Placenta stem/stromal cell-derived extracellular vesicles for potential use in lung repair
|
|
|
- Kathleen O’Neal’
- 2 years ago
- Views:
Transcription
1 Placenta stem/stromal cell-derived extracellular vesicles for potential use in lung repair Sally Yunsun Kim 1,2, Mugdha V. Joglekar 3, Anandwardhan A. Hardikar 3, Thanh Huyen Phan 1,2, Dipesh Khanal 1,2, Priyanka Tharkar 1,2, Christina Limantoro 1,2, Jancy Johnson 4, Bill Kalionis 4 and Wojciech Chrzanowski 1,2 1 The University of Sydney, Faculty of Medicine and Health, Sydney Pharmacy School, New South Wales, Australia. 2 The University of Sydney Nano Institute, New South Wales, Australia. 3 Islet Biology and Diabetes Group, NHMRC CTC, The University of Sydney, 92 Parramatta Road, Camperdown, New South Wales, Australia 4 Department of Maternal-Fetal Medicine Pregnancy Research Centre and University of Melbourne. Department of Obstetrics and Gynaecology, Royal Women s Hospital, Parkville, Victoria, Australia. Corresponding author: Associate Professor Wojciech Chrzanowski The University of Sydney School of Pharmacy Faculty of Medicine and Health Rm 500, Badham Building A16, The University of Sydney NSW 2006 Australia Ph: Fax: This is the author manuscript accepted for publication and has undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: /pmic
2 List of Abbreviations AFM, atomic force microscopy; AFM-IR, atomic force microscope infrared-spectroscopy; CMSC29, chorionic mesenchymal stromal cell line; COPD, chronic obstructive pulmonary disease; DMSC23, decidual mesenchymal stromal cell line; EVs, extracellular vesicles; HBSS(-), Hanks' Balanced Salt Solution; LPS, lipopolysaccharide; MSC, mesenchymal stem/stromal cell; NTA, nanoparticle tracking analysis; SEM, scanning electron microscopy; SFM, serum-free media; TEM, transmission electron microscopy; TRPS, tunable resistive pulse sensing. Keywords: Extracellular vesicles, placenta mesenchymal stromal cells, regenerative potential, lung diseases Total number of words: 7748 Abstract Many acute and chronic lung injuries are incurable and rank as the fourth leading cause of death globally. While stem cell treatment for lung injuries is a promising approach, there is growing evidence that the therapeutic efficacy of stem cells originates from secreted extracellular vesicles (EVs). Consequently, EVs are emerging as next-generation therapeutics. While EVs are extensively researched for diagnostic applications, their therapeutic potential to promote tissue repair is not fully elucidated. By housing and delivering tissue-repairing cargo, EVs refine the cellular microenvironment, modulate inflammation, and ultimately repair injury. Here, we present the potential use of EVs derived from two placental mesenchymal stem/stromal cell (MSC) lines; a chorionic MSC line (CMSC29) and a decidual MSC cell line (DMSC23) for applications in lung diseases. Functional analyses using in vitro models of injury demonstrated that these EVs have a role
3 in ameliorating injuries caused to lung cells. We also showed that EVs promote repair of lung epithelial cells. This study is fundamental to advancing the field of EVs and to unlock the full potential of EVs in regenerative medicine. Statement of significance of the study Current studies provide strong evidence that the therapeutic benefit of stem cells is mainly through the paracrine actions of extracellular vesicles (EVs). Here we demonstrate that EVs derived from placental mesenchymal stem/stromal cell lines ameliorate inflammation and stimulate tissue repair. Direct delivery of EVs to lungs is a promising therapeutic approach to drive lung tissue regeneration after injury. In this study, we present the use of single vesicle characterisation and microrna profiling for identifying types of EVs that contain the appropriate molecular composition for use in lung injuries. We provided an extensive approach to EV characterisation using our highly novel resonance enhanced atomic force microscopy method, which exceeds recommended minimum reporting requirements for EVs, and which enables single vesicle structural and molecular analysis. The use of our approach allows for unequivocal analysis of vesicle heterogeneity and identification of nanoscale differences in EV composition. Both parameters are pivotal for identifying and isolating - EV subtypes that have potential use in ameliorating diseases. Together with microrna profiling, these data are of fundamental importance when designing rational approaches that employ EVs for lung tissue repair. 1. Introduction Acute and chronic respiratory diseases remain incurable and new treatment strategies are urgently required [1-3]. Each year, four million people die prematurely due to respiratory diseases [3]. Acute respiratory distress syndrome, pulmonary fibrosis and chronic obstructive pulmonary disorder (COPD) are amongst the most frequent causes of deaths globally, despite
4 ongoing advances in supportive care [1, 4]. Current strategies for enhancing tissue regeneration are only supportive and do not actively promote tissue repair. Stem cell-based therapy for tissue regeneration emerged as a promising option not only for controlling symptoms of lung diseases but also for its potential as a curative treatment regimen [5, 6]. The efficacy of mesenchymal stem cells (MSCs) in alleviating inflammation, a major hallmark of treatment for many lung diseases, is confirmed in numerous studies [6, 7]. Moreover, cell-free regenerative therapy using MSC-derived extracellular vesicles (EVs) is emerging as a more promising alternative due to the lower risk of an immunogenic response, preservation of biochemical activity upon storage [8] and higher stability in hostile environments compared with MSCs [9]. MSCs respond to their changing microenvironment by selectively packaging EVs with heterogeneous compositions of proteins, lipids, and nucleic acids [10]. The transfer of specific protein and microrna (mirna) cargo to the injured cell is widely accepted as an essential driver of the regenerative process, through its impact on multiple downstream biological pathways, and its ability to regulate the host immune response [11]. We investigated the potential of placenta-derived mesenchymal stem/stromal cell-derived EVs for regenerative medicine. Human placenta is an abundant source of stem cells, yet it is usually discarded at birth. Placenta-derived stem cells grow fast and more robustly compared to stem cells from other tissues while having the lowest osteogenic potential [12]. Our focus was on developing therapeutic EVs for direct delivery to the lung to yield enhanced therapeutic effects. The EVs secreted by the chorionic MSC line (CMSC29) and decidual MSC line (DMSC23) were analysed using a combination of conventional microscopy, nanoparticle tracking analysis, tunable resistive pulse sensing, Nano Flow Analyzer and biological analyses. We also used resonance enhanced atomic force microscope-infrared spectroscopy (AFM-IR) for the characterisation of EV chemical composition and Nano Flow
5 Analyzer for the determination of total nucleic acid content in EVs. Our recently published study shows AFM-IR enables the nanoscale study of structural and molecular differences of individual EVs [13]. In this study we include functional analyses of in vitro models of injury to demonstrate the potential of EVs derived from the DMSC23 cell line in reducing cellular stress, which support progress into pre-clinical studies. Our work is fundamental in advancing the use of placental cell line-derived EVs for future application in regenerative medicine. 2. Materials and Methods 2.1 Cell culture and maintenance Primary MSCs from the fetal chorion and maternal decidual components of the term human placenta were transduced with telomerase reverse transcriptase (htert) to create the CMSC29 and DMSC23 cell lines respectively [14] These two cell lines have extended life span while maintaining stemness, whereas primary MSCs have limited life spans and their stemness are subject to patient-to-patient variation. CMSC29 was cultured in AmnioMAX TM (Invitrogen TM, Thermo Fisher Scientific, North Ryde, NSW, Australia) and DMSC23 was cultured using the Mesencult TM proliferation kit (STEMCELL Technologies, Tullamarine, VIC, Australia). The immortalised human bronchial epithelial cell line (BEAS-2B) and the human monocyte cell line (THP-1) were cultured in Dulbecco's Modified Eagle Medium (DMEM) and Roswell Park Memorial Institute medium (RPMI 1640) respectively (Sigma-Aldrich, Castle Hill, NSW, Australia). Both these lines were supplemented with 10% foetal bovine serum (FBS; French origin, Scientifix, Cheltenham, VIC, Australia) and 1% penicillin-streptomycin 5000 U/ml (PenStrep, Thermo Fisher Scientific). Hanks' Balanced Salt Solution (HBSS(-), Sigma-Aldrich) was used for washing DMSC23 and CMSC29, while phosphate buffer saline without Ca 2+ and Mg 2+ (PBS; Lonza, Bella Vista, NSW, Australia) was used for BEAS-2B and THP-1. TrypLE TM Select Enzyme (Thermo Fisher Scientific) was used as the
6 dissociation reagent. DMSC23 and CMSC29 were used at passages P23-28, while BEAS-2B and THP-1 cells were used at P12-16 and P14-20 respectively. All cells were maintained at 37 C and 5% CO 2 in incubators. 2.2 Isolation of EVs by ultracentrifugation DMSC23 and CMSC29 were cultured until 80% confluence in normal maintenance culture media then cells were washed twice with HBSS(-). Cells were then cultured in EV isolation media (serum-free media containing 0.5% bovine serum albumin and 1% PenStrep) for 48 h. EV-containing media were collected and centrifuged at 500 g for 5 min, then the supernatant was further centrifuged at 2,000 g for 10 min to remove cells and debris. The supernatant was centrifuged at 31,200 rpm (100,000 g) for 60 min at 4 C using a Ti-70 rotor in an Optima LE-80K Ultra Centrifuge (Beckman Coulter, Lane Cove West, NSW, Australia). The supernatant was discarded and the EV pellet was resuspended in 1 ml RNase-free PBS (Lonza). The ultracentrifugation was repeated at 31,200 rpm (100,000 g) for 60 min at 4 C and the supernatant was discarded. The EV pellet was resuspended in 200 µl RNase-free water, transferred to RNase-free microcentrifuge tubes and stored at 80 C if not used immediately. 2.3 Measurement of size and concentration of EVs Nanoparticle tracking analysis (NTA) EV samples were diluted 1:40 with nuclease-free water, vortexed at a moderate speed for 30 secs and then measured using a NanoSight NS300 (Malvern Instruments, ATA Scientific, Australia). Diluted samples (1 ml) were infused using a syringe pump with temperature control set at 25 C and the particles were imaged with an auto-focus camera for 60 secs, and this process was repeated three times per sample. Data were processed using the NTA software using camera level 11 and a detection threshold of 4, which allowed the calculation and comparison of mean particle diameter, mode, D50 values and particle concentration.
7 2.3.2 Tunable Resistive Pulse Sensing (TRPS) For the measurement of particle size and concentration with higher precision and accuracy, TRPS was used using a qnano (IZON Science, Christchurch, New Zealand). Particles were resuspended in electrolytes and passed through an engineered pore (NP100), which provided a direct measurement of size and concentration. All reagents were freshly prepared and filtered (0.22 µm) Protein quantification using Bradford assay The total protein in each EV sample was quantified using the Pierce TM Detergent Compatible Bradford Assay Kit (Thermo Fisher). EV samples were lysed by incubating a 10 µl sample with 290 µl of radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher) in each well of a 96-well plate. After thorough mixing by pipetting, 10 µl from each well was transferred into a new 96-well plate. To each well, 300 µl of Bradford Assay Reagent was added and mixed by pipetting. BSA µg/ml was used to prepare a standard curve. After a 10 min incubation at room temperature in the dark, the absorbance at 600 nm was measured using a plate reader (VICTOR X, Perkin Elmer, Melbourne, VIC, Australia). 2.4 Analysis of size and morphology of EVs Scanning electron microscopy Scanning electron microscopy (SEM) was used to observe the changes in morphology and particle size. Samples were gold sputter-coated using a K550X sputter coater (Quorum Emitech, Kent, UK). Images were obtained using a Zeiss Sigma high definition field emission gun scanning electron microscope (Carl Zeiss AG, Oberkochen, Germany) Transmission electron microscopy The size and morphology of the particles were analysed using transmission electron microscopy (TEM). Samples were mounted on carbon and Formvar-coated copper grids for 30 minutes and fixed with 2.5% glutaraldehyde for 30 min. Following three washes in sterile PBS, they were negative stained with 2% (w/v) ammonium molybdate in water for 5 min.
8 Excess stain was removed by blotting and the samples were air-dried further, before imaging in JEOL JEM-1400 TEM (Tokyo, JPN) Atomic force microscopy For observing particle size and 3D morphology, EV samples were placed onto freshly cleaved mica and dried for atomic force microscopy (AFM) on the nanoir TM AFM-IR instrument (Anasys Instruments, USA). Images were obtained at a scan rate of Hz using a tapping-mode with an EX-T125 probe (resonance frequency khz and spring constant N/m, Anasys Instruments). 2.5 Analysis of EV molecular composition using atomic force microscope-infrared spectroscopy (AFM- IR) and Nano Flow Analyzer Our protocol for using AFM-IR (nanoir TM, Anasys Instruments) for EV characterisation [13] was followed. Each EV sample (5 µl) was placed on a zinc selenide prism and dried overnight. After optimisation of the laser signal, AFM-IR spectra were collected from 1000 to 1800 cm 1 at 1 cm 1 intervals with a scan rate of 0.1 Hz and with a coaverage of 16. A silicon nitride cantilever (EXC450 tips, AppNano, CA, USA) with a nominal spring constant of 0.5 Nm 1 was used for all measurements. The scan sizes were µm for probing one point per EV and 1 1 µm for probing multiple points on individual EVs. Analysis Studio TM software was used for data analyses. Smoothing of the spectra using the Savitzky-Golay function was achieved by using the polynomial function of 2, and 15 numbers of points. The size and nucleic acid content of EVs were measured using Nano Flow Analyser (nanofcm, China). The standard 200 nm polystyrene spheres (100 µl) were used to align the laser. After the laser optimization, the size of silica nanospheres was measured and used as the size standards, followed by the construction of a calibration curve. The size measurement of each EV sample (diluted in 100 µl RNase-free PBS) was acquired directly using the same instrument settings as the silica nanoparticles reference standards. To quantify total nucleic acid content, each EV sample was diluted in 50 µl RNase-free PBS and SYTO RNASelect
9 green fluorescent cell stain 5 mm stock solution in DMSO (Thermofisher) was added to make the final concentration 10 µm. The mixture was incubated at 37 C for 30 minutes. Subsequently, the samples were loaded on the exosome spin column MW 3000 (Thermofisher) to remove all unbound dyes and measured on Nano Flow Analyzer. The control samples were PBS and basal medium without EVs using the same RNA staining process. 2.6 RNA isolation from EVs Total RNA was isolated from EVs using the TRIzol TM reagent (Invitrogen), following the manufacturer s protocol with minor modifications. Briefly, 300 μl TRIzol was added directly to the EV pellet following ultracentrifugation (see 2.2 above) and then this was transferred to an RNase-free microcentrifuge tube. An additional 200 μl TRIzol was added to the ultracentrifuge tube to ensure all the residual pellet was collected, and then samples were stored immediately at 80 C. 2.7 OpenArray for mirna profiling of EVs For interrogating the mirna content in EVs, OpenArray was conducted, with a modified protocol for low sample input. RNA samples were diluted to 10 ng/µl. Reverse Transcriptase (RT) mastermix was prepared using the manufacturer s protocol. Mastermix (4.5 µl) was added to each sample, inverted to mix, centrifuged and incubated on ice for 5 min. For cdna synthesis, the thermocycler program was: 40 cycles (16 C for 2 min, 42 C for 1 min, 50 C for 1 sec), 85 C for 5 min, and then held at 4 C. To generate pre-amplified cdna, PreAmp master mix (TaqMan PreAmp Mastermix) and PreAmp primers (Thermo Fisher Scientific, diluted in nuclease-free water), were added and the thermocycler program was: 95 C for 10 min, 55 C for 2 min, 72 C for 2 min, 16 cycles (95 C for 15 sec, 60 C for 4 min), 99 C for 10 min, and then held at 4 C. Preamplified cdna was diluted 1:20 in 0.1 TE buffer. The diluted preamplified cdna (5 µl) was combined with TaqMan OpenArray real-time PCR
10 master mix (5 µl). The plate was sealed and vortexed to mix. After transferring 5 µl of each sample to each well of the 384-well sample plate, the plate was centrifuged at 490 g for 1 min at 4 C. QuantStudio 12K Flex software was initialised and the qpcr was run. All run files were uploaded to Thermo cloud software for data normalization, which was performed using global normalisation method. Normalised data were exported, analysed and visualised as heatmap, volcano plots and bar graphs. The RStudio software (version ) was used for generating heatmaps and Prism software was used for Volcano plots and bar graphs. Reproducibility of data were confirmed from four replicate samples for CMSC29 and DMSC23 cells and five replicate samples for CMSC29 EVs and DMSC23 EVs. 2.8 Assessment of cell migration and invasion upon EV treatment Cell wound repopulation assay Epithelial cell migration in the presence of DMSC23 EVs or CMSC29 EVs was assessed by comparing the extent of closure of a two-dimensional scratch wound on a monolayer of cells. BEAS-2B cells were seeded at cells/well on 96-well plates and were cultured in normal media until 90% confluent. The cells were then incubated in low serum medium (containing 1% FBS) for 24 h prior to scratching a wound on the midline of the culture well using a 96-pin wound making tool (IncuCyte WoundMaker TM ). After washing with low serum medium twice to remove cell debris, fresh medium was added. EVs were added at particles per well. Wound images were taken every 2 h with 10 magnification using the IncuCyte live cell imaging system and IncuCyte ZOOM software program (Essen BioScience, Inc. Ann Arbor, MI, USA) Cell invasion and migration assay using impedance measurement Cell invasion and migration were monitored using an xcelligence Real-Time Cell Analyzer Dual Purpose (RTCA DP) instrument using the 16-well electronic cell invasion and migration plate (CIM-Plate 16). Following the manufacturer s instructions, the CIM-Plate
11 16 was filled with media and equilibrated at 37 C for 1 h, then background impedance values were recorded. BEAS-2B cells were passaged a day prior to the experiment, kept in serum-free media (SFM) for 24 h. Each plate was seeded with BEAS-2B cells in the upper chamber at 40,000 cells per well in 100 µl serum-free media. DMSC23 EVs and CMSC29 EVs were diluted to particles in 20 µl SFM and added to cells immediately after seeding. Plates were left at room temperature for 30 min to allow the cells to settle at the bottom of the upper chamber. The plates were then loaded onto the RTCA DP instrument inside a 37 C incubator and the impedance was measured every 10 min for 72 h. 2.9 Analysis of cellular responses to EV treatment after LPS injury EVs were diluted in DMEM complete media and transferred to a 24-well plate, with each well containing particles in 100 µl media. The control group was given medium without EVs. Then BEAS-2B were seeded at cells per well on 24-well plate, to incubate with EVs 30 min prior to exposing cells to lipopolysaccharide (LPS) injury used at final concentration of 100 ng/ml (E.Coli O111:B4, Sigma-Aldrich). After 16 h incubation, cells were collected and prepared according to the manufacturer s instructions for the Muse TM Nitric Oxide Assay Kit (Merck Millipore, Frenches Forest, NSW, Australia) Measurement of cytokine release in response to LPS injury after EV treatment THP-1 cells were seeded at cells per well (180 µl of cells/ml) then treated with LPS (E. coli O111:B4, Sigma-Aldrich) at 100 ng/ml final concentration, incubated on a shaker at 37 C, 5% CO 2 for 4 h. EVs were made to or particles/ml and added to THP-1 cells at 5 particles/cell. The enzyme-linked immunosorbent assay (ELISA) was conducted using IL-6 and TNF-α ELISA kits (BD Biosciences, North Ryde, NSW, Australia) following the manufacturer s protocols. Briefly, Nunc MaxiSorp TM plates (Thermo Fisher Scientific) were coated with
12 capture antibodies (100 µl/well; 40 µl in 10 ml for one 96-well plate to give a final dilution of 1:250) and incubated at 4 C for 16 h (20 h LPS exposure in total). Samples were mixed by pipetting, collected (240 µl) and centrifuged to remove cells. The supernatant (220 µl) was transferred to an empty 96-well plate (i.e. the sample plate) and checked using a light microscope to ensure no cells were present. IL-6 and TNF-α standards were prepared in assay diluent. Wash buffer diluted from a 20 stock solution was used to wash away unbound capture antibodies, blocked with assay diluent for 1 h and then the manufacturer s protocol was followed for subsequent washing and incubation steps, using the BD OptEIA TM set (BD Biosciences) Statistical analysis A minimum of triplicate, or quadruplicate samples, were used for reliability in each experimental condition being examined. Data were analysed and presented as a mean ± standard deviation. The differences between the experimental and control groups were analysed using the one-way analysis of variance (ANOVA) test. A p-value of less than 0.05 was considered to be a statistically significant difference. 3. Results and Discussion 3.1 Comparison of EV size and concentration Analysis of 3D images of EVs showed that EVs were spherical with no morphological differences between EVs isolated from CMSC29 or DMSC23 cells. AFM height images revealed the EVs diameters were in a range of nm. EV subpopulations that with larger diameters: 100 to 300 nm were also identified (Fig. 1A). TEM analyses showed primarily smaller vesicles (less than 100 nm), which were characterised by a typical lipid bilayer, and there was evidence of smaller vesicles within larger vesicles (Fig. 1B).
13 I J Fig. 1. A. Three-dimensional atomic force microscopy (AFM) image of DMSC23 EVs; B. Transmission electron micrographs of DMSC23 EVs showing the lipid bilayer structures, scale bars 100 nm; Scanning electron micrographs of: C. DMSC23 EVs; and D. CMSC29 EVs (left: secondary electrons detector, and right: InLens detector, scale bars 200 nm). Comparison of particle size and concentration for DMSC23 EVs and CMSC29 EVs, using: E. nanoparticle tracking analysis (NanoSight); tunable resistive pulse sensing (qnano) of: F. DMSC23 EVs and G. CMSC29 EVs; H. protein quantification (Bradford assay); and size distribution using Nano Flow Analyzer of: I. DMSC23 EVs and J. CMSC29 EVs. Further analyses of morphological changes of the surface were by SEM. Due to the limitation in resolution, only EV subpopulations with sizes of about nm in diameter were analysed (Fig. 1C, D). The use of InLens signal merged with secondary electron detection produced images with higher contrast and detail (Fig. 1C, D (right)). The surface of EVs appeared to be relatively smooth with small protruding patches of a secondary structure on the membrane. These patches could be associated with components of secretome, e.g. proteins (Fig. 1C).
14 Table 1. Summary of averaged particle size and concentration measured using NTA and TRPS methods for CMSC29 EVs and DMSC23 EVs. Measurement Method NTA TRPS Nano Flow Analyzer Mean (nm) Mode (nm) Mean (nm) Mode (nm) Concentration (particles/ ml) Mean (nm) CMSC29 EV ± ± ± ± ± 16.6 DMSC23 EV ± ± ± ± ± 18.4 The size distribution and concentration of EVs were analysed using NTA and TRPS. These methods confirmed a relatively broad size distribution for both CMSC29 EVs and DMSC23 EVs (Table 1, Fig. 1E-G). The averaged mean and mode sizes identified from NTA were slightly larger than those measured by TRPS (Table 1, Fig. 1E-G). The mode diameters for EVs measured by NTA and TRPS were ± 10.1 nm and ± 1.4 nm for DMSC23 EVs respectively, and 65.3 ± 39.5 nm and 75 ± 2.8 nm for CMSC29 EVs respectively. Although NTA and TRPS yielded comparable total particle counts overall, NTA detected more particles with larger diameter (i.e. greater than 110 nm) while TRPS detected more particles in the smaller diameter (less than 110 nm) range. Quantitative assessments using NTA and TRPS were influenced by the type of particle as well as the settings used e.g. camera level and detection threshold for NTA, and nanopore size for TRPS [15],[16]. As a nanopore NP100 was used in this study, the detectable vesicle size for TRPS was in the range of 50 to 330 nm and was less sensitive for detection of EV particles smaller than 50 nm. The size distributions of DMSC23 EVs and CMSC29 EVs were also analysed in Nano Flow Analyzer (Table 1, Fig 1I, J), which is able to detect the smaller diameter particles due to the lower detection threshold. The mean sizes of DMSC23 EVs and CMSC29 EVs were 79.0 ±
15 18.4 nm and 77.7 ± 16.6 nm respectively, which obviously indicate the higher sensitivity of Nano Flow Analyzer compared to NTA and TRPS (Table 1). Protein content analysis Proteins, which are essential building blocks for tissue repair, where quantified using the Bradford assay. Results of the assay showed the total protein content was higher for DMSC23 EVs (99.7 ± 4.8 µg/ml) compared with that of CMSC29 EVs (84.1 ± 9.1 µg/ml) (Fig. 1H). Cumulatively, particles size/concentration and protein analyses demonstrated comparable size distribution, with CMSC29 EVs having a broader size distribution compared to DMSC23 EVs in both analysis methods. CMSC29 EVs appeared to be more aggregated. However, EVs imaged using the AFM did not reveal any aggregates, which could be attributed to differences in sample preparation and the number of particulates that are typically analysed by imaging techniques, when compared to size/concentration analysis using dynamic light scattering or TRPS. It is important to note that there were some discrepancies in data generated by each of the characterisation methods, which emphasised their individual limitations and reinforced the need to use multiple techniques to increase the accuracy of EV size and concentration measurements. The limitations of these techniques must be taken into account when results are interpreted. 3.2 Analysis of EV molecular composition using AFM-IR and Nano Flow Analyzer Heterogeneity and molecular composition of individual CMSC29 EVs and DMSC23 EVs was interrogated using resonance enhanced AFM-IR [13]. For both samples characteristic peaks that corresponded to proteins, nucleic acids, and lipids were identified. There were substantial differences between both groups of EVs (Fig. 2.1). DMSC23 EVs characterised with lower amount of proteins and relatively broad peak with three dominant peaks at 1650,
16 1590 and 1530 cm -1 indicated conformational changes to the protein structure Amide I and Amide II. Additional peak at 1730 cm -1 (C=O stretch) confirmed presence of phospholipids. DMSC23 EVs characterised with very strong peaks at 1105 cm -1 and 1025 cm -1. Whereas, CMSC29 EVs had a dominating amide I peak at 1600 cm -1 and well-defined peaks in the range 1450 and 1350 cm -1 which corresponded to the phosphatidylcholine head group and thymine that reveals presence of RNAs in EVs. Overall these peaks, when analysed in relation to protein peaks, were of higher intensity for DMSC23 EVs. Importantly, we observed two dominating peaks in DMSC23 EVs at 1080 and 1030 cm -1 ; these peaks are associated with glycogen and nucleic acid: desoxyribose, RNA and ribose. In summary, DMSC23 EVs characterised with relatively higher amount of DNA comparing to amount of protein. Whereas, CMSC29 EVs had very well-balanced composition. Qualitative analysis of the peak intensities showed that CMSC29 EVs samples had much stronger peaks with lower noise-to-signal ration which suggests greater concertation of the molecular content. The differences of nucleic acid contents between DMSC23 EVs and CMSC29 EVs were further demonstrated using Nano Flow Analyzer (Fig 2.2). Indeed, the exosomal nucleic acids of DMSC23 (2.6%) are higher (Fig 2.2A) than CMSC29 EVs (1.9%) (Fig 2.2B) (red dots represented for the fluorescence residues). Differences in total RNA and DNA contents were in agreement with quantification using AFM-IR (Fig. 2.1). The control group, which were basal medium and PBS with RNA staining (Fig 2.2C and D), indicated no fluorescence presented in both samples. Total nucleic acids content of EVs comprises of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). It was previously shown that the RNA spectrum has two clear spectral components centered at 1240 and 1220 cm 1, while the ds-dna band in the same region is sharper and peaked at 1226 cm 1 {Zucchiatti, 2016 #1264}. The symmetric stretching band of nucleic acid phosphate is typically centered at 1086 cm 1. Conversely, the
17 ribose nature of the phosphate sugar backbone of RNA is defined in spectral band centered at 1119 cm 1 (stretching vibration of the skeletal structure around the C2 OH group of ribose) (39) and characterise with strong spectral features of the C O are close to each other for ribose (1059 cm 1 ) and deoxyribose (1053 cm 1 ) (24). Analysis of the spectra for EVs isolated from DMSC23 and CMSC29 showed presence of a 1230 cm 1 band which was stronger for DMSC23 EVs, suggesting greater amount of total nucleic acids content. This result agrees with total DNA/RNA content obtained using Nano Flow Analyser. However, CMSC29 EVs showed a band ~1080 cm 1 associated with nucleic acid, which for DMSC23 EVs was shifted to ~1105 cm -1 which could be associated with phospholipids C O stretch. CMSC29 EVs characterised with two peaks ~1120 cm 1 and ~1045 cm 1, which are associated with RNA. Peaks in these positions were identified only for some of the DMSC23 EVs, which suggests that the EVs were heterogenous. AFM-IR does not allow determining mirna content, therefore the comparison to total mirna measured using Open Array is not possible. However, AFM-IR identified major differences in the composition of both populations of EVs in terms of RNA, DNA content, protein content and structure and some differences in lipid structure.
18 Fig Infrared absorption spectra of EV molecular components with corresponding topographical AFM images for: A. DMSC23 EVs; and B. CMSC29 EVs, where the spectra is generated from probing of individual EV particles. A Burst trace Dot-plots B Burst trace Dot-plots
19 C Burst trace Dot-plots D Burst trace Dot-plots Fig The real time burst trace of size scattering (blue) and fluorescence FITC (green) channels with corresponding dot-plots in RNA staining of: A. DMSC23 EVs, B. CMSC29 EVs, C. Basal medium DMSC23 and D. PBS
20 3.3 DMSC23 EVs and CMSC29 EVs increase lung epithelial cell migration Lung epithelial cells (BEAS-2B) treated with DMSC23 and CMSC29 EVs were placed in a chamber with a chemotactic gradient (FBS) to test whether the EVs increase BEAS-2B migration. From the results of the impedance values that measure the translocation of cells as they migrate from the upper chamber to lower chamber (xcelligence CIM-16), an increased number of cells passed through the chamber when treated with EVs (Fig. 3). Both DMSC23 EVs and CMSC29 EVs increased the migration of BEAS-2B, which suggests that lung epithelial cells with increased mobility may be helpful in the microenvironment during the recovery processes from a lung injury. This was further investigated by the wound closure assay. In a simplified wound model using a scratch wound, DMSC23 EVs increased the speed of cells migrating towards the wound. Cell migration-enhancing capability is important in wound healing as it is a feature of natural wound repair processes in the epithelium. The progenitor cells near the injured epithelium migrate and spread to cover the denuded surfaces [17]. Fig. 3. Assessment of cell migration of BEAS-2B in response to EV treatment by: A. Impedance measurement sensing cell translocation towards chemoattractant (xcelligence); B. Scratch wound assay with calculated percentage of wound closure from live cell imaging (IncuCyte); C. Images from the scratch wound assay with highlighted wound area over 24 h, 10 magnification.
21 3.4 DMSC23 EVs reduce cellular stress and the inflammatory response caused by LPS As is the case with DMSC primary cells, DMSC23 cells are resistant to oxidative stress due to the pre-conditioning of the placenta-derived tissue where the cells are isolated from [14] We previously showed that DMSC23 are more resistant to hypoxic stress compared to CMSC29 [18]. Since EVs convey similar properties to the cells which they are secreted from [19], DMSC23 EVs were expected to reduce oxidative stress in an established in vitro model of lung injury; LPS-injured BEAS-2B lung epithelial cells. The measurement of nitric oxide expression allows quantification of cellular stress in response to injury. Nitric oxide is a small molecule generated by the enzyme nitric oxide synthase (NOS) in cells, which participates in many biological functions [20]. In the context of cellular stress, nitric oxide is produced in large amounts in response to inflammatory stimuli, as part of a defense mechanism in oxidative toxicity [21]. The intracellular detection of nitric oxide is important for the detection of apoptotic signal cascades as the cell protects itself from pathological processes including inflammation and apoptosis [22]. The comparison of the nitric oxide profiles after LPS injury demonstrated that the cells treated by EVs had lower nitric oxide expression as shown from the reduction in the average percentage of nitric oxide positive cells from 8.0 ± 0.4 %, to 7.0 ± 0.6 % and 5.5 ± 1.3 %, respectively for DMSC23 and CMSC29 EVs (Fig. 4. A,B). This suggests that the oxidative stress signalling cascade within cells is modified in response to LPS injury after the uptake of CMSC29 EVs (p < 0.05), more obviously than cells treated with DMSC23 EVs. The immunomodulatory effects of DMSC23 EVs on cytokine release by THP-1 were investigated using ELISA. Although the effects of EVs on IL-6 was minimal, both EV types reduced the release of TNF-α (Fig. 4. C, D), a pro-inflammatory cytokine implicated in many diseases and particularly well characterised in pulmonary diseases including acute lung injuries, asthma, emphysema and pulmonary fibrosis [23].
22 Fig. 4. Comparison of cellular responses to CMSC29 EV and DMSC23 EV treatment: A. Nitric oxide expression profiles of LPS-injured BEAS-2B cell line, representative of triplicate samples; B. Comparison of total nitric oxide-positive cell population; ELISA analyses of proinflammatory cytokines released from THP-1 upon treatment with LPS, with or without EVs for: C. IL-6, and D. TNF-α; *p < 0.05; E. Unsupervised hierarchical cluster analysis of microrna profiles in DMSC23 and CMSC29 cells (4 replicates for each type) and EVs (5 replicates for each type) is presented as a heatmap. 3.5 The therapeutic potential of DMSC23 EVs and CMSC29 EVs is attributable to their molecular composition EVs produced by MSCs reprogram tissue-injured cells by delivering mrna and/or mirna to modulate soluble paracrine mediator production, mediate cell de-differentiation and cell-
23 cycle re-entry and thereby contributing to tissue regeneration [24]. Another important rationale for analysing the EV mirna profile is that the EVs released from the injured cells may mediate phenotypic transfer of surrounding MSCs to acquire certain features by delivering mrna and/or mirna to MSCs [24]. Thus, comparative analysis of the mirnas carried in EVs and mirnas contained in the cells secreting those EVs may reveal important mechanisms by which MSCs and their EVs carry out their therapeutic effects. We interrogated the mirna content of CMSC29s and DMSC23s and their EVs, using an OpenArray TM platform, which enabled the analysis of 754 mirnas of which 405 mirnas were, detected (Fig. 4E). Unsupervised hierarchical clustering demonstrated that mirna profiles of CMSC29s and DMSC23s were similar than their respective EVs (Fig. 4E) implying differences in packaging of microrna cargos in EVs from the two cell types. There were more mirnas packaged in CMSC29 EVs compared to DMSC23 EVs, which was in agreement with the AFM-IR results. Furthermore, the CMSC29 EV mirna profile was more similar to the mirna profile of secreting CMSC29 cells, than the mirna profiles of DMSC23 EVs compared with secreting DMSC23 cells (Fig. 4E). Univariate analysis presented in Supplementary Figure 1A identified several mirnas that are significantly differentially expressed (Supplementary Figure 1B,C). Since mirnas are known to be important regulators of cellular repair and tissue regeneration, we assessed our data for the presence of key mirna in EVs and secreting cells that have been previously reported to be involved in lung repair (Table 2). Amongst the known mirnas associated with lung repair (Table 2), our study did not find mir-505 to be localized to EVs released from either of the two cell types. Similarly, mir-30a-3p was also consistently undetectable from the EVs in DMSC23 cell-derived EVs. Intriguingly, some mirnas (mir-548c, mir-548a as well as mir-603) that came up in our discovery analyses were found to be consistently and selectively packaged into EVs assessed from both cell types, whilst remaining to be depleted
24 from the cells that transcribe them. These may potentially be effector molecules targeting specific cells and could also be biomarkers of early response to injury. On the other hand, mirna candidates such as let-7a and mir-181a-2#, although present in both cell types were always excluded from EVs (Suppl. Figure 2). The process of selective packaging of molecular components in EVs by cells may be investigated in future studies to gain an insight into modifying the content of specific mirnas and other biological signals carried inside the EVs for potential therapeutic applications. Our analyses from mirna profiling studies not only demonstrate differences between mirna composition based on the secreting cell type, but also identify candidates that could be included in future clinical studies/trials for screening tissue damage and response. These studies also highlight the need for further investigations to interrogate molecular composition of EVs for potential therapeutic applications [25]. Table 2. Selected mirnas in EVs that may be involved in tissue repair [26].
25 4. Concluding remarks A critical component of the mechanism for MSC-driven tissue regeneration is now widely accepted to be the transfer of genetic information between MSCs and injured cells in tissues. The results from this study demonstrate that DMSC23 EVs and CMSC29 EVs were beneficial in promoting migration and reducing oxidative stress and inflammation, suggesting their potential application in reducing lung injury and enhancing cellular repair. This study leads to further in vivo studies that will be fundamental in advancing the field of EV therapeutics, in particular to improve the regeneration of the lung after injury. Further mirna mir-505 mir-29a/b mir-146a Functions Highly involved in targeting relevant gene functions in repair The master regulators of tissue regeneration Reduce inflammation, increase proliferation, migration, and angiogenesis of endothelial progenitor cells mir-30d mir-133a mir-30c mir-30a-3p mir-30a-5p mir-483-5p mir-130a Important in the late repair phase Regulatory roles in airway inflammation, tumour suppressor Important in the late repair phase Wound healing Tumour suppressor, modulate epithelial-mesenchymal transition Protective role in chronic obstructive pulmonary disease Involved in lung organogenesis development of the single vesicle characterisation technique using AFM-IR as presented in this study will enable the identification of the most therapeutically relevant EV subtypes.
26 Taking this innovative approach together with profiling of mirna and proteins will advance the field of EV-therapies towards clinical translation. Acknowledgments WCh acknowledges The University of Sydney for the SOAR Fellowship. The authors acknowledge the facilities and the scientific and technical assistance of the Bosch Molecular Biology Facility and Australian Microscopy & Microanalysis Research Facility at the Australian Centre for Microscopy & Microanalysis, The University of Sydney. AAH and MVJ acknowledge the support through the JDRF Australia T1DCRN Fellowship and the JDRF International Advanced Post-doctoral fellowship, respectively. Conflicts of interest The authors have declared no conflict of interest. References [1] I. Gangwar, N. Kumar Sharma, G. Panzade, S. Awasthi, A. Agrawal, R. Shankar, Scientific Reports 2017, 7, [2] M. Patrick, J. S. L, Future Prescriber 2013, 14, 13. [3] T. Ferkol, D. Schraufnagel, Annals of the American Thoracic Society 2014, 11, 404. [4] G. Bellani, J. G. Laffey, T. Pham, et al., JAMA 2016, 315, 788; J. B. Soriano, A. A. Abajobir, K. H. Abate, S. F. Abera, A. Agrawal, M. B. Ahmed, A. N. Aichour, I. Aichour, M. T. E. Aichour, K. Alam, N. Alam, J. M. Alkaabi, F. Al-Maskari, N. Alvis-Guzman, A. Amberbir, Y. A. Amoako, M. G. Ansha, J. M. Antó, H. Asayesh, T. M. Atey, E. F. G. A. Avokpaho, A. Barac, S. Basu, N. Bedi, I. M. Bensenor, A. Berhane, A. S. Beyene, Z. A. Bhutta, S. Biryukov, D. J. Boneya, M. Brauer, D. O. Carpenter, D. Casey, D. J. Christopher, L. Dandona, R. Dandona, S. D. Dharmaratne, H. P. Do, F. Fischer, T. T. Gebrehiwot, A. Geleto, A. G. Ghoshal, R. F. Gillum, I. A. M. Ginawi, V. Gupta, S. I. Hay, M. T. Hedayati, N. Horita, H. D. Hosgood, M. B. Jakovljevic, S. L. James, J. B. Jonas, A. Kasaeian, Y. S. Khader, I. A. Khalil, E. A. Khan, Y.-H. Khang, J. Khubchandani, L. D. Knibbs, S. Kosen, P. A. Koul, G. A. Kumar, C. T. Leshargie, X. Liang, H. M. A. El Razek, A. Majeed, D. C. Malta, T. Manhertz, N. Marquez, A. Mehari, G. A. Mensah, T. R. Miller, K. A. Mohammad, K. E. Mohammed, S. Mohammed, A. H. Mokdad, M. Naghavi, C. T. Nguyen, G. Nguyen, Q. Le Nguyen, T. H. Nguyen, D. N. A. Ningrum, V. M. Nong, J. I. Obi, Y. E. Odeyemi, F. A. Ogbo, E. Oren, M. Pa, E.-K. Park, G. C. Patton, K. Paulson, M. Qorbani, R. Quansah, A. Rafay, M. H. U. Rahman, R. K. Rai, S. Rawaf, N. Reinig, S. Safiri, R. Sarmiento-Suarez, B. Sartorius, M. Savic, M. Sawhney, M. Shigematsu, M. Smith, F. Tadese, G. D. Thurston, R. Topor-Madry, B. X. Tran, K. N. Ukwaja, J. F. M. van Boven, V. V. Vlassov, S. E. Vollset, X. Wan, A. Werdecker, S. W. Hanson, Y. Yano, H. H. Yimam, N. Yonemoto, C. Yu, Z. Zaidi, M. El Sayed Zaki, A. D. Lopez, C. J. L.
27 Murray, T. Vos, The Lancet Respiratory Medicine 2017, 5, 691; N. Snell, D. Strachan, R. Hubbard, J. Gibson, E. Limb, R. Gupta, A. Martin, M. Laffan, I. Jarrold, European Respiratory Journal 2016, 48. [5] S. Geiger, D. Hirsch, F. G. Hermann, European Respiratory Review 2017, 26; G. F. Curley, M. Jerkic, S. Dixon, G. Hogan, C. Masterson, D. O Toole, J. Devaney, J. G. Laffey, Critical care medicine 2017, 45, e202; S. H. Mei, S. D. McCarter, Y. Deng, C. H. Parker, W. C. Liles, D. J. Stewart, PLoS medicine 2007, 4, e269. [6] M. Reddy, L. Fonseca, S. Gowda, B. Chougule, A. Hari, S. Totey, International Journal of Stem Cells 2016, 9, 192. [7] E. S. Kim, Y. S. Chang, S. J. Choi, J. K. Kim, H. S. Yoo, S. Y. Ahn, D. K. Sung, S. Y. Kim, Y. R. Park, W. S. Park, Respiratory Research 2011, 12, 108; J. Li, D. Li, X. Liu, S. Tang, F. Wei, Journal of inflammation (London, England) 2012, 9, 33. [8] V. B. Konala, M. K. Mamidi, R. Bhonde, A. K. Das, R. Pochampally, R. Pal, Cytotherapy 2016, 18, 13; L. Sun, R. Xu, X. Sun, Y. Duan, Y. Han, Y. Zhao, H. Qian, W. Zhu, W. Xu, Cytotherapy 2016, 18, 413. [9] C. A. Herberts, M. S. G. Kwa, H. P. H. Hermsen, Journal of Translational Medicine 2011, 9, 29; K. Lu, H.-y. Li, K. Yang, J.-l. Wu, X.-w. Cai, Y. Zhou, C.-q. Li, Stem Cell Research & Therapy 2017, 8, 108. [10] M. C. Deregibus, A. Iavello, C. Tetta, G. Camussi, in Adult Stem Cell Therapies: Alternatives to Plasticity, (Ed: M. Z. Ratajczak), Springer New York, New York, NY 2014, 231. [11] F. Collino, M. Pomatto, S. Bruno, R. S. Lindoso, M. Tapparo, W. Sicheng, P. Quesenberry, G. Camussi, Stem cell reviews 2017, 13, 226; Y. Wu, W. Deng, D. J. Klinke, The Analyst 2015, 140, [12] J. S. Heo, Y. Choi, H.-S. Kim, H. O. Kim, International journal of molecular medicine 2016, 37, 115. [13] S. Y. Kim, D. Khanal, P. Tharkar, B. Kalionis, W. Chrzanowski, Nanoscale Horizons 2018, 3, 430. [14] S. Q. Qin, G. D. Kusuma, B. Al-Sowayan, R. A. Pace, S. Isenmann, M. D. Pertile, S. Gronthos, M. H. Abumaree, S. P. Brennecke, B. Kalionis, Placenta 2016, 39, 134; G. D. Kusuma, M. H. Abumaree, M. D. Pertile, A. V. Perkins, S. P. Brennecke, B. Kalionis, Stem cell reviews 2016, 12, 285. [15] S. L. N. Maas, J. de Vrij, E. J. van der Vlist, B. Geragousian, L. van Bloois, E. Mastrobattista, R. M. Schiffelers, M. H. M. Wauben, M. L. D. Broekman, E. N. M. Nolte-'t Hoen, Journal of Controlled Release 2015, 200, 87. [16] J. C. Akers, V. Ramakrishnan, J. P. Nolan, E. Duggan, C.-C. Fu, F. H. Hochberg, C. C. Chen, B. S. Carter, PLoS ONE 2016, 11, e [17] L. M. Crosby, C. M. Waters, American Journal of Physiology - Lung Cellular and Molecular Physiology 2010, 298, L715. [18] K. Yang, K. G. Leslie, S. Y. Kim, B. Kalionis, W. Chrzanowski, K. A. Jolliffe, E. J. New, Organic & Biomolecular Chemistry 2018, 16, 619. [19] H. Xin, Y. Li, M. Chopp, Frontiers in cellular neuroscience 2014, 8, 377. [20] N. Omer, A. Rohilla, S. Rohilla, A. Kushnoor, International Journal of Pharmaceutical Sciences and Drug Research 2012, 4, 105. [21] C. Bogdan, Nature immunology 2001, 2, 907. [22] P. K. Kim, R. Zamora, P. Petrosko, T. R. Billiar, International immunopharmacology 2001, 1, [23] S. Mukhopadhyay, J. R. Hoidal, T. K. Mukherjee, Respiratory Research 2006, 7, 125. [24] L. Guo, R. C. H. Zhao, Y. Wu, Experimental Hematology 2011, 39, 608. [25] I. M. Bjorge, S. Y. Kim, J. F. Mano, B. Kalionis, W. Chrzanowski, Biomaterials Science 2018, 6, 60. [26] K. S. Tan, H. Choi, X. Jiang, L. Yin, J. E. Seet, V. Patzel, B. P. Engelward, V. T. Chow, BMC Genomics 2014, 15, 587; C. K. Sen, S. Ghatak, The American Journal of Pathology 2015, 185,
28 2629; S. D. Alipoor, E. Mortaz, J. Garssen, M. Movassaghi, M. Mirsaeidi, I. M. Adcock, Mediators of Inflammation 2016, 2016, 11; L. Zhang, Y. Wang, W. Li, P. A. Tsonis, Z. Li, L. Xie, Y. Huang, Scientific Reports 2017, 7, 1117; A. O. Ribeiro, C. R. G. Schoof, A. Izzotti, L. V. Pereira, L. R. Vasques, MicroRNA 2014, 3, 45. [27] A. A. Hardikar, R. J. Farr, M. V. Joglekar, J Am Heart Assoc 2014, 3, e
29 Minerva Access is the Institutional Repository of The University of Melbourne Author/s: Kim, SY; Joglekar, MV; Hardikar, AA; Thanh, HP; Khanal, D; Tharkar, P; Limantoro, C; Johnson, J; Kalionis, B; Chrzanowski, W Title: Placenta Stem/Stromal Cell-Derived Extracellular Vesicles for Potential Use in Lung Repair Date: Citation: Kim, S. Y., Joglekar, M. V., Hardikar, A. A., Thanh, H. P., Khanal, D., Tharkar, P., Limantoro, C., Johnson, J., Kalionis, B. & Chrzanowski, W. (2019). Placenta Stem/Stromal Cell-Derived Extracellular Vesicles for Potential Use in Lung Repair. PROTEOMICS, 19 (17), Persistent Link: File Description: Accepted version
Covalent Conjugation to Cytodiagnostics Carboxylated Gold Nanoparticles Tech Note #105
 Covalent Conjugation to Cytodiagnostics Carboxylated Gold Nanoparticles Tech Note #105 Background Gold nanoparticle conjugates have been widely used in biological research and biosensing applications.
Covalent Conjugation to Cytodiagnostics Carboxylated Gold Nanoparticles Tech Note #105 Background Gold nanoparticle conjugates have been widely used in biological research and biosensing applications.
Human serum albumin (HSA) nanoparticles stabilized with. intermolecular disulfide bonds. Supporting Information
 Human serum albumin (HSA) nanoparticles stabilized with intermolecular disulfide bonds Wentan Wang, Yanbin Huang*, Shufang Zhao, Ting Shao and Yi Cheng* Department of Chemical Engineering, Tsinghua University,
Human serum albumin (HSA) nanoparticles stabilized with intermolecular disulfide bonds Wentan Wang, Yanbin Huang*, Shufang Zhao, Ting Shao and Yi Cheng* Department of Chemical Engineering, Tsinghua University,
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
Instructions. Torpedo sirna. Material. Important Guidelines. Specifications. Quality Control
 is a is a state of the art transfection reagent, specifically designed for the transfer of sirna and mirna into a variety of eukaryotic cell types. is a state of the art transfection reagent, specifically
is a is a state of the art transfection reagent, specifically designed for the transfer of sirna and mirna into a variety of eukaryotic cell types. is a state of the art transfection reagent, specifically
Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides
 Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides Polyacrylamide gel electrophoresis (PAGE) and subsequent analyses are common tools in biochemistry and molecular biology. This Application
Western Blot Analysis with Cell Samples Grown in Channel-µ-Slides Polyacrylamide gel electrophoresis (PAGE) and subsequent analyses are common tools in biochemistry and molecular biology. This Application
Application Note. Single Cell PCR Preparation
 Application Note Single Cell PCR Preparation From Automated Screening to the Molecular Analysis of Single Cells The AmpliGrid system is a highly sensitive tool for the analysis of single cells. In combination
Application Note Single Cell PCR Preparation From Automated Screening to the Molecular Analysis of Single Cells The AmpliGrid system is a highly sensitive tool for the analysis of single cells. In combination
Cell Cycle Phase Determination Kit
 Cell Cycle Phase Determination Kit Item No. 10009349 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Safety
Cell Cycle Phase Determination Kit Item No. 10009349 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 3 Safety
Modulating Glucose Uptake in Skeletal Myotubes:
 icell Skeletal Myoblasts Application Protocol Introduction Modulating Glucose Uptake in Skeletal Myotubes: Insulin Induction with Bioluminescent Glucose Uptake Analysis The skeletal muscle is one of the
icell Skeletal Myoblasts Application Protocol Introduction Modulating Glucose Uptake in Skeletal Myotubes: Insulin Induction with Bioluminescent Glucose Uptake Analysis The skeletal muscle is one of the
CytoSelect 48-Well Cell Adhesion Assay (ECM Array, Fluorometric Format)
 Product Manual CytoSelect 48-Well Cell Adhesion Assay (ECM Array, Fluorometric Format) Catalog Number CBA-071 CBA-071-5 48 assays 5 x 48 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures
Product Manual CytoSelect 48-Well Cell Adhesion Assay (ECM Array, Fluorometric Format) Catalog Number CBA-071 CBA-071-5 48 assays 5 x 48 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures
Application Guide... 2
 Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
MTT Cell Proliferation Assay
 ATCC 30-1010K Store at 4 C This product is intended for laboratory research purposes only. It is not intended for use in humans, animals or for diagnostics. Introduction Measurement of cell viability and
ATCC 30-1010K Store at 4 C This product is intended for laboratory research purposes only. It is not intended for use in humans, animals or for diagnostics. Introduction Measurement of cell viability and
User Manual. CelluLyser Lysis and cdna Synthesis Kit. Version 1.4 Oct 2012 From cells to cdna in one tube
 User Manual CelluLyser Lysis and cdna Synthesis Kit Version 1.4 Oct 2012 From cells to cdna in one tube CelluLyser Lysis and cdna Synthesis Kit Table of contents Introduction 4 Contents 5 Storage 5 Additionally
User Manual CelluLyser Lysis and cdna Synthesis Kit Version 1.4 Oct 2012 From cells to cdna in one tube CelluLyser Lysis and cdna Synthesis Kit Table of contents Introduction 4 Contents 5 Storage 5 Additionally
Agencourt RNAdvance Blood Kit for Free Circulating DNA and mirna/rna Isolation from 200-300μL of Plasma and Serum
 SUPPLEMENTAL PROTOCOL WHITE PAPER Agencourt RNAdvance Blood Kit for Free Circulating DNA and mirna/rna Isolation from 200-300μL of Plasma and Serum Bee Na Lee, Ph.D., Beckman Coulter Life Sciences Process
SUPPLEMENTAL PROTOCOL WHITE PAPER Agencourt RNAdvance Blood Kit for Free Circulating DNA and mirna/rna Isolation from 200-300μL of Plasma and Serum Bee Na Lee, Ph.D., Beckman Coulter Life Sciences Process
Real-time quantitative RT -PCR (Taqman)
 Real-time quantitative RT -PCR (Taqman) Author: SC, Patti Lab, 3/03 This is performed as a 2-step reaction: 1. cdna synthesis from DNase 1-treated total RNA 2. PCR 1. cdna synthesis (Advantage RT-for-PCR
Real-time quantitative RT -PCR (Taqman) Author: SC, Patti Lab, 3/03 This is performed as a 2-step reaction: 1. cdna synthesis from DNase 1-treated total RNA 2. PCR 1. cdna synthesis (Advantage RT-for-PCR
Protein extraction from Tissues and Cultured Cells using Bioruptor Standard & Plus
 Protein extraction from Tissues and Cultured Cells using Bioruptor Standard & Plus Introduction Protein extraction from tissues and cultured cells is the first step for many biochemical and analytical
Protein extraction from Tissues and Cultured Cells using Bioruptor Standard & Plus Introduction Protein extraction from tissues and cultured cells is the first step for many biochemical and analytical
HBV Quantitative Real Time PCR Kit
 Revision No.: ZJ0002 Issue Date: Aug 7 th, 2008 HBV Quantitative Real Time PCR Kit Cat. No.: HD-0002-01 For Use with LightCycler 1.0/LightCycler2.0/LightCycler480 (Roche) Real Time PCR Systems (Pls ignore
Revision No.: ZJ0002 Issue Date: Aug 7 th, 2008 HBV Quantitative Real Time PCR Kit Cat. No.: HD-0002-01 For Use with LightCycler 1.0/LightCycler2.0/LightCycler480 (Roche) Real Time PCR Systems (Pls ignore
Supporting Information
 Supporting Information Simple and Rapid Synthesis of Ultrathin Gold Nanowires, Their Self-Assembly and Application in Surface-Enhanced Raman Scattering Huajun Feng, a Yanmei Yang, a Yumeng You, b Gongping
Supporting Information Simple and Rapid Synthesis of Ultrathin Gold Nanowires, Their Self-Assembly and Application in Surface-Enhanced Raman Scattering Huajun Feng, a Yanmei Yang, a Yumeng You, b Gongping
a Beckman Coulter Life Sciences: White Paper
 a Beckman Coulter Life Sciences: White Paper Violet-Excited nim-da Allows Efficient and Reproducible Cell Cycle Analysis on the Gallios Flow Cytometer Authors: Valdez, Ben 1. Carr, Karen 2. Norman, John
a Beckman Coulter Life Sciences: White Paper Violet-Excited nim-da Allows Efficient and Reproducible Cell Cycle Analysis on the Gallios Flow Cytometer Authors: Valdez, Ben 1. Carr, Karen 2. Norman, John
ArC Amine Reactive Compensation Bead Kit
 ArC Amine Reactive Compensation Bead Kit Catalog no. A1346 Table 1. Contents and storage information. Material Amount Composition Storage Stability ArC reactive beads (Component A) ArC negative beads (Component
ArC Amine Reactive Compensation Bead Kit Catalog no. A1346 Table 1. Contents and storage information. Material Amount Composition Storage Stability ArC reactive beads (Component A) ArC negative beads (Component
protocol handbook 3D cell culture mimsys G hydrogel
 handbook 3D cell culture mimsys G hydrogel supporting real discovery handbook Index 01 Cell encapsulation in hydrogels 02 Cell viability by MTS assay 03 Live/Dead assay to assess cell viability 04 Fluorescent
handbook 3D cell culture mimsys G hydrogel supporting real discovery handbook Index 01 Cell encapsulation in hydrogels 02 Cell viability by MTS assay 03 Live/Dead assay to assess cell viability 04 Fluorescent
Measuring Cell Viability/Cytotoxicity: Cell Counting Kit-F
 Introduction The Cell Counting Kit-F is a fluorometic assay for the determination of viable cell numbers. Calcein-AM in this kit passes through the cell membrane and is hydrolized by the esterase in the
Introduction The Cell Counting Kit-F is a fluorometic assay for the determination of viable cell numbers. Calcein-AM in this kit passes through the cell membrane and is hydrolized by the esterase in the
Islet Viability Assessment by Single Cell Flow Cytometry
 Islet Viability Assessment by Single Cell Flow Cytometry Page 1 of 8 Purpose: To comprehensively assess the viability of the islet cell preparation prior to transplantation. Tissue Samples: A sample containing
Islet Viability Assessment by Single Cell Flow Cytometry Page 1 of 8 Purpose: To comprehensively assess the viability of the islet cell preparation prior to transplantation. Tissue Samples: A sample containing
UltraClean Soil DNA Isolation Kit
 PAGE 1 UltraClean Soil DNA Isolation Kit Catalog # 12800-50 50 preps New improved PCR inhibitor removal solution (IRS) included Instruction Manual (New Alternative Protocol maximizes yields) Introduction
PAGE 1 UltraClean Soil DNA Isolation Kit Catalog # 12800-50 50 preps New improved PCR inhibitor removal solution (IRS) included Instruction Manual (New Alternative Protocol maximizes yields) Introduction
Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity
 Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity Today you analyze the results of your bacterial transformation from last week and determine the efficiency
Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity Today you analyze the results of your bacterial transformation from last week and determine the efficiency
PRODUCT INFORMATION...
 VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
Cell Migration, Chemotaxis and Invasion Assay Protocol
 Cell Migration, Chemotaxis and Invasion Assay Protocol Hilary Sherman, Pilar Pardo and Todd Upton Corning Life Sciences 900 Chelmsford Street Lowell, MA 01851 Introduction Cell migration, the movement
Cell Migration, Chemotaxis and Invasion Assay Protocol Hilary Sherman, Pilar Pardo and Todd Upton Corning Life Sciences 900 Chelmsford Street Lowell, MA 01851 Introduction Cell migration, the movement
DNA Integrity Number (DIN) For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments
 DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
Steatosis Colorimetric Assay Kit
 Steatosis Colorimetric Assay Kit Item No. 10012643 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Steatosis Colorimetric Assay Kit Item No. 10012643 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
DNA Isolation Kit for Cells and Tissues
 DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
Mir-X mirna First-Strand Synthesis Kit User Manual
 User Manual Mir-X mirna First-Strand Synthesis Kit User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories, Inc.
User Manual Mir-X mirna First-Strand Synthesis Kit User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories, Inc.
Biological Stability and Activity of sirna in Ionic Liquids**
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Supporting Information Biological Stability and Activity of sirna in Ionic Liquids** Romiza R.
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2014 Supporting Information Biological Stability and Activity of sirna in Ionic Liquids** Romiza R.
NCL Method ITA-6. Leukocyte Proliferation Assay
 NCL Method ITA-6 Leukocyte Proliferation Assay Nanotechnology Characterization Laboratory National Cancer Institute-Frederick SAIC-Frederick Frederick, MD 21702 (301) 846-6939 ncl@mail.nih.gov December
NCL Method ITA-6 Leukocyte Proliferation Assay Nanotechnology Characterization Laboratory National Cancer Institute-Frederick SAIC-Frederick Frederick, MD 21702 (301) 846-6939 ncl@mail.nih.gov December
Plant Genomic DNA Extraction using CTAB
 Plant Genomic DNA Extraction using CTAB Introduction The search for a more efficient means of extracting DNA of both higher quality and yield has lead to the development of a variety of protocols, however
Plant Genomic DNA Extraction using CTAB Introduction The search for a more efficient means of extracting DNA of both higher quality and yield has lead to the development of a variety of protocols, however
Benchtop Mitochondria Isolation Protocol
 Benchtop Mitochondria Isolation Protocol Note: Specific protocols are available for the following products: MS850 Mitochondria Isolation Kit for Rodent Tissue MS851 Mitochondria Isolation Kit for Rodent
Benchtop Mitochondria Isolation Protocol Note: Specific protocols are available for the following products: MS850 Mitochondria Isolation Kit for Rodent Tissue MS851 Mitochondria Isolation Kit for Rodent
Taqman TCID50 for AAV Vector Infectious Titer Determination
 Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Related topics: Application Note 27 Data Analysis of Tube Formation Assays.
 Tube Formation Assays in µ-slide Angiogenesis Related topics: Application Note 27 Data Analysis of Tube Formation Assays. Contents 1. General Information... 1 2. Material... 2 3. Work Flow Overview...
Tube Formation Assays in µ-slide Angiogenesis Related topics: Application Note 27 Data Analysis of Tube Formation Assays. Contents 1. General Information... 1 2. Material... 2 3. Work Flow Overview...
Radius 24-Well Cell Migration Assay (Laminin Coated)
 Product Manual Radius 24-Well Cell Migration Assay (Laminin Coated) Catalog Number CBA-125-LN 24 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cell migration is a highly
Product Manual Radius 24-Well Cell Migration Assay (Laminin Coated) Catalog Number CBA-125-LN 24 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cell migration is a highly
RNA Extraction and Quantification, Reverse Transcription, and Real-time PCR (q-pcr)
 RNA Extraction and Quantification, Reverse Transcription, and Real-time Preparation of Samples Cells: o Remove media and wash cells 2X with cold PBS. (2 ml for 6 well plate or 3 ml for 6cm plate) Keep
RNA Extraction and Quantification, Reverse Transcription, and Real-time Preparation of Samples Cells: o Remove media and wash cells 2X with cold PBS. (2 ml for 6 well plate or 3 ml for 6cm plate) Keep
Measuring Protein Concentration through Absorption Spectrophotometry
 Measuring Protein Concentration through Absorption Spectrophotometry In this lab exercise you will learn how to homogenize a tissue to extract the protein, and then how to use a protein assay reagent to
Measuring Protein Concentration through Absorption Spectrophotometry In this lab exercise you will learn how to homogenize a tissue to extract the protein, and then how to use a protein assay reagent to
Cell Viability Assays: Microtitration (MTT) Viability Test Live/Dead Fluorescence Assay. Proliferation Assay: Anti-PCNA Staining
 Cell Viability Assays: Microtitration (MTT) Viability Test Live/Dead Fluorescence Assay Proliferation Assay: Anti-PCNA Staining Spring 2008 1 Objectives To determine the viability of cells under different
Cell Viability Assays: Microtitration (MTT) Viability Test Live/Dead Fluorescence Assay Proliferation Assay: Anti-PCNA Staining Spring 2008 1 Objectives To determine the viability of cells under different
Annexin V-FITC Apoptosis Detection Kit
 ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
MystiCq microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C
 microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C Product Description The microrna cdna Synthesis Mix has been designed to easily convert micrornas into cdna templates for qpcr
microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C Product Description The microrna cdna Synthesis Mix has been designed to easily convert micrornas into cdna templates for qpcr
No-wash, no-lyse detection of leukocytes in human whole blood on the Attune NxT Flow Cytometer
 APPLICATION NOTE Attune NxT Flow Cytometer No-wash, no-lyse detection of leukocytes in human whole blood on the Attune NxT Flow Cytometer Introduction Standard methods for isolating and detecting leukocytes
APPLICATION NOTE Attune NxT Flow Cytometer No-wash, no-lyse detection of leukocytes in human whole blood on the Attune NxT Flow Cytometer Introduction Standard methods for isolating and detecting leukocytes
ab185915 Protein Sumoylation Assay Ultra Kit
 ab185915 Protein Sumoylation Assay Ultra Kit Instructions for Use For the measuring in vivo protein sumoylation in various samples This product is for research use only and is not intended for diagnostic
ab185915 Protein Sumoylation Assay Ultra Kit Instructions for Use For the measuring in vivo protein sumoylation in various samples This product is for research use only and is not intended for diagnostic
PROTOCOL. Immunostaining for Flow Cytometry. Background. Materials and equipment required.
 PROTOCOL Immunostaining for Flow Cytometry 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 Rev.0 Background The combination of single cell analysis using flow cytometry and the specificity of antibody-based
PROTOCOL Immunostaining for Flow Cytometry 1850 Millrace Drive, Suite 3A Eugene, Oregon 97403 Rev.0 Background The combination of single cell analysis using flow cytometry and the specificity of antibody-based
CFSE Cell Division Assay Kit
 CFSE Cell Division Assay Kit Item No. 10009853 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 4 Precautions
CFSE Cell Division Assay Kit Item No. 10009853 Customer Service 800.364.9897 * Technical Support 888.526.5351 www.caymanchem.com TABLE OF CONTENTS GENERAL INFORMATION 3 Materials Supplied 4 Precautions
Classic Immunoprecipitation
 292PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Classic Immunoprecipitation Utilizes Protein A/G Agarose for Antibody Binding (Cat.
292PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Classic Immunoprecipitation Utilizes Protein A/G Agarose for Antibody Binding (Cat.
Application Information Bulletin: Set-Up of the CytoFLEX Set-Up of the CytoFLEX* for Extracellular Vesicle Measurement
 Application Information Bulletin: Set-Up of the CytoFLEX Set-Up of the CytoFLEX* for Extracellular Vesicle Measurement Andreas Spittler, MD, Associate Professor for Pathophysiology, Medical University
Application Information Bulletin: Set-Up of the CytoFLEX Set-Up of the CytoFLEX* for Extracellular Vesicle Measurement Andreas Spittler, MD, Associate Professor for Pathophysiology, Medical University
Annexin V-FITC Apoptosis Detection Kit
 ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
ab14085 Annexin V-FITC Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of Apoptosis in living cells (adherent and suspension). This product is for research
QuickZyme Soluble Collagen Assay
 QuickZyme Soluble Collagen Assay Version June 2012 FOR RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC PROCEDURES This package insert must be read in its entirety before using this product. Introduction Collagen
QuickZyme Soluble Collagen Assay Version June 2012 FOR RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC PROCEDURES This package insert must be read in its entirety before using this product. Introduction Collagen
High-quality genomic DNA isolation and sensitive mutation analysis
 Application Note High-quality genomic DNA isolation and sensitive mutation analysis Izabela Safin, Ivonne Schröder-Stumberger and Peter Porschewski Introduction A major objective of cancer research is
Application Note High-quality genomic DNA isolation and sensitive mutation analysis Izabela Safin, Ivonne Schröder-Stumberger and Peter Porschewski Introduction A major objective of cancer research is
LAB 14 ENZYME LINKED IMMUNOSORBENT ASSAY (ELISA)
 STUDENT GUIDE LAB 14 ENZYME LINKED IMMUNOSORBENT ASSAY (ELISA) GOAL The goal of this laboratory lesson is to explain the concepts and technique of enzyme linked immunosorbent assay (ELISA). OBJECTIVES
STUDENT GUIDE LAB 14 ENZYME LINKED IMMUNOSORBENT ASSAY (ELISA) GOAL The goal of this laboratory lesson is to explain the concepts and technique of enzyme linked immunosorbent assay (ELISA). OBJECTIVES
Annexin V-EGFP Apoptosis Detection Kit
 ab14153 Annexin V-EGFP Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of apoptosis in various samples This product is for research use only and is not intended
ab14153 Annexin V-EGFP Apoptosis Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of apoptosis in various samples This product is for research use only and is not intended
Hydrogen Peroxide Cell-Based Assay Kit
 Hydrogen Peroxide Cell-Based Assay Kit Item No. 600050 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Hydrogen Peroxide Cell-Based Assay Kit Item No. 600050 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Chromatin Immunoprecipitation
 Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Chromatin Immunoprecipitation A) Prepare a yeast culture (see the Galactose Induction Protocol for details). 1) Start a small culture (e.g. 2 ml) in YEPD or selective media from a single colony. 2) Spin
Seahorse XF Cell Mito Stress Test Kit
 Seahorse XF Cell Mito Stress Test Kit Part # 103015-100 User Guide For use with Seahorse XF e and XF Extracellular Flux Analyzers For Research Use Only 1 Table of Contents Product Description... 2 Introduction...
Seahorse XF Cell Mito Stress Test Kit Part # 103015-100 User Guide For use with Seahorse XF e and XF Extracellular Flux Analyzers For Research Use Only 1 Table of Contents Product Description... 2 Introduction...
6 Characterization of Casein and Bovine Serum Albumin
 6 Characterization of Casein and Bovine Serum Albumin (BSA) Objectives: A) To separate a mixture of casein and bovine serum albumin B) to characterize these proteins based on their solubilities as a function
6 Characterization of Casein and Bovine Serum Albumin (BSA) Objectives: A) To separate a mixture of casein and bovine serum albumin B) to characterize these proteins based on their solubilities as a function
NimbleGen DNA Methylation Microarrays and Services
 NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
RealLine HCV PCR Qualitative - Uni-Format
 Instructions for use PCR KIT FOR EXTRACTION OF RNA AND REAL TIME PCR DETECTION KIT FOR HEPATITIS C VIRUS RNA Research Use Only Qualitative Uni Format VBD0798 48 tests valid from: December 2013 Rev11122013
Instructions for use PCR KIT FOR EXTRACTION OF RNA AND REAL TIME PCR DETECTION KIT FOR HEPATITIS C VIRUS RNA Research Use Only Qualitative Uni Format VBD0798 48 tests valid from: December 2013 Rev11122013
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
50 g 650 L. *Average yields will vary depending upon a number of factors including type of phage, growth conditions used and developmental stage.
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: techsupport@norgenbiotek.com Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
Genolution Pharmaceuticals, Inc. Life Science and Molecular Diagnostic Products
 Genolution Pharmaceuticals, Inc. Revolution through genes, And Solution through genes. Life Science and Molecular Diagnostic Products www.genolution1.com TEL; 02-3010-8670, 8672 Geno-Serum Hepatitis B
Genolution Pharmaceuticals, Inc. Revolution through genes, And Solution through genes. Life Science and Molecular Diagnostic Products www.genolution1.com TEL; 02-3010-8670, 8672 Geno-Serum Hepatitis B
MISCIBILITY AND INTERACTIONS IN CHITOSAN AND POLYACRYLAMIDE MIXTURES
 MISCIBILITY AND INTERACTIONS IN CHITOSAN AND POLYACRYLAMIDE MIXTURES Katarzyna Lewandowska Faculty of Chemistry Nicolaus Copernicus University, ul. Gagarina 7, 87-100 Toruń, Poland e-mail: reol@chem.umk.pl
MISCIBILITY AND INTERACTIONS IN CHITOSAN AND POLYACRYLAMIDE MIXTURES Katarzyna Lewandowska Faculty of Chemistry Nicolaus Copernicus University, ul. Gagarina 7, 87-100 Toruń, Poland e-mail: reol@chem.umk.pl
Human Adult Mesothelial Cell Manual
 Human Adult Mesothelial Cell Manual INSTRUCTION MANUAL ZBM0025.01 SHIPPING CONDITIONS Human Adult Mesothelial Cells Orders are delivered via Federal Express courier. All US and Canada orders are shipped
Human Adult Mesothelial Cell Manual INSTRUCTION MANUAL ZBM0025.01 SHIPPING CONDITIONS Human Adult Mesothelial Cells Orders are delivered via Federal Express courier. All US and Canada orders are shipped
MLX BCG Buccal Cell Genomic DNA Extraction Kit. Performance Characteristics
 MLX BCG Buccal Cell Genomic DNA Extraction Kit Performance Characteristics Monolythix, Inc. 4720 Calle Carga Camarillo, CA 93012 Tel: (805) 484-8478 monolythix.com Page 2 of 9 MLX BCG Buccal Cell Genomic
MLX BCG Buccal Cell Genomic DNA Extraction Kit Performance Characteristics Monolythix, Inc. 4720 Calle Carga Camarillo, CA 93012 Tel: (805) 484-8478 monolythix.com Page 2 of 9 MLX BCG Buccal Cell Genomic
ab185916 Hi-Fi cdna Synthesis Kit
 ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
Real-time monitoring of rolling circle amplification using aggregation-induced emission: applications for biological detection
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 215 Supplementary Information Real-time monitoring of rolling circle amplification using aggregation-induced
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 215 Supplementary Information Real-time monitoring of rolling circle amplification using aggregation-induced
Z-Stacking and Z-Projection using a Scaffold-based 3D Cell Culture Model
 A p p l i c a t i o n N o t e Z-Stacking and Z-Projection using a Scaffold-based 3D Cell Culture Model Brad Larson and Peter Banks, BioTek Instruments, Inc., Winooski, VT Grant Cameron, TAP Biosystems
A p p l i c a t i o n N o t e Z-Stacking and Z-Projection using a Scaffold-based 3D Cell Culture Model Brad Larson and Peter Banks, BioTek Instruments, Inc., Winooski, VT Grant Cameron, TAP Biosystems
INTERFERin in vitro sirna/mirna transfection reagent PROTOCOL. 1 Standard sirna transfection of adherent cells... 2
 INTERFERin in vitro sirna/mirna transfection reagent PROTOCOL DESCRIPTION INTERFERin is a powerful sirna/mirna transfection reagent that ensures efficient gene silencing and reproducible transfection in
INTERFERin in vitro sirna/mirna transfection reagent PROTOCOL DESCRIPTION INTERFERin is a powerful sirna/mirna transfection reagent that ensures efficient gene silencing and reproducible transfection in
Genomic DNA Extraction Kit INSTRUCTION MANUAL
 Genomic DNA Extraction Kit INSTRUCTION MANUAL Table of Contents Introduction 3 Kit Components 3 Storage Conditions 4 Recommended Equipment and Reagents 4 Introduction to the Protocol 4 General Overview
Genomic DNA Extraction Kit INSTRUCTION MANUAL Table of Contents Introduction 3 Kit Components 3 Storage Conditions 4 Recommended Equipment and Reagents 4 Introduction to the Protocol 4 General Overview
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual
 Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Basic Science in Medicine
 Medical Journal of th e Islamic Republic of Iran Volume 18 Number 3 Fall 1383 November 2004 Basic Science in Medicine ] EXPANSION OF HUMAN CORD BLOOD PRIMITIVE PROGENITORS IN SERUM-FREE MEDIA USING HUMAN
Medical Journal of th e Islamic Republic of Iran Volume 18 Number 3 Fall 1383 November 2004 Basic Science in Medicine ] EXPANSION OF HUMAN CORD BLOOD PRIMITIVE PROGENITORS IN SERUM-FREE MEDIA USING HUMAN
Immunophenotyping peripheral blood cells
 IMMUNOPHENOTYPING Attune Accoustic Focusing Cytometer Immunophenotyping peripheral blood cells A no-lyse, no-wash, no cell loss method for immunophenotyping nucleated peripheral blood cells using the Attune
IMMUNOPHENOTYPING Attune Accoustic Focusing Cytometer Immunophenotyping peripheral blood cells A no-lyse, no-wash, no cell loss method for immunophenotyping nucleated peripheral blood cells using the Attune
Inc. Wuhan. Quantity Pre-coated, ready to use 96-well strip plate 1 Plate sealer for 96 wells 4 Standard (liquid) 2
 Uscn Life Science Inc. Wuhan Website: www.uscnk.com Phone: +86 27 84259552 Fax: +86 27 84259551 E-mail: uscnk@uscnk.com ELISA Kit for Human Prostaglandin E1(PG-E1) Instruction manual Cat. No.: E0904Hu
Uscn Life Science Inc. Wuhan Website: www.uscnk.com Phone: +86 27 84259552 Fax: +86 27 84259551 E-mail: uscnk@uscnk.com ELISA Kit for Human Prostaglandin E1(PG-E1) Instruction manual Cat. No.: E0904Hu
INSTRUCTION Probemaker
 INSTRUCTION Probemaker Instructions for Duolink In Situ Probemaker PLUS (Art. no. 92009-0020) and Duolink In Situ Probemaker MINUS (Art. no. 92010-0020) Table of content 1. Introduction 4 2. Applications
INSTRUCTION Probemaker Instructions for Duolink In Situ Probemaker PLUS (Art. no. 92009-0020) and Duolink In Situ Probemaker MINUS (Art. no. 92010-0020) Table of content 1. Introduction 4 2. Applications
MEF Nucleofector Kit 1 and 2
 page 1 of 7 MEF Nucleofector Kit 1 and 2 for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the they are isolated
page 1 of 7 MEF Nucleofector Kit 1 and 2 for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the they are isolated
EdU Flow Cytometry Kit. User Manual
 User Manual Ordering information: (for detailed kit content see Table 2) EdU Flow Cytometry Kits for 50 assays: Product number EdU Used fluorescent dye BCK-FC488-50 10 mg 6-FAM Azide BCK-FC555-50 10 mg
User Manual Ordering information: (for detailed kit content see Table 2) EdU Flow Cytometry Kits for 50 assays: Product number EdU Used fluorescent dye BCK-FC488-50 10 mg 6-FAM Azide BCK-FC555-50 10 mg
How To Make A Tri Reagent
 TRI Reagent For processing tissues, cells cultured in monolayer or cell pellets Catalog Number T9424 Store at room temperature. TECHNICAL BULLETIN Product Description TRI Reagent is a quick and convenient
TRI Reagent For processing tissues, cells cultured in monolayer or cell pellets Catalog Number T9424 Store at room temperature. TECHNICAL BULLETIN Product Description TRI Reagent is a quick and convenient
FOR REFERENCE PURPOSES
 BIOO LIFE SCIENCE PRODUCTS FOR REFERENCE PURPOSES This manual is for Reference Purposes Only. DO NOT use this protocol to run your assays. Periodically, optimizations and revisions are made to the kit
BIOO LIFE SCIENCE PRODUCTS FOR REFERENCE PURPOSES This manual is for Reference Purposes Only. DO NOT use this protocol to run your assays. Periodically, optimizations and revisions are made to the kit
RealStar HBV PCR Kit 1.0 11/2012
 RealStar HBV PCR Kit 1.0 11/2012 RealStar HBV PCR Kit 1.0 For research use only! (RUO) Product No.: 201003 96 rxns INS-201000-GB-02 Store at -25 C... -15 C November 2012 altona Diagnostics GmbH Mörkenstraße
RealStar HBV PCR Kit 1.0 11/2012 RealStar HBV PCR Kit 1.0 For research use only! (RUO) Product No.: 201003 96 rxns INS-201000-GB-02 Store at -25 C... -15 C November 2012 altona Diagnostics GmbH Mörkenstraße
BD PuraMatrix Peptide Hydrogel
 BD PuraMatrix Peptide Hydrogel Catalog No. 354250 Guidelines for Use FOR RESEARCH USE ONLY NOT FOR CLINICAL, DIAGNOSTIC OR THERAPEUTIC USE SPC-354250-G rev 2.0 1 TABLE OF CONTENTS Intended Use... 2 Materials
BD PuraMatrix Peptide Hydrogel Catalog No. 354250 Guidelines for Use FOR RESEARCH USE ONLY NOT FOR CLINICAL, DIAGNOSTIC OR THERAPEUTIC USE SPC-354250-G rev 2.0 1 TABLE OF CONTENTS Intended Use... 2 Materials
Uses of Flow Cytometry
 Uses of Flow Cytometry 1. Multicolour analysis... 2 2. Cell Cycle and Proliferation... 3 a. Analysis of Cellular DNA Content... 4 b. Cell Proliferation Assays... 5 3. Immunology... 6 4. Apoptosis... 7
Uses of Flow Cytometry 1. Multicolour analysis... 2 2. Cell Cycle and Proliferation... 3 a. Analysis of Cellular DNA Content... 4 b. Cell Proliferation Assays... 5 3. Immunology... 6 4. Apoptosis... 7
CytoSelect Cell Viability and Cytotoxicity Assay Kit
 Product Manual CytoSelect Cell Viability and Cytotoxicity Assay Kit Catalog Number CBA-240 96 assays (96-well plate) FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction The measurement
Product Manual CytoSelect Cell Viability and Cytotoxicity Assay Kit Catalog Number CBA-240 96 assays (96-well plate) FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction The measurement
EVALUATION OF THE EFFECT OF THE COMPOUND RIBOXYL ON THE ATP LEVEL AND CELLULAR RESPIRATION FROM HUMAN DERMAL FIBROBLASTS
 EVALUATION OF THE EFFECT OF THE COMPOUND RIBOXYL ON THE ATP LEVEL AND CELLULAR RESPIRATION FROM HUMAN DERMAL FIBROBLASTS FINAL REPORT OF THE STUDY GT050923 Study promoter: LUCAS MEYER COSMETICS ZA Les
EVALUATION OF THE EFFECT OF THE COMPOUND RIBOXYL ON THE ATP LEVEL AND CELLULAR RESPIRATION FROM HUMAN DERMAL FIBROBLASTS FINAL REPORT OF THE STUDY GT050923 Study promoter: LUCAS MEYER COSMETICS ZA Les
HPLC Analysis of Acetaminophen Tablets with Waters Alliance and Agilent Supplies
 HPLC Analysis of Acetaminophen Tablets with Waters Alliance and Agilent Supplies Application Note Small Molecule Pharmaceuticals Authors Jignesh Shah, Tiantian Li, and Anil Sharma Agilent Technologies,
HPLC Analysis of Acetaminophen Tablets with Waters Alliance and Agilent Supplies Application Note Small Molecule Pharmaceuticals Authors Jignesh Shah, Tiantian Li, and Anil Sharma Agilent Technologies,
AAGPs TM Anti-Aging Glyco Peptides. Enhancing Cell, Tissue and Organ Integrity Molecular and biological attributes of lead AAGP molecule
 AAGPs TM Anti-Aging Glyco Peptides Enhancing Cell, Tissue and Organ Integrity Molecular and biological attributes of lead AAGP molecule 1 Acknowledgements This presentation was prepared by Dr. Samer Hussein
AAGPs TM Anti-Aging Glyco Peptides Enhancing Cell, Tissue and Organ Integrity Molecular and biological attributes of lead AAGP molecule 1 Acknowledgements This presentation was prepared by Dr. Samer Hussein
SmartFlare RNA Detection Probes: Principles, protocols and troubleshooting
 Technical Guide SmartFlare RNA Detection Probes: Principles, protocols and troubleshooting Principles of SmartFlare technology RNA detection traditionally requires transfection, laborious sample prep,
Technical Guide SmartFlare RNA Detection Probes: Principles, protocols and troubleshooting Principles of SmartFlare technology RNA detection traditionally requires transfection, laborious sample prep,
QuickTiter FeLV Core Antigen ELISA Kit (FeLV p27)
 Product Manual QuickTiter FeLV Core Antigen ELISA Kit (FeLV p27) Catalog Numbers VPK-155 96 wells FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Feline leukemia virus (FeLV) is
Product Manual QuickTiter FeLV Core Antigen ELISA Kit (FeLV p27) Catalog Numbers VPK-155 96 wells FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Feline leukemia virus (FeLV) is
Chromatin Immunoprecipitation (ChIP)
 Chromatin Immunoprecipitation (ChIP) Day 1 A) DNA shearing 1. Samples Dissect tissue (One Mouse OBs) of interest and transfer to an eppendorf containing 0.5 ml of dissecting media (on ice) or PBS but without
Chromatin Immunoprecipitation (ChIP) Day 1 A) DNA shearing 1. Samples Dissect tissue (One Mouse OBs) of interest and transfer to an eppendorf containing 0.5 ml of dissecting media (on ice) or PBS but without
The cell lines used in this study were obtained from the American Type Culture
 Supplementary materials and methods Cell culture and drug treatments The cell lines used in this study were obtained from the American Type Culture Collection (ATCC) and grown as previously described.
Supplementary materials and methods Cell culture and drug treatments The cell lines used in this study were obtained from the American Type Culture Collection (ATCC) and grown as previously described.
QUANTITATIVE RT-PCR. A = B (1+e) n. A=amplified products, B=input templates, n=cycle number, and e=amplification efficiency.
 QUANTITATIVE RT-PCR Application: Quantitative RT-PCR is used to quantify mrna in both relative and absolute terms. It can be applied for the quantification of mrna expressed from endogenous genes, and
QUANTITATIVE RT-PCR Application: Quantitative RT-PCR is used to quantify mrna in both relative and absolute terms. It can be applied for the quantification of mrna expressed from endogenous genes, and
Procedure for RNA isolation from human muscle or fat
 Procedure for RNA isolation from human muscle or fat Reagents, all Rnase free: 20% SDS DEPC-H2O Rnase ZAP 75% EtOH Trizol Chloroform Isopropanol 0.8M NaCitrate/1.2M NaCl TE buffer, ph 7.0 1. Homogenizer-probe
Procedure for RNA isolation from human muscle or fat Reagents, all Rnase free: 20% SDS DEPC-H2O Rnase ZAP 75% EtOH Trizol Chloroform Isopropanol 0.8M NaCitrate/1.2M NaCl TE buffer, ph 7.0 1. Homogenizer-probe
Title: Mapping T cell epitopes in PCV2 capsid protein - NPB #08-159. Date Submitted: 12-11-09
 Title: Mapping T cell epitopes in PCV2 capsid protein - NPB #08-159 Investigator: Institution: Carol Wyatt Kansas State University Date Submitted: 12-11-09 Industry summary: Effective circovirus vaccines
Title: Mapping T cell epitopes in PCV2 capsid protein - NPB #08-159 Investigator: Institution: Carol Wyatt Kansas State University Date Submitted: 12-11-09 Industry summary: Effective circovirus vaccines
Optimized Protocol sirna Test Kit for Cell Lines and Adherent Primary Cells
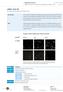 page 1 of 8 sirna Test Kit for Cell Lines and Adherent Primary Cells Kit principle Co-transfection of pmaxgfp, encoding the green fluorescent protein (GFP) from Pontellina p. with an sirna directed against
page 1 of 8 sirna Test Kit for Cell Lines and Adherent Primary Cells Kit principle Co-transfection of pmaxgfp, encoding the green fluorescent protein (GFP) from Pontellina p. with an sirna directed against
Real-time PCR: Understanding C t
 APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
TransIT -2020 Transfection Reagent
 Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/5400 INTRODUCTION TransIT -2020 Transfection Reagent is a high-performance, animal-origin free, broad spectrum reagent
Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/5400 INTRODUCTION TransIT -2020 Transfection Reagent is a high-performance, animal-origin free, broad spectrum reagent
Western Blot Protocol Protein isolation
 Western Blot Protocol Protein isolation A. Preparation of cell lysates. - Preparation of materials: -Dial the microcentrifuge temperature control setting to 4 C -Prepare a bucket of ice -Prepare lysis
Western Blot Protocol Protein isolation A. Preparation of cell lysates. - Preparation of materials: -Dial the microcentrifuge temperature control setting to 4 C -Prepare a bucket of ice -Prepare lysis
use. 3,5 Apoptosis Viability Titration Assays
 Introduction Monitoring cell viability can provide critical insights into the effectiveness of biological protocols and the stability of biological systems including: the efficacy of cell harvesting procedures,
Introduction Monitoring cell viability can provide critical insights into the effectiveness of biological protocols and the stability of biological systems including: the efficacy of cell harvesting procedures,
CompleteⅡ 1st strand cdna Synthesis Kit
 Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Osteoblast Differentiation and Mineralization
 Osteoblast Differentiation and Mineralization Application Note Background Osteoblasts are specialized fibroblasts that secrete and mineralize the bone matrix. They develop from mesenchymal precursors.
Osteoblast Differentiation and Mineralization Application Note Background Osteoblasts are specialized fibroblasts that secrete and mineralize the bone matrix. They develop from mesenchymal precursors.
