as Equation 7.24 shows. Using the experimental data from Section 6.3 we found calls ( m n/m + 1). Experimental Validation We evaluate our proposed cost functions using the observed query times from Section 6.3. Figure 7.6(a) shows the observed query times for reachability queries given a pair of nodes, while Figure 7.6(b) shows the predicted cost. For IO_cost we use 1, for CPU_cost The figures show we are able to determine which algorithm is best suited for a given number of nodes in most cases. Only for very small graphs (less than 500 nodes) we overestimate the cost of the recursive query strategy and thus would never choose this algorithm if indexes are created. Choosing GRIPP does not significantly prolong the execution time of a query, as the difference in terms of query time between querying recursively and GRIPP is not big. Average query time for method pair on graphs Estimated cost for method pair on graphs 1 recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) 1000 cost_rec(s,t,g) cost_gripp(s,t,g) cost_tc(s,t,g) Average query time (sec) Estimated cost e e+06 Number of nodes Number of nodes (a) Average query times. (b) Estimated costs. Figure 7.6.: The figures show the actual query times and estimated costs for different graphs to answer reachability queries for a given pair of nodes. For a given set of start nodes Figure 7.7 shows we correctly estimate that using the recursive query strategy is worst. We underestimate the cost of querying TC compared to querying GRIPP. As already stated in Section 6 it may be due to sizes of the input relation, with TC being over two orders of magnitude larger than GRIPP. We currently do not take this factor in our cost model into account, as from our perspective the proposed functions work sufficiently well to suggest a good algorithm Distance Queries For distance queries we may use three different implementations, namely GRIPP, the recursive query strategy, and querying TC. Again, querying TC for distances is done in constant time as Equation 7.25 shows. To deduce the cost function for the recursive query 129
142 7. GRIcano Query times for method set A on graphs with 1000 nodes Estimated cost for method set A on graphs with 1000 nodes recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) cost_rec_single(s,n,m) cost_gripp_single(s,n,m) cost_tc_single(s,n,m) Avg. query time (sec) Estimated cost Size set A Size set A (a) Average query times. (b) Estimated costs. Figure 7.7.: The figures show the actual query times and estimated costs for different graphs to answer reachability queries for a set of start nodes. strategy we consider the figures in Section 6.3. They show the recursive query strategy requires almost the same time to answer distance queries without length restriction as to answer reachability queries. Thus, Cost Φ(recursive) (s, t, G) = Cost φ(recursive) (s, t, G). For GRIPP the situation is more complicated. The number of recursive calls for GRIPP is (n/m) times the number of calls for the recursive query strategy as we may use knowledge from the GRIPP index structure. On the other hand, in each call we have to invest more work to update the path lengths for all instances found in the current reachable instance set. We assume we have to update on average information for m/2 instances. Equation 7.26 shows our proposed cost function for GRIPP. Cost Φ(TC) (s, t, G) = Cost φ(tc) (s, t, G) (7.25) Cost Φ(GRIPP) (s, t, G) = s t ( m n n m + 1) n m IO_cost + s t ( m n n m + 1) n m m CPU _cost (7.26) 2 Cost Φ(recursive) (s, t, G) = Cost φ(recursive) (s, t, G) (7.27) Set of Start Nodes Equations 7.28, 7.29, and 7.30 show the proposed cost functions for the three algorithms that take a set of start nodes and return the distances for all reachable end nodes. Just like for reachability queries the algorithms perform an exhaustive search. We account for this fact by using ( m n/m) as term to estimate the number of recursive calls. 130
143 7.2. Cost Functions Cost Φ(TCSingle) (s, G) = Cost φ(tcsingle) (s, G) (7.28) ( ) m n Cost Φ(GRIPPSingle) (s, G) = s n m m IO_cost ( ) m n + s n m m m CPU _cost (7.29) 2 Cost Φ(recursiveSingle) (s, G) = Cost φ(recursivesingle) (s, G) (7.30) Distance with Length Restriction In Equations we define cost functions to answer distance queries with length restriction. The cost function for querying TC with length restriction is equal to Cost Φ(TC) (s, t, G). For the recursive query strategy and GRIPP we use function seen_nodes(g, l) developed for cardinality estimates of distance queries with length restriction in Equation 7.3 on page 120 to estimate the number of recursive calls. Both strategies use a bi-directional search. Thus, nodes up to length (l/2 1) are used in forward and reverse direction for recursive calls. For GRIPP we have to perform (n/m) less calls, as we only use tree instances, but in each call we have to perform on average (m/2) updates. Cost Φlow (TC)(s, t, G, l) = Cost Φ(TC) (s, t, G) (7.31) Cost Φlow (GRIPP)(s, t, G, l) = s t 2 seen_nodes (G, l ) 2 1 n m IO_cost + s t 2 seen_nodes (G, l ) 2 1 n m m CPU _cost 2 (7.32) Cost Φlow (recursive)(s, t, G, l) = s t 2 seen_seen (G, l ) 2 1 IO_cost (7.33) For distance queries with length restrictions that have a set of start nodes as input we may use Equations 7.32 and 7.33 in modified form. We omit the terms t and 2 and do not divide l by 2, thus leaving s seen(g, l 1) as first factor. As this step is straight forward, we omit the equations at this point. Experimental Validation We use the experimentally determined query times for answering distance queries given in Section to validate our proposed cost functions. As for reachability queries we assume IO_cost = 1 and CPU _cost = Figures 7.8(a) and 7.8(b) show that we correctly estimate using TC is always best. The cost functions would predict an advantage of GRIPP over the recursive query strategy for vary small graphs, which is not the case as Figure 7.8(a) shows. As the absolute time difference is only marginal it does not significantly prolong the query time. The cost functions also work well for a constant number of nodes and increasing number of edges 131
144 7. GRIcano (data not shown). Figures 7.9(a) and 7.9(b) show the query times for varying numbers of start and end nodes. Here as well the cost estimates are able to predict the fastest algorithm to use. Query times for method pair on graphs Estimated cost for method pair on graphs 1 recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) 1000 cost_rec(s,t,n,m) cost_gripp(s,t,n,m) cost_tc(s,t,n,m) Avg. query time (sec) Estimated cost Number nodes Number of nodes (a) Determined query time. (b) Estimated cost. Figure 7.8.: Comparison of query times and estimated costs to answer distance queries for a pair of nodes on graphs with increasing number of nodes and outdegree = 2. Please note, for the cost estimate of distance queries the cost estimates are identical for random and scale-free graphs as the maximum outdegree and the number of nodes without outgoing edges is not input to the equations. Figure 7.10 shows the experimentally determined query times and predicted cost to answer distance queries with length restriction. Here as well, we are able to predict that using TC is best. When comparing GRIPP and the recursive query strategy we correctly identify the latter to be the better strategy for larger graphs. Only for very small graphs (n 1,000) we prefer to use GRIPP instead of the recursive query strategy. For these graphs the query times for both strategies are in the same order, and thus, choosing the unfavorable algorithm may not have a huge impact on the query time Path Length Queries For answering path length queries we have two different implementations. We either recursively traverse the graph or query GRIPP. For both implementations it is important how many calls are required. In both cases the number is correlated to the number of paths found, for which Equation 7.12 (page 123) gives an estimate for a single start node. Equations 7.34 and 7.35 show the cost functions for the recursive query strategy and GRIPP to answer path length queries. We assume we require (n/m) less calls for GRIPP than for the recursive query strategy. For a given set of end nodes we have to compare the result of each start node with the set of end nodes. This factor is reflected in the second term. 132
145 7.2. Cost Functions Query times for method set AB on random graphs with 1000 nodes recursive (random) GRIPP (random) TC (random) Estimated cost for method set AB on random graphs with 1000 nodes cost_rec(s,t,n,m) cost_gripp(s,t,n,m) cost_tc(s,t,n,m) Avg. query time (sec) Size Set A Size Set B Estimated cost Size Set A Size Set B (a) Determined query time. (b) Estimated cost. Figure 7.9.: Query times and estimated cost to answer distance queries for a varying number of start and end nodes on random graphs with n=1,000, m= 2,000. Query times for method pair low with l<6 Estimated cost for method pair low with l<6 0.1 recursive (random) GRIPP (random) TC (random) recursive (scale-free) GRIPP (scale-free) TC (scale-free) 1000 cost_rec_low(s,t, G,l), random cost_gripp_low(s,t, G, l), random cost_tc_low(s, t, G, l), random cost_rec_low(s, t, G, l), scale-free cost_gripp_low(s,t, G,l), scale-free cost_tc_low(s, t, G, l), scale-free 100 Avg. query time (sec) 0.01 Estimated cost Number of nodes (a) Determined query time (l<6) Number of nodes (b) Estimated cost (l<6). Figure 7.10.: Query times and estimated cost to answer distance queries with length restriction (l<6) on different graphs with outdegree=2. Experimental Validation Cost ψ(recursive) (s, t, G, l) = s seen_paths(g, l) IO_cost + s log t CPU _cost (7.34) Cost ψ(gripp) (s, t, G, l) = s seen_paths(g, l) n m IO_cost + s log t CPU _cost (7.35) To validate Equations 7.34 and 7.35 we use the experimentally determined query times from Chapter 6. Figure 7.11 shows we are able to correctly predict that GRIPP is slightly better to answer path length queries given a pair of nodes. We are also able 133
146 7. GRIcano to model the influence of the graph type, i.e., we correctly predict that answering path length queries on scale-free graphs is more costly than on random graphs of the same size. Figure 7.12 shows the influence of the number of start and end nodes. These figures undermine the statement that the number of end nodes (Size Set B) has no significant influence on the cost of a query. Avg. query time (sec) Query times for method pair on graphs with n = 1,000 and m = 2, recursive (random) GRIPP (random) recursive (scale-free) GRIPP (scale-free) Estimated cost Estimated cost for method pair on graphs with n = 1,000 and m = 2, cost_rec(s,t, G, l) (random) cost_gripp(s,t, G, l) (random) cost_rec(s,t, G, l) (scale-free) cost_gripp (s,t,g, l) (scale-free) (a) Determined query time. (b) Estimated cost. Figure 7.11.: Determined query times and predicted cost for path length queries for a pair of nodes on graphs with n=1.000, m=2.000 and varying path length. Query times for method set AB on graphs with n=100 and l<8 Estimated cost for method set AB on graphs with n=100 and l<8 GRIPP (random) GRIPP (scale-free) cost_gripp(s, t, G, l) (random) cost_gripp(s, t, G, l) (scale-free) Size Set A Size Set B Size Set A Size Set B (a) Determined query time. (b) Estimated cost. Figure 7.12.: Determined query times and predicted cost for path length queries on graphs with n=100, m=200 and varying number of start and end nodes Path Queries In the following we provide cost functions for the different implementations to answer path queries. As for path length queries available implementations are querying recursively or GRIPP. Remember, path queries return all nodes of a path. To account for this fact we add the term Ψ(low)(s, t, G, l) ) CPU _cost in comparison to the equations for path length queries, as we have to process more output. 134
147 7.3. GRIcano Cost Ψ(recursive) (s, t, G, l) = s seen_paths(g, l) IO_cost + (s log t + Ψ(low)(s, t, G, l) ) CPU _cost (7.36) Cost Ψ(GRIPP) (s, t, G, l) = s seen_paths(g, l) n m IO_cost + (s log t + Ψ(low)(s, t, G, l) ) CPU _cost (7.37) Experimental Validation Again, we validate Equation 7.36 and 7.37 using the experimentally determined values from Section 6.3. The figures for path queries are similar to the curves for path length queries and thus omitted Validation on Real World Graphs We validate the cost functions postulated for generated graphs using the presented realworld graphs. Figure 7.13 shows the experimentally determined query times and predicted cost for reachability queries for a pair of nodes on real-world graphs. As the figures show, the proposed cost functions correctly predict that using TC, when available, is always favorable. For small graphs from NetPath the cost function favors GRIPP over the recursive query strategy, while the experimentally determined query times show an advantage of the recursive query strategy over GRIPP. This advantage is marginal and thus may not severely prolong the query time. For distance queries the cost functions correctly predict using GRIPP is always slowest (data not shown). Figure 7.14 shows that the measured query times and predicted cost for path length queries with length < 6 for pairs of nodes on real-world graphs. The figures show we correctly predict that for graphs from NetPath GRIPP is fastest for answering path length queries. For metabolic networks we predict GRIPP is better than the recursive strategy, which is not supported by the experimentally determined query times. We also predict that we have highest cost for the graph BioCyc A. thaliana, while the experimental data show we require most time to answer path length queries on the graph of KEGG. These figures show we may have to rethink about the cost functions for path length and path queries, by possibly also taking other key figures of graphs, such as diameter or radius [ARS + 08], into account GRIcano We implemented a novel prototype of a graph query optimizer, called GRIcano. This prototype is constructed according to Figure 2.3 in Section 2.3 on page 20. The first step of query processing is query parsing. We use SableCC [htt11a] to parse the PQL query into an internal representation. Using this representation we create the input file for the query optimizer. The optimizer is based on the Volcano framework presented in Section 2.4. We extended this framework for the algebraic operators and 135
148 7. GRIcano Avg. query time (sec) GRIPP Recursive TC Query times for pairs of nodes on real world graphs NetPath KitReceptor NetPath Notch NetPath TNF-alpha NetPath TGF-beta KEGG Reactome BioCyc Ecoli BioCyc Athaliana BioCyc Hsapiens Estimated cost GRIPP Recursive TC Estimated costs for pairs of nodes on real world graphs NetPath KitReceptor NetPath Notch NetPath TNF-alpha NetPath TGF-beta KEGG Reactome BioCyc Athaliana BioCyc Ecoli BioCyc Hsapiens Datasources Datasources (a) Determined query time. (b) Estimated cost. Figure 7.13.: Determined query times and predicted cost for reachability queries for a pair of nodes on real-world graphs. Avg. query time (sec) Query times for pairs of nodes (l<6) on real world graphs GRIPP Recursive Estimated cost Estimated costs for pairs of nodes (l<6) on real world graphs GRIPP Recursive NetPath KitReceptor NetPath TNF-alpha NetPath TGF-beta NetPath Notch KEGG Reactome BioCyc Ecoli BioCyc Athaliana BioCyc Hsapiens 10 NetPath KitReceptor NetPath TNF-alpha NetPath TGF-beta NetPath Notch KEGG Reactome BioCyc Athalia BioCyc Ecoli BioCyc Hsapiens Datasources Datasources (a) Determined query time. (b) Estimated cost. Figure 7.14.: Determined query times and predicted cost for path length queries (l< 6) on real-world graphs given a pair of nodes. rewrite rules presented in Chapter 4. An excerpt of the model specification may be found in Appendix E. We use the algorithms presented in Chapter 5 as implementations for our proposed operators. The third input to Volcano is the cost model, for which we have to provide information about the expected cardinalities of operators, the estimated cost, and applicability of an algorithm. In Appendix F we provide the C code to specify this information in Volcano. The output of the query optimizer is then parsed and a stored procedure for the execution in an RDBMS is created. To show how GRIcano works we provide Example 7.1. Example 7.1 (Query optimization in GRIcano). SELECT C, E FROM kegg LET node A, node B, node C, node E, path P, path Q, path R WHERE A.Name = Salicylate 136
149 7.3. GRIcano AND B.Name = Alcohol AND C.Type LIKE Reaction AND P.path = A[->]C AND P.length < 5 AND Q.path = B[->]C AND R.path = C[->]E AND R.length = 1 AND E.Type LIKE Compound ; name= Salidylate A P.length<5 B C R.length=1 type LIKE Reaction type LIKE Compound E name= Alcohol Figure 7.15.: The query graph for the PQL query form Example 7.1 that returns chemical compounds (in node variable E) produced by a reaction (node variable C), which may directly or indirectly be influenced by a combined intake of alcohol and aspirin, as salicylate is the first degradation product of aspirin. The PQL query in Example 7.1, for which Figure 7.15 shows the query graph, returns chemical compounds (in node variable E) produced by a reaction (node variable C), which is directly or indirectly influenced by a combined intake of alcohol and aspirin, as salicylate is the first degradation product of aspirin. It is well known that the combination of those two drugs is harmful [PHH83] and should thus be avoided. Figure 7.16 shows the original, not optimized query plan that is input to the extended Volcano query optimizer. For this plan Volcano estimates the cost of execution to IO plus CPU units using GRIPP as implementation for every path operator. After cost-based query optimization Volcano returns the plan presented in Figure In the optimized plan all path operators are replaced by reachability, distance and path length operators. The optimized plan has with 183, 431 IO plus CPU units much lower estimated cost than the original plan. In the optimized plan Volcano proposes to use implementations of GRIPP for φ start, Φ start, and ψ start. For the distance operator this is surprising, as we determined experimentally in Section 6.3 that the recursive implementation has advantages over GRIPP. The explanation is simple when considering Equations 7.26 and In our cost model IO cost of GRIPP is lower, while the CPU cost is much higher than for the recursive query strategy. As Volcano considers both cost types separately and first considers only IO as decision criteria, GRIPP is chosen over the recursive strategy. In this point 137
150 7. GRIcano Figure 7.16.: Original query tree before optimization. The estimated cost of execution of this plan is IO plus CPU units. The figures in brackets show the estimated cardinality for each step. The path operators are shown in red and Nodes X stands for Node X Label X. we should either rethink our cost model for Φ or alter the cost comparison of Volcano towards a combined cost function. In Figure 7.17 we also show the estimated and actual cardinalities of intermediate result sets. Note, we estimate the cardinalities of φ start, Φ start, and ψ start quite well. On the other hand, our cardinality estimates of and δ do not work well in all cases. The reason is that we did not focus on theses cardinality estimates and used simple functions. Using more elaborate functions should overcome this problem. The optimized expression tree given in Figure 7.17 is used as input to create a stored procedure, which may be used to execute the query inside an RDBMS. The query returns 5, 442 reaction compound pairs in about 5 seconds. In this list we may identify 3, 464 unique compounds. This number may be too large for manual inspection, but, provided a table of toxic or harmful agents we are able to use this result as input to a further query Experimental Evaluation We experimentally evaluate GRIcano using 16 exemplary queries, which are given in Appendix G. We use three queries for each query type, i.e., reachability (r), distance 138
151 7.3. GRIcano Figure 7.17.: Rewritten query tree after cost-based query optimization by GRIcano. Volcano estimates that this plan has costs of 183, 431 IO and CPU units. The path operators are again shown in red. The labels on edges are the actual cardinalities (in brackets the estimated ones) of intermediate results. (d), path length (l), and path (p). The three queries have the following general form. SELECT A, B / P FROM network LET node A, node B, path P WHERE A.Type = value AND B.Type = value AND P.path = A[->]B AND P.length {< =} x For query type 1 we use a condition that selects one node into node variable A, for query type 2 it select a small set (5 nodes), and for query type 3 it selects a larger set (some 500 nodes). For node variable B we use a condition that selects a larger set (some 2, 000 nodes) for all three queries. Volcano correctly identifies the different sizes of the sets. For reachability, distance, and path length queries we use A, B in SELECT, only 139
152 7. GRIcano for path queries we use P. We omit the path length condition for reachability queries, set the length to < n for distance and path queries and = n for path length queries. Queries 1 and 3 use KEGG, Query 2 uses Reactome. In addition, we created 4 complex queries (c). The query from Example 7.1 is labeled c2. For c3 we use a similar query, but return paths between nodes. Query c4 uses KEGG and Intact together. All 16 queries were parsed, optimized in Volcano, embedded into a stored procedure, and then executed inside an RDBMS. The parsing and query optimization in Volcano takes a few milliseconds for queries with up to two path variables. For queries involving three and more path variables, of which the path operator of some path variables may be rewritten to a path length, distance, or reachability operator, the optimization may take up to some seconds. One possibility to improve the optimization time is to consider the addition of dedicated pruning strategies for the newly proposed operators to Volcano. For each query type Volcano exchanges the path operator Ψ of the original plan. For reachability queries r1 r3 Volcano exchanges Ψ to φ start, and for distance queries d1 d3 Φ start(low) is used. For path length queries l1 l3 Ψ is exchanged to ψ start, and for path queries p1 p3 to Ψ start. For the complex queries c1 c4 Ψ is replaced by Volcano to the expected operators. Table 7.2 shows the estimated cost of execution for r1 r3 for the optimized plan using the reachability operator φ start and for the non-optimized plan using Ψ (with a maximum path length of 10 for cost estimation). The table shows the actual execution time of the optimized query. As we do not have an implementation that computes paths of infinite length, we cannot provide an execution time for the plan containing Ψ. The table also shows r3 takes longest, as the cost estimates predict. Query 1 Query 2 Query 3 Estimated cost IO CPU IO CPU IO CPU r: Optimized φ start , Non-optimized Ψ Query time (sec) r: Optimized φ start Table 7.2.: This table shows the estimated cost of execution for r1 r3 for the optimized plan (φ start ) and for the non-optimized plan (Ψ) for reachability queries. In addition, we give the query times for the execution of the optimized plan. Table 7.3 shows the effect of query rewriting on estimated cost and execution time for d1 d3, l1 l3, and p1 p3. The basis for all these queries are plans that contain the path operator Ψ, from which the output is joined with the sets of selected nodes for path variables A and B. This means, we have to compute for each node of the graph all paths of length < n and then discard all paths whose start or end node does not fulfill the condition. For Reactome (Query 2) this takes less than two hours, while for KEGG the computation did not finish within three hours. The figures in Table 7.3 show that the optimizer correctly identifies it is advantageous to rewrite the expression from the non-optimized version (NodesA A=start Φ) to Φ start. 140
153 7.3. GRIcano The query times as well as estimated cost are orders of magnitude lower than those of the original plan. The reason is that we only compute paths starting at selected nodes for node variable A, which is a small subset of all nodes in the graph. The data also show using the path length operator is cheaper than using the path operator, while using the distance operator has lowest estimated cost for the same length restriction. This fact is also supported by the experimentally determined query times. Table 7.3 also shows we underestimate the cost for Query 1 and overestimate the cost for Query 2. The reason is that the cost functions are based on averages, but individual nodes may exceed or fall below this average. For Query 1 we found a node, which exceeds this average, while in the set of input nodes for Query 2 several nodes fall below this average. Query 1 Query 2 Query 3 Estimated cost IO CPU IO CPU IO CPU d: Optimized Φ start l: Optimized ψ start p: Optimized Ψ start Non-optimized Ψ Query time (sec) d: Optimized Φ start l: Optimized ψ start p: Optimized Ψ start Non-optimized Ψ > 10, > 10, 000 Table 7.3.: This table shows the estimated cost of execution of d1 d3, l1 l3, and p1 p3 for the optimized plans (Φ start, ψ start, and Ψ start ) and for the nonoptimized plan (Ψ) for distance, path length, and path queries. In addition, the query times for the execution of the different plans are given. Figure 7.18(a) shows the expected and experimentally determined cardinalities of the 16 exemplary queries. We predict the highest cardinalities for r1 r3, which is also supported by the experimental values. The high difference between expected and actual cardinality is caused by the deficiency of the cardinality estimates for the de-duplication operator δ. Another observation is that the experimentally determined cardinalities for d1 d3, l1 l3, and p1 p3 do not correspond well with the predicted ones. An explanation is that the equations for the cardinality estimates, like the functions for cost estimates, are based on averages. The cardinality estimates for complex queries work quite well, except for Query c2, for which the problem with δ may be accounted. In Figure 7.18(b) we contrast the estimated costs (IO CPU) with the average query time of 10 query executions inside the RDBMS, which are partly already given in Table 7.2 and 7.3. We estimate the cost of execution for c1 c3 quite well and from manual inspection the rewritten plans contain the expected operators. Only for c4 we severely underestimate the cost compared to query execution time. Although the rewritten plan looks as expected, a reason may be that we use KEGG and Intact as data graphs. We assume the estimates for KEGG work well, while for Intact, a network 141
154 7. GRIcano that only contains undirected edges, our cost estimates may be far off as we only used directed graphs as basis for our cost functions. Clearly, this provides potential for further improvement. Cardinality 1e+12 1e+10 1e+08 1e Determined and estimated cardinality for optimized PQL queries Exp. determined Estimated Query times (sec) Determined query times and estimated costs for PQL queries Exp. determined (y) Estimated (y2) Estimated cost (IO CPU) 1 r1 r2 r3 d1 d2 d3 l1 l2 l3 p1 p2 p3 c1 c2 c3 c4 Query Number (a) Estimated and observed cardinalities. 0.1 r1 r2 r3 d1 d2 d3 l1 l2 l3 p1 p2 p3 c1 c2 c3 c4 Query Number (b) Estimated cost and observed query times. Figure 7.18.: Comparison of estimated cardinalities and cost to experimentally determined cardinalities and query times on exemplary PQL queries optimized by GRIcano. To get an insight into the quality of the selected plan we use Query c2 and enforce Volcano to apply certain algorithms or operators when creating the optimized plan. Table 7.4 shows the expected cost and actual query time of different plans. The figures show the execution of only one query plan (Φ start rec) is faster than the best plan chosen by Volcano (labeled as original ). In the plan with the fastest execution time the algorithm for Φ start is the recursive query strategy instead of GRIPP. As we already mentioned, Volcano takes as first decision criteria IO cost. For Φ start the IO cost of the recursive query strategy is higher than the cost for GRIPP, while CPU cost is lower. For the remaining operators using the recursive query strategy is, as predicted by the cost functions, worse than using GRIPP. Table 7.4 also shows the effect of not allowing the optimizer to use certain operators, which was enforced in Volcano by deleting rewrite rules. The query times show none of the alternative plans with different operators beats the original best plan. The plan, for which we disabled the rewrite rules for all operators to exchange an operator op to op start even has an execution time that is two orders of magnitude slower than the execution time of the original plan. This leads us to the assumption that one of the most important rewrite strategies is to restrict the number of start and target nodes early during query execution, i.e., pushing the selection operator down. Although in most cases IO and CPU cost are only marginally higher than for the original plan, we are able to determine the plan that is executed as one of the fastest. And this is the most important criteria when optimizing for query times. Based on the presented experiments we believe the cost-based optimizer created by Volcano, which uses the presented cardinality estimates from Section 7.1 and cost functions from Section 7.2 is able to propose a good execution plan. We showed that the 142
155 7.4. Related Work Query plan Cost IO Cost CPU Query time (sec) original 183, Algorithm exchange φ start rec 183, Φ start rec 186, ψ start rec 183, Operator exchange no φ 183, no Φ 284, no ψ 183, no op start 287, , Table 7.4.: Estimated cost and query execution times for different optimized plans when enforcing Volcano to use certain algorithms or operators or Query c2. optimizer avoids very time consuming plans by rewriting the algebra expression and choosing fast algorithms for the operators Related Work Cost-based query optimization in RDBMS is a well researched topic [SAC + 79, HFLP89, GM93, Ioa96]. There exist cost functions and cardinality estimates for the classic relational algebra operators selection, projection, and join. The situation is different for cost-based query optimization of graph queries. Only few researchers target this topic. In the following we discuss these works Cardinality and Cost Estimates To estimate the result size of reachability queries and general recursive Datalog queries Lipton and Naughton present in [LN89] and refine in [LN90] an adaptive sampling algorithm. They suggest to estimate the result size based on the given query Q only at query time. They assume the input of Q may be partitioned and each of these partitions returns a subset of the result. To estimate the result sizes they partition the input and continuously use partitions to compute the result until a predetermined cost parameter (for example time for sampling) is exceeded. The advantage of this method is that the cardinality estimate is based on the actual input set and thus may produce a better estimate than φ(s, t, G), which we propose in Equation 7.1. The disadvantage is that estimating the result size for input partitions also requires time, which may in some cases even exceed the actual query time Rule-based Query Optimization In [BR86] Bancilhon and Ramakrishnan review query optimization strategies for deductive databases. All strategies under review use a rule-based rewrite strategy. The aim of all those strategies is to push the selection operator as deep as possible. 143
156 7. GRIcano Buneman and colleagues also present in [BDHS96] rule-based optimization strategies for their graph query language UnQL and their proposed calculus. Their main focus also lies on pushing the selection operation as far down as possible. Odonez proposes in [Ord05] a similar strategy for the execution of graph queries in RDBMS using recursive views to answer reachability queries on trees and DAGs. For general graphs a length condition must be given to ensure termination. Their optimization strategy also enforces the pushing of the selection operator as deep as possible. In [Ord10] the authors show the effect of the different optimization strategies on different types of graphs. In general these rule-based strategies should allow for an efficient execution of graph queries. In Section 7.3 we also identified that restricting the set of start and target nodes early during query execution as important rewrite strategy. Although the proposed rule-based strategies may work in many cases, we believe using the cost-based query optimization, which we propose in this work, allows for more flexibility in choosing the order of the different operators Cost-based Query Optimization The first cost-based query optimization methods for graphs have been proposed for XML documents. Frasincar et al. [FHP02] and Cheng et al. [CYD07] optimize the order of the execution of XPath steps. Both approximate the cost through the expected size of an intermediate result, i.e., the larger the result size, the higher the cost. To estimate the cardinality they use frequencies of node labels involved in an XPath step. In [WPJ03b] and [WPJ02] Wu et al. show that frequencies of node labels may not predict the cardinality of an XPath step well enough. Thus, they propose a so called position histogram, which must be created for each unique label in the graph. In [WPJ03a] they use the cardinality and cost estimates that may be computed from these position histograms for cost-based query optimization of XML queries. In addition, their research shows limiting query optimization to left-deep plans may not select the most efficient plan in the XML setting using their algorithms, but full dynamic programming with different pruning strategies should be employed. Volcano does not specifically produce left-deep plans, as it uses what the authors call a directed dynamic programming approach, which prunes more costly sub-plans early during query optimization regardless their position in the tree. McHugh and Widom present in [MW99] the query processor of Lore, an XML database. Their cardinality estimates are based on an index of pre-computed paths of up to length k. This index may become quite large with growing k and growing number of different labels. Thus, in [AAN01] Aboulnaga et al. propose an index, which summarizes XML documents while computing the statistics for paths. All these optimizations and estimates only work if there are few different labels for nodes, which is the case in XML documents. In addition, position histograms and document summarization only work well on tree structured data. As biological networks are general graphs and each node has an individual label the proposed cardinality estimates are not applicable in our setting. In recent years cost-based query optimization is also used to find matching subgraphs 144
157 7.4. Related Work in a given data graph, which we discuss in the following. He and Singh want to find isomorphic subgraphs for a query graph in a data graph [HS08]. They consider each edge of the query graph separately and optimize the order in which edges of the query graph are mapped to edges in the data graph. They approximate the cost through the expected size of an intermediate result. To estimate this size they use the frequency of the edge label divided by the frequencies of their adjacent node labels. Zhao and Han argue it is not efficient to match the query graph edge by edge to the data graph [ZH10]. Their approach is to create subgraphs of the query graph and match these entirely to the data graph. To quickly reduce the number of possible matching points they propose an index structure called neighborhood signature, for which they index the vicinity of each node up to a predefined distance k. Using this index structure a cardinality estimate for each subgraph, i.e., how many possible matches this subgraph has, may be gained. To obtain the complete result all subgraphs of the query graph have to be joined, where the cardinality estimates are basis for the optimization of the join order. In [ZCÖ09] Zou and colleagues do not target the problem of finding isomorphic subgraphs, but assume each edge in the query graph maps to a path of up to length k in the data graph. They argue the frequencies of node and edge labels are not sufficient to estimate the number of possible matches for each edge in the query graph. They propose sampling to estimate connectivity for different distances and use this information for a correction factor of the cardinality estimate. The cardinality estimate is used as input for the cost functions for their proposed algorithms. The goal is to optimize the order in which paths bound to two adjacent edges of the query graph are joined. All three methods to optimize the search for matching subgraphs in a data graph also rely on the assumption that nodes in the data graph share few distinct node labels. As mentioned before, in biological networks each node has a unique label, and thus the presented methods for cardinality estimation may not be applicable. In [Kos09] Koschmieder targets the problem of finding regular path expressions in large graphs. Their approach is to first determine the exact number of node mappings for each node in the regular path expression. Starting with the node with the lowest number of mappings they extend the search along the edges in the regular path expression. A disadvantage of this approach is that it is only possible to extend paths starting from one initial node. In our approach we allow for different start points, whose paths may then be joined according to the cardinality and cost estimates. 145
158
159 8. Conclusion and Outlook This chapter concludes this thesis and provides an outlook on possible improvements for cost-based query optimization for graph queries Summary In this thesis we present GRIcano, a system that efficiently executes graph queries. For GRIcano we assume graphs are stored and queried using relational database management systems (RDBMS). We presented in Chapter 3 the syntax and semantics of an extended version of the pathway query language PQL to express graph queries. We employ ideas from RDBMS to improve the performance of query execution. Thus, the core of GRIcano is a cost-based query optimizer that is created using the Volcano optimizer generator presented in Section 2.4. In this thesis we made contributions to all three required components of the optimizer, the relational algebra, implementations, and cost model. In Chapter 4 we argued that relational algebra operators alone are not sufficient to express graph queries. Thus, we first presented new operators to rewrite PQL queries to algebra expressions. We proposed the reachability φ, distance Φ, path length ψ, and path operator Ψ. In addition, we provided rewrite rules for the newly proposed operators in combination with standard relational algebra operators that may be used to rewrite expressions and exchange operators. Secondly, we provided implementations for each proposed operator in Chapter 5. The main contribution is GRIPP, an index structure that efficiently executes reachability queries on very large graphs, containing directed edges. The advantage of GRIPP over other existing index structures, which we reviewed in that chapter, is that GRIPP is able to answer reachability queries for a given pair of nodes on average in constant time regardless the size or shape of the graph, while the created index is in the size of the graph. For example, using GRIPP to answer reachability queries for a given pair of nodes is on average only about 3 times slower than using the transitive closure (TC). In comparison, GRIPP is up to 30 times faster than the recursive query strategy. We also show we may employ GRIPP and the recursive query strategy, which we also present in Chapter 5, to provide implementations for all four proposed operators. The third component of Volcano is the cost model, which requires cardinality estimates for the proposed operators and cost models for the used implementations. Based on extensive experimental evaluation of the proposed implementations on generated random and scale-free graphs presented in Chapter 6 we present functions to estimate the cardinality of the φ, Φ, ψ, and Ψ operator in Chapter 7. In addition, we deduced functions for the presented algorithms to estimate the cost of execution. The novel approach is that these functions only use key figures of the graph, which are number of nodes 147
160 8. Conclusion and Outlook and edges, degree of the node with highest outdegree, and number of nodes with zero outdegree. We finally present the effectiveness of GRIcano using exemplary graph queries on real biological networks. The experiments conducted show the cost-based query optimizer created by Volcano is able to suggest a very efficient plan for graph query execution Future Work This thesis contains several aspects for which an improvement may be proposed. PQL Extension In Chapter 3 we describe an extended version of PQL. This query language already allows users to express various graph queries, but it lacks the capability to formulate regular expressions for paths. An example for a regular path expression is to look for paths that lead from a chemical compound A to another compound B over two to four reaction-compound edges, including at least one edge, which contains a self-catalyzing reaction R. Using PQL we are not able to express this query. This is a deficiency of the current version of PQL. Thus, regular path expressions may be incorporated in a future version. The proposed operators in Chapter 4 are sufficient to rewrite PQL statements to algebra expressions. Although, if PQL is extended for regular path expressions, an adaption of the path operator Ψ may become necessary. Improved Algorithms for Graph Queries A hugh improvement potential offer implementations for the proposed operators, especially for the distance Φ, path length ψ, and path operator Ψ. For the reachability operator φ we believe our proposed GRIPP index offers a very good compromise between query time and index size. Compared to other query methods for answering reachability queries we still believe using GRIPP is one of the best choices to efficiently answer reachability queries on different graph types and sizes in RDBMS. When considering implementations for the distance operator Φ the situation is different. In this case GRIPP has disadvantages over the competing recursive query strategy. Even the recursive query strategy is up to orders of magnitude slower than querying the transitive closure (TC). From my point of view, an interesting challenge for the future is to develop an index structure that is smaller than TC and less costly to create, but which also allows answering distance queries efficiently, possibly even in constant time. Recently developed algorithms to answer distance queries are interesting [CY09], but we believe there is still potential for improvement. To find efficient implementations for path length and path queries the situation is even more complex and also requires attention. In this work we focused on graphs with directed, unlabeled, and unweighted edges. For other graph types, e.g., for graphs with undirected edges, other types of algorithms may 148
161 8.2. Future Work be favorable. Thus, when developing new algorithms one should also consider graphs with other types of edges. Cardinality Estimates and Cost Functions In Chapter 7 we developed functions to estimate the cardinality of the different operators and costs of the execution of algorithms. These functions are based on extensive measurements on generated scale-free and random graphs with directed edges. Thus, for further validation and improvement of the functions it is inevitable to test them on other graph types and shapes. It may even become necessary to extend the equations for other key figures of the graph, such as size and number of connected components, the network diameter, radius, centralization, heterogeneity, or clustering coefficient [ARS + 08]. The result of such a validation might be that different functions could be needed for different types of graphs, even when employing the same algorithm. GRIcano We presented a prototypic implementation of GRIcano in Chapter 7. This prototype may be improved in several ways. First, it currently does not support the grouping operator, which is essential to express PQL queries that contain a HAVING clause. We believe the prototype already shows the prove of concept, even with this deficiency. Second, the estimates of the classical relational algebra operators may need improvement to better estimate the result size of join, selection, or de-duplication operators. We saw in Section 7.3 the evaluation of graph queries may be very time consuming, especially computing paths between sets of nodes. If we incorrectly estimate the sizes of input sets we may choose an unfavorable plan for query execution. We currently rely on static and predefined selectivity information of relations to estimate the number of selected nodes. A first improvement would be to use the estimations of an RDBMS, which are hopefully better. Still, these estimations may not always be correct. Thus, for a future implementation it may be prudent to revisit the ideas of dynamic query optimization [KD98], adapt them for graph query optimization, and then accommodate them in GRIcano. Currently, GRIcano returns the result of a graph query as relation. A user may expect as answer to a graph query a result graph, i.e., a representation containing nodes and edges. Thus, we may consider to extend an existing bioinformatics package for biological network visualization such as Cytoscape [SOR + 11]. An important feature of GRIcano is its modular design principle that allows for an easy extension of the system to include new implementations for the proposed operators. If new and more efficient algorithms for answering distance, path length or path queries are developed they may easily be included if cost functions are provided, which are designed similar to the cost functions presented in Section 7.2. Overall, we believe GRIcano is an important first step in cost-based graph query optimization and more contributions may and should be made in this area. 149
162
163 A. Strongly Connected Component A.1. Kosaraju s Algorithm Algorithm A.1 presents Kosaraju s algorithm, which performs two depth-first searches. The first depth-first search assigns every node in the reverse graph of G a postorder value. In the second depth-first search on the forward graph we assign every node the number of the strongly connected component. All nodes with the same number are in the same strongly connected component. As Algorithm A.1 only performs two depth-first searches the runtime is O( V + E ). Given a graph G = (V, E) and strongly connected components C 1, C 2,..., C n of G. We may construct the component graph G C by representing every strongly connected component C i as node in G C and add edges between C i and C j, with i j, if at least one node in C i has a directed edge to at least one node in C j. Theorem A.1. The resulting graph G C forms a DAG. Proof A.1. Let us assume G C is not cycle-free. Therefore, there must exist two nodes u, v V for which holds u v and v u. According to Definition 2.10 these two nodes must be contained in one strongly connected component, i.e., no such nodes exist. 151
164 A. Strongly Connected Component Algorithm A.1: Kosaraju s algorithm to compute strongly connected components of G. Data: graph G 1 traversed 2 pp_count 0 3 comp_count 0 4 FUNCTION strongly-connected 5 foreach u V do 6 if u / traversed then 7 depth-first-reverse(u) 8 end 9 end 10 traversed 11 foreach u V, ordered by postorder, descending do 12 if u / traversed then 13 depth-first-forward(u) 14 comp_count comp_count end 16 end 17 end 18 FUNCTION depth-first-reverse(u) 19 traversed traversed u 20 foreach v parents(u) do 21 if v / traversed then 22 depth-first-reverse(v) 23 end 24 end 25 u post pp_count end 27 FUNCTION depth-first-forward(u) 28 traversed traversed u 29 u comp comp_count 30 foreach v children(u) do 31 if v / traversed then 32 depth-first-forward(v) 33 end 34 end 35 end 152
165 B. Rewrite Rules for Operators B.1. Path Operator B.1.1. Restriction on Start and End Node Equations B.1 B.3 show the rewrite rules if restriction on start and end nodes are given. Ψ start cond, start=a.node_id(nodes A, Edge) := σ start=a.node_id (Nodes A Ψ cond (Edge)) = Nodes A A.node_id=start Ψ cond (Edge) (B.1) Ψ end cond, end=b.node_id(edge, Nodes B) := σ end=b.node_id (Ψ cond (Edge) Nodes B) = Ψ cond (Edge) end=b.node_id Nodes B (B.2) Ψ both cond, start=a.node_id AND end=b.node_id(nodes A, Edge, Nodes B) := σ start=a.node_id AND end=b.node_id (Nodes A Ψ cond (Edge) Nodes B) = Nodes A A.node_id=start Ψ cond (Edge) end=b.node_id Nodes B (B.3) B.1.2. Path Operator and Other Operators Equations B.4 B.6 show rewrite rules for Ψ in combination with other operators. σ sel_cond (Ψ start cond(nodes, Edge)) = Ψ start cond(σ sel_cond (Nodes), Edge) if sel_cond contains only conditions on attributes of Nodes (B.4) σ sel_cond (Ψ end cond(edge, Nodes)) = Ψ end cond(edge, σ sel_cond (Nodes)) if sel_cond contains only conditions on attributes of Nodes (B.5) 153
166 B. Rewrite Rules for Operators σ sel_cond (Ψ both cond(nodes A, Edge, Nodes B)) = σ sel_cond_a (σ sel_cond_b (Ψ both cond(nodes A, Edge, Nodes B))) = σ sel_cond_a (Ψ both cond(nodes A, Edge, σ sel_cond_b (Nodes B))) = Ψ both cond(σ sel_cond_a (Nodes A), Edge, σ sel_cond_b (Nodes B)) (B.6) if sel_cond_a contains only conditions on attributes of Nodes A and sel_cond_b contains only conditions on attributes of Nodes B and sel_cond = sel_cond_a AND sel_cond_b Equations B.7 B.10 show the rewrite rules to push the join into Ψ. R R.attr op A.attr Ψ start cond(nodes A, Edge) = Ψ start cond((r R.attr op A.attr Nodes A), Edge) (B.7) R R.attr op B.attr Ψ end cond(edge, Nodes B) = Ψ end cond(edge, (R R.attr op B.attr Nodes B)) (B.8) R R.attr op A.attr Ψ both cond(nodes A, Edge, Nodes B) = Ψ both cond((r R.attr op A.attr Nodes A), Edge, Nodes B) (B.9) R R.attr op B.attr Ψ both cond(nodes A, Edge, Nodes A)) = (Ψ both cond(nodes A, Edge, (R R.attr op B.attr Nodes B)) (B.10) B.2. Path Length Operator B.2.1. Restriction on Start and End Node Equations B.11 B.13 show the rewrite rules if restriction on start and end nodes are given. ψ start cond, start=a.node_id(nodes A, Edge) := σ start=a.node_id (Nodes A ψ cond (Edge)) = Nodes A A.node_id=start ψ cond (Edge) (B.11) 154
167 B.2. Path Length Operator ψ end cond, end=b.node_id(edge, Nodes B) := σ end=b.node_id (ψ cond (Edge) Nodes B) = ψ cond (Edge) end=b.node_id Nodes B (B.12) ψ both cond, start=a.node_id AND end=b.node_id(nodes A, Edge, Nodes B) := σ start=a.node_id AND end=b.node_id (Nodes A ψ cond (Edge) Nodes B) = Nodes A A.node_id=start ψ cond (Edge) end=b.node_id Nodes B (B.13) B.2.2. From Path Operator Ψ to Path Length Operator ψ Equations B.14 and B.15 show the rewrite rule to rewrite an expression containing Ψ to an expression containing ψ. δ(π A (Ψ cond (Edge))) = π A (ψ cond (Edge)) if A = {start, end, length} (B.14) δ(γ A,count(path_id) (Ψ cond (Edge))) = γ A,count(path_id) (ψ cond (Edge)) if A = {path_id, start, end, length} (B.15) Using Equation B.14 and Equations B.11 B.13 we can deduce the rewrite rules for Ψ start to ψ start, Ψ end to ψ end, and Ψ both to ψ both. B.2.3. Path Length Operator and Other Operators Equations B.16 B.18 shows the rewrite rule to push the selection operator to Nodes. σ sel_cond (ψcond(nodes, start Edge)) = ψcond(σ start sel_cond (Nodes), Edge) if sel_cond contains only conditions on attributes of Nodes (B.16) σ sel_cond (ψcond(edge, end Nodes)) = ψcond(edge, end σ sel_cond (Nodes)) if sel_cond contains only conditions on attributes of Nodes (B.17) 155
168 B. Rewrite Rules for Operators σ sel_cond (ψ both cond(nodes A, Edge, Nodes B)) = σ sel_cond_a (σ sel_cond_b (ψ both cond(nodes A, Edge, Nodes B))) = σ sel_cond_a (ψ both cond(nodes A, Edge, σ sel_cond_b (Nodes B))) = ψ both cond(σ sel_cond_a (Nodes A), Edge, σ sel_cond_b (Nodes B)) (B.18) if sel_cond_a contains only conditions on attributes of Nodes A and sel_cond_b contains only conditions on attributes of Nodes B and sel_cond = sel_cond_a AND sel_cond_b Equations B.7 B.10 show the rewrite rules to push the join into ψ. R R.attr op A.attr ψ start cond(nodes A, Edge) = ψ start cond((r R.attr op A.attr Nodes A), Edge) (B.19) R R.attr op B.attr ψ end cond(edge, Nodes B) = ψ end cond(edge, (R R.attr op B.attr Nodes B)) (B.20) R R.attr op A.attr ψ both cond(nodes A, Edge, Nodes B) = ψ both cond((r R.attr op A.attr Nodes A), Edge, Nodes B) (B.21) R R.attr op B.attr ψ both cond(nodes A, Edge, Nodes A)) = (ψ both cond(nodes A, Edge, (R R.attr op B.attr Nodes B)) (B.22) B.3. Distance Operator B.3.1. Restriction on Start and End Node Equations B.23 B.25 show the rewrite rules if restriction on start or end nodes are given. Φ start cond, start=a.node_id(nodes A, Edge) := σ start=a.node_id (Nodes A Φ cond (Edge)) = Nodes A A.node_id=start Φ cond (Edge) (B.23) 156
169 B.3. Distance Operator Φ end cond, end=b.node_id(edge, Nodes B) := σ end=b.node_id (Φ cond (Edge) Nodes B) = Φ cond (Edge) end=b.node_id Nodes B (B.24) Φ both cond, start=a.node_id AND end=b.node_id(nodes A, Edge, Nodes B) := σ start=a.node_id AND end=b.node_id (Nodes A Φ cond (Edge) Nodes B) = Nodes A A.node_id=start Φ cond (Edge) end=b.node_id Nodes B (B.25) B.3.2. From Path Operator Ψ to Distance Operator Φ Equation B.26 shows the rewrite rule to rewrite an expression containing Ψ to an expression containing φ. π A (Ψ cond (Edge)) = π A (φ cond (Edge)) if A = {start, end} and cond = [->] [-] [<->], (length_cond)? with length_cond := length op n (AND length op n)* and op := < (B.26) Using Equation B.26 and Equations B.23 B.25 we can deduce the rewrite rules for Ψ start to φ start, Ψ end to φ end, and Ψ both to φ both. B.3.3. Distance Operator and Other Operators Equations B.27 B.27 show the rewrite rule to push the selection operator to Nodes. σ sel_cond (Φ start cond(nodes, Edge)) = Φ start cond(σ sel_cond (Nodes), Edge) if sel_cond contains only conditions on attributes of Nodes (B.27) σ sel_cond (Φ end cond(edge, Nodes)) = Φ end cond(edge, σ sel_cond (Nodes)) if sel_cond contains only conditions on attributes of Nodes (B.28) 157
170 B. Rewrite Rules for Operators σ sel_cond (Φ both cond(nodes A, Edge, Nodes B)) = σ sel_cond_a (σ sel_cond_b (Φ both cond(nodes A, Edge, Nodes B))) = σ sel_cond_a (Φ both cond(nodes A, Edge, σ sel_cond_b (Nodes B))) = Φ both cond(σ sel_cond_a (Nodes A), Edge, σ sel_cond_b (Nodes B)) (B.29) if sel_cond_a contains only conditions on attributes of Nodes A and sel_cond_b contains only conditions on attributes of Nodes B and sel_cond = sel_cond_a AND sel_cond_b Equations B.30 B.33 show the rewrite rules to push the join into Φ. R R.attr op A.attr Φ start cond(nodes A, Edge) = Φ start cond((r R.attr op A.attr Nodes A), Edge) (B.30) R R.attr op B.attr Φ end cond(edge, Nodes B) = Φ end cond(edge, (R R.attr op B.attr Nodes B)) (B.31) R R.attr op A.attr Φ both cond(nodes A, Edge, Nodes B) = Φ both cond((r R.attr op A.attr Nodes A), Edge, Nodes B) (B.32) R R.attr op B.attr Φ both cond(nodes A, Edge, Nodes A) = Φ both cond(nodes A, Edge, (R R.attr op B.attr Nodes B)) (B.33) B.4. Reachability Operator B.4.1. Restriction on Start and End Node Equations B.34 B.36 show the rewrite rule if restrictions on the start or end node are given. φ start cond, start=a.node_id(nodes A, Edge) := σ start=a.node_id (Nodes A φ cond (Edge)) = Nodes A A.node_id=start φ cond (Edge) (B.34) 158
171 B.4. Reachability Operator φ end cond, end=b.node_id(edge, Nodes B) := σ end=b.node_id (φ cond (Edge) Nodes B) = φ cond (Edge) end=b.node_id Nodes B (B.35) φ both cond, start=a.node_id AND end=b.node_id(nodes A, Edge, Nodes B) := σ start=a.node_id AND end=b.node_id (Nodes A φ cond (Edge) Nodes B) = Nodes A A.node_id=start φ cond (Edge) end=b.node_id Nodes B (B.36) B.4.2. From Path Operator Ψ to Reachability Operator φ Equation B.37 shows the rewrite rule to rewrite an expression containing Ψ to an expression containing φ. δ(π A (Ψ cond (Edge))) = π A (φ cond (Edge)) if A = {start, end} and cond = [->] [-] [<->] (B.37) Using Equation B.37 and Equations B.34 B.36 we can deduce the rewrite rules for Ψ start to φ start, Ψ end to φ end, and Ψ both to φ both. B.4.3. Reachability Operator and Other Operators Equations B.38 B.36 show the rewrite rules to push down the selection operator. σ sel_cond (φ start cond(nodes, Edge)) = φ start cond(σ sel_cond (Nodes), Edge) if sel_cond contains only conditions on attributes of Nodes (B.38) σ sel_cond (φ end cond(edge, Nodes)) = φ end cond(edge, σ sel_cond (Nodes)) if sel_cond contains only conditions on attributes of Nodes (B.39) 159
172 B. Rewrite Rules for Operators σ sel_cond (φ both cond(nodes A, Edge, Nodes B)) = σ sel_cond_a (σ sel_cond_b (φ both cond(nodes A, Edge, Nodes B))) = σ sel_cond_a (φ both cond(nodes A, Edge, σ sel_cond_b (Nodes B))) = φ both cond(σ sel_cond_a (Nodes A), Edge, σ sel_cond_b (Nodes B)) (B.40) if sel_cond_a contains only conditions on attributes of Nodes A and sel_cond_b contains only conditions on attributes of Nodes B and sel_cond = sel_cond_a AND sel_cond_b Equations B.41 B.44 show the rewrite rules to push the join into φ. R R.attr op A.attr φ start cond(nodes A, Edge) = φ start cond((r R.attr op A.attr Nodes A), Edge) (B.41) R R.attr op B.attr φ end cond(edge, Nodes B) = φ end cond(edge, (R R.attr op B.attr Nodes B)) (B.42) R R.attr op A.attr φ both cond(nodes A, Edge, Nodes B) = φ both cond((r R.attr op A.attr Nodes A), Edge, Nodes B) (B.43) R R.attr op B.attr φ both cond(nodes A, Edge, Nodes A) = φ both cond(nodes A, Edge, (R R.attr op B.attr Nodes B)) (B.44) 160
173 C. Additional Algorithms for GRIPP C.1. Relational Schema for Storing GRIPP The GRIPP index as well as the stop nodes are stored in relations. Figure C.1 shows the database schema to store the GRIPP index as well as the stop nodes. The instance type of a node is stored as special attribute in the index table. For stop nodes we store apart from the node_id also the pre- and postorder value and the depth of the tree instance of that stop node. GRIPP node_id preorder postorder depth edge_id instance_type Stop_nodes node_id preorder postorder depth Figure C.1.: The database schema to store the GRIPP index structure and stop nodes. C.2. Stop Node List for GRIPP To create the list of stop nodes we would have to check the reachable instance set of every node. As this is too time consuming we test only selected nodes. We are especially interested in nodes whose reachable instance set covers many instances. Therefore, we only consider child nodes c of the virtual root node as stop node candidates. In addition for every c we compute the size of RIS(c), RIS(c), which is basically c post c pre /2. We only consider c as stop node candidate if RIS(c) t, with t being a cut-off value. For our experiments we use the cut-off value t = max( RIS(c) ), which we determined empirically as tradeoff between the number of nodes we must evaluate during the stop node list generation and the number of stop nodes found. Furthermore, we only consider a node as stop node candidate if it is a potential hop node, i.e., if it has a non-tree instance in IND(G). For a stop node candidate s we check if the tree instance h of any hop node in RIS(s) is also in RIS(s). If that is not the case, h is sibling to s and s is not a stop node; otherwise, s is a stop node and is added to the list of stop nodes. Algorithm C.1 shows the procedure to compute the list of the stop nodes. The child nodes to the root node are retrieved according to the size of their reachable instance sets. 161
174 C. Additional Algorithms for GRIPP Algorithm C.1: The algorithm to compute the stop node list 1 PROCEDURE compute_stop_nodes(root_node) 2 t 0 3 while cand next(children(root_node)) // order by RIS 4 do 5 if t = 0 then 6 t post(cand) pre(cand) 7 end 8 if post( cand) pre( cand) > t 9 AND hasnon-tree(cand) AND stopnodecond(cand) then 10 STOP_NODES STOP_NODES (node(cand), pre(cand), post(cand)); 11 end 12 end 13 end 14 FUNCTION stopnodecond(cand) 15 forall the non_tree_inst RIS(cand) do 16 tree_inst gettree(non_tree_inst) 17 if tree_inst / RIS(cand) then return false 18 end 19 return true 20 end C.3. Reachability for Sets of Nodes The function given in Algorithm C.2 is a variation of Algorithm 5.2 presented on page 79. To find all reachable nodes from u we basically use every possible hop node unless this hop node has already been used or is successor to a used hop node. In addition, we add each hop node to a list of reachable nodes, which initially is empty. To get all reachable nodes we have to add an additional step. This step starts in line 22 of Algorithm C.2. The idea is to find all tree instances in the reachable instance set of the hop node h. To avoid that we retrieve nodes multiple times we skip all intervals of used hop nodes that lie in RIS(h) (line 23). 162
175 C.3. Reachability for Sets of Nodes Algorithm C.2: Function to find all nodes that are reachable from u using GRIPP. 1 used_hops ; used_stops ; reached_nodes 2 FUNCTION reachability(u) RETURNS boolean 3 used_hops used_hops (u) 4 if u STOP_NODES then 5 used_stops used_stops (u) 6 else 7 while non_tree_inst nextstop(ris(u)) do 8 tree_inst gettreeinst(non_tree_inst) 9 reachability(tree_inst) 10 end 11 if isinris(u, used_stops) then return 12 H 1... H n getusedhopsinris(u) // skip ranges 13 non_tree_instances getnontreeinst(ris(u) \ RIS(H 1) \... \ RIS(H n)) 14 foreach non_tree_inst non_tree_instances do 15 tree_inst gettreeinst(non_tree_inst) 16 if!haschildren(tree_inst) then continue // if new hop has been used as hop 17 if tree_inst used_hops then continue // if new hop is in a RIS of a used hop 18 if isinris(tree_inst, used_hops) then continue // otherwise call recursively 19 reachability(tree_inst) 20 end 21 end // now add all tree instances that are reachable min_pre pre(u); reached_nodes reached_nodes (u) 24 foreach inst gettreeinris(u) do 25 if min_pre < pre( inst) then 26 if inst used_hops then 27 min_pre post(inst) 28 else 29 reached_nodes reached_nodes (inst) 30 end 31 end 32 end 33 end 163
176
177 D. Graph Properties No. nodes No. edges Max. degree SD(Max. SD (Zero Zero degree degree) degree) , ,000 2, ,500 5, ,000 10, ,000 20, , ,000 50, , , , , , , , , , ,000 1,000, , ,000,000 2,000, , ,500,000 5,000, , ,000 5, , ,000 7, , ,000 10, ,000 12, ,000 15, ,000 17, ,000 20, ,000 25, ,000 30, ,000 40, ,000 50, Table D.1.: Key figures for different random graphs (Max. degree = outdegree of the nodes with highest outdegree; Zero degree = number of nodes with an outdegree 0). The figures are an average over five generated graphs for each graph type and its standard deviation (SD). 165
178 D. Graph Properties No. nodes No. edges Max. degree SD (Max. SD (Zero Zero degree degree) degree) , ,000 2, ,500 5, ,000 10, ,000 20, , ,000 50, , , , , , , , , , , ,000 1,000, , ,000,000 2,000, , ,500,000 5,000,000 1, , ,000 5, ,000 7, ,000 10, ,000 12, ,000 15, ,000 17, ,000 20, ,000 25, ,000 30, ,000 40, ,000 50, Table D.2.: Key figures for different scale-free graphs (Max. degree = outdegree of the nodes with highest outdegree; Zero degree = number of nodes with an outdegree 0). The figures are an average over five generated graphs for each graph type and its standard deviation (SD). 166
179 E. Model Specification for Volcano In Listing E.1 we show an excerpt of the file model.input, which holds the model specifications for Volcano. We omit the rewrite rules for the standard relational algebra operators SELECT, JOIN, and PROJECT. We also omit the code for operator and algorithm definitions, implementation and transformation rules for the different varieties of LENGTH, DISTANCE, and REACHABILITY operators as these are very similar to the presented code for the PATH operator. Listing E.1: Excerpt from model.input, the model specification for Volcano Define operators and their arities. Set base operators %operator SELECT 1 %operator JOIN 2... and more (PROJECT, DISTINCT) Paths operators %operator PATHS 1 %operator PATHS_START 2 %operator PATHS_END 2 %operator PATHS_BOTH 3... and more (LENGTHS, DISTANCE, REACHABILITY) Define algorithms and their arities. the normal ones %algorithm FILTER 1 %algorithm HASH_JOIN 2 %algorithm MERGE_JOIN 2... and more (PROJECT, DISTINCT, SORT) for the paths %algorithm PATHS_REC 1 %algorithm PATHS_GRIPP 1 %algorithm PATHS_START_REC 2 %algorithm PATHS_START_GRIPP 2 %algorithm PATHS_END_REC 2 %algorithm PATHS_END_GRIPP 2 %algorithm PATHS_BOTH_FW_REC 3 %algorithm PATHS_BOTH_FW_GRIPP 3 %algorithm PATHS_BOTH_RV_REC 3 %algorithm PATHS_BOTH_RV_GRIPP 3... and more (LENGTHS, DISTANCE, REACHABILITY) Implementation rules which algorithm is an implementation for which operator the normal rules 167
180 E. Model Specification for Volcano %impl_rule ( SELECT?op_arg1 (?1) ) > ( FILTER?al_arg1 (?1) ) %impl_rule ( JOIN?op_arg1 (?1?2) ) > ( HASH_JOIN?al_arg1 (?1?2) ) %impl_rule ( JOIN?op_arg1 (?1?2) ) > ( MERGE_JOIN?al_arg1 (?1?2) )... and more (PROJECT, DISTINCT) the path rules %impl_rule ( GRAPH_GET?op_arg1 () ) > ( GRAPH_SCAN?al_arg1() ) %impl_rule ( PATHS?op_arg1 (?1) ) > ( PATHS_REC?al_arg1 (?1) ) %impl_rule ( PATHS?op_arg1 (?1) ) > ( PATHS_GRIPP?al_arg1 (?1) ) %impl_rule ( PATHS_END?op_arg1 (?1?2) ) > ( PATHS_END_REC?al_arg1 (?1?2) ) %impl_rule ( PATHS_END?op_arg1 (?1?2) ) > ( PATHS_END_GRIPP?al_arg1 (?1?2) ) %impl_rule ( PATHS_START?op_arg1 (?1?2) ) > ( PATHS_START_REC?al_arg1 (?1?2) ) %impl_rule ( PATHS_START?op_arg1 (?1?2) ) > ( PATHS_START_GRIPP?al_arg1 (?1?2) ) %impl_rule ( PATHS_BOTH?op_arg1 (?1?2?3) ) > ( PATHS_BOTH_FW_REC?al_arg1 (?1?2?3) ) %impl_rule ( PATHS_BOTH?op_arg1 (?1?2?3) ) > ( PATHS_BOTH_FW_GRIPP?al_arg1 (?1?2?3) ) %impl_rule ( PATHS_BOTH?op_arg1 (?1?2?3) ) > ( PATHS_BOTH_RV_REC?al_arg1 (?1?2?3) ) %impl_rule ( PATHS_BOTH?op_arg1 (?1?2?3) ) > ( PATHS_BOTH_RV_GRIPP?al_arg1 (?1?2?3) )... and more (LENGTHS, DISTANCE, REACHABILITY) Rewrite rules how to rewrite the expressions SELECT and JOIN non interference. %trans_rule (SELECT?op_arg1 ((JOIN?op_arg2 (?1?2)))) > (JOIN?op_arg3 ((SELECT?op_arg4 (?1))?2)) %cond_code {{ LOGICAL_PROPERTY right; eq_class_logical_property(?2, right); if (select_in_set(&(?op_arg1 >select), right)) REJECT; }} %appl_code {{ / Just copy over / copy_operator_arg(?op_arg3,?op_arg2); copy_operator_arg(?op_arg4,?op_arg1); }}... and more (Combinations of SELECT, JOIN, PROJECT, DISTINCT) join and paths can be rewritten to paths start if the join condition contains start_node %trans_rule ( JOIN?op_arg1 (?1 (PATHS?op_arg2 (?2)))) >! ( PATHS_START?op_arg3 (?1?2 ) ) 168
181 %cond_code {{ char str [80]; strcpy( str,?op_arg2 >paths.name); strcat (str, ".start_node"); if ( strcmp(?op_arg1 >join.attr2, str )!=0 && strcmp(?op_arg1 >join.attr1, str )!= 0) REJECT; }} %appl_code {{ //printf("rewrite from paths to paths start \n"); copy_operator_arg(?op_arg3,?op_arg2);?op_arg3 >paths_start.terms =?op_arg2 >paths.terms + 1; strcpy(?op_arg3 >paths_start.cond[?op_arg2 >paths.terms].attr,?op_arg1 >join.attr1); strcpy(?op_arg3 >paths_start.cond[?op_arg2 >paths.terms].attr2,?op_arg1 >join.attr2);?op_arg3 >paths_start.cond[?op_arg2 >paths.terms].op =?op_arg1 >join.op; }} if (?op_arg2 >paths.terms > 0 )?op_arg3 >paths_start.op[?op_arg2 >paths.terms 1] = AND_OP; join and paths can be rewritten to paths end if the join condition contains end_node %trans_rule ( JOIN?op_arg1 (?1 (PATHS?op_arg2 (?2)))) >! ( PATHS_END?op_arg3 (?2?1 ) ) %cond_code {{ char str [80]; strcpy( str,?op_arg2 >paths.name); strcat (str, ".end_node"); if ( strcmp(?op_arg1 >join.attr2, str )!=0 && strcmp(?op_arg1 >join.attr1, str )!= 0 ) REJECT; }} %appl_code {{ //printf("rewrite from paths to paths end\n"); copy_operator_arg(?op_arg3,?op_arg2);?op_arg3 >paths_end.terms =?op_arg2 >paths.terms + 1; strcpy(?op_arg3 >paths_end.cond[?op_arg2 >paths.terms].attr,?op_arg1 >join.attr1); strcpy(?op_arg3 >paths_end.cond[?op_arg2 >paths.terms].attr2,?op_arg1 >join.attr2);?op_arg3 >paths_end.cond[?op_arg2 >paths.terms].op =?op_arg1 >join.op; }} if (?op_arg2 >paths.terms > 0 )?op_arg3 >paths_end.op[?op_arg2 >paths.terms 1] = AND_OP; join, join and paths can be rewritten to paths both if one join condition contains end_node and the other start_node %trans_rule ( JOIN?op_arg1 ( (JOIN?op_arg2(?1 (PATHS?op_arg3 (?2))))?3 ) ) >! ( PATHS_BOTH?op_arg4 (?1?2?3) ) %cond_code 169
182 E. Model Specification for Volcano {{ char start [80]; char end[80]; strcpy( start,?op_arg3 >paths.name); strcat ( start, ".start_node"); strcpy(end,?op_arg3 >paths.name); strcat (end, ".end_node"); if ( NOT (strcmp(?op_arg2 >join.attr2, start) == 0 && strcmp(?op_arg1 >join.attr1, end ) == 0) ) REJECT; }} %appl_code {{ //printf("rewrite from paths to paths both\n"); copy_operator_arg(?op_arg4,?op_arg3);?op_arg4 >paths.terms =?op_arg3 >paths.terms + 2; strcpy(?op_arg4 >paths.cond[?op_arg3 >paths.terms].attr,?op_arg2 >join.attr1); strcpy(?op_arg4 >paths.cond[?op_arg3 >paths.terms].attr2,?op_arg2 >join.attr2);?op_arg4 >paths.cond[?op_arg3 >paths.terms].op =?op_arg2 >join.op; if (?op_arg3 >paths.terms > 0 )?op_arg4 >paths.op[?op_arg3 >paths.terms 1] = AND_OP; strcpy(?op_arg4 >paths.cond[?op_arg3 >paths.terms+1].attr,?op_arg1 >join.attr1); strcpy(?op_arg4 >paths.cond[?op_arg3 >paths.terms+1].attr2,?op_arg1 >join.attr2);?op_arg4 >paths.cond[?op_arg3 >paths.terms+1].op =?op_arg1 >join.op; if (?op_arg3 >paths.terms > 0 )?op_arg4 >paths.op[?op_arg3 >paths.terms] = AND_OP; }}... and more (LENGTHS, DISTANCE, REACHABILITY) rewrite from PATHS to others %trans_rule ( PROJECT?op_arg1 ( (PATHS?op_arg2 (?1)) ) ) >! ( PROJECT?op_arg3 ( (REACHABILITY?op_arg4 (?1)) ) ) %cond_code {{ int i ; int found = 0; char str1 [80]; char str2 [80]; char str3 [80]; strcpy( str1,?op_arg2 >paths.name); strcat (str1, ".node_id"); strcpy( str2,?op_arg2 >paths.name); strcat (str2, ".position"); strcpy( str3,?op_arg2 >paths.name); strcat (str3, ".length"); for ( i = 0; i!=?op_arg1 >project.terms; i++) { if (strcmp(?op_arg1 >project.term[i].attr, str1)==0 170
183 OR strcmp(?op_arg1 >project.term[i].attr, str2)==0 OR strcmp(?op_arg1 >project.term[i].attr, str3)==0 ) { found = 1; } } if (found == 1) { REJECT; } for ( i = 0; i!=?op_arg2 >reachability.terms; i++ ) { if (strcmp(?op_arg2 >reachability.cond[i].attr, str1)==0 OR strcmp(?op_arg2 >reachability.cond[i].attr, str2)==0 OR strcmp(?op_arg2 >reachability.cond[i].attr, str3)==0 ) { found = 1; } } if (found == 1) { REJECT; } }} %appl_code {{ //printf("rewrite from paths to reachability \n"); copy_operator_arg(?op_arg3,?op_arg1); copy_operator_arg(?op_arg4,?op_arg2); }}... and more (PATHS TO LENGTHS, DISTANCE, REACHABILITY for START, END, BOTH) 171
184
185 F. Cost and Cardinality Functions for Volcano In Listing F.1 we show an excerpt of the file dbi.c, which holds C code for Volcano. In dbi.c the user has to specify the expected cardinality as system property and resulting schema as logical property for each operator. In addition, the user has to provide for each algorithm its physical properties (e.g., if the output is sorted), the cost, the applicability, and the input properties. In Listing F.1 we show these for the defined operator REACHABILITY and one of its implementations REACHABILITY_GRIPP. Listing F.1: Excerpt from dbi.c, the C source to specify cardinality of operators and cost and applicability of algorithms for Volcano / dbi.c / #include "../optgen/opt_defs.h" #include "../optgen/global.h" #include "dbi_gen.h" #include "dbi.h" #include "../optgen/opt.h" additional definition and assignments / derive_reachability_sys_prop This routine determines the resulting system properties of getting all paths / STATUS derive_reachability_sys_prop(oa, input_sys_prop, input_log_prop, derived_sys_prop) OPERATOR_ARGUMENT oa; SYSTEM_PROPERTY input_sys_prop[]; LOGICAL_PROPERTY input_log_prop[]; SYSTEM_PROPERTY derived_sys_prop; { double card = 0.0; double exponent = 0.0; derived_sys_prop >record_width = 2 LONG_WIDTH; exponent = (double)(input_log_prop[0] >pql.graph.no_edges input_log_prop[0] >pql.graph.no_nodes)/ (double)input_log_prop[0] >pql.graph.no_nodes; } card = (double)input_log_prop[0] >pql.graph.no_nodes ( 1.0 exp( exponent)); derived_sys_prop >cardinality = card (double)input_log_prop[0] >pql.graph.no_nodes; return (OKAY) ; one derive_operator_sys_prop for each defined operator 173
186 F. Cost and Cardinality Functions for Volcano / derive_reachability_log_prop This routine determines the resulting logical properties (shape of relation ) of reachability / STATUS derive_reachability_log_prop(oa, input_log_prop, derived_log_prop) OPERATOR_ARGUMENT oa; LOGICAL_PROPERTY input_log_prop[]; LOGICAL_PROPERTY derived_log_prop; { / Sanity check insure no null pointers / if (oa == NULL OR derived_log_prop == NULL OR input_log_prop == NULL) return (ERR_NULL_POINTER) ; derived_log_prop = input_log_prop[0]; / Initialize the derived logical property vector / strcpy(derived_log_prop >gen.name, Internal_set); derived_log_prop >gen.object_type = MIXED_SET; // get the schema start_node, end_node derived_log_prop >rel.arity = 2; strcat (strcpy(derived_log_prop >rel.attr[0].name, oa >reachability.name), ".start_node"); derived_log_prop >rel.attr[0].attr_type = INTEGER; derived_log_prop >rel.attr[0].attr_width = LONG_WIDTH; derived_log_prop >rel.attr[0].btree_avail = FALSE; derived_log_prop >rel.attr[0].domain_min.int_value = 0; derived_log_prop >rel.attr[0].domain_max.int_value = input_log_prop[0] >pql.graph.no_nodes; derived_log_prop >rel.attr[0]. selectivity = (double)1.0/(double)input_log_prop[0] >pql.graph.no_nodes; derived_log_prop >rel.attr[0].unique = input_log_prop[0] >pql.graph.no_nodes; strcat (strcpy(derived_log_prop >rel.attr[1].name, oa >reachability.name), ".end_node"); derived_log_prop >rel.attr[1].attr_type = INTEGER; derived_log_prop >rel.attr[1].attr_width = LONG_WIDTH; derived_log_prop >rel.attr[1].btree_avail = FALSE; derived_log_prop >rel.attr[1].domain_min.int_value = 0; derived_log_prop >rel.attr[1].domain_max.int_value = input_log_prop[0] >pql.graph.no_nodes; derived_log_prop >rel.attr[1]. selectivity = (double)1.0/(double)input_log_prop[0] >pql.graph.no_nodes; derived_log_prop >rel.attr[1].unique = input_log_prop[0] >pql.graph.no_nodes; } return (OKAY) ; one derive_operator_log_prop for each defined operator / 174
187 derive_reachability_gripp_phy_prop This routine determines the resulting physical properties (e.g. sort) of a gripp / STATUS derive_reachability_gripp_phy_prop(aa, input_pv, sys_prop, log_prop, input_sys_prop, input_log_prop, needed_pv, derived_pv) ALGORITHM_ARGUMENT aa; PROPERTY_VECTOR input_pv[]; SYSTEM_PROPERTY sys_prop; LOGICAL_PROPERTY log_prop; SYSTEM_PROPERTY input_sys_prop[]; LOGICAL_PROPERTY input_log_prop[]; PROPERTY_VECTOR needed_pv; PROPERTY_VECTOR derived_pv; { / Sanity check / if (aa == NULL OR input_pv == NULL OR input_log_prop == NULL OR derived_pv == NULL) return (ERR_NULL_POINTER); / The property vector matches that of the input / derived_pv = input_pv[0]; return (OKAY) ; } one derive_algorithm_phy_prop for each defined algorithm / cost_reachability_gripp This routine determines the cost of reachability gripp. / STATUS cost_reachability_gripp(aa, sys_prop, log_prop, input_sys_prop, input_log_prop, needed_pv, derived_pv, input_pv, cost) ALGORITHM_ARGUMENT aa; SYSTEM_PROPERTY sys_prop; LOGICAL_PROPERTY log_prop; SYSTEM_PROPERTY input_sys_prop[]; LOGICAL_PROPERTY input_log_prop[]; PROPERTY_VECTOR needed_pv; PROPERTY_VECTOR derived_pv; PROPERTY_VECTOR input_pv[]; COST cost ; { double term; / If derived_pv == NULL, then this is just an "estimated" cost. / if (derived_pv == NULL) { cost = zero_cost; return OKAY; } / if / term = 3.0; cost >set = 1; 175
188 F. Cost and Cardinality Functions for Volcano cost >cost.io = (double)input_log_prop[0] >pql.graph.no_nodes term IO_COST; cost >cost.cpu = (double)input_log_prop[0] >pql.graph.no_nodes term CPU_COST; //printf("cost io for reachability gripp: %4.2f ", cost >cost.io); //printf("cost cpu for reachability gripp: %4.2f\n", cost >cost.cpu); return (OKAY) ; } one cost_algorithm for each defined algorithm / do_any_good_reachability_gripp This routine determines whether the given filter will satisfy the physical property requirements. / STATUS do_any_good_reachability_gripp(current_operator, oa, needed_pv, sys_prop, log_prop, excl_pv, input_operator, ret_value) OPERATOR current_operator; OPERATOR_ARGUMENT oa; PROPERTY_VECTOR needed_pv; SYSTEM_PROPERTY sys_prop; LOGICAL_PROPERTY log_prop; PROPERTY_VECTOR excl_pv; OPERATOR input_operator[]; ALGO_APPL_RETURN ret_value; { // only apply the gripp algorithm if gripp is available if ( log_prop >pql.graph.gripp_fw_avail == TRUE ) { ret_value >apply_truth_value = TRUE; ret_value >reestimate_cost = TRUE; ret_value >algorithm_argument.reachability_gripp = oa >reachability; ret_value >process_input_count = 1; } else { ret_value >apply_truth_value = FALSE; } return (OKAY); } one do_any_good_algorithm for each defined algorithm / get_input_pv_reachability_gripp This routine requests property vectors for the input to a filter operator. / STATUS get_input_pv_reachability_gripp(aa, needed_pv, excl_pv, input_id, iteration, input_pv, input_excl_pv, input_sys_prop, input_log_prop, input_ret_pele, input_ret_cost, fail) ALGORITHM_ARGUMENT aa; PROPERTY_VECTOR needed_pv; PROPERTY_VECTOR excl_pv; int input_id; int iteration ; 176
189 PROPERTY_VECTOR input_pv[]; PROPERTY_VECTOR input_excl_pv[]; SYSTEM_PROPERTY input_sys_prop[]; LOGICAL_PROPERTY input_log_prop[]; PHY_EXPR_LIST_ELEM input_ret_pele[]; COST input_ret_cost[]; BOOLEAN fail ; { / Initialize property vectors / init_property_vec(input_pv[input_id]); init_property_vec(input_excl_pv[input_id]); / Paths doesn t care about input sort property vectors, but passes them / / through from the output. Excluded property vectors are likewise. / copy_property_vec(input_pv[0], needed_pv); copy_property_vec(input_excl_pv[0], excl_pv); return (OKAY); } one get_input_pv_algorithm for each defined algorithm additional code for printing 177
190
191 G. Exemplary Queries for GRIcano Listing G.1: Examplary queries used in Section x reachability get all reactions catalyzed by a given enzyme query r1 SELECT A, B FROM kegg LET node A, node B, path P WHERE A.Name = phenylalanine 4-monooxygenase AND P.path = A[ >]B AND B.Type LIKE Reaction ; which enzymes may be influenced query r2 SELECT A, B FROM reactome LET node A, node B, path P WHERE A.Name = %nitrophenol% AND A.Type LIKE Compound AND P.path = A[ >]B AND B.Type LIKE Enzyme ; query r3 SELECT A, B FROM kegg LET node A, node B, path P WHERE A.Class LIKE Nucleotidyltransferases AND P.path = A[ >]B AND B.Type LIKE Enzyme ; 3 x distance get all reactions catalyzed by a given enzyme query d1 SELECT A, B FROM kegg LET node A, node B, path P WHERE A.Name = phenylalanine 4-monooxygenase AND P.path = A[ >]B AND P.length < 6 AND B.Type LIKE Reaction ; given a single compound, which are the enzymes responsible for degradation query d2 SELECT A, B 179
192 G. Exemplary Queries for GRIcano FROM reactome LET node A, node B, path P WHERE A.Name LIKE %nitrophenol% AND A.Type LIKE Compound AND P.path = A[ >]B AND P.length < 7 AND B.Type LIKE Enzyme ; query d3 SELECT A, B FROM kegg LET node A, node B, path P WHERE A.Class LIKE Nucleotidyltransferases AND P.path = A[ >]B AND P.length < 6 AND B.Type LIKE Enzyme ; 3x path length query l1 SELECT A, B FROM kegg LET node A, node B, path P WHERE A.Name = phenylalanine 4-monooxygenase AND P.path = A[ >]B AND P.length = 5 AND B.Type LIKE Reaction ; given a single compound, which are the enzymes responsible for degradation query l2 SELECT A, B FROM reactome LET node A, node B, path P WHERE A.Name LIKE %nitrophenol% AND A.Type LIKE Compound AND P.path = A[ >]B AND P.length = 6 AND B.Type LIKE Enzyme ; query l3 SELECT A, B FROM kegg LET node A, node B, path P WHERE A.Class LIKE Nucleotidyltransferases AND P.path = A[ >]B AND P.length = 5 AND B.Type LIKE Enzyme ; 3 x paths query p1 SELECT P FROM kegg LET node A, node B, path P 180
193 WHERE A.Name = phenylalanine 4-monooxygenase AND P.path = A[ >]B AND P.length < 6 AND B.Type LIKE Reaction ; given a single compound, which are the enzymes responsible for degradation query p2 SELECT P FROM reactome LET node A, node B, path P WHERE A.Name LIKE %nitrophenol% AND A.Type LIKE Compound AND P.path = A[ >]B AND P.length < 7 AND B.Type LIKE Enzyme ; query p3 SELECT P FROM kegg LET node A, node B, path P WHERE A.Class LIKE Nucleotidyltransferases AND P.path = A[ >]B AND P.length < 6 AND B.Type LIKE Enzyme ; complex queries query c1 SELECT P FROM kegg LET node A, node B, path P WHERE A.name = L-Arginine AND B.name = L-Proline AND P.path = A[ >]B AND P.length < 5; given two compounds, what may be the result after a reaction query c2 SELECT C, E FROM kegg LET node A, node B, node C, node E, path P, path Q, path R WHERE A.Name = Salicylate AND B.Name = Alcohol AND C.Type LIKE Reaction AND P.path = A[ >]C AND P.length < 5 AND Q.path = B[ >]C AND R.path = C[ >]E AND R.length = 1 AND E.Type LIKE Compound ; given two compounds, what may be the result after a reaction, get all paths query c3 181
194 G. Exemplary Queries for GRIcano SELECT P, Q, R FROM kegg LET node A, node B, node C, node E, path P, path Q, path R WHERE A.Name = Salicylate AND B.Name = Alcohol AND C.Type LIKE Reaction AND P.path = A[ >]C AND P.length < 5 AND Q.path = B[ >]C AND Q.length < 5 AND R.path = C[ >]E AND R.length = 1 AND E.Type LIKE Compound ; query c4 SELECT A, D FROM kegg, intact LET node A IN kegg, node B IN kegg, path P IN kegg, path Q IN intact, node C IN intact, node D IN intact WHERE B.Name = fatty-acid synthase AND P.path = A[ >]B AND C.Name = A.Name AND Q.path = C[ >]D AND Q.length < 3; 182
195 Bibliography [AAN01] [ABE09] [Abi97] [ABJ89] [AG05] [Agr88] Aboulnaga, Ashraf; Alameldeen, Alaa R.; Naughton, Jeffrey F.: Estimating the Selectivity of XML Path Expressions for Internet Scale Applications. In: Proceedings of the 27th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Alkhateeb, Faisal; Baget, Jean-François; Euzenat, Jérôme: Extending SPARQL with regular expression patterns (for querying RDF). In: J. Web Sem., volume 7(2):pp , Abiteboul, Serge: Querying Semi-Structured Data. In: Proceedings of the 6th International Conference on Database Theory (ICDT), volume 1186 of Lecture Notes in Computer Science, pp Springer, Agrawal, Rakesh; Borgida, Alexander; Jagadish, H. V.: Efficient Management of Transitive Relationships in Large Data and Knowledge Bases. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Angles, Renzo; Gutierrez, Claudio: Survey of Graph Database Models. Technical Report TR/DCC , Computer Science Department, Universidad de Chile, Agrawal, Rakesh: Alpha: An Extension of Relational Algebra to Express a Class of Recursive Queries. In: IEEE Transactions on Software Engineering, volume 14(7):pp , [Ahl06] Ahl, Stephan: Visuelle Graphanfragen für biologische Netzwerke. Diplomarbeit, Freie Universität zu Berlin, Institut für Informatik, [AJ87] [AJW + 08] [AQM + 97] [ARS + 08] [AS92] Agrawal, Rakesh; Jagadish, H. V.: Direct Algorithms for Computing the Transitive Closure of Database Relations. In: Proceedings of the 13th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Alberts, Bruce; Johnson, Alexander; Walter, Peter; Lewis, Julian; Raff, Martin; Roberts, Keith; Orme, Nigel: Molecular Biology of the Cell. Taylor & Francis, 5th edition, Abiteboul, Serge; Quass, Dallan; McHugh, Jason; Widom, Jennifer; Wiener, Janet L.: The Lorel Query Language for Semistructured Data. In: Int. J. on Digital Libraries, volume 1(1):pp , Assenov, Yassen; Ramírez, Fidel; Schelhorn, Sven-Eric; Lengauer, Thomas; Albrecht, Mario: Computing topological parameters of biological networks. In: Bioinformatics, volume 24(2):pp , Jan Amann, Bernd; Scholl, Michel: Gram: A Graph Data Model and Query Language. In: Proceedings of the European Conference on Hypertext Technology (ECHT), pp ACM Press, [BA99] Barabási, Albert-Lázló; Albert, Réka: Emergence of Scaling in Random Networks. In: Science, volume 286(5439):pp , Oct [Bay72] [BBH03] Bayer, Rudolf: Symmetric Binary B-Trees: Data Structure and Maintenance Algorithms. In: Acta Inf., volume 1:pp , Bader, Gary D; Betel, Doron; Hogue, Christopher W V: BIND: the Biomolecular Interaction Network Database. In: Nucleic Acids Research, volume 31(1):pp , Jan
196 Bibliography [BDHS96] [Bec04] Buneman, Peter; Davidson, Susan B.; Hillebrand, Gerd G.; Suciu, Dan: A Query Language and Optimization Techniques for Unstructured Data. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Beckett, Dave: RDF/XML Syntax Specification (Revised). Feb [BFM06] Bast, Holger; Funke, Stefan; Matijevic, Domagoj: TRANSIT: Ultrafast Shortest-Path Queries with Linear-Time Preprocessing. In: Proceedings of the 9th DIMACS Implementation Challenge Shortest Path. DIMACS, [BFS00] [BKS02] Buneman, Peter; Fernandez, Mary F.; Suciu, Dan: UnQL: A Query Language and Algebra for Semistructured Data Based on Structural Recursion. In: VLDB Journal, volume 9(1):pp , Bruno, Nicolas; Koudas, Nick; Srivastava, Divesh: Holistic Twig Joins: Optimal XML Pattern Matching. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, [BMR99] Beech, David; Malhotra, Ashok; Rys, Michael: A Formal Data Model and Algebra for XML [BN05] [BO04] [BPSM + 06] Borodina, Irina; Nielsen, Jens: From genomes to in silico cells via metabolic networks. In: Current Opinion in Biotechnology, volume 16(3):pp , Jun Barabási, Albert-László; Oltvai, Zoltán N: Network biology: understanding the cell s functional organization. In: Nature Reviews Genetics, volume 5(2):pp , Feb Bray, Tim; Paoli, Jean; Sperberg-McQueen, C. M.; Maler, Eve; Yergeau, François: Extensible Markup Language (XML) 1.0 (Fourth Edition). Sep [BR86] Bancilhon, François; Ramakrishnan, Raghu: An Amateur s Introduction to Recursive Query Processing Strategies. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, [BS03] [BT99] [CCS00] [CF04] Bornholdt, Stefan; Schuster, Heinz Georg: Handbook of Graphs and Networks. Wiley-VCH, Beeri, Catriel; Tzaban, Yariv: SAL: An Algebra for Semistructured Data and XML. In: Informal Proceedings of ACM SIGMOD Workshop on The Web and Databases (WebDB), pp Christophides, Vassilis; Cluet, Sophie; Siméon, Jérôme: On Wrapping Query Languages and Efficient XML Integration. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Cooper, Colin; Frieze, Alan M.: The Size of the Largest Strongly Connected Component of a Random Digraph with a Given Degree Sequence. In: Combinatorics, Probability & Computing, volume 13(3):pp , [CGK05] Chen, Li; Gupta, Amarnath; Kurul, M. Erdem: Stack-based Algorithms for Pattern Matching on DAGs. In: Proceedings of the 31st International Conference on Very Large Data Bases (VLDB), pp ACM Press, [Cha98] Chaudhuri, Surajit: An Overview of Query Optimization in Relational Systems. In: Proceedings of the 17th ACM SIGACT-SIGMOD-SIGART Symposium on Principles of Database Systems (PODS), pp ACM Press, [CHKZ03] Cohen, Edith; Halperin, Eran; Kaplan, Haim; Zwick, Uri: Reachability and Distance Queries via 2-Hop Labels. In: SIAM J. Comput., volume 32(5):pp ,
197 Bibliography [CKNL07] [CLR01] [CM90] [CMS02] [CY09] [CYD07] [CYL + 06] [CYL + 08] [CYY + 07] Cheng, James; Ke, Yiping; Ng, Wilfred; Lu, An: FG-Index: Towards Verification-Free Query Processing on Graph Databases. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Cormen, Thomas H.; Leiserson, Charles E.; Rivest, Ronald L.: Introduction to Algorithms. MIT Press, Consens, Mariano P.; Mendelzon, Alberto O.: GraphLog: a Visual Formalism for Real Life Recursion. In: Proceedings of the Ninth ACM SIGACT-SIGMOD-SIGART Symposium on Principles of Database Systems (PODS), pp ACM Press, Chung, Chin-Wan; Min, Jun-Ki; Shim, Kyuseok: APEX: an adaptive path index for XML data. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Cheng, Jiefeng; Yu, Jeffrey Xu: On-line exact shortest distance query processing. In: Proceedings of the 12th International Conference on Extending Database Technology (EDBT), volume 360 of ACM International Conference Proceeding Series, pp ACM Press, Cheng, Jiefeng; Yu, Jeffrey Xu; Ding, Bolin: Cost-Based Query Optimization for Multi Reachability Joins. In: Proceeding of the 12th International Conference on Database Systems for Advanced Applications (DASFAA), volume 4443 of Lecture Notes in Computer Science, pp Springer, Cheng, Jiefeng; Yu, Jeffrey Xu; Lin, Xuemin; Wang, Haixun; Yu, Philip S.: Fast Computation of Reachability Labeling for Large Graphs. In: Proceedings of the 10th International Conference on Extending Database Technology (EDBT ), volume 3896 of Lecture Notes in Computer Science, pp Springer, Cheng, Jiefeng; Yu, Jeffrey Xu; Lin, Xuemin; Wang, Haixun; Yu, Philip S.: Fast computing reachability labelings for large graphs with high compression rate. In: Proceedings of the 11th International Conference on Extending Database Technology (EDBT), volume 261 of ACM International Conference Proceeding Series, pp ACM Press, Chen, Chen; Yan, Xifeng; Yu, Philip S.; Han, Jiawei; Zhang, Dong-Qing; Gu, Xiaohui: Towards Graph Containment Search and Indexing. In: Proceedings of the 33rd International Conference on Very Large Data Bases (VLDB), pp ACM Press, [DA93] Dar, S.; Agrawal, R.: Extending SQL with Generalized Transitive Closure. In: IEEE Transactions on Knowledge and Data Engineering, volume 5(5):pp , [DAJ91] Dar, Shaul; Agrawal, Rakesh; Jagadish, H. V.: Optimization of Generalized Transitive Closure Queries. In: Proceedings of the Seventh International Conference on Data Engineering (ICDE), pp IEEE Computer Society, [Dat03] Date, Chris J.: An Introduction to Database Systems. Addison-Wesley, 8th edition, [DD97] [DFF + 07] [DJG + 02] Date, Chris J.; Darwen, Hugh: A Guide to the SQL Standard. Addison-Wesley Longman, Draper, Denise; Fankhauser, Peter; Fernández, Mary; Malhotra, Ashok; Rose, Kristoffer; Rys, Michael; Siméon, Jérôme; Wadler, Philip: XQuery 1.0 and XPath 2.0 Formal Semantics. Jan DeRose, Steven; Jr., Ron Daniel; Grosso, Paul; Maler, Eve; Marsh, Jonathan; Walsh, Norman: XML Pointer Language (XPointer). Aug [DMO01] DeRose, Steve; Maler, Eve; Orchard, David: XML Linking Language (XLink) Version June [DNR09] Dries, Anton; Nijssen, Siegfried; Raedt, Luc De: A query language for analyzing networks. In: Proceedings of the 18th ACM Conference on Information and Knowledge Management (CIKM), pp ACM Press,
198 Bibliography [DS87] [DSB08] Dietz, Paul; Sleator, Daniel: Two algorithms for maintaining order in a list. In: Proceedings of the 19th annual ACM Symposium on Theory of computing (STOC), pp ACM Press, Detwiler, Landon T; Suciu, Dan; Brinkley, James F: Regular paths in SparQL: querying the NCI Thesaurus. In: AMIA Annu Symp Proc, pp , [EB06] Eckman, Barbara A.; Brown, P. G.: Graph data management for molecular and cell biology. In: IBM J. Res & Dev., volume 50(6):pp , Nov [ER60] [FFLS97] [FHP02] [FSW00] [GM93] Erdös, P.; Rényi, A.: On the Evolution of Random Graphs. In: Publ. Math. Inst. Hungar. Acad. Sci., volume 5:pp , Fernandez, Mary F.; Florescu, Daniela; Levy, Alon Y.; Suciu, Dan: A Query Language for a Web-Site Management System. In: SIGMOD Record, volume 26(3):pp. 4 11, Frasincar, Flavius; Houben, Geert-Jan; Pau, Cristian: XAL: An Algebra For XML Query Optimization. In: Thirteenth Australasian Database Conference on Database Technologies (ADC), volume 5 of CRPIT. Australian Computer Society, Fernández, Mary F.; Siméon, Jérôme; Wadler, Philip: An Algebra for XML Query. In: Proceedings of the 20th Conference on Foundations of Software Technology and Theoretical Computer Science (FST TCS), volume 1974 of Lecture Notes in Computer Science, pp Springer, Graefe, Goetz; McKenna, William J.: The Volcano Optimizer Generator: Extensibility and Efficient Search. In: Proceedings of the Ninth International Conference on Data Engineering (ICDE), pp IEEE Computer Society, [GMUW02] Garcia-Molina, Hector; Ullman, Jeffrey D.; Widom, Jennifer: Database Systems: The Complete Book. Prentice Hall, [GP99] Gulutzan, Peter; Pelzer, Trudy: SQL-99 Complete, Really. Mcgraw-Hill Professional, [Gra95] [Gru02] [GS02] [Gut84] [Güt94] [GV07] [GvKT03] [GvKT04] [GW97] Graefe, Goetz: The Cascades Framework for Query Optimization. In: IEEE Data Eng. Bull., volume 18(3):pp , Grust, Torsten: Accelerating XPath location steps. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Giugno, Rosalba; Shasha, Dennis: GraphGrep: A Fast and Universal Method for Querying Graphs. In: Proceedings of the 16th International Conference on Pattern Recognition (ICPR) - Volume 2, pp Guttman, Antonin: R-Trees: A Dynamic Index Structure for Spatial Searching. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Güting, Ralf Hartmut: GraphDB: Modeling and Querying Graphs in Databases. In: Proceedings of the 20th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Georgiadis, Haris; Vassalos, Vasilis: Xpath on steroids: exploiting relational engines for xpath performance. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Grust, Torsten; van Keulen, Maurice; Teubner, Jens: Staircase Join: Teach a Relational DBMS to Watch its (Axis) Steps. In: Proceedings of the 29th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Grust, Torsten; van Keulen, Maurice; Teubner, Jens: Accelerating XPath evaluation in any RDBMS. In: ACM Trans. Database Syst., volume 29:pp , Goldman, Roy; Widom, Jennifer: DataGuides: Enabling Query Formulation and Optimization in Semistructured Databases. In: Proceedings of the 23rd International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann,
199 Bibliography [GW02] [Ham82] [HFLP89] [HH07] [HLQR07] [HNM + 00] [HS06] [HS08] [htt08] Groff, James R; Weinberg, Paul N.: SQL: The Complete Reference. McGraw-Hill, second edition edition, Hamilton, A. G.: Numbers, sets and axioms: the apparatus of mathematics. Cambridge University Press, Haas, Laura M.; Freytag, Johann Christoph; Lohman, Guy M.; Pirahesh, Hamid: Extensible Query Processing in Starburst. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Hartig, Olaf; Heese, Ralf: The SPARQL Query Graph Model for Query Optimization. In: Proceedings of the 4th European Semantic Web Conference (ESWC 2007), volume 4519 of Lecture Notes in Computer Science, pp Springer, Heese, Ralf; Leser, Ulf; Quilitz, Bastian; Rothe, Christian: Index Support for SPARQL. Poster Paper at European Semantic Web Conference (ESWC), Helden, J van; Naim, A; Mancuso, R; Eldridge, M; Wernisch, L; Gilbert, D; Wodak, SJ: Representing and analysing molecular and cellular function using the computer. In: Journal of Biological Chemistry, volume 381(9-10):pp , He, Huahai; Singh, Ambuj K.: Closure-Tree: An Index Structure for Graph Queries. In: Proceedings of the 22nd International Conference on Data Engineering (ICDE), p. 38. IEEE Computer Society, He, Huahai; Singh, Ambuj K.: Graphs-at-a-time: query language and access methods for graph databases. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Open Source Native XML Database. Web-Page, [htt11a] SableCC. Web-Page, June [htt11b] [HWYY05] [Ioa86] BioCarta - Charting Pathways oflife. Web- Page, June He, Hao; Wang, Haixun; Yang, Jun; Yu, Philip S.: Compact reachability labeling for graph-structured data. In: Proceedings of the 14th ACM Conference on Information and Knowledge Management (CIKM), pp ACM Press, Ioannidis, Yannis E.: On the Computation of the Transitive Closure of Relational Operators. In: Proceedings of the 12th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, [Ioa96] Ioannidis, Yannis E.: Query Optimization. In: ACM Comput. Surv., volume 28(1):pp , [Ioa03] [JLKH01] [JLST01] [JTGV + 05] Ioannidis, Yannis E.: The History of Histograms (abridged). In: Proceedings of the 29th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Jenssen, TK; Laegreid, A; Komorowski, J; Hovig, E: A literature network of human genes for high-throughput analysis of gene expression. In: Nat Genet, volume 28(1):pp , May Jagadish, H. V.; Lakshmanan, Laks V. S.; Srivastava, Divesh; Thompson, Keith: TAX: A Tree Algebra for XML. In: Proceedings of the 8th International Workshop on Database Programming Languages (DBPL), volume 2397 of Lecture Notes in Computer Science, pp Springer, Joshi-Tope, G.; Gillespie, M.; Vastrik, I.; D Eustachio, P.; Schmidt, E.; de Bono, B.; Jassal, B.; Gopinath, G. R.; Wu, G. R.; Matthews, L.; Lewis, S.; Birney, E.; Stein, L.: Reactome: a knowledgebase of biological pathways. In: Nucleic Acids Research, volume 33(Database issue):pp. D428 32, Jan
200 Bibliography [JXRF09] [JXRW08] [KAC + 02] [KD98] Jin, Ruoming; Xiang, Yang; Ruan, Ning; Fuhry, David: 3-HOP: a high-compression indexing scheme for reachability query. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Jin, Ruoming; Xiang, Yang; Ruan, Ning; Wang, Haixun: Efficiently answering reachability queries on very large directed graphs. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Karvounarakis, Gregory; Alexaki, Sofia; Christophides, Vassilis; Plexousakis, Dimitris; Scholl, Michel: RQL: a declarative query language for RDF. In: Proceedings of the 11th International World Wide Web Conference (WWW), pp Kabra, Navin; DeWitt, David J.: Efficient Mid-Query Re-Optimization of Sub-Optimal Query Execution Plans. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, [KGK + 04] Kanehisa, Minoru; Goto, Susumu; Kavashima, Shuichi; Okuno, Yasushi; Hattori, Masahiro: The KEGG resource for deciphering the genome. In: Nucleic Acids Research, volume 32(Database issue):pp. D277 D280, [KGKN02] [KGS04] Kanehisa, Minoru; Goto, Susumu; Kawashima, Shuichi; Nakaya, Akihiro: The KEGG databases at GenomeNet. In: Nucleic Acids Research, volume 30(1):pp , Jan Koyutürk, Mehmet; Grama, Ananth; Szpankowski, Wojciech: An efficient algorithm for detecting frequent subgraphs in biological networks. In: Bioinformatics, volume 20 Suppl 1:pp. i200 i207, Aug [KJ07] Kochut, Krys; Janik, Maciej: SPARQLeR: Extended Sparql for Semantic Association Discovery. In: Proceedings of the 4th European Semantic Web Conference (ESWC 2007), volume 4519 of Lecture Notes in Computer Science, pp Springer, [KK04] [KMR + 10] Kuramochi, Michihiro; Karypis, George: An Efficient Algorithm for Discovering Frequent Subgraphs. In: IEEE Transactions on Knowledge and Data Engineering, volume 16(9):pp , Kandasamy, Kumaran; Mohan, S. Sujatha; Raju, Rajesh; Keerthikumar, Shivakumar; Kumar, Ghantasala S Sameer; Venugopal, Abhilash K; Telikicherla, Deepthi; Navarro, J. Daniel; Mathivanan, Suresh; Pecquet, Christian; Gollapudi, Sashi Kanth; Tattikota, Sudhir Gopal; Mohan, Shyam; Padhukasahasram, Hariprasad; Subbannayya, Yashwanth; Goel, Renu; Jacob, Harrys K C; Zhong, Jun; Sekhar, Raja; Nanjappa, Vishalakshi; Balakrishnan, Lavanya; Subbaiah, Roopashree; Ramachandra, Y. L.; Rahiman, B. Abdul; Prasad, T. S Keshava; Lin, Jian-Xin; Houtman, Jon C D; Desiderio, Stephen; Renauld, Jean-Christophe; Constantinescu, Stefan N; Ohara, Osamu; Hirano, Toshio; Kubo, Masato; Singh, Sujay; Khatri, Purvesh; Draghici, Sorin; Bader, Gary D; Sander, Chris; Leonard, Warren J; Pandey, Akhilesh: NetPath: a public resource of curated signal transduction pathways. In: Genome Biol, volume 11(1):p. R3, [KOMK + 05] Karp, Peter D; Ouzounis, Christos A; Moore-Kochlacs, Caroline; Goldovsky, Leon; Kaipa, Pallavi; Ahrén, Dag; Tsoka, Sophia; Darzentas, Nikos; Kunin, Victor; López-Bigas, Núria: Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. In: Nucleic Acids Research, volume 33(19):pp , [Kos09] Koschmieder, Andre: Hauptspeicherbasierte Bearbeitung von Pfadanfragen an große Graphen. Diplomarbeit, Humboldt-Universität zu Berlin, Institut für Informatik, [KPV + 06] [KRBV06] Krull, Mathias; Pistor, Susanne; Voss, Nico; Kel, Alexander; Reuter, Ingmar; Kronenberg, Deborah; Michael, Holger; Schwarzer, Knut; Potapov, Anatolij; Choi, Claudia; Kel- Margoulis, Olga; Wingender, Edgar: TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations. In: Nucleic Acids Res, volume 34(Database issue):pp. D546 D551, Jan Kotz, Samuel; Read, Campbell B.; Balakrishnan, N.; Vidakovic, Brani: Encyclopedia of Statistical Sciences. John Wiley & Sons,
201 Bibliography [KSBG02] [KYHJ02] [LAC + 04] Kaushik, Raghav; Shenoy, Pradeep; Bohannon, Philip; Gudes, Ehud: Exploiting Local Similarity for Indexing Paths in Graph-Structured Data. In: Proceedings of the 18th International Conference on Data Engineering (ICDE), pp IEEE Computer Society, Kim, Beom Jun; Yoon, Chang No; Han, Seung Kee; Jeong, Hawoong: Path finding strategies in scale-free networks. In: Physical Review E Statistical, Nonlinear, and Soft Matter Physics, volume 65(2 Pt 2):p , Feb Lemer, Christian; Antezana, Erick; Couche, Fabian; Fays, Frédéric; Santolaria, Xavier; Janky, Rekin s; Deville, Yves; Richelle, Jean; Wodak, Shoshana J: The amaze Light- Bench: a web interface to a relational database of cellular processes. In: Nucleic Acids Research, volume 32 Database issue:pp. D , Jan [Les05a] Leser, Ulf: A Query Language for Biological Networks. In: Bioinformatics, volume 21 Suppl 2:pp. ii33 ii39, Sep [Les05b] [Lis07] [LMR87] [LN89] [LN90] Leser, Ulf: A Query Language for Biological Networks. Technical Report 187, Humboldt- Universität zu Berlin, Institut für Informatik, Liske, Peter: Auswertung regulärer Pfadausdrücke in Graphen. Diplomarbeit, Humboldt- Universität zu Berlin, Institut für Informatik, Lu, Hongjun; Mikkilineni, Krishna P.; Richardson, James P.: Design and Evaluation of Algorithms to Compute the Transitive Closure of a Database Relation. In: Proceedings of the Third International Conference on Data Engineering (ICDE), pp IEEE Computer Society, Lipton, Richard J.; Naughton, Jeffrey F.: Estimating the Size of Generalized Transitive Closures. In: Proceedings of the 15th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Lipton, Richard J.; Naughton, Jeffrey F.: Query Size Estimation by Adaptive Sampling. In: Proceedings of the Ninth ACM SIGACT-SIGMOD-SIGART Symposium on Principles of Database Systems (PODS), pp ACM Press, [LÖ09] Liu, Ling; Özsu, M. Tamer, editors: Encyclopedia of Database Systems. Springer US, [LR78] LaPaugh, Andrea S.; Rivest, Ronald L.: The Subgraph Homeomorphism Problem. In: Conference Record of the Tenth Annual ACM Symposium on Theory of Computing (STOC), pp ACM Press, [LT09] Leser, Ulf; Trißl, Silke: Graph Management in the Life Sciences. In: Liu, Ling; Özsu, M. Tamer, editors, Encyclopedia of Database Systems, pp Springer US, [Lu87] Lu, Hongjun: New Strategies for Computing the Transitive Closure of a Database Relation. In: Proceedings of the 13th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, [Mai83] Maier, David: The Theory of Relational Databases. Computer Science Press, [MS99] Milo, Tova; Suciu, Dan: Index Structures for Path Expressions. In: Proceedings of the 7th International Conference on Database Theory (ICDT), volume 1540 of Lecture Notes in Computer Science, pp Springer, [MW89] Mendelzon, Alberto O.; Wood, Peter T.: Finding Regular Simple Paths in Graph Databases. In: Proceedings of the 15th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, [MW99] [Ord05] McHugh, Jason; Widom, Jennifer: Query Optimization for XML. In: Proceedings of the 25th International Conference on Very Large Data Bases (VLDB), pp Morgan Kaufmann, Ordonez, Carlos: Optimizing recursive queries in SQL. In: Proceedings of the ACM SIG- MOD International Conference on Management of Data, pp ACM Press,
202 Bibliography [Ord10] Ordonez, Carlos: Optimization of Linear Recursive Queries in SQL. In: IEEE Trans. Knowl. Data Eng., volume 22(2):pp , [PBBS10] [PHH83] [PPT95] Przymus, Piotr; Boniewicz, Aleksandra; Burzańska, Marta; Stencel, Krzysztof: Recursive Query Facilities in Relational Databases: A Survey. In: Database Theory and Application, Bio-Science and Bio-Technology, volume 118 of Communications in Computer and Information Science, pp Springer, Puurunen, J.; Huttunen, P.; Hirvonen, J.: Interactions between ethanol and acetylsalicylic acid in damaging the rat gastric mucosa. In: Acta Pharmacol Toxicol (Copenh), volume 52(5):pp , May Paredaens, Jan; Peelman, Peter; Tanca, Letizia: G-Log: A Graph-Based Query Language. In: IEEE Trans. Knowl. Data Eng., volume 7(3):pp , [Pri04] Price, Jason: Oracle Database 10g SQL. Mcgraw-Hill Professional, [PS08] Prud hommeaux, Eric; Seaborne, Andy: SPARQL Query Language for RDF. January [RDM + 06] [RHDM86] [RMT + 04] [SAC + 79] [Sch04] [Sed04] [SH07] [SHZ04] [SL05] [SÖÖ99] [SOR + 11] Rother, Kristian; Dunkel, Mathias; Michalsky, Elke; Trißl, Silke; Goede, Andrean; Preissner, Robert: A structural keystone for drug design. In: Journal of Integrative Bioinformatics, Rosenthal, Arnon; Heiler, Sandra; Dayal, Umeshwar; Manola, Frank: Traversal recursion: a practical approach to supporting recursive applications. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Rother, Kristian; Müller, Heiko; Trißl, Silke; Koch, Ina; Steinke, Thomas; Preissner, Robert; Frömmel, Cornelius; Leser, Ulf: Columba: Multidimensional Data Integration of Protein Annotations. In: Proceedings of the First International Workshop on Data Integration in the Life Sciences (DILS), volume 2994 of Lecture Notes in Computer Science, pp Springer, Selinger, Patricia G.; Astrahan, Morton M.; Chamberlin, Donald D.; Lorie, Raymond A.; Price, Thomas G.: Access Path Selection in a Relational Database Management System. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Schaefer, Carl F: Pathway databases. In: Annals of the New York Academy of Sciences, volume 1020:pp , May Sedgewick, Robert: Algorithms in Java - Part 5. Addison-Wesley, third edition edition, Suderman, Matthew; Hallett, Michael: Tools for visually exploring biological networks. In: Bioinformatics, volume 23(20):pp , Oct Sohler, Florian; Hanisch, Daniel; Zimmer, Ralf: New methods for joint analysis of biological networks and expression data. In: Bioinformatics, volume 20(10):pp , Jul Strömbäck, Lena; Lambrix, Patrick: Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. In: Bioinformatics, volume 21(24):pp , Dec Sheng, Lei; Özsoyoglu, Z. Meral; Özsoyoglu, Gultekin: A Graph Query Language and Its Query Processing. In: Proceedings of the 15th International Conference on Data Engineering (ICDE), pp IEEE Computer Society, Smoot, Michael E; Ono, Keiichiro; Ruscheinski, Johannes; Wang, Peng-Liang; Ideker, Trey: Cytoscape 2.8: new features for data integration and network visualization. In: Bioinformatics, volume 27(3):pp , Feb
203 Bibliography [STW04] [SWG02] [SZ05] [THC + 99] [TL05] [TL06] [TL07] [TL10] [Tri05] [Tri07] [TRM + 05] [TVB + 02] [TZL07] [VDS + 07] [vmjs + 05] [Wag06] Schenkel, Ralf; Theobald, Anja; Weikum, Gerhard: HOPI: An Efficient Connection Index for Complex XML Document Collections. In: Proceedings of the 9th International Conference on Extending Database Technology (EDBT), volume 2992 of Lecture Notes in Computer Science, pp Springer, Shasha, Dennis; Wang, Jason T. L.; Giugno, Rosalba: Algorithmics and applications of tree and graph searching. In: Proceedings of the 21st ACM SIGMOD-SIGACT-SIGART symposium on Principles of database systems, pp ACM Press, Sohler, Florian; Zimmer, Ralf: Identifying active transcription factors and kinases from expression data using pathway queries. In: Bioinformatics, volume 21 Suppl 2:pp. ii115 ii122, Sep Tavazoie, S; Hughes, JD; Campbell, MJ; Cho, RJ; Church, GM: Systematic determination of genetic network architecture. In: Nat Genet, volume 22(3):pp , Jul Trißl, Silke; Leser, Ulf: Querying Ontologies in Relational Database Systems. In: Proceedings of the Second International Workshop on Data Integration in the Life Sciences (DILS), volume 3615 of Lecture Notes in Computer Science, pp Springer, Trißl, Silke; Leser, Ulf: GRIPP - Indexing and Querying Graphs based on Pre- and Postorder Numbering. Technical Report 207, Humboldt-Universität zu Berlin, Institut für Informatik, Trißl, Silke; Leser, Ulf: Fast and Practical Indexing and Querying of Very Large Graphs. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Trißl, Silke; Leser, Ulf: Estimating Result Size and Execution Times for Graph Queries. In: Proceedings of the Workshop on Querying Graph Structured Data (GraphQ) Trißl, Silke: Anfragen in Ontologien in relationalen Datenbanken (German). In: Proceedings of the 17. GI-Workshop über Grundlagen von Datenbanken Trißl, Silke: Cost-based Optimization of Graph Queries. In: Proceedings of the SIGMOD /PODS PhD Workshop on Innovative Database Research (IDAR) Trißl, Silke; Rother, Kristian; Müller, Heiko; Steinke, Thomas; Koch, Ina; Preissner, Robert; Frömmel, Cornelius; Leser, Ulf: Columba: an integrated database of proteins, structures, and annotations. In: BMC Bioinformatics, volume 6(1):p. 81, Mar Tatarinov, Igor; Viglas, Stratis; Beyer, Kevin S.; Shanmugasundaram, Jayavel; Shekita, Eugene J.; Zhang, Chun: Storing and querying ordered XML using a relational database system. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, Trißl, Silke; Zipser, Florian; Leser, Ulf: Applying GRIPP to XML Documents containing XInclude and XLink Elements. In: Berliner XML Tage Vastrik, Imre; D Eustachio, Peter; Schmidt, Esther; Joshi-Tope, Geeta; Gopinath, Gopal; Croft, David; de Bono, Bernard; Gillespie, Marc; Jassal, Bijay; Lewis, Suzanna; Matthews, Lisa; Wu, Guanming; Birney, Ewan; Stein, Lincoln: Reactome: a knowledge base of biologic pathways and processes. In: Genome Biol, volume 8(3):p. R39, von Mering, Christian; Jensen, Lars J; Snel, Berend; Hooper, Sean D; Krupp, Markus; Foglierini, Mathilde; Jouffre, Nelly; Huynen, Martijn A; Bork, Peer: STRING: known and predicted protein-protein associations, integrated and transferred across organisms. In: Nucleic Acids Research, volume 33(Database issue):pp. D433 D437, Jan Wagner, Christoph: Indexstrukturen für Graphen zur Unterstützung von Erreichbarkeitsund Distanzanfragen in Datenbanken (German). Diplomarbeit, Humboldt-Universität zu Berlin, Institut für Informatik, [War62] Warshal, Stephenl: A Theorem on Boolean Matrices. In: Journal of the ACM, volume 9(1):pp ,
204 Bibliography [War75] [WHY + 06] Warren, Henry S.: A Modification of Warshall s Algorithm for the Transitive Closure of Binary Relations. In: Commun. ACM, volume 18(4):pp , Wang, Haixun; He, Hao; Yang, Jun; Yu, Philip S.; Yu, Jeffrey Xu: Dual Labeling: Answering Graph Reachability Queries in Constant Time. In: Proceedings of the 22nd International Conference on Data Engineering (ICDE), p. 75. IEEE Computer Society, [WPJ02] Wu, Yuqing; Patel, Jignesh M.; Jagadish, H. V.: Estimating Answer Sizes for XML Queries. In: Proceedings of the 8th International Conference on Extending Database Technology (EDBT), volume 2287 of Lecture Notes in Computer Science, pp Springer, [WPJ03a] Wu, Yuqing; Patel, Jignesh M.; Jagadish, H. V.: Structural Join Order Selection for XML Query Optimization. In: Proceedings of the 19th International Conference on Data Engineering (ICDE), pp IEEE Computer Society, [WPJ03b] [XSD + 02] [YC10] [YCZ10] [YS07] Wu, Yuqing; Patel, Jignesh M.; Jagadish, H. V.: Using histograms to estimate answer sizes for XML queries. In: Inf. Syst., volume 28(1-2):pp , Xenarios, Ioannis; Salwínski, Lukasz; Duan, Xiaoqun Joyce; Higney, Patrick; Kim, Sul- Min; Eisenberg, David: DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. In: Nucleic Acids Research, volume 30(1):pp , Jan Yu, Jeffrey Xu; Cheng, Jiefeng: Graph Reachability Queries: A Survey. In: Managing and Mining Graph Data, volume 40 of The Kluwer International Series on Advances in Database Systems, pp Springer, Yildirim, Hilmi; Chaoji, Vineet; Zaki, Mohammed Javeed: Grail: Scalable reachability index for large graphs. In: Proceedings of the VLDB Endowment (PVLDB), volume 3(1):pp , Yang, Qingwu; Sze, Sing-Hoi: Path Matching and Graph Matching in Biological Networks. In: Journal of Computational Biology, volume 14(1):pp , [YYH04] Yan, Xifeng; Yu, Philip S.; Han, Jiawei: Graph indexing: a frequent structure-based approach. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp ACM Press, [YYH05] [ZCÖ09] Yan, Xifeng; Yu, Philip S.; Han, Jiawei: Graph indexing based on discriminative frequent structure analysis. In: ACM Trans. Database Syst., volume 30(4):pp , Zou, Lei; Chen, Lei; Özsu, M. Tamer: DistanceJoin: Pattern Match Query In a Large Graph Database. In: Proceedings of the VLDB Endowment (PVLDB), volume 2(1):pp , [ZH10] Zhao, Peixiang; Han, Jiawei: On Graph Query Optimization in Large Networks. In: Proceedings of the VLDB Endowment (PVLDB), volume 3(1):pp , [ZHY07] Zhang, Shijie; Hu, Meng; Yang, Jiong: TreePi: A Novel Graph Indexing Method. In: Proceedings of the 23rd International Conference on Data Engineering (ICDE), pp IEEE, [ZL07] [Zwi98] Zou, Lei; 0002, Lei Chen; Lu, Yansheng: Top-K Subgraph Matching Query in A Large Graph. In: Proceedings of the First Ph.D. Workshop at CIKM (PIKM), pp ACM Press, Zwick, Uri: All Pairs Shortest Paths in Weighted Directed Graphs Exact and Almost Exact Algorithms. In: Proceedings of the FOCS Conference, pp
205 Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit mit dem Titel Cost-based Optimization of Graph Queries in Relational Database Management Systems selbstständig und nur unter Verwendung der angegebenen Literatur und Hilfsmittel angefertigt und sie an keiner anderen Universität eingereicht habe, dass ich keinen Doktorgrad im Fach Informatik besitze, und dass mir die Promotionsordnung der Mathematisch-Naturwissenschaftlichen Fakultät II der Humboldt-Universität zu Berlin vom , zuletzt geändert am , veröffentlicht im Amtlichen Mitteilungsblatt Nr. 34/2006, bekannt ist. Berlin, den Silke Trißl 193
Protein Protein Interaction Networks
 Functional Pattern Mining from Genome Scale Protein Protein Interaction Networks Young-Rae Cho, Ph.D. Assistant Professor Department of Computer Science Baylor University it My Definition of Bioinformatics
Functional Pattern Mining from Genome Scale Protein Protein Interaction Networks Young-Rae Cho, Ph.D. Assistant Professor Department of Computer Science Baylor University it My Definition of Bioinformatics
Bioinformatics: Network Analysis
 Bioinformatics: Network Analysis Graph-theoretic Properties of Biological Networks COMP 572 (BIOS 572 / BIOE 564) - Fall 2013 Luay Nakhleh, Rice University 1 Outline Architectural features Motifs, modules,
Bioinformatics: Network Analysis Graph-theoretic Properties of Biological Networks COMP 572 (BIOS 572 / BIOE 564) - Fall 2013 Luay Nakhleh, Rice University 1 Outline Architectural features Motifs, modules,
Chapter 13: Query Processing. Basic Steps in Query Processing
 Chapter 13: Query Processing! Overview! Measures of Query Cost! Selection Operation! Sorting! Join Operation! Other Operations! Evaluation of Expressions 13.1 Basic Steps in Query Processing 1. Parsing
Chapter 13: Query Processing! Overview! Measures of Query Cost! Selection Operation! Sorting! Join Operation! Other Operations! Evaluation of Expressions 13.1 Basic Steps in Query Processing 1. Parsing
Social Media Mining. Graph Essentials
 Graph Essentials Graph Basics Measures Graph and Essentials Metrics 2 2 Nodes and Edges A network is a graph nodes, actors, or vertices (plural of vertex) Connections, edges or ties Edge Node Measures
Graph Essentials Graph Basics Measures Graph and Essentials Metrics 2 2 Nodes and Edges A network is a graph nodes, actors, or vertices (plural of vertex) Connections, edges or ties Edge Node Measures
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION. Professor Bharat Patel Office: Science 2, 2.36 Email: [email protected].
 Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: [email protected] What is Gene Expression & Gene Regulation? 1. Gene Expression
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: [email protected] What is Gene Expression & Gene Regulation? 1. Gene Expression
Analysis of Algorithms, I
 Analysis of Algorithms, I CSOR W4231.002 Eleni Drinea Computer Science Department Columbia University Thursday, February 26, 2015 Outline 1 Recap 2 Representing graphs 3 Breadth-first search (BFS) 4 Applications
Analysis of Algorithms, I CSOR W4231.002 Eleni Drinea Computer Science Department Columbia University Thursday, February 26, 2015 Outline 1 Recap 2 Representing graphs 3 Breadth-first search (BFS) 4 Applications
Home Page. Data Structures. Title Page. Page 1 of 24. Go Back. Full Screen. Close. Quit
 Data Structures Page 1 of 24 A.1. Arrays (Vectors) n-element vector start address + ielementsize 0 +1 +2 +3 +4... +n-1 start address continuous memory block static, if size is known at compile time dynamic,
Data Structures Page 1 of 24 A.1. Arrays (Vectors) n-element vector start address + ielementsize 0 +1 +2 +3 +4... +n-1 start address continuous memory block static, if size is known at compile time dynamic,
Data Warehousing und Data Mining
 Data Warehousing und Data Mining Multidimensionale Indexstrukturen Ulf Leser Wissensmanagement in der Bioinformatik Content of this Lecture Multidimensional Indexing Grid-Files Kd-trees Ulf Leser: Data
Data Warehousing und Data Mining Multidimensionale Indexstrukturen Ulf Leser Wissensmanagement in der Bioinformatik Content of this Lecture Multidimensional Indexing Grid-Files Kd-trees Ulf Leser: Data
SQL Query Evaluation. Winter 2006-2007 Lecture 23
 SQL Query Evaluation Winter 2006-2007 Lecture 23 SQL Query Processing Databases go through three steps: Parse SQL into an execution plan Optimize the execution plan Evaluate the optimized plan Execution
SQL Query Evaluation Winter 2006-2007 Lecture 23 SQL Query Processing Databases go through three steps: Parse SQL into an execution plan Optimize the execution plan Evaluate the optimized plan Execution
Efficiently Identifying Inclusion Dependencies in RDBMS
 Efficiently Identifying Inclusion Dependencies in RDBMS Jana Bauckmann Department for Computer Science, Humboldt-Universität zu Berlin Rudower Chaussee 25, 12489 Berlin, Germany [email protected]
Efficiently Identifying Inclusion Dependencies in RDBMS Jana Bauckmann Department for Computer Science, Humboldt-Universität zu Berlin Rudower Chaussee 25, 12489 Berlin, Germany [email protected]
Search Engines Chapter 2 Architecture. 14.4.2011 Felix Naumann
 Search Engines Chapter 2 Architecture 14.4.2011 Felix Naumann Overview 2 Basic Building Blocks Indexing Text Acquisition Text Transformation Index Creation Querying User Interaction Ranking Evaluation
Search Engines Chapter 2 Architecture 14.4.2011 Felix Naumann Overview 2 Basic Building Blocks Indexing Text Acquisition Text Transformation Index Creation Querying User Interaction Ranking Evaluation
Datenbanksysteme II: Implementation of Database Systems Implementing Joins
 Datenbanksysteme II: Implementation of Database Systems Implementing Joins Material von Prof. Johann Christoph Freytag Prof. Kai-Uwe Sattler Prof. Alfons Kemper, Dr. Eickler Prof. Hector Garcia-Molina
Datenbanksysteme II: Implementation of Database Systems Implementing Joins Material von Prof. Johann Christoph Freytag Prof. Kai-Uwe Sattler Prof. Alfons Kemper, Dr. Eickler Prof. Hector Garcia-Molina
Control of Gene Expression
 Home Gene Regulation Is Necessary? Control of Gene Expression By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Home Gene Regulation Is Necessary? Control of Gene Expression By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection favoring
Comp 5311 Database Management Systems. 16. Review 2 (Physical Level)
 Comp 5311 Database Management Systems 16. Review 2 (Physical Level) 1 Main Topics Indexing Join Algorithms Query Processing and Optimization Transactions and Concurrency Control 2 Indexing Used for faster
Comp 5311 Database Management Systems 16. Review 2 (Physical Level) 1 Main Topics Indexing Join Algorithms Query Processing and Optimization Transactions and Concurrency Control 2 Indexing Used for faster
Practical Graph Mining with R. 5. Link Analysis
 Practical Graph Mining with R 5. Link Analysis Outline Link Analysis Concepts Metrics for Analyzing Networks PageRank HITS Link Prediction 2 Link Analysis Concepts Link A relationship between two entities
Practical Graph Mining with R 5. Link Analysis Outline Link Analysis Concepts Metrics for Analyzing Networks PageRank HITS Link Prediction 2 Link Analysis Concepts Link A relationship between two entities
IE 680 Special Topics in Production Systems: Networks, Routing and Logistics*
 IE 680 Special Topics in Production Systems: Networks, Routing and Logistics* Rakesh Nagi Department of Industrial Engineering University at Buffalo (SUNY) *Lecture notes from Network Flows by Ahuja, Magnanti
IE 680 Special Topics in Production Systems: Networks, Routing and Logistics* Rakesh Nagi Department of Industrial Engineering University at Buffalo (SUNY) *Lecture notes from Network Flows by Ahuja, Magnanti
Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism )
 Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
SPANNING CACTI FOR STRUCTURALLY CONTROLLABLE NETWORKS NGO THI TU ANH NATIONAL UNIVERSITY OF SINGAPORE
 SPANNING CACTI FOR STRUCTURALLY CONTROLLABLE NETWORKS NGO THI TU ANH NATIONAL UNIVERSITY OF SINGAPORE 2012 SPANNING CACTI FOR STRUCTURALLY CONTROLLABLE NETWORKS NGO THI TU ANH (M.Sc., SFU, Russia) A THESIS
SPANNING CACTI FOR STRUCTURALLY CONTROLLABLE NETWORKS NGO THI TU ANH NATIONAL UNIVERSITY OF SINGAPORE 2012 SPANNING CACTI FOR STRUCTURALLY CONTROLLABLE NETWORKS NGO THI TU ANH (M.Sc., SFU, Russia) A THESIS
Data Structure [Question Bank]
![Data Structure [Question Bank] Data Structure [Question Bank]](/thumbs/40/21355705.jpg) Unit I (Analysis of Algorithms) 1. What are algorithms and how they are useful? 2. Describe the factor on best algorithms depends on? 3. Differentiate: Correct & Incorrect Algorithms? 4. Write short note:
Unit I (Analysis of Algorithms) 1. What are algorithms and how they are useful? 2. Describe the factor on best algorithms depends on? 3. Differentiate: Correct & Incorrect Algorithms? 4. Write short note:
G-SPARQL: A Hybrid Engine for Querying Large Attributed Graphs
 G-SPARQL: A Hybrid Engine for Querying Large Attributed Graphs Sherif Sakr National ICT Australia UNSW, Sydney, Australia [email protected] Sameh Elnikety Microsoft Research Redmond, WA, USA [email protected]
G-SPARQL: A Hybrid Engine for Querying Large Attributed Graphs Sherif Sakr National ICT Australia UNSW, Sydney, Australia [email protected] Sameh Elnikety Microsoft Research Redmond, WA, USA [email protected]
www.gr8ambitionz.com
 Data Base Management Systems (DBMS) Study Material (Objective Type questions with Answers) Shared by Akhil Arora Powered by www. your A to Z competitive exam guide Database Objective type questions Q.1
Data Base Management Systems (DBMS) Study Material (Objective Type questions with Answers) Shared by Akhil Arora Powered by www. your A to Z competitive exam guide Database Objective type questions Q.1
Graph theoretic approach to analyze amino acid network
 Int. J. Adv. Appl. Math. and Mech. 2(3) (2015) 31-37 (ISSN: 2347-2529) Journal homepage: www.ijaamm.com International Journal of Advances in Applied Mathematics and Mechanics Graph theoretic approach to
Int. J. Adv. Appl. Math. and Mech. 2(3) (2015) 31-37 (ISSN: 2347-2529) Journal homepage: www.ijaamm.com International Journal of Advances in Applied Mathematics and Mechanics Graph theoretic approach to
CSE 326, Data Structures. Sample Final Exam. Problem Max Points Score 1 14 (2x7) 2 18 (3x6) 3 4 4 7 5 9 6 16 7 8 8 4 9 8 10 4 Total 92.
 Name: Email ID: CSE 326, Data Structures Section: Sample Final Exam Instructions: The exam is closed book, closed notes. Unless otherwise stated, N denotes the number of elements in the data structure
Name: Email ID: CSE 326, Data Structures Section: Sample Final Exam Instructions: The exam is closed book, closed notes. Unless otherwise stated, N denotes the number of elements in the data structure
Data Lineage and Meta Data Analysis in Data Warehouse Environments
 Department of Informatics, University of Zürich BSc Thesis Data Lineage and Meta Data Analysis in Data Warehouse Environments Martin Noack Matrikelnummer: 09-222-232 Email: [email protected] January
Department of Informatics, University of Zürich BSc Thesis Data Lineage and Meta Data Analysis in Data Warehouse Environments Martin Noack Matrikelnummer: 09-222-232 Email: [email protected] January
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Databases and Information Systems 1 Part 3: Storage Structures and Indices
 bases and Information Systems 1 Part 3: Storage Structures and Indices Prof. Dr. Stefan Böttcher Fakultät EIM, Institut für Informatik Universität Paderborn WS 2009 / 2010 Contents: - database buffer -
bases and Information Systems 1 Part 3: Storage Structures and Indices Prof. Dr. Stefan Böttcher Fakultät EIM, Institut für Informatik Universität Paderborn WS 2009 / 2010 Contents: - database buffer -
V. Adamchik 1. Graph Theory. Victor Adamchik. Fall of 2005
 V. Adamchik 1 Graph Theory Victor Adamchik Fall of 2005 Plan 1. Basic Vocabulary 2. Regular graph 3. Connectivity 4. Representing Graphs Introduction A.Aho and J.Ulman acknowledge that Fundamentally, computer
V. Adamchik 1 Graph Theory Victor Adamchik Fall of 2005 Plan 1. Basic Vocabulary 2. Regular graph 3. Connectivity 4. Representing Graphs Introduction A.Aho and J.Ulman acknowledge that Fundamentally, computer
Discrete Mathematics & Mathematical Reasoning Chapter 10: Graphs
 Discrete Mathematics & Mathematical Reasoning Chapter 10: Graphs Kousha Etessami U. of Edinburgh, UK Kousha Etessami (U. of Edinburgh, UK) Discrete Mathematics (Chapter 6) 1 / 13 Overview Graphs and Graph
Discrete Mathematics & Mathematical Reasoning Chapter 10: Graphs Kousha Etessami U. of Edinburgh, UK Kousha Etessami (U. of Edinburgh, UK) Discrete Mathematics (Chapter 6) 1 / 13 Overview Graphs and Graph
Visualizing Networks: Cytoscape. Prat Thiru
 Visualizing Networks: Cytoscape Prat Thiru Outline Introduction to Networks Network Basics Visualization Inferences Cytoscape Demo 2 Why (Biological) Networks? 3 Networks: An Integrative Approach Zvelebil,
Visualizing Networks: Cytoscape Prat Thiru Outline Introduction to Networks Network Basics Visualization Inferences Cytoscape Demo 2 Why (Biological) Networks? 3 Networks: An Integrative Approach Zvelebil,
Load balancing Static Load Balancing
 Chapter 7 Load Balancing and Termination Detection Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination detection
Chapter 7 Load Balancing and Termination Detection Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination detection
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
Algorithms in Computational Biology (236522) spring 2007 Lecture #1
 Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Algorithms in Computational Biology (236522) spring 2007 Lecture #1 Lecturer: Shlomo Moran, Taub 639, tel 4363 Office hours: Tuesday 11:00-12:00/by appointment TA: Ilan Gronau, Taub 700, tel 4894 Office
Subgraph Patterns: Network Motifs and Graphlets. Pedro Ribeiro
 Subgraph Patterns: Network Motifs and Graphlets Pedro Ribeiro Analyzing Complex Networks We have been talking about extracting information from networks Some possible tasks: General Patterns Ex: scale-free,
Subgraph Patterns: Network Motifs and Graphlets Pedro Ribeiro Analyzing Complex Networks We have been talking about extracting information from networks Some possible tasks: General Patterns Ex: scale-free,
Metabolic Network Analysis
 Metabolic Network nalysis Overview -- modelling chemical reaction networks -- Levels of modelling Lecture II: Modelling chemical reaction networks dr. Sander Hille [email protected] http://www.math.leidenuniv.nl/~shille
Metabolic Network nalysis Overview -- modelling chemical reaction networks -- Levels of modelling Lecture II: Modelling chemical reaction networks dr. Sander Hille [email protected] http://www.math.leidenuniv.nl/~shille
Gene Regulation -- The Lac Operon
 Gene Regulation -- The Lac Operon Specific proteins are present in different tissues and some appear only at certain times during development. All cells of a higher organism have the full set of genes:
Gene Regulation -- The Lac Operon Specific proteins are present in different tissues and some appear only at certain times during development. All cells of a higher organism have the full set of genes:
Application of Graph-based Data Mining to Metabolic Pathways
 Application of Graph-based Data Mining to Metabolic Pathways Chang Hun You, Lawrence B. Holder, Diane J. Cook School of Electrical Engineering and Computer Science Washington State University Pullman,
Application of Graph-based Data Mining to Metabolic Pathways Chang Hun You, Lawrence B. Holder, Diane J. Cook School of Electrical Engineering and Computer Science Washington State University Pullman,
Map-like Wikipedia Visualization. Pang Cheong Iao. Master of Science in Software Engineering
 Map-like Wikipedia Visualization by Pang Cheong Iao Master of Science in Software Engineering 2011 Faculty of Science and Technology University of Macau Map-like Wikipedia Visualization by Pang Cheong
Map-like Wikipedia Visualization by Pang Cheong Iao Master of Science in Software Engineering 2011 Faculty of Science and Technology University of Macau Map-like Wikipedia Visualization by Pang Cheong
Chapter 7 Active Reading Guide Cellular Respiration and Fermentation
 Name: AP Biology Mr. Croft Chapter 7 Active Reading Guide Cellular Respiration and Fermentation Overview: Before getting involved with the details of cellular respiration and photosynthesis, take a second
Name: AP Biology Mr. Croft Chapter 7 Active Reading Guide Cellular Respiration and Fermentation Overview: Before getting involved with the details of cellular respiration and photosynthesis, take a second
Network Analysis. BCH 5101: Analysis of -Omics Data 1/34
 Network Analysis BCH 5101: Analysis of -Omics Data 1/34 Network Analysis Graphs as a representation of networks Examples of genome-scale graphs Statistical properties of genome-scale graphs The search
Network Analysis BCH 5101: Analysis of -Omics Data 1/34 Network Analysis Graphs as a representation of networks Examples of genome-scale graphs Statistical properties of genome-scale graphs The search
Inside the PostgreSQL Query Optimizer
 Inside the PostgreSQL Query Optimizer Neil Conway [email protected] Fujitsu Australia Software Technology PostgreSQL Query Optimizer Internals p. 1 Outline Introduction to query optimization Outline of
Inside the PostgreSQL Query Optimizer Neil Conway [email protected] Fujitsu Australia Software Technology PostgreSQL Query Optimizer Internals p. 1 Outline Introduction to query optimization Outline of
Load Balancing and Termination Detection
 Chapter 7 Load Balancing and Termination Detection 1 Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination detection
Chapter 7 Load Balancing and Termination Detection 1 Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination detection
131-1. Adding New Level in KDD to Make the Web Usage Mining More Efficient. Abstract. 1. Introduction [1]. 1/10
![131-1. Adding New Level in KDD to Make the Web Usage Mining More Efficient. Abstract. 1. Introduction [1]. 1/10 131-1. Adding New Level in KDD to Make the Web Usage Mining More Efficient. Abstract. 1. Introduction [1]. 1/10](/thumbs/25/4963058.jpg) 1/10 131-1 Adding New Level in KDD to Make the Web Usage Mining More Efficient Mohammad Ala a AL_Hamami PHD Student, Lecturer m_ah_1@yahoocom Soukaena Hassan Hashem PHD Student, Lecturer soukaena_hassan@yahoocom
1/10 131-1 Adding New Level in KDD to Make the Web Usage Mining More Efficient Mohammad Ala a AL_Hamami PHD Student, Lecturer m_ah_1@yahoocom Soukaena Hassan Hashem PHD Student, Lecturer soukaena_hassan@yahoocom
Web-Based Genomic Information Integration with Gene Ontology
 Web-Based Genomic Information Integration with Gene Ontology Kai Xu 1 IMAGEN group, National ICT Australia, Sydney, Australia, [email protected] Abstract. Despite the dramatic growth of online genomic
Web-Based Genomic Information Integration with Gene Ontology Kai Xu 1 IMAGEN group, National ICT Australia, Sydney, Australia, [email protected] Abstract. Despite the dramatic growth of online genomic
From DNA to Protein. Proteins. Chapter 13. Prokaryotes and Eukaryotes. The Path From Genes to Proteins. All proteins consist of polypeptide chains
 Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Asking Hard Graph Questions. Paul Burkhardt. February 3, 2014
 Beyond Watson: Predictive Analytics and Big Data U.S. National Security Agency Research Directorate - R6 Technical Report February 3, 2014 300 years before Watson there was Euler! The first (Jeopardy!)
Beyond Watson: Predictive Analytics and Big Data U.S. National Security Agency Research Directorate - R6 Technical Report February 3, 2014 300 years before Watson there was Euler! The first (Jeopardy!)
Binary Trees and Huffman Encoding Binary Search Trees
 Binary Trees and Huffman Encoding Binary Search Trees Computer Science E119 Harvard Extension School Fall 2012 David G. Sullivan, Ph.D. Motivation: Maintaining a Sorted Collection of Data A data dictionary
Binary Trees and Huffman Encoding Binary Search Trees Computer Science E119 Harvard Extension School Fall 2012 David G. Sullivan, Ph.D. Motivation: Maintaining a Sorted Collection of Data A data dictionary
Network (Tree) Topology Inference Based on Prüfer Sequence
 Network (Tree) Topology Inference Based on Prüfer Sequence C. Vanniarajan and Kamala Krithivasan Department of Computer Science and Engineering Indian Institute of Technology Madras Chennai 600036 [email protected],
Network (Tree) Topology Inference Based on Prüfer Sequence C. Vanniarajan and Kamala Krithivasan Department of Computer Science and Engineering Indian Institute of Technology Madras Chennai 600036 [email protected],
Query Processing C H A P T E R12. Practice Exercises
 C H A P T E R12 Query Processing Practice Exercises 12.1 Assume (for simplicity in this exercise) that only one tuple fits in a block and memory holds at most 3 blocks. Show the runs created on each pass
C H A P T E R12 Query Processing Practice Exercises 12.1 Assume (for simplicity in this exercise) that only one tuple fits in a block and memory holds at most 3 blocks. Show the runs created on each pass
Optimization of SQL Queries in Main-Memory Databases
 Optimization of SQL Queries in Main-Memory Databases Ladislav Vastag and Ján Genči Department of Computers and Informatics Technical University of Košice, Letná 9, 042 00 Košice, Slovakia [email protected]
Optimization of SQL Queries in Main-Memory Databases Ladislav Vastag and Ján Genči Department of Computers and Informatics Technical University of Košice, Letná 9, 042 00 Košice, Slovakia [email protected]
Customer Intimacy Analytics
 Customer Intimacy Analytics Leveraging Operational Data to Assess Customer Knowledge and Relationships and to Measure their Business Impact by Francois Habryn Scientific Publishing CUSTOMER INTIMACY ANALYTICS
Customer Intimacy Analytics Leveraging Operational Data to Assess Customer Knowledge and Relationships and to Measure their Business Impact by Francois Habryn Scientific Publishing CUSTOMER INTIMACY ANALYTICS
Why? A central concept in Computer Science. Algorithms are ubiquitous.
 Analysis of Algorithms: A Brief Introduction Why? A central concept in Computer Science. Algorithms are ubiquitous. Using the Internet (sending email, transferring files, use of search engines, online
Analysis of Algorithms: A Brief Introduction Why? A central concept in Computer Science. Algorithms are ubiquitous. Using the Internet (sending email, transferring files, use of search engines, online
[Refer Slide Time: 05:10]
![[Refer Slide Time: 05:10] [Refer Slide Time: 05:10]](/thumbs/29/13447118.jpg) Principles of Programming Languages Prof: S. Arun Kumar Department of Computer Science and Engineering Indian Institute of Technology Delhi Lecture no 7 Lecture Title: Syntactic Classes Welcome to lecture
Principles of Programming Languages Prof: S. Arun Kumar Department of Computer Science and Engineering Indian Institute of Technology Delhi Lecture no 7 Lecture Title: Syntactic Classes Welcome to lecture
General Network Analysis: Graph-theoretic. COMP572 Fall 2009
 General Network Analysis: Graph-theoretic Techniques COMP572 Fall 2009 Networks (aka Graphs) A network is a set of vertices, or nodes, and edges that connect pairs of vertices Example: a network with 5
General Network Analysis: Graph-theoretic Techniques COMP572 Fall 2009 Networks (aka Graphs) A network is a set of vertices, or nodes, and edges that connect pairs of vertices Example: a network with 5
Graph Database Proof of Concept Report
 Objectivity, Inc. Graph Database Proof of Concept Report Managing The Internet of Things Table of Contents Executive Summary 3 Background 3 Proof of Concept 4 Dataset 4 Process 4 Query Catalog 4 Environment
Objectivity, Inc. Graph Database Proof of Concept Report Managing The Internet of Things Table of Contents Executive Summary 3 Background 3 Proof of Concept 4 Dataset 4 Process 4 Query Catalog 4 Environment
Unit 5 Photosynthesis and Cellular Respiration
 Unit 5 Photosynthesis and Cellular Respiration Advanced Concepts What is the abbreviated name of this molecule? What is its purpose? What are the three parts of this molecule? Label each part with the
Unit 5 Photosynthesis and Cellular Respiration Advanced Concepts What is the abbreviated name of this molecule? What is its purpose? What are the three parts of this molecule? Label each part with the
Vector storage and access; algorithms in GIS. This is lecture 6
 Vector storage and access; algorithms in GIS This is lecture 6 Vector data storage and access Vectors are built from points, line and areas. (x,y) Surface: (x,y,z) Vector data access Access to vector
Vector storage and access; algorithms in GIS This is lecture 6 Vector data storage and access Vectors are built from points, line and areas. (x,y) Surface: (x,y,z) Vector data access Access to vector
Quiz! Database Indexes. Index. Quiz! Disc and main memory. Quiz! How costly is this operation (naive solution)?
 Database Indexes How costly is this operation (naive solution)? course per weekday hour room TDA356 2 VR Monday 13:15 TDA356 2 VR Thursday 08:00 TDA356 4 HB1 Tuesday 08:00 TDA356 4 HB1 Friday 13:15 TIN090
Database Indexes How costly is this operation (naive solution)? course per weekday hour room TDA356 2 VR Monday 13:15 TDA356 2 VR Thursday 08:00 TDA356 4 HB1 Tuesday 08:00 TDA356 4 HB1 Friday 13:15 TIN090
Hormones & Chemical Signaling
 Hormones & Chemical Signaling Part 2 modulation of signal pathways and hormone classification & function How are these pathways controlled? Receptors are proteins! Subject to Specificity of binding Competition
Hormones & Chemical Signaling Part 2 modulation of signal pathways and hormone classification & function How are these pathways controlled? Receptors are proteins! Subject to Specificity of binding Competition
Relational Databases
 Relational Databases Jan Chomicki University at Buffalo Jan Chomicki () Relational databases 1 / 18 Relational data model Domain domain: predefined set of atomic values: integers, strings,... every attribute
Relational Databases Jan Chomicki University at Buffalo Jan Chomicki () Relational databases 1 / 18 Relational data model Domain domain: predefined set of atomic values: integers, strings,... every attribute
Load Balancing and Termination Detection
 Chapter 7 slides7-1 Load Balancing and Termination Detection slides7-2 Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination
Chapter 7 slides7-1 Load Balancing and Termination Detection slides7-2 Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination
Data storage Tree indexes
 Data storage Tree indexes Rasmus Pagh February 7 lecture 1 Access paths For many database queries and updates, only a small fraction of the data needs to be accessed. Extreme examples are looking or updating
Data storage Tree indexes Rasmus Pagh February 7 lecture 1 Access paths For many database queries and updates, only a small fraction of the data needs to be accessed. Extreme examples are looking or updating
Twincore - Zentrum für Experimentelle und Klinische Infektionsforschung Institut für Molekulare Bakteriologie
 Twincore - Zentrum für Experimentelle und Klinische Infektionsforschung Institut für Molekulare Bakteriologie 0 HELMHOLTZ I ZENTRUM FÜR INFEKTIONSFORSCHUNG Technische Universität Braunschweig Institut
Twincore - Zentrum für Experimentelle und Klinische Infektionsforschung Institut für Molekulare Bakteriologie 0 HELMHOLTZ I ZENTRUM FÜR INFEKTIONSFORSCHUNG Technische Universität Braunschweig Institut
Big Data and Scripting. Part 4: Memory Hierarchies
 1, Big Data and Scripting Part 4: Memory Hierarchies 2, Model and Definitions memory size: M machine words total storage (on disk) of N elements (N is very large) disk size unlimited (for our considerations)
1, Big Data and Scripting Part 4: Memory Hierarchies 2, Model and Definitions memory size: M machine words total storage (on disk) of N elements (N is very large) disk size unlimited (for our considerations)
DATA STRUCTURES USING C
 DATA STRUCTURES USING C QUESTION BANK UNIT I 1. Define data. 2. Define Entity. 3. Define information. 4. Define Array. 5. Define data structure. 6. Give any two applications of data structures. 7. Give
DATA STRUCTURES USING C QUESTION BANK UNIT I 1. Define data. 2. Define Entity. 3. Define information. 4. Define Array. 5. Define data structure. 6. Give any two applications of data structures. 7. Give
Cell Structure & Function!
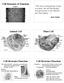 Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
Activity 7.21 Transcription factors
 Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
Database-Supported XML Processors
 Database-Supported XML Processors Prof. Dr. Torsten Grust [email protected] Winter 2008/2009 Torsten Grust (WSI) Database-Supported XML Processors Winter 2008/09 1 Part I Preliminaries Torsten
Database-Supported XML Processors Prof. Dr. Torsten Grust [email protected] Winter 2008/2009 Torsten Grust (WSI) Database-Supported XML Processors Winter 2008/09 1 Part I Preliminaries Torsten
Query Optimization for Distributed Database Systems Robert Taylor Candidate Number : 933597 Hertford College Supervisor: Dr.
 Query Optimization for Distributed Database Systems Robert Taylor Candidate Number : 933597 Hertford College Supervisor: Dr. Dan Olteanu Submitted as part of Master of Computer Science Computing Laboratory
Query Optimization for Distributed Database Systems Robert Taylor Candidate Number : 933597 Hertford College Supervisor: Dr. Dan Olteanu Submitted as part of Master of Computer Science Computing Laboratory
Distance Degree Sequences for Network Analysis
 Universität Konstanz Computer & Information Science Algorithmics Group 15 Mar 2005 based on Palmer, Gibbons, and Faloutsos: ANF A Fast and Scalable Tool for Data Mining in Massive Graphs, SIGKDD 02. Motivation
Universität Konstanz Computer & Information Science Algorithmics Group 15 Mar 2005 based on Palmer, Gibbons, and Faloutsos: ANF A Fast and Scalable Tool for Data Mining in Massive Graphs, SIGKDD 02. Motivation
Name Date Period. 2. When a molecule of double-stranded DNA undergoes replication, it results in
 DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
for High Performance Computing
 Technische Universität München Institut für Informatik Lehrstuhl für Rechnertechnik und Rechnerorganisation Automatic Performance Engineering Workflows for High Performance Computing Ventsislav Petkov
Technische Universität München Institut für Informatik Lehrstuhl für Rechnertechnik und Rechnerorganisation Automatic Performance Engineering Workflows for High Performance Computing Ventsislav Petkov
AP WORLD LANGUAGE AND CULTURE EXAMS 2012 SCORING GUIDELINES
 AP WORLD LANGUAGE AND CULTURE EXAMS 2012 SCORING GUIDELINES Interpersonal Writing: E-mail Reply 5: STRONG performance in Interpersonal Writing Maintains the exchange with a response that is clearly appropriate
AP WORLD LANGUAGE AND CULTURE EXAMS 2012 SCORING GUIDELINES Interpersonal Writing: E-mail Reply 5: STRONG performance in Interpersonal Writing Maintains the exchange with a response that is clearly appropriate
5. A full binary tree with n leaves contains [A] n nodes. [B] log n 2 nodes. [C] 2n 1 nodes. [D] n 2 nodes.
![5. A full binary tree with n leaves contains [A] n nodes. [B] log n 2 nodes. [C] 2n 1 nodes. [D] n 2 nodes. 5. A full binary tree with n leaves contains [A] n nodes. [B] log n 2 nodes. [C] 2n 1 nodes. [D] n 2 nodes.](/thumbs/40/21353779.jpg) 1. The advantage of.. is that they solve the problem if sequential storage representation. But disadvantage in that is they are sequential lists. [A] Lists [B] Linked Lists [A] Trees [A] Queues 2. The
1. The advantage of.. is that they solve the problem if sequential storage representation. But disadvantage in that is they are sequential lists. [A] Lists [B] Linked Lists [A] Trees [A] Queues 2. The
How To Find Local Affinity Patterns In Big Data
 Detection of local affinity patterns in big data Andrea Marinoni, Paolo Gamba Department of Electronics, University of Pavia, Italy Abstract Mining information in Big Data requires to design a new class
Detection of local affinity patterns in big data Andrea Marinoni, Paolo Gamba Department of Electronics, University of Pavia, Italy Abstract Mining information in Big Data requires to design a new class
AP BIOLOGY CHAPTER 7 Cellular Respiration Outline
 AP BIOLOGY CHAPTER 7 Cellular Respiration Outline I. How cells get energy. A. Cellular Respiration 1. Cellular respiration includes the various metabolic pathways that break down carbohydrates and other
AP BIOLOGY CHAPTER 7 Cellular Respiration Outline I. How cells get energy. A. Cellular Respiration 1. Cellular respiration includes the various metabolic pathways that break down carbohydrates and other
Module 3 Questions. 7. Chemotaxis is an example of signal transduction. Explain, with the use of diagrams.
 Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
Module 3 Questions Section 1. Essay and Short Answers. Use diagrams wherever possible 1. With the use of a diagram, provide an overview of the general regulation strategies available to a bacterial cell.
Chapter 7 Load Balancing and Termination Detection
 Chapter 7 Load Balancing and Termination Detection Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination detection
Chapter 7 Load Balancing and Termination Detection Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination detection
Extraction and Visualization of Protein-Protein Interactions from PubMed
 Extraction and Visualization of Protein-Protein Interactions from PubMed Ulf Leser Knowledge Management in Bioinformatics Humboldt-Universität Berlin Finding Relevant Knowledge Find information about Much
Extraction and Visualization of Protein-Protein Interactions from PubMed Ulf Leser Knowledge Management in Bioinformatics Humboldt-Universität Berlin Finding Relevant Knowledge Find information about Much
GENE REGULATION. Teacher Packet
 AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
TIn 1: Lecture 3: Lernziele. Lecture 3 The Belly of the Architect. Basic internal components of the 8086. Pointers and data storage in memory
 Mitglied der Zürcher Fachhochschule TIn 1: Lecture 3 The Belly of the Architect. Lecture 3: Lernziele Basic internal components of the 8086 Pointers and data storage in memory Architektur 8086 Besteht
Mitglied der Zürcher Fachhochschule TIn 1: Lecture 3 The Belly of the Architect. Lecture 3: Lernziele Basic internal components of the 8086 Pointers and data storage in memory Architektur 8086 Besteht
Load Balancing and Termination Detection
 Chapter 7 Slide 1 Slide 2 Load Balancing and Termination Detection Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination
Chapter 7 Slide 1 Slide 2 Load Balancing and Termination Detection Load balancing used to distribute computations fairly across processors in order to obtain the highest possible execution speed. Termination
A Comparison of Dictionary Implementations
 A Comparison of Dictionary Implementations Mark P Neyer April 10, 2009 1 Introduction A common problem in computer science is the representation of a mapping between two sets. A mapping f : A B is a function
A Comparison of Dictionary Implementations Mark P Neyer April 10, 2009 1 Introduction A common problem in computer science is the representation of a mapping between two sets. A mapping f : A B is a function
Chapter 2 Data Storage
 Chapter 2 22 CHAPTER 2. DATA STORAGE 2.1. THE MEMORY HIERARCHY 23 26 CHAPTER 2. DATA STORAGE main memory, yet is essentially random-access, with relatively small differences Figure 2.4: A typical
Chapter 2 22 CHAPTER 2. DATA STORAGE 2.1. THE MEMORY HIERARCHY 23 26 CHAPTER 2. DATA STORAGE main memory, yet is essentially random-access, with relatively small differences Figure 2.4: A typical
A Fast Algorithm For Finding Hamilton Cycles
 A Fast Algorithm For Finding Hamilton Cycles by Andrew Chalaturnyk A thesis presented to the University of Manitoba in partial fulfillment of the requirements for the degree of Masters of Science in Computer
A Fast Algorithm For Finding Hamilton Cycles by Andrew Chalaturnyk A thesis presented to the University of Manitoba in partial fulfillment of the requirements for the degree of Masters of Science in Computer
Chapter 9 Mitochondrial Structure and Function
 Chapter 9 Mitochondrial Structure and Function 1 2 3 Structure and function Oxidative phosphorylation and ATP Synthesis Peroxisome Overview 2 Mitochondria have characteristic morphologies despite variable
Chapter 9 Mitochondrial Structure and Function 1 2 3 Structure and function Oxidative phosphorylation and ATP Synthesis Peroxisome Overview 2 Mitochondria have characteristic morphologies despite variable
Exemplar for Internal Achievement Standard. German Level 1
 Exemplar for Internal Achievement Standard German Level 1 This exemplar supports assessment against: Achievement Standard 90885 Interact using spoken German to communicate personal information, ideas and
Exemplar for Internal Achievement Standard German Level 1 This exemplar supports assessment against: Achievement Standard 90885 Interact using spoken German to communicate personal information, ideas and
Mining Social-Network Graphs
 342 Chapter 10 Mining Social-Network Graphs There is much information to be gained by analyzing the large-scale data that is derived from social networks. The best-known example of a social network is
342 Chapter 10 Mining Social-Network Graphs There is much information to be gained by analyzing the large-scale data that is derived from social networks. The best-known example of a social network is
Interconnection Networks. Interconnection Networks. Interconnection networks are used everywhere!
 Interconnection Networks Interconnection Networks Interconnection networks are used everywhere! Supercomputers connecting the processors Routers connecting the ports can consider a router as a parallel
Interconnection Networks Interconnection Networks Interconnection networks are used everywhere! Supercomputers connecting the processors Routers connecting the ports can consider a router as a parallel
Clustering & Visualization
 Chapter 5 Clustering & Visualization Clustering in high-dimensional databases is an important problem and there are a number of different clustering paradigms which are applicable to high-dimensional data.
Chapter 5 Clustering & Visualization Clustering in high-dimensional databases is an important problem and there are a number of different clustering paradigms which are applicable to high-dimensional data.
Regulation of enzyme activity
 1 Regulation of enzyme activity Regulation of enzyme activity is important to coordinate the different metabolic processes. It is also important for homeostasis i.e. to maintain the internal environment
1 Regulation of enzyme activity Regulation of enzyme activity is important to coordinate the different metabolic processes. It is also important for homeostasis i.e. to maintain the internal environment
