Molecular Dynamics Simulations
|
|
|
- Stephen Horn
- 10 years ago
- Views:
Transcription
1 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH)
2 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular Dynamics Simulations IV. Applications 2
3 I. Introduction 3
4 Evolution of molecular simulations Pre-computer era Computer simulation 4
5 Molecular dynamics simulation Molecular systems Molecular dynamics simulations Properties of the molecular systems Carried out on computers 5
6 Molecular Dynamics simulation Virtual experiment at atomistic scale Direct observation and manipulation of atoms and molecules Statistical Mechanics MD & others Molecular Modeling Computer Simulation 6
7 Molecular dynamics simulation Carried out on computer(s) Inter-atomic interactions MD simulation program output How to describe them? How does an MD program work? What can be obtained? 7
8 II. Molecular Mechanical Force Fields 8
9 Molecular dynamics simulation Inter-atomic interactions MD simulation program output How to describe them? 9
10 Basic ideas If we want to study a protein, a piece of DNA, biological membranes, crystal lattice, nanomaterials, diffusion in liquids, the number of electrons become impossible to handle even with present-day computers. Instead, we replace the nuclei and electrons, and their interactions, by classical atoms and new potential functions. No cumbersome integrals to solve - Enables us to study very large systems ( atoms). 10
11 Molecular mechanics force fields Force fields use simplified functions (potential functions) to describe the interactions between atoms. Force fields are constructed by parameterising the potential functions using either experimental data (Xray and electron diffraction, NMR and IR spectroscopy) or ab initio and semi-empirical quantum mechanical calculations. Despite classical nature, force fields can mimic the behaviour of atomistic systems with an accuracy which approaches the high level of quantum mechanical calculations in a fraction of the time. 11
12 Force Field Function Form V V V V V V V FF bond angle torsion improper vdw elec V bonded V non-bonded Bonded terms bond stretching i j r angle bending i k j 12
13 Bonded terms proper torsional angle i j k l ϕ improper torsional angle i k j d l Non-Bonded terms van der Waals electrostatic q + q + q - 13
14 Widely Used Force Fields OPLS/AA Force Field Empirically fitted charges Electrically neutral subunits AMBER Force Field AMBER94, AMBER99, AMBER03 Charges from RESP (restrained electrostatic potential) fitting CHARMM Force Field Charges based on the solute-water complexes Urey-Bradley term accounting for 1-3 interaction 14
15 Water Models TIP3P 0.834e Å e e TIP4P Å +0.52e +0.52e 1.04e SPC/E e 1.0 Å e e 15
16 III. Molecular Dynamics Simulations 16
17 Molecular dynamics simulation Carried out on computer(s) Inter-atomic interactions MD simulation program output How does an MD program work? 17
18 Molecular dynamics simulation How does an MD program work? Inter-atomic interactions Force acting on each atom Acceleration on each atom Next time step New velocity and position Changes of velocity and position Molecular dynamics simulations simulate the collisions between atoms! 18
19 Equation of Motion and Integrator Newton s Equation of Motion F i m i a i Calculated from inter-atomic interactions Making the changes of velocity, position of atom i 19
20 Leap-frog Algorithm Velocity-Verlet algorithm 20
21 Periodic Boundary Condition Periodic boundary condition is used to simulate an infinite system Treatment of nonbonded Interaction Cut-off radius Minimum image convention Recovery of the electrostatic interactions beyond cut-off: Ewald summation & Particle mesh Ewald 21
22 Molecular dynamics simulation Inter-atomic interactions MD simulation program output What can be obtained? 22
23 What can be obtained from Molecular Dynamics simulation? Cooperative phenomena; Collective properties; MD simulations The effects from: temperature, pressure, solvents, intermolecular interactions, Structures: bulk materials, solutions, aggregates, DNAs, proteins,clusters, molecules, Dynamics properties: diffusion coefficients; relaxation times, 23
24 IV. Applications 24
25 1. Protein adsorption onto TiO 2 Background Titanium is a promising biocompatible material TiO 2 film exists on the titanium surface Water molecules are known to dissociate on TiO 2 surface Dental implant Question What is the effect of water dissociation on the biocompatibility of TiO 2? 25
26 Protein adsorption onto TiO 2 Snapshots on hydroxylated and non-hydroxylated surface (kj/mol) 26
27 Protein adsorption onto TiO 2 Vertical number densities of interfacial water and protein 27
28 Protein adsorption onto TiO 2 What do we obtain? Protein adsorption onto TiO 2 surface is strongly mediated by the interfacial water molecules. Protein affinity of the surface is enhanced by dissociated water molecules. Biocompatibility can be manipulated through appropriate surface modification. 28
29 2. O 2 -induced 19 F NMR Shift Background O 2 -induced 19 F NMR shift can be used to determine the immersion depth of transmembrane protein O 2 is inhomogeneously distributed in membrane 19 F NMR shift is sensitive to O 2 concentration 19 F labels can be conveniently introduced into cysteine residue of protein Question How does O 2 molecule reside around 19 F label and affect the 19 F NMR shift? Hydrophilic Hydrophobic Hydrophilic [O 2 ] 29
30 O 2 -induced 19 F NMR Shift Force fields Fluorinated cysteine General AMBER force field RESP partial atomic charges Water model TIP4P O 2 model Designed to reproduce the experimental quadrupole moment Zasetsky, A. Y., et al. (2001) Chem. Phys. Lett., Vol. 334, pp oxygen virtual oxygen site 30
31 O 2 -induced 19 F NMR Shift Shortest O-F distance: cutoff at 4.0 Å 7.6% Computational method Pennanen, T. O., et al. (2008) Phys. Rev. Lett., Vol. 100, p
32 O 2 -induced 19 F NMR Shift Results 32
33 O 2 -induced 19 F NMR Shift What do we obtain? The paramagnetic shift depends on both the shortest O-F distance and the corresponding F-O-O angle. Positive spin density downfield 19 F shifts; negative spin density upfield 19 F shifts. Downfield shift of 3.38 ± 0.60 ppm is comparable with the experimental data. 33
34 Thank you! 34
What is molecular dynamics (MD) simulation and how does it work?
 What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
1 The water molecule and hydrogen bonds in water
 The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
 Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Introduction Molecular Mechanics or force-field methods use classical type models
Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Introduction Molecular Mechanics or force-field methods use classical type models
NMR SPECTROSCOPY. Basic Principles, Concepts, and Applications in Chemistry. Harald Günther University of Siegen, Siegen, Germany.
 NMR SPECTROSCOPY Basic Principles, Concepts, and Applications in Chemistry Harald Günther University of Siegen, Siegen, Germany Second Edition Translated by Harald Günther JOHN WILEY & SONS Chichester
NMR SPECTROSCOPY Basic Principles, Concepts, and Applications in Chemistry Harald Günther University of Siegen, Siegen, Germany Second Edition Translated by Harald Günther JOHN WILEY & SONS Chichester
Applications of Quantum Chemistry HΨ = EΨ
 Applications of Quantum Chemistry HΨ = EΨ Areas of Application Explaining observed phenomena (e.g., spectroscopy) Simulation and modeling: make predictions New techniques/devices use special quantum properties
Applications of Quantum Chemistry HΨ = EΨ Areas of Application Explaining observed phenomena (e.g., spectroscopy) Simulation and modeling: make predictions New techniques/devices use special quantum properties
CHEM6085: Density Functional Theory Lecture 2. Hamiltonian operators for molecules
 CHEM6085: Density Functional Theory Lecture 2 Hamiltonian operators for molecules C.-K. Skylaris 1 The (time-independent) Schrödinger equation is an eigenvalue equation operator for property A eigenfunction
CHEM6085: Density Functional Theory Lecture 2 Hamiltonian operators for molecules C.-K. Skylaris 1 The (time-independent) Schrödinger equation is an eigenvalue equation operator for property A eigenfunction
Structure Determination
 5 Structure Determination Most of the protein structures described and discussed in this book have been determined either by X-ray crystallography or by nuclear magnetic resonance (NMR) spectroscopy. Although
5 Structure Determination Most of the protein structures described and discussed in this book have been determined either by X-ray crystallography or by nuclear magnetic resonance (NMR) spectroscopy. Although
Introduction to Molecular Dynamics Simulations
 Introduction to Molecular Dynamics Simulations Roland H. Stote Institut de Chimie LC3-UMR 7177 Université Louis Pasteur Strasbourg France 1EA5 Title Native Acetylcholinesterase (E.C. 3.1.1.7) From Torpedo
Introduction to Molecular Dynamics Simulations Roland H. Stote Institut de Chimie LC3-UMR 7177 Université Louis Pasteur Strasbourg France 1EA5 Title Native Acetylcholinesterase (E.C. 3.1.1.7) From Torpedo
2054-2. Structure and Dynamics of Hydrogen-Bonded Systems. 26-27 October 2009. Hydrogen Bonds and Liquid Water
 2054-2 Structure and Dynamics of Hydrogen-Bonded Systems 26-27 October 2009 Hydrogen Bonds and Liquid Water Ruth LYNDEN-BELL University of Cambridge, Department of Chemistry Lensfield Road Cambridge CB2
2054-2 Structure and Dynamics of Hydrogen-Bonded Systems 26-27 October 2009 Hydrogen Bonds and Liquid Water Ruth LYNDEN-BELL University of Cambridge, Department of Chemistry Lensfield Road Cambridge CB2
Molecular Docking. - Computational prediction of the structure of receptor-ligand complexes. Receptor: Protein Ligand: Protein or Small Molecule
 Scoring and Docking Molecular Docking - Computational prediction of the structure of receptor-ligand complexes Receptor: Protein Ligand: Protein or Small Molecule Protein-Protein Docking Protein-Small
Scoring and Docking Molecular Docking - Computational prediction of the structure of receptor-ligand complexes Receptor: Protein Ligand: Protein or Small Molecule Protein-Protein Docking Protein-Small
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005
 Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Plastics and Polymer Business. Properties enhancement for Plastics
 News Letter Vol. 18, issue October-December, 2012 Hyperdispersants and Coupling Agents for Thermoplastics and Thermosets Solplus, Ircolplus and Solsperse hyperdispersants and coupling agents have been
News Letter Vol. 18, issue October-December, 2012 Hyperdispersants and Coupling Agents for Thermoplastics and Thermosets Solplus, Ircolplus and Solsperse hyperdispersants and coupling agents have been
Universitätsstrasse 1, D-40225 Düsseldorf, Germany 3 Current address: Institut für Festkörperforschung,
 Lane formation in oppositely charged colloidal mixtures - supplementary information Teun Vissers 1, Adam Wysocki 2,3, Martin Rex 2, Hartmut Löwen 2, C. Patrick Royall 1,4, Arnout Imhof 1, and Alfons van
Lane formation in oppositely charged colloidal mixtures - supplementary information Teun Vissers 1, Adam Wysocki 2,3, Martin Rex 2, Hartmut Löwen 2, C. Patrick Royall 1,4, Arnout Imhof 1, and Alfons van
- particle with kinetic energy E strikes a barrier with height U 0 > E and width L. - classically the particle cannot overcome the barrier
 Tunnel Effect: - particle with kinetic energy E strikes a barrier with height U 0 > E and width L - classically the particle cannot overcome the barrier - quantum mechanically the particle can penetrated
Tunnel Effect: - particle with kinetic energy E strikes a barrier with height U 0 > E and width L - classically the particle cannot overcome the barrier - quantum mechanically the particle can penetrated
DISTANCE DEGREE PROGRAM CURRICULUM NOTE:
 Bachelor of Science in Electrical Engineering DISTANCE DEGREE PROGRAM CURRICULUM NOTE: Some Courses May Not Be Offered At A Distance Every Semester. Chem 121C General Chemistry I 3 Credits Online Fall
Bachelor of Science in Electrical Engineering DISTANCE DEGREE PROGRAM CURRICULUM NOTE: Some Courses May Not Be Offered At A Distance Every Semester. Chem 121C General Chemistry I 3 Credits Online Fall
Data Visualization for Atomistic/Molecular Simulations. Douglas E. Spearot University of Arkansas
 Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras
 Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O.
 Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O. Melnyk b a National University of life and environmental sciences
Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O. Melnyk b a National University of life and environmental sciences
10.7 Kinetic Molecular Theory. 10.7 Kinetic Molecular Theory. Kinetic Molecular Theory. Kinetic Molecular Theory. Kinetic Molecular Theory
 The first scheduled quiz will be given next Tuesday during Lecture. It will last 5 minutes. Bring pencil, calculator, and your book. The coverage will be pp 364-44, i.e. Sections 0.0 through.4. 0.7 Theory
The first scheduled quiz will be given next Tuesday during Lecture. It will last 5 minutes. Bring pencil, calculator, and your book. The coverage will be pp 364-44, i.e. Sections 0.0 through.4. 0.7 Theory
Section Activity #1: Fill out the following table for biology s most common elements assuming that each atom is neutrally charged.
 LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
Indiana's Academic Standards 2010 ICP Indiana's Academic Standards 2016 ICP. map) that describe the relationship acceleration, velocity and distance.
 .1.1 Measure the motion of objects to understand.1.1 Develop graphical, the relationships among distance, velocity and mathematical, and pictorial acceleration. Develop deeper understanding through representations
.1.1 Measure the motion of objects to understand.1.1 Develop graphical, the relationships among distance, velocity and mathematical, and pictorial acceleration. Develop deeper understanding through representations
Turbulence, Heat and Mass Transfer (THMT 09) Poiseuille flow of liquid methane in nanoscopic graphite channels by molecular dynamics simulation
 Turbulence, Heat and Mass Transfer (THMT 09) Poiseuille flow of liquid methane in nanoscopic graphite channels by molecular dynamics simulation Sapienza Università di Roma, September 14, 2009 M. T. HORSCH,
Turbulence, Heat and Mass Transfer (THMT 09) Poiseuille flow of liquid methane in nanoscopic graphite channels by molecular dynamics simulation Sapienza Università di Roma, September 14, 2009 M. T. HORSCH,
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.
 CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
Chapter 2 Polar Covalent Bonds; Acids and Bases
 John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
Biomaterials in tissue engineering
 Biomaterials in tissue engineering S. Swaminathan Director Centre for Nanotechnology & Advanced Biomaterials School of Chemical & Biotechnology SASTRA University Thanjavur 613 401 Tamil Nadu Page 1 of
Biomaterials in tissue engineering S. Swaminathan Director Centre for Nanotechnology & Advanced Biomaterials School of Chemical & Biotechnology SASTRA University Thanjavur 613 401 Tamil Nadu Page 1 of
Organic Chemistry Tenth Edition
 Organic Chemistry Tenth Edition T. W. Graham Solomons Craig B. Fryhle Welcome to CHM 22 Organic Chemisty II Chapters 2 (IR), 9, 3-20. Chapter 2 and Chapter 9 Spectroscopy (interaction of molecule with
Organic Chemistry Tenth Edition T. W. Graham Solomons Craig B. Fryhle Welcome to CHM 22 Organic Chemisty II Chapters 2 (IR), 9, 3-20. Chapter 2 and Chapter 9 Spectroscopy (interaction of molecule with
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING
 CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
Fluids Confined in Carbon Nanotubes
 Fluids Confined in Carbon Nanotubes Constantine M. Megaridis Micro/Nanoscale Fluid Transport Laboratory Mechanical and Industrial Engineering University of Illinois at Chicago 1 Background and Societal
Fluids Confined in Carbon Nanotubes Constantine M. Megaridis Micro/Nanoscale Fluid Transport Laboratory Mechanical and Industrial Engineering University of Illinois at Chicago 1 Background and Societal
BNG 331 Cell-Tissue Material Interactions. Biomaterial Surfaces
 BNG 331 Cell-Tissue Material Interactions Biomaterial Surfaces Course update Updated syllabus Homework 4 due today LBL 5 Friday Schedule for today: Chapter 8 Biomaterial surface characterization Surface
BNG 331 Cell-Tissue Material Interactions Biomaterial Surfaces Course update Updated syllabus Homework 4 due today LBL 5 Friday Schedule for today: Chapter 8 Biomaterial surface characterization Surface
Infrared Spectroscopy: Theory
 u Chapter 15 Infrared Spectroscopy: Theory An important tool of the organic chemist is Infrared Spectroscopy, or IR. IR spectra are acquired on a special instrument, called an IR spectrometer. IR is used
u Chapter 15 Infrared Spectroscopy: Theory An important tool of the organic chemist is Infrared Spectroscopy, or IR. IR spectra are acquired on a special instrument, called an IR spectrometer. IR is used
Surface Area and Porosity
 Surface Area and Porosity 1 Background Techniques Surface area Outline Total - physical adsorption External Porosity meso micro 2 Length 1 Å 1 nm 1 µm 1 1 1 1 1 mm macro meso micro metal crystallite 1-1
Surface Area and Porosity 1 Background Techniques Surface area Outline Total - physical adsorption External Porosity meso micro 2 Length 1 Å 1 nm 1 µm 1 1 1 1 1 mm macro meso micro metal crystallite 1-1
Boyle s law - For calculating changes in pressure or volume: P 1 V 1 = P 2 V 2. Charles law - For calculating temperature or volume changes: V 1 T 1
 Common Equations Used in Chemistry Equation for density: d= m v Converting F to C: C = ( F - 32) x 5 9 Converting C to F: F = C x 9 5 + 32 Converting C to K: K = ( C + 273.15) n x molar mass of element
Common Equations Used in Chemistry Equation for density: d= m v Converting F to C: C = ( F - 32) x 5 9 Converting C to F: F = C x 9 5 + 32 Converting C to K: K = ( C + 273.15) n x molar mass of element
4.5 Physical Properties: Solubility
 4.5 Physical Properties: Solubility When a solid, liquid or gaseous solute is placed in a solvent and it seems to disappear, mix or become part of the solvent, we say that it dissolved. The solute is said
4.5 Physical Properties: Solubility When a solid, liquid or gaseous solute is placed in a solvent and it seems to disappear, mix or become part of the solvent, we say that it dissolved. The solute is said
Towards Large-Scale Molecular Dynamics Simulations on Graphics Processors
 Towards Large-Scale Molecular Dynamics Simulations on Graphics Processors Joe Davis, Sandeep Patel, and Michela Taufer University of Delaware Outline Introduction Introduction to GPU programming Why MD
Towards Large-Scale Molecular Dynamics Simulations on Graphics Processors Joe Davis, Sandeep Patel, and Michela Taufer University of Delaware Outline Introduction Introduction to GPU programming Why MD
Name Class Date. In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question.
 Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Interpretation of density profiles and pair correlation functions
 Interpretation of density profiles and pair correlation functions Martin Horsch and Cemal Engin Paderborn, 22 nd June 12 IV. Annual Meeting of the Boltzmann Zuse Society Surface tension of nanodroplets
Interpretation of density profiles and pair correlation functions Martin Horsch and Cemal Engin Paderborn, 22 nd June 12 IV. Annual Meeting of the Boltzmann Zuse Society Surface tension of nanodroplets
Physics 9e/Cutnell. correlated to the. College Board AP Physics 1 Course Objectives
 Physics 9e/Cutnell correlated to the College Board AP Physics 1 Course Objectives Big Idea 1: Objects and systems have properties such as mass and charge. Systems may have internal structure. Enduring
Physics 9e/Cutnell correlated to the College Board AP Physics 1 Course Objectives Big Idea 1: Objects and systems have properties such as mass and charge. Systems may have internal structure. Enduring
CHAPTER 6 Chemical Bonding
 CHAPTER 6 Chemical Bonding SECTION 1 Introduction to Chemical Bonding OBJECTIVES 1. Define Chemical bond. 2. Explain why most atoms form chemical bonds. 3. Describe ionic and covalent bonding.. 4. Explain
CHAPTER 6 Chemical Bonding SECTION 1 Introduction to Chemical Bonding OBJECTIVES 1. Define Chemical bond. 2. Explain why most atoms form chemical bonds. 3. Describe ionic and covalent bonding.. 4. Explain
INSPIRE GK12 Lesson Plan. The Chemistry of Climate Change Length of Lesson
 Lesson Title The Chemistry of Climate Change Length of Lesson 180 min Created By David Wilson Subject Physical Science / Chemistry / Organic Chemistry Grade Level 8-12 State Standards 2c, 4d / 2a, 4d /
Lesson Title The Chemistry of Climate Change Length of Lesson 180 min Created By David Wilson Subject Physical Science / Chemistry / Organic Chemistry Grade Level 8-12 State Standards 2c, 4d / 2a, 4d /
Molecular-Orbital Theory
 Molecular-Orbital Theory 1 Introduction Orbitals in molecules are not necessarily localized on atoms or between atoms as suggested in the valence bond theory. Molecular orbitals can also be formed the
Molecular-Orbital Theory 1 Introduction Orbitals in molecules are not necessarily localized on atoms or between atoms as suggested in the valence bond theory. Molecular orbitals can also be formed the
Lecture 24 - Surface tension, viscous flow, thermodynamics
 Lecture 24 - Surface tension, viscous flow, thermodynamics Surface tension, surface energy The atoms at the surface of a solid or liquid are not happy. Their bonding is less ideal than the bonding of atoms
Lecture 24 - Surface tension, viscous flow, thermodynamics Surface tension, surface energy The atoms at the surface of a solid or liquid are not happy. Their bonding is less ideal than the bonding of atoms
SCIENCE. Introducing updated Cambridge International AS & A Level syllabuses for. Biology 9700 Chemistry 9701 Physics 9702
 Introducing updated Cambridge International AS & A Level syllabuses for SCIENCE Biology 9700 Chemistry 9701 Physics 9702 The revised Cambridge International AS & A Level Biology, Chemistry and Physics
Introducing updated Cambridge International AS & A Level syllabuses for SCIENCE Biology 9700 Chemistry 9701 Physics 9702 The revised Cambridge International AS & A Level Biology, Chemistry and Physics
HYDRONMR and Fast-HYDRONMR
 File:hydronmr7c-pub.doc HYDRONMR and Fast-HYDRONMR Index 1. Introduction to HYDRONMR 2. Literature 3. Running HYDRONMR. Input data file 4. Output files 5. Notes and hints 7. Release notes Version 7c, September
File:hydronmr7c-pub.doc HYDRONMR and Fast-HYDRONMR Index 1. Introduction to HYDRONMR 2. Literature 3. Running HYDRONMR. Input data file 4. Output files 5. Notes and hints 7. Release notes Version 7c, September
Chapter 2. Atomic Structure and Interatomic Bonding
 Chapter 2. Atomic Structure and Interatomic Bonding Interatomic Bonding Bonding forces and energies Primary interatomic bonds Secondary bonding Molecules Bonding Forces and Energies Considering the interaction
Chapter 2. Atomic Structure and Interatomic Bonding Interatomic Bonding Bonding forces and energies Primary interatomic bonds Secondary bonding Molecules Bonding Forces and Energies Considering the interaction
DO PHYSICS ONLINE FROM QUANTA TO QUARKS QUANTUM (WAVE) MECHANICS
 DO PHYSICS ONLINE FROM QUANTA TO QUARKS QUANTUM (WAVE) MECHANICS Quantum Mechanics or wave mechanics is the best mathematical theory used today to describe and predict the behaviour of particles and waves.
DO PHYSICS ONLINE FROM QUANTA TO QUARKS QUANTUM (WAVE) MECHANICS Quantum Mechanics or wave mechanics is the best mathematical theory used today to describe and predict the behaviour of particles and waves.
Adsorption at Surfaces
 Adsorption at Surfaces Adsorption is the accumulation of particles (adsorbate) at a surface (adsorbent or substrate). The reverse process is called desorption. fractional surface coverage: θ = Number of
Adsorption at Surfaces Adsorption is the accumulation of particles (adsorbate) at a surface (adsorbent or substrate). The reverse process is called desorption. fractional surface coverage: θ = Number of
PCV Project: Excitons in Molecular Spectroscopy
 PCV Project: Excitons in Molecular Spectroscopy Introduction The concept of excitons was first introduced by Frenkel (1) in 1931 as a general excitation delocalization mechanism to account for the ability
PCV Project: Excitons in Molecular Spectroscopy Introduction The concept of excitons was first introduced by Frenkel (1) in 1931 as a general excitation delocalization mechanism to account for the ability
Size effects. Lecture 6 OUTLINE
 Size effects 1 MTX9100 Nanomaterials Lecture 6 OUTLINE -Why does size influence the material s properties? -How does size influence the material s performance? -Why are properties of nanoscale objects
Size effects 1 MTX9100 Nanomaterials Lecture 6 OUTLINE -Why does size influence the material s properties? -How does size influence the material s performance? -Why are properties of nanoscale objects
Structural Bioinformatics (C3210) Experimental Methods for Macromolecular Structure Determination
 Structural Bioinformatics (C3210) Experimental Methods for Macromolecular Structure Determination Introduction Knowing the exact 3D-structure of bio-molecules is essential for any attempt to understand
Structural Bioinformatics (C3210) Experimental Methods for Macromolecular Structure Determination Introduction Knowing the exact 3D-structure of bio-molecules is essential for any attempt to understand
Study the following diagrams of the States of Matter. Label the names of the Changes of State between the different states.
 Describe the strength of attractive forces between particles. Describe the amount of space between particles. Can the particles in this state be compressed? Do the particles in this state have a definite
Describe the strength of attractive forces between particles. Describe the amount of space between particles. Can the particles in this state be compressed? Do the particles in this state have a definite
4. It is possible to excite, or flip the nuclear magnetic vector from the α-state to the β-state by bridging the energy gap between the two. This is a
 BASIC PRINCIPLES INTRODUCTION TO NUCLEAR MAGNETIC RESONANCE (NMR) 1. The nuclei of certain atoms with odd atomic number, and/or odd mass behave as spinning charges. The nucleus is the center of positive
BASIC PRINCIPLES INTRODUCTION TO NUCLEAR MAGNETIC RESONANCE (NMR) 1. The nuclei of certain atoms with odd atomic number, and/or odd mass behave as spinning charges. The nucleus is the center of positive
Chemical Bonds and Groups - Part 1
 hemical Bonds and Groups - Part 1 ARB SKELETS arbon has a unique role in the cell because of its ability to form strong covalent bonds with other carbon atoms. Thus carbon atoms can join to form chains.
hemical Bonds and Groups - Part 1 ARB SKELETS arbon has a unique role in the cell because of its ability to form strong covalent bonds with other carbon atoms. Thus carbon atoms can join to form chains.
Basic principles and mechanisms of NSOM; Different scanning modes and systems of NSOM; General applications and advantages of NSOM.
 Lecture 16: Near-field Scanning Optical Microscopy (NSOM) Background of NSOM; Basic principles and mechanisms of NSOM; Basic components of a NSOM; Different scanning modes and systems of NSOM; General
Lecture 16: Near-field Scanning Optical Microscopy (NSOM) Background of NSOM; Basic principles and mechanisms of NSOM; Basic components of a NSOM; Different scanning modes and systems of NSOM; General
Proton Nuclear Magnetic Resonance ( 1 H-NMR) Spectroscopy
 Proton Nuclear Magnetic Resonance ( 1 H-NMR) Spectroscopy Theory behind NMR: In the late 1940 s, physical chemists originally developed NMR spectroscopy to study different properties of atomic nuclei,
Proton Nuclear Magnetic Resonance ( 1 H-NMR) Spectroscopy Theory behind NMR: In the late 1940 s, physical chemists originally developed NMR spectroscopy to study different properties of atomic nuclei,
Section 3: Crystal Binding
 Physics 97 Interatomic forces Section 3: rystal Binding Solids are stable structures, and therefore there exist interactions holding atoms in a crystal together. For example a crystal of sodium chloride
Physics 97 Interatomic forces Section 3: rystal Binding Solids are stable structures, and therefore there exist interactions holding atoms in a crystal together. For example a crystal of sodium chloride
The Hofmeister series
 The Hofmeister series a review of monatomic ions near the hydrophobic/water interface Student: Ernst Rösler Student ID: 0105813 Course: Literature Study MSc Chemistry, track Theoretical and Computational
The Hofmeister series a review of monatomic ions near the hydrophobic/water interface Student: Ernst Rösler Student ID: 0105813 Course: Literature Study MSc Chemistry, track Theoretical and Computational
Non-Covalent Bonds (Weak Bond)
 Non-Covalent Bonds (Weak Bond) Weak bonds are those forces of attraction that, in biological situations, do not take a large amount of energy to break. For example, hydrogen bonds are broken by energies
Non-Covalent Bonds (Weak Bond) Weak bonds are those forces of attraction that, in biological situations, do not take a large amount of energy to break. For example, hydrogen bonds are broken by energies
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Free Electron Fermi Gas (Kittel Ch. 6)
 Free Electron Fermi Gas (Kittel Ch. 6) Role of Electrons in Solids Electrons are responsible for binding of crystals -- they are the glue that hold the nuclei together Types of binding (see next slide)
Free Electron Fermi Gas (Kittel Ch. 6) Role of Electrons in Solids Electrons are responsible for binding of crystals -- they are the glue that hold the nuclei together Types of binding (see next slide)
PARTICLE SIMULATION ON MULTIPLE DUST LAYERS OF COULOMB CLOUD IN CATHODE SHEATH EDGE
 PARTICLE SIMULATION ON MULTIPLE DUST LAYERS OF COULOMB CLOUD IN CATHODE SHEATH EDGE K. ASANO, S. NUNOMURA, T. MISAWA, N. OHNO and S. TAKAMURA Department of Energy Engineering and Science, Graduate School
PARTICLE SIMULATION ON MULTIPLE DUST LAYERS OF COULOMB CLOUD IN CATHODE SHEATH EDGE K. ASANO, S. NUNOMURA, T. MISAWA, N. OHNO and S. TAKAMURA Department of Energy Engineering and Science, Graduate School
Introduction, Noncovalent Bonds, and Properties of Water
 Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
NMR and IR spectra & vibrational analysis
 Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Lab 5: NMR and IR spectra & vibrational analysis A brief theoretical background 1 Some of the available chemical quantum methods for calculating NMR chemical shifts are based on the Hartree-Fock self-consistent
Sputtering by Particle Bombardment I
 Sputtering by Particle Bombardment I Physical Sputtering of Single-Element Solids Edited by R. Behrisch With Contributions by H. H. Andersen H. L. Bay R. Behrisch M. T. Robinson H. E. Roosendaal P. Sigmund
Sputtering by Particle Bombardment I Physical Sputtering of Single-Element Solids Edited by R. Behrisch With Contributions by H. H. Andersen H. L. Bay R. Behrisch M. T. Robinson H. E. Roosendaal P. Sigmund
Photoinduced volume change in chalcogenide glasses
 Photoinduced volume change in chalcogenide glasses (Ph.D. thesis points) Rozália Lukács Budapest University of Technology and Economics Department of Theoretical Physics Supervisor: Dr. Sándor Kugler 2010
Photoinduced volume change in chalcogenide glasses (Ph.D. thesis points) Rozália Lukács Budapest University of Technology and Economics Department of Theoretical Physics Supervisor: Dr. Sándor Kugler 2010
Used to determine relative location of atoms within a molecule Most helpful spectroscopic technique in organic chemistry Related to MRI in medicine
 Structure Determination: Nuclear Magnetic Resonance CHEM 241 UNIT 5C 1 The Use of NMR Spectroscopy Used to determine relative location of atoms within a molecule Most helpful spectroscopic technique in
Structure Determination: Nuclear Magnetic Resonance CHEM 241 UNIT 5C 1 The Use of NMR Spectroscopy Used to determine relative location of atoms within a molecule Most helpful spectroscopic technique in
Incorporating Internal Gradient and Restricted Diffusion Effects in Nuclear Magnetic Resonance Log Interpretation
 The Open-Access Journal for the Basic Principles of Diffusion Theory, Experiment and Application Incorporating Internal Gradient and Restricted Diffusion Effects in Nuclear Magnetic Resonance Log Interpretation
The Open-Access Journal for the Basic Principles of Diffusion Theory, Experiment and Application Incorporating Internal Gradient and Restricted Diffusion Effects in Nuclear Magnetic Resonance Log Interpretation
Proton Nuclear Magnetic Resonance Spectroscopy
 Proton Nuclear Magnetic Resonance Spectroscopy Introduction: The NMR Spectrum serves as a great resource in determining the structure of an organic compound by revealing the hydrogen and carbon skeleton.
Proton Nuclear Magnetic Resonance Spectroscopy Introduction: The NMR Spectrum serves as a great resource in determining the structure of an organic compound by revealing the hydrogen and carbon skeleton.
The strength of the interaction
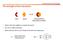 The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
The Four Questions to Ask While Interpreting Spectra. 1. How many different environments are there?
 1 H NMR Spectroscopy (#1c) The technique of 1 H NMR spectroscopy is central to organic chemistry and other fields involving analysis of organic chemicals, such as forensics and environmental science. It
1 H NMR Spectroscopy (#1c) The technique of 1 H NMR spectroscopy is central to organic chemistry and other fields involving analysis of organic chemicals, such as forensics and environmental science. It
Shielding vs. Deshielding:
 Shielding vs. Deshielding: Pre-tutorial: Things we need to know before we start the topic: What does the NMR Chemical shift do? The chemical shift is telling us the strength of the magnetic field that
Shielding vs. Deshielding: Pre-tutorial: Things we need to know before we start the topic: What does the NMR Chemical shift do? The chemical shift is telling us the strength of the magnetic field that
NMR for Physical and Biological Scientists Thomas C. Pochapsky and Susan Sondej Pochapsky Table of Contents
 Preface Symbols and fundamental constants 1. What is spectroscopy? A semiclassical description of spectroscopy Damped harmonics Quantum oscillators The spectroscopic experiment Ensembles and coherence
Preface Symbols and fundamental constants 1. What is spectroscopy? A semiclassical description of spectroscopy Damped harmonics Quantum oscillators The spectroscopic experiment Ensembles and coherence
C o m p u te r M o d e lin g o f M o le c u la r E le c tro n ic S tru c tu re
 C o m p u te r M o d e lin g o f M o le c u la r E le c tro n ic S tru c tu re P e te r P u la y D e p a rtm e n t o f C h e m is try a n d B io c h e m is try, U n iv e rs ity o f A rk a n s a s, F a
C o m p u te r M o d e lin g o f M o le c u la r E le c tro n ic S tru c tu re P e te r P u la y D e p a rtm e n t o f C h e m is try a n d B io c h e m is try, U n iv e rs ity o f A rk a n s a s, F a
Comparative crystal structure determination of griseofulvin: Powder X-ray diffraction versus single-crystal X-ray diffraction
 Article Analytical Chemistry October 2012 Vol.57 No.30: 3867 3871 doi: 10.1007/s11434-012-5245-5 SPECIAL TOPICS: Comparative crystal structure determination of griseofulvin: Powder X-ray diffraction versus
Article Analytical Chemistry October 2012 Vol.57 No.30: 3867 3871 doi: 10.1007/s11434-012-5245-5 SPECIAL TOPICS: Comparative crystal structure determination of griseofulvin: Powder X-ray diffraction versus
Use the Force! Noncovalent Molecular Forces
 Use the Force! Noncovalent Molecular Forces Not quite the type of Force we re talking about Before we talk about noncovalent molecular forces, let s talk very briefly about covalent bonds. The Illustrated
Use the Force! Noncovalent Molecular Forces Not quite the type of Force we re talking about Before we talk about noncovalent molecular forces, let s talk very briefly about covalent bonds. The Illustrated
Laboratorio Regionale LiCryL CNR-INFM
 Laboratorio Regionale LiCryL CNR-INFM c/o Physics Department - University of Calabria, Ponte P. Bucci, Cubo 33B, 87036 Rende (CS) Italy UNIVERSITÀ DELLA CALABRIA Dipartimento di FISICA Researchers Dr.
Laboratorio Regionale LiCryL CNR-INFM c/o Physics Department - University of Calabria, Ponte P. Bucci, Cubo 33B, 87036 Rende (CS) Italy UNIVERSITÀ DELLA CALABRIA Dipartimento di FISICA Researchers Dr.
Viscoelasticity of Polymer Fluids.
 Viscoelasticity of Polymer Fluids. Main Properties of Polymer Fluids. Entangled polymer fluids are polymer melts and concentrated or semidilute (above the concentration c) solutions. In these systems polymer
Viscoelasticity of Polymer Fluids. Main Properties of Polymer Fluids. Entangled polymer fluids are polymer melts and concentrated or semidilute (above the concentration c) solutions. In these systems polymer
Chapter 8 Maxwell relations and measurable properties
 Chapter 8 Maxwell relations and measurable properties 8.1 Maxwell relations Other thermodynamic potentials emerging from Legendre transforms allow us to switch independent variables and give rise to alternate
Chapter 8 Maxwell relations and measurable properties 8.1 Maxwell relations Other thermodynamic potentials emerging from Legendre transforms allow us to switch independent variables and give rise to alternate
Overview of NAMD and Molecular Dynamics
 Overview of NAMD and Molecular Dynamics Jim Phillips Low-cost Linux Clusters for Biomolecular Simulations Using NAMD Outline Overview of molecular dynamics Overview of NAMD NAMD parallel design NAMD config
Overview of NAMD and Molecular Dynamics Jim Phillips Low-cost Linux Clusters for Biomolecular Simulations Using NAMD Outline Overview of molecular dynamics Overview of NAMD NAMD parallel design NAMD config
What is a weak hydrogen bond?
 What is a weak hydrogen bond? Gautam R. Desiraju School of Chemistry University of yderabad yderabad 500 046, India [email protected] http://202.41.85.161/~grd/ What is a hydrogen bond? Under certain
What is a weak hydrogen bond? Gautam R. Desiraju School of Chemistry University of yderabad yderabad 500 046, India [email protected] http://202.41.85.161/~grd/ What is a hydrogen bond? Under certain
Topic 2: Energy in Biological Systems
 Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Chapter Outline. Diffusion - how do atoms move through solids?
 Chapter Outline iffusion - how do atoms move through solids? iffusion mechanisms Vacancy diffusion Interstitial diffusion Impurities The mathematics of diffusion Steady-state diffusion (Fick s first law)
Chapter Outline iffusion - how do atoms move through solids? iffusion mechanisms Vacancy diffusion Interstitial diffusion Impurities The mathematics of diffusion Steady-state diffusion (Fick s first law)
Chapter 8 Concepts of Chemical Bonding
 Chapter 8 Concepts of Chemical Bonding Chemical Bonds Three types: Ionic Electrostatic attraction between ions Covalent Sharing of electrons Metallic Metal atoms bonded to several other atoms Ionic Bonding
Chapter 8 Concepts of Chemical Bonding Chemical Bonds Three types: Ionic Electrostatic attraction between ions Covalent Sharing of electrons Metallic Metal atoms bonded to several other atoms Ionic Bonding
THE DEVELOPMENT LINES OF PROGEO
 THE DEVELOPMENT LINES OF PROGEO ANTONIO LAGANA, University of Perugia ANDREA CAPRICCIOLI, ENEA Frascati PREAMBLE The ICSA (Innovative Computational Science Applications) Association has started several
THE DEVELOPMENT LINES OF PROGEO ANTONIO LAGANA, University of Perugia ANDREA CAPRICCIOLI, ENEA Frascati PREAMBLE The ICSA (Innovative Computational Science Applications) Association has started several
New magnetism of 3d monolayers grown with oxygen surfactant: Experiment vs. ab initio calculations
 New magnetism of 3d monolayers grown with oxygen surfactant: Experiment vs. ab initio calculations 1. Growth and structure 2. Magnetism and MAE 3. Induced magnetism at oxygen Klaus Baberschke Institut
New magnetism of 3d monolayers grown with oxygen surfactant: Experiment vs. ab initio calculations 1. Growth and structure 2. Magnetism and MAE 3. Induced magnetism at oxygen Klaus Baberschke Institut
CFD SIMULATION OF SDHW STORAGE TANK WITH AND WITHOUT HEATER
 International Journal of Advancements in Research & Technology, Volume 1, Issue2, July-2012 1 CFD SIMULATION OF SDHW STORAGE TANK WITH AND WITHOUT HEATER ABSTRACT (1) Mr. Mainak Bhaumik M.E. (Thermal Engg.)
International Journal of Advancements in Research & Technology, Volume 1, Issue2, July-2012 1 CFD SIMULATION OF SDHW STORAGE TANK WITH AND WITHOUT HEATER ABSTRACT (1) Mr. Mainak Bhaumik M.E. (Thermal Engg.)
WJP, PHY381 (2015) Wabash Journal of Physics v4.3, p.1. Cloud Chamber. R.C. Dennis, Tuan Le, M.J. Madsen, and J. Brown
 WJP, PHY381 (2015) Wabash Journal of Physics v4.3, p.1 Cloud Chamber R.C. Dennis, Tuan Le, M.J. Madsen, and J. Brown Department of Physics, Wabash College, Crawfordsville, IN 47933 (Dated: May 7, 2015)
WJP, PHY381 (2015) Wabash Journal of Physics v4.3, p.1 Cloud Chamber R.C. Dennis, Tuan Le, M.J. Madsen, and J. Brown Department of Physics, Wabash College, Crawfordsville, IN 47933 (Dated: May 7, 2015)
Why? Intermolecular Forces. Intermolecular Forces. Chapter 12 IM Forces and Liquids. Covalent Bonding Forces for Comparison of Magnitude
 1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
Numerical Model for the Study of the Velocity Dependence Of the Ionisation Growth in Gas Discharge Plasma
 Journal of Basrah Researches ((Sciences)) Volume 37.Number 5.A ((2011)) Available online at: www.basra-science -journal.org ISSN 1817 2695 Numerical Model for the Study of the Velocity Dependence Of the
Journal of Basrah Researches ((Sciences)) Volume 37.Number 5.A ((2011)) Available online at: www.basra-science -journal.org ISSN 1817 2695 Numerical Model for the Study of the Velocity Dependence Of the
Lesson 3. Chemical Bonding. Molecular Orbital Theory
 Lesson 3 Chemical Bonding Molecular Orbital Theory 1 Why Do Bonds Form? An energy diagram shows that a bond forms between two atoms if the overall energy of the system is lowered when the two atoms approach
Lesson 3 Chemical Bonding Molecular Orbital Theory 1 Why Do Bonds Form? An energy diagram shows that a bond forms between two atoms if the overall energy of the system is lowered when the two atoms approach
ENHANCEMENT OF NATURAL CONVECTION HEAT TRANSFER BY THE EFFECT OF HIGH VOLTAGE D.C. ELECTRIC FIELD
 Int. J. Mech. Eng. & Rob. Res. 014 Amit Kumar and Ritesh Kumar, 014 Research Paper ISSN 78 0149 www.ijmerr.com Vol. 3, No. 1, January 014 014 IJMERR. All Rights Reserved ENHANCEMENT OF NATURAL CONVECTION
Int. J. Mech. Eng. & Rob. Res. 014 Amit Kumar and Ritesh Kumar, 014 Research Paper ISSN 78 0149 www.ijmerr.com Vol. 3, No. 1, January 014 014 IJMERR. All Rights Reserved ENHANCEMENT OF NATURAL CONVECTION
Surface Tension: Liquids Stick Together Teacher Version
 Surface Tension: Liquids Stick Together Teacher Version In this lab you will learn about properties of liquids, specifically cohesion, adhesion, and surface tension. These principles will be demonstrated
Surface Tension: Liquids Stick Together Teacher Version In this lab you will learn about properties of liquids, specifically cohesion, adhesion, and surface tension. These principles will be demonstrated
JOURNAL INTEGRATED CIRCUITS AND SYSTEMS, VOL 1, NO. 3, JULY 2006. 39
 JOURNAL INTEGRATED CIRCUITS AND SYSTEMS, VOL 1, NO. 3, JULY 2006. 39 Self-Assembled Polystyrene Micro-Spheres Applied for Photonic Crystals and Templates Fabrication Daniel S. Raimundo 1, Francisco J.
JOURNAL INTEGRATED CIRCUITS AND SYSTEMS, VOL 1, NO. 3, JULY 2006. 39 Self-Assembled Polystyrene Micro-Spheres Applied for Photonic Crystals and Templates Fabrication Daniel S. Raimundo 1, Francisco J.
Extension of COSMO-RS to interfacial phenomena
 Extension of COSMO-RS to interfacial phenomena Martin P. Andersson et al. Nano-Science Center Department of Chemistry University of Copenhagen Denmark Presentation at the 4th COSMO-RS symposium Mar 2015
Extension of COSMO-RS to interfacial phenomena Martin P. Andersson et al. Nano-Science Center Department of Chemistry University of Copenhagen Denmark Presentation at the 4th COSMO-RS symposium Mar 2015
