SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants
|
|
|
- Violet Wiggins
- 10 years ago
- Views:
Transcription
1 SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants Xiuwen Zheng Department of Biostatistics University of Washington Seattle Dec 28, 2014 Contents 1 Overview 2 2 Preparing Data Data formats used in SeqArray Format conversion from VCF files Export to a VCF File Modification Data Processing Get Data Apply Functions Over Array Margins Apply Functions in Parallel Integration with Other Packages in Bioconductor Examples The performance of seqapply Missing Rates Multi-process Implementation Allele Frequency Multi-process Implementation Principal Component Analysis Multi-process Implementation Individual Inbreeding Coefficient Multi-process Implementation Seamless R and C Integration The 1000 Genomes Project 31 1
2 Variants 2 6 Resources 33 7 Session Info 33 1 Overview In the era of big data, thousands of gigabyte-size data sets are challenging scientists for data management, even on well-equipped hardware. Currently, next-generation sequencing techniques are being adopted to investigate common and rare variants, making the analyses of large-scale genotypic data challenging. For example, the 1000 Genomes Project has identified approximately 38 million single nucleotide polymorphisms (SNPs), 1.4 million short insertions and deletions, and more than 14,000 larger deletions from whole-genome sequencing technologies [1]. In the near future, new technologies, like third-generation whole-genome sequencing, will be enabling data to be generated at an unprecedented scale [2]. The Variant Call Format (VCF) was developed for the 1000 Genomes Project, which is a generic text format for storing DNA polymorphism data such as SNPs, insertions, deletions and structural variants, together with rich annotations [3]. However, this format is less efficient for largescale analyses since numeric data have to be parsed from a text VCF file before further analyses. The computational burden associated with sequencing variants is especially evident with large sample and variant sizes, and it requires efficient numerical implementation and data management. Here I introduce a high-performance C/C computing library CoreArray ( net) for big-data management of genome-wide variants [4]. CoreArray was designed for developing portable and scalable storage technologies for bioinformatics data, allowing parallel computing at the multicore and cluster levels. It provides the genomic data structure (GDS) file format for array-oriented data: this is a universal data format to store multiple data variables in a single file. The CoreArray library modules are demonstrated in Figure 1. A hierarchical data structure is used to store multiple extensible data variables in the GDS format, and all datasets are stored in a single file with chunked storage layout. A document about CoreArray can be found: packages/gdsfmt/vignettes/corearraytutorial.pdf. Here, I focus on the application of CoreArray for statisticians working in the R environment, and developed R packages gdsfmt and SeqArray to address or reduce the computational burden associated with data management of sequencing variants. Gdsfmt provides a general R interface for the CoreArray library, and SeqArray is specifically designed for data management of sequencing variants. The kernels of gdsfmt and SeqArray were written in C/C and highly optimized. Genotypic data and annotations are stored in a binary and array-oriented manner, offering efficient access of genetic variants using the R language. There are six key functions in SeqArray (Table 1), and most of data analyses could be done using these six functions. The 1000 Genomes Project released 39 million genetic variants for 1092 individuals, and a 26G data file was created by SeqArray to store sequencing variants with phasing information, where 2 bits were used as a primitive data type. The file size can be further reduced to 1.5G by compression algorithms without sacrificing access efficiency, since it has a large proportion of rare variants. SeqArray will be of great interest to scientists involved in data analyses of large-scale genomic sequencing data using R environment, particularly those with limited experience of low-level C programming and
3 Variants 3 Figure 1: CoreArray library modules. Function seqvcf2gds seqsummary seqsetfilter seqgetdata seqapply seqparallel Table 1: The key functions in the SeqArray package. Description Imports VCF files Gets the summary of a sequencing GDS file (# of samples, # of variants, INFO/FORMAT variables, etc) Sets a filter to sample or variant (define a subset of data) Gets data from a sequencing GDS file (from a subset of data) Applies a user-defined function over array margins Applies functions in parallel parallel computing. 2 Preparing Data
4 Variants Data formats used in SeqArray To support efficient memory management for genome-wide numerical data, the gdsfmt package provides the genomic data structure (GDS) file format for array-oriented bioinformatic data based on the Core- Array library, which is a container for storing genotypic and annotation data. The GDS format supports data blocking so that only the subset of data that is being processed needs to reside in memory. > # load the R package SeqArray > library(seqarray) Here is a typical GDS file shipped with the SeqArray package: > gds.fn <- seqexamplefilename("gds") > # or gds.fn <- "C:/YourFolder/Your_GDS_File.gds" > gds.fn [1] "/tmp/rtmpbo98st/rinst46302deddfc3/seqarray/extdata/ceu_exon.gds" > seqsummary(gds.fn) File: /tmp/rtmpbo98st/rinst46302deddfc3/seqarray/extdata/ceu_exon.gds Sequence Variant Format: v1.0 The number of samples: 90 The number of variants: 1348 The chromosomes: <NA> The number of alleles per site: Annotation, information variables: AA,., String, Ancestral Allele AC, 1, Integer, total number of alternate alleles in called genotypes AN, 1, Integer, total number of alleles in called genotypes DP, 1, Integer, Total Depth HM2, 0, Flag, HapMap2 membership HM3, 0, Flag, HapMap3 membership OR, 1, String, Previous rs number GP, 1, String, GRCh37 position(s) BN, 1, Integer, First dbsnp build # Annotation, format variables: DP,., Integer, Read Depth from MOSAIK BAM seqexamplefilename returns the file name of a GDS file used as an example in SeqArray, and it is a subset of data from the 1000 Genomes Project. seqsummary summarizes the genotypes and annotations stored in the GDS file.
5 Variants 5 > # open a GDS file > genofile <- seqopen(gds.fn) > # display the contents of the GDS file in a hierarchical structure > genofile Object of class "SeqVarGDSClass" File: /tmp/rtmpbo98st/rinst46302deddfc3/seqarray/extdata/ceu_exon.gds [ ] -- description [ ] * -- sample.id { VStr8 90 ZIP(28.33%) } -- variant.id { Int ZIP(35.39%) } -- position { Int ZIP(86.11%) } -- chromosome { VStr ZIP(2.14%) } -- allele { VStr ZIP(16.86%) } -- genotype [ ] * -- data { Bit2 2x90x1348 ZIP(28.36%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Int16 0 ZIP } -- phase [ ] -- data { Bit1 90x1348 ZIP(0.24%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Bit1 0 ZIP } -- annotation [ ] -- id { VStr ZIP(40.89%) } -- qual { Float ZIP(0.57%) } -- filter { Int32,factor 1348 ZIP(0.56%) } * -- info [ ] -- AA { VStr ZIP(23.55%) } * -- AC { Int ZIP(26.89%) } * -- AN { Int ZIP(20.29%) } * -- DP { Int ZIP(62.24%) } * -- HM2 { Bit ZIP(106.51%) } * -- HM3 { Bit ZIP(106.51%) } * -- OR { VStr ZIP(12.92%) } * -- GP { VStr ZIP(34.24%) } * -- BN { Int ZIP(21.31%) } * -- format [ ] -- DP [ ] * -- data { Int32 90x1348 ZIP(33.83%) } For those who would like to know how variable-length genotypic data and annotations are stored in an array-oriented manner, print(, all=true) displays all contents including hidden structures: > # display all contents of the GDS file > print(genofile, all=true)
6 Variants 6 File: /tmp/rtmpbo98st/rinst46302deddfc3/seqarray/extdata/ceu_exon.gds [ ] -- description [ ] * -- vcf.header [ data.frame ] * -- id { VStr8 1 } -- value { VStr8 1 } -- sample.id { VStr8 90 ZIP(28.33%) } -- variant.id { Int ZIP(35.39%) } -- position { Int ZIP(86.11%) } -- chromosome { VStr ZIP(2.14%) } -- allele { VStr ZIP(16.86%) } -- genotype [ ] * -- data { Bit2 2x90x1348 ZIP(28.36%) } { Int ZIP(0.56%) } * -- extra.index { Int32 3x0 ZIP } * -- extra { Int16 0 ZIP } -- phase [ ] -- data { Bit1 90x1348 ZIP(0.24%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Bit1 0 ZIP } -- annotation [ ] -- id { VStr ZIP(40.89%) } -- qual { Float ZIP(0.57%) } -- filter { Int32,factor 1348 ZIP(0.56%) } * -- info [ ] -- AA { VStr ZIP(23.55%) } * { Int ZIP(0.56%) } * -- AC { Int ZIP(26.89%) } * -- AN { Int ZIP(20.29%) } * -- DP { Int ZIP(62.24%) } * -- HM2 { Bit ZIP(106.51%) } * -- HM3 { Bit ZIP(106.51%) } * -- OR { VStr ZIP(12.92%) } * -- GP { VStr ZIP(34.24%) } * -- BN { Int ZIP(21.31%) } * -- format [ ] -- DP [ ] * -- data { Int32 90x1348 ZIP(33.83%) } { Int ZIP(0.56%) } * > # close the GDS file > seqclose(genofile) The output lists all variables stored in the GDS file. At the first level, it stores variables sample.id, variant.id, etc. The additional information are displayed in the square brackets indicating data type, size, compressed or not compression ratio, where Bit2 indicates that each byte encodes up to
7 Variants 7 four alleles since one byte consists of eight bits, and VStr8 indicates variable-length character. The annotations are stored in the directory annotation, which includes the fields of ID, QUAL, FILTER, INFO and FORMAT corresponding to the original VCF file(s). All of the functions in SeqArray require a minimum set of variables in the annotation data: ˆ sample.id, a unique identifier for each sample. ˆ variant.id, a unique identifier for each variant. ˆ position, integer, the base position of each variant on the chromosome, and 0 or NA for unknown position. ˆ chromosome, character, the chromosome code, e.g., 1-22 for autosomes, X, Y, XY (the pseudoautosomal region), M (the mitochondrial probes), and (a blank string) for probes with unknown chromosome. ˆ allele, character, reference and alternative alleles using comma as a separator. ˆ genotype, a folder, data, a 3-dimensional array for genotypic data, the first dimension refers to the number of ploidy, the second is sample and the third is the index for the variable data, and the is used to indicate the index. It should be equal-size as variant.id, which is used to specify the data size of each variant. extra.index, an index (3-by- ) matrix for triploid call (look like 0/0/1 in the VCF file). E.g., for 0/0/1, the first two alleles are stored in data, and the last allele is saved in the variable extra. For each column of extra.index, the first value is the index of sample (starting from 1), the second value is the index of variant (starting from 1), and the last value is how many alleles remain (usually it is 1 since the first two alleles are stored in data) that indicates how many alleles stored in extra contiguously. extra, one-dimensional array, the additional data for triploid call, each allele block corresponds to each column in extra.index. The optional variables include phase (phasing information) and annotation. includes: The variable phase ˆ data, a matrix or 3-dimensional array for phasing information. 0 for unphased status and 1 for phased status. If it is a matrix, the first dimension is sample and the second is variant, corresponding to diploid genotypic data. If it is a 3-dimensional array, the first dimension refers to the number of ploidy minus one. More details about / and in a VCF file can be founded: genomes.org/wiki/Analysis/VariantCallFormat/vcf-variant-call-format-version-41. ˆ extra.index, an index (3-by- ) matrix for triploid call (look like 0/0/1 in the VCF file). E.g., for 0/0/1, the first separator ( / here) is stored in data, and the last separator is saved in the variable extra. For each column of extra.index, the first value is the index of sample (starting from 1), the second value is the index of variant (starting from 1), and the last value is how many separators remain (usually it is 1 since the first separator is stored in data) that indicates how many separator stored in extra contiguously. ˆ extra, one-dimensional array, the additional data of separator indicator for triploid call, each separator block corresponds to each column in extra.index. The variable annotation includes: ˆ id, ID semi-colon separated list of unique identifiers where available. If this is a dbsnp variant
8 Variants 8 it is encouraged to use the rs number(s). No identifier should be present in more than one data record. If there is no identifier available, then a blank string is used. ˆ qual, phred-scaled quality score for the assertion made in ALT. ˆ filter, PASS if this position has passed all filters, i.e. a call is made at this position. ˆ info, additional information for each variant, according to the INFO field in a VCF file, VARIABLE_NAME, variable. If it is fixed-length, missing value indicates that there is no entry for that variant in the VCF optional if VARIABLE_NAME is variable-length, one-dimensional array. The is used to indicate the index data. It should be equal-size as variant.id, which is used to specify the data size of each variant. OTHER_VARIABLES,... ˆ format, additional information for each variant and sample, according to the FORMAT field in a VCF file, VARIABLE_NAME, a folder, * data, a n samp -by- matrix. one-dimensional array, the index data for the variable data, and the is used to indicate the index data. It should be equal-size as variant.id, which is used to specify the data size of each variant. OTHER_VARIABLES, Format conversion from VCF files The SeqArray package provides a function seqvcf2gds to reformat a VCF file, and it allows merging multiple VCF files during format conversion. The genotypic and annotation data are stored in a compressed manner. > # the VCF file, using the example in the SeqArray package > vcf.fn <- seqexamplefilename("vcf") > # or vcf.fn <- "C:/YourFolder/Your_VCF_File.vcf" > vcf.fn [1] "/tmp/rtmpbo98st/rinst46302deddfc3/seqarray/extdata/ceu_exon.vcf.gz" > # parse the header > seqvcf.header(vcf.fn) $fileformat [1] "VCFv4.0" $info ID Number Type Description 1 AA. String Ancestral Allele 2 AC 1 Integer total number of alternate alleles in called genotypes 3 AN 1 Integer total number of alleles in called genotypes 4 DP 1 Integer Total Depth 5 HM2 0 Flag HapMap2 membership
9 Variants 9 6 HM3 0 Flag HapMap3 membership 7 OR 1 String Previous rs number 8 GP 1 String GRCh37 position(s) 9 BN 1 Integer First dbsnp build # $filter NULL $format ID Number Type Description 1 GT 1 String Genotype 2 DP. Integer Read Depth from MOSAIK BAM $alt NULL $contig NULL $assembly NULL $header id value 13 reference human_b36_both.fasta $num.ploidy [1] 2 attr(,"class") [1] "SeqVCFHeaderClass" The columns Number, Type and Description are defined by the 1000 Genomes Project: Briefly, the Number entry is an Integer that describes the number of values that can be included with the INFO field. For example, if the INFO field contains a single number, then this value should be 1; if the INFO field describes a pair of numbers, then this value should be 2 and so on. If the field has one value per alternate allele then this value should be A ; if the field has one value for each possible genotype then this value should be G. If the number of possible values varies, is unknown, or is unbounded, then this value should be.. The Flag type indicates that the INFO field does not contain a Value entry, and hence the Number should be 0 in this case. Possible Types for FORMAT fields are: Integer, Float, Character, and String (this field is otherwise defined precisely as the INFO field). > # convert, save in "tmp.gds" > seqvcf2gds(vcf.fn, "tmp.gds")
10 Variants 10 The Variant Call Format (VCF) header: file format: VCFv4.0 the number of sets of chromosomes (ploidy): 2 the number of samples: 90 Parsing "/tmp/rtmpbo98st/rinst46302deddfc3/seqarray/extdata/ceu_exon.vcf.gz"... genotype/data { Bit2 2x90x1348 ZIP } Done. Optimize the access efficiency... Clean up the fragments of GDS file: open the file "tmp.gds" (size: ). # of fragments in total: 92. save it to "tmp.gds.tmp". rename "tmp.gds.tmp" (size: ). # of fragments in total: 67. > seqsummary("tmp.gds") File: /tmp/rtmpbo98st/rbuild dd0/seqarray/vignettes/tmp.gds Sequence Variant Format: v1.0 The number of samples: 90 The number of variants: 1348 The chromosomes: <NA> The number of alleles per site: Annotation, information variables: AA,., String, Ancestral Allele AC, 1, Integer, total number of alternate alleles in called genotypes AN, 1, Integer, total number of alleles in called genotypes DP, 1, Integer, Total Depth HM2, 0, Flag, HapMap2 membership HM3, 0, Flag, HapMap3 membership OR, 1, String, Previous rs number GP, 1, String, GRCh37 position(s) BN, 1, Integer, First dbsnp build # Annotation, format variables: DP,., Integer, Read Depth from MOSAIK BAM
11 Variants Export to a VCF File The SeqArray package provides a function seqgds2vcf to export data to a VCF file. The arguments info.var and fmt.var in seqgds2vcf allow users to specify the variables listed in the INFO and FORMAT fields of VCF format, or remove the INFO and FORMAT information. seqsetfilter can be used to define a subset of data for the export. > # the file of GDS > gds.fn <- seqexamplefilename("gds") > # or gds.fn <- "C:/YourFolder/Your_GDS_File.gds" > > # open a GDS file > genofile <- seqopen(gds.fn) > # convert > seqgds2vcf(genofile, "tmp.vcf.gz") Output: tmp.vcf.gz The INFO field: AA, AC, AN, DP, HM2, HM3, OR, GP, BN The FORMAT field: DP Done. > # read > z <- readlines("tmp.vcf.gz", n=20) > for (i in z) cat(substr(i, 1, 76), "...\n", sep="") ##fileformat=vcfv ##filedate= ##source=seqarray_rpackage_v ##INFO=<ID=AA,Number=.,Type=String,Description="Ancestral Allele">... ##INFO=<ID=AC,Number=1,Type=Integer,Description="total number of alternate a... ##INFO=<ID=AN,Number=1,Type=Integer,Description="total number of alleles in... ##INFO=<ID=DP,Number=1,Type=Integer,Description="Total Depth">... ##INFO=<ID=HM2,Number=0,Type=Flag,Description="HapMap2 membership">... ##INFO=<ID=HM3,Number=0,Type=Flag,Description="HapMap3 membership">... ##INFO=<ID=OR,Number=1,Type=String,Description="Previous rs number">... ##INFO=<ID=GP,Number=1,Type=String,Description="GRCh37 position(s)">... ##INFO=<ID=BN,Number=1,Type=Integer,Description="First dbsnp build #">... ##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">... ##FORMAT=<ID=DP,Number=.,Type=Integer,Description="Read Depth from MOSAIK BA... ##reference=human_b36_both.fasta... #CHROM POS ID REF ALT QUAL FILTER INFO rs T C. PASS AA=T;AC= rs G A. PASS AA=G;AC= rs G A. PASS AA=A;AC=6;A rs T C. PASS AA=C;AC=128
12 Variants 12 > # output BN,GP,AA,HM2 in INFO (the variables are in this order), no FORMAT > seqgds2vcf(genofile, "tmp2.vcf.gz", info.var=c("bn","gp","aa","hm2"), fmt.var=character Output: tmp2.vcf.gz The INFO field: BN, GP, AA, HM2 The FORMAT field: Done. > # read > z <- readlines("tmp2.vcf.gz", n=15) > for (i in z) cat(substr(i, 1, 56), "...\n", sep="") ##fileformat=vcfv ##filedate= ##source=seqarray_rpackage_v ##INFO=<ID=BN,Number=1,Type=Integer,Description="First d... ##INFO=<ID=GP,Number=1,Type=String,Description="GRCh37 p... ##INFO=<ID=AA,Number=.,Type=String,Description="Ancestra... ##INFO=<ID=HM2,Number=0,Type=Flag,Description="HapMap2 m... ##FORMAT=<ID=GT,Number=1,Type=String,Description="Genoty... ##reference=human_b36_both.fasta... #CHROM POS ID REF ALT QUAL FILTER INFO rs T C. PASS BN=132;GP rs G A. PASS BN=132;GP rs G A. PASS BN=88;GP=1: rs T C. PASS BN=100;GP= rs G C. PASS BN=100;GP=1 > # close the GDS file > seqclose(genofile) Users can use diff, a command line tool in Unix-like systems, to compare files line by line, in order to confirm data consistency. # assuming the original VCF file is old.vcf.gz, # call "seqvcf2gds" for the import and "seqgds2vcf" for the export to create a new VCF fi $ diff <(zcat old.vcf.gz) <(zcat new.vcf.gz) # OR $ diff <(gunzip -c old.vcf.gz) <(gunzip -c new.vcf.gz) 1a2,3 > ##filedate= > ##source=seqarray_rpackage_v1.0 # LOOK GOOD! There are only two lines different, and both are in the header. > # delete temporary files > unlink(c("tmp.vcf.gz", "tmp1.vcf.gz", "tmp2.vcf.gz"))
13 Variants Modification The SeqArray package provides a function seqdelete to remove data annotations in the INFO and FORMAT fields. It is suggested to use cleanup.gds in the gdsfmt package after calling seqdelete to reduce the file size. For example, > # the file of VCF > vcf.fn <- seqexamplefilename("vcf") > # or vcf.fn <- "C:/YourFolder/Your_VCF_File.vcf" > > # convert > seqvcf2gds(vcf.fn, "tmp.gds", verbose=false) > # make sure that open with "readonly=false" > genofile <- seqopen("tmp.gds", readonly=false) > # display the original structure > genofile Object of class "SeqVarGDSClass" File: /tmp/rtmpbo98st/rbuild dd0/seqarray/vignettes/tmp.gds [ ] * -- description [ ] * -- sample.id { VStr8 90 ZIP(28.77%) } -- variant.id { Int ZIP(35.39%) } -- position { Int ZIP(86.11%) } -- chromosome { VStr ZIP(2.22%) } -- allele { VStr ZIP(16.90%) } -- genotype [ ] * -- data { Bit2 2x90x1348 ZIP(28.36%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Int16 0 ZIP } -- phase [ ] -- data { Bit1 90x1348 ZIP(0.24%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Bit1 0 ZIP } -- annotation [ ] -- id { VStr ZIP(40.89%) } -- qual { Float ZIP(0.57%) } -- filter { Int32,factor 1348 ZIP(0.56%) } * -- info [ ] -- AA { VStr ZIP(23.60%) } * -- AC { Int ZIP(26.89%) } * -- AN { Int ZIP(20.29%) } * -- DP { Int ZIP(62.24%) } * -- HM2 { Bit ZIP(106.51%) } * -- HM3 { Bit ZIP(106.51%) } * -- OR { VStr ZIP(12.90%) } *
14 Variants GP { VStr ZIP(34.26%) } * -- BN { Int ZIP(21.31%) } * -- format [ ] -- DP [ ] * -- data { Int32 90x1348 ZIP(33.83%) } > # delete "HM2", "HM3", "AA", "OR" and "DP" > seqdelete(genofile, info.varname=c("hm2", "HM3", "AA", "OR"), format.varname="dp") > # display > genofile Object of class "SeqVarGDSClass" File: /tmp/rtmpbo98st/rbuild dd0/seqarray/vignettes/tmp.gds [ ] * -- description [ ] * -- sample.id { VStr8 90 ZIP(28.77%) } -- variant.id { Int ZIP(35.39%) } -- position { Int ZIP(86.11%) } -- chromosome { VStr ZIP(2.22%) } -- allele { VStr ZIP(16.90%) } -- genotype [ ] * -- data { Bit2 2x90x1348 ZIP(28.36%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Int16 0 ZIP } -- phase [ ] -- data { Bit1 90x1348 ZIP(0.24%) } -- extra.index { Int32 3x0 ZIP } * -- extra { Bit1 0 ZIP } -- annotation [ ] -- id { VStr ZIP(40.89%) } -- qual { Float ZIP(0.57%) } -- filter { Int32,factor 1348 ZIP(0.56%) } * -- info [ ] -- AA { VStr ZIP(23.60%) } * -- AC { Int ZIP(26.89%) } * -- AN { Int ZIP(20.29%) } * -- DP { Int ZIP(62.24%) } * -- HM2 { Bit ZIP(106.51%) } * -- HM3 { Bit ZIP(106.51%) } * -- OR { VStr ZIP(12.90%) } * -- GP { VStr ZIP(34.26%) } * -- BN { Int ZIP(21.31%) } * -- format [ ] -- DP [ ] * -- data { Int32 90x1348 ZIP(33.83%) }
15 Variants 15 > # close the GDS file > seqclose(genofile) > # clean up the fragments, reduce the file size > cleanup.gds("tmp.gds") Clean up the fragments of GDS file: open the file "tmp.gds" (size: ). # of fragments in total: 67. save it to "tmp.gds.tmp". rename "tmp.gds.tmp" (size: ). # of fragments in total: Data Processing 3.1 Get Data > # open a GDS file > gds.fn <- seqexamplefilename("gds") > genofile <- seqopen(gds.fn) It is suggested to use seqgetdata to take out data from the GDS file since this function can take care of variable-length data and multi-allelic genotypes, although users could also use read.gdsn in the gdsfmt package to read data. > # take out sample id > head(samp.id <- seqgetdata(genofile, "sample.id")) [1] "NA06984" "NA06985" "NA06986" "NA06989" "NA06994" "NA07000" > # take out variant id > head(variant.id <- seqgetdata(genofile, "variant.id")) [1] > # get "chromosome" > table(seqgetdata(genofile, "chromosome")) > # get "allele" > head(seqgetdata(genofile, "allele")) [1] "T,C" "G,A" "G,A" "T,C" "G,C" "C,T" > # get "annotation/info/gp" > head(seqgetdata(genofile, "annotation/info/gp")) [1] "1: " "1: " "1: " "1: " "1: " "1: "
16 Variants 16 Users can set a filter to sample and/or variant by seqsetfilter. For example, we consider a data subset consisting of three samples and four variants: > # set sample and variant filters > seqsetfilter(genofile, sample.id=samp.id[c(2,4,6)]) # of selected samples: 3 > set.seed(100) > seqsetfilter(genofile, variant.id=sample(variant.id, 4)) # of selected variants: 4 > # get "allele" > seqgetdata(genofile, "allele") [1] "T,A" "G,A" "G,C" "A,G" Get genotypic data, it is a 3-dimensional array with respect to allele, sample and variant. 0 refers to the reference allele (or the first allele in the variable allele), 1 for the second allele, and so on, while NA is missing allele. > # get genotypic data > seqgetdata(genofile, "genotype"),, 1 sample allele [,1] [,2] [,3] [1,] [2,] 0 0 0,, 2 sample allele [,1] [,2] [,3] [1,] [2,] 0 0 0,, 3 sample allele [,1] [,2] [,3] [1,] [2,] 0 0 0,, 4 sample
17 Variants 17 allele [,1] [,2] [,3] [1,] [2,] Now let us take a look at a variable-length dataset annotation/info/aa, which corresponds to the INFO column in the original VCF file. There are four variants, each variant has data with size ONE ($length), and data are saved in $data contiguously. $length could be ZERO indicating no data for that variant. > # get "annotation/info/aa", a variable-length dataset > seqgetdata(genofile, "annotation/info/aa") $length [1] $data [1] "T" "G" "G" "A" Another variable-length dataset is annotation/format/dp corresponding to the FORMAT column in the original VCF file. Again, $length refers to the size of each variant, and data are saved in $data contiguously with respect to the dimension variant. $length could be ZERO indicating no data for that variant. > # get "annotation/format/dp", a variable-length dataset > seqgetdata(genofile, "annotation/format/dp") $length [1] $data variant sample [,1] [,2] [,3] [,4] [1,] [2,] [3,] Apply Functions Over Array Margins SeqArray provides seqapply to apply a user-defined function over array margins, which is coded in C/C. It is suggested to use seqapply instead of apply.gdsn in the gdsfmt package, since this function can take care of variable-length data and multi-allelic genotypes. For example, read two variables genotype and annotation/id variant by variant: > # set sample and variant filters > set.seed(100) > seqsetfilter(genofile, sample.id=samp.id[c(2,4,6)], variant.id=sample(variant.id, 4))
18 Variants 18 # of selected samples: 3 # of selected variants: 4 > # read multiple variables variant by variant > seqapply(genofile, c(geno="genotype", id="annotation/id"), FUN=function(x) print(x), margin="by.variant", as.is="none") $geno sample allele [,1] [,2] [,3] [1,] [2,] $id [1] "rs " $geno sample allele [,1] [,2] [,3] [1,] [2,] $id [1] "rs12347" $geno sample allele [,1] [,2] [,3] [1,] [2,] $id [1] "rs " $geno sample allele [,1] [,2] [,3] [1,] [2,] $id [1] "rs " > # remove the sample and variant filters > seqsetfilter(genofile)
19 Variants 19 # of selected samples: 90 # of selected variants: 1348 > # get the numbers of alleles per variant > z <- seqapply(genofile, "allele", FUN=function(x) length(unlist(strsplit(x,","))), as.is="integer") > table(z) z Another example is to use the argument var.index in the function seqapply to include external information in the analysis, where the variable index in the user-defined FUN is an index of the specified dimension starting from 1 (e.g., variant). > HM3 <- seqgetdata(genofile, "annotation/info/hm3") > # Now HM3 is a global variable > # print out RS id if the frequency of reference allele is less than 0.5% and it is HM3 > seqapply(genofile, c(geno="genotype", id="annotation/id"), FUN = function(index, x) { p <- mean(x$geno == 0, na.rm=true) # the frequency of reference allele if ((p < 0.005) & HM3[index]) print(x$id) }, as.is="none", var.index="relative", margin="by.variant") [1] "rs " [1] "rs939777" [1] "rs698673" [1] "rs943542" [1] "rs " [1] "rs " [1] "rs322965" [1] "rs " 3.3 Apply Functions in Parallel Now, let us consider an example of calculating the frequency of reference allele, and this calculation can be done using seqapply and seqparallel. Let s try the uniprocessor implementation first. > # calculate the frequency of reference allele, > afreq <- seqapply(genofile, "genotype", FUN=function(x) mean(x==0, na.rm=true), as.is="double", margin="by.variant") > length(afreq) [1] 1348 > summary(afreq)
20 Variants 20 Min. 1st Qu. Median Mean 3rd Qu. Max A multi-process implementation: > # load the "parallel" package > library(parallel) > # Use option cl.core to choose an appropriate cluster size (or # of cores) > cl <- makecluster(getoption("cl.cores", 2)) > # run in parallel > afreq <- seqparallel(cl, genofile, FUN = function(gdsfile) { seqapply(gdsfile, "genotype", as.is="double", FUN=function(x) mean(x==0, na.rm=true)) }, split = "by.variant") > length(afreq) [1] 1348 > summary(afreq) Min. 1st Qu. Median Mean 3rd Qu. Max > # Since we created the cluster manually, we should stop it: > stopcluster(cl) > seqclose(genofile) 3.4 Integration with Other Packages in Bioconductor An R/Bioconductor package SeqVarTools is available on Bioconductor, which defines S4 classes and methods for other common operations and analyses on SeqArray datasets. 4 Examples In this section, a GDS file shipped with the package is used as an example: > # open a GDS file > gds.fn <- seqexamplefilename("gds") > genofile <- seqopen(gds.fn) 4.1 The performance of seqapply Let us try three approaches to export unphased genotypes: 1) the for loop in R; 2) vectorize the function in R; 3) the for loop in seqapply. The function seqapply has been highly optimized by blocking the
21 Variants 21 computations to exploit the high-speed memory instead of disk. The results of running times (listed as follows) indicate that seqapply works well and is comparable with vectorization in R. 1) the for loop in R: > system.time({ > geno <- seqgetdata(genofile, "genotype") > gc <- matrix("", nrow=dim(geno)[2], ncol=dim(geno)[3]) > for (i in 1:dim(geno)[3]) > { > for (j in 1:dim(geno)[2]) > gc[j,i] <- paste(geno[1,j,i], geno[2,j,i], sep="/") > } > gc[gc == "NA/NA"] <- NA > gc > }) user system elapsed <- the function takes 2.4 seconds > dim(gc) [1] > table(c(gc)) 0/0 0/1 1/0 1/1 <NA> ) Vectorize the function in R: > system.time({ > geno <- seqgetdata(genofile, "genotype") > gc <- matrix(paste(geno[1,,], geno[2,,], sep="/"), nrow=dim(geno)[2], ncol=dim(geno > gc[gc == "NA/NA"] <- NA > gc > }) user system elapsed <- the function takes 0.15 seconds 3) the for loop in seqapply: > system.time({ > gc <- seqapply(genofile, "genotype", function(x) { paste(x[1,], x[2,], sep="/") }, > margin="by.variant", as.is="list") > gc2 <- matrix(unlist(gc), ncol=length(gc)) > gc2[gc2 == "NA/NA"] <- NA > gc2 > })
22 Variants 22 user system elapsed <- the function takes 0.17 seconds 4.2 Missing Rates 1) Calculate the missing rate per variant: > m.variant <- local({ # get ploidy, the number of samples and variants z <- seqsummary(genofile, "genotype") # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- z$seldim # ploidy ploidy <- z$dim[1] # apply the function marginally m <- seqapply(genofile, "genotype", function(x) { sum(is.na(x)) }, margin="by.variant", as.is="integer") # output m / (ploidy * dm[1]) }) > length(m.variant) [1] 1348 > summary(m.variant) Min. 1st Qu. Median Mean 3rd Qu. Max ) Calculate the missing rate per sample: > m.sample <- local({ # get ploidy, the number of samples and variants z <- seqsummary(genofile, "genotype") # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- z$seldim # ploidy ploidy <- z$dim[1] # need a global variable (only available in the bracket of "local") n <- integer(dm[1]) # apply the function marginally # use "<<-" operator to find "n" in the parent environment seqapply(genofile, "genotype", function(x) { n <<- n colsums(is.na(x)) },
23 Variants 23 margin="by.variant", as.is="none") # output n / (ploidy * dm[2]) }) > length(m.sample) [1] 90 > summary(m.sample) Min. 1st Qu. Median Mean 3rd Qu. Max Multi-process Implementation > # load the "parallel" package > library(parallel) > # Use option cl.core to choose an appropriate cluster size (or # of cores) > cl <- makecluster(getoption("cl.cores", 2)) 1) Calculate the missing rate per variant: > # run in parallel > m.variant <- local({ # get ploidy, the number of samples and variants z <- seqsummary(genofile, "genotype") # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- z$seldim # ploidy ploidy <- z$dim[1] # run in parallel m <- seqparallel(cl, genofile, FUN = function(gdsfile) { # apply the function marginally seqapply(gdsfile, "genotype", function(x) { sum(is.na(x)) }, margin="by.variant", as.is="integer") }, split = "by.variant") # output m / (ploidy * dm[1]) }) > summary(m.variant) Min. 1st Qu. Median Mean 3rd Qu. Max ) Calculate the missing rate per sample:
24 Variants 24 > m.sample <- local({ # run in parallel m <- seqparallel(cl, genofile, FUN = function(gdsfile) { # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- seqsummary(gdsfile, "genotype")$seldim # need a global variable (only available in the bracket of "local") n <- integer(dm[1]) # apply the function marginally # use "<<-" operator to find "n" in the parent environment seqapply(gdsfile, "genotype", function(x) { n <<- n colsums(is.na(x)) }, margin="by.variant", as.is="none") # output n },.combine = "", # sum all variables "n" of different processes split = "by.variant") # get ploidy, the number of samples and variants z <- seqsummary(genofile, "genotype") # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- z$seldim # ploidy ploidy <- z$dim[1] # output m / (ploidy * dm[2]) }) > summary(m.sample) Min. 1st Qu. Median Mean 3rd Qu. Max > # Since we created the cluster manually, we should stop it: > stopcluster(cl) 4.3 Allele Frequency Calculate the frequency of reference allele: > # apply the function variant by variant > afreq <- seqapply(genofile, "genotype", function(x) { mean(x==0, na.rm=true) }, as.is="double", margin="by.variant")
25 Variants 25 > length(afreq) [1] 1348 > summary(afreq) Min. 1st Qu. Median Mean 3rd Qu. Max Multi-process Implementation > # load the "parallel" package > library(parallel) > # Use option cl.core to choose an appropriate cluster size (or # of cores) > cl <- makecluster(getoption("cl.cores", 2)) > # run in parallel > afreq <- seqparallel(cl, genofile, FUN = function(gdsfile) { seqapply(gdsfile, "genotype", as.is="double", FUN=function(x) mean(x==0, na.rm=true)) }, split = "by.variant") > length(afreq) [1] 1348 > summary(afreq) Min. 1st Qu. Median Mean 3rd Qu. Max > # Since we created the cluster manually, we should stop it: > stopcluster(cl) 4.4 Principal Component Analysis In the principal component analysis, we employ the dosage of reference alleles to avoid confusion of multiple alleles. The genetic correlation matrix is defined as M = [m j,j ]: m j,j = 1 L L l=1 (g j,l 2p l )(g j,l 2p l ) p l (1 p l ) where g j,l is a genotype of individual j at locus l ( {0, 1, 2}, # of reference allele), p l is the frequency of reference allele and there are L loci in total. > # genetic correlation matrix > genmat <- local({ # get the number of samples and variants # dm[1] -- # of selected samples, dm[2] -- # of selected variants
26 Variants 26 dm <- seqsummary(genofile, "genotype")$seldim # need a global variable (only available in the bracket of "local") s <- matrix(0.0, nrow=dm[1], ncol=dm[1]) # apply the function variant by variant # use "<<-" operator to find "s" in the parent environment seqapply(genofile, "genotype", function(x) { g <- (x==0) # indicator of reference allele p <- mean(g, na.rm=true) # allele frequency g2 <- colsums(g) - 2*p # genotypes adjusted by allele frequency m <- (g2 %o% g2) / (p*(1-p)) # genetic correlation matrix m[!is.finite(m)] <- 0 # correct missing values s <<- s m # output to the global variable "s" }, margin="by.variant", as.is="none") # output, scaled by the trace of matrix "s" over the number of samples s / (sum(diag(s)) / dm[1]) }) > # eigen-decomposition > eig <- eigen(genmat) > # eigenvalues > head(eig$value) [1] > # eigenvectors > plot(eig$vectors[,1], eig$vectors[,2], xlab="pc 1", ylab="pc 2")
27 Variants Multi-process Implementation > # load the "parallel" package > library(parallel) > # Use option cl.core to choose an appropriate cluster size (or # of cores) > cl <- makecluster(getoption("cl.cores", 2)) > genmat <- seqparallel(cl, genofile, FUN = function(gdsfile) { # get the number of samples and variants # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- seqsummary(gdsfile, "genotype")$seldim # need a global variable (only available in the bracket of "local") s <- matrix(0.0, nrow=dm[1], ncol=dm[1])
28 Variants 28 # apply the function variant by variant # use "<<-" operator to find "s" in the parent environment seqapply(gdsfile, "genotype", function(x) { g <- (x==0) # indicator of reference allele p <- mean(g, na.rm=true) # allele frequency g2 <- colsums(g) - 2*p # genotypes adjusted by allele frequency m <- (g2 %o% g2) / (p*(1-p)) # genetic correlation matrix m[!is.finite(m)] <- 0 # correct missing values s <<- s m # output to the global variable "s" }, margin="by.variant", as.is="none") # output s },.combine = "", # sum all variables "s" of different processes split = "by.variant") > # finally, scaled by the trace of matrix "s" over the number of samples > dm <- seqsummary(genofile, "genotype")$seldim > genmat <- genmat / (sum(diag(genmat)) / dm[1]) > # eigen-decomposition > eig <- eigen(genmat) > # eigenvalues > head(eig$value) [1] > # Since we created the cluster manually, we should stop it: > stopcluster(cl) 4.5 Individual Inbreeding Coefficient To calculate an individual inbreeding coefficient using SNP genotype data, I demonstrate how to use seqapply to calculate Visscher s estimator described in Yang et al. (2010) [5]. The estimator of individual inbreeding coefficient is defined as ˆθ = 1 L L l=1 g 2 l g l (1 2p l ) 2p 2 l 2p l (1 p l ) where g l is a SNP genotype at locus l ( {0, 1, 2}, # of reference allele), p l is the frequency of reference allele and there are L loci in total. > coeff <- local({ # get the number of samples and variants # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- seqsummary(genofile, "genotype")$seldim
29 Variants 29 # need global variables (only available in the bracket of "local") n <- integer(dm[1]) s <- double(dm[1]) # apply the function variant by variant # use "<<-" operator to find "n" and "s" in the parent environment seqapply(genofile, "genotype", function(x) { p <- mean(x==0, na.rm=true) # allele frequency g <- colsums(x==0) # genotypes: # of reference allele d <- (g*g - g*(1 2*p) 2*p*p) / (2*p*(1-p)) n <<- n is.finite(d) # output to the global variable "n" d[!is.finite(d)] <- 0 s <<- s d # output to the global variable "s" }, margin="by.variant", as.is="none") # output s / n }) > length(coeff) [1] 90 > summary(coeff) Min. 1st Qu. Median Mean 3rd Qu. Max e e e e e e Multi-process Implementation > # load the "parallel" package > library(parallel) > # Use option cl.core to choose an appropriate cluster size (or # of cores) > cl <- makecluster(getoption("cl.cores", 2)) > coeff <- seqparallel(cl, genofile, FUN = function(gdsfile) { # get the number of samples and variants # dm[1] -- # of selected samples, dm[2] -- # of selected variants dm <- seqsummary(gdsfile, "genotype")$seldim # need global variables (only available in the bracket) n <- integer(dm[1]) s <- double(dm[1]) # apply the function variant by variant # use "<<-" operator to find "n" and "s" in the parent environment
30 Variants 30 seqapply(gdsfile, "genotype", function(x) { p <- mean(x==0, na.rm=true) # allele frequency g <- colsums(x==0) # genotypes: # of reference allele d <- (g*g - g*(1 2*p) 2*p*p) / (2*p*(1-p)) n <<- n is.finite(d) # output to the global variable "n" d[!is.finite(d)] <- 0 s <<- s d # output to the global variable "s" }, margin="by.variant", as.is="none") # output list(s=s, n=n) }, # sum all variables "s" and "n" of different processes.combine = function(x1,x2) { list(s = x1$s x2$s, n = x1$n x2$n) }, split = "by.variant") > # finally, average! > coeff <- coeff$s / coeff$n > summary(coeff) Min. 1st Qu. Median Mean 3rd Qu. Max e e e e e e-02 > # Since we created the cluster manually, we should stop it: > stopcluster(cl) 4.6 Seamless R and C Integration The Rcpp package provides R functions as well as a C library which facilitate the integration of R and C. > library(rcpp) The user can dynamically define an inline C/C function in R. > cppfunction('double RefAlleleFreq(IntegerMatrix x) { int nrow = x.nrow(), ncol = x.ncol(); int cnt=0, zero_cnt=0, g; for (int i = 0; i < nrow; i) { for (int j = 0; j < ncol; j) { if ((g = x(i, j))!= NA_INTEGER) { cnt ; if (g == 0) zero_cnt ; }
31 Variants 31 } } return double(zero_cnt) / cnt; }') Now, RefAlleleFreq is a function in R. > RefAlleleFreq > afreq <- seqapply(genofile, "genotype", RefAlleleFreq, as.is="double", margin="by.variant") > summary(afreq) > # close the GDS file > seqclose(genofile) 5 The 1000 Genomes Project The 1000 Genomes Project datasets consist of 39,706,715 variants and 1,092 study samples. The original VCF data files for the 1000 Genomes Project were downloaded from ebi.ac.uk/vol1/ftp/phase1/analysis_results/integrated_call_sets/, and all compressed VCF files are 180G in total. The function seqvcf2gds in the R package SeqArray was used to convert and merge all VCF files, and all 39M variants and annotations were extracted from the VCF files without losing any information. A single GDS file of 140G was created, and all data variables were stored in a compressed manner. The detailed information are listed here: > seqsummary("g1000-release.gds") Sequence Variant Format: v1.0 The number of samples: 1092 The number of variants: The chromosomes: X <NA> The number of alleles per site: Annotation, information variables: LDAF, 1, Float, MLE Allele Frequency Accounting for LD AVGPOST, 1, Float, Average posterior probability from MaCH/Thunder RSQ, 1, Float, Genotype imputation quality from MaCH/Thunder
32 Variants 32 ERATE, 1, Float, Per-marker Mutation rate from MaCH/Thunder THETA, 1, Float, Per-marker Transition rate from MaCH/Thunder CIEND, 2, Integer, Confidence interval around END for imprecise variants CIPOS, 2, Integer, Confidence interval around POS for imprecise variants END, 1, Integer, End position of the variant described in this record HOMLEN,., Integer, Length of base pair identical micro-homology at event breakpoints HOMSEQ,., String, Sequence of base pair identical micro-homology at event breakpoints SVLEN, 1, Integer, Difference in length between REF and ALT alleles SVTYPE, 1, String, Type of structural variant AC,., Integer, Alternate Allele Count AN, 1, Integer, Total Allele Count AA, 1, String, Ancestral Allele, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/pilot_data/te AF, 1, Float, Global Allele Frequency based on AC/AN AMR_AF, 1, Float, Allele Frequency for samples from AMR based on AC/AN ASN_AF, 1, Float, Allele Frequency for samples from ASN based on AC/AN AFR_AF, 1, Float, Allele Frequency for samples from AFR based on AC/AN EUR_AF, 1, Float, Allele Frequency for samples from EUR based on AC/AN VT, 1, String, indicates what type of variant the line represents SNPSOURCE,., String, indicates if a snp was called when analysing the low coverage or Annotation, format variables: DS, 1, Float, Genotype dosage from MaCH/Thunder GL,., Float, Genotype Likelihoods [ ] -- description [ ] * -- sample.id [ VStr ZIP(26.02%) ] -- variant.id [ Int ZIP(34.58%) ] -- position [ Int ZIP(45.97%) ] -- chromosome [ VStr ZIP(0.10%) ] -- allele [ VStr ZIP(20.59%) ] -- genotype [ ] * -- data [ Bit2 2x1092x ZIP(5.61%) ] -- extra.index [ Int32 3x0 ZIP ] * -- extra [ Int16 0 ZIP ] -- phase [ ] -- data [ Bit1 1092x ZIP(0.11%) ] -- extra.index [ Int32 3x0 ZIP ] * -- extra [ Bit1 0 ZIP ] -- annotation [ ] -- id [ VStr ZIP(38.26%) ] -- qual [ Float ZIP(3.44%) ] -- filter [ Int32,factor ZIP(0.10%) ] * -- info [ ] -- LDAF [ Float ZIP(47.36%) ] * -- AVGPOST [ Float ZIP(28.40%) ] *
33 Variants RSQ [ Float ZIP(58.30%) ] * -- ERATE [ Float ZIP(13.38%) ] * -- THETA [ Float ZIP(20.83%) ] * -- CIEND [ Int32 2x ZIP(14.41%) ] * -- CIPOS [ Int32 2x ZIP(14.41%) ] * -- END [ Int ZIP(26.38%) ] * -- HOMLEN [ Int ZIP(20.11%) ] * -- HOMSEQ [ VStr ZIP(27.39%) ] * -- SVLEN [ Int ZIP(26.38%) ] * -- SVTYPE [ VStr ZIP(1.70%) ] * -- AC [ Int ZIP(29.30%) ] * -- AN [ Int ZIP(0.10%) ] * -- AA [ VStr ZIP(20.51%) ] * -- AF [ Float ZIP(22.77%) ] * -- AMR_AF [ Float ZIP(21.79%) ] * -- ASN_AF [ Float ZIP(22.93%) ] * -- AFR_AF [ Float ZIP(23.61%) ] * -- EUR_AF [ Float ZIP(23.07%) ] * -- VT [ VStr ZIP(1.31%) ] * -- SNPSOURCE [ VStr ZIP(0.35%) ] * -- format [ ] -- DS [ ] * -- data [ Float x ZIP(2.88%) ] -- GL [ ] * -- data [ Float x ZIP(27.61%) ] 6 Resources 1. CoreArray project: 2. gdsfmt R package: 3. SeqArray an R/Bioconductor package: SeqArray/ 7 Session Info > tolatex(sessioninfo()) ˆ R version ( ), x86_64-unknown-linux-gnu ˆ Locale: LC_CTYPE=en_US.UTF-8, LC_NUMERIC=C, LC_TIME=en_US.UTF-8, LC_COLLATE=C, LC_MONETARY=en_US.UTF-8, LC_MESSAGES=en_US.UTF-8, LC_PAPER=en_US.UTF-8, LC_NAME=C, LC_ADDRESS=C, LC_TELEPHONE=C, LC_MEASUREMENT=en_US.UTF-8, LC_IDENTIFICATION=C
34 Variants 34 ˆ Base packages: base, datasets, grdevices, graphics, methods, parallel, stats, utils ˆ Other packages: SeqArray 1.6.1, gdsfmt ˆ Loaded via a namespace (and not attached): AnnotationDbi , BBmisc 1.8, BSgenome , BatchJobs 1.5, Biobase , BiocGenerics , BiocParallel 1.0.0, BiocStyle 1.4.1, Biostrings , DBI 0.3.1, GenomeInfoDb 1.2.4, GenomicAlignments 1.2.1, GenomicFeatures , GenomicRanges , IRanges 2.0.1, RCurl , RSQLite 1.0.0, Rsamtools , S4Vectors 0.4.0, VariantAnnotation , XML , XVector 0.6.0, base64enc 0.1-2, biomart , bitops 1.0-6, brew 1.0-6, checkmate 1.5.1, codetools 0.2-9, digest 0.6.8, fail 1.2, foreach 1.4.2, iterators 1.0.7, rtracklayer , sendmailr 1.2-1, stats , stringr 0.6.2, tools 3.1.2, zlibbioc References Genomes Project Consortium, Abecasis, G. R., Auton, A., Brooks, L. D., DePristo, M. A., Durbin, R. M., Handsaker, R. E., Kang, H. M., Marth, G. T., and McVean, G. A. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 7422 (Nov 2012), Schadt, E. E., Linderman, M. D., Sorenson, J., Lee, L., and Nolan, G. P. Computational solutions to large-scale data management and analysis. Nat Rev Genet 11, 9 (Sep 2010), Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., Handsaker, R. E., Lunter, G., Marth, G. T., Sherry, S. T., McVean, G., Durbin, R., and 1000 Genomes Project Analysis Group. The variant call format and vcftools. Bioinformatics 27, 15 (Aug 2011), Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A High-performance Computing Toolset for Relatedness and Principal Component Analysis of SNP Data. Bioinformatics Oct Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 42(7): Epub 2010 Jun 20.
SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants
 SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants Xiuwen Zheng Department of Biostatistics University of Washington Seattle Jan 14, 2015 Contents 1 Overview
SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants Xiuwen Zheng Department of Biostatistics University of Washington Seattle Jan 14, 2015 Contents 1 Overview
SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants
 SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants Xiuwen Zheng Department of Biostatistics University of Washington Seattle Introduction Thousands of gigabyte
SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequencing Variants Xiuwen Zheng Department of Biostatistics University of Washington Seattle Introduction Thousands of gigabyte
SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequence Variants
 SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequence Variants 1 Dr. Xiuwen Zheng Department of Biostatistics University of Washington Seattle Introduction Thousands of gigabyte
SeqArray: an R/Bioconductor Package for Big Data Management of Genome-Wide Sequence Variants 1 Dr. Xiuwen Zheng Department of Biostatistics University of Washington Seattle Introduction Thousands of gigabyte
Creating a New Annotation Package using SQLForge
 Creating a New Annotation Package using SQLForge Marc Carlson, Herve Pages, Nianhua Li February 4, 2016 1 Introduction The AnnotationForge package provides a series of functions that can be used to build
Creating a New Annotation Package using SQLForge Marc Carlson, Herve Pages, Nianhua Li February 4, 2016 1 Introduction The AnnotationForge package provides a series of functions that can be used to build
A High-performance Computing Toolset for Big Data Analysis of Genome-Wide Variants
 A High-performance Computing Toolset for Big Data Analysis of Genome-Wide Variants Xiuwen Zheng Jan 02, 2014 Contents 1 Overview 1 2 Background 2 2.1 SNP Data in Genome-wide Association Studies................
A High-performance Computing Toolset for Big Data Analysis of Genome-Wide Variants Xiuwen Zheng Jan 02, 2014 Contents 1 Overview 1 2 Background 2 2.1 SNP Data in Genome-wide Association Studies................
GWAS Data Cleaning. GENEVA Coordinating Center Department of Biostatistics University of Washington. January 13, 2016.
 GWAS Data Cleaning GENEVA Coordinating Center Department of Biostatistics University of Washington January 13, 2016 Contents 1 Overview 2 2 Preparing Data 3 2.1 Data formats used in GWASTools............................
GWAS Data Cleaning GENEVA Coordinating Center Department of Biostatistics University of Washington January 13, 2016 Contents 1 Overview 2 2 Preparing Data 3 2.1 Data formats used in GWASTools............................
The Variant Call Format (VCF) Version 4.2 Specification
 The Variant Call Format (VCF) Version 4.2 Specification 26 Jan 2015 The master version of this document can be found at https://github.com/samtools/hts-specs. This printing is version cbd60fe from that
The Variant Call Format (VCF) Version 4.2 Specification 26 Jan 2015 The master version of this document can be found at https://github.com/samtools/hts-specs. This printing is version cbd60fe from that
Estimating batch effect in Microarray data with Principal Variance Component Analysis(PVCA) method
 Estimating batch effect in Microarray data with Principal Variance Component Analysis(PVCA) method Pierre R. Bushel, Jianying Li April 16, 2015 Contents 1 Introduction 1 2 Installation 2 3 An example run
Estimating batch effect in Microarray data with Principal Variance Component Analysis(PVCA) method Pierre R. Bushel, Jianying Li April 16, 2015 Contents 1 Introduction 1 2 Installation 2 3 An example run
HowTo: Querying online Data
 HowTo: Querying online Data Jeff Gentry and Robert Gentleman May 3, 2016 1 Overview This article demonstrates how you can make use of the tools that have been provided for on-line querying of data resources.
HowTo: Querying online Data Jeff Gentry and Robert Gentleman May 3, 2016 1 Overview This article demonstrates how you can make use of the tools that have been provided for on-line querying of data resources.
FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem
 FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem Elsa Bernard Laurent Jacob Julien Mairal Jean-Philippe Vert May 3, 2016 Abstract FlipFlop implements a fast method for de novo transcript
FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem Elsa Bernard Laurent Jacob Julien Mairal Jean-Philippe Vert May 3, 2016 Abstract FlipFlop implements a fast method for de novo transcript
SeattleSNPs Interactive Tutorial: Web Tools for Site Selection, Linkage Disequilibrium and Haplotype Analysis
 SeattleSNPs Interactive Tutorial: Web Tools for Site Selection, Linkage Disequilibrium and Haplotype Analysis Goal: This tutorial introduces several websites and tools useful for determining linkage disequilibrium
SeattleSNPs Interactive Tutorial: Web Tools for Site Selection, Linkage Disequilibrium and Haplotype Analysis Goal: This tutorial introduces several websites and tools useful for determining linkage disequilibrium
Text file One header line meta information lines One line : variant/position
 Software Calling: GATK SAMTOOLS mpileup Varscan SOAP VCF format Text file One header line meta information lines One line : variant/position ##fileformat=vcfv4.1! ##filedate=20090805! ##source=myimputationprogramv3.1!
Software Calling: GATK SAMTOOLS mpileup Varscan SOAP VCF format Text file One header line meta information lines One line : variant/position ##fileformat=vcfv4.1! ##filedate=20090805! ##source=myimputationprogramv3.1!
Package hoarder. June 30, 2015
 Type Package Title Information Retrieval for Genetic Datasets Version 0.1 Date 2015-06-29 Author [aut, cre], Anu Sironen [aut] Package hoarder June 30, 2015 Maintainer Depends
Type Package Title Information Retrieval for Genetic Datasets Version 0.1 Date 2015-06-29 Author [aut, cre], Anu Sironen [aut] Package hoarder June 30, 2015 Maintainer Depends
Combining Data from Different Genotyping Platforms. Gonçalo Abecasis Center for Statistical Genetics University of Michigan
 Combining Data from Different Genotyping Platforms Gonçalo Abecasis Center for Statistical Genetics University of Michigan The Challenge Detecting small effects requires very large sample sizes Combined
Combining Data from Different Genotyping Platforms Gonçalo Abecasis Center for Statistical Genetics University of Michigan The Challenge Detecting small effects requires very large sample sizes Combined
UKB_WCSGAX: UK Biobank 500K Samples Genotyping Data Generation by the Affymetrix Research Services Laboratory. April, 2015
 UKB_WCSGAX: UK Biobank 500K Samples Genotyping Data Generation by the Affymetrix Research Services Laboratory April, 2015 1 Contents Overview... 3 Rare Variants... 3 Observation... 3 Approach... 3 ApoE
UKB_WCSGAX: UK Biobank 500K Samples Genotyping Data Generation by the Affymetrix Research Services Laboratory April, 2015 1 Contents Overview... 3 Rare Variants... 3 Observation... 3 Approach... 3 ApoE
Towards Integrating the Detection of Genetic Variants into an In-Memory Database
 Towards Integrating the Detection of Genetic Variants into an 2nd International Workshop on Big Data in Bioinformatics and Healthcare Oct 27, 2014 Motivation Genome Data Analysis Process DNA Sample Base
Towards Integrating the Detection of Genetic Variants into an 2nd International Workshop on Big Data in Bioinformatics and Healthcare Oct 27, 2014 Motivation Genome Data Analysis Process DNA Sample Base
Version 5.0 Release Notes
 Version 5.0 Release Notes 2011 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074 (fax) www.genecodes.com
Version 5.0 Release Notes 2011 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074 (fax) www.genecodes.com
netresponse probabilistic tools for functional network analysis
 netresponse probabilistic tools for functional network analysis Leo Lahti 1,2, Olli-Pekka Huovilainen 1, António Gusmão 1 and Juuso Parkkinen 1 (1) Dpt. Information and Computer Science, Aalto University,
netresponse probabilistic tools for functional network analysis Leo Lahti 1,2, Olli-Pekka Huovilainen 1, António Gusmão 1 and Juuso Parkkinen 1 (1) Dpt. Information and Computer Science, Aalto University,
ASSIsT: An Automatic SNP ScorIng Tool for in and out-breeding species Reference Manual
 ASSIsT: An Automatic SNP ScorIng Tool for in and out-breeding species Reference Manual Di Guardo M, Micheletti D, Bianco L, Koehorst-van Putten HJJ, Longhi S, Costa F, Aranzana MJ, Velasco R, Arús P, Troggio
ASSIsT: An Automatic SNP ScorIng Tool for in and out-breeding species Reference Manual Di Guardo M, Micheletti D, Bianco L, Koehorst-van Putten HJJ, Longhi S, Costa F, Aranzana MJ, Velasco R, Arús P, Troggio
Tutorial for Windows and Macintosh. Preparing Your Data for NGS Alignment
 Tutorial for Windows and Macintosh Preparing Your Data for NGS Alignment 2015 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) 1.734.769.7249
Tutorial for Windows and Macintosh Preparing Your Data for NGS Alignment 2015 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) 1.734.769.7249
DNA Sequencing Data Compression. Michael Chung
 DNA Sequencing Data Compression Michael Chung Problem DNA sequencing per dollar is increasing faster than storage capacity per dollar. Stein (2010) Data 3 billion base pairs in human genome Genomes are
DNA Sequencing Data Compression Michael Chung Problem DNA sequencing per dollar is increasing faster than storage capacity per dollar. Stein (2010) Data 3 billion base pairs in human genome Genomes are
Practical Guideline for Whole Genome Sequencing
 Practical Guideline for Whole Genome Sequencing Disclosure Kwangsik Nho Assistant Professor Center for Neuroimaging Department of Radiology and Imaging Sciences Center for Computational Biology and Bioinformatics
Practical Guideline for Whole Genome Sequencing Disclosure Kwangsik Nho Assistant Professor Center for Neuroimaging Department of Radiology and Imaging Sciences Center for Computational Biology and Bioinformatics
Step by Step Guide to Importing Genetic Data into JMP Genomics
 Step by Step Guide to Importing Genetic Data into JMP Genomics Page 1 Introduction Data for genetic analyses can exist in a variety of formats. Before this data can be analyzed it must imported into one
Step by Step Guide to Importing Genetic Data into JMP Genomics Page 1 Introduction Data for genetic analyses can exist in a variety of formats. Before this data can be analyzed it must imported into one
R4RNA: A R package for RNA visualization and analysis
 R4RNA: A R package for RNA visualization and analysis Daniel Lai May 3, 2016 Contents 1 R4RNA 1 1.1 Reading Input............................................ 1 1.2 Basic Arc Diagram..........................................
R4RNA: A R package for RNA visualization and analysis Daniel Lai May 3, 2016 Contents 1 R4RNA 1 1.1 Reading Input............................................ 1 1.2 Basic Arc Diagram..........................................
org.rn.eg.db December 16, 2015 org.rn.egaccnum is an R object that contains mappings between Entrez Gene identifiers and GenBank accession numbers.
 org.rn.eg.db December 16, 2015 org.rn.egaccnum Map Entrez Gene identifiers to GenBank Accession Numbers org.rn.egaccnum is an R object that contains mappings between Entrez Gene identifiers and GenBank
org.rn.eg.db December 16, 2015 org.rn.egaccnum Map Entrez Gene identifiers to GenBank Accession Numbers org.rn.egaccnum is an R object that contains mappings between Entrez Gene identifiers and GenBank
MORPHEUS. http://biodev.cea.fr/morpheus/ Prediction of Transcription Factors Binding Sites based on Position Weight Matrix.
 MORPHEUS http://biodev.cea.fr/morpheus/ Prediction of Transcription Factors Binding Sites based on Position Weight Matrix. Reference: MORPHEUS, a Webtool for Transcripton Factor Binding Analysis Using
MORPHEUS http://biodev.cea.fr/morpheus/ Prediction of Transcription Factors Binding Sites based on Position Weight Matrix. Reference: MORPHEUS, a Webtool for Transcripton Factor Binding Analysis Using
SAP HANA Enabling Genome Analysis
 SAP HANA Enabling Genome Analysis Joanna L. Kelley, PhD Postdoctoral Scholar, Stanford University Enakshi Singh, MSc HANA Product Management, SAP Labs LLC Outline Use cases Genomics review Challenges in
SAP HANA Enabling Genome Analysis Joanna L. Kelley, PhD Postdoctoral Scholar, Stanford University Enakshi Singh, MSc HANA Product Management, SAP Labs LLC Outline Use cases Genomics review Challenges in
Using the generecommender Package
 Using the generecommender Package Greg Hather May 3, 2016 1 What is generecommender? generecommender is a package used to identify coexpressed genes in microarray data. In particular, the package ranks
Using the generecommender Package Greg Hather May 3, 2016 1 What is generecommender? generecommender is a package used to identify coexpressed genes in microarray data. In particular, the package ranks
Statistical Analysis for Genetic Epidemiology (S.A.G.E.) Version 6.2 Graphical User Interface (GUI) Manual
 Statistical Analysis for Genetic Epidemiology (S.A.G.E.) Version 6.2 Graphical User Interface (GUI) Manual Department of Epidemiology and Biostatistics Wolstein Research Building 2103 Cornell Rd Case Western
Statistical Analysis for Genetic Epidemiology (S.A.G.E.) Version 6.2 Graphical User Interface (GUI) Manual Department of Epidemiology and Biostatistics Wolstein Research Building 2103 Cornell Rd Case Western
HIGH DENSITY DATA STORAGE IN DNA USING AN EFFICIENT MESSAGE ENCODING SCHEME Rahul Vishwakarma 1 and Newsha Amiri 2
 HIGH DENSITY DATA STORAGE IN DNA USING AN EFFICIENT MESSAGE ENCODING SCHEME Rahul Vishwakarma 1 and Newsha Amiri 2 1 Tata Consultancy Services, India [email protected] 2 Bangalore University, India ABSTRACT
HIGH DENSITY DATA STORAGE IN DNA USING AN EFFICIENT MESSAGE ENCODING SCHEME Rahul Vishwakarma 1 and Newsha Amiri 2 1 Tata Consultancy Services, India [email protected] 2 Bangalore University, India ABSTRACT
EDASeq: Exploratory Data Analysis and Normalization for RNA-Seq
 EDASeq: Exploratory Data Analysis and Normalization for RNA-Seq Davide Risso Modified: May 22, 2012. Compiled: October 14, 2013 1 Introduction In this document, we show how to conduct Exploratory Data
EDASeq: Exploratory Data Analysis and Normalization for RNA-Seq Davide Risso Modified: May 22, 2012. Compiled: October 14, 2013 1 Introduction In this document, we show how to conduct Exploratory Data
Package GEOquery. August 18, 2015
 Type Package Package GEOquery August 18, 2015 Title Get data from NCBI Gene Expression Omnibus (GEO) Version 2.34.0 Date 2014-09-28 Author Maintainer BugReports
Type Package Package GEOquery August 18, 2015 Title Get data from NCBI Gene Expression Omnibus (GEO) Version 2.34.0 Date 2014-09-28 Author Maintainer BugReports
Delivering the power of the world s most successful genomics platform
 Delivering the power of the world s most successful genomics platform NextCODE Health is bringing the full power of the world s largest and most successful genomics platform to everyday clinical care NextCODE
Delivering the power of the world s most successful genomics platform NextCODE Health is bringing the full power of the world s largest and most successful genomics platform to everyday clinical care NextCODE
Globally, about 9.7% of cancers in men are prostate cancers, and the risk of developing the
 Chapter 5 Analysis of Prostate Cancer Association Study Data 5.1 Risk factors for Prostate Cancer Globally, about 9.7% of cancers in men are prostate cancers, and the risk of developing the disease has
Chapter 5 Analysis of Prostate Cancer Association Study Data 5.1 Risk factors for Prostate Cancer Globally, about 9.7% of cancers in men are prostate cancers, and the risk of developing the disease has
DNA Copy Number and Loss of Heterozygosity Analysis Algorithms
 DNA Copy Number and Loss of Heterozygosity Analysis Algorithms Detection of copy-number variants and chromosomal aberrations in GenomeStudio software. Introduction Illumina has developed several algorithms
DNA Copy Number and Loss of Heterozygosity Analysis Algorithms Detection of copy-number variants and chromosomal aberrations in GenomeStudio software. Introduction Illumina has developed several algorithms
Introduction. Overview of Bioconductor packages for short read analysis
 Overview of Bioconductor packages for short read analysis Introduction General introduction SRAdb Pseudo code (Shortread) Short overview of some packages Quality assessment Example sequencing data in Bioconductor
Overview of Bioconductor packages for short read analysis Introduction General introduction SRAdb Pseudo code (Shortread) Short overview of some packages Quality assessment Example sequencing data in Bioconductor
Single Nucleotide Polymorphisms (SNPs)
 Single Nucleotide Polymorphisms (SNPs) Additional Markers 13 core STR loci Obtain further information from additional markers: Y STRs Separating male samples Mitochondrial DNA Working with extremely degraded
Single Nucleotide Polymorphisms (SNPs) Additional Markers 13 core STR loci Obtain further information from additional markers: Y STRs Separating male samples Mitochondrial DNA Working with extremely degraded
Image Compression through DCT and Huffman Coding Technique
 International Journal of Current Engineering and Technology E-ISSN 2277 4106, P-ISSN 2347 5161 2015 INPRESSCO, All Rights Reserved Available at http://inpressco.com/category/ijcet Research Article Rahul
International Journal of Current Engineering and Technology E-ISSN 2277 4106, P-ISSN 2347 5161 2015 INPRESSCO, All Rights Reserved Available at http://inpressco.com/category/ijcet Research Article Rahul
Scalable Data Analysis in R. Lee E. Edlefsen Chief Scientist UserR! 2011
 Scalable Data Analysis in R Lee E. Edlefsen Chief Scientist UserR! 2011 1 Introduction Our ability to collect and store data has rapidly been outpacing our ability to analyze it We need scalable data analysis
Scalable Data Analysis in R Lee E. Edlefsen Chief Scientist UserR! 2011 1 Introduction Our ability to collect and store data has rapidly been outpacing our ability to analyze it We need scalable data analysis
SQL Server Array Library 2010-11 László Dobos, Alexander S. Szalay
 SQL Server Array Library 2010-11 László Dobos, Alexander S. Szalay The Johns Hopkins University, Department of Physics and Astronomy Eötvös University, Department of Physics of Complex Systems http://voservices.net/sqlarray,
SQL Server Array Library 2010-11 László Dobos, Alexander S. Szalay The Johns Hopkins University, Department of Physics and Astronomy Eötvös University, Department of Physics of Complex Systems http://voservices.net/sqlarray,
PaRFR : Parallel Random Forest Regression on Hadoop for Multivariate Quantitative Trait Loci Mapping. Version 1.0, Oct 2012
 PaRFR : Parallel Random Forest Regression on Hadoop for Multivariate Quantitative Trait Loci Mapping Version 1.0, Oct 2012 This document describes PaRFR, a Java package that implements a parallel random
PaRFR : Parallel Random Forest Regression on Hadoop for Multivariate Quantitative Trait Loci Mapping Version 1.0, Oct 2012 This document describes PaRFR, a Java package that implements a parallel random
09336863931 : provid.ir
 provid.ir 09336863931 : NET Architecture Core CSharp o Variable o Variable Scope o Type Inference o Namespaces o Preprocessor Directives Statements and Flow of Execution o If Statement o Switch Statement
provid.ir 09336863931 : NET Architecture Core CSharp o Variable o Variable Scope o Type Inference o Namespaces o Preprocessor Directives Statements and Flow of Execution o If Statement o Switch Statement
Hands-On Data Science with R Dealing with Big Data. [email protected]. 27th November 2014 DRAFT
 Hands-On Data Science with R Dealing with Big Data [email protected] 27th November 2014 Visit http://handsondatascience.com/ for more Chapters. In this module we explore how to load larger datasets
Hands-On Data Science with R Dealing with Big Data [email protected] 27th November 2014 Visit http://handsondatascience.com/ for more Chapters. In this module we explore how to load larger datasets
SQL Server An Overview
 SQL Server An Overview SQL Server Microsoft SQL Server is designed to work effectively in a number of environments: As a two-tier or multi-tier client/server database system As a desktop database system
SQL Server An Overview SQL Server Microsoft SQL Server is designed to work effectively in a number of environments: As a two-tier or multi-tier client/server database system As a desktop database system
Final Project Report
 CPSC545 by Introduction to Data Mining Prof. Martin Schultz & Prof. Mark Gerstein Student Name: Yu Kor Hugo Lam Student ID : 904907866 Due Date : May 7, 2007 Introduction Final Project Report Pseudogenes
CPSC545 by Introduction to Data Mining Prof. Martin Schultz & Prof. Mark Gerstein Student Name: Yu Kor Hugo Lam Student ID : 904907866 Due Date : May 7, 2007 Introduction Final Project Report Pseudogenes
Logistic Regression (1/24/13)
 STA63/CBB540: Statistical methods in computational biology Logistic Regression (/24/3) Lecturer: Barbara Engelhardt Scribe: Dinesh Manandhar Introduction Logistic regression is model for regression used
STA63/CBB540: Statistical methods in computational biology Logistic Regression (/24/3) Lecturer: Barbara Engelhardt Scribe: Dinesh Manandhar Introduction Logistic regression is model for regression used
GAIA: Genomic Analysis of Important Aberrations
 GAIA: Genomic Analysis of Important Aberrations Sandro Morganella Stefano Maria Pagnotta Michele Ceccarelli Contents 1 Overview 1 2 Installation 2 3 Package Dependencies 2 4 Vega Data Description 2 4.1
GAIA: Genomic Analysis of Important Aberrations Sandro Morganella Stefano Maria Pagnotta Michele Ceccarelli Contents 1 Overview 1 2 Installation 2 3 Package Dependencies 2 4 Vega Data Description 2 4.1
An example of bioinformatics application on plant breeding projects in Rijk Zwaan
 An example of bioinformatics application on plant breeding projects in Rijk Zwaan Xiangyu Rao 17-08-2012 Introduction of RZ Rijk Zwaan is active worldwide as a vegetable breeding company that focuses on
An example of bioinformatics application on plant breeding projects in Rijk Zwaan Xiangyu Rao 17-08-2012 Introduction of RZ Rijk Zwaan is active worldwide as a vegetable breeding company that focuses on
EndNote Beyond the Basics
 IOE Library Guide EndNote Beyond the Basics These notes assume that you know EndNote basics and are using it regularly. Additional tips and instruction is contained within the guides and FAQs available
IOE Library Guide EndNote Beyond the Basics These notes assume that you know EndNote basics and are using it regularly. Additional tips and instruction is contained within the guides and FAQs available
Database schema documentation for SNPdbe
 Database schema documentation for SNPdbe Changes 02/27/12: seqs_containingsnps.taxid removed dbsnp_snp.tax_id renamed to dbsnp_snp.taxid General information: Data in SNPdbe is organized on several levels.
Database schema documentation for SNPdbe Changes 02/27/12: seqs_containingsnps.taxid removed dbsnp_snp.tax_id renamed to dbsnp_snp.taxid General information: Data in SNPdbe is organized on several levels.
Copy Number Variation: available tools
 Copy Number Variation: available tools Jeroen F. J. Laros Leiden Genome Technology Center Department of Human Genetics Center for Human and Clinical Genetics Introduction A literature review of available
Copy Number Variation: available tools Jeroen F. J. Laros Leiden Genome Technology Center Department of Human Genetics Center for Human and Clinical Genetics Introduction A literature review of available
LifeScope Genomic Analysis Software 2.5
 USER GUIDE LifeScope Genomic Analysis Software 2.5 Graphical User Interface DATA ANALYSIS METHODS AND INTERPRETATION Publication Part Number 4471877 Rev. A Revision Date November 2011 For Research Use
USER GUIDE LifeScope Genomic Analysis Software 2.5 Graphical User Interface DATA ANALYSIS METHODS AND INTERPRETATION Publication Part Number 4471877 Rev. A Revision Date November 2011 For Research Use
BAPS: Bayesian Analysis of Population Structure
 BAPS: Bayesian Analysis of Population Structure Manual v. 6.0 NOTE: ANY INQUIRIES CONCERNING THE PROGRAM SHOULD BE SENT TO JUKKA CORANDER (first.last at helsinki.fi). http://www.helsinki.fi/bsg/software/baps/
BAPS: Bayesian Analysis of Population Structure Manual v. 6.0 NOTE: ANY INQUIRIES CONCERNING THE PROGRAM SHOULD BE SENT TO JUKKA CORANDER (first.last at helsinki.fi). http://www.helsinki.fi/bsg/software/baps/
Journal of Statistical Software
 JSS Journal of Statistical Software October 2006, Volume 16, Code Snippet 3. http://www.jstatsoft.org/ LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria between Single Nucleotide
JSS Journal of Statistical Software October 2006, Volume 16, Code Snippet 3. http://www.jstatsoft.org/ LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria between Single Nucleotide
KEGGgraph: a graph approach to KEGG PATHWAY in R and Bioconductor
 KEGGgraph: a graph approach to KEGG PATHWAY in R and Bioconductor Jitao David Zhang and Stefan Wiemann October 13, 2015 Abstract We demonstrate the capabilities of the KEGGgraph package, an interface between
KEGGgraph: a graph approach to KEGG PATHWAY in R and Bioconductor Jitao David Zhang and Stefan Wiemann October 13, 2015 Abstract We demonstrate the capabilities of the KEGGgraph package, an interface between
LRmix tutorial, version 4.1
 LRmix tutorial, version 4.1 Hinda Haned Netherlands Forensic Institute, The Hague, The Netherlands May 2013 Contents 1 What is LRmix? 1 2 Installation 1 2.1 Install the R software...........................
LRmix tutorial, version 4.1 Hinda Haned Netherlands Forensic Institute, The Hague, The Netherlands May 2013 Contents 1 What is LRmix? 1 2 Installation 1 2.1 Install the R software...........................
Fast Analytics on Big Data with H20
 Fast Analytics on Big Data with H20 0xdata.com, h2o.ai Tomas Nykodym, Petr Maj Team About H2O and 0xdata H2O is a platform for distributed in memory predictive analytics and machine learning Pure Java,
Fast Analytics on Big Data with H20 0xdata.com, h2o.ai Tomas Nykodym, Petr Maj Team About H2O and 0xdata H2O is a platform for distributed in memory predictive analytics and machine learning Pure Java,
Using Illumina BaseSpace Apps to Analyze RNA Sequencing Data
 Using Illumina BaseSpace Apps to Analyze RNA Sequencing Data The Illumina TopHat Alignment and Cufflinks Assembly and Differential Expression apps make RNA data analysis accessible to any user, regardless
Using Illumina BaseSpace Apps to Analyze RNA Sequencing Data The Illumina TopHat Alignment and Cufflinks Assembly and Differential Expression apps make RNA data analysis accessible to any user, regardless
Your Best Next Business Solution Big Data In R 24/3/2010
 Your Best Next Business Solution Big Data In R 24/3/2010 Big Data In R R Works on RAM Causing Scalability issues Maximum length of an object is 2^31-1 Some packages developed to help overcome this problem
Your Best Next Business Solution Big Data In R 24/3/2010 Big Data In R R Works on RAM Causing Scalability issues Maximum length of an object is 2^31-1 Some packages developed to help overcome this problem
Chapter 13. Disk Storage, Basic File Structures, and Hashing
 Chapter 13 Disk Storage, Basic File Structures, and Hashing Chapter Outline Disk Storage Devices Files of Records Operations on Files Unordered Files Ordered Files Hashed Files Dynamic and Extendible Hashing
Chapter 13 Disk Storage, Basic File Structures, and Hashing Chapter Outline Disk Storage Devices Files of Records Operations on Files Unordered Files Ordered Files Hashed Files Dynamic and Extendible Hashing
SeqScape Software Version 2.5 Comprehensive Analysis Solution for Resequencing Applications
 Product Bulletin Sequencing Software SeqScape Software Version 2.5 Comprehensive Analysis Solution for Resequencing Applications Comprehensive reference sequence handling Helps interpret the role of each
Product Bulletin Sequencing Software SeqScape Software Version 2.5 Comprehensive Analysis Solution for Resequencing Applications Comprehensive reference sequence handling Helps interpret the role of each
Biomedical Big Data and Precision Medicine
 Biomedical Big Data and Precision Medicine Jie Yang Department of Mathematics, Statistics, and Computer Science University of Illinois at Chicago October 8, 2015 1 Explosion of Biomedical Data 2 Types
Biomedical Big Data and Precision Medicine Jie Yang Department of Mathematics, Statistics, and Computer Science University of Illinois at Chicago October 8, 2015 1 Explosion of Biomedical Data 2 Types
Microsoft Office 2010: Access 2010, Excel 2010, Lync 2010 learning assets
 Microsoft Office 2010: Access 2010, Excel 2010, Lync 2010 learning assets Simply type the id# in the search mechanism of ACS Skills Online to access the learning assets outlined below. Titles Microsoft
Microsoft Office 2010: Access 2010, Excel 2010, Lync 2010 learning assets Simply type the id# in the search mechanism of ACS Skills Online to access the learning assets outlined below. Titles Microsoft
Cloud-Based Big Data Analytics in Bioinformatics
 Cloud-Based Big Data Analytics in Bioinformatics Presented By Cephas Mawere Harare Institute of Technology, Zimbabwe 1 Introduction 2 Big Data Analytics Big Data are a collection of data sets so large
Cloud-Based Big Data Analytics in Bioinformatics Presented By Cephas Mawere Harare Institute of Technology, Zimbabwe 1 Introduction 2 Big Data Analytics Big Data are a collection of data sets so large
Memory Allocation Technique for Segregated Free List Based on Genetic Algorithm
 Journal of Al-Nahrain University Vol.15 (2), June, 2012, pp.161-168 Science Memory Allocation Technique for Segregated Free List Based on Genetic Algorithm Manal F. Younis Computer Department, College
Journal of Al-Nahrain University Vol.15 (2), June, 2012, pp.161-168 Science Memory Allocation Technique for Segregated Free List Based on Genetic Algorithm Manal F. Younis Computer Department, College
Bioinformatics Resources at a Glance
 Bioinformatics Resources at a Glance A Note about FASTA Format There are MANY free bioinformatics tools available online. Bioinformaticists have developed a standard format for nucleotide and protein sequences
Bioinformatics Resources at a Glance A Note about FASTA Format There are MANY free bioinformatics tools available online. Bioinformaticists have developed a standard format for nucleotide and protein sequences
Accelerating variant calling
 Accelerating variant calling Mauricio Carneiro GSA Broad Institute Intel Genomic Sequencing Pipeline Workshop Mount Sinai 12/10/2013 This is the work of many Genome sequencing and analysis team Mark DePristo
Accelerating variant calling Mauricio Carneiro GSA Broad Institute Intel Genomic Sequencing Pipeline Workshop Mount Sinai 12/10/2013 This is the work of many Genome sequencing and analysis team Mark DePristo
RevoScaleR Speed and Scalability
 EXECUTIVE WHITE PAPER RevoScaleR Speed and Scalability By Lee Edlefsen Ph.D., Chief Scientist, Revolution Analytics Abstract RevoScaleR, the Big Data predictive analytics library included with Revolution
EXECUTIVE WHITE PAPER RevoScaleR Speed and Scalability By Lee Edlefsen Ph.D., Chief Scientist, Revolution Analytics Abstract RevoScaleR, the Big Data predictive analytics library included with Revolution
Copyright 2007 Ramez Elmasri and Shamkant B. Navathe. Slide 13-1
 Slide 13-1 Chapter 13 Disk Storage, Basic File Structures, and Hashing Chapter Outline Disk Storage Devices Files of Records Operations on Files Unordered Files Ordered Files Hashed Files Dynamic and Extendible
Slide 13-1 Chapter 13 Disk Storage, Basic File Structures, and Hashing Chapter Outline Disk Storage Devices Files of Records Operations on Files Unordered Files Ordered Files Hashed Files Dynamic and Extendible
CASSI: Genome-Wide Interaction Analysis Software
 CASSI: Genome-Wide Interaction Analysis Software 1 Contents 1 Introduction 3 2 Installation 3 3 Using CASSI 3 3.1 Input Files................................... 4 3.2 Options....................................
CASSI: Genome-Wide Interaction Analysis Software 1 Contents 1 Introduction 3 2 Installation 3 3 Using CASSI 3 3.1 Input Files................................... 4 3.2 Options....................................
Differential privacy in health care analytics and medical research An interactive tutorial
 Differential privacy in health care analytics and medical research An interactive tutorial Speaker: Moritz Hardt Theory Group, IBM Almaden February 21, 2012 Overview 1. Releasing medical data: What could
Differential privacy in health care analytics and medical research An interactive tutorial Speaker: Moritz Hardt Theory Group, IBM Almaden February 21, 2012 Overview 1. Releasing medical data: What could
Web Document Clustering
 Web Document Clustering Lab Project based on the MDL clustering suite http://www.cs.ccsu.edu/~markov/mdlclustering/ Zdravko Markov Computer Science Department Central Connecticut State University New Britain,
Web Document Clustering Lab Project based on the MDL clustering suite http://www.cs.ccsu.edu/~markov/mdlclustering/ Zdravko Markov Computer Science Department Central Connecticut State University New Britain,
Normalization of RNA-Seq
 Normalization of RNA-Seq Davide Risso Modified: April 27, 2012. Compiled: April 27, 2012 1 Retrieving the data Usually, an RNA-Seq data analysis from scratch starts with a set of FASTQ files (see e.g.
Normalization of RNA-Seq Davide Risso Modified: April 27, 2012. Compiled: April 27, 2012 1 Retrieving the data Usually, an RNA-Seq data analysis from scratch starts with a set of FASTQ files (see e.g.
Extensible Sequence (XSQ) File Format Specification 1.0.1
 Extensible Sequence (XSQ) File Format Specification 1.0.1 Table of Contents 1 INTRODUCTION... 1 2 FILE FORMAT... 1 3 GENERALIZATIONS AND EXTENDED SPECIFICATION... 11 4 FIGURES... 13 1 Introduction This
Extensible Sequence (XSQ) File Format Specification 1.0.1 Table of Contents 1 INTRODUCTION... 1 2 FILE FORMAT... 1 3 GENERALIZATIONS AND EXTENDED SPECIFICATION... 11 4 FIGURES... 13 1 Introduction This
IBM SPSS Direct Marketing 23
 IBM SPSS Direct Marketing 23 Note Before using this information and the product it supports, read the information in Notices on page 25. Product Information This edition applies to version 23, release
IBM SPSS Direct Marketing 23 Note Before using this information and the product it supports, read the information in Notices on page 25. Product Information This edition applies to version 23, release
G563 Quantitative Paleontology. SQL databases. An introduction. Department of Geological Sciences Indiana University. (c) 2012, P.
 SQL databases An introduction AMP: Apache, mysql, PHP This installations installs the Apache webserver, the PHP scripting language, and the mysql database on your computer: Apache: runs in the background
SQL databases An introduction AMP: Apache, mysql, PHP This installations installs the Apache webserver, the PHP scripting language, and the mysql database on your computer: Apache: runs in the background
Chapter 13 Disk Storage, Basic File Structures, and Hashing.
 Chapter 13 Disk Storage, Basic File Structures, and Hashing. Copyright 2004 Pearson Education, Inc. Chapter Outline Disk Storage Devices Files of Records Operations on Files Unordered Files Ordered Files
Chapter 13 Disk Storage, Basic File Structures, and Hashing. Copyright 2004 Pearson Education, Inc. Chapter Outline Disk Storage Devices Files of Records Operations on Files Unordered Files Ordered Files
Package illuminaio. R topics documented: December 15, 2015. Version 0.13.0 Title Parsing Illumina Microarray Output Files
 Version 0.13.0 Title Parsing Illumina Microarray Output Files Package illuminaio December 15, 2015 Tools for parsing Illumina's microarray output s, including IDAT. Suggests RUnit, BiocGenerics, IlluminaDataTestFiles
Version 0.13.0 Title Parsing Illumina Microarray Output Files Package illuminaio December 15, 2015 Tools for parsing Illumina's microarray output s, including IDAT. Suggests RUnit, BiocGenerics, IlluminaDataTestFiles
HETEROGENEOUS DATA INTEGRATION FOR CLINICAL DECISION SUPPORT SYSTEM. Aniket Bochare - [email protected]. CMSC 601 - Presentation
 HETEROGENEOUS DATA INTEGRATION FOR CLINICAL DECISION SUPPORT SYSTEM Aniket Bochare - [email protected] CMSC 601 - Presentation Date-04/25/2011 AGENDA Introduction and Background Framework Heterogeneous
HETEROGENEOUS DATA INTEGRATION FOR CLINICAL DECISION SUPPORT SYSTEM Aniket Bochare - [email protected] CMSC 601 - Presentation Date-04/25/2011 AGENDA Introduction and Background Framework Heterogeneous
Leading Genomics. Diagnostic. Discove. Collab. harma. Shanghai Cambridge, MA Reykjavik
 Leading Genomics Diagnostic harma Discove Collab Shanghai Cambridge, MA Reykjavik Global leadership for using the genome to create better medicine WuXi NextCODE provides a uniquely proven and integrated
Leading Genomics Diagnostic harma Discove Collab Shanghai Cambridge, MA Reykjavik Global leadership for using the genome to create better medicine WuXi NextCODE provides a uniquely proven and integrated
Package retrosheet. April 13, 2015
 Type Package Package retrosheet April 13, 2015 Title Import Professional Baseball Data from 'Retrosheet' Version 1.0.2 Date 2015-03-17 Maintainer Richard Scriven A collection of tools
Type Package Package retrosheet April 13, 2015 Title Import Professional Baseball Data from 'Retrosheet' Version 1.0.2 Date 2015-03-17 Maintainer Richard Scriven A collection of tools
Tutorial on gplink. http://pngu.mgh.harvard.edu/~purcell/plink/gplink.shtml. PLINK tutorial, December 2006; Shaun Purcell, [email protected].
 Tutorial on gplink http://pngu.mgh.harvard.edu/~purcell/plink/gplink.shtml Basic gplink analyses Data management Summary statistics Association analysis Population stratification IBD-based analysis gplink
Tutorial on gplink http://pngu.mgh.harvard.edu/~purcell/plink/gplink.shtml Basic gplink analyses Data management Summary statistics Association analysis Population stratification IBD-based analysis gplink
Optimization of sampling strata with the SamplingStrata package
 Optimization of sampling strata with the SamplingStrata package Package version 1.1 Giulio Barcaroli January 12, 2016 Abstract In stratified random sampling the problem of determining the optimal size
Optimization of sampling strata with the SamplingStrata package Package version 1.1 Giulio Barcaroli January 12, 2016 Abstract In stratified random sampling the problem of determining the optimal size
Data Analysis Tools. Tools for Summarizing Data
 Data Analysis Tools This section of the notes is meant to introduce you to many of the tools that are provided by Excel under the Tools/Data Analysis menu item. If your computer does not have that tool
Data Analysis Tools This section of the notes is meant to introduce you to many of the tools that are provided by Excel under the Tools/Data Analysis menu item. If your computer does not have that tool
Factors for success in big data science
 Factors for success in big data science Damjan Vukcevic Data Science Murdoch Childrens Research Institute 16 October 2014 Big Data Reading Group (Department of Mathematics & Statistics, University of Melbourne)
Factors for success in big data science Damjan Vukcevic Data Science Murdoch Childrens Research Institute 16 October 2014 Big Data Reading Group (Department of Mathematics & Statistics, University of Melbourne)
Cache Configuration Reference
 Sitecore CMS 6.2 Cache Configuration Reference Rev: 2009-11-20 Sitecore CMS 6.2 Cache Configuration Reference Tips and Techniques for Administrators and Developers Table of Contents Chapter 1 Introduction...
Sitecore CMS 6.2 Cache Configuration Reference Rev: 2009-11-20 Sitecore CMS 6.2 Cache Configuration Reference Tips and Techniques for Administrators and Developers Table of Contents Chapter 1 Introduction...
IBM SPSS Data Preparation 22
 IBM SPSS Data Preparation 22 Note Before using this information and the product it supports, read the information in Notices on page 33. Product Information This edition applies to version 22, release
IBM SPSS Data Preparation 22 Note Before using this information and the product it supports, read the information in Notices on page 33. Product Information This edition applies to version 22, release
GEMMA User Manual. Xiang Zhou. May 18, 2016
 GEMMA User Manual Xiang Zhou May 18, 2016 Contents 1 Introduction 4 1.1 What is GEMMA...................................... 4 1.2 How to Cite GEMMA................................... 4 1.3 Models............................................
GEMMA User Manual Xiang Zhou May 18, 2016 Contents 1 Introduction 4 1.1 What is GEMMA...................................... 4 1.2 How to Cite GEMMA................................... 4 1.3 Models............................................
Introduction to the data.table package in R
 Introduction to the data.table package in R Revised: September 18, 2015 (A later revision may be available on the homepage) Introduction This vignette is aimed at those who are already familiar with creating
Introduction to the data.table package in R Revised: September 18, 2015 (A later revision may be available on the homepage) Introduction This vignette is aimed at those who are already familiar with creating
Molecular typing of VTEC: from PFGE to NGS-based phylogeny
 Molecular typing of VTEC: from PFGE to NGS-based phylogeny Valeria Michelacci 10th Annual Workshop of the National Reference Laboratories for E. coli in the EU Rome, November 5 th 2015 Molecular typing
Molecular typing of VTEC: from PFGE to NGS-based phylogeny Valeria Michelacci 10th Annual Workshop of the National Reference Laboratories for E. coli in the EU Rome, November 5 th 2015 Molecular typing
Clustering through Decision Tree Construction in Geology
 Nonlinear Analysis: Modelling and Control, 2001, v. 6, No. 2, 29-41 Clustering through Decision Tree Construction in Geology Received: 22.10.2001 Accepted: 31.10.2001 A. Juozapavičius, V. Rapševičius Faculty
Nonlinear Analysis: Modelling and Control, 2001, v. 6, No. 2, 29-41 Clustering through Decision Tree Construction in Geology Received: 22.10.2001 Accepted: 31.10.2001 A. Juozapavičius, V. Rapševičius Faculty
Data Mining Applications in Manufacturing
 Data Mining Applications in Manufacturing Dr Jenny Harding Senior Lecturer Wolfson School of Mechanical & Manufacturing Engineering, Loughborough University Identification of Knowledge - Context Intelligent
Data Mining Applications in Manufacturing Dr Jenny Harding Senior Lecturer Wolfson School of Mechanical & Manufacturing Engineering, Loughborough University Identification of Knowledge - Context Intelligent
Data Tool Platform SQL Development Tools
 Data Tool Platform SQL Development Tools ekapner Contents Setting SQL Development Preferences...5 Execution Plan View Options Preferences...5 General Preferences...5 Label Decorations Preferences...6
Data Tool Platform SQL Development Tools ekapner Contents Setting SQL Development Preferences...5 Execution Plan View Options Preferences...5 General Preferences...5 Label Decorations Preferences...6
Big Data & Scripting Part II Streaming Algorithms
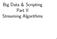 Big Data & Scripting Part II Streaming Algorithms 1, Counting Distinct Elements 2, 3, counting distinct elements problem formalization input: stream of elements o from some universe U e.g. ids from a set
Big Data & Scripting Part II Streaming Algorithms 1, Counting Distinct Elements 2, 3, counting distinct elements problem formalization input: stream of elements o from some universe U e.g. ids from a set
IRanges, GenomicRanges, and Biostrings
 IRanges, GenomicRanges, and Biostrings Bioconductor Infrastructure Packages for Sequence Analysis Patrick Aboyoun Fred Hutchinson Cancer Research Center 7-9 June, 2010 Outline Introduction Genomic Intervals
IRanges, GenomicRanges, and Biostrings Bioconductor Infrastructure Packages for Sequence Analysis Patrick Aboyoun Fred Hutchinson Cancer Research Center 7-9 June, 2010 Outline Introduction Genomic Intervals
