S1. Sample applications of the PRS method
|
|
|
- Abel Tate
- 8 years ago
- Views:
Transcription
1 S1. Sample applications of the PRS method In this section we provide results from the PRS method on different protein systems. Each example illustrates a different situation linked to mechanical changes in the system. A. Stabilization of Closed Conformation by Ligand Binding: K- R- Orn- Binding Protein The periplasmic lysine/arginine/ornithine binding protein (LAO) is a 238 residue long chain that contains two lobes moving relative to each other upon binding the substrate. Its apo- and holo (lysine bound) structures have been determined at 1.8 and 1.9 Å resolution, respectively, by x-ray crystallography [1]. It is one of the earlier examples whereby it was claimed that the role of the ligand is to stabilize the closed conformation upon binding, as opposed to the previous view of a direct conformational change induced by the ligand. We have applied the PRS method to both the apo and the holo forms of this protein (PDB codes 2lao and 1lst, respectively; the overall RMSD between the two structures is 4.7 Å; r c = 9 Å). The results are summarized in Figure S1. In the apo form, a large number of residues in different parts of the protein yield high correlations (C i in equation 19) with the displacements obtained from x- ray structures (102 residues have C i 0.8). The holo form, on the other hand, is stabilized by the ligand, and perturbing only those residues in close proximity to the ligand (shown in orange in the figure) lead to higher C i values. Unlike FBP, long range contacts whose perturbations manipulate the ligand-binding region are not detected in LAO. Figure A. (Left panel) The correlation coefficient between the displacement profiles from PRS and x-ray structures, for the apo (black curve), and the holo form (gray curve) as the initial structure; this is the counterpart of figures 4a and 4b for LAO. (Right panel) The x-ray structure of the holo form; the ligand lysine is colored in olive. The residues whose perturbation leads to higher correlations in the holo form are shown in orange in both panels, and all belong to the ligand binding site.
2 B. Allosteric Control in CheY The chemotaxis protein Y (CheY) is a response regulator in two-component signal transduction systems. Its apo and holo (Mg +2 ion bound) structures have been determined at 1.7 and 1.8 Å resolution, respectively [2,3]. As a simple example of protein allostery, the activation mechanism in CheY was studied by molecular simulation [4]. Therein, it was proposed that the 4 4 loop acts as a gating element that remotely regulates the population of the active loop configuration. We have studied the apo-form of CheY with PRS methodology (PDB code of apo and holo structures are 3chy and 1chn, respectively; the overall RMSD between the two structures is 1.4 Å; r c = 8 Å). The results are displayed in figure S2. The Mg +2 ion in the holo form coordinates residues 13, 57 and 59 (highlighted in olive in figure S2) along with three water molecules. The 4 4 loop is highlighted in orange. In conformity with the above-mentioned allosteric role of this loop, we find that forces exerted on this flexible region induce displacements on the order of 1 Å around the binding site. Figure B. (Bottom panel) The correlation coefficient between the displacement profiles from PRS and x-ray structures. The CheY apo-structure is shown in the inset. The residues directly contacting the Mg +2 ion in the bound form and the 4 4 loop residues are highlighted in the colors olive and orange, respectively. (Top panel) A sample displacement profile, as a result of a perturbation on residue 91 which resides in the 4 4 loop is exhibited. The displacements from the x-ray coordinates are also shown for comparison; residue 91 gives the highest displacement similarity to those recorded in the x-ray structures (the peak in the bottom panel curve). C. Open Closed Form Conformational Change in Bacteriorhodopsin in the Absence of Illumination: Bacteriorhodopsin functions as a light-driven proton pump residing in the cell membrane of halobacterium salinarum. It contains the chromophore retinal, which isomerizes to 13-cis retinal by absorbing a photon. This is followed by protein conformational changes occurring in the dark, resulting in the translocation of one or more protons through the membrane [5]. It was shown by
3 electron microscopy and later confirmed by x-ray crystallography that, even in the absence of illumination, selected mutations could be used to partially destabilize the native conformation towards the open conformation [6,7]. In the extreme, it has even been possible to obtain the full conformational change using the triple mutant D96G/F171C/F219L, without any further significant changes upon illumination. We have scanned the native conformation of bacteriorhodopsin to understand the extent to which singly inserted perturbations on the structure may drive it towards the open form. The results are displayed in figure S3 (PDB code of native and open conformations are 1fbb and 1fbk, respectively; the overall RMSD between the two structures is 0.95 Å; r c = 9 Å). We find that several residue residing on the cytoplasmic side of the F and G helices may be perturbed to yield average correlations of with the experimentally determined conformational change. In particular, perturbations on residue T170 lead to the average correlation of ca Note that two of the residues in the experimentally determined triple mutant reside in the cytoplasmic side of helices F and G [8]. Figure C. (Bottom left panel) The correlation coefficient between the displacement profiles of the native and open conformations of bacteriorhodopsin from PRS and experimental structures. The residues yielding the highest correlations with the open conformation reside in the cytoplasmic side F and G helices, and are colored in purple and cyan, respectively. (Top left panel) A displacement profile, as a result of a perturbation on T170 which resides in the F helix. The displacements from the x-ray coordinates are also shown for comparison; this residue gives the highest displacement similarity to those recorded in the x-ray structures. (Right panel) The native conformation. The seven transmembrane helices are each colored differently and labeled. Retinal is shown in red ball-and-stick representation. Residues leading to the largest correlation with the experimental overall displacements upon singly inserted perturbations are colored in purple and cyan, in accord with the curve in the bottom left panel. D. Binding affinity of an antibody to different epitopes. In this example, we use the PRS methodology for a different purpose, namely, to differentiate the binding affinities of different ligands to the same protein. This is based on the argument that we expect tighter binding to be associated with inducing larger correlated motions in the overall protein. To test this idea, we use the interesting example of promiscuity in the primary immune response, that has been presented on the germline antibody [9]. In this system, four distinct conformations are displayed in the unliganded state; yet, a common one of these is stabilized upon complexation with three different dodecapeptides. Interestingly, the positions of the peptides are different in the combining site, sharing only two Tyr residues. The structure of this
4 common conformation is shown in figure S4, and the two paratope Tyr residues (Y50 and Y106) that reside in the variable domain are shown in space filling representation. We have scanned this common conformation (PDB code 2a6j) using the PRS method (r c = 11 Å); results are shown in figure S4. The three peptides whose bound states are studied are RLLIADPPSPRE, KLASIPTHTSPL and SLGDNLTNHNLR (PDB codes 2a6d, 2a6i and 2a6k, respectively), and are labeled by the first three residues. The RMSD between the unbound and bound structures of the variable domain (residues 1-128) are 0.98, 1.1 and 1.1 Å, respectively. Since the structural differences in the constant domain are very small, the forces inserted on residues in that region do not yield discriminating results. More interesting findings are obtained by perturbing the variable domain residues, whereby the degree of achieving higher correlations is in the order SLG > RLL > KLA; this is in accord with the measured [10] binding affinities of these peptides (0.21, 0.15 and 0.12 M, respectively). Figure D. (Top panel) The x-ray structure of the common peptide-bound conformation of the antibody heavy (H) chain. The variable (containing V H and C H1 ) and constant domain residues (containing C H2 and C H3 ) are colored in gray and purple, respectively. (Bottom panel) The correlation coefficient between the PRS and x-ray structure displacement profiles of the unbound and bound structures for the three peptides. The higher correlations obtained by perturbing the variable domain residues are in accord with the binding affinities of the peptides. E. N-lobe of human transferrin As discussed at length in the Introduction, human transferrin (ht) has many structural features common to bacterial FBP. The three-dimensional structure of only the apo form of full length ht is available. However, the x-ray structures of the N-lobe of apo- and holo-ht have been determined to 1.6 and 2.2 Å, respectively (PDB codes 1BP5 [11] and 1A8E [12]). Sequence identity of N-lobe ht with FBP is 12 % and sequence similarity is ca. 62 %; ht has 337 residues as opposed to FBP having 309. Structural alignment using MultiProt [13] of bacterial FBP and human transferrin N-lobe, both in the Fe bound form, yields 187 residues aligned at an RMSD of 3.48 Å. Although the overall structure with the two domains enclosing the Fe ion persists in N- lobe ht with many of the secondary structural elements having corresponding locations, overall
5 there are structural shifts as well as loop insertions in ht. In particular, the Fe ion is located deeper in the groove between the two domains. As a result, there are 16 residues whose C atoms reside within 8 Å of the ion. Six of these (L62, D63, A64, G65, S248, H249) are located in the moving domain and the others (R124, S125, A126, G127, Y188, V205, K206, V246) are in the fixed domain. The structure of holo-ht is shown in the top panel of Figure S5, and the residues in contact with Fe ion are marked on the figure. By applying PRS on the apo form (using r c = 8 Å as in FBP), we find that 60 % of the residues yield correlations better than 0.9. However, only five residues (A10, N183, K278, A299, H300) yield such good correlations when the holo form is perturbed. These results are shown in orange on Figure S5. Of these five residues, K278, which is 25 Å away from the ion, is found to organize the moving domain residues in contact with the ligand, as shown in the inset to the bottom panel of Figure S5. All six residues move parallel to each other when K278 is perturbed, with the angle between any two pairs (equation 21) being in the interval Moreover, the response is collected along a line on all the six residues (i.e. the fractional contribution of the highest eigenvalue p 1 of the response sub-matrix (equation 20) is in the range for these residues). Such a collectivity, along with the correct displacement profile of the residues, is obtained only when this distant residue is perturbed. Note that K278 has also been identified as an allosteric kinetically significant anion binding site in MD/molecular mechanics simulations in the presence of Cl - [14]. Figure E. (Top) The x-ray structure of the N-lobe of ht, holo form. Fe ion is shown in red. Residues whose C atoms are within 8 Å of the ion are shown in green (moving domain) and pink (fixed domain), respectively. (Bottom) The correlation coefficient between the displacement profiles from PRS and x-ray structures, for the apo (upper curve), and the holo form (lower curve) as the initial structure, sorted from largest to smallest value (counterpart of figure 4c for ht). Residues A10, N183, K278, A299, H300 (marked in orange on the x-ray structure) give the highest correlation (C i > 0.9) in the holo form; conversely, 60 % of the perturbations lead to the large correlations in the apo form. (Inset) Magnified response of the Fecontacting residues to random perturbations on K278 induce a correlated response in the moving domain residues, all six having a preferred direction (green), whereas the displacements in the fixed domain residues (pink) are uncorrelated.
6 References 1. Oh BH, Pandit J, Kang CH, Nikaido K, Gokcen S, et al. (1993) 3-Dimensional Structures of the Periplasmic Lysine Arginine Ornithine-Binding Protein with and without a Ligand. Journal of Biological Chemistry 268: Bellsolell L, Prieto J, Serrano L, Coll M (1994) Magnesium Binding to the Bacterial Chemotaxis Protein Chey Results in Large Conformational-Changes Involving Its Functional Surface. Journal of Molecular Biology 238: Volz K, Matsumura P (1991) Crystal-Structure of Escherichia-Coli Chey Refined at 1.7-Å Resolution. Journal of Biological Chemistry 266: Formaneck MS, Ma L, Cui Q (2006) Reconciling the "old" and "new" views of protein allostery: A molecular simulation study of chemotaxis Y protein (CheY). Proteins-Structure Function and Bioinformatics 63: Kaulen AD (2000) Electrogenic processes and protein conformational changes accompanying the bacteriorhodopsin photocycle. Biochimica Et Biophysica Acta-Bioenergetics 1460: Hirai T, Subramaniam S (2009) Protein Conformational Changes in the Bacteriorhodopsin Photocycle: Comparison of Findings from Electron and X-Ray Crystallographic Analyses. PLoS One 4: e Subramaniam S, Lindahl I, Bullough P, Faruqi AR, Tittor J, et al. (1999) Protein conformational changes in the bacteriorhodopsin photocycle. Journal of Molecular Biology 287: Subramaniam S, Hirai T, Henderson R (2002) From structure to mechanism: electron crystallographic studies of bacteriorhodopsin. Philosophical Transactions of the Royal Society of London Series a- Mathematical Physical and Engineering Sciences 360: Sethi DK, Agarwal A, Manivel V, Rao KVS, Salunke DM (2006) Differential epitope positioning within the germline antibody paratope enhances promiscuity in the primary immune response. Immunity 24: Manivel V, Bayiroglu F, Siddiqui Z, Salunke DM, Rao KVS (2002) The primary antibody repertoire represents a linked network of degenerate antigen specificities. Journal of Immunology 169: Jeffrey PD, Bewley MC, MacGillivray RTA, Mason AB, Woodworth RC, et al. (1998) Ligandinduced conformational change in transferrins: Crystal structure of the open form of the N- terminal half-molecule of human transferrin. Biochemistry 37: MacGillivray RTA, Moore SA, Chen J, Anderson BF, Baker H, et al. (1998) Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release. Biochemistry 37: Shatsky M, Nussinov R, Wolfson HJ (2004) A method for simultaneous alignment of multiple protein structures. Proteins-Structure Function and Bioinformatics 56: Amin EA, Harris WR, Welsh WJ (2004) Identification of possible kinetically significant anionbinding sites in human serum transferrin using molecular modeling strategies. Biopolymers 73:
Analysis of structural dynamics by H/D-exchange coupled to mass spectrometry HDX-MS
 Analysis of structural dynamics by H/D-exchange coupled to mass spectrometry () New Approaches in Drug Design & Discovery 2014 25 th of March 2014 Introduction What are the challenges in structure-based
Analysis of structural dynamics by H/D-exchange coupled to mass spectrometry () New Approaches in Drug Design & Discovery 2014 25 th of March 2014 Introduction What are the challenges in structure-based
VTT TECHNICAL RESEARCH CENTRE OF FINLAND
 Figure from: http://www.embl.de/nmr/sattler/teaching Why NMR (instead of X ray crystallography) a great number of macromolecules won't crystallize) natural environmant (water) ligand binding and inter
Figure from: http://www.embl.de/nmr/sattler/teaching Why NMR (instead of X ray crystallography) a great number of macromolecules won't crystallize) natural environmant (water) ligand binding and inter
Supplementary Figures S1 - S11
 1 Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations Hang Yu 1,2, Klaus Schulten 1,2,3, 1 Beckman Institute, University of Illinois, Urbana, Illinois, USA 2 Center of Biophysics
1 Membrane Sculpting by F-BAR Domains Studied by Molecular Dynamics Simulations Hang Yu 1,2, Klaus Schulten 1,2,3, 1 Beckman Institute, University of Illinois, Urbana, Illinois, USA 2 Center of Biophysics
Refinement of a pdb-structure and Convert
 Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
Refinement of a pdb-structure and Convert A. Search for a pdb with the closest sequence to your protein of interest. B. Choose the most suitable entry (or several entries). C. Convert and resolve errors
PROTEINS THE PEPTIDE BOND. The peptide bond, shown above enclosed in the blue curves, generates the basic structural unit for proteins.
 Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
Ca 2+ The contents of this module were developed under grant award # P116B-001338 from the Fund for the Improvement of Postsecondary Education (FIPSE), United States Department of Education. However, those
CHAPTER 8 IMMUNOLOGICAL IMPLICATIONS OF PEPTIDE CARBOHYDRATE MIMICRY
 CHAPTER 8 IMMUNOLOGICAL IMPLICATIONS OF PEPTIDE CARBOHYDRATE MIMICRY Immunological Implications of Peptide-Carbohydrate Mimicry 8.1 Introduction The two chemically dissimilar molecules, a peptide (12mer)
CHAPTER 8 IMMUNOLOGICAL IMPLICATIONS OF PEPTIDE CARBOHYDRATE MIMICRY Immunological Implications of Peptide-Carbohydrate Mimicry 8.1 Introduction The two chemically dissimilar molecules, a peptide (12mer)
Helices From Readily in Biological Structures
 The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The α Helix and the β Sheet Are Common Folding Patterns Although the overall conformation each protein is unique, there are only two different folding patterns are present in all proteins, which are α
The immune response Antibodies Antigens Epitopes (antigenic determinants) the part of a protein antigen recognized by an antibody Haptens small
 The immune response Antibodies Antigens Epitopes (antigenic determinants) the part of a protein antigen recognized by an antibody Haptens small molecules that can elicit an immune response when linked
The immune response Antibodies Antigens Epitopes (antigenic determinants) the part of a protein antigen recognized by an antibody Haptens small molecules that can elicit an immune response when linked
Myoglobin and Hemoglobin
 Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Myoglobin and Hemoglobin Myoglobin and hemoglobin are hemeproteins whose physiological importance is principally related to their ability to bind molecular oxygen. Myoglobin (Mb) The oxygen storage protein
Biochemistry 462a Hemoglobin Structure and Function Reading - Chapter 7 Practice problems - Chapter 7: 1-6; Proteins extra problems
 Biochemistry 462a Hemoglobin Structure and Function Reading - Chapter 7 Practice problems - Chapter 7: 1-6; Proteins extra problems Myoglobin and Hemoglobin Oxygen is required for oxidative metabolism
Biochemistry 462a Hemoglobin Structure and Function Reading - Chapter 7 Practice problems - Chapter 7: 1-6; Proteins extra problems Myoglobin and Hemoglobin Oxygen is required for oxidative metabolism
Guidance for Industry
 Guidance for Industry Interpreting Sameness of Monoclonal Antibody Products Under the Orphan Drug Regulations U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation
Guidance for Industry Interpreting Sameness of Monoclonal Antibody Products Under the Orphan Drug Regulations U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation
Therapeutic Systems Immunology
 Therapeutic Systems Immunology Coordination: Robert Preissner, Charité, Berlin 13 Partners from Berlin (7) Göttingen (1) Heidelberg (1) Rostock (2) Tübingen (1) Overview Intro / Mission Partners Systems
Therapeutic Systems Immunology Coordination: Robert Preissner, Charité, Berlin 13 Partners from Berlin (7) Göttingen (1) Heidelberg (1) Rostock (2) Tübingen (1) Overview Intro / Mission Partners Systems
Bioinformatics for Biologists. Protein Structure
 Bioinformatics for Biologists Comparative Protein Analysis: Part III. Protein Structure Prediction and Comparison Robert Latek, PhD Sr. Bioinformatics Scientist Whitehead Institute for Biomedical Research
Bioinformatics for Biologists Comparative Protein Analysis: Part III. Protein Structure Prediction and Comparison Robert Latek, PhD Sr. Bioinformatics Scientist Whitehead Institute for Biomedical Research
Papers listed: Cell2. This weeks papers. Chapt 4. Protein structure and function
 Papers listed: Cell2 During the semester I will speak of information from several papers. For many of them you will not be required to read these papers, however, you can do so for the fun of it (and it
Papers listed: Cell2 During the semester I will speak of information from several papers. For many of them you will not be required to read these papers, however, you can do so for the fun of it (and it
Amino Acids. Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain. Alpha Carbon. Carboxyl. Group.
 Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Protein Structure Amino Acids Amino acids are the building blocks of proteins. All AA s have the same basic structure: Side Chain Alpha Carbon Amino Group Carboxyl Group Amino Acid Properties There are
Chapter 3. Protein Structure and Function
 Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Structure Determination
 5 Structure Determination Most of the protein structures described and discussed in this book have been determined either by X-ray crystallography or by nuclear magnetic resonance (NMR) spectroscopy. Although
5 Structure Determination Most of the protein structures described and discussed in this book have been determined either by X-ray crystallography or by nuclear magnetic resonance (NMR) spectroscopy. Although
AP BIOLOGY 2008 SCORING GUIDELINES
 AP BIOLOGY 2008 SCORING GUIDELINES Question 1 1. The physical structure of a protein often reflects and affects its function. (a) Describe THREE types of chemical bonds/interactions found in proteins.
AP BIOLOGY 2008 SCORING GUIDELINES Question 1 1. The physical structure of a protein often reflects and affects its function. (a) Describe THREE types of chemical bonds/interactions found in proteins.
Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert Karakaş, Phoebe L. Stewart, and Jens Meiler
 Structure 17 Supplemental Data EM-Fold: De Novo Folding of α-helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert
Structure 17 Supplemental Data EM-Fold: De Novo Folding of α-helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps Steffen Lindert, René Staritzbichler, Nils Wötzel, Mert
Photosystems I and II
 Photosystems I and II March 17, 2003 Bryant Miles Within the thylakoid membranes of the chloroplast, are two photosystems. Photosystem I optimally absorbs photons of a wavelength of 700 nm. Photosystem
Photosystems I and II March 17, 2003 Bryant Miles Within the thylakoid membranes of the chloroplast, are two photosystems. Photosystem I optimally absorbs photons of a wavelength of 700 nm. Photosystem
Protein engineering for structural biology
 Protein engineering for structural biology J. Michael Sauder, Ph.D. Lilly Biotechnology Center Eli Lilly and Company San Diego, CA 92121 michael.sauder@lilly.com PSDI 2012, Nov 12-13 Structural Biology
Protein engineering for structural biology J. Michael Sauder, Ph.D. Lilly Biotechnology Center Eli Lilly and Company San Diego, CA 92121 michael.sauder@lilly.com PSDI 2012, Nov 12-13 Structural Biology
2007 7.013 Problem Set 1 KEY
 2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
2007 7.013 Problem Set 1 KEY Due before 5 PM on FRIDAY, February 16, 2007. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Where in a eukaryotic cell do you
Transmembrane proteins span the bilayer. α-helix transmembrane domain. Multiple transmembrane helices in one polypeptide
 Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
Transmembrane proteins span the bilayer α-helix transmembrane domain Hydrophobic R groups of a.a. interact with fatty acid chains Multiple transmembrane helices in one polypeptide Polar a.a. Hydrophilic
8/20/2012 H C OH H R. Proteins
 Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
Proteins Rubisco monomer = amino acids 20 different amino acids polymer = polypeptide protein can be one or more polypeptide chains folded & bonded together large & complex 3-D shape hemoglobin Amino acids
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon. V. Polypeptides and Proteins
 IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbon A. Acid/Base properties 1. carboxyl group is proton donor! weak acid 2. amino group is proton acceptor! weak base 3. At physiological ph: H
Review of Chemical Equilibrium 7.51 September 1999. free [A] (µm)
![Review of Chemical Equilibrium 7.51 September 1999. free [A] (µm) Review of Chemical Equilibrium 7.51 September 1999. free [A] (µm)](/thumbs/40/21435474.jpg) Review of Chemical Equilibrium 7.51 September 1999 Equilibrium experiments study how the concentration of reaction products change as a function of reactant concentrations and/or reaction conditions. For
Review of Chemical Equilibrium 7.51 September 1999 Equilibrium experiments study how the concentration of reaction products change as a function of reactant concentrations and/or reaction conditions. For
Increasing the Multiplexing of High Resolution Targeted Peptide Quantification Assays
 Increasing the Multiplexing of High Resolution Targeted Peptide Quantification Assays Scheduled MRM HR Workflow on the TripleTOF Systems Jenny Albanese, Christie Hunter AB SCIEX, USA Targeted quantitative
Increasing the Multiplexing of High Resolution Targeted Peptide Quantification Assays Scheduled MRM HR Workflow on the TripleTOF Systems Jenny Albanese, Christie Hunter AB SCIEX, USA Targeted quantitative
Combinatorial Biochemistry and Phage Display
 Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
Combinatorial Biochemistry and Phage Display Prof. Valery A. Petrenko Director - Valery Petrenko Instructors Galina Kouzmitcheva and I-Hsuan Chen Auburn 2006, Spring semester COMBINATORIAL BIOCHEMISTRY
6.5 Periodic Variations in Element Properties
 324 Chapter 6 Electronic Structure and Periodic Properties of Elements 6.5 Periodic Variations in Element Properties By the end of this section, you will be able to: Describe and explain the observed trends
324 Chapter 6 Electronic Structure and Periodic Properties of Elements 6.5 Periodic Variations in Element Properties By the end of this section, you will be able to: Describe and explain the observed trends
Blood-Based Cancer Diagnostics
 The Biotechnology Education Company Blood-Based Cancer Diagnostics EDVO-Kit 141 Store entire experiment at room temperature. EXPERIMENT OBJECTIVE: The objective of this experiment is to learn and understand
The Biotechnology Education Company Blood-Based Cancer Diagnostics EDVO-Kit 141 Store entire experiment at room temperature. EXPERIMENT OBJECTIVE: The objective of this experiment is to learn and understand
Biomolecular Modelling
 Biomolecular Modelling Carmen Domene Physical & Theoretical Chemistry Laboratory University of Oxford, UK THANKS Dr Joachim Hein Dr Iain Bethune Dr Eilidh Grant & Qi Huangfu 2 EPSRC Grant, Simulations
Biomolecular Modelling Carmen Domene Physical & Theoretical Chemistry Laboratory University of Oxford, UK THANKS Dr Joachim Hein Dr Iain Bethune Dr Eilidh Grant & Qi Huangfu 2 EPSRC Grant, Simulations
PHYSIOLOGY AND MAINTENANCE Vol. II - On The Determination of Enzyme Structure, Function, and Mechanism - Glumoff T.
 ON THE DETERMINATION OF ENZYME STRUCTURE, FUNCTION, AND MECHANISM University of Oulu, Finland Keywords: enzymes, protein structure, X-ray crystallography, bioinformatics Contents 1. Introduction 2. Structure
ON THE DETERMINATION OF ENZYME STRUCTURE, FUNCTION, AND MECHANISM University of Oulu, Finland Keywords: enzymes, protein structure, X-ray crystallography, bioinformatics Contents 1. Introduction 2. Structure
Structure Check. Authors: Eduard Schreiner Leonardo G. Trabuco. February 7, 2012
 University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop Structure Check Authors: Eduard Schreiner Leonardo
18.2 Protein Structure and Function: An Overview
 18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
18.2 Protein Structure and Function: An Overview Protein: A large biological molecule made of many amino acids linked together through peptide bonds. Alpha-amino acid: Compound with an amino group bonded
Antibody Function & Structure
 Antibody Function & Structure Specifically bind to antigens in both the recognition phase (cellular receptors) and during the effector phase (synthesis and secretion) of humoral immunity Serology: the
Antibody Function & Structure Specifically bind to antigens in both the recognition phase (cellular receptors) and during the effector phase (synthesis and secretion) of humoral immunity Serology: the
Role of Hydrogen Bonding on Protein Secondary Structure Introduction
 Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
Role of Hydrogen Bonding on Protein Secondary Structure Introduction The function and chemical properties of proteins are determined by its three-dimensional structure. The final architecture of the protein
(c) How would your answers to problem (a) change if the molecular weight of the protein was 100,000 Dalton?
 Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Problem 1. (12 points total, 4 points each) The molecular weight of an unspecified protein, at physiological conditions, is 70,000 Dalton, as determined by sedimentation equilibrium measurements and by
Some terms: An antigen is a molecule or pathogen capable of eliciting an immune response
 Overview of the immune system We continue our discussion of protein structure by considering the structure of antibodies. All organisms are continually subject to attack by microorganisms and viruses.
Overview of the immune system We continue our discussion of protein structure by considering the structure of antibodies. All organisms are continually subject to attack by microorganisms and viruses.
博 士 論 文 ( 要 約 ) A study on enzymatic synthesis of. stable cyclized peptides which. inhibit protein-protein interactions
 博 士 論 文 ( 要 約 ) 論 文 題 目 A study on enzymatic synthesis of stable cyclized peptides which inhibit protein-protein interactions ( 蛋 白 質 間 相 互 作 用 を 阻 害 する 安 定 な 環 状 化 ペプチドの 酵 素 合 成 に 関 する 研 究 ) 氏 名 張 静 1
博 士 論 文 ( 要 約 ) 論 文 題 目 A study on enzymatic synthesis of stable cyclized peptides which inhibit protein-protein interactions ( 蛋 白 質 間 相 互 作 用 を 阻 害 する 安 定 な 環 状 化 ペプチドの 酵 素 合 成 に 関 する 研 究 ) 氏 名 張 静 1
KMS-Specialist & Customized Biosimilar Service
 KMS-Specialist & Customized Biosimilar Service 1. Polyclonal Antibody Development Service KMS offering a variety of Polyclonal Antibody Services to fit your research and production needs. we develop polyclonal
KMS-Specialist & Customized Biosimilar Service 1. Polyclonal Antibody Development Service KMS offering a variety of Polyclonal Antibody Services to fit your research and production needs. we develop polyclonal
Hydrogen Bonds The electrostatic nature of hydrogen bonds
 Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Hydrogen Bonds Hydrogen bonds have played an incredibly important role in the history of structural biology. Both the structure of DNA and of protein a-helices and b-sheets were predicted based largely
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7. 4. Which of the following weak acids would make the best buffer at ph = 5.0?
 Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Paper: 6 Chemistry 2.130 University I Chemistry: Models Page: 2 of 7 4. Which of the following weak acids would make the best buffer at ph = 5.0? A) Acetic acid (Ka = 1.74 x 10-5 ) B) H 2 PO - 4 (Ka =
Phase determination methods in macromolecular X- ray Crystallography
 Phase determination methods in macromolecular X- ray Crystallography Importance of protein structure determination: Proteins are the life machinery and are very essential for the various functions in the
Phase determination methods in macromolecular X- ray Crystallography Importance of protein structure determination: Proteins are the life machinery and are very essential for the various functions in the
ENZYMES. Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin. Principle of Enzyme Catalysis
 ENZYMES Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin Principle of Enzyme Catalysis Linus Pauling (1946) formulated the first basic principle of enzyme catalysis Enzyme increase the rate
ENZYMES Serine Proteases Chymotrypsin, Trypsin, Elastase, Subtisisin Principle of Enzyme Catalysis Linus Pauling (1946) formulated the first basic principle of enzyme catalysis Enzyme increase the rate
NO CALCULATORS OR CELL PHONES ALLOWED
 Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Biol 205 Exam 1 TEST FORM A Spring 2008 NAME Fill out both sides of the Scantron Sheet. On Side 2 be sure to indicate that you have TEST FORM A The answers to Part I should be placed on the SCANTRON SHEET.
Lecture 19: Proteins, Primary Struture
 CPS260/BGT204.1 Algorithms in Computational Biology November 04, 2003 Lecture 19: Proteins, Primary Struture Lecturer: Pankaj K. Agarwal Scribe: Qiuhua Liu 19.1 The Building Blocks of Protein [1] Proteins
CPS260/BGT204.1 Algorithms in Computational Biology November 04, 2003 Lecture 19: Proteins, Primary Struture Lecturer: Pankaj K. Agarwal Scribe: Qiuhua Liu 19.1 The Building Blocks of Protein [1] Proteins
Ch. 9 - Electron Organization. The Bohr Model [9.4] Orbitals [9.5, 9.6] Counting Electrons, configurations [9.7]
![Ch. 9 - Electron Organization. The Bohr Model [9.4] Orbitals [9.5, 9.6] Counting Electrons, configurations [9.7] Ch. 9 - Electron Organization. The Bohr Model [9.4] Orbitals [9.5, 9.6] Counting Electrons, configurations [9.7]](/thumbs/27/11797291.jpg) Ch. 9 - Electron Organization The Bohr Model [9.4] Orbitals [9.5, 9.6] Counting Electrons, configurations [9.7] Predicting ion charges from electron configurations. CHEM 100 F07 1 Organization of Electrons
Ch. 9 - Electron Organization The Bohr Model [9.4] Orbitals [9.5, 9.6] Counting Electrons, configurations [9.7] Predicting ion charges from electron configurations. CHEM 100 F07 1 Organization of Electrons
Oxygen-Binding Proteins
 Oxygen-Binding Proteins Myoglobin, Hemoglobin, Cytochromes bind O 2. Oxygen is transported from lungs to various tissues via blood in association with hemoglobin In muscle, hemoglobin gives up O 2 to myoglobin
Oxygen-Binding Proteins Myoglobin, Hemoglobin, Cytochromes bind O 2. Oxygen is transported from lungs to various tissues via blood in association with hemoglobin In muscle, hemoglobin gives up O 2 to myoglobin
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Molecular Dynamics Simulations Yaoquan Tu Division of Theoretical Chemistry and Biology, Royal Institute of Technology (KTH) 2011-06 1 Outline I. Introduction II. Molecular Mechanics Force Field III. Molecular
Peptide Bonds: Structure
 Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Peptide Bonds: Structure Peptide primary structure The amino acid sequence, from - to C-terminus, determines the primary structure of a peptide or protein. The amino acids are linked through amide or peptide
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK
 Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
Advanced Medicinal & Pharmaceutical Chemistry CHEM 5412 Dept. of Chemistry, TAMUK Dai Lu, Ph.D. dlu@tamhsc.edu Tel: 361-221-0745 Office: RCOP, Room 307 Drug Discovery and Development Drug Molecules Medicinal
CSC 2427: Algorithms for Molecular Biology Spring 2006. Lecture 16 March 10
 CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
CSC 2427: Algorithms for Molecular Biology Spring 2006 Lecture 16 March 10 Lecturer: Michael Brudno Scribe: Jim Huang 16.1 Overview of proteins Proteins are long chains of amino acids (AA) which are produced
1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA
 1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA Tomáš Kubař Institute of Organic Chemistry and Biochemistry Praha, Czech Republic Thermodynamic Integration Alchemical change
1. Free energy with controlled uncertainty 2. The modes of ligand binding to DNA Tomáš Kubař Institute of Organic Chemistry and Biochemistry Praha, Czech Republic Thermodynamic Integration Alchemical change
Chapter 8. Low energy ion scattering study of Fe 4 N on Cu(100)
 Low energy ion scattering study of 4 on Cu(1) Chapter 8. Low energy ion scattering study of 4 on Cu(1) 8.1. Introduction For a better understanding of the reconstructed 4 surfaces one would like to know
Low energy ion scattering study of 4 on Cu(1) Chapter 8. Low energy ion scattering study of 4 on Cu(1) 8.1. Introduction For a better understanding of the reconstructed 4 surfaces one would like to know
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis? O2, NADP+
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? Na+, H+ 2. (4 pts) What is the terminal electron
REMOTE CONTROL by DNA as a Bio-sensor -antenna.
 REMOTE CONTROL by DNA as a Bio-sensor -antenna. "Piezoelectric quantum transduction is a fundamental property of at- distance induction of genetic control " Paolo Manzelli: pmanzelli@gmail.com ; www.edscuola.it/lre.html;www.egocreanet.it
REMOTE CONTROL by DNA as a Bio-sensor -antenna. "Piezoelectric quantum transduction is a fundamental property of at- distance induction of genetic control " Paolo Manzelli: pmanzelli@gmail.com ; www.edscuola.it/lre.html;www.egocreanet.it
B Cell Generation, Activation & Differentiation. B cell maturation
 B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
3D structure visualization and high quality imaging. Chimera
 3D structure visualization and high quality imaging. Chimera Vincent Zoete 2008 Contact : vincent.zoete@isb sib.ch 1/27 Table of Contents Presentation of Chimera...3 Exercise 1...4 Loading a structure
3D structure visualization and high quality imaging. Chimera Vincent Zoete 2008 Contact : vincent.zoete@isb sib.ch 1/27 Table of Contents Presentation of Chimera...3 Exercise 1...4 Loading a structure
Built from 20 kinds of amino acids
 Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
Built from 20 kinds of amino acids Each Protein has a three dimensional structure. Majority of proteins are compact. Highly convoluted molecules. Proteins are folded polypeptides. There are four levels
This class deals with the fundamental structural features of proteins, which one can understand from the structure of amino acids, and how they are
 This class deals with the fundamental structural features of proteins, which one can understand from the structure of amino acids, and how they are put together. 1 A more detailed view of a single protein
This class deals with the fundamental structural features of proteins, which one can understand from the structure of amino acids, and how they are put together. 1 A more detailed view of a single protein
Lecture 8. Protein Trafficking/Targeting. Protein targeting is necessary for proteins that are destined to work outside the cytoplasm.
 Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Recap. Lecture 2. Protein conformation. Proteins. 8 types of protein function 10/21/10. Proteins.. > 50% dry weight of a cell
 Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Lecture 2 Protein conformation ecap Proteins.. > 50% dry weight of a cell ell s building blocks and molecular tools. More important than genes A large variety of functions http://www.tcd.ie/biochemistry/courses/jf_lectures.php
Consensus alignment server for reliable comparative modeling with distant templates
 W50 W54 Nucleic Acids Research, 2004, Vol. 32, Web Server issue DOI: 10.1093/nar/gkh456 Consensus alignment server for reliable comparative modeling with distant templates Jahnavi C. Prasad 1, Sandor Vajda
W50 W54 Nucleic Acids Research, 2004, Vol. 32, Web Server issue DOI: 10.1093/nar/gkh456 Consensus alignment server for reliable comparative modeling with distant templates Jahnavi C. Prasad 1, Sandor Vajda
Molecular Models in Biology
 Molecular Models in Biology Objectives: After this lab a student will be able to: 1) Understand the properties of atoms that give rise to bonds. 2) Understand how and why atoms form ions. 3) Model covalent,
Molecular Models in Biology Objectives: After this lab a student will be able to: 1) Understand the properties of atoms that give rise to bonds. 2) Understand how and why atoms form ions. 3) Model covalent,
Section I Using Jmol as a Computer Visualization Tool
 Section I Using Jmol as a Computer Visualization Tool Jmol is a free open source molecular visualization program used by students, teachers, professors, and scientists to explore protein structures. Section
Section I Using Jmol as a Computer Visualization Tool Jmol is a free open source molecular visualization program used by students, teachers, professors, and scientists to explore protein structures. Section
High flexibility of DNA on short length scales probed by atomic force microscopy
 High flexibility of DNA on short length scales probed by atomic force microscopy Wiggins P. A. et al. Nature Nanotechnology (2006) presented by Anja Schwäger 23.01.2008 Outline Theory/Background Elasticity
High flexibility of DNA on short length scales probed by atomic force microscopy Wiggins P. A. et al. Nature Nanotechnology (2006) presented by Anja Schwäger 23.01.2008 Outline Theory/Background Elasticity
Peptide bonds: resonance structure. Properties of proteins: Peptide bonds and side chains. Dihedral angles. Peptide bond. Protein physics, Lecture 5
 Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational
Proteins the primary biological macromolecules of living organisms
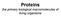 Proteins the primary biological macromolecules of living organisms Protein structure and folding Primary Secondary Tertiary Quaternary structure of proteins Structure of Proteins Protein molecules adopt
Proteins the primary biological macromolecules of living organisms Protein structure and folding Primary Secondary Tertiary Quaternary structure of proteins Structure of Proteins Protein molecules adopt
Chapter 18: Applications of Immunology
 Chapter 18: Applications of Immunology 1. Vaccinations 2. Monoclonal vs Polyclonal Ab 3. Diagnostic Immunology 1. Vaccinations What is Vaccination? A method of inducing artificial immunity by exposing
Chapter 18: Applications of Immunology 1. Vaccinations 2. Monoclonal vs Polyclonal Ab 3. Diagnostic Immunology 1. Vaccinations What is Vaccination? A method of inducing artificial immunity by exposing
Functional Architecture of RNA Polymerase I
 Cell, Volume 131 Supplemental Data Functional Architecture of RNA Polymerase I Claus-D. Kuhn, Sebastian R. Geiger, Sonja Baumli, Marco Gartmann, Jochen Gerber, Stefan Jennebach, Thorsten Mielke, Herbert
Cell, Volume 131 Supplemental Data Functional Architecture of RNA Polymerase I Claus-D. Kuhn, Sebastian R. Geiger, Sonja Baumli, Marco Gartmann, Jochen Gerber, Stefan Jennebach, Thorsten Mielke, Herbert
Chem 106 Thursday Feb. 3, 2011
 Chem 106 Thursday Feb. 3, 2011 Chapter 13: -The Chemistry of Solids -Phase Diagrams - (no Born-Haber cycle) 2/3/2011 1 Approx surface area (Å 2 ) 253 258 Which C 5 H 12 alkane do you think has the highest
Chem 106 Thursday Feb. 3, 2011 Chapter 13: -The Chemistry of Solids -Phase Diagrams - (no Born-Haber cycle) 2/3/2011 1 Approx surface area (Å 2 ) 253 258 Which C 5 H 12 alkane do you think has the highest
Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O.
 Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O. Melnyk b a National University of life and environmental sciences
Features of the formation of hydrogen bonds in solutions of polysaccharides during their use in various industrial processes. V.Mank a, O. Melnyk b a National University of life and environmental sciences
Modelling Compounds. 242 MHR Unit 2 Atoms, Elements, and Compounds
 6.3 Figure 6.26 To build the Michael Lee-Chin Crystal at the Royal Ontario Museum, models were used at different stages to convey different types of information. Modelling Compounds The Michael Lee-Chin
6.3 Figure 6.26 To build the Michael Lee-Chin Crystal at the Royal Ontario Museum, models were used at different stages to convey different types of information. Modelling Compounds The Michael Lee-Chin
Clustering & Visualization
 Chapter 5 Clustering & Visualization Clustering in high-dimensional databases is an important problem and there are a number of different clustering paradigms which are applicable to high-dimensional data.
Chapter 5 Clustering & Visualization Clustering in high-dimensional databases is an important problem and there are a number of different clustering paradigms which are applicable to high-dimensional data.
The Scheduled MRM Algorithm Enables Intelligent Use of Retention Time During Multiple Reaction Monitoring
 The Scheduled MRM Algorithm Enables Intelligent Use of Retention Time During Multiple Reaction Monitoring Delivering up to 2500 MRM Transitions per LC Run Christie Hunter 1, Brigitte Simons 2 1 AB SCIEX,
The Scheduled MRM Algorithm Enables Intelligent Use of Retention Time During Multiple Reaction Monitoring Delivering up to 2500 MRM Transitions per LC Run Christie Hunter 1, Brigitte Simons 2 1 AB SCIEX,
Assessing Checking the the reliability of protein-ligand structures
 Assessing Checking the the reliability of protein-ligand structures Aim A common task in structure-based drug design is the validation of protein-ligand structures. This process needs to be quick, visual
Assessing Checking the the reliability of protein-ligand structures Aim A common task in structure-based drug design is the validation of protein-ligand structures. This process needs to be quick, visual
Chapter 8: An Introduction to Metabolism
 Chapter 8: An Introduction to Metabolism Name Period Concept 8.1 An organism s metabolism transforms matter and energy, subject to the laws of thermodynamics 1. Define metabolism. The totality of an organism
Chapter 8: An Introduction to Metabolism Name Period Concept 8.1 An organism s metabolism transforms matter and energy, subject to the laws of thermodynamics 1. Define metabolism. The totality of an organism
Computational Systems Biology. Lecture 2: Enzymes
 Computational Systems Biology Lecture 2: Enzymes 1 Images from: David L. Nelson, Lehninger Principles of Biochemistry, IV Edition, Freeman ed. or under creative commons license (search for images at http://search.creativecommons.org/)
Computational Systems Biology Lecture 2: Enzymes 1 Images from: David L. Nelson, Lehninger Principles of Biochemistry, IV Edition, Freeman ed. or under creative commons license (search for images at http://search.creativecommons.org/)
Antibody Structure, and the Generation of B-cell Diversity CHAPTER 4 04/05/15. Different Immunoglobulins
 Antibody Structure, and the Generation of B-cell Diversity B cells recognize their antigen without needing an antigen presenting cell CHAPTER 4 Structure of Immunoglobulin G Different Immunoglobulins Differences
Antibody Structure, and the Generation of B-cell Diversity B cells recognize their antigen without needing an antigen presenting cell CHAPTER 4 Structure of Immunoglobulin G Different Immunoglobulins Differences
T cell Epitope Prediction
 Institute for Immunology and Informatics T cell Epitope Prediction EpiMatrix Eric Gustafson January 6, 2011 Overview Gathering raw data Popular sources Data Management Conservation Analysis Multiple Alignments
Institute for Immunology and Informatics T cell Epitope Prediction EpiMatrix Eric Gustafson January 6, 2011 Overview Gathering raw data Popular sources Data Management Conservation Analysis Multiple Alignments
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
BCHM 32200 Analytical Biochemistry Syllabus Spring, 2013
 INSTRUCTOR: Dr. Mark Hall office: BCHM 214 TEL: 494-0714 e-mail: mchall@purdue.edu DEPARTMENT OF BIOCHEMISTRY BCHM 32200 Analytical Biochemistry Syllabus Spring, 2013 Office hours: By appointment only
INSTRUCTOR: Dr. Mark Hall office: BCHM 214 TEL: 494-0714 e-mail: mchall@purdue.edu DEPARTMENT OF BIOCHEMISTRY BCHM 32200 Analytical Biochemistry Syllabus Spring, 2013 Office hours: By appointment only
APh/BE161: Physical Biology of the Cell Winter 2009 Recap on Photosynthesis Rob Phillips
 APh/BE161: Physical Biology of the Cell Winter 2009 Recap on Photosynthesis Rob Phillips Big picture: why are we doing this? A) photosynthesis will explain shortly, b) more generally, interaction of light
APh/BE161: Physical Biology of the Cell Winter 2009 Recap on Photosynthesis Rob Phillips Big picture: why are we doing this? A) photosynthesis will explain shortly, b) more generally, interaction of light
MCAT Organic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins
 MCAT rganic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins Question No. 1 of 10 Question 1. Which amino acid does not contain a chiral center? Question #01 (A) Serine (B) Proline (C)
MCAT rganic Chemistry - Problem Drill 23: Amino Acids, Peptides and Proteins Question No. 1 of 10 Question 1. Which amino acid does not contain a chiral center? Question #01 (A) Serine (B) Proline (C)
CHEM 451 BIOCHEMISTRY I. SUNY Cortland Fall 2010
 CHEM 451 BIOCHEMISTRY I SUNY Cortland Fall 2010 Instructor: Dr. Frank Rossi Office: Bowers 135 Office Hours: Mon. 2:30-4:00, Wed. 4:00-5:30, Friday 2:30-3:00, or by appointment. Extra evening office hours
CHEM 451 BIOCHEMISTRY I SUNY Cortland Fall 2010 Instructor: Dr. Frank Rossi Office: Bowers 135 Office Hours: Mon. 2:30-4:00, Wed. 4:00-5:30, Friday 2:30-3:00, or by appointment. Extra evening office hours
1 The water molecule and hydrogen bonds in water
 The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
The Physics and Chemistry of Water 1 The water molecule and hydrogen bonds in water Stoichiometric composition H 2 O the average lifetime of a molecule is 1 ms due to proton exchange (catalysed by acids
Two-photon FCS Tutorial. Berland Lab Department of Physics Emory University
 Two-photon FCS Tutorial Berland Lab Department of Physics Emory University What is FCS? FCS : Fluorescence Correlation Spectroscopy FCS is a technique for acquiring dynamical information from spontaneous
Two-photon FCS Tutorial Berland Lab Department of Physics Emory University What is FCS? FCS : Fluorescence Correlation Spectroscopy FCS is a technique for acquiring dynamical information from spontaneous
Supporting Information. Fast and Efficient Fragment-Based Lead Generation. by Fully Automated Processing and Analysis of
 Supporting Information Fast and Efficient Fragment-Based Lead Generation by Fully Automated Processing and Analysis of Ligand-Observed NMR Binding Data Chen Peng, Alexandra Frommlet, Manuel Perez, Carlos
Supporting Information Fast and Efficient Fragment-Based Lead Generation by Fully Automated Processing and Analysis of Ligand-Observed NMR Binding Data Chen Peng, Alexandra Frommlet, Manuel Perez, Carlos
Module 2 overview SPRING BREAK
 1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
1 Module 2 overview lecture lab 1. Introduction to the module 1. Start-up protein eng. 2. Rational protein design 2. Site-directed mutagenesis 3. Fluorescence and sensors 3. DNA amplification 4. Protein
CUSTOM ANTIBODIES. Fully customised services: rat and murine monoclonals, rat and rabbit polyclonals, antibody characterisation, antigen preparation
 CUSTOM ANTIBODIES Highly competitive pricing without compromising quality. Rat monoclonal antibodies for the study of gene expression and proteomics in mice and in mouse models of human diseases available.
CUSTOM ANTIBODIES Highly competitive pricing without compromising quality. Rat monoclonal antibodies for the study of gene expression and proteomics in mice and in mouse models of human diseases available.
SPECIAL PERTURBATIONS UNCORRELATED TRACK PROCESSING
 AAS 07-228 SPECIAL PERTURBATIONS UNCORRELATED TRACK PROCESSING INTRODUCTION James G. Miller * Two historical uncorrelated track (UCT) processing approaches have been employed using general perturbations
AAS 07-228 SPECIAL PERTURBATIONS UNCORRELATED TRACK PROCESSING INTRODUCTION James G. Miller * Two historical uncorrelated track (UCT) processing approaches have been employed using general perturbations
A Short Introduction to Computer Graphics
 A Short Introduction to Computer Graphics Frédo Durand MIT Laboratory for Computer Science 1 Introduction Chapter I: Basics Although computer graphics is a vast field that encompasses almost any graphical
A Short Introduction to Computer Graphics Frédo Durand MIT Laboratory for Computer Science 1 Introduction Chapter I: Basics Although computer graphics is a vast field that encompasses almost any graphical
Chapter 6: Antigen-Antibody Interactions
 Chapter 6: Antigen-Antibody Interactions I. Strength of Ag-Ab interactions A. Antibody Affinity - strength of total noncovalent interactions between single Ag-binding site on an Ab and a single epitope
Chapter 6: Antigen-Antibody Interactions I. Strength of Ag-Ab interactions A. Antibody Affinity - strength of total noncovalent interactions between single Ag-binding site on an Ab and a single epitope
Publication of small-unit-cell structures in Acta Crystallographica Michael Hoyland ECM28 University of Warwick, 2013
 Publication of small-unit-cell structures in Acta Crystallographica Michael Hoyland ECM28 University of Warwick, 2013 International Union of Crystallography 5 Abbey Square Chester CH1 2HU UK International
Publication of small-unit-cell structures in Acta Crystallographica Michael Hoyland ECM28 University of Warwick, 2013 International Union of Crystallography 5 Abbey Square Chester CH1 2HU UK International
EXPERIMENT 1: Survival Organic Chemistry: Molecular Models
 EXPERIMENT 1: Survival Organic Chemistry: Molecular Models Introduction: The goal in this laboratory experience is for you to easily and quickly move between empirical formulas, molecular formulas, condensed
EXPERIMENT 1: Survival Organic Chemistry: Molecular Models Introduction: The goal in this laboratory experience is for you to easily and quickly move between empirical formulas, molecular formulas, condensed
Amino Acids, Peptides, Proteins
 Amino Acids, Peptides, Proteins Functions of proteins: Enzymes Transport and Storage Motion, muscle contraction Hormones Mechanical support Immune protection (Antibodies) Generate and transmit nerve impulses
Amino Acids, Peptides, Proteins Functions of proteins: Enzymes Transport and Storage Motion, muscle contraction Hormones Mechanical support Immune protection (Antibodies) Generate and transmit nerve impulses
Characterization of monoclonal antibody epitope specificity using Biacore s SPR technology
 A P P L I C A T I O N N O T E 1 Characterization of monoclonal antibody epitope specificity using Biacore s SPR technology Abstract Biacore s SPR technology based on surface plasmon resonance technology
A P P L I C A T I O N N O T E 1 Characterization of monoclonal antibody epitope specificity using Biacore s SPR technology Abstract Biacore s SPR technology based on surface plasmon resonance technology
Making the switch to a safer CAR-T cell therapy
 Making the switch to a safer CAR-T cell therapy HaemaLogiX 2015 Technical Journal Club May 24 th 2016 Christina Müller - chimeric antigen receptor = CAR - CAR T cells are generated by lentiviral transduction
Making the switch to a safer CAR-T cell therapy HaemaLogiX 2015 Technical Journal Club May 24 th 2016 Christina Müller - chimeric antigen receptor = CAR - CAR T cells are generated by lentiviral transduction
BSC 2010 - Exam I Lectures and Text Pages. The Plasma Membrane Structure and Function. Phospholipids. I. Intro to Biology (2-29) II.
 BSC 2010 - Exam I Lectures and Text Pages I. Intro to Biology (2-29) II. Chemistry of Life Chemistry review (30-46) Water (47-57) Carbon (58-67) Macromolecules (68-91) III. Cells and Membranes Cell structure
BSC 2010 - Exam I Lectures and Text Pages I. Intro to Biology (2-29) II. Chemistry of Life Chemistry review (30-46) Water (47-57) Carbon (58-67) Macromolecules (68-91) III. Cells and Membranes Cell structure
Lecture 15: Enzymes & Kinetics Mechanisms
 ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
ROLE OF THE TRANSITION STATE Lecture 15: Enzymes & Kinetics Mechanisms Consider the reaction: H-O-H + Cl - H-O δ- H Cl δ- HO - + H-Cl Reactants Transition state Products Margaret A. Daugherty Fall 2004
