Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint
|
|
|
- Owen Griffin
- 8 years ago
- Views:
Transcription
1 The EMBO Journal Vol. 22 No. 5 pp. 1075±1087, 2003 Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint Satoko Yamaguchi 1, Anabelle Decottignies 2 and Paul Nurse 3 Cell Cycle Laboratory, Cancer Research UK, 44 Lincoln's Inn Fields, London WC2A 3PX, UK 1 Present address: Department of Biology, New York University, 100 Washington Square East, New York, NY 10003, USA 2 Present address: Catholic University of Louvain, Faculty of Medicine, 74 Avenue Hippocrate, 1200 Brussels, Belgium 3 Corresponding author paul.nurse@cancer.org.uk S.Yamaguchi and A.Decottignies contributed equally to this work Cdc2p is a cyclin-dependent kinase (CDK) essential for both mitotic and meiotic cell cycle progression in ssion yeast. We have found that the spindle checkpoint kinase Bub1p becomes phosphorylated by Cdc2p during spindle damage in mitotic cells. Cdc2p directly phosphorylates Bub1p in vitro at the CDK consensus sites. A Bub1p mutant that cannot be phosphorylated by Cdc2p is checkpoint defective, indicating that Cdc2p-dependent Bub1p phosphorylation is required to activate the checkpoint after spindle damage. The kinase activity of Bub1p is required, but is not suf cient, for complete spindle checkpoint function. The role of Bub1p in maintaining centromeric localization of Rec8p during meiosis I is entirely dependent upon its kinase activity, suggesting that Bub1p kinase activity is essential for establishing proper kinetochore function. Finally, we show that there is a Bub1p-dependent meiotic checkpoint, which is activated in recombination mutants. Keywords: Bub1p kinase/cdc2p/meiotic checkpoint/ Rec8p/spindle checkpoint Introduction The spindle checkpoint plays an essential role in eukaryotic cells to inhibit onset of anaphase until proper attachment of chromosomes to the spindle (reviewed by Amon, 1999; Millband et al., 2002). One of the proteins involved in this checkpoint is the conserved Bub1p protein kinase that localizes to kinetochores during mitosis (reviewed by Straight, 1997). In this paper, we investigate the roles that Bub1p phosphorylation and kinase activity play in spindle checkpoints during the mitotic and meiotic cell cycles of ssion yeast. Bub1p was initially identi ed in budding yeast as being required for pre-anaphase delay in response to spindle damage (Hoyt et al., 1991; Roberts et al., 1994). It plays a role in maintaining chromosomal stability in a number of eukaryotes, and mutations in the bub1 gene have been found in some cancers (reviewed by Cahill et al., 1998; Jallepalli and Lengauer, 2001). Bub1p localizes at the kinetochores during prophase and prometaphase of mitosis (Taylor and McKeon, 1997; Basu et al., 1999) and is thought to be recruited to unattached kinetochores. Bub1p physically associates with Bub3p (Roberts et al., 1994) and Mad1p (Brady and Hardwick, 2000), and is required for the kinetochore localization of Bub3p (Basu et al., 1999; Sharp-Baker and Chen, 2001). Bub1, Bub3p, Mad1p, Mad2p and Mad3p probably form a complex at the unattached kinetochore (reviewed by Amon, 1999). Bub1p has kinase activity in vitro and can phosphorylate itself as well as Bub3p (Roberts et al., 1994) and Mad1p (Seeley et al., 1999) in vitro. Other spindle checkpoint components such as Mad1p, Mad2p and Mad3p interact with Cdc20±APC (the anaphase-promoting complex/ cyclosome), which inhibits the metaphase±anaphase transition (reviewed by Amon, 1999; Millband et al., 2002). In budding yeast, a point mutation in the conserved lysine residue of the kinase domain abolished in vitro kinase activity, and such mutants appeared to be checkpoint defective (Roberts et al., 1994). In Xenopus laevis, Bub1p recruits other spindle checkpoint components (Mad1p, Mad2p and Bub3p) to kinetochores independently of its kinase activity (Sharp-Baker and Chen, 2001). The role of Bub1p during meiosis I (MI) differs from mitosis and meiosis II (MII) because chromosomal segregation is reductional, with sister chromatids remaining attached and moving together to one pole (reviewed by Kleckner, 1996). The spindle checkpoint proteins are recruited to kinetochores during meiosis (Yu et al., 1999; Kallio et al., 2000), and both Bub1p and Mad2p prevent non-disjunction events during MI in yeast (Shonn et al., 2000; Bernard et al., 2001). In ssion yeast, Bub1p has been reported to play a role in ensuring reductional segregation during MI (Bernard et al., 2001). The meiotic cohesin Rec8p also plays an essential role in establishing reductional segregation (Watanabe and Nurse, 1999; Buonomo et al., 2000), and its association with the inner centromeres is important to establish monopolar orientation (Watanabe et al., 2001). Fission yeast Bub1p is associated with the kinetochores during both meiosis and mitosis (Bernard et al., 1998, 2001), and is required for the centromeric localization of Rec8p during anaphase I. How Bub1p is regulated is not well understood. Sequence analysis of Bub1p reveals four putative CDK phosphorylation sites in the amino acid sequence of Bub1p. Cdc2p is a cyclin-dependent kinase that plays multiple roles during both mitosis and meiosis (reviewed by Nigg, 1995; Nurse, 2000). Cdc2p has been shown to be localized at centromeres during meiosis in ssion yeast (Decottignies et al., 2001), and so we have investigated whether Cdc2p plays a role in regulating Bub1p during the cell cycle. We show that at least one of the four putative CDK sites in Bub1p is phosphorylated by Cdc2p, and have ã European Molecular Biology Organization 1075
2 S.Yamaguchi, A.Decottignies and P.Nurse investigated the roles this phosphorylation and the Bub1p kinase domain play in the spindle checkpoint during mitosis and the rst meiotic division. Results Phosphorylation of Bub1p during mitosis To determine whether Bub1p phosphorylation changes during the mitotic cell cycle, cells were synchronized in G 2 by incubating a cdc25±22 mutant strain in which the endogenous bub1 + was tagged with HA at 36 C for 4 h before being released into mitosis at 25 C (Figure 1A). Western blot analysis using anti-ha antibodies revealed multiple bands of Bub1-HAp in exponentially growing cells, but these remained unchanged throughout the cell cycle (Figure 1A). Next, we investigated Bub1p phosphorylation during spindle damage, which is known to involve Bub1p function (reviewed by Amon, 1999). To disrupt the mitotic spindle, cold-sensitive b-tubulin mutant nda3-km311 cells were incubated at 18 C (Hiraoka et al., 1984). Upon shifting the cells to 18 C, slower migrating forms of Bub1-HAp appeared on the gel in addition to the multiple bands present in asynchronously growing cells (Figure 1B). All the bands that appeared in the nda3 block were sensitive to phosphatase treatment (Figure 1B). These results indicate that Bub1p becomes hyperphosphorylated in an nda3 block. Sequence analysis revealed four putative CDK consensus sites (S/T-P-X-K/R) (Maller et al., 1989) in the middle region of Bub1p (Figure 1C). To investigate whether hyperphosphorylation of Bub1p in a mitotic block occurs at these CDK sites, we constructed an `unphosphorylatable' mutant (bub1-p*) of Bub1p by changing the four S/T residues T340, T423, T455 and S466 to alanine (Figure 1C). Bub1p hyperphosphorylation was eliminated in this bub1-p*-ha strain in an nda3 block (Figure 1C). Phosphatase treatment revealed that the other bands observed in the bub1-p*-ha strain were also due to phosphorylation (Figure 1C), suggesting that Bub1p may be additionally phosphorylated by other kinases. These results demonstrate that Bub1p becomes hyperphosphorylated at the CDK consensus sites when spindle formation is disrupted. To determine whether Cdc2p kinase activity is required for Bub1p hyperphosphorylation, we destabilized microtubules (MTs) with drugs in a thermosensitive (ts) cdc2 mutant strain (Figure 1D). Cdc2-M26 cells were incubated for 3 h at 36 C, arresting most cells in late G 2 (Carr et al., 1989). The cells were released to 25 C in the presence of 50 mg/ml carbendazim (MBC), and after 40 min, both bub1-ha and bub1-p*-ha cells were analysed by western blotting (Figure 1D). Slower migrating forms of Bub1- HAp were detected in addition to the normal multiple bands (Figure 1D, left panel, 3h40). In contrast, Fig. 1. Phosphorylation of Bub1p during mitosis. (A) Percentage of cells with 1or 2 nuclei (left panel) and western blotting (right panel) of G 2 block and release experiments. Cdc25-22 bub1-ha cells were incubated at 36 C for 4 h, then released at 25 C for the indicated time to proceed through synchronous mitosis. Anti-HA antibody was used to detect Bub1-HAp. (B) Bub1p became hyperphosphorylated in an nda3 block. Nda3-km311 cells were incubated at 18 C for the indicated time in hours (left panel). Extracts were prepared from nda3-km311 cells blocked at 18 C for 6 h (pre) and incubated with l phosphatase in the absence (lpp) or presence (lpp+inh) of phosphatase inhibitors (right panel). (C) Hyperphosphorylation was abolished in nda3-km311 bub1-p*-ha strain. The four CDK consensus sites were mutated to alanine in the bub1-p* mutant (left panel). Hyperphosphorylation in an nda3 block was eliminated in bub1-p*-ha strain (centre panel). Phosphatase treatment of extracts from bub1-p*-ha cells (right panel). (D) Hyperphosphorylation of Bub1p was dependent on Cdc2p kinase activity. Cdc2-M26 bub1-ha and cdc2-m26 bub1-p*-ha cells were incubated at 36 C for 3 h, released to 25 C in the presence of 50 mg/ml MBC for 40 min until cells blocked in mitosis, and then shifted back to 36 C in the presence of 100 mg/ml MBC. 1076
3 Bub1p kinase is regulated by Cdc2p (Figure 2A). This was not due to autophosphorylation of Bub1p, as GST±Bub1-Dkinasep alone did not give any signal (Figure 2A). We concluded that Cdc2p can phosphorylate Bub1p in vitro. To investigate whether this phosphorylation by Cdc2p occurs at the CDK consensus sites, we puri ed unphosphorylatable mutant of Bub1p from bacteria, in which four CDK consensus sites are mutated (GST±Bub1-P*- Dkinasep). Incubation with either S.pombe or H.sapiens Cdc2p revealed that the [ 32 P]Bub1p signal was much reduced in the GST±Bub1-P*-Dkinasep mutant protein (Figure 2B), indicating that Cdc2p phosphorylates Bub1p at the CDK consensus sites in vitro. Fig. 2. Cdc2p phosphorylates Bub1p at CDK consensus sites in vitro. (A) Left panel: GST±Bub1-Dkinasep or GST was incubated either alone (substrate alone) or with S.pombe Cdc2p (Sp Cdc2p) or H.sapiens Cdc2p (Hs Cdc2p). The band just below GST±Bub1- Dkinasep is a degradation product, as shown by the western blotting. Top right panel: GST±Bub1-Dkinasep was incubated either with Hs Cdc2p alone or with Hs Cdc2p and l phosphatase (lpp). Bottom right panel: Coomassie Blue staining and anti-gst western blotting of GST±Bub1-Dkinase and GST. (B) GST±Bub1-Dkinasep (WT) or GST±Bub1-P*-Dkinasep (Bub1-P*) was incubated with either Sp Cdc2p (left panel) or Hs Cdc2p (right panel). Coomassie Blue staining is shown as loading control. hyperphosphorylation of Bub1-P*-HAp was not observed in the bub1-p*-ha cells (Figure 1D, right panel, 3h40). The upper bands of Bub1-HAp disappeared when cells were shifted back to 36 C (Figure 1D, left panel, 4h20), establishing that Cdc2p kinase activity is continuously required to maintain the MBC-induced hyperphosphorylation of Bub1p. Cdc2p can phosphorylate Bub1p in vitro To investigate whether this Cdc2p-dependent phosphorylation of Bub1p is direct or not, we performed an in vitro kinase assay. Since Bub1p is able to autophosphorylate (Roberts et al., 1994), bacterially-produced glutathione S-transferase (GST) fusion protein of Bub1p lacking the kinase domain was used as a substrate for Cdc2p. When GST±Bub1-Dkinasep was incubated with either Schizosaccharomyces pombe or Homo sapiens Cdc2p, we detected the phosphorylation of a 115 kda band Cdc2p-dependent hyperphosphorylation of Bub1p and the spindle checkpoint We next tested whether Cdc2p-mediated hyperphosphorylation of Bub1p plays a role in the spindle checkpoint. Bub1-P* cells were more sensitive to thiabendazole (TBZ) than wild-type (WT) cells, but more resistant than bub1d cells (Figure 3A). The TBZ sensitivity of bub1-p* cells was comparable with that of mad2d cells (Figure 3A), which have been shown to be sensitive to MT drugs (Li and Murray, 1991; Kim et al., 1998). To quantify these effects, the number of cells that could proceed through mitosis to septation in the presence of MBC was scored. Cdc2-M26 cells were synchronized in late G 2 by incubation at 36 C and subsequently released to 25 C in the presence of 75 mg/ml MBC. Bub1 + cells became blocked in mitosis before septation, and 80 min after release only 3% of the cells were found to be septated (Figure 3B). In contrast, the spindle checkpoint is defective in bub1d cells and 45% of the cells were septated at 80 min. In the bub1-p* mutant, 20% of the cells were septated, suggesting that phosphorylation is required to completely block cells in mitosis. The inability of Bub1-P*p to completely activate the checkpoint was not due to a defect in kinetochore localization because Bub1±GFPp and Bub1-P*±GFPp both localized similarly to kinetochores in the presence of MBC (Figure 3C). Bub1p kinase domain and the spindle checkpoint The kinase domain is at the C-terminus of the Bub1p protein (Figure 1C) and includes the conserved K762 residue in the ATP-binding site (Figure 4A). To investigate the role of Bub1p kinase activity in the spindle checkpoint, we constructed two different mutants, one with the K762R mutation, which abolishes the kinase activity of Saccharomyces cerevisiae Bub1p (Roberts et al., 1994), and a second that lacked the entire kinase domain (bub1-dkinase) (Figure 4B). Both mutants were more sensitive to TBZ than WT cells, but not as sensitive as bub1d cells (Figure 4C). We quanti ed these effects using synchronized cdc2-m26 cells. Eighty minutes after release from G 2 in the presence of MBC, 21% of the bub1- Dkinase cells and 22% of the bub1-k762r cells were septated compared with 3% of WT cells and 45% of bub1d cells (Figure 4D). These observations suggest that the kinase activity of Bub1p is necessary for complete spindle checkpoint function. However, there must also be an additional role for Bub1p that is independent of its kinase activity and is necessary for full operation of the checkpoint. Fusion of the Bub1-Dkinase protein to green 1077
4 S.Yamaguchi, A.Decottignies and P.Nurse Fig. 3. Bub1-P* mutant cells are checkpoint defective. (A) WT, bub1-p*, bub1d and mad2d cells were grown to exponential phase, spotted onto YE5S plates containing 0, 10 or 15 mg/ml TBZ and incubated for 2±3 days. (B) Cdc2-M26 (WT), cdc2-m26 bub1-p* (bub1-p*), cdc2-m26 bub1d (bub1d) strains were incubated at 36 C for 3 h, released to 25 C in the presence of 75mg/ml MBC and incubated for the indicated time in minutes. Cdc2-M26 strain released in the absence of MBC (WT, ±MBC) is indicated as a control. Septated cells were scored by calco uor staining. (C) Bub1±GFPp and Bub1-P*±GFPp were observed by live uorescence microscopy after 40 min of release at 25 C in the presence of MBC. uorescent protein (GFP) revealed that the mutant protein could still localize to the kinetochores of MBC-treated cells (Figure 4E). The defective spindle checkpoint in kinase mutants was not due to a reduction in protein level, as the protein amount of Bub1p in bub1-k762r, bub1- Dkinase and WT cells was comparable (Figure 4B, right panel). Because the bub1-p* and bub1-dkinase mutants are both partially defective in the spindle checkpoint response, we investigated the phenotype of the bub1-p*- Dkinase double mutant. TBZ resistance tests revealed that the bub1-p*-dkinase mutant was more sensitive to the drug than the single mutants, but was still less sensitive than a bub1d mutant (Figure 4C). Quantitative experiments in liquid medium gave similar results (Figure 4D). Therefore, we conclude that although both CDK phosphorylation of Bub1p and the kinase domain of Bub1p contribute to the spindle checkpoint, other aspects of Bub1p function must also be required for a full response. We next investigated the relationship between Mad2p and Bub1p kinase activity, by testing to see if the double mutant of bub1-dkinase and mad2d would give the same phenotype as the single mutants. This was not the case: bub1-dkinase mad2d double mutant cells were more sensitive to TBZ than the single mutants (Figure 4C), indicating that Mad2p is not the only target of Bub1p kinase activity. In a mad2d bub1-p*-d kinase mutant, TBZ sensitivity was similar to bub1d cells (Figure 4C), establishing that CDK phosphorylation, Bub1p kinase activity and the Mad2p-dependent pathway have additive effects. The mad2d bub1d double mutant showed similar TBZ sensitivity to that of bub1d single mutant (Figure 4C). However, we found that Mad2p is still recruited to the kinetochores during spindle damage in the absence of Bub1p (data not shown), suggesting that Bub1p is not required for Mad2p localization at the kinetochore. Bub1p becomes hyperphosphorylated at CDK sites during meiosis Bub1p is required for reductional segregation of sister chromatids during MI (Bernard et al., 2001), and both Cdc2p and Bub1p localize to the centromeres during meiosis (Bernard et al., 2001; Decottignies et al., 2001). Therefore we examined the phosphorylation state of Bub1p in meiotic cells and the relevance of the CDK consensus sites for this phosphorylation. Pat1-114/ pat1-114 mutant cells were used to induce synchronous 1078
5 Bub1p kinase is regulated by Cdc2p Fig. 4. Kinase activity of Bub1p is required but not suf cient for spindle checkpoint. (A) Conservation of K762 residue in the kinase domain of Bub1p. (B) Left panel: kinase mutants. bub1-k762r carries a point mutation and bub1-dkinase lacks the entire kinase domain. Right panel: cell extracts from exponentially growing bub1-ha, bub1-k762r-ha and bub1-dkinase-ha strains were analysed by western blotting using anti-ha antibody. Cdc2p is shown as loading control. (C) See Figure 3 legend for details. The concentrations of TBZ used are indicated in mg/ml. (D) Cdc2-M26 (WT), cdc2-m26 bub1-k762r (bub1-k762r), cdc2-m26 bub1dkinase (bub1dkinase), cdc2-m26 bub1-p*-dkinase (bub1-p*-dkinase) and cdc2- M26 bub1d (bub1d) strains were used for the experiment described in Figure 3B. (E) The experiment described in Figure 3C was performed using kinase mutants. meiotic divisions (BaÈhler et al., 1991). Pre-meiotic DNA synthesis occurred 2 h after the shift (Figure 5A, left panel), followed by MI and MII (Figure 5A, right panel). Western blot analysis revealed that Bub1-HAp becomes hyperphosphorylated 4 h after the shift, corresponding to the time of the meiotic nuclear divisions (Figure 5B). This hyperphosphorylation is maintained for 1h and was not present when the Cdc2p consensus sites were mutated (Figure 5C). In mes1 mutant cells, which block between MI and MII (Figure 6A; Shimoda et al, 1985), Bub1-HAp became hyperphosphorylated at the rst meiotic division (Figure 6B) and dephosphorylated at MI exit (Figure 6B). In mei4 mutant cells, which arrest at meiotic prophase (Figure 6A; Shimoda et al., 1985; BaÈhler et al., 1993; Horie et al., 1998), Bub1-HAp did not become hyperphosphorylated (Figure 6C). These data show that Bub1p hyperphosphorylation at Cdc2p consensus sites occurs during MI, after meiotic prophase. Bub1p phosphorylation and sister chromatid segregation To determine whether Cdc2p-dependent phosphorylation of Bub1p is required for reductional segregation, we scored cen1-gfp distribution during MI (Nabeshima et al., 1998; Watanabe and Nurse, 1999). In bub1 + cells, only 3% of the cells showed equational distribution. In contrast, in 1079
6 S.Yamaguchi, A.Decottignies and P.Nurse Fig. 5. Bub1p becomes hyperphosphorylated at CDK consensus sites during meiotic divisions. (A) FACS analysis (left panel) and percentage of cells with 1, 2 or 4 nuclei (right panel) of pat1-induced synchronous meiosis using pat1-114/pat1-114 bub1-ha/bub1ha strain (pat1-114 bub1-ha). Time after induction is indicated in hours. (B) Western blotting of cell extracts from the time-course experiment described in (A) using anti-ha and anti- Cdc13p antibodies. Cdc13p level is shown as an indication of Cdc13p-associated Cdc2p kinase activity. (C) Western blotting of cell extracts from synchronous meiosis using pat1-114 bub1-ha and pat1-114 bub1-p*-ha strains. Fig. 6. Bub1p becomes phosphorylated during meiosis I. (A) mei4 is required for exit from prophase I, and mes1 is required for entry into meiosis II. (B) Percentage of cells with 1or 2 nuclei at each time point as scored by DAPI staining (left panel) and western blotting of cell extracts (right panel) from synchronous meiosis using pat1-114 mes1d strain. (C) Western blotting of extracts from pat1-114 and pat1-114 mei4d strains. Time after induction is indicated in hours. 1080
7 Bub1p kinase is regulated by Cdc2p Fig. 7. Bub1p kinase activity is required for maintenance of centromeric Rec8p during anaphase I and reductional segregation. (A) Spore viability (%) and equational segregation frequency (%) of WT, bub1d, bub1-p*, bub1-dkinase, bub1-k762r and mad2d cells. (B) In vivo localization of Bub1±GFPp (WT), Bub1-P*±GFPp (bub1-p*), Bub1-Dkinase±GFPp (bub1-dkinase) and Bub1-K762R±GFPp (bub1-k762r) during meiosis. (C) Rec8±GFPp localization during anaphase I in WT, bub1d, bub1-p*, bub1-dkinase and bub1-k762r cells. the absence of Bub1p, cells underwent an equational division in 45% of MI events (Figure 6A), in agreement with data published previously (Bernard et al., 2001). Spore viability was also reduced to 60% (Figure 7A). In the bub1-p* mutant (90% spore viability), equational divisions occurred in 14% of MI events (Figure 7A). The Bub1-P*±GFP protein had a localization similar to WT Bub1±GFPp (Figure 7B). We conclude that although phosphorylation has little effect on spore viability, it does have some role in ensuring reductional division during MI. Reductional segregation requires Bub1p kinase activity To examine the role of the Bub1p kinase domain in sister chromatid cohesion during MI, cen1-gfp distribution in both the bub1dkinase and the bub1-k762r mutants was examined. In the bub1dkinase mutant, equational segregation of cen1-gfp occurred in 43% of MI events, a value similar to the 45% observed in bub1d cells (Figure 7A). In the bub1-k762r mutant, 34% of the MI cells underwent equational segregation, whilst in the mad2d cells, equational segregation was observed in only 2% of MI cells (Figure 7A; Bernard et al., 2001). These results show that the kinase activity of Bub1p is required for complete reductional segregation of sister chromatids during MI. The failure of Bub1p kinase mutants to maintain ef cient sister chromatid cohesion during MI may be due to mislocalization of the mutant protein. We examined the localization of Bub1p kinase mutants during meiotic divisions (Figure 7B). In zygotes undergoing meiotic divisions, WT Bub1±GFPp became associated with the centromeres after meiotic prophase and remained associated until anaphase I, con rming previous reports using xed cells (Bernard et al., 2001; Figure 7B, WT, f±h). Both the Bub1Dkinase±GFPp and Bub1-K762R±GFPp proteins were associated with centromeres during anaphase I (Figure 7B, bub1-dkinase, d±g and bub1- K762R). These data suggest that the effect of Bub1p kinase domain on sister chromatid segregation in MI is not due to a failure of mutant proteins to localize to the centromeres, although the localization of kinase mutant proteins within the centromeric region may be modi ed. Watanabe and Nurse (1999) have shown that maintenance of Rec8p at the centromeric regions during anaphase I is required for reductional segregation of sister chromatids. In MII, centromeric Rec8p is degraded, allowing separation of sister chromatids. In vivo centromeric staining of Rec8±GFPp was detected as bright dots in dividing nuclei 1081
8 S.Yamaguchi, A.Decottignies and P.Nurse (Figure 7Ca, arrows), but in the absence of Bub1p, centromeric Rec8±GFPp was no longer maintained during anaphase I (Figure 7Cb), in agreement with the results using xed cells (Bernard et al., 2001). As expected from cen1±gfp segregation in MI (Figure 7A), the centromeric localization of Rec8±GFPp was lost during anaphase I in the majority of bub1dkinase and bub1-k762r cells (Figure 7Cd and e). These results show that even though Bub1p kinase mutants localize at the centromeric region during anaphase I, they are no longer capable of maintaining Rec8p at this location. A Bub1p-dependent meiotic spindle checkpoint To investigate the existence of a meiotic spindle checkpoint in ssion yeast, we used recombination-de cient (rec) mutants, which exhibit delays in MI (Molnar et al., 2001). We used the GFP±a2-tubulin allele (Yamamoto et al., 1999) to measure the length of MI. In WT cells, the time-course experiment shows that MI spindle elongation occurs in three steps: the spindle elongates slowly between 0 and 10 min (phase I), there is no elongation between 10 and 20 min (phase II) and, nally, the spindle elongates rapidly between 20 and 40 min (phase III) (Figure 8Aa, upper panel; Mallavarapu et al., 1999). Rec7 + is an early meiotic recombination gene, deletion of which leads to non-disjunction of homologous chromosomes and low spore viability (Ponticelli and Smith, 1989; Lin et al., 1992; Molnar et al., 2001). In rec7-146 cells, we found that, after the rst step of slow elongation between 0 and 10 min (phase I), the spindle continues to elongate slowly and then undergoes regression for ~60 min (phase II) before it starts to elongate rapidly until 70 min (phase III) 1082
9 Bub1p kinase is regulated by Cdc2p (Figure 8Ac, upper panel). We found that this MI delay is dependent upon Bub1p because MI spindle elongation time is reduced to 40 min in rec7-146 bub1d cells (Figure 8Ad, upper panel). In this case, however, cells seem to lack phase II and spindle elongates continuously between 0 and 40 min (Figure 8Ad, upper panel). Next, we tested whether the delay is dependent on Bub1p kinase activity. In a rec7-146 bub1dkinase mutant, the MI delay occurs, although reduced by ~20 min compared with rec7-146 bub1 + cells; no spindle regression was observed during phase II (Figure 8Ae, upper panel). This Bub1pdependent delay was also observed in a rec14d mutant (data not shown; Evans et al., 1997; Cervantes et al., 2000). Our data suggest the existence of a Bub1p-dependent spindle checkpoint in MI. To investigate whether CDK activity would be a target of Bub1p-dependent meiotic spindle checkpoint activation, we looked at the association of Cdc2±YFPp (Decottignies et al., 2001) with MI spindle since CDKs from different species are known to regulate MT dynamics in mitosis by direct interaction with Fig. 8. Bub1p-dependent delay during meiosis I in rec7 mutants. (A) a-tubulin±gfpp (top panel) and Cdc2±YFPp on MI spindles (bottom panel) were observed in the following strains: (a) WT, (b) bub1d, (c) rec7-146, (d) rec7-146 bub1d, and (e) rec7-146 bub1dkinase. (f) Cdc2±YFP was observed on MI spindle in bub1 + -overexpressing cells (nmt1-bub1). MI spindle elongation time and the time of Cdc2±YFPp uorescence association with MI spindle (Cdc2±YFP on the MI spindle) in each strain are shown in the table. (B) Association time of Cdc2±YFPp uorescence with MI spindle was measured in rec7-146 cells combined with (a) bub1 +, (b) bub1d, (c) bub1-p*, (d) bub1-dkinase, (e) bub1-p*-dkinase and (f) mad2d. The association times were grouped into ve categories (I: 0±10, II: 10±20, III: 20±30, IV: 30±40, V: 40±50 mins). Histograms show the number of cells falling into each category. MT-associated proteins (reviewed by Andersen, 2000). In WT cells, Cdc2±YFPp uorescence stays on the MI spindle for <10 min (Figure 8Aa, lower panel). In rec7-146 cells, we found that Cdc2±YFPp remained at the spindle for ~40 min (Figure 8Ac, lower panel and Ba), persisting on the elongated spindle after separation of the nucleus into two DNA masses (Figure 8Ac, lower panel). This is never observed in WT cells. In agreement with GFP±a2- tubulin data, the association of Cdc2±YFPp with the MI spindle is reduced to <10 min in rec7-146 bub1d cells (Figure 8Ad, lower panel and Bb). The same reduction was observed in rec7-146 mad2d cells (Figure 8Bf). In rec7-146 bub1dkinase cells, the MI delay was reduced by ~20 min (Figure 8Ae, lower panel and Bd) and Cdc2±YFPp was never found on the elongated spindles. As CDK-dependent phosphorylation of Bub1p occurs during the rst meiotic division (Figures 5 and 6), we tested whether this phosphorylation plays any role in the meiotic checkpoint. However, in a rec7-146 bub1-p* mutant, the delay in MI was still present (Figure 8Bc), indicating that CDK-dependent phosphorylation does not play a signi cant role in this checkpoint. These observations suggest that the Bub1p-dependent delay in MI acts on Cdc2p association with the spindle and occurs at two distinct stages, an early stage with one nuclear mass, and a later stage with separated nuclei which requires kinase activity. We also observed a similar early MI delay in cells overexpressing bub1 + (Figure 8Af). Discussion We have found that Bub1p becomes hyperphosphorylated at CDK consensus sites in a Cdc2p-dependent manner when the ssion yeast mitotic spindle is destabilized using drugs or a b-tubulin mutant. Our in vitro results have shown that Cdc2p directly phosphorylates Bub1p at these CDK consensus sites. A Bub1-P*p mutant, which cannot be phosphorylated on the four CDK consensus sites, has a spindle checkpoint defect, although the defect is not as pronounced as in the bub1d mutant. In vivo observation of Bub1-P*±GFPp revealed that Cdc2p-dependent phosphorylation of Bub1p is not required to maintain the kinetochore localization of the enzyme during spindle damage. It has also been shown that human Bub1p is phosphorylated when the spindle is disrupted, but not during normal mitosis (Taylor et al., 2001). There are putative CDK sites present in Bub1p from human, mouse, y, budding yeast and Xenopus laevis, although the position of the sites is not conserved. Therefore it is possible that there may be a role for CDK phosphorylation of Bub1p in the mitotic spindle of eukaryotes generally. In this respect, Xenopus Bub1p has been shown to be directly phosphorylated by Cdc2p in vitro (Schwab et al., 2001). However, the function of this phosphorylation is still unclear, as the kinase activity of Bub1p was not affected by this phosphorylation (Schwab et al., 2001). In ssion yeast, the main function affected by Cdc2p-dependent phosphorylation is unlikely to be Bub1p kinase activity, as the effects of mutations in phosphorylation sites and kinase activity regarding sensitivity to MT-destabilizing drugs are additive. One hypothesis would be that Cdc2pdependent phosphorylation of Bub1p regulates signal transmission to downstream effectors like Mad3p 1083
10 S.Yamaguchi, A.Decottignies and P.Nurse Table I. Schizosaccharomyces pombe strains used in this study PN4252 bub1::bub1-ha[kanr] leu1-32 h- <bub1-ha> PN4262 cdc25±22 bub1-ha leu1-32 h- PN3822 nda3-km311 leu1 ± his2 ± h+ T.Toda PN4270 bub1::bub1-p*-ha[kanr] leu1-32 ura4-d18 h+ <bub1-p*-ha> PN4465 nda3-km311 bub1-ha his2 ± leu1-32 h+ PN4163 nda3-km311 bub1-p*-ha leu1-32 h+ PN72 cdc2-m26 leu1-32 h- PN4253 bub1-ha cdc2-m26 leu1-32 h+ PN4273 bub1-p*-ha cdc2-m26 leu1-32 h+ PN4255 bub1d::ura4 + ura4-d18 leu1-32 h- Bernard et al. (1998) PN1 972 h- PN4320 bub1::bub1-p*[ura4 + ] ura4-d18 leu1-32 h+ PN2647 mad2d::ura4 + ura4-d18 leu1-32 h- Kim et al. (1998) PN4294 bub1d::ura4 + cdc2-m26 leu1-32 ura4-d18 h- PN4329 cdc2-m26 bub1-p*[ura4 + ] ura4-d18 leu1-32 h- PN4091 bub1::bub1-gfp[kanr] leu1-32 ura4-d18 ade6-704 h- PN4464 bub1::bub1-p*-gfp[kanr] ura4-d18 leu1-32 h- PN4157 bub1::bub1dkinase-gfp[kanr] ura4-d18 leu1-32 ade6-704 h- PN4298 bub1::bub1dkinase[kanr] leu1-32 h- PN4260 bub1d::ura4 + mad2d::ura4 + ura4-d18 leu1-32 h+ PN4472 bub1::bub1dkinase[kanr] mad2d::ura4 + ura4-d18 leu1-32 h- PN4506 mad2d::ura4 + ura4-d18 bub1-p*-dkinase[kanr] leu1-32 PN4353 bub1::bub1-k762r leu1-32 h- PN4628 bub1::bub1-k762r-ha[kanr] ura4-d18 leu1-32 PN4156 bub1::bub1dkinase-ha[kanr] ura4-d18 leu1-32 ade6-704 h- PN4347 bub1::bub1-k762r-gfp[kanr] ura4-d18 leu1-32 h- PN4466 bub1::bub1-k762r cdc2-m26 leu1-32 h+ PN4330 bub1::bub1dkinase[kanr] cdc2-m26 leu1-32 h- PN4340 cdc2-m26 bub1-p*-dkinase[kanr] leu1-32 PN4259 bub1-ha/bub1-ha pat1-114/pat1-114 leu1-32/leu1-32 ade6-m210/ade6-m216 PN4297 pat1-114/pat1-114 bub1-p*-ha/bub1-p*-ha leu1-32/leu1-32 PN4295 pat1-114/pat1-114 bub1-ha/bub1-ha mei4d::ura4+/mei4d::ura4+ leu1-32/leu1-32 ura4-d18/ura4-d18 ade6-m210/ade6-m216 PN4274 pat1-114/pat1-114 bub1-ha/bub1-ha mes1d::leu2/ mes1d::leu2 leu1-32/ leu1-32 ade6-m210/ade6-m216 MKY7A-4 LacO[lys1 + ] GFP-LacI-NLS[his7 + ] ura4 ± leu1 ± h+ <cen1-gfp> Nabeshima et al. (1998) PN4095 bub1d::ura4+ ura4-d18 leu1-32 ade6-m210 cen1-gfp h+ PN4468 bub1-p*[ura4 + ] cen1-gfp leu1-32 ura4-d18 h+ PN4303 bub1dkinase[kanr] cen1-gfp leu1-32 h+ PN4354 bub1-k762r cen1-gfp ura4-d18 leu1-32 h+ PN4144 mad2d::ura4+ ura4-d18 leu1-32 cen1-gfp ade6± h+ PN2377 rec8::rec8-gfp[kanr] h- <rec8-gfp> Watanabe and Nurse (1999) PN4106 rec8-gfp bub1d::ura4 + ura4-d18 ade6-m210 h+ PN4309 rec8-gfp bub1dkinase[kanr] h+ PN4467 rec8-gfp bub1-k762r h+ PN4469 rec8-gfp bub1-p*[ura4 + ] ura4 ± h+ PN4145 rec8d::kanr bub1-gfp[kanr] ura4-d18 h+ GP985 rec7-146::tn10llk ade6-m26 leu1-32 ura4-294 h+ <rec7-146> G.Smith PN4068 cdc2-yfp[sup3-5] Dnmt1-cdc2::kanR leu1-32 ura4-d18 ade6-704 h-<cdc2-yfp> Decottignies et al. (2001) PN4470 rec7-146 cdc2-yfp ura4 ± leu1-32 h- PN4474 rec7-146 cdc2-yfp bub1d::ura4+ ura4 ± leu1-32 h- PN4473 rec7-146 cdc2-yfp bub1dkinase[kanr] ura4 ± leu1-32 h- PN4476 rec7-146 cdc2-yfp bub1-p*[ura4+] ura4 ± leu1-32 h- PN4357 bub1::nmt1-bub1[kanr] cdc2-yfp leu1-32 ura4-d18 h- PN4617 rec7-146::tn10llk cdc2-yfp bub1-p*-dkinase[kanr] leu1-32 ura4-d18 PN4502 rec7-146::tn10llk cdc2-yfp mad2d::ura4 + ura4-d18 leu1-32 GP2004 rec14d::leu2 leu1-32 ade6-52 h- G.Smith PN3774 leu1-32 ura4d18 nmt1::gfp-a-tubulin integrant h- <GFP-tub> Yamamoto et al. (1999) PN4622 GFP-tub bub1d::ura4 + ura4-d18 leu1-32 PN4623 GFP-tub rec7-146::tn10llk leu1-32 PN4624 GFP-tub rec7-146::tn10llk bub1d::ura4+ leu1-32 ura4-d18 PN4626 GFP-tub rec7-146::tn10llk bub1-dkinase[kanr] leu1-32 ura4-d18 MAG071 mad2::mad2-gfp[kanr] leu1-32 ura4 ± h- Garcia et al. (2001) PN4619 mad2-gfp[kanr] cdc2-m26 leu1-32 PN4620 mad2-gfp[kanr] bub1d::ura4 + cdc2-m26 ura4-d18 leu1-32 (Millband and Hardwick, 2002). Alternatively, CDK may regulate Bub1p interaction with other spindle checkpoint components at the kinetochore, as previously shown for the budding yeast Mps1p kinase, which is required for formation of the Mad1p/Bub1p/Bub3p complex (Brady and Hardwick, 2000). Strains lacking the kinase domain of Bub1p (bub1- Dkinase) or with a point mutation in the kinase domain 1084
11 Bub1p kinase is regulated by Cdc2p (bub1-k762r) are partially checkpoint defective. In Xenopus, it has been reported that Bub1p kinase activity is required for metaphase arrest (Tunquist et al., 2002), although recruitment of other spindle checkpoint proteins does not require Bub1p kinase activity (Sharp-Baker and Chen, 2001). Bub1-Dkinase±GFPp and Bub1-K762R± GFPp are able to localize to kinetochores during spindle damage, indicating that Bub1p kinase activity is not required for Bub1p recruitment to the kinetochores. When both the CDK sites and the kinase domain of Bub1p are mutated, cells are more sensitive to MT-destabilizing drugs, but are less sensitive than bub1d cells. We conclude that both CDK phosphorylation and kinase activity are important for Bub1p function, and that Bub1p may also have a role in the structural maintenance of the mitotic kinetochore. Unlike the situation in mitosis, we found that Bub1p becomes hyperphosphorylated at CDK sites during meiosis. This hyperphosphorylation occurs during MI, after meiotic prophase. Sister chromatids of chromosome I separate in 14% of the bub1-p* cells during MI compared with 3% in WT cells, indicating that Cdc2p-dependent phosphorylation of Bub1p plays some role in chromosome segregation during MI. Xenopus Bub1p has also been reported to be regulated by phosphorylation through the MAPK pathway during MI and MII, and it has been suggested that phosphorylation may be required for metaphase arrest in oocytes (Schwab et al., 2001). Deletion of the kinase domain of Bub1p has a greater effect on chromosome segregation in MI, with equational segregation of chromosome I occurring in 43% of the bub1-dkinase cells. There is also a premature loss of centromeric Rec8±GFPp in this mutant, although the Bub1-Dkinase±GFPp protein is still localized at the kinetochores. Bub1D cells show 45% equational segregation, indicating that Bub1p function in MI is largely dependent on its kinase domain. The inability of Bub1- Dkinasep to maintain centromeric Rec8p suggests that the role of Bub1p in meiosis may be to phosphorylate kinetochore components that speci cally protect centromeric cohesin from being degraded or delocalized. The maintenance of centromeric cohesion may re ect a `local' effect of Bub1p kinase activity since the majority of Rec8p, found along the chromatid arms, is degraded in anaphase I (Watanabe and Nurse, 1999). Live observation of GFP±a-tubulin in rec7-146 mutants revealed that progression through MI was delayed (Molnar et al., 2001). We found that MI spindle elongation time was increased by ~30 min compared with WT cells, with the spindle undergoing slow elongation followed by regression before the nal elongation step. Deletion of the bub1 gene abolishes the MI delay in rec7 mutants, demonstrating that there is a bub1-dependent delay during meiotic progression. The delay in meiotic progression may be analogous to the spindle checkpoint during mitosis because defects in homologous chromosome pairing are likely to reduce the ef ciency of kinetochore capture by MTs. Observation of CDK Cdc2±YFPp in rec7-146 and rec14d diploids revealed that, like in mitosis, CDK is likely to be a target of the spindle checkpoint in meiosis, since Cdc2±YFPp remains associated with the spindle of separating nuclei, in contrast to WT cells, where the CDK leaves the metaphase spindle before anaphase I (Decottignies et al., 2001). The increased association time of Cdc2±YFPp with the spindle was no longer observed in a rec7-146 bub1d (or rec14d bub1d) diploid, suggesting that, in recombination-de cient mutants, a bub1- dependent checkpoint signal is sent, resulting in Cdc2p remaining on the spindle. Like the mitotic spindle checkpoint, some checkpoint function remains when the kinase domain of Bub1p is deleted. On the other hand, Cdc2p-dependent phosphorylation of Bub1p plays only a minor role in MI delay. We have identi ed a new role for Cdc2p in the Bub1pdependent spindle checkpoint control in ssion yeast. Phosphorylation by a CDK is required for complete function of Bub1p during spindle damage, and may also have a role during meiosis I. Cdc2p-dependent regulation of Bub1p and the kinase activity of Bub1p appear to act in different pathways, and Bub1p kinase activity is required to establish proper kinetochore function. The conservation of Bub1p kinases from yeast to human suggests that similar controls may operate in higher eukaryotes. Materials and methods Fission yeast strains and methods All experiments were carried out in Edinburgh minimal media (EMM2) or rich media (YE5S), and growth conditions are as described previously (Moreno et al., 1991). Strains used in this study are shown in Table I. Temperature-sensitive strains were grown exponentially at 25 C and shifted to 36 C, the nda3-km311 strain was grown at 32 C and shifted to 18 C, and other strains were grown at 32 C. Yeast transformation was performed as described previously (BaÈhler et al., 1998). Septation was scored by calco uor staining and nuclear division by DAPI staining, after xing cells in 70% ethanol. Western blotting and phosphatase treatment Cells were boiled for 6 min after washing with STOP buffer (150 mm NaCl, 50 mm NaF, 1mM NaN 3, 10 mm EDTA ph 8.0). Protein extracts were made by vortexing with glass beads in HB buffer (Moreno et al., 1991) and protein concentration was determined with the BCA assay kit (Sigma). Cell extracts were boiled in 23 sample buffer for 3 min and 50 mg of each sample were loaded onto an 8% SDS±polyacrylamide gel (Laemmli, 1970). Visualization of Bub1p phosphorylation was achieved using acrylamide containing low bis concentration (200:1) (Lindsay et al., 1998). Proteins were transferred onto ImmobilonÔ-P membrane (Millipore) and detected using ECL (Amersham). The antibodies used were anti-ha monoclonal antibody 12CA5 (1:1000; Babco), anti-cdc13p monoclonal antibody 6F (1:500; Hayles and Steel), anti-cdc2p monoclonal antibody Y63 (1:200; Yamano et al., 1996) and anti-gst polyclonal antibody GST1(1:2000; Sigma). l phosphatase treatments were performed as described previously (Yamaguchi et al., 2000), using l phosphatase (New England Biolab). Site-directed mutagenesis of CDK phosphorylation consensus sites and kinase domain The serine and threonine residues of the four CDK phosphorylation consensus sites S/T-P-X-K/R (T340, T423, T455 and S466) and K762 were changed to alanine and arginine respectively, using the Quikchange site-directed mutagenesis kit (Stratagene). The mutated clones were con rmed by sequencing of the open reading frame. The genomic bub1 fragment carrying the mutations was then transformed into bub1d::ura4 + cells and integrants were selected for 5-FOA resistance and con rmed by colony PCR. Construction of GST±Bub1-Dkinasep The 4.5 kb KpnI/DraIII fragment containing bub1 + ORF was isolated from cosmid SPCC1322 (kindly provided by the Sanger Center). The puri ed 4.5 kb was subsequently digested by BamHI and cloned into BamHI/KpnI-cleaved psk. The bub1dkinase gene was isolated from EcoRI/BamHI-cleaved psk::bub1 + and cloned into EcoRI/BamHIcleaved pgex-kt (Hakes and Dixon, 1992). The 1.2 kb BamHI/SacI fragment of pgex-kt::bub1dkinase was replaced with a 0.8 kb PCR 1085
12 S.Yamaguchi, A.Decottignies and P.Nurse fragment ampli ed on S.pombe genomic DNA using the 5 -CGGGAT- CCATGTCCGATTGGCGG and 5 -CGAGCTCTTATCATAGAGC- AGG primers. The resulting pgex-kt::short bub1dkinase plasmid allows bacterial expression of GST±Bub1Dkinasep. The same strategy was used to get pgex-kt::short bub1-p*-dkinase. Puri cation of GST±Bub1-Dkinasep Bacteria cells (JM109) carrying pgex-kt::short bub1dkinase or pgex- KT::short bub1-p*-dkinase plasmid were grown to OD 595 = 0.3 and protein expression was induced by adding 1mM IPTG for 4 h at 25 C. Extracts were puri ed using glutathione±sepharose 4 fast ow (Amersham Bioscience). Protein concentration was estimated on gel by Coomassie Blue staining. In vitro phosphorylation Five microliters of anti-cdc2p polyclonal antibody C2 (Simanis and Nurse, 1986) were incubated in the presence of Sepharose bead-coupled protein A in HB buffer (Moreno et al., 1991) for 30 min at room temperature. After three washes with HB buffer, 5 mg of S.pombe cell extracts were added to the beads and rotated for 3 h at 4 C. Immunoprecipitates (S.pombe Cdc2p) were washed three times with HB and incubated with 200 mm ATP, 40 mci/ml [g- 32 P]ATP, and GST±Bub1-Dkinasep, GST±Bub1-P*-Dkinasep or GST (Sigma) at 30 C for 30 min. In vitro phosphorylation by H.sapiens Cdc2p (New England Biolab) was performed in the same conditions. The reactions were stopped by boiling for 3 min in the presence of 23 SDS sample buffer. Samples were analysed by SDS±polyacrylamide gel. pat1 ts -induced meiosis To induce meiosis, pat1-114/pat1-114 ade6-m216/m210 leu1-32/leu1-32 cells were grown to mid-exponential phase at 25 C in minimal medium containing 1% glucose, 5 g/l NH 4 Cl and 0.1g/l leucine. Cells were washed three times, shifted to minimal medium containing 1% glucose and 0.05 g/l leucine and incubated for 16 h at 25 C to arrest the cells in G 1. Cells were then shifted to 34 C to inactivate Pat1p and at the same time 0.5 g/l NH 4 Cl and 0.05 g/l leucine were added. Samples were taken every 30 min for 8 h and processed for western blot analysis, FACS analysis and DAPI staining. Cell methods FACS analysis was performed as described previously (Sazer and Sherwood, 1990) (Becton Dickinson). The sister chromatid segregation pattern during MI was scored by using cen1±gfp strains (Nabeshima et al., 1998). Cells of opposite mating types, one of which carried cen1±gfp, were crossed and cells in anaphase I were scored. Spore viability was scored using tetrad analysis. Live uorescence microscopy was performed as described in Decottignies et al. (2001). For Figure 8B, cells with MI spindles were observed every 10 min for 50 min. GFP- and HA-C-terminus tagging of bub1 was perfomed as described previously (BaÈhler et al., 1998). For mitotic block experiments, cdc2-m26 strains were grown to exponential phase in rich medium at 25 C, shifted to 36 C and incubated for 3 h. Cells were then released to 25 C in the presence of 50 mg/ml MBC for 40 min. When the cells were shifted back to 36 C, additional MBC was added to bring the nal concentration to 100 mg/ml. For the TBZ sensitivity test, TBZ was dissolved in DMSO at 10 mg/ml and added to YE5S plates at a nal concentration of 0 (DMSO only), 10, 15or20mg/ml. Cells were grown to cells/ml in rich medium and serial dilutions (1:10) of the cells were spotted onto the plates. Acknowledgements We would like to thank Drs Jean-Paul Javerzat, Tomohiro Matsumoto, Gerald Smith, Yasushi Hiraoka, Takashi Toda, Hiroyuki Yamano and Mitsuhiro Yanagida for providing us with strains and antibodies. We would like to thank Drs Heidi Browning, Rafael Carazo-Salas, Naomi Fersht, Damien Hermand, Klaus Leonhard, Frank Uhlmann and Takashi Toda for critical reading of the manuscript. We would like to thank Drs Annie Borgne, Yoshinori Watanabe and Stefan Weitzer for technical advice. We would like to thank all the people in the Cell Cycle Laboratory for their help and support. This work was supported by Cancer Research UK. S.Y. was supported by a Japan Society for the Promotion of Science predoctoral fellowship. A.D. was supported by the European Commission, Biotechnology Programme and Cancer Research UK. References Amon,A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69±75. Anderson,S.S. (2000) Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol., 10, 261±267. BaÈhler,J., Schuchert,P., Grimm,C. and Kohli,J. (1991) Synchronized meiosis and recombination in ssion yeast: observations with pat1-114 diploid cells. Curr. Genet., 19, 445±451. BaÈhler,J., Wyler,T., Loidl,J. and Kohli,J. (1993) Unusual nuclear structures in meiotic prophase of ssion yeast: a cytological analysis. J. Cell Biol., 121, 241±256. BaÈhler,J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever, A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for ef cient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943±951. Basu,J., Bousbaa,H., Logarinho,E., Li,Z., Williams,B.C., Lopes,C., Sunkel,C.E. and Goldberg,M.L. (1999) Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol., 146, 13±28. Bernard,P., Hardwick,K. and Javerzat,J.P. (1998) Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol., 143, 1775±1787. Bernard,P., Maure,J.F. and Javerzat,J.P. (2001) Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat. Cell Biol., 3, 522±526. Brady,D.M. and Hardwick,K.G. (2000) Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol., 10, 675±678. Buonomo,S.B., Clyne,R.K., Fuchs,J., Loidl,J., Uhlmann,F. and Nasmyth,K. (2000) Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell, 103, 387±398. Cahill,D.P., Lengauer,C., Yu,J., Riggins,G.J., Willson,J.K., Markowitz, S.D., Kinzler,K.W. and Vogelstein,B. (1998) Mutations of mitotic checkpoint genes in human cancers. Nature, 392, 300±303. Carr,A.M., MacNeill,S.A., Hayles,J. and Nurse,P. (1989) Molecular cloning and sequence analysis of mutant alleles of the ssion yeast cdc2 protein kinase gene: implications for cdc2+ protein structure and function. Mol. Gen. Genet., 218, 41±49. Cervantes,M.D., Farah,J.A. and Smith,G.R. (2000) Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell, 5, 883±888. Decottignies,A., Zarzov,P. and Nurse,P. (2001) In vivo localisation of ssion yeast cyclin-dependent kinase cdc2p and cyclin B cdc13p during mitosis and meiosis. J. Cell Sci., 114, 2627±2640. Evans,D.H., Li,Y.F., Fox,M.E. and Smith,G.R. (1997) A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics, 146, 1253±1264. Garcia,M.A., Vardy,L., Koonrugsa,N. and Toda,T. (2001) Fission yeast ch-tog/xmap215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J., 20, 3389±3401. Hakes,D.J. and Dixon,J.E. (1992) New vectors for high level expression of recombinant proteins in bacteria. Anal. Biochem., 202, 293±298. Hiraoka,Y., Toda,T. and Yanagida,M. (1984) The NDA3 gene of ssion yeast encodes b-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell, 39, 349±358. Horie,S., Watanabe,Y., Tanaka,K., Nishiwaki,S., Fujioka,H., Abe,H., Yamamoto,M. and Shimoda,C. (1998) The Schizosaccharomyces pombe mei4 + gene encodes a meiosis-speci c transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol., 18, 2118±2129. Hoyt,M.A., Totis,L. and Roberts,B.T. (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507±517. Jallepalli,P.V. and Lengauer,C. (2001) Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer, 1, 109±117. Kallio,M., Eriksson,J.E. and Gorbsky,G.J. (2000) Differences in spindle association of the mitotic checkpoint protein Mad2 in mammalian spermatogenesis and oogenesis. Dev. Biol., 225, 112±123. Kim,S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045±
13 Bub1p kinase is regulated by Cdc2p Kleckner,N. (1996) Meiosis: how could it work? Proc. Natl Acad. Sci. USA, 93, 8167±8174. Laemmli,U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680±685. Li,R. and Murray,A.W. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519±531. Lin,Y., Larson,K.L., Dorer,R. and Smith,G.R. (1992) Meiotically induced rec7 and rec8 genes of Schizosaccharomyces pombe. Genetics, 132, 75±85. Lindsay,H.D., Grif ths,d.j., Edwards,R.J., Christensen,P.U., Murray, J.M., Osman,F., Walworth,N. and Carr,A.M. (1998) S-phase-speci c activation of Cds1kinase de nes a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev., 12, 382±395. Mallavarapu,A., Sawin,K. and Mitchison,T. (1999) A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol., 9, 1423±1426. Maller,J., Gautier,J., Langan,T.A., Lohka,M.J., Shenoy,S., Shalloway,D. and Nurse,P. (1989) Maturation-promoting factor and the regulation of the cell cycle. J. Cell Sci. (Suppl.), 12, 53±63. Millband,D.N. and Hardwick,K.G. (2002) Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p- and Mph1p-dependent manner. Mol. Cell. Biol., 22, 2728±2742. Millband,D.N., Campbell,L. and Hardwick,K.G. (2002) The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol., 12, 205±209. Molnar,M., Parisi,S., Kakihara,Y., Nojima,H., Yamamoto,A., Hiraoka,Y., Bozsik,A., Sipiczki,M. and Kohli,J. (2001) Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics, 157, 519±532. Moreno,S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of ssion yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795±823. Nabeshima,K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase±anaphase transition in ssion yeast: Dis1is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211±3225. Nigg,E.A. (1995) Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays, 17, 471±480. Nurse,P. (2000) A long twentieth century of the cell cycle and beyond. Cell, 100, 71±78. Ponticelli,A.S. and Smith,G.R. (1989) Meiotic recombination-de cient mutants of Schizosaccharomyces pombe. Genetics, 123, 45±54. Roberts,B.T., Farr,K.A. and Hoyt,M.A. (1994) The Saccharomyces cerevisiae checkpoint gene BUB1encodes a novel protein kinase. Mol. Cell. Biol., 14, 8282±8291. Sazer,S. and Sherwood,S.W. (1990) Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in ssion yeast. J. Cell Sci., 97, 509±516. Schwab,M.S., Roberts,B.T., Gross,S.D., Tunquist,B.J., Taieb,F.E., Lewellyn,A.L. and Maller,J.L. (2001) Bub1 is activated by the protein kinase p90(rsk) during Xenopus oocyte maturation. Curr. Biol., 11, 141±150. Seeley,T.W., Wang,L. and Zhen,J.Y. (1999) Phosphorylation of human MAD1by the BUB1kinase in vitro. Biochem. Biophys. Res. Commun., 257, 589±595. Sharp-Baker,H. and Chen,R.H. (2001) Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3 and CENP-E, independently of its kinase activity. J. Cell Biol., 153, 1239±1250. Shimoda,C., Hirata,A., Kishida,M., Hashida,T. and Tanaka,K. (1985) Characterization of meiosis-de cient mutants by electron microscopy and mapping of four essential genes in the ssion yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 200, 252±257. Shonn,M.A., McCarroll,R. and Murray,A.W. (2000) Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science, 289, 300±303. Simanis,V. and Nurse,P. (1986) The cell cycle control gene cdc2 + of ssion yeast encodes a protein kinase potentially regulated by phosphorylation. Cell, 45, 261±268. Straight,A.F. (1997) Cell cycle: checkpoint proteins and kinetochores. Curr. Biol., 7, R613±R616. Taylor,S.S., Hussein,D., Wang,Y., Elderkin,S. and Morrow,C.J. (2001) Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1and BubR1are differentially regulated by spindle events in human cells. J. Cell Sci., 114, 4385±4395. Taylor,S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727±735. Tunquist,B.J., Schwab,M.S., Chen,L.G. and Mallar,J.L. (2002) The spindle checkpoint kinase Bub1and Cyclin E/Cdk2 both contribute to the establishment of meitoc metaphase arrest by cytostatic factor. Curr. Biol., 12, 1027±1033. Watanabe,Y. and Nurse,P. (1999) Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature, 400, 461±464. Watanabe,Y., Yokobayashi,S., Yamamoto,M. and Nurse,P. (2001) Premeiotic S phase is linked to reductional chromosome segregation and recombination. Nature, 409, 359±363. Yamaguchi,S., Okayama,H. and Nurse,P. (2000) Fission yeast Fizzyrelated protein srw1p is a G 1 -speci c promoter of mitotic cyclin B degradation. EMBO J., 19, 3968±3977. Yamamoto,A., West,R.R., McIntosh,J.R. and Hiraoka,Y. (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and ef cient meiotic recombination in ssion yeast. J. Cell Biol., 145, 1233±1249. Yamano,H., Gannon,J. and Hunt,T. (1996) The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J., 15, 5268±5279. Yu,H.G., Muszynski,M.G. and Kelly Dawe,R. (1999) The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol., 145, 425±435. Received July 29, 2002; revised November 14, 2002; accepted January 7,
by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains was previously described
 Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
CYCLIN DEPENDENT KINASES AND CELL CYCLE CONTROL
 CYCLIN DEPENDENT KINASES AND CELL CYCLE CONTROL Nobel Lecture, December 9, 2001 by PAUL M. NURSE Cancer Research UK, PO Box 123, 61 Lincoln s Inn Fields, London WC2A 3PK, United Kingdom. The cell is the
CYCLIN DEPENDENT KINASES AND CELL CYCLE CONTROL Nobel Lecture, December 9, 2001 by PAUL M. NURSE Cancer Research UK, PO Box 123, 61 Lincoln s Inn Fields, London WC2A 3PK, United Kingdom. The cell is the
List, describe, diagram, and identify the stages of meiosis.
 Meiosis and Sexual Life Cycles In this topic we will examine a second type of cell division used by eukaryotic cells: meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis
Meiosis and Sexual Life Cycles In this topic we will examine a second type of cell division used by eukaryotic cells: meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis
Lecture 7 Mitosis & Meiosis
 Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
Lecture 7 Mitosis & Meiosis Cell Division Essential for body growth and tissue repair Interphase G 1 phase Primary cell growth phase S phase DNA replication G 2 phase Microtubule synthesis Mitosis Nuclear
Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA
 Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes?
 Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
Chapter 13: Meiosis and Sexual Life Cycles 1. Why is mitosis alone insufficient for the life cycle of sexually reproducing eukaryotes? 2. Define: gamete zygote meiosis homologous chromosomes diploid haploid
The cell cycle, mitosis and meiosis
 The cell cycle, mitosis and meiosis Learning objective This learning material is about the life cycle of a cell and the series of stages by which genetic materials are duplicated and partitioned to produce
The cell cycle, mitosis and meiosis Learning objective This learning material is about the life cycle of a cell and the series of stages by which genetic materials are duplicated and partitioned to produce
If and when cancer cells stop dividing, they do so at random points, not at the normal checkpoints in the cell cycle.
 Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Cancer cells have escaped from cell cycle controls Cancer cells divide excessively and invade other tissues because they are free of the body s control mechanisms. Cancer cells do not stop dividing when
Sample Questions for Exam 3
 Sample Questions for Exam 3 1. All of the following occur during prometaphase of mitosis in animal cells except a. the centrioles move toward opposite poles. b. the nucleolus can no longer be seen. c.
Sample Questions for Exam 3 1. All of the following occur during prometaphase of mitosis in animal cells except a. the centrioles move toward opposite poles. b. the nucleolus can no longer be seen. c.
Lecture 2: Mitosis and meiosis
 Lecture 2: Mitosis and meiosis 1. Chromosomes 2. Diploid life cycle 3. Cell cycle 4. Mitosis 5. Meiosis 6. Parallel behavior of genes and chromosomes Basic morphology of chromosomes telomere short arm
Lecture 2: Mitosis and meiosis 1. Chromosomes 2. Diploid life cycle 3. Cell cycle 4. Mitosis 5. Meiosis 6. Parallel behavior of genes and chromosomes Basic morphology of chromosomes telomere short arm
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Reproduction Growth and development Tissue removal Example
Lecture 11 The Cell Cycle and Mitosis
 Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
Lecture 11 The Cell Cycle and Mitosis In this lecture Cell division Chromosomes The cell cycle Mitosis PPMAT Apoptosis What is cell division? Cells divide in order to reproduce themselves The cell cycle
Chapter 13: Meiosis and Sexual Life Cycles
 Name Period Chapter 13: Meiosis and Sexual Life Cycles Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know.
Name Period Chapter 13: Meiosis and Sexual Life Cycles Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know.
Chapter 12: The Cell Cycle
 Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Example 2. What is meant by the cell cycle? Concept 12.1
Name Period Chapter 12: The Cell Cycle Overview: 1. What are the three key roles of cell division? State each role, and give an example. Key Role Example 2. What is meant by the cell cycle? Concept 12.1
The Somatic Cell Cycle
 The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
Anti-ATF6 α antibody, mouse monoclonal (1-7)
 Anti-ATF6 α antibody, mouse monoclonal (1-7) 73-500 50 ug ATF6 (activating transcription factor 6) is an endoplasmic reticulum (ER) membrane-bound transcription factor activated in response to ER stress.
Anti-ATF6 α antibody, mouse monoclonal (1-7) 73-500 50 ug ATF6 (activating transcription factor 6) is an endoplasmic reticulum (ER) membrane-bound transcription factor activated in response to ER stress.
LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS
 LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS Los Angeles Mission College Biology 3 Name: Date: INTRODUCTION BINARY FISSION: Prokaryotic cells (bacteria) reproduce asexually by binary fission. Bacterial
LAB 8 EUKARYOTIC CELL DIVISION: MITOSIS AND MEIOSIS Los Angeles Mission College Biology 3 Name: Date: INTRODUCTION BINARY FISSION: Prokaryotic cells (bacteria) reproduce asexually by binary fission. Bacterial
CHROMOSOME STRUCTURE CHROMOSOME NUMBERS
 CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
CHROMOSOME STRUCTURE 1. During nuclear division, the DNA (as chromatin) in a Eukaryotic cell's nucleus is coiled into very tight compact structures called chromosomes. These are rod-shaped structures made
Angelo Peschiaroli, N. Valerio Dorrello, Daniele Guardavaccaro, Monica Venere, Thanos Halazonetis, Nicholas E. Sherman, and Michele Pagano
 Molecular Cell, Volume 23 Supplemental Data SCF βtrcp -Mediated Degradation of Claspin Regulates Recovery from the DNA Replication Checkpoint Response Angelo Peschiaroli, N. Valerio Dorrello, Daniele Guardavaccaro,
Molecular Cell, Volume 23 Supplemental Data SCF βtrcp -Mediated Degradation of Claspin Regulates Recovery from the DNA Replication Checkpoint Response Angelo Peschiaroli, N. Valerio Dorrello, Daniele Guardavaccaro,
1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells
 Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Division CELL DIVISION. Mitosis. Designation of Number of Chromosomes. Homologous Chromosomes. Meiosis
 Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
5. The cells of a multicellular organism, other than gametes and the germ cells from which it develops, are known as
 1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
1. True or false? The chi square statistical test is used to determine how well the observed genetic data agree with the expectations derived from a hypothesis. True 2. True or false? Chromosomes in prokaryotic
Cell Division Mitosis and the Cell Cycle
 Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
REVIEW ARTICLE DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts
 Biochem. J. (2000) 349, 1 12 (Printed in Great Britain) 1 REVIEW ARTICLE DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts Hiroshi MURAKAMI 1 and
Biochem. J. (2000) 349, 1 12 (Printed in Great Britain) 1 REVIEW ARTICLE DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts Hiroshi MURAKAMI 1 and
Discovery of MPF (M-Phase Promoting Factor)
 Discovery of MPF (M-Phase Promoting Factor) transfert de cytoplasme après GVBD Dettlaff, 1964 Bufo sp. GVBD GVBD Masui & Markert, 1971 ; Smith & Ecker, 1971 Rana pipiens GVBD GVBD Kishimoto, 1976 Etoile
Discovery of MPF (M-Phase Promoting Factor) transfert de cytoplasme après GVBD Dettlaff, 1964 Bufo sp. GVBD GVBD Masui & Markert, 1971 ; Smith & Ecker, 1971 Rana pipiens GVBD GVBD Kishimoto, 1976 Etoile
PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY
 Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Cell Growth and Reproduction Module B, Anchor 1
 Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Cell Growth and Reproduction Module B, Anchor 1 Key Concepts: - The larger a cell becomes, the more demands the cell places on its DNA. In addition, a larger cell is less efficient in moving nutrients
Cellular Reproduction
 9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
9 Cellular Reproduction section 1 Cellular Growth Before You Read Think about the life cycle of a human. On the lines below, write some of the stages that occur in the life cycle of a human. In this section,
Bio EOC Topics for Cell Reproduction: Bio EOC Questions for Cell Reproduction:
 Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
Bio EOC Topics for Cell Reproduction: Asexual vs. sexual reproduction Mitosis steps, diagrams, purpose o Interphase, Prophase, Metaphase, Anaphase, Telophase, Cytokinesis Meiosis steps, diagrams, purpose
Chromatin Immunoprecipitation
 Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
pcas-guide System Validation in Genome Editing
 pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION
 Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION Susan Carson Heather B. Miller D.Scott Witherow ELSEVIER AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN
Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION Susan Carson Heather B. Miller D.Scott Witherow ELSEVIER AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN
CHAPTER 10 CELL CYCLE AND CELL DIVISION
 CHAPTER 10 CELL CYCLE AND CELL DIVISION Cell division is an inherent property of living organisms. It is a process in which cells reproduce their own kind. The growth, differentiation, reproduction and
CHAPTER 10 CELL CYCLE AND CELL DIVISION Cell division is an inherent property of living organisms. It is a process in which cells reproduce their own kind. The growth, differentiation, reproduction and
Biology 3A Laboratory MITOSIS Asexual Reproduction
 Biology 3A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Biology 3A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Recombinant DNA Technology
 Recombinant DNA Technology Dates in the Development of Gene Cloning: 1965 - plasmids 1967 - ligase 1970 - restriction endonucleases 1972 - first experiments in gene splicing 1974 - worldwide moratorium
Recombinant DNA Technology Dates in the Development of Gene Cloning: 1965 - plasmids 1967 - ligase 1970 - restriction endonucleases 1972 - first experiments in gene splicing 1974 - worldwide moratorium
PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI,
 Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
www.njctl.org PSI Biology Mitosis & Meiosis
 Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Mitosis and Meiosis Mitosis Classwork 1. Identify two differences between meiosis and mitosis. 2. Provide an example of a type of cell in the human body that would undergo mitosis. 3. Does cell division
Recombinant DNA Unit Exam
 Recombinant DNA Unit Exam Question 1 Restriction enzymes are extensively used in molecular biology. Below are the recognition sites of two of these enzymes, BamHI and BclI. a) BamHI, cleaves after the
Recombinant DNA Unit Exam Question 1 Restriction enzymes are extensively used in molecular biology. Below are the recognition sites of two of these enzymes, BamHI and BclI. a) BamHI, cleaves after the
Protein transfer from SDS-PAGE to nitrocellulose membrane using the Trans-Blot SD cell (Western).
 Western Blot SOP Protein transfer from SDS-PAGE to nitrocellulose membrane using the Trans-Blot SD cell (Western). Date: 8/16/05, 10/31/05, 2/6/06 Author: N.Oganesyan, R. Kim Edited by: R. Kim Summary:
Western Blot SOP Protein transfer from SDS-PAGE to nitrocellulose membrane using the Trans-Blot SD cell (Western). Date: 8/16/05, 10/31/05, 2/6/06 Author: N.Oganesyan, R. Kim Edited by: R. Kim Summary:
Genetics Lecture Notes 7.03 2005. Lectures 1 2
 Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
Genetics Lecture Notes 7.03 2005 Lectures 1 2 Lecture 1 We will begin this course with the question: What is a gene? This question will take us four lectures to answer because there are actually several
mirnaselect pep-mir Cloning and Expression Vector
 Product Data Sheet mirnaselect pep-mir Cloning and Expression Vector CATALOG NUMBER: MIR-EXP-C STORAGE: -80ºC QUANTITY: 2 vectors; each contains 100 µl of bacterial glycerol stock Components 1. mirnaselect
Product Data Sheet mirnaselect pep-mir Cloning and Expression Vector CATALOG NUMBER: MIR-EXP-C STORAGE: -80ºC QUANTITY: 2 vectors; each contains 100 µl of bacterial glycerol stock Components 1. mirnaselect
Monopolar Attachment of Sister Kinetochores at Meiosis I Requires Casein Kinase 1
 Monopolar Attachment of Sister Kinetochores at Meiosis I Requires Casein Kinase 1 Mark Petronczki, 1,6 Joao Matos, 2,6 Saori Mori, 3 Juraj Gregan, 1 Aliona Bogdanova, 2 Martin Schwickart, 2,4 Karl Mechtler,
Monopolar Attachment of Sister Kinetochores at Meiosis I Requires Casein Kinase 1 Mark Petronczki, 1,6 Joao Matos, 2,6 Saori Mori, 3 Juraj Gregan, 1 Aliona Bogdanova, 2 Martin Schwickart, 2,4 Karl Mechtler,
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes.
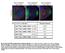 Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
LAB 09 Cell Division
 LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
The Huntington Library, Art Collections, and Botanical Gardens
 The Huntington Library, Art Collections, and Botanical Gardens Rooting for Mitosis Overview Students will fix, stain, and make slides of onion root tips. These slides will be examined for the presence
The Huntington Library, Art Collections, and Botanical Gardens Rooting for Mitosis Overview Students will fix, stain, and make slides of onion root tips. These slides will be examined for the presence
A and B are not absolutely linked. They could be far enough apart on the chromosome that they assort independently.
 Name Section 7.014 Problem Set 5 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68-120 by 5:00pm on Friday
Name Section 7.014 Problem Set 5 Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in to the box outside 68-120 by 5:00pm on Friday
From DNA to Protein
 Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
Nucleus Control center of the cell contains the genetic library encoded in the sequences of nucleotides in molecules of DNA code for the amino acid sequences of all proteins determines which specific proteins
Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network
 Research Article 843 Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network Sandra Dischinger 1,2, Andrea Krapp 1,2, Linfeng Xie 3, James R. Paulson 3 and
Research Article 843 Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network Sandra Dischinger 1,2, Andrea Krapp 1,2, Linfeng Xie 3, James R. Paulson 3 and
How to construct transgenic mice
 How to construct transgenic mice Sandra Beer-Hammer Autumn School 2012 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
How to construct transgenic mice Sandra Beer-Hammer Autumn School 2012 Bad Schandau Pharmakologie und Experimentelle Therapie (APET) Overview History Generation of embryonic stem (ES) cell lines Generation
BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis
 BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis Introduction - Fields of Genetics To answer the following question, review the three traditional subdivisions of
BioSci 2200 General Genetics Problem Set 1 Answer Key Introduction and Mitosis/ Meiosis Introduction - Fields of Genetics To answer the following question, review the three traditional subdivisions of
Expression and Purification of Recombinant Protein in bacteria and Yeast. Presented By: Puspa pandey, Mohit sachdeva & Ming yu
 Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
Expression and Purification of Recombinant Protein in bacteria and Yeast Presented By: Puspa pandey, Mohit sachdeva & Ming yu DNA Vectors Molecular carriers which carry fragments of DNA into host cell.
2.1.2 Characterization of antiviral effect of cytokine expression on HBV replication in transduced mouse hepatocytes line
 i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
1.1 Introduction. 1.2 Cells CHAPTER. 1.2.1 Prokaryotic Cells. 1.2.2 Eukaryotic Cells
 C HAPTER 1CELLS AND CELL DIVISION CHAPTER 1.1 Introduction In genetics, we view cells as vessels for the genetic material. Our main interest is in the chromosomes and their environment. This being said,
C HAPTER 1CELLS AND CELL DIVISION CHAPTER 1.1 Introduction In genetics, we view cells as vessels for the genetic material. Our main interest is in the chromosomes and their environment. This being said,
4.2 Meiosis. Meiosis is a reduction division. Assessment statements. The process of meiosis
 4.2 Meiosis Assessment statements State that meiosis is a reduction division of a diploid nucleus to form haploid nuclei. Define homologous chromosomes. Outline the process of meiosis, including pairing
4.2 Meiosis Assessment statements State that meiosis is a reduction division of a diploid nucleus to form haploid nuclei. Define homologous chromosomes. Outline the process of meiosis, including pairing
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
Protein Expression. A Practical Approach J. HIGGIN S
 Protein Expression A Practical Approach S. J. HIGGIN S B. D. HAMES List of contributors Abbreviations xv Xvi i 1. Protein expression in mammalian cell s Marlies Otter-Nilsson and Tommy Nilsso n 1. Introduction
Protein Expression A Practical Approach S. J. HIGGIN S B. D. HAMES List of contributors Abbreviations xv Xvi i 1. Protein expression in mammalian cell s Marlies Otter-Nilsson and Tommy Nilsso n 1. Introduction
Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage
 The EMBO Journal Vol.18 No.10 pp.2707 2721, 1999 Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage Gabriela Alexandru, Wolfgang
The EMBO Journal Vol.18 No.10 pp.2707 2721, 1999 Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage Gabriela Alexandru, Wolfgang
Design of conditional gene targeting vectors - a recombineering approach
 Recombineering protocol #4 Design of conditional gene targeting vectors - a recombineering approach Søren Warming, Ph.D. The purpose of this protocol is to help you in the gene targeting vector design
Recombineering protocol #4 Design of conditional gene targeting vectors - a recombineering approach Søren Warming, Ph.D. The purpose of this protocol is to help you in the gene targeting vector design
Protein Expression and Analysis. Vijay Yajnik, MD, PhD GI Unit MGH
 Protein Expression and Analysis Vijay Yajnik, MD, PhD GI Unit MGH Identify your needs Antigen production Biochemical studies Cell Biology Protein interaction studies including proteomics Structural studies
Protein Expression and Analysis Vijay Yajnik, MD, PhD GI Unit MGH Identify your needs Antigen production Biochemical studies Cell Biology Protein interaction studies including proteomics Structural studies
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257
 CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
CHAPTER 9 CELLULAR REPRODUCTION P. 243-257 SECTION 9-1 CELLULAR GROWTH Page 244 ESSENTIAL QUESTION Why is it beneficial for cells to remain small? MAIN IDEA Cells grow until they reach their size limit,
The chromosomes are structures in living cells that contain
 Brooker Widmaier Graham Stiling: III. Nucleic Acid Structure and DNA Replication 15. Eukaryotic Chromosomes, Mitosis, 47 EUKARYOTIC CHROMOSOMES, MITOSIS, AND MEIOSIS C HAPTER O UTLINE 15.1 Molecular Structure
Brooker Widmaier Graham Stiling: III. Nucleic Acid Structure and DNA Replication 15. Eukaryotic Chromosomes, Mitosis, 47 EUKARYOTIC CHROMOSOMES, MITOSIS, AND MEIOSIS C HAPTER O UTLINE 15.1 Molecular Structure
THE His Tag Antibody, mab, Mouse
 THE His Tag Antibody, mab, Mouse Cat. No. A00186 Technical Manual No. TM0243 Update date 01052011 I Description.... 1 II Key Features. 2 III Storage 2 IV Applications.... 2 V Examples - ELISA..... 2 VI
THE His Tag Antibody, mab, Mouse Cat. No. A00186 Technical Manual No. TM0243 Update date 01052011 I Description.... 1 II Key Features. 2 III Storage 2 IV Applications.... 2 V Examples - ELISA..... 2 VI
Chapter 13: Meiosis and Sexual Life Cycles
 Name Period Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know. Define: gene locus gamete male gamete female
Name Period Concept 13.1 Offspring acquire genes from parents by inheriting chromosomes 1. Let s begin with a review of several terms that you may already know. Define: gene locus gamete male gamete female
hydrocortisone (5 mg/ml), EGF (10 µg/ml) and Heparin (5000 U/ml). Antibodies against the N-terminal peptide of MEK1 (MEK1-N) and against Flotillin 1
 SUPPLEMENTARY METHODS Cells HUVEC (Human Umbilical Vein Endothelial Cells) were grown in complete Medium 199 (Gibco) supplemented with glutamax, 10% foetal calf serum, BBE (9 mg/ml), hydrocortisone (5
SUPPLEMENTARY METHODS Cells HUVEC (Human Umbilical Vein Endothelial Cells) were grown in complete Medium 199 (Gibco) supplemented with glutamax, 10% foetal calf serum, BBE (9 mg/ml), hydrocortisone (5
Pharmaceutical Biotechnology. Recombinant DNA technology Western blotting and SDS-PAGE
 Pharmaceutical Biotechnology Recombinant DNA technology Western blotting and SDS-PAGE Recombinant DNA Technology Protein Synthesis Western Blot Western blots allow investigators to determine the molecular
Pharmaceutical Biotechnology Recombinant DNA technology Western blotting and SDS-PAGE Recombinant DNA Technology Protein Synthesis Western Blot Western blots allow investigators to determine the molecular
J. Cell Sci. 128: doi:10.1242/jcs.164566: Supplementary Material
 Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Appendix C DNA Replication & Mitosis
 K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
K.Muma Bio 6 Appendix C DNA Replication & Mitosis Study Objectives: Appendix C: DNA replication and Mitosis 1. Describe the structure of DNA and where it is found. 2. Explain complimentary base pairing:
Notch 1 -dependent regulation of cell fate in colorectal cancer
 Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Germ cell formation / gametogenesis And Fertilisation
 Developmental Biology BY1101 P. Murphy Lecture 3 The first steps to forming a new organism Descriptive embryology I Germ cell formation / gametogenesis And Fertilisation Why bother with sex? In terms of
Developmental Biology BY1101 P. Murphy Lecture 3 The first steps to forming a new organism Descriptive embryology I Germ cell formation / gametogenesis And Fertilisation Why bother with sex? In terms of
Chapter 18: Applications of Immunology
 Chapter 18: Applications of Immunology 1. Vaccinations 2. Monoclonal vs Polyclonal Ab 3. Diagnostic Immunology 1. Vaccinations What is Vaccination? A method of inducing artificial immunity by exposing
Chapter 18: Applications of Immunology 1. Vaccinations 2. Monoclonal vs Polyclonal Ab 3. Diagnostic Immunology 1. Vaccinations What is Vaccination? A method of inducing artificial immunity by exposing
Understanding the immune response to bacterial infections
 Understanding the immune response to bacterial infections A Ph.D. (SCIENCE) DISSERTATION SUBMITTED TO JADAVPUR UNIVERSITY SUSHIL KUMAR PATHAK DEPARTMENT OF CHEMISTRY BOSE INSTITUTE 2008 CONTENTS Page SUMMARY
Understanding the immune response to bacterial infections A Ph.D. (SCIENCE) DISSERTATION SUBMITTED TO JADAVPUR UNIVERSITY SUSHIL KUMAR PATHAK DEPARTMENT OF CHEMISTRY BOSE INSTITUTE 2008 CONTENTS Page SUMMARY
For non-us, non-canada, non-uk healthcare professionals only Plk inhibition Mechanism of Action Slide kit
 Procedure ID: 3386 - October 2014 For non-us, non-canada, non-uk healthcare professionals only Plk inhibition Mechanism of Action Slide kit Polo-like kinases (Plks) are an emerging focus for the haemato-oncology
Procedure ID: 3386 - October 2014 For non-us, non-canada, non-uk healthcare professionals only Plk inhibition Mechanism of Action Slide kit Polo-like kinases (Plks) are an emerging focus for the haemato-oncology
Practice Problems 4. (a) 19. (b) 36. (c) 17
 Chapter 10 Practice Problems Practice Problems 4 1. The diploid chromosome number in a variety of chrysanthemum is 18. What would you call varieties with the following chromosome numbers? (a) 19 (b) 36
Chapter 10 Practice Problems Practice Problems 4 1. The diploid chromosome number in a variety of chrysanthemum is 18. What would you call varieties with the following chromosome numbers? (a) 19 (b) 36
HCS604.03 Exercise 1 Dr. Jones Spring 2005. Recombinant DNA (Molecular Cloning) exercise:
 HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
Exploiting science for engineering: BRCA2 targeted therapies
 20.109 MOD1 DNA ENGINEERING Fall 2010 Exploiting science for engineering: BRCA2 targeted therapies Orsi Kiraly Engelward lab Homologous recombination is important No HR chromosomal aberrations cell death
20.109 MOD1 DNA ENGINEERING Fall 2010 Exploiting science for engineering: BRCA2 targeted therapies Orsi Kiraly Engelward lab Homologous recombination is important No HR chromosomal aberrations cell death
Chapter 3. Cell Division. Laboratory Activities Activity 3.1: Mock Mitosis Activity 3.2: Mitosis in Onion Cells Activity 3.
 Chapter 3 Cell Division Laboratory Activities Activity 3.1: Mock Mitosis Activity 3.2: Mitosis in Onion Cells Activity 3.3: Mock Meiosis Goals Following this exercise students should be able to Recognize
Chapter 3 Cell Division Laboratory Activities Activity 3.1: Mock Mitosis Activity 3.2: Mitosis in Onion Cells Activity 3.3: Mock Meiosis Goals Following this exercise students should be able to Recognize
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE Q5B
 INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
INTERNATIONAL CONFERENCE ON HARMONISATION OF TECHNICAL REQUIREMENTS FOR REGISTRATION OF PHARMACEUTICALS FOR HUMAN USE ICH HARMONISED TRIPARTITE GUIDELINE QUALITY OF BIOTECHNOLOGICAL PRODUCTS: ANALYSIS
Workshop: Cellular Reproduction via Mitosis & Meiosis
 Workshop: Cellular Reproduction via Mitosis & Meiosis Introduction In this workshop you will examine how cells divide, including how they partition their genetic material (DNA) between the two resulting
Workshop: Cellular Reproduction via Mitosis & Meiosis Introduction In this workshop you will examine how cells divide, including how they partition their genetic material (DNA) between the two resulting
European Medicines Agency
 European Medicines Agency July 1996 CPMP/ICH/139/95 ICH Topic Q 5 B Quality of Biotechnological Products: Analysis of the Expression Construct in Cell Lines Used for Production of r-dna Derived Protein
European Medicines Agency July 1996 CPMP/ICH/139/95 ICH Topic Q 5 B Quality of Biotechnological Products: Analysis of the Expression Construct in Cell Lines Used for Production of r-dna Derived Protein
cdc2 and the Regulation of Mitosis: Six Interacting mcs Genes
 Copyright 0 1989 by the Genetics Society of America cdc2 and the Regulation of Mitosis: Six Interacting mcs Genes Lisa Molz, Robert Booher, Paul Young and David Beach Cold Spring Harbor Laboratory, Cold
Copyright 0 1989 by the Genetics Society of America cdc2 and the Regulation of Mitosis: Six Interacting mcs Genes Lisa Molz, Robert Booher, Paul Young and David Beach Cold Spring Harbor Laboratory, Cold
Cell Cycle Vignettes Irreversible Transitions in the Cell Cycle
 Cell Cycle Vignettes Irreversible Transitions in the Cell Cycle John J. Tyson & Bela Novak Department of Biological Sciences Virginia Polytechnic Institute & State University Blacksburg, Virginia USA &
Cell Cycle Vignettes Irreversible Transitions in the Cell Cycle John J. Tyson & Bela Novak Department of Biological Sciences Virginia Polytechnic Institute & State University Blacksburg, Virginia USA &
Bio 101 Section 001: Practice Questions for First Exam
 Do the Practice Exam under exam conditions. Time yourself! MULTIPLE CHOICE: 1. The substrate fits in the of an enzyme: (A) allosteric site (B) active site (C) reaction groove (D) Golgi body (E) inhibitor
Do the Practice Exam under exam conditions. Time yourself! MULTIPLE CHOICE: 1. The substrate fits in the of an enzyme: (A) allosteric site (B) active site (C) reaction groove (D) Golgi body (E) inhibitor
Test Two Study Guide
 Test Two Study Guide 1. Describe what is happening inside a cell during the following phases (pictures may help but try to use words): Interphase: : Consists of G1 / S / G2. Growing stage, cell doubles
Test Two Study Guide 1. Describe what is happening inside a cell during the following phases (pictures may help but try to use words): Interphase: : Consists of G1 / S / G2. Growing stage, cell doubles
In vitro analysis of pri-mirna processing. by Drosha-DGCR8 complex. (Narry Kim s lab)
 In vitro analysis of pri-mirna processing by Drosha-DGCR8 complex (Narry Kim s lab) 1-1. Preparation of radiolabeled pri-mirna transcript The RNA substrate for a cropping reaction can be prepared by in
In vitro analysis of pri-mirna processing by Drosha-DGCR8 complex (Narry Kim s lab) 1-1. Preparation of radiolabeled pri-mirna transcript The RNA substrate for a cropping reaction can be prepared by in
DNA Fingerprinting. Unless they are identical twins, individuals have unique DNA
 DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
DNA Fingerprinting Unless they are identical twins, individuals have unique DNA DNA fingerprinting The name used for the unambiguous identifying technique that takes advantage of differences in DNA sequence
Cell Cycle in Onion Root Tip Cells (IB)
 Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Forensic DNA Testing Terminology
 Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
CURRICULUM VITAE Rosella Visintin. Assistant professor of SEMM (European School of Molecular Medicine). IFOM-IEO Campus Milan - Italy
 CURRICULUM VITAE Rosella Visintin Positions: Since 2006 Since July 2005 Assistant professor of SEMM (European School of Molecular Medicine). IFOM-IEO Campus Milan - Italy Junior group leader at the European
CURRICULUM VITAE Rosella Visintin Positions: Since 2006 Since July 2005 Assistant professor of SEMM (European School of Molecular Medicine). IFOM-IEO Campus Milan - Italy Junior group leader at the European
Interphase (A) and mitotic (B) D16 cells were plated on concanavalin A and processed for
 E07-10-1069 Rogers SUPPLEMENTARY DATA Supplemental Figure 1. Interphase day-7 control and γ-tubulin23c RNAi-treated S2 cells stained for MTs (green), γ-tubulin (red), and DNA (blue). Scale, 5μm. Supplemental
E07-10-1069 Rogers SUPPLEMENTARY DATA Supplemental Figure 1. Interphase day-7 control and γ-tubulin23c RNAi-treated S2 cells stained for MTs (green), γ-tubulin (red), and DNA (blue). Scale, 5μm. Supplemental
(1-p) 2. p(1-p) From the table, frequency of DpyUnc = ¼ (p^2) = #DpyUnc = p^2 = 0.0004 ¼(1-p)^2 + ½(1-p)p + ¼(p^2) #Dpy + #DpyUnc
 Advanced genetics Kornfeld problem set_key 1A (5 points) Brenner employed 2-factor and 3-factor crosses with the mutants isolated from his screen, and visually assayed for recombination events between
Advanced genetics Kornfeld problem set_key 1A (5 points) Brenner employed 2-factor and 3-factor crosses with the mutants isolated from his screen, and visually assayed for recombination events between
QUANTITATIVE RT-PCR. A = B (1+e) n. A=amplified products, B=input templates, n=cycle number, and e=amplification efficiency.
 QUANTITATIVE RT-PCR Application: Quantitative RT-PCR is used to quantify mrna in both relative and absolute terms. It can be applied for the quantification of mrna expressed from endogenous genes, and
QUANTITATIVE RT-PCR Application: Quantitative RT-PCR is used to quantify mrna in both relative and absolute terms. It can be applied for the quantification of mrna expressed from endogenous genes, and
Protocol for Western Blotting
 Protocol for Western Blotting Materials Materials used on Day 3 Protease inhibitor stock: 1 μg/μl pepstatin A in DMSO 200 μm leupeptin in OG Buffer 200 mm PMSF: Freshly made. Ex) 34.8 mg PMSF in 1 ml isopropanol
Protocol for Western Blotting Materials Materials used on Day 3 Protease inhibitor stock: 1 μg/μl pepstatin A in DMSO 200 μm leupeptin in OG Buffer 200 mm PMSF: Freshly made. Ex) 34.8 mg PMSF in 1 ml isopropanol
Biology Final Exam Study Guide: Semester 2
 Biology Final Exam Study Guide: Semester 2 Questions 1. Scientific method: What does each of these entail? Investigation and Experimentation Problem Hypothesis Methods Results/Data Discussion/Conclusion
Biology Final Exam Study Guide: Semester 2 Questions 1. Scientific method: What does each of these entail? Investigation and Experimentation Problem Hypothesis Methods Results/Data Discussion/Conclusion
I. Genes found on the same chromosome = linked genes
 Genetic recombination in Eukaryotes: crossing over, part 1 I. Genes found on the same chromosome = linked genes II. III. Linkage and crossing over Crossing over & chromosome mapping I. Genes found on the
Genetic recombination in Eukaryotes: crossing over, part 1 I. Genes found on the same chromosome = linked genes II. III. Linkage and crossing over Crossing over & chromosome mapping I. Genes found on the
Genetics Test Biology I
 Genetics Test Biology I Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Avery s experiments showed that bacteria are transformed by a. RNA. c. proteins.
Genetics Test Biology I Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Avery s experiments showed that bacteria are transformed by a. RNA. c. proteins.
The Need for a PARP in vivo Pharmacodynamic Assay
 The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
The CRL4 Cdt2 Ubiquitin Ligase Mediates the Proteolysis of Cyclin-Dependent Kinase Inhibitor Xic1 through a Direct Association with PCNA
 MOLECULAR AND CELLULAR BIOLOGY, Sept. 2010, p. 4120 4133 Vol. 30, No. 17 0270-7306/10/$12.00 doi:10.1128/mcb.01135-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The CRL4 Cdt2
MOLECULAR AND CELLULAR BIOLOGY, Sept. 2010, p. 4120 4133 Vol. 30, No. 17 0270-7306/10/$12.00 doi:10.1128/mcb.01135-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The CRL4 Cdt2
STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS
 STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS THESIS SUBMITTED FOR THE DEGREB OF DOCTOR OF PHILOSOPHY (SCIENCE) OF THE UNIVERSITY OF CALCUTTA 1996 NRISINHA DE, M.Sc DEPARTMENT OF BIOCHEMISTRY
STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS THESIS SUBMITTED FOR THE DEGREB OF DOCTOR OF PHILOSOPHY (SCIENCE) OF THE UNIVERSITY OF CALCUTTA 1996 NRISINHA DE, M.Sc DEPARTMENT OF BIOCHEMISTRY
SUPPLEMENTARY DATA 1
 SUPPLEMENTARY DATA 1 Supplementary Figure S1. Overexpression of untagged Ago2 inhibits the nuclear transport of the dotted foci of GFP-signal of myc-gfp-tnrc6a-nes-mut. (A C) HeLa cells expressing myc-gfp-tnrc6a-nes-mut
SUPPLEMENTARY DATA 1 Supplementary Figure S1. Overexpression of untagged Ago2 inhibits the nuclear transport of the dotted foci of GFP-signal of myc-gfp-tnrc6a-nes-mut. (A C) HeLa cells expressing myc-gfp-tnrc6a-nes-mut
