Molecular Dynamics Simulation
|
|
|
- Matilda Barker
- 8 years ago
- Views:
Transcription
1 Molecular Dynamics Simulation A Short Introduction Michel Cuendet Molecular Dynamics Simulation - Michel Cuendet - EMBL
2 Plan Introduction The classical force field Setting up a simulation Connection to statistical mechanics Usage of MD simulation Molecular Dynamics Simulation - Michel Cuendet - EMBL
3 Why we do simulation In some cases, experiment is : 1. impossible Inside of stars Weather forecast 2. too dangerous Flight simulation Explosion simulation 3. expensive High pressure simulation Windchannel simulation 4. blind Some properties cannot be observed on very short time-scales and very small space-scales Simulation is a useful complement, because it can : replace experiment provoke experiment explain experiment aid in establishing intellectual property Molecular Dynamics Simulation - Michel Cuendet - EMBL
4 Molecular modeling What is Molecular Modeling? Molecular Modeling is concerned with the description of the atomic and molecular interactions that govern microscopic and macroscopic behaviors of physical systems What is it good for? The essence of molecular modeling resides in the connection between the macroscopic world and the microscopic world provided by the theory of statistical mechanics Macroscopic observable (Solvation energy, affinity between two proteins, H-H distance, conformation,... ) Average of observable over selected microscopic states Molecular Dynamics Simulation - Michel Cuendet - EMBL
5 Computational tools Quantum Mechanics (QM) Electronic structure (Schrödinger) ACCURATE EXPENSIVE small system atoms ps Classical Molecular Mechanics (MM) Empirical forces (Newton) LESS ACCURATE FAST atoms ns Mixed Quantum/Classical (QM/MM) MM QM atoms ps Molecular Dynamics Simulation - Michel Cuendet - EMBL
6 Types of phenomena Goal : simulate/predict processes such as 1. polypeptide folding 2. biomolecular association 3. partitioning between solvents 4. membrane/micelle formation 5. chemical reactions, enzyme catalysis 6. enzyme catalysis 7. photochemical ractions, electron transfer thermodynamic equilibria governed by weak (non-bonded) forces chemical transformations governed by strong forces characteristics (1-4): - degrees of freedom: atomic (solute + solvent) - equations of motion: classical dynamics - governing theory: statistical mechanics characteristics (5-7): - degrees of freedom: electronic, nuclear - equations of motion: quantum dynamics - governing theory: quantum statistical mechanics classical MD quantum MD Molecular Dynamics Simulation - Michel Cuendet - EMBL
7 Processes: Thermodynamic Equilibria Folding Micelle Formation folded/native denatured micelle mixture Complexation Partitioning bound unbound in membrane in water in mixtures Molecular Dynamics Simulation - Michel Cuendet - EMBL
8 Plan Introduction The classical force field Setting up a simulation Connection to statistical mechanics Usage of MD simulation Molecular Dynamics Simulation - Michel Cuendet - EMBL
9 Definition of a model for molecular simulation Every molecule consists of atoms that are very strongly bound to each other Forces or interactions between atoms Degrees of freedom: atoms as elementary particles + topology Boundary conditions MOLECULAR MODEL Force field = physico-chemical knowledge Methods to generate configurations of atoms: Newton system temperature pressure walls external forces Molecular Dynamics Simulation - Michel Cuendet - EMBL
10 Classical force fields Goals of classical (semi-empirical) force fields - Definition of empirical potential energy functions V(r) to model the molecular interactions - These functions need to be differentiable in order to compute the ( F=- V(r forces acting on each atom: Implementation of calssical potential energy functions 1. Theoretical functional forms are derived for the potential energy V(r). 2. Definition of atom types that differ by their atomic number and chemical environment, e.g. the carbons in C=O or C-C are of different types. 3. Parameters are determined so as to reproduce the interactions between the various atom types by fitting procedures ( CHARMM ) - experimental enthalpies ( AMBER - experimental free energies (GROMOS, Parametrization available for proteins, lipids, sugars, ADN,... Molecular Dynamics Simulation - Michel Cuendet - EMBL
11 Covalent bonds and angles Bonds Type: CT1-CT1 r E bond = K b (r " r 0 ) 2 r 0 r 0 Angles Type: CT1-CT1-CT3 E angle = K " (" #" 0 ) 2 θ 0 Molecular Dynamics Simulation - Michel Cuendet - EMBL
12 Dihedrals and Improper torsions Dihedral angles Type: X-CT1-CT1-X E dihedral = K " [ 1+ cos( n" #$)] Improper angles O H 2 N C C O R E improper = K " (# $# 0 ) 2 Type: OC-OC-CT1-CC ψ 0 Molecular Dynamics Simulation - Michel Cuendet - EMBL
13 Van der Waals interactions Lennard -Jones potential : + $ E VdW = 4" -& #,% r ' ) ( 12 $ *& # % r ' ) ( 6. + $ r 0 = " -& m /,% r ' ) ( 12 $ * 2 r ' m & ) % r ( 6. 0 / σ : collision parameter ε : well depth r m : distance at min r m = " Combination rule for two different atoms i, j : σ ε Type: CT3-CT3 r m E VdW r m = r m,i + r m, j " = " i " j Repulsive : Pauli exclusion principle " 1 r 12 Attractive: induced dipole / induced dipole "# 1 r 6 Molecular Dynamics Simulation - Michel Cuendet - EMBL
14 Electrostatic interactions Coulomb law E elec = q i q j 4"# 0 # r ij where ε is the dielectric constant : 1 for vacuum, 4-20 for protein core, 80 for water The Coulomb energy decreases only as 1/r Despite dielectric shielding effects, it is a long range interaction Special techniques to deal with this : - PME : for stystems with periodic boundary conditions - Reaction Field : suppose homogeneous dielectric outside cutoff Molecular Dynamics Simulation - Michel Cuendet - EMBL
15 Derived Interactions Some interactions are often referred to as particular interactions, but they result from the two interactions previously described, i.e. the electrostatic and the van der Waals interactions. ( Hb ) 1) Hydrogen bonds φ δ A - Interaction of the type D-H A - The origin of this interaction is a dipole-dipole attraction - Typical ranges for distance and angle: 2.4 < d < 4.5Å and 180º < φ < 90º 2) Hydrophobic effect δ D H d +δ - Collective effect resulting from the energetically unfavorable surface of contact between the water ( Hb and an apolar medium (loss of water-water - The apolar medium reorganizes to minimize the water exposed surface (compaction, association... ) Water Oil Molecular Dynamics Simulation - Michel Cuendet - EMBL
16 The total potential energy function E = # K b ( r " r 0 ) 2 + # K $ ( $ "$ 0 ) 2 + # K % [ 1+ cos( n% "&)] + # K ' ' "' 0 bonds angles / + ( r 12 ), m +. " 2 r 6 ), 2 m # * r - * r - i> j 3 # i> j q i q j 45( 0 ( r dihedrals impropers ( ) 2 For a system with 1500 atoms ~ 10 6 pairs of interacting atoms Introduction of cutoff Molecular Dynamics Simulation - Michel Cuendet - EMBL
17 Cutoff for non-bonded interactions For an atom A, only non-bonded interactions with atoms within δ Å are calculated Non-bonded neighbour lists A Generally, δ = 8 to 14 Å δ Three cutoff schemes: strict, shift, switch Shift and switch: E'(r) = E(r) " S(r) S(r) differentiable cutofnb cutonnb cutnb Molecular Dynamics Simulation - Michel Cuendet - EMBL
18 Effect of cutoff No cutoff 8 Å cutoff Elec VdW Total Elec VdW Total Molecular Dynamics Simulation - Michel Cuendet - EMBL
19 Force field parametrization E = # K b ( r " r 0 ) 2 + # K $ ( $ "$ 0 ) 2 + # K % [ 1+ cos( n% "&)] + # K ' (' "' 0 ) 2 + 4( 0 ) 12 * -, + r / " ) 6 * - 3 q # 2, / 5 + i q # j. + bonds angles dihedrals impropers 1 r. i> j 4 i> j 46( 0 ( r Type of data structural data (exp.) spectroscopic data (exp.) quantum-chemical calculations : energy profiles (theor.) quantum-chemical calculations : electron densities (theor.) Type of system small molecules small molecules small molecules small molecules Phase crystalline solid phase gas phase gas phase gas phase Type of properties molecular geometry: bond lengths, angles intra-molecular vibrations: force constants torsional-angle rotational profiles atom charges Force field parameters r 0, θ 0, ψ 0 K b, K θ, K ψ K φ, δ, n charges q i (initial) thermodynamic data (exp.) molecules in solution, mixtures condensed phase heat of vaporisation, density, free energy of solvation v. d. Waals : σ i, ε i charges q i (final) dielectric data (exp.) small molecules condensed phase dielectric permittivity, relaxation charges q i transport data (exp.) small molecules condensed phase transport coefficients: diffusion, viscosity v. d. Waals : σ i, ε i charges q i Molecular Dynamics Simulation - Michel Cuendet - EMBL
20 Solvation Fundamental influence on the structure, dynamics and thermodynamics of biological molecules Effect through: Solvation of charge Screening of charge - charge interactions q E elec = i q j 4"# 0 #(r) r ε=1 - + ε=80 Hydrogen bonds between water molecules and polar functions of the solute Taken into account via: Explicit solvation. Water molecules are included. Stochastic boudary conditions Periodic boundary conditions Implicit solvation. Water effect is modeled. Screening constant Implicit solvation models (Poisson Boltzmann, Generalized Born) Molecular Dynamics Simulation - Michel Cuendet - EMBL
21 Explicit solvation schemes Stochastic boundary conditions The region of interest is solvated in a water sphere at 1atm. The water molecules are submitted to an additional force field that restrain them in the sphere while maintaining a strong semblance to bulk water. Periodic boundary conditions The fully solvated central cell is simulated, in the environment produced by the repetition of this cell in all directions. Molecular Dynamics Simulation - Michel Cuendet - EMBL
22 Implicit solvation Screening constant " = Nr N=4,8. The dielectric constant is a function of atom distance. Mimic screening effect of solvent. Simple, unphysical but efficient. ε=1 ε=80 Implicit solvent models In CHARMM: NBOND RDIE EPS 4.0 Poisson Boltzmann (PB) equation. " #(r)"$(r) { } %&'sinh[ $(r) ] = %4'((r) f(r) : electrostatic potential, Equation solved numerically. Very time consuming. In CHARMM: PBEQ module. Generalized born (GB) equation. q G elec = $ i q j 4"# 0 r % 1 & 2 1% 1 ) ( ' # + * i> j j r(r) : charge density q i q j r 2 + a i a j exp (%r 2 4a i a j ) $ $ a i : Born radius i Others: EEF1, SASA, etc... solvation energy Explicit hydrogen bonds with water molecules are lost Molecular Dynamics Simulation - Michel Cuendet - EMBL
23 Introduction to molecular surfaces Hydrophobic effect : Simply modelled by a non-polar solvation (free) energy term, proportional to the solvent accessible surface area (SASA) : "G np, solv = #SASA + b γ = kcal mol 1 Å 2 b = 0.92 kcal mol -1 Rotating probe: radius 1.4 Å Connolly Surface Solvent accessible Surface (SASA) Definitions: Van der Waals Surface - Van der Waals: ensemble of van der Waals sphere centered at each atom - Connolly: ensemble of contact points between probe and vdw spheres - Solvent: ensemble of probe sphere centers Molecular Dynamics Simulation - Michel Cuendet - EMBL
24 Examples of molecular surfaces Van der Waals Connolly ( Contact ) Solvent accessible Molecular Dynamics Simulation - Michel Cuendet - EMBL
25 Limitations of classical MD Problems Solutions 1) Fixed set of atom types 2) No electronic polarization: - fixed partial charges allow for conformational polarization but not electronic polarization Fluctuating charges treated as dynamical parameters Charges on springs representing e - clouds QM-MM Full QM simulations 3) Quadratic form of potentials: - problematic far from equilibrium values - no bond creation or deletion r QM-MM Full QM simulations Molecular Dynamics Simulation - Michel Cuendet - EMBL
26 Coarse grain models All-atom model 16 (CH 2 or CH 3 ) atoms Map to all-atom configurations Coarse-grained model 4 atoms Centre of mass A 1 A 4 A Centre of mass B 1 B 4 B Centre of mass C 1 C 4 C Centre of mass D 1 D 4 D Centre of mass W 1 W 4 W Molecular Dynamics Simulation - Michel Cuendet - EMBL
27 Molecular dynamics software Package name CHARMM Amber amber.scripps.edu GROMOS Gromacs NAMD E = explicit solvent I = implicit solvent supported force fields CHARMM (E / I; AA / UA), Amber Amber (E / I ; AA) Gromos (E / vacuum ; UA) Amber, Gromos, OPLS - (all E) CHARMM, Amber, Gromos,... AA = all atom UA = united atom (apolar H omitted) Molecular Dynamics Simulation - Michel Cuendet - EMBL
28 Plan Introduction The classical force field Setting up a simulation Connection to statistical mechanics Usage of MD simulation Molecular Dynamics Simulation - Michel Cuendet - EMBL
29 Minimal input for MM 1) Topological properties: description of the covalent connectivity of the molecules to be modeled 2) Structural properties: the starting conformation of the molecule, provided by an X-ray structure, NMR data or a theoretical model 3) Energetical properties: a force field describing the force acting on each atom of the molecules 4) Thermodynamical properties: a sampling algorithm that generates the thermodynamical ensemble that matchese experimental conditions for the system, e.g. N,V,T, N,P,T,... NV T 0 Molecular Dynamics Simulation - Michel Cuendet - EMBL
30 MD flowchart Initial coordinates (X-ray, NMR) Structure minimization (release strain) Solvation (if explicit solvent) Initial velocities assignment Heating dynamics (Temp. to 300K) Equilibration dynamics (control of Temp. and structure) Rescale velocities Temp. Ok? Structure Ok? Production dynamics (NVE, NVT, NPT) Analysis of trajectory Calculation of macroscopic values Molecular Dynamics Simulation - Michel Cuendet - EMBL
31 Coordinate files Cartesian coordinates (x,y,z) ATOM 1 N VAL PEP ATOM 2 HT1 VAL PEP ATOM 3 HT2 VAL PEP ATOM 4 HT3 VAL PEP ATOM 5 CA VAL PEP ATOM 6 HA VAL PEP ATOM 7 CB VAL PEP Internal coordinates (IC) Given 4 consecutive atoms A-B-C-D, the IC are: A B C D R AB, θ ABC, φ ABCD, θ BCD, R CD 1 1 N 1 C 1 *CA 1 CB N 1 C 1 *CA 1 HA N 1 CA 1 CB 1 CG CG1 1 CA 1 *CB 1 CG CG1 1 CA 1 *CB 1 HB CA 1 CB 1 CG1 1 HG HG11 1 CB 1 *CG1 1 HG HG11 1 CB 1 *CG1 1 HG It is possible to calculate missing cartesian coordinates from the existing ones and the IC Molecular Dynamics Simulation - Michel Cuendet - EMBL
32 Definition of atom types The residue topology file (RTF) contains the atom types and the standard topology of residues. Example: the CHARMM force field, version 22 file : top_all22_prot.inp Atom types section : MASS 1 H H polar H MASS 2 HC H N-ter H MASS 3 HA H nonpolar H MASS 4 HT H TIPS3P WATER HYDROGEN MASS 5 HP H aromatic H MASS 6 HB H backbone H [...] MASS 20 C C carbonyl C, peptide backbone MASS 21 CA C aromatic C MASS 22 CT C aliphatic sp3 C for CH MASS 23 CT C aliphatic sp3 C for CH2 MASS 24 CT C aliphatic sp3 C for CH3 MASS 25 CPH C his CG and CD2 carbons 90 atom types No. atom type mass atom Molecular Dynamics Simulation - Michel Cuendet - EMBL
33 Decomposition into residues In CHARMM, molecules are decomposed into residues. A molecule may be composed of one to several hundreds or thousands of residues. Residues correspond to amino acids for proteins or to nucleotides for DNA. Topologies for individual residues are pre-defined in CHARMM Easy construction of the protein topology from the sequence. Only informations about 20 amino acids needed to construct the topology of all proteins. NH R O NH R O NH R O NH R O NH R O Residue definition in CHARMM Molecular Dynamics Simulation - Michel Cuendet - EMBL
34 Residue topology definition file : top_all22_prot.inp RESI ALA 0.00 GROUP ATOM N NH ATOM HN H 0.31 HN-N ATOM CA CT HB1 ATOM HA HB 0.09 / GROUP HA-CA--CB-HB2 ATOM CB CT \ ATOM HB1 HA 0.09 HB3 ATOM HB2 HA 0.09 O=C ATOM HB3 HA 0.09 GROUP ATOM C C 0.51 ATOM O O BOND CB CA N HN N CA BOND C CA C +N CA HA CB HB1 CB HB2 CB HB3 DOUBLE O C IMPR N -C CA HN C CA +N O DONOR HN N ACCEPTOR O C IC -C CA *N HN IC -C N CA C IC N CA C +N IC +N CA *C O IC CA C +N +CA IC N C *CA CB IC N C *CA HA IC C CA CB HB IC HB1 CA *CB HB IC HB1 CA *CB HB ATOM N NH Atom name Angles and dihedrals can be generated automatically from this. Total charge Atom type Atom charge Bond Improper angle Previous residue Next residue Molecular Dynamics Simulation - Michel Cuendet - EMBL IC
35 Patching residues Treat protein special features. To make N- and C-termini: PRES NTER 1.00 standard N-terminus GROUP use in generate statement ATOM N NH ATOM HT1 HC 0.33 HT1 ATOM HT2 HC 0.33 (+)/ ATOM HT3 HC CA--N--HT2 ATOM CA CT \ ATOM HA HB 0.10 HA HT3 DELETE ATOM HN BOND HT1 N HT2 N HT3 N DONOR HT1 N DONOR HT2 N DONOR HT3 N IC HT1 N CA C IC HT2 CA *N HT IC HT3 CA *N HT To make disulfide bridges: PRES DISU patch for disulfides. Patch must be 1-CYS and 2-CYS. use in a patch statement follow with AUTOgenerate ANGLes DIHEdrals command GROUP ATOM 1CB CT ATOM 1SG SM SG--2CB-- GROUP / ATOM 2SG SM CB--1SG ATOM 2CB CT DELETE ATOM 1HG1 DELETE ATOM 2HG1 BOND 1SG 2SG IC 1CA 1CB 1SG 2SG IC 1CB 1SG 2SG 2CB IC 1SG 2SG 2CB 2CA Etc... Molecular Dynamics Simulation - Michel Cuendet - EMBL
36 The parameter file Contains force field parameters. file : par_all22_prot.inp Bonds: BONDS V(bond) = Kb(b - b0)**2 Kb: kcal/mole/a**2 b0: A atom type Kb b0 Carbon Dioxide CST OST JES Heme to Sulfate (PSUL) link SS FE force constant a guess equilbrium bond length optimized to reproduce CSD survey values of 2.341pm0.01 (mean, standard error) adm jr., 7/01 C C ALLOW ARO HEM Heme vinyl substituent (KK, from propene (JCS)) CA CA ALLOW ARO benzene, JES 8/25/89 CE1 CE for butene; from propene, yin/adm jr., 12/95 CE1 CE for propene, yin/adm jr., 12/95 CE1 CT for butene; from propene, yin/adm jr., 12/95 CE1 CT for butene, yin/adm jr., 12/95 CE2 CE K b r 0 Idem for angles, dihedrals, impropers,... Molecular Dynamics Simulation - Michel Cuendet - EMBL
37 Setting up a system in CHARMM $ charmm < input_file.inp > output_file.out * Standard Topology and Parameters OPEN UNIT 1 CARD READ NAME top_all22_prot.inp READ RTF CARD UNIT 1 CLOSE UNIT 1 OPEN UNIT 1 CARD READ NAME par_all22_prot.inp READ PARA CARD UNIT 1 CLOSE UNIT 1 Generate actual topology OPEN UNIT 1 READ CARD NAME 1aho-xray.pdb READ SEQUENCE PDB UNIT 1 GENE 1S SETUP FIRST NTER LAST CTER REWIND UNIT 1 READ COOR PDB UNIT 1 CLOSE UNIT 1 Make disulfide bridges PATCH DISU 1S 12 1S 63 PATCH DISU 1S 16 1S 36 PATCH DISU 1S 22 1S 46 PATCH DISU 1S 26 1S 48 AUTOgenerate ANGLes DIHEdrals Fill IC table and build missing coordinates IC FILL IC PARA ALL IC BUILD Build better coordinates for hydrogens HBUILD SELE TYPE H* END Write coordinates in CHARMM format OPEN UNIT 1 WRITE CARD NAME 1aho.crd WRITE COOR CARD UNIT 1 CLOSE UNIT 1 Write coordinates in PDB format OPEN UNIT 1 WRITE CARD NAME 1aho.pdb WRITE COOR PDB UNIT 1 CLOSE UNIT 1 Write Protein Structure File for subsequent use OPEN UNIT 1 WRITE CARD NAME 1aho.psf WRITE PSF CARD UNIT 1 CLOSE UNIT 1 PSFSUM> Summary of the structure file counters : Number of segments = 1 Number of residues = 64 Number of atoms = 962 Number of groups = 298 Number of bonds = 980 Number of angles = 1753 Number of dihedrals = 2591 Number of impropers = 169 Number of cross-terms = 0 Number of HB acceptors = 98 Number of HB donors = 118 Number of NB exclusions = 0 Total charge = Molecular Dynamics Simulation - Michel Cuendet - EMBL
38 The protein structure file (PSF) The PSF is generated by CHARMM from the sequence of the proteins, the ligands, the water molecules, etc..., using the information present in the residue topology file (RTF) It contains all the information needed for future simulations : Residues and segments. How the system is divided into residues and segments. Atom information. Names, types, charges, masses. Bond, angle, dihedral and improper dihedral lists Electrostatic groupings. How some numbers of atoms are grouped for the purpose of calculating long range electrostatic A segment is a group of molecules, for example: one single protein a collection of water molecules a collection of ions a ligand Molecular Dynamics Simulation - Michel Cuendet - EMBL
39 Performing a calculation in CHARMM Second step : perform calculation, e.g. energy evaluation * input Standard Topology and Parameters OPEN UNIT 1 CARD READ NAME top_all22_prot.inp READ RTF CARD UNIT 1 CLOSE UNIT 1 OPEN UNIT 1 CARD READ NAME par_all22_prot.inp READ PARA CARD UNIT 1 CLOSE UNIT Actual topology OPEN UNIT 1 READ CARD NAME 1aho.psf READ PSF CARD UNIT 1 CLOSE UNIT Coordinates OPEN UNIT 1 READ CARD NAME 1aho.pdb READ COOR PDB UNIT 1 CLOSE UNIT Energy calculation ENERGY End of input file STOP CHARMM> ENERGY output NONBOND OPTION FLAGS: ELEC VDW ATOMs CDIElec SHIFt VATOm VSWItch BYGRoup NOEXtnd NOEWald CUTNB = CTEXNB = CTONNB = CTOFNB = WMIN = WRNMXD = E14FAC = EPS = NBXMOD = 5 There are 0 atom pairs and 0 atom exclusions. There are 0 group pairs and 0 group exclusions. <MAKINB> with mode 5 found 2733 exclusions and 2534 interactions(1-4) <MAKGRP> found 886 group exclusions. Generating nonbond list with Exclusion mode = 5 == PRIMARY == SPACE FOR ATOM PAIRS AND 0 GROUP PAIRS General atom nonbond list generation found: ATOM PAIRS WERE FOUND FOR ATOM LIST 9439 GROUP PAIRS REQUIRED ATOM SEARCHES ENER ENR: Eval# ENERgy Delta-E GRMS ENER INTERN: BONDs ANGLes UREY-b DIHEdrals IMPRopers ENER EXTERN: VDWaals ELEC HBONds ASP USER ENER> ENER INTERN> ENER EXTERN> Molecular Dynamics Simulation - Michel Cuendet - EMBL
40 Energy landscape Landscape for φ/ψ plane of dialanine Complex landscape for a protein E φ ψ 3N Spatial coordinates Molecular Dynamics Simulation - Michel Cuendet - EMBL
41 Minimization Landscape for ϕ/ψ plane of dialanine Finding minimum energy conformations given a potential energy function: "E = 0 " 2 E and 2 "x i "x > 0 i Used to: relieve strain in experimental conformations find (energetically) stable states ϕ ψ Huge number of degrees of freedom in macromolecular systems. Huge amount of local minima Impossible to find true global minimum Different minimization methods available: Steepest descent (SD) relieve strain, find close local minimum Conjugated gradient (CONJ) Adopted Basis Newton Raphson (ABNR) Find lower energy mimima Molecular Dynamics Simulation - Michel Cuendet - EMBL
42 CHARMM input for minimization * Standard Topology and Parameters OPEN UNIT 1 CARD READ NAME top_all22_prot.inp READ RTF CARD UNIT 1 CLOSE UNIT 1 OPEN UNIT 1 CARD READ NAME par_all22_prot.inp READ PARA CARD UNIT 1 CLOSE UNIT Actual topology OPEN UNIT 1 READ CARD NAME val.psf READ PSF CARD UNIT 1 CLOSE UNIT Coordinates OPEN UNIT 1 READ CARD NAME val.pdb READ COOR PDB UNIT 1 CLOSE UNIT ABNR minimization MINI ABNR NSTEP End of input file STOP ABNER> An energy minimization has been requested. EIGRNG = MINDIM = 5 NPRINT = 50 NSTEP = 200 PSTRCT = SDSTP = STPLIM = STRICT = TOLFUN = TOLGRD = TOLITR = 100 TOLSTP = FMEM = MINI MIN: Cycle ENERgy Delta-E GRMS Step-size MINI INTERN: BONDs ANGLes UREY-b DIHEdrals IMPRopers MINI EXTERN: VDWaals ELEC HBONds ASP USER MINI> MINI INTERN> MINI EXTERN> MINI> MINI INTERN> MINI EXTERN> MINI> MINI INTERN> MINI EXTERN> UPDECI: Nonbond update at step 103 Generating nonbond list with Exclusion mode = 5 == PRIMARY == SPACE FOR 172 ATOM PAIRS AND 0 GROUP PAIRS General atom nonbond list generation found: 120 ATOM PAIRS WERE FOUND FOR ATOM LIST 0 GROUP PAIRS REQUIRED ATOM SEARCHES MINI> MINI INTERN> MINI EXTERN> MINI> MINI INTERN> MINI EXTERN> ABNER> Minimization exiting with number of steps limit ( 200) exceeded. ABNR MIN: Cycle ENERgy Delta-E GRMS Step-size ABNR INTERN: BONDs ANGLes UREY-b DIHEdrals IMPRopers ABNR EXTERN: VDWaals ELEC HBONds ASP USER ABNR> ABNR INTERN> ABNR EXTERN> Molecular Dynamics Simulation - Michel Cuendet - EMBL
43 Simple Molecular dynamics Newton s law of motion: F i = m i a i = " de i dr i MD algorithm calculate forces F(t) In discete time : integration algorithm. Example: Verlet algrorithm r(t + "t) = r(t) + v(t) #"t a(t) #"t 2 t = t + "t update r(t + "t) r(t "#t) = r(t) " v(t) $#t a(t) $#t 2 r(t + "t) = 2r(t) # r(t #"t) + F(t) m $"t 2 δt ~ 1 fs = s Propagation of time: position at time t+dt is a determined by position at time t and t-dt, and by the acceleration at time t (i.e., the forces at time t) The equations of motion are deterministic, e.g., the positions and the velocities at time zero determine the positions and velocities at all other times, t. Molecular Dynamics Simulation - Michel Cuendet - EMBL
44 CHARMM input file for MD simulation * Standard Topology and Parameters OPEN UNIT 1 CARD READ NAME top_all22_prot.inp READ RTF CARD UNIT 1 CLOSE UNIT 1 OPEN UNIT 1 CARD READ NAME par_all22_prot.inp READ PARA CARD UNIT 1 CLOSE UNIT Actual topology OPEN UNIT 1 READ CARD NAME val.psf READ PSF CARD UNIT 1 CLOSE UNIT Coordinates OPEN UNIT 1 READ CARD NAME val.pdb READ COOR PDB UNIT 1 CLOSE UNIT MD simulation OPEN WRITE UNIT 31 CARD NAME traj/dyna.rst Restart file OPEN WRITE UNIT 32 FILE NAME traj/dyna.dcd Coordinates file OPEN WRITE UNIT 33 CARD NAME traj/dyna.ene Energy file DYNA VERLET START NSTEP 1000 TIMESTEP IUNWRI 31 IUNCRD 32 KUNIT 33 - IPRFRQ 100 NPRINT 100 NSAVC 100 NSAVV 100 ISVFRQ FIRSTT FINALT ICHEW 1 - IEQFRQ 10 IASORS 0 ISCVEL 0 IASVEL 1 ISEED INBFRQ End of input file STOP Generated files: Restart file (rst). To restart after crash or to continue MD sim. Collection of instantaneous coordinates along trajectory (dcd) Control propagation of time. Timestep in ps (i.e. 1fs here) Control output Control temperature Control non bonded list update frequency Molecular Dynamics Simulation - Michel Cuendet - EMBL
45 Output of MD simulation DYNA DYN: Step Time TOTEner TOTKe ENERgy TEMPerature DYNA PROP: GRMS HFCTote HFCKe EHFCor VIRKe DYNA INTERN: BONDs ANGLes UREY-b DIHEdrals IMPRopers DYNA EXTERN: VDWaals ELEC HBONds ASP USER DYNA PRESS: VIRE VIRI PRESSE PRESSI VOLUme DYNA> DYNA PROP> DYNA INTERN> DYNA EXTERN> DYNA PRESS> [...] DYNA DYN: Step Time TOTEner TOTKe ENERgy TEMPerature DYNA PROP: GRMS HFCTote HFCKe EHFCor VIRKe DYNA INTERN: BONDs ANGLes UREY-b DIHEdrals IMPRopers DYNA EXTERN: VDWaals ELEC HBONds ASP USER DYNA PRESS: VIRE VIRI PRESSE PRESSI VOLUme DYNA> DYNA PROP> DYNA INTERN> DYNA EXTERN> DYNA PRESS> UPDECI: Nonbond update at step 10 [...] Molecular Dynamics Simulation - Michel Cuendet - EMBL
46 Newtonian dynamics in practice In theory, Newtonian dynamics conserves the total energy (isolated system) : r = p m p = F(r) H(r,p) = p i 2 " +V(r) = cste i 2m i In practice, constant energy dynamics is not often used for two reasons : 1) Inaccuracies of the MD algorithm tend to heat up the system We can couple the system to a heat reservoir to absorb the excess heat Integrator cutoffs constraints barostat System : Ε reservoir 2) The constant energy dynamics (NVE) does rearely represents the experimental conditions for the system simulated. Molecular Dynamics Simulation - Michel Cuendet - EMBL
47 Plan Introduction The classical force field Setting up a simulation Connection to statistical mechanics Usage of MD simulation Molecular Dynamics Simulation - Michel Cuendet - EMBL
48 Thermodynamic ensembles A macroscopic state is described by : - number of particles : N - chemical potential : µ - volume : V - pressure : P - energy : E - temperature : T Definition: a thermodynamical ensemble is a collection of microscopic states that all realize an identical macroscopic state A microscopic state of the system is given by a point (r, p) of the phase space of the system, where r= (r 1,..., r N ) and p= (p 1,..., p N ) are the positions and the momenta of the N atoms of the system. Examples of thermodynamical ensembles: - Microcanonical: fixed N, V, E - Canonical: fixed N, V, T often used in MD - Constant P-T: fixed N, P, T often used in MD - Grand Canonical: fixed µ, P, T Molecular Dynamics Simulation - Michel Cuendet - EMBL
49 Bolzmann (canonical) distribution Boltzmann showed that the canonical probability of the microstate i is given by P i = 1 Z e"#e i β = 1/K B T K B = Boltzmann constant Z is the partition function, Z = $ j e "#Ej such that : " P i =1 i The partition function is a very complex function to compute, because it represents a measure of the whole space accessible to the system. Illustration: If a system can have two unique states, state (1) and state (2), then the ratio of systems in state (1) and (2) is ( 1 ) P 1 = e E 1 = e E 1_ E _ 2 = e E at 300K, a ΔE of 1.3 kcal/mol P 2 e E 2 results in a P1/P2 of 1 log 10. Cave: if state (1) and (2) are composed of several microscopic states, ΔE ΔG ( 2 ) Molecular Dynamics Simulation - Michel Cuendet - EMBL
50 The partition function The determination of the macroscopic behavior of a system from a thermodynamical point of view is tantamount to computing the partition function, Z, from which all the properties can be derived. Z = $ e "#E i i O = 1 Z " O i e #$E i i... Expectation Value E = " "# ln Z ( ) =U Internal Energy ( ) # p = kt % "ln Z $ "V Pressure & ( ' N,T A = "kt ln(z) Helmoltz free energy Molecular Dynamics Simulation - Michel Cuendet - EMBL
51 The ensemble average Macroscopic Microscopic E 1, P 1 ~ e -βe 1 Expectation value E 2, P 2 ~ e -βe 2 O = 1 Z " O i e #$E i E 3, P 3 ~ e -βe 3 Where Z = e "#E i $ is the partition function E 4, P 4 ~ e -βe 4 E 5, P 5 ~ e -βe 5 Molecular Dynamics Simulation - Michel Cuendet - EMBL
52 Ergodic Hypothesis The ergodic hypothesis is that the ensemble averages used to compute expectation values can be replaced by time averages over the simulation. Ergodicity O ensemble = O time The microstates sampled by molecular dynamics are usually a small subset of the entire thermodynamical ensemble. 1 Z O(r, p)e"#e(r,p ) $ drdp = 1 % The validity of this hypothesis depends on the quality of the sampling produced by the molecular modelling technique. The sampling should reach all important minima and explore them with the correct probability, - NVE simulations Microcanonic ensemble P = cst. - NVT simulations Canonical ensemble P(E) = e -βe - NPT simulations Isothermic-isobaric ensemble ( β(e+pv - P(E) = e e "#E O(t)dt Note that the Bolzmann weight is not present in the time average because it is assumed that conformations are sampled from the right probability. % $ 0 Molecular Dynamics Simulation - Michel Cuendet - EMBL
53 Ergodic hypothesis : intuitive view MD Trajectory E NVE simulation in a local minimum ϕ O ensemble = 1 Z ψ? ) $ O(r, p)e"#e(r,p drdp = % 1 $ O(t)dt = O % time 0 3N Spatial coordinates Two main requirements for MD simulation : 1) Accurate energy function E(r,p) 2) Appropriate algorithm, which - generates the right ensemble - samples efficiently Molecular Dynamics Simulation - Michel Cuendet - EMBL
54 Sampling of the various ensembles ( N,V,E 1) Microcanonical ensemble (constant sampling is obtained by simple integration of the Newtonian dynamics: - Verlet, Leap-Frog, Velocity Verlet, Gear ( N,V,T 2) Canonical ensemble (constant sampling is obtained using thermostats : 1) Berendsen: scaling of velocities to obtain an exponential relaxation of the temperature to T 2) Nose-Hoover: additional degree of freedom coupled to the physical system acts as heat bath. ( N,P,T 3) Isothermic-isobaric ensemble (constant In addition to the thermostat, the volume of the system is allowed to fluctuate, and is regulated by barostat algorithms. NV N NVE T (infinite E ( reservoir fixed P T (infinite E ( reservoir Molecular Dynamics Simulation - Michel Cuendet - EMBL
55 The Nosé-Hoover thermostat Phase space extended by two extra variables : physical variables Newton friction term temperature regulation One can demonstrate that the canonical distribution is reproduced for the physical variables Conserved quantity : Non-Hamiltonian dynamics... Molecular Dynamics Simulation - Michel Cuendet - EMBL
56 Other sampling methods I ( LD ) Langevin Dynamics In Langevin Dynamics, two additional forces are added to the standard force field: - a friction force: -γ i p i whose direction is opposed to the velocity of atom i - a stochastic (random) force: ζ(t) such that <ζ(t)> = 0. This leads to the following equation for the motion of atom i: This equation can for example simulate the friction and stochastic effect of the solvent in implicit solvent simulations. The temperature is adjusted via γ and ζ, using the dissipation-fluctuation theorem. The stochastic term can improve barrier crossing and hence sampling. r i = p i m i p i = F i (r) + "p i + # (t) LD does not reproduce dynamical properties Molecular Dynamics Simulation - Michel Cuendet - EMBL
57 Other sampling methods II Monte Carlo Simulations and the Metropolis criterion In this sampling method, instead of computing the forces on each atom to solve its time evolution, random movements are assigned to the system and the potential energy of the resulting conformer is evaluated. To insure Boltzmann sampling, additional criteria need to be applied on the new conformer. Let C be the initial conformer and C' the randomly modified: - if V(C') < V(C), the new conformer is kept and C' becomes C for next step - if V(C') > V(C), a random number, ρ, in the [0,1] interval is generated and if e -β(v'-v) > ρ, the new conformer is kept and C' becomes C for next step Using this algorithm, one insures Boltzmann statistics, P( C ") P(C) = e#$ " ( V #V ) Molecular Dynamics Simulation - Michel Cuendet - EMBL
58 Plan Introduction The classical force field Setting up a simulation Connection to statistical mechanics Usage of MD simulation Molecular Dynamics Simulation - Michel Cuendet - EMBL
59 The importance of entropy Mechanics: Statistical mechanics: A state is characterised by one minimum energy structure (global minimum) A state is characterised by an ensemble of structures Very small energy differences between states (~k B T = 2.5 kj/mol) resulting from summation over very many contributions Entropic effects : energy U(x) Not only energy minima are of importance but whole range of x-values with energies ~k B T The free energy (F) governs the system ½ k B T S(x 1 ) S(x 2 ) F = U - TS U(x 2 ) x 2 may have higher energy but lower free energy than x 1 U(x 1 ) x 1 x 2 coordinate x Energy (U) entropy (S) compensation at finite temperature T Molecular Dynamics Simulation - Michel Cuendet - EMBL
60 Dynamical behavior of proteins Biological molecules exhibit a wide range of time scales over which specific processes occur; for example: Local Motions (0.01 to 5 Å, to 10-1 s) Atomic fluctuations Sidechain Motions Loop Motions Rigid Body Motions (1 to 10 Å, 10-9 to 1 s) Helix Motions Domain Motions Subunit motions Large-Scale Motions (> 5 Å, 10-7 to 10 4 s) Helix coil transitions Dissociation/Association Folding and Unfolding Molecular Dynamics Simulation - Michel Cuendet - EMBL
61 Types of problems Molecular dynamics simulations permit the study of complex, dynamic processes that occur in biological systems. These include, for example: Protein stability Conformational changes Protein folding Molecular recognition: proteins, DNA, membranes, complexes Ion transport in biological systems and provide the mean to carry out the following studies, Drug Design Structure determination: X-ray and NMR Molecular Dynamics Simulation - Michel Cuendet - EMBL
62 Historical perspective Theoretical milestones: Newton ( ): Boltzmann( ): Schrödinger ( ): Classical equations of motion: F(t)=m a(t) Foundations of statistical mechanics Quantum mechanical eq. of motion: -ih t Ψ(t)=H(t) Ψ(t) Molecular mechanics milestones: Metropolis (1953): First Monte Carlo (MC) simulation of a liquid (hard spheres) Wood (1957): First MC simulation with Lennard-Jones potential Alder (1957): First Molecular Dynamics (MD) simulation of a liquid (hard spheres) Rahman (1964): First MD simulation with Lennard-Jones potential Karplus (1977) & First MD simulation of proteins McCammon (1977) Karplus (1983): The CHARMM general purpose FF & MD program Kollman(1984): The AMBER general purpose FF & MD program Car-Parrinello(1985): First full QM simulations Kollmann(1986): First QM-MM simulations Molecular Dynamics Simulation - Michel Cuendet - EMBL Liquids Proteins
63 System sizes Simulations in explicit solvent BPTI (VAC), bovine pancreatic trypsin inhibitor without solvent BPTI, bovine pancreatic trypsin inhibitor with solvent RHOD, photosynthetic reaction center of Rhodopseudomonas viridis HIV-1, HIV-1 protease ES, estrogen DNA STR, streptavidin DOPC, DOPC lipid bilayer RIBO, ribosome Solid curve, Moore s law doubling every 28.2 months. Dashed curve, Moore s law doubling every 39.6 months. K.Y. Sanbonmatsu, C.-S. Tung, Journal of Structural Biology 157(3) : 470, 2007 Molecular Dynamics Simulation - Michel Cuendet - EMBL
64 Largest system 2006 satellite tobacco mosaic virus 1 million atoms Simulation time : 50 ns system size : 220 A P. L. Freddolino, A. S. Arkhipov, S. B. Larson, A. McPherson, and K. Schulten, Molecular dynamics simulations of the complete satellite tobacco mosaic virus, Structure 14 (2006), 437. Molecular Dynamics Simulation - Michel Cuendet - EMBL
65 The tradeoff we can afford Molecular Dynamics Simulation - Michel Cuendet - EMBL
66 Exemple : Aquaporin Selective translocation of water across a membrane S. Hub and Bert L. de Groot. Mechanism of selectivity in aquaporins and aquaglyceroporins PNAS. 105: (2008) Molecular Dynamics Simulation - Michel Cuendet - EMBL
67 Exemple : Aquaporin Selective translocation of water across a membrane Propose a model for selectivity at the atomic level S. Hub and Bert L. de Groot. Mechanism of selectivity in aquaporins and aquaglyceroporins, PNAS 105: (2008) Molecular Dynamics Simulation - Michel Cuendet - EMBL
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Topic 2: Energy in Biological Systems
 Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Topic 2: Energy in Biological Systems Outline: Types of energy inside cells Heat & Free Energy Energy and Equilibrium An Introduction to Entropy Types of energy in cells and the cost to build the parts
Introduction to Molecular Dynamics Simulations
 Introduction to Molecular Dynamics Simulations Roland H. Stote Institut de Chimie LC3-UMR 7177 Université Louis Pasteur Strasbourg France 1EA5 Title Native Acetylcholinesterase (E.C. 3.1.1.7) From Torpedo
Introduction to Molecular Dynamics Simulations Roland H. Stote Institut de Chimie LC3-UMR 7177 Université Louis Pasteur Strasbourg France 1EA5 Title Native Acetylcholinesterase (E.C. 3.1.1.7) From Torpedo
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005
 Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Hands-on exercises on solvent models & electrostatics EMBnet - Molecular Modeling Course 2005 Exercise 1. The purpose of this exercise is to color the solvent accessible surface of a protein according
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7. 4. Which of the following weak acids would make the best buffer at ph = 5.0?
 Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Language: English Lecturer: Gianni de Fabritiis. Teaching staff: Language: English Lecturer: Jordi Villà i Freixa
 MSI: Molecular Simulations Descriptive details concerning the subject: Name of the subject: Molecular Simulations Code : MSI Type of subject: Optional ECTS: 5 Total hours: 125.0 Scheduling: 11:00-13:00
MSI: Molecular Simulations Descriptive details concerning the subject: Name of the subject: Molecular Simulations Code : MSI Type of subject: Optional ECTS: 5 Total hours: 125.0 Scheduling: 11:00-13:00
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon. V. Polypeptides and Proteins
 IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
Peptide bonds: resonance structure. Properties of proteins: Peptide bonds and side chains. Dihedral angles. Peptide bond. Protein physics, Lecture 5
 Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
What is molecular dynamics (MD) simulation and how does it work?
 What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
What is molecular dynamics (MD) simulation and how does it work? A lecture for CHM425/525 Fall 2011 The underlying physical laws necessary for the mathematical theory of a large part of physics and the
Introduction, Noncovalent Bonds, and Properties of Water
 Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
Lecture 1 Introduction, Noncovalent Bonds, and Properties of Water Reading: Berg, Tymoczko & Stryer: Chapter 1 problems in textbook: chapter 1, pp. 23-24, #1,2,3,6,7,8,9, 10,11; practice problems at end
1 The water molecule and hydrogen bonds in water
 The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
Overview of NAMD and Molecular Dynamics
 Overview of NAMD and Molecular Dynamics Jim Phillips Low-cost Linux Clusters for Biomolecular Simulations Using NAMD Outline Overview of molecular dynamics Overview of NAMD NAMD parallel design NAMD config
Overview of NAMD and Molecular Dynamics Jim Phillips Low-cost Linux Clusters for Biomolecular Simulations Using NAMD Outline Overview of molecular dynamics Overview of NAMD NAMD parallel design NAMD config
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK
 Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
The strength of the interaction
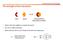 The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
The strength of the interaction Host Guest Supramolecule (host-guest complex) When is the host capable to recognize the guest? How do we define selectivity Which element will we use to design the host
Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology
 Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Introduction Molecular Mechanics or force-field methods use classical type models
Introduction to Molecular Mechanics C. David Sherrill School of Chemistry and Biochemistry Georgia Institute of Technology Introduction Molecular Mechanics or force-field methods use classical type models
INTERMOLECULAR FORCES
 INTERMOLECULAR FORCES Intermolecular forces- forces of attraction and repulsion between molecules that hold molecules, ions, and atoms together. Intramolecular - forces of chemical bonds within a molecule
INTERMOLECULAR FORCES Intermolecular forces- forces of attraction and repulsion between molecules that hold molecules, ions, and atoms together. Intramolecular - forces of chemical bonds within a molecule
Protein Dynamics Intro
 Protein Dynamics Intro From rigid structures to motions on energy landscapes Do you all remember Anfinsen? What concept now associated with his name made Anfinsen famous? Right, it is the concept that
Protein Dynamics Intro From rigid structures to motions on energy landscapes Do you all remember Anfinsen? What concept now associated with his name made Anfinsen famous? Right, it is the concept that
1 Peptide bond rotation
 1 Peptide bond rotation We now consider an application of data mining that has yielded a result that links the quantum scale with the continnum level electrostatic field. In other cases, we have considered
1 Peptide bond rotation We now consider an application of data mining that has yielded a result that links the quantum scale with the continnum level electrostatic field. In other cases, we have considered
Indiana's Academic Standards 2010 ICP Indiana's Academic Standards 2016 ICP. map) that describe the relationship acceleration, velocity and distance.
 .1.1 Measure the motion of objects to understand.1.1 Develop graphical, the relationships among distance, velocity and mathematical, and pictorial acceleration. Develop deeper understanding through representations
.1.1 Measure the motion of objects to understand.1.1 Develop graphical, the relationships among distance, velocity and mathematical, and pictorial acceleration. Develop deeper understanding through representations
Non-Covalent Bonds (Weak Bond)
 Non-Covalent Bonds (Weak Bond) Weak bonds are those forces of attraction that, in biological situations, do not take a large amount of energy to break. For example, hydrogen bonds are broken by energies
Non-Covalent Bonds (Weak Bond) Weak bonds are those forces of attraction that, in biological situations, do not take a large amount of energy to break. For example, hydrogen bonds are broken by energies
Carbohydrates, proteins and lipids
 Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Carbohydrates, proteins and lipids Chapter 3 MACROMOLECULES Macromolecules: polymers with molecular weights >1,000 Functional groups THE FOUR MACROMOLECULES IN LIFE Molecules in living organisms: proteins,
Section Activity #1: Fill out the following table for biology s most common elements assuming that each atom is neutrally charged.
 LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
LS1a Fall 2014 Section Week #1 I. Valence Electrons and Bonding The number of valence (outer shell) electrons in an atom determines how many bonds it can form. Knowing the number of valence electrons present
Structure of proteins
 Structure of proteins Primary structure: is amino acids sequence or the covalent structure (50-2500) amino acids M.Wt. of amino acid=110 Dalton (56 110=5610 Dalton). Single chain or more than one polypeptide
Structure of proteins Primary structure: is amino acids sequence or the covalent structure (50-2500) amino acids M.Wt. of amino acid=110 Dalton (56 110=5610 Dalton). Single chain or more than one polypeptide
Lecture Overview. Hydrogen Bonds. Special Properties of Water Molecules. Universal Solvent. ph Scale Illustrated. special properties of water
 Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Lecture Overview special properties of water > water as a solvent > ph molecules of the cell > properties of carbon > carbohydrates > lipids > proteins > nucleic acids Hydrogen Bonds polarity of water
Structure Check. Authors: Eduard Schreiner Leonardo G. Trabuco. February 7, 2012
 University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
Role of Hydrogen Bonding on Protein Secondary Structure Introduction
 Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
H 2O gas: molecules are very far apart
 Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Non-Covalent Molecular Forces 2/27/06 3/1/06 How does this reaction occur: H 2 O (liquid) H 2 O (gas)? Add energy H 2O gas: molecules are very far apart H 2O liquid: bonding between molecules Use heat
Chemical Bonds. Chemical Bonds. The Nature of Molecules. Energy and Metabolism < < Covalent bonds form when atoms share 2 or more valence electrons.
 The Nature of Molecules Chapter 2 Energy and Metabolism Chapter 6 Chemical Bonds Molecules are groups of atoms held together in a stable association. Compounds are molecules containing more than one type
The Nature of Molecules Chapter 2 Energy and Metabolism Chapter 6 Chemical Bonds Molecules are groups of atoms held together in a stable association. Compounds are molecules containing more than one type
Physics 9e/Cutnell. correlated to the. College Board AP Physics 1 Course Objectives
 Physics 9e/Cutnell correlated to the College Board AP Physics 1 Course Objectives Big Idea 1: Objects and systems have properties such as mass and charge. Systems may have internal structure. Enduring
Physics 9e/Cutnell correlated to the College Board AP Physics 1 Course Objectives Big Idea 1: Objects and systems have properties such as mass and charge. Systems may have internal structure. Enduring
PROTEINS THE PEPTIDE BOND. The peptide bond, shown above enclosed in the blue curves, generates the basic structural unit for proteins.
 Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
8/20/2012 H C OH H R. Proteins
 Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Chapter 2 Polar Covalent Bonds; Acids and Bases
 John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
John E. McMurry http://www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds; Acids and Bases Javier E. Horta, M.D., Ph.D. University of Massachusetts Lowell Polar Covalent Bonds: Electronegativity
Name Class Date. In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question.
 Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Assessment Chapter Test A Chapter: States of Matter In the space provided, write the letter of the term or phrase that best completes each statement or best answers each question. 1. The kinetic-molecular
Helices From Readily in Biological Structures
 The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
Chapter 13 - LIQUIDS AND SOLIDS
 Chapter 13 - LIQUIDS AND SOLIDS Problems to try at end of chapter: Answers in Appendix I: 1,3,5,7b,9b,15,17,23,25,29,31,33,45,49,51,53,61 13.1 Properties of Liquids 1. Liquids take the shape of their container,
Chapter 13 - LIQUIDS AND SOLIDS Problems to try at end of chapter: Answers in Appendix I: 1,3,5,7b,9b,15,17,23,25,29,31,33,45,49,51,53,61 13.1 Properties of Liquids 1. Liquids take the shape of their container,
18.2 Protein Structure and Function: An Overview
 18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
Sidechain Torsional Potentials and Motion of Amino Acids in Proteins:
 Proc. Nat. Acad. Sci. USA Vol. 72, No. 6, pp. 2002-2006, June 1975 Sidechain Torsional Potentials and Motion of Amino Acids in Proteins: Bovine Pancreatic Trypsin Inhibitor (nuclear magnetic resonance
Proc. Nat. Acad. Sci. USA Vol. 72, No. 6, pp. 2002-2006, June 1975 Sidechain Torsional Potentials and Motion of Amino Acids in Proteins: Bovine Pancreatic Trypsin Inhibitor (nuclear magnetic resonance
Protein Physics. A. V. Finkelstein & O. B. Ptitsyn LECTURE 1
 Protein Physics A. V. Finkelstein & O. B. Ptitsyn LECTURE 1 PROTEINS Functions in a Cell MOLECULAR MACHINES BUILDING BLOCKS of a CELL ARMS of a CELL ENZYMES - enzymatic catalysis of biochemical reactions
Protein Physics A. V. Finkelstein & O. B. Ptitsyn LECTURE 1 PROTEINS Functions in a Cell MOLECULAR MACHINES BUILDING BLOCKS of a CELL ARMS of a CELL ENZYMES - enzymatic catalysis of biochemical reactions
Molecular Docking. - Computational prediction of the structure of receptor-ligand complexes. Receptor: Protein Ligand: Protein or Small Molecule
 Scoring and Docking Molecular Docking - Computational prediction of the structure of receptor-ligand complexes Receptor: Protein Ligand: Protein or Small Molecule Protein-Protein Docking Protein-Small
Scoring and Docking Molecular Docking - Computational prediction of the structure of receptor-ligand complexes Receptor: Protein Ligand: Protein or Small Molecule Protein-Protein Docking Protein-Small
Statistical Mechanics, Kinetic Theory Ideal Gas. 8.01t Nov 22, 2004
 Statistical Mechanics, Kinetic Theory Ideal Gas 8.01t Nov 22, 2004 Statistical Mechanics and Thermodynamics Thermodynamics Old & Fundamental Understanding of Heat (I.e. Steam) Engines Part of Physics Einstein
Statistical Mechanics, Kinetic Theory Ideal Gas 8.01t Nov 22, 2004 Statistical Mechanics and Thermodynamics Thermodynamics Old & Fundamental Understanding of Heat (I.e. Steam) Engines Part of Physics Einstein
Peptide Bonds: Structure
 Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
(c) How would your answers to problem (a) change if the molecular weight of the protein was 100,000 Dalton?
 Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
A disaccharide is formed when a dehydration reaction joins two monosaccharides. This covalent bond is called a glycosidic linkage.
 CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
CH 5 Structure & Function of Large Molecules: Macromolecules Molecules of Life All living things are made up of four classes of large biological molecules: carbohydrates, lipids, proteins, and nucleic
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Chapter 6 An Overview of Organic Reactions
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 6 An Overview of Organic Reactions Why this chapter? To understand organic and/or biochemistry, it is necessary to know: -What occurs -Why and
Scoring Functions and Docking. Keith Davies Treweren Consultants Ltd 26 October 2005
 Scoring Functions and Docking Keith Davies Treweren Consultants Ltd 26 October 2005 Overview Applications Docking Algorithms Scoring Functions Results Demonstration Docking Applications Drug Design Lead
Scoring Functions and Docking Keith Davies Treweren Consultants Ltd 26 October 2005 Overview Applications Docking Algorithms Scoring Functions Results Demonstration Docking Applications Drug Design Lead
CHEM 120 Online Chapter 7
 CHEM 120 Online Chapter 7 Date: 1. Which of the following statements is not a part of kinetic molecular theory? A) Matter is composed of particles that are in constant motion. B) Particle velocity increases
CHEM 120 Online Chapter 7 Date: 1. Which of the following statements is not a part of kinetic molecular theory? A) Matter is composed of particles that are in constant motion. B) Particle velocity increases
Why? Intermolecular Forces. Intermolecular Forces. Chapter 12 IM Forces and Liquids. Covalent Bonding Forces for Comparison of Magnitude
 1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
1 Why? Chapter 1 Intermolecular Forces and Liquids Why is water usually a liquid and not a gas? Why does liquid water boil at such a high temperature for such a small molecule? Why does ice float on water?
Lecture 15: Enzymes & Kinetics Mechanisms
 ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
2054-2. Structure and Dynamics of Hydrogen-Bonded Systems. 26-27 October 2009. Hydrogen Bonds and Liquid Water
 2054-2 Structure and Dynamics of Hydrogen-Bonded Systems 26-27 October 2009 Hydrogen Bonds and Liquid Water Ruth LYNDEN-BELL University of Cambridge, Department of Chemistry Lensfield Road Cambridge CB2
2054-2 Structure and Dynamics of Hydrogen-Bonded Systems 26-27 October 2009 Hydrogen Bonds and Liquid Water Ruth LYNDEN-BELL University of Cambridge, Department of Chemistry Lensfield Road Cambridge CB2
CSC 2427: Algorithms for Molecular Biology Spring 2006. Lecture 16 March 10
 CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
7. 1.00 atm = 760 torr = 760 mm Hg = 101.325 kpa = 14.70 psi. = 0.446 atm. = 0.993 atm. = 107 kpa 760 torr 1 atm 760 mm Hg = 790.
 CHATER 3. The atmosphere is a homogeneous mixture (a solution) of gases.. Solids and liquids have essentially fixed volumes and are not able to be compressed easily. have volumes that depend on their conditions,
CHATER 3. The atmosphere is a homogeneous mixture (a solution) of gases.. Solids and liquids have essentially fixed volumes and are not able to be compressed easily. have volumes that depend on their conditions,
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
The peptide bond is rigid and planar
 Level Description Bonds Primary Sequence of amino acids in proteins Covalent (peptide bonds) Secondary Structural motifs in proteins: α- helix and β-sheet Hydrogen bonds (between NH and CO groups in backbone)
Level Description Bonds Primary Sequence of amino acids in proteins Covalent (peptide bonds) Secondary Structural motifs in proteins: α- helix and β-sheet Hydrogen bonds (between NH and CO groups in backbone)
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 4 ENZYMATIC CATALYSIS
 ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 4 ENZYMATIC CATALYSIS We will continue today our discussion on enzymatic catalysis
ENZYME SCIENCE AND ENGINEERING PROF. SUBHASH CHAND DEPARTMENT OF BIOCHEMICAL ENGINEERING AND BIOTECHNOLOGY IIT DELHI LECTURE 4 ENZYMATIC CATALYSIS We will continue today our discussion on enzymatic catalysis
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush α-keratins, bundles of α- helices Contain polypeptide chains organized approximately parallel along a single axis: Consist
Chapter 2 Polar Covalent Bonds: Acids and Bases
 John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds: Acids and Bases Modified by Dr. Daniela R. Radu Why This Chapter? Description of basic ways chemists account for chemical
John E. McMurry www.cengage.com/chemistry/mcmurry Chapter 2 Polar Covalent Bonds: Acids and Bases Modified by Dr. Daniela R. Radu Why This Chapter? Description of basic ways chemists account for chemical
Amino Acids, Peptides, Proteins
 Amino Acids, Peptides, Proteins Functions of proteins: Enzymes Transport and Storage Motion, muscle contraction Hormones Mechanical support Immune protection (Antibodies) Generate and transmit nerve impulses
Amino Acids, Peptides, Proteins Functions of proteins: Enzymes Transport and Storage Motion, muscle contraction Hormones Mechanical support Immune protection (Antibodies) Generate and transmit nerve impulses
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING
 CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
CHEMISTRY STANDARDS BASED RUBRIC ATOMIC STRUCTURE AND BONDING Essential Standard: STUDENTS WILL UNDERSTAND THAT THE PROPERTIES OF MATTER AND THEIR INTERACTIONS ARE A CONSEQUENCE OF THE STRUCTURE OF MATTER,
AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts
 AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts 8.1 Types of Chemical Bonds A. Ionic Bonding 1. Electrons are transferred 2. Metals react with nonmetals 3. Ions paired have lower energy
AP Chemistry A. Allan Chapter 8 Notes - Bonding: General Concepts 8.1 Types of Chemical Bonds A. Ionic Bonding 1. Electrons are transferred 2. Metals react with nonmetals 3. Ions paired have lower energy
Myoglobin and Hemoglobin
 Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Type of Chemical Bonds
 Type of Chemical Bonds Covalent bond Polar Covalent bond Ionic bond Hydrogen bond Metallic bond Van der Waals bonds. Covalent Bonds Covalent bond: bond in which one or more pairs of electrons are shared
Type of Chemical Bonds Covalent bond Polar Covalent bond Ionic bond Hydrogen bond Metallic bond Van der Waals bonds. Covalent Bonds Covalent bond: bond in which one or more pairs of electrons are shared
Transmembrane proteins span the bilayer. α-helix transmembrane domain. Multiple transmembrane helices in one polypeptide
 Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT
 BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
BIOLOGICAL MEMBRANES: FUNCTIONS, STRUCTURES & TRANSPORT UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY BMLS II / B Pharm II / BDS II VJ Temple
Biomolecular Modelling
 Biomolecular Modelling Carmen Domene Physical & Theoretical Chemistry Laboratory University of Oxford, UK THANKS Dr Joachim Hein Dr Iain Bethune Dr Eilidh Grant & Qi Huangfu 2 EPSRC Grant, Simulations
Biomolecular Modelling Carmen Domene Physical & Theoretical Chemistry Laboratory University of Oxford, UK THANKS Dr Joachim Hein Dr Iain Bethune Dr Eilidh Grant & Qi Huangfu 2 EPSRC Grant, Simulations
Built from 20 kinds of amino acids
 Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Test Review # 9. Chemistry R: Form TR9.13A
 Chemistry R: Form TR9.13A TEST 9 REVIEW Name Date Period Test Review # 9 Collision theory. In order for a reaction to occur, particles of the reactant must collide. Not all collisions cause reactions.
Chemistry R: Form TR9.13A TEST 9 REVIEW Name Date Period Test Review # 9 Collision theory. In order for a reaction to occur, particles of the reactant must collide. Not all collisions cause reactions.
Physics 176 Topics to Review For the Final Exam
 Physics 176 Topics to Review For the Final Exam Professor Henry Greenside May, 011 Thermodynamic Concepts and Facts 1. Practical criteria for identifying when a macroscopic system is in thermodynamic equilibrium:
Physics 176 Topics to Review For the Final Exam Professor Henry Greenside May, 011 Thermodynamic Concepts and Facts 1. Practical criteria for identifying when a macroscopic system is in thermodynamic equilibrium:
EXIT TIME PROBLEMS AND ESCAPE FROM A POTENTIAL WELL
 EXIT TIME PROBLEMS AND ESCAPE FROM A POTENTIAL WELL Exit Time problems and Escape from a Potential Well Escape From a Potential Well There are many systems in physics, chemistry and biology that exist
EXIT TIME PROBLEMS AND ESCAPE FROM A POTENTIAL WELL Exit Time problems and Escape from a Potential Well Escape From a Potential Well There are many systems in physics, chemistry and biology that exist
KINETIC THEORY OF MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature
 1 KINETIC TERY F MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature TE STATES F MATTER 1. Gas a) ideal gas - molecules move freely - molecules have
1 KINETIC TERY F MATTER - molecules in matter are always in motion - speed of molecules is proportional to the temperature TE STATES F MATTER 1. Gas a) ideal gas - molecules move freely - molecules have
The strength of the hydrogen bond in the linking of protein
 Energetics of hydrogen bonds in peptides Sheh-Yi Sheu*, Dah-Yen Yang, H. L. Selzle, and E. W. Schlag *Department of Life Science, National Yang-Ming University, Taipei 112, Taiwan; Institute of Atomic
Energetics of hydrogen bonds in peptides Sheh-Yi Sheu*, Dah-Yen Yang, H. L. Selzle, and E. W. Schlag *Department of Life Science, National Yang-Ming University, Taipei 112, Taiwan; Institute of Atomic
Chapter 7: Polarization
 Chapter 7: Polarization Joaquín Bernal Méndez Group 4 1 Index Introduction Polarization Vector The Electric Displacement Vector Constitutive Laws: Linear Dielectrics Energy in Dielectric Systems Forces
Chapter 7: Polarization Joaquín Bernal Méndez Group 4 1 Index Introduction Polarization Vector The Electric Displacement Vector Constitutive Laws: Linear Dielectrics Energy in Dielectric Systems Forces
Exam 4 Practice Problems false false
 Exam 4 Practice Problems 1 1. Which of the following statements is false? a. Condensed states have much higher densities than gases. b. Molecules are very far apart in gases and closer together in liquids
Exam 4 Practice Problems 1 1. Which of the following statements is false? a. Condensed states have much higher densities than gases. b. Molecules are very far apart in gases and closer together in liquids
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras
 Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Modern Construction Materials Prof. Ravindra Gettu Department of Civil Engineering Indian Institute of Technology, Madras Module - 2 Lecture - 2 Part 2 of 2 Review of Atomic Bonding II We will continue
Boyle s law - For calculating changes in pressure or volume: P 1 V 1 = P 2 V 2. Charles law - For calculating temperature or volume changes: V 1 T 1
 Common Equations Used in Chemistry Equation for density: d= m v Converting F to C: C = ( F - 32) x 5 9 Converting C to F: F = C x 9 5 + 32 Converting C to K: K = ( C + 273.15) n x molar mass of element
Common Equations Used in Chemistry Equation for density: d= m v Converting F to C: C = ( F - 32) x 5 9 Converting C to F: F = C x 9 5 + 32 Converting C to K: K = ( C + 273.15) n x molar mass of element
Biological Membranes. Impermeable lipid bilayer membrane. Protein Channels and Pores
 Biological Membranes Impermeable lipid bilayer membrane Protein Channels and Pores 1 Biological Membranes Are Barriers for Ions and Large Polar Molecules The Cell. A Molecular Approach. G.M. Cooper, R.E.
Biological Membranes Impermeable lipid bilayer membrane Protein Channels and Pores 1 Biological Membranes Are Barriers for Ions and Large Polar Molecules The Cell. A Molecular Approach. G.M. Cooper, R.E.
A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys
 Questions- Proteins & Enzymes A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys Reaction of the intact peptide
Questions- Proteins & Enzymes A. A peptide with 12 amino acids has the following amino acid composition: 2 Met, 1 Tyr, 1 Trp, 2 Glu, 1 Lys, 1 Arg, 1 Thr, 1 Asn, 1 Ile, 1 Cys Reaction of the intact peptide
http://faculty.sau.edu.sa/h.alshehri
 http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
http://faculty.sau.edu.sa/h.alshehri Definition: Proteins are macromolecules with a backbone formed by polymerization of amino acids. Proteins carry out a number of functions in living organisms: - They
Molecular Visualization. Introduction
 Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
LECTURE 11 : GLASSY DYNAMICS - Intermediate scattering function - Mean square displacement and beyond - Dynamic heterogeneities - Isoconfigurational
 LECTURE 11 : GLASSY DYNAMICS - Intermediate scattering function - Mean square displacement and beyond - Dynamic heterogeneities - Isoconfigurational Ensemble - Energy landscapes A) INTERMEDIATE SCATTERING
LECTURE 11 : GLASSY DYNAMICS - Intermediate scattering function - Mean square displacement and beyond - Dynamic heterogeneities - Isoconfigurational Ensemble - Energy landscapes A) INTERMEDIATE SCATTERING
Invariant residue-a residue that is always conserved. It is assumed that these residues are essential to the structure or function of the protein.
 Chapter 6 The amino acid side chains have polar and nonpolar properties, and the relative hydrophobicity of the amino acid side chains is critical for the folding and stability of a protein. The more hydrophobic
Chapter 6 The amino acid side chains have polar and nonpolar properties, and the relative hydrophobicity of the amino acid side chains is critical for the folding and stability of a protein. The more hydrophobic
The Role of Electric Polarization in Nonlinear optics
 The Role of Electric Polarization in Nonlinear optics Sumith Doluweera Department of Physics University of Cincinnati Cincinnati, Ohio 45221 Abstract Nonlinear optics became a very active field of research
The Role of Electric Polarization in Nonlinear optics Sumith Doluweera Department of Physics University of Cincinnati Cincinnati, Ohio 45221 Abstract Nonlinear optics became a very active field of research
Data Visualization for Atomistic/Molecular Simulations. Douglas E. Spearot University of Arkansas
 Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Data Visualization for Atomistic/Molecular Simulations Douglas E. Spearot University of Arkansas What is Atomistic Simulation? Molecular dynamics (MD) involves the explicit simulation of atomic scale particles
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Two Forms of Energy
 Module 2D - Energy and Metabolism Objective # 19 All living organisms require energy for survival. In this module we will examine some general principles about chemical reactions and energy usage within
Module 2D - Energy and Metabolism Objective # 19 All living organisms require energy for survival. In this module we will examine some general principles about chemical reactions and energy usage within
Chemistry Diagnostic Questions
 Chemistry Diagnostic Questions Answer these 40 multiple choice questions and then check your answers, located at the end of this document. If you correctly answered less than 25 questions, you need to
Chemistry Diagnostic Questions Answer these 40 multiple choice questions and then check your answers, located at the end of this document. If you correctly answered less than 25 questions, you need to
Amino Acids and Proteins
 Amino Acids and Proteins Proteins are composed of amino acids. There are 20 amino acids commonly found in proteins. All have: N2 C α R COO Amino acids at neutral p are dipolar ions (zwitterions) because
Amino Acids and Proteins Proteins are composed of amino acids. There are 20 amino acids commonly found in proteins. All have: N2 C α R COO Amino acids at neutral p are dipolar ions (zwitterions) because
States of Matter CHAPTER 10 REVIEW SECTION 1. Name Date Class. Answer the following questions in the space provided.
 CHAPTER 10 REVIEW States of Matter SECTION 1 SHORT ANSWER Answer the following questions in the space provided. 1. Identify whether the descriptions below describe an ideal gas or a real gas. ideal gas
CHAPTER 10 REVIEW States of Matter SECTION 1 SHORT ANSWER Answer the following questions in the space provided. 1. Identify whether the descriptions below describe an ideal gas or a real gas. ideal gas
Intermolecular Forces
 Intermolecular Forces: Introduction Intermolecular Forces Forces between separate molecules and dissolved ions (not bonds) Van der Waals Forces 15% as strong as covalent or ionic bonds Chapter 11 Intermolecular
Intermolecular Forces: Introduction Intermolecular Forces Forces between separate molecules and dissolved ions (not bonds) Van der Waals Forces 15% as strong as covalent or ionic bonds Chapter 11 Intermolecular
VAPORIZATION IN MORE DETAIL. Energy needed to escape into gas phase GAS LIQUID. Kinetic energy. Average kinetic energy
 30 VAPORIZATION IN MORE DETAIL GAS Energy needed to escape into gas phase LIQUID Kinetic energy Average kinetic energy - For a molecule to move from the liquid phase to the gas phase, it must acquire enough
30 VAPORIZATION IN MORE DETAIL GAS Energy needed to escape into gas phase LIQUID Kinetic energy Average kinetic energy - For a molecule to move from the liquid phase to the gas phase, it must acquire enough
1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA
 1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA Tomáš Kubař Institute of Organic Chemistry and Biochemistry Praha, Czech Republic Thermodynamic Integration Alchemical change
1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA Tomáš Kubař Institute of Organic Chemistry and Biochemistry Praha, Czech Republic Thermodynamic Integration Alchemical change
7. Gases, Liquids, and Solids 7.1 Kinetic Molecular Theory of Matter
 7. Gases, Liquids, and Solids 7.1 Kinetic Molecular Theory of Matter Kinetic Molecular Theory of Matter The Kinetic Molecular Theory of Matter is a concept that basically states that matter is composed
7. Gases, Liquids, and Solids 7.1 Kinetic Molecular Theory of Matter Kinetic Molecular Theory of Matter The Kinetic Molecular Theory of Matter is a concept that basically states that matter is composed
CASSANDRA. Computational Atomistic Simulation Software At Notre Dame for Research Advances
 1 CASSANDRA Computational Atomistic Simulation Software At Notre Dame for Research Advances User Manual 1.0 Written by: Edward J. Maginn, Jindal K. Shah, Eliseo MarinRimoldi, Sandip Khan, Neeraj Rai, Thomas
1 CASSANDRA Computational Atomistic Simulation Software At Notre Dame for Research Advances User Manual 1.0 Written by: Edward J. Maginn, Jindal K. Shah, Eliseo MarinRimoldi, Sandip Khan, Neeraj Rai, Thomas
Chapter 5: Diffusion. 5.1 Steady-State Diffusion
 : Diffusion Diffusion: the movement of particles in a solid from an area of high concentration to an area of low concentration, resulting in the uniform distribution of the substance Diffusion is process
: Diffusion Diffusion: the movement of particles in a solid from an area of high concentration to an area of low concentration, resulting in the uniform distribution of the substance Diffusion is process
MOLECULAR MODELING: MOLECULAR MECHANICS
 An Introduction to Computational Biochemistry. C. Stan Tsai Copyright 2002 by Wiley-Liss, Inc. ISBN: 0-471-40120-X 14 MOLECULAR MODELING: MOLECULAR MECHANICS Molecular modeling can be defined as an application
An Introduction to Computational Biochemistry. C. Stan Tsai Copyright 2002 by Wiley-Liss, Inc. ISBN: 0-471-40120-X 14 MOLECULAR MODELING: MOLECULAR MECHANICS Molecular modeling can be defined as an application
Chem 1A Exam 2 Review Problems
 Chem 1A Exam 2 Review Problems 1. At 0.967 atm, the height of mercury in a barometer is 0.735 m. If the mercury were replaced with water, what height of water (in meters) would be supported at this pressure?
Chem 1A Exam 2 Review Problems 1. At 0.967 atm, the height of mercury in a barometer is 0.735 m. If the mercury were replaced with water, what height of water (in meters) would be supported at this pressure?
Molecular Spectroscopy
 Molecular Spectroscopy UV-Vis Spectroscopy Absorption Characteristics of Some Common Chromophores UV-Vis Spectroscopy Absorption Characteristics of Aromatic Compounds UV-Vis Spectroscopy Effect of extended
Molecular Spectroscopy UV-Vis Spectroscopy Absorption Characteristics of Some Common Chromophores UV-Vis Spectroscopy Absorption Characteristics of Aromatic Compounds UV-Vis Spectroscopy Effect of extended
1.1.2. thebiotutor. AS Biology OCR. Unit F211: Cells, Exchange & Transport. Module 1.2 Cell Membranes. Notes & Questions.
 thebiotutor AS Biology OCR Unit F211: Cells, Exchange & Transport Module 1.2 Cell Membranes Notes & Questions Andy Todd 1 Outline the roles of membranes within cells and at the surface of cells. The main
thebiotutor AS Biology OCR Unit F211: Cells, Exchange & Transport Module 1.2 Cell Membranes Notes & Questions Andy Todd 1 Outline the roles of membranes within cells and at the surface of cells. The main
