Multiple Pathways Used for the Targeting of Thylakoid Proteins in Chloroplasts
|
|
|
- Gavin Alan Martin
- 8 years ago
- Views:
Transcription
1 Traffic : Copyright C Munksgaard 2001 Munksgaard International Publishers ISSN Review Multiple Pathways Used for the Targeting of Thylakoid Proteins in Chloroplasts Colin Robinson*, Simon J. Thompson and Cheryl Woolhead Department of Biological Sciences, University of Warwick, Coventry CV4 7AL, United Kingdom *Corresponding author: Crobinson@bio.warwick.ac.uk The assembly of the chloroplast thylakoid membrane requires the import of numerous proteins from the cytosol and their targeting into or across the thylakoid membrane. It is now clear that multiple pathways are involved in the thylakoid-targeting stages, depending on the type of protein substrate. Two very different pathways are used by thylakoid lumen proteins; one is the Sec pathway which has been well-characterised in bacteria, and which involves the threading of the substrate through a narrow channel. In contrast, the more recently characterised twin-arginine translocation (Tat) system is able to translocate fully folded proteins across this membrane. Recent advances on bacterial Tat systems shed further light on the structure and function of this system. Membrane proteins, on the other hand, use two further pathways. One is the signal recognition particle-dependent pathway, involving a complex interplay between many different factors, whereas other proteins insert without the assistance of any known apparatus. This article reviews advances in the study of these pathways and considers the rationale behind the surprising complexity. Key words: chloroplast, membrane, protein transport, tat, thylakoid Received and accepted for publication 5 February 2001 The chloroplast is well known as the site of photosynthesis in plants but it also carries out many other critical functions and consequently contains a vast range of protein types probably well over Some of these proteins, especially those involved in photosynthetic functions, are also present at very high concentrations, and the total protein concentration in the organelle is in the order of 300 mg/ml. A small proportion of these proteins are synthesised within the organelle, but the vast majority are encoded by nuclear genes and synthesised in the cytosol, with the net result that chloroplast biogenesis requires the import of a colossal amount of protein in a relatively short time. To complicate matters, this organelle is particularly complex in structural terms, comprising three separate membranes, which enclose three distinct soluble phases. The chloroplast is bounded by a doublemembrane envelope, which encloses an intermembrane space, about which rather little is known. The major soluble phase is the stroma, which is the site of carbon fixation, amino acid synthesis and many other pathways, and the dominant membrane is the extensive interconnecting thylakoid network, where light is captured and ATP synthesised. Finally, the thylakoid membrane encloses a further soluble phase, the thylakoid lumen, which houses a number of extrinsic photosynthetic proteins as well as many others. Imported proteins are targeted into all six chloroplast subcompartments and the underlying mechanisms have been the focus of considerable attention over the past 25 years. Perhaps surprisingly, most proteins appear to be imported across the envelope membranes by a basically similar mechanism. This topic has been covered in a recent review (1) and will not be dealt with in detail here. Rather, the aim of this article is to review recent progress on the intraorganellar sorting of thylakoid proteins, where it is now clear that numerous pathways are utilised for the targeting of thylakoid lumen and membrane proteins. One of the lumen-targeting pathways, the Tat pathway, has been the centre of much interest since its discovery in bacteria, and the same applies to the Alb3- mediated pathway for membrane proteins. This review will focus on the mainstream pathways used for thylakoid proteins with an emphasis on providing a rationale for the surprising multiplicity. Two Very Different Pathways for the Targeting of Thylakoid Lumen Proteins Nearly all thylakoid lumen proteins are synthesised in the cytosol, and all of them appear to undergo a two-stage import pathway via stromal intermediate forms. They are initially synthesised in the cytosol as precursor proteins bearing bipartite N-terminal presequences which contain two targeting signals in tandem, after which they are transported across the envelope at contact sites between the two membranes. This import process requires ATP and probably GTP, and a key feature of the translocation mechanism is that the protein is transported in a largely unfolded form (reviewed in (1)). Once in the stroma, the first envelope transit domain of the presequence is removed by a stromal processing peptidase. The intermediate form is then transported across the thylakoid membrane by one of two distinct mechanisms. A subset 245
2 Robinson et al. of lumenal proteins, including the 33-kDa photosystem II protein (33K), plastocyanin and others, are transported by a Sec pathway which closely resembles the well-characterised Sec-dependent export pathways in bacteria (reviewed in (2)). Many proteins are exported across the plasma membrane to the periplasm (or further) in bacteria, and until recently it was believed that this process was mediated almost exclusively by the Sec pathway. In this process, the substrate protein is synthesised with a signal peptide comprising three domains: a basic N-terminal (N-) domain, hydrophobic core (H-) domain and more polar C-terminal (C-) domain. The export mechanism involves the threading of the protein through a SecYEG translocon, driven by the ATPase activity of a peripheral protein, SecA. Plant chloroplasts contain SecY, SecE and SecA homologues which are involved in targeting of thylakoid lumen proteins by a very similar mechanism (3 5), and this finding comes as no surprise since it is generally accepted that chloroplasts evolved from cyanobacterial-type organisms. The discovery of a second mainstream pathway, on the other hand, did come as a surprise because at that time there were no indications of a second export system in bacteria. It was shown some time ago that other lumen proteins are transported by a mechanism that does not require SecA or nucleoside triphosphates (NTPs), but which relies totally on the DpH across the thylakoid membrane (6,7). Substrates for this pathway are also synthesised with bipartite presequences and the second, thylakoid-targeting domains resemble those of Sec substrates in many respects. However, these signals contain all of the information specifying translocation by the DpH-dependent pathway and are able to direct foreign proteins with high efficiency by this route (8,9). Closer examination of these signals showed that they contain an invariant twin-arginine motif that is critical for DpH-dependent targeting (10); a further important determinant is the presence of a highly hydrophobic residue two or three residues after this motif (11). These features are illustrated in Figure 1, which lists Sec- and DpH-dependent thylakoid-targeting signals of known lumenal proteins. Discovery of a Related, Sec-independent Export Pathway in Bacteria The major breakthrough in this field came with the characterisation of a maize mutant, termed hcf106, that was shown to be specifically defective in the DpH-dependent targeting pathway (12). Substrates for this pathway were shown to accumulate as intermediates in the stroma and the hcf106 gene was later cloned, sequenced and shown to encode a thylakoid membrane protein (13). This work provided the next serious surprise because homologues of the hcf106 gene were present in most sequenced bacterial genomes, all as unassigned open reading frames. In fact, bacteria usually contain two or more hcf106 homologues and the gene organisation in Escherichia coli is shown Figure 1: Targeting signals for thylakoid lumen proteins. Signal peptides specifying translocation by the Tat and Sec pathways in chloroplasts. The signals are shown for representative substrates from spinach (Sp), wheat (Wh), maize (Ma), Arabidopsis (Ara), barley (Bar) and Cotton (Cot). The list includes: proteins that have been experimentally shown to depend on the Tat-system and several that specify targeting by the Sec pathway. Hydrophobic (H-) domains are underlined, charged residues are shown in bold and twin-arginine motifs are shown in bold italics. in Figure 2, together with predicted basic structures and orientations of the encoded proteins. As discussed below, these genes do indeed encode components of second export pathway that is related to the thylakoid DpH-dependent pathway, and the genes have been termed tat genes (for twin-arginine translocase). The E. coli genome contains three genes homologous to hcf106, two of which (tata and tatb) form the first two genes of a four-gene operon. A third hcf106 homologue, tate, is monocistronic. Knockout studies have shown that disruption of either tatb or tata/e leads to a complete block in the export of a range of periplasmic proteins (14,15). While all three genes are sequence-related to each other and to hcf106, they encode proteins with two distinct functions, and it is now clear that the export system basically requires TatB and either TatA or TatE. TatA is the more important of the latter two and its disruption has a far greater effect, but the DtatA cells can be complemented by overexpression of TatE (16). These Tat components (and Hcf106) are all strongly predicted to be singlespan proteins with a very short N-terminal region in the periplasm/thylakoid lumen and a short domain in the cytoplasm/stroma (that of TatB being larger than TatA). The next gene in the tat operon, tatc is also critical for Tatdependent export and E. coli cells disrupted in this gene are completely defective in the Tat export pathway (17). The TatC protein is predicted to span the plasma membrane six times with the N- and C- termini in the cytoplasm. However, the tatd gene appears not be involved (or to only a minor extent). This gene encodes a soluble cytoplasmic protein and DtatD cells are unaffected in Tat-dependent export (18). The Tat 246 Traffic 2001; 2:
3 Multiple Pathways for Targeting Thylakoid Proteins Speciality of the Tat Pathway: the Translocation of Fully Folded Proteins Figure 2: tat gene organisation in E. coli and the predicted structures of Tat proteins. A: Gene organisation. The primary known genes are organised in a tatabcd operon as shown. The tatabc genes encode membrane-bound components whereas the tatd gene product is a cytoplasmic protein which does not appear to be involved in Tat-dependent export. The monocistronic tate gene encodes a TatA-type protein but this gene is expressed at low levels and the function of TatE is relatively unimportant. B: Predicted protein structure. TatA and TatB almost certainly contain a short periplasmic N-terminal domain and a single membrane-spanning region followed by a predicted short amphipathic region. TatA, TatE and TatB contain small cytoplasmic domains. TatC is predicted to contain 6 transmembrane helices and short interconnecting loop regions, with the N- and C-termini located in the cytoplasm. Chloroplasts contain homologues of all three Tat proteins. pathway in bacteria has thus so far been shown to require the activities of three membrane-bound proteins, although the existence of further Tat subunits cannot be ruled out since the system has yet to be purified in a functional form. All of the indications are that the thylakoidal system is essentially identical; Hcf106 is believed to be a TatB homologue and homologues of TatA and TatC have been identified in higher plants (19,20). Although this article is primarily concerned with thylakoid protein targeting, it is of passing interest to note that the targeting signals for bacterial Tat substrates are generally similar to thylakoid lumen proteins and they likewise contain the invariant RR-motif (21,22). They are furthermore able to direct proteins across the chloroplast thylakoid membrane with high efficiency (23 25), indicating that some features of the mechanism have remained very highly conserved. However, Gram-negative bacterial RR-signal peptides contain additional conserved features that are generally absent from thylakoid Tat signals; most contain a phenylalanine two residues after the RR-motif and many contain a lysine two residues later (21,22). The consensus motif for these bacterial signals is thus RRxFxK, but the precise significance of the conserved Phe and Lys residues is uncertain since they are not strictly required for efficient translocation (22). Traffic 2001; 2: The targeting signals for the Sec- and Tat-type translocation pathways may be surprisingly similar but their mechanisms could hardly be more different, because there is now considerable evidence that the Tat system has the unique ability to translocate folded proteins across tightly sealed biological membranes. Two studies on the thylakoidal system have addressed this point directly using in vitro import assays. In the first (26), it was found that bovine pancreatic trypsin inhibitor could be transported across the thylakoid membrane by the Tat pathway using a Tat-specific signal, even after internal cross-linking to block any unfolding. The second study (27) showed that dihdyrofolate reductase could be transported by this pathway after binding a folate analogue in the active site, and that the ligand was almost certainly transported across the membrane with the protein, implying transport in a folded state. The two contrasting pathways used for the targeting of thylakoid lumen proteins are shown diagrammatically in Figure 3. There is also a wealth of circumstantial, but equally compelling evidence for a similar transport mechanism in bacteria. Studies of the substrate specificity of the Tat system (21,28) indicate that the main substrates are those periplasmic proteins that bind any of a range of redox cofactors, including molybdopterin, FeS and others. There is good evidence that these cofactors can only be inserted in the cytoplasm and hence these proteins can only be exported after folding to a substantial degree. This is impossible by the Sec pathway, which can only transport proteins in an unfolded state, and hence one primary role of the Tat pathway is to transport proteins that are simply obliged to fold prior to translocation. However, there is good evidence that some Tat substrates do not bind cofactors. This applies particularly to substrates of the thylakoid Tat system, where only polyphenol oxidase of the group shown in Figure 1 is believed to bind such a cofactor (a multicopper centre (29)). Thus the second group of substrates is likely to comprise those proteins that either fold too quickly and/or too tightly for the Sec pathway to handle. The Tat system is clearly capable of carrying out some remarkable translocation reactions. The largest known substrates in bacteria include TMAO reductase and formate dehydrogenase N, which have molecular masses of 83 and 132 kda, respectively. If, as seems likely, these proteins are exported in a fully folded state, the Tat system must have a quite unique translocation mechanism because the transport of these relatively huge molecules must occur without substantial leakage of protons, since the system operates in energy-transducing membranes. All other protein translocases in such tightly sealed membranes (for example, the endoplasmic reticulum (ER) and bacterial Sec systems, and the chloroplast and mitochondrial envelope-localised translocases) operate by a threading mechanism and it appears that the Tat system must have evolved to overcome the limitations of these systems. At present, however, we know very little 247
4 Robinson et al. Figure 3: The Tat and Sec pathways for the targeting of thylakoid lumen proteins. Imported thylakoid lumen proteins such as plastocyanin (PC; a Sec substrate) and 23K (a Tat substrate) are synthesised with bipartite presequences containing envelope transit signals (red boxes) and thylakoid-targeting signals in tandem. Sec- and Tat-type thylakoid-targeting signals are depicted by green and black boxes, respectively. The envelope transit peptides of both precursors are recognised by a protein transport system in the envelope membranes which facilitates translocation across the envelope membranes in an unfolded state. The envelope transit signals are usually removed in the stroma. Proteins such as 23K are believed to refold in the stroma before being transported in a folded form by a DpH-driven Tat system; the translocation of Sec substrates involves stromal SecA, ATP hydrolysis and a membrane-bound Sec complex believed to be similar to prokaryotic SecYEG complexes. It is presently unclear whether these proteins fold in the stroma, but the later stages involve SecA/ATPdriven translocation of the intermediate in an unfolded state through the Sec translocon. After translocation, substrates on both pathways are processed to the mature forms by the thylakoidal processing peptidase. about the structure or mechanism of this system. TatA and TatB have been found to co-immunoprecipitate with each other in a solubilized E. coli complex of about 600 kda (30). One artificial Tat substrate has been found to lodge in a thylakoid Tat complex of about the same size (31), but the system has yet to be purified in a functional state or otherwise characterised in any detail. Signal Recognition Particle-Dependent Insertion of Thylakoid Membrane Proteins: Another Inherited Pathway The thylakoid membrane houses integral membrane proteins of many types, from simple single-span proteins to complex multispanning, pigment binding proteins. Once again, most of these proteins are imported from the cytosol prior to being inserted into the thylakoid membrane, and the underlying mechanisms have been analysed in some detail, primarily using in vitro reconstitution assays. Much of the work has centred on a single model thylakoid membrane protein, the major light-harvesting chlorophyll-binding protein, Lhcb1. This protein spans the membrane three times, with the N- terminus located on the stromal side and the C-terminus in the lumen. As with almost all imported chloroplast proteins, Lhcb1 is synthesised with an N-terminal envelope transit peptide which directs import into the organelle, but unlike the lumenal proteins discussed above there is no second cleavable thylakoid-targeting peptide. Instead, the thylakoid- targeting information is located within the mature Lhcb1 protein (32,33). Initial reconstitution assays showed that this protein undergoes a complex insertion pathway that requires a stromal signal recognition particle (SRP), FtsY, GTP and initially unidentified thylakoid-bound translocation machinery (34 36). These data clearly point to a prokaryotic origin for this pathway because SRP and FtsY have been shown to be involved in the insertion of several membrane proteins in bacteria (reviewed in (37,38)). Both of these factors hydrolyse GTP during their functioning and the emerging picture in bacteria suggests that SRP binds to highly hydrophobic regions, and hence selects mostly membrane proteins, whereas SecA is used for the targeting of soluble periplasmic proteins. Both pathways, however, appear to converge at the SecYEG translocon, at least in some cases (reviewed in (37)). A similar situation may well apply in chloroplasts but this important point has yet to be resolved. Although the bacterial and chloroplastic SRP-dependent targeting systems are similar in several respects, it is interesting that the chloroplast SRP differs from other known SRPs. The eukaryotic SRP involved in targeting to the ER is a complex particle containing an RNA molecule and six protein subunits, while the E. coli particle is a simpler version containing an RNA molecule and one protein subunit, homologous to the 54-kDa signal peptide-binding subunit of the eukaryotic SRP (reviewed in (37,38)). In contrast, the chloroplast SRP contains the 54-kDa subunit together with another 43-kDa subunit that is completely novel (39). This arrangement may 248 Traffic 2001; 2:
5 Multiple Pathways for Targeting Thylakoid Proteins have evolved to cope with the obligatorily post-translational mode of insertion for imported thylakoid membrane proteins, since the eukaryotic and bacterial SRPs may well bind to their cognate substrates in a cotranslational manner. However, further work is required before we can appreciate the global role of SRP in thylakoid membrane biogenesis, since Lhcb1 is the only cytoplasmically synthesised substrate identified to date. Moreover, there is evidence that the 43-kDa subunit specifically recognises a peptide sequence (denoted L18) that is present only in members of the light-harvesting chlorophyll-binding proteins family (40), which raises the possibility that SRP (or at least SRP43-containing SRP) is essentially dedicated to the biogenesis of this type of protein. Other studies strongly suggest a role for SRP in the insertion of one major chloroplast-encoded protein, D1 (41) but it is unclear whether the 43-kDa subunit is involved in this process. The picture has also become more complex with the recent emergence of a critical new targeting factor that is used for the biogenesis of membrane proteins in many membrane systems. The Oxa1 protein is involved in the biogenesis of a range of mitochondrial inner membrane proteins, specifically those that insert from the matrix side of the membrane (42). An Oxa1 homologue, termed Alb3, is also essential for the insertion of Lhcb1 in thylakoids (43), and recent work has shown an E. coli homologue to be essential for the insertion of a range of membrane proteins in this organism (44). The latter work is particularly important because in this study it was found that both SRP-dependent and -independent proteins depend on the Oxa1 homologue, YidC. This raises the question as to how YidC (and Alb3) fit into the SRP-dependent and -independent insertion pathways. Some YidC is found tightly bound to the SecYEG translocon in bacteria (45), suggesting that it might participate in the insertion reaction either before or after SRP and FtsY deliver the membrane protein to the SecYEG translocon. However, YidC is also involved in SRP/Sec-independent insertion reactions, which suggests that it may form a separate translocon on its own or with other, as yet unidentified components. The precise role of Alb3 in thylakoid protein insertion is similarly unclear, but Figure 4 shows one possible model for the SRP-dependent targeting pathway in chloroplasts, in which Alb3 is involved in releasing the inserted protein from the translocon. Note, however, that the thylakoid SecYEG complex has not been shown to be involved in Lhcb1 insertion and this model is based partly on its involvement in the bacterial SRP-dependent pathway. A Novel Insertion Mechanism for Other Thylakoid Membrane Proteins The Sec-, Tat- and SRP-dependent targeting pathways described above were all apparently inherited from the cyanobacterial-type progenitors of higher plant chloroplasts, and this conservation of translocation machinery is of course logical in most respects. However, a further pathway has been iden- Traffic 2001; 2: tified for the targeting of thylakoid membrane proteins, and this particular pathway is not only virtually unique but also clearly more recent in origin. Initial studies showed that a range of single-span membrane proteins, including CF o II, PsbW and PsbX, differ from other imported membrane proteins in that they are synthesised with clear bipartite presequences that strongly resemble those of imported lumenal proteins. However, these proteins insert into the thylakoid membrane in the absence of SRP, NTPs or a functional Sec machinery, and proteolysis of thylakoids, which destroys all of the known translocation apparatus, has no effect on their insertion (46 49). More recent studies showed that a further protein on this pathway, PsbY, is even more remarkable since this protein contains two cleavable signal peptides, which are cleaved after insertion to yield two small single-span mature proteins (50). In the absence of any identifiable insertion machinery or energy requirements, it has been proposed that these proteins may insert spontaneously, but the possible involvement of unknown machinery (in particular Alb3) cannot yet be ruled out. If this scenario is correct, however, it may be that the role of the signal peptide is to provide a second hydrophobic region which, in concert with that in the mature protein, may partition into the membrane and drive the overall insertion process. This pathway is shown in Figure 4. Irrespective of the precise insertion mechanism, this is one pathway that appears not to have been inherited from cyanobacteria. Genes encoding proteins homologous to CF o II, PsbX and PsbY are present in cyanobacteria and also in the plastid genomes of some eukaryotic algae, and in no case is the encoded protein synthesised with a signal peptide. In these organisms, the mature protein is clearly directed into the thylakoid membrane, which suggests that the requirement for cleavable signal peptides coincided with the transfer of the genes to the nucleus. Possibly, the signal peptides are needed for a rather different insertion mechanism that results from the much more complex pathway from cytosol to thylakoid. Summarising Remarks The biogenesis of thylakoids has turned out to be much more complex than originally anticipated, and even more targeting pathways may emerge in future studies. The underlying mechanisms are now understood in some detail and most of the known components have been identified and cloned. We also now appreciate the basic distinctions between the Secand Tat-dependent lumen-targeting pathways and the rationale behind their existence. However, many aspects of thylakoid proteins remain poorly understood. The Tat components have only recently been identified, we have essentially no idea how this remarkable system works, and the central reactions in the SRP pathway remain to be elucidated in real detail (this applies to bacteria too). Finally, the spontaneous insertion pathway needs further study because it seems inherently unlikely that membrane-spanning proteins could re- 249
6 Robinson et al. Figure 4: Models for the SRP-dependent and spontaneous targeting pathways for membrane proteins. The SRP pathway: Lhcb1 is a chlorophyll binding protein that spans the membrane three times (membrane-spanning regions depicted as grey boxes). It is synthesised in the cytosol as a precursor containing a single envelope transit peptide. This directs the protein across the envelope membranes in an unfolded form, after which the presequence is removed and the protein is bound by stromal signal recognition particle, SRP (not necessarily in that order). SRP then delivers the protein to translocation apparatus in the thylakoid membrane (probably the SecYEG translocon, but this is not certain) and, together with FtsY, facilitates insertion by a GTP-dependent process. A further factor, Alb3, is required for full insertion. Spontaneous pathway: PsbW is a single-span protein synthesised in the cytosol with a bipartite presequence comprising an envelope transit peptide followed by a hydrophobic signal-type peptide. Translocation across the envelope probably takes place by the standard route, after which the transit peptide is removed and the protein then inserts into the thylakoid membrane in a loop conformation. The thylakoidal processing peptidase cleaves on the lumenal face of the membrane, generating the mature PsbW protein. main soluble and insertion-competent for so long in the stroma. Nevertheless, the availability of so many genes encoding these components, together with the ability of thylakoids to import proteins so efficiently, means that we can look forward to continued progress in this field. This will be good for an understanding of this remarkable membrane system and also for an appreciation of the bacterial membranes from which it originated. References 1. Heins L, Collinson IR, Soll J. The protein transport apparatus of chloroplast envelopes. Trends Plant Sci 1998;3: Manting EH, Driessen AJ. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol 2000;37: Yuan J, Henry R, McCaffery M, Cline K. SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science 1994;266: Laidler V, Chaddock AM, Knott TG, Walker D, Robinson C. A SecY homolog in Arabidopsis thaliana. Sequence of a full-length cdna clone and import of the precursor protein into chloroplasts. J Biol Chem 1995;270: Schuenemann D, Amin P, Hartmann E, Hoffman NE. Chloroplast SecY is complexed to SecE and involved in the translocation of the 33-kDa, but not the 23-kDa subunit of the oxygen-evolving complex. J Biol Chem 1999;274: Mould RM, Robinson C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J Biol Chem 1991;266: Cline K, Ettinger WF, Theg SM. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J Biol Chem 1992;267: Robinson C, Cai D, Hulford A, Brock IW, Michl D, Hazell L, Schmidt I, Herrmann RG, Klösgen RB. The presequence of a chimeric construct dictates which of two mechanisms are utilized for translocation across the thylakoid membrane: evidence for the existence of two distinct translocation systems. EMBO J 1994;13: Henry R, Kapazoglou A, McCaffery M, Cline K. Differences between lumen targeting domains of chloroplast transit peptides determine pathway specificity for thylakoid transport. J Biol Chem 1994;269: Chaddock AM, Mant A, Karnauchov I, Brink S, Herrmann RG, Klösgen RB, Robinson C. A new type of signal peptide: central role of a twinarginine motif in transfer signals for the DpH-dependent thylakoidal protein translocase. EMBO J 1995;14: Brink S, Bogsch EG, Edwards WR, Hynds PJ, Robinson C. Targeting of thylakoid proteins by the DpH-driven twin-arginine translocation pathway requires a specific signal in the hydrophobic domain in conjunction with the twin-arginine motif. FEBS Lett 1998;434: Voelker R, Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J 1995;14: Settles MA, Yonetani A, Baron A, Bush DR, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science 1997;278: Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 1998;93: Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. Overlapping functions of components of a bacterial Secindependent export pathway. EMBO J 1998;17: Traffic 2001; 2:
7 Multiple Pathways for Targeting Thylakoid Proteins 16. Sargent F, Stanley NR, Berks BC, Palmer T. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J Biol Chem 1999;274: Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem 1998;273: Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in Sec-independent protein export. J Biol Chem 2000;275: Walker MB, Roy LM, Coleman E, Voelker R, Barkan A. The maize tha4 gene functions in Sec-independent protein transport in chloroplasts and is related to hcf106, tata and tatb. J Cell Biol 1999;147: Mori H, Summer EJ, Ma X, Cline K. Component specificity for the thylakoidal Sec and delta ph-dependent protein transport pathways. J Cell Biol 1999;146: Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 1996;22: Stanley NR, Palmer T, Berks BC. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem 2000;275: Mori H, Cline K. A signal peptide that directs non-sec transport in bacteria also directs efficient and exclusive transport on the thylakoid delta ph pathway. J Biol Chem 1998;273: Wexler M, Bogsch EG, Palmer T, Robinson C, Berks BC. Targeting signals for a bacterial Sec-independent export system direct plant thylakoid import by the DpH pathway. FEBS Lett 1998;431: Halbig D, Hou B, Freudl R, Sprenger GA, Klösgen RB. Bacterial proteins carrying twin-r signal peptides are specifically targeted by the DpH-dependent transport machinery of the thylakoid membrane system. FEBS Lett 1999;447: Clark SA, Theg SM. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol Biol Cell 1997;8: Hynds PJ, Robinson D, Robinson C. The Sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J Biol Chem 1998;273: Santini CL, Ize B, Chanal A, Müller M, Giordano G, Wu LF. A novel Secindependent periplasmic protein translocation pathway in Escherichia coli. EMBO J 1998;17: Koussevitzky S, Ne eman E, Sommer A, Steffens JC, Harel E. Purification and properties of a novel chloroplast stromal peptidase. Processing of polyphenol oxidase and other imported precursors. J Biol Chem 1998;273: Bolhuis A, Bogsch EG, Robinson C. Subunit interactions in the twinarginine translocase complex of Escherichia coli. FEBS Lett 2000;472: Berghöfer J, Klösgen RB Two distinct translocation intermediates can be distinguished during protein transport by the TAT (delta ph) pathway across the thylakoid. FEBS Letters 1999;460: Lamppa GK. The chlorophyll a/b -binding protein inserts into the thylakoids independent of its cognate transit peptide. J Biol Chem 1988;263: Viitanen PV, Doran ER, Dunsmuir P. What is the role of the transit peptide in thylakoid integration of the light-harvesting chlorophyll a/b protein? J Biol Chem 1988;263: Li X, Henry R, Yuan J, Cline K, Hoffman NE. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the post-translational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA 1995;92: Tu CJ, Schuenemann D, Hoffman NE. Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J Biol Chem 1999;274: Kogata N, Nishio K, Hirohashi T, Kikuchi S, Nakai M. Involvement of a chloroplast homologue of the signal recognition particle receptor protein, FtsY, in protein targeting to thylakoids. FEBS Lett 1999;329: Dalbey RE, Robinson C. Protein translocation into and across the bacterial plasma membrane and the plant thylakoid membrane. Trends Biochem Sci 1999;24: De Gier J-WL, Valent QA, von Heijne G, Luirink J. The E. coli SRP: preferences of a targeting factor. FEBS Lett 1997;408: Schuenemann D, Gupta S, Persello-Cartieaux F, Klimyuk VI, Jones JDG, Nussaume L, Hoffman NE. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci USA 1998;95: DeLille J, Peterson EC, Johnson T, Moore M, Kight A, Henry R. A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc Natl Acad Sci USA 2000;97: Nilsson R, Brunner J, Hoffman NE, van Wijk KJ. Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpsrp54. EMBO J 1999;18: Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J 1997;16: Moore M, Harrison MS, Peterson EC, Henry R. Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J Biol Chem 2000;275: Samuelson JC, Chen M, Jiang F, Moeller I, Wiedmann M, Phillips G, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature 2000;406: Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJM, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J 2000;19:in press. 46. Michl D, Robinson C, Shackleton JB, Herrmann RG, Klösgen RB. Targeting of proteins to the thylakoids by bipartite presequences: CF 0 II is imported by a novel, third pathway. EMBO J 1994;13: Kim SJ, Robinson C, Mant A. Sec/SRP-independent insertion of two thylakoid membrane proteins bearing cleavable signal peptides. FEBS Lett 1998;424: Lorkovic ZJ, Schröder WP, Pakrasi HB, Irrgang K-D, Herrmann RG, Oelmüller R. Molecular characterisation of PSII-W, the only nuclearencoded component of the photosystem II reaction centre. Proc Natl Acad Sci USA 1995;92: Robinson D, Karnauchov I, Herrmann RG, Klösgen RB, Robinson C. Protease-sensitive thylakoidal import machinery for the Sec-, DpHand signal recognition particle-dependent protein targeting pathways, but not for CF o II integration. Plant J 1996;10: Thompson SJ, Robinson C, Mant A. Dual signal peptides mediate the Sec/SRP-independent insertion of a thylakoid membrane polyprotein, PsbY. J Biol Chem 1999; 274: Traffic 2001; 2:
Transport of proteins into and across the thylakoid membrane
 Journal of Experimental Botany, Vol. 51, GMP Special Issue, pp. 369 374, February 2000 Transport of proteins into and across the thylakoid membrane Colin Robinson1, Cheryl Woolhead and Wayne Edwards Department
Journal of Experimental Botany, Vol. 51, GMP Special Issue, pp. 369 374, February 2000 Transport of proteins into and across the thylakoid membrane Colin Robinson1, Cheryl Woolhead and Wayne Edwards Department
Mechanism of ph/tat-dependent protein transport at the thylakoid membrane
 Mechanism of ph/tat-dependent protein transport at the thylakoid membrane Dissertation zur Erlangung des akademischen Grades doctor rerum naturalium (Dr.rer.nat) vorgelegt der Mathematisch-Naturwissenschaftlich-Technischen
Mechanism of ph/tat-dependent protein transport at the thylakoid membrane Dissertation zur Erlangung des akademischen Grades doctor rerum naturalium (Dr.rer.nat) vorgelegt der Mathematisch-Naturwissenschaftlich-Technischen
Lecture 8. Protein Trafficking/Targeting. Protein targeting is necessary for proteins that are destined to work outside the cytoplasm.
 Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
PROTEIN TARGETING TO THE THYLAKOID MEMBRANE
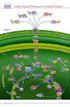 Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998. 49:97 126 Copyright c 1998 by Annual Reviews. All rights reserved PROTEIN TARGETING TO THE THYLAKOID MEMBRANE Danny J. Schnell Department of Biological
Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998. 49:97 126 Copyright c 1998 by Annual Reviews. All rights reserved PROTEIN TARGETING TO THE THYLAKOID MEMBRANE Danny J. Schnell Department of Biological
Eukaryotes have organelles
 Energy-transducing Eukaryotes have organelles membrane systems An organelle is a discrete membrane bound cellular structure specialized functions. An organelle is to the cell what an organ is to the body
Energy-transducing Eukaryotes have organelles membrane systems An organelle is a discrete membrane bound cellular structure specialized functions. An organelle is to the cell what an organ is to the body
BIOGENESIS OF GREEN PLANT THYLAKOID MEMBRANES
 1 BIOGENESIS OF GREEN PLANT THYLAKOID MEMBRANES Kenneth Cline Horticultural Sciences and Plant Molecular and Cellular Biology University of Florida Horticultural Sciences and Plant Molecular and Cellular
1 BIOGENESIS OF GREEN PLANT THYLAKOID MEMBRANES Kenneth Cline Horticultural Sciences and Plant Molecular and Cellular Biology University of Florida Horticultural Sciences and Plant Molecular and Cellular
Two Very Different Pathways of Thylakanthan One
 Post-translational protein translocation into thylakoids by the Sec and DpH-dependent pathways Hiroki Mori and Kenneth line Horticultural Sciences and Plant Molecular and ellular Biology, University of
Post-translational protein translocation into thylakoids by the Sec and DpH-dependent pathways Hiroki Mori and Kenneth line Horticultural Sciences and Plant Molecular and ellular Biology, University of
Biological energy cycle BIOMASS
 Photosynthesis Global carbon cycle Biological energy cycle BIOMASS Photosynthetic Reaction equation light CO 2 + 2H 2 O (CH 2 O) + O 2 + H 2 O Beware! This is not what s going on! The crucial equation
Photosynthesis Global carbon cycle Biological energy cycle BIOMASS Photosynthetic Reaction equation light CO 2 + 2H 2 O (CH 2 O) + O 2 + H 2 O Beware! This is not what s going on! The crucial equation
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
Electron Transport Generates a Proton Gradient Across the Membrane
 Electron Transport Generates a Proton Gradient Across the Membrane Each of respiratory enzyme complexes couples the energy released by electron transfer across it to an uptake of protons from water in
Electron Transport Generates a Proton Gradient Across the Membrane Each of respiratory enzyme complexes couples the energy released by electron transfer across it to an uptake of protons from water in
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
Chapter 2: Cell Structure and Function pg. 70-107
 UNIT 1: Biochemistry Chapter 2: Cell Structure and Function pg. 70-107 Organelles are internal structures that carry out specialized functions, interacting and complementing each other. Animal and plant
UNIT 1: Biochemistry Chapter 2: Cell Structure and Function pg. 70-107 Organelles are internal structures that carry out specialized functions, interacting and complementing each other. Animal and plant
APh/BE161: Physical Biology of the Cell Winter 2009 Recap on Photosynthesis Rob Phillips
 APh/BE161: Physical Biology of the Cell Winter 2009 Recap on Photosynthesis Rob Phillips Big picture: why are we doing this? A) photosynthesis will explain shortly, b) more generally, interaction of light
APh/BE161: Physical Biology of the Cell Winter 2009 Recap on Photosynthesis Rob Phillips Big picture: why are we doing this? A) photosynthesis will explain shortly, b) more generally, interaction of light
Photosystems I and II
 Photosystems I and II March 17, 2003 Bryant Miles Within the thylakoid membranes of the chloroplast, are two photosystems. Photosystem I optimally absorbs photons of a wavelength of 700 nm. Photosystem
Photosystems I and II March 17, 2003 Bryant Miles Within the thylakoid membranes of the chloroplast, are two photosystems. Photosystem I optimally absorbs photons of a wavelength of 700 nm. Photosystem
Chem 465 Biochemistry II
 Chem 465 Biochemistry II Name: 2 points Multiple choice (4 points apiece): 1. Formation of the ribosomal initiation complex for bacterial protein synthesis does not require: A) EF-Tu. B) formylmethionyl
Chem 465 Biochemistry II Name: 2 points Multiple choice (4 points apiece): 1. Formation of the ribosomal initiation complex for bacterial protein synthesis does not require: A) EF-Tu. B) formylmethionyl
AP Bio Photosynthesis & Respiration
 AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
AP Bio Photosynthesis & Respiration Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. What is the term used for the metabolic pathway in which
CELLS: PLANT CELLS 20 FEBRUARY 2013
 CELLS: PLANT CELLS 20 FEBRUARY 2013 Lesson Description In this lesson we will discuss the following: The Cell Theory Terminology Parts of Plant Cells: Organelles Difference between plant and animal cells
CELLS: PLANT CELLS 20 FEBRUARY 2013 Lesson Description In this lesson we will discuss the following: The Cell Theory Terminology Parts of Plant Cells: Organelles Difference between plant and animal cells
* Is chemical energy potential or kinetic energy? The position of what is storing energy?
 Biology 1406 Exam 2 - Metabolism Chs. 5, 6 and 7 energy - capacity to do work 5.10 kinetic energy - energy of motion : light, electrical, thermal, mechanical potential energy - energy of position or stored
Biology 1406 Exam 2 - Metabolism Chs. 5, 6 and 7 energy - capacity to do work 5.10 kinetic energy - energy of motion : light, electrical, thermal, mechanical potential energy - energy of position or stored
The Cell Teaching Notes and Answer Keys
 The Cell Teaching Notes and Answer Keys Subject area: Science / Biology Topic focus: The Cell: components, types of cells, organelles, levels of organization Learning Aims: describe similarities and differences
The Cell Teaching Notes and Answer Keys Subject area: Science / Biology Topic focus: The Cell: components, types of cells, organelles, levels of organization Learning Aims: describe similarities and differences
Cell Structure and Function. Eukaryotic Cell: Neuron
 Cell Structure and Function Eukaryotic Cell: Neuron Cell Structure and Function Eukaryotic Cells: Blood Cells Cell Structure and Function Prokaryotic Cells: Bacteria Cell Structure and Function All living
Cell Structure and Function Eukaryotic Cell: Neuron Cell Structure and Function Eukaryotic Cells: Blood Cells Cell Structure and Function Prokaryotic Cells: Bacteria Cell Structure and Function All living
SOME Important Points About Cellular Energetics by Dr. Ty C.M. Hoffman
 SOME Important Points About Cellular Energetics by Dr. Ty C.M. Hoffman An Introduction to Metabolism Most biochemical processes occur as biochemical pathways, each individual reaction of which is catalyzed
SOME Important Points About Cellular Energetics by Dr. Ty C.M. Hoffman An Introduction to Metabolism Most biochemical processes occur as biochemical pathways, each individual reaction of which is catalyzed
Lecture Series 7. From DNA to Protein. Genotype to Phenotype. Reading Assignments. A. Genes and the Synthesis of Polypeptides
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Plasma Membrane hydrophilic polar heads
 The Parts of the Cell 3 main parts in ALL cells: plasma membrane, cytoplasm, genetic material this is about the parts of a generic eukaryotic cell Plasma Membrane -is a fluid mosaic model membrane is fluid
The Parts of the Cell 3 main parts in ALL cells: plasma membrane, cytoplasm, genetic material this is about the parts of a generic eukaryotic cell Plasma Membrane -is a fluid mosaic model membrane is fluid
A. Incorrect! No, while this statement is correct, it is not the best answer to the question.
 Biochemistry - Problem Drill 18: Photosynthesis No. 1 of 10 1. What is photosynthesis? Select the best answer. (A) Photosynthesis happens in the chloroplasts. (B) Light absorption by chlorophyll induces
Biochemistry - Problem Drill 18: Photosynthesis No. 1 of 10 1. What is photosynthesis? Select the best answer. (A) Photosynthesis happens in the chloroplasts. (B) Light absorption by chlorophyll induces
Chapter 9 Mitochondrial Structure and Function
 Chapter 9 Mitochondrial Structure and Function 1 2 3 Structure and function Oxidative phosphorylation and ATP Synthesis Peroxisome Overview 2 Mitochondria have characteristic morphologies despite variable
Chapter 9 Mitochondrial Structure and Function 1 2 3 Structure and function Oxidative phosphorylation and ATP Synthesis Peroxisome Overview 2 Mitochondria have characteristic morphologies despite variable
Chapter 5 Organelles. Lesson Objectives List the organelles of the cell and their functions. Distinguish between plant and animal cells.
 Chapter 5 Organelles Lesson Objectives List the organelles of the cell and their functions. Distinguish between plant and animal cells. Check Your Understanding What is a cell? How do we visualize cells?
Chapter 5 Organelles Lesson Objectives List the organelles of the cell and their functions. Distinguish between plant and animal cells. Check Your Understanding What is a cell? How do we visualize cells?
PROTEIN IMPORT INTO CHLOROPLASTS
 ROTEIN IMORT INTO CHLOROLASTS Jürgen Soll and Enrico Schleiff Chloroplasts are organelles of endosymbiotic origin, and they transferred most of their genetic information to the host nucleus during this
ROTEIN IMORT INTO CHLOROLASTS Jürgen Soll and Enrico Schleiff Chloroplasts are organelles of endosymbiotic origin, and they transferred most of their genetic information to the host nucleus during this
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Name Date Period. 2. When a molecule of double-stranded DNA undergoes replication, it results in
 DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
DNA, RNA, Protein Synthesis Keystone 1. During the process shown above, the two strands of one DNA molecule are unwound. Then, DNA polymerases add complementary nucleotides to each strand which results
CHAPTER 40 The Mechanism of Protein Synthesis
 CHAPTER 40 The Mechanism of Protein Synthesis Problems: 2,3,6,7,9,13,14,15,18,19,20 Initiation: Locating the start codon. Elongation: Reading the codons (5 3 ) and synthesizing protein amino carboxyl.
CHAPTER 40 The Mechanism of Protein Synthesis Problems: 2,3,6,7,9,13,14,15,18,19,20 Initiation: Locating the start codon. Elongation: Reading the codons (5 3 ) and synthesizing protein amino carboxyl.
BCH401G Lecture 39 Andres
 BCH401G Lecture 39 Andres Lecture Summary: Ribosome: Understand its role in translation and differences between translation in prokaryotes and eukaryotes. Translation: Understand the chemistry of this
BCH401G Lecture 39 Andres Lecture Summary: Ribosome: Understand its role in translation and differences between translation in prokaryotes and eukaryotes. Translation: Understand the chemistry of this
Photosynthesis January 23 Feb 1, 2013 WARM-UP JAN 23/24. Mr. Stephens, IB Biology III 1
 WARM-UP JAN 23/24 Mr. Stephens, IB Biology III 1 Photosynthesis & Cellular Respiration What is the connection between Photosynthesis and Cellular Respiration? Energy Production Inorganic Molecules Specialized
WARM-UP JAN 23/24 Mr. Stephens, IB Biology III 1 Photosynthesis & Cellular Respiration What is the connection between Photosynthesis and Cellular Respiration? Energy Production Inorganic Molecules Specialized
Cytology. Living organisms are made up of cells. Either PROKARYOTIC or EUKARYOTIC cells.
 CYTOLOGY Cytology Living organisms are made up of cells. Either PROKARYOTIC or EUKARYOTIC cells. A. two major cell types B. distinguished by structural organization See table on handout for differences.
CYTOLOGY Cytology Living organisms are made up of cells. Either PROKARYOTIC or EUKARYOTIC cells. A. two major cell types B. distinguished by structural organization See table on handout for differences.
Cell Structure & Function!
 Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
Cell Structure & Function! Chapter 3! The most exciting phrase to hear in science, the one that heralds new discoveries, is not 'Eureka!' but 'That's funny.! -- Isaac Asimov Animal Cell Plant Cell Cell
Effects of altered TatC proteins on protein secretion efficiency via the twin-arginine translocation pathway of Bacillus subtilis
 Microbiology (2009), 155, 1776 1785 DOI 10.1099/mic.0.027987-0 Effects of altered TatC proteins on protein secretion efficiency via the twin-arginine translocation pathway of Bacillus subtilis Robyn T.
Microbiology (2009), 155, 1776 1785 DOI 10.1099/mic.0.027987-0 Effects of altered TatC proteins on protein secretion efficiency via the twin-arginine translocation pathway of Bacillus subtilis Robyn T.
Visualizing Cell Processes
 Visualizing Cell Processes A Series of Five Programs produced by BioMEDIA ASSOCIATES Content Guide for Program 3 Photosynthesis and Cellular Respiration Copyright 2001, BioMEDIA ASSOCIATES www.ebiomedia.com
Visualizing Cell Processes A Series of Five Programs produced by BioMEDIA ASSOCIATES Content Guide for Program 3 Photosynthesis and Cellular Respiration Copyright 2001, BioMEDIA ASSOCIATES www.ebiomedia.com
Lecture 4. Polypeptide Synthesis Overview
 Initiation of Protein Synthesis (4.1) Lecture 4 Polypeptide Synthesis Overview Polypeptide synthesis proceeds sequentially from N Terminus to C terminus. Amino acids are not pre-positioned on a template.
Initiation of Protein Synthesis (4.1) Lecture 4 Polypeptide Synthesis Overview Polypeptide synthesis proceeds sequentially from N Terminus to C terminus. Amino acids are not pre-positioned on a template.
Photosynthesis. Photosynthesis: Converting light energy into chemical energy. Photoautotrophs capture sunlight and convert it to chemical energy
 Photosynthesis: Converting light energy into chemical energy Photosynthesis 6 + 12H 2 O + light energy Summary Formula: C 6 H 12 O 6 + 6O 2 + 6H 2 O 6 + 6H 2 O C 6 H 12 O 6 + 6O 2 Photosythesis provides
Photosynthesis: Converting light energy into chemical energy Photosynthesis 6 + 12H 2 O + light energy Summary Formula: C 6 H 12 O 6 + 6O 2 + 6H 2 O 6 + 6H 2 O C 6 H 12 O 6 + 6O 2 Photosythesis provides
The Cell: Organelle Diagrams
 The Cell: Organelle Diagrams Fig 7-4. A prokaryotic cell. Lacking a true nucleus and the other membrane-enclosed organelles of the eukaryotic cell, the prokaryotic cell is much simpler in structure. Only
The Cell: Organelle Diagrams Fig 7-4. A prokaryotic cell. Lacking a true nucleus and the other membrane-enclosed organelles of the eukaryotic cell, the prokaryotic cell is much simpler in structure. Only
Organelles and Their Functions
 Organelles and Their Functions The study of cell organelles and their functions is a fascinating part of biology. The current article provides a brief description of the structure of organelles and their
Organelles and Their Functions The study of cell organelles and their functions is a fascinating part of biology. The current article provides a brief description of the structure of organelles and their
PLANT PHYSIOLOGY. Az Agrármérnöki MSc szak tananyagfejlesztése TÁMOP-4.1.2-08/1/A-2009-0010
 PLANT PHYSIOLOGY Az Agrármérnöki MSc szak tananyagfejlesztése TÁMOP-4.1.2-08/1/A-2009-0010 The light reactions of the photosynthesis Photosynthesis inhibiting herbicides Overview 1. Photosynthesis, general
PLANT PHYSIOLOGY Az Agrármérnöki MSc szak tananyagfejlesztése TÁMOP-4.1.2-08/1/A-2009-0010 The light reactions of the photosynthesis Photosynthesis inhibiting herbicides Overview 1. Photosynthesis, general
Photosynthesis takes place in three stages:
 Photosynthesis takes place in three stages: Light-dependent reactions Light-independent reactions The Calvin cycle 1. Capturing energy from sunlight 2. Using energy to make ATP and NADPH 3. Using ATP and
Photosynthesis takes place in three stages: Light-dependent reactions Light-independent reactions The Calvin cycle 1. Capturing energy from sunlight 2. Using energy to make ATP and NADPH 3. Using ATP and
Chapter 19a Oxidative Phosphorylation and Photophosphorylation. Multiple Choice Questions
 Chapter 19a Oxidative Phosphorylation and Photophosphorylation Multiple Choice Questions 1. Electron-transfer reactions in mitochondria Page: 707 Difficulty: 1 Ans: E Almost all of the oxygen (O 2 ) one
Chapter 19a Oxidative Phosphorylation and Photophosphorylation Multiple Choice Questions 1. Electron-transfer reactions in mitochondria Page: 707 Difficulty: 1 Ans: E Almost all of the oxygen (O 2 ) one
Chloroplasts and Mitochondria
 Name: KEY Period: Chloroplasts and Mitochondria Plant cells and some Algae contain an organelle called the chloroplast. The chloroplast allows plants to harvest energy from sunlight to carry on a process
Name: KEY Period: Chloroplasts and Mitochondria Plant cells and some Algae contain an organelle called the chloroplast. The chloroplast allows plants to harvest energy from sunlight to carry on a process
Biological cell membranes
 Unit 14: Cell biology. 14 2 Biological cell membranes The cell surface membrane surrounds the cell and acts as a barrier between the cell s contents and the environment. The cell membrane has multiple
Unit 14: Cell biology. 14 2 Biological cell membranes The cell surface membrane surrounds the cell and acts as a barrier between the cell s contents and the environment. The cell membrane has multiple
Microscopes. Eukaryotes Eukaryotic cells are characterized by having: DNA in a nucleus that is bounded by a membranous nuclear envelope
 CH 6 The Cell Microscopy Scientists use microscopes to visualize cells too small to see with the naked eye. In a light microscope (LM), visible light is passed through a specimen and then through glass
CH 6 The Cell Microscopy Scientists use microscopes to visualize cells too small to see with the naked eye. In a light microscope (LM), visible light is passed through a specimen and then through glass
Evolution of Metabolism. Introduction. Introduction. Introduction. How Cells Harvest Energy. Chapter 7 & 8
 How ells Harvest Energy hapter 7 & 8 Evolution of Metabolism A hypothetical timeline for the evolution of metabolism - all in prokaryotic cells!: 1. ability to store chemical energy in ATP 2. evolution
How ells Harvest Energy hapter 7 & 8 Evolution of Metabolism A hypothetical timeline for the evolution of metabolism - all in prokaryotic cells!: 1. ability to store chemical energy in ATP 2. evolution
pathway that involves taking in heat from the environment at each step. C.
 Study Island Cell Energy Keystone Review 1. Cells obtain energy by either capturing light energy through photosynthesis or by breaking down carbohydrates through cellular respiration. In both photosynthesis
Study Island Cell Energy Keystone Review 1. Cells obtain energy by either capturing light energy through photosynthesis or by breaking down carbohydrates through cellular respiration. In both photosynthesis
ecture 16 Oct 7, 2005
 Lecture utline ecture 16 ct 7, 005 hotosynthesis 1 I. Reactions 1. Importance of Photosynthesis to all life on earth - primary producer, generates oxygen, ancient. What needs to be accomplished in photosynthesis
Lecture utline ecture 16 ct 7, 005 hotosynthesis 1 I. Reactions 1. Importance of Photosynthesis to all life on earth - primary producer, generates oxygen, ancient. What needs to be accomplished in photosynthesis
Introduction to the Cell: Plant and Animal Cells
 Introduction to the Cell: Plant and Animal Cells Tissues, Organs, and Systems of Living Things Cells, Cell Division, and Animal Systems and Plant Systems Cell Specialization Human Systems All organisms
Introduction to the Cell: Plant and Animal Cells Tissues, Organs, and Systems of Living Things Cells, Cell Division, and Animal Systems and Plant Systems Cell Specialization Human Systems All organisms
CELL/ PHOTOSYNTHESIS/ CELLULAR RESPIRATION Test 2011 ANSWER 250 POINTS ANY WAY IN WHICH YOU WANT
 CELL/ PHOTOSYNTHESIS/ CELLULAR RESPIRATION Test 2011 ANSWER 250 POINTS ANY WAY IN WHICH YOU WANT Completion: complete each statement. (1 point each) 1. All cells arise from. 2. The basic unit of structure
CELL/ PHOTOSYNTHESIS/ CELLULAR RESPIRATION Test 2011 ANSWER 250 POINTS ANY WAY IN WHICH YOU WANT Completion: complete each statement. (1 point each) 1. All cells arise from. 2. The basic unit of structure
1. f. Students know usable energy is captured from sunlight by chloroplasts and is stored through the synthesis of sugar from carbon dioxide.
 1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
1. The fundamental life processes of plants and animals depend on a variety of chemical reactions that occur in specialized areas of the organism s cells. As a basis for understanding this concept: 1.
Given these characteristics of life, which of the following objects is considered a living organism? W. X. Y. Z.
 Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
Cell Structure and Organization 1. All living things must possess certain characteristics. They are all composed of one or more cells. They can grow, reproduce, and pass their genes on to their offspring.
ISTEP+: Biology I End-of-Course Assessment Released Items and Scoring Notes
 ISTEP+: Biology I End-of-Course Assessment Released Items and Scoring Notes Page 1 of 22 Introduction Indiana students enrolled in Biology I participated in the ISTEP+: Biology I Graduation Examination
ISTEP+: Biology I End-of-Course Assessment Released Items and Scoring Notes Page 1 of 22 Introduction Indiana students enrolled in Biology I participated in the ISTEP+: Biology I Graduation Examination
Lecture 7 Outline (Ch. 10)
 Lecture 7 Outline (Ch. 10) I. Photosynthesis overview A. Purpose B. Location II. The light vs. the dark reaction III. Chloroplasts pigments A. Light absorption B. Types IV. Light reactions A. Photosystems
Lecture 7 Outline (Ch. 10) I. Photosynthesis overview A. Purpose B. Location II. The light vs. the dark reaction III. Chloroplasts pigments A. Light absorption B. Types IV. Light reactions A. Photosystems
Cells & Cell Organelles
 Cells & Cell Organelles The Building Blocks of Life H Biology Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
Cells & Cell Organelles The Building Blocks of Life H Biology Types of cells bacteria cells Prokaryote - no organelles Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell
2. PHOTOSYNTHESIS. The general equation describing photosynthesis is light + 6 H 2 O + 6 CO 2 C 6 H 12 O 6 + 6 O 2
 2. PHOTOSYNTHESIS Photosynthesis is the process by which light energy is converted to chemical energy whereby carbon dioxide and water are converted into organic molecules. The process occurs in most algae,
2. PHOTOSYNTHESIS Photosynthesis is the process by which light energy is converted to chemical energy whereby carbon dioxide and water are converted into organic molecules. The process occurs in most algae,
Transcription in prokaryotes. Elongation and termination
 Transcription in prokaryotes Elongation and termination After initiation the σ factor leaves the scene. Core polymerase is conducting the elongation of the chain. The core polymerase contains main nucleotide
Transcription in prokaryotes Elongation and termination After initiation the σ factor leaves the scene. Core polymerase is conducting the elongation of the chain. The core polymerase contains main nucleotide
Comparing Plant And Animal Cells
 Comparing Plant And Animal Cells http://khanacademy.org/video?v=hmwvj9x4gny Plant Cells shape - most plant cells are squarish or rectangular in shape. amyloplast (starch storage organelle)- an organelle
Comparing Plant And Animal Cells http://khanacademy.org/video?v=hmwvj9x4gny Plant Cells shape - most plant cells are squarish or rectangular in shape. amyloplast (starch storage organelle)- an organelle
Copyright 2000-2003 Mark Brandt, Ph.D. 54
 Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
Pyruvate Oxidation Overview of pyruvate metabolism Pyruvate can be produced in a variety of ways. It is an end product of glycolysis, and can be derived from lactate taken up from the environment (or,
PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY
 Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Name PRESTWICK ACADEMY NATIONAL 5 BIOLOGY CELL BIOLOGY SUMMARY Cell Structure Identify animal, plant, fungal and bacterial cell ultrastructure and know the structures functions. Plant cell Animal cell
Sickle cell anemia: Altered beta chain Single AA change (#6 Glu to Val) Consequence: Protein polymerizes Change in RBC shape ---> phenotypes
 Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Protein Structure Polypeptide: Protein: Therefore: Example: Single chain of amino acids 1 or more polypeptide chains All polypeptides are proteins Some proteins contain >1 polypeptide Hemoglobin (O 2 binding
Bacterial (Prokaryotic) Cell. Common features of all cells. Tour of the Cell. Eukaryotic Cell. Plasma Membrane defines inside from outside
 www.denniskunkel.com Tour of the Cell www.denniskunkel.com Today s Topics Properties of all cells Prokaryotes and Eukaryotes Functions of Major Cellular Organelles Information, Synthesis&Transport,, Vesicles
www.denniskunkel.com Tour of the Cell www.denniskunkel.com Today s Topics Properties of all cells Prokaryotes and Eukaryotes Functions of Major Cellular Organelles Information, Synthesis&Transport,, Vesicles
Compartmentalization of the Cell. Objectives. Recommended Reading. Professor Alfred Cuschieri. Department of Anatomy University of Malta
 Compartmentalization of the Cell Professor Alfred Cuschieri Department of Anatomy University of Malta Objectives By the end of this session the student should be able to: 1. Identify the different organelles
Compartmentalization of the Cell Professor Alfred Cuschieri Department of Anatomy University of Malta Objectives By the end of this session the student should be able to: 1. Identify the different organelles
AP BIOLOGY 2006 SCORING GUIDELINES. Question 1
 AP BIOLOGY 2006 SCORING GUIDELINES Question 1 A major distinction between prokaryotes and eukaryotes is the presence of membrane-bound organelles in eukaryotes. (a) Describe the structure and function
AP BIOLOGY 2006 SCORING GUIDELINES Question 1 A major distinction between prokaryotes and eukaryotes is the presence of membrane-bound organelles in eukaryotes. (a) Describe the structure and function
CONCEPTS OF BIOLOGY - BIOL115 Dr. SG Saupe; Fall 2006 Exam #2
 CONCEPTS OF BIOLOGY - BIOL115 Dr. SG Saupe; Fall 2006 Exam #2 name And he gave it for his opinion, that whoever could make two ears of corn or two blades of grass to grow upon a spot where only one grew
CONCEPTS OF BIOLOGY - BIOL115 Dr. SG Saupe; Fall 2006 Exam #2 name And he gave it for his opinion, that whoever could make two ears of corn or two blades of grass to grow upon a spot where only one grew
Photosynthesis (Life from Light)
 Photosynthesis Photosynthesis (Life from Light) Energy needs of life All life needs a constant input of energy o Heterotrophs (consumers) Animals, fungi, most bacteria Get their energy from other organisms
Photosynthesis Photosynthesis (Life from Light) Energy needs of life All life needs a constant input of energy o Heterotrophs (consumers) Animals, fungi, most bacteria Get their energy from other organisms
Photosynthesis. Name. Light reactions Calvin cycle Oxidation Reduction Electronegativity Photosystem Electron carrier NADP+ Concentration gradient
 Vocabulary Terms Photoautotroph Chemoautotroph Electromagnetic spectrum Wavelength Chloroplast Thylakoid Stroma Chlorophyll Absorption spectrum Photosynthesis Light reactions Calvin cycle Oxidation Reduction
Vocabulary Terms Photoautotroph Chemoautotroph Electromagnetic spectrum Wavelength Chloroplast Thylakoid Stroma Chlorophyll Absorption spectrum Photosynthesis Light reactions Calvin cycle Oxidation Reduction
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Chapter 18 Regulation of Gene Expression 18.1. Gene Regulation Is Necessary By switching genes off when they are not needed, cells can prevent resources from being wasted. There should be natural selection
Summary of Metabolism. Mechanism of Enzyme Action
 Summary of Metabolism Mechanism of Enzyme Action 1. The substrate contacts the active site 2. The enzyme-substrate complex is formed. 3. The substrate molecule is altered (atoms are rearranged, or the
Summary of Metabolism Mechanism of Enzyme Action 1. The substrate contacts the active site 2. The enzyme-substrate complex is formed. 3. The substrate molecule is altered (atoms are rearranged, or the
Photosynthesis and Cellular Respiration. Stored Energy
 Photosynthesis and Cellular Respiration Stored Energy What is Photosynthesis? plants convert the energy of sunlight into the energy in the chemical bonds of carbohydrates sugars and starches. SUMMARY EQUATION:
Photosynthesis and Cellular Respiration Stored Energy What is Photosynthesis? plants convert the energy of sunlight into the energy in the chemical bonds of carbohydrates sugars and starches. SUMMARY EQUATION:
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT
 BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
Like The Guy From Krypton Photosynthesis: Energy from Sunlight What Is Photosynthesis?
 Like The Guy From Krypton Photosynthesis: Energy from Sunlight What Is Photosynthesis? Photosynthesis: synthesis from light The broad outline: Plants take in CO 2 and release water and O 2 Light is required
Like The Guy From Krypton Photosynthesis: Energy from Sunlight What Is Photosynthesis? Photosynthesis: synthesis from light The broad outline: Plants take in CO 2 and release water and O 2 Light is required
CHAPTER 9 IMMUNOGLOBULIN BIOSYNTHESIS
 CHAPTER 9 IMMUNOGLOBULIN BIOSYNTHESIS Although the process by which a functional gene for immunoglobulin HEAVY and LIGHT CHAINS is formed is highly unusual, the SYNTHESIS, POST- TRANSLATIONAL PROCESSING
CHAPTER 9 IMMUNOGLOBULIN BIOSYNTHESIS Although the process by which a functional gene for immunoglobulin HEAVY and LIGHT CHAINS is formed is highly unusual, the SYNTHESIS, POST- TRANSLATIONAL PROCESSING
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION. Professor Bharat Patel Office: Science 2, 2.36 Email: b.patel@griffith.edu.
 Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: b.patel@griffith.edu.au What is Gene Expression & Gene Regulation? 1. Gene Expression
Lecture 1 MODULE 3 GENE EXPRESSION AND REGULATION OF GENE EXPRESSION Professor Bharat Patel Office: Science 2, 2.36 Email: b.patel@griffith.edu.au What is Gene Expression & Gene Regulation? 1. Gene Expression
Rubisco; easy Purification and Immunochemical Determination
 Rubisco; easy Purification and Immunochemical Determination Ulrich Groß Justus-Liebig-Universität Gießen, Institute of Plant Nutrition, Department of Tissue Culture, Südanlage 6, D-35390 Giessen e-mail:
Rubisco; easy Purification and Immunochemical Determination Ulrich Groß Justus-Liebig-Universität Gießen, Institute of Plant Nutrition, Department of Tissue Culture, Südanlage 6, D-35390 Giessen e-mail:
Which regions of the electromagnetic spectrum do plants use to drive photosynthesis?
 Which regions of the electromagnetic spectrum do plants use to drive photosynthesis? Green Light: The Forgotten Region of the Spectrum. In the past, plant physiologists used green light as a safe light
Which regions of the electromagnetic spectrum do plants use to drive photosynthesis? Green Light: The Forgotten Region of the Spectrum. In the past, plant physiologists used green light as a safe light
7.2 Cells: A Look Inside
 CHAPTER 7 CELL STRUCTURE AND FUNCTION 7.2 Cells: A Look Inside Imagine a factory that makes thousands of cookies a day. Ingredients come into the factory, get mixed and baked, then the cookies are packaged.
CHAPTER 7 CELL STRUCTURE AND FUNCTION 7.2 Cells: A Look Inside Imagine a factory that makes thousands of cookies a day. Ingredients come into the factory, get mixed and baked, then the cookies are packaged.
Bio 101 Section 001: Practice Questions for First Exam
 Do the Practice Exam under exam conditions. Time yourself! MULTIPLE CHOICE: 1. The substrate fits in the of an enzyme: (A) allosteric site (B) active site (C) reaction groove (D) Golgi body (E) inhibitor
Do the Practice Exam under exam conditions. Time yourself! MULTIPLE CHOICE: 1. The substrate fits in the of an enzyme: (A) allosteric site (B) active site (C) reaction groove (D) Golgi body (E) inhibitor
Lecture 4 Cell Membranes & Organelles
 Lecture 4 Cell Membranes & Organelles Structure of Animal Cells The Phospholipid Structure Phospholipid structure Encases all living cells Its basic structure is represented by the fluidmosaic model Phospholipid
Lecture 4 Cell Membranes & Organelles Structure of Animal Cells The Phospholipid Structure Phospholipid structure Encases all living cells Its basic structure is represented by the fluidmosaic model Phospholipid
mrna EDITING Watson et al., BIOLOGIA MOLECOLARE DEL GENE, Zanichelli editore S.p.A. Copyright 2005
 mrna EDITING mrna EDITING http://dbb.urmc.rochester.edu/labs/smith/research_2.htm The number of A to I sites in the human transcriptome >15;000 the vast majority of these sites occurring in Alu repeats
mrna EDITING mrna EDITING http://dbb.urmc.rochester.edu/labs/smith/research_2.htm The number of A to I sites in the human transcriptome >15;000 the vast majority of these sites occurring in Alu repeats
Review Questions Photosynthesis
 Review Questions Photosynthesis 1. Describe a metabolic pathway. In a factory, labor is divided into small individual jobs. A carmaker, for example, will have one worker install the front windshield, another
Review Questions Photosynthesis 1. Describe a metabolic pathway. In a factory, labor is divided into small individual jobs. A carmaker, for example, will have one worker install the front windshield, another
Methods for Protein Analysis
 Methods for Protein Analysis 1. Protein Separation Methods The following is a quick review of some common methods used for protein separation: SDS-PAGE (SDS-polyacrylamide gel electrophoresis) separates
Methods for Protein Analysis 1. Protein Separation Methods The following is a quick review of some common methods used for protein separation: SDS-PAGE (SDS-polyacrylamide gel electrophoresis) separates
BCOR 011 Exam 2, 2004
 BCOR 011 Exam 2, 2004 Name: Section: MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1. According to the first law of thermodynamics, A. the universe
BCOR 011 Exam 2, 2004 Name: Section: MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1. According to the first law of thermodynamics, A. the universe
Chapter-21b: Hormones and Receptors
 1 hapter-21b: Hormones and Receptors Hormone classes Hormones are classified according to the distance over which they act. 1. Autocrine hormones --- act on the same cell that released them. Interleukin-2
1 hapter-21b: Hormones and Receptors Hormone classes Hormones are classified according to the distance over which they act. 1. Autocrine hormones --- act on the same cell that released them. Interleukin-2
Transmembrane proteins span the bilayer. α-helix transmembrane domain. Multiple transmembrane helices in one polypeptide
 Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Recent advances in the understanding of membrane protein assembly and structure
 Quarterly Reviews of Biophysics 32, 4 (1999), pp. 285 307 1999 Cambridge University Press Printed in the United Kingdom 285 Recent advances in the understanding of membrane protein assembly and structure
Quarterly Reviews of Biophysics 32, 4 (1999), pp. 285 307 1999 Cambridge University Press Printed in the United Kingdom 285 Recent advances in the understanding of membrane protein assembly and structure
Synthesis, assembly and degradation of thylakoid membrane proteins
 Biochimie 82 (2000) 615 634 2000 Société française de biochimie et biologie moléculaire / Éditions scientifiques et médicales Elsevier SAS. All rights reserved. S030090840000609X/FLA Synthesis, assembly
Biochimie 82 (2000) 615 634 2000 Société française de biochimie et biologie moléculaire / Éditions scientifiques et médicales Elsevier SAS. All rights reserved. S030090840000609X/FLA Synthesis, assembly
Biology. Slide 1of 51. End Show. Copyright Pearson Prentice Hall
 Biology 1of 51 8-3 The Reactions of Photosynthesis 2of 51 Inside a Chloroplast Inside a Chloroplast In plants, photosynthesis takes place inside chloroplasts. Plant Chloroplast Plant cells 3of 51 Inside
Biology 1of 51 8-3 The Reactions of Photosynthesis 2of 51 Inside a Chloroplast Inside a Chloroplast In plants, photosynthesis takes place inside chloroplasts. Plant Chloroplast Plant cells 3of 51 Inside
--not necessarily a protein! (all proteins are polypeptides, but the converse is not true)
 00Note Set 5b 1 PEPTIDE BONDS AND POLYPEPTIDES OLIGOPEPTIDE: --chain containing only a few amino acids (see tetrapaptide, Fig 5.9) POLYPEPTIDE CHAINS: --many amino acids joined together --not necessarily
00Note Set 5b 1 PEPTIDE BONDS AND POLYPEPTIDES OLIGOPEPTIDE: --chain containing only a few amino acids (see tetrapaptide, Fig 5.9) POLYPEPTIDE CHAINS: --many amino acids joined together --not necessarily
Keystone Review Practice Test Module A Cells and Cell Processes. 1. Which characteristic is shared by all prokaryotes and eukaryotes?
 Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Keystone Review Practice Test Module A Cells and Cell Processes 1. Which characteristic is shared by all prokaryotes and eukaryotes? a. Ability to store hereditary information b. Use of organelles to control
Chapter 5: Organization and Expression of Immunoglobulin Genes
 Chapter 5: Organization and Expression of Immunoglobulin Genes I. Genetic Model Compatible with Ig Structure A. Two models for Ab structure diversity 1. Germ-line theory: maintained that the genome contributed
Chapter 5: Organization and Expression of Immunoglobulin Genes I. Genetic Model Compatible with Ig Structure A. Two models for Ab structure diversity 1. Germ-line theory: maintained that the genome contributed
Hormones: Classification. Hormones: Classification. Peptide Hormone Synthesis, Packaging, and Release
 Hormones: Classification Hormones: Classification Be able to give types and example. Compare synthesis, half-life and location of receptor 1. Peptide or protein hormones Insulin from amino acids 2. Steroid
Hormones: Classification Hormones: Classification Be able to give types and example. Compare synthesis, half-life and location of receptor 1. Peptide or protein hormones Insulin from amino acids 2. Steroid
7.2 Cell Structure. Lesson Objectives. Lesson Summary. Cell Organization Eukaryotic cells contain a nucleus and many specialized structures.
 7.2 Cell Structure Lesson Objectives Describe the structure and function of the cell nucleus. Describe the role of vacuoles, lysosomes, and the cytoskeleton. Identify the role of ribosomes, endoplasmic
7.2 Cell Structure Lesson Objectives Describe the structure and function of the cell nucleus. Describe the role of vacuoles, lysosomes, and the cytoskeleton. Identify the role of ribosomes, endoplasmic
1 Mutation and Genetic Change
 CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
CHAPTER 14 1 Mutation and Genetic Change SECTION Genes in Action KEY IDEAS As you read this section, keep these questions in mind: What is the origin of genetic differences among organisms? What kinds
Unit I: Introduction To Scientific Processes
 Unit I: Introduction To Scientific Processes This unit is an introduction to the scientific process. This unit consists of a laboratory exercise where students go through the QPOE2 process step by step
Unit I: Introduction To Scientific Processes This unit is an introduction to the scientific process. This unit consists of a laboratory exercise where students go through the QPOE2 process step by step
Biology 101 Chapter 4 Cells as the Basic Unit of Life. The Cell Theory Major Contributors: Galileo = first observations made with a microscope
 Biology 101 Chapter 4 Cells as the Basic Unit of Life The Cell Theory Major Contributors: Galileo = first observations made with a microscope Robert Hooke = first to observe small compartments in dead
Biology 101 Chapter 4 Cells as the Basic Unit of Life The Cell Theory Major Contributors: Galileo = first observations made with a microscope Robert Hooke = first to observe small compartments in dead
Q: How are proteins (amino acid chains) made from the information in mrna? A: Translation Ribosomes translate mrna into protein
 ranslation (written lesson) Q: How are proteins (amino acid chains) made from the information in mrn? : ranslation Ribosomes translate mrn into protein ranslation has 3 steps also! 1. ranslation Initiation:
ranslation (written lesson) Q: How are proteins (amino acid chains) made from the information in mrn? : ranslation Ribosomes translate mrn into protein ranslation has 3 steps also! 1. ranslation Initiation:
Translation. Translation: Assembly of polypeptides on a ribosome
 Translation Translation: Assembly of polypeptides on a ribosome Living cells devote more energy to the synthesis of proteins than to any other aspect of metabolism. About a third of the dry mass of a cell
Translation Translation: Assembly of polypeptides on a ribosome Living cells devote more energy to the synthesis of proteins than to any other aspect of metabolism. About a third of the dry mass of a cell
