Chapter 11 Preparation of FFPE Tissue Slides for Solid Tumor FISH Analysis
|
|
|
- Hillary Richard
- 9 years ago
- Views:
Transcription
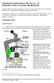
1 Chapter 11 Preparation of FFPE Tissue Slides for Solid Tumor FISH Analysis Sven Müller PhD, Steen H. Matthiesen PhD and Kirsten V. Nielsen MS Tumor tissue analysis by fluorescence in situ hybridization (FISH) is a well-established method for identification of genetic aberrations like gene copy number changes and translocations. The method has been used as a research tool for more than 20 years and due to the prognostic and predictive value of some FISH markers, e.g. TOP2A and HER2, the method has been implemented in the clinic. In clinical pathology, the quantitative nature of analysis results has made FISH an attractive supplement or alternative to IHC-based analysis. Although promising substitutes have been identified (1, 2), tissue preservation practice is still largely dominated by the classic method of fixation in neutral-buffered formalin, followed by paraffin embedding (FFPE). The FFPE tissue preservation method is fully compatible with FISH, but technically valid FISH results require that tissue preparation match the fixation history of the individual tissue sample. This article focuses on preparation of FFPE tissue slides for FISH analysis and how sub-optimal stains can be identified. Composition of a Solid Tumor FISH Probe Mixture The optimal composition of a FISH probe mixture for detection of changes in gene copy number in solid tumors includes two labeled probes. One probe directed towards the gene of interest and one probe directed towards a reference sequence for the chromosome on which the target gene is located. Traditionally, the reference probes are directed towards the non-coding centromere sequences. 8q24 CEN-8 Gene-directed probes are based on genomic clones typically COS or BAC clones and cover the genomic area where the coding sequences of the gene of interest are located. Labeling of the genomic clones, e.g. by nick translation, includes labeling of introns and gene flanking sequences. Thereby the repetitive sequences interspersed in the human genome are labeled as well. The resulting unspecific staining can be blocked by addition of unlabeled competitive sequences. The first consistently efficient blocker identified was the Cot-1 DNA fraction (3). It has, subsequently, been demonstrated that a mixture of unlabeled peptide nucleic acid (PNA) oligonucleotides directed towards the dominating Alu repeat sequences is comparable to or better than Cot-1 DNA fraction for blocking (4). Tissue Sample Preparation Specimens from biopsies, excisions or resections must be handled as soon as possible to preserve the tissue for FISH. Specimens should be preserved in 10% neutral-buffered formalin (NBF), preferably as 3-4 mm blocks fixed for hours followed by dehydration and embedment in paraffin. Sections should be cut into 4-6 µm, mounted on positively charged slides (e.g. SuperFrost Plus, Mentel-Gläser, Thermo Scientific) and adhered to the slide by baking at 60 C for approximately 1 hour. Fixation in formalin is suitable because the induced protein-protein and protein-nucleic acid cross-links preserve the tissue efficiently while retaining morphology relatively intact. However, the macromolecular network introduced by formalin significantly reduces the access of FISH probes to target DNA. Consequently, the initial steps in a FISH staining must address suitable breakdown of this network. FFPE Slide Preparation MYC CEN kb Figure 1. Example of a FISH probe mixture. A 291 kb long Texas Red-labeled probe directed towards MYC and a fluorescein-labeled probe towards CEN-8 reference sequences on the same chromosome. Deparaffinization The need for deparaffinization of FFPE slides for FISH analysis does not differ from standard preparation of slides for histological staining methods like H&E, special stains or IHC. Following mounting by baking, the slides are deparaffinized in xylene (or substitutes) and rehydrated in a series of ethanol. IHC Staining Methods, Fifth Edition 67
2 Pre-treatment The aim of pre-treatment is to ease the subsequent protease digestion by breaking the formalin-induced disulphide bonds (6). Correct pre-treatment, therefore, has a huge impact on final FISH staining quality. Inadequate pre-treatment results in increased autofluorescence level due to intact proteins and ultimately reduced FISH probe signal intensity, whereas excessive pre-treatment distorts tissue morphology. It is important to adjust pre-treatment methods to tissues that have been fixed in weaker fixatives than NBF, otherwise pre-treatment may induce significant harm to the nuclear membrane. Degraded nuclear membranes will show as a blurred membrane demarcation (Figure 3.D). Degradation of the nuclear membrane and areas close to the membrane may reduce the presence of the non-coding (centromere) reference target sequences as non-coding sequences have a preference for localization at the nuclear envelope (7). Some of the more intense pre-treatment methods combine incubation in acidic solution (e.g. HCl) with exposure to chaotropic agents (e.g. sodium thiocyanate). A pre-treatment which can be harsh for both morphology and tissue slide attachment. However, very efficient results can be obtained solely by incubation in 2-(N-morpholino) ethanesulfonic acid (MES) at C for 10 minutes. The MES-based pre-treatment is relatively tolerant to variations in NBF fixation times, but in order for this simple approach to bring consistent results, it is crucial that incubation is performed at no less than 95 C. Enzymatic digestion Proper protease digestion is by far the most decisive step in order to obtain technically valid FISH results. The breaking of peptide bindings by protease digestion directly affects signal quality as it eases access of the FISH probes to the genomic target DNA and reduces autofluorescence generated by intact proteins. Some protocols are based on enzymatic digestion by the very active serine protease, proteinase K, but gentler and still adequate digestion can be achieved by use of a carboxyl protease called pepsin. Enzymatic digestion time must be adapted to the tissue fixation time. For tissues that have been fixed in NBF for 6-72 hours, the enzymatic digestion time can be between 30 seconds and 12 minutes at 37 C using Dako Ready-to-Use Pepsin. The digestion can also be performed at room temperature (20-25 C), but will require 3-4 times longer incubation time. In general terms, longer fixation time requires longer enzymatic digestion time. Typically, 2-3 minutes at 37 C will be an optimal enzymatic digestion time for the majority of routine FFPE samples fixed in NFB for hours. However, present hospital routines, in accordance with which fixation time can vary significantly or even be unknown to the FISH laboratory, present a challenge for proper digestion. It is important that the FISH laboratory recognizes when a sample is unsuitable for analysis and accordingly reprocess the sample using a digestion intensity-adapted protocol. Identification of Sub-optimal Digestion To verify the digestion quality of a stained slide several evaluation parameters must be examined, the most important being inspection of: Autofluorescence level DAPI staining pattern Signal distribution Autofluorescence Heavily under-digested samples may be identified by inspection of the sample in the relevant double filter (e.g. Texas Red/FITC), see Figure 2.A. Under-digested samples are often characterized by apparent green autofluorescence in the cytosol as well as the extracellular matrix (Figure 2.A and C). 68 IHC Staining Methods, Fifth Edition
3 Figure 2. FFPE breast tissue stained with MYC/CEN-8 (Y5504, Dako). (a) Under-digested with excessive green cytosolic and extracellular autofluorescence background staining. No red gene signals (MYC) and some green reference signals (CEN-8). The arrow points to a green signal. (b) DAPI stain of A. (c) Slightly under-digested with green cytosolic and extracellular autofluorescence background staining (long arrow). Some red gene signals and green reference signals are seen (short arrow). (d) DAPI stain of C. (e) Acceptable digestion time. Short arrow: Green and red signals; long arrow: Low level of cytosolic and extracellular autofluorescence. (f) DAPI stain of E. IHC Staining Methods, Fifth Edition 69
4 When under-digestion is pronounced, probe hybridization is hampered by the high presence of proteins and peptide chains. This reduces signal intensity as a consequence of the higher autofluorescence and lower probe hybridization, thereby reducing the signal to noise ratio. The use of labeled PNA oligos or DNA as reference probes towards highly repetitive sequences (e.g. centromeric sequences) may result in the presence of green signals despite sub-optimal (under-) digestion of the sample (Figure 2.A). FISH stains with under-digestion of tissue can be recognized due to autofluorescence in the cytosol and extracellular matrix and potential lack or reduced presence and intensity of red signals from the gene of interest (single locus). In properly digested cells, the autofluorescence is mainly restricted to the nuclei, and the level does not disturb red and green signals (Figure 2.E). Visual inspection of autofluorescence level may help identify under-digested samples, but for over-digested samples the autofluorescence level does not differentiate properly-digested samples from over-digested samples. DAPI staining pattern DAPI (4,-6-diamidino-2-phenylindole) forms fluorescent complexes with double-stranded DNA and this ability makes it possible to use the DAPI staining pattern as a tool to evaluate digestion status. Insufficient enzymatic digestion impedes the DNA DAPI complex formation and makes the nuclei staining appear heterogeneous. This is seen when the staining pattern of an optimally digested sample (Figure 3.A) is compared to an under-digested sample (Figure 3.B). Over-digestion can also be seen in the DAPI staining pattern as doughnut formation or damaged nuclei membranes (Figure 3.C and Figure 3.D, respectively). Figure 3. Nuclei DAPI counter stains of FFPE breast tissue at different digestion times. (a) Optimal digestion time, homogenous staining (arrow). (b) Underdigested, heterogenous staining (arrow). (c) Over digested, some nuclei with doughnut formation (arrow). (d) Heavily over-digested, ghost nuclei (long arrow) and damaged nuclei membranes (short arrow). However, using the DAPI staining pattern for evaluation of underdigested samples is very coarse as many lightly fixed tissues appear homogenously stained despite insufficient digestion. A well digested tissue will often morphologically, depending of tissue type, result in a more spherical nucleus with a light swollen appearance compared to an under-digested tissue. 70 IHC Staining Methods, Fifth Edition
5 Over-digestion of a tissue section results in destroyed tissue, cell and nuclei morphology as well as loss of DNA. As binding of DAPI to double-stranded DNA results in an approximately 20-fold fluorescence enhancement (8), the reduced presence or absence of DNA is reflected in the DAPI staining pattern. Partly over-digested nuclei may be identified by the absence of DAPI staining in the center of the nucleus, giving rise to a doughnut-like appearance (Figure 3.C). In case of intense over-digestion, degradation of the nuclei becomes quite easy to spot with almost empty ghost nuclei and destroyed nuclear morphology (Figure 3.D). Caution should be taken not to conclude cases of under-digestion as over-digested based solely on a DAPI staining pattern. In underdigested samples, the DAPI molecule will only slowly enter the core of the nuclei (Figure 2.B and D) and this effect may result in lack of core staining and the doughnut-like appearance seen for overdigested samples. Signal distribution The digestion pattern as shown by DAPI staining (Figure 3.A-D) indicates that enzymatic digestion of material in the nucleus is heterogeneous. According to the DAPI staining pattern, the center of the nucleus is the most affected area. Loss of genomic DNA as a consequence of over-digestion compromises the technical validity of a FISH staining as the number of probe target sequences may be reduced. A study by Bolzer, et al. (7) showed that localization of genomic DNA in human nuclei correlates to gene density. Genepoor domains form a layer beneath the nuclear envelope and genedense domains are enriched in the nuclear interior. The centromerebased reference probes used in solid tumor FISH probe mixes are per definition targeted to gene-poor domains and should according to the observations by Bolzer, et al. have a preference for localization close to the nuclear envelope. This is supported by the signal distribution that can be seen when centromere sequence-based probes are used for interphase stains, see Figure 4. Figure 4. Staining with CEN-18 FITC-labeled PNA probe (green signals). Arrows illustrate the localization preference close to the membrane of gene-poor DNA domains. FISH probe target genes are often located in gene-dense domains enriched in the nuclear interior, thus a heterogeneous enzymatic degradation affecting the center of the nucleus may bias probe signals towards a reduced presence of gene targeted signals versus centromere probes closer to the envelope. A possible outcome of over-digestion is shown in Figure 5. IHC Staining Methods, Fifth Edition 71
6 relative humidity) as evaporation during hybridization can change the probe composition or even dry out the solution thereby compromising signal quality. Unspecific hybridization events are washed away at stringent conditions prior to mounting. Optimal temperature of the posthybridization wash depends mainly on the salt concentration in the buffer. High salt concentration decreases DNA backbone repulsion and thereby increases the DNA duplex melting temperature. Wash using a saline-sodium citrate (SSC) buffer with a detergent for 10 minutes at 65 C efficiently removes mismatch hybrids, but if this temperature is exceeded, it may result in a reduction of signal intensity or loss of the specific signals. Mounting and Visualization Figure 5. Over-digested (8-minute enzymatic digestion) FFPE breast tissue stained with gene probe (red), a centromere reference PNA probe (green) and DAPI (blue). The image shows doughnut formation and uneven DNA counter stain (blue). The arrow points to a nuclei with loss of red signal. The tissue specimen has a normal 1:1 gene to reference ratio at its optimal digestion time (3-minute enzymatic digestion data not shown). After mounting, the slides should be left in the dark for at least 15 minutes for DAPI to stain the nuclear material. Slides are inspected in a fluorescence microscope. It is important to use the recommended filter sets since use of sub-optimal filters significantly reduces signal intensity (see Chapter 12, Filters for FISH Imaging). Table 1. Possible Staining Patterns* Probe Hybridization and Stringency Wash Following pre-treatment and digestion, a two-step process is required to ensure efficient marking of the target sequences. First the genomic target DNA and the labeled probes (when double-stranded) must be denaturated by heating. Then the temperature is lowered to start a hybridization competition in which unlabeled repetitive sequences in the probe mix hybridize to complimentary sequences in the target DNA to block unspecific signals. Simultaneously, the labeled probe sequences hybridize to complimentary sequences in the target DNA and thereby mark the target sequences. The denaturation and hybridization events take place in the presence of formamide, and the optimal temperature profile depends mainly on the concentration of this chaotrope. In a 45% formamide solution, optimal results are obtained by denaturation at 82 C for 5 minutes followed by hybridization at 45 C over night (14-20 hours). The reaction should be covered and sealed and the humidity kept at saturation (100% Preparation Autofluorescence DAPI Pattern Possible Effects Underdigestion Overdigestion Optimal digestion excessive autofluorescence autofluorescence level does not differentiate properly digested samples from over-digested samples non-disturbing autofluorescence heterogeneous staining some doughnut nuclei doughnut nuclei ghost nuclei destroyed tissue morphology intact nuclei intact tissue morphology homogeneous DAPI staining *These effects may not be seen in all cells or in all slides. reduced target gene signal in some cells loss of target gene signal in some cells all signals will show reference signals, typically, near membrane target gene signals, typically, in interior 72 IHC Staining Methods, Fifth Edition
7 Conclusion Correct pre-treatment of FFPE slides is very important to obtain valid FISH staining results. The key factor is to adjust the enzymatic digestion to the fixation history of the tissue. Inspection of autofluorescence level, DAPI staining pattern and nuclear morphology are informative tools for an initial evaluation of sample digestion. The signal distribution pattern may add valuable information to assess, if digestion has been optimal. An optimally processed sample is characterized by intact nuclear membranes, non-disturbing autofluorescence and homogeneous DAPI staining. Reference signals are expected to be present near the nuclear membrane, whereas the gene targets are predominantly expected in the nuclear interior when arising from gene-dense domains. Finally, areas of normal cells may serve as a control for proper tissue treatment. In normal cells, the target/reference signal ratio should match the ratio which is expected in a normal diploid cell. A prerequisite for use of normal cell areas as tissue treatment control is that the impact of fixation is similar in the normal and the malignant areas. Acknowledgement We wish to thank Dr. Martina Schmidt, Scientist, and Dr. Oliver Stoss, Project Manager, Biomarker Analytics and Bioinformatics, Targos Molecular Pathology Gmbh, Kassel, Germany, for providing the FISH images used in Figure 2-3. References 1. Morales A R, Nassiri M, Kanhoush R, Vicek V, Nadji M. Experience with an automated microwave-assisted rapid tissue processing method. Validation of histologic quality and impact on the timeliness of diagnostic surgical pathology. Am J Clin Pathol 121: , Nassiri M, Ramos S, Zohourian H, Vinvek V, Morales A R, Nadji M. Preservation of biomolecules in breast cancer tissue by a formalin-free histology system. BMC Clinical Pathology 8:1, Landegent J E, Jansen in de Wal N, Dirks R W, Baas F and van der Ploeg M. Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet 77: , Nielsen K V, Müller S, Poulsen T S, Gabs S and Schønau A. Combined Use of PNA and DNA for Fluorescence In Situ Hybridization (FISH). In PEPTIDE NUCLEIC ACIDS Protocols and Applications. 2 nd Edition. Nielsen P E, eds. Horizon Bioscience, Nielsen K V, Müller S, Agerholm I, Poulsen T S, Matthiesen S H, Witton C J, Schønau A. PNA suppression method combined with fluorescence in situ hybridization (FISH) technique. In PRINS and PNA Technologies in Chromosomal Investigation. Frank Pellestor, eds. NOVA Publishers, Hopman A H N, Poddighe P, Moesker O and Ramaekers F C S. Interphase cytogenetics: an approach to the detection of genetic aberration in tumours. In Diagnostic molecular pathology: A practical approach. M J O and H C S, eds. IRL Press, NY, Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher M R, Cremer T. Three-Dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes. PLoS Biology 3, Barcellona M L, Cardiel G, Gratton E. Time-resolved fluorescence of DAPI in solution and bound to polydeoxynucleotides. Biochem Biophys Res Commun. 170: , IHC Staining Methods, Fifth Edition 73
HER2 FISH pharmdx. Assay Kit
 PATHOLOGY HER2 FISH pharmdx Assay Kit HER2 FISH pharmdx Clarity You Can Count On Reliable Results at a Glance The robust HER2 FISH pharmdx assay offers bright and distinct signals for easy and fast reading
PATHOLOGY HER2 FISH pharmdx Assay Kit HER2 FISH pharmdx Clarity You Can Count On Reliable Results at a Glance The robust HER2 FISH pharmdx assay offers bright and distinct signals for easy and fast reading
March 19, 2014. Dear Dr. Duvall, Dr. Hambrick, and Ms. Smith,
 Dr. Daniel Duvall, Medical Officer Center for Medicare, Hospital and Ambulatory Policy Group Centers for Medicare and Medicaid Services 7500 Security Boulevard Baltimore, Maryland 21244 Dr. Edith Hambrick,
Dr. Daniel Duvall, Medical Officer Center for Medicare, Hospital and Ambulatory Policy Group Centers for Medicare and Medicaid Services 7500 Security Boulevard Baltimore, Maryland 21244 Dr. Edith Hambrick,
CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA
 CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA Cytogenetics is the study of chromosomes and their structure, inheritance, and abnormalities. Chromosome abnormalities occur in approximately:
CHROMOSOMES Dr. Fern Tsien, Dept. of Genetics, LSUHSC, NO, LA Cytogenetics is the study of chromosomes and their structure, inheritance, and abnormalities. Chromosome abnormalities occur in approximately:
micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved
 microrna 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions of target mrnas Act as negative
microrna 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions of target mrnas Act as negative
INSTRUCTION Probemaker
 INSTRUCTION Probemaker Instructions for Duolink In Situ Probemaker PLUS (Art. no. 92009-0020) and Duolink In Situ Probemaker MINUS (Art. no. 92010-0020) Table of content 1. Introduction 4 2. Applications
INSTRUCTION Probemaker Instructions for Duolink In Situ Probemaker PLUS (Art. no. 92009-0020) and Duolink In Situ Probemaker MINUS (Art. no. 92010-0020) Table of content 1. Introduction 4 2. Applications
What is special about a stain, and why do we call some stains
 Technical Articles Evolution of Use of Special Stains Alton D. Floyd, PhD Independent Consultant Edwardsburg, MI, USA What is special about a stain, and why do we call some stains special? We all think
Technical Articles Evolution of Use of Special Stains Alton D. Floyd, PhD Independent Consultant Edwardsburg, MI, USA What is special about a stain, and why do we call some stains special? We all think
ab176915 StayBlue/AP Plus Stain Kit Instructions for Use An immunohistochemical chromogen substrate for staining tissue sections
 ab176915 StayBlue/AP Plus Stain Kit Instructions for Use An immunohistochemical chromogen substrate for staining tissue sections This product is for research use only and is not intended for diagnostic
ab176915 StayBlue/AP Plus Stain Kit Instructions for Use An immunohistochemical chromogen substrate for staining tissue sections This product is for research use only and is not intended for diagnostic
MLX BCG Buccal Cell Genomic DNA Extraction Kit. Performance Characteristics
 MLX BCG Buccal Cell Genomic DNA Extraction Kit Performance Characteristics Monolythix, Inc. 4720 Calle Carga Camarillo, CA 93012 Tel: (805) 484-8478 monolythix.com Page 2 of 9 MLX BCG Buccal Cell Genomic
MLX BCG Buccal Cell Genomic DNA Extraction Kit Performance Characteristics Monolythix, Inc. 4720 Calle Carga Camarillo, CA 93012 Tel: (805) 484-8478 monolythix.com Page 2 of 9 MLX BCG Buccal Cell Genomic
Technical Note. Roche Applied Science. No. LC 18/2004. Assay Formats for Use in Real-Time PCR
 Roche Applied Science Technical Note No. LC 18/2004 Purpose of this Note Assay Formats for Use in Real-Time PCR The LightCycler Instrument uses several detection channels to monitor the amplification of
Roche Applied Science Technical Note No. LC 18/2004 Purpose of this Note Assay Formats for Use in Real-Time PCR The LightCycler Instrument uses several detection channels to monitor the amplification of
IKDT Laboratory. IKDT as Service Lab (CRO) for Molecular Diagnostics
 Page 1 IKDT Laboratory IKDT as Service Lab (CRO) for Molecular Diagnostics IKDT lab offer is complete diagnostic service to all external customers. We could perform as well single procedures or complex
Page 1 IKDT Laboratory IKDT as Service Lab (CRO) for Molecular Diagnostics IKDT lab offer is complete diagnostic service to all external customers. We could perform as well single procedures or complex
protocol handbook 3D cell culture mimsys G hydrogel
 handbook 3D cell culture mimsys G hydrogel supporting real discovery handbook Index 01 Cell encapsulation in hydrogels 02 Cell viability by MTS assay 03 Live/Dead assay to assess cell viability 04 Fluorescent
handbook 3D cell culture mimsys G hydrogel supporting real discovery handbook Index 01 Cell encapsulation in hydrogels 02 Cell viability by MTS assay 03 Live/Dead assay to assess cell viability 04 Fluorescent
Fluorescence in situ hybridisation (FISH)
 Fluorescence in situ hybridisation (FISH) rarechromo.org Fluorescence in situ hybridization (FISH) Chromosomes Chromosomes are structures that contain the genetic information (DNA) that tells the body
Fluorescence in situ hybridisation (FISH) rarechromo.org Fluorescence in situ hybridization (FISH) Chromosomes Chromosomes are structures that contain the genetic information (DNA) that tells the body
Outline. 1. Experiment. 2. Sample analysis and storage. 3. Image analysis and presenting data. 4. Probemaker
 Tips and tricks Note: this is just an informative document with general recommendations. Please contact [email protected] should you have any queries. Document last reviewed 2011-11-17 Outline 1. Experiment
Tips and tricks Note: this is just an informative document with general recommendations. Please contact [email protected] should you have any queries. Document last reviewed 2011-11-17 Outline 1. Experiment
Forensic DNA Testing Terminology
 Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Ms. Campbell Protein Synthesis Practice Questions Regents L.E.
 Name Student # Ms. Campbell Protein Synthesis Practice Questions Regents L.E. 1. A sequence of three nitrogenous bases in a messenger-rna molecule is known as a 1) codon 2) gene 3) polypeptide 4) nucleotide
Name Student # Ms. Campbell Protein Synthesis Practice Questions Regents L.E. 1. A sequence of three nitrogenous bases in a messenger-rna molecule is known as a 1) codon 2) gene 3) polypeptide 4) nucleotide
Expression and Purification of Recombinant Protein in bacteria and Yeast. Presented By: Puspa pandey, Mohit sachdeva & Ming yu
 Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
ab183294 TripleStain IHC Kit: M&M&R on rodent tissue (DAB, DAB/Ni & AP/Red)
 ab183294 TripleStain IHC Kit: M&M&R on rodent tissue (DAB, DAB/Ni & AP/Red) Instructions for Use For the detection of Rabbit and Mouse Primary antibodies on Rodent Tissue. This product is for research
ab183294 TripleStain IHC Kit: M&M&R on rodent tissue (DAB, DAB/Ni & AP/Red) Instructions for Use For the detection of Rabbit and Mouse Primary antibodies on Rodent Tissue. This product is for research
PROTOCOL. Immunocytochemistry (ICC) MATERIALS AND EQUIPMENT REQUIRED
 PROTOCOL Immunocytochemistry (ICC) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 11-07 MATERIALS AND EQUIPMENT REQUIRED Materials: MitoSciences primary monoclonal antibody/antibodies Fluorophore-conjugated
PROTOCOL Immunocytochemistry (ICC) 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 11-07 MATERIALS AND EQUIPMENT REQUIRED Materials: MitoSciences primary monoclonal antibody/antibodies Fluorophore-conjugated
High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B. PEPperPRINT GmbH Heidelberg, 06/2014
 High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B PEPperPRINT GmbH Heidelberg, 06/2014 Introduction This report describes the epitope mapping of autoimmune sera
High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B PEPperPRINT GmbH Heidelberg, 06/2014 Introduction This report describes the epitope mapping of autoimmune sera
DNA SPOOLING 1 ISOLATION OF DNA FROM ONION
 DNA SPOOLING 1 ISOLATION OF DNA FROM ONION INTRODUCTION This laboratory protocol will demonstrate several basic steps required for isolation of chromosomal DNA from cells. To extract the chromosomal DNA,
DNA SPOOLING 1 ISOLATION OF DNA FROM ONION INTRODUCTION This laboratory protocol will demonstrate several basic steps required for isolation of chromosomal DNA from cells. To extract the chromosomal DNA,
Fluorescent dyes for use with the
 Detection of Multiple Reporter Dyes in Real-time, On-line PCR Analysis with the LightCycler System Gregor Sagner, Cornelia Goldstein, and Rob van Miltenburg Roche Molecular Biochemicals, Penzberg, Germany
Detection of Multiple Reporter Dyes in Real-time, On-line PCR Analysis with the LightCycler System Gregor Sagner, Cornelia Goldstein, and Rob van Miltenburg Roche Molecular Biochemicals, Penzberg, Germany
Application Guide... 2
 Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
HER2 Testing in Breast Cancer
 HER2 Testing in Breast Cancer GAIL H. VANCE, M.D. AGT MEETING JUNE 13, 2014 LOUISVILLE, KENTUCKY No Conflict of Interest to Report Human Epidermal Growth Factor Receptor 2-HER2 Human epidermal growth factor
HER2 Testing in Breast Cancer GAIL H. VANCE, M.D. AGT MEETING JUNE 13, 2014 LOUISVILLE, KENTUCKY No Conflict of Interest to Report Human Epidermal Growth Factor Receptor 2-HER2 Human epidermal growth factor
Co Extra (GM and non GM supply chains: Their CO EXistence and TRAceability) Outcomes of Co Extra
 GM and non GM supply chains: Their CO EXistence and TRAceability Outcomes of Co Extra Comparison of different real time PCR chemistries and their suitability for detection and quantification of genetically
GM and non GM supply chains: Their CO EXistence and TRAceability Outcomes of Co Extra Comparison of different real time PCR chemistries and their suitability for detection and quantification of genetically
Recombinant DNA & Genetic Engineering. Tools for Genetic Manipulation
 Recombinant DNA & Genetic Engineering g Genetic Manipulation: Tools Kathleen Hill Associate Professor Department of Biology The University of Western Ontario Tools for Genetic Manipulation DNA, RNA, cdna
Recombinant DNA & Genetic Engineering g Genetic Manipulation: Tools Kathleen Hill Associate Professor Department of Biology The University of Western Ontario Tools for Genetic Manipulation DNA, RNA, cdna
PNA BRAF Mutation Detection Kit
 - PNA BRAF Mutation Detection Kit Catalog Number KA2102 50 tests/kit Version: 01 Intended for research use only www.abnova.com Introduction and Background Intended use The PNA BRAF Mutation Detection Kit
- PNA BRAF Mutation Detection Kit Catalog Number KA2102 50 tests/kit Version: 01 Intended for research use only www.abnova.com Introduction and Background Intended use The PNA BRAF Mutation Detection Kit
Chapter 3 Contd. Western blotting & SDS PAGE
 Chapter 3 Contd. Western blotting & SDS PAGE Western Blot Western blots allow investigators to determine the molecular weight of a protein and to measure relative amounts of the protein present in different
Chapter 3 Contd. Western blotting & SDS PAGE Western Blot Western blots allow investigators to determine the molecular weight of a protein and to measure relative amounts of the protein present in different
Comparative genomic hybridization Because arrays are more than just a tool for expression analysis
 Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
Nucleic Acid Techniques in Bacterial Systematics
 Nucleic Acid Techniques in Bacterial Systematics Edited by Erko Stackebrandt Department of Microbiology University of Queensland St Lucia, Australia and Michael Goodfellow Department of Microbiology University
Nucleic Acid Techniques in Bacterial Systematics Edited by Erko Stackebrandt Department of Microbiology University of Queensland St Lucia, Australia and Michael Goodfellow Department of Microbiology University
HER2. Improved Quality and Efficiency FISH. HER2 IQFISH pharmdx
 WHI T E PAP E R FISH HER2 IQFISH pharmdx HER2 IQFISH pharmdx HER2 IQFISH pharmdx Improved Quality and Efficiency Improved Quality and Efficiency Debra S. Cohen, BS, CG(ASCAP)CM & Sharon Alsobrook, CG(ASCP)CM,
WHI T E PAP E R FISH HER2 IQFISH pharmdx HER2 IQFISH pharmdx HER2 IQFISH pharmdx Improved Quality and Efficiency Improved Quality and Efficiency Debra S. Cohen, BS, CG(ASCAP)CM & Sharon Alsobrook, CG(ASCP)CM,
Mitosis in Onion Root Tip Cells
 Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
DNA Integrity Number (DIN) For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments
 DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
Sample Questions for Exam 3
 Sample Questions for Exam 3 1. All of the following occur during prometaphase of mitosis in animal cells except a. the centrioles move toward opposite poles. b. the nucleolus can no longer be seen. c.
Sample Questions for Exam 3 1. All of the following occur during prometaphase of mitosis in animal cells except a. the centrioles move toward opposite poles. b. the nucleolus can no longer be seen. c.
Analysis of the DNA Methylation Patterns at the BRCA1 CpG Island
 Analysis of the DNA Methylation Patterns at the BRCA1 CpG Island Frédérique Magdinier 1 and Robert Dante 2 1 Laboratory of Molecular Biology of the Cell, Ecole Normale Superieure, Lyon, France 2 Laboratory
Analysis of the DNA Methylation Patterns at the BRCA1 CpG Island Frédérique Magdinier 1 and Robert Dante 2 1 Laboratory of Molecular Biology of the Cell, Ecole Normale Superieure, Lyon, France 2 Laboratory
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ncounter Gene Expression Assay Manual Total RNA and Cell Lysate Protocols
 ncounter Gene Expression Assay Manual Total RNA and Cell Lysate Protocols v.20090807 For research use only. Not for use in diagnostic procedures. Limited License Subject to the terms and conditions of
ncounter Gene Expression Assay Manual Total RNA and Cell Lysate Protocols v.20090807 For research use only. Not for use in diagnostic procedures. Limited License Subject to the terms and conditions of
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT
 BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
Protein immunoblotting
 Protein immunoblotting (Western blotting) Dr. Serageldeen A. A. Sultan Lecturer of virology Dept. of Microbiology SVU, Qena, Egypt [email protected] Western blotting -It is an analytical technique used to
Protein immunoblotting (Western blotting) Dr. Serageldeen A. A. Sultan Lecturer of virology Dept. of Microbiology SVU, Qena, Egypt [email protected] Western blotting -It is an analytical technique used to
Real-time PCR: Understanding C t
 APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
EXTRACTION OF DNA FROM CALF THYMUS CELLS Revised 2/1/96 Introduction
 Revised 2/1/96 Introduction Cells may be classified into two primary types depending on whether they have a discrete nucleus (eukaryotic) or do not (prokaryotic). Prokaryotes include bacteria, such as
Revised 2/1/96 Introduction Cells may be classified into two primary types depending on whether they have a discrete nucleus (eukaryotic) or do not (prokaryotic). Prokaryotes include bacteria, such as
Principles of Immunohistochemistry Queen s Laboratory For Molecular Pathology
 Principles of Immunohistochemistry Queen s Laboratory For Molecular Pathology Table of Contents POLYCLONAL VERSUS MONOCLONAL ANTIBODIES...PAGE 3 DIRECT & INDIRECT ASSAYS.PAGE 4 LABELS..PAGE 5 DETECTION...PAGE
Principles of Immunohistochemistry Queen s Laboratory For Molecular Pathology Table of Contents POLYCLONAL VERSUS MONOCLONAL ANTIBODIES...PAGE 3 DIRECT & INDIRECT ASSAYS.PAGE 4 LABELS..PAGE 5 DETECTION...PAGE
ISOLATION AND PROPERTIES OF SECRETORY GRANULES FROM RAT ISLETS OF LANGERHANS. II. Ultrastructure of the Beta Granule
 ISOLATION AND PROPERTIES OF SECRETORY GRANULES FROM RAT ISLETS OF LANGERHANS II Ultrastructure of the Beta Granule MARIE H GREIDER, S L HOWELL, and P E LACY From the Department of Pathology, Washington
ISOLATION AND PROPERTIES OF SECRETORY GRANULES FROM RAT ISLETS OF LANGERHANS II Ultrastructure of the Beta Granule MARIE H GREIDER, S L HOWELL, and P E LACY From the Department of Pathology, Washington
WESTERN BLOTTING TIPS AND TROUBLESHOOTING GUIDE TROUBLESHOOTING GUIDE
 WESTERN BLOTTING TIPS AND TROUBLESHOOTING GUIDE TIPS FOR SUCCESSFUL WESTERB BLOTS TROUBLESHOOTING GUIDE 1. Suboptimal protein transfer. This is the most common complaint with western blotting and could
WESTERN BLOTTING TIPS AND TROUBLESHOOTING GUIDE TIPS FOR SUCCESSFUL WESTERB BLOTS TROUBLESHOOTING GUIDE 1. Suboptimal protein transfer. This is the most common complaint with western blotting and could
Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA
 Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Bio 3A Lab: DNA Isolation and the Polymerase Chain Reaction
 Bio 3A Lab: DNA Isolation and the Polymerase Chain Reaction Objectives Understand the process of DNA isolation Perform DNA isolation using cheek cells Use thermal cycler and Taq polymerase to perform DNA
Bio 3A Lab: DNA Isolation and the Polymerase Chain Reaction Objectives Understand the process of DNA isolation Perform DNA isolation using cheek cells Use thermal cycler and Taq polymerase to perform DNA
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs)
 Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE
 AP Biology Date SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE LEARNING OBJECTIVES Students will gain an appreciation of the physical effects of sickle cell anemia, its prevalence in the population,
AP Biology Date SICKLE CELL ANEMIA & THE HEMOGLOBIN GENE TEACHER S GUIDE LEARNING OBJECTIVES Students will gain an appreciation of the physical effects of sickle cell anemia, its prevalence in the population,
50 g 650 L. *Average yields will vary depending upon a number of factors including type of phage, growth conditions used and developmental stage.
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
Plant Genomic DNA Extraction using CTAB
 Plant Genomic DNA Extraction using CTAB Introduction The search for a more efficient means of extracting DNA of both higher quality and yield has lead to the development of a variety of protocols, however
Plant Genomic DNA Extraction using CTAB Introduction The search for a more efficient means of extracting DNA of both higher quality and yield has lead to the development of a variety of protocols, however
IBCSG Tissue Bank Policy
 THE INTERNATIONAL BREAST CANCER STUDY GROUP IBCSG Tissue Bank Policy Accepted by the IBCSG Executive Committee on March 24, 2006 Accepted by the IBCSG Ethics Committee on March 24, 2006 Accepted by the
THE INTERNATIONAL BREAST CANCER STUDY GROUP IBCSG Tissue Bank Policy Accepted by the IBCSG Executive Committee on March 24, 2006 Accepted by the IBCSG Ethics Committee on March 24, 2006 Accepted by the
Protease Peptide Microarrays Ready-to-use microarrays for protease profiling
 Protocol Protease Peptide Microarrays Ready-to-use microarrays for protease profiling Contact us: InfoLine: +49-30-97893-117 Order per fax: +49-30-97893-299 Or e-mail: [email protected] www: www.jpt.com
Protocol Protease Peptide Microarrays Ready-to-use microarrays for protease profiling Contact us: InfoLine: +49-30-97893-117 Order per fax: +49-30-97893-299 Or e-mail: [email protected] www: www.jpt.com
1. The diagram below represents a biological process
 1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
1. The diagram below represents a biological process 5. The chart below indicates the elements contained in four different molecules and the number of atoms of each element in those molecules. Which set
REAL TIME PCR USING SYBR GREEN
 REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
REAL TIME PCR USING SYBR GREEN 1 THE PROBLEM NEED TO QUANTITATE DIFFERENCES IN mrna EXPRESSION SMALL AMOUNTS OF mrna LASER CAPTURE SMALL AMOUNTS OF TISSUE PRIMARY CELLS PRECIOUS REAGENTS 2 THE PROBLEM
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
P4 Distribution of Cetuximab in Models of Human Lung Cancer
 Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, University of Tübingen, Robert Bosch Foundation Stuttgart, Germany FORSYS Partner Project Predictive Cancer Therapy Project Meeting 14.10.09
Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, University of Tübingen, Robert Bosch Foundation Stuttgart, Germany FORSYS Partner Project Predictive Cancer Therapy Project Meeting 14.10.09
The following chapter is called "Preimplantation Genetic Diagnosis (PGD)".
 Slide 1 Welcome to chapter 9. The following chapter is called "Preimplantation Genetic Diagnosis (PGD)". The author is Dr. Maria Lalioti. Slide 2 The learning objectives of this chapter are: To learn the
Slide 1 Welcome to chapter 9. The following chapter is called "Preimplantation Genetic Diagnosis (PGD)". The author is Dr. Maria Lalioti. Slide 2 The learning objectives of this chapter are: To learn the
VLLM0421c Medical Microbiology I, practical sessions. Protocol to topic J10
 Topic J10+11: Molecular-biological methods + Clinical virology I (hepatitis A, B & C, HIV) To study: PCR, ELISA, your own notes from serology reactions Task J10/1: DNA isolation of the etiological agent
Topic J10+11: Molecular-biological methods + Clinical virology I (hepatitis A, B & C, HIV) To study: PCR, ELISA, your own notes from serology reactions Task J10/1: DNA isolation of the etiological agent
Annexin V-EGFP Apoptosis Detection Kit
 ab14153 Annexin V-EGFP Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of apoptosis in various samples This product is for research use only and is not intended
ab14153 Annexin V-EGFP Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of apoptosis in various samples This product is for research use only and is not intended
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
MagExtractor -Genome-
 Instruction manual MagExtractor-Genome-0810 F0981K MagExtractor -Genome- NPK-101 100 preparations Store at 4 C Contents [1] Introduction [2] Components [3] Materials required [4] Protocol 1. Purification
Instruction manual MagExtractor-Genome-0810 F0981K MagExtractor -Genome- NPK-101 100 preparations Store at 4 C Contents [1] Introduction [2] Components [3] Materials required [4] Protocol 1. Purification
Summary of Discussion on Non-clinical Pharmacology Studies on Anticancer Drugs
 Provisional Translation (as of January 27, 2014)* November 15, 2013 Pharmaceuticals and Bio-products Subcommittees, Science Board Summary of Discussion on Non-clinical Pharmacology Studies on Anticancer
Provisional Translation (as of January 27, 2014)* November 15, 2013 Pharmaceuticals and Bio-products Subcommittees, Science Board Summary of Discussion on Non-clinical Pharmacology Studies on Anticancer
Microarray Technology
 Microarrays And Functional Genomics CPSC265 Matt Hudson Microarray Technology Relatively young technology Usually used like a Northern blot can determine the amount of mrna for a particular gene Except
Microarrays And Functional Genomics CPSC265 Matt Hudson Microarray Technology Relatively young technology Usually used like a Northern blot can determine the amount of mrna for a particular gene Except
Southern Blot Analysis (from Baker lab, university of Florida)
 Southern Blot Analysis (from Baker lab, university of Florida) DNA Prep Prepare DNA via your favorite method. You may find a protocol under Mini Yeast Genomic Prep. Restriction Digest 1.Digest DNA with
Southern Blot Analysis (from Baker lab, university of Florida) DNA Prep Prepare DNA via your favorite method. You may find a protocol under Mini Yeast Genomic Prep. Restriction Digest 1.Digest DNA with
ChIP TROUBLESHOOTING TIPS
 ChIP TROUBLESHOOTING TIPS Creative Diagnostics Abstract ChIP dissects the spatial and temporal dynamics of the interactions between chromatin and its associated factors CD Creative Diagnostics info@creative-
ChIP TROUBLESHOOTING TIPS Creative Diagnostics Abstract ChIP dissects the spatial and temporal dynamics of the interactions between chromatin and its associated factors CD Creative Diagnostics info@creative-
RT31-020 20 rxns. RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl
 Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
DNA Isolation Kit for Cells and Tissues
 DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
MUCICARMINE STAIN - SOUTHGATE S - MUCIN PURPOSE:
 SURGICAL PATHOLOGY - HISTOLOGY Date: STAINING MANUAL - CARBOHYDRATES Page: 1 of 2 MUCICARMINE STAIN - SOUTHGATE S - MUCIN PURPOSE: To stain mucin which is a secretion produced by a variety of epithelial
SURGICAL PATHOLOGY - HISTOLOGY Date: STAINING MANUAL - CARBOHYDRATES Page: 1 of 2 MUCICARMINE STAIN - SOUTHGATE S - MUCIN PURPOSE: To stain mucin which is a secretion produced by a variety of epithelial
Quantifiler Human DNA Quantification Kit Quantifiler Y Human Male DNA Quantification Kit
 Product Bulletin Human Identification Quantifiler Human DNA Quantification Kit Quantifiler Y Human Male DNA Quantification Kit The Quantifiler kits produce reliable and reproducible results, helping to
Product Bulletin Human Identification Quantifiler Human DNA Quantification Kit Quantifiler Y Human Male DNA Quantification Kit The Quantifiler kits produce reliable and reproducible results, helping to
Genomic DNA Extraction Kit INSTRUCTION MANUAL
 Genomic DNA Extraction Kit INSTRUCTION MANUAL Table of Contents Introduction 3 Kit Components 3 Storage Conditions 4 Recommended Equipment and Reagents 4 Introduction to the Protocol 4 General Overview
Genomic DNA Extraction Kit INSTRUCTION MANUAL Table of Contents Introduction 3 Kit Components 3 Storage Conditions 4 Recommended Equipment and Reagents 4 Introduction to the Protocol 4 General Overview
Annexin V-FITC Apoptosis Detection Kit
 ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY
 MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
Annexin V-FITC Apoptosis Detection Kit
 ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
PREPARED FOR: U.S. Army Medical Research and Materiel CommandFort Detrick, Maryland 21702 5012
 AD Award Number: W81XWH 07 1 0542 TITLE: Is Nuclear Structure Altered in Breast Cancer Cells? PRINCIPAL INVESTIGATOR: Han Htun, Ph.D. CONTRACTING ORGANIZATION: University of California Los Angeles, CA
AD Award Number: W81XWH 07 1 0542 TITLE: Is Nuclear Structure Altered in Breast Cancer Cells? PRINCIPAL INVESTIGATOR: Han Htun, Ph.D. CONTRACTING ORGANIZATION: University of California Los Angeles, CA
ELUTION OF DNA FROM AGAROSE GELS
 ELUTION OF DNA FROM AGAROSE GELS OBTECTIVE: To isolate specific bands or regions of agarose-separated DNA for use in subsequent experiments and/or procedures. INTRODUCTION: It is sometimes necessary to
ELUTION OF DNA FROM AGAROSE GELS OBTECTIVE: To isolate specific bands or regions of agarose-separated DNA for use in subsequent experiments and/or procedures. INTRODUCTION: It is sometimes necessary to
Gene Mapping Techniques
 Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Replication Study Guide
 Replication Study Guide This study guide is a written version of the material you have seen presented in the replication unit. Self-reproduction is a function of life that human-engineered systems have
Replication Study Guide This study guide is a written version of the material you have seen presented in the replication unit. Self-reproduction is a function of life that human-engineered systems have
inform Advanced Image Analysis Software For Accurately Quantifying Biomarkers in Tissue Sections
 P R O D U C T N O T E inform Advanced Image Analysis Key Features Quantitative analysis of biomarker epression in tissue sections and TMAs Separation of weakly epressing and overlapping markers Cellular
P R O D U C T N O T E inform Advanced Image Analysis Key Features Quantitative analysis of biomarker epression in tissue sections and TMAs Separation of weakly epressing and overlapping markers Cellular
Laboratory Testing for Her2 Status in Breast Cancer
 Laboratory Testing for Her2 Status in Breast Cancer Katherine Geiersbach, M.D. Assistant Professor, Department of Pathology May 28, 2015 Overview Clinical relevance of Her2 status for treatment of breast
Laboratory Testing for Her2 Status in Breast Cancer Katherine Geiersbach, M.D. Assistant Professor, Department of Pathology May 28, 2015 Overview Clinical relevance of Her2 status for treatment of breast
OBJECTIVES PROCEDURE. Lab 2- Bio 160. Name:
 Lab 2- Bio 160 Name: Prokaryotic and Eukaryotic Cells OBJECTIVES To explore cell structure and morphology in prokaryotes and eukaryotes. To gain more experience using the microscope. To obtain a better
Lab 2- Bio 160 Name: Prokaryotic and Eukaryotic Cells OBJECTIVES To explore cell structure and morphology in prokaryotes and eukaryotes. To gain more experience using the microscope. To obtain a better
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
How To Get A Cell Print
 QUICK CELL CAPTURE AND CHARACTERIZATION GUIDE FOR CELLSEARCH CUSTOMERS CellSave EDTA Blood sample Rare cell capture Enumeration Single protein marker Cell capture for molecular characterization CELLSEARCH
QUICK CELL CAPTURE AND CHARACTERIZATION GUIDE FOR CELLSEARCH CUSTOMERS CellSave EDTA Blood sample Rare cell capture Enumeration Single protein marker Cell capture for molecular characterization CELLSEARCH
Optimal Conditions for F(ab ) 2 Antibody Fragment Production from Mouse IgG2a
 Optimal Conditions for F(ab ) 2 Antibody Fragment Production from Mouse IgG2a Ryan S. Stowers, 1 Jacqueline A. Callihan, 2 James D. Bryers 2 1 Department of Bioengineering, Clemson University, Clemson,
Optimal Conditions for F(ab ) 2 Antibody Fragment Production from Mouse IgG2a Ryan S. Stowers, 1 Jacqueline A. Callihan, 2 James D. Bryers 2 1 Department of Bioengineering, Clemson University, Clemson,
Validation of BRAF Mutational Analysis in Thyroid Fine Needle Aspirations: A Morphologic- Molecular Approach
 Validation of BRAF Mutational Analysis in Thyroid Fine Needle Aspirations: A Morphologic- Molecular Approach Kerry C. Councilman, MD Assistant Professor University of Colorado Denver Goals: BRAF Mutation
Validation of BRAF Mutational Analysis in Thyroid Fine Needle Aspirations: A Morphologic- Molecular Approach Kerry C. Councilman, MD Assistant Professor University of Colorado Denver Goals: BRAF Mutation
www.gbo.com/bioscience Tissue Culture 1 Cell/ Microplates 2 HTS- 3 Immunology/ HLA 4 Microbiology/ Bacteriology Purpose Beakers 5 Tubes/Multi-
 11 Cryo 5 Tubes/Multi 2 HTS 3 Immunology / Immunology Technical Information 3 I 2 96 Well ELISA 3 I 4 96 Well ELISA Strip Plates 3 I 6 8 Well Strip Plates 3 I 7 12 Well Strip Plates 3 I 8 16 Well Strip
11 Cryo 5 Tubes/Multi 2 HTS 3 Immunology / Immunology Technical Information 3 I 2 96 Well ELISA 3 I 4 96 Well ELISA Strip Plates 3 I 6 8 Well Strip Plates 3 I 7 12 Well Strip Plates 3 I 8 16 Well Strip
Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides
 Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides Polyacrylamide gel electrophoresis (PAGE) and subsequent analyses are common tools in biochemistry and molecular biology. This Application
Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides Polyacrylamide gel electrophoresis (PAGE) and subsequent analyses are common tools in biochemistry and molecular biology. This Application
The RNA strategy. RNA as a tool and target in human disease diagnosis and therapy.
 The RNA strategy RNA as a tool and target in human disease diagnosis and therapy. The Laboratory of RNA Biology and Biotechnology at the Centre for Integrative Biology (CIBIO) of the University of Trento,
The RNA strategy RNA as a tool and target in human disease diagnosis and therapy. The Laboratory of RNA Biology and Biotechnology at the Centre for Integrative Biology (CIBIO) of the University of Trento,
Genomic DNA Clean & Concentrator Catalog Nos. D4010 & D4011
 Page 0 INSTRUCTION MANUAL Catalog Nos. D4010 & D4011 Highlights Quick (5 minute) spin column recovery of large-sized DNA (e.g., genomic, mitochondrial, plasmid (BAC/PAC), viral, phage, (wga)dna, etc.)
Page 0 INSTRUCTION MANUAL Catalog Nos. D4010 & D4011 Highlights Quick (5 minute) spin column recovery of large-sized DNA (e.g., genomic, mitochondrial, plasmid (BAC/PAC), viral, phage, (wga)dna, etc.)
Pharmaceutical Biotechnology. Recombinant DNA technology Western blotting and SDS-PAGE
 Pharmaceutical Biotechnology Recombinant DNA technology Western blotting and SDS-PAGE Recombinant DNA Technology Protein Synthesis Western Blot Western blots allow investigators to determine the molecular
Pharmaceutical Biotechnology Recombinant DNA technology Western blotting and SDS-PAGE Recombinant DNA Technology Protein Synthesis Western Blot Western blots allow investigators to determine the molecular
Single Nucleotide Polymorphisms (SNPs)
 Single Nucleotide Polymorphisms (SNPs) Additional Markers 13 core STR loci Obtain further information from additional markers: Y STRs Separating male samples Mitochondrial DNA Working with extremely degraded
Single Nucleotide Polymorphisms (SNPs) Additional Markers 13 core STR loci Obtain further information from additional markers: Y STRs Separating male samples Mitochondrial DNA Working with extremely degraded
How To Make A Tri Reagent
 TRI Reagent For processing tissues, cells cultured in monolayer or cell pellets Catalog Number T9424 Store at room temperature. TECHNICAL BULLETIN Product Description TRI Reagent is a quick and convenient
TRI Reagent For processing tissues, cells cultured in monolayer or cell pellets Catalog Number T9424 Store at room temperature. TECHNICAL BULLETIN Product Description TRI Reagent is a quick and convenient
Macromolecules in my food!!
 Macromolecules in my food!! Name Notes/Background Information Food is fuel: All living things need to obtain fuel from something. Whether it is self- made through the process of photosynthesis, or by ingesting
Macromolecules in my food!! Name Notes/Background Information Food is fuel: All living things need to obtain fuel from something. Whether it is self- made through the process of photosynthesis, or by ingesting
Biopharmaceuticals and Biotechnology Unit 2 Student Handout. DNA Biotechnology and Enzymes
 DNA Biotechnology and Enzymes 35 Background Unit 2~ Lesson 1 The Biotechnology Industry Biotechnology is a process (or a technology) that is used to create products like medicines by using micro-organisms,
DNA Biotechnology and Enzymes 35 Background Unit 2~ Lesson 1 The Biotechnology Industry Biotechnology is a process (or a technology) that is used to create products like medicines by using micro-organisms,
Overview of Genetic Testing and Screening
 Integrating Genetics into Your Practice Webinar Series Overview of Genetic Testing and Screening Genetic testing is an important tool in the screening and diagnosis of many conditions. New technology is
Integrating Genetics into Your Practice Webinar Series Overview of Genetic Testing and Screening Genetic testing is an important tool in the screening and diagnosis of many conditions. New technology is
Immunohistochemistry. Services & Products. Indivumed GmbH Falkenried 88, Bldg. D 20251 Hamburg Germany
 Services & Products Indivumed GmbH Falkenried 88, Bldg. D 20251 Hamburg Germany Phone Europe: +49-40-41 33 83-0 Fax Europe: +49-40-41 33 83-14 Phone USA: +1-301-588-4650 [email protected] www.indivumed.com
Services & Products Indivumed GmbH Falkenried 88, Bldg. D 20251 Hamburg Germany Phone Europe: +49-40-41 33 83-0 Fax Europe: +49-40-41 33 83-14 Phone USA: +1-301-588-4650 [email protected] www.indivumed.com
Objectives: Vocabulary:
 Introduction to Agarose Gel Electrophoresis: A Precursor to Cornell Institute for Biology Teacher s lab Author: Jennifer Weiser and Laura Austen Date Created: 2010 Subject: Molecular Biology and Genetics
Introduction to Agarose Gel Electrophoresis: A Precursor to Cornell Institute for Biology Teacher s lab Author: Jennifer Weiser and Laura Austen Date Created: 2010 Subject: Molecular Biology and Genetics
6 Characterization of Casein and Bovine Serum Albumin
 6 Characterization of Casein and Bovine Serum Albumin (BSA) Objectives: A) To separate a mixture of casein and bovine serum albumin B) to characterize these proteins based on their solubilities as a function
6 Characterization of Casein and Bovine Serum Albumin (BSA) Objectives: A) To separate a mixture of casein and bovine serum albumin B) to characterize these proteins based on their solubilities as a function
Liquid-based cytology
 Liquid-based cytology Claire Bourgain UZBrussel Introduction LBC for gynaecological cytology has been introduced in Belgium a decade ago There is no consensus On performance compared to conventional cytology
Liquid-based cytology Claire Bourgain UZBrussel Introduction LBC for gynaecological cytology has been introduced in Belgium a decade ago There is no consensus On performance compared to conventional cytology
