GIPZ Lentiviral shrna
|
|
|
- Clyde Gardner
- 9 years ago
- Views:
Transcription
1 GE Healthcare GIPZ Lentiviral shrna Technical Manual Product Description: The Dharmacon GIPZ Lentiviral shrna Library was developed in collaboration with Dr. Greg Hannon of Cold Spring Harbor Laboratory [CSHL] and Dr. Steve Elledge of Harvard Medical School. This library combines the design advantages of Dharmacon microrna-adapted shrna with the pgipz lentiviral vector to create a powerful RNA tool capable of producing RNA interference (RNAi) in most cell types including primary and non-dividing cells. Note: GIPZ shrna is available in glycerol stock or viral particle format. If viral particle format is purchased, begin work with Protocol IX Determining Relative Transduction Efficiency. Important Safety Note: Please follow the safety guidelines for use and production of vector-based lentivirus as set by your institution s biosafety committee. For glycerol stocks of E. coli containing lentiviral plasmids, BSL1 guidelines should be followed For handling and use of lentiviral products to produce lentiviral particles, BSL2 or BSL2+ guidelines should be followed For handling and use of lentiviral particle products, BSL2 or BSL2+ guidelines should be followed Additional information on the safety features incorporated in the pgipz lentiviral vector and the Dharmacon Trans-Lentiviral Packaging System can be found on page 3. Note: Dharmacon GIPZ shrna vectors are not compatible with third generation packaging systems, due to the requirement of the expression of tat, which third generation systems do not contain. We recommend the Trans-Lentiviral Packaging System for use with our vectors. Design Information: Unique microrna-30 Based Hairpin Design Short hairpin RNA (shrna) constructs are expressed as human microrna-30 (mir-30) primary transcripts. This design adds a Drosha processing site to the hairpin construct and has been shown to greatly increase gene silencing efficiency (Boden 2004). The hairpin stem consists of 22 nucleotides (nt) of dsrna and a 19 nucleotides (nt) loop from human mir-30. Adding the mir-30 loop and 125 nucleotides (nt) of mir-30 flanking sequence on either side of the hairpin results in greater than 10-fold increase in Drosha and Dicer processing of the expressed hairpins when compared with conventional shrna designs (Silva 2005). Increased Drosha and Dicer processing translates into greater shrna production and greater potency for expressed hairpins. Use of the mir-30 design also allowed the use of rules-based designs for target sequence selection. One such rule is the destabilizing of the 5' end of the antisense strand, which results in strand specific incorporation of microrna/sirnas into RISC. The proprietary design algorithm targets sequences in coding regions and the 3' UTR with the additional requirement that they contain greater than 3 mismatches to any other sequence in the human or mouse genomes. Each shrna construct has been bioinformatically verified to match NCBI sequence data. To assure the highest possibility of modulating the gene expression level, each gene is represented by multiple shrna constructs, each covering a unique region of the target gene. Vector Information: Versatile Vector Design Features of the pgipz lentiviral vector (Figures 1 and 2) that make it a versatile tool for RNAi studies include: Ability to perform transfections or transductions using the replication incompetent lentivirus (Shimada 1995) TurboGFP (Evrogen, Moscow, Russia) and shrna are part of a bicistronic transcript allowing the visual marking of shrna expressing cells. Amenable to in vitro and in vivo applications. Puromycin drug resistance marker for selecting stable cell lines. Molecular barcodes enable multiplexed screening in pools.
2 Revised versions From PowerPoint (R&D) tgfp IRES Puro R WPRE tgfp IRES Puro R WPRE Ψ 5' LTR Amp R pgipz puc ori shrna SV40 ori 3' SIN LTR Ψ 5ʹ LTR Figure 1. pgipz lentiviral vector. pgipz shrna Amp R pucori SV40 ori 3ʹ SIN LTR Table 1. Features of the pgipz vector. Vector Element tgfp 5' LTR Ψ Puro R IRES shrna 5' LTR Ψ Vector Map: 5' LTR Ψ RSV/5' LTR Ψ TRE trfp Amp R Utility UBC rtta3 IRES Puro R WPRE Human cytomegalovirus promoter drives strong transgene expression Puromycin ptripz resistance permits antibiotic-selective pressure and propagation of stable integrants ptripz Internal ribosomal entry site allows expression of TurboGFP and puromycin resistance genes in a single transcript psmart 2.0 plko.1 pgipz 11.8 kb tgfp IRES Puro R microrna WPRE psmart 2.0 Amp R puc ori SV40 ori Amp R pucori SV40 ori Ψ TurboGFP shrna reporter for visual tracking of transduction and expression puc ori 5' LTR 5' long terminal repeat SV40 ori Amp R puc ori SV40 ori U6 hpgk shrna Puro R puc ori Amp R 3' SIN LTR 3' SIN LTR 3' SIN LTR 3' SIN LTR 5ʹ LTR microrna-adapted shrna (based on mir-30) for gene knockdown 5ʹ LTR Ψ RSV/5ʹ LTR Ψ 5ʹ LTR Ψ TRE UBC trfp rtta3 IRES Puro R WPRE shrna Amp R pucori SV40 ori 3' SIN LTR 3' self-inactivating long terminal repeat for increased lentivirus safety Ψ Psi packaging sequence allows viral genome packaging using lentiviral packaging systems Rev response element enhances titer by increasing packaging efficiency of full-length viral genomes tgfp IRES Puro R shrna WPRE cppt/cts tgfp IRES Puro R shrna WPRE WPRE Woodchuck hepatitis posttranscriptional regulatory element enhances transgene expression in the target cells psmart 2.0 Amp R pucori SV40 ori U6 hpgk shrna Puro R plko.1 pucori Amp R tgfp IRES Puro R microrna WPRE psmart 2.0 3ʹ SIN LTR 3ʹ SIN LTR 3ʹ SIN LTR 3ʹ SIN LTR 5' LTR Ψ Figure 2. Detailed vector map of pgipz lentiviral vector. ORF IRES Multi Tag Antibiotic Resistance: Cloning Site -2atGFPnuc Blast R pgipz contains three antibiotic resistance ploc markers (Table 2). Table 2. Antibiotic resistances conveyed by pgipz. WPRE 3' SIN LTR Drawing was created using Geneious ( Antibiotic Amp R puc ori SV40 ori Concentration Utility Amp R pucori SV40 ori Ampicillin (carbenicillin) 100 μg/ml Bacterial selection marker (outside LTRs) Ψ 5ʹ LTR ORF IRES Multi Tag Cloning Site ploc Zeocin 25 μg/ml Bacterial selection marker (inside LTRs) Puromycin Variable Mammalian selection marker tgfp nuc -2a- Blast R WPRE 3ʹ SIN LTR SMARTchoice promoters mcmv hef1α mef1α CAG PGK UBC SMARTchoice shrna SMARTchoice promoters mcmv hef1α mef1α CAG PGK UBC 5' LTR Ψ tgfp IRES Puro R WPRE or trfp SMARTvector universal scaffold 3' SIN LTR 5ʹ LTR Ψ tgfp or trfp IRES Puro R WPRE 3ʹ SIN LTR SMARTvector2.0 universal scaffold
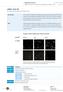
3 Quality Control: The GIPZ Lentiviral shrna Library has passed through internal QC processes to ensure high quality and low recombination (Figures 3 and 4). Figure 3. Representative GIPZ Lentiviral shrna clones grown for 16 hours at 30 C. Plasmid was isolated and normalized to a standard concentration. Clones were then digested with SacII and run on an agarose gel with uncut plasmid. The expected band sizes are 1259 bp, 2502 bp, and 7927 bp. No recombinant products are visible. 10 kb molecular weight ladder (10 kb, 7 kb, 5 kb, 4 kb, 3 kb, 2.5 kb, 2 kb, 1.5 kb, 1 kb). Figure 4. Gel image of a restriction digest of clones from the GIPZ shrna library cultured for 10 successive generations in an attempt to determine the tendency of the pgipz vector to recombine. Each generation was thawed, replicated and incubated overnight for 16 hours at 30 C then frozen, thawed and replicated. This process was repeated for 10 growth cycles. After the 10th growth cycle, plasmid was isolated and normalized to a standard concentration. Clones were digested with SacII and run on an agarose gel. The pgipz vector appears stable without showing any recombination. Expected band sizes: 1259 bp, 2502 bp, and 7927 bp. 10 kb molecular weight ladder (10 kb, 7 kb, 5 kb, 4 kb, 3 kb, 2.5 kb, 2 kb, 1.5 kb, 1 kb). Additional Safety Information: Historically, the greatest safety risk associated with a lentiviral delivery platform stems from the potential generation of recombinant viruses that are capable of autonomous replication. The pgipz Lentiviral shrna platform minimizes these hazards to the greatest degree by combining a disabled viral genome with the proprietary Trans-Lentiviral packaging process. Starting with the HXB2 clone of HIV-1 (GenBank, Accession #K03455), the lentiviral backbone has been modified to eliminate all but the most essential genetic elements necessary for packaging and integration (such as 5' LTR, Psi sequences, polypurine tracts, Rev responsive elements and 3' LTR). The resultant self-inactivating (SIN) vector greatly reduces the probability of producing recombinant particles and limits cellular toxicity often associated with expression of HIV genes. Additional safety features can be incorporated by the packaging process itself. Generation of pgipz Lentiviral shrna particles requires a packaging step during which the expression construct containing the silencing sequence is enclosed in a viral capsid. Gene functions that facilitate this process (such as those encoded by the structural genes gag, pol, env, etc.) are distributed amongst multiple helper plasmids which do not contain significant regions of homology. This tactic further minimizes the probability of recombination events that might otherwise generate viruses capable of autonomous replication. Among commercially available lentiviral vector systems, the Trans-Lentiviral Packaging System offers a superior safety profile as the packaging components are separated onto five plasmids. Additionally, expression of gag-pro and tat-rev are under the control of the conditional tetracycline-responsive promoter element (TRE), limiting expression of these viral components strictly to the packaging cell line. A detailed description of the Trans-Lentiviral Packaging System can be found in (Wu 2000). With these safety measures in place, GIPZ lentiviral shrna particles can be employed in standard Biosafety Level 2 tissue culture facilities. Any investigator who purchases Dharmacon Viral Vector Products is responsible for consulting with their institution s health and biosafety group for specific guidelines on the handling of lentiviral vector particles. Further, each investigator is fully responsible for obtaining the required permissions for the acceptance of lentiviral particles into their local geography and institution. In the U.S., download the U.S. Department of Health and Human Services Centers for Disease Control and Prevention and National Institutes of Health, Biosafety in Microbiological and Biomedical Laboratories (BMBL), Fifth Edition, Feb 2007 here. See also: NIH Guidelines For Research Involving Recombinant DNA Molecules (NIH Guidelines), September 2009, downloadable here. For Biosafety Considerations for Research with Lentiviral Vectors, see.
4 Protocol I Replication: For archive replication, grow all GIPZ shrna clones at 30 C in 2x LB broth (low salt)* medium plus 25 μg/ml Zeocin and 100 μg/ml carbenicillin in order to provide maximum stability of the clones. Prepare medium with 8% glycerol** and the appropriate antibiotics. 2x LB broth (low-salt) medium preparation LB-Broth-Lennox 20 g/l Peptone 10 g/l Yeast Extract 5 g/l Appropriate antibiotic(s) at recommended concentration(s). Glycerol 8% for long-term storage. *1x LB medium can be used instead of 2x LB broth medium. **Glycerol can be omitted from the medium if you are culturing for plasmid preparation. If making copies of the constructs for long-term storage at -80 C, 8% glycerol is required. Table 3. Materials for plate replication. Item Vendor Cat # 2x LB Broth (low salt) Fisher Scientific BP Peptone, granulated, 2 kg Difco Fisher Scientific BP Yeast Extract, 500 g, granulated Fisher Scientific BP NaCl Fisher Scientific BP3581 Glycerol Fisher Scientific BP2291 Carbenicillin Fisher Scientific BP Zeocin Invitrogen ant-zn-5p Puromycin Fisher Scientific BP well microplates Fisher Scientific Aluminum seals Fisher Scientific Disposable replicators Fisher Scientific NC Replication of Plates Prepare target plates by dispensing ~ 160 μl of 2x LB broth (low salt) medium supplemented with 8% glycerol** and appropriate antibiotic (25 μg/ml Zeocin and 100 μg/ml carbenicillin). Prepare Source Plates 1. Remove foil seals from the source plates while they are still frozen. This minimizes cross-contamination. 2. Thaw the source plates with the lid on. Wipe any condensation underneath the lid with a paper wipe soaked in ethanol. Replicate 1. Gently place a disposable replicator in the thawed source plate and lightly move the replicator around inside the well to mix the culture. Make sure to scrape the bottom of the plate of the well. 2. Gently remove the replicator from the source plate and gently place in the target plate and mix in the same manner to transfer cells. 3. Dispose of the replicator. 4. Place the lids back on the source plates and target plates. 5. Repeat steps 1-4 until all plates have been replicated. 6. Return the source plates to the -80 C freezer. 7. Place the inoculated target plates in a 30 C incubator for hours. Freeze at -80 C for long-term storage. Avoid long periods of storage at room temperature or higher in order to control background recombination products. Note: Due to the tendency of viral vectors to recombine, we recommend keeping the incubation times as short as possible and avoid subculturing. Return to your glycerol stock for each plasmid preparation. Protocol II Plasmid Preparation: Culture Conditions for Individual Plasmid Preparations For plasmid preparation, grow all GIPZ shrna clones at 37 C in 2x LB broth (low salt) medium plus 100 μg/ml carbenicillin only. 1. Upon receiving your glycerol stock(s) containing the shrna of interest, store at -80 C until ready to begin. 2. To prepare plasmid DNA, first thaw your glycerol stock culture and pulse vortex to resuspend any E. coli that may have settled to the bottom of the tube. 3. Take a 10 μl inoculum from the glycerol stock into 3-5 ml of 2x LB broth (low salt) medium with 100 μg/ml carbenicillin. Return the glycerol stock(s) to -80 C. Note: If a large culture volume is desired, incubate the 3-5 ml culture for 8 hours at 37 C with shaking and use as a starter inoculum. Dilute the starter culture 1:500-1:1000 into the larger volume. 4. Incubate at 37 C for hours with vigorous shaking. 5. Pellet the culture and begin preparation of plasmid DNA. Plasmid DNA can be isolated using Thermo Scientific GeneJET Plasmid Miniprep Kit (Cat #K0502) or similar. 6. Run μg of the plasmid DNA on a 1% agarose gel. pgipz with shrna is bp. Note: Due to the tendency of viral vectors to recombine, we recommend keeping the incubation times as short as possible and avoid subculturing. Return to your original glycerol stock for each plasmid preparation.
5 Culture Conditions For 96-Well Bio-Block Plasmid Preparation Inoculate a 96-well bio-block containing 1 ml per well of 2x LB broth (low salt) medium with 100 μg/ml carbenicillin with 1 µl of the glycerol stock culture. Incubate at 37 C with shaking (~ rpm). We have observed that incubation times between hours produce good plasmid yield. For plasmid preparation, follow the protocols recommended by the plasmid isolation kit manufacturer. Note: For the library collection, we use the above 96-well bio-block plasmid preparation protocol in conjunction with a Qiagen Turbo Kit (Cat #27191). We use 2 bio-blocks combined. Do not perform the optional wash and elute the DNA in molecular grade water. Protocol III Restriction Digest: The following is a sample protocol for restriction enzyme digestion using Thermo Scientific FastDigest Restriction Enzymes KpnI (Cat #FD0524), SalI (Cat #FD0644), XhoI (Cat #FD0694) and/or NotI (Cat #FD0594) and Thermo Scientific Restriction Enzyme SalI (Cat #ER0205) for diagnostic quality control of pgipz lentiviral vectors. 1. Add the following components (Table 4), in the order stated, to a sterile PCR thin-wall tube. 2. Mix gently by pipetting. 3. Incubate in a thermal cycler at 37 C for 5 minutes for FastDigest enzymes or as suggested by the manufacturer. 4. Load the gel with 10 µl of each of the digested samples, KpnI, SacII, SalI, XhoI and/or NotI on a 1% agarose gel. Run uncut sample alongside the digested samples. (Figure 5). Table 4. Restriction Digest Components. Component Water, nuclease-free (Cat #R0581) 10x FastDigest buffer DNA sample (up to 1 µg) in water FastDigest enzyme Final Volume Amount X μl 2 μl X μl 1 μl 20 μl Lane 1 - Lane 2 - Lane 3 - Lane 4 - Lane 5 - Lane 6-10 kb molecular weight ladder (10 kb, 7 kb, 5 kb, 4 kb, 3 kb, 2.5 kb, 2 kb, 1.5 kb, 1 kb) Uncut pgipz vector KpnI digested pgipz produces two bands at 1917 bp and 9857 bp. SacII digest produces three bands at 1345 bp, 2502 bp and 7927 bp SalI produces three bands at 2188 bp, 4465 bp and 5121 bp XhoI, NotI double digest produces two bands at 1291 bp and 10,483 bp Figure 5. Restriction digests with pgipz. Protocol IV Transfection: Quantities and volumes should be scaled-up according to the number of cells/wells to be transfected (Table 5). This example is for 24-well plate format. 1. In each well, seed ~ adherent cells or ~ suspension cells in 1 ml of growth medium 24 hours prior to transfection. Note: The recommended confluency for adherent cells on the day of transfection is 70-90%. Suspension cells should be in logarithmic growth phase at the time of transfection. 2. Dilute 1 µg of DNA in 100 µl of DMEM or other serum-free growth medium. Note: The transfection efficiency with Thermo Scientific TurboFect Transfection Reagent (Cat #R0531) is equally high in the presence of serum. This is not the case with many other transfection reagents. 3. Briefly vortex TurboFect Transfection Reagent and add 2 µl to the diluted DNA. Mix immediately by pipetting or vortexing. 4. Incubate minutes at room temperature. Note: Prepare immediately prior to transfection. We recommend starting with 1 µg of DNA and 2 µl of TurboFect reagent per well in a 24-well plate (see scale-up Table 5). Subsequent optimization may further increase transfection efficiency depending on the cell line and transgene used. 5. Add 100 µl of the TurboFect reagent/dna mixture drop-wise to each well. Do not remove the growth medium from the cells. 6. Gently rock the plate to achieve even distribution of the complexes. 7. Incubate at 37 C in a CO 2 incubator. 8. Analyze transgene expression hours later. For stable transfection, cells should be grown in selective medium for days (see Protocol V Puromycin Selection). Table 5. Scale-up ratios for transfection of adherent and suspension cells with TurboFect Transfection Reagent. Tissue Culture Vessel Growth area, cm 2 /well Volume of medium, ml Adherent (suspension) cells to seed the day before transfection* Amount of DNA Volume of TurboFect µg** µl*** Recommended Range 96-well plate ( ) well plate ( ) well plate ( ) well plate ( ) well plate ( ) mm plate ( ) * These numbers were determined using HeLa and Jurkat cells. Actual values depend on the cell type. ** Amount of DNA and TurboFect Transfection Reagent used may require optimization. *** The volume of DNA should be 1/10 of the volume of the culture medium used for dilution of the DNA.
6 Cells Successfully Transfected with Dharmacon TurboFect Transfection Reagents Include: Permanently growing cell lines Primary cell cultures Cos-7 african green monkey kidney cells Rat fibroblasts HeLa human cervix adenocarcinoma cells Mouse bone marrow derived dendritic cells CHO chinese hamster ovary cells Mouse bone marrow derived macrophages HEK293 human embryonic kidney cells HLF human lung fibroblasts B50 rat nervous tissue neuronal cells Calu1 human lung epidermoid carcinoma cells RAW264 mouse leukaemic monocyte-macrophage cells WEHI mouse B cell lymphoma cells MDCK Madin Darby Canine Kidney cells Raji human Burkitt s lymphoma cells COLO human colon adenocarcinoma cells Jurkat human leukaemic T cells Sp2/Ag14 mouse myeloma cells HeLa S3 human cervix carcinoma cells Hep2C human larynx carcinoma cells L929 mouse connective tissue fibroblasts NIH3T3 mouse embryo fibroblasts Protocol V Puromycin Selection: Determining Antibiotic Dose Response (Kill Curve) In order to generate stable cell lines expressing the transgene of interest, it is important to determine the minimum amount of antibiotic required to kill non-transfected cells. A simple procedure to test this is as follows: 1. Day 1: Using the same cell type and relative cell densities to be used in subsequent transfection or transduction procedures, plate cells and culture overnight. 2. Day 2: Replace complete growth medium with growth medium supplemented with a range of puromycin concentrations (0-15 μg/ml), including untreated control cells with no antibiotic added. 3. Day 4: Refresh medium and assess viability. 4. Replace medium with fresh medium supplemented with the appropriate concentration of puromycin every 2-3 days depending on the growth of cells. 5. Examine cells daily and identify the minimal concentration of antibiotic that efficiently kills all non-transfected/transduced cells between 3-6 days following addition of puromycin. Puromycin Selection If adding antibiotic for selection, use the appropriate concentration as determined based on the above kill curve. 1. Add medium containing antibiotic 24 or hours post-transfection or post-transduction, respectively, to begin selection Note: It is important to wait at least 24 hours after transfection before beginning selection. 2. Cells can be harvested for transgene expression hours after starting selection. 3. If longer selection is required for cells to be confluent, replace selective medium approximately every 2-3 days. 4. Monitor the cells daily and observe the percentage of surviving cells. Cells surviving selection will be expressing the transgene. 5. If generating stable cell lines (optional), select and grow for days. 6. Once non-transfected cells are eliminated and/or you have selected for stably transfected cell lines if desired, you can proceed to assay for transgene expression. RT-qPCR, Western blot analysis or other appropriate functional assay can be used; compare treated samples to untreated, reporter alone, non-silencing control, or other controls as appropriate. Protocol VI Packaging Lentivirus: The pgipz vector is tat dependant, so you must use a packaging system that expresses the tat gene. For packaging our lentiviral shrna constructs, we recommend the Trans-Lentiviral shrna Packaging Kit (Cat #TLP5912 or TLP5917). The Trans-Lentiviral shrna Packaging System allows creation of a replication-incompetent, HIV-1-based lentivirus which can be used to deliver and express your shrna of interest in either dividing or non-dividing mammalian cells. The Trans-Lentiviral shrna Packaging System uses a replication-incompetent lentivirus based on the trans-lentiviral system developed by Kappes (Kappes 2001). For protocols and information on packaging pgipz with our Trans-Lentiviral shrna Packaging System, please see the product manual available on our website. Protocol VII Titering: Viral Titering Follow the procedure below to determine the titer of your lentiviral stock using the mammalian cell line of choice IF YOU HAVE PRODUCED VIRAL PARTICLES YOURSELF. This protocol uses the HEK293T cell line that is available as part of our Trans-Lentiviral shrna Packaging. Kit (Cat #TLP5918) or separately (Cat #HCL4517). You can use a standard HEK293T cell line as an alternative. Note: If you have generated a lentiviral stock of the expression control (such as GIPZ non-silencing construct), we recommend titering this stock as well. Virus stock 1 Virus stock 2 Virus stock 3 Virus stock 4 A B C D E F G H Figure 6. Five-fold serial dilutions of virus stock. Dilution Plate
7 1. The day before transduction, seed a 24-well tissue culture plate with HEK293T Cells at cells per well in DMEM (10% FBS, 1% pen-strep). Note: The following day, each well should be no more than 40-50% confluent. 2. Make dilutions of the viral stock in a round bottom 96-well plate using serum-free media. Utilize the plate as shown in (Figure 6) using one row for each virus stock to be tested. Use the procedure below (starting at step 4) for dilution of the viral stocks. The goal is to produce a series of 5-fold dilutions to reach a final dilution of 390,625-fold. 3. Add 80 µl of serum-free media to each well. 4. Add 20 µl of thawed virus stock to each corresponding well in column 1 (five-fold dilution). Note: Pipette contents of well up and down times. Discard pipette tip. 5. With new pipette tips, transfer 20 µl from each well of column 1 to the corresponding well in column 2. Note: Pipette up and down times and discard pipette tip. 6. With new pipette tips, transfer 20 µl from each well of column 2 to the corresponding well in column 3. Note: Pipette up and down times and discard pipette tip. 7. Repeat transfers of 20 µl from columns 3 through 8, pipetting up and down times and changing pipette tips between each dilution. Note: It is strongly recommended that you use a high quality multichannel pipettor when performing multiple dilutions. Pre-incubate the dilutions of the virus stock for 5 minutes at room temperature. 8. Label 24-well plate as shown in (Figure 7) using one row for each virus stock to be tested. 9. Remove culture media from the cells in the 24-well plate. 10. Add 225 µl of serum-free media to each well Virus stock 1 A Virus stock 2 B Virus stock 3 C Virus stock 4 D 11. Transduce cells by adding 25 µl of diluted virus from the original 96-well plate (Figure 6) to a well on the 24-well destination plate (Figure 7) containing the cells. For example, transfer 25 µl from well A2 of the 96-well plate into well A1 in the 24-well plate (Table 6). 12. Incubate transduced cultures at 37 C for 4 hours. 13. Remove transduction mix from cultures and add 1 ml of DMEM (10% FBS, 1% Pen-Strep). 14. Culture cells for 48 hours. 15. Count the TurboGFP expressing cells or colonies of cells (Figure 8). Note: Count each multi-cell colony as 1 transduced cell, as the cells will be dividing over the 48 hour culture period. Figure 8 illustrates this principle of cell counting. 16. Transducing units per ml (TU/mL) can be determined using the following formula: # of TurboGFP positive colonies counted dilution factor 40 = # TU/mL: Example: 55 TurboGFP positive colonies counted in well A3 55 (TurboGFP positive colonies) 625 (dilution factor) 40 = TU/mL Table 6. Example of set up for dilutions. Originating (96-well plate) Figure 7. Twenty four well tissue culture plate, seeded with HEK293T cells, used to titer the virus. Well (Row A, B, C, or D) Destination (24-well plate) Volume Diluted Virus Used Dilution Factor A1 25 μl 5 * A2 A1 25 μl 25 A3 A2 25 μl 125 A4 A3 25 μl 625 A5 A4 25 μl 3125 A6 A5 25 μl A7 A6 25 μl A8 25 μl * *Please note that when expecting very high or very low titers, it would be advisable to include either well 8 or well 1 respectively.
8 A B Figure 8. Examples of individual colonies. Once you have generated a lentiviral stock with a suitable titer, you are ready to transduce the lentiviral vector into the mammalian cell line of choice and assay for gene silencing Multiplicity of Infection (MOI) To obtain optimal silencing of your shrna, you will need to transduce the lentiviral vector into your mammalian cell line of choice using a suitable MOI. MOI is defined as the number of transducing units per cell. Determining the Optimal MOI A number of factors can influence determination of an optimal MOI including the nature of your mammalian cell (actively dividing versus non-dividing), its transduction efficiency, your application of interest, and the nature of your gene of interest. If you are transducing your lentiviral construct into the mammalian cell line of choice for the first time, after you have titered the lentiviral particles, we recommend using a range of MOIs (for example, 0, 0.5, 1, 2, 5, 10, 20) to determine the MOI required to obtain optimal expression for your particular application. It should be noted that to achieve single copy knockdown, an MOI of 0.3 is generally used, as less than 4% of your cells will have more than one insert. Protocol VIII Transduction: Transduction of Target Cells The protocol below is optimized for transduction of the lentiviral particles into HEK293T, OVCAR-8 or MCF7 cells in a 24-well plate using serum-free media. If a different culture dish is used, adjust the number of cells, volumes, and reagent quantities in proportion to the change in surface area (Table 7). It is strongly recommended that you optimize transduction conditions to suit your target cell line to provide for the highest transduction efficiency possible. It is preferable that transduction be carried out in medium that is serum free and antibiotic free. A reduction in transduction efficiency occurs in the presence of serum; however it is possible to carry out successful transductions with serum present; you will have to optimize the protocol according to your cells of interest. 1. On day 0, plate cells per well in a 24-well plate. Incubate overnight. You will be using full growth medium (with serum) at this stage. 2. The next day (day 1), remove the medium and add an appropriate amount of the virus to acheive the MOI you wish to use. Set up all desired experiments and controls in a similar fashion. Bring the total volume of liquid up so that it just covers the cells efficiently with serum-free media (see Table 7 for guidelines). If you are using concentrated virus you are likely to use a small volume of virus if you are using unconcentrated virus, you will find you need more virus volume. Table 7. Suggested volumes of media per surface area per well of adherent cells. Tissue Culture Dish Surface Area Per Well (cm 2 ) Suggested Total Serum-free Medium Volume Per Well (ml) 100 mm mm mm well well well well Approximately 4-6 hours post-transduction, add an additional 1 ml of full medium (serum plus Pen-Strep if you are using it) to your cells and incubate overnight. Note: We have experienced low toxicity with transduction in the cell lines tested, therefore removal of virus is not required for many cell lines. In our experience, higher transduction efficiencies have been achieved if the virus is not removed after 6 hours. However, if toxicity is a problem, aspirate the mixture after 4-6 hours and replace with fresh growth medium. Additionally, fresh growth medium should be replenished as required for continued cell growth. 4. At 48 hours post-transduction examine the cells microscopically for the presence of reporter gene (TurboGFP) expression as this will be your first indication as to the efficiency of your transduction. Note: When visualizing TurboGFP expression, if less than 90% of all cells are green, it is recommended in these cases to utilize puromycin selection in order to reduce background expression of your gene of interest from untransduced cells.
9 a. If adding puromycin, use the appropriate concentration as determined based on the kill curve (see Protocol V). Incubate cells with the selection medium. b. Approximately every 2-3 days replace with freshly prepared selective medium. c. Monitor the cells daily and observe the percentage of surviving cells. Once the non-transduced control cells are dead, the surviving cells in the transduced wells will be expressing the shrna. Optimum effectiveness of the puromycin selection should be reached in 4-6 days with puromycin dependent upon the concentration of puromycin chosen from the kill curve. Note: The higher the MOI you have chosen, the more copies of the shrna and puromycin resistance gene you will have per cell. When selecting with puromycin, it is worth remembering that at higher MOIs, cells containing multiple copies of the resistance gene can withstand higher puromycin concentrations than those at lower MOIs. Adjust the concentration of puromycin to a level that will select for the population of transduced cells you require for your application without going below the minimum antibiotic concentration you have established in your kill curve. 5. Once your transduction efficiency is at an acceptable level (with or without puromycin selection performed post-transduction), you can proceed to assay cells for reduction in gene expression or fluorescent reporter activity by reverse transcription quantitative/real-time PCR (RT-qPCR), Western blot analysis or other appropriate functional assay. Compare target gene to untreated, reporter alone (empty vector), non-silencing shrna, or other negative controls. Note: Optimal length of incubation from the start of transfection to analysis is dependent on cell type, gene of interest, and the stability of the mrna and/or protein being analyzed. RT-qPCR generally gives the best indication of mrna expression and gene silencing. The use of Western blot analysis to determine knockdown is dependent on quantity and quality of the protein, its half-life, and the sensitivity of the antibody and detection systems used. Protocol IX Determining Relative Transduction Efficiency Follow the procedure below to determine the relative transduction efficiency of purchased GIPZ lentiviral particles (Cat #VGH5518, VGM5520, VGH5526). This protocol should only be used with purchased GIPZ shrna individual clones in viral particle format. Prior to transducing with purchased GIPZ shrna individual clones in viral particle format, we recommend determining the relative transduction efficiency of your cell type. Lentiviral titers provided with purchased GIPZ lentiviral particles have been calculated by transducing HEK293T cells. Transduction efficiencies vary significantly by cell type. The relative transduction efficiency of your cells may be estimated by determining the functional titer of a control virus such as GIPZ Non-silencing control viral particles (Cat #RHS4348) in your cells of interest. Follow the procedure below to determine the functional titer of the GIPZ Non-silencing control shrna viral stock in the mammalian cell line of your choice. The following conditions have been optimized for transduction of HEK293T cells. When determining the relative transduction efficiency of your cell type, use the transduction conditions that have been optimized for your cells of interest. 1. The day before transduction (day 0), seed a 24-well tissue culture plate with your cells at cells per well in their respective medium. Note: The following day, each well should be no more than 40-50% confluent. 2. Make dilutions of the Non-silencing control shrna viral stock in a round bottom 96-well plate using serum-free medium. Utilize the plate as shown in Figure 9 with one row for each replicate (we recommend performing at least two replicates). Use the procedure below for dilution of the viral stock. The goal is to produce a series of five-fold dilutions to reach a final dilution of 390, 625-fold. Virus stock 1 Virus stock 2 Virus stock 3 Virus stock 4 A B C D E F G H Figure 9. Five-fold serial dilutions of virus stock. Dilution Plate 3. Add 40 ul of serum-free media to each well in column. 4. Add 80 ul of serum-free media to each well of columns 2-8. Note: If desired, include 8 μg/ml polybrene in the dilution medium. 5. Add 10 µl of thawed control shrna virus stock to each well in column 1 (five-fold dilution). Note: Pipette contents of well up and down times. Discard pipette tip. Post dox 24 hours Post dox 72 hours
10 Virus stock 1 A Virus stock 2 B Virus stock 3 C Virus stock 4 D Figure 10. Twenty-four well tissue culture plate, seeded with HEK293T cells, used totiter the virus. 6. With new pipette tips, transfer 20 µl from each well of column 1 to the corresponding well in column 2. Pipette up and down times and discard pipette tip. 7. With new pipette tips, transfer 20 µl from each well of column 2 to the corresponding well in column 3. Pipette up and down times and discard pipette tip. 8. Repeat transfers of 20 µl from columns 3 through 8, pipetting up and down times and changing pipette tips between each dilution. It is strongly recommended that you use a high-quality multichannel pipettor when performing multiple dilutions. Incubate the dilutions of the virus stock for 5 minutes at room temperature. 9. Label a 24-well plate as shown in Figure 10 using one row for each replicate. 10. Remove culture medium from the cells in the 24-well plate. 11. Add 225 µl of serum-free medium to each well. 12. Transduce cells by adding 25 µl of diluted control shrna lentivirus from the original 96-well plate (Figure 9) to a well on the 24-well destination plate (Figure 10) containing the cells. For example, transfer 25 µl from well A2 of the 96-well plate into well A1in the 24-well plate (Table 8). 13. Incubate transduced cultures at 37 C for 4-6 hours. 14. Add 1 ml of your medium (normal serum concentration). 15. Culture cells for 72 hours. 16. Count the TurboGFP expressing cells or colonies of cells (Figure 11). Count each multi-cell colony as 1 transduced cell, as the cells will be dividing over the 72 hour culture period. Figure 11 illustrates this principle of cell counting. Count the number of TurboGFP expressing colonies in wells corresponding to at least two viral dilutions. Table 8. Example of set up for dilutions. Well (Row A, B, C, or D) Originating (96-well plate) Destination (24-well plate) Volume Diluted Virus Used Dilution Factor A1 25 μl 5 * A2 A1 25 μl 25 A3 A2 25 μl 125 A4 A3 25 μl 625 A5 A4 25 μl 3125 A6 A5 25 μl A7 A6 25 μl A8 25 μl * *Please note that when expecting very high or very low titers, it would be advisable to include either well 8 or well 1 respectively. A. B. Figure 11. Examples of individual colonies.
11 17. Transducing units per ml (TU/mL) can be determined using the following formula: # of TurboGFP positive colonies counted dilution factor 40 = # TU/mL Note: 25 μlof diluted virus was added to the cells. This is 1/40th of a ml. Example: 55 TurboGFP positive colonies counted in well A3 55 (TurboGFP positive colonies) 625 (dilution factor) 40 = TU/mL. 18. The functional titer calculated for your cell line under your experimental conditions can be used to determine the relative transduction efficiency of your cell type by using the following formula: Functional titer of Non-silencing control shrna virus stock in your cell line Titer of Non-silencing control shrna virus stock as calculated in HEK293T = Relative transduction efficiency For example, if the titer of the Non-silencing control shrna virus stock in HEK293T (as provided on the product specification sheet) is TU/mL and the functional titer of the control shrna virus stock in your cell line is TU/mL, the relative transduction efficiency of your cell type is 0.2. To extrapolate the average functional titer of the provided GIPZ viral particles, multiply the average titer of each plate as provided on the product specification sheet by the relative transduction efficiency of your cell type. In our example, if the titer of the GIPZ viral particles in HEK293T cells is TU/mL and the relative transduction efficiency of your cell type is 0.2, the extrapolated average functional titer of that plate your cell type is TU/mL. Once the relative transduction efficiency of the GIPZ virus has been established in your cell line, use the optimized transduction conditions determined in Protocol VIII to transduce your cell line with the purchased GIPZ shrna individual clones in viral particle format. If the titer of the non-silencing control shrna virus is not satisfactory in your cell line you might consider choosing a different cell line more permissive to transduction by lentivirus before proceeding. Protocol X PCR QPCR Experimental Recommendations One of the biggest challenges of any qpcr experiment is to obtain reproducible and reliable data. Due to the sensitivity of this multi-step technique, care must be taken to ensure results obtained are accurate and trustworthy (see Bustin et al., 2009). 1. Experimental samples should be run at least in duplicate. It should be noted that with duplicate experiments it will not be possible to assign error bars to indicate consistency from experimental sample to experimental sample. Using triplicate samples or higher will enable error bars to be assigned indicating the level of experimental variation. 2. Reverse Transcriptase reactions for cdna synthesis should always include a No Template Control (NTC) and No Reverse Transcriptase (no RT) control to check for reagents contamination and the presence of contaminating DNA, respectively. Use a robust reverse transcriptase enzyme for cdna synthesis such as the Thermo Scientific Maxima cdna Synthesis Kit for RT-qPCR (Cat #K1641). 3. We have found that normalizing the RNA concentration prior to cdna synthesis will increase consistency downstream. 4. qpcr should be done at least in triplicate. Again, it should be noted that with duplicate reactions it will not be possible to assign error bars to indicate the consistency in your qpcr reactions. Using triplicate samples or higher will enable error bars to be assigned indicating the level of variation between qpcr reactions. Use validated primer sets for SYBR-based assays or primers/probe for probe-based assays. 5. Make sure the mrna you are using as your internal reference control for qpcr is expressed at a level higher than your target gene's message. 6. Use only high-quality calibrated pipettes, in conjunction with well fitting barrier tips. 7. When pipetting, take the time to visually inspect the fluid in the pipette tip(s) for accuracy and lack of bubbles, especially when using a multi-channel pipette. 8. Be sure to spin your qpcr plate prior to loading in the real-time instrument in order to collect the sample at the bottom of the well and eliminate any bubbles that may have developed. 9. With regard to knockdown experiments using shrna, it is vitally important that you greatly reduce if not eliminate entirely those cells which are not transduced or transfected from the population. This can be done in several ways: increase the efficiency of your transfection, use a higher mulitplicity of infection (MOI) for your transduction, utilize the puromycin selection marker and select against those cells that do not contain the shrna or utilize fluorescent sorting to select against those cells that do not contain the shrna. 10. Always utilize the non-silencing control as a reference for target gene expression, as opposed to an untreated sample. The non-silencing treated samples will most accurately reproduce the conditions in your experimental samples. The non-silencing best controls for changes in qpcr internal reference gene expression. 11. You may also use an untreated sample to indicate substantial changes in target gene expression as seen in the non-silencing control due to generic consequences of viral transduction and transgene expression. However, it should be noted that small changes in expression levels between an untreated sample and the non-silencing control are to be expected. 12. Cq values greater than 35 should be avoided as they tend to be more variable. Samples with such high Cq values should be repeated at higher cdna concentrations and with a lower expressing qpcr internal reference control (such as TBP). 13. Cq values less than 11 for the qpcr internal reference control should be avoided as it is difficult to determine a proper background subtraction using these values. If this occurs, use Cq values from both your internal reference control as well as your experimental target to determine an optimum cdna concentration. 14. It may be necessary to change internal reference controls if conditions in steps 12 and 13 cannot be simultaneously met. Controls and Validation: RNAintro shrna Starter Kits The use of vector-based RNAi for gene silencing is a powerful and versatile tool. Successful gene silencing in vitro is dependent on several variables including 1) The target cell line being studied, 2) transfection and transduction efficiency, 3) abundance of the mrna or protein of interest in the target cell line, 4) half life of the protein, and 5) robust experimental protocols. For all these reasons, it is important to run controlled experiments where the transfection and transduction efficiencies are as high as possible and measurable.
12 Controls are a critical part of a gene silencing experiment. They enable accurate representation of knockdown data and provide confidence in the specificity of the response. Changes in the mrna or protein levels in cells treated with negative or non-silencing controls reflect non-specific responses in cells and can be used as a baseline against which specific knockdown can be measured. Positive controls are useful to demonstrate that your experimental system is functional. 140% 120% 80% 60% 40% 20% 0% 113% 88% Residual Gene Activity 93% Control GAPDH EG5 PP1B Figure 12. Non-silencing lentiviral shrna control does not knockdown common endogenous genes. The above data represents the baseline amount of GAPDH, EG5 or PP1B mrna set at in the control. The relative amounts of each of these mrnas are then represented after treatment with non-silencing shrna of these genes. 120% 80% 60% 40% 20% 0% Untransduced Control 34% Residual GAPDH Expression 28% Figure 13. HEK293T cells were transduced with lentiviral particles expressing GAPDH or Non-silencing shrna at variable MOIs ranging from The graph depicts the residual levels of GAPDH relative to Non-silencing control. 12% NS#1 NS#1 NS#1 GAPDH GAPDH GAPDH Controls The EG5 and GAPDH GIPZ lentiviral shrna vectors have been validated as positive controls for RNAi experiments performed using the GIPZ shrna-containing lentiviral vectors. These shrna have been tested in transduction based experiments and have shown efficient knockdown at both mrna and protein levels. The EG5 control has been validated to knockdown human EG5 by RT-qPCR (Figure 14 and 15) and in situ hybridization of cells in tissue culture. The GAPDH control has been validated to knockdown human and mouse GAPDH by RT-qPCR (Figure 13). The GIPZ Non-silencing lentiviral shrna vector has been validated as a negative control for RNAi experiments performed using the GIPZ shrna-containing lentiviral vectors (Figure 12). 120% Residual EG5 Expression OVERLAY ANTI-TUBULIN 80% 60% ANTI-EG5 DAPI 40% 20% 0% NS#1 MOI 3.5 NS#1 MOI 8.5 NS#1 MOI 17 29% EG5 MOI % EG5 MOI % EG5 MOI 17 Figure 14. HEK293T cells were transduced with lentiviral particles expressing EG5 or non-silencing shrna at MOIs of 3.5, 8.5 and 17. The graph depicts the residual levels of EG5 relative to its non-silencing control Figure 15. The characteristic phenotype observed by the targeting of the EG5 (KIF11) gene results in the formation of half spindles, mitotic arrest and monoastral microtubular arrays (green, see the cell on the left). By contrast, normal cells show bipolar spindles and microtubule networks in mitosis and in interphase (see the cell on the right). The comparative expression of EG5 (red) between the cell on the left and the right shows the extensive knockdown of EG5 in the cell displaying the phenotype (left). The cells were visualized at 100x magnification using aleica DMIRB fluorescence microscope. HEK293T cells were stained for tubulin (anti-tubulin, green), DNA (DAPI, blue) and EG5 (anti-eg5, red). Table 9. Related Reagents. Related Reagents Vendor Cat # GAPDH Verified Positive Control* Dharmacon RHS4371 EG5 Verified Positive control* Dharmacon RHS4480 Non-silencing Verified Negative Control* Dharmacon RHS4346 TurboFECT Transfection Reagent 1 ml-5 mls* Dharmacon R0531, R0532 GIPZ shrna Empty Vector Dharmacon RHS4349 Trans-Lentiviral shrna Packaging System Dharmacon TLP5912 Trans-Lentiviral shrna Packaging System with HEK293T Cells Dharmacon TLP5917 *These items also available in the GIPZ lentiviral transfection starter kit (Cat #RHS4287).
13 Frequently Asked Questions (FAQs): What clones are part of my library collection? A CD containing the data for this collection will be shipped with each collection. This file contains the location and accession number for each construct in the collection. Where can I find the sequence of an individual shrna construct? If you are looking for the sequence of an individual shrna construct, you can search for the clone on our website (gelifesciences.com/dharmacon). Enter the catalog number or clone ID of your construct into the search at the top of the page. You should see your product in the catalog number section of the results. Click on the plus sign to expand the details for this clone and select the Sequence tab. Which antibiotic should I use? You should grow all GIPZ shrna constructs in 2x LB broth medium with both 25 μg/ml zeocin and 100 μg/ml carbenicillin for archive replication. For plasmid preparations, grow the constructs in 2x LB broth medium containing only 100 ug/ml carbenicillin. What packaging cell line should I use for making lentivirus? The GIPZ shrna vector is tat dependant, so a packaging system that expresses the tat gene. For packaging our lentiviral shrna constructs, we recommend the Trans-Lentiviral shrna Packaging Kit (TLP5912, TLP5917). The Trans-Lentiviral Packaging Kit allows creation of a replication-incompetent (Shimada, et al. 1995), HIV-1-based lentivirus which can be used to deliver and express your gene or shrna of interest in either dividing or non-dividing mammalian cells. The Trans-Lentiviral Packaging Kit uses a replication-incompetent lentivirus based on the trans-lentiviral system developed by Kappes (Kappes and Wu et al. 2001). For protocols and information on packaging GIPZ shrna with our Trans-Lentiviral shrna Packaging Kit, please see the product manual available at here. Can I use any 2 nd generation packaging system with the pgipz vector? The pgipz vector is tat dependant, so you must use a packaging system that expresses the tat gene. What does the number 40 refer to in the formula for the calculation of titer? The titer units are given in transducing units (TU) per ml, so the number 40 is used to convert the 25 µl used in the titration ( volume of diluted virus used, Table 6) to one milliliter. What is the sequencing primer for pgipz? The pgipz sequencing primer is 5' - GCATTAAAGCAGCGTATC - 3' Note: The binding site lies from base and runs in the reverse complement direction. The melting temperature of this 18mer = 52.7 C. Where do you purchase puromycin? We purchase puromycin from Fisher Scientific Cellgro (Cat #BP ). How many transfections are available in each volume size of TurboFect Transfection Reagent? The number of transfections that can be performed depends on the size of the culture dish used and the volume size of TurboFect purchased. Troubleshooting: For help with transfection or transduction of your lentiviral constructs, please technical support at [email protected] with the answers to the questions below, your sales order or purchase order number and the catalog number or clone ID of the construct with which you are having trouble. 1. Are you using direct transfection or transduction into your cell line? 2. What was the 260/280 ratio of DNA? Over 1.8? 3. What was the transfection efficiency if you used direct transfection? What transfection reagent was used? 4. Were positive and negative knockdown controls used (such as our GAPDH or EG5 validated positive controls and the validated non-silencing negative control)? 5. What were the results of the controlled experiments? 6. How was knockdown measured (for example real-time RT-qPCR or western blot analysis)? 7. What is the abundance and the half-life of the protein? Does the protein have many isoforms? 8. What packaging cell line was used if you are using transduction rather than transfection? 9. What was your viral titer? 10. What was your MOI? 11. Did you maintain the cells in puromycin selection media after transfection or transduction? 12. How much time elapsed from transfection/transduction to puromycin selection? If transfection into your cell line is unsuccessful, you may need to consider the following list of factors influencing successful transfection. 1. Concentration and purity of plasmid DNA and nucleic acids determine the concentration of your DNA using 260 nm absorbance. Avoid cytotoxic effects by using pure preparations of nucleic acids. 2. Insufficient mixing of transfection reagent or transfection complexes. 3. Transfection in serum containing or serum-free media our studies indicate that the transfection efficiency with Thermo Scientific TurboFect Transfection Reagent is equally high in the presence of serum. This is not the case with many other transfection reagents. 4. Presence of antibiotics in transfection medium the presence of antibiotics do not interfere with both DNA/TurboFect complex formation and cell transfection. This is not the case for other transfection reagents. 5. Cell history, density, and passage number it is very important to use healthy cells that are regularly passaged and in growth phase. The highest transfection efficiencies are achieved if cells are plated the day before; however, adequate time should be given to allow the cells to recover from the passaging (generally > 12 hours). Plate cells at a consistent density to minimize experimental variation. If transfection efficiencies are low or reduction occurs over time, thawing a new batch of cells or using cells with a lower passage number may improve the results.
14 If transduction into your cell line is unsuccessful, you may need to consider the following list of factors influencing successful transduction. 1. Transduction efficiency is integrally related to the quality and the quantity of the virus you have produced. Factors to consider when transducing include MOI (related to accurate titer in the target cell line), the presence of serum in the media, the use of polybrene in the media, length of exposure to virus, and viral toxicity to your particular cells. 2. High quality transfer vector DNA and the appropriate and efficient viral packaging are required to make high quality virus able to transduce cells effectively. 3. See also suggestions 3-5 for factors influencing successful transfection (above). References: Cited References and Additional Suggested Reading 1. Bartel, D. P. (2004). micrornas: genomics, biogenesis, mechanism, and function. Cell 116(2): Boden, D., O. Pusch, et al. (2004). Enhanced gene silencing of HIV-1 specific sirna using microrna designed hairpins Nucleic Acids Res 32(3): Chendrimada, T. P., R. I. Gregory, et al. (2005). TRBP recruits the Dicer complex to Ago2 for microrna processing and gene silencing. Nature 436(7051): Cleary, M. A., K. Kilian, et al. (2004). Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods 1(3): Cullen, B. R. (2004). Transcription and processing of human microrna precursors. Mol Cell 16(6): Cullen, B. R. (2005). RNAi the natural way. Nat Genet 37(11): Dickins, R. A., M. T. Hemann, et al. (2005). Probing tumor phenotypes using stable and regulated synthetic microrna precursors. Nat Genet 37(11): Editors of Nature Cell Biology (2003). Whither RNAi? Nat Cell Biol 5(6): Elbashir, S. M., J. Harborth, et al. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836): Fire, A., S. Xu, et al. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669): Gregory, R. I., T. P. Chendrimada, et al. (2005). Human RISC couples microrna biogenesis and posttranscriptional gene silencing. Cell 123(4): Kappes, J. C. and X. Wu (2001). Safety considerations in vector development. Somat Cell Mol Genet 26(1-6): Kappes, J. C., X. Wu, et al. (2003). Production of trans-lentiviral vector with predictable safety. Methods Mol Med 76: Paddison, P. J., J. M. Silva, et al. (2004). A resource for large-scale RNA-interference-based screens in mammals. Nature 428(6981): Shimada, T., et.al. (1995). Development of vectors utilized for gene therapy for AIDS. AIDS Silva, J. M., M. Z. Li, et al. (2005). Second-generation shrna libraries covering the mouse and human genomes. Nat Genet 37(11): Limited Use Licenses: Limited Use Label License: Evrogen Fluorescent Proteins This product contains a proprietary nucleic acid(s) coding for a proprietary fluorescent protein being, including its derivatives or modifications, the subject of pending U.S. and foreign patent applications and/or patents owned by Evrogen JSC (hereinafter Evrogen Fluorescent Proteins ). The purchase of this product conveys to the buyer the non-transferable right to use Evrogen Fluorescent Proteins for (i) not-for-profit internal Research conducted by the buyer (whether the buyer is an academic or for-profit entity), where Research means non-commercial uses or activities which (or the results of which) do not generate revenue, and (ii) evaluation of Evrogen Fluorescent Proteins for the purpose of testing its appropriateness for development of a therapeutic, clinical diagnostic, vaccine or prophylactic product, provided that Evrogen Fluorescent Proteins are not used in the development or manufacture of such product. Offer of Evrogen Fluorescent Proteins for resale; distribution, transfer, or otherwise providing access to Evrogen Fluorescent Proteins to any third party for any purpose, or any use of Evrogen Fluorescent Proteins other than for Research is strictly prohibited. The buyer cannot sell or otherwise transfer materials made by the employment of Evrogen Fluorescent Proteins to a third party or otherwise use Evrogen Fluorescent Proteins for Commercial Purposes. The buyer may transfer information made through the use of this product solely for research and not for Commercial Purposes. Commercial Purposes means any activity by a party for consideration and may include, but is not limited to: (1) use of Evrogen Fluorescent Proteins in manufacturing; (2) use of Evrogen Fluorescent Proteins to provide a service, information, or data; (3) use of Evrogen Fluorescent Proteins for therapeutic, diagnostic or prophylactic purposes. The purchase of this product does not convey a license under any method claims in the foregoing patents or patentapplications. For information on the foregoing patents or patent applications or on purchasing a license to use Evrogen Fluorescent Proteins for purposes other than those permitted above, contact Licensing Department, Evrogen JSC, Miklukho-Maklaya street 16/10, Moscow, , Russian Federation license@evrogen. com. shrna Limited Use Agreement Permitted Use. Portions of this product are covered by published patent applications (the shrna IP Rights ) owned by Cold Spring Harbor Laboratory, referred to herein as CSHL. This product, and/or any components or derivatives of this product, and/or any materials made using this product or its components, are referred to herein as the Product. Subject to the terms of this Limited Use Agreement, sale of the Product by ( OBS ) conveys the limited, nonexclusive, nontransferable right to any company or other entity that orders, pays for and takes delivery of the Product (the Customer ) from OBS to use the Product solely for its internal research and only at such Customer s facility where the Product is delivered by OBS. Restrictions. Customer obtains no right to transfer, assign, or sublicense its use rights, or to transfer, resell, package, or otherwise distribute the Product, or to use the Product for the benefit of any third party or for any commercial purpose. The Product or components of the Product may not be used in vitro or in vivo for any diagnostic, preventative, therapeutic or vaccine application, or in the manufacture or testing of a product therefore, or used (directly or indirectly) in humans for any purpose. Compliance. Customer may only use the Product in compliance with applicable local, state and federal laws, regulations and rules, including without limitation (for uses in the United States) EPA, FDA, USDA and NIH guidelines. Customer may not (directly or indirectly) use the Product, or allow the transfer, transmission, export or re-export of all or any part of the Product, in violation of any export control law or regulation of the United States or any other relevant jurisdiction.
15 Other Uses. Except for the permitted use expressly specified above and subject to the restrictions set forth above, no other use is permitted. CSHL shall retain all right, title and interest in and to the shrna IP Rights. Nothing herein confers to Customer (by implication, estoppel or otherwise) any right or license under any patent of CSHL, including, for avoidance of doubt, under any patent that may issue from or relate to the shrna IP Rights. For information on obtaining an agreement to use this Product for purposes other than those expressly permitted by this Limited Use Agreement, or to practice under CSHL patent rights, please contact the CSHL Office of Technology Transfer at (516) Termination of Rights. Upon issuance of any patent covering any portion of the Product, any and all rights conveyed to any Customer (other than a research or educational institution that is a tax-exempt organization under section 501(c)(3) of the U.S. IRS code) under this Limited Use Agreement shall terminate and such Customer shall need a license from CSHL to continue to use any Product that was previously purchased, or to purchase from OBS or any other vendor or use any product claimed by such issued patent. At such time, such Customer may contact the CSHL Office of Technology Transfer at (516) and seek a license to use this Product under CSHL s patent rights. YOUR USE OF THIS PRODUCT CONSTITUTES ACCEPTANCE OF THE TERMS OF THIS LIMITED USE AGREEMENT. If you are not willing to accept the limitations of this agreement, OBS is willing to accept return of the Product for a full refund. Limited Label License: Benitec This product is sold solely for use for research purposes in fields other than plants. This product is not transferable. If the purchaser is not willing to accept the conditions of this label license, supplier is willing to accept the return of the unopened product and provide the purchaser with a full refund. However, if the product is opened, then the purchaser agrees to be bound by the conditions of this limited use statement. This product is sold by supplier under license from Benitec Australia Ltd and CSIRO as co-owners of U.S. Patent No. 6,573,099 and foreign counterparts. For information regarding licenses to these patents for use of ddrnai as a therapeutic agent or as a method to treat/prevent human disease, please contact Benitec at [email protected]. For the use of ddrnai in other fields, please contact CSIRO at Fluorescent Protein and TurboGFP are trademarks of Evrogen. HXB2 is a trademark of GenBank. Zeocin, FastDigest, GeneJET, and Maxima are trademarks of Thermo Fisher Scientific, Inc. GE, imagination at work and GE monogram are trademarks of General Electric Company. Dharmacon, Inc., a General Electric company doing business as GE Healthcare. All other trademarks are the property of General Electric Company or one of its subsidiaries General Electric Company All rights reserved. Version published September GE Healthcare UK Limited, Amersham Place, Little Chalfont, Buckinghamshire, HP7 9NA, UK. Orders can be placed at: gelifesciences.com/dharmacon Customer Support: [email protected] Technical Support: [email protected] or ; if you have any questions. V2-0915
MGC premier Expression-Ready cdna clones TCH1103, TCM1104, TCR1105, TCB1106, TCH1203, TCM1204, TCR1205, TCB1206, TCH1303, TCM1304, TCR1305
 MGC premier Expression-Ready cdna clones TCH1103, TCM1104, TCR1105, TCB1106, TCH1203, TCM1204, TCR1205, TCB1206, TCH1303, TCM1304, TCR1305 The MGC premier Expression-Ready collection has the high quality,
MGC premier Expression-Ready cdna clones TCH1103, TCM1104, TCR1105, TCB1106, TCH1203, TCM1204, TCR1205, TCB1206, TCH1303, TCM1304, TCR1305 The MGC premier Expression-Ready collection has the high quality,
TransIT -2020 Transfection Reagent
 Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/5400 INTRODUCTION TransIT -2020 Transfection Reagent is a high-performance, animal-origin free, broad spectrum reagent
Quick Reference Protocol, MSDS and Certificate of Analysis available at mirusbio.com/5400 INTRODUCTION TransIT -2020 Transfection Reagent is a high-performance, animal-origin free, broad spectrum reagent
Product: Expression Arrest TM egfp control shrna vector
 Product: Expression Arrest TM egfp control vector Catalog #: RHS1702 Product Description The laboratory of Dr. Greg Hannon at Cold Spring Harbor Laboratory (CSHL) has created an RNAi Clone Library comprised
Product: Expression Arrest TM egfp control vector Catalog #: RHS1702 Product Description The laboratory of Dr. Greg Hannon at Cold Spring Harbor Laboratory (CSHL) has created an RNAi Clone Library comprised
EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES
 C GUGAAG EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES MicroRNA-adapted shrna (shrna mir ) for increased, specific and consistent knockdown. MicroRNA PROCESSING PATHWAY UTILIZED FOR shrna mir Developed
C GUGAAG EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES MicroRNA-adapted shrna (shrna mir ) for increased, specific and consistent knockdown. MicroRNA PROCESSING PATHWAY UTILIZED FOR shrna mir Developed
Manual for: sirna-trans Maximo Reagent
 Manual for: sirna-trans Maximo Reagent Contact: [email protected]; www.geneon.net GeneON GmbH Germany GeneON GmbH / Protocol sirna-trans reagent vs. 1.3 / www.geneon.net / - 0 - sirna-trans reagent Instruction
Manual for: sirna-trans Maximo Reagent Contact: [email protected]; www.geneon.net GeneON GmbH Germany GeneON GmbH / Protocol sirna-trans reagent vs. 1.3 / www.geneon.net / - 0 - sirna-trans reagent Instruction
Instructions. Torpedo sirna. Material. Important Guidelines. Specifications. Quality Control
 is a is a state of the art transfection reagent, specifically designed for the transfer of sirna and mirna into a variety of eukaryotic cell types. is a state of the art transfection reagent, specifically
is a is a state of the art transfection reagent, specifically designed for the transfer of sirna and mirna into a variety of eukaryotic cell types. is a state of the art transfection reagent, specifically
RNA Extraction and Quantification, Reverse Transcription, and Real-time PCR (q-pcr)
 RNA Extraction and Quantification, Reverse Transcription, and Real-time Preparation of Samples Cells: o Remove media and wash cells 2X with cold PBS. (2 ml for 6 well plate or 3 ml for 6cm plate) Keep
RNA Extraction and Quantification, Reverse Transcription, and Real-time Preparation of Samples Cells: o Remove media and wash cells 2X with cold PBS. (2 ml for 6 well plate or 3 ml for 6cm plate) Keep
Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity
 Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity Today you analyze the results of your bacterial transformation from last week and determine the efficiency
Lab 10: Bacterial Transformation, part 2, DNA plasmid preps, Determining DNA Concentration and Purity Today you analyze the results of your bacterial transformation from last week and determine the efficiency
First Strand cdna Synthesis
 380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 [email protected] A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
380PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 [email protected] A Geno Technology, Inc. (USA) brand name First Strand cdna Synthesis (Cat. # 786 812) think proteins! think G-Biosciences
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual
 Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Thermo Scientific DyNAmo cdna Synthesis Kit for qrt-pcr Technical Manual F- 470S 20 cdna synthesis reactions (20 µl each) F- 470L 100 cdna synthesis reactions (20 µl each) Table of contents 1. Description...
Taqman TCID50 for AAV Vector Infectious Titer Determination
 Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
Page 1 of 8 Purpose: To determine the concentration of infectious particles in an AAV vector sample. This process involves serial dilution of the vector in a TCID50 format and endpoint determination through
ab185915 Protein Sumoylation Assay Ultra Kit
 ab185915 Protein Sumoylation Assay Ultra Kit Instructions for Use For the measuring in vivo protein sumoylation in various samples This product is for research use only and is not intended for diagnostic
ab185915 Protein Sumoylation Assay Ultra Kit Instructions for Use For the measuring in vivo protein sumoylation in various samples This product is for research use only and is not intended for diagnostic
RT31-020 20 rxns. RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl
 Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
Components RT31-020 20 rxns RT31-050 50 rxns RT31-100 100 rxns TRANSCRIPTME Enzyme Mix (1) 40 µl 2 x 50 µl 5 x 40 µl 2x RT Master Mix (2) 200 µl 2 x 250 µl 5 x 200 µl RNase H (E. coli) 20 µl 2 x 25 µl
ab185916 Hi-Fi cdna Synthesis Kit
 ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
ab185916 Hi-Fi cdna Synthesis Kit Instructions for Use For cdna synthesis from various RNA samples This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated 1
MEF Starter Nucleofector Kit
 page 1 of 7 MEF Starter Nucleofector Kit for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the mice they are isolated
page 1 of 7 MEF Starter Nucleofector Kit for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the mice they are isolated
qstar mirna qpcr Detection System
 qstar mirna qpcr Detection System Table of Contents Table of Contents...1 Package Contents and Storage Conditions...2 For mirna cdna synthesis kit...2 For qstar mirna primer pairs...2 For qstar mirna qpcr
qstar mirna qpcr Detection System Table of Contents Table of Contents...1 Package Contents and Storage Conditions...2 For mirna cdna synthesis kit...2 For qstar mirna primer pairs...2 For qstar mirna qpcr
pcas-guide System Validation in Genome Editing
 pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
PRODUCT INFORMATION...
 VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
VIROMER RED In vitro plasmid DNA and mrna Standard Transfection PRODUCT INFORMATION... 2 GENERAL... 2 RED OR YELLOW?... 3 PROTOCOL GUIDELINES... 4 GENERAL REMARKS... 4 CELL CULTURE AND PLATING... 4 FORWARD/REVERSE
Optimized Protocol sirna Test Kit for Cell Lines and Adherent Primary Cells
 page 1 of 8 sirna Test Kit for Cell Lines and Adherent Primary Cells Kit principle Co-transfection of pmaxgfp, encoding the green fluorescent protein (GFP) from Pontellina p. with an sirna directed against
page 1 of 8 sirna Test Kit for Cell Lines and Adherent Primary Cells Kit principle Co-transfection of pmaxgfp, encoding the green fluorescent protein (GFP) from Pontellina p. with an sirna directed against
MystiCq microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C
 microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C Product Description The microrna cdna Synthesis Mix has been designed to easily convert micrornas into cdna templates for qpcr
microrna cdna Synthesis Mix Catalog Number MIRRT Storage Temperature 20 C Product Description The microrna cdna Synthesis Mix has been designed to easily convert micrornas into cdna templates for qpcr
TransformAid Bacterial Transformation Kit
 Home Contacts Order Catalog Support Search Alphabetical Index Numerical Index Restriction Endonucleases Modifying Enzymes PCR Kits Markers Nucleic Acids Nucleotides & Oligonucleotides Media Transfection
Home Contacts Order Catalog Support Search Alphabetical Index Numerical Index Restriction Endonucleases Modifying Enzymes PCR Kits Markers Nucleic Acids Nucleotides & Oligonucleotides Media Transfection
CompleteⅡ 1st strand cdna Synthesis Kit
 Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
Instruction Manual CompleteⅡ 1st strand cdna Synthesis Kit Catalog # GM30401, GM30402 Green Mountain Biosystems. LLC Web: www.greenmountainbio.com Tel: 800-942-1160 Sales: Sales@ greenmountainbio.com Support:
User Manual. CelluLyser Lysis and cdna Synthesis Kit. Version 1.4 Oct 2012 From cells to cdna in one tube
 User Manual CelluLyser Lysis and cdna Synthesis Kit Version 1.4 Oct 2012 From cells to cdna in one tube CelluLyser Lysis and cdna Synthesis Kit Table of contents Introduction 4 Contents 5 Storage 5 Additionally
User Manual CelluLyser Lysis and cdna Synthesis Kit Version 1.4 Oct 2012 From cells to cdna in one tube CelluLyser Lysis and cdna Synthesis Kit Table of contents Introduction 4 Contents 5 Storage 5 Additionally
OriGene Technologies, Inc. MicroRNA analysis: Detection, Perturbation, and Target Validation
 OriGene Technologies, Inc. MicroRNA analysis: Detection, Perturbation, and Target Validation -Optimal strategies to a successful mirna research project Optimal strategies to a successful mirna research
OriGene Technologies, Inc. MicroRNA analysis: Detection, Perturbation, and Target Validation -Optimal strategies to a successful mirna research project Optimal strategies to a successful mirna research
GRS Plasmid Purification Kit Transfection Grade GK73.0002 (2 MaxiPreps)
 1 GRS Plasmid Purification Kit Transfection Grade GK73.0002 (2 MaxiPreps) (FOR RESEARCH ONLY) Sample : Expected Yield : Endotoxin: Format : Operation Time : Elution Volume : 50-400 ml of cultured bacterial
1 GRS Plasmid Purification Kit Transfection Grade GK73.0002 (2 MaxiPreps) (FOR RESEARCH ONLY) Sample : Expected Yield : Endotoxin: Format : Operation Time : Elution Volume : 50-400 ml of cultured bacterial
Transfection-Transfer of non-viral genetic material into eukaryotic cells. Infection/ Transduction- Transfer of viral genetic material into cells.
 Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
Transfection Key words: Transient transfection, Stable transfection, transfection methods, vector, plasmid, origin of replication, reporter gene/ protein, cloning site, promoter and enhancer, signal peptide,
Bacterial Transformation with Green Fluorescent Protein. Table of Contents Fall 2012
 Bacterial Transformation with Green Fluorescent Protein pglo Version Table of Contents Bacterial Transformation Introduction..1 Laboratory Exercise...3 Important Laboratory Practices 3 Protocol...... 4
Bacterial Transformation with Green Fluorescent Protein pglo Version Table of Contents Bacterial Transformation Introduction..1 Laboratory Exercise...3 Important Laboratory Practices 3 Protocol...... 4
INTERFERin in vitro sirna/mirna transfection reagent PROTOCOL. 1 Standard sirna transfection of adherent cells... 2
 INTERFERin in vitro sirna/mirna transfection reagent PROTOCOL DESCRIPTION INTERFERin is a powerful sirna/mirna transfection reagent that ensures efficient gene silencing and reproducible transfection in
INTERFERin in vitro sirna/mirna transfection reagent PROTOCOL DESCRIPTION INTERFERin is a powerful sirna/mirna transfection reagent that ensures efficient gene silencing and reproducible transfection in
MEF Nucleofector Kit 1 and 2
 page 1 of 7 MEF Nucleofector Kit 1 and 2 for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the they are isolated
page 1 of 7 MEF Nucleofector Kit 1 and 2 for Mouse Embryonic Fibroblasts (MEF) MEF display significant phenotypic variations which depend on the strain, the genetic background of the they are isolated
Application Guide... 2
 Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
Protocol for GenomePlex Whole Genome Amplification from Formalin-Fixed Parrafin-Embedded (FFPE) tissue Application Guide... 2 I. Description... 2 II. Product Components... 2 III. Materials to be Supplied
FOR REFERENCE PURPOSES
 BIOO LIFE SCIENCE PRODUCTS FOR REFERENCE PURPOSES This manual is for Reference Purposes Only. DO NOT use this protocol to run your assays. Periodically, optimizations and revisions are made to the kit
BIOO LIFE SCIENCE PRODUCTS FOR REFERENCE PURPOSES This manual is for Reference Purposes Only. DO NOT use this protocol to run your assays. Periodically, optimizations and revisions are made to the kit
NIH Mammalian Gene Collection (MGC)
 USER GUIDE NIH Mammalian Gene Collection (MGC) Catalog number FL1002 Revision date 28 November 2011 Publication Part number 25-0610 MAN0000351 For Research Use Only. Not for diagnostic procedures. ii Table
USER GUIDE NIH Mammalian Gene Collection (MGC) Catalog number FL1002 Revision date 28 November 2011 Publication Part number 25-0610 MAN0000351 For Research Use Only. Not for diagnostic procedures. ii Table
RevertAid Premium First Strand cdna Synthesis Kit
 RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
RevertAid Premium First Strand cdna Synthesis Kit #K1651, #K1652 CERTIFICATE OF ANALYSIS #K1651 Lot QUALITY CONTROL RT-PCR using 100 fg of control GAPDH RNA and GAPDH control primers generated a prominent
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
HiPer RT-PCR Teaching Kit
 HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
HiPer RT-PCR Teaching Kit Product Code: HTBM024 Number of experiments that can be performed: 5 Duration of Experiment: Protocol: 4 hours Agarose Gel Electrophoresis: 45 minutes Storage Instructions: The
Mir-X mirna First-Strand Synthesis Kit User Manual
 User Manual Mir-X mirna First-Strand Synthesis Kit User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories, Inc.
User Manual Mir-X mirna First-Strand Synthesis Kit User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories, Inc.
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis
 ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
ab139418 Propidium Iodide Flow Cytometry Kit for Cell Cycle Analysis Instructions for Use To determine cell cycle status in tissue culture cell lines by measuring DNA content using a flow cytometer. This
mirnaselect pep-mir Cloning and Expression Vector
 Product Data Sheet mirnaselect pep-mir Cloning and Expression Vector CATALOG NUMBER: MIR-EXP-C STORAGE: -80ºC QUANTITY: 2 vectors; each contains 100 µl of bacterial glycerol stock Components 1. mirnaselect
Product Data Sheet mirnaselect pep-mir Cloning and Expression Vector CATALOG NUMBER: MIR-EXP-C STORAGE: -80ºC QUANTITY: 2 vectors; each contains 100 µl of bacterial glycerol stock Components 1. mirnaselect
HiPerFect Transfection Reagent Handbook
 Fifth Edition October 2010 HiPerFect Transfection Reagent Handbook For transfection of eukaryotic cells with sirna and mirna Sample & Assay Technologies QIAGEN Sample and Assay Technologies QIAGEN is the
Fifth Edition October 2010 HiPerFect Transfection Reagent Handbook For transfection of eukaryotic cells with sirna and mirna Sample & Assay Technologies QIAGEN Sample and Assay Technologies QIAGEN is the
Proto col. GoClone Repor ter Construc ts: Sample Protocol for Adherent Cells. Tech support: 877-994-8240. Luciferase Assay System
 Luciferase Assay System Proto col GoClone Repor ter Construc ts: Sample Protocol for Adherent Cells LightSwitch Luciferase Assay System GoClone Reporter Assay Workflow Step 1: Seed cells in plate format.
Luciferase Assay System Proto col GoClone Repor ter Construc ts: Sample Protocol for Adherent Cells LightSwitch Luciferase Assay System GoClone Reporter Assay Workflow Step 1: Seed cells in plate format.
Sequencing Library qpcr Quantification Guide
 Sequencing Library qpcr Quantification Guide FOR RESEARCH USE ONLY Introduction 3 Quantification Workflow 4 Best Practices 5 Consumables and Equipment 6 Select Control Template 8 Dilute qpcr Control Template
Sequencing Library qpcr Quantification Guide FOR RESEARCH USE ONLY Introduction 3 Quantification Workflow 4 Best Practices 5 Consumables and Equipment 6 Select Control Template 8 Dilute qpcr Control Template
Classic Immunoprecipitation
 292PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 [email protected] A Geno Technology, Inc. (USA) brand name Classic Immunoprecipitation Utilizes Protein A/G Agarose for Antibody Binding (Cat.
292PR 01 G-Biosciences 1-800-628-7730 1-314-991-6034 [email protected] A Geno Technology, Inc. (USA) brand name Classic Immunoprecipitation Utilizes Protein A/G Agarose for Antibody Binding (Cat.
Effects of Antibiotics on Bacterial Growth and Protein Synthesis: Student Laboratory Manual
 Effects of Antibiotics on Bacterial Growth and Protein Synthesis: Student Laboratory Manual I. Purpose...1 II. Introduction...1 III. Inhibition of Bacterial Growth Protocol...2 IV. Inhibition of in vitro
Effects of Antibiotics on Bacterial Growth and Protein Synthesis: Student Laboratory Manual I. Purpose...1 II. Introduction...1 III. Inhibition of Bacterial Growth Protocol...2 IV. Inhibition of in vitro
All-in-One First-Strand cdna Synthesis Kit
 All-in-One First-Strand cdna Synthesis Kit For reliable first-strand cdna synthesis from all RNA sources Cat. No. AORT-0020 (20 synthesis reactions) Cat. No. AORT-0050 (50 synthesis reactions) User Manual
All-in-One First-Strand cdna Synthesis Kit For reliable first-strand cdna synthesis from all RNA sources Cat. No. AORT-0020 (20 synthesis reactions) Cat. No. AORT-0050 (50 synthesis reactions) User Manual
MLX BCG Buccal Cell Genomic DNA Extraction Kit. Performance Characteristics
 MLX BCG Buccal Cell Genomic DNA Extraction Kit Performance Characteristics Monolythix, Inc. 4720 Calle Carga Camarillo, CA 93012 Tel: (805) 484-8478 monolythix.com Page 2 of 9 MLX BCG Buccal Cell Genomic
MLX BCG Buccal Cell Genomic DNA Extraction Kit Performance Characteristics Monolythix, Inc. 4720 Calle Carga Camarillo, CA 93012 Tel: (805) 484-8478 monolythix.com Page 2 of 9 MLX BCG Buccal Cell Genomic
All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna
 All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna All-in-One TM mirna First-Strand cdna Synthesis Kit AMRT-0020 (20 RT reactions), AMRT-0060 (60 RT reactions) Used in combination
All-in-One mirna qrt-pcr Reagent Kits For quantitative detection of mature mirna All-in-One TM mirna First-Strand cdna Synthesis Kit AMRT-0020 (20 RT reactions), AMRT-0060 (60 RT reactions) Used in combination
ExpressArt Bacterial H-TR cdna synthesis kit. With extreme selectivity against rrnas
 ExpressArt Bacterial H-TR cdna synthesis kit With extreme selectivity against rrnas suitable for 2 to 4 µg total RNA Catalogue No. 8004-A30 (30 rxns) Reagents Materials are provided for 30 cdna synthesis
ExpressArt Bacterial H-TR cdna synthesis kit With extreme selectivity against rrnas suitable for 2 to 4 µg total RNA Catalogue No. 8004-A30 (30 rxns) Reagents Materials are provided for 30 cdna synthesis
The RNAi Consortium (TRC) Broad Institute
 TRC Laboratory Protocols Protocol Title: One Step PCR Preparation of Samples for Illumina Sequencing Current Revision Date: 11/10/2012 RNAi Platform,, [email protected] Brief Description: This
TRC Laboratory Protocols Protocol Title: One Step PCR Preparation of Samples for Illumina Sequencing Current Revision Date: 11/10/2012 RNAi Platform,, [email protected] Brief Description: This
The fastest spin-column based procedure for purifying up to 10 mg of ultra-pure endotoxin-free transfection-grade plasmid DNA.
 INSTRUCTION MANUAL ZymoPURE Plasmid Gigaprep Kit Catalog Nos. D4204 (Patent Pending) Highlights The fastest spin-column based procedure for purifying up to 10 mg of ultra-pure endotoxin-free transfection-grade
INSTRUCTION MANUAL ZymoPURE Plasmid Gigaprep Kit Catalog Nos. D4204 (Patent Pending) Highlights The fastest spin-column based procedure for purifying up to 10 mg of ultra-pure endotoxin-free transfection-grade
sirna Duplexes & RNAi Explorer
 P r o d u c t S p e c i f i c a t i o n s Guaranteed RNAi Explorer kit with Fl/Dabcyl Molecular Beacon Catalog No.:27-6402-01 Guaranteed RNAi Explorer kit with Fluorescein/Tamra TaqMan Catalog No.:27-6402-01
P r o d u c t S p e c i f i c a t i o n s Guaranteed RNAi Explorer kit with Fl/Dabcyl Molecular Beacon Catalog No.:27-6402-01 Guaranteed RNAi Explorer kit with Fluorescein/Tamra TaqMan Catalog No.:27-6402-01
Transformation Protocol
 To make Glycerol Stocks of Plasmids ** To be done in the hood and use RNase/DNase free tips** 1. In a 10 ml sterile tube add 3 ml autoclaved LB broth and 1.5 ul antibiotic (@ 100 ug/ul) or 3 ul antibiotic
To make Glycerol Stocks of Plasmids ** To be done in the hood and use RNase/DNase free tips** 1. In a 10 ml sterile tube add 3 ml autoclaved LB broth and 1.5 ul antibiotic (@ 100 ug/ul) or 3 ul antibiotic
BacReady TM Multiplex PCR System
 BacReady TM Multiplex PCR System Technical Manual No. 0191 Version 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI Detailed Experimental
BacReady TM Multiplex PCR System Technical Manual No. 0191 Version 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 2 V Simplified Procedures. 2 VI Detailed Experimental
Path-ID Multiplex One-Step RT-PCR Kit
 USER GUIDE Path-ID Multiplex One-Step RT-PCR Kit TaqMan probe-based multiplex one-step real-time RT-PCR detection of RNA targets Catalog Numbers 4428206, 4428207, 4440022 Publication Part Number 4440907
USER GUIDE Path-ID Multiplex One-Step RT-PCR Kit TaqMan probe-based multiplex one-step real-time RT-PCR detection of RNA targets Catalog Numbers 4428206, 4428207, 4440022 Publication Part Number 4440907
Green Fluorescent Protein (GFP): Genetic Transformation, Synthesis and Purification of the Recombinant Protein
 Green Fluorescent Protein (GFP): Genetic Transformation, Synthesis and Purification of the Recombinant Protein INTRODUCTION Green Fluorescent Protein (GFP) is a novel protein produced by the bioluminescent
Green Fluorescent Protein (GFP): Genetic Transformation, Synthesis and Purification of the Recombinant Protein INTRODUCTION Green Fluorescent Protein (GFP) is a novel protein produced by the bioluminescent
TECHNICAL BULLETIN. MISSION! shrna Bacterial Glycerol Stock. Catalog Number SHCLNG Storage Temperature 70 C
 MISSION! shrna Bacterial Glycerol Stock Catalog Number SHCLNG Storage Temperature 70 C TECHNICAL BULLETIN Product Description Small interfering RNAs (sirnas) processed from short hairpin RNAs (shrnas)
MISSION! shrna Bacterial Glycerol Stock Catalog Number SHCLNG Storage Temperature 70 C TECHNICAL BULLETIN Product Description Small interfering RNAs (sirnas) processed from short hairpin RNAs (shrnas)
User Manual/Hand book. qpcr mirna Arrays ABM catalog # MA003 (human) and MA004 (mouse)
 User Manual/Hand book qpcr mirna Arrays ABM catalog # MA003 (human) and MA004 (mouse) Kit Components Cat. No. MA003...Human Whole Genome mirna qpcr Profiling Kit (-20 C) The following components are sufficient
User Manual/Hand book qpcr mirna Arrays ABM catalog # MA003 (human) and MA004 (mouse) Kit Components Cat. No. MA003...Human Whole Genome mirna qpcr Profiling Kit (-20 C) The following components are sufficient
Validating Microarray Data Using RT 2 Real-Time PCR Products
 Validating Microarray Data Using RT 2 Real-Time PCR Products Introduction: Real-time PCR monitors the amount of amplicon as the reaction occurs. Usually, the amount of product is directly related to the
Validating Microarray Data Using RT 2 Real-Time PCR Products Introduction: Real-time PCR monitors the amount of amplicon as the reaction occurs. Usually, the amount of product is directly related to the
Transformation of the bacterium E. coli. using a gene for Green Fluorescent Protein
 Transformation of the bacterium E. coli using a gene for Green Fluorescent Protein Background In molecular biology, transformation refers to a form of genetic exchange in which the genetic material carried
Transformation of the bacterium E. coli using a gene for Green Fluorescent Protein Background In molecular biology, transformation refers to a form of genetic exchange in which the genetic material carried
PicoMaxx High Fidelity PCR System
 PicoMaxx High Fidelity PCR System Instruction Manual Catalog #600420 (100 U), #600422 (500 U), and #600424 (1000 U) Revision C Research Use Only. Not for Use in Diagnostic Procedures. 600420-12 LIMITED
PicoMaxx High Fidelity PCR System Instruction Manual Catalog #600420 (100 U), #600422 (500 U), and #600424 (1000 U) Revision C Research Use Only. Not for Use in Diagnostic Procedures. 600420-12 LIMITED
Mouse ES Cell Nucleofector Kit
 page 1 of 7 Mouse ES Cell Nucleofector Kit for Mouse Embryonic Stem Cells Cell type Origin Cells derived from mouse blastocysts. Morphology Round cells growing in clumps. Important remarks! 1. This protocol
page 1 of 7 Mouse ES Cell Nucleofector Kit for Mouse Embryonic Stem Cells Cell type Origin Cells derived from mouse blastocysts. Morphology Round cells growing in clumps. Important remarks! 1. This protocol
Troubleshooting Guide for DNA Electrophoresis
 Troubleshooting Guide for Electrophoresis. ELECTROPHORESIS Protocols and Recommendations for Electrophoresis electrophoresis problem 1 Low intensity of all or some bands 2 Smeared bands 3 Atypical banding
Troubleshooting Guide for Electrophoresis. ELECTROPHORESIS Protocols and Recommendations for Electrophoresis electrophoresis problem 1 Low intensity of all or some bands 2 Smeared bands 3 Atypical banding
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
TOOLS sirna and mirna. User guide
 TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
TOOLS sirna and mirna User guide Introduction RNA interference (RNAi) is a powerful tool for suppression gene expression by causing the destruction of specific mrna molecules. Small Interfering RNAs (sirnas)
50 g 650 L. *Average yields will vary depending upon a number of factors including type of phage, growth conditions used and developmental stage.
 3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
3430 Schmon Parkway Thorold, ON, Canada L2V 4Y6 Phone: 866-667-4362 (905) 227-8848 Fax: (905) 227-1061 Email: [email protected] Phage DNA Isolation Kit Product # 46800, 46850 Product Insert
Cloning GFP into Mammalian cells
 Protocol for Cloning GFP into Mammalian cells Studiepraktik 2013 Molecular Biology and Molecular Medicine Aarhus University Produced by the instructors: Tobias Holm Bønnelykke, Rikke Mouridsen, Steffan
Protocol for Cloning GFP into Mammalian cells Studiepraktik 2013 Molecular Biology and Molecular Medicine Aarhus University Produced by the instructors: Tobias Holm Bønnelykke, Rikke Mouridsen, Steffan
All-in-One mirna qrt-pcr Detection System Handbook
 All-in-One mirna qrt-pcr Detection System Handbook For quantitative detection of mature mirna All-in-One mirna First-Strand cdna Synthesis Kit Cat. No. AMRT-0020 (20 mirna reverse transcription reactions)
All-in-One mirna qrt-pcr Detection System Handbook For quantitative detection of mature mirna All-in-One mirna First-Strand cdna Synthesis Kit Cat. No. AMRT-0020 (20 mirna reverse transcription reactions)
Amaxa Mouse T Cell Nucleofector Kit
 Amaxa Mouse T Cell Nucleofector Kit For T cells isolated from C57BL/6 & BALB/c mice Evaluated for murine T cells isolated from C57BL/6 & BALB/c mice This protocol is designed for murine lymphocytes or
Amaxa Mouse T Cell Nucleofector Kit For T cells isolated from C57BL/6 & BALB/c mice Evaluated for murine T cells isolated from C57BL/6 & BALB/c mice This protocol is designed for murine lymphocytes or
UltraClean Forensic DNA Isolation Kit (Single Prep Format)
 UltraClean Forensic DNA Isolation Kit (Single Prep Format) Catalog No. Quantity 14000-10 10 preps 14000-S 1 prep Instruction Manual Please recycle Version: 10302012 1 Table of Contents Introduction...
UltraClean Forensic DNA Isolation Kit (Single Prep Format) Catalog No. Quantity 14000-10 10 preps 14000-S 1 prep Instruction Manual Please recycle Version: 10302012 1 Table of Contents Introduction...
Technical Manual No. 0173 Update Date 10112010
 TissueDirect TM Multiplex PCR System Technical Manual No. 0173 Update Date 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 3 V Simplified Procedures. 3 VI Detailed
TissueDirect TM Multiplex PCR System Technical Manual No. 0173 Update Date 10112010 I Description.. 1 II Applications 2 III Key Features.. 2 IV Shipping and Storage. 3 V Simplified Procedures. 3 VI Detailed
RT-PCR: Two-Step Protocol
 RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
RT-PCR: Two-Step Protocol We will provide both one-step and two-step protocols for RT-PCR. We recommend the twostep protocol for this class. In the one-step protocol, the components of RT and PCR are mixed
GENETIC TRANSFORMATION OF BACTERIA WITH THE GENE FOR GREEN FLUORESCENT PROTEIN (GFP)
 GENETIC TRANSFORMATION OF BACTERIA WITH THE GENE FOR GREEN FLUORESCENT PROTEIN (GFP) LAB BAC3 Adapted from "Biotechnology Explorer pglo Bacterial Transformation Kit Instruction Manual". (Catalog No. 166-0003-EDU)
GENETIC TRANSFORMATION OF BACTERIA WITH THE GENE FOR GREEN FLUORESCENT PROTEIN (GFP) LAB BAC3 Adapted from "Biotechnology Explorer pglo Bacterial Transformation Kit Instruction Manual". (Catalog No. 166-0003-EDU)
MTT Cell Proliferation Assay
 ATCC 30-1010K Store at 4 C This product is intended for laboratory research purposes only. It is not intended for use in humans, animals or for diagnostics. Introduction Measurement of cell viability and
ATCC 30-1010K Store at 4 C This product is intended for laboratory research purposes only. It is not intended for use in humans, animals or for diagnostics. Introduction Measurement of cell viability and
UltraClean Soil DNA Isolation Kit
 PAGE 1 UltraClean Soil DNA Isolation Kit Catalog # 12800-50 50 preps New improved PCR inhibitor removal solution (IRS) included Instruction Manual (New Alternative Protocol maximizes yields) Introduction
PAGE 1 UltraClean Soil DNA Isolation Kit Catalog # 12800-50 50 preps New improved PCR inhibitor removal solution (IRS) included Instruction Manual (New Alternative Protocol maximizes yields) Introduction
Transfection reagent for visualizing lipofection with DNA. For ordering information, MSDS, publications and application notes see www.biontex.
 METAFECTENE FluoR Transfection reagent for visualizing lipofection with DNA For ordering information, MSDS, publications and application notes see www.biontex.com Description Cat. No. Size METAFECTENE
METAFECTENE FluoR Transfection reagent for visualizing lipofection with DNA For ordering information, MSDS, publications and application notes see www.biontex.com Description Cat. No. Size METAFECTENE
2.1.2 Characterization of antiviral effect of cytokine expression on HBV replication in transduced mouse hepatocytes line
 i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
DNA Isolation Kit for Cells and Tissues
 DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
DNA Isolation Kit for Cells and Tissues for 10 isolations of 00 mg each for tissue or 5 x 10 7 cultured cells Cat. No. 11 81 770 001 Principle Starting material Application Time required Results Benefits
Arcturus PicoPure RNA Isolation Kit. User Guide
 Arcturus PicoPure RNA Isolation Kit User Guide For Research Use Only. Not intended for any animal or human therapeutic or diagnostic use. Information in this document is subject to change without notice.
Arcturus PicoPure RNA Isolation Kit User Guide For Research Use Only. Not intended for any animal or human therapeutic or diagnostic use. Information in this document is subject to change without notice.
One Shot TOP10 Competent Cells
 USER GUIDE One Shot TOP10 Competent Cells Catalog Numbers C4040-10, C4040-03, C4040-06, C4040-50, and C4040-52 Document Part Number 280126 Publication Number MAN0000633 Revision A.0 For Research Use Only.
USER GUIDE One Shot TOP10 Competent Cells Catalog Numbers C4040-10, C4040-03, C4040-06, C4040-50, and C4040-52 Document Part Number 280126 Publication Number MAN0000633 Revision A.0 For Research Use Only.
NimbleGen DNA Methylation Microarrays and Services
 NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
NimbleGen DNA Methylation Microarrays and Services Sample Preparation Instructions Outline This protocol describes the process for preparing samples for NimbleGen DNA Methylation microarrays using the
Essentials of Real Time PCR. About Sequence Detection Chemistries
 Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
Essentials of Real Time PCR About Real-Time PCR Assays Real-time Polymerase Chain Reaction (PCR) is the ability to monitor the progress of the PCR as it occurs (i.e., in real time). Data is therefore collected
AxyPrep TM Mag PCR Clean-up Protocol
 AxyPrep TM Mag PCR Clean-up Protocol Intro The AxyPrep Mag PCR Clean-up kit utilizes a unique paramagnetic bead technology for rapid, high-throughput purification of PCR amplicons. Using this kit, PCR
AxyPrep TM Mag PCR Clean-up Protocol Intro The AxyPrep Mag PCR Clean-up kit utilizes a unique paramagnetic bead technology for rapid, high-throughput purification of PCR amplicons. Using this kit, PCR
Global MicroRNA Amplification Kit
 Global MicroRNA Amplification Kit Store kit at -20 C on receipt (ver. 3-060901) A limited-use label license covers this product. By use of this product, you accept the terms and conditions outlined in
Global MicroRNA Amplification Kit Store kit at -20 C on receipt (ver. 3-060901) A limited-use label license covers this product. By use of this product, you accept the terms and conditions outlined in
Agencourt RNAdvance Blood Kit for Free Circulating DNA and mirna/rna Isolation from 200-300μL of Plasma and Serum
 SUPPLEMENTAL PROTOCOL WHITE PAPER Agencourt RNAdvance Blood Kit for Free Circulating DNA and mirna/rna Isolation from 200-300μL of Plasma and Serum Bee Na Lee, Ph.D., Beckman Coulter Life Sciences Process
SUPPLEMENTAL PROTOCOL WHITE PAPER Agencourt RNAdvance Blood Kit for Free Circulating DNA and mirna/rna Isolation from 200-300μL of Plasma and Serum Bee Na Lee, Ph.D., Beckman Coulter Life Sciences Process
Protocol 001298v001 Page 1 of 1 AGENCOURT RNACLEAN XP IN VITRO PRODUCED RNA AND CDNA PURIFICATION
 Page 1 of 1 AGENCOURT RNACLEAN XP IN VITRO PRODUCED RNA AND CDNA PURIFICATION Please refer to http://www.agencourt.com/technical for updated protocols and refer to MSDS instructions when handling or shipping
Page 1 of 1 AGENCOURT RNACLEAN XP IN VITRO PRODUCED RNA AND CDNA PURIFICATION Please refer to http://www.agencourt.com/technical for updated protocols and refer to MSDS instructions when handling or shipping
Amaxa 4D-Nucleofector Protocol for Mouse Embryonic Stem [ES] Cells For 4D-Nucleofector X Unit Transfection in suspension
![Amaxa 4D-Nucleofector Protocol for Mouse Embryonic Stem [ES] Cells For 4D-Nucleofector X Unit Transfection in suspension Amaxa 4D-Nucleofector Protocol for Mouse Embryonic Stem [ES] Cells For 4D-Nucleofector X Unit Transfection in suspension](/thumbs/30/14702458.jpg) For 4D-Nucleofector X Unit Transfection in suspension Cells derived from mouse blastocysts; round cells, growing in clumps s 1. This protocol is meant to provide an outline for the handling and the Nucleofection
For 4D-Nucleofector X Unit Transfection in suspension Cells derived from mouse blastocysts; round cells, growing in clumps s 1. This protocol is meant to provide an outline for the handling and the Nucleofection
RealLine HCV PCR Qualitative - Uni-Format
 Instructions for use PCR KIT FOR EXTRACTION OF RNA AND REAL TIME PCR DETECTION KIT FOR HEPATITIS C VIRUS RNA Research Use Only Qualitative Uni Format VBD0798 48 tests valid from: December 2013 Rev11122013
Instructions for use PCR KIT FOR EXTRACTION OF RNA AND REAL TIME PCR DETECTION KIT FOR HEPATITIS C VIRUS RNA Research Use Only Qualitative Uni Format VBD0798 48 tests valid from: December 2013 Rev11122013
Quantitative Real Time PCR Protocol. Stack Lab
 Quantitative Real Time PCR Protocol Stack Lab Overview Real-time quantitative polymerase chain reaction (qpcr) differs from regular PCR by including in the reaction fluorescent reporter molecules that
Quantitative Real Time PCR Protocol Stack Lab Overview Real-time quantitative polymerase chain reaction (qpcr) differs from regular PCR by including in the reaction fluorescent reporter molecules that
DNA Integrity Number (DIN) For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments
 DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
Recombinant DNA and Biotechnology
 Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
HCS604.03 Exercise 1 Dr. Jones Spring 2005. Recombinant DNA (Molecular Cloning) exercise:
 HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
HCS604.03 Exercise 1 Dr. Jones Spring 2005 Recombinant DNA (Molecular Cloning) exercise: The purpose of this exercise is to learn techniques used to create recombinant DNA or clone genes. You will clone
RAPAd mirna Adenoviral Expression System Catalog #: VPK-253
 DATENBLATT RAPAd mirna Adenoviral Expression System Catalog #: VPK-253 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction MicroRNAs (mirnas) are 18 24 nucleotide RNA molecules that
DATENBLATT RAPAd mirna Adenoviral Expression System Catalog #: VPK-253 FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction MicroRNAs (mirnas) are 18 24 nucleotide RNA molecules that
FastLine cell cdna Kit
 1. FastLine cell cdna Kit For high-speed preparation of first-strand cdna directly from cultured cells without RNA purification www.tiangen.com RT100701 FastLine cell cdna Kit Cat. no. KR105 Kit Contents
1. FastLine cell cdna Kit For high-speed preparation of first-strand cdna directly from cultured cells without RNA purification www.tiangen.com RT100701 FastLine cell cdna Kit Cat. no. KR105 Kit Contents
Genolution Pharmaceuticals, Inc. Life Science and Molecular Diagnostic Products
 Genolution Pharmaceuticals, Inc. Revolution through genes, And Solution through genes. Life Science and Molecular Diagnostic Products www.genolution1.com TEL; 02-3010-8670, 8672 Geno-Serum Hepatitis B
Genolution Pharmaceuticals, Inc. Revolution through genes, And Solution through genes. Life Science and Molecular Diagnostic Products www.genolution1.com TEL; 02-3010-8670, 8672 Geno-Serum Hepatitis B
Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr
 User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
User Manual Mir-X mirna First-Strand Synthesis and SYBR qrt-pcr User Manual United States/Canada 800.662.2566 Asia Pacific +1.650.919.7300 Europe +33.(0)1.3904.6880 Japan +81.(0)77.543.6116 Clontech Laboratories,
Human Free Testosterone(F-TESTO) ELISA Kit
 Human Free Testosterone(F-TESTO) ELISA Kit Catalog Number. MBS700040 For the quantitative determination of human free testosterone(f-testo) concentrations in serum, plasma. This package insert must be
Human Free Testosterone(F-TESTO) ELISA Kit Catalog Number. MBS700040 For the quantitative determination of human free testosterone(f-testo) concentrations in serum, plasma. This package insert must be
Mouse glycated hemoglobin A1c(GHbA1c) ELISA Kit
 Mouse glycated hemoglobin A1c(GHbA1c) ELISA Kit Catalog Number. CSB-E08141m For the quantitative determination of mouse glycated hemoglobin A1c(GHbA1c) concentrations in lysate for RBC. This package insert
Mouse glycated hemoglobin A1c(GHbA1c) ELISA Kit Catalog Number. CSB-E08141m For the quantitative determination of mouse glycated hemoglobin A1c(GHbA1c) concentrations in lysate for RBC. This package insert
HighPure Maxi Plasmid Kit
 HighPure Maxi Plasmid Kit For purification of high pure plasmid DNA with high yields www.tiangen.com PP120109 HighPure Maxi Plasmid Kit Kit Contents Storage Cat.no. DP116 Contents RNaseA (100 mg/ml) Buffer
HighPure Maxi Plasmid Kit For purification of high pure plasmid DNA with high yields www.tiangen.com PP120109 HighPure Maxi Plasmid Kit Kit Contents Storage Cat.no. DP116 Contents RNaseA (100 mg/ml) Buffer
LAB 7 DNA RESTRICTION for CLONING
 BIOTECHNOLOGY I DNA RESTRICTION FOR CLONING LAB 7 DNA RESTRICTION for CLONING STUDENT GUIDE GOALS The goals of this lab are to provide the biotech student with experience in DNA digestion with restriction
BIOTECHNOLOGY I DNA RESTRICTION FOR CLONING LAB 7 DNA RESTRICTION for CLONING STUDENT GUIDE GOALS The goals of this lab are to provide the biotech student with experience in DNA digestion with restriction
High-quality genomic DNA isolation and sensitive mutation analysis
 Application Note High-quality genomic DNA isolation and sensitive mutation analysis Izabela Safin, Ivonne Schröder-Stumberger and Peter Porschewski Introduction A major objective of cancer research is
Application Note High-quality genomic DNA isolation and sensitive mutation analysis Izabela Safin, Ivonne Schröder-Stumberger and Peter Porschewski Introduction A major objective of cancer research is
