Applying the power of MetaCore to
|
|
|
- Calvin Reynolds
- 8 years ago
- Views:
Transcription
1 Applying the power of MetaCore to Next Generation Sequencing Data: Uncovering potential mechanisms for Interferon alpha induced Thyroiditis case study Introduction Interferon (IFN) alpha was the first cytokine to be reproduced by recombinant DNA technology and has emerged as a major therapeutic intervention for several disease including melanoma, renal cell carcinomas, hairy cell leukemia, hepatitis B and most commonly, Hepatitis C (Baron S, et al., 1991). However, despite its successful therapeutic actions, IFN alpha therapy is also associated with a common side effect profile that includes the development of autoimmune disease and reports of the development of type I diabetes mellitus, psoriasis, rheumatoid arthritis, systemic lupus-like syndromes, and thyroid disease. In particular IFN alpha induced thyroiditis (IIT) is a major clinical problem and has been reported to develop in a range from percent of patients receiving IFN alpha therapy (Reviewed in Oppenheim Y et al., 2003). A new classification for IIT has been suggested based on the epidemiological characteristics of IIT where IIT is described as either Autoimmune IIT or Nonautoimmune IIT. Autoimmune IIT can manifest by the development of thyroid antibodies with or without clinical disease. Non-autoimmune IIT can manifest as destructive thyroiditis or as hypothyroidism with negative thyroid antibodies. (Reviewed in Menconi and Tomer, 2009). This suggests IFN alpha induces tissue inflammation through atypical immune as well as non-immune mechanisms however, neither of these mechanisms are well understood. To further elucidate the immune and non-immune mechanisms induced by IFN alpha, the current study applied a pathway analysis approach using Thomson Reuters MetaCore to Next Generation Sequencing (RNAseq) data generated from an in vivo mouse model of IFN alpha induced Thyroiditis (IIT). The current study demonstrates one of the logical and powerful workflows available using Thomson Reuters Systems Biology Solutions. Through the application of unique tools and manually curated content, Next Generation Sequencing data was analyzed and interpreted to generate hypothesis based on strong experimentally derived information. These hypotheses could save valuable time and money as they provide a strong foundation to potentially drive fast and accurate decisions for the direction for future research and significant findings. Materials and Methods Next generation sequencing data used for this study was obtained from the transcriptome analysis (using RNAseq) of an in vivo IIT mouse model (Akeno et al., 2011). Briefly, thyroid tissue RNA was dissected, pooled and isolated from the thyroid tissues of 10 transgenic mice with thyroid specific overexpression of IFN alpha at 8 months of age (TG) and a pool of control thyroid tissues obtained from 8 litter mate controls (WT) (Akeno et al., 2011). For the two samples (TG and WT) mrna was extracted, fragmented and used to prepare a cdna library compatible with the Illumina Next Generation Sequencing technology and sequencing was performed on an Illumina Genome Analyzer IIx as described (Akeno et al., 2011). Gene expression in each sample is proportional to the number of reads aligning to a specific gene sequence and reads were aligned to the mouse (mm9) genome where the resulting counts were calculated and normalized to relative abundance in Fragment per kilobase of exon model per Million (FPKM). This data was obtained from the public GEO database under Series GSE25115 ( nlm.nih.gov/geo/query/acc.cgi?acc=gse25115). The FPKM values for samples GSM (mouse_wt_ RNA-seq) and GSM (mouse-tg_rna-seq) were used to generate fold change values for the differentially expressed genes of the mouse-tg_rnaseq sample relative to the mouse_wt_rna-seq sample (DEGs TGvsWT) applying similar methodology outlined in the corresponding publication (Akeno et al., 2011). The samples were read into R and in order to enable quantitative comparisons between the samples, only those transcripts which had values available in both samples were compared. Samples were normalized by scaling to have the same total mean expression value and only genes with > 50 FPKM in at least 1 sample were retained. These settings were slightly more stringent than the authors who accepted genes where one sample had 50 FPKM. The differential expression analysis was then performed using the DEGseq package and the MA method with random sampling. Genes were filtered where a fold change +/- 2 was retained. The fold change values for the resulting 927 mouse RefSeq IDs were converted to a log2 ratio and uploaded into MetaCore for pathway analysis. Of the 927 RefSeq IDs, 926 were recognized by MetaCore.
2 Figure 1: (b) Metacore Analysis Workflow for mouse Thyroid tissue DEGs TGvs WT. MetaCore is an integrated software suite for functional analysis of experimental data and gene lists including resulting originating from Next Generation Sequencing. MetaCore is based on a high-quality, manually-curated database providing accurate systems biology content and provides an array of sophisticated functional tools to interpret and analyze high throughput data. The current study applies a logical workflow incorporating a selection of these tools to analyze the Next Generation Sequencing data set and elucidate potential mechanisms of IFN alpha in Thyroiditis (refer to Figure 1 for an overview of this workflow). RESULTS Figure 2 MetaCore Canonical Pathway Maps histograms after enrichment analysis with the DEGs from thryoid tissue of Mouse TGvsWT filtered for a fold change +/- 2. The top ten canonical maps for enriched genes are reported as histogram plots as shown. 1. Enrichment analysis: MetaCore Pathway Maps Ontology The manually curated MetaCore Pathway Maps ontology was used to identify canonical Pathway Maps significantly enriched by the TGvsWT dataset. Using the MetaCore variation of the Fisher s exact test and adjusting for multiple sample testing using (FDR) pathway enrichments were more stringent and ensured more accurate results (Figure 2). Consistent with the over expression of IFN alpha in the thyroid of TG mice, the most significantly enriched canonical pathways included Immune response_antiviral actions of interferons and Immune response_ifnalpha/ beta signaling pathway. Other significantly enriched maps included immunological responses such as Antigen presentation by MHC class I and complement system pathways (Figure 2).
3 2. Interactions by Protein Function When exploring transcriptional data it is important to also consider that important regulatory elements may be altered at biological levels other than the transcriptional, which can easily be missed when looking exclusively at transcriptional data. For example, transcription factors often have no detectable change in transcription but change in activity, promoter binding affinity, etc. To further extend the current analysis and try to capture such hidden regulators, the Interactome workflow in MetaCore was utilized using the log2 ratios of the DEGs from mouse thyroid tissue in TGvsWT. The resulting list of 138 potential regulators (Figure 3) was then also used in combination with previous results to get a larger multi-level approach to the analysis. 3. Enrichment analysis: using results from Interactions by Protein Function tool & Pathway Maps Ontology MetaCore allows easy export of all results to either external software such as Excel or as a new file within the tool for further analysis. By saving the interactome analysis results as an experimental list, it could then be used in enrichment analysis of the MetaCore Pathway Maps ontology. This additional analysis compliments the initial enrichment analysis of the DEGs TGvsWT and can also highlight additional canonical pathways that may further elucidate the mechanisms of IFN alpha in inducing Thyroiditis in the current IIT model. Results shown in Figure 4 include the same three Immune pathways identified in Figure 2. And the two lists (1) Figure 3 Results of Interactome analysis for network objects associated with DEGS from thyroid tissue of mouse TGvsWT. The top results for each protein class are shown following filtering by by z-value. Network objects significantly overconnected and also that appears in the original DEG list are highlighted in orange. Columns are as follows: Actual - number of targets in the activated datasets regulated by the chosen network object, n - number of network objects in the activated datasets, R - number of targets in the complete database or background list regulated by the chosen network object N - total number of gene-based objects in the background list, Expected - mean value for hypergeometric distribution (n*r/n), Ratio- connectivity ratio (Actual/Expected), p-value - probability (from the hypergeometric distribution) of seeing the observed value of r or higher, z-score - ((r-mean)/ sqrt(variance)). Figure 4 MetaCore Canonical Pathway Maps histograms after enrichment analysis with Network objects significantly overconnected with DEGs from thryoid tissue of Mouse TGvsWT filtered for a fold change +/- 2. The top thirteen canonical maps for enriched genes are reported as histogram plots as shown.
4 Applying the power of MetaCore to Next Generation Sequencing Data: Uncovering potential mechanisms DEGs and (2) Overconnected network objects can be overlaid onto the MetaCore Canonical Pathway Maps and highlight continuous mechanistic pathways (Figure 5 and 6). Significant up-regulation of the canonical pathway Immune response_antigen presentation by MHC class I demonstrates increased proteosome activity (which is commonly correlated with disease activity in autoimmunity) and antigen processing and presentation by MHC class I (Figure 5). IFN alpha is well known to increase MHC class I antigen expression on cells, including thyroid epithelial cells (Roti, E et al., 1996) and is further confirmed by the Figure 5 MetaCore pathway map Immune response_antigen presentation by MHC class I. The thermometers represent the different datasets overlayed on map: (1) DEGs from thyroid tissue of Mouse TGvsWT (ref). (2) Proteins significantly overconnected to the list of DEGs from thyroid tissue of Mouse TGvsWT. The vertical thermometers represent the log2 ratios for the DEGs (red represents up-regulation) and overconnected network objects are given a default value of +1. Figure 6 MetaCore pathway map Immune response_antiviral actions of Interferons. The thermometers represent the different datasets overlayed on map: (1) DEGs from thyroid tissue of Mouse TGvsWT (ref). (2) Proteins significantly overconnected to the list of DEGs from thyroid tissue of Mouse TGvsWT. The vertical thermometers represent the log2 ratios for the DEGs (red represents up-regulation) and overconnected network objects are given a default value of +1.
5 significantly enriched Immune response _Antiviral actions of interferons pathway in the mouse thyroid tissue of TGvsWT (Figure 6). Results reported from the publication in which the current data set was taken (Akeno et al., 2011) also reported a significant increase in thyroid specific proteins (Tg) when a rat thyroid cell line (PCCL3) is induced with IFN alpha in vitro (data not shown). Combined with an increase in antigen processing and presentation by MHC class I it is reasonable to hypothesis that thyroid self antigens could be the antigens presented to the adaptive immune system in the thyroid tissue of TG mice and potentially be a immune modulated mechanism in which IFN alpha induces autoimmune thyroiditis. Enrichment analysis of the proteins significantly overconnected to the original data set can also highlight additional canonical pathways that may further elucidate the mechanisms of IFN alpha in inducing thyroiditis. As shown from the enrichment results in Figure 4 the IL-6 signaling pathway is also highlighted where IL-6 is known to be induced by IFN alpha and is also associated with autoimmune Thyroiditis (Ajjan et al., 1996). An additional immune response pathway Immune response-il-7 signaling in T lymphocytes is highlighted and is involved in the is an essential cytokine pathway for proliferation, maintenance and survival of T-lymphocytes (Kittipatarin C et al., 2001). 4. Enrichment analysis: Pathway Map Folders Ontology The MetaCore Pathway Map Folders ontology is a collection of manually created pathway maps, grouped hierarchically into folders according to main biological processes. This ontology allows the exploration of the data at a higher level of the hierarchical tree and often can elucidate significant changes to larger categories that can be overlooked when looking at the individual pathways themselves. Using this ontology allows identification of key biological processes from the DEGs of TGvsWT in a single click. This analysis also confirmed a predominant enrichment of Immune system responses (Figure 7). Observations originally published with the current dataset also revealed thyroid cell death both in vivo (during thyroid dissection of the current animals analyzed) and in vitro (from IFN alpha stimulated Rat thyroid cell line (PCCL3)). The authors excluded apoptosis from their in vitro studies by a Vybrant apoptosis assay kit (Akeno et Figure 7 MetaCore Canonical Pathway Map Folders histograms after enrichment analysis with the DEGs from thryoid tissue of Mouse TGvsWT filtered for a fold change +/- 2. The top ten Pathway Map Folders representing biological processes are reported as histogram plots as shown. Bold and faded Orange bars show if the pathway map folder passed or failed the FDR analysis filer, respectively. al., 2011). A similar result for the in vivo IIT model can be hypothesized by the current pathway analysis approach as there is a distinct absence of enrichment in any apoptopic canonical pathway map (Figure 2 and 4) and the apoptoptic pathway map folder (Figure 7). Collectively these results suggest the observed cell death is not mediated by mechanistic pathways of apoptosis. 5. Creating a custom ontology representing toxic pathology types In order to identify potential toxic pathologies associated with IIT in the current model, the DEGs from the thyroid tissue of TGvsWT were enriched using custom built networks based on specific toxic pathologies. The Thomson Reuters Systems Toxicology Module adds manually curated content to MetaCore focusing on specific toxic pathology types annotated specifically to 12 major organs. As many toxic pathology mechanisms and biomarkers overlap between organs (Cobb et al., 1996), manually curated biomarkers associated with the different toxic pathology types were pooled in this study from the 12 major organs annotated. To this end the advanced search tool in MetaCore was utilized and biomarkers associated with different toxic pathology types were extracted from the content knowledgebase add on provided with the Systems Toxicology knowledgebase. These were then used to build custom networks representing biomarkers for 12 main toxic pathology types. This was quickly achieved using the MetaCore network building algorithm: Direct interactions with canonical pathways. The resulting networks were saved and enrichment of the DEGs TGvsWT was performed from the new custom Toxic Pathology types ontology generated from manually curated toxicology biomarkers. 6. Perform Enrichment Analysis using custom ontology for toxic pathology types Inflammation was the top toxic pathology type enriched by the DEGs of thyroid tissue of mouse TGvsWT (Figure 8) and is consistent with non-immune IIT (destructive thyroiditis) (Tomer, Y & Menconi, F. 2009) and is secondary to necrotic cell death. Necrosis was the second most enriched toxic pathology while apoptosis was the least furthering strengthening the findings that thyroid cell death observed in the current IIT in vivo model is by necrosis rather than apoptosis.
6 Figure 8 Custom toxicology pathology networks histograms after enrichment analysis with Network objects significantly overconnected with DEGs from thryoid tissue of Mouse TGvsWT filtered for a fold change +/- 2. Twelve networks representing manually curated biomarkers for 12 well annotated toxic pathology types were generartd and used for enrichment in the create your own ontology feature of MetaCore. Bold and faded Orange bars show if the pathway map folder passed or failed the FDR analysis filer, respectively. SUMMARY & CONCLUSION The current study used Next Generation Sequencing data from an in vivo IFN alpha Induced Thyroiditis (IIT) mouse model. The data set was obtained from the GEO database (GSE series 25115) and read counts from the thyroid tissue of transgenic mice overexpressing IFN alpha (TG) and wildtype mice (WT) were processed and uploaded into MetaCore. Various tools within MetaCore were applied including enrichment analysis of the log2 ratios from the DEGs of TGvsWT using the manually curated ontologies, canonical pathway maps and pathway map folders as well as a custom ontology built from networks of various toxic pathology types. A summary of the results is provided in figure 9. Figure 9 Summary of findings for the current case study highlighting the original aim and Metacore tools applied for analysis. STUDY AIM 1. To identify key canonical pathways induced by IFN alpha in the in vivo IIT model THOMSON REUTERS SYSTEMS BIOLOGY SOLUTIONS APPROACH Metacore Pathway Maps Ontology: One click enrichment analysis KEY RESULTING FINDINGS IN THIS STUDY 1. Top significantly enriched pathway maps include: Immune response_antiviral actions of interferons Immune response_ifnalpha/ beta signaling pathway Immune response _Antigen presentation by MHC class I 2. Increased proteosome activity and upregulation of antigen processing and presentation by MHC class I A list of 138 network objects that are significantly overconnected to DEGs TGvsWT that were used to expand enrichment analysis and visualised on Pathway Maps. 1. Further validated results from (1) where same three maps were significantly enriched 2. Highlighted additional pathway maps relevant to in vivo ITT model Immune response-il-7 signaling in T lymphocytes Immune response_il-6 signaling pathway 1. Main biological process enriched was Immunological Responses. 2. Absence of the pathway map folder for Apoptosis. 1. Inflammation was the most significant toxic pathology type enriched by the DEGs of TGvsWT. 2. Necrosis was the second most significant toxic pathology while Apoptosis was the least significant. 2. Identify regulatory components responsible for the biological events induced by IFN alpha in the in vivo IIT model Interactome Analysis 3. To extend analysis and perform enrichment using results from step 2 and further elucidate potential mechanisms of IFN alpha in inducing Thyroiditis in the current IIT model. One click enrichment analysis using results from 2 with Metacore Pathway Maps Ontology 4. Identify the main biological processes induced by IFN alpha in the in vivo IIT model One click enrichment analysis using Pathway Map Folders Ontology 5. Identify key toxic pathology types induced by IFN alpha in the in vivo ITT model One click enrichment analysis using custom Toxic Pathology Ontology consisting of 12 manually built networks from extracted from Thomson Reuters Systems Toxicology Module Knowledge base
7 Overall the results drew similar findings to the authors where IFN alpha upregulated several immune response pathways in the Thyroid of TG mice consistent with increased IFN alpha levels. There was no significant enrichment of any apoptopic pathway maps or map folders in the in vivo IIT model and inflammation and necrosis were the most significantly enriched toxic pathology types. Apoptosis was the least enriched toxic pathology and reflects similar results found by the authors in vitro where observed cell death was necrosis and not apoptosis. Interestingly, in the current analysis there was no significant upregulation of the Granzyme B signaling pathway referred to during the enrichment analysis by the authors (refer Akeno et al., 2011). Granzyme B is responsible for rapid induction of caspase-dependent apoptosis (Bots et al., 2006) and the 20 percent upregulation of the Granzyme B signaling pathway (upregulated genes were Granzyme B and LaminB1) demonstrated this pathway as significantly upregulated in the authors analysis. In the current analysis Granzyme B was not a significant DEG due to the more stringent FPKM cut off-of >50 rather than a minimum of 50 in one of the two samples. (Granzyme B s FPKM = 50 and 2 for the TG and WT, respectively). Robust methodology in processing read counts is necessary to account for the pooling of RNA tissue and demonstrates the importance of processing FPKM prior to pathway analysis upload. In the current analysis, two DEGs were up-regulated in the MetaCore Granzyme B signaling pathway (Caspase-7 and LaminB1); however this pathway was not significantly enriched and did not appear in the initial and extended enrichment analysis conducted in MetaCore (Refer to figures 2 and 4). Collectively the results of the current study suggest that IFN alpha therapy could be inducing thyroid cell death (by the way of necrosis) and the up-regulation of antigen processing and presentation by the MHC class I molecule events. These results offer a mechanism in which thyroid self antigens are released following necrotic cell death and the increase in proteolysis activity and MHC class I expression could suggest the self antigens are being presented to the adaptive immune system causing the autoimmune response. The current study applied a unique pathway analysis workflow and produce potentially important, robust and actionable conclusions that are a direct result of the high quality, 100 percent manually curated content of the Thomson Reuters Systems Biology Solutions. The combination of a biological interactions knowledgebase built on robust interaction information (quality controlled at the experimental design level) with intuitive yet powerful bioinformatic algorithms allows researchers to make better decisions as to next steps, faster. Using MetaCore, high throughput Next Generation Sequencing data was analyzed and identified key biological pathways and processes involved in IFN alpha Induced Thyroiditis. In addition, when used in combination with the Thomson Reuters Systems Toxicology Module, MetaCore also identified significant toxic pathologies potentially induced by IFN alpha in vivo and were consistent with previous findings in vitro.
8 REFERENCES 1) Baron S, Tyring SK, Fleischmann WR Jr, Coppenhaver DH, Niesel DW, Klimpel GR, Stanton GJ, Hughes TK (1991). The interferons. Mechanisms of action and clinical applications. JAMA. 266(10): ) Oppenheim Y, Ban Y, Tomer Y (2004). Interferon induced Autoimmune Thyroid Disease (AITD): a model for human autoimmunity. Autoimmun Rev. Jul; 3(5): ) Tomer, Y and Menconi, F (2009). Interferon induced thyroiditis. Best Pract Res Clin Endocrinol Metab. 23(6): 703 4) Akeno N., Smith, E.P., Stefan, M., Amanda K. Huber, A.K., Zhang, W., Keddache, M., and Tomer, Y. (2011). Interferon-alpha mediates the development of Autoimmunity both by direct tissue toxicity and through immune-cell recruitment mechanisms. J Immunol. 186(8): ) Roti E, Minelli R, Giuberti T, Marchelli S, Schianchi C, Gardini E, Salvi M, Fiaccadori F, Ugolotti G, Neri TM, Braverman LE. (1996) Multiple changes in thyroid function in patients with chronic active HCV hepatitis treated with recombinant interferon-alpha. Am J Med;101(5): ) Ajjan RA, Watson PF, McIntosh RS, Weetman AP (1996) Intrathyroidal cytokine gene expression in Hashimoto s thyroiditis. Clin Exp Immunol ;105(3): ) Kittipatarin., C. And Khaled AR (2007) Interlinking interleukin-7. Cytokine;39(1): ) Cobb JP, Hotchkiss RS, Karl IE, Buchman TG (1996) Mechanisms of cell injury and death, Buchman Br J Anaesth. 77(1): ) Bots M, Medema JP Granzymes at a glance. Journal of cell science 2006 Dec 15;119(Pt 24): THOMSON REUTERS Regional Offices Contact us to find out more about MetaCore or visit thomsonreuters.com/diseaseinsight North America Philadelphia Latin America Europe, Middle East and Africa Barcelona London Asia Pacific Singapore Tokyo For a complete office list visit: ip-science.thomsonreuters.com/ contact LS SCI-NGSCS Copyright 2012 Thomson Reuters
Thomson Reuters Biomarker Solutions: Hepatitis C Treatment Biomarkers and special considerations in patients with Asthma
 : Hepatitis C Treatment Biomarkers and special considerations in patients with Asthma Abstract This case study aims to demonstrate the process of biomarker identification and validation utilizing Thomson
: Hepatitis C Treatment Biomarkers and special considerations in patients with Asthma Abstract This case study aims to demonstrate the process of biomarker identification and validation utilizing Thomson
Understanding West Nile Virus Infection
 Understanding West Nile Virus Infection The QIAGEN Bioinformatics Solution: Biomedical Genomics Workbench (BXWB) + Ingenuity Pathway Analysis (IPA) Functional Genomics & Predictive Medicine, May 21-22,
Understanding West Nile Virus Infection The QIAGEN Bioinformatics Solution: Biomedical Genomics Workbench (BXWB) + Ingenuity Pathway Analysis (IPA) Functional Genomics & Predictive Medicine, May 21-22,
How to create and interpret the predictive analysis of a compound
 How to create and interpret the predictive analysis of a compound Platform with suite of tools Predict & understand biological effects of small molecules & compounds Predict targets and metabolites, potential
How to create and interpret the predictive analysis of a compound Platform with suite of tools Predict & understand biological effects of small molecules & compounds Predict targets and metabolites, potential
Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation
 Identification of rheumatoid arthritis and osterthritis patients by transcriptome-based rule set generation Bering Limited Report generated on September 19, 2014 Contents 1 Dataset summary 2 1.1 Project
Identification of rheumatoid arthritis and osterthritis patients by transcriptome-based rule set generation Bering Limited Report generated on September 19, 2014 Contents 1 Dataset summary 2 1.1 Project
Autoimmunity and immunemediated. FOCiS. Lecture outline
 1 Autoimmunity and immunemediated inflammatory diseases Abul K. Abbas, MD UCSF FOCiS 2 Lecture outline Pathogenesis of autoimmunity: why selftolerance fails Genetics of autoimmune diseases Therapeutic
1 Autoimmunity and immunemediated inflammatory diseases Abul K. Abbas, MD UCSF FOCiS 2 Lecture outline Pathogenesis of autoimmunity: why selftolerance fails Genetics of autoimmune diseases Therapeutic
AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE
 ACCELERATING PROGRESS IS IN OUR GENES AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE GENESPRING GENE EXPRESSION (GX) MASS PROFILER PROFESSIONAL (MPP) PATHWAY ARCHITECT (PA) See Deeper. Reach Further. BIOINFORMATICS
ACCELERATING PROGRESS IS IN OUR GENES AGILENT S BIOINFORMATICS ANALYSIS SOFTWARE GENESPRING GENE EXPRESSION (GX) MASS PROFILER PROFESSIONAL (MPP) PATHWAY ARCHITECT (PA) See Deeper. Reach Further. BIOINFORMATICS
Tutorial for proteome data analysis using the Perseus software platform
 Tutorial for proteome data analysis using the Perseus software platform Laboratory of Mass Spectrometry, LNBio, CNPEM Tutorial version 1.0, January 2014. Note: This tutorial was written based on the information
Tutorial for proteome data analysis using the Perseus software platform Laboratory of Mass Spectrometry, LNBio, CNPEM Tutorial version 1.0, January 2014. Note: This tutorial was written based on the information
The role of IBV proteins in protection: cellular immune responses. COST meeting WG2 + WG3 Budapest, Hungary, 2015
 The role of IBV proteins in protection: cellular immune responses COST meeting WG2 + WG3 Budapest, Hungary, 2015 1 Presentation include: Laboratory results Literature summary Role of T cells in response
The role of IBV proteins in protection: cellular immune responses COST meeting WG2 + WG3 Budapest, Hungary, 2015 1 Presentation include: Laboratory results Literature summary Role of T cells in response
A Genetic Analysis of Rheumatoid Arthritis
 A Genetic Analysis of Rheumatoid Arthritis Introduction to Rheumatoid Arthritis: Classification and Diagnosis Rheumatoid arthritis is a chronic inflammatory disorder that affects mainly synovial joints.
A Genetic Analysis of Rheumatoid Arthritis Introduction to Rheumatoid Arthritis: Classification and Diagnosis Rheumatoid arthritis is a chronic inflammatory disorder that affects mainly synovial joints.
Analyzing microrna Data and Integrating mirna with Gene Expression Data in Partek Genomics Suite 6.6
 Analyzing microrna Data and Integrating mirna with Gene Expression Data in Partek Genomics Suite 6.6 Overview This tutorial outlines how microrna data can be analyzed within Partek Genomics Suite. Additionally,
Analyzing microrna Data and Integrating mirna with Gene Expression Data in Partek Genomics Suite 6.6 Overview This tutorial outlines how microrna data can be analyzed within Partek Genomics Suite. Additionally,
Gene Therapy. The use of DNA as a drug. Edited by Gavin Brooks. BPharm, PhD, MRPharmS (PP) Pharmaceutical Press
 Gene Therapy The use of DNA as a drug Edited by Gavin Brooks BPharm, PhD, MRPharmS (PP) Pharmaceutical Press Contents Preface xiii Acknowledgements xv About the editor xvi Contributors xvii An introduction
Gene Therapy The use of DNA as a drug Edited by Gavin Brooks BPharm, PhD, MRPharmS (PP) Pharmaceutical Press Contents Preface xiii Acknowledgements xv About the editor xvi Contributors xvii An introduction
Guide for Data Visualization and Analysis using ACSN
 Guide for Data Visualization and Analysis using ACSN ACSN contains the NaviCell tool box, the intuitive and user- friendly environment for data visualization and analysis. The tool is accessible from the
Guide for Data Visualization and Analysis using ACSN ACSN contains the NaviCell tool box, the intuitive and user- friendly environment for data visualization and analysis. The tool is accessible from the
Frequently Asked Questions Next Generation Sequencing
 Frequently Asked Questions Next Generation Sequencing Import These Frequently Asked Questions for Next Generation Sequencing are some of the more common questions our customers ask. Questions are divided
Frequently Asked Questions Next Generation Sequencing Import These Frequently Asked Questions for Next Generation Sequencing are some of the more common questions our customers ask. Questions are divided
specific B cells Humoral immunity lymphocytes antibodies B cells bone marrow Cell-mediated immunity: T cells antibodies proteins
 Adaptive Immunity Chapter 17: Adaptive (specific) Immunity Bio 139 Dr. Amy Rogers Host defenses that are specific to a particular infectious agent Can be innate or genetic for humans as a group: most microbes
Adaptive Immunity Chapter 17: Adaptive (specific) Immunity Bio 139 Dr. Amy Rogers Host defenses that are specific to a particular infectious agent Can be innate or genetic for humans as a group: most microbes
2.1.2 Characterization of antiviral effect of cytokine expression on HBV replication in transduced mouse hepatocytes line
 i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
i 1 INTRODUCTION 1.1 Human Hepatitis B virus (HBV) 1 1.1.1 Pathogenesis of Hepatitis B 1 1.1.2 Genome organization of HBV 3 1.1.3 Structure of HBV virion 5 1.1.4 HBV life cycle 5 1.1.5 Experimental models
Analysis of gene expression data. Ulf Leser and Philippe Thomas
 Analysis of gene expression data Ulf Leser and Philippe Thomas This Lecture Protein synthesis Microarray Idea Technologies Applications Problems Quality control Normalization Analysis next week! Ulf Leser:
Analysis of gene expression data Ulf Leser and Philippe Thomas This Lecture Protein synthesis Microarray Idea Technologies Applications Problems Quality control Normalization Analysis next week! Ulf Leser:
Analysis of Illumina Gene Expression Microarray Data
 Analysis of Illumina Gene Expression Microarray Data Asta Laiho, Msc. Tech. Bioinformatics research engineer The Finnish DNA Microarray Centre Turku Centre for Biotechnology, Finland The Finnish DNA Microarray
Analysis of Illumina Gene Expression Microarray Data Asta Laiho, Msc. Tech. Bioinformatics research engineer The Finnish DNA Microarray Centre Turku Centre for Biotechnology, Finland The Finnish DNA Microarray
Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS)
 Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS) A typical RNA Seq experiment Library construction Protocol variations Fragmentation methods RNA: nebulization,
Introduction to transcriptome analysis using High Throughput Sequencing technologies (HTS) A typical RNA Seq experiment Library construction Protocol variations Fragmentation methods RNA: nebulization,
New Technologies for Sensitive, Low-Input RNA-Seq. Clontech Laboratories, Inc.
 New Technologies for Sensitive, Low-Input RNA-Seq Clontech Laboratories, Inc. Outline Introduction Single-Cell-Capable mrna-seq Using SMART Technology SMARTer Ultra Low RNA Kit for the Fluidigm C 1 System
New Technologies for Sensitive, Low-Input RNA-Seq Clontech Laboratories, Inc. Outline Introduction Single-Cell-Capable mrna-seq Using SMART Technology SMARTer Ultra Low RNA Kit for the Fluidigm C 1 System
B Cells and Antibodies
 B Cells and Antibodies Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School Lecture outline Functions of antibodies B cell activation; the role of helper T cells in antibody production
B Cells and Antibodies Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School Lecture outline Functions of antibodies B cell activation; the role of helper T cells in antibody production
Dr Alexander Henzing
 Horizon 2020 Health, Demographic Change & Wellbeing EU funding, research and collaboration opportunities for 2016/17 Innovate UK funding opportunities in omics, bridging health and life sciences Dr Alexander
Horizon 2020 Health, Demographic Change & Wellbeing EU funding, research and collaboration opportunities for 2016/17 Innovate UK funding opportunities in omics, bridging health and life sciences Dr Alexander
Non-clinical development of biologics
 Aurigon Life Science GmbH Non-clinical development of biologics Requirements, challenges and case studies Committed to Life. Sigrid Messemer vet. med. M4 Seminar March 10 th 2014 Aurigon - your full service
Aurigon Life Science GmbH Non-clinical development of biologics Requirements, challenges and case studies Committed to Life. Sigrid Messemer vet. med. M4 Seminar March 10 th 2014 Aurigon - your full service
Microarray Data Analysis. A step by step analysis using BRB-Array Tools
 Microarray Data Analysis A step by step analysis using BRB-Array Tools 1 EXAMINATION OF DIFFERENTIAL GENE EXPRESSION (1) Objective: to find genes whose expression is changed before and after chemotherapy.
Microarray Data Analysis A step by step analysis using BRB-Array Tools 1 EXAMINATION OF DIFFERENTIAL GENE EXPRESSION (1) Objective: to find genes whose expression is changed before and after chemotherapy.
Gene Expression Analysis
 Gene Expression Analysis Jie Peng Department of Statistics University of California, Davis May 2012 RNA expression technologies High-throughput technologies to measure the expression levels of thousands
Gene Expression Analysis Jie Peng Department of Statistics University of California, Davis May 2012 RNA expression technologies High-throughput technologies to measure the expression levels of thousands
PreciseTM Whitepaper
 Precise TM Whitepaper Introduction LIMITATIONS OF EXISTING RNA-SEQ METHODS Correctly designed gene expression studies require large numbers of samples, accurate results and low analysis costs. Analysis
Precise TM Whitepaper Introduction LIMITATIONS OF EXISTING RNA-SEQ METHODS Correctly designed gene expression studies require large numbers of samples, accurate results and low analysis costs. Analysis
Shouguo Gao Ph. D Department of Physics and Comprehensive Diabetes Center
 Computational Challenges in Storage, Analysis and Interpretation of Next-Generation Sequencing Data Shouguo Gao Ph. D Department of Physics and Comprehensive Diabetes Center Next Generation Sequencing
Computational Challenges in Storage, Analysis and Interpretation of Next-Generation Sequencing Data Shouguo Gao Ph. D Department of Physics and Comprehensive Diabetes Center Next Generation Sequencing
Expression Quantification (I)
 Expression Quantification (I) Mario Fasold, LIFE, IZBI Sequencing Technology One Illumina HiSeq 2000 run produces 2 times (paired-end) ca. 1,2 Billion reads ca. 120 GB FASTQ file RNA-seq protocol Task
Expression Quantification (I) Mario Fasold, LIFE, IZBI Sequencing Technology One Illumina HiSeq 2000 run produces 2 times (paired-end) ca. 1,2 Billion reads ca. 120 GB FASTQ file RNA-seq protocol Task
岑 祥 股 份 有 限 公 司 技 術 專 員 費 軫 尹 20100803
 技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
技 術 專 員 費 軫 尹 20100803 Overview of presentation Basic Biology of RNA interference Application of sirna for gene function? How to study mirna? How to deliver sirna and mirna? New prospects on RNAi research
Rheumatoid arthritis: an overview. Christine Pham MD
 Rheumatoid arthritis: an overview Christine Pham MD RA prevalence Chronic inflammatory disease affecting approximately 0.5 1% of the general population Prevalence is higher in North America (approaching
Rheumatoid arthritis: an overview Christine Pham MD RA prevalence Chronic inflammatory disease affecting approximately 0.5 1% of the general population Prevalence is higher in North America (approaching
ANIMALS FORM & FUNCTION BODY DEFENSES NONSPECIFIC DEFENSES PHYSICAL BARRIERS PHAGOCYTES. Animals Form & Function Activity #4 page 1
 AP BIOLOGY ANIMALS FORM & FUNCTION ACTIVITY #4 NAME DATE HOUR BODY DEFENSES NONSPECIFIC DEFENSES PHYSICAL BARRIERS PHAGOCYTES Animals Form & Function Activity #4 page 1 INFLAMMATORY RESPONSE ANTIMICROBIAL
AP BIOLOGY ANIMALS FORM & FUNCTION ACTIVITY #4 NAME DATE HOUR BODY DEFENSES NONSPECIFIC DEFENSES PHYSICAL BARRIERS PHAGOCYTES Animals Form & Function Activity #4 page 1 INFLAMMATORY RESPONSE ANTIMICROBIAL
Special report. Chronic Lymphocytic Leukemia (CLL) Genomic Biology 3020 April 20, 2006
 Special report Chronic Lymphocytic Leukemia (CLL) Genomic Biology 3020 April 20, 2006 Gene And Protein The gene that causes the mutation is CCND1 and the protein NP_444284 The mutation deals with the cell
Special report Chronic Lymphocytic Leukemia (CLL) Genomic Biology 3020 April 20, 2006 Gene And Protein The gene that causes the mutation is CCND1 and the protein NP_444284 The mutation deals with the cell
The Most Common Autoimmune Disease: Rheumatoid Arthritis. Bonita S. Libman, M.D.
 The Most Common Autoimmune Disease: Rheumatoid Arthritis Bonita S. Libman, M.D. Disclosures Two googled comics The Normal Immune System Network of cells and proteins that work together Goal: protect against
The Most Common Autoimmune Disease: Rheumatoid Arthritis Bonita S. Libman, M.D. Disclosures Two googled comics The Normal Immune System Network of cells and proteins that work together Goal: protect against
Chapter 43: The Immune System
 Name Period Our students consider this chapter to be a particularly challenging and important one. Expect to work your way slowly through the first three concepts. Take particular care with Concepts 43.2
Name Period Our students consider this chapter to be a particularly challenging and important one. Expect to work your way slowly through the first three concepts. Take particular care with Concepts 43.2
Introduction To Real Time Quantitative PCR (qpcr)
 Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
Introduction To Real Time Quantitative PCR (qpcr) SABiosciences, A QIAGEN Company www.sabiosciences.com The Seminar Topics The advantages of qpcr versus conventional PCR Work flow & applications Factors
BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS
 BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS 1. The Technology Strategy sets out six areas where technological developments are required to push the frontiers of knowledge
BBSRC TECHNOLOGY STRATEGY: TECHNOLOGIES NEEDED BY RESEARCH KNOWLEDGE PROVIDERS 1. The Technology Strategy sets out six areas where technological developments are required to push the frontiers of knowledge
Using Illumina BaseSpace Apps to Analyze RNA Sequencing Data
 Using Illumina BaseSpace Apps to Analyze RNA Sequencing Data The Illumina TopHat Alignment and Cufflinks Assembly and Differential Expression apps make RNA data analysis accessible to any user, regardless
Using Illumina BaseSpace Apps to Analyze RNA Sequencing Data The Illumina TopHat Alignment and Cufflinks Assembly and Differential Expression apps make RNA data analysis accessible to any user, regardless
Natalia Taborda Vanegas. Doc. Sci. Student Immunovirology Group Universidad de Antioquia
 Pathogenesis of Dengue Natalia Taborda Vanegas Doc. Sci. Student Immunovirology Group Universidad de Antioquia Infection process Epidermis keratinocytes Dermis Archives of Medical Research 36 (2005) 425
Pathogenesis of Dengue Natalia Taborda Vanegas Doc. Sci. Student Immunovirology Group Universidad de Antioquia Infection process Epidermis keratinocytes Dermis Archives of Medical Research 36 (2005) 425
2 Year (2013 15) CLL/SLL Research Initiative CLL/SLL Collaborative Grant
 2 Year (2013 15) CLL/SLL Research Initiative CLL/SLL Collaborative Grant Guidelines & General Instructions for Application KEY DATES: Application Release Date: December 12, 2012 Application Deadline: February
2 Year (2013 15) CLL/SLL Research Initiative CLL/SLL Collaborative Grant Guidelines & General Instructions for Application KEY DATES: Application Release Date: December 12, 2012 Application Deadline: February
Analysis of ChIP-seq data in Galaxy
 Analysis of ChIP-seq data in Galaxy November, 2012 Local copy: https://galaxy.wi.mit.edu/ Joint project between BaRC and IT Main site: http://main.g2.bx.psu.edu/ 1 Font Conventions Bold and blue refers
Analysis of ChIP-seq data in Galaxy November, 2012 Local copy: https://galaxy.wi.mit.edu/ Joint project between BaRC and IT Main site: http://main.g2.bx.psu.edu/ 1 Font Conventions Bold and blue refers
Basic Overview of Preclinical Toxicology Animal Models
 Basic Overview of Preclinical Toxicology Animal Models Charles D. Hebert, Ph.D., D.A.B.T. December 5, 2013 Outline Background In Vitro Toxicology In Vivo Toxicology Animal Models What is Toxicology? Background
Basic Overview of Preclinical Toxicology Animal Models Charles D. Hebert, Ph.D., D.A.B.T. December 5, 2013 Outline Background In Vitro Toxicology In Vivo Toxicology Animal Models What is Toxicology? Background
CD22 Antigen Is Broadly Expressed on Lung Cancer Cells and Is a Target for Antibody-Based Therapy
 CD22 Antigen Is Broadly Expressed on Lung Cancer Cells and Is a Target for Antibody-Based Therapy Joseph M. Tuscano, Jason Kato, David Pearson, Chengyi Xiong, Laura Newell, Yunpeng Ma, David R. Gandara,
CD22 Antigen Is Broadly Expressed on Lung Cancer Cells and Is a Target for Antibody-Based Therapy Joseph M. Tuscano, Jason Kato, David Pearson, Chengyi Xiong, Laura Newell, Yunpeng Ma, David R. Gandara,
Medical Therapies Limited EGM Presentation
 Medical Therapies Limited EGM Presentation Maria Halasz Chief Executive Officer 5 May 2009 1 Agenda 1. Company information 2. Recent developments 3. Business strategy 4. Key value inflection points for
Medical Therapies Limited EGM Presentation Maria Halasz Chief Executive Officer 5 May 2009 1 Agenda 1. Company information 2. Recent developments 3. Business strategy 4. Key value inflection points for
CUSTOM ANTIBODIES. Fully customised services: rat and murine monoclonals, rat and rabbit polyclonals, antibody characterisation, antigen preparation
 CUSTOM ANTIBODIES Highly competitive pricing without compromising quality. Rat monoclonal antibodies for the study of gene expression and proteomics in mice and in mouse models of human diseases available.
CUSTOM ANTIBODIES Highly competitive pricing without compromising quality. Rat monoclonal antibodies for the study of gene expression and proteomics in mice and in mouse models of human diseases available.
Modelling and analysis of T-cell epitope screening data.
 Modelling and analysis of T-cell epitope screening data. Tim Beißbarth 2, Jason A. Tye-Din 1, Gordon K. Smyth 1, Robert P. Anderson 1 and Terence P. Speed 1 1 WEHI, 1G Royal Parade, Parkville, VIC 3050,
Modelling and analysis of T-cell epitope screening data. Tim Beißbarth 2, Jason A. Tye-Din 1, Gordon K. Smyth 1, Robert P. Anderson 1 and Terence P. Speed 1 1 WEHI, 1G Royal Parade, Parkville, VIC 3050,
Analysis and Integration of Big Data from Next-Generation Genomics, Epigenomics, and Transcriptomics
 Analysis and Integration of Big Data from Next-Generation Genomics, Epigenomics, and Transcriptomics Christopher Benner, PhD Director, Integrative Genomics and Bioinformatics Core (IGC) idash Webinar,
Analysis and Integration of Big Data from Next-Generation Genomics, Epigenomics, and Transcriptomics Christopher Benner, PhD Director, Integrative Genomics and Bioinformatics Core (IGC) idash Webinar,
Ingenuity Pathway Analysis (IPA )
 ProductProfile Ingenuity Pathway Analysis (IPA ) For the analysis and interpretation of omics data IPA is a web-based software application for the analysis, integration, and interpretation of data derived
ProductProfile Ingenuity Pathway Analysis (IPA ) For the analysis and interpretation of omics data IPA is a web-based software application for the analysis, integration, and interpretation of data derived
dixa a data infrastructure for chemical safety Jos Kleinjans Dept of Toxicogenomics Maastricht University
 dixa a data infrastructure for chemical safety Jos Kleinjans Dept of Toxicogenomics Maastricht University Current protocol for chemical safety testing Short Term Tests for Genetic Toxicity Bacterial Reverse
dixa a data infrastructure for chemical safety Jos Kleinjans Dept of Toxicogenomics Maastricht University Current protocol for chemical safety testing Short Term Tests for Genetic Toxicity Bacterial Reverse
ALLIANCE FOR LUPUS RESEARCH AND PFIZER S CENTERS FOR THERAPEUTIC INNOVATION CHALLENGE GRANT PROGRAM PROGRAM GUIDELINES
 ALLIANCE FOR LUPUS RESEARCH AND PFIZER S CENTERS FOR THERAPEUTIC INNOVATION CHALLENGE GRANT PROGRAM PROGRAM GUIDELINES DESCRIPTION OF GRANT MECHANISM The Alliance for Lupus Research (ALR) is an independent,
ALLIANCE FOR LUPUS RESEARCH AND PFIZER S CENTERS FOR THERAPEUTIC INNOVATION CHALLENGE GRANT PROGRAM PROGRAM GUIDELINES DESCRIPTION OF GRANT MECHANISM The Alliance for Lupus Research (ALR) is an independent,
B Cell Generation, Activation & Differentiation. B cell maturation
 B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
B Cell Generation, Activation & Differentiation Naïve B cells- have not encountered Ag. Have IgM and IgD on cell surface : have same binding VDJ regions but different constant region leaves bone marrow
Core Facility Genomics
 Core Facility Genomics versatile genome or transcriptome analyses based on quantifiable highthroughput data ascertainment 1 Topics Collaboration with Harald Binder and Clemens Kreutz Project: Microarray
Core Facility Genomics versatile genome or transcriptome analyses based on quantifiable highthroughput data ascertainment 1 Topics Collaboration with Harald Binder and Clemens Kreutz Project: Microarray
Gene expression analysis. Ulf Leser and Karin Zimmermann
 Gene expression analysis Ulf Leser and Karin Zimmermann Ulf Leser: Bioinformatics, Wintersemester 2010/2011 1 Last lecture What are microarrays? - Biomolecular devices measuring the transcriptome of a
Gene expression analysis Ulf Leser and Karin Zimmermann Ulf Leser: Bioinformatics, Wintersemester 2010/2011 1 Last lecture What are microarrays? - Biomolecular devices measuring the transcriptome of a
SYMPOSIUM. June 15, 2013. Photonics Center Colloquium Room (906) 8 Saint Mary's St., Boston, MA 02215 Boston University
 SYMPOSIUM June 15, 2013 Photonics Center Colloquium Room (906) 8 Saint Mary's St., Boston, MA 02215 Bette Korber Los Alamos National Labs HIV evolution and the neutralizing antibody response We have recently
SYMPOSIUM June 15, 2013 Photonics Center Colloquium Room (906) 8 Saint Mary's St., Boston, MA 02215 Bette Korber Los Alamos National Labs HIV evolution and the neutralizing antibody response We have recently
Reprogramming, Screening and Validation of ipscs and Terminally Differentiated Cells using the qbiomarker PCR Array System
 Reprogramming, Screening and Validation of ipscs and Terminally Differentiated Cells using the qbiomarker PCR Array System Outline of Webinar What are induced pluripotent Stem Cells (ips Cells or ipscs)?
Reprogramming, Screening and Validation of ipscs and Terminally Differentiated Cells using the qbiomarker PCR Array System Outline of Webinar What are induced pluripotent Stem Cells (ips Cells or ipscs)?
RNA-seq. Quantification and Differential Expression. Genomics: Lecture #12
 (2) Quantification and Differential Expression Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #12 Today (2) Gene Expression per Sources of bias,
(2) Quantification and Differential Expression Institut für Medizinische Genetik und Humangenetik Charité Universitätsmedizin Berlin Genomics: Lecture #12 Today (2) Gene Expression per Sources of bias,
Quantitative proteomics background
 Proteomics data analysis seminar Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post transcriptional regulation of key cellular processes de Groot, M., Daran
Proteomics data analysis seminar Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post transcriptional regulation of key cellular processes de Groot, M., Daran
RETRIEVING SEQUENCE INFORMATION. Nucleotide sequence databases. Database search. Sequence alignment and comparison
 RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
RETRIEVING SEQUENCE INFORMATION Nucleotide sequence databases Database search Sequence alignment and comparison Biological sequence databases Originally just a storage place for sequences. Currently the
Standards, Guidelines and Best Practices for RNA-Seq V1.0 (June 2011) The ENCODE Consortium
 Standards, Guidelines and Best Practices for RNA-Seq V1.0 (June 2011) The ENCODE Consortium I. Introduction: Sequence based assays of transcriptomes (RNA-seq) are in wide use because of their favorable
Standards, Guidelines and Best Practices for RNA-Seq V1.0 (June 2011) The ENCODE Consortium I. Introduction: Sequence based assays of transcriptomes (RNA-seq) are in wide use because of their favorable
CONTRACTING ORGANIZATION: University of Alabama at Birmingham Birmingham, AL 35294
 AD Award Number: W81XWH-08-1-0030 TITLE: Regulation of Prostate Cancer Bone Metastasis by DKK1 PRINCIPAL INVESTIGATOR: Gregory A. Clines, M.D., Ph.D. CONTRACTING ORGANIZATION: University of Alabama at
AD Award Number: W81XWH-08-1-0030 TITLE: Regulation of Prostate Cancer Bone Metastasis by DKK1 PRINCIPAL INVESTIGATOR: Gregory A. Clines, M.D., Ph.D. CONTRACTING ORGANIZATION: University of Alabama at
Molecular Genetics: Challenges for Statistical Practice. J.K. Lindsey
 Molecular Genetics: Challenges for Statistical Practice J.K. Lindsey 1. What is a Microarray? 2. Design Questions 3. Modelling Questions 4. Longitudinal Data 5. Conclusions 1. What is a microarray? A microarray
Molecular Genetics: Challenges for Statistical Practice J.K. Lindsey 1. What is a Microarray? 2. Design Questions 3. Modelling Questions 4. Longitudinal Data 5. Conclusions 1. What is a microarray? A microarray
HuCAL Custom Monoclonal Antibodies
 HuCAL Custom Monoclonal HuCAL Custom Monoclonal Antibodies Highly Specific, Recombinant Antibodies in 8 Weeks Highly Specific Monoclonal Antibodies in Just 8 Weeks HuCAL PLATINUM (Human Combinatorial Antibody
HuCAL Custom Monoclonal HuCAL Custom Monoclonal Antibodies Highly Specific, Recombinant Antibodies in 8 Weeks Highly Specific Monoclonal Antibodies in Just 8 Weeks HuCAL PLATINUM (Human Combinatorial Antibody
From Reads to Differentially Expressed Genes. The statistics of differential gene expression analysis using RNA-seq data
 From Reads to Differentially Expressed Genes The statistics of differential gene expression analysis using RNA-seq data experimental design data collection modeling statistical testing biological heterogeneity
From Reads to Differentially Expressed Genes The statistics of differential gene expression analysis using RNA-seq data experimental design data collection modeling statistical testing biological heterogeneity
TREATING AUTOIMMUNE DISEASES WITH HOMEOPATHY. Dr. Stephen A. Messer, MSEd, ND, DHANP Professor and Chair of Homeopathic Medicine
 TREATING AUTOIMMUNE DISEASES WITH HOMEOPATHY Dr. Stephen A. Messer, MSEd, ND, DHANP Professor and Chair of Homeopathic Medicine AUTOIMMUNE DISEASES An autoimmune disorder occurs when the body s immune
TREATING AUTOIMMUNE DISEASES WITH HOMEOPATHY Dr. Stephen A. Messer, MSEd, ND, DHANP Professor and Chair of Homeopathic Medicine AUTOIMMUNE DISEASES An autoimmune disorder occurs when the body s immune
Lecture 11 Data storage and LIMS solutions. Stéphane LE CROM lecrom@biologie.ens.fr
 Lecture 11 Data storage and LIMS solutions Stéphane LE CROM lecrom@biologie.ens.fr Various steps of a DNA microarray experiment Experimental steps Data analysis Experimental design set up Chips on catalog
Lecture 11 Data storage and LIMS solutions Stéphane LE CROM lecrom@biologie.ens.fr Various steps of a DNA microarray experiment Experimental steps Data analysis Experimental design set up Chips on catalog
CCR Biology - Chapter 9 Practice Test - Summer 2012
 Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
Name: Class: Date: CCR Biology - Chapter 9 Practice Test - Summer 2012 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Genetic engineering is possible
IKDT Laboratory. IKDT as Service Lab (CRO) for Molecular Diagnostics
 Page 1 IKDT Laboratory IKDT as Service Lab (CRO) for Molecular Diagnostics IKDT lab offer is complete diagnostic service to all external customers. We could perform as well single procedures or complex
Page 1 IKDT Laboratory IKDT as Service Lab (CRO) for Molecular Diagnostics IKDT lab offer is complete diagnostic service to all external customers. We could perform as well single procedures or complex
Molecule Shapes. support@ingenuity.com www.ingenuity.com 1
 IPA 8 Legend This legend provides a key of the main features of Network Explorer and Canonical Pathways, including molecule shapes and colors as well as relationship labels and types. For a high-resolution
IPA 8 Legend This legend provides a key of the main features of Network Explorer and Canonical Pathways, including molecule shapes and colors as well as relationship labels and types. For a high-resolution
Protein Protein Interaction Networks
 Functional Pattern Mining from Genome Scale Protein Protein Interaction Networks Young-Rae Cho, Ph.D. Assistant Professor Department of Computer Science Baylor University it My Definition of Bioinformatics
Functional Pattern Mining from Genome Scale Protein Protein Interaction Networks Young-Rae Cho, Ph.D. Assistant Professor Department of Computer Science Baylor University it My Definition of Bioinformatics
A disease and antibody biology approach to antibody drug discovery
 A disease and antibody biology approach to antibody drug discovery Björn Frendéus, PhD VP, Preclinical research Presenter: Björn Frendéus Date: 2011-11-08 1 Antibodies have revolutionized Cancer Treatment!
A disease and antibody biology approach to antibody drug discovery Björn Frendéus, PhD VP, Preclinical research Presenter: Björn Frendéus Date: 2011-11-08 1 Antibodies have revolutionized Cancer Treatment!
FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem
 FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem Elsa Bernard Laurent Jacob Julien Mairal Jean-Philippe Vert September 24, 2013 Abstract FlipFlop implements a fast method for de novo transcript
FlipFlop: Fast Lasso-based Isoform Prediction as a Flow Problem Elsa Bernard Laurent Jacob Julien Mairal Jean-Philippe Vert September 24, 2013 Abstract FlipFlop implements a fast method for de novo transcript
THOMSON REUTERS CORTELLIS FOR INFORMATICS. REUTERS/ Aly Song
 THOMSON REUTERS CORTELLIS FOR INFORMATICS REUTERS/ Aly Song THOMSON REUTERS CORTELLIS FOR INFORMATICS 1 Table of Contents Table of Contents...1 The challenge... 2 The solution... 2 WHAT CAN YOU DO WITH
THOMSON REUTERS CORTELLIS FOR INFORMATICS REUTERS/ Aly Song THOMSON REUTERS CORTELLIS FOR INFORMATICS 1 Table of Contents Table of Contents...1 The challenge... 2 The solution... 2 WHAT CAN YOU DO WITH
Thyroid pathology in the Presence of antiviral treatment of chronic hepatitis C. Professor Nikitin Igor G Russian State Medical University MOSCOW
 Thyroid pathology in the Presence of antiviral treatment of chronic hepatitis C Professor Nikitin Igor G Russian State Medical University MOSCOW The structure of the side effects associated with antiviral
Thyroid pathology in the Presence of antiviral treatment of chronic hepatitis C Professor Nikitin Igor G Russian State Medical University MOSCOW The structure of the side effects associated with antiviral
EMA and Progressive Multifocal Leukoencephalopathy.
 EMA and Progressive Multifocal Leukoencephalopathy. ENCePP Plenary, London 23 November 2011 Presented by: Henry Fitt Head of Coordination & Networking, Pharmacovigilance & Risk Management An agency of
EMA and Progressive Multifocal Leukoencephalopathy. ENCePP Plenary, London 23 November 2011 Presented by: Henry Fitt Head of Coordination & Networking, Pharmacovigilance & Risk Management An agency of
DeCyder Extended Data Analysis module Version 1.0
 GE Healthcare DeCyder Extended Data Analysis module Version 1.0 Module for DeCyder 2D version 6.5 User Manual Contents 1 Introduction 1.1 Introduction... 7 1.2 The DeCyder EDA User Manual... 9 1.3 Getting
GE Healthcare DeCyder Extended Data Analysis module Version 1.0 Module for DeCyder 2D version 6.5 User Manual Contents 1 Introduction 1.1 Introduction... 7 1.2 The DeCyder EDA User Manual... 9 1.3 Getting
Vitamin D deficiency exacerbates ischemic cell loss and sensory motor dysfunction in an experimental stroke model
 Vitamin D deficiency exacerbates ischemic cell loss and sensory motor dysfunction in an experimental stroke model Robyn Balden & Farida Sohrabji Texas A&M Health Science Center- College of Medicine ISC
Vitamin D deficiency exacerbates ischemic cell loss and sensory motor dysfunction in an experimental stroke model Robyn Balden & Farida Sohrabji Texas A&M Health Science Center- College of Medicine ISC
Notch 1 -dependent regulation of cell fate in colorectal cancer
 Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
Notch 1 -dependent regulation of cell fate in colorectal cancer Referees: PD Dr. Tobias Dick Prof. Dr. Wilfried Roth http://d-nb.info/1057851272 CONTENTS Summary 1 Zusammenfassung 2 1 INTRODUCTION 3 1.1
The immune system. Bone marrow. Thymus. Spleen. Bone marrow. NK cell. B-cell. T-cell. Basophil Neutrophil. Eosinophil. Myeloid progenitor
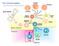 The immune system Basophil Neutrophil Bone marrow Eosinophil Myeloid progenitor Dendritic cell Pluripotent Stem cell Lymphoid progenitor Platelets Bone marrow Thymus NK cell T-cell B-cell Spleen Cancer
The immune system Basophil Neutrophil Bone marrow Eosinophil Myeloid progenitor Dendritic cell Pluripotent Stem cell Lymphoid progenitor Platelets Bone marrow Thymus NK cell T-cell B-cell Spleen Cancer
PrimePCR Assay Validation Report
 Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID papillary renal cell carcinoma (translocation-associated) PRCC Human This gene
Gene Information Gene Name Gene Symbol Organism Gene Summary Gene Aliases RefSeq Accession No. UniGene ID Ensembl Gene ID papillary renal cell carcinoma (translocation-associated) PRCC Human This gene
Pathway Analysis : An Introduction
 Pathway Analysis : An Introduction Experiments Literature and other KB Data Knowledge Structure in Data through statistics Structure in Knowledge through GO and other Ontologies Gain insight into Data
Pathway Analysis : An Introduction Experiments Literature and other KB Data Knowledge Structure in Data through statistics Structure in Knowledge through GO and other Ontologies Gain insight into Data
ALPHA (TNFa) IN OBESITY
 THE ROLE OF TUMOUR NECROSIS FACTOR ALPHA (TNFa) IN OBESITY Alison Mary Morris, B.Sc (Hons) A thesis submitted to Adelaide University for the degree of Doctor of Philosophy Department of Physiology Adelaide
THE ROLE OF TUMOUR NECROSIS FACTOR ALPHA (TNFa) IN OBESITY Alison Mary Morris, B.Sc (Hons) A thesis submitted to Adelaide University for the degree of Doctor of Philosophy Department of Physiology Adelaide
A Primer of Genome Science THIRD
 A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
A Primer of Genome Science THIRD EDITION GREG GIBSON-SPENCER V. MUSE North Carolina State University Sinauer Associates, Inc. Publishers Sunderland, Massachusetts USA Contents Preface xi 1 Genome Projects:
ELITE Custom Antibody Services
 ELITE Custom Antibody Services ELITE Custom Antibody Services Experience, confidence, and understanding As a manufacturer and service provider, we have the experience, confidence, and understanding to
ELITE Custom Antibody Services ELITE Custom Antibody Services Experience, confidence, and understanding As a manufacturer and service provider, we have the experience, confidence, and understanding to
HuCAL Custom Monoclonal Antibodies
 HuCAL Custom Monoclonal Antibodies Highly Specific Monoclonal Antibodies in just 8 Weeks PROVEN, HIGHLY SPECIFIC, HIGH AFFINITY ANTIBODIES IN 8 WEEKS WITHOUT HuCAL PLATINUM IMMUNIZATION (Human Combinatorial
HuCAL Custom Monoclonal Antibodies Highly Specific Monoclonal Antibodies in just 8 Weeks PROVEN, HIGHLY SPECIFIC, HIGH AFFINITY ANTIBODIES IN 8 WEEKS WITHOUT HuCAL PLATINUM IMMUNIZATION (Human Combinatorial
High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B. PEPperPRINT GmbH Heidelberg, 06/2014
 High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B PEPperPRINT GmbH Heidelberg, 06/2014 Introduction This report describes the epitope mapping of autoimmune sera
High Resolution Epitope Mapping of Human Autoimmune Sera against Antigens CENPA and KDM6B PEPperPRINT GmbH Heidelberg, 06/2014 Introduction This report describes the epitope mapping of autoimmune sera
SUMMARY AND CONCLUSIONS
 SUMMARY AND CONCLUSIONS Summary and Conclusions This study has attempted to document the following effects of targeting protein antigens to the macrophage scavenger receptors by maleylation. 1. Modification
SUMMARY AND CONCLUSIONS Summary and Conclusions This study has attempted to document the following effects of targeting protein antigens to the macrophage scavenger receptors by maleylation. 1. Modification
Gene Expression Assays
 APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
APPLICATION NOTE TaqMan Gene Expression Assays A mpl i fic ationef ficienc yof TaqMan Gene Expression Assays Assays tested extensively for qpcr efficiency Key factors that affect efficiency Efficiency
The Need for a PARP in vivo Pharmacodynamic Assay
 The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
The Need for a PARP in vivo Pharmacodynamic Assay Jay George, Ph.D., Chief Scientific Officer, Trevigen, Inc., Gaithersburg, MD For further infomation, please contact: William Booth, Ph.D. Tel: +44 (0)1235
What s New in Pathway Studio Web 11.1
 1 1 What s New in Pathway Studio Web 11.1 Elseiver is pleased to announce the release of Pathway Studio Web 11.1 for all database subscriptions (Mammal, Mammal+ChemEffect+DiseaseFx, Plant). This release
1 1 What s New in Pathway Studio Web 11.1 Elseiver is pleased to announce the release of Pathway Studio Web 11.1 for all database subscriptions (Mammal, Mammal+ChemEffect+DiseaseFx, Plant). This release
InSyBio BioNets: Utmost efficiency in gene expression data and biological networks analysis
 InSyBio BioNets: Utmost efficiency in gene expression data and biological networks analysis WHITE PAPER By InSyBio Ltd Konstantinos Theofilatos Bioinformatician, PhD InSyBio Technical Sales Manager August
InSyBio BioNets: Utmost efficiency in gene expression data and biological networks analysis WHITE PAPER By InSyBio Ltd Konstantinos Theofilatos Bioinformatician, PhD InSyBio Technical Sales Manager August
HUNTINGTON S DISEASE THERAPIES RESEARCH UPDATE
 HUNTINGTON S DISEASE MULTIDISCIPLINARY CLINIC HUNTINGTON S DISEASE THERAPIES RESEARCH UPDATE From gene to treatments The gene that causes Huntington s disease (HD) was discovered in 1993. Since then, enormous
HUNTINGTON S DISEASE MULTIDISCIPLINARY CLINIC HUNTINGTON S DISEASE THERAPIES RESEARCH UPDATE From gene to treatments The gene that causes Huntington s disease (HD) was discovered in 1993. Since then, enormous
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland 21702-5012
 AD Award Number: W81XWH-05-1-0584 TITLE: Early Host Responses to Prion Infection and Development of an In Vitro Bioassay PRINCIPAL INVESTIGATOR: George A. Carlson, Ph.D. Leroy E. Hood, M.D., Ph.D. CONTRACTING
AD Award Number: W81XWH-05-1-0584 TITLE: Early Host Responses to Prion Infection and Development of an In Vitro Bioassay PRINCIPAL INVESTIGATOR: George A. Carlson, Ph.D. Leroy E. Hood, M.D., Ph.D. CONTRACTING
BioMmune Technologies Inc. Corporate Presentation 2015
 BioMmune Technologies Inc Corporate Presentation 2015 * Harnessing the body s own immune system to fight cancer & other autoimmune diseases BioMmune Technologies Inc. (IMU) ABOUT A public biopharmaceutical
BioMmune Technologies Inc Corporate Presentation 2015 * Harnessing the body s own immune system to fight cancer & other autoimmune diseases BioMmune Technologies Inc. (IMU) ABOUT A public biopharmaceutical
Frequently Asked Questions (FAQ)
 Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
Frequently Asked Questions (FAQ) Why screen your (therapeutic) antibody for cross-reactivity? Cross-reactivity of therapeutic antibodies leads to adverse effects and might render the antibody unsuitable
RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial
 RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial Samuel J. Rulli, Jr., Ph.D. qpcr-applications Scientist Samuel.Rulli@QIAGEN.com Pathway Focused Research from Sample Prep to Data Analysis! -2-
RT 2 Profiler PCR Array: Web-Based Data Analysis Tutorial Samuel J. Rulli, Jr., Ph.D. qpcr-applications Scientist Samuel.Rulli@QIAGEN.com Pathway Focused Research from Sample Prep to Data Analysis! -2-
Activity 7.21 Transcription factors
 Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
Purpose To consolidate understanding of protein synthesis. To explain the role of transcription factors and hormones in switching genes on and off. Play the transcription initiation complex game Regulation
Autoimmunity. Autoimmunity. Genetic Contributions to Autoimmunity. Targets of Autoimmunity
 Autoimmunity Factors predisposing an individual to autoimmune disease Mechanisms of initiation of autoimmunity Pathogenesis of particular autoimmune disease Animal models of autoimmune disease Treatment
Autoimmunity Factors predisposing an individual to autoimmune disease Mechanisms of initiation of autoimmunity Pathogenesis of particular autoimmune disease Animal models of autoimmune disease Treatment
How To Expand Hematopoietic Stem Cells
 Purification and Expansion of Hematopoietic Stem Cells Based on Proteins Expressed by a Novel Stromal Cell Population Our bodies are constantly killing old, nonfunctional, and unneeded cells and making
Purification and Expansion of Hematopoietic Stem Cells Based on Proteins Expressed by a Novel Stromal Cell Population Our bodies are constantly killing old, nonfunctional, and unneeded cells and making
COMMITTEE FOR MEDICINAL PRODUCTS FOR HUMAN USE (CHMP) REFLECTION PAPER
 European Medicines Agency London 23 April 2009 EMEA/CHMP/BMWP/102046/2006 COMMITTEE FOR MEDICINAL PRODUCTS FOR HUMAN USE (CHMP) REFLECTION PAPER NON-CLINICAL AND CLINICAL DEVELOPMENT OF SIMILAR MEDICINAL
European Medicines Agency London 23 April 2009 EMEA/CHMP/BMWP/102046/2006 COMMITTEE FOR MEDICINAL PRODUCTS FOR HUMAN USE (CHMP) REFLECTION PAPER NON-CLINICAL AND CLINICAL DEVELOPMENT OF SIMILAR MEDICINAL
REUTERS/TIM WIMBORNE SCHOLARONE MANUSCRIPTS COGNOS REPORTS
 REUTERS/TIM WIMBORNE SCHOLARONE MANUSCRIPTS COGNOS REPORTS 28-APRIL-2015 TABLE OF CONTENTS Select an item in the table of contents to go to that topic in the document. USE GET HELP NOW & FAQS... 1 SYSTEM
REUTERS/TIM WIMBORNE SCHOLARONE MANUSCRIPTS COGNOS REPORTS 28-APRIL-2015 TABLE OF CONTENTS Select an item in the table of contents to go to that topic in the document. USE GET HELP NOW & FAQS... 1 SYSTEM
Single-Cell DNA Sequencing with the C 1. Single-Cell Auto Prep System. Reveal hidden populations and genetic diversity within complex samples
 DATA Sheet Single-Cell DNA Sequencing with the C 1 Single-Cell Auto Prep System Reveal hidden populations and genetic diversity within complex samples Single-cell sensitivity Discover and detect SNPs,
DATA Sheet Single-Cell DNA Sequencing with the C 1 Single-Cell Auto Prep System Reveal hidden populations and genetic diversity within complex samples Single-cell sensitivity Discover and detect SNPs,
Recognition of T cell epitopes (Abbas Chapter 6)
 Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
Recognition of T cell epitopes (Abbas Chapter 6) Functions of different APCs (Abbas Chapter 6)!!! Directon Routes of antigen entry (Abbas Chapter 6) Flow of Information Barrier APCs LNs Sequence of Events
