App. Note # FTMS-47+MT-113 MALDI Imaging with Single Cell Resolution at 10 µm Pixel Size
|
|
|
- Monica Osborne
- 10 years ago
- Views:
Transcription
1 pp. Note # FTMS-47+MT-113 MLDI Imaging with Single Cell Resolution at 10 µm Pixel Size bstract Key parameters for MLDI imaging are spatial resolution and mass spectral quality. In this application note, we explore the feasibility of single cell imaging. Typically, high spatial resolution requires matrix preparation that is drier than usual and often is limited to high abundant lipid species. For the high resolution imaging presented here, we utilized a dry sublimation device, which enabled us to perform MLDI imaging at a spatial resolution of 10µm and to resolve individual cells in tissue images. The 10 µm pixel size was achieved on both solarix FTMS and autoflexspeed and ultraflextreme platforms. uthors Michael ecker, Eckhard elau, Sören-Oliver Deininger, Jens Fuchser ruker Daltonik GmbH, remen, Germany Shannon Cornett ruker Daltonics, illerica, M, US Keywords MLDI Imaging FTMS MLDI-TOF Instrumentation and Software solarix MLDI-TOF
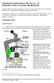
2 Introduction Interest in MLDI imaging has grown significantly in recent years with applications in the fields of histology, histopathology, drug development and others. Several performance factors are relevant to mass spectrometric imaging techniques, such as spatial resolution, analysis speed, and mass spectral quality. Usually the optimization of one of these factors is detrimental to the others, so in real-life, experiments are often not done at the limits of spatial resolution. Nevertheless, in this application we focus on the optimization of spatial resolution. Sample preparation is an important factor for achieving high resolution MLDI imaging. Often, the MLDI matrix is applied as solution in the form of an aerosol. The solvent droplets extract the compounds of interest which are subsequently incorporated in the matrix for the measurement. For many compounds, such as drugs or proteins, a wet sample preparation (longer aerosol) leads to a better spectral quality, but at the same time leads to larger matrix crystals, thereby limiting the maximum spatial resolution. Especially for highly abundant lipids, it has been shown that matrix can be applied in a dry fashion [1], which leads to small matrix crystal size and therefore allows a higher spatial resolution. In this study, we used sublimation to apply matrix onto the prepared slides. This allows a very high spatial resolution because no solvent is involved [2] and is especially suited to assess the instrument-based limits of the lateral resolution. Spatial resolution in imaging mass spectrometry Within the field of mass spectrometric imaging spatial resolution can be defined differently. Often, the term spatial resolution is used synonymously with the achievable raster width (i.e. pixel size ) or the geometric distance between spectra. In these cases, technical features such as minimum laser focus diameter and precise sample stage movement are considered the limiting factors (in the absence of large matrix crystals). To determine the highest spatial resolution that we could achieve on biological tissue we selected rat cerebellum and testis tissue as test specimens. In each testis section there are a number of structures of different sizes including tubuli, interstitial spaces and differing layers of cells on the outside of each tubule as well as in the lumen. In the cerebellum organized layers of individual large neurons, the Purkinje cells, have a soma diameter of 30µm. The goal of this study was to prepare and analyse these samples such that the smallest of these features was well resolved in the ion images. Experimental Cryosections of fresh-frozen rat cerebellum and testis were cut at 10µm thickness and thaw-mounted onto indium-tin-oxide (ITO, ruker) coated glass slides. Sample slides were not washed and directly desiccated in a vacuum desiccator for 1 hour. Sublimation was performed in a custom built device using a 152 mm inner diameter sublimation apparatus (ce Glass, NJ, US) exactly as described in Chaurand et al. [2] using 2,5-dihydroxybencoid acid as matrix. MLDI imaging measurements were performed on commercially available Ultraflextreme, utoflex Speed and Solarix (all ruker) instruments equipped with a smartbeam II laser. The standard laser setting of minimum diameter was used exclusively for all measurements. FlexImaging 3.0 software (ruker) was used to set up the imaging measurements and to generate ion images from the acquired data. MLDI-TOF images were normalized to total ion count (TIC), MLDI-FTMS images were normalized to root mean square (RMS) intensity. The intensity thresholds for the reconstructed MLDI images were adjusted to result in bright images and low noise. fter image collection, slides were removed and washed in ethanol to remove matrix and stained using a standard Hematoxylin and Eosin (H&E) protocol. Microscopic images were acquired on a Mirax desk slide scanner (Zeiss). Results MLDI-TOF imaging Figure 1 shows an illustration of the main features of rat cerebellum, including white matter, granular layer, Purkinje cell layer and molecular layer. Purkinje cell soma have an average diameter of ~30 µm in rat which are well separated from each other and therefore are ideal structures to test MLDI Imaging spatial resolution. Figures 2-4 show a selection of MLDI images of lipids from rat cerebellum at increasing magnification. Figure 2 shows two reconstructed MLDI images of five selected mass signals and an optical image of the measured area. In Figure 2a, the mass signal at Da (displayed in red) is localized to the cerebellar white matter and the signal at Da (in blue) is found in the granular layer. The mass signal at is seen as a line of green dots that correspond to individual Purkinje neurons. The signal at is seen in Figure 2b as a well-structured image of several anatomical features.
3 Simplified anatomy of rat cerebellum Figure 1: : The main structure in the cerebellum is comprised of cerebellar white matter (WM) surrounded by the granular layer (GL). The granular layer is surrounded by a single layer of Purkinje cells (PCL), which is in turn surrounded by the molecular layer (ML). : Enlarged area as marked, the Purkinje cells are large cells with irregular spacing. In mouse cerebellum, the diameter of Purkinje cells is ~30 µm. MLDI images of rat cerebellum WM GL ML C PCL Da Da Figure 2: MLDI images from part of rat cerebellum acquired at 10µm pixel size. : Three selected masses which show cellular localization: the mass signal in green highlights the individual Purkinje cells; the mass signal Da in red is found in the cerebellar white matter; and the mass signal Da is found in the granular layer. : Two selected mass signals that show strong differentiation, the mass signal at also exhibits spatial differentiation according to intensity range. C: Optical image of the measured cerebellum with measured region shown for context. ll scale bars: 200 µm. WM Enlarged figure on the back side GL Purkinje Cells ML Figures 3a-b show, in progressively greater detail, that the mass signal at is indeed localized to the Purkinje cell layer. fter the MLDI acquisition, an H&E stained image of the section was acquired and is shown in Figure 3c. Individual Purkinje cells are indicated by arrows on both the molecular and the microscopic images. That the somata are represented by multiple pixels suggests that the spatial resolution is higher than 30 µm. The images in Figure 4 show expanded regions of Figures 3b-c that contains a single Purkinje cell. Here, the molecular image is not interpolated so each square pixel represents data from a single MS spectrum (at 10µm raster width/ spatial resolution). The Purkinje cell soma has a diameter of ~30 µm, and is represented by 3 3 mass spectrometric pixels, clearly demonstrating an actual mass spectrometric pixel size of 10µm. Testis consists of a number of well-defined anatomical structures that span a range of sizes and make it another good model system for evaluating mass spectrometric imaging performance. The main structures are seminiferous tubules which have a diameter ~200 µm. Within each tubular lumen the stage of spermatogenesis determines the internal structure. The interstitial space between tubules can range in size from a few µm up to 40 µm in width. Finally, the tubules are lined on the outside with layer of smooth muscle cells. Whether or not the interstitial space and the muscle layer surrounding the tubules can be clearly resolved is therefore a good test for evaluating spatial resolution in a mass spectrometric imaging experiment. small region of testis was imaged by MLDI-TOF and the distributions of three mass signals are shown in Figure 5 along with an H&E image of the analysis region taken after the MLDI experiment. The main anatomical structures are visible, especially a layered structure inside the seminiferous tubules highlighted by the ions shown in green and blue. The interstitial space (denoted by the ion in red) is also clearly resolved. MLDI-FTMS imaging FTMS instruments are especially suited for imaging of small molecules such as lipids because of their unsurpassed mass resolution and measurement accuracy. In the case of complex samples such as tissue, there are typically many isobaric compounds at each nominal m/z and a more accurate description of the sample makeup can be obtained when these are well resolved. Moreover, the precise mass measurement often allows an unambiguous determination of the molecular formulas of the observed compounds. Combining such high MS performance with a high spatial resolution promises to generate datasets rich with information.
4 Purkinje cells single Purkinje cell C Figure 4: Expanded view of the region highlighted in Fig. 3b, surrounding a single Purkinje cell. : The mass spectrometric image shown without interpolation. Each pixel represents one mass spectrum from a 10 µm area. : H&E image of the same section. The the dimension bar shows the Purkinje cell to have a diameter of 30 µm. Rat testis imaged at 10 µm pixel size with MLDI-TOF Figure 3: n example of a mass signal which is localized to the Purkinje cells. : Wide-field view showing the distinctive cellular layer surrounding the granular layer. : Expanded view of the highlighted region. C: H&E image of the same tissue section. In C individual Purkinje cells are designated with arrows. The corresponding Purkinje cells are annotated with arrows in Da Da Da Here, we applied MLDI-FTMS imaging analysis to the rat testis sample. Five structurally localized ions are shown in Figure 6 without display interpolation so that every visible square pixel represents one mass spectrum. From the three selected mass signals in Figure 6a we see that a pixel size of 10µm is sufficient to clearly distinguish the layer surrounding the tubules, the interstitial space and the lumen of the tubules. Figure 6b shows the distribution of two mass signals that are only 3 mda apart and exhibit very different distribution in the tissue. One mass signal is detected only from the interstitial space while the other is Da detected in the lumen of the tubules Da Da Figure 5: : Image which shows localization of three selected mass signals to different structures: seminiferous tubules (in green and blue) and interstitial space (in red). : H&E image of the same section.
5 The two peaks (as detected from the pixels highlighted in Figure 6b) are shown in Figure 6c. In this broadband (wide m/z range) FTMS measurement the average mass resolving power was approximately 190,000, which is sufficient to generate the image because those signals are also spatially resolved. If these peaks were in the same spectrum the resolution necessary to resolve them at a nominal mass of 808 Da would need to be greater than 300,000 which can be achieved using CSI. The signal of Da is not fully resolved from a lower-intensity, co-localized signal at Da. However: the peaks have sufficient spatial differentiation that does not contribute significant interference to the ion image of mass signal Da. Further, the measurement accuracy of the FTMS measurement allows the direct estimation of the molecular formulas labeled in Figure 6c. The mass signal at Da likely corresponds to the molecular formula [C 44 H 84 NNaO 8 P] + (a calculated mass of Da, a 0.94 ppm deviation from the measured mass). The signal at Da most likely corresponds to the molecular formula [C 46 H 83 NO 8 P] + (a calculated mass of Da, a 0.14 ppm deviation from the measured mass). Conclusion Sample preparation and instrumental parameters compete to be the limiting factor for achieving high spatial resolution in MLDI imaging. Sublimation of matrix offers an ideal dry preparation for MLDI imaging of lipids in which matrix crystal size and analyte migration do not limit high spatial resolution. Using this preparation strategy we were able to readily achieve a spatial resolution of 10µm using standard commercial ultraflextreme, autoflex speed and solarix instruments with the smartbeam II laser. Rat testis imaged at 10 µm pixel size with MLDI-FTMS C Δ = 3.05 mda m/z = [ C ] + 46 H 83 NO 8 P 50 µm m/z = [C 44H ] + 84 NNaO 8 P Da, Da, Da m/z = m/z 50 µm Da, Da Figure 6: : Image which shows three peaks localized to: tubule lumen (blue), basal layer (green) and interstitial space (red). : Image of two peaks which have a mass difference of 3 mda differentiated to tubule lumen and interstitial space. C: Mass signals from designated pixels in ) with suggested molecular formulas
6 ruker Daltonics is continually improving its products and reserves the right to change specifications without notice. ruker Daltonics , MLDI images of rat cerebellum C Da Da Reproduced from inside: Figure 2: MLDI images from part of rat cerebellum acquired at 10µm pixel size. : Three selected masses which show cellular localization: the mass signal in green highlights the individual Purkinje cells; the mass signal Da in red is found in the cerebellar white matter; and the mass signal Da is found in the granular layer. : Two selected mass signals that show strong differentiation, the mass signal at also exhibits spatial differentiation according to intensity range. C: Optical image of the measured cerebellum with measured region shown for context. ll scale bars: 200 µm. References For research use only. Not for use in diagnostic procedures. [1] Puolitaival SM, urnum KE, Cornett DS, Caprioli RM Solvent-free matrix dry-coating for MLDI imaging of phospholipids J m Soc Mass Spectrom Jun;19(6):882-6 [2] Chaurand P, Cornett DS, ngel PM, Caprioli RM From whole-body sections down to cellular level, multiscale imaging of phospholipids by MLDI mass spectrometry Mol Cell Proteomics Feb;10(2):O ruker Daltonik GmbH remen Germany Phone +49 (0) Fax +49 (0) [email protected] ruker Daltonics Inc. illerica, M US Phone +1 (978) Fax +1 (978) [email protected] Fremont, C US Phone +1 (510) Fax +1 (510) [email protected]
Bruker ToxScreener TM. Innovation with Integrity. A Comprehensive Screening Solution for Forensic Toxicology UHR-TOF MS
 Bruker ToxScreener TM A Comprehensive Screening Solution for Forensic Toxicology Innovation with Integrity UHR-TOF MS ToxScreener - Get the Complete Picture Forensic laboratories are frequently required
Bruker ToxScreener TM A Comprehensive Screening Solution for Forensic Toxicology Innovation with Integrity UHR-TOF MS ToxScreener - Get the Complete Picture Forensic laboratories are frequently required
Increasing the Multiplexing of High Resolution Targeted Peptide Quantification Assays
 Increasing the Multiplexing of High Resolution Targeted Peptide Quantification Assays Scheduled MRM HR Workflow on the TripleTOF Systems Jenny Albanese, Christie Hunter AB SCIEX, USA Targeted quantitative
Increasing the Multiplexing of High Resolution Targeted Peptide Quantification Assays Scheduled MRM HR Workflow on the TripleTOF Systems Jenny Albanese, Christie Hunter AB SCIEX, USA Targeted quantitative
Application Note # LCMS-81 Introducing New Proteomics Acquisiton Strategies with the compact Towards the Universal Proteomics Acquisition Method
 Application Note # LCMS-81 Introducing New Proteomics Acquisiton Strategies with the compact Towards the Universal Proteomics Acquisition Method Introduction During the last decade, the complexity of samples
Application Note # LCMS-81 Introducing New Proteomics Acquisiton Strategies with the compact Towards the Universal Proteomics Acquisition Method Introduction During the last decade, the complexity of samples
Copyright 2007 Casa Software Ltd. www.casaxps.com. ToF Mass Calibration
 ToF Mass Calibration Essentially, the relationship between the mass m of an ion and the time taken for the ion of a given charge to travel a fixed distance is quadratic in the flight time t. For an ideal
ToF Mass Calibration Essentially, the relationship between the mass m of an ion and the time taken for the ion of a given charge to travel a fixed distance is quadratic in the flight time t. For an ideal
Pesticide Analysis by Mass Spectrometry
 Pesticide Analysis by Mass Spectrometry Purpose: The purpose of this assignment is to introduce concepts of mass spectrometry (MS) as they pertain to the qualitative and quantitative analysis of organochlorine
Pesticide Analysis by Mass Spectrometry Purpose: The purpose of this assignment is to introduce concepts of mass spectrometry (MS) as they pertain to the qualitative and quantitative analysis of organochlorine
Tutorial for Proteomics Data Submission. Katalin F. Medzihradszky Robert J. Chalkley UCSF
 Tutorial for Proteomics Data Submission Katalin F. Medzihradszky Robert J. Chalkley UCSF Why Have Guidelines? Large-scale proteomics studies create huge amounts of data. It is impossible/impractical to
Tutorial for Proteomics Data Submission Katalin F. Medzihradszky Robert J. Chalkley UCSF Why Have Guidelines? Large-scale proteomics studies create huge amounts of data. It is impossible/impractical to
Accurate Mass Screening Workflows for the Analysis of Novel Psychoactive Substances
 Accurate Mass Screening Workflows for the Analysis of Novel Psychoactive Substances TripleTOF 5600 + LC/MS/MS System with MasterView Software Adrian M. Taylor AB Sciex Concord, Ontario (Canada) Overview
Accurate Mass Screening Workflows for the Analysis of Novel Psychoactive Substances TripleTOF 5600 + LC/MS/MS System with MasterView Software Adrian M. Taylor AB Sciex Concord, Ontario (Canada) Overview
The Olympus stereology system. The Computer Assisted Stereological Toolbox
 The Olympus stereology system The Computer Assisted Stereological Toolbox CAST is a Computer Assisted Stereological Toolbox for PCs running Microsoft Windows TM. CAST is an interactive, user-friendly,
The Olympus stereology system The Computer Assisted Stereological Toolbox CAST is a Computer Assisted Stereological Toolbox for PCs running Microsoft Windows TM. CAST is an interactive, user-friendly,
Accurate calibration of on-line Time of Flight Mass Spectrometer (TOF-MS) for high molecular weight combustion product analysis
 Accurate calibration of on-line Time of Flight Mass Spectrometer (TOF-MS) for high molecular weight combustion product analysis B. Apicella*, M. Passaro**, X. Wang***, N. Spinelli**** [email protected]
Accurate calibration of on-line Time of Flight Mass Spectrometer (TOF-MS) for high molecular weight combustion product analysis B. Apicella*, M. Passaro**, X. Wang***, N. Spinelli**** [email protected]
Care and Use of the Compound Microscope
 Revised Fall 2011 Care and Use of the Compound Microscope Objectives After completing this lab students should be able to 1. properly clean and carry a compound and dissecting microscope. 2. focus a specimen
Revised Fall 2011 Care and Use of the Compound Microscope Objectives After completing this lab students should be able to 1. properly clean and carry a compound and dissecting microscope. 2. focus a specimen
AB SCIEX TOF/TOF 4800 PLUS SYSTEM. Cost effective flexibility for your core needs
 AB SCIEX TOF/TOF 4800 PLUS SYSTEM Cost effective flexibility for your core needs AB SCIEX TOF/TOF 4800 PLUS SYSTEM It s just what you expect from the industry leader. The AB SCIEX 4800 Plus MALDI TOF/TOF
AB SCIEX TOF/TOF 4800 PLUS SYSTEM Cost effective flexibility for your core needs AB SCIEX TOF/TOF 4800 PLUS SYSTEM It s just what you expect from the industry leader. The AB SCIEX 4800 Plus MALDI TOF/TOF
Choices, choices, choices... Which sequence database? Which modifications? What mass tolerance?
 Optimization 1 Choices, choices, choices... Which sequence database? Which modifications? What mass tolerance? Where to begin? 2 Sequence Databases Swiss-prot MSDB, NCBI nr dbest Species specific ORFS
Optimization 1 Choices, choices, choices... Which sequence database? Which modifications? What mass tolerance? Where to begin? 2 Sequence Databases Swiss-prot MSDB, NCBI nr dbest Species specific ORFS
Tutorial 9: SWATH data analysis in Skyline
 Tutorial 9: SWATH data analysis in Skyline In this tutorial we will learn how to perform targeted post-acquisition analysis for protein identification and quantitation using a data-independent dataset
Tutorial 9: SWATH data analysis in Skyline In this tutorial we will learn how to perform targeted post-acquisition analysis for protein identification and quantitation using a data-independent dataset
# LCMS-35 esquire series. Application of LC/APCI Ion Trap Tandem Mass Spectrometry for the Multiresidue Analysis of Pesticides in Water
 Application Notes # LCMS-35 esquire series Application of LC/APCI Ion Trap Tandem Mass Spectrometry for the Multiresidue Analysis of Pesticides in Water An LC-APCI-MS/MS method using an ion trap system
Application Notes # LCMS-35 esquire series Application of LC/APCI Ion Trap Tandem Mass Spectrometry for the Multiresidue Analysis of Pesticides in Water An LC-APCI-MS/MS method using an ion trap system
In-Depth Qualitative Analysis of Complex Proteomic Samples Using High Quality MS/MS at Fast Acquisition Rates
 In-Depth Qualitative Analysis of Complex Proteomic Samples Using High Quality MS/MS at Fast Acquisition Rates Using the Explore Workflow on the AB SCIEX TripleTOF 5600 System A major challenge in proteomics
In-Depth Qualitative Analysis of Complex Proteomic Samples Using High Quality MS/MS at Fast Acquisition Rates Using the Explore Workflow on the AB SCIEX TripleTOF 5600 System A major challenge in proteomics
Application Note #503 Comparing 3D Optical Microscopy Techniques for Metrology Applications
 Screw thread image generated by WLI Steep PSS angles WLI color imaging Application Note #503 Comparing 3D Optical Microscopy Techniques for Metrology Applications 3D optical microscopy is a mainstay metrology
Screw thread image generated by WLI Steep PSS angles WLI color imaging Application Note #503 Comparing 3D Optical Microscopy Techniques for Metrology Applications 3D optical microscopy is a mainstay metrology
Micromass LCT User s Guide
 Micromass LCT User s Guide 1) Log on to MassLynx with your username & password. 2) After you have logged in, the MassLynx software will automatically run. 3) After MassLynx has come up, open your project
Micromass LCT User s Guide 1) Log on to MassLynx with your username & password. 2) After you have logged in, the MassLynx software will automatically run. 3) After MassLynx has come up, open your project
Fast, Reproducible LC-MS/MS Analysis of Dextromethorphan and Dextrorphan
 Fast, Reproducible LC-MS/MS Analysis of and Kimberly Phipps, Thermo Fisher Scientific, Runcorn, Cheshire, UK Application Note 685 Key Words Accucore C18, dextromethorphan, dextrorphan, SOLA CX Abstract
Fast, Reproducible LC-MS/MS Analysis of and Kimberly Phipps, Thermo Fisher Scientific, Runcorn, Cheshire, UK Application Note 685 Key Words Accucore C18, dextromethorphan, dextrorphan, SOLA CX Abstract
Alignment and Preprocessing for Data Analysis
 Alignment and Preprocessing for Data Analysis Preprocessing tools for chromatography Basics of alignment GC FID (D) data and issues PCA F Ratios GC MS (D) data and issues PCA F Ratios PARAFAC Piecewise
Alignment and Preprocessing for Data Analysis Preprocessing tools for chromatography Basics of alignment GC FID (D) data and issues PCA F Ratios GC MS (D) data and issues PCA F Ratios PARAFAC Piecewise
Overview. Introduction. AB SCIEX MPX -2 High Throughput TripleTOF 4600 LC/MS/MS System
 Investigating the use of the AB SCIEX TripleTOF 4600 LC/MS/MS System for High Throughput Screening of Synthetic Cannabinoids/Metabolites in Human Urine AB SCIEX MPX -2 High Throughput TripleTOF 4600 LC/MS/MS
Investigating the use of the AB SCIEX TripleTOF 4600 LC/MS/MS System for High Throughput Screening of Synthetic Cannabinoids/Metabolites in Human Urine AB SCIEX MPX -2 High Throughput TripleTOF 4600 LC/MS/MS
ALLEN Mouse Brain Atlas
 TECHNICAL WHITE PAPER: QUALITY CONTROL STANDARDS FOR HIGH-THROUGHPUT RNA IN SITU HYBRIDIZATION DATA GENERATION Consistent data quality and internal reproducibility are critical concerns for high-throughput
TECHNICAL WHITE PAPER: QUALITY CONTROL STANDARDS FOR HIGH-THROUGHPUT RNA IN SITU HYBRIDIZATION DATA GENERATION Consistent data quality and internal reproducibility are critical concerns for high-throughput
Enhancing GCMS analysis of trace compounds using a new dynamic baseline compensation algorithm to reduce background interference
 Enhancing GCMS analysis of trace compounds using a new dynamic baseline compensation algorithm to reduce background interference Abstract The advantages of mass spectrometry (MS) in combination with gas
Enhancing GCMS analysis of trace compounds using a new dynamic baseline compensation algorithm to reduce background interference Abstract The advantages of mass spectrometry (MS) in combination with gas
Aiping Lu. Key Laboratory of System Biology Chinese Academic Society [email protected]
 Aiping Lu Key Laboratory of System Biology Chinese Academic Society [email protected] Proteome and Proteomics PROTEin complement expressed by genome Marc Wilkins Electrophoresis. 1995. 16(7):1090-4. proteomics
Aiping Lu Key Laboratory of System Biology Chinese Academic Society [email protected] Proteome and Proteomics PROTEin complement expressed by genome Marc Wilkins Electrophoresis. 1995. 16(7):1090-4. proteomics
Compartmentalization of the Cell. Objectives. Recommended Reading. Professor Alfred Cuschieri. Department of Anatomy University of Malta
 Compartmentalization of the Cell Professor Alfred Cuschieri Department of Anatomy University of Malta Objectives By the end of this session the student should be able to: 1. Identify the different organelles
Compartmentalization of the Cell Professor Alfred Cuschieri Department of Anatomy University of Malta Objectives By the end of this session the student should be able to: 1. Identify the different organelles
HRMS in Clinical Research: from Targeted Quantification to Metabolomics
 A sponsored whitepaper. HRMS in Clinical Research: from Targeted Quantification to Metabolomics By: Bertrand Rochat Ph. D., Research Project Leader, Faculté de Biologie et de Médecine of the Centre Hospitalier
A sponsored whitepaper. HRMS in Clinical Research: from Targeted Quantification to Metabolomics By: Bertrand Rochat Ph. D., Research Project Leader, Faculté de Biologie et de Médecine of the Centre Hospitalier
Bio and Polymer Analytics. RD Instrumental Analytical Chemistry. Organic Trace Analytics. RD Environmental & Process Analytics
 MOLEKULARE IMAGING MASSENSPEKTROMETRIE VON GEWEBSOBERFLÄCHEN und BIOPOLYMERANALTIK Günter Allmaier RESEARCH GROUP BIO- AND POLYMER ANALYSIS Vienna University of Technology, Institute of Chemical Technologies
MOLEKULARE IMAGING MASSENSPEKTROMETRIE VON GEWEBSOBERFLÄCHEN und BIOPOLYMERANALTIK Günter Allmaier RESEARCH GROUP BIO- AND POLYMER ANALYSIS Vienna University of Technology, Institute of Chemical Technologies
Application Note # LCMS-62 Walk-Up Ion Trap Mass Spectrometer System in a Multi-User Environment Using Compass OpenAccess Software
 Application Note # LCMS-62 Walk-Up Ion Trap Mass Spectrometer System in a Multi-User Environment Using Compass OpenAccess Software Abstract Presented here is a case study of a walk-up liquid chromatography
Application Note # LCMS-62 Walk-Up Ion Trap Mass Spectrometer System in a Multi-User Environment Using Compass OpenAccess Software Abstract Presented here is a case study of a walk-up liquid chromatography
Appendix 5 Overview of requirements in English
 Appendix 5 Overview of requirements in English This document is a translation of Appendix 4 (Bilag 4) section 2. This translation is meant as a service for the bidder and in case of any differences between
Appendix 5 Overview of requirements in English This document is a translation of Appendix 4 (Bilag 4) section 2. This translation is meant as a service for the bidder and in case of any differences between
Explain the role of blood and bloodstain patterns in forensics science. Analyze and identify bloodstain patterns by performing bloodstain analysis
 Lab 4 Blood Learning Objectives Explain the role of blood and bloodstain patterns in forensics science Analyze and identify bloodstain patterns by performing bloodstain analysis Introduction Blood, a
Lab 4 Blood Learning Objectives Explain the role of blood and bloodstain patterns in forensics science Analyze and identify bloodstain patterns by performing bloodstain analysis Introduction Blood, a
MRMPilot Software: Accelerating MRM Assay Development for Targeted Quantitative Proteomics
 MRMPilot Software: Accelerating MRM Assay Development for Targeted Quantitative Proteomics With Unique QTRAP and TripleTOF 5600 System Technology Targeted peptide quantification is a rapidly growing application
MRMPilot Software: Accelerating MRM Assay Development for Targeted Quantitative Proteomics With Unique QTRAP and TripleTOF 5600 System Technology Targeted peptide quantification is a rapidly growing application
Functional Data Analysis of MALDI TOF Protein Spectra
 Functional Data Analysis of MALDI TOF Protein Spectra Dean Billheimer [email protected]. Department of Biostatistics Vanderbilt University Vanderbilt Ingram Cancer Center FDA for MALDI TOF
Functional Data Analysis of MALDI TOF Protein Spectra Dean Billheimer [email protected]. Department of Biostatistics Vanderbilt University Vanderbilt Ingram Cancer Center FDA for MALDI TOF
Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides
 Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides Polyacrylamide gel electrophoresis (PAGE) and subsequent analyses are common tools in biochemistry and molecular biology. This Application
Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides Polyacrylamide gel electrophoresis (PAGE) and subsequent analyses are common tools in biochemistry and molecular biology. This Application
MASCOT Search Results Interpretation
 The Mascot protein identification program (Matrix Science, Ltd.) uses statistical methods to assess the validity of a match. MS/MS data is not ideal. That is, there are unassignable peaks (noise) and usually
The Mascot protein identification program (Matrix Science, Ltd.) uses statistical methods to assess the validity of a match. MS/MS data is not ideal. That is, there are unassignable peaks (noise) and usually
Effects of Intelligent Data Acquisition and Fast Laser Speed on Analysis of Complex Protein Digests
 Effects of Intelligent Data Acquisition and Fast Laser Speed on Analysis of Complex Protein Digests AB SCIEX TOF/TOF 5800 System with DynamicExit Algorithm and ProteinPilot Software for Robust Protein
Effects of Intelligent Data Acquisition and Fast Laser Speed on Analysis of Complex Protein Digests AB SCIEX TOF/TOF 5800 System with DynamicExit Algorithm and ProteinPilot Software for Robust Protein
ProteinPilot Report for ProteinPilot Software
 ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Pow erful mass spectrometers like
ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Pow erful mass spectrometers like
MIRAX SCAN The new way of looking at pathology
 Microscopy from Carl Zeiss MIRAX SCAN The new way of looking at pathology Greater reliability. Greater efficiency. Virtual microscopy for research & analysis. Better. More efficient. Quality as the basis
Microscopy from Carl Zeiss MIRAX SCAN The new way of looking at pathology Greater reliability. Greater efficiency. Virtual microscopy for research & analysis. Better. More efficient. Quality as the basis
www.gbo.com/bioscience Tissue Culture 1 Cell/ Microplates 2 HTS- 3 Immunology/ HLA 4 Microbiology/ Bacteriology Purpose Beakers 5 Tubes/Multi-
 11 Cryo 5 Tubes/Multi 2 HTS 3 Immunology / Immunology Technical Information 3 I 2 96 Well ELISA 3 I 4 96 Well ELISA Strip Plates 3 I 6 8 Well Strip Plates 3 I 7 12 Well Strip Plates 3 I 8 16 Well Strip
11 Cryo 5 Tubes/Multi 2 HTS 3 Immunology / Immunology Technical Information 3 I 2 96 Well ELISA 3 I 4 96 Well ELISA Strip Plates 3 I 6 8 Well Strip Plates 3 I 7 12 Well Strip Plates 3 I 8 16 Well Strip
Mass Spectrometry Signal Calibration for Protein Quantitation
 Cambridge Isotope Laboratories, Inc. www.isotope.com Proteomics Mass Spectrometry Signal Calibration for Protein Quantitation Michael J. MacCoss, PhD Associate Professor of Genome Sciences University of
Cambridge Isotope Laboratories, Inc. www.isotope.com Proteomics Mass Spectrometry Signal Calibration for Protein Quantitation Michael J. MacCoss, PhD Associate Professor of Genome Sciences University of
LC-MS/MS Method for the Determination of Docetaxel in Human Serum for Clinical Research
 LC-MS/MS Method for the Determination of Docetaxel in Human Serum for Clinical Research J. Jones, J. Denbigh, Thermo Fisher Scientific, Runcorn, Cheshire, UK Application Note 20581 Key Words SPE, SOLA,
LC-MS/MS Method for the Determination of Docetaxel in Human Serum for Clinical Research J. Jones, J. Denbigh, Thermo Fisher Scientific, Runcorn, Cheshire, UK Application Note 20581 Key Words SPE, SOLA,
泛 用 蛋 白 質 體 學 之 質 譜 儀 資 料 分 析 平 台 的 建 立 與 應 用 Universal Mass Spectrometry Data Analysis Platform for Quantitative and Qualitative Proteomics
 泛 用 蛋 白 質 體 學 之 質 譜 儀 資 料 分 析 平 台 的 建 立 與 應 用 Universal Mass Spectrometry Data Analysis Platform for Quantitative and Qualitative Proteomics 2014 Training Course Wei-Hung Chang ( 張 瑋 宏 ) ABRC, Academia
泛 用 蛋 白 質 體 學 之 質 譜 儀 資 料 分 析 平 台 的 建 立 與 應 用 Universal Mass Spectrometry Data Analysis Platform for Quantitative and Qualitative Proteomics 2014 Training Course Wei-Hung Chang ( 張 瑋 宏 ) ABRC, Academia
MultiQuant Software 2.0 for Targeted Protein / Peptide Quantification
 MultiQuant Software 2.0 for Targeted Protein / Peptide Quantification Gold Standard for Quantitative Data Processing Because of the sensitivity, selectivity, speed and throughput at which MRM assays can
MultiQuant Software 2.0 for Targeted Protein / Peptide Quantification Gold Standard for Quantitative Data Processing Because of the sensitivity, selectivity, speed and throughput at which MRM assays can
Guide to Reverse Phase SpinColumns Chromatography for Sample Prep
 Guide to Reverse Phase SpinColumns Chromatography for Sample Prep www.harvardapparatus.com Contents Introduction...2-3 Modes of Separation...4-6 Spin Column Efficiency...7-8 Fast Protein Analysis...9 Specifications...10
Guide to Reverse Phase SpinColumns Chromatography for Sample Prep www.harvardapparatus.com Contents Introduction...2-3 Modes of Separation...4-6 Spin Column Efficiency...7-8 Fast Protein Analysis...9 Specifications...10
TECHNICAL BULLETIN. ProteoMass Peptide & Protein MALDI-MS Calibration Kit. Catalog Number MSCAL1 Store at Room Temperature
 ProteoMass Peptide & Protein MALDI-MS Calibration Kit Catalog Number MSCAL1 Store at Room Temperature TECHNICAL BULLETIN Description This kit provides a range of standard peptides and proteins for the
ProteoMass Peptide & Protein MALDI-MS Calibration Kit Catalog Number MSCAL1 Store at Room Temperature TECHNICAL BULLETIN Description This kit provides a range of standard peptides and proteins for the
Simultaneous qualitative and quantitative analysis using the Agilent 6540 Accurate-Mass Q-TOF
 Simultaneous qualitative and quantitative analysis using the Agilent 654 Accurate-Mass Q-TOF Technical Overview Authors Pat Perkins Anabel Fandino Lester Taylor Agilent Technologies, Inc. Santa Clara,
Simultaneous qualitative and quantitative analysis using the Agilent 654 Accurate-Mass Q-TOF Technical Overview Authors Pat Perkins Anabel Fandino Lester Taylor Agilent Technologies, Inc. Santa Clara,
A Streamlined Workflow for Untargeted Metabolomics
 A Streamlined Workflow for Untargeted Metabolomics Employing XCMS plus, a Simultaneous Data Processing and Metabolite Identification Software Package for Rapid Untargeted Metabolite Screening Baljit K.
A Streamlined Workflow for Untargeted Metabolomics Employing XCMS plus, a Simultaneous Data Processing and Metabolite Identification Software Package for Rapid Untargeted Metabolite Screening Baljit K.
What Do We Learn about Hepatotoxicity Using Coumarin-Treated Rat Model?
 What Do We Learn about Hepatotoxicity Using Coumarin-Treated Rat Model? authors M. David Ho 1, Bob Xiong 1, S. Stellar 2, J. Proctor 2, J. Silva 2, H.K. Lim 2, Patrick Bennett 1, and Lily Li 1 Tandem Labs,
What Do We Learn about Hepatotoxicity Using Coumarin-Treated Rat Model? authors M. David Ho 1, Bob Xiong 1, S. Stellar 2, J. Proctor 2, J. Silva 2, H.K. Lim 2, Patrick Bennett 1, and Lily Li 1 Tandem Labs,
Mass Frontier 7.0 Quick Start Guide
 Mass Frontier 7.0 Quick Start Guide The topics in this guide briefly step you through key features of the Mass Frontier application. Editing a Structure Working with Spectral Trees Building a Library Predicting
Mass Frontier 7.0 Quick Start Guide The topics in this guide briefly step you through key features of the Mass Frontier application. Editing a Structure Working with Spectral Trees Building a Library Predicting
Preprocessing, Management, and Analysis of Mass Spectrometry Proteomics Data
 Preprocessing, Management, and Analysis of Mass Spectrometry Proteomics Data M. Cannataro, P. H. Guzzi, T. Mazza, and P. Veltri Università Magna Græcia di Catanzaro, Italy 1 Introduction Mass Spectrometry
Preprocessing, Management, and Analysis of Mass Spectrometry Proteomics Data M. Cannataro, P. H. Guzzi, T. Mazza, and P. Veltri Università Magna Græcia di Catanzaro, Italy 1 Introduction Mass Spectrometry
Image Lab Software for the GS-900 Densitometer
 Image Lab Software for the GS-900 Densitometer Quick Start Guide Catalog # 170-9690 Bio-Rad Technical Support For help and technical advice, please contact the Bio-Rad Technical Support department. In
Image Lab Software for the GS-900 Densitometer Quick Start Guide Catalog # 170-9690 Bio-Rad Technical Support For help and technical advice, please contact the Bio-Rad Technical Support department. In
The Scheduled MRM Algorithm Enables Intelligent Use of Retention Time During Multiple Reaction Monitoring
 The Scheduled MRM Algorithm Enables Intelligent Use of Retention Time During Multiple Reaction Monitoring Delivering up to 2500 MRM Transitions per LC Run Christie Hunter 1, Brigitte Simons 2 1 AB SCIEX,
The Scheduled MRM Algorithm Enables Intelligent Use of Retention Time During Multiple Reaction Monitoring Delivering up to 2500 MRM Transitions per LC Run Christie Hunter 1, Brigitte Simons 2 1 AB SCIEX,
MICROSCOPY. To demonstrate skill in the proper utilization of a light microscope.
 MICROSCOPY I. OBJECTIVES To demonstrate skill in the proper utilization of a light microscope. To demonstrate skill in the use of ocular and stage micrometers for measurements of cell size. To recognize
MICROSCOPY I. OBJECTIVES To demonstrate skill in the proper utilization of a light microscope. To demonstrate skill in the use of ocular and stage micrometers for measurements of cell size. To recognize
aurora ICP-MS Innovation with Integrity The Science of Sensitivity ICP-MS
 aurora ICP-MS The Science of Sensitivity Innovation with Integrity ICP-MS Powers the Business of Quantification aurora ICP-MS enhanced value package Bruker s ICP-MS is famous for its innovative and unique
aurora ICP-MS The Science of Sensitivity Innovation with Integrity ICP-MS Powers the Business of Quantification aurora ICP-MS enhanced value package Bruker s ICP-MS is famous for its innovative and unique
INFITEC - A NEW STEREOSCOPIC VISUALISATION TOOL BY WAVELENGTH MULTIPLEX IMAGING
 INFITEC - A NEW STEREOSCOPIC VISUALISATION TOOL BY WAVELENGTH MULTIPLEX IMAGING Helmut Jorke, Markus Fritz INFITEC GmbH, Lise-Meitner-Straße 9, 89081 Ulm [email protected] Phone +49 731 550299 56 Fax _
INFITEC - A NEW STEREOSCOPIC VISUALISATION TOOL BY WAVELENGTH MULTIPLEX IMAGING Helmut Jorke, Markus Fritz INFITEC GmbH, Lise-Meitner-Straße 9, 89081 Ulm [email protected] Phone +49 731 550299 56 Fax _
amazon SL Innovation with Integrity Setting New Standards in Performance, Simplicity and Value Ion Trap MS
 amazon SL Setting New Standards in Performance, Simplicity and Value Innovation with Integrity Ion Trap Best-In-Class Ion Trap Mass Spectrometer for Routine Analysis The amazon SL entry-level system is
amazon SL Setting New Standards in Performance, Simplicity and Value Innovation with Integrity Ion Trap Best-In-Class Ion Trap Mass Spectrometer for Routine Analysis The amazon SL entry-level system is
Per Andrén Dept. of Pharmaceutical Biosciences Medical Mass Spectrometry, Uppsala University, Uppsala, Sweden
 Lung Imaging in Inhaled Product Development Novartis, Horsham, UK, 2011-11-09 Mapping Drug Deposition using Imaging Mass Spectrometry Per Andrén Dept. of Pharmaceutical Biosciences Medical Mass Spectrometry,
Lung Imaging in Inhaled Product Development Novartis, Horsham, UK, 2011-11-09 Mapping Drug Deposition using Imaging Mass Spectrometry Per Andrén Dept. of Pharmaceutical Biosciences Medical Mass Spectrometry,
CELERY LAB - Structure and Function of a Plant
 CELERY LAB - Structure and Function of a Plant READ ALL INSTRUCTIONS BEFORE BEGINNING! YOU MAY WORK WITH A PARTNER ON THIS ACTIVITY, BUT YOU MUST COMPLETE YOUR OWN LAB SHEET! Look at the back of this paper
CELERY LAB - Structure and Function of a Plant READ ALL INSTRUCTIONS BEFORE BEGINNING! YOU MAY WORK WITH A PARTNER ON THIS ACTIVITY, BUT YOU MUST COMPLETE YOUR OWN LAB SHEET! Look at the back of this paper
Fast and Accurate Analysis of Vitamin D Metabolites Using Novel Chromatographic Selectivity and Sample Preparation
 Fast and Accurate Analysis of Vitamin D Metabolites Using Novel Chromatographic Selectivity and Sample Preparation Craig Aurand 1, David Bell 1, Anders Fridstrom 2, and Klaus Buckendahl 2 1 Supelco, Div.
Fast and Accurate Analysis of Vitamin D Metabolites Using Novel Chromatographic Selectivity and Sample Preparation Craig Aurand 1, David Bell 1, Anders Fridstrom 2, and Klaus Buckendahl 2 1 Supelco, Div.
MarkerView Software 1.2.1 for Metabolomic and Biomarker Profiling Analysis
 MarkerView Software 1.2.1 for Metabolomic and Biomarker Profiling Analysis Overview MarkerView software is a novel program designed for metabolomics applications and biomarker profiling workflows 1. Using
MarkerView Software 1.2.1 for Metabolomic and Biomarker Profiling Analysis Overview MarkerView software is a novel program designed for metabolomics applications and biomarker profiling workflows 1. Using
ProteinScape. Innovation with Integrity. Proteomics Data Analysis & Management. Mass Spectrometry
 ProteinScape Proteomics Data Analysis & Management Innovation with Integrity Mass Spectrometry ProteinScape a Virtual Environment for Successful Proteomics To overcome the growing complexity of proteomics
ProteinScape Proteomics Data Analysis & Management Innovation with Integrity Mass Spectrometry ProteinScape a Virtual Environment for Successful Proteomics To overcome the growing complexity of proteomics
Application Note. Single Cell PCR Preparation
 Application Note Single Cell PCR Preparation From Automated Screening to the Molecular Analysis of Single Cells The AmpliGrid system is a highly sensitive tool for the analysis of single cells. In combination
Application Note Single Cell PCR Preparation From Automated Screening to the Molecular Analysis of Single Cells The AmpliGrid system is a highly sensitive tool for the analysis of single cells. In combination
Chapter 4. Microscopy, Staining, and Classification. Lecture prepared by Mindy Miller-Kittrell North Carolina State University
 Chapter 4 Microscopy, Staining, and Classification 2012 Pearson Education Inc. Lecture prepared by Mindy Miller-Kittrell North Carolina State University Microscopy and Staining 2012 Pearson Education Inc.
Chapter 4 Microscopy, Staining, and Classification 2012 Pearson Education Inc. Lecture prepared by Mindy Miller-Kittrell North Carolina State University Microscopy and Staining 2012 Pearson Education Inc.
Q-TOF User s Booklet. Enter your username and password to login. After you login, the data acquisition program will automatically start.
 Q-TOF User s Booklet Enter your username and password to login. After you login, the data acquisition program will automatically start. ~ 1 ~ This data acquisition window will come up after the program
Q-TOF User s Booklet Enter your username and password to login. After you login, the data acquisition program will automatically start. ~ 1 ~ This data acquisition window will come up after the program
Application of TargetView software within the food industry - the identi cation of pyrazine compounds in potato crisps
 Application Note: ANTV10 Application of TargetView software within the food industry - the identi cation of pyrazine compounds in potato crisps Abstract In order to regulate product safety and quality
Application Note: ANTV10 Application of TargetView software within the food industry - the identi cation of pyrazine compounds in potato crisps Abstract In order to regulate product safety and quality
CONFOCAL LASER SCANNING MICROSCOPY TUTORIAL
 CONFOCAL LASER SCANNING MICROSCOPY TUTORIAL Robert Bagnell 2006 This tutorial covers the following CLSM topics: 1) What is the optical principal behind CLSM? 2) What is the spatial resolution in X, Y,
CONFOCAL LASER SCANNING MICROSCOPY TUTORIAL Robert Bagnell 2006 This tutorial covers the following CLSM topics: 1) What is the optical principal behind CLSM? 2) What is the spatial resolution in X, Y,
Technical Bulletin. Threshold and Analysis of Small Particles on the BD Accuri C6 Flow Cytometer
 Threshold and Analysis of Small Particles on the BD Accuri C6 Flow Cytometer Contents 2 Thresholds 2 Setting the Threshold When analyzing small particles, defined as particles smaller than 3.0 µm, on the
Threshold and Analysis of Small Particles on the BD Accuri C6 Flow Cytometer Contents 2 Thresholds 2 Setting the Threshold When analyzing small particles, defined as particles smaller than 3.0 µm, on the
Scanners and How to Use Them
 Written by Jonathan Sachs Copyright 1996-1999 Digital Light & Color Introduction A scanner is a device that converts images to a digital file you can use with your computer. There are many different types
Written by Jonathan Sachs Copyright 1996-1999 Digital Light & Color Introduction A scanner is a device that converts images to a digital file you can use with your computer. There are many different types
New drugs of abuse? A case study on Phenazepam & Methoxetamine
 New drugs of abuse? A case study on Phenazepam & Methoxetamine Presenter: Nadia Wong Co authors: Dr Yao Yi Ju & Alex Low Xuan Kai Analytical Toxicology Laboratory Clinical & Forensic Toxicology Unit Applied
New drugs of abuse? A case study on Phenazepam & Methoxetamine Presenter: Nadia Wong Co authors: Dr Yao Yi Ju & Alex Low Xuan Kai Analytical Toxicology Laboratory Clinical & Forensic Toxicology Unit Applied
AMD Analysis & Technology AG
 AMD Analysis & Technology AG Application Note 120419 Author: Karl-Heinz Maurer APCI-MS Trace Analysis of volatile organic compounds in ambient air A) Introduction Trace analysis of volatile organic compounds
AMD Analysis & Technology AG Application Note 120419 Author: Karl-Heinz Maurer APCI-MS Trace Analysis of volatile organic compounds in ambient air A) Introduction Trace analysis of volatile organic compounds
Name Date Hour. Nerve Histology Microscope Lab
 Name Date Hour Nerve Histology Microscope Lab PRE-LAB: Answer the following questions using your reading and class notes before starting the microscope lab. 1. What is the difference between the functions
Name Date Hour Nerve Histology Microscope Lab PRE-LAB: Answer the following questions using your reading and class notes before starting the microscope lab. 1. What is the difference between the functions
Technical Considerations Detecting Transparent Materials in Particle Analysis. Michael Horgan
 Technical Considerations Detecting Transparent Materials in Particle Analysis Michael Horgan Who We Are Fluid Imaging Technology Manufacturers of the FlowCam series of particle analyzers FlowCam HQ location
Technical Considerations Detecting Transparent Materials in Particle Analysis Michael Horgan Who We Are Fluid Imaging Technology Manufacturers of the FlowCam series of particle analyzers FlowCam HQ location
LIST OF FIGURES. Figure 1: Diagrammatic representation of electromagnetic wave. Figure 2: A representation of the electromagnetic spectrum.
 LIST OF FIGURES Figure 1: Diagrammatic representation of electromagnetic wave. Figure 2: A representation of the electromagnetic spectrum. Figure 3: Picture depicting the internal circuit of mobile phone
LIST OF FIGURES Figure 1: Diagrammatic representation of electromagnetic wave. Figure 2: A representation of the electromagnetic spectrum. Figure 3: Picture depicting the internal circuit of mobile phone
Advantages of the LTQ Orbitrap for Protein Identification in Complex Digests
 Application Note: 386 Advantages of the LTQ Orbitrap for Protein Identification in Complex Digests Rosa Viner, Terry Zhang, Scott Peterman, and Vlad Zabrouskov, Thermo Fisher Scientific, San Jose, CA,
Application Note: 386 Advantages of the LTQ Orbitrap for Protein Identification in Complex Digests Rosa Viner, Terry Zhang, Scott Peterman, and Vlad Zabrouskov, Thermo Fisher Scientific, San Jose, CA,
Physics 441/2: Transmission Electron Microscope
 Physics 441/2: Transmission Electron Microscope Introduction In this experiment we will explore the use of transmission electron microscopy (TEM) to take us into the world of ultrasmall structures. This
Physics 441/2: Transmission Electron Microscope Introduction In this experiment we will explore the use of transmission electron microscopy (TEM) to take us into the world of ultrasmall structures. This
[ Care and Use Manual ]
![[ Care and Use Manual ] [ Care and Use Manual ]](/thumbs/26/8588921.jpg) PREP Calibration Mix DIOS Low i. Introduction Pre-packaged PREP Calibration Mixtures eliminate the need to purchase and store large quantities of the component calibration reagents, simplifying sample
PREP Calibration Mix DIOS Low i. Introduction Pre-packaged PREP Calibration Mixtures eliminate the need to purchase and store large quantities of the component calibration reagents, simplifying sample
Protease Peptide Microarrays Ready-to-use microarrays for protease profiling
 Protocol Protease Peptide Microarrays Ready-to-use microarrays for protease profiling Contact us: InfoLine: +49-30-97893-117 Order per fax: +49-30-97893-299 Or e-mail: [email protected] www: www.jpt.com
Protocol Protease Peptide Microarrays Ready-to-use microarrays for protease profiling Contact us: InfoLine: +49-30-97893-117 Order per fax: +49-30-97893-299 Or e-mail: [email protected] www: www.jpt.com
AppNote 6/2003. Analysis of Flavors using a Mass Spectral Based Chemical Sensor KEYWORDS ABSTRACT
 AppNote 6/2003 Analysis of Flavors using a Mass Spectral Based Chemical Sensor Vanessa R. Kinton Gerstel, Inc., 701 Digital Drive, Suite J, Linthicum, MD 21090, USA Kevin L. Goodner USDA, Citrus & Subtropical
AppNote 6/2003 Analysis of Flavors using a Mass Spectral Based Chemical Sensor Vanessa R. Kinton Gerstel, Inc., 701 Digital Drive, Suite J, Linthicum, MD 21090, USA Kevin L. Goodner USDA, Citrus & Subtropical
Thermo Scientific SIEVE Software for Differential Expression Analysis
 m a s s s p e c t r o m e t r y Thermo Scientific SIEVE Software for Differential Expression Analysis Automated, label-free, semi-quantitative analysis of proteins, peptides, and metabolites based on comparisons
m a s s s p e c t r o m e t r y Thermo Scientific SIEVE Software for Differential Expression Analysis Automated, label-free, semi-quantitative analysis of proteins, peptides, and metabolites based on comparisons
1. Examine the metric ruler. This ruler is 1 meter long. The distance between two of the lines with numbers on this ruler is 1 centimeter.
 Nano Scale How small is small? It depends on your point of reference. A human is very small compared to the earth. A grain of salt is very small compared to a human. However, a grain of salt is very large
Nano Scale How small is small? It depends on your point of reference. A human is very small compared to the earth. A grain of salt is very small compared to a human. However, a grain of salt is very large
Calibration of Volumetric Glassware
 Chemistry 119: Experiment 2 Calibration of Volumetric Glassware For making accurate measurements in analytical procedures, next in importance to the balance is volumetric equipment. In this section volumetric
Chemistry 119: Experiment 2 Calibration of Volumetric Glassware For making accurate measurements in analytical procedures, next in importance to the balance is volumetric equipment. In this section volumetric
DRUG METABOLISM. Drug discovery & development solutions FOR DRUG METABOLISM
 DRUG METABLISM Drug discovery & development solutions FR DRUG METABLISM Fast and efficient metabolite identification is critical in today s drug discovery pipeline. The goal is to achieve rapid structural
DRUG METABLISM Drug discovery & development solutions FR DRUG METABLISM Fast and efficient metabolite identification is critical in today s drug discovery pipeline. The goal is to achieve rapid structural
Electromagnetic Radiation (EMR) and Remote Sensing
 Electromagnetic Radiation (EMR) and Remote Sensing 1 Atmosphere Anything missing in between? Electromagnetic Radiation (EMR) is radiated by atomic particles at the source (the Sun), propagates through
Electromagnetic Radiation (EMR) and Remote Sensing 1 Atmosphere Anything missing in between? Electromagnetic Radiation (EMR) is radiated by atomic particles at the source (the Sun), propagates through
Fluorescence Microscopy for an NMR- Biosensor Project
 Fluorescence Microscopy for an NMR- Biosensor Project Ole Hirsch Physikalisch-Technische Bundesanstalt Medical Optics Abbestr. -1, 10587 Berlin, Germany Overview NMR Sensor Project Dimensions in biological
Fluorescence Microscopy for an NMR- Biosensor Project Ole Hirsch Physikalisch-Technische Bundesanstalt Medical Optics Abbestr. -1, 10587 Berlin, Germany Overview NMR Sensor Project Dimensions in biological
Integration of a fin experiment into the undergraduate heat transfer laboratory
 Integration of a fin experiment into the undergraduate heat transfer laboratory H. I. Abu-Mulaweh Mechanical Engineering Department, Purdue University at Fort Wayne, Fort Wayne, IN 46805, USA E-mail: [email protected]
Integration of a fin experiment into the undergraduate heat transfer laboratory H. I. Abu-Mulaweh Mechanical Engineering Department, Purdue University at Fort Wayne, Fort Wayne, IN 46805, USA E-mail: [email protected]
Fractional Distillation and Gas Chromatography
 Fractional Distillation and Gas Chromatography Background Distillation The previous lab used distillation to separate a mixture of hexane and toluene based on a difference in boiling points. Hexane boils
Fractional Distillation and Gas Chromatography Background Distillation The previous lab used distillation to separate a mixture of hexane and toluene based on a difference in boiling points. Hexane boils
Thermo Scientific GC-MS Data Acquisition Instructions for Cerno Bioscience MassWorks Software
 Thermo Scientific GC-MS Data Acquisition Instructions for Cerno Bioscience MassWorks Software Mark Belmont and Alexander N. Semyonov, Thermo Fisher Scientific, Austin, TX, USA Ming Gu, Cerno Bioscience,
Thermo Scientific GC-MS Data Acquisition Instructions for Cerno Bioscience MassWorks Software Mark Belmont and Alexander N. Semyonov, Thermo Fisher Scientific, Austin, TX, USA Ming Gu, Cerno Bioscience,
SELDI-TOF Mass Spectrometry Protein Data By Huong Thi Dieu La
 SELDI-TOF Mass Spectrometry Protein Data By Huong Thi Dieu La References Alejandro Cruz-Marcelo, Rudy Guerra, Marina Vannucci, Yiting Li, Ching C. Lau, and Tsz-Kwong Man. Comparison of algorithms for pre-processing
SELDI-TOF Mass Spectrometry Protein Data By Huong Thi Dieu La References Alejandro Cruz-Marcelo, Rudy Guerra, Marina Vannucci, Yiting Li, Ching C. Lau, and Tsz-Kwong Man. Comparison of algorithms for pre-processing
using ms based proteomics
 quantification using ms based proteomics lennart martens Computational Omics and Systems Biology Group Department of Medical Protein Research, VIB Department of Biochemistry, Ghent University Ghent, Belgium
quantification using ms based proteomics lennart martens Computational Omics and Systems Biology Group Department of Medical Protein Research, VIB Department of Biochemistry, Ghent University Ghent, Belgium
ab176915 StayBlue/AP Plus Stain Kit Instructions for Use An immunohistochemical chromogen substrate for staining tissue sections
 ab176915 StayBlue/AP Plus Stain Kit Instructions for Use An immunohistochemical chromogen substrate for staining tissue sections This product is for research use only and is not intended for diagnostic
ab176915 StayBlue/AP Plus Stain Kit Instructions for Use An immunohistochemical chromogen substrate for staining tissue sections This product is for research use only and is not intended for diagnostic
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY
 MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
DETECTION OF SUBSURFACE DAMAGE IN OPTICAL TRANSPARENT MATERIALSS USING SHORT COHERENCE TOMOGRAPHY. Rainer Boerret, Dominik Wiedemann, Andreas Kelm
 URN (Paper): urn:nbn:de:gbv:ilm1-2014iwk-199:0 58 th ILMENAU SCIENTIFIC COLLOQUIUM Technische Universität Ilmenau, 08 12 September 2014 URN: urn:nbn:de:gbv:ilm1-2014iwk:3 DETECTION OF SUBSURFACE DAMAGE
URN (Paper): urn:nbn:de:gbv:ilm1-2014iwk-199:0 58 th ILMENAU SCIENTIFIC COLLOQUIUM Technische Universität Ilmenau, 08 12 September 2014 URN: urn:nbn:de:gbv:ilm1-2014iwk:3 DETECTION OF SUBSURFACE DAMAGE
Digital Remote Sensing Data Processing Digital Remote Sensing Data Processing and Analysis: An Introduction and Analysis: An Introduction
 Digital Remote Sensing Data Processing Digital Remote Sensing Data Processing and Analysis: An Introduction and Analysis: An Introduction Content Remote sensing data Spatial, spectral, radiometric and
Digital Remote Sensing Data Processing Digital Remote Sensing Data Processing and Analysis: An Introduction and Analysis: An Introduction Content Remote sensing data Spatial, spectral, radiometric and
ForensicDB: A Web-accessible Spectral Database
 ForensicDB: A Web-accessible Spectral Database Table of Contents 1. Introduction to ForensicDB... 1 2. Accessing ForensicDB... 1 3. Setting System Properties... 5 Spectrum Properties... 5 Frame Properties...
ForensicDB: A Web-accessible Spectral Database Table of Contents 1. Introduction to ForensicDB... 1 2. Accessing ForensicDB... 1 3. Setting System Properties... 5 Spectrum Properties... 5 Frame Properties...
Spectrophotometry and the Beer-Lambert Law: An Important Analytical Technique in Chemistry
 Spectrophotometry and the Beer-Lambert Law: An Important Analytical Technique in Chemistry Jon H. Hardesty, PhD and Bassam Attili, PhD Collin College Department of Chemistry Introduction: In the last lab
Spectrophotometry and the Beer-Lambert Law: An Important Analytical Technique in Chemistry Jon H. Hardesty, PhD and Bassam Attili, PhD Collin College Department of Chemistry Introduction: In the last lab
AxioCam MR The All-round Camera for Biology, Medicine and Materials Analysis Digital Documentation in Microscopy
 Microscopy from Carl Zeiss AxioCam MR The All-round Camera for Biology, Medicine and Materials Analysis Digital Documentation in Microscopy New Dimensions in Performance AxioCam MR from Carl Zeiss Both
Microscopy from Carl Zeiss AxioCam MR The All-round Camera for Biology, Medicine and Materials Analysis Digital Documentation in Microscopy New Dimensions in Performance AxioCam MR from Carl Zeiss Both
Nuclear Magnetic Resonance notes
 Reminder: These notes are meant to supplement, not replace, the laboratory manual. Nuclear Magnetic Resonance notes Nuclear Magnetic Resonance (NMR) is a spectrometric technique which provides information
Reminder: These notes are meant to supplement, not replace, the laboratory manual. Nuclear Magnetic Resonance notes Nuclear Magnetic Resonance (NMR) is a spectrometric technique which provides information
WebEx Sharing Resources
 WebEx Sharing Resources OTS PUBLICATION: WX0 REVISED: 4/8/06 04 TOWSON UNIVERSITY OFFICE OF TECHNOLOGY SERVICES =Shortcut =Advice =Caution Introduction During a WebEx session, the host has the ability
WebEx Sharing Resources OTS PUBLICATION: WX0 REVISED: 4/8/06 04 TOWSON UNIVERSITY OFFICE OF TECHNOLOGY SERVICES =Shortcut =Advice =Caution Introduction During a WebEx session, the host has the ability
Measuring Line Edge Roughness: Fluctuations in Uncertainty
 Tutor6.doc: Version 5/6/08 T h e L i t h o g r a p h y E x p e r t (August 008) Measuring Line Edge Roughness: Fluctuations in Uncertainty Line edge roughness () is the deviation of a feature edge (as
Tutor6.doc: Version 5/6/08 T h e L i t h o g r a p h y E x p e r t (August 008) Measuring Line Edge Roughness: Fluctuations in Uncertainty Line edge roughness () is the deviation of a feature edge (as
Master course KEMM03 Principles of Mass Spectrometric Protein Characterization. Exam
 Exam Master course KEMM03 Principles of Mass Spectrometric Protein Characterization 2010-10-29 kl 08.15-13.00 Use a new paper for answering each question! Write your name on each paper! Aids: Mini calculator,
Exam Master course KEMM03 Principles of Mass Spectrometric Protein Characterization 2010-10-29 kl 08.15-13.00 Use a new paper for answering each question! Write your name on each paper! Aids: Mini calculator,
