Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation
|
|
|
- Juniper Thompson
- 8 years ago
- Views:
Transcription
1 Identification of rheumatoid arthritis and osterthritis patients by transcriptome-based rule set generation Bering Limited Report generated on September 19, 2014 Contents 1 Dataset summary Project description Array processing and normalization 3 3 Quality Outlier detection Principal Component Analysis Signal density and box plots Array similarity heatmap and hierarchical clustering Batch correction Quality summary Differential expression analysis 8 5 Gene Ontology enrichment Biological Process Cellular Component Molecular Function Reactome pathway enrichment 11 1
2 1 Dataset summary Number of samples: 30 Number of chip identifiers: Comparison: ra vs. 1.1 Project description ArrayExpression accession number: E-GEO Discrimination of rheumatoid arthritis (RA) patients from patients with other inflammatory/degenerative joint diseases or healthy individuals purely on the basis of genes differentially expressed in high-throughput data has proven very difficult. Thus, the present study sought to achieve such discrimination by employing a novel unbiased apprch using rule-based classifiers. Three multi-center genome-wide transcriptomic data sets (Affymetrix HG- U133 A/B) from a total of 79 individuals, including 20 healthy s ( group - CG), as well as 26 osterthritis (OA) and 33 RA patients. Reference: Woetzel D., et al. Identification of rheumatoid arthritis and osterthritis patients by transcriptome-based rule set generation. Arthritis Research & Therapy 2014, 16:R84 file.name GSM ND 1 S... GSM ND 2 S... GSM ND 3 S... GSM ND 4 S... GSM ND 5 S... GSM ND 6 S... GSM ND 7 S... GSM ND 8 S... GSM ND 9 S... GSM ND GSM OA 1 S... GSM OA 2 S... GSM OA 3 S... GSM OA 4 S... GSM OA 5 S... GSM OA 6 S... GSM OA 7 S... GSM OA 8 S... GSM OA 9 S... GSM OA phenotype
3 GSM RA 1 S... ra GSM RA 2 S... ra GSM RA 3 S... ra GSM RA 4 S... ra GSM RA 5 S... ra GSM RA 6 S... ra GSM RA 7 S... ra GSM RA 8 S... ra GSM RA 9 S... ra GSM RA ra Table 1: Sample-data relationships 2 Array processing and normalization After array normalization and detection of present probes, probes were retained. 3 Quality GeneProfiler pipeline aims to identify outliers, batch effects, and overly noisy experiments. Automated quality is carried out using the arrayqualitymetrics Bioconductor package. 3.1 Outlier detection Outlier detection is carried out using three distinct apprches: Box plot: Each box corresponds to one array. Typically, one expects the boxes to have similar positions and widths. If the distribution of an array is very different from the others, this may indicate an experimental problem. Outlier detection is performed by computing the Kolmogorov-Smirnov statistic Ka between each array s distribution and the distribution of the pooled data. Signal Density plot: Typically, the distributions of the arrays should have similar shapes and ranges. Arrays whose distributions are very different from the others should be considered for possible problems. Inter-sample correlation heatmap: Patterns in this plot can indicate clustering of the arrays either because of intended biological or unintended experimental factors (batch effects). The distance between two arrays is computed as the mean absolute difference between the data of the arrays (using the data from all probes without filtering). Outlier detection is performed by
4 looking for arrays for which the sum of the distances to all other arrays is exceptionally large. Results of outlier detection are shown in Table 2. Table columns contain results of a specific outlier detection test. FALSE value indicates that an array is not an outlier, while TRUE value indiciates that an array is an outlier. An array will be considered an outlier, and labeled so in a column Vote, if it is called an outlier by at least two methods. Boxplot Density Heatmap Vote GSM ND 1 S... FALSE FALSE FALSE FALSE GSM ND 2 S... TRUE FALSE FALSE FALSE GSM ND 3 S... FALSE FALSE FALSE FALSE GSM ND 4 S... FALSE FALSE FALSE FALSE GSM ND 5 S... FALSE FALSE FALSE FALSE GSM ND 6 S... FALSE FALSE FALSE FALSE GSM ND 7 S... TRUE FALSE FALSE FALSE GSM ND 8 S... TRUE FALSE FALSE FALSE GSM ND 9 S... FALSE FALSE FALSE FALSE GSM ND FALSE FALSE FALSE FALSE GSM OA 1 S... FALSE FALSE FALSE FALSE GSM OA 2 S... FALSE FALSE FALSE FALSE GSM OA 3 S... FALSE FALSE FALSE FALSE GSM OA 4 S... FALSE FALSE FALSE FALSE GSM OA 5 S... FALSE FALSE FALSE FALSE GSM OA 6 S... FALSE FALSE FALSE FALSE GSM OA 7 S... FALSE FALSE FALSE FALSE GSM OA 8 S... FALSE FALSE FALSE FALSE GSM OA 9 S... FALSE FALSE FALSE FALSE GSM OA TRUE FALSE FALSE FALSE GSM RA 1 S... FALSE FALSE FALSE FALSE GSM RA 2 S... FALSE FALSE FALSE FALSE GSM RA 3 S... FALSE FALSE FALSE FALSE GSM RA 4 S... FALSE FALSE FALSE FALSE GSM RA 5 S... FALSE FALSE FALSE FALSE GSM RA 6 S... FALSE FALSE FALSE FALSE GSM RA 7 S... FALSE FALSE FALSE FALSE GSM RA 8 S... FALSE FALSE FALSE FALSE GSM RA 9 S... FALSE FALSE FALSE FALSE GSM RA FALSE FALSE FALSE FALSE Table 2: Outlying arrays.
5 3.1.1 Principal Component Analysis Figure 1: Scatterplot visualising Principal Component Analysis for 30 arrays. Outliers (if any) are shown in red. Principal Components Analysis (PCA) plots were used to visualize the overall quality of a micrrray dataset. Each point in the PCA plots corresponds to an array. Dissimilar arrays are further apart.
6 3.1.2 Signal density and box plots Figure 2: Boxplots and signal intensity densities for 30 arrays. Outliers (if any) are shown in red Array similarity heatmap and hierarchical clustering Hiearachical clustering was used to determine if sample clusters correspond to the experimental sample groups, rather than to technical sources of variation.
7 Figure 3: Array similarity heatmap for 30 arrays. The color scale is chosen to cover the range of distances encountered in the dataset. There were 0 outlying arrays. 3.2 Batch correction If batches are specified, they are corrected. 3.3 Quality summary Of probes, passed quality protocols. 30 samples passed outlier detection criteria.
8 4 Differential expression analysis Differential expression analysis was carried out comparing ra vs.. There were 692 up-regulated and 801 down-regulated genes (p value 0.05, FDR-correction: No). Top 10 differentially expressed genes are shown in Table 3. Symbol Name logfc P.Value CXCL13 Chemokine (C-X-C motif) ligand 1.1E E SLAMF8 SLAM family member 8 7.4E E-12 TPD52L1 Tumor protein D52-like 1-4.8E E-11 ADAMDEC1 ADAM-like, decysin 1 4.7E E-10 SERPINA1 Serpin peptidase inhibitor, 4.6E E-09 clade A (alpha-1 antiproteinase, antitrypsin), member 1 NOVA1 Neuro-oncological ventral -3.0E E-10 antigen 1 CCL13 Chemokine (C-C motif) ligand 4.5E E ISG20 Interferon stimulated 2.3E E-10 exonuclease gene 20kDa CD27 CD27 molecule 2.9E E-09 CRLF1 Cytokine receptor-like factor 1-4.7E E-07 Table 3: Top 10 differentially expressed genes.
9 Figure 4: Volcano plot of all differentially expressed genes in ra vs.. Top 5 differentially expressed genes are labeled. 5 Gene Ontology enrichment 1493 differentially expressed genes were enriched for Gene Ontology (GO) Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) terms. All micrrray genes (n=8934) were used as background. Headers in Tables 4, 5, and 6, Significant and P.Value reffer to number of significant genes annotated by a term and corresponding significance p-values respectively.
10 5.1 Biological Process Term Significant P.Value Immune Response E-30 Immune System Process E-25 Defense Response E-20 Regulation Of Immune System Process E-19 Regulation Of Immune Response E-19 Positive Regulation Of Immune System Process E-18 Positive Regulation Of Response To Stimulus E-17 Regulation Of Response To Stimulus E-17 Signal Transduction E-17 Signaling E-16 Table 4: Top enriched Gene Ontology Biological Process terms. 5.2 Cellular Component Term Significant P.Value Cell Periphery E-18 Plasma Membrane E-18 Membrane E-17 Extracellular Region E-13 Membrane Part E-11 Intrinsic Component Of Membrane E-11 Integral Component Of Membrane E-10 Extracellular Region Part E-10 Extracellular Space E-10 Side Of Membrane E-10 Table 5: Top enriched Gene Ontology Cellular Component terms. 5.3 Molecular Function Term Significant P.Value Receptor Activity E-12 Signal Transducer Activity E-09 Molecular Transducer Activity E-09 Receptor Binding E-09 Transmembrane Signaling Receptor Activity E-08 Signaling Receptor Activity E-08 Antigen Binding E-08 Sulfur Compound Binding E-07 Heparin Binding E-06 Chemokine Activity E-06 Table 6: Top enriched Gene Ontology Molecular Function terms.
11 6 Reactome pathway enrichment 1493 differentially expressed genes were enriched for Reactome pathways. Top 10 enriched pathways are shown in Table 7. Description P.Value Count Activity.Score Immune System 1.50E Adaptive Immune System 5.40E Phosphorylation of CD3 and TCR zeta 1.90E chains Lipid and lipoprotein metabolism 1.90E Hemostasis 2.00E TCR signaling 2.00E Cytokine Signaling in Immune system 2.60E Platelet activation, signaling and 2.70E aggregation Antigen Activates B Cell Receptor Leading 3.00E to Generation of Second Messengers Alternative complement activation 3.10E Table 7: Top enriched Reactome Pathways. Column P.Value refers to raw enrichment significance p-values. Column Count highlights the total number of differentially expressed genes assigned to a specific pathway. Column Activity.Score corresponds to the average pathway fold change in ra vs. comparison.
Analysis of Illumina Gene Expression Microarray Data
 Analysis of Illumina Gene Expression Microarray Data Asta Laiho, Msc. Tech. Bioinformatics research engineer The Finnish DNA Microarray Centre Turku Centre for Biotechnology, Finland The Finnish DNA Microarray
Analysis of Illumina Gene Expression Microarray Data Asta Laiho, Msc. Tech. Bioinformatics research engineer The Finnish DNA Microarray Centre Turku Centre for Biotechnology, Finland The Finnish DNA Microarray
Microarray Data Analysis. A step by step analysis using BRB-Array Tools
 Microarray Data Analysis A step by step analysis using BRB-Array Tools 1 EXAMINATION OF DIFFERENTIAL GENE EXPRESSION (1) Objective: to find genes whose expression is changed before and after chemotherapy.
Microarray Data Analysis A step by step analysis using BRB-Array Tools 1 EXAMINATION OF DIFFERENTIAL GENE EXPRESSION (1) Objective: to find genes whose expression is changed before and after chemotherapy.
Tutorial for proteome data analysis using the Perseus software platform
 Tutorial for proteome data analysis using the Perseus software platform Laboratory of Mass Spectrometry, LNBio, CNPEM Tutorial version 1.0, January 2014. Note: This tutorial was written based on the information
Tutorial for proteome data analysis using the Perseus software platform Laboratory of Mass Spectrometry, LNBio, CNPEM Tutorial version 1.0, January 2014. Note: This tutorial was written based on the information
The Advantages and Disadvantages of Using Gene Ontology
 Extracting Biological Information from Gene Lists Simon Andrews, Laura Biggins, Boo Virk simon.andrews@babraham.ac.uk laura.biggins@babraham.ac.uk boo.virk@babraham.ac.uk v1.0 Biological material Sample
Extracting Biological Information from Gene Lists Simon Andrews, Laura Biggins, Boo Virk simon.andrews@babraham.ac.uk laura.biggins@babraham.ac.uk boo.virk@babraham.ac.uk v1.0 Biological material Sample
Analyzing the Effect of Treatment and Time on Gene Expression in Partek Genomics Suite (PGS) 6.6: A Breast Cancer Study
 Analyzing the Effect of Treatment and Time on Gene Expression in Partek Genomics Suite (PGS) 6.6: A Breast Cancer Study The data for this study is taken from experiment GSE848 from the Gene Expression
Analyzing the Effect of Treatment and Time on Gene Expression in Partek Genomics Suite (PGS) 6.6: A Breast Cancer Study The data for this study is taken from experiment GSE848 from the Gene Expression
B Cell Generation, Activation & Differentiation. B cell maturation
 B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
Hormones & Chemical Signaling
 Hormones & Chemical Signaling Part 2 modulation of signal pathways and hormone classification & function How are these pathways controlled? Receptors are proteins! Subject to Specificity of binding Competition
Hormones & Chemical Signaling Part 2 modulation of signal pathways and hormone classification & function How are these pathways controlled? Receptors are proteins! Subject to Specificity of binding Competition
Course on Functional Analysis. ::: Gene Set Enrichment Analysis - GSEA -
 Course on Functional Analysis ::: Madrid, June 31st, 2007. Gonzalo Gómez, PhD. ggomez@cnio.es Bioinformatics Unit CNIO ::: Contents. 1. Introduction. 2. GSEA Software 3. Data Formats 4. Using GSEA 5. GSEA
Course on Functional Analysis ::: Madrid, June 31st, 2007. Gonzalo Gómez, PhD. ggomez@cnio.es Bioinformatics Unit CNIO ::: Contents. 1. Introduction. 2. GSEA Software 3. Data Formats 4. Using GSEA 5. GSEA
Hierarchical Clustering Analysis
 Hierarchical Clustering Analysis What is Hierarchical Clustering? Hierarchical clustering is used to group similar objects into clusters. In the beginning, each row and/or column is considered a cluster.
Hierarchical Clustering Analysis What is Hierarchical Clustering? Hierarchical clustering is used to group similar objects into clusters. In the beginning, each row and/or column is considered a cluster.
TEMA 10. REACCIONES INMUNITARIAS MEDIADAS POR CÉLULAS.
 TEMA 10. REACCIONES INMUNITARIAS MEDIADAS POR CÉLULAS. The nomenclature of cytokines partly reflects their first-described function and also the order of their discovery. There is no single unified nomenclature,
TEMA 10. REACCIONES INMUNITARIAS MEDIADAS POR CÉLULAS. The nomenclature of cytokines partly reflects their first-described function and also the order of their discovery. There is no single unified nomenclature,
Analysis of the colorectal tumor microenvironment using integrative bioinformatic tools
 MLECNIK Bernhard & BINDEA Gabriela Analysis of the colorectal tumor microenvironment using integrative bioinformatic tools INSERM U872, Jérôme Galon Team15: Integrative Cancer Immunology Cordeliers Research
MLECNIK Bernhard & BINDEA Gabriela Analysis of the colorectal tumor microenvironment using integrative bioinformatic tools INSERM U872, Jérôme Galon Team15: Integrative Cancer Immunology Cordeliers Research
ProteinQuest user guide
 ProteinQuest user guide 1. Introduction... 3 1.1 With ProteinQuest you can... 3 1.2 ProteinQuest basic version 4 1.3 ProteinQuest extended version... 5 2. ProteinQuest dictionaries... 6 3. Directions for
ProteinQuest user guide 1. Introduction... 3 1.1 With ProteinQuest you can... 3 1.2 ProteinQuest basic version 4 1.3 ProteinQuest extended version... 5 2. ProteinQuest dictionaries... 6 3. Directions for
Molecule Shapes. support@ingenuity.com www.ingenuity.com 1
 IPA 8 Legend This legend provides a key of the main features of Network Explorer and Canonical Pathways, including molecule shapes and colors as well as relationship labels and types. For a high-resolution
IPA 8 Legend This legend provides a key of the main features of Network Explorer and Canonical Pathways, including molecule shapes and colors as well as relationship labels and types. For a high-resolution
BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS
 BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS 1. The Technology Strategy sets out six areas where technological developments are required to push the frontiers of knowledge
BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS 1. The Technology Strategy sets out six areas where technological developments are required to push the frontiers of knowledge
specific B cells Humoral immunity lymphocytes antibodies B cells bone marrow Cell-mediated immunity: T cells antibodies proteins
 Adaptive Immunity Chapter 17: Adaptive (specific) Immunity Bio 139 Dr. Amy Rogers Host defenses that are specific to a particular infectious agent Can be innate or genetic for humans as a group: most microbes
Adaptive Immunity Chapter 17: Adaptive (specific) Immunity Bio 139 Dr. Amy Rogers Host defenses that are specific to a particular infectious agent Can be innate or genetic for humans as a group: most microbes
Dr Alexander Henzing
 Horizon 2020 Health, Demographic Change & Wellbeing EU funding, research and collaboration opportunities for 2016/17 Innovate UK funding opportunities in omics, bridging health and life sciences Dr Alexander
Horizon 2020 Health, Demographic Change & Wellbeing EU funding, research and collaboration opportunities for 2016/17 Innovate UK funding opportunities in omics, bridging health and life sciences Dr Alexander
Guide for Data Visualization and Analysis using ACSN
 Guide for Data Visualization and Analysis using ACSN ACSN contains the NaviCell tool box, the intuitive and user- friendly environment for data visualization and analysis. The tool is accessible from the
Guide for Data Visualization and Analysis using ACSN ACSN contains the NaviCell tool box, the intuitive and user- friendly environment for data visualization and analysis. The tool is accessible from the
Analyzing microrna Data and Integrating mirna with Gene Expression Data in Partek Genomics Suite 6.6
 Analyzing microrna Data and Integrating mirna with Gene Expression Data in Partek Genomics Suite 6.6 Overview This tutorial outlines how microrna data can be analyzed within Partek Genomics Suite. Additionally,
Analyzing microrna Data and Integrating mirna with Gene Expression Data in Partek Genomics Suite 6.6 Overview This tutorial outlines how microrna data can be analyzed within Partek Genomics Suite. Additionally,
Gene expression analysis. Ulf Leser and Karin Zimmermann
 Gene expression analysis Ulf Leser and Karin Zimmermann Ulf Leser: Bioinformatics, Wintersemester 2010/2011 1 Last lecture What are microarrays? - Biomolecular devices measuring the transcriptome of a
Gene expression analysis Ulf Leser and Karin Zimmermann Ulf Leser: Bioinformatics, Wintersemester 2010/2011 1 Last lecture What are microarrays? - Biomolecular devices measuring the transcriptome of a
Molecular Genetics: Challenges for Statistical Practice. J.K. Lindsey
 Molecular Genetics: Challenges for Statistical Practice J.K. Lindsey 1. What is a Microarray? 2. Design Questions 3. Modelling Questions 4. Longitudinal Data 5. Conclusions 1. What is a microarray? A microarray
Molecular Genetics: Challenges for Statistical Practice J.K. Lindsey 1. What is a Microarray? 2. Design Questions 3. Modelling Questions 4. Longitudinal Data 5. Conclusions 1. What is a microarray? A microarray
Quantitative proteomics background
 Proteomics data analysis seminar Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post transcriptional regulation of key cellular processes de Groot, M., Daran
Proteomics data analysis seminar Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post transcriptional regulation of key cellular processes de Groot, M., Daran
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
Chapter 43: The Immune System
 Name Period Our students consider this chapter to be a particularly challenging and important one. Expect to work your way slowly through the first three concepts. Take particular care with Concepts 43.2
Name Period Our students consider this chapter to be a particularly challenging and important one. Expect to work your way slowly through the first three concepts. Take particular care with Concepts 43.2
Network Webinar Series
 Undergraduate Educator Network Series Sponsored by Undergraduate Education Subcommittee SOT Education Committee June 4, 2015 12:00 Noon ET (c) SOT2015 Welcome Kristine Willett, PhD Co-Chair, C Undergraduate
Undergraduate Educator Network Series Sponsored by Undergraduate Education Subcommittee SOT Education Committee June 4, 2015 12:00 Noon ET (c) SOT2015 Welcome Kristine Willett, PhD Co-Chair, C Undergraduate
Exiqon Array Software Manual. Quick guide to data extraction from mircury LNA microrna Arrays
 Exiqon Array Software Manual Quick guide to data extraction from mircury LNA microrna Arrays March 2010 Table of contents Introduction Overview...................................................... 3 ImaGene
Exiqon Array Software Manual Quick guide to data extraction from mircury LNA microrna Arrays March 2010 Table of contents Introduction Overview...................................................... 3 ImaGene
AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE
 ACCELERATING PROGRESS IS IN OUR GENES AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE GENESPRING GENE EXPRESSION (GX) MASS PROFILER PROFESSIONAL (MPP) PATHWAY ARCHITECT (PA) See Deeper. Reach Further. BIOINFORMATICS
ACCELERATING PROGRESS IS IN OUR GENES AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE GENESPRING GENE EXPRESSION (GX) MASS PROFILER PROFESSIONAL (MPP) PATHWAY ARCHITECT (PA) See Deeper. Reach Further. BIOINFORMATICS
B Cells and Antibodies
 B Cells and Antibodies Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School Lecture outline Functions of antibodies B cell activation; the role of helper T cells in antibody production
B Cells and Antibodies Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School Lecture outline Functions of antibodies B cell activation; the role of helper T cells in antibody production
Lecture 11 Data storage and LIMS solutions. Stéphane LE CROM lecrom@biologie.ens.fr
 Lecture 11 Data storage and LIMS solutions Stéphane LE CROM lecrom@biologie.ens.fr Various steps of a DNA microarray experiment Experimental steps Data analysis Experimental design set up Chips on catalog
Lecture 11 Data storage and LIMS solutions Stéphane LE CROM lecrom@biologie.ens.fr Various steps of a DNA microarray experiment Experimental steps Data analysis Experimental design set up Chips on catalog
Recognition of T cell epitopes (Abbas Chapter 6)
 Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
HUMORAL IMMUNE RE- SPONSES: ACTIVATION OF B CELLS AND ANTIBODIES JASON CYSTER SECTION 13
 SECTION 13 HUMORAL IMMUNE RE- SPONSES: ACTIVATION OF B CELLS AND ANTIBODIES CONTACT INFORMATION Jason Cyster, PhD (Email) READING Basic Immunology: Functions and Disorders of the Immune System. Abbas,
SECTION 13 HUMORAL IMMUNE RE- SPONSES: ACTIVATION OF B CELLS AND ANTIBODIES CONTACT INFORMATION Jason Cyster, PhD (Email) READING Basic Immunology: Functions and Disorders of the Immune System. Abbas,
Analysis of gene expression data. Ulf Leser and Philippe Thomas
 Analysis of gene expression data Ulf Leser and Philippe Thomas This Lecture Protein synthesis Microarray Idea Technologies Applications Problems Quality control Normalization Analysis next week! Ulf Leser:
Analysis of gene expression data Ulf Leser and Philippe Thomas This Lecture Protein synthesis Microarray Idea Technologies Applications Problems Quality control Normalization Analysis next week! Ulf Leser:
A Streamlined Workflow for Untargeted Metabolomics
 A Streamlined Workflow for Untargeted Metabolomics Employing XCMS plus, a Simultaneous Data Processing and Metabolite Identification Software Package for Rapid Untargeted Metabolite Screening Baljit K.
A Streamlined Workflow for Untargeted Metabolomics Employing XCMS plus, a Simultaneous Data Processing and Metabolite Identification Software Package for Rapid Untargeted Metabolite Screening Baljit K.
(A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors.
 Legends of supplemental figures and tables Figure 1: Overview of study design and results. (A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors. After raw data gene expression
Legends of supplemental figures and tables Figure 1: Overview of study design and results. (A) Microarray analysis was performed on ATM and MDM isolated from 4 obese donors. After raw data gene expression
Statistical Analysis. NBAF-B Metabolomics Masterclass. Mark Viant
 Statistical Analysis NBAF-B Metabolomics Masterclass Mark Viant 1. Introduction 2. Univariate analysis Overview of lecture 3. Unsupervised multivariate analysis Principal components analysis (PCA) Interpreting
Statistical Analysis NBAF-B Metabolomics Masterclass Mark Viant 1. Introduction 2. Univariate analysis Overview of lecture 3. Unsupervised multivariate analysis Principal components analysis (PCA) Interpreting
Autoimmunity and immunemediated. FOCiS. Lecture outline
 1 Autoimmunity and immunemediated inflammatory diseases Abul K. Abbas, MD UCSF FOCiS 2 Lecture outline Pathogenesis of autoimmunity: why selftolerance fails Genetics of autoimmune diseases Therapeutic
1 Autoimmunity and immunemediated inflammatory diseases Abul K. Abbas, MD UCSF FOCiS 2 Lecture outline Pathogenesis of autoimmunity: why selftolerance fails Genetics of autoimmune diseases Therapeutic
Actions of Hormones on Target Cells Page 1. Actions of Hormones on Target Cells Page 2. Goals/ What You Need to Know Goals What You Need to Know
 Actions of Hormones on Target Cells Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com) Page 1. Actions of Hormones on Target Cells Hormones
Actions of Hormones on Target Cells Graphics are used with permission of: Pearson Education Inc., publishing as Benjamin Cummings (http://www.aw-bc.com) Page 1. Actions of Hormones on Target Cells Hormones
Factors for success in big data science
 Factors for success in big data science Damjan Vukcevic Data Science Murdoch Childrens Research Institute 16 October 2014 Big Data Reading Group (Department of Mathematics & Statistics, University of Melbourne)
Factors for success in big data science Damjan Vukcevic Data Science Murdoch Childrens Research Institute 16 October 2014 Big Data Reading Group (Department of Mathematics & Statistics, University of Melbourne)
Exploratory data analysis for microarray data
 Eploratory data analysis for microarray data Anja von Heydebreck Ma Planck Institute for Molecular Genetics, Dept. Computational Molecular Biology, Berlin, Germany heydebre@molgen.mpg.de Visualization
Eploratory data analysis for microarray data Anja von Heydebreck Ma Planck Institute for Molecular Genetics, Dept. Computational Molecular Biology, Berlin, Germany heydebre@molgen.mpg.de Visualization
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology
 Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology The Master Degree in Medical Laboratory Sciences / Clinical Microbiology, Immunology or
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Microbiology, Immunology and Serology The Master Degree in Medical Laboratory Sciences / Clinical Microbiology, Immunology or
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
Hapten - a small molecule that is antigenic but not (by itself) immunogenic.
 Chapter 3. Antigens Terminology: Antigen: Substances that can be recognized by the surface antibody (B cells) or by the TCR (T cells) when associated with MHC molecules Immunogenicity VS Antigenicity:
Chapter 3. Antigens Terminology: Antigen: Substances that can be recognized by the surface antibody (B cells) or by the TCR (T cells) when associated with MHC molecules Immunogenicity VS Antigenicity:
DeCyder Extended Data Analysis module Version 1.0
 GE Healthcare DeCyder Extended Data Analysis module Version 1.0 Module for DeCyder 2D version 6.5 User Manual Contents 1 Introduction 1.1 Introduction... 7 1.2 The DeCyder EDA User Manual... 9 1.3 Getting
GE Healthcare DeCyder Extended Data Analysis module Version 1.0 Module for DeCyder 2D version 6.5 User Manual Contents 1 Introduction 1.1 Introduction... 7 1.2 The DeCyder EDA User Manual... 9 1.3 Getting
Predictive Gene Signature Selection for Adjuvant Chemotherapy in Non-Small Cell Lung Cancer Patients
 Predictive Gene Signature Selection for Adjuvant Chemotherapy in Non-Small Cell Lung Cancer Patients by Li Liu A practicum report submitted to the Department of Public Health Sciences in conformity with
Predictive Gene Signature Selection for Adjuvant Chemotherapy in Non-Small Cell Lung Cancer Patients by Li Liu A practicum report submitted to the Department of Public Health Sciences in conformity with
Exercise with Gene Ontology - Cytoscape - BiNGO
 Exercise with Gene Ontology - Cytoscape - BiNGO This practical has material extracted from http://www.cbs.dtu.dk/chipcourse/exercises/ex_go/goexercise11.php In this exercise we will analyze microarray
Exercise with Gene Ontology - Cytoscape - BiNGO This practical has material extracted from http://www.cbs.dtu.dk/chipcourse/exercises/ex_go/goexercise11.php In this exercise we will analyze microarray
CyTOF2. Mass cytometry system. Unveil new cell types and function with high-parameter protein detection
 CyTOF2 Mass cytometry system Unveil new cell types and function with high-parameter protein detection DISCOVER MORE. IMAGINE MORE. MASS CYTOMETRY. THE FUTURE OF CYTOMETRY TODAY. Mass cytometry resolves
CyTOF2 Mass cytometry system Unveil new cell types and function with high-parameter protein detection DISCOVER MORE. IMAGINE MORE. MASS CYTOMETRY. THE FUTURE OF CYTOMETRY TODAY. Mass cytometry resolves
Understanding West Nile Virus Infection
 Understanding West Nile Virus Infection The QIAGEN Bioinformatics Solution: Biomedical Genomics Workbench (BXWB) + Ingenuity Pathway Analysis (IPA) Functional Genomics & Predictive Medicine, May 21-22,
Understanding West Nile Virus Infection The QIAGEN Bioinformatics Solution: Biomedical Genomics Workbench (BXWB) + Ingenuity Pathway Analysis (IPA) Functional Genomics & Predictive Medicine, May 21-22,
T Cell Maturation,Activation and Differentiation
 T Cell Maturation,Activation and Differentiation Positive Selection- In thymus, permits survival of only those T cells whose TCRs recognize self- MHC molecules (self-mhc restriction) Negative Selection-
T Cell Maturation,Activation and Differentiation Positive Selection- In thymus, permits survival of only those T cells whose TCRs recognize self- MHC molecules (self-mhc restriction) Negative Selection-
ANIMALS FORM & FUNCTION BODY DEFENSES NONSPECIFIC DEFENSES PHYSICAL BARRIERS PHAGOCYTES. Animals Form & Function Activity #4 page 1
 AP BIOLOGY ANIMALS FORM & FUNCTION ACTIVITY #4 NAME DATE HOUR BODY DEFENSES NONSPECIFIC DEFENSES PHYSICAL BARRIERS PHAGOCYTES Animals Form & Function Activity #4 page 1 INFLAMMATORY RESPONSE ANTIMICROBIAL
AP BIOLOGY ANIMALS FORM & FUNCTION ACTIVITY #4 NAME DATE HOUR BODY DEFENSES NONSPECIFIC DEFENSES PHYSICAL BARRIERS PHAGOCYTES Animals Form & Function Activity #4 page 1 INFLAMMATORY RESPONSE ANTIMICROBIAL
Deep profiling of multitube flow cytometry data Supplemental information
 Deep profiling of multitube flow cytometry data Supplemental information Kieran O Neill et al December 19, 2014 1 Table S1: Markers in simulated multitube data. The data was split into three tubes, each
Deep profiling of multitube flow cytometry data Supplemental information Kieran O Neill et al December 19, 2014 1 Table S1: Markers in simulated multitube data. The data was split into three tubes, each
The immune system. Bone marrow. Thymus. Spleen. Bone marrow. NK cell. B-cell. T-cell. Basophil Neutrophil. Eosinophil. Myeloid progenitor
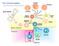 The immune system Basophil Neutrophil Bone marrow Eosinophil Myeloid progenitor Dendritic cell Pluripotent Stem cell Lymphoid progenitor Platelets Bone marrow Thymus NK cell T-cell B-cell Spleen Cancer
The immune system Basophil Neutrophil Bone marrow Eosinophil Myeloid progenitor Dendritic cell Pluripotent Stem cell Lymphoid progenitor Platelets Bone marrow Thymus NK cell T-cell B-cell Spleen Cancer
Statistical issues in the analysis of microarray data
 Statistical issues in the analysis of microarray data Daniel Gerhard Institute of Biostatistics Leibniz University of Hannover ESNATS Summerschool, Zermatt D. Gerhard (LUH) Analysis of microarray data
Statistical issues in the analysis of microarray data Daniel Gerhard Institute of Biostatistics Leibniz University of Hannover ESNATS Summerschool, Zermatt D. Gerhard (LUH) Analysis of microarray data
Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation
 Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation Woetzel et al. Woetzel et al. Arthritis Research & Therapy 2014, 16:R84 Woetzel et al. Arthritis
Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation Woetzel et al. Woetzel et al. Arthritis Research & Therapy 2014, 16:R84 Woetzel et al. Arthritis
Comparing Methods for Identifying Transcription Factor Target Genes
 Comparing Methods for Identifying Transcription Factor Target Genes Alena van Bömmel (R 3.3.73) Matthew Huska (R 3.3.18) Max Planck Institute for Molecular Genetics Folie 1 Transcriptional Regulation TF
Comparing Methods for Identifying Transcription Factor Target Genes Alena van Bömmel (R 3.3.73) Matthew Huska (R 3.3.18) Max Planck Institute for Molecular Genetics Folie 1 Transcriptional Regulation TF
 Blood Sticky, opaque fluid with a metallic taste (Fe 2+ ) Varies from scarlet (P O2 = 100) to dark red (P O2 = 40) ph is between 7.35 and 7.45 Average volume in an adult is 5 L (7% of body weight) 2 L
Blood Sticky, opaque fluid with a metallic taste (Fe 2+ ) Varies from scarlet (P O2 = 100) to dark red (P O2 = 40) ph is between 7.35 and 7.45 Average volume in an adult is 5 L (7% of body weight) 2 L
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Biochemistry
 Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Biochemistry The Master Degree in Medical Laboratory Sciences /Clinical Biochemistry, is awarded by the Faculty of Graduate Studies
Course Curriculum for Master Degree in Medical Laboratory Sciences/Clinical Biochemistry The Master Degree in Medical Laboratory Sciences /Clinical Biochemistry, is awarded by the Faculty of Graduate Studies
ALLEN Mouse Brain Atlas
 TECHNICAL WHITE PAPER: QUALITY CONTROL STANDARDS FOR HIGH-THROUGHPUT RNA IN SITU HYBRIDIZATION DATA GENERATION Consistent data quality and internal reproducibility are critical concerns for high-throughput
TECHNICAL WHITE PAPER: QUALITY CONTROL STANDARDS FOR HIGH-THROUGHPUT RNA IN SITU HYBRIDIZATION DATA GENERATION Consistent data quality and internal reproducibility are critical concerns for high-throughput
Quality Assessment of Exon and Gene Arrays
 Quality Assessment of Exon and Gene Arrays I. Introduction In this white paper we describe some quality assessment procedures that are computed from CEL files from Whole Transcript (WT) based arrays such
Quality Assessment of Exon and Gene Arrays I. Introduction In this white paper we describe some quality assessment procedures that are computed from CEL files from Whole Transcript (WT) based arrays such
Lecture 8. Protein Trafficking/Targeting. Protein targeting is necessary for proteins that are destined to work outside the cytoplasm.
 Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex
Class time required: Two 40-minute class periods + homework. Part 1 may be done as pre-lab homework
 Diagnosing Diabetes Teacher information Summary: Students analyze simulated blood plasma samples collected during a glucose tolerance test for diabetes. They test glucose and insulin levels to determine
Diagnosing Diabetes Teacher information Summary: Students analyze simulated blood plasma samples collected during a glucose tolerance test for diabetes. They test glucose and insulin levels to determine
Chapter-21b: Hormones and Receptors
 1 hapter-21b: Hormones and Receptors Hormone classes Hormones are classified according to the distance over which they act. 1. Autocrine hormones --- act on the same cell that released them. Interleukin-2
1 hapter-21b: Hormones and Receptors Hormone classes Hormones are classified according to the distance over which they act. 1. Autocrine hormones --- act on the same cell that released them. Interleukin-2
Microarray Data Mining: Puce a ADN
 Microarray Data Mining: Puce a ADN Recent Developments Gregory Piatetsky-Shapiro KDnuggets EGC 2005, Paris 2005 KDnuggets EGC 2005 Role of Gene Expression Cell Nucleus Chromosome Gene expression Protein
Microarray Data Mining: Puce a ADN Recent Developments Gregory Piatetsky-Shapiro KDnuggets EGC 2005, Paris 2005 KDnuggets EGC 2005 Role of Gene Expression Cell Nucleus Chromosome Gene expression Protein
Activation and effector functions of HMI
 Activation and effector functions of HMI Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 August 2015 ว ตถ ประสงค หล งจากช วโมงบรรยายน แล วน กศ กษาสามารถ
Activation and effector functions of HMI Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 August 2015 ว ตถ ประสงค หล งจากช วโมงบรรยายน แล วน กศ กษาสามารถ
Validated Cell-Based Assays for Rapid Screening and Functional Characterization of Therapeutic Monoclonal Antibodies
 Validated Cell-Based Assays for Rapid Screening and Functional Characterization of Therapeutic Monoclonal Antibodies Abhishek Saharia, Ph. D. Senior Product Manager Validation Assays Drug Discovery Evolution
Validated Cell-Based Assays for Rapid Screening and Functional Characterization of Therapeutic Monoclonal Antibodies Abhishek Saharia, Ph. D. Senior Product Manager Validation Assays Drug Discovery Evolution
Visualization of the Phosphoproteomic Data from AfCS with the Google Motion Chart Gadget
 Visualization of the Phosphoproteomic Data from AfCS with the Google Motion Chart Gadget Huilei Xu 1, and Avi Ma ayan 1,* 1 Department of Pharmacology and Systems Therapeutics, Mount Sinai School of Medicine,
Visualization of the Phosphoproteomic Data from AfCS with the Google Motion Chart Gadget Huilei Xu 1, and Avi Ma ayan 1,* 1 Department of Pharmacology and Systems Therapeutics, Mount Sinai School of Medicine,
Chapter 8. Summary and Perspectives
 Chapter 8 Summary and Perspectives 131 Chapter 8 Summary Overexpression of the multidrug resistance protein MRP1 confer multidrug resistance (MDR) to cancer cells. The contents of this thesis describe
Chapter 8 Summary and Perspectives 131 Chapter 8 Summary Overexpression of the multidrug resistance protein MRP1 confer multidrug resistance (MDR) to cancer cells. The contents of this thesis describe
Master BioMedical Sciences (BMS) Track Cell Biology and Advanced Microscopy
 Master BioMedical Sciences (BMS) Track Cell Biology and Advanced Microscopy The five tracks offered in the Medical Biology cluster are: Biochemistry and Metabolic Diseases Cell Biology and Advanced Microscopy
Master BioMedical Sciences (BMS) Track Cell Biology and Advanced Microscopy The five tracks offered in the Medical Biology cluster are: Biochemistry and Metabolic Diseases Cell Biology and Advanced Microscopy
Comparison of Non-linear Dimensionality Reduction Techniques for Classification with Gene Expression Microarray Data
 CMPE 59H Comparison of Non-linear Dimensionality Reduction Techniques for Classification with Gene Expression Microarray Data Term Project Report Fatma Güney, Kübra Kalkan 1/15/2013 Keywords: Non-linear
CMPE 59H Comparison of Non-linear Dimensionality Reduction Techniques for Classification with Gene Expression Microarray Data Term Project Report Fatma Güney, Kübra Kalkan 1/15/2013 Keywords: Non-linear
Measuring gene expression (Microarrays) Ulf Leser
 Measuring gene expression (Microarrays) Ulf Leser This Lecture Gene expression Microarrays Idea Technologies Problems Quality control Normalization Analysis next week! 2 http://learn.genetics.utah.edu/content/molecules/transcribe/
Measuring gene expression (Microarrays) Ulf Leser This Lecture Gene expression Microarrays Idea Technologies Problems Quality control Normalization Analysis next week! 2 http://learn.genetics.utah.edu/content/molecules/transcribe/
Correlation of microarray and quantitative real-time PCR results. Elisa Wurmbach Mount Sinai School of Medicine New York
 Correlation of microarray and quantitative real-time PCR results Elisa Wurmbach Mount Sinai School of Medicine New York Microarray techniques Oligo-array: Affymetrix, Codelink, spotted oligo-arrays (60-70mers)
Correlation of microarray and quantitative real-time PCR results Elisa Wurmbach Mount Sinai School of Medicine New York Microarray techniques Oligo-array: Affymetrix, Codelink, spotted oligo-arrays (60-70mers)
A truly robust Expression analyzer
 Genowiz A truly robust Expression analyzer Abstract Gene expression profiles of 10,000 tumor samples, disease classification, novel gene finding, linkage analysis, clinical profiling of diseases, finding
Genowiz A truly robust Expression analyzer Abstract Gene expression profiles of 10,000 tumor samples, disease classification, novel gene finding, linkage analysis, clinical profiling of diseases, finding
MarkerView Software 1.2.1 for Metabolomic and Biomarker Profiling Analysis
 MarkerView Software 1.2.1 for Metabolomic and Biomarker Profiling Analysis Overview MarkerView software is a novel program designed for metabolomics applications and biomarker profiling workflows 1. Using
MarkerView Software 1.2.1 for Metabolomic and Biomarker Profiling Analysis Overview MarkerView software is a novel program designed for metabolomics applications and biomarker profiling workflows 1. Using
Gene Expression Analysis
 Gene Expression Analysis Jie Peng Department of Statistics University of California, Davis May 2012 RNA expression technologies High-throughput technologies to measure the expression levels of thousands
Gene Expression Analysis Jie Peng Department of Statistics University of California, Davis May 2012 RNA expression technologies High-throughput technologies to measure the expression levels of thousands
Name (print) Name (signature) Period. (Total 30 points)
 AP Biology Worksheet Chapter 43 The Immune System Lambdin April 4, 2011 Due Date: Thurs. April 7, 2011 You may use the following: Text Notes Power point Internet One other person in class "On my honor,
AP Biology Worksheet Chapter 43 The Immune System Lambdin April 4, 2011 Due Date: Thurs. April 7, 2011 You may use the following: Text Notes Power point Internet One other person in class "On my honor,
Making the switch to a safer CAR-T cell therapy
 Making the switch to a safer CAR-T cell therapy HaemaLogiX 2015 Technical Journal Club May 24 th 2016 Christina Müller - chimeric antigen receptor = CAR - CAR T cells are generated by lentiviral transduction
Making the switch to a safer CAR-T cell therapy HaemaLogiX 2015 Technical Journal Club May 24 th 2016 Christina Müller - chimeric antigen receptor = CAR - CAR T cells are generated by lentiviral transduction
GENEGOBI : VISUAL DATA ANALYSIS AID TOOLS FOR MICROARRAY DATA
 COMPSTAT 2004 Symposium c Physica-Verlag/Springer 2004 GENEGOBI : VISUAL DATA ANALYSIS AID TOOLS FOR MICROARRAY DATA Eun-kyung Lee, Dianne Cook, Eve Wurtele, Dongshin Kim, Jihong Kim, and Hogeun An Key
COMPSTAT 2004 Symposium c Physica-Verlag/Springer 2004 GENEGOBI : VISUAL DATA ANALYSIS AID TOOLS FOR MICROARRAY DATA Eun-kyung Lee, Dianne Cook, Eve Wurtele, Dongshin Kim, Jihong Kim, and Hogeun An Key
Thomson Reuters Biomarker Solutions: Hepatitis C Treatment Biomarkers and special considerations in patients with Asthma
 : Hepatitis C Treatment Biomarkers and special considerations in patients with Asthma Abstract This case study aims to demonstrate the process of biomarker identification and validation utilizing Thomson
: Hepatitis C Treatment Biomarkers and special considerations in patients with Asthma Abstract This case study aims to demonstrate the process of biomarker identification and validation utilizing Thomson
Graduate and Postdoctoral Affairs School of Biomedical Sciences College of Medicine. Graduate Certificate. Metabolic & Nutritional Medicine
 Graduate and Postdoctoral Affairs School of Biomedical Sciences College of Medicine Graduate Certificate in Metabolic & Nutritional Medicine Graduate Certificate Metabolic & Nutritional Medicine Purpose
Graduate and Postdoctoral Affairs School of Biomedical Sciences College of Medicine Graduate Certificate in Metabolic & Nutritional Medicine Graduate Certificate Metabolic & Nutritional Medicine Purpose
LESSON 3: ANTIBODIES/BCR/B-CELL RESPONSES
 Introduction to immunology. LESSON 3: ANTIBODIES/BCR/B-CELL RESPONSES Today we will get to know: The antibodies How antibodies are produced, their classes and their maturation processes Antigen recognition
Introduction to immunology. LESSON 3: ANTIBODIES/BCR/B-CELL RESPONSES Today we will get to know: The antibodies How antibodies are produced, their classes and their maturation processes Antigen recognition
CNV Univariate Analysis Tutorial
 CNV Univariate Analysis Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Overview 2 2. CNAM Optimal Segmenting 4 A. Performing CNAM Optimal Segmenting..................................
CNV Univariate Analysis Tutorial Release 8.1 Golden Helix, Inc. March 18, 2014 Contents 1. Overview 2 2. CNAM Optimal Segmenting 4 A. Performing CNAM Optimal Segmenting..................................
BSc in Medical Sciences with PHARMACOLOGY
 BSc in Medical Sciences with PHARMACOLOGY Course Director Dr Christopher John Module Leaders Dr Robert Dickinson (Module 1) Dr Anabel Varela Carver (Module 2) Dr Sohag Saleh (Module 3) Course Administrator
BSc in Medical Sciences with PHARMACOLOGY Course Director Dr Christopher John Module Leaders Dr Robert Dickinson (Module 1) Dr Anabel Varela Carver (Module 2) Dr Sohag Saleh (Module 3) Course Administrator
OplAnalyzer: A Toolbox for MALDI-TOF Mass Spectrometry Data Analysis
 OplAnalyzer: A Toolbox for MALDI-TOF Mass Spectrometry Data Analysis Thang V. Pham and Connie R. Jimenez OncoProteomics Laboratory, Cancer Center Amsterdam, VU University Medical Center De Boelelaan 1117,
OplAnalyzer: A Toolbox for MALDI-TOF Mass Spectrometry Data Analysis Thang V. Pham and Connie R. Jimenez OncoProteomics Laboratory, Cancer Center Amsterdam, VU University Medical Center De Boelelaan 1117,
Lecture 2: Descriptive Statistics and Exploratory Data Analysis
 Lecture 2: Descriptive Statistics and Exploratory Data Analysis Further Thoughts on Experimental Design 16 Individuals (8 each from two populations) with replicates Pop 1 Pop 2 Randomly sample 4 individuals
Lecture 2: Descriptive Statistics and Exploratory Data Analysis Further Thoughts on Experimental Design 16 Individuals (8 each from two populations) with replicates Pop 1 Pop 2 Randomly sample 4 individuals
Introduction to Flow Cytometry
 Outline Introduction to Flow Cytometry Basic Concept of Flow Cytometry Introduction to Instrument Subsystems Daisy Kuo Assistant Product Manager E-mail: daisy_kuo@bd.com BDBiosciences Application Examples
Outline Introduction to Flow Cytometry Basic Concept of Flow Cytometry Introduction to Instrument Subsystems Daisy Kuo Assistant Product Manager E-mail: daisy_kuo@bd.com BDBiosciences Application Examples
岑 祥 股 份 有 限 公 司 技 術 專 員 費 軫 尹 20100803
 技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
MultiExperiment Viewer Quickstart Guide
 MultiExperiment Viewer Quickstart Guide Table of Contents: I. Preface - 2 II. Installing MeV - 2 III. Opening a Data Set - 2 IV. Filtering - 6 V. Clustering a. HCL - 8 b. K-means - 11 VI. Modules a. T-test
MultiExperiment Viewer Quickstart Guide Table of Contents: I. Preface - 2 II. Installing MeV - 2 III. Opening a Data Set - 2 IV. Filtering - 6 V. Clustering a. HCL - 8 b. K-means - 11 VI. Modules a. T-test
Time series experiments
 Time series experiments Time series experiments Why is this a separate lecture: The price of microarrays are decreasing more time series experiments are coming Often a more complex experimental design
Time series experiments Time series experiments Why is this a separate lecture: The price of microarrays are decreasing more time series experiments are coming Often a more complex experimental design
User Manual. Transcriptome Analysis Console (TAC) Software. For Research Use Only. Not for use in diagnostic procedures. P/N 703150 Rev.
 User Manual Transcriptome Analysis Console (TAC) Software For Research Use Only. Not for use in diagnostic procedures. P/N 703150 Rev. 1 Trademarks Affymetrix, Axiom, Command Console, DMET, GeneAtlas,
User Manual Transcriptome Analysis Console (TAC) Software For Research Use Only. Not for use in diagnostic procedures. P/N 703150 Rev. 1 Trademarks Affymetrix, Axiom, Command Console, DMET, GeneAtlas,
Inflammatory Cytokine-induced Expression of Vasohibin-1 by Rheumatoid Synovial Fibroblasts
 9 6 6 958 Inflammatory Cytokine-induced Expression of Vasohibin- by Rheumatoid Synovial Fibroblasts a b c a a a a d e a* a a b c d e 5 6 6 9 5 56 67 7 ʼ ʼ ʼ ʼ ʼ ʼ ʼ ʼ 5 6 6 ʼ 5 75 989 6 69 8 95 56 56896
9 6 6 958 Inflammatory Cytokine-induced Expression of Vasohibin- by Rheumatoid Synovial Fibroblasts a b c a a a a d e a* a a b c d e 5 6 6 9 5 56 67 7 ʼ ʼ ʼ ʼ ʼ ʼ ʼ ʼ 5 6 6 ʼ 5 75 989 6 69 8 95 56 56896
Frequently Asked Questions (FAQ)
 Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
Antibody Function & Structure
 Antibody Function & Structure Specifically bind to antigens in both the recognition phase (cellular receptors) and during the effector phase (synthesis and secretion) of humoral immunity Serology: the
Antibody Function & Structure Specifically bind to antigens in both the recognition phase (cellular receptors) and during the effector phase (synthesis and secretion) of humoral immunity Serology: the
Session 1. Course Presentation: Mass spectrometry-based proteomics for molecular and cellular biologists
 Program Overview Session 1. Course Presentation: Mass spectrometry-based proteomics for molecular and cellular biologists Session 2. Principles of Mass Spectrometry Session 3. Mass spectrometry based proteomics
Program Overview Session 1. Course Presentation: Mass spectrometry-based proteomics for molecular and cellular biologists Session 2. Principles of Mass Spectrometry Session 3. Mass spectrometry based proteomics
Vitamin D deficiency exacerbates ischemic cell loss and sensory motor dysfunction in an experimental stroke model
 Vitamin D deficiency exacerbates ischemic cell loss and sensory motor dysfunction in an experimental stroke model Robyn Balden & Farida Sohrabji Texas A&M Health Science Center- College of Medicine ISC
Vitamin D deficiency exacerbates ischemic cell loss and sensory motor dysfunction in an experimental stroke model Robyn Balden & Farida Sohrabji Texas A&M Health Science Center- College of Medicine ISC
Pulling the Plug on Cancer Cell Communication. Stephen M. Ansell, MD, PhD Mayo Clinic
 Pulling the Plug on Cancer Cell Communication Stephen M. Ansell, MD, PhD Mayo Clinic Why do Waldenstrom s cells need to communicate? Waldenstrom s cells need activating signals to stay alive. WM cells
Pulling the Plug on Cancer Cell Communication Stephen M. Ansell, MD, PhD Mayo Clinic Why do Waldenstrom s cells need to communicate? Waldenstrom s cells need activating signals to stay alive. WM cells
ProteinPilot Report for ProteinPilot Software
 ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Pow erful mass spectrometers like
ProteinPilot Report for ProteinPilot Software Detailed Analysis of Protein Identification / Quantitation Results Automatically Sean L Seymour, Christie Hunter SCIEX, USA Pow erful mass spectrometers like
Comparative genomic hybridization Because arrays are more than just a tool for expression analysis
 Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
Microarray Data Analysis Workshop MedVetNet Workshop, DTU 2008 Comparative genomic hybridization Because arrays are more than just a tool for expression analysis Carsten Friis ( with several slides from
Biochemistry. Entrance Requirements. Requirements for Honours Programs. 148 Bishop s University 2015/2016
 148 Bishop s University 2015/2016 Biochemistry The Biochemistry program at Bishop s is coordinated through an interdisciplinary committee of chemists, biochemists and biologists, providing students with
148 Bishop s University 2015/2016 Biochemistry The Biochemistry program at Bishop s is coordinated through an interdisciplinary committee of chemists, biochemists and biologists, providing students with
Objectives. Immunologic Methods. Objectives. Immunology vs. Serology. Cross Reactivity. Sensitivity and Specificity. Definitions
 Immunologic Methods Part One Definitions Part Two Antigen-Antibody Reactions CLS 420 Clinical Immunology and Molecular Diagnostics Kathy Trudell MLS (ASCP) CM SBB CM ktrudell@nebraskamed.com Discuss the
Immunologic Methods Part One Definitions Part Two Antigen-Antibody Reactions CLS 420 Clinical Immunology and Molecular Diagnostics Kathy Trudell MLS (ASCP) CM SBB CM ktrudell@nebraskamed.com Discuss the
What s New in Pathway Studio Web 11.1
 1 1 What s New in Pathway Studio Web 11.1 Elseiver is pleased to announce the release of Pathway Studio Web 11.1 for all database subscriptions (Mammal, Mammal+ChemEffect+DiseaseFx, Plant). This release
1 1 What s New in Pathway Studio Web 11.1 Elseiver is pleased to announce the release of Pathway Studio Web 11.1 for all database subscriptions (Mammal, Mammal+ChemEffect+DiseaseFx, Plant). This release
Discovery & Modeling of Genomic Regulatory Networks with Big Data
 Discovery & Modeling of Genomic Regulatory Networks with Big Data Hamid Bolouri Division of Human Biology Fred Hutchinson Cancer Research Center labs.fhcrc.org/bolouri I have no financial relationships
Discovery & Modeling of Genomic Regulatory Networks with Big Data Hamid Bolouri Division of Human Biology Fred Hutchinson Cancer Research Center labs.fhcrc.org/bolouri I have no financial relationships
Microarray analysis of viral infections
 Microarray analysis of viral infections Dipl. Biol. Department of Virology Bernhard Nocht Institute for Tropical Medicine Berhard Nocht Institut für Tropenmedizin Possible investigations of viral infections
Microarray analysis of viral infections Dipl. Biol. Department of Virology Bernhard Nocht Institute for Tropical Medicine Berhard Nocht Institut für Tropenmedizin Possible investigations of viral infections
