Supplementary Information - PCR amplification PCR amplification reactions for the partial mitochondrial cytochrome oxidase subunit I (COI), the
|
|
|
- Jodie Hodges
- 8 years ago
- Views:
Transcription
1 Supplementary Information - PCR amplification PCR amplification reactions for the partial mitochondrial cytochrome oxidase subunit I (COI), the ribosomal 16S rdna gene and a fragment of the nuclear single locus recombination activating gene 1 (RAG1) spanning the highly variable 5`-end of the gene were performed in 20 µl reactions containing 2 µl 10x reaction buffer, mm MgCl 2, 0.2 mm dntps, 10 mm of forward and reverse primers, 0.5 U of AmpliTaq and ng template. Denaturation in 95 o C (10 mins) was followed by 35 cycles of a denaturation step of 94 o C (30 s), an annealing step of 50 o C, 54 o C or 57 o C for 16S, COI and RAG1 respectively for 30 s, an extension step of 72 o C (1 min) and a final extension of 72 o C (7 min). PCR products were quantified by agarose gel electrophoresis on a 1% TAE agarose gel using known standards. Amplicons were purified with AMPure XP (Agencourt) following manufacturer s recommendations and sequenced using the ABI Prism BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). 1
2 Table S1 Specimens used in this study; accession numbers of sequences and collection information. Details of the COI haplotypes are also given for the specimens of Neotrygon annotata, N. kuhlii and N. ningalooensis. Species Sample # COI 16s RAG1 Collection location Lat Long COI Haplotype Neotrygon annotata A11229 KC KC KC Australia, QLD, Torres Strait Hap35 Neotrygon annotata A11230 KC KC KC Australia, QLD, Torres Strait Hap35 Neotrygon annotata A11231 KC KC KC Australia, QLD, Torres Strait Hap37 Neotrygon cf annotata A2643 EU Australia, QLD, Torres Strait Hap36 Neotrygon cf annotata A2644 EU Indonesia, West Java, Muara Angke Hap36 Neotrygon cf annotata A2645 EU KC Indonesia, West Java, Muara Angke Hap36 Neotrygon cf annotata A2646 EU KC KC Indonesia, West Java, Muara Angke Hap36 Neotrygon cf annotata A2647 EU KC KC Indonesia, West Java, Muara Angke Hap36 Neotrygon kuhlii clade 1 A2584 EU KC KC Taiwan, Penghu Islands Hap1 Neotrygon kuhlii clade 1 A2585 EU KC Taiwan, Penghu Islands Hap3 Neotrygon kuhlii clade 1 A2586 EU KC Taiwan, Penghu Islands Hap1 Neotrygon kuhlii clade 1 A2587 KC KC Taiwan, Penghu Islands Hap1 Neotrygon kuhlii clade 1 A6218 GU KC KC Thailand, Gulf Coast Hap1 Neotrygon kuhlii clade 1 A6219 GU KC Thailand, Gulf Coast Hap1 Neotrygon kuhlii clade 1 A6276 GU KC KC Vietnam, Con Son Island fish market Hap2 Neotrygon kuhlii clade 1 A7817 HM KC Taiwan, Penghu Islands Hap1 Neotrygon kuhlii clade 1 A7818 KC Taiwan, Penghu Islands Neotrygon kuhlii clade 1 A7819 HM KC Taiwan, Penghu Islands Hap1 Neotrygon kuhlii clade 1 A7820 KC Taiwan, Penghu Islands Neotrygon kuhlii clade 1 BO409 KC KC KC Malaysia, Sarawak Hap2 Neotrygon kuhlii clade 2 A2575 EU KC KC Indonesia, West Java, Muara Angke Hap6 Neotrygon kuhlii clade 2 A2576 EU KC KC Indonesia, West Java, Muara Angke Hap4 Neotrygon kuhlii clade 2 A2577 EU KC Indonesia, West Java, Muara Angke Hap9 Neotrygon kuhlii clade 2 A2578 EU KC Indonesia, West Java, Muara Angke Hap7 Neotrygon kuhlii clade 2 A2579 EU KC KC Indonesia, West Java, Muara Angke Hap8 Neotrygon kuhlii clade 2 A6221 GU KC KC Thailand, Gulf Coast Hap5 Neotrygon kuhlii clade 2 A7737 GU KC Indonesia, West Java, Muara Angke Hap9 Neotrygon kuhlii clade 2 BO424 KC KC Malaysia, Sarawak Tanjung Manis Hap5 2
3 Neotrygon kuhlii clade 2 BO473 KC KC KC Malaysia, Sarawak Mukah Hap5 Neotrygon kuhlii clade 3 A6220 GU KC KC Thailand, Gulf Coast Hap10 Neotrygon kuhlii clade 3 BO423 KC KC Malaysia, Sarawak Tanjung Manis Hap10 Neotrygon kuhlii clade 4 P AB Japan, Okinawa, Ishigaki Is Hap11 Neotrygon kuhlii clade 5 A208 DQ KC KC Australia, QLD, Gulf of Carpentaria Hap14 Neotrygon kuhlii clade 5 A5649 KC KC Australia, QLD, Off Turtle Head Is Hap14 Neotrygon kuhlii clade 5 A5650 KC KC Australia, QLD, Torres Straits Hap14 Neotrygon kuhlii clade 5 A5960 KC KC Indonesia, Lombok, Tanjung Luar Hap16 Neotrygon kuhlii clade 5 A5961 KC KC Indonesia, Lombok, Tanjung Luar Hap16 Neotrygon kuhlii clade 5 A6198 KC KC Australia, WA, Shark Bay Hap12 Neotrygon kuhlii clade 5 A6199 GU KC Australia, WA, Shark Bay Hap15 Neotrygon kuhlii clade 5 A6849 KC KC KC Australia, QLD, off Weipa Hap13 Neotrygon kuhlii clade 5 A6850 KC KC Australia, QLD, off Weipa Hap14 Neotrygon kuhlii clade 5 A7794 HM Australia, QLD, off Weipa Hap14 Neotrygon kuhlii clade 6 A2571 EU KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A2572 EU KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A2573 EU Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A2574 EU KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A2580 EF KC KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A2582 KC KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A2583 EU KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 6 A5731 KC KC KC Indonesia, Central Java, Sadang Hap17 Neotrygon kuhlii clade 6 A5737 KC KC KC Indonesia, Central Java, Sadang Hap17 Neotrygon kuhlii clade 6 A5738 KC KC Indonesia, Central Java, Sadang Hap17 Neotrygon kuhlii clade 6 A5739 KC KC KC Indonesia, Central Java, Sadang Hap17 Neotrygon kuhlii clade 6 A7812 HM KC KC Indonesia, Bali, Kedonganan Hap17 Neotrygon kuhlii clade 7 A6222 GU KC Thailand, Andaman Coast Hap18 Neotrygon kuhlii clade 7 A6223 GU KC KC Thailand, Andaman Coast Hap20 Neotrygon kuhlii clade 7 A6224 GU KC Thailand, Andaman Coast Hap20 Neotrygon kuhlii clade 7 A6225 GU KC KC Thailand, Andaman Coast Hap19 Neotrygon kuhlii clade 7 A6226 GU KC KC Thailand, Andaman Coast Hap20 Neotrygon kuhlii clade 8 CHN 156 HM HM India, Kochi, Kerala Hap21 Neotrygon kuhlii clade KC KC KC Tanzania, Tanga Hap22 Neotrygon kuhlii clade 9 A6217 GU KC KC New Caledonia, Magenta Beach Hap24 3
4 Neotrygon kuhlii clade 9 A7791 HM KC KC Australia, NSW, off Yamba Hap25 Neotrygon kuhlii clade 9 A7792 HM KC Australia, NSW Hap25 Neotrygon kuhlii clade 9 A7793 HM KC Australia, NSW Hap25 Neotrygon kuhlii clade 9 A7808 HM KC KC Australia, QLD, Gladstone Hap28 Neotrygon kuhlii clade 9 A7809 HM KC Australia, QLD, Gladstone Hap25 Neotrygon kuhlii clade 9 A7810 HM KC Australia, QLD, Gladstone Hap26 Neotrygon kuhlii clade 9 A7811 KC Australia, QLD, Gladstone Neotrygon kuhlii clade 9 A7813 HM KC KC Australia, QLD, Moreton Bay Hap25 Neotrygon kuhlii clade 9 A7814 HM KC Australia, QLD, Moreton Bay Hap27 Neotrygon kuhlii clade 9 A7815 HM KC Australia, QLD, Moreton Bay Hap25 Neotrygon kuhlii clade 9 A7816 HM KC Australia, QLD, Moreton Bay Hap25 Neotrygon kuhlii clade 9 UGA008 KC Australia, QLD, Lizard Island Hap23 Neotrygon leylandi A2917 EU KC KC Australia, WA, Exmouth Neotrygon leylandi A2935 EU KC KC Australia, WA, Shark Bay Neotrygon leylandi A6235 KC KC Australia, WA, Coral Bay Neotrygon picta A5653 KC KC KC Australia, QLD, Torres Strait Neotrygon picta A5659 KC KC KC Australia, QLD, Torres Strait Neotrygon picta A5661 KC KC Australia, QLD, Torres Strait Neotrygon ningalooensis A6233 HQ KC KC Australia, WA, Coral Bay Hap34 Neotrygon ningalooensis A6234 KC KC KC Australia, WA, Coral Bay Hap34 Neotrygon ningalooensis A6205 GU KC KC Australia, WA, Shark Bay Hap32 Neotrygon ningalooensis A6215 GU KC Australia, WA, Shark Bay Hap30 Neotrygon ningalooensis A6216 GU KC Australia, WA, Shark Bay Hap29 Neotrygon ningalooensis A6204 GU Australia, WA, Shark Bay Hap32 Neotrygon ningalooensis A6206 GU Australia, WA, Shark Bay Hap31 Neotrygon ningalooensis A6207 GU Australia, WA, Shark Bay Hap31 Neotrygon ningalooensis A6212 GU Australia, WA, Shark Bay Hap33 Outgroups Dasyatis brevicaudata EU EU Dasyatis parvonigra A3755 EU KC KC Dasyatis ushiei BW-2176 EU KC KC Dasyatis zugei A3765 EU Dasyatis zugei A3766 EU KC Himantura astra A225 DQ KC
5 Himantura walga EU KC KC Pastinachus atrus A2161 EU KC KC Pteroplatytrygon violacea HM HM HM Taeniura lymma UG0012 KC KC KC Taeniura lymma UG0713 KC KC KC Taeniurops meyeni A6227 GU KC KC Urogymnus asperrimus A4563 KC KC KC Rhynchobatus australiae A2380 EU KC Rhynchobatus australiae A2925 EU Carcharhinus leuca U62645 *All Axxxx numbers refer to corresponding Barcode of Life Database specimen numbers BW-Axxxx Table S2 Kimura 2-Parameter (K2P) corrected Neighbour Joining distance matrix with minimum and maximum values (in brackets) for the Neotrygon kuhlii mitochondrial COI clades 1 to 9. Clade ( ) ( ) 0.58 (0-0.93) ( ) 1.33 ( ) 0 (0-0) ( ) 2.54 ( ) 2.48 ( ) N/A ( ) 3.16 ( ) 3.10 ( ) 3.47 ( ) 0.53 (0-1.4) ( ) 2.61 ( ) 2.53 ( ) 3.17 ( ) 2.68 ( ) 0 (0-0) ( ) 2.35 ( ) 2.33 ( ) 3.31 ( ) 2.84 ( ) 1.69 ( ) 0.16 ( ) ( ) 2.56 ( ) 2.36 ( ) 3.17 ( ) 2.53 ( ) 1.41 ( ) 1.58 ( ) 1.56 ( ) ( ) 4.05 ( ) 4.00 ( ) 3.57 ( ) 4.53 ( ) 3.97 ( ) 4.07 ( ) 3.8 ( ) 0.3 ( ) 5
6 Table S3 K2P-corrected neighbour-joining distance matrix of Neotrygon species calculated from the COI alignment. Genetic distance is along the lower diagonal and standard error along the upper diagonal. N. annotata N. cf annotata N. ningalooensis N. cf ningalooensis N. leylandi N. picta N. kuhlii clade N. kuhlii clade N. kuhlii clade N. kuhlii clade N. kuhlii clade N. kuhlii clade N. kuhlii clade N. kuhlii clade N. kuhlii clade
7 Table S4 Mean divergence times and their 95% HPD confidence intervals based on fossil calibrations and published COI mutation rates (0.58% / My and 0.9% / My) calculated in the spotted eagle ray (A. narinari) from the isthmus of panama closure for nodes 1 to 10. Fossil Mutation rate 0.58% / My 0.9% / My Node Age (95% CI) Age (95% CI) Age (95% CI) ( ) 7.9 ( ) 11.6 ( ) ( ) 3.9 ( ) 9.0 ( ) ( ) 1.2 ( ) 7.3 ( ) ( ) 8.9 ( ) 5.8 ( ) ( ) 2.9 ( ) 1.9 ( ) ( ) 2.0 ( ) 1.4 ( ) ( ) 2.3 ( ) 1.5 ( ) ( ) 2.9 ( ) 1.9 ( ) ( ) 3.0 ( ) 2.0 ( ) ( ) 0.8 ( ) 0.5 ( ) Mutation rates from the present study COI = 0.229% per My 16s = % per My RAG1 = % per My 7
8 Figure S1 Substitution frequencies of transitions (cross) and transversions (triangle) for: a) the 1 st, b) 2 nd and c) 3 rd codon positions of the partial COI gene. Observed frequencies are calculated with the K2P genetic distance. a b a 8
9 Figure S2 Substitution frequencies of transitions (cross) and transversions (triangle) for: a) the 1 st, b) 2 nd and c) 3 rd codon positions of the partial 16s gene. Observed frequencies are calculated with the K2P genetic distance. a b c 9
10 Figure S3 Substitution frequencies of transitions (cross) and transversions (triangle) for: a) the 1 st, b) 2 nd and c) 3 rd codon positions of the partial RAG1 gene. Observed frequencies are calculated with the K2P genetic distance. a b c 10
11 Figure S4 Mismatch distributions for pairwise comparisons of Neotrygon kuhlii COI mitochondrial clades. Monomorphic clades 3, 4 and 6 were excluded from the calculation. The frequencies of expected and observed differences are given as a solid and dashed line, respectively. Figure: Ts-Tv_ratio C1 C7 Substitution frequencies of transitions (cross) and transversions (triangle) for the 1 st, 2 nd and 3 rd codon positions for the COI, 16s and RAG1 gene. Observed frequencies are calculated with the K2P genetic distance. τ = τ = C2 C8 τ = τ = C5 C9 τ = τ =
Protocols. Internal transcribed spacer region (ITS) region. Niklaus J. Grünwald, Frank N. Martin, and Meg M. Larsen (2013)
 Protocols Internal transcribed spacer region (ITS) region Niklaus J. Grünwald, Frank N. Martin, and Meg M. Larsen (2013) The nuclear ribosomal RNA (rrna) genes (small subunit, large subunit and 5.8S) are
Protocols Internal transcribed spacer region (ITS) region Niklaus J. Grünwald, Frank N. Martin, and Meg M. Larsen (2013) The nuclear ribosomal RNA (rrna) genes (small subunit, large subunit and 5.8S) are
Troubleshooting Sequencing Data
 Troubleshooting Sequencing Data Troubleshooting Sequencing Data No recognizable sequence (see page 7-10) Insufficient Quantitate the DNA. Increase the amount of DNA in the sequencing reactions. See page
Troubleshooting Sequencing Data Troubleshooting Sequencing Data No recognizable sequence (see page 7-10) Insufficient Quantitate the DNA. Increase the amount of DNA in the sequencing reactions. See page
ID kit. imegen Anchovies II. and E. japonicus) DNA detection by. User manual. Anchovies species (E. encrasicolus. sequencing.
 User manual imegen Anchovies II ID kit Anchovies species (E. encrasicolus and E. japonicus) DNA detection by sequencing Reference: Made in Spain The information in this guide is subject to change without
User manual imegen Anchovies II ID kit Anchovies species (E. encrasicolus and E. japonicus) DNA detection by sequencing Reference: Made in Spain The information in this guide is subject to change without
A Fast, Accurate, and Automated Workflow for Multi Locus Sequence Typing of Bacterial Isolates
 Application Note MLST A Fast, Accurate, and Automated Workflow for Multi Locus Sequence Typing of Bacterial Isolates Using Applied Biosystems 3130 and 3730 Series Capillary Electrophoresis Systems and
Application Note MLST A Fast, Accurate, and Automated Workflow for Multi Locus Sequence Typing of Bacterial Isolates Using Applied Biosystems 3130 and 3730 Series Capillary Electrophoresis Systems and
Real-time quantitative RT -PCR (Taqman)
 Real-time quantitative RT -PCR (Taqman) Author: SC, Patti Lab, 3/03 This is performed as a 2-step reaction: 1. cdna synthesis from DNase 1-treated total RNA 2. PCR 1. cdna synthesis (Advantage RT-for-PCR
Real-time quantitative RT -PCR (Taqman) Author: SC, Patti Lab, 3/03 This is performed as a 2-step reaction: 1. cdna synthesis from DNase 1-treated total RNA 2. PCR 1. cdna synthesis (Advantage RT-for-PCR
PrimeSTAR HS DNA Polymerase
 Cat. # R010A For Research Use PrimeSTAR HS DNA Polymerase Product Manual Table of Contents I. Description...3 II. III. IV. Components...3 Storage...3 Features...3 V. General Composition of PCR Reaction
Cat. # R010A For Research Use PrimeSTAR HS DNA Polymerase Product Manual Table of Contents I. Description...3 II. III. IV. Components...3 Storage...3 Features...3 V. General Composition of PCR Reaction
360 Master Mix. , and a supplementary 360 GC Enhancer.
 Product Bulletin AmpliTaq Gold 360 Master Mix and 360 DNA Polymerase AmpliTaq Gold 360 Master Mix AmpliTaq Gold 360 DNA Polymerase 360 Coverage for a Full Range of Targets AmpliTaq Gold 360 Master Mix
Product Bulletin AmpliTaq Gold 360 Master Mix and 360 DNA Polymerase AmpliTaq Gold 360 Master Mix AmpliTaq Gold 360 DNA Polymerase 360 Coverage for a Full Range of Targets AmpliTaq Gold 360 Master Mix
Sequencing Guidelines Adapted from ABI BigDye Terminator v3.1 Cycle Sequencing Kit and Roswell Park Cancer Institute Core Laboratory website
 Biomolecular Core Facility AI Dupont Hospital for Children, Rockland Center One, Room 214 Core: (302) 651-6712, Office: (302) 651-6707, mbcore@nemours.org Katia Sol-Church, Ph.D., Director Jennifer Frenck
Biomolecular Core Facility AI Dupont Hospital for Children, Rockland Center One, Room 214 Core: (302) 651-6712, Office: (302) 651-6707, mbcore@nemours.org Katia Sol-Church, Ph.D., Director Jennifer Frenck
The Central Dogma of Molecular Biology
 Vierstraete Andy (version 1.01) 1/02/2000 -Page 1 - The Central Dogma of Molecular Biology Figure 1 : The Central Dogma of molecular biology. DNA contains the complete genetic information that defines
Vierstraete Andy (version 1.01) 1/02/2000 -Page 1 - The Central Dogma of Molecular Biology Figure 1 : The Central Dogma of molecular biology. DNA contains the complete genetic information that defines
PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI,
 Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
Supplemental Text/Tables PCR Amplification and Sequencing PCR was carried out in a reaction volume of 20 µl using the ABI AmpliTaq GOLD kit (ABI, Foster City, CA). Each PCR reaction contained 20 ng genomic
FOR REFERENCE PURPOSES
 BIOO LIFE SCIENCE PRODUCTS FOR REFERENCE PURPOSES This manual is for Reference Purposes Only. DO NOT use this protocol to run your assays. Periodically, optimizations and revisions are made to the kit
BIOO LIFE SCIENCE PRODUCTS FOR REFERENCE PURPOSES This manual is for Reference Purposes Only. DO NOT use this protocol to run your assays. Periodically, optimizations and revisions are made to the kit
Extensive Cryptic Diversity in Indo-Australian Rainbowfishes Revealed by DNA Barcoding
 Extensive Cryptic Diversity in Indo-Australian Rainbowfishes Revealed by DNA Barcoding Kadarusman, Hubert N, Hadiaty R.K #, Sudarto, Paradis E., Pouyaud L. Akademi Perikanan Sorong, Papua Barat, Indonesia
Extensive Cryptic Diversity in Indo-Australian Rainbowfishes Revealed by DNA Barcoding Kadarusman, Hubert N, Hadiaty R.K #, Sudarto, Paradis E., Pouyaud L. Akademi Perikanan Sorong, Papua Barat, Indonesia
Intended Use: The kit is designed to detect the 5 different mutations found in Asian population using seven different primers.
 Unzipping Genes MBPCR014 Beta-Thalassemia Detection Kit P r o d u c t I n f o r m a t i o n Description: Thalassemia is a group of genetic disorders characterized by quantitative defects in globin chain
Unzipping Genes MBPCR014 Beta-Thalassemia Detection Kit P r o d u c t I n f o r m a t i o n Description: Thalassemia is a group of genetic disorders characterized by quantitative defects in globin chain
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR
 Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
RT-PCR: Two-Step Protocol
 RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
Procedures For DNA Sequencing
 Procedures For DNA Sequencing Plant-Microbe Genomics Facility (PMGF) Ohio State University 420 Biological Sciences Building 484 W. 12th Ave., Columbus OH 43210 Telephone: 614/247-6204 FAX: 614/292-6337
Procedures For DNA Sequencing Plant-Microbe Genomics Facility (PMGF) Ohio State University 420 Biological Sciences Building 484 W. 12th Ave., Columbus OH 43210 Telephone: 614/247-6204 FAX: 614/292-6337
Data Analysis for Ion Torrent Sequencing
 IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
IFU022 v140202 Research Use Only Instructions For Use Part III Data Analysis for Ion Torrent Sequencing MANUFACTURER: Multiplicom N.V. Galileilaan 18 2845 Niel Belgium Revision date: August 21, 2014 Page
ab185916 Hi-Fi cdna Synthesis Kit
 ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
Real-Time PCR Vs. Traditional PCR
 Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Real-Time PCR Vs. Traditional PCR Description This tutorial will discuss the evolution of traditional PCR methods towards the use of Real-Time chemistry and instrumentation for accurate quantitation. Objectives
Isolation and characterization of nine microsatellite loci in the Pale Pitcher Plant. MARGARET M. KOOPMAN*, ELIZABETH GALLAGHER, and BRYAN C.
 Page 1 of 28 1 1 2 3 PERMANENT GENETIC RESOURCES Isolation and characterization of nine microsatellite loci in the Pale Pitcher Plant Sarracenia alata (Sarraceniaceae). 4 5 6 MARGARET M. KOOPMAN*, ELIZABETH
Page 1 of 28 1 1 2 3 PERMANENT GENETIC RESOURCES Isolation and characterization of nine microsatellite loci in the Pale Pitcher Plant Sarracenia alata (Sarraceniaceae). 4 5 6 MARGARET M. KOOPMAN*, ELIZABETH
CompleteⅡ 1st strand cdna Synthesis Kit
 Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Troubleshooting for PCR and multiplex PCR
 Page 1 of 5 Page designed and maintained by Octavian Henegariu (Email: Tavi's Yale email or Tavi's Yahoo email). As I am currently pursuing a new junior faculty position, the Yale URL and email may change
Page 1 of 5 Page designed and maintained by Octavian Henegariu (Email: Tavi's Yale email or Tavi's Yahoo email). As I am currently pursuing a new junior faculty position, the Yale URL and email may change
STA DARD OPERATI G PROCEDURE FOR THE DETECTIO OF AFRICA SWI E FEVER VIRUS (ASFV) BY CO VE TIO AL POLYMERASE CHAI REACTIO (PCR)
 STA DARD OPERATI G PROCEDURE FOR THE DETECTIO OF AFRICA SWI E FEVER VIRUS (ASFV) BY CO VE TIO AL POLYMERASE CHAI REACTIO (PCR) jmvizcaino@vet.ucm.es Av/ Puerta de Hierro s/n. 28040 Madrid. Tel: (34) 913944082
STA DARD OPERATI G PROCEDURE FOR THE DETECTIO OF AFRICA SWI E FEVER VIRUS (ASFV) BY CO VE TIO AL POLYMERASE CHAI REACTIO (PCR) jmvizcaino@vet.ucm.es Av/ Puerta de Hierro s/n. 28040 Madrid. Tel: (34) 913944082
quantitative real-time PCR, grain, simplex DNA extraction: PGS0426 RT-PCR: PGS0494 & PGS0476
 BioScience quantitative real-time PCR, grain, simplex DNA extraction: PGS0426 RT-PCR: PGS0494 & PGS0476 This method describes a Real-time semi-quantitative TaqMan PCR procedure for the determination of
BioScience quantitative real-time PCR, grain, simplex DNA extraction: PGS0426 RT-PCR: PGS0494 & PGS0476 This method describes a Real-time semi-quantitative TaqMan PCR procedure for the determination of
Castillo et al. Rice Archaeogenetics electronic supplementary information
 Castillo et al. Rice Archaeogenetics electronic supplementary information Figure S1: PCR amplification for the nuclear genome region Sh4; and the sequence analysis. (a) Electrophoresis of 10 PCR products
Castillo et al. Rice Archaeogenetics electronic supplementary information Figure S1: PCR amplification for the nuclear genome region Sh4; and the sequence analysis. (a) Electrophoresis of 10 PCR products
ABSTRACT. Promega Corporation, Updated September 2008. http://www.promega.com/pubhub. 1 Campbell-Staton, S.
 A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA... A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA from Shed Reptile Skin ABSTRACT
A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA... A Modified Wizard SV Genomic DNA Purification System Protocol to Purify Genomic DNA from Shed Reptile Skin ABSTRACT
HiPer RT-PCR Teaching Kit
 HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
SOP Title: Multiplex-PCR check of genomic DNA isolated from FFPE tissue for its usability in array CGH analysis
 SOP Title: Multiplex-PCR check of genomic DNA isolated from FFPE tissue for its usability in array CGH analysis The STORE processing methods were shown to be fit-for purpose for DNA, RNA and protein extraction
SOP Title: Multiplex-PCR check of genomic DNA isolated from FFPE tissue for its usability in array CGH analysis The STORE processing methods were shown to be fit-for purpose for DNA, RNA and protein extraction
First Strand cdna Synthesis
 380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
PCR Instruments and Consumables
 Index Page GeneAmp PCR Instrument Systems 2 GeneAmp PCR System 9700 Components 3 Accessories and Software for GeneAmp PCR Instrument Systems 4 Disposables 5 PCR Enzymes 7 GeneAmp PCR Kits 12 RNA PCR Kits
Index Page GeneAmp PCR Instrument Systems 2 GeneAmp PCR System 9700 Components 3 Accessories and Software for GeneAmp PCR Instrument Systems 4 Disposables 5 PCR Enzymes 7 GeneAmp PCR Kits 12 RNA PCR Kits
SYBR Green PCR Master Mix and SYBR Green RT-PCR Reagents Kit
 USER GUIDE SYBR Green PCR Master Mix and SYBR Green RT-PCR Reagents Kit Catalog Number 4309155 (Master Mix) and 4306736 (RT-PCR Reagents Kit) Publication Part Number 4310251 Rev. G Revision Date September
USER GUIDE SYBR Green PCR Master Mix and SYBR Green RT-PCR Reagents Kit Catalog Number 4309155 (Master Mix) and 4306736 (RT-PCR Reagents Kit) Publication Part Number 4310251 Rev. G Revision Date September
Improved methods for site-directed mutagenesis using Gibson Assembly TM Master Mix
 CLONING & MAPPING DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Improved methods for site-directed mutagenesis using Gibson Assembly TM Master Mix LIBRARY PREP FOR NET GEN SEQUENCING PROTEIN
CLONING & MAPPING DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Improved methods for site-directed mutagenesis using Gibson Assembly TM Master Mix LIBRARY PREP FOR NET GEN SEQUENCING PROTEIN
mircute mirna qpcr Detection Kit (SYBR Green)
 mircute mirna qpcr Detection Kit (SYBR Green) For detection of mirna using real-time RT-PCR (SYBR Green I) www.tiangen.com QP110302 mircute mirna qpcr Detection Kit (SYBR Green) Kit Contents Cat. no. FP401
mircute mirna qpcr Detection Kit (SYBR Green) For detection of mirna using real-time RT-PCR (SYBR Green I) www.tiangen.com QP110302 mircute mirna qpcr Detection Kit (SYBR Green) Kit Contents Cat. no. FP401
A quick and simple method for the identi cation of meat species and meat products by PCR assay
 Meat Science 51 (1999) 143±148 A quick and simple method for the identi cation of meat species and meat products by PCR assay T. Matsunaga a, K. Chikuni b *, R. Tanabe b, S. Muroya b, K. Shibata a, J.
Meat Science 51 (1999) 143±148 A quick and simple method for the identi cation of meat species and meat products by PCR assay T. Matsunaga a, K. Chikuni b *, R. Tanabe b, S. Muroya b, K. Shibata a, J.
DNA Sequencing Handbook
 Genomics Core 147 Biotechnology Building Ithaca, New York 14853-2703 Phone: (607) 254-4857; Fax (607) 254-4847 Web: http://cores.lifesciences.cornell.edu/brcinfo/ Email: DNA_Services@cornell.edu DNA Sequencing
Genomics Core 147 Biotechnology Building Ithaca, New York 14853-2703 Phone: (607) 254-4857; Fax (607) 254-4847 Web: http://cores.lifesciences.cornell.edu/brcinfo/ Email: DNA_Services@cornell.edu DNA Sequencing
Power SYBR Green PCR Master Mix and Power SYBR Green RT-PCR Reagents Kit
 USER GUIDE Power SYBR Green PCR Master Mix and Power SYBR Green RT-PCR Reagents Kit Catalog Number 4368577, 4367659, 4367660, 4368706, 4368702, 4368708 (Master Mix) and 4368711 (RT-PCR Reagents Kit) Publication
USER GUIDE Power SYBR Green PCR Master Mix and Power SYBR Green RT-PCR Reagents Kit Catalog Number 4368577, 4367659, 4367660, 4368706, 4368702, 4368708 (Master Mix) and 4368711 (RT-PCR Reagents Kit) Publication
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual
 Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
PyroPhage 3173 DNA Polymerase, Exonuclease Minus (Exo-)
 PyroPhage 3173 DNA Polymerase, Exonuclease Minus (Exo-) FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608)
PyroPhage 3173 DNA Polymerase, Exonuclease Minus (Exo-) FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608)
1/12 Dideoxy DNA Sequencing
 1/12 Dideoxy DNA Sequencing Dideoxy DNA sequencing utilizes two steps: PCR (polymerase chain reaction) amplification of DNA using dideoxy nucleoside triphosphates (Figures 1 and 2)and denaturing polyacrylamide
1/12 Dideoxy DNA Sequencing Dideoxy DNA sequencing utilizes two steps: PCR (polymerase chain reaction) amplification of DNA using dideoxy nucleoside triphosphates (Figures 1 and 2)and denaturing polyacrylamide
Evidence from Mitochondrial DNA That Head Lice and Body Lice of Humans (Phthiraptera: Pediculidae) are Conspecific
 SHORT COMMUNICATION Evidence from Mitochondrial DNA That Head Lice and Body Lice of Humans (Phthiraptera: Pediculidae) are Conspecific N. P. LEO, 1,2 N.J.H. CAMPBELL, 2 X. YANG, 3 K. MUMCUOGLU, 4 AND S.
SHORT COMMUNICATION Evidence from Mitochondrial DNA That Head Lice and Body Lice of Humans (Phthiraptera: Pediculidae) are Conspecific N. P. LEO, 1,2 N.J.H. CAMPBELL, 2 X. YANG, 3 K. MUMCUOGLU, 4 AND S.
Forensic DNA Testing Terminology
 Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Forensic DNA Testing Terminology ABI 310 Genetic Analyzer a capillary electrophoresis instrument used by forensic DNA laboratories to separate short tandem repeat (STR) loci on the basis of their size.
Taq98 Hot Start 2X Master Mix
 Taq98 Hot Start 2X Master Mix Optimized for 98C Denaturation Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608) 831-9011 FAX: (608) 831-9012 lucigen@lucigen.com
Taq98 Hot Start 2X Master Mix Optimized for 98C Denaturation Lucigen Corporation 2905 Parmenter St, Middleton, WI 53562 USA Toll Free: (888) 575-9695 (608) 831-9011 FAX: (608) 831-9012 lucigen@lucigen.com
BigDye Terminator v3.1 Cycle Sequencing Kit. Protocol. DRAFT August 27, 2002 12:32 pm, 4337035A_v3.1Title.fm
 BigDye Terminator v3.1 Cycle Sequencing Kit Protocol August 27, 2002 12:32 pm, 4337035A_v3.1Title.fm Copyright 2002, Applied Biosystems. All rights reserved. For Research Use Only. Not for use in diagnostic
BigDye Terminator v3.1 Cycle Sequencing Kit Protocol August 27, 2002 12:32 pm, 4337035A_v3.1Title.fm Copyright 2002, Applied Biosystems. All rights reserved. For Research Use Only. Not for use in diagnostic
RevertAid Premium First Strand cdna Synthesis Kit
 RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
GENOTYPING ASSAYS AT ZIRC
 GENOTYPING ASSAYS AT ZIRC A. READ THIS FIRST - DISCLAIMER Dear ZIRC user, We now provide detailed genotyping protocols for a number of zebrafish lines distributed by ZIRC. These protocols were developed
GENOTYPING ASSAYS AT ZIRC A. READ THIS FIRST - DISCLAIMER Dear ZIRC user, We now provide detailed genotyping protocols for a number of zebrafish lines distributed by ZIRC. These protocols were developed
Troubleshooting the Single-step PCR Site-directed Mutagenesis Procedure Intended to Create a Non-functional rop Gene in the pbr322 Plasmid
 Troubleshooting the Single-step PCR Site-directed Mutagenesis Procedure Intended to Create a Non-functional rop Gene in the pbr322 Plasmid Lina Jew Department of Microbiology & Immunology, University of
Troubleshooting the Single-step PCR Site-directed Mutagenesis Procedure Intended to Create a Non-functional rop Gene in the pbr322 Plasmid Lina Jew Department of Microbiology & Immunology, University of
Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541
 PRODUCT INFORMATION Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541 Lot _ Store at -20 C Expiry Date _ www.thermoscientific.com/onebio CERTIFICATE OF ANALYSIS The Phusion Site-Directed Mutagenesis
PRODUCT INFORMATION Thermo Scientific Phusion Site-Directed Mutagenesis Kit #F-541 Lot _ Store at -20 C Expiry Date _ www.thermoscientific.com/onebio CERTIFICATE OF ANALYSIS The Phusion Site-Directed Mutagenesis
Genetic Analysis. Phenotype analysis: biological-biochemical analysis. Genotype analysis: molecular and physical analysis
 Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
Genetic Analysis Phenotype analysis: biological-biochemical analysis Behaviour under specific environmental conditions Behaviour of specific genetic configurations Behaviour of progeny in crosses - Genotype
Multiple Losses of Flight and Recent Speciation in Steamer Ducks Tara L. Fulton, Brandon Letts, and Beth Shapiro
 Supplementary Material for: Multiple Losses of Flight and Recent Speciation in Steamer Ducks Tara L. Fulton, Brandon Letts, and Beth Shapiro 1. Supplementary Tables Supplementary Table S1. Sample information.
Supplementary Material for: Multiple Losses of Flight and Recent Speciation in Steamer Ducks Tara L. Fulton, Brandon Letts, and Beth Shapiro 1. Supplementary Tables Supplementary Table S1. Sample information.
Gene Mapping Techniques
 Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Gene Mapping Techniques OBJECTIVES By the end of this session the student should be able to: Define genetic linkage and recombinant frequency State how genetic distance may be estimated State how restriction
Hepatitis B Virus Genemer Mix
 Product Manual Hepatitis B Virus Genemer Mix Primer Pair for amplification of HBV Specific DNA Fragment Includes Internal Negative Control Primers and Template Catalog No.: 60-2007-12 Store at 20 o C For
Product Manual Hepatitis B Virus Genemer Mix Primer Pair for amplification of HBV Specific DNA Fragment Includes Internal Negative Control Primers and Template Catalog No.: 60-2007-12 Store at 20 o C For
All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna
 All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna All-in-One TM mirna First-Strand cdna Synthesis Kit AMRT-0020 (20 RT reactions), AMRT-0060 (60 RT reactions) Used in combination
All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna All-in-One TM mirna First-Strand cdna Synthesis Kit AMRT-0020 (20 RT reactions), AMRT-0060 (60 RT reactions) Used in combination
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
Cloning Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems
 Promega Notes Number 71, 1999, p. 10 Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems By Kimberly Knoche, Ph.D., and Dan Kephart, Ph.D. Promega Corporation Corresponding
Promega Notes Number 71, 1999, p. 10 Blunt-End Pfu DNA Polymerase- Generated PCR Fragments into pgem -T Vector Systems By Kimberly Knoche, Ph.D., and Dan Kephart, Ph.D. Promega Corporation Corresponding
TaqMan Fast Advanced Master Mix. Protocol
 TaqMan Fast Advanced Master Mix Protocol For Research Use Only. Not intended for any animal or human therapeutic or diagnostic use. Information in this document is subject to change without notice. APPLIED
TaqMan Fast Advanced Master Mix Protocol For Research Use Only. Not intended for any animal or human therapeutic or diagnostic use. Information in this document is subject to change without notice. APPLIED
Introduction. Preparation of Template DNA
 Procedures and Recommendations for DNA Sequencing at the Plant-Microbe Genomics Facility Ohio State University Biological Sciences Building Room 420, 484 W. 12th Ave., Columbus OH 43210 Telephone: 614/247-6204;
Procedures and Recommendations for DNA Sequencing at the Plant-Microbe Genomics Facility Ohio State University Biological Sciences Building Room 420, 484 W. 12th Ave., Columbus OH 43210 Telephone: 614/247-6204;
The RNAi Consortium (TRC) Broad Institute
 TRC Laboratory Protocols Protocol Title: One Step PCR Preparation of Samples for Illumina Sequencing Current Revision Date: 11/10/2012 RNAi Platform,, trc_info@broadinstitute.org Brief Description: This
TRC Laboratory Protocols Protocol Title: One Step PCR Preparation of Samples for Illumina Sequencing Current Revision Date: 11/10/2012 RNAi Platform,, trc_info@broadinstitute.org Brief Description: This
Fungal DNA sequencing from laser capture microdissection samples
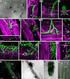 APPLICATION NOTE Laser capture microdissection and Sanger sequencing Fungal DNA sequencing from laser capture microdissection samples Introduction The direct identification of human fungal pathogens by
APPLICATION NOTE Laser capture microdissection and Sanger sequencing Fungal DNA sequencing from laser capture microdissection samples Introduction The direct identification of human fungal pathogens by
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Table of Contents. I. Description... 2. II. Kit Components... 2. III. Storage... 2. IV. 1st Strand cdna Synthesis Reaction... 3
 Table of Contents I. Description... 2 II. Kit Components... 2 III. Storage... 2 IV. 1st Strand cdna Synthesis Reaction... 3 V. RT-PCR, Real-time RT-PCR... 4 VI. Application... 5 VII. Preparation of RNA
Table of Contents I. Description... 2 II. Kit Components... 2 III. Storage... 2 IV. 1st Strand cdna Synthesis Reaction... 3 V. RT-PCR, Real-time RT-PCR... 4 VI. Application... 5 VII. Preparation of RNA
ABI PRISM BigDye Primer Cycle Sequencing Ready Reaction Kit With AmpliTaq DNA Polymerase, FS
 ABI PRISM BigDye Primer Cycle Sequencing Ready Reaction Kit With AmpliTaq DNA Polymerase, FS Protocol Copyright 2003, 2010 Applied Biosystems. Printed in the U.S.A. For Research Use Only. Not for use in
ABI PRISM BigDye Primer Cycle Sequencing Ready Reaction Kit With AmpliTaq DNA Polymerase, FS Protocol Copyright 2003, 2010 Applied Biosystems. Printed in the U.S.A. For Research Use Only. Not for use in
Development of a Workflow to Detect Sequence Variants in the BRCA1 and BRCA2 Genes
 Your Innovative Research BRCA1 and BRCA2 Variant Detection Development of a Workflow to Detect Sequence Variants in the BRCA1 and BRCA2 Genes The oncogenetics group in the DNA Diagnostics division of the
Your Innovative Research BRCA1 and BRCA2 Variant Detection Development of a Workflow to Detect Sequence Variants in the BRCA1 and BRCA2 Genes The oncogenetics group in the DNA Diagnostics division of the
For information regarding shipping specifications, please refer to the following link: http://dna.macrogen.com/eng/help/orderg.jsp
 Macrogen Sample Submission Guide Macrogen has served over 10 years in sequencing field using the cutting edge technology and delivering fast and reliable results. We use high throughput Applied Biosystems
Macrogen Sample Submission Guide Macrogen has served over 10 years in sequencing field using the cutting edge technology and delivering fast and reliable results. We use high throughput Applied Biosystems
United States Department of Agriculture Food Safety and Inspection Service, Office of Public Health Science
 CLG-FPCR.01 Page 1 of 14 Contents A. INTRODUCTION... 2 B. EQUIPMENT... 2 C. REAGENTS AND SOLUTIONS... 3 D. STANDARD(S)... 7 E. SAMPLE PREPARATION... 7 F. ANALYTICAL PROCEDURE... 7 G. CALCULATIONS / IDENTIFICATION...
CLG-FPCR.01 Page 1 of 14 Contents A. INTRODUCTION... 2 B. EQUIPMENT... 2 C. REAGENTS AND SOLUTIONS... 3 D. STANDARD(S)... 7 E. SAMPLE PREPARATION... 7 F. ANALYTICAL PROCEDURE... 7 G. CALCULATIONS / IDENTIFICATION...
Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA
 Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Page 1 of 5 Biology Behind the Crime Scene Week 4: Lab #4 Genetics Exercise (Meiosis) and RFLP Analysis of DNA Genetics Exercise: Understanding how meiosis affects genetic inheritance and DNA patterns
Terra PCR Direct Polymerase Mix User Manual
 Clontech Laboratories, Inc. Terra PCR Direct Polymerase Mix User Manual Cat. Nos. 639269, 639270, 639271 PT5126-1 (031416) Clontech Laboratories, Inc. A Takara Bio Company 1290 Terra Bella Avenue, Mountain
Clontech Laboratories, Inc. Terra PCR Direct Polymerase Mix User Manual Cat. Nos. 639269, 639270, 639271 PT5126-1 (031416) Clontech Laboratories, Inc. A Takara Bio Company 1290 Terra Bella Avenue, Mountain
AffinityScript QPCR cdna Synthesis Kit
 AffinityScript QPCR cdna Synthesis Kit INSTRUCTION MANUAL Catalog #600559 Revision C.01 For In Vitro Use Only 600559-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this
AffinityScript QPCR cdna Synthesis Kit INSTRUCTION MANUAL Catalog #600559 Revision C.01 For In Vitro Use Only 600559-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this
DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
 Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2014 DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
Electronic Supplementary Material (ESI) for Lab on a Chip. This journal is The Royal Society of Chemistry 2014 DNA-functionalized hydrogels for confined membrane-free in vitro transcription/translation
SeqScape Software Version 2.5 Comprehensive Analysis Solution for Resequencing Applications
 Product Bulletin Sequencing Software SeqScape Software Version 2.5 Comprehensive Analysis Solution for Resequencing Applications Comprehensive reference sequence handling Helps interpret the role of each
Product Bulletin Sequencing Software SeqScape Software Version 2.5 Comprehensive Analysis Solution for Resequencing Applications Comprehensive reference sequence handling Helps interpret the role of each
DNA: A Person s Ultimate Fingerprint
 A partnership between the UAB Center for Community Outreach Development and McWane Center DNA: A Person s Ultimate Fingerprint This project is supported by a Science Education Partnership Award (SEPA)
A partnership between the UAB Center for Community Outreach Development and McWane Center DNA: A Person s Ultimate Fingerprint This project is supported by a Science Education Partnership Award (SEPA)
Absolute Quantification Getting Started Guide
 5 cdna Reverse Primer Oligo d(t) or random hexamer Synthesis of 1st cdna strand 3 5 cdna Applied Biosystems 7300/7500/7500 Fast Real-Time PCR System Absolute Quantification Getting Started Guide Introduction
5 cdna Reverse Primer Oligo d(t) or random hexamer Synthesis of 1st cdna strand 3 5 cdna Applied Biosystems 7300/7500/7500 Fast Real-Time PCR System Absolute Quantification Getting Started Guide Introduction
SEQUENCING SERVICES. McGill University and Génome Québec Innovation Centre JANUARY 21, 2016. Version 3.4
 JANUARY 21, 2016 SEQUENCING SERVICES McGill University and Génome Québec Innovation Centre Sanger Sequencing User Guide Version 3.4 Copyright 2010 McGill University and Génome Québec Innovation Centre
JANUARY 21, 2016 SEQUENCING SERVICES McGill University and Génome Québec Innovation Centre Sanger Sequencing User Guide Version 3.4 Copyright 2010 McGill University and Génome Québec Innovation Centre
DNA Sequence Analysis
 DNA Sequence Analysis Two general kinds of analysis Screen for one of a set of known sequences Determine the sequence even if it is novel Screening for a known sequence usually involves an oligonucleotide
DNA Sequence Analysis Two general kinds of analysis Screen for one of a set of known sequences Determine the sequence even if it is novel Screening for a known sequence usually involves an oligonucleotide
Essentials of Real Time PCR. About Sequence Detection Chemistries
 Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
SYBR Green Realtime PCR Master Mix -Plus-
 Instruction manual SYBR Green Realtime PCR Master Mix -Plus- 0810 F0925K SYBR Green Realtime PCR Master Mix -Plus- Contents QPK-212T 1mLx1 QPK-212 1mLx5 Store at -20 C, protected from light [1] Introduction
Instruction manual SYBR Green Realtime PCR Master Mix -Plus- 0810 F0925K SYBR Green Realtime PCR Master Mix -Plus- Contents QPK-212T 1mLx1 QPK-212 1mLx5 Store at -20 C, protected from light [1] Introduction
Crime Scenes and Genes
 Glossary Agarose Biotechnology Cell Chromosome DNA (deoxyribonucleic acid) Electrophoresis Gene Micro-pipette Mutation Nucleotide Nucleus PCR (Polymerase chain reaction) Primer STR (short tandem repeats)
Glossary Agarose Biotechnology Cell Chromosome DNA (deoxyribonucleic acid) Electrophoresis Gene Micro-pipette Mutation Nucleotide Nucleus PCR (Polymerase chain reaction) Primer STR (short tandem repeats)
HISTO SPOT SSO System
 HISTO SPOT SSO System for HLA Typing HISTO SPOT Kits MR.SPOT Processor HISTO MATCH Software Complete system certified for IvD use HISTO SPOT Features The HISTO SPOT SSO assay in combination with the MR.SPOT
HISTO SPOT SSO System for HLA Typing HISTO SPOT Kits MR.SPOT Processor HISTO MATCH Software Complete system certified for IvD use HISTO SPOT Features The HISTO SPOT SSO assay in combination with the MR.SPOT
All-in-One mirna qrt-pcr Detection System Handbook
 All-in-One mirna qrt-pcr Detection System Handbook For quantitative detection of mature mirna All-in-One mirna First-Strand cdna Synthesis Kit Cat. No. AMRT-0020 (20 mirna reverse transcription reactions)
All-in-One mirna qrt-pcr Detection System Handbook For quantitative detection of mature mirna All-in-One mirna First-Strand cdna Synthesis Kit Cat. No. AMRT-0020 (20 mirna reverse transcription reactions)
Amplicon Template Preparation and Sequencing
 Please note: the shared protocols described herein may not have been validated by Pacific Biosciences and are provided as-is and without any warranty. Use of these protocols is offered to those customers
Please note: the shared protocols described herein may not have been validated by Pacific Biosciences and are provided as-is and without any warranty. Use of these protocols is offered to those customers
DNA Sequencing Setup and Troubleshooting
 DNA Sequencing Setup and Troubleshooting Lara Cullen, PhD Scientific Applications Specialist Australia and New Zealand Reviewing Sequencing Data Review the Electropherogram Review the Raw Data (Signal
DNA Sequencing Setup and Troubleshooting Lara Cullen, PhD Scientific Applications Specialist Australia and New Zealand Reviewing Sequencing Data Review the Electropherogram Review the Raw Data (Signal
FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE.
 Instruction manual for KOD FX Neo 1103 F1100K KOD FX Neo KFX-201 200 U 200 reactions Store at -20 C Contents [1] Introduction [2] Components [3] Quality testing [4] Primer design [5] Cloning of PCR products
Instruction manual for KOD FX Neo 1103 F1100K KOD FX Neo KFX-201 200 U 200 reactions Store at -20 C Contents [1] Introduction [2] Components [3] Quality testing [4] Primer design [5] Cloning of PCR products
Mitochondrial DNA Analysis
 Mitochondrial DNA Analysis Lineage Markers Lineage markers are passed down from generation to generation without changing Except for rare mutation events They can help determine the lineage (family tree)
Mitochondrial DNA Analysis Lineage Markers Lineage markers are passed down from generation to generation without changing Except for rare mutation events They can help determine the lineage (family tree)
Sanger Sequencing and Quality Assurance. Zbigniew Rudzki Department of Pathology University of Melbourne
 Sanger Sequencing and Quality Assurance Zbigniew Rudzki Department of Pathology University of Melbourne Sanger DNA sequencing The era of DNA sequencing essentially started with the publication of the enzymatic
Sanger Sequencing and Quality Assurance Zbigniew Rudzki Department of Pathology University of Melbourne Sanger DNA sequencing The era of DNA sequencing essentially started with the publication of the enzymatic
Reverse Transcription System
 TECHNICAL BULLETIN Reverse Transcription System Instruc ons for use of Product A3500 Revised 1/14 TB099 Reverse Transcription System All technical literature is available on the Internet at: www.promega.com/protocols/
TECHNICAL BULLETIN Reverse Transcription System Instruc ons for use of Product A3500 Revised 1/14 TB099 Reverse Transcription System All technical literature is available on the Internet at: www.promega.com/protocols/
DNA Integrity Number (DIN) For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments
 DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
NimbleGen DNA Methylation Microarrays and Services
 NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
June 09, 2009 Random Mutagenesis
 Why Mutagenesis? Analysis of protein function June 09, 2009 Random Mutagenesis Analysis of protein structure Protein engineering Analysis of structure-function relationship Analysis of the catalytic center
Why Mutagenesis? Analysis of protein function June 09, 2009 Random Mutagenesis Analysis of protein structure Protein engineering Analysis of structure-function relationship Analysis of the catalytic center
Sanger Sequencing: Sample Preparation Guide
 Sanger Sequencing: Sample Preparation Guide Use this as a guide to prepare your samples for Sanger sequencing at AGRF CONTENTS 1 Overview... 2 1.1 Capillary Separation (CS) or electrophoretic separation
Sanger Sequencing: Sample Preparation Guide Use this as a guide to prepare your samples for Sanger sequencing at AGRF CONTENTS 1 Overview... 2 1.1 Capillary Separation (CS) or electrophoretic separation
Aurora Forensic Sample Clean-up Protocol
 Aurora Forensic Sample Clean-up Protocol 106-0008-BA-D 2015 Boreal Genomics, Inc. All rights reserved. All trademarks are property of their owners. http://www.borealgenomics.com support@borealgenomics.com
Aurora Forensic Sample Clean-up Protocol 106-0008-BA-D 2015 Boreal Genomics, Inc. All rights reserved. All trademarks are property of their owners. http://www.borealgenomics.com support@borealgenomics.com
ZR DNA Sequencing Clean-up Kit
 INSTRUCTION MANUAL ZR DNA Sequencing Clean-up Kit Catalog Nos. D40 & D4051 Highlights Simple 2 Minute Bind, Wash, Elute Procedure Flexible 6-20 µl Elution Volumes Allow for Direct Loading of Samples with
INSTRUCTION MANUAL ZR DNA Sequencing Clean-up Kit Catalog Nos. D40 & D4051 Highlights Simple 2 Minute Bind, Wash, Elute Procedure Flexible 6-20 µl Elution Volumes Allow for Direct Loading of Samples with
APOT - Assay. Protocol for HPV16 and 18. Amplification of Papilloma Virus Oncogene Transcripts HPV. E6 E7 E1 Zelluläre DNA poly(a)
 E5 E2 E1 APOT - Assay Amplification of Papilloma Virus Oncogene Transcripts URR E6 E7 L1 HPV L2 E4 E6 E7 E1 Zelluläre DNA poly(a) Protocol for HPV16 and 18 Brief summary of the APOT assay Fig.1A shows
E5 E2 E1 APOT - Assay Amplification of Papilloma Virus Oncogene Transcripts URR E6 E7 L1 HPV L2 E4 E6 E7 E1 Zelluläre DNA poly(a) Protocol for HPV16 and 18 Brief summary of the APOT assay Fig.1A shows
FDA SOP for Generating DNA Barcodes Suitable for Species Identification of an Unknown Fish Tissue Sample:
 FDA SOP for Generating DNA Barcodes Suitable for Species Identification of an Unknown Fish Tissue Sample: The process of generating DNA barcodes suitable for species identification from an unknown fish
FDA SOP for Generating DNA Barcodes Suitable for Species Identification of an Unknown Fish Tissue Sample: The process of generating DNA barcodes suitable for species identification from an unknown fish
GenScript BloodReady TM Multiplex PCR System
 GenScript BloodReady TM Multiplex PCR System Technical Manual No. 0174 Version 20040915 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI
GenScript BloodReady TM Multiplex PCR System Technical Manual No. 0174 Version 20040915 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI
Authentication of Basmati rice using SSR-PCR and QIAxcel Advanced
 Application Note Authentication of Basmati rice using SSR-PCR and QIAxcel Advanced R. Cassier ADGENE Laboratoire, Thury Harcourt, France Introduction Basmati is one of the most popular types of rice in
Application Note Authentication of Basmati rice using SSR-PCR and QIAxcel Advanced R. Cassier ADGENE Laboratoire, Thury Harcourt, France Introduction Basmati is one of the most popular types of rice in
Work carried out in partnership with: LGL Environmental Research Associates, BC Ministry of Environment. Coquitlam nerkid genetics 1
 1 Assessment of the genetic relationship among sockeye salmon and kokanee populations of the lower Mainland of British Columbia: Implications for establishment of an anadromous sockeye run in the Coquitlam
1 Assessment of the genetic relationship among sockeye salmon and kokanee populations of the lower Mainland of British Columbia: Implications for establishment of an anadromous sockeye run in the Coquitlam
ANNEX TABLES Table 1. Revenue by ICT Type Revenue * Share to Total Revenues Growth Rate ICT Type 2004 2005 2004 2005 (in percent)
 ANNEX TABLES Table 1. Revenue by Revenue * Share to Revenues 2004 2005 2004 2005 Contact Centers 32,904.1 54,295.1 44.4 49.4 65.0 Medical Transcription 236.7 466.2 0.3 0.4 97.0 Animation 694.2 939.1 0.9
ANNEX TABLES Table 1. Revenue by Revenue * Share to Revenues 2004 2005 2004 2005 Contact Centers 32,904.1 54,295.1 44.4 49.4 65.0 Medical Transcription 236.7 466.2 0.3 0.4 97.0 Animation 694.2 939.1 0.9
Welcome to Pacific Biosciences' Introduction to SMRTbell Template Preparation.
 Introduction to SMRTbell Template Preparation 100 338 500 01 1. SMRTbell Template Preparation 1.1 Introduction to SMRTbell Template Preparation Welcome to Pacific Biosciences' Introduction to SMRTbell
Introduction to SMRTbell Template Preparation 100 338 500 01 1. SMRTbell Template Preparation 1.1 Introduction to SMRTbell Template Preparation Welcome to Pacific Biosciences' Introduction to SMRTbell
Brief Communication. B. Freeman, 1 N. Smith, 1 C. Curtis, 1 L. Huckett, 1 J. Mill, 1 and I. W. Craig 1,2
 Behavior Genetics, Vol. 33, No. 1, January 2003 ( 2003) Brief Communication DNA from Buccal Swabs Recruited by Mail: Evaluation of Storage Effects on Long-term Stability and Suitability for Multiplex Polymerase
Behavior Genetics, Vol. 33, No. 1, January 2003 ( 2003) Brief Communication DNA from Buccal Swabs Recruited by Mail: Evaluation of Storage Effects on Long-term Stability and Suitability for Multiplex Polymerase
Description: Molecular Biology Services and DNA Sequencing
 Description: Molecular Biology s and DNA Sequencing DNA Sequencing s Single Pass Sequencing Sequence data only, for plasmids or PCR products Plasmid DNA or PCR products Plasmid DNA: 20 100 ng/μl PCR Product:
Description: Molecular Biology s and DNA Sequencing DNA Sequencing s Single Pass Sequencing Sequence data only, for plasmids or PCR products Plasmid DNA or PCR products Plasmid DNA: 20 100 ng/μl PCR Product:
Sanger Sequencing. Troubleshooting Guide. Failed sequence
 Sanger Sequencing Troubleshooting Guide Below are examples of the main problems experienced in ABI Sanger sequencing. Possible causes for failure and their solutions are listed below each example. The
Sanger Sequencing Troubleshooting Guide Below are examples of the main problems experienced in ABI Sanger sequencing. Possible causes for failure and their solutions are listed below each example. The
