Genetics, genomics and gene discovery in the auditory system
|
|
|
- Cecily Cameron
- 8 years ago
- Views:
Transcription
1 # 2002 Oxford University Press Human Molecular Genetics, 2002, Vol. 11, No Genetics, genomics and gene discovery in the auditory system Cynthia C. Morton * Departments of Obstetrics, Gynecology, and Reproductive Biology and Pathology, Brigham and Women s Hospital and Harvard Medical School, Boston, MA, USA Received April 5, 2002; Accepted April 10, 2002 The sounds of silence have forever been broken as genetics and genomics approaches in human and model organisms have provided a powerful and rapid entry into gene discovery in the auditory system. An understanding of the complexities and beauty of the biological process of hearing itself is unfolding as genes underlying hereditary hearing impairment are identified. Genes involved in modifying hearing are also being found, and will be critical to a full comprehension of genotype phenotype relationships. Investigations in the auditory system will provide important insight into how the nervous system decodes molecular information. Deafness represents a common sensory disorder that can interfere dramatically in the acquisition of speech and language in children, and in the quality of life for a growing aged population. As newborn screening for hearing impairment is being implemented in many birth hospitals, the prospects for precise clinical diagnosis, appropriate genetic counseling and proper medical management for auditory disorders has never been at a more exciting crossroad. INTRODUCTION It has long been recognized that heredity plays a major role in hearing impairment. Despite the fact that understanding the genetic basis of hearing loss has fascinated human and medical geneticists for decades, only within the past few years have the genes and molecular mechanisms underlying deafness begun to be discovered. In part, this results from various obstacles to investigation of the auditory system including: inaccessibility of the sensory end organ for hearing, the cochlea, within the dense temporal bone; length of time to direct pathologic observation of the deaf ear due to an otherwise normal lifespan of individuals with hearing loss; unparalleled genetic heterogeneity; and assortative mating. The history of the genetics of deafness has had a sordid past of blatant discrimination of the deaf (1), and there is mistrust about genetics among some members of the deaf population. Nevertheless, the past few years have witnessed an explosion of discoveries that are providing fundamental insight into the biology of hearing. The frequency of hearing loss is estimated at one per thousand newborns and half of all cases are attributed to genetic causes. In addition to being a common etiology of congenital deafness, mutations in genes are responsible for progressive hearing loss, and no doubt will be found to play an important role in progressive hearing loss with ageing (presbycusis). Environmental etiologies of hearing loss are likely to represent a declining proportion of cases as better therapies for bacterial and viral infections (e.g. vaccines) are implemented, acoustic trauma in the workplace is recognized and prevented, and ototoxic drugs (e.g. aminoglycosides) are avoided in genetically susceptible individuals. GENETICS OF DEAFNESS The study of the genetics of deafness is unique among inherited disorders for several reasons and illustrates well various concepts in human genetics. Notably, there is incomparable genetic heterogeneity with over 90% of matings among the deaf resulting in all hearing offspring. This reflects matings among the deaf with mutations in different genes as well as matings of couples in which one individual is deaf due to a genetic mechanism and the other due to an environmental etiology. Matings among the deaf are well recognized and, with the exception of assortative mating for stature, may represent one of the most common genetic traits on which an altered mating structure occurs in human populations. Furthermore, the hearing offspring of deaf couples who themselves can be native signers are more likely than random members of the population to have a partner who is deaf due to a shared language and culture. Both genetic heterogeneity and assortative mating confound gene discovery using traditional methods of genetic linkage analysis. *To whom correspondence should be addressed at: Department of Pathology, Brigham and Women s Hospital, 75 Francis Street, Boston, MA 02115, USA. Tel: ; Fax: ; cmorton@partners.org
2 1230 Human Molecular Genetics, 2002, Vol. 11, No. 10 Table 1. Gene discovery in the auditory system No. Gene Protein or RNA Map position Syndromic or Disorders Reference nonsyndromic hearing loss 1 ATP6B1 2cen q13 SHL Distal renal tubular acidosis (76) associated with sensorineural deafness 2 BSND barttin 1p32.3 SHL Bartter syndrome (77) 3 CDH23 cadherin 23 10q21 q22 NSHL þ SHL Usher syndrome type 1D (10) (USH1D) Usher syndrome type 1D (11) (USH1D) þ DFNB12 4 CLDN14 claudin 14 21q22 NSHL DFNB29 (78) 5 COCH cochlin 14q12 q13 NSHL DFNA9 (22) 6 COL1A2 collagen type 1 a2 7q22.1 SHL Osteogenesis imperfecta (79) 7 COL2A1 collagen type 2 a1 12q13.1 q13.2 SHL Stickler syndrome (STL1) (80) 8 COL4A3 collagen type 4 a3 2q36 q37 SHL Alport syndrome (23) 9 COL4A4 collagen type 4 a4 2q36 q37 SHL Alport syndrome (23) 10 COL4A5 collagen type 4 a5 Xq22 SHL Alport syndrome (20) 11 COL11A1 collagen type 11 a1 1p21 SHL Stickler syndrome (STL2) (81) 12 COL11A2 collagen type 11 a2 6p21.3 NSHL þ SHL Stickler syndrome (STL3) (82) DFNA13 (83) 13 DDP Xq22 SHL Mohr-Tranebjaerg syndrome (16) 14 DFNA5 7p15 NSHL DFNA5 (19) 15 DIAPH1 diaphanous 5q31 NSHL DFNA1 (84) 16 DSPP dentin sialophosphoprotein 4q21.3 SHL Dentinogenesis imperfecta 1 (85) (DGI1), DFNA39 17 EDN3 endothelin 3 20q13.2 q13.3 SHL Waardenburg Shah syndrome (86) (WS4) 18 EDNRB endothelin receptor B 13q22 SHL Waardenburg Shah syndrome (87) (WS4) 19 EYA1 8q13.3 SHL Branchio-oto-renal syndrome (88) 20 EYA4 6q22 q23 NSHL DFNA10 (89) 21 GJA1 connexin 43 6q21 q23.2 NSHL Autosomal recessive deafness (67) 22 GJB2 connexin 26 13q12 NSHL DFNB1 (24) DFNA3 (25) 23 GJB3 connexin 31 1p34 NSHL DFNA2 (26) Autosomal recessive deafness (90) 24 GJB6 connexin 30 13q12 NSHL DFNA3 (66) 25 KCNE1 21q22.1 q22.2 SHL Jervell and Lange Nielsen (91,92) syndrome (JLNS2) 26 KCNQ4 1p34 NSHL DFNA2 (93) 27 KVLQT1 11p15.5 SHL Jervell and Lange-Nielsen (94) syndrome (JLNS1) 28 MITF 3p12.3 p14.1 SHL Waardenburg syndrome type II (28) (WS2) Tietz syndrome (95) 29 MYH9 myosin heavy chain 9 22q13 NSHL þ SHL May Hegglin and Fechtner (96) syndromes DFNA17 (97) 30 MYO6 myosin 6 6q13 NSHL DFNA22 (49) 31 MYO7A myosin 7A 11q12.3 q21 NSHL þ SHL Usher syndrome type 1B (7) (USH1B) DFNA11 (6) DFNB2 (8,9) Atypical Usher syndrome (98) 32 MYO15A myosin 15A 17p11.2 NSHL DFNB3 (99) 33 NDP norrin Xp11.3 SHL Norrie disease (14,100) 34 OTOF otoferlin 2p22 p23 NSHL DFNB9 (29) 35 PAX3 2q35 SHL Waardenburg syndrome type I (27) (WS1) Waardenburg syndrome types (101) I þ III (WS1 þ WS3) Craniofacial dysmorphism, (102) hand abnormalities, profound sensorineural deafness (CDHS) 36 PCDH15 protocadherin 15 10q21 q22 SHL Usher syndrome type 1F (103,104) (USH1F) 37 POU3F4 Xq21.1 NSHL DFN3 (105)
3 Human Molecular Genetics, 2002, Vol. 11, No Table 1. (Continued) 38 POU4F3 5q31 NSHL DFNA15 (69) 39 PMP22 peripheral myelin 17p11.2 SHL Charcot Marie Tooth disease (106) 40 12S rrna ribosomal RNA mitochondrial NSHL Sensorineural deafness (74) 41 SLC19A2 1q23.3 SHL Thiamine-responsive megaloblastic (107,108) anemia with diabetes mellitus and deafness 42 SLC26A4 pendrin 7q31; 7q21 q34 NSHL þ SHL Pendred syndrome (PDS) (17) DFNB4 (109) 43 SOX9 17q24.3 q25.1 SHL Campomelic dysplasia (110) 44 SOX10 22q13 SHL Waardenburg Shah syndrome (WS4) (111) 45 STRC stereocilin 15q21 q22 NSHL DFNB16 (31) 46 TCOF1 treacle 5q32 q33.1 SHL Treacher Collins syndrome (15) 47 TECTA alpha tectorin 11q22 q24 NSHL DFNA8, DFNA12 (21) DFNB21 (112) 48 TMC1 9q13 q21 NSHL DFNA36 (12) DFNB7/B11 49 TMPRSS3 21q22.3 NSHL DFNB8, DFNB10 (113) 50 trna-glu transfer RNA mitochondrial SHL Diabetes and deafness (114) 51 trna-leu transfer RNA mitochondrial SHL Myopathy, encephalopathy, lactic (115) acidosis and stroke-like episodes (MELAS) Diabetes mellitis and deafness (116) 52 trna-lys transfer RNA mitochondrial SHL Myoclonic epilepsy and ragged-red fiber (117) disease (MERRF) 53 trna-ser transfer RNA mitochondrial SHL Retinitis pigmentosum and progressive (118) sensorineural hearing loss 54 trna-ser(unc) transfer RNA mitochondrial NSHL þ SHL Sensorineural deafness (119) Progressive myoclonic epilepsy, ataxia (120) and hearing loss Palmoplantar keratoderma and deafness (121) 55 USH1C harmonin 11p15.1 SHL Usher syndrome type 1C (USH1C) (30) (122) 56 USH2A usherin 1q41 SHL Usher syndrome type 2A (USH2A) (18) 57 USH3 3q21 q25 SHL Usher syndrome type 3 (USH3) (123) 58 WFS1 wolframin 4p16 SHL Wolfram syndrome (124,125) NSHL DFNA6/A14 (126) DFNA38 (127) Hundreds of syndromic forms of deafness have been described (2 4) and the underlying genetic mutation identified for many of the more common forms, but only 30% of genetic cases are estimated to be part of a heritable syndrome. Thus, the vast majority of genetic deafness is designated as nonsyndromic and to date over 65 loci have been mapped. Nonsyndromic hearing impairment is categorized further by mode of inheritance: approximately 77% of cases are autosomal recessive; 22% autosomal dominant; 1% X-linked; and < 1% due to mitochondrial inheritance (5). Dominant loci are designated with the prefix DFNA, recessive loci DFNB, X-linked loci DFN and modifying loci with DFNM. Generally, patients with autosomal recessive hearing impairment have prelingual and congenital deafness and patients with autosomal dominant hearing impairment have postlingual and progressive hearing impairment. This observation may be explained by the complete absence of functional protein in recessive disorders, while in autosomal dominant disorders, dominant mutations may be consistent with initial function and subsequent hearing impairment due to a cumulative, degenerative process. A recent tally of nonsyndromic hearing loss disorders reveals 32 autosomal dominant, 27 autosomal recessive, and 4 X-linked forms (4). It remains to be shown whether each of these loci will correspond to a unique gene. In fact, various deafness disorders have been found already to be the result of the same genetic etiology (e.g. DFNA8 and A12; DFNA6, A14 and A38). In addition, at least 58 auditory genes have been identified: 16 for autosomal dominant and 11 for autosomal recessive loci, and 1 for an X-linked locus; 6 mitochondrial genes and at least 38 genes for syndromic hearing loss have also been discovered (n.b. some genes cause multiple forms of deafness) (Table 1). Although this magnitude of progress is remarkable and significant advances have been made, it is clear that many more genes for hearing await detection. Mutations within the same gene have been found to result in a variety of clinical phenotypes with different modes of inheritance. For example, mutations in MYO7A are pathogenetic in the autosomal recessive deafness and blindness disorder Usher syndrome type 1B (USH1B), and in two nonsyndromic hearing disorders, DFNB2 and DFNA11, displaying autosomal recessive and dominant segregation, respectively. It has been suggested that the mutation in DFNA11, a 9 bp deletion in the coiled-coil domain of MYO7A which is involved in dimerization, could have a dominant negative effect (6) as compared to splicing and missense mutations observed in recessive USH1B and DFNB2 (7 9). Another example of phenotypic heterogeneity also involving Usher syndrome is seen in mutations in CDH23 detected in USH1D and DFNB12 (10,11). Mutations in PDS cause Pendred syndrome and nonsyndromic autosomal recessive
4 1232 Human Molecular Genetics, 2002, Vol. 11, No. 10 hearing loss, DFNB4. Similarly, mutations in WFS1 cause Wolfram syndrome and also account for an autosomal dominant nonsyndromic deafness, DFNA6/A14/A38. Most recently, DFNA36 and DFNB7/B11 were determined to be the result of mutations in TMC1 (12). GENOMIC APPROACHES TO GENE DISCOVERY IN THE AUDITORY SYSTEM Traditional methods for mapping disease genes, such as genetic linkage analysis, have a less than totally optimal use in gene discovery efforts for hearing disorders, mainly because of the complex genetic nature of deafness. Successful use of genetic linkage for mapping hearing disorders, especially for autosomal recessive nonsyndromic loci, has been restricted largely to consanguineous kindreds or populations in which there has been limited immigration. Even in families in which a heritable hearing disorder is successfully mapped, there is often an insufficient number of recombination events to narrow a chromosomal interval, resulting in a candidate region consisting of megabases of genomic DNA. Positional cloning has been productive for a modest number of human deafness genes including NDP (13,14), TCOF1 (15), DDP (16), SLC26A4 (17), USH2A (18) and DFNA5 (19). Positional candidate genes from human (e.g. COL4A5 (20), TECTA (21), COCH (22), COL4A and 4A4 (23), GJB2 (24,25), GJB3 (26)) and mouse (e.g. PAX3 (27), MITF (28), OTOF (29), USH1C (30), STRC (31)) among others have been the primary method for gene identification. Organ and tissue-specific methods for auditory candidate genes A complementary method to genetic linkage analysis for gene identification is one that utilizes tissue or organ-specific cdna libraries to provide candidate genes (32 34). A transcript map of the inner ear provides a ready source of positional candidate genes for mutation screens in gene discovery efforts. To this end, cochlear cdna libraries constructed from human (35,36) and mouse (37) have provided precious biological tools for gene discovery in the mammalian cochlea. A cdna library from an analogous organ in chicken, the basilar papillae, has also been of great value (38). Almost human (Morton fetal cochlear cdna library) and 1600 mouse (Soares mouse NMIE cdna library) inner ear ESTs are currently available in GenBank and the sequences of an additional 9000 mouse ESTs will soon be deposited there. ESTs derived from two sequencing projects from human cochlear cdna clones (39,40) have elucidated thousands of potential positional candidate genes for hearing disorders (41) in addition to providing a snapshot of gene expression in the week gestational age fetal cochlea. BLAST analysis of 8153 human ESTs revealed that about 50% (n ¼ 4040) had sequence similarity to a total of 1449 known human genes. The most abundantly expressed gene was COL1A2, and two other collagens, COL3A1 and COL1A1, were among the most highly represented transcripts. In total, 10 different collagen genes were present in the cochlear ESTs. Forty-three human homologs of nonhuman mammalian genes were also identified, and among them are ESTs for membrane proteins, extracellular proteins and trafficking proteins. Of the remaining 4055 ESTs, Table 2. Web resources for genetic and genomic studies of the auditory system Web page name Summary of contents URL Connexin-deafness homepage Information on connexin mutations Corey lab Microarray expression data from mouse inner ear coreylab/index.html Harvard Medical School Center Resources for families, clinicians and hearing.harvard.edu for Hereditary Deafness researchers Hereditary hearing impairment in mice Results from a large scale mouse screening program Hereditary hearing loss homepage Syndromic and nonsyndromic disorders, mitochondrial disorders, otosclerosis Morton hearing research group Human cochlear ESTs summary and map hearing.bwh.harvard.edu locations MRC Institute of Hearing Research inner ear mutant table Mouse mutants with various inner ear defects MutantsTable.shtml MRC Institute of Hearing Research inner ear developmental gene expression table Developmental gene expression in the inner ear in a variety of species genetable/index.shtml National Institute on Deafness and Other Research from the Laboratory of Molecular Communication Disorders Genetics and other resources OMIM Human heritable disorders query.fcgi?db ¼ OMIM Otobase Repository of candidate genes for inner ear Hudspeth-sgi.rockefeller.edu disorders in human, mouse and zebrafish Tour de l oreille, Université Montpellier Anatomy, physiology and pathophysiology of the auditory system audition/english/start.htm Univ. of WI Dept. of Neurophysiology Hearing Educational and research resources and Balance Washington Univ. inner ear protein database Catalog of biochemical characterization of tissues and fluids of the inner ear oto.wustl.edu/thc/innerear2d.htm
5 Human Molecular Genetics, 2002, Vol. 11, No Table 3. Mouse hearing loss mutants and their human homologs Mouse mutant Gene Human disorder(s) References Ames waltzer (av) Pcdh15 Usher syndrome type 1F (USH1F) (103,104,128) Beethoven (Bth) and deafness (dn) Tmc1 DFNA36, DFNB7/B11 (12,50) chondrodysplasia(cho) Col11a1 Stickler syndrome type 2 (STL2) (81,129,130) Col1a1 transgene disruption Col1a1 Osteogenesis imperfecta (OI) ( ) Col11a2 targeted null Col11a2 Stickler syndrome type 3 (STL3), DFNA13 (82,83) Col4a3 targeted null Col4a3 Alport syndrome (23,135) Disproportionate micromelia (Dmm) Col2a1 Stickler syndrome type 1 (STL1) (80,136,137) and mutant transgenes Dominant megacolon (Dom) Sox10 Waardenburg Shah syndrome (WS4) (111,138,139) dominant spotting (W) Kit Piebald trait (PBT) ( ) Eya1 bor and targeted null Eya1 Branchio-oto-renal syndrome (BOR) (88,143,144) Fgfr3 targeted null Fgfr3 Craniosynostosis (145,146) Gata targeted null Gata3 Hypoparathyroidism, sensorineural deafness (147,148) and renal dysplasia syndrome (HDR) Kcne1 targeted null and Kcne1 pkr Kcne1 Jervell and Lange Nielsen syndrome (JLNS2) (91,92,149,150) Kcnq1 targeted null Kcnq1 Jervell and Lange Nielsen syndrome (JLNS1) (94,151,152) lethal spotting (ls) Edn3 Waardenburg Shah syndrome (WS4) (86,153) microphthalmia (mi) Mitf Waardenburg syndrome type 2 (WS2), (28,95,154,155) Tietz syndrome Ndph targeted null Ndph Norrie disease (ND) (13,14,156) Pax2 targeted null Pax2 Renal-coloboma syndrome (157,158) piebald (s) Ednrb Waardenburg Shah syndrome (WS4) (87,159) Pou3f4 targeted null and sex-linked fidget (slf) Pou3f4 DFN3 (105, ) Pou4f3 targeted null and dreidel (ddl) Pou4f3 DFNA15 (69,163,164) quivering (qv) Spnb4 Charcot-Marie-Tooth disease type 4F (CMT4F) (165) shaker-1 (sh1) Myo7a Usher syndrome type 1B (USH1B), DFNB2, (7,9,98,166,167) DFNA11, atypical Usher syndrome shaker-2 (sh2) Myo15a DFNB3 (99,168) Slc26a4 targeted null Slc26a4 Pendred syndrome (PDS), DFNB4 (17,109,169) Snell s waltzer (sv) Myo6 DFNA22 (48,49) splotch (Sp) Pax3 Waardenburg syndromes types 1 and 3 (WS1, WS3), (27,101,102,170,171) Craniofacial dysmorphism, hand abnormalities, profound sensorineural deafness (CDHS) Tecta targeted null Tecta DFNA8/A12, DFNB21 (21,112,172) Thrd targeted null Thrb Thyroid hormone resistance (173,174) tremble (Tr) Pmp22 Charcot-Marie-Tooth disease type 1A (CMT1A) (106,175,176) waltzer (v) Cdh23 Usher syndrome type 1D (USH1D), DFNB12 (10,11,177) 3277 had sequence similarity to other ESTs representing 2266 unique clusters; 778 categorized into 700 clusters had no sequence similarity to known genes or ESTs and can be considered to be cochlear-specific. Identification of additional known genes, ESTs and cochlear-specific ESTs provides new candidate genes for both syndromic and nonsyndromic deafness disorders. A variety of web resources have been developed for genetic and genomic studies of the auditory system that facilitate candidate gene approaches and are listed in Table 2. In addition to sequence-based approaches, gene expression chips provide an important means to explore the repertoire of cochlear messages in the normal and diseased state. Data from gene chip assays are available on the web for normal mouse cochlea for ages P2 and P32 (42). Several preferentially expressed cochlear genes, namely COCH (43) and OTOR (44,45), have been identified from the human fetal cochlear cdna library; COCH was further shown to be responsible for a sensorineural deafness and vestibular disorder, DFNA9 (22). Various genes have been identified from a similar approach using mouse inner ear transcripts and include Otog (37), Ocn95 (46), Ush1c (30), Fdp (mouse homolog of OTOR) (47) and Strc (31); the human homolog of Ush1c was found to underlie mutations in USH1C and of Strc to be etiologic in DFNB16. Mouse models for discovery of hearing genes Identification of mouse models of specific forms of deafness is of great interest as the mouse is clearly the model organism of choice for the study of hearing loss in humans. The mouse cochlea is highly similar in structure to that of the human. Studies of mouse mutants from fetal to adult ages makes possible investigation into the pathology at developmental time points simply not possible in humans. Of particular relevance to understanding the pathobiology and underlying molecular mechanisms of genetic mutations is the ability to evaluate early developmental stages in mouse mutants because hair cell defects may result from degenerative processes secondary to a primary abnormality in another cell type. Although there has been tremendous progress in identifying genes underlying deafness, there are still relatively few mouse models (Table 3). In some instances, identification of the mouse mutation has greatly preceded detection of the human disorder (48,49),
6 1234 Human Molecular Genetics, 2002, Vol. 11, No. 10 whereas in other cases discovery of the genetic basis for deafness has occurred concurrently (12,50). Positional cloning of deafness genes in the mouse is facilitated by the ability to breed large numbers of mice with the same mutation to narrow the interval for study. A large screen of inbred mouse strains by ABR threshold analyses at The Jackson Laboratory is currently underway, and likely to identify mutants that will lead to discovery of new genes and modifying genes for deafness (51). In addition, large numbers of new mouse mutants for investigating mammalian gene function including deafness are being generated rapidly through ENU mutagenesis (52). This chemical mutagenesis program also makes possible genedriven approaches to mouse mutants and using this approach, two missense and one stop mutation were recently identified in Gjb2, the most common cause of nonsyndromic deafness in the human population (53). Besides the spontaneous deaf mouse mutants and those generated from mutagenesis programs, a number of gene targeting experiments have been performed following identification of the human gene, and have provided valuable mouse mutants for investigation. As the human and mouse DNA sequencing projects are finished, the mouse human synteny maps will also become better defined and it will become increasingly easier to locate potential mouse mutants for mapped human deafness disorders. GENE DISCOVERY IN THE AUDITORY SYSTEM: THE PATH TO IMPROVED DIAGNOSIS AND CLINICAL CARE Inspection of the genes identified in hearing disorders and those among the gene lists from the EST sequencing projects reveals a great diversity of transcripts, perhaps not surprising due to the large variety of cells and complexity of the inner ear. The finding of a large percentage of cochlear ESTs not identified in any other tissue may indicate the existence of genes that are unique to the cochlea. Certainly, the great degree of genetic heterogeneity reflected in the many different syndromes involving hearing loss and mapped loci is indicative of a large number of genes orchestrating the hearing process. Grouping the genes discovered to be etiologic in deafness disorders into functional categories begins the process of understanding their role in hearing. Knowledge of the pathways in which many of these genes function will be an exciting journey in hearing science; no doubt pathways exist that are not yet imagined. Another important aspect of gene discovery for deafness disorders is that it makes possible the development of diagnostic tests and accurate genetic counseling. Appropriate medical management and therapeutic options may be based on an understanding of the specific disorder. Gap junction proteins: the connexins Prominent among the group of genes are those encoding gap junctions. A somewhat surprising finding in the field has been the prevalence of mutations in a single gene encoding the gapjunction protein connexin 26, GJB2, accounting for up to 50% of all cases of autosomal recessive prelingual deafness in tested populations (54 65). Connexin 26 gap junctions are believed to play a critical role in the recycling of potassium ions from their entry into hair cells during sensory transduction from the endolymph through to the stria vascularis, where other potassium channels pump potassium back into the endolymph. The gap junction itself, the connexon, is formed from six monomers of connexin and forms a pore between cells by binding with a connexon on an adjacent cell. Several recurrent mutations have been found in GJB2 (e.g. 35delG, 167delT, and 235delC), some with ethnic predilections. In addition to the recurrent mutations, the gene is small (681 bp) making it especially amenable for genetic testing. Screening for mutations in GJB2 has already emerged as the cornerstone of genetic testing for hearing loss and has been incorporated in some centers into the clinical work-up of infants who fail newborn hearing tests. The connexin-deafness homepage provides information on connexin mutations in deafness (Table 2). Besides GJB2, genes for other gap junction proteins have been found to be associated with hearing loss including GJB3 (26), GJB6 (66) and GJA1 (67). A curious finding in genetic testing of the deaf for GJB2 has been the frequent observation of heterozygosity for a mutation. Various explanations have been proposed including the possibility that the deafness was due to another gene or that there was a mutation in a non-coding region of GJB2 not evaluated in the test. Recent identification of a 342 kb deletion in the GJB6 gene (D(GJB6 D13S1830)) as the second most frequent mutation causing prelingual deafness in the Spanish population may provide the sought after answer in many cases (68). GJB6 maps within the same chromosomal region as GJB2, and these recently published data suggest that mutations in the DFNB1 locus can result in a monogenic or digenic pattern of inheritance. Of note, the typical mutationdetection assays commonly in use may miss such large deletions. Genes for maintenance of hair cell function Another group of genes of intense interest are those required for survival of sensory hair cells. The POU domain transcription factor gene POU4F3 is required for terminal differentiation and maintenance of inner hair cells and an 8 bp deletion in the POU homeodomain results in progressive hearing loss in DFNA15 (69). Studies of such genes may result in valuable insight into the molecular triggers for hair cell degeneration. Loss of hair cells is presumed to be a fundamental cause of progressive age-related hearing loss (presbycusis) and an understanding of this degenerative process might provide the basis for therapeutic intervention. The recent finding of the transmembrane cochlear-expressed gene TMC1 uncovers a common cause of nonsyndromic recessive deafness in Pakistan and India at the DFNB7/B11 locus on chromosome 9 in bands q13 q21; mutations in TMC1 account for the deafness phenotype in % of 230 families screened (12). Cloning of TMC1 resulted from an interesting genomicsbased approach that first involved identification of a predicted gene (subsequently designated TMC2) on chromosome 20 with sequence similarity to query sequences in a tblastx analysis of a BAC from the linked genetic interval. TMC1 mutations were also found to be etiologic in DFNA36 (12) and in the mouse mutants deafness (dn) (12) and Beethoven (Bth) (50). It is predicted that TMC1 protein may mediate an ion-transport or channel function required for the normal function of hair cells.
7 Human Molecular Genetics, 2002, Vol. 11, No The recessive dn mutant displays no auditory response and has secondary hair cell degeneration and the dominant Bth mutant appears to have normally functioning hair cells prior to degeneration. Modifier genes Molecular analyses of the auditory system have already yielded a number of genes in mice and humans that influence the expression or function of other genes. Studies of these genes are certain to provide insight into the interaction of their gene products. Notable among the mouse genes are tub (tubby) and moth1 (modifier of tubby hearing) (70), and dfw (deafwaddler) and mdfw (modifier of deaf waddler) (71). Moth1 can worsen or prevent the hearing impairment in tubby, depending upon the type of moth1 allele and whether one or both copies of the allele are functional. One allele of Mdfw can protect dfw heterozygotes from hearing loss, whereas another is permissive for hearing loss in dfw heterozygotes. In humans, the locus for a modifier gene (DFNM1) for the deafness haplotype in DFNB26 has been identified on chromosome 1 in band q24 (72). Mitochondrial genes A variety of mitochondrial disorders have been found to involve hearing loss, perhaps reflecting the highly metabolic state of the hearing process (73). Of particular interest has been the A1555!G mutation in 12S rrna that is recognized as the most frequent cause of aminoglycoside-induced deafness and as the etiology of a nonsyndromic deafness (74). In a recent study a nearly identical degree of mitochondrial dysfunction was observed in enucleated lymphoblastoid cells derived from both symptomatic and asymptomatic individuals from the same kindred (75) supporting the possibility of a nuclear gene in modifying the effect of the mutation. Investigations in hereditary deafness have revealed many lessons in genetics, foremost among them a sensory system with profound genotypic and phenotypic heterogeneity. Despite the recent tremendous successes in the genetics of deafness, our knowledge remains woefully incomplete. An astonishing number of deafness loci have been mapped in humans and mice, yet the genetic basis of many disorders remains unknown. Genomic approaches using a combination of methods of positional cloning, candidate genes and mouse models continue to yield new and novel genes providing valuable insight into the molecular basis of the process of hearing. REFERENCES 1. Ruben, R.J. (1991) The history of the genetics of hearing impairment. Ann. N. Y. Acad. Sci., 630, Gorlin, R.J., Toriello, H.V. and Cohen, M.M. (1995) Hereditary Hearing Loss and Its Syndromes. Oxford University Press, New York. 3. OMIM Online Mendelian Inheritance in Man (OMIM). 4. Van Camp, G. and Smith, R.J.H. Hereditary hearing loss homepage. 5. Morton, N.E. (1991) Genetic epidemiology of hearing impairment. Ann. N. Y. Acad. Sci., 630, Liu, X.Z., Walsh, J., Tamagawa, Y., Kitamura, K., Nishizawa, M., Steel, K.P. and Brown, S.D. (1997) Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat. Genet., 17, Weil, D., Blanchard, S., Kaplan, J., Guilford, P., Gibson, F., Walsh, J., Mburu, P., Varela, A., Levilliers, J., Weston, M.D. et al. (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature, 374, Weil, D., Kussel, P., Blanchard, S., Levy, G., Levi-Acobas, F., Drira, M., Ayadi, H. and Petit, C. (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin- VIIA gene. Nat. Genet., 16, Liu, X.-Z., Walsh, J., Mburu, P., Kendrick-Jones, J., Cope, M.J.T.V., Steel, K.P. and Brown, S.D.M. (1997) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat. Genet., 16, Bolz, H., von Brederlow, B., Ramirez, A., Bryda, E.C., Kutsche, K., Nothwang, H.G., Seeliger, M., del, C.S.C.M., Vila, M.C., Molina, O.P. et al. (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet., 27, Bork, J.M., Peters, L.M., Riazuddin, S., Bernstein, S.L., Ahmed, Z.M., Ness, S.L., Polomeno, R., Ramesh, A., Schloss, M., Srisailpathy, C.R. et al. (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherinlike gene CDH23. Am. J. Hum. Genet., 68, Kurima, K., Peters, L.M., Yang, Y., Riazuddin, S., Ahmed, Z.M., Naz, S., Arnaud, D., Drury, S., Mo, J., Makishima, T. et al. (2002) Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet., 30, Berger, W., Meindl, A., van de Pol, T.J.R., Cremers, F.P.M., Ropers, H.H., Doerner, C., Monaco, A., Bergen, A.A.B., Lebo, R., Warburg, M. et al. (1992) Isolation of a candidate gene for Norrie disease by positional cloning. Nat. Genet., 1, Chen, Z.-Y., Hendriks, R.W., Jobling, M.A., Powell, J.F., Breakefield, X.O., Sims, K.B. and Craig, I.W. (1992) Isolation and characterization of a candidate gene for Norrie disease. Nat. Genet., 1, Dixon, M.J. (1996) Treacher Collins syndrome. Hum. Mol. Genet., 5, Jin, H., May, M., Tranebjaerg, L., Kendall, E., Fontan, G., Jackson, J., Subramony, S.H., Arena, F., Lubs, H., Smith, S. et al. (1996) A novel X- "linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat. Genet., 14, Everett, L.A., Glaser, B., Beck, J.C., Idol, J.R., Buchs, A., Heyman, M., Adawi, F., Hazani, E., Nassir, E., Baxevanis, A.D. et al. (1997) Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat. Genet., 17, Eudy, J.D., Weston, M.D., Yao, S., Hoover, D.M., Rehm, H.L., Ma-Edmonds, M., Yan, D., Ahmad, I., Cheng, J.J., Ayuso, C. et al. (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science, 280, Van Laer, L., Huizing, E.H., Verstreken, M., van Zuijlen, D., Wauters, J.G., Bossuyt, P.J., Van de Heyning, P., McGuirt, W.T., Smith, R.J., Willems, P.J. et al. (1998) Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat. Genet., 20, Barker, D.F., Hostikka, S.L., Chow, L.T., Oliphant, A.R., Gerken, S.C., Gregory, M.C., Skolnick, M.H., Atkin, C.L. and Tryggvason, K. (1990) Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science, 248, Verhoeven, K., Van Laer, L., Kirschhofer, K., Legan, P.K., Hughes, D.C., Schatteman, I., Verstreken, M., Van Hauwe, P., Coucke, P., Chen, A. et al. (1998) Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat. Genet., 19, Robertson, N.G., Lu, L., Heller, S., Merchant, S.N., Eavey, R.D., McKenna, M., Nadol, J.B., Jr., Miyamoto, R.T., Linthicum, F.H., Jr., Lubianca Neto, J.F. et al. (1998) Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat. Genet., 20, Mochizuki, T., Lemmink, H.H., Mariyama, M., Antignac, C., Gubler, M.C., Pirson, Y., Verellen-Dumoulin, C., Chan, B., Schroder, C.H., Smeets, H.J. et al. (1994) Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat. Genet., 8, Kelsell, D.P., Dunlop, J., Stevens, H.P., Lench, N.J., Liang, J.N., Parry, G., Mueller, R.F. and Leigh, I.M. (1997) Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature, 387, Denoyelle, F., Lina-Granade, G., Plauchu, H., Bruzzone, R., Chaib, H., Levi-Acobas, F., Weil, D. and Petit, C. (1998) Connexin 26 gene linked to a dominant deafness. Nature, 393,
8 1236 Human Molecular Genetics, 2002, Vol. 11, No Xia, J.H., Liu, C.Y., Tang, B.S., Pan, Q., Huang, L., Dai, H.P., Zhang, B.R., Xie, W., Hu, D.X., Zheng, D. et al. (1998) Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment. Nat. Genet., 20, Tassabehji, M., Read, A.P., Newton, V.E., Harris, P., Balling, R., Gruss, P. and Strachan, T. (1992) Waardenburg s syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature, 355, Tassabehji, M., Newton, V.E. and Read, A.P. (1994) Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet., 8, Yasunaga, S., Grati, M., Cohen-Salmon, M., El-Amraoui, A., Mustapha, M., Salem, N., El-Zir, E., Loiselet, J. and Petit, C. (1999) A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat. Genet., 21, Verpy, E., Leibovici, M., Zwaenepoel, I., Liu, X.Z., Gal, A., Salem, N., Mansour, A., Blanchard, S., Kobayashi, I., Keats, B.J. et al. (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat. Genet., 26, Verpy, E., Masmoudi, S., Zwaenepoel, I., Leibovici, M., Hutchin, T.P., Del Castillo, I., Nouaille, S., Blanchard, S., Laine, S., Popot, J.L. et al. (2001) Mutations in a new gene encoding a protein of the hair bundle cause nonsyndromic deafness at the DFNB16 locus. Nat. Genet., 29, Hedrick, S.M., Cohen, D.I., Nielsen, E.A. and Davis, M.M. (1984) Isolation of cdna clones encoding T cell-specific membrane-associated proteins. Nature, 308, Jones, D.T. and Reed, R.R. (1989) G olf : an olfactory neuron specific- G protein involved in odorant signal transduction. Science, 244, Gurish, M.F., Bell, A.F., Smith, T.J., Ducharme, L.A., Wang, R.K. and Weis, J.H. (1992) Expression of murine beta 7, alpha 4, and beta 1 integrin genes by rodent mast cells. J. Immunol., 149, Robertson, N.G., Khetarpal, U., Gutiérrez-Espeleta, G.A., Bieber, F.R. and Morton, C.C. (1994) Isolation of novel and known genes from a human fetal cochlear cdna library using subtractive hybridization and differential screening. Genomics, 23, Jacob, A.N., Baskaran, N., Kandpal, G., Narayan, D., Bhargava, A.K. and Kandpal, R.P. (1997) Isolation of human ear specific cdnas and construction of cdna libraries from surgically removed small amounts of inner ear tissues. Somat. Cell Mol. Genet., 23, Cohen-Salmon, M., El-Amraoui, A., Leibovici, M. and Petit, C. (1997) Otogelin: a glycoprotein specific to the acellular membranes of the inner ear. Proc. Natl Acad. Sci. USA, 94, Heller, S., Sheane, C.A., Javed, Z. and Hudspeth, A.J. (1998) Molecular markers for cell types of the inner ear and candidate genes for hearing disorders. Proc. Natl Acad. Sci. USA, 95, Skvorak, A.B., Weng, Z., Yee, A.J., Robertson, N.G. and Morton, C.C. (1999) Human cochlear expressed sequence tags provide insight into cochlear gene expression and identify candidate genes for deafness. Hum. Mol. Genet., 8, Resendes, B.L., Robertson, N.G., Szustakowski, J.D., Resendes, R.J., Weng, Z. and Morton, C.C. (2002) Gene discovery in the auditory system: Characterization of additional cochlear sequences. JARO, 3, Morton, C.C. Morton Hearing Research Group Corey, D. The Corey Lab Robertson, N.G., Skvorak, A.B., Yin, Y., Weremowicz, S., Johnson, K.R., Kovatch, K.A., Battey, J.F., Bieber, F.R. and Morton, C.C. (1997) Mapping and characterization of a novel cochlear gene in human and in mouse: a positional candidate gene for a deafness disorder, DFNA9. Genomics, 46, Robertson, N.G., Heller, S., Lin, J.S., Resendes, B.L., Weremowicz, S., Denis, C.S., Bell, A.M., Hudspeth, A.J. and Morton, C.C. (2000) A novel conserved cochlear gene, OTOR: identification, expression analysis, and chromosomal mapping. Genomics, 66, Rendtorff, N.D., Frodin, M., Attie-Bitach, T., Vekemans, M. and Tommerup, N. (2001) Identification and characterization of an inner ear-expressed human melanoma inhibitory activity (MIA)-like gene (MIAL) with a frequent polymorphism that abolishes translation. Genomics, 71, Verpy, E., Leibovici, M. and Petit, C. (1999) Characterization of otoconin- 95, the major protein of murine otoconia, provides insights into the formation of these inner ear biominerals. Proc. Natl Acad. Sci. USA, 96, Cohen-Salmon, M., Frenz, D., Liu, W., Verpy, E., Voegeling, S. and Petit, C. (2000) Fdp, a new fibrocyte-derived protein related to MIA/CD-RAP, has an in vitro effect on the early differentiation of the inner ear mesenchyme. J. Biol. Chem., 275, Avraham, K.B., Hasson, T., Steel, K.P., Kingsley, D.M., Russell, L.B., Mooseker, M.S., Copeland, N.G. and Jenkins, N.A. (1995) The mouse Snell s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat. Genet., 11, Melchionda, S., Ahituv, N., Bisceglia, L., Sobe, T., Glaser, F., Rabionet, R., Arbones, M.L., Notarangelo, A., Di Iorio, E., Carella, M. et al. (2001) MYO6, the human homologue of the gene responsible for deafness in Snell s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am. J. Hum. Genet., 69, Vreugde, S., Erven, A., Kros, C.J., Marcotti, W., Fuchs, H., Kurima, K., Wilcox, E.R., Friedman, T.B., Griffith, A.J., Balling, R. et al. (2002) Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet., 30, Zheng, Q.Y., Johnson, K.R. and Erway, L.C. (1999) Assessment of hearing in 80 strains of mice by ABR threshold analyses. Hear. Res., 130, Nolan, P.M., Peters, J., Strivens, M., Rogers, D., Hagan, J., Spurr, N., Gray, I.C., Vizor, L., Brooker, D., Whitehill, E. et al. (2000) A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet., 25, Coghill, E.L., Hugill, A., Parkinson, N., Davison, C., Glenister, P., Clements, S., Hunter, J., Cox, R.D. and Brown, S.D. (2002) A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet., 19, Estivill, X., Fortina, P., Surrey, S., Rabionet, R., Melchionda, S., D Agruma, L., Mansfield, E., Rappaport, E., Govea, N., Mila, M. et al. (1998) Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet, 351, Kelley, P.M., Harris, D.J., Comer, B.C., Askew, J.W., Fowler, T., Smith, S.D. and Kimberling, W.J. (1998) Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am. J. Hum. Genet., 62, Lench, N., Houseman, M., Newton, V., Van Camp, G. and Mueller, R. (1998) Connexin-26 mutations in sporadic non-syndromal sensorineural deafness. Lancet, 351, Scott, D.A., Kraft, M.L., Carmi, R., Ramesh, A., Elbedour, K., Yairi, Y., Srisailapathy, C.R., Rosengren, S.S., Markham, A.F., Mueller, R.F. et al. (1998) Identification of mutations in the connexin 26 gene that cause autosomal recessive nonsyndromic hearing loss. Hum. Mutat., 11, Denoyelle, F., Marlin, S., Weil, D., Moatti, L., Chauvin, P., Garabedian, E.N. and Petit, C. (1999) Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: implications for genetic counselling. Lancet, 353, Murgia, A., Orzan, E., Polli, R., Martella, M., Vinanzi, C., Leonardi, E., Arslan, E. and Zacchello, F. (1999) Cx26 deafness: mutation analysis and clinical variability. J. Med. Genet., 36, Abe, S., Usami, S., Shinkawa, H., Kelley, P.M. and Kimberling, W.J. (2000) Prevalent connexin 26 gene (GJB2) mutations in Japanese. J. Med. Genet., 37, Rabionet, R., Zelante, L., Lopez-Bigas, N., D Agruma, L., Melchionda, S., Restagno, G., Arbones, M.L., Gasparini, P. and Estivill, X. (2000) Molecular basis of childhood deafness resulting from mutations in the GJB2 (connexin 26) gene. Hum. Genet., 106, Sobe, T., Vreugde, S., Shahin, H., Berlin, M., Davis, N., Kanaan, M., Yaron, Y., Orr-Urtreger, A., Frydman, M., Shohat, M. et al. (2000) The prevalence and expression of inherited connexin 26 mutations associated with nonsyndromic hearing loss in the Israeli population. Hum. Genet., 106, Wilcox, S.A., Saunders, K., Osborn, A.H., Arnold, A., Wunderlich, J., Kelly, T., Collins, V., Wilcox, L.J., McKinlay Gardner, R.J., Kamarinos, M. et al. (2000) High frequency hearing loss correlated with mutations in the GJB2 gene. Hum. Genet., 106,
9 Human Molecular Genetics, 2002, Vol. 11, No Gabriel, H., Kupsch, P., Sudendey, J., Winterhager, E., Jahnke, K. and Lautermann, J. (2001) Mutations in the connexin26/gjb2 gene are the most common event in non- syndromic hearing loss among the German population. Hum. Mutat., 17, Loffler, J., Nekahm, D., Hirst-Stadlmann, A., Gunther, B., Menzel, H.J., Utermann, G. and Janecke, A.R. (2001) Sensorineural hearing loss and the incidence of Cx26 mutations in Austria. Eur. J. Hum. Genet., 9, Grifa, A., Wagner, C.A., D Ambrosio, L., Melchionda, S., Bernardi, F., Lopez-Bigas, N., Rabionet, R., Arbones, M., Monica, M.D., Estivill, X. et al. (1999) Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nat. Genet., 23, Liu, X.Z., Xia, X.J., Adams, J., Chen, Z.Y., Welch, K.O., Tekin, M., Ouyang, X.M., Kristiansen, A., Pandya, A., Balkany, T. et al. (2001) Mutations in GJA1 (connexin 43) are associated with non-syndromic autosomal recessive deafness. Hum. Mol. Genet., 10, del Castillo, I., Villamar, M., Moreno-Pelayo, M.A., del Castillo, F.J., Alvarez, A., Telleria, D., Menendez, I. and Moreno, F. (2002) A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. New Engl. J. Med., 346, Vahava, O., Morell, R., Lynch, E.D., Weiss, S., Kagan, M.E., Ahituv, N., Morrow, J.E., Lee, M.K., Skvorak, A.B., Morton, C.C. et al. (1998) Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science, 279, Ikeda, A., Zheng, Q.Y., Rosenstiel, P., Maddatu, T., Zuberi, A.R., Roopenian, D.C., North, M.A., Naggert, J.K., Johnson, K.R. and Nishina, P.M. (1999) Genetic modification of hearing in tubby mice: evidence for the existence of a major gene (moth1) which protects tubby mice from hearing loss. Hum. Mol. Genet., 8, Noben-Trauth, K., Zheng, Q.Y., Johnson, K.R. and Nishina, P.M. (1997) mdfw: a deafness susceptibility locus that interacts with deaf waddler (dfw). Genomics, 44, Riazuddin, S., Castelein, C.M., Ahmed, Z.M., Lalwani, A.K., Mastroianni, M.A., Naz, S., Smith, T.N., Liburd, N.A., Friedman, T.B., Griffith, A.J. et al. (2000) Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat. Genet., 26, Fischel-Ghodsian, N. (1999) Mitochondrial deafness mutations reviewed. Hum. Mutat., 13, Prezant, T.R., Agapian, J.V., Bohlman, M.C., Bu, X., Oztas, S., Qiu, W.Q., Arnos, K.S., Cortopassi, G.A., Jaber, L., Rotter, J.I. et al. (1993) Mitochondrial ribosomal RNA mutation associated with both antibioticinduced and non-syndromic deafness. Nat. Genet., 4, Guan, M.X., Fischel-Ghodsian, N. and Attardi, G. (2001) Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rrna mutation. Hum. Mol. Genet., 10, Sykes, B., Ogilvie, D., Wordsworth, P., Anderson and Jones, N. (1986) Osteogenesis imperfecta is linked to both type I collagen structural genes. Lancet, 2, Goto, Y., Nonaka, I. and Horai, S. (1990) A mutation in the trna(leu)(uur) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature, 348, van den Ouweland, J.M.W., Lemkes, H.H.P.J., Ruitenbeek, W., Sandkuijl, L.A., de Vijlder, M.F., Struyvenberg, P.A.A., van de Kamp, J.J.P. and Maassen, J.A. (1992) Mutation in mitochondrial trna Leu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet., 1, Shoffner, J.M., Lott, M.T., Lezza, A.M., Seibel, P., Ballinger, S.W. and Wallace, D.C. (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA trna(lys) mutation. Cell, 61, Hoth, C.F., Milunsky, A., Lipsky, N., Sheffer, R., Clarren, S.K. and Baldwin, C.T. (1993) Mutations in the paired domain of the human PAX3 gene cause Klein- Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I). Am. J. Hum. Genet., 52, Asher, J.H., Jr., Sommer, A., Morell, R. and Friedman, T.B. (1996) Missense mutation in the paired domain of PAX3 causes craniofacialdeafness-hand syndrome. Hum. Mutat., 7, Berger, W., van de Pol, D., Warburg, M., Gal, A., Bleeker-Wagemakers, L., de Silva, H., Meindl, A., Meitinger, T., Cremers, F. and Ropers, H.H. (1992) Mutations in the candidate gene for Norrie disease. Hum. Mol. Genet., 1, Reid, F.M., Vernham, G.A. and Jacobs, H.T. (1994) A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum. Mutat., 3, Tiranti, V., Chariot, P., Carella, F., Toscano, A., Soliveri, P., Girlanda, P., Carrara, F., Fratta, G.M., Reid, F.M., Mariotti, C. et al. (1995) Maternally inherited hearing loss, ataxia and myoclonus associated with a novel point mutation in mitochondrial trnaser(ucn) gene. Hum. Mol. Genet., 4, Sevior, K.B., Hatamochi, A., Stewart, I.A., Bykhovskaya, Y., Allen-Powell, D.R., Fischel-Ghodsian, N. and Maw, M.A. (1998) Mitochondrial A7445G mutation in two pedigrees with palmoplantar keratoderma and deafness. Am. J. Med. Genet., 75, Smith, S.D., Kelley, P.M., Kenyon, J.B. and Hoover, D. (2000) Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J. Med. Genet., 37, Foster, J.W., Dominguez-Steglich, M.A., Guioli, S., Kowk, G., Weller, P.A., Stevanovic, M., Weissenbach, J., Mansour, S., Young, I.D., Goodfellow, P.N. et al. (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature, 372, Vikkula, M., Mariman, E.C., Lui, V.C., Zhidkova, N.I., Tiller, G.E., Goldring, M.B., van Beersum, S.E., de Waal Malefijt, M.C., van den Hoogen, F.H., Ropers, H.H. et al. (1995) Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell, 80, McGuirt, W.T., Prasad, S.D., Griffith, A.J., Kunst, H.P., Green, G.E., Shpargel, K.B., Runge, C., Huybrechts, C., Mueller, R.F., Lynch, E. et al. (1999) Mutations in COL11A2 cause non-syndromic hearing loss (DFNA13). Nat. Genet., 23, de Kok, Y.J.M., van der Maarel, S.M., Bitner-Glindzicz, M., Huber, I., Monaco, A.P., Malcolm, S., Pembrey, M.E., Ropers, H.-H. and Cremers, F.P.M. (1995) Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science, 267, Liu, X.Z., Hope, C., Walsh, J., Newton, V., Ke, X.M., Liang, C.Y., Xu, L.R., Zhou, J.M., Trump, D., Steel, K.P. et al. (1998) Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am. J. Hum. Genet., 63, Hao, H., Bonilla, E., Manfredi, G., DiMauro, S. and Moraes, C.T. (1995) Segregation patterns of a novel mutation in the mitochondrial trna glutamic acid gene associated with myopathy and diabetes mellitus. Am. J. Hum. Genet., 56, Attie, T., Till, M., Pelet, A., Amiel, J., Edery, P., Boutrand, L., Munnich, A. and Lyonnet, S. (1995) Mutation of the endothelin-receptor B gene in Waardenburg-Hirschsprung disease. Hum. Mol. Genet., 4, Edery, P., Attie, T., Amiel, J., Pelet, A., Eng, C., Hofstra, R.M., Martelli, H., Bidaud, C., Munnich, A. and Lyonnet, S. (1996) Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah- Waardenburg syndrome). Nat. Genet., 12, Williams, C.J., Ganguly, A., Considine, E., McCarron, S., Prockop, D.J., Walsh-Vockley, C. and Michels, V.V. (1996) A-2 > G transition at the 3 0 acceptor splice site of IVS17 characterizes the COL2A1 gene mutation in the original Stickler syndrome kindred. Am. J. Med. Genet., 63, Richards, A.J., Yates, J.R., Williams, R., Payne, S.J., Pope, F.M., Scott, J.D. and Snead, M.P. (1996) A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum. Mol. Genet., 5, Abdelhak, S., Kalatzis, V., Heilig, R., Compain, S., Samson, D., Vincent, C., Weil, D., Cruaud, C., Sahly, I., Leibovici, M. et al. (1997) A human homologue of the Drosophila eyes absent gene underlies Branchio-Oto-Renal (BOR) syndrome and identifies a novel gene family. Nat. Genet., 15, Neyroud, N., Tesson, F., Denjoy, I., Leibovici, M., Donger, C., Barhanin, J., Fauré, S., Gary, F., Coumel, P., Petit, C. et al. (1997) A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange- Nielsen cardioauditory syndrome. Nat. Genet., 15, Lynch, E.D., Lee, M.K., Morrow, J.E., Welcsh, P.L., Leon, P.E. and King, M.-C. (1997) Non-syndromic deafness DFNA1 is associated with mutation in the human homolog of Drosophila diaphanous, a profilin ligand and target of Rho that regulates actin polymerization. Science, 278, Tyson, J., Tranebjaerg, L., Bellman, S., Wren, C., Taylor, J.F., Bathen, J., Aslaksen, B., Sorland, S.J., Lund, O., Malcolm, S. et al. (1997) IsK and KvLQT1: mutation in either of the two subunits of the slow component of the delayed rectifier potassium channel can cause Jervell and Lange- Nielsen syndrome. Hum. Mol. Genet., 6,
10 1238 Human Molecular Genetics, 2002, Vol. 11, No Schulze-Bahr, E., Wang, Q., Wedekind, H., Haverkamp, W., Chen, Q., Sun, Y., Rubie, C., Hördt, M., Towbin, J.A., Borggrefe, M. et al. (1997) KCNE1 mutations cause Jervell and Lange-Nielsen syndrome. Nat. Genet., 17, Li, X.C., Everett, L.A., Lalwani, A.K., Desmukh, D., Friedman, T.B., Green, E.D. and Wilcox, E.R. (1998) A mutation in PDS causes non-syndromic recessive deafness. Nat. Genet., 18, Pingault, V., Bondurand, N., Kuhlbrodt, K., Goerich, D.E., Prehu, M.O., Puliti, A., Herbarth, B., Hermans-Borgmeyer, I., Legius, E., Matthijs, G. et al. (1998) SOX10 mutations in patients with Waardenburg- Hirschsprung disease. Nat. Genet., 18, Mustapha, M., Weil, D., Chardenoux, S., Elias, S., El-Zir, E., Beckmann, J.S., Loiselet, J. and Petit, C. (1999) An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum. Mol. Genet., 8, Wang, A., Liang, Y., Fridell, R.A., Probst, F.J., Wilcox, E.R., Touchman, J.W., Morton, C.C., Morell, R.J., Noben-Trauth, K., Camper, S.A. et al. (1998) Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science, 280, Karet, F.E., Finberg, K.E., Nelson, R.D., Nayir, A., Mocan, H., Sanjad, S.A., Rodriguez-Soriano, J., Santos, F., Cremers, C.W., Di Pietro, A. et al. (1999) Mutations in the gene encoding B1 subunit of H þ - ATPase cause renal tubular acidosis with sensorineural deafness. [see comments]. Nat. Genet., 21, Kubisch, C., Schroeder, B.C., Friedrich, T., Lutjohann, B., El-Amraoui, A., Marlin, S., Petit, C. and Jentsch, T.J. (1999) KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell, 96, Mansergh, F.C., Millington-Ward, S., Kennan, A., Kiang, A.S., Humphries, M., Farrar, G.J., Humphries, P. and Kenna, P.F. (1999) Retinitis pigmentosa and progressive sensorineural hearing loss caused by a C12258A mutation in the mitochondrial MTTS2 gene. Am. J. Hum. Genet., 64, Kovach, M.J., Lin, J.P., Boyadjiev, S., Campbell, K., Mazzeo, L., Herman, K., Rimer, L.A., Frank, W., Llewellyn, B., Wang Jabs, E. et al. (1999) A unique point mutation in the PMP22 gene is associated with Charcot- Marie-Tooth disease and deafness. Am. J. Hum. Genet., 64, Labay, V., Raz, T., Baron, D., Mandel, H., Williams, H., Barrett, T., Szargel, R., McDonald, L., Shalata, A., Nosaka, K. et al. (1999) Mutations in SLC19A2 cause thiamine-responsive megaloblastic anaemia associated with diabetes mellitus and deafness. Nat. Genet., 22, Fleming, J.C., Tartaglini, E., Steinkamp, M.P., Schorderet, D.F., Cohen, N. and Neufeld, E.J. (1999) The gene mutated in thiamine-responsive anaemia with diabetes and deafness (TRMA) encodes a functional thiamine transporter. Nat. Genet., 22, Bitner-Glindzicz, M., Lindley, K.J., Rutland, P., Blaydon, D., Smith, V.V., Milla, P.J., Hussain, K., Furth-Lavi, J., Cosgrove, K.E., Shepherd, R.M. et al. (2000) A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat. Genet., 26, Seri, M., Cusano, R., Gangarossa, S., Caridi, G., Bordo, D., Lo Nigro, C., Ghiggeri, G.M., Ravazzolo, R., Savino, M., Del Vecchio, M. et al. (2000) Mutations in MYH9 result in the May-Hegglin anomaly, and Fechtner and Sebastian syndromes. The May-Heggllin/Fechtner Syndrome Consortium. Nat. Genet., 26, Lalwani, A.K., Goldstein, J.A., Kelley, M.J., Luxford, W., Castelein, C.M. and Mhatre, A.N. (2000) Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am. J. Hum. Genet., 67, Wilcox, E.R., Burton, Q.L., Naz, S., Riazuddin, S., Smith, T.N., Ploplis, B., Belyantseva, I., Ben-Yosef, T., Liburd, N.A., Morell, R.J. et al. (2001) Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell, 104, Scott, H.S., Kudoh, J., Wattenhofer, M., Shibuya, K., Berry, A., Chrast, R., Guipponi, M., Wang, J., Kawasaki, K., Asakawa, S. et al. (2001) Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet., 27, Wayne, S., Robertson, N.G., DeClau, F., Chen, N., Verhoeven, K., Prasad, S., Tranebjarg, L., Morton, C.C., Ryan, A.F., Van Camp, G. et al. (2001) Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet., 10, Xiao, S., Yu, C., Chou, X., Yuan, W., Wang, Y., Bu, L., Fu, G., Qian, M., Yang, J., Shi, Y. et al. (2001) Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet., 27, Ahmed, Z.M., Riazuddin, S., Bernstein, S.L., Ahmed, Z., Khan, S., Griffith, A.J., Morell, R.J., Friedman, T.B. and Wilcox, E.R. (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet., 69, Alagramam, K.N., Yuan, H., Kuehn, M.H., Murcia, C.L., Wayne, S., Srisailpathy, C.R., Lowry, R.B., Knaus, R., Van Laer, L., Bernier, F.P. et al. (2001) Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum. Mol. Genet., 10, Joensuu, T., Hamalainen, R., Yuan, B., Johnson, C., Tegelberg, S., Gasparini, P., Zelante, L., Pirvola, U., Pakarinen, L., Lehesjoki, A.E. et al. (2001) Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am. J. Hum. Genet., 69, Inoue, H., Tanizawa, Y., Wasson, J., Behn, P., Kalidas, K., Bernal-Mizrachi, E., Mueckler, M., Marshall, H., Donis-Keller, H., Crock, P. et al. (1998) A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat. Genet., 20, Strom, T.M., Hortnagel, K., Hofmann, S., Gekeler, F., Scharfe, C., Rabl, W., Gerbitz, K.D. and Meitinger, T. (1998) Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum. Mol. Genet., 7, Bespalova, I.N., Van Camp, G., Bom, S.J., Brown, D.J., Cryns, K., DeWan, A.T., Erson, A.E., Flothmann, K., Kunst, H.P., Kurnool, P. et al. (2001) Mutations in the Wolfram syndrome 1 gene (WFS1) are a common cause of low frequency sensorineural hearing loss. Hum. Mol. Genet., 10, Young, T.L., Ives, E., Lynch, E., Person, R., Snook, S., MacLaren, L., Cator, T., Griffin, A., Fernandez, B., Lee, M.K. et al. (2001) Nonsyndromic progressive hearing loss DFNA38 is caused by heterozygous missense mutation in the Wolfram syndrome gene WFS1. Hum. Mol. Genet., 10, Birkenhager, R., Otto, E., Schurmann, M.J., Vollmer, M., Ruf, E.M., Maier-Lutz, I., Beekmann, F., Fekete, A., Omran, H., Feldmann, D. et al. (2001) Mutation of BSND causes Bartter syndrome with sensorineural deafness and kidney failure. Nat. Genet., 29, Alagramam, K.N., Murcia, C.L., Kwon, H.Y., Pawlowski, K.S., Wright, C.G. and Woychik, R.P. (2001) The mouse Ames waltzer hearingloss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet., 27, Cho, H., Yamada, Y. and Yoo, T.J. (1991) Ultrastructural changes of cochlea in mice with hereditary chondrodysplasia (cho/cho). Ann. N. Y. Acad. Sci., 630, Li, Y., Lacerda, D.A., Warman, M.L., Beier, D.R., Yoshioka, H., Ninomiya, Y., Oxford, J.T., Morris, N.P., Andrikopoulos, K., Ramirez, F. et al. (1995) A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell, 80, Barsh, G.S., David, K.E. and Byers, P.H. (1982) Type I osteogenesis imperfecta: a nonfunctional allele for pro alpha 1 (I) chains of type I procollagen. Proc. Natl Acad. Sci. USA, 79, Shapiro, J.R., Pikus, A., Weiss, G. and Rowe, D.W. (1982) Hearing and middle ear function in osteogenesis imperfecta. JAMA, 247, Bonadio, J., Saunders, T.L., Tsai, E., Goldstein, S.A., Morris-Wiman, J., Brinkley, L., Dolan, D.F., Altschuler, R.A., Hawkins, J.E., Jr., Bateman, J.F. et al. (1990) Transgenic mouse model of the mild dominant form of osteogenesis imperfecta. Proc. Natl Acad. Sci. USA, 87, Altschuler, R.A., Dolan, D.F., Ptok, M., Gholizadeh, G., Bonadio, J. and Hawkins, J.E. (1991) An evaluation of otopathology in the MOV-13 transgenic mutant mouse. Ann. N. Y. Acad. Sci., 630, Cosgrove, D., Samuelson, G., Meehan, D.T., Miller, C., McGee, J., Walsh, E.J. and Siegel, M. (1998) Ultrastructural, physiological, and molecular defects in the inner ear of a gene-knockout mouse model for autosomal Alport syndrome. Hear. Res., 121, Berggren, D., Frenz, D., Galinovic-Schwartz, V. and Van de Water, T.R. (1997) Fine structure of extracellular matrix and basal laminae in two types of abnormal collagen production: L-proline analog-treated otocyst
11 Human Molecular Genetics, 2002, Vol. 11, No cultures and disproportionate micromelia (Dmm/Dmm) mutants. Hear. Res., 107, Maddox, B.K., Garofalo, S., Horton, W.A., Richardson, M.D. and Trune, D.R. (1998) Craniofacial and otic capsule abnormalities in a transgenic mouse strain with a Col2a1 mutation. J. Craniofac. Genet. Dev. Biol., 18, Herbarth, B., Pingault, V., Bondurand, N., Kuhlbrodt, K., Hermans- Borgmeyer, I., Puliti, A., Lemort, N., Goossens, M. and Wegner, M. (1998) Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc. Natl Acad. Sci. USA, 95, Southard-Smith, E.M., Kos, L. and Pavan, W.J. (1998) Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet., 18, Geissler, E.N., Ryan, M.A. and Housman, D.E. (1988) The dominantwhite spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell, 55, Steel, K.P. and Barkway, C. (1989) Another role for melanocytes: their importance for normal stria vascularis development in the mammalian inner ear. Development, 107, Giebel, L.B. and Spritz, R.A. (1991) Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc. Natl Acad. Sci. USA, 88, Johnson, K.R., Cook, S.A., Erway, L.C., Matthews, A.N., Sanford, L.P., Paradies, N.E. and Friedman, R.A. (1999) Inner ear and kidney anomalies caused by IAP insertion in an intron of the Eya1 gene in a mouse model of BOR syndrome. Hum. Mol. Genet., 8, Xu, P.X., Adams, J., Peters, H., Brown, M.C., Heaney, S. and Maas, R. (1999) Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet., 23, Colvin, J.S., Bohne, B.A., Harding, G.W., McEwen, D.G. and Ornitz, D.M. (1996) Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat. Genet., 12, Hollway, G.E., Suthers, G.K., Battese, K.M., Turner, A.M., David, D.J. and Mulley, J.C. (1998) Deafness due to Pro250Arg mutation of FGFR3. Lancet, 351, Van Esch, H., Groenen, P., Nesbit, M.A., Schuffenhauer, S., Lichtner, P., Vanderlinden, G., Harding, B., Beetz, R., Bilous, R.W., Holdaway, I. et al. (2000) GATA3 haplo-insufficiency causes human HDR syndrome. Nature, 406, Karis, A., Pata, I., van Doorninck, J.H., Grosveld, F., de Zeeuw, C.I., de Caprona, D. and Fritzsch, B. (2001) Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J. Comp. Neurol., 429, Vetter, D.E., Mann, J.R., Wangemann, P., Liu, J., McLaughlin, K.J., Lesage, F., Marcus, D.C., Lazdunski, M., Heinemann, S.F. and Barhanin, J. (1996) Inner ear defects induced by null mutation of the isk gene. Neuron, 17, Letts, V.A., Valenzuela, A., Dunbar, C., Zheng, Q.Y., Johnson, K.R. and Frankel, W.N. (2000) A new spontaneous mouse mutation in the Kcne1 gene. Mamm. Genome., 11, Lee, M.P., Hu, R.-J., Johnson, L.A. and Feinberg, A.P. (1997) Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat. Genet., 15, Casimiro, M.C., Knollmann, B.C., Ebert, S.N., Vary, J.C., Jr., Greene, A.E., Franz, M.R., Grinberg, A., Huang, S.P. and Pfeifer, K. (2001) Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange-Nielsen Syndrome. Proc. Natl Acad. Sci. USA, 98, Baynash, A.G., Hosoda, K., Giaid, A., Richardson, J.A., Emoto, N., Hammer, R.E. and Yanagisawa, M. (1994) Interaction of endothelin-3 with endothelin-b receptor is essential for development of epidermal melanocytes and enteric neurons. Cell, 79, Hodgkinson, C.A., Moore, K.J., Nakayama, A., Steingrimsson, E., Copeland, N.G., Jemkins, N.A. and Arnheiter, H. (1993) Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell, 74, Hughes, A.E., Newton, V.E., Liu, X.Z. and Read, A.P. (1994) A gene for Waardenburg syndrome type 2 maps close to the human homologue of the microphthalmia gene at chromosome 3p12-p14.1. Nat. Genet., 7, Berger, W., van de Pol, D., Bachner, D., Oerlemans, F., Winkens, H., Hameister, H., Wieringa, B., Hendriks, W. and Ropers, H.H. (1996) An animal model for Norrie disease (ND): gene targeting of the mouse ND gene. Hum. Mol. Genet., 5, Torres, M., Gomez-Pardo, E. and Gruss, P. (1996) Pax2 contributes to inner ear patterning and optic nerve trajectory. Development, 122, Eccles, M.R. and Schimmenti, L.A. (1999) Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clin. Genet., 56, Hosoda, K., Hammer, R.E., Richardson, J.A., Baynash, A.G., Cheung, J.C., Giaid, A. and Yanagisawa, M. (1994) Targeted and natural (piebald-lethal) mutations of endothelin-b receptor gene produce megacolon associated with spotted coat color in mice. Cell, 79, Minowa, O., Ikeda, K., Sugitani, Y., Oshima, T., Nakai, S., Katori, Y., Suzuki, M., Furukawa, M., Kawase, T., Zheng, Y. et al. (1999) Altered cochlear fibrocytes in a mouse model of DFN3 nonsyndromic deafness. Science, 285, Phippard, D., Lu, L., Lee, D., Saunders, J.C. and Crenshaw, E.B., 3rd (1999) Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. J. Neurosci., 19, Phippard, D., Boyd, Y., Reed, V., Fisher, G., Masson, W.K., Evans, E.P., Saunders, J.C. and Crenshaw, E.B., 3rd (2000) The sex-linked fidget mutation abolishes Brn4/Pou3f4 gene expression in the embryonic inner ear. Hum. Mol. Genet., 9, Erkman, L., McEvilly, R.J., Luo, L., Ryan, A.K., Hooshmand, F., O Connell, S.M., Keithley, E.M., Rapaport, D.H., Ryan, A.F. and Rosenfeld, M.G. (1996) Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature, 381, Xiang, M., Gan, L., Li, D., Chen, Z.Y., Zhou, L., O Malley, B.W., Jr., Klein, W. and Nathans, J. (1997) Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc. Natl Acad. Sci. USA, 94, Parkinson, N.J., Olsson, C.L., Hallows, J.L., McKee-Johnson, J., Keogh, B.P., Noben-Trauth, K., Kujawa, S.G. and Tempel, B.L. (2001) Mutant beta-spectrin 4 causes auditory and motor neuropathies in quivering mice. Nat. Genet., 29, Gibson, F., Walsh, J., Mburu, P., Varela, A., Brown, K.A., Antonio, M., Beisel, K.W., Steel, K.P. and Brown, S.D.M. (1995) A type VII myosin encoded by the mouse deafness gene shaker-1. Nature, 374, Liu, X.Z., Newton, V.E., Steel, K.P. and Brown, S.D. (1997) Identification of a new mutation of the myosin VII head region in Usher syndrome type 1. Hum. Mutat., 10, Probst, F.J., Fridell, R.A., Raphael, Y., Saunders, T.L., Wang, A., Liang, Y., Morell, R.J., Touchman, J.W., Lyons, R.H., Noben-Trauth, K. et al. (1998) Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science, 280, Everett, L.A., Belyantseva, I.A., Noben-Trauth, K., Cantos, R., Chen, A., Thakkar, S.I., Hoogstraten-Miller, S.L., Kachar, B., Wu, D.K. and Green, E.D. (2001) Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum. Mol. Genet., 10, Epstein, D.J., Vekemans, M. and Gros, P. (1991) splotch (Sp 2H ), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell, 67, Steel, K.P. and Smith, R.J. (1992) Normal hearing in Splotch (Sp/ þ ), the mouse homologue of Waardenburg syndrome type 1. Nat. Genet., 2, Legan, P.K., Lukashkina, V.A., Goodyear, R.J., Kossi, M., Russell, I.J. and Richardson, G.P. (2000) A targeted deletion in alpha-tectorin reveals that the tectorial membrane is required for the gain and timing of cochlear feedback. Neuron, 28, Sakurai, A., Takeda, K., Ain, K., Ceccarelli, P., Nakai, A., Seino, S., Bell, G.I., Refetoff, S. and DeGroot, L.J. (1989) Generalized resistance to thyroid hormone associated with a mutation in the ligand-binding domain of the human thyroid hormone receptor beta. Proc. Natl Acad. Sci. USA, 86, Forrest, D., Erway, L.C., Ng, L., Altschuler, R. and Curran, T. (1996) Thyroid hormone receptor beta is essential for development of auditory function. Nat. Genet., 13,
12 1240 Human Molecular Genetics, 2002, Vol. 11, No Suter, U., Welcher, A.A., Ozcelik, T., Snipes, G.J., Kosaras, B., Francke, U., Billings-Gagliardi, S., Sidman, R.L. and Shooter, E.M. (1992) Trembler mouse carries a point mutation in a myelin gene. Nature, 356, Zhou, R., Assouline, J.G., Abbas, P.J., Messing, A. and Gantz, B.J. (1995) Anatomical and physiological measures of auditory system in mice with peripheral myelin deficiency. Hear. Res., 88, Di Palma, F., Holme, R.H., Bryda, E.C., Belyantseva, I.A., Pellegrino, R., Kachar, B., Steel, K.P. and Noben-Trauth, K. (2001) Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat. Genet., 27,
Corporate Medical Policy Genetic Testing for Hereditary Hearing Loss
 Corporate Medical Policy Genetic Testing for Hereditary Hearing Loss File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_hereditary_hearing_loss 10/2013 8/2015 8/2016
Corporate Medical Policy Genetic Testing for Hereditary Hearing Loss File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_hereditary_hearing_loss 10/2013 8/2015 8/2016
The Genetics of Hearing Loss. Heidi L. Rehm, Ph.D. Harvard Medical School Boston, MA
 The Genetics of Hearing Loss Heidi L. Rehm, Ph.D. Harvard Medical School Boston, MA Faculty Disclosure Information In the past 12 months, I have not had a significant financial interest or other relationship
The Genetics of Hearing Loss Heidi L. Rehm, Ph.D. Harvard Medical School Boston, MA Faculty Disclosure Information In the past 12 months, I have not had a significant financial interest or other relationship
G. Shashidhar Pai, MD MUSC Children s Hospital Department of Pediatrics Division of Genetics
 G. Shashidhar Pai, MD MUSC Children s Hospital Department of Pediatrics Division of Genetics One of every 500 newborns has bilateral permanent sensorineural hearing loss 40 db which makes it the most common
G. Shashidhar Pai, MD MUSC Children s Hospital Department of Pediatrics Division of Genetics One of every 500 newborns has bilateral permanent sensorineural hearing loss 40 db which makes it the most common
Jennifer A. Defant, M.S., C.G.C. Certified Genetic Counselor Division of Genetics and Metabolism University of Florida
 Jennifer A. Defant, M.S., C.G.C. Certified Genetic Counselor Division of Genetics and Metabolism University of Florida 60% of childhood hearing loss is genetic Syndromic Nonsyndromic 40% of childhood hearing
Jennifer A. Defant, M.S., C.G.C. Certified Genetic Counselor Division of Genetics and Metabolism University of Florida 60% of childhood hearing loss is genetic Syndromic Nonsyndromic 40% of childhood hearing
Single Gene and NextGen Panels
 Molecular Testing for Hearing Loss: Single Gene and NextGen Panels Honey V Reddi, PhD, FACMG Clinical Molecular Geneticist Prevention Genetics, Marshfield, WI www.preventiongenetics.com Outline of Presentation
Molecular Testing for Hearing Loss: Single Gene and NextGen Panels Honey V Reddi, PhD, FACMG Clinical Molecular Geneticist Prevention Genetics, Marshfield, WI www.preventiongenetics.com Outline of Presentation
Title: Genetics and Hearing Loss: Clinical and Molecular Characteristics
 Session # : 46 Day/Time: Friday, May 1, 2015, 1:00 4:00 pm Title: Genetics and Hearing Loss: Clinical and Molecular Characteristics Presenter: Kathleen S. Arnos, PhD, Gallaudet University This presentation
Session # : 46 Day/Time: Friday, May 1, 2015, 1:00 4:00 pm Title: Genetics and Hearing Loss: Clinical and Molecular Characteristics Presenter: Kathleen S. Arnos, PhD, Gallaudet University This presentation
Victims Compensation Claim Status of All Pending Claims and Claims Decided Within the Last Three Years
 Claim#:021914-174 Initials: J.T. Last4SSN: 6996 DOB: 5/3/1970 Crime Date: 4/30/2013 Status: Claim is currently under review. Decision expected within 7 days Claim#:041715-334 Initials: M.S. Last4SSN: 2957
Claim#:021914-174 Initials: J.T. Last4SSN: 6996 DOB: 5/3/1970 Crime Date: 4/30/2013 Status: Claim is currently under review. Decision expected within 7 days Claim#:041715-334 Initials: M.S. Last4SSN: 2957
Goals for Today. Clinical and Molecular Aspects of Genetic Hearing Loss. Case presentation. Case presentation continued. Case presentation continued
 Clinical and Molecular Aspects of Genetic Hearing Loss Kathleen Arnos, PhD Department of Science, Technology, & Mathematics Gallaudet University Washington, DC 20002 kathleen.arnos@gallaudet.edu Goals
Clinical and Molecular Aspects of Genetic Hearing Loss Kathleen Arnos, PhD Department of Science, Technology, & Mathematics Gallaudet University Washington, DC 20002 kathleen.arnos@gallaudet.edu Goals
Objectives Role of Medical Genetics in Hearing Loss Evaluation. 5 y.o. boy with severe SNHL
 Objectives Role of Medical Genetics in Hearing Loss Evaluation Millan Patel, MD UBC Dept. of Medical Genetics October 22, 2010 Case presentation to illustrate importance of defining syndromic hearing loss
Objectives Role of Medical Genetics in Hearing Loss Evaluation Millan Patel, MD UBC Dept. of Medical Genetics October 22, 2010 Case presentation to illustrate importance of defining syndromic hearing loss
Genetic Testing for Hereditary Hearing Loss
 MEDICAL POLICY POLICY RELATED POLICIES POLICY GUIDELINES DESCRIPTION SCOPE BENEFIT APPLICATION RATIONALE REFERENCES CODING APPENDIX HISTORY Genetic Testing for Hereditary Hearing Loss Number 12.04.87 Effective
MEDICAL POLICY POLICY RELATED POLICIES POLICY GUIDELINES DESCRIPTION SCOPE BENEFIT APPLICATION RATIONALE REFERENCES CODING APPENDIX HISTORY Genetic Testing for Hereditary Hearing Loss Number 12.04.87 Effective
A Common Genetic Disorder Deafness
 A Common Genetic Disorder Deafness Shahid Hussain Department of Bioinformatics, Mohammad Ali Jinnah University Islamabad, Pakistan Abstract: Deafness is a complete or partial loss of hearing, caused due
A Common Genetic Disorder Deafness Shahid Hussain Department of Bioinformatics, Mohammad Ali Jinnah University Islamabad, Pakistan Abstract: Deafness is a complete or partial loss of hearing, caused due
Genetics of Hearing Loss
 2 Genetics of Hearing Loss Ella Shalit and Karen B. Avraham 1. Introduction The revolution in genetics in the past decades has enabled identification of many of the genes associated with human hereditary
2 Genetics of Hearing Loss Ella Shalit and Karen B. Avraham 1. Introduction The revolution in genetics in the past decades has enabled identification of many of the genes associated with human hereditary
Genetic Testing for Hearing Loss
 Genetic Testing for Hearing Loss S H O B A N A K U B E N D R A N, M B B S, M S, C G C G E N E T I C C O U N S E L O R A S S T P R O F E S S O R D E P T O F P E D I A T R I C S K U S M - W Objectives Indications
Genetic Testing for Hearing Loss S H O B A N A K U B E N D R A N, M B B S, M S, C G C G E N E T I C C O U N S E L O R A S S T P R O F E S S O R D E P T O F P E D I A T R I C S K U S M - W Objectives Indications
Patient Information. for Childhood
 Patient Information Genetic Testing for Childhood Hearing Loss Introduction This document describes the most common genetic cause of childhood hearing loss and explains the role of genetic testing. Childhood
Patient Information Genetic Testing for Childhood Hearing Loss Introduction This document describes the most common genetic cause of childhood hearing loss and explains the role of genetic testing. Childhood
Cochlear Implantation for Children With GJB2-Related Deafness
 The Laryngoscope Lippincott Williams & Wilkins, Inc. 2004 The American Laryngological, Rhinological and Otological Society, Inc. Cochlear Implantation for Children With GJB2-Related Deafness Robert D.
The Laryngoscope Lippincott Williams & Wilkins, Inc. 2004 The American Laryngological, Rhinological and Otological Society, Inc. Cochlear Implantation for Children With GJB2-Related Deafness Robert D.
About The Causes of Hearing Loss
 About 1 in 500 infants is born with or develops hearing loss during early childhood. Hearing loss has many causes: some are genetic (that is, caused by a baby s genes) or non-genetic (such as certain infections
About 1 in 500 infants is born with or develops hearing loss during early childhood. Hearing loss has many causes: some are genetic (that is, caused by a baby s genes) or non-genetic (such as certain infections
Genetic Bases of Hearing Loss: Future Treatment Implications
 Genetic Bases of Hearing Loss: Future Treatment Implications Luis F. Escobar, MD Medical Director Medical Genetics & Neurodevelopmental Pediatrics of Indiana Peyton Manning Children s Hospital St. Vincent
Genetic Bases of Hearing Loss: Future Treatment Implications Luis F. Escobar, MD Medical Director Medical Genetics & Neurodevelopmental Pediatrics of Indiana Peyton Manning Children s Hospital St. Vincent
Genetic Testing for Hereditary Hearing Loss. Populations Interventions Comparators Outcomes Interventions of interest are: Genetic testing
 Protocol Genetic Testing for Hereditary Hearing Loss (20487) Medical Benefit Effective Date: 01/01/15 Next Review Date: 11/16 Preauthorization Yes Review Dates: 01/14, 11/14, 11/15 Preauthorization is
Protocol Genetic Testing for Hereditary Hearing Loss (20487) Medical Benefit Effective Date: 01/01/15 Next Review Date: 11/16 Preauthorization Yes Review Dates: 01/14, 11/14, 11/15 Preauthorization is
Chromosomes, Mapping, and the Meiosis Inheritance Connection
 Chromosomes, Mapping, and the Meiosis Inheritance Connection Carl Correns 1900 Chapter 13 First suggests central role for chromosomes Rediscovery of Mendel s work Walter Sutton 1902 Chromosomal theory
Chromosomes, Mapping, and the Meiosis Inheritance Connection Carl Correns 1900 Chapter 13 First suggests central role for chromosomes Rediscovery of Mendel s work Walter Sutton 1902 Chromosomal theory
Supervisors Prof.Dr.Ahmad Sameh Farid Professor of ENT and Audiology Faculty of Medicine Cairo University
 Connexin 26 gene in non-syndromic hearing loss Thesis Submitted for partial fulfillment of MD degree in Audiology by Mona Ahmed El-Akkad M.B, B.CH., MSc. Supervisors Prof.Dr.Ahmad Sameh Farid Professor
Connexin 26 gene in non-syndromic hearing loss Thesis Submitted for partial fulfillment of MD degree in Audiology by Mona Ahmed El-Akkad M.B, B.CH., MSc. Supervisors Prof.Dr.Ahmad Sameh Farid Professor
CONGENITAL HEARING LOSS
 CONGENITAL HEARING LOSS Congenital hearing loss, for the purposes of this fact sheet, is defined as permanent and is bilateral or unilateral, is sensory or conductive, and averages 30 db or more in the
CONGENITAL HEARING LOSS Congenital hearing loss, for the purposes of this fact sheet, is defined as permanent and is bilateral or unilateral, is sensory or conductive, and averages 30 db or more in the
Deafness and Hereditary Hearing Loss Overview
 Page 1 of 25 NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health. Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviews [Internet]. Seattle (WA): University
Page 1 of 25 NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health. Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviews [Internet]. Seattle (WA): University
Just the Facts: A Basic Introduction to the Science Underlying NCBI Resources
 1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
1 of 8 11/7/2004 11:00 AM National Center for Biotechnology Information About NCBI NCBI at a Glance A Science Primer Human Genome Resources Model Organisms Guide Outreach and Education Databases and Tools
Medical Policy Manual. Date of Origin: October 2014. Topic: Genetic Testing for Hereditary Hearing Loss. Last Reviewed Date: December 2015
 Medical Policy Manual Topic: Genetic Testing for Hereditary Hearing Loss Section: Genetic Testing Policy No: 36 Date of Origin: October 2014 Last Reviewed Date: December 2015 Effective Date: February 1,
Medical Policy Manual Topic: Genetic Testing for Hereditary Hearing Loss Section: Genetic Testing Policy No: 36 Date of Origin: October 2014 Last Reviewed Date: December 2015 Effective Date: February 1,
DURATION OF HEARING LOSS
 When your child is diagnosed with a hearing loss, it may be very overwhelming. This may be a difficult time for you and your family. However, gaining a greater knowledge in this area is crucial in helping
When your child is diagnosed with a hearing loss, it may be very overwhelming. This may be a difficult time for you and your family. However, gaining a greater knowledge in this area is crucial in helping
Gene mutation and molecular medicine Chapter 15
 Gene mutation and molecular medicine Chapter 15 Lecture Objectives What Are Mutations? How Are DNA Molecules and Mutations Analyzed? How Do Defective Proteins Lead to Diseases? What DNA Changes Lead to
Gene mutation and molecular medicine Chapter 15 Lecture Objectives What Are Mutations? How Are DNA Molecules and Mutations Analyzed? How Do Defective Proteins Lead to Diseases? What DNA Changes Lead to
Mouse Models for Deafness: Lessons for the Human Inner Ear and Hearing Loss
 Mouse Models for Deafness: Lessons for the Human Inner Ear and Hearing Loss Karen B. Avraham In the field of hearing research, recent advances using the mouse as a model for human hearing loss have brought
Mouse Models for Deafness: Lessons for the Human Inner Ear and Hearing Loss Karen B. Avraham In the field of hearing research, recent advances using the mouse as a model for human hearing loss have brought
Genetic Mutations Cause Many Birth Defects:
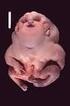 Genetic Mutations Cause Many Birth Defects: What We Learned from the FORGE Canada Project Jan M. Friedman, MD, PhD University it of British Columbia Vancouver, Canada I have no conflicts of interest related
Genetic Mutations Cause Many Birth Defects: What We Learned from the FORGE Canada Project Jan M. Friedman, MD, PhD University it of British Columbia Vancouver, Canada I have no conflicts of interest related
Sickle cell anemia: Altered beta chain Single AA change (#6 Glu to Val) Consequence: Protein polymerizes Change in RBC shape ---> phenotypes
 Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Mendelian inheritance and the
 Mendelian inheritance and the most common genetic diseases Cornelia Schubert, MD, University of Goettingen, Dept. Human Genetics EUPRIM-Net course Genetics, Immunology and Breeding Mangement German Primate
Mendelian inheritance and the most common genetic diseases Cornelia Schubert, MD, University of Goettingen, Dept. Human Genetics EUPRIM-Net course Genetics, Immunology and Breeding Mangement German Primate
Connexin 26 and autosomal recessive non-syndromic hearing loss
 40 Review Article Connexin 26 and autosomal recessive non-syndromic hearing loss Monisha Mukherjee, S. R. Phadke, B. Mittal Department of Medical Genetics, Sanja y Gandhi Postgraduate Institute of Medical
40 Review Article Connexin 26 and autosomal recessive non-syndromic hearing loss Monisha Mukherjee, S. R. Phadke, B. Mittal Department of Medical Genetics, Sanja y Gandhi Postgraduate Institute of Medical
Hearing loss is an etiologically heterogeneous trait with
 The new england journal of medicine review article Current Concepts Newborn Hearing Screening A Silent Revolution Cynthia C. Morton, Ph.D., and Walter E. Nance, M.D., Ph.D. Hearing loss is an etiologically
The new england journal of medicine review article Current Concepts Newborn Hearing Screening A Silent Revolution Cynthia C. Morton, Ph.D., and Walter E. Nance, M.D., Ph.D. Hearing loss is an etiologically
BEST PRACTICES Pediatric Bilateral Sensorineural Hearing Loss
 BEST PRACTICES Pediatric Bilateral Sensorineural Hearing Loss Christina L. Runge, PhD, CCC-A Associate Professor Chief, Division of Communication Sciences Director, Koss Cochlear Implant Program Resources
BEST PRACTICES Pediatric Bilateral Sensorineural Hearing Loss Christina L. Runge, PhD, CCC-A Associate Professor Chief, Division of Communication Sciences Director, Koss Cochlear Implant Program Resources
Biological Sciences Initiative. Human Genome
 Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
Biological Sciences Initiative HHMI Human Genome Introduction In 2000, researchers from around the world published a draft sequence of the entire genome. 20 labs from 6 countries worked on the sequence.
Understanding the Genetics of Deafness
 Understanding the Genetics of Deafness A Guide for Patients and Families Harvard Medical School Center For Hereditary Deafness Understanding the Genetics of Deafness Understanding the Genetics of Deafness
Understanding the Genetics of Deafness A Guide for Patients and Families Harvard Medical School Center For Hereditary Deafness Understanding the Genetics of Deafness Understanding the Genetics of Deafness
The correct answer is c A. Answer a is incorrect. The white-eye gene must be recessive since heterozygous females have red eyes.
 1. Why is the white-eye phenotype always observed in males carrying the white-eye allele? a. Because the trait is dominant b. Because the trait is recessive c. Because the allele is located on the X chromosome
1. Why is the white-eye phenotype always observed in males carrying the white-eye allele? a. Because the trait is dominant b. Because the trait is recessive c. Because the allele is located on the X chromosome
Medical Policy Genetic Testing for Hereditary Hearing Loss
 Medical Policy Genetic Testing for Hereditary Hearing Loss Table of Contents Policy: Commercial Coding Information Information Pertaining to All Policies Policy: Medicare Description References Authorization
Medical Policy Genetic Testing for Hereditary Hearing Loss Table of Contents Policy: Commercial Coding Information Information Pertaining to All Policies Policy: Medicare Description References Authorization
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs)
 Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Lecture 6: Single nucleotide polymorphisms (SNPs) and Restriction Fragment Length Polymorphisms (RFLPs) Single nucleotide polymorphisms or SNPs (pronounced "snips") are DNA sequence variations that occur
Mouse models to study inner ear development and hereditary hearing loss
 Int. J. Dev. Biol. 51: 609-631 (2007) doi: 10.1387/ijdb.072365lf Mouse models to study inner ear development and hereditary hearing loss LILACH M. FRIEDMAN, AMIEL A. DROR and KAREN B. AVRAHAM* Department
Int. J. Dev. Biol. 51: 609-631 (2007) doi: 10.1387/ijdb.072365lf Mouse models to study inner ear development and hereditary hearing loss LILACH M. FRIEDMAN, AMIEL A. DROR and KAREN B. AVRAHAM* Department
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
The Genomics of Hearing Loss
 The Genomics of Hearing Loss Ian Krantz, M.D. The Children s Hospital of Philadelphia and the Perelman School of Medicine at the University of Pennsylvania Objectives 1. Understand the clinical and genetic
The Genomics of Hearing Loss Ian Krantz, M.D. The Children s Hospital of Philadelphia and the Perelman School of Medicine at the University of Pennsylvania Objectives 1. Understand the clinical and genetic
Gene Mapping Techniques
 Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Congenital Hearing Loss
 Congenital Hearing Loss A resident asks... Why would a primary care physician want to know about the genetics of hearing loss? Key Points: Newborn screening for hearing loss is being implemented in many
Congenital Hearing Loss A resident asks... Why would a primary care physician want to know about the genetics of hearing loss? Key Points: Newborn screening for hearing loss is being implemented in many
Molecular Genetic Testing in Public Health and Clinical Settings
 Molecular Genetic Testing in Public Health and Clinical Settings Ira M. Lubin, PhD, FACMG Division of Laboratory Systems NCPDCID, CCID Centers for Disease Control and Prevention Atlanta, Georgia Disclaimers
Molecular Genetic Testing in Public Health and Clinical Settings Ira M. Lubin, PhD, FACMG Division of Laboratory Systems NCPDCID, CCID Centers for Disease Control and Prevention Atlanta, Georgia Disclaimers
The future role of genetic screening to detect newborns at risk of childhood-onset hearing loss
 International Journal of Audiology 2013; 52: 124 133 Discussion Article The future role of genetic screening to detect newborns at risk of childhood-onset hearing loss Luan Linden Phillips *, Maria Bitner-Glindzicz,
International Journal of Audiology 2013; 52: 124 133 Discussion Article The future role of genetic screening to detect newborns at risk of childhood-onset hearing loss Luan Linden Phillips *, Maria Bitner-Glindzicz,
Breast cancer and the role of low penetrance alleles: a focus on ATM gene
 Modena 18-19 novembre 2010 Breast cancer and the role of low penetrance alleles: a focus on ATM gene Dr. Laura La Paglia Breast Cancer genetic Other BC susceptibility genes TP53 PTEN STK11 CHEK2 BRCA1
Modena 18-19 novembre 2010 Breast cancer and the role of low penetrance alleles: a focus on ATM gene Dr. Laura La Paglia Breast Cancer genetic Other BC susceptibility genes TP53 PTEN STK11 CHEK2 BRCA1
Diagnostic Scoring System for LQTS
 Medical Coverage Policy Genetic Testing: Congenital Long QT Syndrome Device/Equipment Drug Medical Surgery Test Other Effective Date: 2/15/2011 Policy Last Updated: 2/21/2012 Prospective review is recommended/required.
Medical Coverage Policy Genetic Testing: Congenital Long QT Syndrome Device/Equipment Drug Medical Surgery Test Other Effective Date: 2/15/2011 Policy Last Updated: 2/21/2012 Prospective review is recommended/required.
Usher Syndrome Genetics
 Usher Syndrome Genetics October 2012 Page 1 of 20 Introduction Usher syndrome is a genetic or inherited condition that affects hearing, vision and balance The sight loss is caused by an eye condition known
Usher Syndrome Genetics October 2012 Page 1 of 20 Introduction Usher syndrome is a genetic or inherited condition that affects hearing, vision and balance The sight loss is caused by an eye condition known
Sensorineural Hearing Loss
 Sensorineural Hearing Loss 1. Review the patterns of hearing loss in hereditary hearing impairment. AL First, we must understand that genetic hearing loss seems to breach all categories of hearing loss,
Sensorineural Hearing Loss 1. Review the patterns of hearing loss in hereditary hearing impairment. AL First, we must understand that genetic hearing loss seems to breach all categories of hearing loss,
commentary Genetics IN Medicine 521 November/December 2004 Vol. 6 No. 6
 November/December 2004 Vol. 6 No. 6 commentary Genetic testing as part of the Early Hearing Detection and Intervention (EHDI) process Lisa A. Schimmenti, MD 1, Ariadna Martinez, MS, MS 2, Michelle Fox,
November/December 2004 Vol. 6 No. 6 commentary Genetic testing as part of the Early Hearing Detection and Intervention (EHDI) process Lisa A. Schimmenti, MD 1, Ariadna Martinez, MS, MS 2, Michelle Fox,
Gene Therapy and Genetic Counseling. Chapter 20
 Gene Therapy and Genetic Counseling Chapter 20 What is Gene Therapy? Treating a disease by replacing, manipulating or supplementing a gene The act of changing an individual s DNA sequence to fix a non-functional
Gene Therapy and Genetic Counseling Chapter 20 What is Gene Therapy? Treating a disease by replacing, manipulating or supplementing a gene The act of changing an individual s DNA sequence to fix a non-functional
A bout 1 in 1000 British children are born deaf.
 1of5 ELECTRONIC LETTER Genetic information but not termination: pregnant women s attitudes and willingness to pay for carrier screening for deafness genes M Ryan, Z Miedzybrodzka, L Fraser, M Hall... J
1of5 ELECTRONIC LETTER Genetic information but not termination: pregnant women s attitudes and willingness to pay for carrier screening for deafness genes M Ryan, Z Miedzybrodzka, L Fraser, M Hall... J
Genetics Module B, Anchor 3
 Genetics Module B, Anchor 3 Key Concepts: - An individual s characteristics are determines by factors that are passed from one parental generation to the next. - During gamete formation, the alleles for
Genetics Module B, Anchor 3 Key Concepts: - An individual s characteristics are determines by factors that are passed from one parental generation to the next. - During gamete formation, the alleles for
A pproximately one child in 500 650 is born deaf,
 147 ORIGINAL ARTICLE A genotype-phenotype correlation for GJB2 (connexin 26) deafness K Cryns*, E Orzan*, A Murgia*, P L M Huygen, F Moreno, I del Castillo, G Parker Chamberlin, H Azaiez, S Prasad, R A
147 ORIGINAL ARTICLE A genotype-phenotype correlation for GJB2 (connexin 26) deafness K Cryns*, E Orzan*, A Murgia*, P L M Huygen, F Moreno, I del Castillo, G Parker Chamberlin, H Azaiez, S Prasad, R A
Genetic Hearing Loss
 Genetic Hearing Loss Stephanie Cordes, MD Faculty Advisor: Norman Friedman, MD The University of Texas Medical Branch Department of Otolaryngology Grand Rounds Presentation April 2000 Introduction Deafness
Genetic Hearing Loss Stephanie Cordes, MD Faculty Advisor: Norman Friedman, MD The University of Texas Medical Branch Department of Otolaryngology Grand Rounds Presentation April 2000 Introduction Deafness
Attitudes toward genetic testing
 DOI 10.1515/jbcpp-2013-0063 J Basic Clin Physiol Pharmacol 2013; 24(3): 165 170 Mini Review Natali Idan, Zippora Brownstein, Shaked Shivatzki and Karen B. Avraham* Advances in genetic diagnostics for hereditary
DOI 10.1515/jbcpp-2013-0063 J Basic Clin Physiol Pharmacol 2013; 24(3): 165 170 Mini Review Natali Idan, Zippora Brownstein, Shaked Shivatzki and Karen B. Avraham* Advances in genetic diagnostics for hereditary
The Need for a PARP in vivo Pharmacodynamic Assay
 The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
CONNEXINS IN HEARING LOSS: A COMPREHENSIVE OVERVIEW
 CONNEXINS IN HEARING LOSS: A COMPREHENSIVE OVERVIEW Adi D. Sabag, Orit Dagan and Karen B. Avraham Department of Human Genetics and Molecular Medicine, Sackler School of Medicine, Tel Aviv University, Tel
CONNEXINS IN HEARING LOSS: A COMPREHENSIVE OVERVIEW Adi D. Sabag, Orit Dagan and Karen B. Avraham Department of Human Genetics and Molecular Medicine, Sackler School of Medicine, Tel Aviv University, Tel
Von Mäusen und Menschen E - 1
 Von Mäusen und Menschen E - 1 Mus musculus: Genetic Portrait of the House Mouse E - 3 Outline Mouse genome Mouse life cycle Transgenic protocols Addition of genes by nuclear injection Removal of genes
Von Mäusen und Menschen E - 1 Mus musculus: Genetic Portrait of the House Mouse E - 3 Outline Mouse genome Mouse life cycle Transgenic protocols Addition of genes by nuclear injection Removal of genes
Usefulness of polymorphic markers in exclusion of BRCA1/BRCA2 mutations in families with aggregation of breast/ovarian cancers
 J. Appl. Genet. 44(3), 2003, pp. 419-423 Short communication Usefulness of polymorphic markers in exclusion of BRCA1/BRCA2 mutations in families with aggregation of breast/ovarian cancers Bohdan GÓRSKI,
J. Appl. Genet. 44(3), 2003, pp. 419-423 Short communication Usefulness of polymorphic markers in exclusion of BRCA1/BRCA2 mutations in families with aggregation of breast/ovarian cancers Bohdan GÓRSKI,
Rare Diseases: Genetic Testing and Targeted Therapy
 Rare Diseases: Genetic Testing and Targeted Therapy Meral Özgüç Hacettepe University Faculty of Medicine Department of Medical Biology & DNA/Cell Bank RDs Definition! Low prevalence (Less than 5/10000)!
Rare Diseases: Genetic Testing and Targeted Therapy Meral Özgüç Hacettepe University Faculty of Medicine Department of Medical Biology & DNA/Cell Bank RDs Definition! Low prevalence (Less than 5/10000)!
Heritability: Twin Studies. Twin studies are often used to assess genetic effects on variation in a trait
 TWINS AND GENETICS TWINS Heritability: Twin Studies Twin studies are often used to assess genetic effects on variation in a trait Comparing MZ/DZ twins can give evidence for genetic and/or environmental
TWINS AND GENETICS TWINS Heritability: Twin Studies Twin studies are often used to assess genetic effects on variation in a trait Comparing MZ/DZ twins can give evidence for genetic and/or environmental
UNIT 13 (OPTION) Genetic Abnormalities
 Unit 13 Genetic Abnormailities 1 UNIT 13 (OPTION) Genetic Abnormalities Originally developed by: Hildur Helgedottir RN, MN Revised (2000) by: Marlene Reimer RN, PhD, CCN (C) Associate Professor Faculty
Unit 13 Genetic Abnormailities 1 UNIT 13 (OPTION) Genetic Abnormalities Originally developed by: Hildur Helgedottir RN, MN Revised (2000) by: Marlene Reimer RN, PhD, CCN (C) Associate Professor Faculty
TERATOGENESIS ONTOGENESIS
 TERATOGENESIS ONTOGENESIS Inborn developmental defects Occured during prenatal development Are present by delivery At about 3-5 % newborns are affected. Inborn developmental defects 1. CHROMOSOMAL ABERRATIONS
TERATOGENESIS ONTOGENESIS Inborn developmental defects Occured during prenatal development Are present by delivery At about 3-5 % newborns are affected. Inborn developmental defects 1. CHROMOSOMAL ABERRATIONS
Becker Muscular Dystrophy
 Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
Control of Gene Expression
 Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Control of Gene Expression What is Gene Expression? Gene expression is the process by which informa9on from a gene is used in the synthesis of a func9onal gene product. What is Gene Expression? Figure
Chapter 4 Pedigree Analysis in Human Genetics. Chapter 4 Human Heredity by Michael Cummings 2006 Brooks/Cole-Thomson Learning
 Chapter 4 Pedigree Analysis in Human Genetics Mendelian Inheritance in Humans Pigmentation Gene and Albinism Fig. 3.14 Two Genes Fig. 3.15 The Inheritance of Human Traits Difficulties Long generation time
Chapter 4 Pedigree Analysis in Human Genetics Mendelian Inheritance in Humans Pigmentation Gene and Albinism Fig. 3.14 Two Genes Fig. 3.15 The Inheritance of Human Traits Difficulties Long generation time
The world of non-coding RNA. Espen Enerly
 The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
The world of non-coding RNA Espen Enerly ncrna in general Different groups Small RNAs Outline mirnas and sirnas Speculations Common for all ncrna Per def.: never translated Not spurious transcripts Always/often
So, how do we hear? outer middle ear inner ear
 The ability to hear is critical to understanding the world around us. The human ear is a fully developed part of our bodies at birth and responds to sounds that are very faint as well as sounds that are
The ability to hear is critical to understanding the world around us. The human ear is a fully developed part of our bodies at birth and responds to sounds that are very faint as well as sounds that are
SENSORINEURAL HEARING LOSS INVESTIGATION & REFERRAL
 Epidemiology NZ Hearing Screening Referral Pathway Important Services When to Investigate Initial Assessment Referrals Investigations Indications for Cochlear Transplant Acknowledgements References Epidemiology
Epidemiology NZ Hearing Screening Referral Pathway Important Services When to Investigate Initial Assessment Referrals Investigations Indications for Cochlear Transplant Acknowledgements References Epidemiology
Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iplex technology
 Svidnicki et al. BMC Medical Genetics (2015) 16:85 DOI 10.1186/s12881-015-0232-8 RESEARCH ARTICLE Open Access Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iplex
Svidnicki et al. BMC Medical Genetics (2015) 16:85 DOI 10.1186/s12881-015-0232-8 RESEARCH ARTICLE Open Access Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iplex
AP BIOLOGY 2008 SCORING GUIDELINES
 AP BIOLOGY 2008 SCORING GUIDELINES Question 1 1. The physical structure of a protein often reflects and affects its function. (a) Describe THREE types of chemical bonds/interactions found in proteins.
AP BIOLOGY 2008 SCORING GUIDELINES Question 1 1. The physical structure of a protein often reflects and affects its function. (a) Describe THREE types of chemical bonds/interactions found in proteins.
Worksheet - COMPARATIVE MAPPING 1
 Worksheet - COMPARATIVE MAPPING 1 The arrangement of genes and other DNA markers is compared between species in Comparative genome mapping. As early as 1915, the geneticist J.B.S Haldane reported that
Worksheet - COMPARATIVE MAPPING 1 The arrangement of genes and other DNA markers is compared between species in Comparative genome mapping. As early as 1915, the geneticist J.B.S Haldane reported that
Systematic discovery of regulatory motifs in human promoters and 30 UTRs by comparison of several mammals
 Systematic discovery of regulatory motifs in human promoters and 30 UTRs by comparison of several mammals Xiaohui Xie 1, Jun Lu 1, E. J. Kulbokas 1, Todd R. Golub 1, Vamsi Mootha 1, Kerstin Lindblad-Toh
Systematic discovery of regulatory motifs in human promoters and 30 UTRs by comparison of several mammals Xiaohui Xie 1, Jun Lu 1, E. J. Kulbokas 1, Todd R. Golub 1, Vamsi Mootha 1, Kerstin Lindblad-Toh
AP Biology Essential Knowledge Student Diagnostic
 AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
AP Biology Essential Knowledge Student Diagnostic Background The Essential Knowledge statements provided in the AP Biology Curriculum Framework are scientific claims describing phenomenon occurring in
Chapter 9 Patterns of Inheritance
 Bio 100 Patterns of Inheritance 1 Chapter 9 Patterns of Inheritance Modern genetics began with Gregor Mendel s quantitative experiments with pea plants History of Heredity Blending theory of heredity -
Bio 100 Patterns of Inheritance 1 Chapter 9 Patterns of Inheritance Modern genetics began with Gregor Mendel s quantitative experiments with pea plants History of Heredity Blending theory of heredity -
Lecture 3: Mutations
 Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
Lecture 3: Mutations Recall that the flow of information within a cell involves the transcription of DNA to mrna and the translation of mrna to protein. Recall also, that the flow of information between
The Developing Person Through the Life Span 8e by Kathleen Stassen Berger
 The Developing Person Through the Life Span 8e by Kathleen Stassen Berger Chapter 3 Heredity and Environment PowerPoint Slides developed by Martin Wolfger and Michael James Ivy Tech Community College-Bloomington
The Developing Person Through the Life Span 8e by Kathleen Stassen Berger Chapter 3 Heredity and Environment PowerPoint Slides developed by Martin Wolfger and Michael James Ivy Tech Community College-Bloomington
BioBoot Camp Genetics
 BioBoot Camp Genetics BIO.B.1.2.1 Describe how the process of DNA replication results in the transmission and/or conservation of genetic information DNA Replication is the process of DNA being copied before
BioBoot Camp Genetics BIO.B.1.2.1 Describe how the process of DNA replication results in the transmission and/or conservation of genetic information DNA Replication is the process of DNA being copied before
A CASE FOR ETIOLOGIC FOCUS IN AUDIOLOGY GENETIC TESTING. Thesis
 A CASE FOR ETIOLOGIC FOCUS IN AUDIOLOGY GENETIC TESTING Thesis Presented in Partial Fulfillment of the Requirements for The Masters in Speech and Hearing Science in the Graduate School of The Ohio State
A CASE FOR ETIOLOGIC FOCUS IN AUDIOLOGY GENETIC TESTING Thesis Presented in Partial Fulfillment of the Requirements for The Masters in Speech and Hearing Science in the Graduate School of The Ohio State
1.- L a m e j o r o p c ió n e s c l o na r e l d i s co ( s e e x p li c a r á d es p u é s ).
 PROCEDIMIENTO DE RECUPERACION Y COPIAS DE SEGURIDAD DEL CORTAFUEGOS LINUX P ar a p od e r re c u p e ra r nu e s t r o c o rt a f u e go s an t e un d es a s t r e ( r ot u r a d e l di s c o o d e l a
PROCEDIMIENTO DE RECUPERACION Y COPIAS DE SEGURIDAD DEL CORTAFUEGOS LINUX P ar a p od e r re c u p e ra r nu e s t r o c o rt a f u e go s an t e un d es a s t r e ( r ot u r a d e l di s c o o d e l a
Human Cloning The Science and Ethics of Nuclear Transplantation
 Human Cloning The Science and Ethics of Transplantation Rudolf Jaenisch, M.D. In addition to the moral argument against the use of somatic-cell nuclear for the creation of a child ( reproductive cloning
Human Cloning The Science and Ethics of Transplantation Rudolf Jaenisch, M.D. In addition to the moral argument against the use of somatic-cell nuclear for the creation of a child ( reproductive cloning
RETRIEVING SEQUENCE INFORMATION. Nucleotide sequence databases. Database search. Sequence alignment and comparison
 RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
Heredity - Patterns of Inheritance
 Heredity - Patterns of Inheritance Genes and Alleles A. Genes 1. A sequence of nucleotides that codes for a special functional product a. Transfer RNA b. Enzyme c. Structural protein d. Pigments 2. Genes
Heredity - Patterns of Inheritance Genes and Alleles A. Genes 1. A sequence of nucleotides that codes for a special functional product a. Transfer RNA b. Enzyme c. Structural protein d. Pigments 2. Genes
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
Structure and Function of DNA
 Structure and Function of DNA DNA and RNA Structure DNA and RNA are nucleic acids. They consist of chemical units called nucleotides. The nucleotides are joined by a sugar-phosphate backbone. The four
Structure and Function of DNA DNA and RNA Structure DNA and RNA are nucleic acids. They consist of chemical units called nucleotides. The nucleotides are joined by a sugar-phosphate backbone. The four
The Human Genome Project
 The Human Genome Project Brief History of the Human Genome Project Physical Chromosome Maps Genetic (or Linkage) Maps DNA Markers Sequencing and Annotating Genomic DNA What Have We learned from the HGP?
The Human Genome Project Brief History of the Human Genome Project Physical Chromosome Maps Genetic (or Linkage) Maps DNA Markers Sequencing and Annotating Genomic DNA What Have We learned from the HGP?
Bio 102 Practice Problems Genetic Code and Mutation
 Bio 102 Practice Problems Genetic Code and Mutation Multiple choice: Unless otherwise directed, circle the one best answer: 1. Beadle and Tatum mutagenized Neurospora to find strains that required arginine
Bio 102 Practice Problems Genetic Code and Mutation Multiple choice: Unless otherwise directed, circle the one best answer: 1. Beadle and Tatum mutagenized Neurospora to find strains that required arginine
Validation and Replication
 Validation and Replication Overview Definitions of validation and replication Difficulties and limitations Working examples from our group and others Why? False positive results still occur. even after
Validation and Replication Overview Definitions of validation and replication Difficulties and limitations Working examples from our group and others Why? False positive results still occur. even after
Genomes and SNPs in Malaria and Sickle Cell Anemia
 Genomes and SNPs in Malaria and Sickle Cell Anemia Introduction to Genome Browsing with Ensembl Ensembl The vast amount of information in biological databases today demands a way of organising and accessing
Genomes and SNPs in Malaria and Sickle Cell Anemia Introduction to Genome Browsing with Ensembl Ensembl The vast amount of information in biological databases today demands a way of organising and accessing
12.1 The Role of DNA in Heredity
 12.1 The Role of DNA in Heredity Only in the last 50 years have scientists understood the role of DNA in heredity. That understanding began with the discovery of DNA s structure. In 1952, Rosalind Franklin
12.1 The Role of DNA in Heredity Only in the last 50 years have scientists understood the role of DNA in heredity. That understanding began with the discovery of DNA s structure. In 1952, Rosalind Franklin
CHROMOSOMES AND INHERITANCE
 SECTION 12-1 REVIEW CHROMOSOMES AND INHERITANCE VOCABULARY REVIEW Distinguish between the terms in each of the following pairs of terms. 1. sex chromosome, autosome 2. germ-cell mutation, somatic-cell
SECTION 12-1 REVIEW CHROMOSOMES AND INHERITANCE VOCABULARY REVIEW Distinguish between the terms in each of the following pairs of terms. 1. sex chromosome, autosome 2. germ-cell mutation, somatic-cell
Mutation Research/Reviews in Mutation Research
 Mutation Research 681 (2009) 189 196 Contents lists available at ScienceDirect Mutation Research/Reviews in Mutation Research journal homepage: www.elsevier.com/locate/reviewsmr Community address: www.elsevier.com/locate/mutres
Mutation Research 681 (2009) 189 196 Contents lists available at ScienceDirect Mutation Research/Reviews in Mutation Research journal homepage: www.elsevier.com/locate/reviewsmr Community address: www.elsevier.com/locate/mutres
How to construct transgenic mice
 How to construct transgenic mice Sandra Beer-Hammer Autumn School 2012 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
How to construct transgenic mice Sandra Beer-Hammer Autumn School 2012 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
Influence of Sex on Genetics. Chapter Six
 Influence of Sex on Genetics Chapter Six Humans 23 Autosomes Chromosomal abnormalities very severe Often fatal All have at least one X Deletion of X chromosome is fatal Males = heterogametic sex XY Females
Influence of Sex on Genetics Chapter Six Humans 23 Autosomes Chromosomal abnormalities very severe Often fatal All have at least one X Deletion of X chromosome is fatal Males = heterogametic sex XY Females
CCR Biology - Chapter 7 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 7 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. A person who has a disorder caused
Name: Class: Date: CCR Biology - Chapter 7 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. A person who has a disorder caused
Causes of Birth Defects
 Causes of Birth Defects Some medical / genetic terms: congenital defects: visible defects present at birth (due to any cause (genetic, developmental error ). syndrome: the symptoms that characterize any
Causes of Birth Defects Some medical / genetic terms: congenital defects: visible defects present at birth (due to any cause (genetic, developmental error ). syndrome: the symptoms that characterize any
Antigenic variation in Plasmodium falciparum : Erythrocyte invasion and immune escape mechanisms
 Antigenic variation in Plasmodium falciparum : Erythrocyte invasion and immune escape mechanisms Introduction Why does immunity to malaria take so long to develop? The parasite s survival depends on its
Antigenic variation in Plasmodium falciparum : Erythrocyte invasion and immune escape mechanisms Introduction Why does immunity to malaria take so long to develop? The parasite s survival depends on its
1/26/2011. 50% of deafness and hearing impairment is avoidable through prevention, early diagnosis, and management.
 Hearing Impairment Roseann Mulligan, DDS, MS Herman Ostrow School of Dentistry of the University of Southern California 1 JAMA, July 4, 2007 Vol 298, No. 1 2 278 million - moderate to profound bilateral
Hearing Impairment Roseann Mulligan, DDS, MS Herman Ostrow School of Dentistry of the University of Southern California 1 JAMA, July 4, 2007 Vol 298, No. 1 2 278 million - moderate to profound bilateral
