H3.1 H3.3 ] H3-GFP. Loading control
|
|
|
- Colleen Hill
- 4 years ago
- Views:
Transcription
1 A B TR/ TR1.1 TR4/TR5. C ene name Promoter size (bp) Atg27 TR 251 At4g00 TR4 28 At4g0 TR At5g90 TR1 81 TR TR1 TR4 TR5..1 D ene ID E Mw l-0 Co R R5 R4 R1 T T T T 55 ] Loading control Supplemental Figure 1. Characterization of fluorescently-labeled proteins (A) enomic organization of.1 (TR and TR1) and. (TR4 and TR5) genes used in this study. The genomic sequence was fused to appropriate tags and expressed in transgenic Arabidopsis plants under their endogenous promoters, as indicated in the main text and Experimental Procedures. (B) Size of promoters used to express each TR gene. (C) Absolute amount of endogenous and transgene expression. Copy number of each endogenous gene (grey bars) and each - transgene (dark green) was determined in wild type Col-0 and tagged lines, respectively. Data are the mean values ア s.d., n =. (D) Representative examples of the localization pattern of proteins used in this study in the root meristem nuclei of plants described in panel A. (E) Nuclear extract of Col-0 and transgenic plants expressing the indicated - proteins were fractionated and the - proteins detected with anti- antibodies. The line correspond to a line expressing alone. Note that the slight different mobility of - is due to the different vectors used that provide slightly different linkers between the and the moieties. 1
2 TR + TR5.1-.-mR A cell wall B B1 A1 D C1 D1.1- TR1 TR4.- TR5 Cortex TR C F I J K L M N O P QC plane Epidermis E Supplemental Figure 2. Expression pattern of various fluorescently labeled lines used in this study. (A-D) Embryos of plants expressing both.1 (TR-) and. (TR5-mR) at various stages: globular (A), early (B) and late (C) heart, and torpedo (D). Insets A1-D1 correspond to the sections highlighted in the upper panels. Arrowheads point to the QC. (E-P Examples of the patchy and constitutive expression patterns of.1(tr- and TR1-; E,F,I,J,M,N) and. (TR4- and TR5-;,,K,L,O,P), respectively, in the cortex (A-D), epidermis (I-L) and middle plane (M-P) of the root apical meristem. Note that panels I and L are also shown in Figs. 2A and 2B, respectively. 2
3 0 Atrichoblasts 0 Trichoblasts Supplemental Figure. Relative position of mitotic figures in trichoblast and atrichoblast cell layers. Mitotic figures labeled with.(tr5-mr) only or with both.1(tr-) and.(tr5-mr) were scored along the RAM as described in Fig. 2F. Atrichoblasts, n=27; trichoblasts, n=26. The two populations were statistically different (p=0.05, t-test) in the case of atrichoblasts.
4 A differentiated wt ( -). (TR5- +).1 (TR1- +) B apical wt ( -). (TR4- +).1 (TR- +) differentiated wt ( -) Supplemental Figure 4. Analysis of ploidy levels of -tagged nuclei (A) Analysis of ploidy level of wild type (Wt, ),.(TR5)- and.1(tr1)- nuclei of the differentiated part of the root (the entire root except the 5 mm apical part) by two-channel flow cytometry. was used for nuclear staining and determination of DNA content (2-16C). Wild type Col-0 and. (TR5)- nuclei were used to define the (left panel) and + (mid and right panels) gates, respectively. (B) Analysis of ploidy level of wild type (Wt, ),.(TR4)- and.1(tr)- nuclei of the 5 mm apical (upper panels) and differentiated (lower panels; as defined in panel a) parts of the root. was used for nuclear staining and determination of DNA content (2-16C). Wild type Col-0 and.(tr4)- nuclei were used to define the (left panels) and + (mid and right panels) gates, respectively. 4
5 Stage I Stage IV Supplemental Figure labeling during initiation of lateral roots. Cell cycle reactivation of lateral root founder cells is associated with incorporation of.1, since stage I. At later stages, e.g. stage IV, nuclei show different levels of.1- labeling, indicative of cells at different phases of the cell cycle. Note that, frequently, cells of central cylinder appear -labeled. The exact nature of these nuclei remains to be determined. 5
6 Supplemental Table 1. Chromatin and cell cycle genes downregulated from slice to slice 4 (fold change fc > 1.5) ene ID AT4g0 AT5g19 AT4g08 ATg226 AT122 AT AT161 AT AT11 AT275 AT4299 AT558 AT2167 AT259 AT2164 AT5491 AT AT5488 AT21270 AT2000 AT AT427 AT84 AT561 AT507 AT185 AT48 AT15150 AT2451 AT AT518 AT2550 AT0 AT5146 AT5254 AT5167 AT11 AT190 AT1765 AT5649 ene name SWINER SUV4 SD4 RDM1 RBR RFC RFC2 MCM7 ORC4 ORC2 ORC5 MSI1 MSI2 MAD2 MCM4 MET1 KU70 ICK6 CDT1A OBBIT UB1 TA2 DA08 DA6 D2C DA15 D1 PY2 AM1 AMMA-2AX FZR ET1 DEL/E2FF DRM2 DNMT2 DOT2/MDF CDKD1; CDKB2;2 CDKB2;1 CDKC2 AT50070 AT5672 AT11 AT176 AT456 AT2176 AT144 AT2267 AT115 AT170 AT1441 AT AT24 AT519 AT1156 AT016 AT670 AT AT AT48150 AT AT AT5 AT54990 AT1496 AT248 AT170 AT7790 AT2195 AT55090 AT1447 AT8 AT8590 AT1578 AT59550 CYCD; CYCD;2 CYCB;1 CYCB2;4 CYCB2;2 CYCB2;1 CYCB1;5 CYCB1;4 CYCB1; CYCA2;4 CYCA1;1 CMT CR CR2 CENP-C CDC48C CDC48B CAK4 ATXR5 APC8 APC6 APC ARP6 SUVR2 ICK5 ICK TR12 TB1 MSI4 ISTONE ISTONE CAPERONE DOMAIN ISTONE CAPERONE DOMAIN VIM6 VIM1 VIM 6
7 Chromatin and cell cycle genes downregulated from slice 2 + to slice 4 (fold change fc > 1.5) ene ID AT59550 AT AT5049 AT5159 AT8 AT122 AT5546 AT AT161 AT AT11 AT275 AT1268 AT4299 AT558 AT45050 AT5491 AT AT21270 AT427 AT15 AT185 AT5228 AT5567 AT1798 AT AT0 AT5167 AT5649 AT47 AT5 AT25 AT52 AT AT170 AT7790 AT88 AT55090 AT400 AT50 ene name VIM ULT1 SUV1 SMC5 SD4 RBR TA1 RPA2 RFC2 MCM7 ORC4 ORC2 ORC6 ORC5 MSI1 MSI MET1 KU70 CDT1A TA2 TA DA15 TB2 A2 L2 AMMA 2AX DEL/E2FF DOT2/MDF CDKC2 CYCD5;1 CYCA;1 CDC45 ATXR6 APC TR12 TB1 TA5 ISTONE ISTONE ISTONE AT590 AT27470 AT4590 AT27 ISTONE ISTONE ISTONE ISTONE 7
8 Supplemental Table 2. Primers used in this study Cloning of TR genes TR genes were cloned using the following primers: TR-W-F: 5 -ACAATTTTACAAAAAACACTATCCATCTCTTACTTTCCTA- TR-W-R: 5 -ACCACTTTTACAAAAACTTCACCCTCTCACCTCTAATCC- TR4-W-F: 5 -ACAATTTTACAAAAAACACTAAACTCCCTTTCCAA- TR4-W-R: 5 -ACCACTTTTACAAAAACTTCACCTTCACCTCTATAC- TR5-W-F: 5 -ACAATTTTACAAAAAACACTCATTTACTCAAAACAATA- TR5-W-R: 5 -ACCACTTTTACAAAAACTTCACACTTCTCCTCTATCCT- TR1-W-F: 5 -ACAATTTTACAAAAAACACTATTCTTCTTCATCTCCAT- TR1-W-R: 5 -ACCACTTTTACAAAAACTTCACTCTTTCTCCTCTATTCTCC- qpcr measurements The expression of the TR genes in Col-0 plants was monitored using the following primers: TR-endo-F 5 -TCAAACACACATCTCAACAA- TR-endo-R 5 -CTACTTTCCTACAATCT- TR4-endo-F 5 -CCTTAACCAACCAAAAT- TR4-endo-R 5 -ATTAAAAAATTCCAACACAC- TR5-endo-F 5 -CTTAATTTATTAA- TR5-endo-R 5 -CACCATTTTTCCAATCTCC- TR1-endo-F 5 -CAAAATCAAAA- TR1-endo-R 5 -CCAAATCAAAATCCAAAA- The expression of the TR- tagged genes was measured using the following primers: TR--F 5 -TATCATCCACTCTTC- TR--R 5 -TTTTACAAAAACTTCA- TR4--F 5 -CTAATTCTCAAATTTCA- TR4--R 5 -TTTTACAAAAACTTCA- TR5/1--F 5 -ACAACAAAAAACCATC- TR5/1--R 5 -AAACAATTTTAC- 8
Mitotic Membrane Turnover Coordinates Differential Induction of the Heart Progenitor Lineage
 Developmental Cell Supplemental Information Mitotic Membrane Turnover Coordinates Differential Induction of the Heart Progenitor Lineage Christina D. Cota and Brad Davidson Supplemental Figures S1-S7 S1.
Developmental Cell Supplemental Information Mitotic Membrane Turnover Coordinates Differential Induction of the Heart Progenitor Lineage Christina D. Cota and Brad Davidson Supplemental Figures S1-S7 S1.
by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains was previously described
 Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
Table S1. Fission yeast strains used in this study. Gene disruption was performed by the PCR-mediated method (Krawchuk and Wahls, 1999). The construction of Ams2-null and conditional ams2-shut-off strains
OriGene Technologies, Inc. MicroRNA analysis: Detection, Perturbation, and Target Validation
 OriGene Technologies, Inc. MicroRNA analysis: Detection, Perturbation, and Target Validation -Optimal strategies to a successful mirna research project Optimal strategies to a successful mirna research
OriGene Technologies, Inc. MicroRNA analysis: Detection, Perturbation, and Target Validation -Optimal strategies to a successful mirna research project Optimal strategies to a successful mirna research
pcas-guide System Validation in Genome Editing
 pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
pcas-guide System Validation in Genome Editing Tagging HSP60 with HA tag genome editing The latest tool in genome editing CRISPR/Cas9 allows for specific genome disruption and replacement in a flexible
Supplementary Figure 1 Characterization of 5xFAD derived hippocampal cultures. A-B) Average total numbers of cells per hippocampus as well as average
 Supplementary Figure 1 Characterization of 5xFAD derived hippocampal cultures. A-B) Average total numbers of cells per hippocampus as well as average total numbers of cells per field of view were similar
Supplementary Figure 1 Characterization of 5xFAD derived hippocampal cultures. A-B) Average total numbers of cells per hippocampus as well as average total numbers of cells per field of view were similar
Plant Growth & Development. Growth Stages. Differences in the Developmental Mechanisms of Plants and Animals. Development
 Plant Growth & Development Plant body is unable to move. To survive and grow, plants must be able to alter its growth, development and physiology. Plants are able to produce complex, yet variable forms
Plant Growth & Development Plant body is unable to move. To survive and grow, plants must be able to alter its growth, development and physiology. Plants are able to produce complex, yet variable forms
Supplemental Information. McBrayer et al. Supplemental Data
 1 Supplemental Information McBrayer et al. Supplemental Data 2 Figure S1. Glucose consumption rates of MM cell lines exceed that of normal PBMC. (A) Normal PBMC isolated from three healthy donors were
1 Supplemental Information McBrayer et al. Supplemental Data 2 Figure S1. Glucose consumption rates of MM cell lines exceed that of normal PBMC. (A) Normal PBMC isolated from three healthy donors were
Interphase (A) and mitotic (B) D16 cells were plated on concanavalin A and processed for
 E07-10-1069 Rogers SUPPLEMENTARY DATA Supplemental Figure 1. Interphase day-7 control and γ-tubulin23c RNAi-treated S2 cells stained for MTs (green), γ-tubulin (red), and DNA (blue). Scale, 5μm. Supplemental
E07-10-1069 Rogers SUPPLEMENTARY DATA Supplemental Figure 1. Interphase day-7 control and γ-tubulin23c RNAi-treated S2 cells stained for MTs (green), γ-tubulin (red), and DNA (blue). Scale, 5μm. Supplemental
Cell Division CELL DIVISION. Mitosis. Designation of Number of Chromosomes. Homologous Chromosomes. Meiosis
 Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
Cell Division CELL DIVISION Anatomy and Physiology Text and Laboratory Workbook, Stephen G. Davenport, Copyright 2006, All Rights Reserved, no part of this publication can be used for any commercial purpose.
J. Cell Sci. 128: doi:10.1242/jcs.164566: Supplementary Material
 Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Figure S1. Microtubule and microfilament drug treatments severely interfere with mitotic spindle morphology, length and positioning. (A) GFP-fused lamin-a/c, LAP2 or BAF1 localizes to the spindle and all
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
Cell, Tissue and Tumor Kinetics
 Cell, issue and umor Kinetics Proliferation Kinetics: rate of growth of a population, change in total cell number. Adult tissues are in homeostasis. Children (and tumors) grow. I. Quantitative Assessment
Cell, issue and umor Kinetics Proliferation Kinetics: rate of growth of a population, change in total cell number. Adult tissues are in homeostasis. Children (and tumors) grow. I. Quantitative Assessment
Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION
 Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION Susan Carson Heather B. Miller D.Scott Witherow ELSEVIER AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN
Molecular Biology Techniques: A Classroom Laboratory Manual THIRD EDITION Susan Carson Heather B. Miller D.Scott Witherow ELSEVIER AMSTERDAM BOSTON HEIDELBERG LONDON NEW YORK OXFORD PARIS SAN DIEGO SAN
Real-time PCR: Understanding C t
 APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
APPLICATION NOTE Real-Time PCR Real-time PCR: Understanding C t Real-time PCR, also called quantitative PCR or qpcr, can provide a simple and elegant method for determining the amount of a target sequence
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
www.sciencesignaling.org/cgi/content/full/7/339/ra80/dc1 Supplementary Materials for Manipulation of receptor oligomerization as a strategy to inhibit signaling by TNF superfamily members Julia T. Warren,
Recognition of T cell epitopes (Abbas Chapter 6)
 Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
Introduction to SAGEnhaft
 Introduction to SAGEnhaft Tim Beissbarth October 13, 2015 1 Overview Serial Analysis of Gene Expression (SAGE) is a gene expression profiling technique that estimates the abundance of thousands of gene
Introduction to SAGEnhaft Tim Beissbarth October 13, 2015 1 Overview Serial Analysis of Gene Expression (SAGE) is a gene expression profiling technique that estimates the abundance of thousands of gene
Superior TrueMAB TM monoclonal antibodies for the recognition of proteins native epitopes
 Superior TrueMAB TM monoclonal antibodies for the recognition of proteins native epitopes Outlines Brief introduction of OriGene s mission on gene-centric product solution. TrueMAB monoclonal antibody
Superior TrueMAB TM monoclonal antibodies for the recognition of proteins native epitopes Outlines Brief introduction of OriGene s mission on gene-centric product solution. TrueMAB monoclonal antibody
The Huntington Library, Art Collections, and Botanical Gardens
 The Huntington Library, Art Collections, and Botanical Gardens Rooting for Mitosis Overview Students will fix, stain, and make slides of onion root tips. These slides will be examined for the presence
The Huntington Library, Art Collections, and Botanical Gardens Rooting for Mitosis Overview Students will fix, stain, and make slides of onion root tips. These slides will be examined for the presence
Supplementary Figure 1.
 Supplementary Figure 1. (A) MicroRNA 212 enhances IS from pancreatic β-cells. INS-1 832/3 β-cells were transfected with precursors for mirnas 212, 375, or negative control oligonucleotides. 48 hrs after
Supplementary Figure 1. (A) MicroRNA 212 enhances IS from pancreatic β-cells. INS-1 832/3 β-cells were transfected with precursors for mirnas 212, 375, or negative control oligonucleotides. 48 hrs after
SUPPLEMENTARY DATA 1
 SUPPLEMENTARY DATA 1 Supplementary Figure S1. Overexpression of untagged Ago2 inhibits the nuclear transport of the dotted foci of GFP-signal of myc-gfp-tnrc6a-nes-mut. (A C) HeLa cells expressing myc-gfp-tnrc6a-nes-mut
SUPPLEMENTARY DATA 1 Supplementary Figure S1. Overexpression of untagged Ago2 inhibits the nuclear transport of the dotted foci of GFP-signal of myc-gfp-tnrc6a-nes-mut. (A C) HeLa cells expressing myc-gfp-tnrc6a-nes-mut
Chapter 3. Cell Division. Laboratory Activities Activity 3.1: Mock Mitosis Activity 3.2: Mitosis in Onion Cells Activity 3.
 Chapter 3 Cell Division Laboratory Activities Activity 3.1: Mock Mitosis Activity 3.2: Mitosis in Onion Cells Activity 3.3: Mock Meiosis Goals Following this exercise students should be able to Recognize
Chapter 3 Cell Division Laboratory Activities Activity 3.1: Mock Mitosis Activity 3.2: Mitosis in Onion Cells Activity 3.3: Mock Meiosis Goals Following this exercise students should be able to Recognize
LAB 09 Cell Division
 LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
LAB 09 Cell Division Introduction: One of the characteristics of living things is the ability to replicate and pass on genetic information to the next generation. Cell division in individual bacteria and
Cell Cycle in Onion Root Tip Cells (IB)
 Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Cell Cycle in Onion Root Tip Cells (IB) A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules,
Lab 3: Testing Hypotheses about Mitosis
 Lab 3: Testing Hypotheses about Mitosis Why do cells divide? Lab today focuses on cellular division, also known as cellular reproduction. To become more familiar with why cells divide, the types of cell
Lab 3: Testing Hypotheses about Mitosis Why do cells divide? Lab today focuses on cellular division, also known as cellular reproduction. To become more familiar with why cells divide, the types of cell
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes.
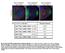 Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Fig. S1. The tail of KHC is important for Gurken localization. (A-C) Gurken in st9 khc 27 mutant oocytes (GLC) containing KHCGFP transgenes. A) KHC1-975 rescues Gurken localization, to a crescent at the
Design high specificity CRISPR-Cas9 grnas: principles and tools. Heidi Huang, PhD
 Design high specificity CRISPR-Cas9 grnas: principles and tools Heidi Huang, PhD Webinar Agenda 1 2 3 4 Introduction of CRISPR-Cas9 grna Design Resources and Services Q&A 2 What is CRISPR? CRISPR Clustered
Design high specificity CRISPR-Cas9 grnas: principles and tools Heidi Huang, PhD Webinar Agenda 1 2 3 4 Introduction of CRISPR-Cas9 grna Design Resources and Services Q&A 2 What is CRISPR? CRISPR Clustered
GeneCopoeia Genome Editing Tools for Safe Harbor Integration in. Mice and Humans. Ed Davis, Liuqing Qian, Ruiqing li, Junsheng Zhou, and Jinkuo Zhang
 G e n e C o p o eia TM Expressway to Discovery APPLICATION NOTE Introduction GeneCopoeia Genome Editing Tools for Safe Harbor Integration in Mice and Humans Ed Davis, Liuqing Qian, Ruiqing li, Junsheng
G e n e C o p o eia TM Expressway to Discovery APPLICATION NOTE Introduction GeneCopoeia Genome Editing Tools for Safe Harbor Integration in Mice and Humans Ed Davis, Liuqing Qian, Ruiqing li, Junsheng
Cells, tissues and organs
 Chapter 8: Cells, tissues and organs Cells: building blocks of life Living things are made of cells. Many of the chemical reactions that keep organisms alive (metabolic functions) take place in cells.
Chapter 8: Cells, tissues and organs Cells: building blocks of life Living things are made of cells. Many of the chemical reactions that keep organisms alive (metabolic functions) take place in cells.
Integrated Protein Services
 Integrated Protein Services Custom protein expression & purification Version DC04-0012 Expression strategy The first step in the recombinant protein generation process is to design an appropriate expression
Integrated Protein Services Custom protein expression & purification Version DC04-0012 Expression strategy The first step in the recombinant protein generation process is to design an appropriate expression
Primary evaluation of induced pluripotent stem cells using flow cytometry
 3 Special Issue: Mesenchymal stem cells Original Article Primary evaluation of induced pluripotent stem cells using flow cytometry Daisuke Araki 1,2, ), Yoshimi Kawamura 2, ), Kunimichi Niibe 1,2), Sadafumi
3 Special Issue: Mesenchymal stem cells Original Article Primary evaluation of induced pluripotent stem cells using flow cytometry Daisuke Araki 1,2, ), Yoshimi Kawamura 2, ), Kunimichi Niibe 1,2), Sadafumi
Fig. S1. Identification of CRMs with activity in Ci-Tbx2/3 expression domains outside the notochord. B-F
 Fig. S1. Identification of CRMs with activity in Ci-Tbx2/3 expression domains outside the notochord. (A) Top: map of the Ci- Tbx2/3 locus. Teal rectangles and lines denote exons and introns, respectively.
Fig. S1. Identification of CRMs with activity in Ci-Tbx2/3 expression domains outside the notochord. (A) Top: map of the Ci- Tbx2/3 locus. Teal rectangles and lines denote exons and introns, respectively.
Current Motif Discovery Tools and their Limitations
 Current Motif Discovery Tools and their Limitations Philipp Bucher SIB / CIG Workshop 3 October 2006 Trendy Concepts and Hypotheses Transcription regulatory elements act in a context-dependent manner.
Current Motif Discovery Tools and their Limitations Philipp Bucher SIB / CIG Workshop 3 October 2006 Trendy Concepts and Hypotheses Transcription regulatory elements act in a context-dependent manner.
Cell Division Mitosis and the Cell Cycle
 Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
Cell Division Mitosis and the Cell Cycle A Chromosome and Sister Chromatids Key Points About Chromosome Structure A chromosome consists of DNA that is wrapped around proteins (histones) and condensed Each
CD3/TCR stimulation and surface detection Determination of specificity of intracellular detection of IL-7Rα by flow cytometry
 CD3/TCR stimulation and surface detection Stimulation of HPB-ALL cells with the anti-cd3 monoclonal antibody OKT3 was performed as described 3. In brief, antibody-coated plates were prepared by incubating
CD3/TCR stimulation and surface detection Stimulation of HPB-ALL cells with the anti-cd3 monoclonal antibody OKT3 was performed as described 3. In brief, antibody-coated plates were prepared by incubating
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID papillary renal cell carcinoma (translocation-associated) PRCC Human This gene
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID papillary renal cell carcinoma (translocation-associated) PRCC Human This gene
Frequently Asked Questions (FAQ)
 Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
Introduction to flow cytometry
 Introduction to flow cytometry Flow cytometry is a popular laser-based technology. Discover more with our introduction to flow cytometry. Flow cytometry is now a widely used method for analyzing the expression
Introduction to flow cytometry Flow cytometry is a popular laser-based technology. Discover more with our introduction to flow cytometry. Flow cytometry is now a widely used method for analyzing the expression
1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells
 Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
Cell Growth and Reproduction 1. When new cells are formed through the process of mitosis, the number of chromosomes in the new cells A. is half of that of the parent cell. B. remains the same as in the
The Somatic Cell Cycle
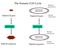 The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
The Somatic Cell Cycle Maternal chromosome Diploid Zygote Diploid Zygote Paternal chromosome MITOSIS MITOSIS Maternal chromosome Diploid organism Diploid organism Paternal chromosome Int terpha ase The
EXPERIMENT 9 - IDENTIFYING FEATURES OF MUTANT EMBRYO USING NOMARSKI MICROSCOPY (GENE TWO)
 EXPERIMENT 9 - IDENTIFYING FEATURES OF MUTANT EMBRYO USING NOMARSKI MICROSCOPY (GENE TWO) Purpose: To introduce Differential Interference Contrast (DIC) or Nomarski Interference Contrast (NIC) microscopy
EXPERIMENT 9 - IDENTIFYING FEATURES OF MUTANT EMBRYO USING NOMARSKI MICROSCOPY (GENE TWO) Purpose: To introduce Differential Interference Contrast (DIC) or Nomarski Interference Contrast (NIC) microscopy
123count ebeads Catalog Number: 01-1234 Also known as: Absolute cell count beads GPR: General Purpose Reagents. For Laboratory Use.
 Page 1 of 1 Catalog Number: 01-1234 Also known as: Absolute cell count beads GPR: General Purpose Reagents. For Laboratory Use. Normal human peripheral blood was stained with Anti- Human CD45 PE (cat.
Page 1 of 1 Catalog Number: 01-1234 Also known as: Absolute cell count beads GPR: General Purpose Reagents. For Laboratory Use. Normal human peripheral blood was stained with Anti- Human CD45 PE (cat.
Sox10 gain-of-function causes XX sex reversal in mice: Implications for human 22q-linked disorders of sex development
 Supplementary information for: Sox10 gain-of-function causes XX sex reversal in mice: Implications for human 22q-linked disorders of sex development Juan Carlos Polanco 1, Dagmar Wilhelm 1, Tara-Lynne
Supplementary information for: Sox10 gain-of-function causes XX sex reversal in mice: Implications for human 22q-linked disorders of sex development Juan Carlos Polanco 1, Dagmar Wilhelm 1, Tara-Lynne
Becker Muscular Dystrophy
 Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Muscular Dystrophy A Case Study of Positional Cloning Described by Benjamin Duchenne (1868) X-linked recessive disease causing severe muscular degeneration. 100 % penetrance X d Y affected male Frequency
Gene Models & Bed format: What they represent.
 GeneModels&Bedformat:Whattheyrepresent. Gene models are hypotheses about the structure of transcripts produced by a gene. Like all models, they may be correct, partly correct, or entirely wrong. Typically,
GeneModels&Bedformat:Whattheyrepresent. Gene models are hypotheses about the structure of transcripts produced by a gene. Like all models, they may be correct, partly correct, or entirely wrong. Typically,
Ubiquitin-conjugated degradation of Golden 2-Like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato
 New Phytologist Supporting Information Ubiquitin-conjugated degradation of Golden 2-Like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato Xiaofeng Tang, Min Miao, Xiangli
New Phytologist Supporting Information Ubiquitin-conjugated degradation of Golden 2-Like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato Xiaofeng Tang, Min Miao, Xiangli
Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice
 Manuscript EMBO-2012-82888 Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice Saya Kagiwada, Kazuki Kurimoto, Takayuki Hirota, Masashi Yamaji, and Mitinori Saitou
Manuscript EMBO-2012-82888 Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice Saya Kagiwada, Kazuki Kurimoto, Takayuki Hirota, Masashi Yamaji, and Mitinori Saitou
A novel polarized cell culture model to study HCV infection
 A novel polarized cell culture model to study HCV infection Sandrine Belouzard Centre d Infection et d Immunité de Lille 15 ième réunion du réseau national hépatites Paris- 30 janvier 2015 HCV entry and
A novel polarized cell culture model to study HCV infection Sandrine Belouzard Centre d Infection et d Immunité de Lille 15 ième réunion du réseau national hépatites Paris- 30 janvier 2015 HCV entry and
MAB Solut. MABSolys Génopole Campus 1 5 rue Henri Desbruères 91030 Evry Cedex. www.mabsolut.com. is involved at each stage of your project
 Mabsolus-2015-UK:Mise en page 1 03/07/15 14:13 Page1 Services provider Department of MABSolys from conception to validation MAB Solut is involved at each stage of your project Creation of antibodies Production
Mabsolus-2015-UK:Mise en page 1 03/07/15 14:13 Page1 Services provider Department of MABSolys from conception to validation MAB Solut is involved at each stage of your project Creation of antibodies Production
How to construct transgenic mice
 How to construct transgenic mice Sandra Beer-Hammer Autumn School 2010 Bad Schandau Methods additional genetic information transgenic mouse line gene inactivation gene-deficient knockout mouse line Jak2
How to construct transgenic mice Sandra Beer-Hammer Autumn School 2010 Bad Schandau Methods additional genetic information transgenic mouse line gene inactivation gene-deficient knockout mouse line Jak2
Supplementary Material. Free-radical production after post-thaw incubation of ram spermatozoa is related to decreased in vivo fertility
 10.1071/RD14043_AC CSIRO 2015 Supplementary Material: Reproduction, Fertility and Development, 2015, 27(8), 1187 1196. Supplementary Material Free-radical production after post-thaw incubation of ram spermatozoa
10.1071/RD14043_AC CSIRO 2015 Supplementary Material: Reproduction, Fertility and Development, 2015, 27(8), 1187 1196. Supplementary Material Free-radical production after post-thaw incubation of ram spermatozoa
Mitosis in Onion Root Tip Cells
 Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
Mitosis in Onion Root Tip Cells A quick overview of cell division The genetic information of plants, animals and other eukaryotic organisms resides in several (or many) individual DNA molecules, or chromosomes.
PrimePCR Assay Validation Report
 Gene Information Gene Name sorbin and SH3 domain containing 2 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SORBS2 Human Arg and c-abl represent the mammalian
Gene Information Gene Name sorbin and SH3 domain containing 2 Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID SORBS2 Human Arg and c-abl represent the mammalian
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR
 Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Rapid Acquisition of Unknown DNA Sequence Adjacent to a Known Segment by Multiplex Restriction Site PCR BioTechniques 25:415-419 (September 1998) ABSTRACT The determination of unknown DNA sequences around
Biotechnology: DNA Technology & Genomics
 Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Chapter 20. Biotechnology: DNA Technology & Genomics 2003-2004 The BIG Questions How can we use our knowledge of DNA to: diagnose disease or defect? cure disease or defect? change/improve organisms? What
Title: Surveying Genome to Identify Origins of DNA Replication In Silico
 Title: Surveying Genome to Identify Origins of DNA Replication In Silico Abstract: DNA replication origins are the bases to realize the process of chromosome replication and analyze the progress of cell
Title: Surveying Genome to Identify Origins of DNA Replication In Silico Abstract: DNA replication origins are the bases to realize the process of chromosome replication and analyze the progress of cell
The illustrations below reflect other scientists results in identifying and counting the stages of the onion root tip and the whitefish blastula.
 Abstract: The purpose of this laboratory experiment was to identify in what stage of mitosis viewed cells were in. The stages of mitosis include prophase, metaphase, anaphase and telophase. Although the
Abstract: The purpose of this laboratory experiment was to identify in what stage of mitosis viewed cells were in. The stages of mitosis include prophase, metaphase, anaphase and telophase. Although the
Aviva Systems Biology
 Aviva Custom Antibody Service and Price Mouse Monoclonal Antibody Service Package Number Description Package Contents Time Price Customer provides antigen protein $6,174 Monoclonal package1 (From protein
Aviva Custom Antibody Service and Price Mouse Monoclonal Antibody Service Package Number Description Package Contents Time Price Customer provides antigen protein $6,174 Monoclonal package1 (From protein
Arabidopsis. A Practical Approach. Edited by ZOE A. WILSON Plant Science Division, School of Biological Sciences, University of Nottingham
 Arabidopsis A Practical Approach Edited by ZOE A. WILSON Plant Science Division, School of Biological Sciences, University of Nottingham OXPORD UNIVERSITY PRESS List of Contributors Abbreviations xv xvu
Arabidopsis A Practical Approach Edited by ZOE A. WILSON Plant Science Division, School of Biological Sciences, University of Nottingham OXPORD UNIVERSITY PRESS List of Contributors Abbreviations xv xvu
Difficult DNA Templates Sequencing. Primer Walking Service
 Difficult DNA Templates Sequencing Primer Walking Service Result 16/18s (ITS 5.8s) rrna Sequencing Phylogenetic tree 16s rrna Region ITS rrna Region ITS and 26s rrna Region Order and Result Cloning Service
Difficult DNA Templates Sequencing Primer Walking Service Result 16/18s (ITS 5.8s) rrna Sequencing Phylogenetic tree 16s rrna Region ITS rrna Region ITS and 26s rrna Region Order and Result Cloning Service
Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1
 Supporting Online Material for Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1 Birgit Schulze, Tobias Mentzel, Anna Jehle,
Supporting Online Material for Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1 Birgit Schulze, Tobias Mentzel, Anna Jehle,
Recombinant DNA and Biotechnology
 Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
Recombinant DNA and Biotechnology Chapter 18 Lecture Objectives What Is Recombinant DNA? How Are New Genes Inserted into Cells? What Sources of DNA Are Used in Cloning? What Other Tools Are Used to Study
OBJECTIVES PROCEDURE. Lab 2- Bio 160. Name:
 Lab 2- Bio 160 Name: Prokaryotic and Eukaryotic Cells OBJECTIVES To explore cell structure and morphology in prokaryotes and eukaryotes. To gain more experience using the microscope. To obtain a better
Lab 2- Bio 160 Name: Prokaryotic and Eukaryotic Cells OBJECTIVES To explore cell structure and morphology in prokaryotes and eukaryotes. To gain more experience using the microscope. To obtain a better
Lecture 13: DNA Technology. DNA Sequencing. DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology
 Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
Lecture 13: DNA Technology DNA Sequencing Genetic Markers - RFLPs polymerase chain reaction (PCR) products of biotechnology DNA Sequencing determine order of nucleotides in a strand of DNA > bases = A,
STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS
 STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS THESIS SUBMITTED FOR THE DEGREB OF DOCTOR OF PHILOSOPHY (SCIENCE) OF THE UNIVERSITY OF CALCUTTA 1996 NRISINHA DE, M.Sc DEPARTMENT OF BIOCHEMISTRY
STUDIES ON SEED STORAGE PROTEINS OF SOME ECONOMICALLY MINOR PLANTS THESIS SUBMITTED FOR THE DEGREB OF DOCTOR OF PHILOSOPHY (SCIENCE) OF THE UNIVERSITY OF CALCUTTA 1996 NRISINHA DE, M.Sc DEPARTMENT OF BIOCHEMISTRY
quantitative real-time PCR, grain, simplex DNA extraction: PGS0426 RT-PCR: PGS0494 & PGS0476
 BioScience quantitative real-time PCR, grain, simplex DNA extraction: PGS0426 RT-PCR: PGS0494 & PGS0476 This method describes a Real-time semi-quantitative TaqMan PCR procedure for the determination of
BioScience quantitative real-time PCR, grain, simplex DNA extraction: PGS0426 RT-PCR: PGS0494 & PGS0476 This method describes a Real-time semi-quantitative TaqMan PCR procedure for the determination of
EDF Extended Depth of Field
 EDF Extended Depth of Field An upgrade for the ImageStream system Think outside the dot. Break the classic depth of field barrier Improve precision Enhance discrimination Simplify analysis Increase resolution
EDF Extended Depth of Field An upgrade for the ImageStream system Think outside the dot. Break the classic depth of field barrier Improve precision Enhance discrimination Simplify analysis Increase resolution
Introduction to Bioinformatics 3. DNA editing and contig assembly
 Introduction to Bioinformatics 3. DNA editing and contig assembly Benjamin F. Matthews United States Department of Agriculture Soybean Genomics and Improvement Laboratory Beltsville, MD 20708 matthewb@ba.ars.usda.gov
Introduction to Bioinformatics 3. DNA editing and contig assembly Benjamin F. Matthews United States Department of Agriculture Soybean Genomics and Improvement Laboratory Beltsville, MD 20708 matthewb@ba.ars.usda.gov
Rubisco; easy Purification and Immunochemical Determination
 Rubisco; easy Purification and Immunochemical Determination Ulrich Groß Justus-Liebig-Universität Gießen, Institute of Plant Nutrition, Department of Tissue Culture, Südanlage 6, D-35390 Giessen e-mail:
Rubisco; easy Purification and Immunochemical Determination Ulrich Groß Justus-Liebig-Universität Gießen, Institute of Plant Nutrition, Department of Tissue Culture, Südanlage 6, D-35390 Giessen e-mail:
BioMmune Technologies Inc. Corporate Presentation 2015
 BioMmune Technologies Inc Corporate Presentation 2015 * Harnessing the body s own immune system to fight cancer & other autoimmune diseases BioMmune Technologies Inc. (IMU) ABOUT A public biopharmaceutical
BioMmune Technologies Inc Corporate Presentation 2015 * Harnessing the body s own immune system to fight cancer & other autoimmune diseases BioMmune Technologies Inc. (IMU) ABOUT A public biopharmaceutical
HPI in-memory-based database system in Task 2b of BioASQ
 CLEF 2014 Conference and Labs of the Evaluation Forum BioASQ workshop HPI in-memory-based database system in Task 2b of BioASQ Mariana Neves September 16th, 2014 Outline 2 Overview of participation Architecture
CLEF 2014 Conference and Labs of the Evaluation Forum BioASQ workshop HPI in-memory-based database system in Task 2b of BioASQ Mariana Neves September 16th, 2014 Outline 2 Overview of participation Architecture
Data Mining Analysis (breast-cancer data)
 Data Mining Analysis (breast-cancer data) Jung-Ying Wang Register number: D9115007, May, 2003 Abstract In this AI term project, we compare some world renowned machine learning tools. Including WEKA data
Data Mining Analysis (breast-cancer data) Jung-Ying Wang Register number: D9115007, May, 2003 Abstract In this AI term project, we compare some world renowned machine learning tools. Including WEKA data
FGF-1 as Cosmetic Supplement
 FGF-1 as Cosmetic Supplement Ing-Ming Chiu, Ph.D. Professor, Internal Medicine and Molecular and Cellular Biochemistry The Ohio State University Columbus, Ohio, U.S.A. GENTEON USA Fibroblast Growth Factor
FGF-1 as Cosmetic Supplement Ing-Ming Chiu, Ph.D. Professor, Internal Medicine and Molecular and Cellular Biochemistry The Ohio State University Columbus, Ohio, U.S.A. GENTEON USA Fibroblast Growth Factor
Integrated Protein Services
 Integrated Protein Services Custom protein expression & purification Last date of revision June 2015 Version DC04-0013 www.iba-lifesciences.com Expression strategy The first step in the recombinant protein
Integrated Protein Services Custom protein expression & purification Last date of revision June 2015 Version DC04-0013 www.iba-lifesciences.com Expression strategy The first step in the recombinant protein
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY
 MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
MITOSIS IN ONION ROOT TIP CELLS: AN INTRODUCTION TO LIGHT MICROSCOPY Adapted from Foundations of Biology I; Lab 6 Introduction to Microscopy Dr. John Robertson, Westminster College Biology Department,
CyTOF2. Mass cytometry system. Unveil new cell types and function with high-parameter protein detection
 CyTOF2 Mass cytometry system Unveil new cell types and function with high-parameter protein detection DISCOVER MORE. IMAGINE MORE. MASS CYTOMETRY. THE FUTURE OF CYTOMETRY TODAY. Mass cytometry resolves
CyTOF2 Mass cytometry system Unveil new cell types and function with high-parameter protein detection DISCOVER MORE. IMAGINE MORE. MASS CYTOMETRY. THE FUTURE OF CYTOMETRY TODAY. Mass cytometry resolves
An Overview of Cells and Cell Research
 An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
An Overview of Cells and Cell Research 1 An Overview of Cells and Cell Research Chapter Outline Model Species and Cell types Cell components Tools of Cell Biology Model Species E. Coli: simplest organism
CCR Biology - Chapter 9 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Biogenesis, Size and Function of Small RNAs
 srnas of Plants Small, Non-coding RNAs of Plants Regulatory RNAs that act through gene silencing Two classes of small RNAs (srnas) o microrna (mirnas) Encoded by genes in the genome o small interfering
srnas of Plants Small, Non-coding RNAs of Plants Regulatory RNAs that act through gene silencing Two classes of small RNAs (srnas) o microrna (mirnas) Encoded by genes in the genome o small interfering
MCB41: Second Midterm Spring 2009
 MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
MCB41: Second Midterm Spring 2009 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 7 pages including this page. You will have 50 minutes for
Ontology-based Expansion of Genomic Metadata and Genometric Queries
 Ontology-based Expansion of Genomic Metadata and Genometric Queries Joint work with Javier Fernandez and Maurizio Lenzerini (Università Sapienza, Roma) Motivation. Genomic Metadata Genomic experiment meta-information:
Ontology-based Expansion of Genomic Metadata and Genometric Queries Joint work with Javier Fernandez and Maurizio Lenzerini (Università Sapienza, Roma) Motivation. Genomic Metadata Genomic experiment meta-information:
DECLARATION OF PERFORMANCE NO. HU-DOP_TN-212-25_001
 NO. HU-DOP_TN-212-25_001 Product type 212 (TN) 3,5x25 mm EN 14566:2008+A1:2009 NO. HU-DOP_TN-212-35_001 Product type 212 (TN) 3,5x35 mm EN 14566:2008+A1:2009 NO. HU-DOP_TN-212-45_001 Product type 212 (TN)
NO. HU-DOP_TN-212-25_001 Product type 212 (TN) 3,5x25 mm EN 14566:2008+A1:2009 NO. HU-DOP_TN-212-35_001 Product type 212 (TN) 3,5x35 mm EN 14566:2008+A1:2009 NO. HU-DOP_TN-212-45_001 Product type 212 (TN)
EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES
 C GUGAAG EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES MicroRNA-adapted shrna (shrna mir ) for increased, specific and consistent knockdown. MicroRNA PROCESSING PATHWAY UTILIZED FOR shrna mir Developed
C GUGAAG EXPRESSION ARREST shrna mir GENOME- WIDE LIBRARIES MicroRNA-adapted shrna (shrna mir ) for increased, specific and consistent knockdown. MicroRNA PROCESSING PATHWAY UTILIZED FOR shrna mir Developed
DNA Integrity Number (DIN) For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments
 DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
DNA Integrity Number () For the Assessment of Genomic DNA Samples in Real-Time Quantitative PCR (qpcr) Experiments Application Note Nucleic Acid Analysis Author Arunkumar Padmanaban Agilent Technologies,
Core Facility Genomics
 Core Facility Genomics versatile genome or transcriptome analyses based on quantifiable highthroughput data ascertainment 1 Topics Collaboration with Harald Binder and Clemens Kreutz Project: Microarray
Core Facility Genomics versatile genome or transcriptome analyses based on quantifiable highthroughput data ascertainment 1 Topics Collaboration with Harald Binder and Clemens Kreutz Project: Microarray
ChIP TROUBLESHOOTING TIPS
 ChIP TROUBLESHOOTING TIPS Creative Diagnostics Abstract ChIP dissects the spatial and temporal dynamics of the interactions between chromatin and its associated factors CD Creative Diagnostics info@creative-
ChIP TROUBLESHOOTING TIPS Creative Diagnostics Abstract ChIP dissects the spatial and temporal dynamics of the interactions between chromatin and its associated factors CD Creative Diagnostics info@creative-
qstar mirna qpcr Detection System
 qstar mirna qpcr Detection System Table of Contents Table of Contents...1 Package Contents and Storage Conditions...2 For mirna cdna synthesis kit...2 For qstar mirna primer pairs...2 For qstar mirna qpcr
qstar mirna qpcr Detection System Table of Contents Table of Contents...1 Package Contents and Storage Conditions...2 For mirna cdna synthesis kit...2 For qstar mirna primer pairs...2 For qstar mirna qpcr
CELERY LAB - Structure and Function of a Plant
 CELERY LAB - Structure and Function of a Plant READ ALL INSTRUCTIONS BEFORE BEGINNING! YOU MAY WORK WITH A PARTNER ON THIS ACTIVITY, BUT YOU MUST COMPLETE YOUR OWN LAB SHEET! Look at the back of this paper
CELERY LAB - Structure and Function of a Plant READ ALL INSTRUCTIONS BEFORE BEGINNING! YOU MAY WORK WITH A PARTNER ON THIS ACTIVITY, BUT YOU MUST COMPLETE YOUR OWN LAB SHEET! Look at the back of this paper
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial
 RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial Samuel J. Rulli, Jr., Ph.D. qpcr-applications Scientist Samuel.Rulli@QIAGEN.com Pathway Focused Research from Sample Prep to Data Analysis! -2-
RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial Samuel J. Rulli, Jr., Ph.D. qpcr-applications Scientist Samuel.Rulli@QIAGEN.com Pathway Focused Research from Sample Prep to Data Analysis! -2-
Guide for Data Visualization and Analysis using ACSN
 Guide for Data Visualization and Analysis using ACSN ACSN contains the NaviCell tool box, the intuitive and user- friendly environment for data visualization and analysis. The tool is accessible from the
Guide for Data Visualization and Analysis using ACSN ACSN contains the NaviCell tool box, the intuitive and user- friendly environment for data visualization and analysis. The tool is accessible from the
About Our Products. Blood Products. Purified Infectious/Inactivated Agents. Native & Recombinant Viral Proteins. DNA Controls and Primers for PCR
 About Our Products Purified Infectious/Inactivated Agents ABI produces a variety of specialized reagents, allowing researchers to choose the best preparations for their studies. Available reagents include
About Our Products Purified Infectious/Inactivated Agents ABI produces a variety of specialized reagents, allowing researchers to choose the best preparations for their studies. Available reagents include
DECLARATION OF PERFORMANCE NO. HU-DOP_TD-25_001
 NO. HU-DOP_TD-25_001 Product type TD 3,5x25 mm EN 14566:2008+A1:2009 NO. HU-DOP_TD-35_001 Product type TD 3,5x35 mm EN 14566:2008+A1:2009 NO. HU-DOP_TD-45_001 Product type TD 3,5x45 mm EN 14566:2008+A1:2009
NO. HU-DOP_TD-25_001 Product type TD 3,5x25 mm EN 14566:2008+A1:2009 NO. HU-DOP_TD-35_001 Product type TD 3,5x35 mm EN 14566:2008+A1:2009 NO. HU-DOP_TD-45_001 Product type TD 3,5x45 mm EN 14566:2008+A1:2009
NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS
 NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS Orly Alter (a) *, Gene H. Golub (b), Patrick O. Brown (c)
NOVEL GENOME-SCALE CORRELATION BETWEEN DNA REPLICATION AND RNA TRANSCRIPTION DURING THE CELL CYCLE IN YEAST IS PREDICTED BY DATA-DRIVEN MODELS Orly Alter (a) *, Gene H. Golub (b), Patrick O. Brown (c)
Manufacturing process of biologics
 Manufacturing process of biologics K. Ho Afssaps, France 2011 ICH International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use 2011 ICH 1 Disclaimer:
Manufacturing process of biologics K. Ho Afssaps, France 2011 ICH International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use 2011 ICH 1 Disclaimer:
Laboratory Observing the Cell Cycle of Onion Root Tip Cells
 Laboratory Observing the Cell Cycle of Onion Root Tip Cells Background: Because of their rapid growth, the cells of the root tips of plants undergo rapid cell division. Ornamental onion root tips cells
Laboratory Observing the Cell Cycle of Onion Root Tip Cells Background: Because of their rapid growth, the cells of the root tips of plants undergo rapid cell division. Ornamental onion root tips cells
How many of you have checked out the web site on protein-dna interactions?
 How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
How many of you have checked out the web site on protein-dna interactions? Example of an approximately 40,000 probe spotted oligo microarray with enlarged inset to show detail. Find and be ready to discuss
Introduction to Proteomics
 Introduction to Proteomics Why Proteomics? Same Genome Different Proteome Black Swallowtail - larvae and butterfly Biological Complexity Yeast - a simple proteome 6,113 proteins = 344,855 tryptic peptides
Introduction to Proteomics Why Proteomics? Same Genome Different Proteome Black Swallowtail - larvae and butterfly Biological Complexity Yeast - a simple proteome 6,113 proteins = 344,855 tryptic peptides
Multicolor Bead Flow Cytometry Standardization Heba Degheidy MD, PhD, QCYM DB/OSEL/CDRH/FDA Manager of MCM Flow Cytometry Facility
 Multicolor Bead Flow Cytometry Standardization Heba Degheidy MD, PhD, QCYM DB/OSEL/CDRH/FDA Manager of MCM Flow Cytometry Facility The mention of commercial products, their sources, or their use in connection
Multicolor Bead Flow Cytometry Standardization Heba Degheidy MD, PhD, QCYM DB/OSEL/CDRH/FDA Manager of MCM Flow Cytometry Facility The mention of commercial products, their sources, or their use in connection
Cell Biology Questions and Learning Objectives
 Cell Biology Questions and Learning Objectives (with hypothetical learning materials that might populate the objective) The topics and central questions listed here are typical for an introductory undergraduate
Cell Biology Questions and Learning Objectives (with hypothetical learning materials that might populate the objective) The topics and central questions listed here are typical for an introductory undergraduate
