The Application of Distributed Computing to the Investigation of Protein Conformational Change
|
|
|
- Julius Robertson
- 8 years ago
- Views:
Transcription
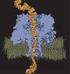
1 The Application of Distributed Computing to the Investigation of Protein Conformational Change C. J. Woods, J. G. Frey, J. W. Essex School of Chemistry, University of Southampton, SO17 1BJ, UK Abstract Distributed computing is a potentially very powerful approach for accessing large amounts of computational power. Under the umbrella of the comb-e-chem project we have examined distributed computing software and applied it to the problem of investigating protein conformational change. These investigations required the development of protein simulations that were suited to distributed computing. Each simulation was split into many coupled, parallel parts. These proved challenging to schedule on the flexible and unreliable distributed computing resource. Scheduling algorithms were thus written that identified which parts of the simulation were likely to impact the overall efficiency. These parts were then rescheduled to be caught-up via a fast and dedicated cluster. 1. Background Distributed computing is a potentially powerful approach for accessing large amounts of computational power. Cycle stealers, which allow a PC user to donate the spare power of their computer, are now used in a wide range of scientific projects, e.g. the SETI@home study, 1 the CAN-DDO cancer screening project 2 and folding@home. 3 While the use of cycle stealers can provide supercomputer-like resources, their use is limited to calculations that may be split into many independently parallel parts (i.e. coarsely parallel simulations). The distributed and unreliable nature of this resource makes it unsuitable for closely coupled parallel calculations. For these calculations, the speed and latency of inter-processor communication are a bottleneck that cannot be overcome simply through the addition of more nodes. Unfortunately, a large number of chemical simulations require closely coupled parallel calculations, and are thus not suitable for deployment over a distributed computing cluster. An example of such a simulation is the investigation of protein conformational change. These simulations are typically performed using molecular dynamics (MD), 4 where the motions of the atoms are integrated over time using Newton s laws. These simulations cannot be broken up into multiple independent parts, as each nanosecond of MD must be run in series and in sequence. The investigation of protein conformational change is important as it lies at the heart of many biological processes, e.g. cell signaling. Some bacteria regulate nitrogen metabolism Figure 1. The native (blue) and phosphorylated (red) conformations of NTRC. The site of phosphorylation (Asp54) is shown as spheres. using one such signalling pathway. Nitrogen Regulatory Protein C (NTRC) 5 plays a key role in this pathway. Changes in nitrogen concentration activate the kinase NTRB. This phosphorylates an aspartate residue in NTRC, causing it to change conformation (figure 1). 5 This change in conformation allows the NTRC to join together to form oligomers, which then help promote the transcription of genes. These genes are used to produce proteins that are used in nitrogen metabolism. 5 A key stage of this pathway is the change in conformation that occurs in NTRC when it is phosphorylated. It is difficult to study this conformational change experimentally as the phosphorylated form of NTRC has a very short lifetime. 6 It is thus desirable to simulate the NTRC protein and
2 Figure 2. Progress of the simulation at each temperature as a function of simulation time. Iterations are run at each temperature; odd iterations are shown in blue and even iterations are shown in green. Our scheduler has to cope with extreme events, e.g. the complete failure of the distributed cluster after about 200 and 270 hours of simulation. The distributed cluster contains both fast and slow nodes. Some iterations can thus take a lot longer than others (visible here as longer bars). In addition, the owners of the PCs will also wish to use them (shown here as red dots). This will interrupt the calculation that temperature, again slowing it down relative to the other temperatures. Because of this, neighbouring temperatures will be ready to test at different times. This can lead to a loss of efficiency as completed temperatures wait for their neighbours. In the worst case this waiting can propagate, as occurs for temperatures around 310 K after 360 hours of simulation. To help prevent this, a catch-up cluster is used that identifies and reschedules slow temperatures (use of the catch-up cluster is shown in yellow). encourage the conformational change on computer. 1.1 The Replica Exchange Method We can use a distributed computing cluster to investigate protein conformational change via Replica Exchange simulations. 7,8 Multiple simulations of the protein are run in parallel, each running under a different condition, e.g. temperature. Periodically the simulations running at neighbouring temperatures are tested and swapped. This enables simulations at high temperatures, where there is rapid conformational change, to rain down to biologically relevant temperatures where conformational change occurs more slowly. The testing of neighbouring temperatures introduces a light coupling to the simulation, meaning that it no longer fits the archetypal coarsely parallel distributed computing model. This light coupling introduces inefficiencies to the scheduling of the simulation, as any delay in the calculation of one temperature can propagate out to delay the calculation of all temperatures. To help overcome this, a catch-up cluster has been developed that monitors the simulation for temperatures that are taking too long to complete, and that are likely to negatively impact the overall efficiency of the simulation. Once identified, the calculation of these temperatures is rescheduled onto a small, yet fast and dedicated, computational resource so that they can catch-up with the other temperatures (figure 2). 2. Experimental Details NMR structures of the phosphorylated (1DC8) and unphosphorylated (1DC7) conformations of the NTRC protein were obtained from the protein databank. 9 Polar hydrogen atoms were added via WhatIf 10. The proteins were solvated in 60 3 Å 3 boxes of TIP3P water and sodium ions were added via the XLEAP module of AMBER to neutralise the system. The CHARMM27 forcefield 12 was used, and the systems minimised, then annealed from 100 K to 300 K.
3 Figure 3. The number of nodes in use for the replica exchange simulation over just the distributed cluster (left), and over the distributed and catchup clusters (right). The systems were finally equilibrated for 100 ps at constant temperature (300 K) and pressure (1 atm). The final structures from equilibration were used as the starting structures for all of the replicas. The temperatures for each replica were chosen using a custom program that optimised the temperature distribution such that a replica exchange move was accepted with a probability of 20%. The temperatures chosen ranged from several replicas below the target temperature of 300 K (290.1 K) to a maximum of 400 K. In total 64 replicas were used for each of the two proteins. The simulations were conducted using NAMD A replica exchange move was attempted between neighbouring temperatures every 2 ps, after the initial 20 ps of sampling that was used to equilibrate each replica to its initial temperature. A Langevin thermostat 14 and a Nose-Hoover Langevin piston barostat 15 were used to sample at constant temperature and pressure, while SHAKE 16 was used to constrain hydrogen bond lengths to equilibrium values. A 1 fs time step was used for the molecular dynamics integrator, and the non-bonded interactions were evaluated using a 12 Å cutoff and the particle mesh ewald sum Details of the Distributed Cluster The simulations were run over the condor 18 cluster provided by the University of Southampton. This cluster uses condor 18 to make available the spare cycles of approximately 450 desktop computers running Microsoft Windows NT 5.1. The two replica exchange simulations were run simultaneously on this cluster. There was little competition between the two simulations for nodes as they each required a maximum of 64 nodes out of the available Implementation of the Catchup Cluster The catchup cluster was implemented via dedicated dual Xeon 2.8 GHz nodes running Linux. Each Xeon processor was able to provide 2 virtual processors, allowing NAMD to run in parallel over 4 virtual processors per node. This meant that each dual Xeon could complete 2 ps of MD approximately 3.5 times quicker than a typical node on the condor cluster. The catchup cluster consisted of two dual Xeon nodes, thus allowing it to catch up two replicas simultaneously. To test the utility of the catchup cluster, it was only made available to the replica exchange simulation on the phosphorylated conformation of the protein (1DC8). As both replica exchange simulations were running simultaneously, any differences in efficiency should thus be wholly attributable to use of the catchup cluster. 3. Results Figure 3 shows the number of nodes in use during the replica exchange simulations on the phosphorylated and unphosphorylated conformations of NTRC. The initial phase of the simulation involved the 20 ps of equilibration of each replica to its initial temperature. This was broken down into 10 iterations of 2 ps. As there were no replica exchange moves during these first 10 iterations the replicas were all independent and thus the maximum number of 64 nodes were in use. However, there were efficiency problems during this phase of the simulation, as the unreliable nature of the distributed cluster caused several short periods of downtime that stopped both simulations. Frequent periods of downtime were common throughout the rest of replica exchange simulations.
4 The second stage of the simulations occurred when the replicas begun to complete their 10 th iteration. At this point each replica had to wait for its partner to complete 10 iterations such that the pair of replicas could be tested and potentially swapped. Owing to the range of processors available in the distributed cluster and the different impacts of downtime on each of the replicas, there was a large spread of times over which each replica completed 10 iterations. This meant that a large number of replicas were left waiting for a significant time for their partner to complete, and thus the number of nodes in use for each simulation dropped from the maximum of 64 down to approximately 30. If the efficiency is defined as the ratio of the number of nodes in use compared to the theoretical maximum, then the efficiency dropped from 100% down to about 47%. After this dip in efficiency, the simulations then moved towards the final stage, which was a steady state, where the number of replicas running and the number of replicas waiting for their partner to complete reached a consistent range of values. This steady state was periodically disrupted by failure of the condor cluster, but was always quickly recovered once the disruption was over. The steady state for the simulation that used the catchup cluster had significantly more replicas running, and significantly fewer replicas waiting compared to the simulation that did not use the catchup cluster. The catchup cluster clearly improved the steady state number of nodes in use to approximately 50, compared to approximately 40 for the simulation that did not use the catchup cluster. This is an improvement in efficiency from 63% to 78%. 3.1 The Heterogenous Distributed Cluster The distributed condor cluster consisted of a range of desktop computers with varying processor speeds. To investigate the effect of running the simulation on this heterogeneous cluster, the total simulation time for each iteration was histogrammed. The histogram of replica completion times for the phosphorylated form of the protein is shown in figure 4. This figure shows that while the majority of iterations completed in under ten thousand seconds (2.8 hours), there was a significant spread of replica completion times up to twenty thousand seconds (5.6 hours). This spread of completion times means that fewer iterations have been completed than would have been expected based on the speed of the processors used. Figure 4. Histogram of the times to complete each iteration of the replica exchange simulation on the phosphorylated form of NTRC. 3.2 Comparison to Normal MD The use of dual Xeons in the catchup cluster aiding the efficiency of a replica exchange simulation was compared to their use as dedicated nodes running a normal Molecular Dynamics simulation. The phosphorylated conformation of NTRC was simulated at 300 K using a standard MD simulation with an identical starting structure as the replica exchange simulation and identical simulation conditions. Over the same period of time as the replica exchange simulations were running, this MD simulation on a single dual Xeon node completed 1.9 ns of dynamics. This compares to a total of 10.5 ns of dynamics generated by the corresponding replica exchange simulation. However, the 1.9 ns of dynamics generated via the MD simulation forms a single, selfconsistent trajectory. In comparison, the 10.5 ns of sampling from the replica exchange simulations was formed over 64 individual trajectories of only 0.16 ns in length. The dedicated node has produced a single trajectory over ten times the length of those produced via the distributed condor cluster with catchup cluster. This is despite the dedicated node only running the MD approximately 3.5 times the speed of a typical node in the distributed cluster. There are two reasons for this discrepancy; firstly, as demonstrated in figure 4, the heterogeneous nature of the distributed cluster meant that there was a large spread in the amount of time needed to complete each iteration of the replica exchange simulation. This could be mitigated against by running each iteration twice at the same time on the distributed cluster and using the results from the first node that completed the calculation. The second reason for the discrepancy is that the
5 not travel far in temperature. Instead each replica tended to oscillate around its initial temperature. No replicas from high temperature swapped down to room temperature. This shows that the replica exchange simulations need to be continued for many more iterations before the improved sampling of high temperature is able to be of use in enhancing the rate of sampling at room temperature. Figure 5. Temperature of each replica as a function of iteration from the replica exchange simulation on the unphosphorylated conformation of NTRC. Four randomly chosen replicas are highlighted. distributed cluster was very unreliable, leading to large periods of time when the simulation was not running. The replica exchange simulations presented here were run at a time when the condor cluster was experiencing a higher than normal amount of downtime. It is anticipated that during normal operation the condor cluster would be more reliable, and that the steady state efficiency of the replica exchange simulations would be maintained throughout the majority of the simulation. However, the experience of running these simulations demonstrate that applications that use distributed clusters need to include estimates of downtime and the range of available resources when predicting how long a particular simulation will take to run. These results also demonstrate that a distributed computing resource is, unsurprisingly, not efficient compared to a dedicated computing resource. However, distributed computing typically provides resources that would otherwise not be available. 3.3 Effectiveness of Replica Exchange The primary aim of running these simulations over the distributed computing resource was to sample the conformational change induced by phosphorylating NTRC. The aim was to use replica exchange to swap simulations running at high temperature, where the conformational change occurs more rapidly, down to room temperature, where the simulation statistics are collected. Figure 5 shows the temperature for each replica of the unphosphorylated simulation as a function of iteration. Four of the replicas are highlighted. This figure shows that while the replica exchange moves were accepted with the desired frequency, the replicas themselves did 4. Conclusion Distributed computing provides a resource that is not ideally suited to a wide range of chemistry problems. The investigation of protein conformational change is one such problem. The replica exchange algorithm was used in an attempt to fit this chemistry problem to the distributed computing resource. The coupled nature of replica exchange simulations caused problems for the scheduling of the computation that were partially solved through the development of a dedicated catchup cluster. This cluster improved the efficiency of the replica exchange simulation from 63% to 78%. Acknowledgements We thank R. Gledhill, A. Wiley and L. Fenu for discussions and the EPSRC for funding comb-echem. References Leach, A.R., Molecular Modelling, Principals and Applications, Longman, Harlow, UK, Pelton, J.G., Kustu, S., Wemmer, D.E., J. Mol. Biol., 292, 1095, Kern, D., Volkman, B.F., Luginbuhl, P., Nohaile, M.J., Kustu, S., Wemmer, D.E., Nature, 402, 894, Sugita, Y., Kitao, A., Okamoto, Y., J. Chem. Phys., 113, , Hansmann, U.H.E., Chem. Phys. Lett., 281, 140, Vriend, G., Hooft, R.W.W., Van Aalten, D., WhatIf, Pearlman, D.A., Case, D.A., Caldwell, J.W., Ross, W.S., Cheatham, T.E., Debolt, S., Ferguson, D., Seibel, G., Kollman, P., Comput. Phys. Commun., 91, 1, Mackerrell, A.D., Bashford, D., Bellot, M., Dunbrack, L., Evanseck, M., Fischer, S., Gao, J., Guo, H., Ha, S., Joseph-Mccarthy, D., Kuchnir, L., Kuczera, K., Lau, F.T.K., Mattos, C., Michnick, S., Ngo, T., Nguyen,
6 D.T., Prodhom, B., Reiher, W.E., Roux, B., Sclenkrich, M., Smith, J.C., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz- Kuczera, J., Yin, D., Karplus, M., J. Phys. Chem. B., 102, 3586, Kale, L., Skeel, R., Bhandarker, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K., Schulten, K., J. Comp. Phys., 151, 283, NAMD was developed by the Theoretical Biophysics Group in the Beckman Institute at Urbana-Champaign. 14. Paterlini, M.G., Ferguson, D.M., Chem. Phys., 236, 243, Feller, S.E., Zhang, Y.H., Pastor, R.W., Brooks, B.R., J. Chem. Phys., 103, 4613, Ryckaert, J.P., Ciccotti, G., Berendsen, J.C., 23, 327, Darden, T., York, D., Pedersen, L., J. Chem. Phys., 98, 10089,
Atsushi Matsumoto. Hisashi Ishida 1, Kei Yura 1, Takuma Kano 1 and Atsushi Matsumoto 1. 1. Introduction
 Chapter 3 Epoch Making Simulation Analysis of the Function of a Large-scale Supra-biomolecule System by Molecular Dynamics Simulation System, (Simulation Codes for huge Biomolecular Assembly) Project Representative
Chapter 3 Epoch Making Simulation Analysis of the Function of a Large-scale Supra-biomolecule System by Molecular Dynamics Simulation System, (Simulation Codes for huge Biomolecular Assembly) Project Representative
Overview of NAMD and Molecular Dynamics
 Overview of NAMD and Molecular Dynamics Jim Phillips Low-cost Linux Clusters for Biomolecular Simulations Using NAMD Outline Overview of molecular dynamics Overview of NAMD NAMD parallel design NAMD config
Overview of NAMD and Molecular Dynamics Jim Phillips Low-cost Linux Clusters for Biomolecular Simulations Using NAMD Outline Overview of molecular dynamics Overview of NAMD NAMD parallel design NAMD config
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
NAMD2- Greater Scalability for Parallel Molecular Dynamics. Presented by Abel Licon
 NAMD2- Greater Scalability for Parallel Molecular Dynamics Laxmikant Kale, Robert Steel, Milind Bhandarkar,, Robert Bunner, Attila Gursoy,, Neal Krawetz,, James Phillips, Aritomo Shinozaki, Krishnan Varadarajan,,
NAMD2- Greater Scalability for Parallel Molecular Dynamics Laxmikant Kale, Robert Steel, Milind Bhandarkar,, Robert Bunner, Attila Gursoy,, Neal Krawetz,, James Phillips, Aritomo Shinozaki, Krishnan Varadarajan,,
Supplementary Figures S1 - S11
 1 Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations Hang Yu 1,2, Klaus Schulten 1,2,3, 1 Beckman Institute, University of Illinois, Urbana, Illinois, USA 2 Center of Biophysics
1 Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations Hang Yu 1,2, Klaus Schulten 1,2,3, 1 Beckman Institute, University of Illinois, Urbana, Illinois, USA 2 Center of Biophysics
Structure Check. Authors: Eduard Schreiner Leonardo G. Trabuco. February 7, 2012
 University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation
 J. Chem. Theory Comput. 2008, 4, 435-447 435 GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation Berk Hess* Max-Planck Institute for Polymer Research, Ackermannweg
J. Chem. Theory Comput. 2008, 4, 435-447 435 GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation Berk Hess* Max-Planck Institute for Polymer Research, Ackermannweg
Biomolecular Modelling
 Biomolecular Modelling Carmen Domene Physical & Theoretical Chemistry Laboratory University of Oxford, UK THANKS Dr Joachim Hein Dr Iain Bethune Dr Eilidh Grant & Qi Huangfu 2 EPSRC Grant, Simulations
Biomolecular Modelling Carmen Domene Physical & Theoretical Chemistry Laboratory University of Oxford, UK THANKS Dr Joachim Hein Dr Iain Bethune Dr Eilidh Grant & Qi Huangfu 2 EPSRC Grant, Simulations
Chapter 8: Energy and Metabolism
 Chapter 8: Energy and Metabolism 1. Discuss energy conversions and the 1 st and 2 nd law of thermodynamics. Be sure to use the terms work, potential energy, kinetic energy, and entropy. 2. What are Joules
Chapter 8: Energy and Metabolism 1. Discuss energy conversions and the 1 st and 2 nd law of thermodynamics. Be sure to use the terms work, potential energy, kinetic energy, and entropy. 2. What are Joules
Recommended hardware system configurations for ANSYS users
 Recommended hardware system configurations for ANSYS users The purpose of this document is to recommend system configurations that will deliver high performance for ANSYS users across the entire range
Recommended hardware system configurations for ANSYS users The purpose of this document is to recommend system configurations that will deliver high performance for ANSYS users across the entire range
ANSWER KEY : BUILD AN ATOM PART I: ATOM SCREEN Build an Atom simulation ( http://phet.colorado.edu/en/simulation/build an atom )
 ANSWER KEY : PART I: ATOM SCREEN Build an Atom simulation ( http://phet.colorado.edu/en/simulation/build an atom ) 1. Explore the Build an Atom simulation with your group. As you explore, talk about what
ANSWER KEY : PART I: ATOM SCREEN Build an Atom simulation ( http://phet.colorado.edu/en/simulation/build an atom ) 1. Explore the Build an Atom simulation with your group. As you explore, talk about what
1 The water molecule and hydrogen bonds in water
 The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
Towards Large-Scale Molecular Dynamics Simulations on Graphics Processors
 Towards Large-Scale Molecular Dynamics Simulations on Graphics Processors Joe Davis, Sandeep Patel, and Michela Taufer University of Delaware Outline Introduction Introduction to GPU programming Why MD
Towards Large-Scale Molecular Dynamics Simulations on Graphics Processors Joe Davis, Sandeep Patel, and Michela Taufer University of Delaware Outline Introduction Introduction to GPU programming Why MD
NMR SPECTROSCOPY A N I N T R O D U C T I O N T O... Self-study booklet NUCLEAR MAGNETIC RESONANCE. 4 3 2 1 0 δ PUBLISHING
 A N I N T R O D U T I O N T O... NMR SPETROSOPY NULEAR MAGNETI RESONANE 4 3 1 0 δ Self-study booklet PUBLISING NMR Spectroscopy NULEAR MAGNETI RESONANE SPETROSOPY Origin of Spectra Theory All nuclei possess
A N I N T R O D U T I O N T O... NMR SPETROSOPY NULEAR MAGNETI RESONANE 4 3 1 0 δ Self-study booklet PUBLISING NMR Spectroscopy NULEAR MAGNETI RESONANE SPETROSOPY Origin of Spectra Theory All nuclei possess
Composition of the Atmosphere. Outline Atmospheric Composition Nitrogen and Oxygen Lightning Homework
 Molecules of the Atmosphere The present atmosphere consists mainly of molecular nitrogen (N2) and molecular oxygen (O2) but it has dramatically changed in composition from the beginning of the solar system.
Molecules of the Atmosphere The present atmosphere consists mainly of molecular nitrogen (N2) and molecular oxygen (O2) but it has dramatically changed in composition from the beginning of the solar system.
Chapter 6 An Overview of Organic Reactions
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
Sample Exercise 8.1 Magnitudes of Lattice Energies
 Sample Exercise 8.1 Magnitudes of Lattice Energies Without consulting Table 8.2, arrange the ionic compounds NaF, CsI, and CaO in order of increasing lattice energy. Analyze From the formulas for three
Sample Exercise 8.1 Magnitudes of Lattice Energies Without consulting Table 8.2, arrange the ionic compounds NaF, CsI, and CaO in order of increasing lattice energy. Analyze From the formulas for three
AP* Atomic Structure & Periodicity Free Response Questions KEY page 1
 AP* Atomic Structure & Periodicity ree Response Questions KEY page 1 1980 a) points 1s s p 6 3s 3p 6 4s 3d 10 4p 3 b) points for the two electrons in the 4s: 4, 0, 0, +1/ and 4, 0, 0, - 1/ for the three
AP* Atomic Structure & Periodicity ree Response Questions KEY page 1 1980 a) points 1s s p 6 3s 3p 6 4s 3d 10 4p 3 b) points for the two electrons in the 4s: 4, 0, 0, +1/ and 4, 0, 0, - 1/ for the three
Chapter 2: The Chemical Context of Life
 Chapter 2: The Chemical Context of Life Name Period This chapter covers the basics that you may have learned in your chemistry class. Whether your teacher goes over this chapter, or assigns it for you
Chapter 2: The Chemical Context of Life Name Period This chapter covers the basics that you may have learned in your chemistry class. Whether your teacher goes over this chapter, or assigns it for you
What is molecular dynamics (MD) simulation and how does it work?
 What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
Language: English Lecturer: Gianni de Fabritiis. Teaching staff: Language: English Lecturer: Jordi Villà i Freixa
 MSI: Molecular Simulations Descriptive details concerning the subject: Name of the subject: Molecular Simulations Code : MSI Type of subject: Optional ECTS: 5 Total hours: 125.0 Scheduling: 11:00-13:00
MSI: Molecular Simulations Descriptive details concerning the subject: Name of the subject: Molecular Simulations Code : MSI Type of subject: Optional ECTS: 5 Total hours: 125.0 Scheduling: 11:00-13:00
CHAPTER 4: Enzyme Structure ENZYMES
 CHAPTER 4: ENZYMES Enzymes are biological catalysts. There are about 40,000 different enzymes in human cells, each controlling a different chemical reaction. They increase the rate of reactions by a factor
CHAPTER 4: ENZYMES Enzymes are biological catalysts. There are about 40,000 different enzymes in human cells, each controlling a different chemical reaction. They increase the rate of reactions by a factor
ZooKeeper. Table of contents
 by Table of contents 1 ZooKeeper: A Distributed Coordination Service for Distributed Applications... 2 1.1 Design Goals...2 1.2 Data model and the hierarchical namespace...3 1.3 Nodes and ephemeral nodes...
by Table of contents 1 ZooKeeper: A Distributed Coordination Service for Distributed Applications... 2 1.1 Design Goals...2 1.2 Data model and the hierarchical namespace...3 1.3 Nodes and ephemeral nodes...
A Simultaneous Solution for General Linear Equations on a Ring or Hierarchical Cluster
 Acta Technica Jaurinensis Vol. 3. No. 1. 010 A Simultaneous Solution for General Linear Equations on a Ring or Hierarchical Cluster G. Molnárka, N. Varjasi Széchenyi István University Győr, Hungary, H-906
Acta Technica Jaurinensis Vol. 3. No. 1. 010 A Simultaneous Solution for General Linear Equations on a Ring or Hierarchical Cluster G. Molnárka, N. Varjasi Széchenyi István University Győr, Hungary, H-906
Load Balancing in the Cloud Computing Using Virtual Machine Migration: A Review
 Load Balancing in the Cloud Computing Using Virtual Machine Migration: A Review 1 Rukman Palta, 2 Rubal Jeet 1,2 Indo Global College Of Engineering, Abhipur, Punjab Technical University, jalandhar,india
Load Balancing in the Cloud Computing Using Virtual Machine Migration: A Review 1 Rukman Palta, 2 Rubal Jeet 1,2 Indo Global College Of Engineering, Abhipur, Punjab Technical University, jalandhar,india
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
H 2O gas: molecules are very far apart
 Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Windows Server Performance Monitoring
 Spot server problems before they are noticed The system s really slow today! How often have you heard that? Finding the solution isn t so easy. The obvious questions to ask are why is it running slowly
Spot server problems before they are noticed The system s really slow today! How often have you heard that? Finding the solution isn t so easy. The obvious questions to ask are why is it running slowly
green B 1 ) into a single unit to model the substrate in this reaction. enzyme
 Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
Teacher Key Objectives You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
CHEM 120 Online Chapter 7
 CHEM 120 Online Chapter 7 Date: 1. Which of the following statements is not a part of kinetic molecular theory? A) Matter is composed of particles that are in constant motion. B) Particle velocity increases
CHEM 120 Online Chapter 7 Date: 1. Which of the following statements is not a part of kinetic molecular theory? A) Matter is composed of particles that are in constant motion. B) Particle velocity increases
Study the following diagrams of the States of Matter. Label the names of the Changes of State between the different states.
 Describe the strength of attractive forces between particles. Describe the amount of space between particles. Can the particles in this state be compressed? Do the particles in this state have a definite
Describe the strength of attractive forces between particles. Describe the amount of space between particles. Can the particles in this state be compressed? Do the particles in this state have a definite
2038-20. Conference: From DNA-Inspired Physics to Physics-Inspired Biology. 1-5 June 2009. How Proteins Find Their Targets on DNA
 2038-20 Conference: From DNA-Inspired Physics to Physics-Inspired Biology 1-5 June 2009 How Proteins Find Their Targets on DNA Anatoly B. KOLOMEISKY Rice University, Department of Chemistry Houston TX
2038-20 Conference: From DNA-Inspired Physics to Physics-Inspired Biology 1-5 June 2009 How Proteins Find Their Targets on DNA Anatoly B. KOLOMEISKY Rice University, Department of Chemistry Houston TX
5. Structure, Geometry, and Polarity of Molecules
 5. Structure, Geometry, and Polarity of Molecules What you will accomplish in this experiment This experiment will give you an opportunity to draw Lewis structures of covalent compounds, then use those
5. Structure, Geometry, and Polarity of Molecules What you will accomplish in this experiment This experiment will give you an opportunity to draw Lewis structures of covalent compounds, then use those
Name Class Date. What is ionic bonding? What happens to atoms that gain or lose electrons? What kinds of solids are formed from ionic bonds?
 CHAPTER 1 2 Ionic Bonds SECTION Chemical Bonding BEFORE YOU READ After you read this section, you should be able to answer these questions: What is ionic bonding? What happens to atoms that gain or lose
CHAPTER 1 2 Ionic Bonds SECTION Chemical Bonding BEFORE YOU READ After you read this section, you should be able to answer these questions: What is ionic bonding? What happens to atoms that gain or lose
Scaling from 1 PC to a super computer using Mascot
 Scaling from 1 PC to a super computer using Mascot 1 Mascot - options for high throughput When is more processing power required? Options: Multiple Mascot installations Using a large Unix server Clusters
Scaling from 1 PC to a super computer using Mascot 1 Mascot - options for high throughput When is more processing power required? Options: Multiple Mascot installations Using a large Unix server Clusters
Review - After School Matter Name: Review - After School Matter Tuesday, April 29, 2008
 Name: Review - After School Matter Tuesday, April 29, 2008 1. Figure 1 The graph represents the relationship between temperature and time as heat was added uniformly to a substance starting at a solid
Name: Review - After School Matter Tuesday, April 29, 2008 1. Figure 1 The graph represents the relationship between temperature and time as heat was added uniformly to a substance starting at a solid
Studying an Organic Reaction. How do we know if a reaction can occur? And if a reaction can occur what do we know about the reaction?
 Studying an Organic Reaction How do we know if a reaction can occur? And if a reaction can occur what do we know about the reaction? Information we want to know: How much heat is generated? How fast is
Studying an Organic Reaction How do we know if a reaction can occur? And if a reaction can occur what do we know about the reaction? Information we want to know: How much heat is generated? How fast is
Molecular Modelling of DNA. Charlie Laughton University of Nottingham
 Molecular Modelling of DNA Charlie Laughton University of Nottingham Why model DNA? Sequences/structures you can t crystallise or get by NMR Drug design Understanding protein-dna recognition Quantitating
Molecular Modelling of DNA Charlie Laughton University of Nottingham Why model DNA? Sequences/structures you can t crystallise or get by NMR Drug design Understanding protein-dna recognition Quantitating
Q.1 Classify the following according to Lewis theory and Brønsted-Lowry theory.
 Acid-base 2816 1 Acid-base theories ACIDS & BASES - IONIC EQUILIBRIA LEWIS acid electron pair acceptor H +, AlCl 3 base electron pair donor NH 3, H 2 O, C 2 H 5 OH, OH e.g. H 3 N: -> BF 3 > H 3 N + BF
Acid-base 2816 1 Acid-base theories ACIDS & BASES - IONIC EQUILIBRIA LEWIS acid electron pair acceptor H +, AlCl 3 base electron pair donor NH 3, H 2 O, C 2 H 5 OH, OH e.g. H 3 N: -> BF 3 > H 3 N + BF
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Two Forms of Energy
 Module 2D - Energy and Metabolism Objective # 19 All living organisms require energy for survival. In this module we will examine some general principles about chemical reactions and energy usage within
Module 2D - Energy and Metabolism Objective # 19 All living organisms require energy for survival. In this module we will examine some general principles about chemical reactions and energy usage within
States of Matter CHAPTER 10 REVIEW SECTION 1. Name Date Class. Answer the following questions in the space provided.
 CHAPTER 10 REVIEW States of Matter SECTION 1 SHORT ANSWER Answer the following questions in the space provided. 1. Identify whether the descriptions below describe an ideal gas or a real gas. ideal gas
CHAPTER 10 REVIEW States of Matter SECTION 1 SHORT ANSWER Answer the following questions in the space provided. 1. Identify whether the descriptions below describe an ideal gas or a real gas. ideal gas
VTT TECHNICAL RESEARCH CENTRE OF FINLAND
 Figure from: http://www.embl.de/nmr/sattler/teaching Why NMR (instead of X ray crystallography) a great number of macromolecules won't crystallize) natural environmant (water) ligand binding and inter
Figure from: http://www.embl.de/nmr/sattler/teaching Why NMR (instead of X ray crystallography) a great number of macromolecules won't crystallize) natural environmant (water) ligand binding and inter
Which substance contains positive ions immersed in a sea of mobile electrons? A) O2(s) B) Cu(s) C) CuO(s) D) SiO2(s)
 BONDING MIDTERM REVIEW 7546-1 - Page 1 1) Which substance contains positive ions immersed in a sea of mobile electrons? A) O2(s) B) Cu(s) C) CuO(s) D) SiO2(s) 2) The bond between hydrogen and oxygen in
BONDING MIDTERM REVIEW 7546-1 - Page 1 1) Which substance contains positive ions immersed in a sea of mobile electrons? A) O2(s) B) Cu(s) C) CuO(s) D) SiO2(s) 2) The bond between hydrogen and oxygen in
Energy & Enzymes. Life requires energy for maintenance of order, growth, and reproduction. The energy living things use is chemical energy.
 Energy & Enzymes Life requires energy for maintenance of order, growth, and reproduction. The energy living things use is chemical energy. 1 Energy exists in two forms - potential and kinetic. Potential
Energy & Enzymes Life requires energy for maintenance of order, growth, and reproduction. The energy living things use is chemical energy. 1 Energy exists in two forms - potential and kinetic. Potential
Name Class Date. In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question.
 Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Determination of Equilibrium Constants using NMR Spectrscopy
 CHEM 331L Physical Chemistry Laboratory Revision 1.0 Determination of Equilibrium Constants using NMR Spectrscopy In this laboratory exercise we will measure a chemical equilibrium constant using key proton
CHEM 331L Physical Chemistry Laboratory Revision 1.0 Determination of Equilibrium Constants using NMR Spectrscopy In this laboratory exercise we will measure a chemical equilibrium constant using key proton
The Action Potential Graphics are used with permission of: adam.com (http://www.adam.com/) Benjamin Cummings Publishing Co (http://www.awl.
 The Action Potential Graphics are used with permission of: adam.com (http://www.adam.com/) Benjamin Cummings Publishing Co (http://www.awl.com/bc) ** If this is not printed in color, it is suggested you
The Action Potential Graphics are used with permission of: adam.com (http://www.adam.com/) Benjamin Cummings Publishing Co (http://www.awl.com/bc) ** If this is not printed in color, it is suggested you
Sample Exercise 8.1 Magnitudes of Lattice Energies
 Sample Exercise 8.1 Magnitudes of Lattice Energies Without consulting Table 8.2, arrange the following ionic compounds in order of increasing lattice energy: NaF, CsI, and CaO. Analyze: From the formulas
Sample Exercise 8.1 Magnitudes of Lattice Energies Without consulting Table 8.2, arrange the following ionic compounds in order of increasing lattice energy: NaF, CsI, and CaO. Analyze: From the formulas
Electron Configurations, Isoelectronic Elements, & Ionization Reactions. Chemistry 11
 Electron Configurations, Isoelectronic Elements, & Ionization Reactions Chemistry 11 Note: Of the 3 subatomic particles, the electron plays the greatest role in determining the physical and chemical properties
Electron Configurations, Isoelectronic Elements, & Ionization Reactions Chemistry 11 Note: Of the 3 subatomic particles, the electron plays the greatest role in determining the physical and chemical properties
Distributed Dynamic Load Balancing for Iterative-Stencil Applications
 Distributed Dynamic Load Balancing for Iterative-Stencil Applications G. Dethier 1, P. Marchot 2 and P.A. de Marneffe 1 1 EECS Department, University of Liege, Belgium 2 Chemical Engineering Department,
Distributed Dynamic Load Balancing for Iterative-Stencil Applications G. Dethier 1, P. Marchot 2 and P.A. de Marneffe 1 1 EECS Department, University of Liege, Belgium 2 Chemical Engineering Department,
Efficient Parallel Execution of Sequence Similarity Analysis Via Dynamic Load Balancing
 Efficient Parallel Execution of Sequence Similarity Analysis Via Dynamic Load Balancing James D. Jackson Philip J. Hatcher Department of Computer Science Kingsbury Hall University of New Hampshire Durham,
Efficient Parallel Execution of Sequence Similarity Analysis Via Dynamic Load Balancing James D. Jackson Philip J. Hatcher Department of Computer Science Kingsbury Hall University of New Hampshire Durham,
THREE DIMENSIONAL REPRESENTATION OF AMINO ACID CHARAC- TERISTICS
 THREE DIMENSIONAL REPRESENTATION OF AMINO ACID CHARAC- TERISTICS O.U. Sezerman 1, R. Islamaj 2, E. Alpaydin 2 1 Laborotory of Computational Biology, Sabancı University, Istanbul, Turkey. 2 Computer Engineering
THREE DIMENSIONAL REPRESENTATION OF AMINO ACID CHARAC- TERISTICS O.U. Sezerman 1, R. Islamaj 2, E. Alpaydin 2 1 Laborotory of Computational Biology, Sabancı University, Istanbul, Turkey. 2 Computer Engineering
List the 3 main types of subatomic particles and indicate the mass and electrical charge of each.
 Basic Chemistry Why do we study chemistry in a biology course? All living organisms are composed of chemicals. To understand life, we must understand the structure, function, and properties of the chemicals
Basic Chemistry Why do we study chemistry in a biology course? All living organisms are composed of chemicals. To understand life, we must understand the structure, function, and properties of the chemicals
Basic Scientific Principles that All Students Should Know Upon Entering Medical and Dental School at McGill
 Fundamentals of Medicine and Dentistry Basic Scientific Principles that All Students Should Know Upon Entering Medical and Dental School at McGill Students entering medical and dental training come from
Fundamentals of Medicine and Dentistry Basic Scientific Principles that All Students Should Know Upon Entering Medical and Dental School at McGill Students entering medical and dental training come from
CHEM 1211K Test IV. MULTIPLE CHOICE (3 points each)
 CEM 1211K Test IV MULTIPLE COICE (3 points each) 1) ow many single covalent bonds must a silicon atom form to have a complete octet in its valence shell? A) 4 B) 3 C) 1 D) 2 E) 0 2) What is the maximum
CEM 1211K Test IV MULTIPLE COICE (3 points each) 1) ow many single covalent bonds must a silicon atom form to have a complete octet in its valence shell? A) 4 B) 3 C) 1 D) 2 E) 0 2) What is the maximum
Consensus Scoring to Improve the Predictive Power of in-silico Screening for Drug Design
 Consensus Scoring to Improve the Predictive Power of in-silico Screening for Drug Design Masato Okada Faculty of Science and Technology, Masato Tsukamoto Faculty of Pharmaceutical Sciences, Hayato Ohwada
Consensus Scoring to Improve the Predictive Power of in-silico Screening for Drug Design Masato Okada Faculty of Science and Technology, Masato Tsukamoto Faculty of Pharmaceutical Sciences, Hayato Ohwada
Proton Nuclear Magnetic Resonance ( 1 H-NMR) Spectroscopy
 Proton Nuclear Magnetic Resonance ( 1 H-NMR) Spectroscopy Theory behind NMR: In the late 1940 s, physical chemists originally developed NMR spectroscopy to study different properties of atomic nuclei,
Proton Nuclear Magnetic Resonance ( 1 H-NMR) Spectroscopy Theory behind NMR: In the late 1940 s, physical chemists originally developed NMR spectroscopy to study different properties of atomic nuclei,
A New Nature-inspired Algorithm for Load Balancing
 A New Nature-inspired Algorithm for Load Balancing Xiang Feng East China University of Science and Technology Shanghai, China 200237 Email: xfeng{@ecusteducn, @cshkuhk} Francis CM Lau The University of
A New Nature-inspired Algorithm for Load Balancing Xiang Feng East China University of Science and Technology Shanghai, China 200237 Email: xfeng{@ecusteducn, @cshkuhk} Francis CM Lau The University of
Ionization energy _decreases from the top to the bottom in a group. Electron affinity increases from the left to the right within a period.
 hem 150 Answer Key roblem et 2 1. omplete the following phrases: Ionization energy _decreases from the top to the bottom in a group. Electron affinity increases from the left to the right within a period.
hem 150 Answer Key roblem et 2 1. omplete the following phrases: Ionization energy _decreases from the top to the bottom in a group. Electron affinity increases from the left to the right within a period.
Chapter 1 Student Reading
 Chapter 1 Student Reading Chemistry is the study of matter You could say that chemistry is the science that studies all the stuff in the entire world. A more scientific term for stuff is matter. So chemistry
Chapter 1 Student Reading Chemistry is the study of matter You could say that chemistry is the science that studies all the stuff in the entire world. A more scientific term for stuff is matter. So chemistry
1A Rate of reaction. AS Chemistry introduced the qualitative aspects of rates of reaction. These include:
 1A Rate of reaction AS Chemistry introduced the qualitative aspects of rates of reaction. These include: Collision theory Effect of temperature Effect of concentration Effect of pressure Activation energy
1A Rate of reaction AS Chemistry introduced the qualitative aspects of rates of reaction. These include: Collision theory Effect of temperature Effect of concentration Effect of pressure Activation energy
Multi-GPU Load Balancing for In-situ Visualization
 Multi-GPU Load Balancing for In-situ Visualization R. Hagan and Y. Cao Department of Computer Science, Virginia Tech, Blacksburg, VA, USA Abstract Real-time visualization is an important tool for immediately
Multi-GPU Load Balancing for In-situ Visualization R. Hagan and Y. Cao Department of Computer Science, Virginia Tech, Blacksburg, VA, USA Abstract Real-time visualization is an important tool for immediately
Optimization of Preventive Maintenance Scheduling in Processing Plants
 18 th European Symposium on Computer Aided Process Engineering ESCAPE 18 Bertrand Braunschweig and Xavier Joulia (Editors) 2008 Elsevier B.V./Ltd. All rights reserved. Optimization of Preventive Maintenance
18 th European Symposium on Computer Aided Process Engineering ESCAPE 18 Bertrand Braunschweig and Xavier Joulia (Editors) 2008 Elsevier B.V./Ltd. All rights reserved. Optimization of Preventive Maintenance
Molecular Visualization. Introduction
 Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Cluster Scalability of ANSYS FLUENT 12 for a Large Aerodynamics Case on the Darwin Supercomputer
 Cluster Scalability of ANSYS FLUENT 12 for a Large Aerodynamics Case on the Darwin Supercomputer Stan Posey, MSc and Bill Loewe, PhD Panasas Inc., Fremont, CA, USA Paul Calleja, PhD University of Cambridge,
Cluster Scalability of ANSYS FLUENT 12 for a Large Aerodynamics Case on the Darwin Supercomputer Stan Posey, MSc and Bill Loewe, PhD Panasas Inc., Fremont, CA, USA Paul Calleja, PhD University of Cambridge,
The strategic role and objectives of operations
 The strategic role and objectives of operations Source: Honda Motor Company What is the role of the Operations function? Operations as implementer Operations as supporter Operations as driver Operations
The strategic role and objectives of operations Source: Honda Motor Company What is the role of the Operations function? Operations as implementer Operations as supporter Operations as driver Operations
Name: Date: Period: DNA Unit: DNA Webquest
 Name: Date: Period: DNA Unit: DNA Webquest Part 1 History, DNA Structure, DNA Replication DNA History http://www.dnaftb.org/dnaftb/1/concept/index.html Read the text and answer the following questions.
Name: Date: Period: DNA Unit: DNA Webquest Part 1 History, DNA Structure, DNA Replication DNA History http://www.dnaftb.org/dnaftb/1/concept/index.html Read the text and answer the following questions.
Canonical sampling through velocity rescaling
 THE JOURNAL OF CHEMICAL PHYSICS 126, 014101 2007 Canonical sampling through velocity rescaling Giovanni Bussi, a Davide Donadio, and Michele Parrinello Computational Science, Department of Chemistry and
THE JOURNAL OF CHEMICAL PHYSICS 126, 014101 2007 Canonical sampling through velocity rescaling Giovanni Bussi, a Davide Donadio, and Michele Parrinello Computational Science, Department of Chemistry and
The Empirical Formula of a Compound
 The Empirical Formula of a Compound Lab #5 Introduction A look at the mass relationships in chemistry reveals little order or sense. The ratio of the masses of the elements in a compound, while constant,
The Empirical Formula of a Compound Lab #5 Introduction A look at the mass relationships in chemistry reveals little order or sense. The ratio of the masses of the elements in a compound, while constant,
Bonding Practice Problems
 NAME 1. When compared to H 2 S, H 2 O has a higher 8. Given the Lewis electron-dot diagram: boiling point because H 2 O contains stronger metallic bonds covalent bonds ionic bonds hydrogen bonds 2. Which
NAME 1. When compared to H 2 S, H 2 O has a higher 8. Given the Lewis electron-dot diagram: boiling point because H 2 O contains stronger metallic bonds covalent bonds ionic bonds hydrogen bonds 2. Which
I. The SMART Project - Status Report and Plans. G. Salton. The SMART document retrieval system has been operating on a 709^
 1-1 I. The SMART Project - Status Report and Plans G. Salton 1. Introduction The SMART document retrieval system has been operating on a 709^ computer since the end of 1964. The system takes documents
1-1 I. The SMART Project - Status Report and Plans G. Salton 1. Introduction The SMART document retrieval system has been operating on a 709^ computer since the end of 1964. The system takes documents
Converting A High Performance Application to an Elastic Cloud Application
 Converting A High Performance Application to an Elastic Cloud Application Dinesh Rajan, Anthony Canino, Jesus A Izaguirre, and Douglas Thain Department of Computer Science and Engineering University of
Converting A High Performance Application to an Elastic Cloud Application Dinesh Rajan, Anthony Canino, Jesus A Izaguirre, and Douglas Thain Department of Computer Science and Engineering University of
Test Bank - Chapter 4 Multiple Choice
 Test Bank - Chapter 4 The questions in the test bank cover the concepts from the lessons in Chapter 4. Select questions from any of the categories that match the content you covered with students. The
Test Bank - Chapter 4 The questions in the test bank cover the concepts from the lessons in Chapter 4. Select questions from any of the categories that match the content you covered with students. The
An Implementation Of Multiprocessor Linux
 An Implementation Of Multiprocessor Linux This document describes the implementation of a simple SMP Linux kernel extension and how to use this to develop SMP Linux kernels for architectures other than
An Implementation Of Multiprocessor Linux This document describes the implementation of a simple SMP Linux kernel extension and how to use this to develop SMP Linux kernels for architectures other than
Protein Protein Interaction Networks
 Functional Pattern Mining from Genome Scale Protein Protein Interaction Networks Young-Rae Cho, Ph.D. Assistant Professor Department of Computer Science Baylor University it My Definition of Bioinformatics
Functional Pattern Mining from Genome Scale Protein Protein Interaction Networks Young-Rae Cho, Ph.D. Assistant Professor Department of Computer Science Baylor University it My Definition of Bioinformatics
Data Visualization for Atomistic/Molecular Simulations. Douglas E. Spearot University of Arkansas
 Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Chemistry 122 Mines, Spring 2014
 Chemistry 122 Mines, Spring 2014 Answer Key, Problem Set 9 1. 18.44(c) (Also indicate the sign on each electrode, and show the flow of ions in the salt bridge.); 2. 18.46 (do this for all cells in 18.44
Chemistry 122 Mines, Spring 2014 Answer Key, Problem Set 9 1. 18.44(c) (Also indicate the sign on each electrode, and show the flow of ions in the salt bridge.); 2. 18.46 (do this for all cells in 18.44
Parallels Virtuozzo Containers
 Parallels Virtuozzo Containers White Paper Virtual Desktop Infrastructure www.parallels.com Version 1.0 Table of Contents Table of Contents... 2 Enterprise Desktop Computing Challenges... 3 What is Virtual
Parallels Virtuozzo Containers White Paper Virtual Desktop Infrastructure www.parallels.com Version 1.0 Table of Contents Table of Contents... 2 Enterprise Desktop Computing Challenges... 3 What is Virtual
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.
 CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
CHAPTER 10: INTERMOLECULAR FORCES: THE UNIQUENESS OF WATER Problems: 10.2, 10.6,10.15-10.33, 10.35-10.40, 10.56-10.60, 10.101-10.102 10.1 INTERACTIONS BETWEEN IONS Ion-ion Interactions and Lattice Energy
Q.1 Classify the following according to Lewis theory and Brønsted-Lowry theory.
 Acid-base A4 1 Acid-base theories ACIDS & BASES - IONIC EQUILIBRIA 1. LEWIS acid electron pair acceptor H, AlCl 3 base electron pair donor NH 3, H 2 O, C 2 H 5 OH, OH e.g. H 3 N: -> BF 3 > H 3 N BF 3 see
Acid-base A4 1 Acid-base theories ACIDS & BASES - IONIC EQUILIBRIA 1. LEWIS acid electron pair acceptor H, AlCl 3 base electron pair donor NH 3, H 2 O, C 2 H 5 OH, OH e.g. H 3 N: -> BF 3 > H 3 N BF 3 see
Brønsted-Lowry Acids and Bases
 Brønsted-Lowry Acids and Bases 1 According to Brønsted and Lowry, an acid-base reaction is defined in terms of a proton transfer. By this definition, the reaction of Cl in water is: Cl(aq) + Cl (aq) +
Brønsted-Lowry Acids and Bases 1 According to Brønsted and Lowry, an acid-base reaction is defined in terms of a proton transfer. By this definition, the reaction of Cl in water is: Cl(aq) + Cl (aq) +
Principles of Soccer
 What criteria can we use to tell how well our team is playing? Analysis of soccer always starts out with the same question, Who has the ball? Principle #1: Possession If our team has possession of the
What criteria can we use to tell how well our team is playing? Analysis of soccer always starts out with the same question, Who has the ball? Principle #1: Possession If our team has possession of the
Chapter 8 Concepts of Chemical Bonding
 Chapter 8 Concepts of Chemical Bonding Chemical Bonds Three types: Ionic Electrostatic attraction between ions Covalent Sharing of electrons Metallic Metal atoms bonded to several other atoms Ionic Bonding
Chapter 8 Concepts of Chemical Bonding Chemical Bonds Three types: Ionic Electrostatic attraction between ions Covalent Sharing of electrons Metallic Metal atoms bonded to several other atoms Ionic Bonding
Oracle Database Scalability in VMware ESX VMware ESX 3.5
 Performance Study Oracle Database Scalability in VMware ESX VMware ESX 3.5 Database applications running on individual physical servers represent a large consolidation opportunity. However enterprises
Performance Study Oracle Database Scalability in VMware ESX VMware ESX 3.5 Database applications running on individual physical servers represent a large consolidation opportunity. However enterprises
Chemistry 13: States of Matter
 Chemistry 13: States of Matter Name: Period: Date: Chemistry Content Standard: Gases and Their Properties The kinetic molecular theory describes the motion of atoms and molecules and explains the properties
Chemistry 13: States of Matter Name: Period: Date: Chemistry Content Standard: Gases and Their Properties The kinetic molecular theory describes the motion of atoms and molecules and explains the properties
Chemical Basis of Life Module A Anchor 2
 Chemical Basis of Life Module A Anchor 2 Key Concepts: - Water is a polar molecule. Therefore, it is able to form multiple hydrogen bonds, which account for many of its special properties. - Water s polarity
Chemical Basis of Life Module A Anchor 2 Key Concepts: - Water is a polar molecule. Therefore, it is able to form multiple hydrogen bonds, which account for many of its special properties. - Water s polarity
Technology Update White Paper. High Speed RAID 6. Powered by Custom ASIC Parity Chips
 Technology Update White Paper High Speed RAID 6 Powered by Custom ASIC Parity Chips High Speed RAID 6 Powered by Custom ASIC Parity Chips Why High Speed RAID 6? Winchester Systems has developed High Speed
Technology Update White Paper High Speed RAID 6 Powered by Custom ASIC Parity Chips High Speed RAID 6 Powered by Custom ASIC Parity Chips Why High Speed RAID 6? Winchester Systems has developed High Speed
PORTrockIT. Spectrum Protect : faster WAN replication and backups with PORTrockIT
 1 PORTrockIT 2 Executive summary IBM Spectrum Protect, previously known as IBM Tivoli Storage Manager or TSM, is the cornerstone of many large companies data protection strategies, offering a wide range
1 PORTrockIT 2 Executive summary IBM Spectrum Protect, previously known as IBM Tivoli Storage Manager or TSM, is the cornerstone of many large companies data protection strategies, offering a wide range
QuickSpecs. Models PC3-10600 (DDR3-1333 MHz) DIMMs
 Overview Models PC3-10600 (DDR3-1333 MHz) DIMMs HP 1-GB PC3-10600 (DDR3-1333 MHz) DIMM HP 2-GB PC3-10600 (DDR3-1333 MHz) DIMM HP 4-GB PC3-10600 (DDR3-1333 MHz) DIMM PC3-10600 (DDR3-1333 MHz) SODIMMs HP
Overview Models PC3-10600 (DDR3-1333 MHz) DIMMs HP 1-GB PC3-10600 (DDR3-1333 MHz) DIMM HP 2-GB PC3-10600 (DDR3-1333 MHz) DIMM HP 4-GB PC3-10600 (DDR3-1333 MHz) DIMM PC3-10600 (DDR3-1333 MHz) SODIMMs HP
Chapter 2 Polar Covalent Bonds; Acids and Bases
 John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
Enzymes and Metabolic Pathways
 Enzymes and Metabolic Pathways Enzyme characteristics Made of protein Catalysts: reactions occur 1,000,000 times faster with enzymes Not part of reaction Not changed or affected by reaction Used over and
Enzymes and Metabolic Pathways Enzyme characteristics Made of protein Catalysts: reactions occur 1,000,000 times faster with enzymes Not part of reaction Not changed or affected by reaction Used over and
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING
 CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
BackupEnabler: Virtually effortless backups for VMware Environments
 White Paper BackupEnabler: Virtually effortless backups for VMware Environments Contents Abstract... 3 Why Standard Backup Processes Don t Work with Virtual Servers... 3 Agent-Based File-Level and Image-Level
White Paper BackupEnabler: Virtually effortless backups for VMware Environments Contents Abstract... 3 Why Standard Backup Processes Don t Work with Virtual Servers... 3 Agent-Based File-Level and Image-Level
PLEASE SCROLL DOWN FOR ARTICLE
 This article was downloaded by:[new York University] [New York University] On: 16 July 2007 Access Details: [subscription number 769426389] Publisher: Taylor & Francis Informa Ltd Registered in England
This article was downloaded by:[new York University] [New York University] On: 16 July 2007 Access Details: [subscription number 769426389] Publisher: Taylor & Francis Informa Ltd Registered in England
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005
 Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
MOLECULAR DYNAMICS SIMULATION OF DYNAMIC RESPONSE OF BERYLLIUM
 MOLECULAR DYNAMICS SIMULATION OF DYNAMIC RESPONSE OF BERYLLIUM Aidan P. Thompson, J. Matthew D. Lane and Michael P. Desjarlais Sandia National Laboratories, Albuquerque, New Mexico 87185, USA Abstract.
MOLECULAR DYNAMICS SIMULATION OF DYNAMIC RESPONSE OF BERYLLIUM Aidan P. Thompson, J. Matthew D. Lane and Michael P. Desjarlais Sandia National Laboratories, Albuquerque, New Mexico 87185, USA Abstract.
Mulliken suggested to split the shared density 50:50. Then the electrons associated with the atom k are given by:
 1 17. Population Analysis Population analysis is the study of charge distribution within molecules. The intention is to accurately model partial charge magnitude and location within a molecule. This can
1 17. Population Analysis Population analysis is the study of charge distribution within molecules. The intention is to accurately model partial charge magnitude and location within a molecule. This can
APPLICATION NOTE TESTING PV MICRO INVERTERS USING A FOUR QUADRANT CAPABLE PROGRAMMABLE AC POWER SOURCE FOR GRID SIMULATION. Abstract.
 TESTING PV MICRO INVERTERS USING A FOUR QUADRANT CAPABLE PROGRAMMABLE AC POWER SOURCE FOR GRID SIMULATION Abstract This application note describes the four quadrant mode of operation of a linear AC Power
TESTING PV MICRO INVERTERS USING A FOUR QUADRANT CAPABLE PROGRAMMABLE AC POWER SOURCE FOR GRID SIMULATION Abstract This application note describes the four quadrant mode of operation of a linear AC Power
Why ClearCube Technology for VDI?
 Why ClearCube Technology for VDI? January 2014 2014 ClearCube Technology, Inc. All Rights Reserved 1 Why ClearCube for VDI? There are many VDI platforms to choose from. Some have evolved inefficiently
Why ClearCube Technology for VDI? January 2014 2014 ClearCube Technology, Inc. All Rights Reserved 1 Why ClearCube for VDI? There are many VDI platforms to choose from. Some have evolved inefficiently
REMOTE CONTROL by DNA as a Bio-sensor -antenna.
 REMOTE CONTROL by DNA as a Bio-sensor -antenna. "Piezoelectric quantum transduction is a fundamental property of at- distance induction of genetic control " Paolo Manzelli: pmanzelli@gmail.com ; www.edscuola.it/lre.html;www.egocreanet.it
REMOTE CONTROL by DNA as a Bio-sensor -antenna. "Piezoelectric quantum transduction is a fundamental property of at- distance induction of genetic control " Paolo Manzelli: pmanzelli@gmail.com ; www.edscuola.it/lre.html;www.egocreanet.it
