CellLine, a stochastic cell lineage simulator: Manual
|
|
|
- Aubrey Griffith
- 9 years ago
- Views:
Transcription
1 CellLine, a stochastic cell lineage simulator: Manual Andre S. Ribeiro, Daniel A. Charlebois, Jason Lloyd-Price July 23, Introduction This document explains how to work with the CellLine modules and the functionality provided. CellLineconsists of 3 modules and requires an initial reactions file. The reactions file defines the initial concentrations of all the substances and the reactions that can occur inside a single cell. Given an initial cell, defined in a file containing the list of possible reactions and initial reactants concentrations, one can use CellLineGen to generate a cell line from the initial cell. If desired, the third module NCellsGen can be used to generate a set of N dynamically independent cell copies of the initial cell or some cell of the lineage. CellLineGen takes the initial reactions file, calls DynSim.exe to run its dynamics for a user defined time interval, and creates two new cells afterwards. The user also defines the number of cell generations desired. The two new cells will have the same set of reactions as the mother cell. Each new cell s initial concentrations are equal to the concentrations of the same elements in the mother cell, at the moment of division. Importantly, elements of the waitlist still not released in the system will also be passed on to the two daughter cells. This is critical since, for example, transcription and translation involve time delays, and the promoters could be in the waitlist at the moment of division. Once their reactions files are generated, the two daughter cells dynamics are then simulated by DynSim.exe. The cells life time and sampling interval are the same as the mother cell. When the dynamics simulation is finished, DynSim.exe outputs the two cells time series and divides each of them in two more daughter cells and so on. CellLineGen provides each cell with a unique random seed and appends it to each cell s reactions files. NCellsGen takes the user defined reactions file and makes N copies of it. The cell copies will have the same set of reactions but can be as distinct as desired on every other aspect such as concentrations, rate constants, and delays. For example, the same reactions in different cells can have different rate constants, for one or more reactions. Initial chemical populations and time delays can also differ. Each reactions file has a different random seed, thus the dynamics of each cell copy will be unique. The seed is appended to each cell s reactions file, so that experiments can be repeated when desired. The results will always be the same as long as no other random variables are defined (such as random rate constants). After making N cell copies and creating an independent reactions file for each cell, NCellsGen calls DynSim.exe, which simulates the dynamics for a user defined time interval and outputs the resulting times series of each cell in independent files. The dynamics resulting from the reactions files generated by NCellsGen or CellLineGen is simulated by DynSim.exe, which is an extension of a previously existing simulator, SGN Sim [4]. This simulator s dynamics is driven by the delayed SSA [5], which consists of the original SSA [2, 1], but allows multiple time delayed reactions to be modelled. CellLine uses the Mersenne Twister pseudo-random number generator [3]. This generator has a period of We now describe each of the program components. 1
2 2 Cell line generator module CellLineGen generates a cell lineage from an initial reactions file provided by the user where the mother cell s system of chemical reactions is defined. It then runs its dynamics for a user-defined amount of time and outputs the system state at a specified sampling interval. When it completes this first step, the program takes a snap-shot of the mother cell s state at the time of division (the concentrations of all chemical elements including those in the waitlist) and it creates two new daughter cells. The daughter cells inherit the set of reactions and the concentrations of the chemical species of the mother cell at the moment of division. The reactions file of each cell in the lineage is named Gener x cell y.g, where x is the generation number and y is the cell number within that generation. The mother cell s reactions file (a copy of the reactions file provided by the user) is named Gener 01 cell 01.g. After simulating the dynamics of any cell of the lineage, the program outputs a results file. This file, named Gener x cell y out.xls, contains the time series of each of the chemical species involved. Additionally, the program also outputs a snapshot file, called Gener x cell y.snap.g with each cell s final state. CellLineGen.exe can be called by invoking the following command: cellline <filename> [number of cell generations] [simulation time] [sampling interval] Filename is the reactions filename provided by the user. Number of cell generations is an integer specifying the number of generations spawned from the original cell. Simulation time is a positive real variable specifying the run time of each cell dynamics. Sampling interval is a positive real variable specifying the time series sampling interval. Number of cell generations, Simulation time, and Sampling interval are all default to 1, when not provided. CellLineGen appends each cell s random seed to the cell s reactions file and therefore all cells will differ in their dynamics. 3 NCellsGen generator module Once the reactions file is constructed (either by the user or resulting from a cell lineage simulation), NCellsGen can be invoked to produce a number of independent cell copies that will be simulated for a specific length of time. This is done by executing the following command: NCellsGen <filename> [number of cells] [simulation time] [sampling interval] Filename is the reactions filename. Number of cells is an integer variable that specifies the number of independent cell copies of the original cell to simulate (from 1 to N). Simulation time is a real positive variable specifying the simulation time of the dynamics of each cell (from 0 to infinite). Sampling interval is a real positive variable specifying the sampling interval of the cell s dynamics. Number of cells, Simulation time, and Sampling interval are all default to 1, when not provided. After the command line is correctly invoked, NCellsGen generates a single reactions file for each cell and names them cell [cell index].g. After running each cell s dynamics, an output file of each cell is created that contains the time series of all species and is named out [cell index].xls. Since each cell has a different random seed, their dynamics will differ. The seed is appended to each cell s reactions file, so that experiments can be repeated when desired. Yet, one may which to vary some parameter from cell to cell, like a rate constant, an element s initial concentration, or a time delay value. How to do this is explained in the following sections describing how to setup a reactions file. Fourier spectrums of time series can only be obtained for single cells (i.e. one cell at a time). For example, to calculate the fourier spectrum of the time series of cell x, open the reactions file from which cell x is created and introduce a new line, before the main lua block, with the following: fourier file fourier filename; Then run NCellsGen, using the reactions file as input and specifying that only a single cell is to be simulated with the following command: 2
3 Ncells <cell filename> 1 [simulation time] [sampling interval] The program will output a results file out 01.xls with the time series of the system, simulation time seconds long, sampled every sampling interval seconds, and a second file, named fourier filename, with the fourier spectrum of the time series of out 01.xls. Both files are tab delimited text files. 4 Delayed Stochastic Simulation Algorithm CellLine dynamics module, DynSim, is driven by the delayed SSA [5]. Unlike the original SSA [2], the delayed SSA stores delayed output events on a waitlist, handling multiple delayed output events for each input event. The waitlist contains the list of elements to be released some time delay after the reaction that produced them occurred (this delay is also stored for each element on the waitlist). The algorithm proceeds as follows [5]: 1) Set t 0, t stop stop time, read initial number of molecules and reactions, create empty waiting list L for delayed generating events. 2) Do an SSA step for reacting events to get the next reacting event R 1 and corresponding occurrence time t + t 1. 3) Compare t + t 1 with t min, the least time in L. If t + t 1 < t min or L is empty, set t t + t 1. Update number of molecules by performing R 1, adding any delayed products to L with a time of t + t 1 + τ, where τ is drawn from an appropriate distribution. 4) If L is not empty and if t + t 1 t min, set t t min. Update number of molecules and L by releasing the first element in L. 5) If t < t stop, go to step 2, else stop. 5 Input Reactions file The initial reactions file must always be provided to the simulator. It consists of a list of the initial quantities of chemical species and the set of reactions (including rate constants and time delays) that each cell consists of. In the following sections we describe the syntax of the reactions file. If the initial concentration of some substance is not defined, the simulator assumes it is zero at t = 0 s. In general, the initial cell can contain any set of reactions. For example, it could consist of a toggle switch model, a repressilator, or the P53-Mdm2 feedback loop chemical system. To input a reactions system into the simulator the reactions have to be listed in a file. The reaction system is specified by a series of identifier-data pairs. The identifier determines how to interpret the data. The next sections of this document defines the set of identifiers and the format of their data. We provide several reactions files. These should be kept intact and any changes should be made in a new file. 5.1 Format Information specified in a file follows the following formats: 1. identifier data; 2. identifier { (data;)* } 3. identifier!{ data }! The first format is used to give simple information such as the initial random seed and the filename of the file to which the fourier spectrum should be output. The second format is used to input a block of similar data and is equivalent to repeating the first form many times. Lists of reactions and initial populations of molecules can then be added quickly. The following example demonstrates how a set of initial population declarations (see section 5.5) for a Toggle Switch system between genes A and B can be condensed into a single block: 3
4 Format 1 Format 2 population A = 0; population { population B = 0; A = 0; population ProA = 1; B = 0; population ProB = 1; ProA = 1; ProB = 1; } The third and final format is designed to allow blocks of data which may contain semicolons or braces inside them to be input. This is primarily useful for writing a chunk of Lua code (see section 6). 5.2 Comments Comments can be placed in an input file to allow inline documentation of the reaction system. Comments follow the C++ comment format, where // can be used to comment a line, and /* */ can be used to comment a block of text. 5.3 Species Names Species names follow the C identifier rules. They must begin with a letter or an underscore, followed by a string of letters, numbers or underscores. That is, they must satisfy the regular expression [a-za-z ][a-za-z0-9 ]*. 5.4 Multimers Often, molecules will bind to one another to form complexes. Names of complexes can be formed by concatenating the names of the elementary species with.. The complexes are treated internally as a completely separate species. That is, A, B and A.B are completely separate species and will be reported as such, rather than have A.B contribute to the count of the number of As and Bs. 5.5 Initial Populations The initial populations of species are given by the population identifier. Populations are usually specified as speciesname = population. Possible uses of the population identifier are illustrated below: Data Description A = 5 Set the initial concentration of A to 5. A += 3 Add 3 to the previously declared initial population of A. A -= 2 Subtract 2 from the previously declared initial population of A. Initial populations cannot be negative. A Equivalent to A += 0. If a species name is encountered that has not yet been seen before in the input, its initial concentration is initialized to 0. This applies to species in reactions and the waiting list as well as the += and -= forms of population. By default, all species are output in the readout file. This can produce a lot of data that may not be necessary. Readout of a species can be explicitly enabled or disabled by placing a! or # before the species name, respectively. Readout of newly-discovered species, from any identifier that adds molecular species (population, reaction and queue) can be enabled or disabled by using the molecule readout identifier with data show or hide. When using the NCellsGen module, the initial populations can vary among cells by adding a call to Lua s math.random here. For example, to initialize some substance X between 10 and 30, from a uniform distribution, write: 4
5 population X = math.random (10,30); 5.6 Reactions Reactions are input with the reaction identifier in an intuitive and human-readable format: substrate-list --[rate-constant]--> product-list For example, the reaction A + B C with a rate constant of 2 can be input as: A + B --[2]--> C Zero-order reactions (no substrates) and decay reactions (no products) can be input by simply omitting the substrate-list or the product-list. For example: --[2]--> A A --[2]--> If more than one of a substrate is consumed in the reaction, such as the reaction 2Y Z, then the number should be prepended to the substrate name, such as: 2Y --[2]--> Z Additionally, the reaction rate is Lua-enabled (see section 6), allowing any formula that can be interpreted by Lua to be inserted, such as: A + B --[math.sqrt(5) math.random(4)]--> C This could be used to adjust the reactions rate constants to variations in the system properties (e.g. volume or temperature). This can also be used to introduce variation in the properties of the cells created by NCellsGen. Lua can also be inserted to calculate the number of reactants consumed or products produced by a reaction by placing the Lua code in square brackets before the species name. This is mostly useful to set these values in global parameters. For example, the reaction A + B --[c react]--> [n]c allows easy manipulation of both the reaction s rate constant and number of C s produced from a central Lua block. 5.7 Delayed Reactions By default, products of a reaction are released immediately. However, in several cases it has been shown that the reaction is better modelled by delaying the release by a certain amount of time. The delay for a given product is specified by placing the delay time in parentheses after the product. This can be used to efficiently simulate multi-step reactions without including all intermediate reactions. For example, the time it takes for a gene to be transcribed by an RNA Polymerase, spliced, translated by a Ribosome, and folded can be simplified into: Promoter + RNAp --[k]--> Promoter(2) + RNAp(20) + P(2000) In this example, the time it takes for the RNAp to transcribe the entire gene and be available to bind to another gene is 20 seconds, while it only takes 2 seconds for the gene s Promoter to be released so other RNAps can bind and begin transcribing. The end product of the reaction is a protein and is released long after the RNAp finishes transcribing the gene to account for the time it needs to be translated and folded. If nothing but a number is specified as the delay, the time delay will be assumed to be constant. However, many delays are modelled best when sampled from a distribution, especially when modeling multi-step reactions. For example, when transcribing a gene, RNA Polymerase molecules do not always move at a constant speed across the gene, resulting in a distribution of time delays for the release of the proteins. In this case, the time delay for a given product can be generated from several available distributions. The distribution is specified in the form (distribution:parameters). The distribution can be any of the following: 5
6 Name Parameters Description delta, const d A constant delay of d gaussian, gaus, normal µ, σ A normal distribution with mean µ and standard deviation σ exponential, exp λ An exponential distribution with a mean of 1/λ For example, the reaction A + B C where C s release delay follows a Gaussian distribution with mean 10 and standard deviation 2 would be input as: A + B --[k]--> C(gaussian:10,2) Additionally, distributions that allow negative delays to be generated (such as the gaussian distribution) will have their negative part truncated, so that negative delays are impossible. Since the time delay parameters are Lua enabled, they can, similar to rates constants, be defined by global variables as well as vary from cell to cell. 5.8 Virtual Substrates and Reaction Rates Sometimes, especially when building a reaction system from a system of ODEs, the propensities of the reactions must vary by some non-linear function of the populations of some other species. For example, when the production of one species is suppressed by another. CellLine like its predecessor, SGN Sim calculates a reaction s propensity from the following formula: a i = c i s S i r s ([s]) (1) Where c i is the reaction s stochastic rate constant, S i is the set of substrates of the reaction, and r s is the substrate s rate function (described below). That is, the propensity of the reaction is the product of the reaction s constant rate and all the rates calculated from the substrates rate functions. A rate function can be assigned to a substrate in a manner similar to assigning a delay distribution to a product. It is specified in the form (function:parameters). Unlike delay distributions, there is no default rate function if no function is given. If a parameter is omitted, it is assumed to be 1. Available rate functions (where a and b are the parameters, and X is the population of the substrate) are: Name Function a j=1 gilh const a linear ax square, sqr ax 2 cube ax 3 pow bx a hill X b a b +X b a b a b +X b X j+1 j invhill max b max(a, X) min b min(a, X) If no special rate function is given, substrates are assumed to have the gilh rate where a is the number of molecules of the substrate that are consumed. gilh corresponds to the calculation of H given on p. 5 of [1]. Sometimes, it is necessary to not have any molecules of a given substrate consumed by a reaction. In this case, the substrate becomes a catalyst or inhibitor of the reaction without itself being affected by it, and becomes a virtual substrate. This scenario can be input either by specifying 0 as the number of molecules consumed, or by placing a * before the species name. Virtual substrates that are not explicitly given a rate are assumed to have the linear rate function. For example, the ODEs representing a toggle switch with cooperative binding, 6
7 can be represented with the following reactions: *B(invhill:1,c) --[k p ]--> A *A(invhill:1,c) --[k p ]--> B A --[k d ]--> B --[k d ]--> d[a] k p = dt 1 + [B] c k d[a] (2) d[b] k p = dt 1 + [A] c k d[b] (3) While the ability to override the usual rate functions greatly enhances the flexibility of the simulator, it is necessary to use them with caution so that populations do not become negative. In this case, the simulation behavior is undefined. 6 Lua Most numeric fields allow for Lua code to be inserted. Lua code encountered where a number is expected is compiled as though return was prepended to it. All Lua code is compiled and run immediately when it is encountered, in the order it appears. A block of generic Lua code can be executed using the lua identifier. It is recommended that the data block be delimited with!{ }! so that semicolons and braces can be used inside the code block without having the block split into several chunks and compiled separately. A global function parse is also provided to feed a string from Lua into the parsing system. The function takes one or two parameters. When two parameters are given, the first is interpreted as a string containing the identifier, and the second is the data. A single string passed to the function will be parsed as though it was from a file. Documentation for the Lua 5.1 language can be found at Generating random numbers from a Poisson distribution We have included a random number generator that follows a Poisson distribution, that is implemented in a file called poisson.lua (provided by us). It can be called at runtime from within a Lua block. Here, we use this to create DNA double strand breaks. To do this, first poisson.lua file must be in the same directory as the reactions file, and it must be included by executing require ("poisson");. Poisson.lua uses math.random to produce uniform random numbers. Therefore math.randomseed should be used to seed this generator. Poisson.lua defines a single function called poisson. This function takes a single parameter: λ, the desired mean of the Poisson distribution. 7 Output The simulation will produce an output file that is tab-delimited spreadsheet in text format. Each row corresponds to a sampling of the system at a given time and contains the sample time (Time), number of times the system time was updated (Steps), number of reactions completed (Num Reactions), number of elements in the waiting list (WaitList Length) and the populations of all the molecular species at that time. Species are output in the order that they are discovered in the input. The following columns will invariantly appear in the output file (X i represent the various species): Time, Steps, Num Reactions, WaitList Length, X 1,..., X n The fourier spectrum of the resulting time series can also be obtained as mentioned above. The Fourier.xls is a tab delimited text file, which can be viewed in EXCEL. 7
8 8 Runtime Complexity Module DynSim.exe algorithm s best-case runtime is Ω(R logr) and worst-case runtime is O(R 2 ), where R is the number of reactions in the system. The worst case occurs when the most commonly used substrate is found in O(R) reactions. Depending on the number of cells one desires to model with NCellsGen, this time grows linearly with N. CellLineGen grows exponentially with the number of generations. References [1] D.T. Gillespie. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. [2] D.T. Gillespie. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys., 22: , [3] M. Matsumoto and T. Nishimura. Mersenne twister: A 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Transactions on Modeling and Computer Simulation, 8(1):3 30, January [4] Andre S. Ribeiro and Jason Lloyd-Price. Sgn sim, a stochastic gene networks simulator. Bioinformatics, /bioinformatics/btm004, [5] M.R. Roussel and Rui Zhu. Validation of an algorithm for delay stochastic simulation of transcription and translation in prokaryotic gene expression. Phys Biol., 3(4):274 84,
Supplement to Call Centers with Delay Information: Models and Insights
 Supplement to Call Centers with Delay Information: Models and Insights Oualid Jouini 1 Zeynep Akşin 2 Yves Dallery 1 1 Laboratoire Genie Industriel, Ecole Centrale Paris, Grande Voie des Vignes, 92290
Supplement to Call Centers with Delay Information: Models and Insights Oualid Jouini 1 Zeynep Akşin 2 Yves Dallery 1 1 Laboratoire Genie Industriel, Ecole Centrale Paris, Grande Voie des Vignes, 92290
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION. Professor Bharat Patel Office: Science 2, 2.36 Email: [email protected].
 Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: [email protected] What is Gene Expression & Gene Regulation? 1. Gene Expression
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: [email protected] What is Gene Expression & Gene Regulation? 1. Gene Expression
Visual Logic Instructions and Assignments
 Visual Logic Instructions and Assignments Visual Logic can be installed from the CD that accompanies our textbook. It is a nifty tool for creating program flowcharts, but that is only half of the story.
Visual Logic Instructions and Assignments Visual Logic can be installed from the CD that accompanies our textbook. It is a nifty tool for creating program flowcharts, but that is only half of the story.
ESPResSo Summer School 2012
 ESPResSo Summer School 2012 Introduction to Tcl Pedro A. Sánchez Institute for Computational Physics Allmandring 3 D-70569 Stuttgart Germany http://www.icp.uni-stuttgart.de 2/26 Outline History, Characteristics,
ESPResSo Summer School 2012 Introduction to Tcl Pedro A. Sánchez Institute for Computational Physics Allmandring 3 D-70569 Stuttgart Germany http://www.icp.uni-stuttgart.de 2/26 Outline History, Characteristics,
SPSS for Windows importing and exporting data
 Guide 86 Version 3.0 SPSS for Windows importing and exporting data This document outlines the procedures to follow if you want to transfer data from a Windows application like Word 2002 (Office XP), Excel
Guide 86 Version 3.0 SPSS for Windows importing and exporting data This document outlines the procedures to follow if you want to transfer data from a Windows application like Word 2002 (Office XP), Excel
Introduction to Statistical Computing in Microsoft Excel By Hector D. Flores; [email protected], and Dr. J.A. Dobelman
 Introduction to Statistical Computing in Microsoft Excel By Hector D. Flores; [email protected], and Dr. J.A. Dobelman Statistics lab will be mainly focused on applying what you have learned in class with
Introduction to Statistical Computing in Microsoft Excel By Hector D. Flores; [email protected], and Dr. J.A. Dobelman Statistics lab will be mainly focused on applying what you have learned in class with
Arena 9.0 Basic Modules based on Arena Online Help
 Arena 9.0 Basic Modules based on Arena Online Help Create This module is intended as the starting point for entities in a simulation model. Entities are created using a schedule or based on a time between
Arena 9.0 Basic Modules based on Arena Online Help Create This module is intended as the starting point for entities in a simulation model. Entities are created using a schedule or based on a time between
Chapter 3 RANDOM VARIATE GENERATION
 Chapter 3 RANDOM VARIATE GENERATION In order to do a Monte Carlo simulation either by hand or by computer, techniques must be developed for generating values of random variables having known distributions.
Chapter 3 RANDOM VARIATE GENERATION In order to do a Monte Carlo simulation either by hand or by computer, techniques must be developed for generating values of random variables having known distributions.
Introduction to Matlab
 Introduction to Matlab Social Science Research Lab American University, Washington, D.C. Web. www.american.edu/provost/ctrl/pclabs.cfm Tel. x3862 Email. [email protected] Course Objective This course provides
Introduction to Matlab Social Science Research Lab American University, Washington, D.C. Web. www.american.edu/provost/ctrl/pclabs.cfm Tel. x3862 Email. [email protected] Course Objective This course provides
From DNA to Protein. Proteins. Chapter 13. Prokaryotes and Eukaryotes. The Path From Genes to Proteins. All proteins consist of polypeptide chains
 Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Proteins From DNA to Protein Chapter 13 All proteins consist of polypeptide chains A linear sequence of amino acids Each chain corresponds to the nucleotide base sequence of a gene The Path From Genes
Name: Class: Date: 9. The compiler ignores all comments they are there strictly for the convenience of anyone reading the program.
 Name: Class: Date: Exam #1 - Prep True/False Indicate whether the statement is true or false. 1. Programming is the process of writing a computer program in a language that the computer can respond to
Name: Class: Date: Exam #1 - Prep True/False Indicate whether the statement is true or false. 1. Programming is the process of writing a computer program in a language that the computer can respond to
A Mathematical Model of a Synthetically Constructed Genetic Toggle Switch
 BENG 221 Mathematical Methods in Bioengineering Project Report A Mathematical Model of a Synthetically Constructed Genetic Toggle Switch Nick Csicsery & Ricky O Laughlin October 15, 2013 1 TABLE OF CONTENTS
BENG 221 Mathematical Methods in Bioengineering Project Report A Mathematical Model of a Synthetically Constructed Genetic Toggle Switch Nick Csicsery & Ricky O Laughlin October 15, 2013 1 TABLE OF CONTENTS
6 Scalar, Stochastic, Discrete Dynamic Systems
 47 6 Scalar, Stochastic, Discrete Dynamic Systems Consider modeling a population of sand-hill cranes in year n by the first-order, deterministic recurrence equation y(n + 1) = Ry(n) where R = 1 + r = 1
47 6 Scalar, Stochastic, Discrete Dynamic Systems Consider modeling a population of sand-hill cranes in year n by the first-order, deterministic recurrence equation y(n + 1) = Ry(n) where R = 1 + r = 1
Eventia Log Parsing Editor 1.0 Administration Guide
 Eventia Log Parsing Editor 1.0 Administration Guide Revised: November 28, 2007 In This Document Overview page 2 Installation and Supported Platforms page 4 Menus and Main Window page 5 Creating Parsing
Eventia Log Parsing Editor 1.0 Administration Guide Revised: November 28, 2007 In This Document Overview page 2 Installation and Supported Platforms page 4 Menus and Main Window page 5 Creating Parsing
E x c e l 2 0 1 0 : Data Analysis Tools Student Manual
 E x c e l 2 0 1 0 : Data Analysis Tools Student Manual Excel 2010: Data Analysis Tools Chief Executive Officer, Axzo Press: Series Designer and COO: Vice President, Operations: Director of Publishing Systems
E x c e l 2 0 1 0 : Data Analysis Tools Student Manual Excel 2010: Data Analysis Tools Chief Executive Officer, Axzo Press: Series Designer and COO: Vice President, Operations: Director of Publishing Systems
GENE REGULATION. Teacher Packet
 AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
AP * BIOLOGY GENE REGULATION Teacher Packet AP* is a trademark of the College Entrance Examination Board. The College Entrance Examination Board was not involved in the production of this material. Pictures
( ) FACTORING. x In this polynomial the only variable in common to all is x.
 FACTORING Factoring is similar to breaking up a number into its multiples. For example, 10=5*. The multiples are 5 and. In a polynomial it is the same way, however, the procedure is somewhat more complicated
FACTORING Factoring is similar to breaking up a number into its multiples. For example, 10=5*. The multiples are 5 and. In a polynomial it is the same way, however, the procedure is somewhat more complicated
MICROSOFT ACCESS STEP BY STEP GUIDE
 IGCSE ICT SECTION 11 DATA MANIPULATION MICROSOFT ACCESS STEP BY STEP GUIDE Mark Nicholls ICT Lounge P a g e 1 Contents Task 35 details Page 3 Opening a new Database. Page 4 Importing.csv file into the
IGCSE ICT SECTION 11 DATA MANIPULATION MICROSOFT ACCESS STEP BY STEP GUIDE Mark Nicholls ICT Lounge P a g e 1 Contents Task 35 details Page 3 Opening a new Database. Page 4 Importing.csv file into the
7-1. This chapter explains how to set and use Event Log. 7.1. Overview... 7-2 7.2. Event Log Management... 7-2 7.3. Creating a New Event Log...
 7-1 7. Event Log This chapter explains how to set and use Event Log. 7.1. Overview... 7-2 7.2. Event Log Management... 7-2 7.3. Creating a New Event Log... 7-6 7-2 7.1. Overview The following are the basic
7-1 7. Event Log This chapter explains how to set and use Event Log. 7.1. Overview... 7-2 7.2. Event Log Management... 7-2 7.3. Creating a New Event Log... 7-6 7-2 7.1. Overview The following are the basic
Bob Jesberg. Boston, MA April 3, 2014
 DNA, Replication and Transcription Bob Jesberg NSTA Conference Boston, MA April 3, 2014 1 Workshop Agenda Looking at DNA and Forensics The DNA, Replication i and Transcription i Set DNA Ladder The Double
DNA, Replication and Transcription Bob Jesberg NSTA Conference Boston, MA April 3, 2014 1 Workshop Agenda Looking at DNA and Forensics The DNA, Replication i and Transcription i Set DNA Ladder The Double
How To Understand Enzyme Kinetics
 Chapter 12 - Reaction Kinetics In the last chapter we looked at enzyme mechanisms. In this chapter we ll see how enzyme kinetics, i.e., the study of enzyme reaction rates, can be useful in learning more
Chapter 12 - Reaction Kinetics In the last chapter we looked at enzyme mechanisms. In this chapter we ll see how enzyme kinetics, i.e., the study of enzyme reaction rates, can be useful in learning more
Tutorial on Using Excel Solver to Analyze Spin-Lattice Relaxation Time Data
 Tutorial on Using Excel Solver to Analyze Spin-Lattice Relaxation Time Data In the measurement of the Spin-Lattice Relaxation time T 1, a 180 o pulse is followed after a delay time of t with a 90 o pulse,
Tutorial on Using Excel Solver to Analyze Spin-Lattice Relaxation Time Data In the measurement of the Spin-Lattice Relaxation time T 1, a 180 o pulse is followed after a delay time of t with a 90 o pulse,
Translation Study Guide
 Translation Study Guide This study guide is a written version of the material you have seen presented in the replication unit. In translation, the cell uses the genetic information contained in mrna to
Translation Study Guide This study guide is a written version of the material you have seen presented in the replication unit. In translation, the cell uses the genetic information contained in mrna to
Minitab Tutorials for Design and Analysis of Experiments. Table of Contents
 Table of Contents Introduction to Minitab...2 Example 1 One-Way ANOVA...3 Determining Sample Size in One-way ANOVA...8 Example 2 Two-factor Factorial Design...9 Example 3: Randomized Complete Block Design...14
Table of Contents Introduction to Minitab...2 Example 1 One-Way ANOVA...3 Determining Sample Size in One-way ANOVA...8 Example 2 Two-factor Factorial Design...9 Example 3: Randomized Complete Block Design...14
Access Queries (Office 2003)
 Access Queries (Office 2003) Technical Support Services Office of Information Technology, West Virginia University OIT Help Desk 293-4444 x 1 oit.wvu.edu/support/training/classmat/db/ Instructor: Kathy
Access Queries (Office 2003) Technical Support Services Office of Information Technology, West Virginia University OIT Help Desk 293-4444 x 1 oit.wvu.edu/support/training/classmat/db/ Instructor: Kathy
Using EXCEL Solver October, 2000
 Using EXCEL Solver October, 2000 2 The Solver option in EXCEL may be used to solve linear and nonlinear optimization problems. Integer restrictions may be placed on the decision variables. Solver may be
Using EXCEL Solver October, 2000 2 The Solver option in EXCEL may be used to solve linear and nonlinear optimization problems. Integer restrictions may be placed on the decision variables. Solver may be
Scicos is a Scilab toolbox included in the Scilab package. The Scicos editor can be opened by the scicos command
 7 Getting Started 7.1 Construction of a Simple Diagram Scicos contains a graphical editor that can be used to construct block diagram models of dynamical systems. The blocks can come from various palettes
7 Getting Started 7.1 Construction of a Simple Diagram Scicos contains a graphical editor that can be used to construct block diagram models of dynamical systems. The blocks can come from various palettes
Random Variate Generation (Part 3)
 Random Variate Generation (Part 3) Dr.Çağatay ÜNDEĞER Öğretim Görevlisi Bilkent Üniversitesi Bilgisayar Mühendisliği Bölümü &... e-mail : [email protected] [email protected] Bilgisayar Mühendisliği
Random Variate Generation (Part 3) Dr.Çağatay ÜNDEĞER Öğretim Görevlisi Bilkent Üniversitesi Bilgisayar Mühendisliği Bölümü &... e-mail : [email protected] [email protected] Bilgisayar Mühendisliği
AMATH 352 Lecture 3 MATLAB Tutorial Starting MATLAB Entering Variables
 AMATH 352 Lecture 3 MATLAB Tutorial MATLAB (short for MATrix LABoratory) is a very useful piece of software for numerical analysis. It provides an environment for computation and the visualization. Learning
AMATH 352 Lecture 3 MATLAB Tutorial MATLAB (short for MATrix LABoratory) is a very useful piece of software for numerical analysis. It provides an environment for computation and the visualization. Learning
Configuring Simulator 3 for IP
 Configuring Simulator 3 for IP Application Note AN405 Revision v1.0 August 2012 AN405 Configuring Simulator 3 for IP 1 Overview This Application Note is designed to help to configure a Sim 3 to run in
Configuring Simulator 3 for IP Application Note AN405 Revision v1.0 August 2012 AN405 Configuring Simulator 3 for IP 1 Overview This Application Note is designed to help to configure a Sim 3 to run in
Time Clock Import Setup & Use
 Time Clock Import Setup & Use Document # Product Module Category CenterPoint Payroll Processes (How To) This document outlines how to setup and use of the Time Clock Import within CenterPoint Payroll.
Time Clock Import Setup & Use Document # Product Module Category CenterPoint Payroll Processes (How To) This document outlines how to setup and use of the Time Clock Import within CenterPoint Payroll.
Dynamical Systems Analysis II: Evaluating Stability, Eigenvalues
 Dynamical Systems Analysis II: Evaluating Stability, Eigenvalues By Peter Woolf [email protected]) University of Michigan Michigan Chemical Process Dynamics and Controls Open Textbook version 1.0 Creative
Dynamical Systems Analysis II: Evaluating Stability, Eigenvalues By Peter Woolf [email protected]) University of Michigan Michigan Chemical Process Dynamics and Controls Open Textbook version 1.0 Creative
The Kinetics of Enzyme Reactions
 The Kinetics of Enzyme Reactions This activity will introduce you to the chemical kinetics of enzyme-mediated biochemical reactions using an interactive Excel spreadsheet or Excelet. A summarized chemical
The Kinetics of Enzyme Reactions This activity will introduce you to the chemical kinetics of enzyme-mediated biochemical reactions using an interactive Excel spreadsheet or Excelet. A summarized chemical
Exercise 4 Learning Python language fundamentals
 Exercise 4 Learning Python language fundamentals Work with numbers Python can be used as a powerful calculator. Practicing math calculations in Python will help you not only perform these tasks, but also
Exercise 4 Learning Python language fundamentals Work with numbers Python can be used as a powerful calculator. Practicing math calculations in Python will help you not only perform these tasks, but also
VISUAL GUIDE to. RX Scripting. for Roulette Xtreme - System Designer 2.0
 VISUAL GUIDE to RX Scripting for Roulette Xtreme - System Designer 2.0 UX Software - 2009 TABLE OF CONTENTS INTRODUCTION... ii What is this book about?... iii How to use this book... iii Time to start...
VISUAL GUIDE to RX Scripting for Roulette Xtreme - System Designer 2.0 UX Software - 2009 TABLE OF CONTENTS INTRODUCTION... ii What is this book about?... iii How to use this book... iii Time to start...
Data Integrator. Pervasive Software, Inc. 12365-B Riata Trace Parkway Austin, Texas 78727 USA
 Data Integrator Event Management Guide Pervasive Software, Inc. 12365-B Riata Trace Parkway Austin, Texas 78727 USA Telephone: 888.296.5969 or 512.231.6000 Fax: 512.231.6010 Email: [email protected]
Data Integrator Event Management Guide Pervasive Software, Inc. 12365-B Riata Trace Parkway Austin, Texas 78727 USA Telephone: 888.296.5969 or 512.231.6000 Fax: 512.231.6010 Email: [email protected]
What happens to the food we eat? It gets broken down!
 Enzymes Essential Questions: What is an enzyme? How do enzymes work? What are the properties of enzymes? How do they maintain homeostasis for the body? What happens to the food we eat? It gets broken down!
Enzymes Essential Questions: What is an enzyme? How do enzymes work? What are the properties of enzymes? How do they maintain homeostasis for the body? What happens to the food we eat? It gets broken down!
Getting off the ground when creating an RVM test-bench
 Getting off the ground when creating an RVM test-bench Rich Musacchio, Ning Guo Paradigm Works [email protected],[email protected] ABSTRACT RVM compliant environments provide
Getting off the ground when creating an RVM test-bench Rich Musacchio, Ning Guo Paradigm Works [email protected],[email protected] ABSTRACT RVM compliant environments provide
Simply Accounting Intelligence Tips and Tricks Booklet Vol. 1
 Simply Accounting Intelligence Tips and Tricks Booklet Vol. 1 1 Contents Accessing the SAI reports... 3 Running, Copying and Pasting reports... 4 Creating and linking a report... 5 Auto e-mailing reports...
Simply Accounting Intelligence Tips and Tricks Booklet Vol. 1 1 Contents Accessing the SAI reports... 3 Running, Copying and Pasting reports... 4 Creating and linking a report... 5 Auto e-mailing reports...
Math Review. for the Quantitative Reasoning Measure of the GRE revised General Test
 Math Review for the Quantitative Reasoning Measure of the GRE revised General Test www.ets.org Overview This Math Review will familiarize you with the mathematical skills and concepts that are important
Math Review for the Quantitative Reasoning Measure of the GRE revised General Test www.ets.org Overview This Math Review will familiarize you with the mathematical skills and concepts that are important
MAS 500 Intelligence Tips and Tricks Booklet Vol. 1
 MAS 500 Intelligence Tips and Tricks Booklet Vol. 1 1 Contents Accessing the Sage MAS Intelligence Reports... 3 Copying, Pasting and Renaming Reports... 4 To create a new report from an existing report...
MAS 500 Intelligence Tips and Tricks Booklet Vol. 1 1 Contents Accessing the Sage MAS Intelligence Reports... 3 Copying, Pasting and Renaming Reports... 4 To create a new report from an existing report...
Computer Science 210: Data Structures. Searching
 Computer Science 210: Data Structures Searching Searching Given a sequence of elements, and a target element, find whether the target occurs in the sequence Variations: find first occurence; find all occurences
Computer Science 210: Data Structures Searching Searching Given a sequence of elements, and a target element, find whether the target occurs in the sequence Variations: find first occurence; find all occurences
1 Description of The Simpletron
 Simulating The Simpletron Computer 50 points 1 Description of The Simpletron In this assignment you will write a program to simulate a fictional computer that we will call the Simpletron. As its name implies
Simulating The Simpletron Computer 50 points 1 Description of The Simpletron In this assignment you will write a program to simulate a fictional computer that we will call the Simpletron. As its name implies
RNA Structure and folding
 RNA Structure and folding Overview: The main functional biomolecules in cells are polymers DNA, RNA and proteins For RNA and Proteins, the specific sequence of the polymer dictates its final structure
RNA Structure and folding Overview: The main functional biomolecules in cells are polymers DNA, RNA and proteins For RNA and Proteins, the specific sequence of the polymer dictates its final structure
Oracle Database: SQL and PL/SQL Fundamentals
 Oracle University Contact Us: 1.800.529.0165 Oracle Database: SQL and PL/SQL Fundamentals Duration: 5 Days What you will learn This course is designed to deliver the fundamentals of SQL and PL/SQL along
Oracle University Contact Us: 1.800.529.0165 Oracle Database: SQL and PL/SQL Fundamentals Duration: 5 Days What you will learn This course is designed to deliver the fundamentals of SQL and PL/SQL along
Creating Basic Excel Formulas
 Creating Basic Excel Formulas Formulas are equations that perform calculations on values in your worksheet. Depending on how you build a formula in Excel will determine if the answer to your formula automatically
Creating Basic Excel Formulas Formulas are equations that perform calculations on values in your worksheet. Depending on how you build a formula in Excel will determine if the answer to your formula automatically
6.170 Tutorial 3 - Ruby Basics
 6.170 Tutorial 3 - Ruby Basics Prerequisites 1. Have Ruby installed on your computer a. If you use Mac/Linux, Ruby should already be preinstalled on your machine. b. If you have a Windows Machine, you
6.170 Tutorial 3 - Ruby Basics Prerequisites 1. Have Ruby installed on your computer a. If you use Mac/Linux, Ruby should already be preinstalled on your machine. b. If you have a Windows Machine, you
grep, awk and sed three VERY useful command-line utilities Matt Probert, Uni of York grep = global regular expression print
 grep, awk and sed three VERY useful command-line utilities Matt Probert, Uni of York grep = global regular expression print In the simplest terms, grep (global regular expression print) will search input
grep, awk and sed three VERY useful command-line utilities Matt Probert, Uni of York grep = global regular expression print In the simplest terms, grep (global regular expression print) will search input
MBA Jump Start Program
 MBA Jump Start Program Module 2: Mathematics Thomas Gilbert Mathematics Module Online Appendix: Basic Mathematical Concepts 2 1 The Number Spectrum Generally we depict numbers increasing from left to right
MBA Jump Start Program Module 2: Mathematics Thomas Gilbert Mathematics Module Online Appendix: Basic Mathematical Concepts 2 1 The Number Spectrum Generally we depict numbers increasing from left to right
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Exam Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) The JDK command to compile a class in the file Test.java is A) java Test.java B) java
Exam Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) The JDK command to compile a class in the file Test.java is A) java Test.java B) java
Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism )
 Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
Biology 1406 Exam 3 Notes Structure of DNA Ch. 10 Genetic information (DNA) determines structure of proteins DNA RNA proteins cell structure 3.11 3.15 enzymes control cell chemistry ( metabolism ) Proteins
IBM Operational Decision Manager Version 8 Release 5. Getting Started with Business Rules
 IBM Operational Decision Manager Version 8 Release 5 Getting Started with Business Rules Note Before using this information and the product it supports, read the information in Notices on page 43. This
IBM Operational Decision Manager Version 8 Release 5 Getting Started with Business Rules Note Before using this information and the product it supports, read the information in Notices on page 43. This
SMock A Test Platform for the Evaluation of Monitoring Tools
 SMock A Test Platform for the Evaluation of Monitoring Tools User Manual Ruth Mizzi Faculty of ICT University of Malta June 20, 2013 Contents 1 Introduction 3 1.1 The Architecture and Design of SMock................
SMock A Test Platform for the Evaluation of Monitoring Tools User Manual Ruth Mizzi Faculty of ICT University of Malta June 20, 2013 Contents 1 Introduction 3 1.1 The Architecture and Design of SMock................
CHAPTER 6: Continuous Uniform Distribution: 6.1. Definition: The density function of the continuous random variable X on the interval [A, B] is.
![CHAPTER 6: Continuous Uniform Distribution: 6.1. Definition: The density function of the continuous random variable X on the interval [A, B] is. CHAPTER 6: Continuous Uniform Distribution: 6.1. Definition: The density function of the continuous random variable X on the interval [A, B] is.](/thumbs/40/21160284.jpg) Some Continuous Probability Distributions CHAPTER 6: Continuous Uniform Distribution: 6. Definition: The density function of the continuous random variable X on the interval [A, B] is B A A x B f(x; A,
Some Continuous Probability Distributions CHAPTER 6: Continuous Uniform Distribution: 6. Definition: The density function of the continuous random variable X on the interval [A, B] is B A A x B f(x; A,
Exercise 1: Python Language Basics
 Exercise 1: Python Language Basics In this exercise we will cover the basic principles of the Python language. All languages have a standard set of functionality including the ability to comment code,
Exercise 1: Python Language Basics In this exercise we will cover the basic principles of the Python language. All languages have a standard set of functionality including the ability to comment code,
S1 Text. Modeling deterministic single-cell microrna-p53-mdm2 network Figure 2 Figure 2
 S1 Text. Modeling deterministic single-cell microrna-p53-mdm2 network The schematic diagram of the microrna-p53-mdm2 oscillator is illustrated in Figure 2. The interaction scheme among the mrnas and the
S1 Text. Modeling deterministic single-cell microrna-p53-mdm2 network The schematic diagram of the microrna-p53-mdm2 oscillator is illustrated in Figure 2. The interaction scheme among the mrnas and the
MODULE 7: FINANCIAL REPORTING AND ANALYSIS
 MODULE 7: FINANCIAL REPORTING AND ANALYSIS Module Overview Businesses running ERP systems capture lots of data through daily activity. This data, which reflects such things as the organization's sales
MODULE 7: FINANCIAL REPORTING AND ANALYSIS Module Overview Businesses running ERP systems capture lots of data through daily activity. This data, which reflects such things as the organization's sales
Gamma Distribution Fitting
 Chapter 552 Gamma Distribution Fitting Introduction This module fits the gamma probability distributions to a complete or censored set of individual or grouped data values. It outputs various statistics
Chapter 552 Gamma Distribution Fitting Introduction This module fits the gamma probability distributions to a complete or censored set of individual or grouped data values. It outputs various statistics
Welcome to the topic on the Import from Excel utility.
 Welcome to the topic on the Import from Excel utility. 1 In this topic, you will see how to import business partner master data, item master data, and price lists using the Import from Excel utility. 2
Welcome to the topic on the Import from Excel utility. 1 In this topic, you will see how to import business partner master data, item master data, and price lists using the Import from Excel utility. 2
VB.NET Programming Fundamentals
 Chapter 3 Objectives Programming Fundamentals In this chapter, you will: Learn about the programming language Write a module definition Use variables and data types Compute with Write decision-making statements
Chapter 3 Objectives Programming Fundamentals In this chapter, you will: Learn about the programming language Write a module definition Use variables and data types Compute with Write decision-making statements
Appendix: Tutorial Introduction to MATLAB
 Resampling Stats in MATLAB 1 This document is an excerpt from Resampling Stats in MATLAB Daniel T. Kaplan Copyright (c) 1999 by Daniel T. Kaplan, All Rights Reserved This document differs from the published
Resampling Stats in MATLAB 1 This document is an excerpt from Resampling Stats in MATLAB Daniel T. Kaplan Copyright (c) 1999 by Daniel T. Kaplan, All Rights Reserved This document differs from the published
KITES TECHNOLOGY COURSE MODULE (C, C++, DS)
 KITES TECHNOLOGY 360 Degree Solution www.kitestechnology.com/academy.php [email protected] [email protected] Contact: - 8961334776 9433759247 9830639522.NET JAVA WEB DESIGN PHP SQL, PL/SQL
KITES TECHNOLOGY 360 Degree Solution www.kitestechnology.com/academy.php [email protected] [email protected] Contact: - 8961334776 9433759247 9830639522.NET JAVA WEB DESIGN PHP SQL, PL/SQL
Financial Econometrics MFE MATLAB Introduction. Kevin Sheppard University of Oxford
 Financial Econometrics MFE MATLAB Introduction Kevin Sheppard University of Oxford October 21, 2013 2007-2013 Kevin Sheppard 2 Contents Introduction i 1 Getting Started 1 2 Basic Input and Operators 5
Financial Econometrics MFE MATLAB Introduction Kevin Sheppard University of Oxford October 21, 2013 2007-2013 Kevin Sheppard 2 Contents Introduction i 1 Getting Started 1 2 Basic Input and Operators 5
Formulas & Functions in Microsoft Excel
 Formulas & Functions in Microsoft Excel Theresa A Scott, MS Biostatistician II Department of Biostatistics Vanderbilt University [email protected] Table of Contents 1 Introduction 1 1.1 Using
Formulas & Functions in Microsoft Excel Theresa A Scott, MS Biostatistician II Department of Biostatistics Vanderbilt University [email protected] Table of Contents 1 Introduction 1 1.1 Using
Building and Using Spreadsheet Decision Models
 Chapter 9 Building and Using Spreadsheet Decision Models Models A model is an abstraction or representation of a real system, idea, or object. Models could be pictures, spreadsheets, or mathematical relationships
Chapter 9 Building and Using Spreadsheet Decision Models Models A model is an abstraction or representation of a real system, idea, or object. Models could be pictures, spreadsheets, or mathematical relationships
Chemical reactions allow living things to grow, develop, reproduce, and adapt.
 Section 2: Chemical reactions allow living things to grow, develop, reproduce, and adapt. K What I Know W What I Want to Find Out L What I Learned Essential Questions What are the parts of a chemical reaction?
Section 2: Chemical reactions allow living things to grow, develop, reproduce, and adapt. K What I Know W What I Want to Find Out L What I Learned Essential Questions What are the parts of a chemical reaction?
Translating Stochastic CLS into Maude
 Translating Stochastic CLS into Maude (ONGOING WORK) Thomas Anung Basuki 1 Antonio Cerone 1 Paolo Milazzo 2 1. Int. Institute for Software Technology, United Nations Univeristy, Macau SAR, China 2. Dipartimento
Translating Stochastic CLS into Maude (ONGOING WORK) Thomas Anung Basuki 1 Antonio Cerone 1 Paolo Milazzo 2 1. Int. Institute for Software Technology, United Nations Univeristy, Macau SAR, China 2. Dipartimento
Reaction diffusion systems and pattern formation
 Chapter 5 Reaction diffusion systems and pattern formation 5.1 Reaction diffusion systems from biology In ecological problems, different species interact with each other, and in chemical reactions, different
Chapter 5 Reaction diffusion systems and pattern formation 5.1 Reaction diffusion systems from biology In ecological problems, different species interact with each other, and in chemical reactions, different
Retrieving Data Using the SQL SELECT Statement. Copyright 2006, Oracle. All rights reserved.
 Retrieving Data Using the SQL SELECT Statement Objectives After completing this lesson, you should be able to do the following: List the capabilities of SQL SELECT statements Execute a basic SELECT statement
Retrieving Data Using the SQL SELECT Statement Objectives After completing this lesson, you should be able to do the following: List the capabilities of SQL SELECT statements Execute a basic SELECT statement
Modeling with Python
 H Modeling with Python In this appendix a brief description of the Python programming language will be given plus a brief introduction to the Antimony reaction network format and libroadrunner. Python
H Modeling with Python In this appendix a brief description of the Python programming language will be given plus a brief introduction to the Antimony reaction network format and libroadrunner. Python
AP Computer Science Java Subset
 APPENDIX A AP Computer Science Java Subset The AP Java subset is intended to outline the features of Java that may appear on the AP Computer Science A Exam. The AP Java subset is NOT intended as an overall
APPENDIX A AP Computer Science Java Subset The AP Java subset is intended to outline the features of Java that may appear on the AP Computer Science A Exam. The AP Java subset is NOT intended as an overall
Sources: On the Web: Slides will be available on:
 C programming Introduction The basics of algorithms Structure of a C code, compilation step Constant, variable type, variable scope Expression and operators: assignment, arithmetic operators, comparison,
C programming Introduction The basics of algorithms Structure of a C code, compilation step Constant, variable type, variable scope Expression and operators: assignment, arithmetic operators, comparison,
Assignment 2: Option Pricing and the Black-Scholes formula The University of British Columbia Science One CS 2015-2016 Instructor: Michael Gelbart
 Assignment 2: Option Pricing and the Black-Scholes formula The University of British Columbia Science One CS 2015-2016 Instructor: Michael Gelbart Overview Due Thursday, November 12th at 11:59pm Last updated
Assignment 2: Option Pricing and the Black-Scholes formula The University of British Columbia Science One CS 2015-2016 Instructor: Michael Gelbart Overview Due Thursday, November 12th at 11:59pm Last updated
Macros allow you to integrate existing Excel reports with a new information system
 Macro Magic Macros allow you to integrate existing Excel reports with a new information system By Rick Collard Many water and wastewater professionals use Microsoft Excel extensively, producing reports
Macro Magic Macros allow you to integrate existing Excel reports with a new information system By Rick Collard Many water and wastewater professionals use Microsoft Excel extensively, producing reports
Real Time Scheduling Basic Concepts. Radek Pelánek
 Real Time Scheduling Basic Concepts Radek Pelánek Basic Elements Model of RT System abstraction focus only on timing constraints idealization (e.g., zero switching time) Basic Elements Basic Notions task
Real Time Scheduling Basic Concepts Radek Pelánek Basic Elements Model of RT System abstraction focus only on timing constraints idealization (e.g., zero switching time) Basic Elements Basic Notions task
To be able to describe polypeptide synthesis including transcription and splicing
 Thursday 8th March COPY LO: To be able to describe polypeptide synthesis including transcription and splicing Starter Explain the difference between transcription and translation BATS Describe and explain
Thursday 8th March COPY LO: To be able to describe polypeptide synthesis including transcription and splicing Starter Explain the difference between transcription and translation BATS Describe and explain
Object Oriented Software Design
 Object Oriented Software Design Introduction to Java - II Giuseppe Lipari http://retis.sssup.it/~lipari Scuola Superiore Sant Anna Pisa September 14, 2011 G. Lipari (Scuola Superiore Sant Anna) Introduction
Object Oriented Software Design Introduction to Java - II Giuseppe Lipari http://retis.sssup.it/~lipari Scuola Superiore Sant Anna Pisa September 14, 2011 G. Lipari (Scuola Superiore Sant Anna) Introduction
Importing Data from a Dat or Text File into SPSS
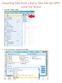 Importing Data from a Dat or Text File into SPSS 1. Select File Open Data (Using Text Wizard) 2. Under Files of type, choose Text (*.txt,*.dat) 3. Select the file you want to import. The dat or text file
Importing Data from a Dat or Text File into SPSS 1. Select File Open Data (Using Text Wizard) 2. Under Files of type, choose Text (*.txt,*.dat) 3. Select the file you want to import. The dat or text file
Chapter 5 Functions. Introducing Functions
 Chapter 5 Functions 1 Introducing Functions A function is a collection of statements that are grouped together to perform an operation Define a function Invoke a funciton return value type method name
Chapter 5 Functions 1 Introducing Functions A function is a collection of statements that are grouped together to perform an operation Define a function Invoke a funciton return value type method name
Confidence Intervals for the Difference Between Two Means
 Chapter 47 Confidence Intervals for the Difference Between Two Means Introduction This procedure calculates the sample size necessary to achieve a specified distance from the difference in sample means
Chapter 47 Confidence Intervals for the Difference Between Two Means Introduction This procedure calculates the sample size necessary to achieve a specified distance from the difference in sample means
Activity 7.21 Transcription factors
 Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
agucacaaacgcu agugcuaguuua uaugcagucuua
 RNA Secondary Structure Prediction: The Co-transcriptional effect on RNA folding agucacaaacgcu agugcuaguuua uaugcagucuua By Conrad Godfrey Abstract RNA secondary structure prediction is an area of bioinformatics
RNA Secondary Structure Prediction: The Co-transcriptional effect on RNA folding agucacaaacgcu agugcuaguuua uaugcagucuua By Conrad Godfrey Abstract RNA secondary structure prediction is an area of bioinformatics
Data Tool Platform SQL Development Tools
 Data Tool Platform SQL Development Tools ekapner Contents Setting SQL Development Preferences...5 Execution Plan View Options Preferences...5 General Preferences...5 Label Decorations Preferences...6
Data Tool Platform SQL Development Tools ekapner Contents Setting SQL Development Preferences...5 Execution Plan View Options Preferences...5 General Preferences...5 Label Decorations Preferences...6
Perl in a nutshell. First CGI Script and Perl. Creating a Link to a Script. print Function. Parsing Data 4/27/2009. First CGI Script and Perl
 First CGI Script and Perl Perl in a nutshell Prof. Rasley shebang line tells the operating system where the Perl interpreter is located necessary on UNIX comment line ignored by the Perl interpreter End
First CGI Script and Perl Perl in a nutshell Prof. Rasley shebang line tells the operating system where the Perl interpreter is located necessary on UNIX comment line ignored by the Perl interpreter End
Computer Programming C++ Classes and Objects 15 th Lecture
 Computer Programming C++ Classes and Objects 15 th Lecture 엄현상 (Eom, Hyeonsang) School of Computer Science and Engineering Seoul National University Copyrights 2013 Eom, Hyeonsang All Rights Reserved Outline
Computer Programming C++ Classes and Objects 15 th Lecture 엄현상 (Eom, Hyeonsang) School of Computer Science and Engineering Seoul National University Copyrights 2013 Eom, Hyeonsang All Rights Reserved Outline
Title. Syntax. stata.com. odbc Load, write, or view data from ODBC sources. List ODBC sources to which Stata can connect odbc list
 Title stata.com odbc Load, write, or view data from ODBC sources Syntax Menu Description Options Remarks and examples Also see Syntax List ODBC sources to which Stata can connect odbc list Retrieve available
Title stata.com odbc Load, write, or view data from ODBC sources Syntax Menu Description Options Remarks and examples Also see Syntax List ODBC sources to which Stata can connect odbc list Retrieve available
Enzymes and Metabolism
 Enzymes and Metabolism Enzymes and Metabolism Metabolism: Exergonic and Endergonic Reactions Chemical Reactions: Activation Every chemical reaction involves bond breaking and bond forming A chemical reaction
Enzymes and Metabolism Enzymes and Metabolism Metabolism: Exergonic and Endergonic Reactions Chemical Reactions: Activation Every chemical reaction involves bond breaking and bond forming A chemical reaction
Engineering Problem Solving and Excel. EGN 1006 Introduction to Engineering
 Engineering Problem Solving and Excel EGN 1006 Introduction to Engineering Mathematical Solution Procedures Commonly Used in Engineering Analysis Data Analysis Techniques (Statistics) Curve Fitting techniques
Engineering Problem Solving and Excel EGN 1006 Introduction to Engineering Mathematical Solution Procedures Commonly Used in Engineering Analysis Data Analysis Techniques (Statistics) Curve Fitting techniques
Industry Environment and Concepts for Forecasting 1
 Table of Contents Industry Environment and Concepts for Forecasting 1 Forecasting Methods Overview...2 Multilevel Forecasting...3 Demand Forecasting...4 Integrating Information...5 Simplifying the Forecast...6
Table of Contents Industry Environment and Concepts for Forecasting 1 Forecasting Methods Overview...2 Multilevel Forecasting...3 Demand Forecasting...4 Integrating Information...5 Simplifying the Forecast...6
Oracle Database: SQL and PL/SQL Fundamentals NEW
 Oracle University Contact Us: + 38516306373 Oracle Database: SQL and PL/SQL Fundamentals NEW Duration: 5 Days What you will learn This Oracle Database: SQL and PL/SQL Fundamentals training delivers the
Oracle University Contact Us: + 38516306373 Oracle Database: SQL and PL/SQL Fundamentals NEW Duration: 5 Days What you will learn This Oracle Database: SQL and PL/SQL Fundamentals training delivers the
Excel 2003: Ringtones Task
 Excel 2003: Ringtones Task 1. Open up a blank spreadsheet 2. Save the spreadsheet to your area and call it Ringtones.xls 3. Add the data as shown here, making sure you keep to the cells as shown Make sure
Excel 2003: Ringtones Task 1. Open up a blank spreadsheet 2. Save the spreadsheet to your area and call it Ringtones.xls 3. Add the data as shown here, making sure you keep to the cells as shown Make sure
How To Use Excel With A Calculator
 Functions & Data Analysis Tools Academic Computing Services www.ku.edu/acs Abstract: This workshop focuses on the functions and data analysis tools of Microsoft Excel. Topics included are the function
Functions & Data Analysis Tools Academic Computing Services www.ku.edu/acs Abstract: This workshop focuses on the functions and data analysis tools of Microsoft Excel. Topics included are the function
Reaction Rates and Chemical Kinetics. Factors Affecting Reaction Rate [O 2. CHAPTER 13 Page 1
 CHAPTER 13 Page 1 Reaction Rates and Chemical Kinetics Several factors affect the rate at which a reaction occurs. Some reactions are instantaneous while others are extremely slow. Whether a commercial
CHAPTER 13 Page 1 Reaction Rates and Chemical Kinetics Several factors affect the rate at which a reaction occurs. Some reactions are instantaneous while others are extremely slow. Whether a commercial
CS 2112 Spring 2014. 0 Instructions. Assignment 3 Data Structures and Web Filtering. 0.1 Grading. 0.2 Partners. 0.3 Restrictions
 CS 2112 Spring 2014 Assignment 3 Data Structures and Web Filtering Due: March 4, 2014 11:59 PM Implementing spam blacklists and web filters requires matching candidate domain names and URLs very rapidly
CS 2112 Spring 2014 Assignment 3 Data Structures and Web Filtering Due: March 4, 2014 11:59 PM Implementing spam blacklists and web filters requires matching candidate domain names and URLs very rapidly
Performance Workload Design
 Performance Workload Design The goal of this paper is to show the basic principles involved in designing a workload for performance and scalability testing. We will understand how to achieve these principles
Performance Workload Design The goal of this paper is to show the basic principles involved in designing a workload for performance and scalability testing. We will understand how to achieve these principles
Search and Replace in SAS Data Sets thru GUI
 Search and Replace in SAS Data Sets thru GUI Edmond Cheng, Bureau of Labor Statistics, Washington, DC ABSTRACT In managing data with SAS /BASE software, performing a search and replace is not a straight
Search and Replace in SAS Data Sets thru GUI Edmond Cheng, Bureau of Labor Statistics, Washington, DC ABSTRACT In managing data with SAS /BASE software, performing a search and replace is not a straight
